Abstract
The ability of bacteria to sense environmental cues and adapt is essential for their survival. The use of second-messenger signaling molecules to translate these cues into a physiological response is a common mechanism employed by bacteria. The second messenger 3’-5’-cyclic diadenosine monophosphate (c-di-AMP) has been linked to a diverse set of biological processes involved in maintaining cell viability and homeostasis, as well as pathogenicity. A complex network of both protein and RNA receptors inside the cell activate specific pathways and mediate phenotypic outputs in response to c-di-AMP. Structural analysis of these RNA and protein receptors has revealed the different recognition elements employed by these effectors to bind the same small molecule. Herein, using a series of c-di-AMP analogs, we probed the interactions made with a riboswitch and a phosphodiesterase protein to identify the features important for c-di-AMP binding and recognition. We found that the ydaO riboswitch binds c-di-AMP in two discrete sites with near identical affinity and a Hill coefficient of 1.6. The ydaO riboswitch distinguishes between c-di-AMP and structurally related second messengers by discriminating against an amine at the C2 position, more than a carbonyl at the C6 position. We also identified phosphate-modified analogs that bind both the ydaO RNA and GdpP protein with high affinity, while symmetrically-modified ribose analogs exhibited a substantial decrease in ydaO affinity, but retained high affinity for GdpP. These ligand modifications resulted in increased resistance to enzyme-catalyzed hydrolysis by the GdpP enzyme. Together, these data suggest that these c-di-AMP analogs could be useful as chemical tools to specifically target subsections of the second-messenger signaling pathways.
INTRODUCTION
Bacteria rely upon signaling molecules to adapt to changing environments and respond to extracellular inputs.1 Small-molecule second messengers are commonly used to relay stimuli from external receptors to effectors within the cell.2–4 One such molecule, cyclic diadenosine monophosphate (c-di-AMP), has recently been identified as a second messenger in a wide variety of bacteria.5,6 C-di-AMP signaling has been implicated in a diverse set of processes including sporulation7, peptidoglycan homeostasis8,9, cell size8,10, biofilm formation8,10,11, virulence10,12–16, and cell viability.8,9,15–21 In addition, c-di-AMP has been shown to activate an innate immune response via the human host proteins STING and DDX41.22–25
Differential gene expression in these pathways is correlated to changes in c-di-AMP concentration in many species, including pathogens such as Bacillus subtilis, Staphylococcus aureus, and Listeria monocytogenes.5,8,16,22,25,26 Intracellular levels of c-di-AMP are controlled by diadenylate cyclases (DACs) that synthesize c-di-AMP from two ATP or ADP molecules5,26, and phosphodiesterases (PDEs) that degrade c-di-AMP to pApA or AMP.16,20,27,28 Despite the important role of c-di-AMP signaling, only a small number of protein targets have been identified that respond directly to this small molecule24,29–37.
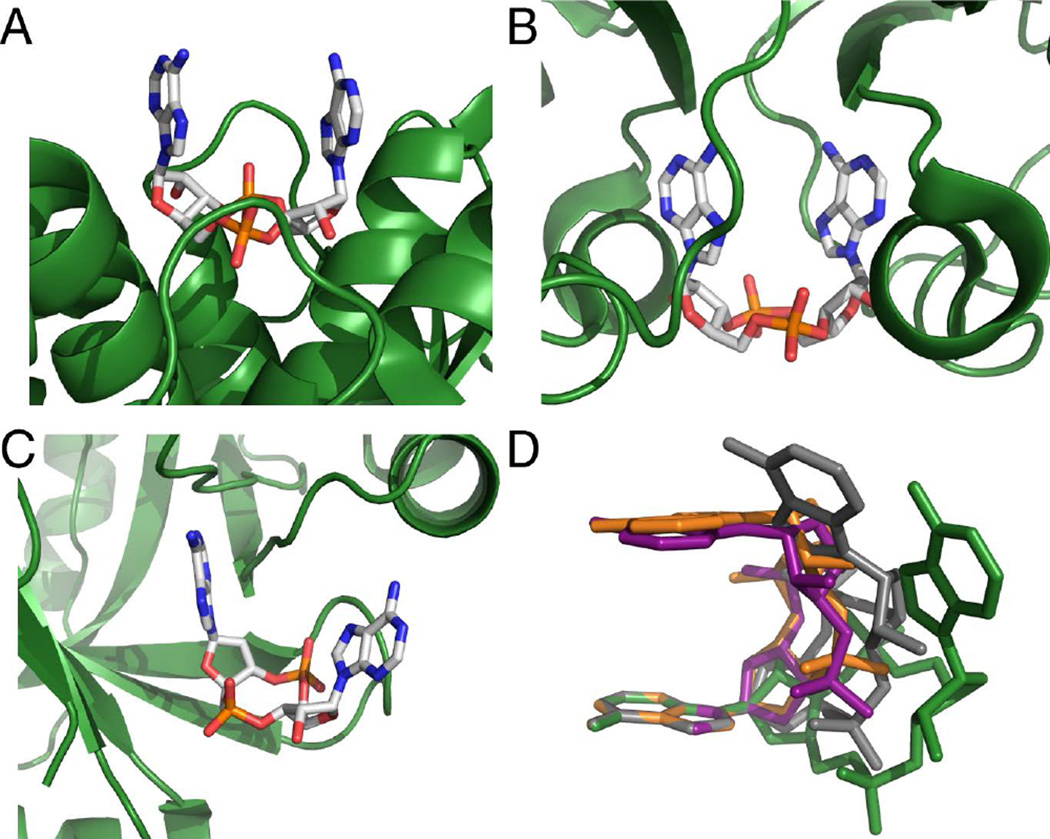
Crystal structures of c-di-AMP bound to these protein receptors reveal unique interactions to recognize and bind the same ligand. Ligand recognition can function through the phosphodiester backbone,32 both adenine bases,37 or a single base (Figure 1a,b,c).33–36 In addition to protein receptors in bacteria, the enzymes that bind and degrade c-di-AMP also employ specific recognition elements. The recent crystal structure of c-di-AMP bound to a PDE protein from L. monocytogenes shows asymmetric contacts between ligand and protein.36 While the phosphate backbone that undergoes cleavage is heavily recognized, the remaining functional groups are less contacted by the protein. These different ligand specificities have begun to uncover how c-di-AMP is recognized by its macromolecular receptors and affects physiological responses in bacteria.
Figure 1.
c-di-AMP bound to macromolecular receptors (A) LmPC, (B) KtrA, and (C) PstASA (ref. 32,33,37). c-di-AMP is colored by atom with carbon in white, oxygen in red, nitrogen in blue, and phosphorous in orange. Protein binding sites are shown as green cartoons. (D) Conformations adopted by c-di-AMP when bound to downstream targets (ref. 32,33,37,45–47). C-di-AMP is colored in green for ydaO, grey for PstASA, orange for LmPC, and purple for KtrA. The structures are superimposed at a single adenosine, with the remaining atoms floating.
C-di-AMP is part of a broader signaling network that also includes riboswitches. Riboswitches are structured RNA domains that bind small-molecule effectors with high affinity and specificity, and control gene expression.38–41 Typically, effector binding induces structural changes that lead to modulation of transcription termination or translation initiation.39,42 C-di-AMP has recently been identified as the ligand for the ydaO riboswitch (Figure 2a), originally discovered in 200443 but whose ligand was unidentified for almost a decade. This RNA motif provides a clear mechanism for gene control by this signaling molecule.44 The presence of these riboswitches in human pathogens, as well as the influence that c-di-AMP has on cellular homeostasis and sporulation, make them attractive antibiotic targets.
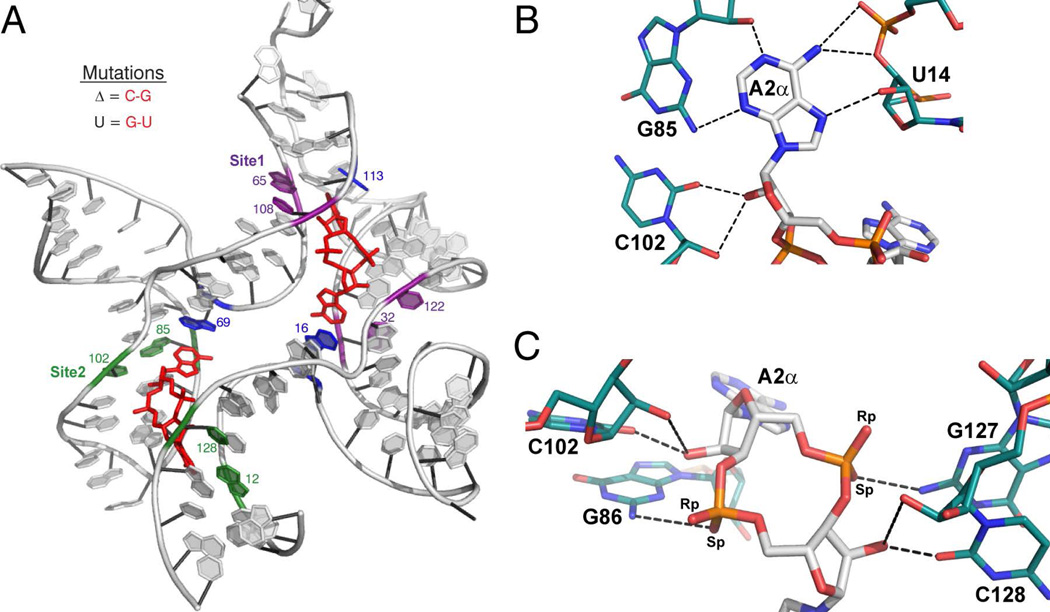
Figure 2.
c-di-AMP recognition by the ydaO riboswitch (ref. 45–47). (A) Structure of the ydaO aptamer from B. subtilis bound to two molecules of c-di-AMP. c-di-AMP is colored in red. Conserved Watson-Crick G-C pairs involved in type-I A-minor interactions with c-di-AMP are shown in purple for Site 1 and green for Site 2. Bases that stack with the adenines are shown in blue. Important nucleotides are labeled. (B and C) c-di-AMP-A2α recognition by the ydaO motif. c-di-AMP is colored by atom with carbon in white, oxygen in red, nitrogen in blue, and phosphorous in orange. Hydrogen-bonding nucleotides of ydaO are colored as described, with carbons in teal. Hydrogen bonds are shown as black dashed lines. (B) Recognition of the Watson-Crick and Hoogsteen faces of the c-di-AMP nucleobases featuring the type-1 A-minor interaction. (C) Backbone recognition of c-di-AMP by the ydaO riboswitch. These c-di-AMP recognition motifs are observed with each adenosine of the ligand.
The crystal structure of the ydaO riboswitch bound to c-di-AMP was recently reported.45–47 The riboswitch adopts a pseudo-symmetric architecture and two discrete binding pockets were observed (Figure 2a). Both binding sites, which are related by pseudo-two-fold symmetry, recognize a single ligand molecule and employ similar stacking and hydrogen bonding interactions. The four adenines contact a conserved Watson-Crick G-C pair to form type-I A-minor interactions (Figure 2b).45–48 The c-di-AMP phosphodiester backbone and ribose 2’-hydroxyls are also highly involved in binding to the riboswitch (Figure 2c).
C-di-AMP recognition by the ydaO RNA is distinct from what is observed for c-di-AMP-binding proteins. Most notably, c-di-AMP adopts an extended conformation in the ydaO riboswitch, while protein targets bind c-di-AMP in a more compact, U-like orientation (Figure 1d).28,32–37 Furthermore, adenine recognition by an A-minor interaction is specific to RNA.
The c-di-AMP recognition motifs of protein and RNA targets suggest that they may differentially bind to chemically modified versions of the ligand. We have previously reported analogs of c-di-GMP, a related second messenger employed in bacterial signaling, that can selectively target the two identified classes of the c-di-GMP riboswitch.49 Furthermore, it has been shown that modifications of a c-di-GMP phosphate can decrease affinity for the class I riboswitch without affecting affinity for the PDE, RocR.50,51 This suggests that it may be possible to specifically target downstream effectors of c-di-AMP using synthetic analogs that take advantage of the different recognition strategies utilized by the RNA and proteins.
Here, we confirmed that both binding sites of the ydaO riboswitch are physiologically relevant and recognize c-di-AMP with similar affinities. We performed a structure-activity relationship study of c-di-AMP binding to RNA and protein targets. We also tested for modifications that render the second messenger resistant to PDE-catalyzed hydrolysis. We used a series of c-di-AMP analogs to systematically perturb the interactions made between the target molecules and the bases and ribosyl-phosphate backbone of the ligand. We identified the functional elements of c-di-AMP that differentially affect binding to the ydaO riboswitch and the GdpP (formerly YybT) phosphodiesterase protein. Furthermore, we discovered modifications that improve the resistance of c-di-AMP derivatives to enzymatic degradation. These second-messenger analogs could potentially be used as tools to differentially activate c-di-AMP signaling pathways.
MATERIALS AND METHODS
Materials
C-di-AMP was purchased from Biolog or synthesized enzymatically as previously described.52 Nucleotide analogs of c-di-AMP were chemically synthesized on solid support using phosphoramidites purchased from either ChemGenes or Glen Research. The controlled pore glass (CPG) solid support, 3-(4,4’-dimethoxytrityloxy)-2,2-(dicarboxymethylamido)propyl-1-O-succinoyl long chain alkylamino-CPG (3’-CPR-II CPG), was purchased from Glen Research. DNA/RNA synthesis grade acetonitrile (ACN), anhydrous pyridine, triethylamine (TEA), 1-(mesitylene-2-sulfonyl)-3-nitro-1,2,4-triazole (MSNT), and triethylamine trihydrofluoride (HF-TEA) were purchased from Sigma-Aldrich. The oxidation reagent tert-butyl hydroperoxide/toluene was prepared following published procedures.53 All RNA molecules were cloned and transcribed in vitro using T7 RNA polymerase.52
Chemical Synthesis of c-di-AMP Analogs
Phosphorothioate-modified analogs were synthesized as described.54 All analogs containing a standard phosphodiester backbone were synthesized using solid phase chemistry (Scheme S1) on a 2 µmol scale as previously described49 with the following adaptations. 5’-DMTr-2’-OTBDMS cyanoethyl phosphate-protected adenosine phosphoramidites (100 mM in acetonitrile (ACN)) were coupled to the solid support using 5-benzylmercaptotetrazole (125 mM in ACN) as the activator. All molecules were purified by high-performance liquid chromatography (HPLC) on a C18 reverse-phase column using a gradient of 0–10% ACN in 50 mM triethylammonium acetate (TEA Ac), pH 6.0. The identity of all compounds was confirmed by Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) in negative ion mode (Table S1), and purity determined by analytical HPLC.
Kd measurements of c-di-AMP by gel-shift and Kd measurements of analogs by competition gel-shift
Radiolabeled c-di-AMP was enzymatically synthesized using the purified DisA diadenylate cyclase protein as previously described.52 The wild-type ydaO aptamer from Bacillus subtilis was transcribed in vitro using T7 RNA polymerase. RNA conformation was checked by fast protein liquid chromatography (FPLC) and ydaO was shown to form monomer and dimer (Figure S1). To achieve a homogenous population of monomer, RNA was folded in the presence of trace amounts of radiolabeled c-di-AMP by heating to 70°C for 3 minutes and slow cooling in folding buffer (10 mM NaCl, 10 mM MgCl2, and 10 mM sodium cacodylate, pH 6.8). Binding reactions were incubated at room temperature (22°C) until equilibrium was reached (at least 12 hours). The Kd of c-di-AMP for the riboswitch was then measured by separating free c-di-AMP from RNA-bound c-di-AMP by native polyacrylamide gel electrophoresis (PAGE) (100 mM Tris/HEPES pH 7.5, 10 mM MgCl2, 0.1 mM EDTA) at 4°C and the fraction of c-di-AMP bound (FB) at each RNA concentration was determined. Gels were scanned using a STORM phosphorimager (GE Healthcare) and ImageQuant (GE Healthcare) was used to quantify bands. Data were fit to the following equation to determine Kd values:
| (1) |
with FB∞= fraction of c-di-AMP bound at RNA saturation, RT= total RNA concentration, and n = Hill coefficient.
Competition experiments to determine the Kd of analogs were performed under similar conditions. In this case, radiolabeled c-di-AMP and unlabeled competitor analog were premixed in folding buffer before adding RNA to a final concentration of 25 nM. RNA was heated to 70°C for 3 min and slow cooled in the presence of both labeled and unlabeled ligand and incubated at room temperature for 12–24 hours before resolving free and bound c-di-AMP by native PAGE. We observed no changes in the measured binding affinities at incubation times longer than 12 hours, indicating that equilibrium had been achieved. The fraction bound (FB) of labeled c-di-AMP was quantified and the Kd of the unlabeled competitor analog was determined from the following equation as previously described49:
| (2) |
where FB∞= fraction bound of c-di-AMP at saturating concentrations of competitor analog, FB0= fraction bound in the absence of competitor, CcdiA= concentration of labeled c-di-AMP (estimated as 0.025 nM based on the efficiency of the enzymatic labeling reaction), KdcdiA= affinity of c-di-AMP for the riboswitch, CT= concentration of unlabeled competitor analog, KdC= affinity of competitor analog, and RT=total concentration of riboswitch RNA.
To determine the change in binding energy (ΔΔGbind), the binding energy (ΔGbind) of each analog was first obtained from the following equation:
| (3) |
Where R = universal gas constant and T = temperature. ΔΔGbind was then calculated according to:
| (4) |
Expression and Purification of Phosphodiesterase Protein GdpP from Bacillus subtilis
Six-histidine-tagged phosphodiesterase protein GdpP (formerly YybT) was purified by affinity chromatography using Ni-NTA agarose (Qiagen) as previously described.27 Briefly, overnight cultures of Escherichia coli BL21 cells harboring the expression plasmid pET-28(a+) encoding the cytoplasmic portion of GdpP (AA84–659) were grown at 37°C. The overnight culture was diluted into 1 liter of fresh LB medium, and cells were grown to an OD600 of 0.8 before inducing with 0.8 mM isopropyl β-D-thiogalactopyranoside (IPTG). The culture was grown at 16°C for 12 hours before it was pelleted by centrifugation. The cells were lysed in 20 mL of lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol, 6 mM BME, and 0.2 mg/mL lysozyme) via sonication. The lysate was cleared by centrifugation (25000 rpm for 30 min) and filtered before incubating with Ni-NTA agarose for 1 hour at 4°C. The resin was washed with 50 mL of W1 buffer (lysis buffer with 20 mM imidazole) and 20 mL of W2 buffer (lysis buffer with 50 mM imidazole). The proteins were eluted using a step gradient method with elution buffers containing 50 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol, and 200, 300, or 500 mM imidazole. After analysis by SDS-PAGE, pure fractions were pooled and dialyzed into 50 mM Tris, pH 8.0, 150 mM NaCl, and 5% glycerol. The proteins were stored at −80°C after flash freezing, and concentration measurement was by the Bradford assay method.
Phosphodiesterase Assays for Evaluating Degradation Rates of c-di-AMP and Its Analogs
Phosphodiesterase activity was evaluated under steady-state reaction conditions. Pure GdpP protein (1 µM) was incubated with 100 µM c-di-AMP (or analog) in PDE buffer (100 mM Tris, pH 8.3, 20 mM KCl, 500 µM MnCl2) at 37°C. Aliquots were removed at each time point and the reaction was quenched with 1 mM EDTA. Samples were diluted in 50 mM TEA Ac buffer (pH 6.0), and compound degradation was analyzed by HPLC on a reverse-phase C8 column using a gradient from 0–4% ACN in 50 mM TEA Ac (pH 6.0) over 15 min for most compounds. A steeper gradient was used for the following compounds as indicated: 0–15% ACN for c-di-(RpRp)-AMPPS, and 0–10% ACN for c-di-(RpSp)-AMPPS. Degradation was monitored in the range where the reaction was still linear to measure the initial velocity. The area under peaks corresponding to linear and cyclic compounds was determined by integration (HPChem Software), and the fraction of linear (FL) product formed was calculated using the following equation:
| (5) |
where areal is the area of the linear product and areac is the area of the cyclic compound. FL was multiplied by the substrate concentration to obtain the amount of product formed, which was plotted against time. The data were fit to a line, and the slope of the line was used to determine the initial rate.
Km Determination of c-di-AMP
The Km of c-di-AMP was determined using the HPLC assay described above. C-di-AMP was incubated with 200 nM GdpP in 1x PDE buffer. The initial rate of degradation was measured for substrate concentrations ranging from 3.1 µM to 800 µM. The initial velocity vs. substrate concentration was plotted and data were fit to the following equation to determine the Km:
| (6) |
where V0 = initial velocity, Vmax = maximum reaction velocity, and [c-di-AMP] = concentration of c-di-AMP substrate.
Competition Phosphodiesterase Assays with GdpP-Resistant Analogs
The degradation of radiolabeled c-di-AMP (*c-di-AMP) to pApA was monitored by determining the fraction of pApA formed by separating cyclic and linear products by polyethyleneimine-cellulose thin layer chromatography (PEI-cellulose TLC). For competition assays, GdpP (1 µM) was incubated with trace amounts of *c-di-AMP, unlabeled c-di-AMP (100 µM), and an excess of unlabeled competitor analog (500 µM) at 37°C in PDE buffer. Aliquots were removed at each time point and reactions quenched by the addition of an equal volume of 0.5 M EDTA. Reaction mixtures were extracted with a phenol/chloroform/isoamyl alcohol mixture (25:24:1) to remove the protein before analysis by PEI-cellulose TLC. PEI-cellulose TLC plates were run in a 1:1.5 (v/v) mixture of saturated NH4SO4 and 1.5 M KH2PO4 (pH 3.6). The plates were scanned and the amounts of c-di-AMP and pApA were quantified as detailed above. The fraction of pApA was plotted against time and the data were fit to a line to determine the initial velocity. The following equation was used to determine the affinity (Ki) of analogs for GdpP.
| (7) |
where V0 = initial velocity, Vmax = maximum reaction velocity (15.6 min−1), [c-di-AMP] = concentration of c-di-AMP substrate (100 µM), and [I] = concentration of analog (500 µM).
RESULTS AND DISCUSSION
Affinity Measurements of c-di-AMP for the Discrete Binding Sites of the ydaO Riboswitch
The crystal structure of the ydaO riboswitch identified two pseudo-symmetric c-di-AMP binding sites. This was further supported by isothermal titration calorimetry that showed a 2:1 binding stoichiometry.45–47 While it is possible that both sites are relevant, the experimental conditions employed required large quantities of both ligand and RNA. Mutant constructs have also been reported that disrupt the A-minor interactions in the individual binding pockets and reduce binding stoichiometry to 1:1.45 The G-C pairs that contact the adenine bases of c-di-AMP were mutated to C-G pairs to perturb either Site 1 or 2, while preserving helices and maintaining the overall RNA structure (Figure 2a). No measurable binding to Site 2 was reported when Site 1 was mutated in this way (ΔSite1); further suggesting that Site 2 may have a non-physiologically relevant binding affinity.45
We set out to measure ligand binding for each site of the ydaO riboswitch at lower, more physiological RNA concentrations, using a variation of a previously reported gel-shift assay used to study the c-di-GMP binding riboswitches.49,52 We found that the binding curve of c-di-AMP for the wild-type ydaO riboswitch is best fit to a cooperative binding model with a Kd of 0.43 nM and a Hill coefficient of 1.6 (Table 1; Figure 3a,b). This is in agreement with previously published values ranging from mid-picomolar to low nanomolar.44–47 The two binding-site mutants, described above, were also tested to determine the affinity of c-di-AMP for the individual sites in isolation. The measured affinity of the ΔSite1 mutant was 5.5 nM, while the ΔSite2 mutant affinity was 3.3 nM, with Hill coefficients of 1 (Table 2; Figure S2). This suggests that only one binding site is functional and that both binding sites of the ydaO riboswitch recognize c-di-AMP with near identical affinity, in contrast to previously published reports. Mutating both binding sites in a single construct disrupted c-di-AMP affinity for the riboswitch.
Table 1.
Binding affinities of analogs measured for the ydaO riboswitch
| Modification | Analog | Kd (nM) | Fold Loss | ΔΔGbind (kcal/mol) |
|---|---|---|---|---|
| c-di-AMP | 0.43 ± 0.02 | |||
| Base | c-AMP-GMP | > 2.3 × 106a | > 4.2 × 106 | > 9.0 |
| c-di-GMP | > 1.1 × 107 | > 2.0 × 107 | > 10 | |
| c-AMP-IMP | 750 ± 43 | 1400 | 4.3 | |
| c-di-IMP | 2000 ± 460 | 3600 | 4.9 | |
| c-AMP-Purine | 18 ± 1.6 | 33 | 2.1 | |
| c-di-Purine | 2900 ± 270 | 5300 | 5.1 | |
| c-di-N6m,N6mAMP | > 5 × 105 | > 9.1 × 105 | > 8.1 | |
| c-AMP-DAP | 360 ± 38 | 650 | 3.8 | |
| c-di-DAP | > 1.5 × 106 | > 2.7 × 106 | > 8.8 | |
| c-di-2AP | > 2.5 × 106 | > 4.5 × 106 | > 9.1 | |
| c-di-c7AMP | 6.4 ± 0.6 | 12 | 1.5 | |
| Ribose | c-dA-AMP | 11 ± 1.6 | 20 | 1.8 |
| c-di-dAMP | 160 ± 13 | 290 | 3.4 | |
| c-2’F-A-AMP | 26 ± 2.4 | 47 | 2.3 | |
| c-di-2’F-AMP | 2800 ± 330 | 5100 | 5.1 | |
| c-dA-2’F-A | 290 ± 23 | 530 | 3.7 | |
| c-2’OMe-A-AMP | 340 ± 41 | 620 | 3.8 | |
| c-di-2’OMe-AMP | > 1.6 × 106 | > 2.9 × 106 | > 8.8 | |
| Phosphate | c-(RpRp)-di-Aps | 1.0 ± 0.1 | 1.8 | 0.4 |
| c-(RpSp)-di-Aps | 1.6 ± 0.4 | 2.9 | 0.6 |
No detectable binding at the highest ligand concentration tested (≥200 µM). Kd values were estimated based on a 5% detection limit.
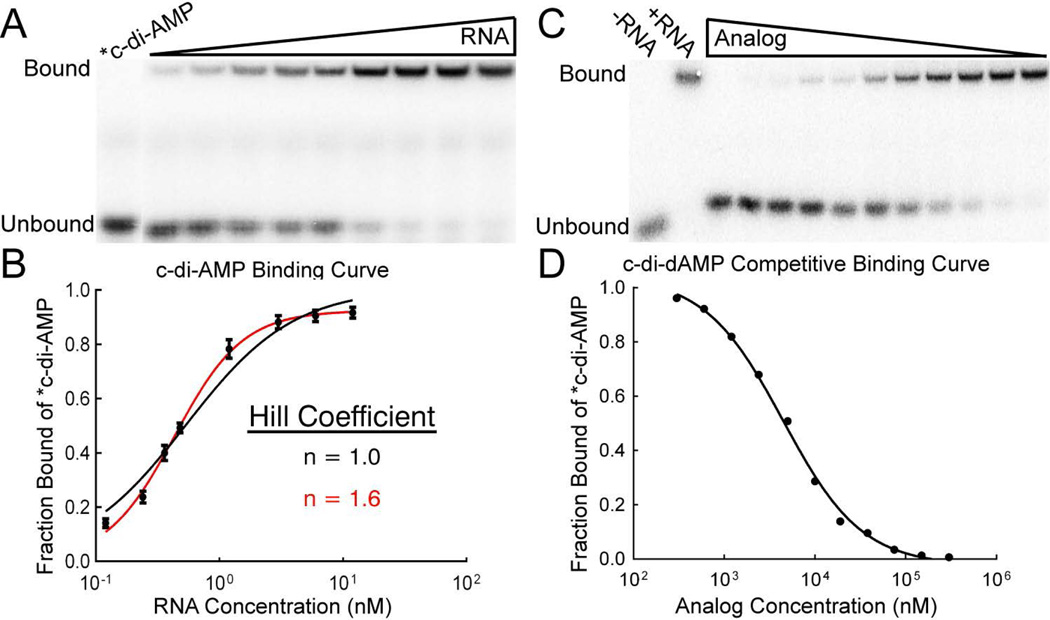
Figure 3.
Kd measurements of c-di-AMP by gel-shift and its various analogs using the competition gel-shift assay with radiolabeled c-di-AMP. (A) Representative gel-shift experiment for measuring the Kd of c-di-AMP for the ydaO RNA by direct binding. (B) c-di-AMP binding curve for the ydaO riboswitch. (C) Representative competition gel-shift experiment. RNA, radiolabeled c-di-AMP, and increasing concentrations of competitor analog are incubated until equilibrium is achieved. Free c-di-AMP is separated from RNA-bound c-di-AMP by native PAGE. (D) Sample binding curve from the competition gel-shift assay with c-di-dAMP. Data are fit to an equation for competitive binding to determine the analog Kd.
Table 2.
Binding affinities of c-di-AMP and dithiophosphate analogs measured for ydaO binding site mutants.
| Ligand Kd (nM) | |||
|---|---|---|---|
| Constructa | c-di-AMP | c-(RpRp)-di-Aps | c-(RpSp)-di-Aps |
| WT144 | 0.43 ± 0.02 | 1.0 ± 0.1 | 1.6 ± 0.4 |
| ΔSite1 | 5.5 ± 0.3 | 11.3 ± 0.8 | 9.4 ± 1.1 |
| ΔSite2 | 3.3 ± 0.5 | 6.2 ± 0.5 | 5.4 ± 0.4 |
Construct sequences are described in Figure 1.
The adenine bases of c-di-AMP are recognized via a type-I A-minor interaction, a well characterized and ubiquitous RNA tertiary element.48 Substantial literature precedent has shown that a type-I A-minor motif is able to form with all canonical Watson-Crick base pairs (a function that is critical for successful recognition of the correct tRNA in the ribosomal A site), as the minor grooves are isosteric.48,59–62 Additionally, the phylogenetic conservation of the four G-C base pairs is not absolute. In 7–11% of constructs the G-C pairs in helices P3 and P7 are observed as C-G pairs, while in these same helices G-U pairs appear very infrequently, <0.2% of the time. The P1 and P5 helices are more highly conserved as G-C pairs, with <2% C-G and <0.8% G-U. We hypothesized that mutation of the conserved G-C pairs to noncanonical G-U wobbles would be more detrimental to the A-minor interaction and provide a better indication of individual binding site affinity. The affinity of c-di-AMP for these constructs, Site 1U and Site 2U, was less than 10-fold lower than the ΔSite mutants, and also had Hill coefficients of 1 (Figure S2). The larger effect on affinity suggests that the G-U mutations do have a larger disruptive effect on the binding sites. Taken together, these data support the crystallographic identification of two binding sites and suggest that both are functional at physiologically relevant c-di-AMP concentrations.
Effects of Base Modifications on Ligand Affinity for the ydaO Riboswitch
Bacteria use a variety of nucleotide-based signaling molecules in addition to c-di-AMP, including the structurally similar second messengers c-di-GMP and c-AMP-GMP, to regulate cellular function.3,4,63 It is critical for riboswitch control that the RNA can distinguish between its cognate ligand and structurally similar molecules. Nelson and colleagues have demonstrated that the ydaO riboswitch selectively binds c-di-AMP over c-di-GMP and c-AMP-GMP by more than 100,000-fold.44
The crystal structure has identified interactions between c-di-AMP and the ydaO riboswitch, but the basis for how this receptor differentiates between other cyclic dinucleotides remains unexplored. To perturb individual interactions between the RNA and ligand, we designed, and chemically synthesized, a series of c-di-AMP analogs, systematically modifying the bases (Figure 4a), ribose rings, and phosphate backbone (Figure 4b) of the second messenger. We prepared both symmetric (modification of both AMP units) and asymmetric (modification of a single AMP unit) versions of the analogs. Radiolabeled c-di-AMP (*c-di-AMP) was prepared enzymatically, but it is not feasible to prepare cyclic dinucleotide analogs using enzymatic synthesis. To measure the affinities of the unlabeled analogs, we used a variation of a previously reported competition gel-shift assay with *c-di-AMP.49,52 RNA and *c-di-AMP were incubated in the presence of increasing concentrations of the unlabeled competitor analog, and the fraction of free *c-di-AMP was monitored to determine the affinity of the competitor (Figure 3c,d). As both RNA binding sites had similar affinities for c-di-AMP and are structurally symmetrical, we expected no difference in analog recognition between the two sites. We confirmed this for a subset of the analogs described below. Therefore, we tested analog binding to the wild-type ydaO riboswitch, with both binding pockets intact, to measure the most relevant affinities.
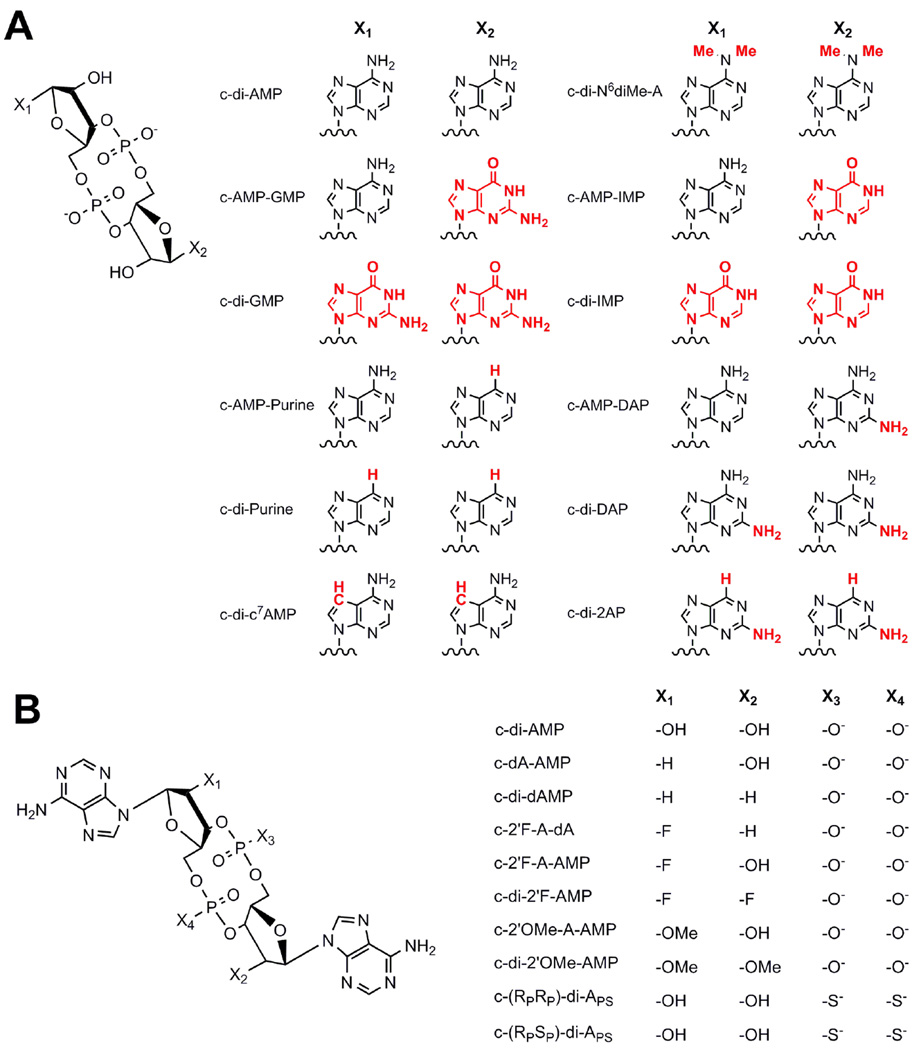
Figure 4.
Structures of c-di-AMP analogs used in this study. X indicates where modifications were made in each series. (A) Base, (B) Ribose, and Phosphate-modified analogs.
The crystal structure revealed an extensive hydrogen-bonding network to the adenine bases of c-di-AMP on both the Watson-Crick and Hoogsteen faces. The Watson-Crick edge of the adenines is buried in the minor groove of a riboswitch G-C base pair. A single hydrogen bond is observed between both the N1 and N3 of c-di-AMP and the O2’ and N2 of the guanine, respectively. Furthermore, the guanine N3 is in close proximity to the C2 of adenine (Figure 2b).45–47 Modifications to the adenine base that make it more similar to guanine would likely affect these contacts and decrease binding affinity. We wanted to explore how well the ydaO riboswitch can discriminate against other cyclic-dinucleotide second analogs.44–47 Consistent with previous reports, we observed no detectable binding to c-AMP-GMP or c-di-GMP in these assays.44–46
The exocyclic amine of c-di-AMP is predicted to make two hydrogen bonds to the backbone of the riboswitch (Figure 2b).45–47 This suggests that it is functionally important for RNA-ligand recognition. To test this prediction, we measured the affinities of inosine (c-AMP-IMP, c-di-IMP), purine (c-AMP-Purine, c-di-Purine), and N6-methyl (c-di-N6diMe-AMP) adenosine analogs. While inosine substitutions measure the negative effect on binding of having a carbonyl at the C6 position, purine analogs would identify the positive effect of the presence of the exocyclic amine. Modification of both bases to either purine or inosine resulted in a loss in binding energy of 5.1 and 4.9 kcal/mol, respectively, suggesting that the riboswitch binds equally poorly to these two molecules (Table 1). To deconvolute the importance of these modifications, we tested the ligand affinity of the asymmetric analogs c-AMP-Purine and c-AMP-IMP. Interestingly, twice the amount of binding energy was lost for c-AMP-IMP (4.3 kcal/mol), as compared to c-AMP-Purine (2.1 kcal/mol, Table 1). The structural similarity between inosine and guanine could explain this larger decrease in binding affinity, given that the riboswitch must be able to functionally distinguish between the carbonyl and amine at this position. We expected the N6-methyl analog to have a larger impact on affinity than the purine and inosine analogs due to the increased steric bulk. As predicted, no detectable binding was observed for c-di-N6diMe-AMP (Table 1).
To identify the effect of introducing an exocyclic amine at the C2 position, comparable to that of guanosine, we tested the base analogs 2-aminopurine (c-di-2AP) and diaminopurine (c-AMP-DAP, c-di-DAP). We had expected that the 2AP analog would have weaker affinity than DAP because the inclusion of the C6 amine would rescue some binding. However, no detectable binding was observed for either c-di-2AP or c-di-DAP at the highest analog concentration tested, representing a loss in binding energy of more than 8.8 kcal/mol for both analogs (Table 1). This suggests that added steric bulk at the C2 position, regardless of the presence of the native exocyclic amine, hinders binding to the riboswitch. This is most likely due to clashes with the nearby guanine base of the riboswitch. Such clashes would disrupt the type-I A-minor interaction and tight packing of the ligand base into the minor groove of the RNA helix.48 Approximately 3.8 kcal/mol of binding energy was lost for c-AMP-DAP, indicating that added steric bulk can be tolerated on one base but heavily destabilizes the RNA-ligand interaction. These data suggest selection against the C2 amine is a stronger determinant of base specificity than selection for the C6 carbonyl (Table 1).
We wanted to demonstrate that all modifications to the base were not equally detrimental by disrupting a contact that was non-specific to adenine or guanine. A single hydrogen bond is predicted between the N7 nitrogen of the ligand and the RNA base that also contacts the exocyclic amine (Figure 2b). We hypothesized that a 7-deazaadenosine analog would disrupt this interaction, while not being structurally characteristic of either purine nucleobase. We observed a 1.5 kcal/mol loss in binding energy for c-di-c7AMP (Table 1). This modest effect suggests that modifications that do not disrupt functional groups on the Watson-Crick face are less detrimental to binding. Additionally, these results indicate that the Watson-Crick edge is the primary means of ligand recognition for the ydaO riboswitch selecting against moieties that mimic c-AMP-GMP or c-di-GMP.
Effects of Modifications to the Ribose Rings on Ligand Affinity for the ydaO Riboswitch
We also examined the effects of modifying the ribose rings of c-di-AMP on ligand affinity for the ydaO riboswitch. In the crystal structure, two hydrogen bonds are observed between the ligand 2’-OHs and the RNA backbone, as part of the A-minor motif (Figure 2c).45–47 To determine the energetic contribution of these interactions to ligand recognition and binding, we tested 2’-deoxy, 2’-fluoro, and 2’-methoxy c-di-AMP analogs (Figure 4b).
A loss in ligand affinity of up to 100-fold for c-di-dAMP was previously reported.44,46 Consistent with these reports, we measure a Kd of 160 nM for the di-deoxy analog. The loss in binding energy was 1.8 kcal/mol for c-dA-AMP and twice that for c-di-dAMP (3.4 kcal/mol, Table 1). This suggests that the ydaO riboswitch recognizes the 2’-OH of c-di-AMP with structural symmetry, consistent with the geometries observed in the crystal structure.45–47
Adenosine analogs with deoxyribose substitutions have been shown to shift the equilibrium of sugar pucker to the 2’-endo conformation.64 C-di-AMP is shown to bind ydaO in the 3’-endo conformation. It is possible the loss in binding affinity for the 2’-deoxy analog may include contributions from altering the ribose structure.45–47,49 To parse out hydrogen bonding effects from ribose conformational effects, we tested 2’-fluoro analogs, a substitution that causes the ribose sugar to preferentially adopt the 3’-endo conformation.64,65 This substitution has been observed in several cases to rescue 2’-deoxy effects that result from conformational changes to the ribose ring.65,66 If a shift in the sugar pucker of the deoxyribose substitution contributes to the loss in binding affinity, we expected the 2’-fluoro analogs to bind with stronger affinity. Instead, we observed the opposite effect with the 2’-fluoro substitutions being more destabilizing than the 2’-deoxy analogs. The binding energy of c-2’F-A-AMP, which contains a single modification, was nearly 0.5 kcal/mol weaker than the corresponding deoxy variant (c-dA-AMP), while the symmetrically modified c-di-2’F-AMP was more than 1.7 kcal/mol weaker than c-di-dAMP (Table 1). To further examine the effect on ligand affinity of 2’-deoxy and 2’-fluoro substituents, the chimeric analog c-dA-2’F-A was synthesized and tested. An improvement in binding energy of nearly 1.4 kcal/mol was measured by replacing a 2’-fluoro substituent with a 2’-deoxy group.
These observations suggest that hydrogen-bonding effects, not the conformation of the ribose sugar, dominate the RNA-ligand interaction and explain the differential loss in ligand affinity. In the ydaO crystal structures, the 3’-Os of c-di-AMP are within hydrogen bond distance of the 2’-OHs of neighboring RNA nucleotides. The 2’-deoxy and 2’-fluoro substitutions would disrupt a putative H-bond with the 2’-OHs of c-di-AMP, which could be compensated by the 3’Os. A conformational shift in c-di-dAMP could enhance these hydrogen bonds as compared to c-di-2’F-AMP and may explain the greater affinity observed for the 2’-deoxy substitutions.
The 2’-methoxy substitutions are expected to create significant steric clashes given the close approach to the 2’-OH in a type I A-minor interaction. The singly substituted analog, c-2’OMe-A-AMP, showed weak binding, with nearly a 3.8 kcal/mol loss in binding energy, while c-di-2’OMe-AMP showed no detectable affinity for the RNA (Table 1). The introduction of a 2’-methoxy was one of the most detrimental single modifications we tested, indicating that the binding pocket is highly selective against additional bulk at the 2’-OH.
Effects of Modifications to the Phosphate Backbone on Ligand Affinity for the ydaO Riboswitch
C-di-AMP binding to the riboswitch also involves contacts to the phosphate. There is a hydrogen bond between the pro-SP non-bridging oxygen and the exocyclic amine of a neighboring guanine (Figure 2c).45–47 Since the covalent radius of sulfur is larger than oxygen and the charge distribution is asymmetric for phosphorothioates, with the negative charge primarily localized on the sulfur, thiophosphate c-di-AMP variants may produce unfavorable electrostatic interactions within the binding pocket.70 To assess this, we measured the affinities of the dithiophosphate analogs [c-(RpRp)-di-Aps and c-(RpSp)-di-Aps, Figure 4b]. We observed only a 0.4 kcal/mol effect on binding energy for c-(RpRp)-di-Aps, and only a 0.6 kcal/mol effect for c-(RpSp)-di-Aps (Table 1). The minimal effect of both the RpRp and RpSp modifications are consistent with the crystal structures as contact between ydaO and the phosphate backbone of c-di-AMP is formed by a single hydrogen bond, and not mediated by metals.45–47,71 Sulfur-containing hydrogen bonds are approximately 0.5 kcal/mol weaker than traditional hydrogen bonds, which is consistent with the magnitude of the effects we measured for both analogs.72,73
To determine if there is differential analog recognition by the discrete binding sites, we tested the affinity of the dithiophosphates for the ydaO binding pocket mutants, ΔSite1 and ΔSite2 (Figure 2a). We measured a 2-fold loss in affinity to both binding site constructs for the RpRp diastereomers when compared to native ligand. A comparable 2-fold effect was observed for the RpSp diastereomer as well. The 10-fold and 6-fold loss in affinities for the ΔSite1 and ΔSite2 mutants, compared with wild-type ydaO, are also mimicked by the dithiophosphate analogs (Table 2). These findings further indicate that c-di-AMP recognition in the individual binding sites is nearly indistinguishable, and ligand modifications result in similar effects in both sites.
C-di-AMP Analog Modifications Effect Enzymatic Hydrolysis Rates
The intracellular concentration of c-di-AMP is directly linked to the phenotypic response induced by downstream targets.5,8,16,22,24–32 Understanding the metabolism of c-di-AMP may allow us to vary the second-messenger concentration and manipulate these phenotypic responses. To determine which modifications alter the specificity of PDE enzymes for c-di-AMP, we measured the rate of hydrolysis for this series of c-di-AMP derivatives by the PDE GdpP from Bacillus subtilis.27
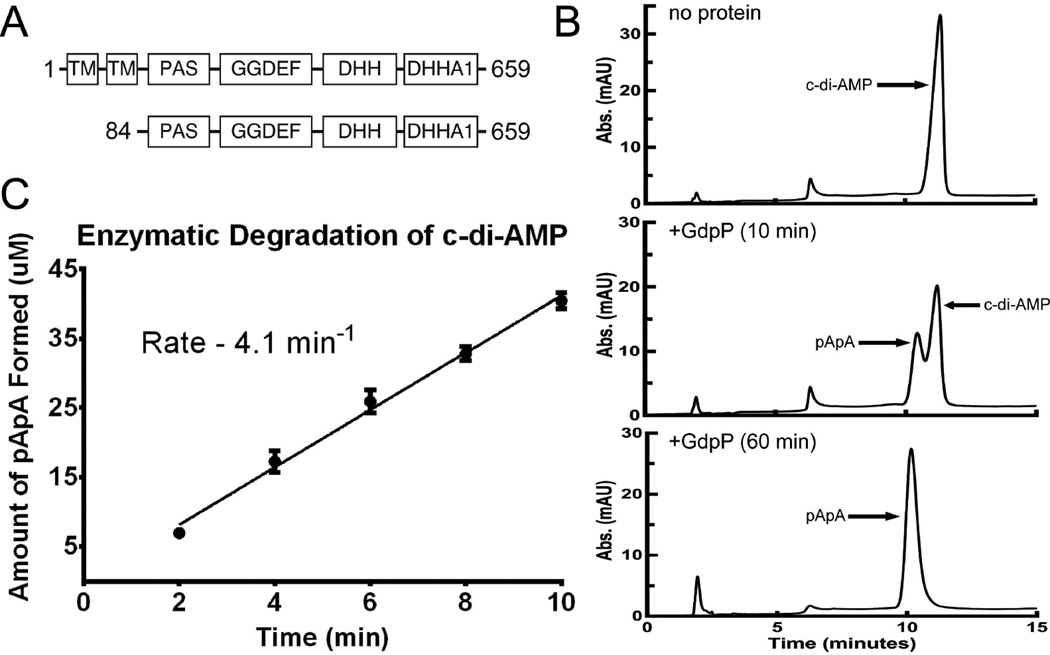
GdpP-family proteins contain two N-terminal transmembrane helices, a heme-responsive PAS domain, a degenerate GGDEF domain, and a DHH/DHHA1 domain (Figure 5a). Sequence alignments and domain-specific constructs showed the DHH/DHHA1 domain possessed phosphodiesterase activity, while the highly modified GGDEF domain exhibited ATPase activity. GdpP has been shown to hydrolyze c-di-AMP and c-di-GMP to pApA and pGpG, respectively. Although GdpP can degrade c-di-GMP, it has a significantly greater Km and is unlikely to be a physiologically relevant substrate.20,27
Figure 5.
Monitoring the enzymatic hydrolysis of c-di-AMP by HPLC. (A) Domain architecture of GdpP-family proteins (1–659) and the protein construct used for enzymatic assay (84–659). (B) HPLC traces showing elution of pApA and c-di-AMP. Peaks corresponding to each of these compounds are labeled. After 60 min under the experimental conditions used, c-di-AMP is completely degraded. (C) The amount of pApA formed was measured to determine the initial velocity of c-di-AMP degradation. Analog degradation was measured and analyzed in the same manner.
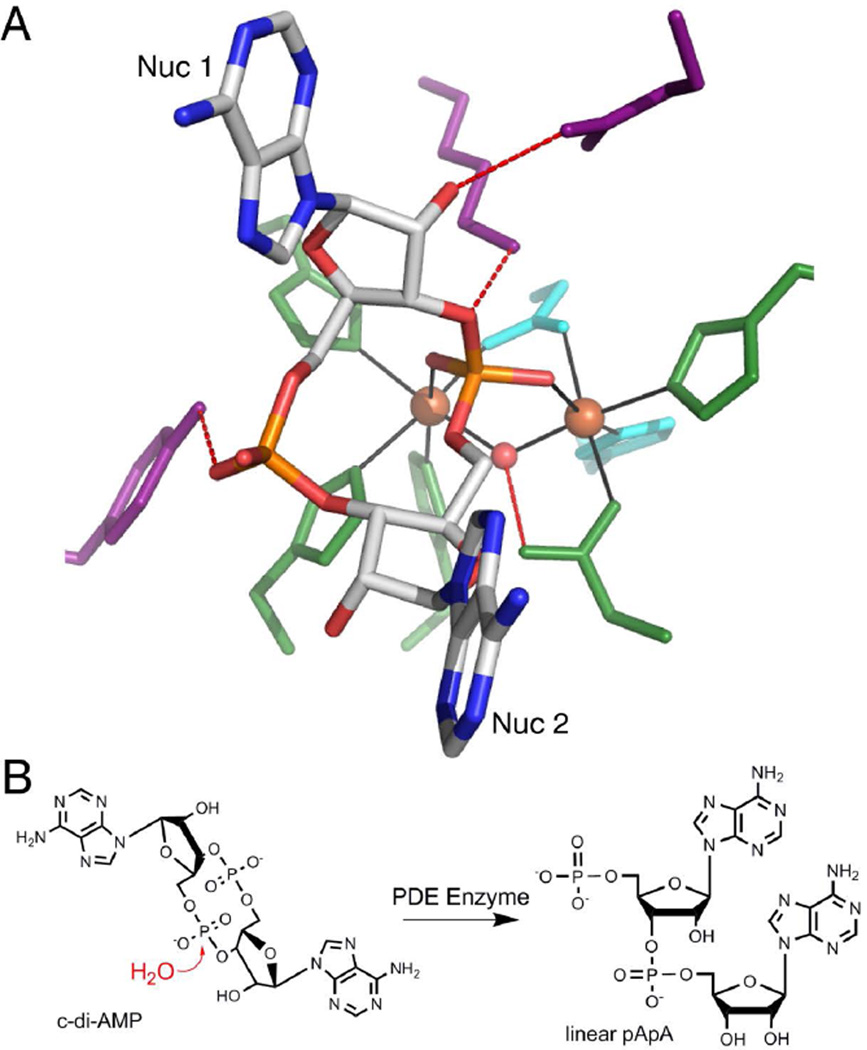
While the structure of GdpP has not been solved, a His-Asp (HD) domain protein from L. monocytogenes, PgpH, has been structurally and biochemically characterized as a c-di-AMP phosphodiesterase.28 The non-bridging oxygens of the active phosphate are coordinated to two Fe metal ions, which in turn coordinate the His and Asp residues, as well as a bridging water (Figure 6a).28 The proposed hydrolysis mechanism involves nucleophilic attack by the activated water on the scissile phosphate to yield linear pApA (Figure 6b). Although the GdpP-family proteins have not been structurally characterized, they are also two-metal Mn-dependent proteins and are predicted to act via a similar mechanism to HD-domain PDEs.6,28
Figure 6.
Recognition and enzymatic hydrolysis of c-di-AMP by PDE proteins (ref. 28). (A) c-di-AMP bound to the active site of HD domain protein PgpH from L. monocytogenes (PDB entry 4S1B). c-di-AMP is colored by atom with carbon in white, oxygen in red, nitrogen in blue, and phosphorous in orange. Ligand binding residues are colored in purple, metal coordinating residues are in green, and the catalytic histidine and aspartic acid are in cyan. Hydrogen-bonding interactions are shown as dashed lines (red) and metal-coordinating interactions are displayed as solid lines (black). Iron metals are shown as orange spheres and the nucleophilic water is a red sphere. (B) Proposed mechanism of c-di-AMP cleavage. In the presence of a PDE protein, a nucleophilic water molecule attacks one of the c-di-AMP phosphodiester bonds to produce the linear 5’-pApA.
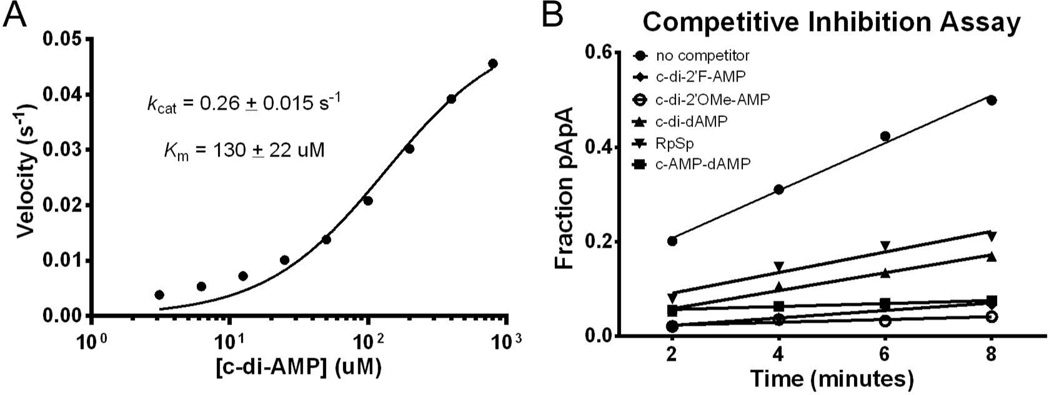
We utilized an HPLC-based assay to measure the rate of degradation for the series of analogs with single and double substitutions by the cytoplasmic portion of GdpP (GdpP84–659, Figure 5a). As a control, c-di-AMP linearization was observed via HPLC by monitoring the emergence of a peak corresponding to pApA (retention time – 9.8 min) and the loss of the peak for c-di-AMP (retention time – 10.9 min) (Figure 5b). Co-injection of a chemically synthesized standard of pApA confirmed that the degradation peak was the correct breaKdown product (Figure S3). The initial rate of c-di-AMP degradation was measured under steady-state conditions with increasing amounts of substrate (3 to 800µM). The initial velocity was plotted against substrate concentration to estimate kcat. Under these conditions, the rate of c-di-AMP hydrolysis was 0.26 ± 0.02 s−1 (Figure 7a). This value is in reasonable agreement with the previously published value (0.55 ± 0.02 s−1).27 The rate of hydrolysis of c-di-AMP analogs was also measured under steady-state conditions using the HPLC assay. However, due to limited analog stocks, we were unable to confirm that the rates measured were under saturating conditions. Therefore, we could not in all cases determine if a reduction in degradation is due to a kcat or a Km effect. All experiments were run using 100 µM substrate and the initial rates were compared to that measured for 100 µM c-di-AMP (4.1 min−1, Table 3 and Figure 5c).
Figure 7.
Degradation of c-di-AMP by GdpP. (A) Steady-state kinetic analysis of the enzyme-catalyzed hydrolysis of c-di-AMP by GdpP84-659. (B) Fraction of pApA formed from degradation of radiolabeled c-di-AMP over time in the presence of unlabeled c-di-AMP analogs.
Table 3.
Rates of enzymatic hydrolysis for c-di-AMP and its nucleotide analogs by phosphodiesterase protein GdpPa
| Modification | Analog | Rate [mol min−1 (mol of enzyme)−1]b |
Fold Change |
|---|---|---|---|
| c-di-AMP | 4.1 ± 0.2 | - | |
| Base | c-AMP-GMP | 2.4 ± 0.3 | 1.7 |
| c-di-GMP | 0.8 ± 0.1 | 5.1 | |
| c-AMP-IMP | 1.3 ± 0.2 | 3.3 | |
| c-di-IMP | ≤ 0.001 c | ≥ 4100 | |
| c-AMP-Purine | 1.1 ± 0.1 | 3.8 | |
| c-di-Purine | 0.11 ± 0.01 | 36 | |
| c-di-N6m,N6mAMP | ≤ 0.001 | ≥ 4100 | |
| c-AMP-DAP | 1.8 ± 0.3 | 2.2 | |
| c-di-DAP | ≤ 0.001 | ≥ 4100 | |
| c-di-2AP | 1.7 ± 0.2 | 2.4 | |
| c-di-c7AMP | 2.3 ± 0.2 | 1.8 | |
| Ribose | c-dA-AMP | 3.5 ± 0.3 | 1.2 |
| c-di-dAMP | ≤ 0.001 | ≥ 4100 | |
| c-2’F-A-AMP | 0.17 ± 0.01 | 24 | |
| c-di-2’F-AMP | ≤ 0.001 | ≥ 4100 | |
| c-dA-2’F-A | ≤ 0.001 | ≥ 4100 | |
| c-2’OMe-A-AMP | 0.14 ± 0.01 | 30 | |
| c-di-2’OMe-AMP | ≤ 0.001 | ≥ 4100 | |
| Phosphate | c-(RpRp)-di-Aps | ≤ 0.001 | ≥ 4100 |
| c-(RpSp)-di-Aps | ≤ 0.001 | ≥ 4100 |
All data are the average of at least three independent trials.
Rates measured under multiple-turnover conditions with 100 µM substrate and 1 µM enzyme.
No degradation under the conditions tested after incubation with GdpP for 24 h. Rates were estimated using a detection limit of 2%.
Phosphate modifications are expected to affect analog linearization as they directly disrupt the site of enzyme catalysis. No detectable degradation was observed upon incubation of c-(RpRp)-di-Aps and c-(RpSp)-di-Aps with GdpP, even after 24 hours (Table 3 and Figure S4). Under these conditions, GdpP is still active with a modest decrease in activity (data not shown). For both dithiophosphate analogs, only the mass of the cyclic product was detected via electrospray ionization mass spectrometry (ESI-MS). This confirmed that the linear and cyclic products were not coeluting in the single peak observed on HPLC. We have previously demonstrated that dithiophosphate analogs of c-di-GMP are resistant to enzymatic degradation by EAL domain proteins.66. Together, these data suggest that replacing both of the phosphodiester bonds with phosphorothioate linkages results in a PDE-resistant analog.
Based on previous structural analysis with c-di-GMP PDEs, the 2’-OH groups are not expected to be critical for recognition.74–76 Furthermore, the PgpH structure only predicts one hydrogen bond to the 2’-hydroxyl of nucleotide 2 (Figure 6a).28 However, the spatial proximity of this functional group to the scissile phosphates may affect the rate of catalysis by interacting with moieties in the active site. To test this, we measured the rate of linearization for the 2’-deoxy, 2’-fluoro, and 2’-O-methyl-modified analogs (Figure 4b). Surprisingly, no degradation was detected after 24-hour incubation with GdpP for the disubstituted 2’OH analogs (Table 3). This indicates that modification of both ribose rings renders the second messenger resistant to enzyme-catalyzed hydrolysis. It is possible that the 2’-OHs play a role in positioning c-di-AMP in the catalytic site and in product release, which could explain the observed resistance.
The rate of analog degradation is influenced by successful binding of the ligand, as well as enzyme-mediated chemistry. Given the limit of detection of the HPLC assay, the rate of hydrolysis of both varieties of nuclease-resistant analogs can be estimated to be no greater than 0.001 min−1. This represents a reduction of the rate of cleavage of at least 4000-fold, compared to c-di-AMP. These linearization deficits could also be explained by a weaker affinity for GdpP. In order to distinguish between these two possibilities, we used a competitive inhibition assay.66 GdpP and c-di-AMP, with trace *c-di-AMP, were incubated in the presence of excess competitor analog. If the analog is able to bind GdpP in the active site, the initial velocity of c-di-AMP hydrolysis should decrease. In the presence of c-dA-AMP, which is hydrolyzed at nearly the same rate, implying it binds to GdpP, there is a measurable deficiency in linearization. This establishes that molecules capable of binding GdpP can limit c-di-AMP degradation (Figure 7b). All nuclease-resistant analogs were tested at concentrations from 50 µM to 5 mM in order to estimate Ki at several inhibitor concentrations.
All resistant analogs are able to bind GdpP. We found that all four ribose-modified analogs (c-di-dAMP, c-di-2’F-AMP, c-di-2’OMe-AMP, and c-dA-2’F-A) reduced degradation velocity when incubated with the enzyme. The Ki was determined to be 200 µM for c-di-dAMP (Table 4), which represents only a 1.5-fold decrease in affinity compared to the Km of c-di-AMP (measured as 130 µM). Alternatively, the binding energies of the 2’-fluoro, 2’-methoxy, and 2’-deoxy/fluoro analogs all showed better binding than c-di-AMP (Table 4). At the concentrations tested, the analogs bind the enzyme, suggesting that the observed resistance to GdpP linearization was because of catalytic rather than binding effects.
Table 4.
Binding affinities of nuclease-resistant analogs measured for the phosphodiesterase protein GdpPa
| Modification | Analog | Ki(µM) | Fold Change |
|---|---|---|---|
| Ribose | c-di-dAMP | 200 ± 18 | 1.5 |
| c-di-2’F-AMP | 26 ± 3.6 | 0.2 | |
| c-dA-2’F-A | 9.4 ± 0.8 | 0.07 | |
| c-di-2’OMe-AMP | 15 ± 2.5 | 0.12 | |
| Phosphate | c-(RpRp)-di-Aps | 260 ± 2.7 | 2.0 |
| c-(RpSp)-di-Aps | 210 ± 17 | 1.6 |
Data are the average of at least three independent trials of inhibitor concentration from 50 µM – 5 mM.
For the dithiophosphate derivatives, we found that both diastereomers were able to slow the rate of c-di-AMP degradation (Figure 7b) consistent with efficient binding to the protein. The Ki of the RpRp and RpSp analogs were 260 µM and 210 µM, respectively. These affinities are within 2-fold of the Km (less than 0.4 kcal/mol), and mimic the binding loss observed between these analogs and the ydaO riboswitch (Table 1).
All asymmetrically substituted analogs were susceptible to PDE degradation (Table 3). No significant effect was observed for the c-dA-AMP analog (1.2-fold), while all other ribose-modified analogs showed a dramatic reduction in the rate of hydrolysis. The monosubstituted 2’-fluoro and 2’-methoxy analogs were degraded 24-fold and 30-fold slower than c-di-AMP, respectively. Modification of a single adenine to a purine, inosine, guanine, or diaminopurine resulted in a hydrolysis rate within 4-fold of that measured for c-di-AMP. This suggests that a single adenine is sufficient for recognition and degradation of the second messenger by this enzyme. Additionally, only a single product peak was observed by HPLC for all asymmetric analogs. This suggests that GdpP preferentially binds the analogs in one orientation. The c-di-AMP binding pocket of the HD-domain protein, PgpH, provides structural insight into these observations. The 5’ phosphate group of nucleotide 2 is bound in the active site. This nucleotide is contacted on one face by a α-helix and solvent exposed on the other face, while nucleotide 1 is inserted into a pocket between two α-helices.28 Recognition of the 2’-hydroxyls is also unequal, with the 2’-OH of nucleotide 1 taking part in a hydrogen bond with a glutamic acid residue, while the nucleotide 2 hydroxyl is not contacted.28 The asymmetric modifications may bind into the more promiscuous nucleotide 2 position while not disrupting catalysis in the active site (Figure 6a).
CONCLUSIONS
C-di-AMP signaling has been connected with a diverse set of processes involved in maintaining cell viability.3,4,6 To date, several proteins and a riboswitch have been identified that respond directly to this small molecule.24,28,36–44 We have shown that the discrete binding sites of the ydaO riboswitch recognize c-di-AMP with low nanomolar affinity. When both sites are active in wild-type RNA, the Kd is in the mid-picomolar range with a Hill coefficient greater than one. The affinities we observed are 100-fold tighter than those previously measured by ITC. This discrepancy is likely because our method involves refolding RNA in the presence of ligand, while c-di-AMP is titrated into prefolded RNA for ITC experiments.
We have also demonstrated that analogs of c-di-AMP have differential selectivity for two receptors in the c-di-AMP signaling pathway. We have described modifications to base, ribose, and phosphate moieties of c-di-AMP that yielded analogs that were resistant to a c-di-AMP-specific phosphodiesterase. Additionally, a subset of these analogs differentially bound a protein and RNA target of this second messenger. Dithiophosphate analogs retained high affinity for both the ydaO riboswitch and GdpP protein and displayed enhanced resistance to enzyme-mediated degradation. In contrast, nuclease-resistant ribose-modified analogs showed a substantial loss in affinity for the riboswitch, while retaining high affinity for the protein. These results suggest that ligand modifications can selectively disrupt recognition elements of specific c-di-AMP receptors, while not affecting binding to other macromolecular targets.
Crystal structures of c-di-AMP bound to several protein targets reveal that unique contacts are made between the ligand and various receptors (Figure 1). The different conformations adopted by c-di-AMP when bound to certain macromolecules could also influence the affinity of analogs. These different binding motifs suggest that discrete analogs, which disrupt unique interactions, may selectively bind specific proteins as well, though this remains to be further explored.
Few examples exist of ligand analogs being used to explicitly target different classes of either RNA or protein.49–51 Given our results, the analogs described could be utilized to selectively target downstream effectors in the c-di-AMP signaling pathway to elucidate which receptors trigger distinct physiological responses. This could be especially useful for distinguishing between the effects of RNA-mediated and protein-mediated second-messenger signaling.
Moreover, the wide distribution of diadenylate cyclase and diguanylate cyclase domains among bacteria suggests that many organisms have the potential to use both c-di-AMP and c-di-GMP as second messengers.3,5,6,27 This has been further supported by the identification of proteins and RNAs that respond to these second messengers in the same bacterial species. We have previously published a series of c-di-GMP analogs that specifically distinguish between the two classes of c-di-GMP riboswitch while being resistant to nuclease degradation.49,62 By combining these two sets of second-messenger analogs, it may be possible to manipulate the diverse biological processes controlled by these ubiquitous signaling molecules. Identifying analogs that are absolutely selective may provide a useful tool for manipulating second-messenger signaling networks.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by National Institutes of Health Grant GM022778 to S.A.S and National Institutes of Health Grant GM79760 to R.A.J.
We would like to thank Dave Hiller, Katie Smith, Brian Dunican, and Michelle Legaspi for helpful discussion and critical comments on the manuscript, and Michelle Legaspi for preliminary work on the analog degradation assays. We would also like to thank the West Campus Analytical Core at Yale University for use of their NMR facility
ABBREVIATIONS
- c-di-AMP
(3’-5’)-cyclic diadenosine monophosphate
- c-di-GMP
(3’-5’)-cyclic diguanosine monophosphate
- c-AMP-GMP
(3’-5’)-cyclic adenosine monophosphate-guanosine monophosphate
- DAC
diadenylate cyclase
- PDE
phosphodiesterase
- PDB
Protein Data Bank
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supplemental results, including four figures, two tables, and one scheme as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Römling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 3.Romling U. Great Times for Small Molecules: c-di-AMP, a Second Messenger Candidate in Bacteria and Archaea. Sci. Signal. 2008;1:e39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- 4.Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013;42:305. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 5.Witte G, Hartung S, Büttner K, Hopfner K-P. Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates. Mol. Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Corrigan RM, Gründling A. Cyclic di-AMP: another second messenger enters the fray. Nat. Rev. Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 7.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP Is a New Second Messenger in Staphylococcus aureus with a Role in Controlling Cell Size and Envelope Stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis σM in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol. 2012;83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O’Gara JP. Methicillin Resistance Alters the Biofilm Phenotype and Attenuates Virulence in Staphylococcus aureus Device-Associated Infections. PLoS Pathog. 2012;8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du B, Ji W, An H, Shi Y, Huang Q, Cheng Y, Fu Q, Wang H, Yan Y, Sun J. Functional analysis of c-di-AMP phosphodiesterase, GdpP, in Streptococcus suis serotype 2. Microbiol. Res. 2014;169:749–758. doi: 10.1016/j.micres.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Cron LE, Stol K, Burghout P, van Selm S, Simonetti ER, Bootsma HJ, Hermans PWM. Two DHH Subfamily 1 Proteins Contribute to Pneumococcal Virulence and Confer Protection against Pneumococcal Disease. Infect. Immun. 2011;79:3697–3710. doi: 10.1128/IAI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz KT, Carleton JD, Quillin SJ, Rollins SD, Portnoy DA, Leber JH. Hyperinduction of Host Beta Interferon by a Listeria monocytogenes Strain Naturally Overexpressing the Multidrug Efflux Pump MdrT. Infect. Immun. 2012;80:1537–1545. doi: 10.1128/IAI.06286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T, Hara H, Tsuchiya K, Sakai S, Fang R, Matsuura M, Nomura T, Sato F, Mitsuyama M, Kawamura I. Listeria monocytogenes Strain-Specific Impairment of the TetR Regulator Underlies the Drastic Increase in Cyclic di-AMP Secretion and Beta Interferon-Inducing Ability. Infect. Immun. 2012;80:2323–2332. doi: 10.1128/IAI.06162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witte CE, Whiteley AT, Burke TP, Sauer J-D, Portnoy DA, Woodward JJ. Cyclic di-AMP Is Critical for Listeria monocytogenes Growth, Cell Wall Homeostasis, and Establishment of Infection. mBio. 2013;4:e00282–e00213. doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. Two DHH Subfamily 1 Proteins in Streptococcus pneumoniae Possess Cyclic Di-AMP Phosphodiesterase Activity and Affect Bacterial Growth and Virulence. J. Bacteriol. 2013;195:5123–5132. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. A mecA-Negative Strain of Methicillin-Resistant Staphylococcus aureus with High-Level β-Lactam Resistance Contains Mutations in Three Genes. Antimicrob. Agents Chemother. 2010;54:4900–4902. doi: 10.1128/AAC.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths JM, O’Neill AJ. Loss of Function of the GdpP Protein Leads to Joint β-Lactam/Glycopeptide Tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012;56:579–581. doi: 10.1128/AAC.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehne FMP, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. Cyclic Di-AMP Homeostasis in Bacillus subtilis BOTH LACK AND HIGH LEVEL ACCUMULATION OF THE NUCLEOTIDE ARE DETRIMENTAL FOR CELL GROWTH. J. Biol. Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye M, Zhang J-J, Fang X, Lawlis GB, Troxell B, Zhou Y, Gomelsky M, Lou Y, Yang XF. DhhP, a Cyclic di-AMP Phosphodiesterase of Borrelia burgdorferi, Is Essential for Cell Growth and Virulence. Infect. Immun. 2014;82:1840–1849. doi: 10.1128/IAI.00030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gándara C, Alonso JC. DisA and c-di-AMP act at the intersection between DNA-damage response and stress homeostasis in exponentially growing Bacillus subtilis cells. DNA Repair. 2015;27:1–8. doi: 10.1016/j.dnarep.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP Secreted by Intracellular Listeria monocytogenes Activates a Host Type I Interferon Response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud A-L, Cambier JC, Lenz LL. MPYS Is Required for IFN Response Factor 3 Activation and Type I IFN Production in the Response of Cultured Phagocytes to Bacterial Second Messengers Cyclic-di-AMP and Cyclic-di-GMP. J. Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu Z-J, Modlin RL, Liu Y, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-Dependent Recognition of Cyclic di-AMP Mediates Type I Interferon Responses during Chlamydia trachomatis Infection. mBio. 2013;4:e00018–e00013. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. Mycobacterium tuberculosis Rv3586 (DacA) Is a Diadenylate Cyclase That Converts ATP or ADP into c-di-AMP. PLoS ONE. 2012;7:e35206. doi: 10.1371/journal.pone.0035206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang Z-X. YybT Is a Signaling Protein That Contains a Cyclic Dinucleotide Phosphodiesterase Domain and a GGDEF Domain with ATPase Activity. J. Biol. Chem. 2010;285:473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh TN, Luo S, Pensinger D, Sauer J-D, Tong L, Woodward JJ. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. 2015;112:E747–E756. doi: 10.1073/pnas.1416485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Li W, He Z-G. DarR, a TetR-like Transcriptional Factor, Is a Cyclic Di-AMP-responsive Repressor in Mycobacterium smegmatis. J. Biol. Chem. 2013;288:3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Gründling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. 2013 doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai Y, Yang J, Zarrella TM, Zhang Y, Metzger DW, Bai G. Cyclic Di-AMP Impairs Potassium Uptake Mediated by a Cyclic Di-AMP Binding Protein in Streptococcus pneumoniae. J. Bacteriol. 2014;196:614–623. doi: 10.1128/JB.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer J-D, Tong L, Woodward JJ. The Cyclic Dinucleotide c-di-AMP Is an Allosteric Regulator of Metabolic Enzyme Function. Cell. 2014;158:1389–1401. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campeotto I, Zhang Y, Mladenov MG, Freemont PS, Gründling A. Complex Structure and Biochemical Characterization of the Staphylococcus aureus Cyclic Diadenylate Monophosphate (c-di-AMP)-binding Protein PstA, the Founding Member of a New Signal Transduction Protein Family. J. Biol. Chem. 2015;290:2888–2901. doi: 10.1074/jbc.M114.621789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller M, Hopfner K-P, Witte G. c-di-AMP recognition by Staphylococcus aureus PstA. FEBS Lett. 2015;589:45–51. doi: 10.1016/j.febslet.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 35.Gundlach J, Dickmanns A, Schröder-Tittmann K, Neumann P, Kaesler J, Kampf J, Herzberg C, Hammer E, Schwede F, Kaever V, Tittmann K, Stülke J, Ficner R. Identification, Characterization, and Structure Analysis of the Cyclic di-AMP-binding PII-like Signal Transduction Protein DarA. J. Biol. Chem. 2015;290:3069–3080. doi: 10.1074/jbc.M114.619619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi PH, Sureka K, Woodward JJ, Tong L. Molecular basis for the recognition of cyclic-di-AMP by PstA, a PII-like signal transduction protein. MicrobiologyOpen. 2015;4:361–374. doi: 10.1002/mbo3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H, Youn S-J, Kim SO, Ko J, Lee J-O, Choi B-S. Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP) J. Biol. Chem. 2015 doi: 10.1074/jbc.M115.641340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 39.Winkler WC, Breaker RR. Regulation of Bacterial Gene Expression by Riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 40.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breaker RR. Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao A, Serganov A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat. Chem. Biol. 2014;10:787–792. doi: 10.1038/nchembio.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren A, Patel DJ. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat. Chem. Biol. 2014;10:780–786. doi: 10.1038/nchembio.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones CP, Ferré-D’Amaré AR. Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA. EMBO J. 2014;33:2692–2703. doi: 10.15252/embj.201489209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shanahan CA, Gaffney BL, Jones RA, Strobel SA. Differential analogue binding by two classes of c-di-GMP riboswitches. J. Am. Chem. Soc. 2011;133:15578–15592. doi: 10.1021/ja204650q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Zhou J, Donaldson GP, Nakayama S, Yan L, Lam Y, Lee VT, Sintim HO. Conservative Change to the Phosphate Moiety of Cyclic Diguanylic Monophosphate Remarkably Affects Its Polymorphism and Ability To Bind DGC, PDE, and PilZ Proteins. J. Am. Chem. Soc. 2011;133:9320–9330. doi: 10.1021/ja1112029. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y, Zhou J, Watt SK, Lee VT, Dayie TK, Sintim HO. Differential binding of 2′ -biotinylated analogs of c-di-GMP with c-di-GMP riboswitches and binding proteins. Mol. Biosyst. 2012;8:772–778. doi: 10.1039/c2mb05338a. [DOI] [PubMed] [Google Scholar]
- 52.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scaringe SA. RNA Oligonucleotide Synthesis via 5′ -Silyl-2′ -Orthoester Chemistry. Methods. 2001;23:206–217. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- 54.Gaffney BL, Veliath E, Zhao J, Jones RA. One-Flask Syntheses of c-di-GMP and the [Rp,Rp] and [Rp,Sp] Thiophosphate Analogues. Org. Lett. 2010;12:3269–3271. doi: 10.1021/ol101236b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill A. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol Lond. 1910;40:4–7. [Google Scholar]
- 56.Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, Smith TM, Yang M, Kurecki T, Gollnick P. The structure of trp RNA-binding attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 57.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A Glycine-Dependent Riboswitch That Uses Cooperative Binding to Control Gene Expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 58.Trausch JJ, Ceres P, Reyes FE, Batey RT. The Structure of a Tetrahydrofolate-Sensing Riboswitch Reveals Two Ligand Binding Sites in a Single Aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the Codon-Anticodon Helix by Ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- 60.Lescoute A, Westhof E. The A-minor motifs in the decoding recognition process. Biochimie. 2006;88:993–999. doi: 10.1016/j.biochi.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 61.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. U. S. A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated Regulation of Accessory Genetic Elements Produces Cyclic Di-Nucleotides for V. cholerae Virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uesugi S, Miki H, Ikehara M, Iwahashi H, Kyogoku Y. A linear relationship between electronegativity of 2′ -substituents and conformation of adenine nucleosides. Tetrahedron Lett. 1979;20:4073–4076. [Google Scholar]
- 65.Micklefield J. Backbone modification of nucleic acids: synthesis, structure and therapeutic applications. Curr. Med. Chem. 2001;8:1157–1179. doi: 10.2174/0929867013372391. [DOI] [PubMed] [Google Scholar]
- 66.Shanahan CA, Gaffney BL, Jones RA, Strobel SA. Identification of c-di-GMP Derivatives Resistant to an EAL Domain Phosphodiesterase. Biochemistry (Mosc.) 2013;52:365–377. doi: 10.1021/bi301510v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frederick CA, Coll M, Van der Marel GA, Van Boom JH, Wang AHJ. Molecular structure of cyclic deoxydiadenylic acid at atomic resolution. Biochemistry (Mosc.) 1988;27:8350–8361. doi: 10.1021/bi00422a010. [DOI] [PubMed] [Google Scholar]
- 68.Blommers MJ, Haasnoot CA, Walters JA, van der Marel GA, van Boom JH, Hilbers CW. Solution structure of the 3’-5’ cyclic dinucleotide d(pApA). A combined NMR, UV melting, and molecular mechanics study. Biochemistry (Mosc.) 1988;27:8361–8369. doi: 10.1021/bi00422a011. [DOI] [PubMed] [Google Scholar]
- 69.Blommers MJJ, Slot HJB, van der Marel GA, van Boom JH, Hilbers CW. Conformational Analysis of the 3′ −5′ -Cyclic Dinucleotide d by Means of Molecular Mechanics. J. Biomol. Struct. Dyn. 1990;8:233–251. doi: 10.1080/07391102.1990.10507804. [DOI] [PubMed] [Google Scholar]
- 70.Frey PA, Sammons RD. Bond order and charge localization in nucleoside phosphorothioates. Science. 1985;228:541–545. doi: 10.1126/science.2984773. [DOI] [PubMed] [Google Scholar]
- 71.Forconi M, Herschlag D. Chapter 15 - Use of Phosphorothioates to Identify Sites of Metal-Ion Binding in RNA. In: Herschlag D, editor. Methods in Enzymology. Academic Press; 2009. pp. 311–333. [DOI] [PubMed] [Google Scholar]
- 72.Wennmohs F, Staemmler V, Schindler M. Theoretical investigation of weak hydrogen bonds to sulfur. J. Chem. Phys. 2003;119:3208–3218. [Google Scholar]
- 73.Zhou P, Tian F, Lv F, Shang Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins Struct. Funct. Bioinforma. 2009;76:151–163. doi: 10.1002/prot.22327. [DOI] [PubMed] [Google Scholar]
- 74.Barends TRM, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 75.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Baslé A, Massa C, Collart FR, Schirmer T, Anderson WF. Crystal Structures of YkuI and Its Complex with Second Messenger Cyclic Di-GMP Suggest Catalytic Mechanism of Phosphodiester Bond Cleavage by EAL Domains. J. Biol. Chem. 2009;284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF. Structural Insight into the Mechanism of c-di-GMP Hydrolysis by EAL Domain Phosphodiesterases. J. Mol. Biol. 2010;402:524–538. doi: 10.1016/j.jmb.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.