Abstract
N,N-bis-(2-chloroethyl)-phosphorodiamidic acid (phosphoramide mustard, PM) and N,N-bis-(2-chloroethyl)-ethylamine (nornitrogen mustard, NOR) are the two biologically active metabolites of cyclophosphamide, a DNA alkylating drug commonly used to treat lymphoma, breast cancer, certain brain cancers, and autoimmune diseases. PM and NOR are reactive bis-electrophiles capable of cross-linking cellular biomolecules to form covalent DNA-DNA and DNA-protein cross-links (DPCs). In the present work, a mass spectrometry-based proteomics approach was employed to characterize PM- and NOR-mediated DNA-protein cross-linking in human cells. Following treatment of human fibrosarcoma cells (HT1080) with cytotoxic concentrations of PM, over 130 proteins were found to be covalently trapped to DNA, including those involved in transcriptional regulation, RNA splicing/processing, chromatin organization, and protein transport. HPLC-ESI+-MS/MS analysis of proteolytic digests of DPC-containing DNA from NOR-treated cells revealed a concentration-dependent formation of N-[2-[cysteinyl]ethyl]-N-[2-(guan-7-yl)ethyl]amine (Cys-NOR-N7G) conjugates, confirming that it cross-links cysteine thiols of proteins to the N-7 position of guanines in DNA. Cys-NOR-N7G adduct numbers were higher in NER-deficient Xeroderma pigmentosum cells (XPA) as compared with repair proficient cells. Furthermore, both XPA and FANCD2 deficient cells were sensitized to NOR treatment as compared to wild type cells, suggesting that Fanconi Anemia and nucleotide excision repair pathways are involved in the removal of cyclophosphamide-induced DNA damage.
Keywords: cyclophosphamide, nitrogen mustard, phosphoramide mustard, human cell culture, mass spectrometry based proteomics, DNA repair, toxicity
TOC image

Introduction
DNA-protein cross-links (DPCs) are bulky DNA lesions that are formed as a result of irreversible trapping of cellular proteins on DNA strands.1 DPCs can form endogenously or may be induced by exposure to transition metal ions,2,3 ionizing radiation,4 and anti-cancer drugs such as nitrogen mustards,5–7 platinum compounds,8–11 and haloethylnitrosureas.12 As a result of their size, DPC lesions are thought to interfere with chromatin folding, DNA replication, transcription, and repair, potentially contributing to toxicity and mutagenicity of cross-linking agents.13
Phosphoramide mustard (PM, Scheme 1) is a biologically active metabolite of cyclophosphamide (CP), a chemotherapeutic agent commonly used to treat lymphomas, breast cancers, certain brain cancers, and autoimmune diseases.14–16 Under physiological conditions, PM spontaneously dephosphoramidates to form another DNA alkylating agent, nornitrogen mustard (NOR) (Scheme 1).17–19 Both PM and NOR can modify N-7-guanine of DNA to yield N-(2-chloroethyl)-N-[2-(7-guaninyl)ethyl] amine, N-(2-hydroxyethyl)-N-[2-(7-guaninyl)ethyl] amine, and N,N-bis-[2-(7-guaninyl)ethyl] amine adducts,17–19 and also produce covalent DNA-protein conjugates (Scheme 1).
Scheme 1.

Metabolism of cyclophosphamide to phosphoramide mustard and nornitrogen mustard and the formation of DNA-protein crosslinks.
DNA-protein cross-linking by nitrogen mustards was first observed by Ewig et al.7 These authors utilized the alkaline elution approach to detect DPCs in mouse leukemia cells (L1210) treated with nitrogen mustard.7 Hansson et al used a similar methodology to demonstrate DPC formation in human melanoma cells upon exposure to NOR and melphalan.20 When compared to other bifunctional lesions such as DNA interstrand and intrastrand cross-links, DPCs accounted for 60 – 70% of total cross-linked lesions.20 Our group initially investigated DNA-protein cross-linking by nitrogen mustards using a model protein, O6-alkylguanine DNA alkyltransferase (AGT).21 Covalent AGT-DNA conjugates formed in a concentration-dependent manner following treatment with mechlorethamine and chlorambucil21 and involved the N7 position of guanine in DNA and AGT active site cysteine residues, C145 and C150.21 More recently, a mass spectrometry-based proteomics approach was employed to identify mechlorethamine-induced DPCs in human fibrosarcoma (HT1080) cells.5 A total of 38 proteins including were identified, including those participating in DNA damage response/repair, RNA processing/mRNA splicing, and transcriptional regulation/translation.5 Unfortunately, many of the cross-linked proteins remained undetected due to the limitations of mass spectrometry methodologies used at the time (ion trap MS).5
In the present study, highly sensitive and accurate Orbitrap methodology was used to characterize DNA-protein cross-linking in HT1080 cells treated with PM and NOR. As mentioned above (Scheme 1), NOR forms upon spontaneous dephosphoramidation of PM, and both mustards produce structurally identical DNA and protein adducts. Proteins covalently bound to chromosomal DNA following PM treatment were isolated using a modified phenol/chloroform extraction methodology developed by our group.5,22 Neutral thermal hydrolysis was used to release PM-induced DPCs from the DNA backbone in the form of guanine-protein cross-links, which were subsequently resolved by SDS-PAGE and identified by mass spectrometry-based proteomics on an Orbitrap Velos mass spectrometer (Scheme 2). A total of 134 proteins were found to form DPCs in the presence of PM, including gene products that function in DNA repair, transcriptional regulation, apoptosis, and cell signaling. Isotope dilution HPLC-ESI+-MS/MS analyses of N-[2-[cysteinyl]ethyl]-N-[2-(guan-7-yl)ethyl]amine (Cys-NOR-N7G) conjugates with 15N5-labeled internal standard were used to quantify DPC formation in NOR-treated cells. Finally, DPC numbers were compared between human wild type fibroblasts and the corresponding cells deficient in nucleotide excision repair (NER) and Fanconi Anemia (FA) repair pathways to gain insight into potential mechanisms of DPC repair in humans.
Scheme 2.

Experimental strategy for isolation of DNA-protein cross-links from phosphoramide mustard-treated cells and their characterization by mass spectrometry.
Experimental
Chemicals and Reagents
Ammonium bicarbonate, ammonium acetate, diepoxybutane, bis-(2-chloroethyl)amine hydrochloride (nornitrogen mustard), mechlorethamine, d,l -1,2,3,4-diepoxybutane, phenylmethanesulfonyl fluoride (PMSF), Boc-L-cysteine (Boc-Cys-OH), triflouroacetic acid (TFA), leupeptin, pepstatin, aprotinin, methoxyamine, UCN-01, dithiothreitol (DTT), iodoacetamide, chloroform, ribonuclease A, deoxyribonuclease I, and alkaline phosphatase were purchased from Sigma (St. Louis, MO). Phosphodiesterase I and phosphodiesterase II were obtained from Worthington Biochemical Corporation (Lakewood, NJ). UltraPure buffer-saturated phenol was obtained from Invitrogen (Carlsbad,CA). Mass spectrometry grade trypsin was purchased from Promega (Madison, WI). Proteinase K was obtained from New England Biolabs (Beverly, MA). Cell Lysis Solution and Protein Precipitation solution were purchased from Qiagen (Valencia, Ca). Phosphoramide mustard was obtained from iTT GmbH/Niomech (Bielefeld, Germany).
N-(2-chloroethyl)-N-[2-(guan-7-yl)ethyl]amine (N7G-NOR-Cl)
2′-Deoxyguanosine (500 mg, 1.87 mmol) was reacted with nornitrogen mustard (3.34 g, 18.7 mmol) in trifluoroethanol (25 mL) at 37 °C for 72 h under anhydrous conditions. The reaction mixture containing N7G-NOR-Cl was dried under argon, and the resulting solid was washed with 1 mL of anhydrous ether three times to remove unreacted NOR. The presence of N7G-NOR-Cl was confirmed by UV spectrophotometry and mass spectrometry. UV: λmax = 246 nm, λmin = 275 nm (pH 4.9); ESI+-MS/MS: m/z 257.7 [M + H], m/z 178.2 [M + H – NH(CH2)2Cl]+, m/z 107.1 [M + H – Gua]+. 15N5-N7G-NOR-Cl was prepared analogously starting with 15N5–2’-deoxyguanosine, and its structure was confirmed by UV and mass spectrometry. UV: λmax 246 nm, λmin 275 nm (pH 4.9); ESI+-MS/MS: m/z 262.7 [M + H]+, m/z 183.1 [M + H − NH(CH2)2Cl]+, m/z 107.1 [M + H − Gua]+.
N-[2-[cysteinyl]ethyl]-N-[2-(guan-7-yl)ethyl]amine (Cys-NOR-N7G)
Boc-Cys-OH (167 mg, 0.7541 mmol) was combined with N7G-NOR-Cl (335 mg, 0.9438 mmol) in 7 mL DMSO, and the reaction mixture was stirred at 37 °C for 72 h. The insoluble product was isolated by filtration and separated by semi-preparative HPLC on a Supelcosil LC-18-DB column (25 cm × 10 mm, 5 μm) eluted with a linear gradient of acetonitrile (B) in 20 mM ammonium acetate, pH 4.9 (A). Solvent composition was changed linearly from 0 to 24% B in 24 min and further to 60% in 6 min. Under these conditions, Boc-Cys-NOR-N7G eluted as a sharp peak at 20.9 min. ESI+-MS/MS: m/z 443.5 [M + H]+ → m/z 343.5 [M + H −Boc]+ and m/z 193.3 [M + H − Boc – Gua]+. The Boc group was removed by incubating Boc- Cys-NOR-N7G with 50% TFA at room temperature for 45 min, and the resulting deprotected product (Cys-NOR-N7G) was purified using the same HPLC method (retention time = 10.4 min). Cys-NOR-N7G: ESI+-MS/MS: m/z 342.1 [M + H]+ → m/z 191.1 [M + H − Gua]+. Cys-NOR-[15N5]-N7G was synthesized analogously starting with 15N57G-NOR-Cl. ESI+-MS/MS: m/z 347.1 [M + H]+ → m/z 191.1 [M + H − Gua]+.
Cell Culture
Human fibrosarcoma (HT1080) cells were obtained from the American Type Culture Collection (Camden, NJ). Human Xeroderma Pigmentosum Complementation Group A (XPA, XPA deficient) cells were obtained from NIGMS Human Genetic Cell Repository (Camden, NJ). Prof. Alexandra Sobeck (University of Minnesota) kindly provided FA-D2 cells (PD20, FANCD2 deficient), and FA-D2-derivative cells stably expressing FANCD2 (PD20 + FANCD2). The cells were maintained as exponentially growing monolayer cultures in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), in a humidified incubator at 37 °C with 5% CO2.
Cytotoxicity Experiments
HT1080, XPA, PD20, and PD20 corrected cells were plated in Dulbecco’s modified Eagle’s medium containing 10% FBS at a density of 3 ×104 cells/dish and permitted to adhere for 24 hours. Cells (1.0 × 105, in triplicate) were treated with 0 – 2000 μM PM, mechlorethamine, or DEB for 3 h at 37 °C. Following treatment, cell media was replaced, and the cells were grown for an additional 48 h at 37 °C. Cell viability was determined using an Alamar Blue assay23 using a Synergy HI Microplate reader (BioTek, Winooski, VT).
Cell Treatment with PM and Isolation of DNA-Protein Crosslinks
HT1080 cells in culture (~ 1 × 107, in triplicate) were treated with increasing concentrations of PM (0, 50, 100, 250, and 500 μM) for 3 h at 37 °C. Following treatment, the cells were washed with phosphate-buffered saline (PBS) and re-suspended in 3 mL PBS buffer. DPC-containing DNA was isolated by a modified phenol-chloroform procedure as described previously.5,22 In brief, the cells were lysed by adding an equal volume of 2X cell lysis buffer (20 mM Tris-HCl/10 mM MgCl2/2% Triton-X100/0.65 M Sucrose), incubated on ice for 5 min, and centrifuged at 2000 g for 10 min at 4 °C. The nuclear pellets were re-suspended in saline-EDTA solution (75 mM NaCl/24 mM EDTA/1% sodium dodecyl sulfate) containing RNase A (10 μg/mL) and a protease inhibitor cocktail (1 mM PMSF; 1 μg/mL pepstatin; 0.5 μg/mL leupeptin; 1.5 μg/mL aprotinin). The resulting nuclei were incubated for 2 h at 37 °C with gentle shaking. To the nuclear lysates, an equal amount of tris-saturated phenol was added, and the solutions were mixed for 5 minutes. Following centrifugation at 1500 RPM for 10 minutes, the aqueous layer containing DNA and the DPC-containing interface was collected. This material was re-extracted with phenol/chloroform as described above two additional times, and DPC-containing DNA was precipitated with cold isopropanol. DNA amounts and its purity were estimated using UV spectrophotometry (A260) and subsequently determined by quantitation of dG in enzymatic hydrolysates as described below. Typical DNA yields from 10 million cells were 40–60 μg.
DNA Quantitation by dG Analysis
To quantify DNA extracted from cells, 5 μg aliquots of DNA were taken and subjected to neutral thermal hydrolysis (1 h at 70 °C) to release protein−guanine conjugates from the DNA backbone. The resulting partially depurinated DNA was digested to 2′-deoxynucleosides in the presence of phosphodiesterase I (120 mU), phosphodiesterase II (105 mU), DNase (35 U) and alkaline phosphatase (22 U) in 20 μL 10 mM Tris-HCl/15 mM ZnCl2 (pH 7.0) for 18 h at 37 °C. Quantitative analysis of dG in enzymatic digests was conducted by HPLC-UV on an Agilent Technologies 1100 HPLC system equipped with a diode array UV detector and an autosampler. The samples were loaded on an Xterra MS C18 column (2.1 × 250 mm, 5 μm, from Waters Corporation, Milford, MA) and eluted with a gradient of 150 mM ammonium acetate (A) and acetonitrile (B). Solvent composition was changed linearly from 0 to 20% B over 30 min and further to 75% B over 3 min, then returned to 0% B over 3 min, where it was kept for the final 13 min of the HPLC run. UV absorbance was monitored at 260 nm. With this method, dG eluted as a sharp peak at 19.9 min. dG amounts were determined by comparing HPLC peak areas to a calibration curve constructed by injecting known dG amounts. No RNA contamination was detected as demonstrated by the absence of ribonucloside peaks in enzymatic digests.
Mass Spectrometric Identification of Cross-Linked Proteins
To identify cellular proteins that become covalently trapped on DNA following exposure to PM, HT1080 cells (1 × 107, in triplicate) were treated with 100 μM PM for 3 h at 37 °C. Chromosomal DNA containing any covalently attached proteins was isolated by the modified phenol/chloroform extraction method described above.5,22 DPC-containing DNA (30 μg) was subjected to neutral thermal hydrolysis to release protein-guanine conjugates (Scheme 2), dried under vacuum, and reconstituted in 1 × NuPAGE Sample Buffer (Invitrogen, Carlsbad, CA). Proteins were resolved by NuPAGE Novex 12% Bis-Tris Gels (Invitrogen, Carlsbad, CA) and stained with SimplyBlue Safe stain (Invitrogen, Carlsbad, CA).
Gel lanes were divided into five sections encompassing the molecular weight range of 5 – 250 kDa. Each section of the gel (260 – 110 kDa, 110 – 80 kDa, 80 – 50 kDa, 50 – 30 kDa, 30 – 5 kDa) was excised and further diced into 1 mm pieces. The proteins present within the gel pieces were subjected to in-gel tryptic digestion by standard methods. In brief, gel pieces were washed with 25 mM ammonium bicarbonate, protein disulfide linkages were reduced with DTT (300 mM), and the resulting thiols were alkylated with saturated iodoacetamide. Gel pieces were dehydrated by incubation with acetonitrile, dried under vacuum, and reconstituted in 25 mM ammonium bicarbonate buffer (pH 9.0). Mass spectrometry grade trypsin (1 μg) was added, and the samples were digested for 18 hours at 37 °C. The resulting tryptic peptides were extracted with 60% acetonitrile containing 0.1% aqueous formic acid, evaporated to dryness, and desalted using C18 ZipTips (Millipore, Temecula, CA). Samples were reconstituted in 0.1% formic acid for HPLC−ESI+−MS/MS analysis.
HPLC-ESI+-MS/MS analyses of tryptic peptides were conducted using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA) equipped with an Eksigent nanoLC 2D HPLC pump, a nanospray source, and Xcalibur 2.1.0 software for instrument control. Peptide mixtures were loaded onto a Symmetry C18 trapping column (180 μm ×20 mm, Waters, Milford, MA) using 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) at a solvent composition of 95% A and 5% B at 5 μL/min for 3 min. Typical injection volume was 8 μL. Following trapping, the HPLC flow was decreased to 0.3 μL/min and reversed to elute the peptides. The peptides were separated on a nanoHPLC column (75 μm i.d.,10 cm packed bed, 15 μm orifice) created by hand packing a commercially purchased fused-silica emitter (New Objective, Woburn MA) with Zorbax SB-C18 5 μm separation media (Agilent, Santa Clara, CA). The gradient program started at 5% B, followed by a linear increase to 60% B over 60 min and further to 95% B in 5 min. Liquid chromatography was carried out at an ambient temperature. The mass spectrometer was calibrated prior to each analysis, and the spray voltage was adjusted to ensure a stable spray. Typically, the MS tune parameters were as follows: spray voltage of 1.6 kV, a capillary temperature of 275 °C, and an S-lens RF level of 50%. MS/MS spectra were collected using data-dependent scanning, in which one full scan mass spectrum acquired in the Orbitrap detector (R = 30,000) was followed by eight MS/MS spectra acquired in the Orbitrap detector (R = 7,500) with an isolation width of 2.5 m/z, activation time of 30 ms, activation Q of 0.25, 35% normalized CID collision energy, 1 microscan, with an AGC setting of 2 × 105 and a maximum injection time of 100 ms. Dynamic exclusion was enabled for 60 s, and singly charged species were excluded from MS/MS analysis.
Spectral data were analyzed using an in-house developed software pipeline “TINT” that linked raw data extraction, database searching, and probability scoring. Raw data were extracted and converted to the mzXML format using ReadW. Spectra that contained fewer than 6 peaks or had measured total ion current (TIC) < 20 were excluded. Data was processed using the SEQUEST v.27 algorithm24 on a high speed, multiprocessor Linux cluster in the Minnesota Supercomputing Institute at the University of Minnesota. Peptide spectra were searched against the UniProt Human Protein Database. Cysteine carboxamidomethylation (+57.0215 Da) was set as a fixed modification, and methionine oxidation (+15.9949 Da) was selected as a variable modification. Precursor mass tolerance was set to 10 ppm within the calculated mass, and fragment ion mass tolerance was set to 10 mmu of their monoisotopic mass. The identified peptides were filtered using Scaffold 3 software (Proteome Software, INC., Portland, OR), to a target false discovery rate (FDR) of 5%. The FDR was calculated with the following expression: FDR = (2R)/(R + F) × 100, where R is the number of passing reversed peptide identifications and F is the number of passing forward (normal orientation) peptide identifications. The second round of filtering removed proteins supported by less than two unique peptide fragments in the analyses. Any proteins also found in control (untreated) samples were excluded from the final list.
Western Blot Analysis
HT1080 cells (~1 × 107) were treated with NOR (0, 50, 100, 250, or 500 μM) for 3 h at 37 °C. Chromosomal DNA, along with any covalent DPCs, was extracted and quantified as described above. From each sample, 50 μg of DNA was subjected to neutral thermal hydrolysis (1 h at 70 °C) to release protein-guanine conjugates from the DNA backbone. The proteins were resolved by NuPAGE Novex 12% Bis-Tris gels (Invitrogen, Carlsbad, CA) and transferred to Invitrolon PVDF filter paper membranes (0.45 μm pore size, Life Technologies, Carlsbad, CA). The membranes were blocked for 4 h in Tris-buffered saline-Tween 20 (TBST) containing 5% (w/v) bovine serum albumin. Following blocking, the membranes were incubated with the primary antibodies against vimentin, nucleophosmin, prohibitin-2, matrin-3, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), poly-(ADP-ribose) polymerase 1 (PARP), and histone-H4 overnight at 4 °C. The membranes were rinsed with TBST and incubated with the corresponding alkaline phosphatase-conjugated antibody for 1 h at room temperature. The gels were developed using SIGMA Fast BCIP/NBT (Sigma, St. Louis, MO) according to manufacturer’s protocol.
Quantitation of Cys-NOR-N7G in NOR-Exposed Cells
HT1080 cells (1 × 107) were treated with increasing concentrations of NOR (0, 50, 100, 250, and 500 μM) for 3 h at 37 °C. Following treatment, the cells were washed with PBS, re-suspended in cell lysis solution (2 mL, Qiagen, Valencia, CA) containing 10 μg RNAse A, and incubated for 16 h at room temperature with gentle inversion. Proteinase K (10 μg) was added, and the samples were incubated for an additional 16 h at room temperature with gentle inversion. The lysates were mixed with 700 μL of protein precipitation solution (Qiagen, Valencia, CA), and the samples were vortexed for one minute. Following centrifugation at 1500 RPM for 10 minutes, the supernatant was collected, and DPC-containing genomic DNA was precipitated with cold isopropanol. DNA was quantified by dG analysis as described above.
DNA (100 μg) was subjected to neutral thermal hydrolysis (1 h at 70 °C) to release protein−guanine conjugates from the DNA backbone. Proteins were cleaved with trypsin (10 μg protein in 25 mM ammonium bicarbonate, overnight at 37 °C), and the resulting peptides were further digested to amino acids in the presence of proteinase K (10 μg in 100 μL water, overnight at 37 °C). The digests were spiked with Cys-[15N5]-NOR-N7G (internal standard for mass spectrometry, 200 fmol), followed by offline HPLC purification as follows. An Agilent Technologies HPLC system (1100 model) was used incorporating a diode array detector, an autosampler, and a fraction collector. A Synergi 4μ Hydro RP (4.6 × 250 mm, 5 μm) column (Sigma-Aldrich, St. Louis, MO) was eluted at a flow rate of 1 mL/min using a gradient of 15 mM ammonium formate, pH 4.9 (A) and acetonitrile (B). Fractions containing Cys-NOR-Gua and its 15N-labeled internal standard (9.7 – 11.2 min) were collected, dried under vacuum, and reconstituted in 0.1% formic acid (20 μL).
Quantitative analyses of Cys-NOR-N7G were conducted with a Dionex UltiMate 3000 RSLC nanoHPLC system (Thermo Scientific, Waltham, MA) interfaced to a TSQ Vantage mass spectrometer (Thermo Scientific, Waltham, MA). Chromatographic separation was accomplished with a Hypercarb HPLC column (100 mm × 0.3 mm, 3 μm) eluted with a gradient of 0.1% formic acid (A) and acetonitrile (B) at a flow rate of 12 μL/min. Solvent composition was linearly changed from 4% to 30% in 13 min and further to 90% 1 min, kept at 90% for 1 min, then brought back to 4% in 1 min. Under these conditions, Cys-NOR-N7G and its internal standard (Cys-NOR-[15N5] N7G) eluted at ~ 9.6 min. Electrospray ionization was achieved at a spray voltage of 3200 V and a capillary temperature of 270 °C. Collision induced dissociation was performed with Ar as a collision gas (1.5 mTorr) at a collision energy of 23 V. Instrument parameters were optimized for maximum response during infusion of a standard solution of Cys-NOR-N7G.
HPLC-ESI+-MS/MS analysis of Cys-NOR-N7G was performed in the selected reaction monitoring mode by following the neutral loss of guanine from protonated molecules of the analyte (m/z 342.1 [M + H]+ → 191.0 [M + H − Gua]+) and the corresponding mass transitions corresponding to 15N5- labeled internal standard (m/z 347.1 [M + H]+ → 191.0 [M + H − 15N5 − Gua]+). Analyte concentrations were determined using the relative response ratios calculated from HPLC-ESI+-MS/MS peak areas in extracted ion chromatograms corresponding to Cys-NOR-N7G and its internal standard.
Cys-NOR-N7G Quantitation in Wild Type and DNA Repair-Deficient Fibroblasts
Human fibrosarcoma cells (HT1080), NER-deficient Xeroderma pigmentosum cells (XPA), FANCD2-deficient cells (PD20), and the corresponding FANCD2-corrected cells (PD20-corrected, 1×107) were treated with 250 μM NOR for 3 h at 37 °C. Following treatment, cell media was replaced, and the cells were allowed to recover at 37 °C for either 4 h (n = 6) or 0, 4, 12, or 24 h (n = 3) to allow for repair. DNA was isolated using Qiagen Cell Lysis Solution extraction procedure and quantified by dG analysis as described above. DNA samples (100 μg) were subjected to neutral thermal hydrolysis (1 h at 70 °C) to release protein-guanine conjugates. Conjugated peptides were further digested to amino acids in the presence of trypsin (10 μg protein in 25 mM ammonium bicarbonate, overnight at 37 ° C), followed by proteinase K (10 μg in 250 μL water, overnight at 37 °C). The digests were spiked with Cys-NOR-[15N5]-N7G internal standard (200 fmol), followed by off-line HPLC purification as described above. Cys-NOR-N7G conjugates were quantified by isotope dilution HPLC-ESI+-MS/MS as described above.
Results
Cytoxicity Experiments in Human Cell Culture
To characterize the toxicity of PM in human fibrosarcoma cell cultures, HT1080 cells (in triplicate) were treated with increasing amounts of PM, and their viability was determined by the Alamar Blue assay.23 We found that PM was significantly less toxic than mechlorethamine, a structurally related nitrogen mustard investigated in our earlier publication (EC50 = 317 ± 43 and 20.2 ± 4.59, respectively),5 but had similar toxicity to 1,2,3,4-diepoxybutane EC50 = 480.8 ± 41.9 (Supplementary Figure S-1). The fractions of live cells following treatment with 50, 100, 250, 500, 750, 1000, 1500, and 2000 μM PM were 87 ± 2.6, 73.0 ± 3.4, 60 ± 3.7, 43 ± 3, 30 ± 3.6, 18 ± 2, 7 ± 0.3, and 8.5 ± 0.3 %, respectively (Figure 1).
Figure 1.

Cytoxicity of phosphoramide mustard in human fibrosarcoma (HT1080) cells. Cells (in triplicate) were treated with 0 – 2000 μM PM for 3 h. Following exposure, the cells were allowed to recover in fresh media for 48 h. Cell viability was measured by the Alamar Blue assay using a Synergi H1 Microplate reader.
Concentration-Dependent Formation of DPCs in PM-Treated Human Cells
To monitor PM-mediated DPC formation, human fibrosarcoma cells (HT1080) were treated with increasing concentrations of PM (0, 50, 100, 250, and 500 μM) for 3 h. DNA was extracted under mild conditions that preserved covalent DNA-protein conjugates. The samples were subjected to thermal hydrolysis to release proteins in the form of protein-guanine conjugates, which were resolved by gel electrophoresis (Scheme 2 and Figure 2A). Although small amounts of endogenous DPCs were present in untreated cells (Lane 3 in Figure 2A), the amounts of cross-linked proteins increased in a concentration dependent manner (Lanes 5, 7, 9, 11 in Figure 2A, Figure 2B). In our previous studies, a 20-fold greater concentration of 1,2,3,4-diepoxybutane (DEB, 2 mM) was required to produce similar numbers of DPCs in HT1080 cells,22 while comparable cross-linking efficiency was observed upon treatment with mechlorethamine.5 This suggests that PM and NOR are equally efficient at forming DPCs, while DEB is less efficient than the two nitrogen mustards.
Figure 2.

(A) Concentration-dependent formation of DPCs in HT1080 cells treated with 0 – 500 μM PM for 3 h. Chromosomal DNA containing drug-induced DPCs was extracted using a modified phenol/chloroform extraction method described in the Experimental section. The proteins were released from the DNA backbone by neutral thermal hydrolysis to yield guanine-protein conjugates (Scheme 2), which were resolved by 12% SDS-PAGE and visualized by staining with SimplyBlue SafeStain. Lane 1: molecular weight marker. Lanes 2, 4, 6, 8, 10 are blank. Lane 3: 50 μM PM. Lane 5: 100 μM PM. Lane 7: 250 μM PM. Lane 9: 500 μM PM. (B) Densitometry measurements of the band intensity form Figure 2A, normalized to the untreated control.
Identification of Cross-linked Proteins by Mass Spectrometry-Based Proteomics
To identify the proteins participating in DPC formation following exposure to PM, HT1080 cells (in triplicate) were treated with 100 μM PM for 3 h. Control cells were incubated with PM-free cell culture media. DPC-containing DNA was extracted as described above, and equal amounts of DNA from each sample (30 μg) were subjected to neutral thermal hydrolysis to release protein-guanine conjugates (Scheme 2), which were detected by gel electrophoresis.
SDS-PAGE analysis of protein-guanine conjugates isolated from PM-treated samples (Lanes 9–11 in Figure 3) revealed intense protein bands, while control samples contained considerably weaker bands corresponding to endogenous DPCs (Lanes 3 – 5 in Figure 3). The gel lanes from both experiments were divided into five sections covering the molecular weight range of 5 – 260 kDa (A – E in Figure 3) and cut out of the gel. Individual gel pieces of each region were subjected to in-gel tryptic digestion, and the resulting peptides were extracted from the gel, desalted, and subjected to HPLC-ESI+-MS/MS analysis for protein identification.
Figure 3.

DNA-protein cross-linking in human fibroblasts used for proteomics experiments. HT1080 cells (~5.0 × 106, in triplicate) were treated with 100 μM PM (lanes 9 – 11) or buffer only (lanes 3 – 5) for 3 h. Following DPC extraction and thermal hydrolysis to release guanine-protein conjugates, the cross-linked proteins were resolved by 12% SDS-PAGE and visualized by staining with SimplyBlue SafeStain. Proteins within the molecular weight ranges of 30 – 5 kDa (A), 50 – 30 kDa (B), 80 – 50 kDa (C), 110 – 80 kDa (D), and 260 – 110 kD (E) were excised from the gels, subjected to in-gel trypsin digestion, and analyzed by HPLC-ESI+-MS/MS.
MS/MS analysis of tryptic peptides from cross-linked proteins yielded characteristic b and y series ions, which were used to determine their amino acid sequences and to identify the corresponding proteins (see example in Supplementary Figure S-2). Protein identification was based on the sequence of at least two unique peptides. Any proteins that were also detected in control samples or were not present in all three treated samples were omitted from the final list. Using these criteria, a total of 134 proteins were identified (Table 1).
Table 1.
Proteins forming covalent cross-links to chromosomal DNA following exposure to PM.
| Swiss-Prot ID | Identified Protein | % Coverage | No. of Unique Peptides | Total Spectra | Primary Biological Function | Protein MW (Da) | |
|---|---|---|---|---|---|---|---|
| 1 | P53999 | Activated RNA polymerase II transcriptional coactivator p15 | 37 | 4 | 4 | Transcriptional Regulation | 14395.9 |
| 2 | O15347 | High mobility group protein B3 | 19 | 3 | 3 | 22980.7 | |
| 3 | P39687 | Acidic leucine-rich nuclear phosphoprotein 32 family member A | 21 | 5 | 7 | 28586.1 | |
| 4 | Q99623 | Prohibitin-2 | 10 | 2 | 2 | 33297.6 | |
| 5 | P16989 | DNA-binding protein A | 8 | 2 | 2 | 40089.4 | |
| 6 | Q14320 | Protein FAM50A | 8 | 2 | 2 | 40242.8 | |
| 7 | Q5BKZ1 | Zinc finger protein 326 | 11 | 4 | 4 | 65653.5 | |
| 8 | P11142 | Heat shock cognate 71 kDa protein | 8 | 3 | 3 | 70899.8 | |
| 9 | Q8WVC0 | RNA polymerase-associated protein LEO1 | 4 | 2 | 2 | 75405.3 | |
| 10 | Q9Y3T9 | Nucleolar complex protein 2 | 3 | 2 | 2 | 84907.1 | |
| 11 | P43243 | Matrin-3 | 13 | 10 | 11 | 94626.7 | |
| 12 | P55884 | Eukaryotic translation initiation factor 3 subunit B | 22 | 15 | 18 | 92482.8 | |
| 13 | Q8TDD1 | ATP-dependent RNA helicase DDX54 | 2 | 2 | 2 | 98597.9 | |
| 14 | Q9BXP5 | Serrate RNA effector molecule homolog | 3 | 3 | 3 | 100669.7 | |
| 15 | Q9Y5B9 | FACT complex subunit SPT16 | 16 | 15 | 17 | 119917.4 | |
| 16 | Q03701 | CCAAT/enhancer binding protein zeta | 2 | 2 | 2 | 120992.2 | |
| 17 | Q9BQG0 | Myb-binding Protein 1A | 2 | 2 | 2 | 148858.3 | |
| 18 | P51532 | Transcription activator BRG1 | 2 | 3 | 3 | 184649.6 | |
| 19 | O60216 | Double-strand-break repair protein rad21 homolog | 6 | 3 | 3 | DNA Repair | 71691.6 |
| 20 | P12956 | X-ray repair cross-complementing protein 5 | 22 | 10 | 12 | 69846.4 | |
| 21 | P84103 | Splicing factor, arginine/serine-rich 3 | 30 | 5 | 5 | RNA Splicing/Processing | 19330.0 |
| 22 | Q07955 | Splicing factor, arginine/serine-rich 1 | 12 | 3 | 3 | 27745.1 | |
| 23 | Q32P51 | Heterogeneous nuclear ribonucleoprotein A1-like 2 | 15 | 4 | 5 | 34225.4 | |
| 24 | P22626 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 10 | 3 | 3 | 37430.3 | |
| 25 | P08621 | U1 small nuclear ribonucleoprotein 70 kDa | 5 | 2 | 2 | 51558.4 | |
| 26 | O00566 | U3 small nucleolar ribonucleoprotein protein MPP10 | 7 | 3 | 3 | 78866.8 | |
| 27 | Q15029 | 116 kDa U5 small nuclear ribonucleoprotein component | 4 | 3 | 3 | 109438.1 | |
| 28 | Q15393 | Splicing factor 3B subunit 3 | 2 | 2 | 2 | 135579.4 | |
| 29 | O75643 | U5 small nuclear ribonucleoprotein 200 kDa helicase | 2 | 4 | 4 | 244513.8 | |
| 30 | P51665 | 26S proteasome non-ATPase regulatory subunit 7 | 18 | 3 | 3 | Apoptosis | 37025.6 |
| 31 | P04083 | 26S proteasome non-ATPase regulatory subunit 3 | 19 | 9 | 9 | 60979.6 | |
| 32 | Q9NY61 | Protein AATF | 22 | 7 | 8 | 63135.0 | |
| 33 | P25205 | DNA replication licensing factor MCM3 | 6 | 4 | 4 | DNA Replication | 90982.6 |
| 34 | Q14566 | DNA replication licensing factor MCM6 | 3 | 2 | 2 | 92890.2 | |
| 35 | P33778 | Histone H2B type 1-B | 20 | 3 | 14 | Chromatin Organization | 13950.8 |
| 36 | Q92522 | Histone H1× | 10 | 2 | 2 | 22487.9 | |
| 37 | Q9P0M6 | Core histone macro-H2A.2 | 7 | 2 | 3 | 40060.1 | |
| 38 | Q969G3 | SWI/SNF-related matrix associated actin dependent regulator of chromatin subfamily E member 1 | 13 | 4 | 5 | 46649.7 | |
| 39 | Q14839 | Chromodomain-helicase-DNA-binding protein 4 | 4 | 6 | 6 | 217994.6 | |
| 40 | Q12873 | Chromodomain-helicase-DNA-binding protein 3 | 1 | 2 | 2 | 226597.2 | |
| 41 | P63173 | 60S ribosomal protein L38 | 29 | 2 | 4 | Translation | 8218.5 |
| 42 | P62979 | 40S ribosomal protein S27a | 40 | 2 | 3 | 9418.4 | |
| 43 | Q6NW1 | Putative 60S ribosomal protein L13a-like MGC87657 | 29 | 3 | 3 | 12135.0 | |
| 44 | P83881 | 60S ribosomal protein L36a | 30 | 4 | 4 | 12441.1 | |
| 45 | Q5JNZ5 | Putative 40S ribosomal protein S26-like 1 | 27 | 3 | 10 | 13002.2 | |
| 46 | P60866 | 40S ribosomal protein S20 | 19 | 2 | 3 | 13373.0 | |
| 47 | P62899 | 60S ribosomal protein L31 | 30 | 4 | 11 | 14463.2 | |
| 48 | P35268 | 60S ribosomal protein L22 | 19 | 2 | 3 | 14787.3 | |
| 49 | P62244 | 40S ribosomal protein S15a | 31 | 4 | 4 | 14840.0 | |
| 50 | P62829 | 60S ribosomal protein L23 | 32 | 3 | 4 | 14865.9 | |
| 51 | P62847 | 40S ribosomal protein S24 | 29 | 4 | 9 | 15423.8 | |
| 52 | P08708 | 40S ribosomal protein S17 | 16 | 2 | 3 | 15550.5 | |
| 53 | P62266 | 40S ribosomal protein S23 | 15 | 2 | 3 | 15807.7 | |
| 54 | P62263 | 40S ribosomal protein S14 | 34 | 4 | 4 | 16272.9 | |
| 55 | P62249 | 40S ribosomal protein S16 | 28 | 4 | 6 | 16445.9 | |
| 56 | P46776 | 60S ribosomal protein L27a | 20 | 3 | 3 | 16561.4 | |
| 57 | P62841 | 40S ribosomal protein S15 | 40 | 3 | 6 | 17040.8 | |
| 58 | P62277 | 40S ribosomal protein S13 | 36 | 6 | 7 | 17223.3 | |
| 59 | P30050 | 60S ribosomal protein L12 | 24 | 3 | 4 | 17819.1 | |
| 60 | P62280 | 40S ribosomal protein S11 | 35 | 5 | 5 | 18431.3 | |
| 61 | P46778 | 60S ribosomal protein L21 | 21 | 3 | 8 | 18565.0 | |
| 62 | P46783 | 40S ribosomal protein S10 | 15 | 2 | 4 | 18898.3 | |
| 63 | Q9NQ39 | Putative 40S ribosomal protein S10-like | 13 | 2 | 3 | 20120.6 | |
| 64 | Q02543 | 60S ribosomal protein L18a | 21 | 3 | 4 | 20762.6 | |
| 65 | T1W3K1 | 60S ribosomal protein L17 | 41 | 6 | 11 | 21397.4 | |
| 66 | P32969 | 60S ribosomal protein L9 | 18 | 3 | 4 | 21863.7 | |
| 67 | P62081 | 40S ribosomal protein S7 | 47 | 6 | 10 | 22127.5 | |
| 68 | P46781 | 40S ribosomal protein S9 | 45 | 12 | 21 | 22592.5 | |
| 69 | P23396 | 40S ribosomal protein S3 | 27 | 5 | 5 | 26688.6 | |
| 70 | P62753 | 40S ribosomal protein S6 | 18 | 4 | 5 | 28681.7 | |
| 71 | P18124 | 60S ribosomal protein L7 | 38 | 12 | 16 | 29227.7 | |
| 72 | P15880 | 40S ribosomal protein S2 | 24 | 6 | 6 | 31325.2 | |
| 73 | P05198 | Eukaryotic translation initiation factor 2 subunit 1 | 10 | 2 | 2 | 36112.7 | |
| 74 | P20042 | Eukaryotic translation initiation factor 2 subunit 2-like protein | 16 | 5 | 5 | 37788.3 | |
| 75 | P60842 | Eukaryotic initiation factor 4A-I | 22 | 7 | 7 | 46155.3 | |
| 76 | O15371 | Eukaryotic translation initiation factor 3 subunit D | 11 | 5 | 5 | 63973.8 | |
| 77 | P55884 | Eukaryotic translation initiation factor 3 subunit B | 22 | 15 | 18 | 92482.8 | |
| 78 | Q14152 | Eukaryotic translation initiation factor 3 subunit A | 12 | 15 | 18 | 166574.9 | |
| 79 | Q04637 | Eukaryotic translation initiation factor 4 gamma 1 | 9 | 13 | 14 | 175494.2 | |
| 80 | P05109 | Protein S100 A8 | 32 | 3 | 4 | Inflammatory Response | 10835.0 |
| 81 | P06702 | Protein S100 A9 | 45 | 4 | 6 | 13242.3 | |
| 82 | P66905 | Hemoglobin subunit alpha | 22 | 2 | 3 | Oxygen Transport | 15257.6 |
| 83 | Q13765 | Nascent polypeptide-associated complex subunit alpha | 26 | 4 | 5 | Protein Transport | 23383.3 |
| 84 | Q96FZ7 | Charged multivesicular body protein 6 | 10 | 2 | 2 | 23485.3 | |
| 85 | O76021 | Ribosomal L1 domain-containing protein 1 | 11 | 4 | 4 | 54974.7 | |
| 86 | O00203 | AP-3 complex subunit beta-1 | 5 | 4 | 4 | 121324.0 | |
| 87 | Q9P2E9 | Ribosome-binding protein 1 | 14 | 16 | 21 | 152470.6 | |
| 88 | Q08378 | Golgin subfamily A member 3 | 3 | 3 | 3 | 167356.0 | |
| 89 | Q00610 | Clathrin heavy chain 1 | 11 | 15 | 15 | 191618.9 | |
| 90 | Q13268 | Dehydrogenase/reductase SDR family member 2, mitochondrial | 11 | 3 | 3 | Cellular Homeostasis | 27438.4 |
| 91 | Q15006 | Tetratricopeptide repeat protein 35 | 18 | 4 | 4 | 34834.8 | |
| 92 | P00338 | L-lactate dehydrogenase A chain | 13 | 4 | 5 | 36689.2 | |
| 93 | P11201 | 78 kDa glucose-regulated protein | 17 | 8 | 8 | 72334.7 | |
| 94 | P04083 | Annexin A1 | 27 | 6 | 8 | 38715.9 | |
| 95 | Q96A33 | Coiled-coil domain-containing protein 47 | 9 | 3 | 3 | 55874.9 | |
| 96 | P25705 | ATP synthase subunit alpha, mitochondrial | 12 | 6 | 6 | 59752.1 | |
| 97 | Q9NR30 | Nucleolar RNA helicase 2 | 6 | 4 | 4 | 87346.0 | |
| 98 | P36776 | Lon protease homolog, mitochondrial | 6 | 5 | 6 | 106491.3 | |
| 99 | P63104 | 14-3-3 protein zeta/delta | 17 | 3 | 4 | Cellular Signaling | 27745.9 |
| 100 | P31946 | 14-3-3 protein beta/alpha | 9 | 2 | 5 | 28083.1 | |
| 101 | P62258 | 14-3-3 protein epsilon | 12 | 3 | 3 | 29175.0 | |
| 102 | Q9UBS4 | DnaJ homolog subfamily B member 11 | 14 | 3 | 3 | 40514.6 | |
| 103 | Q3TIV5 | Zinc finger CCCH domain-containing protein 15 | 10 | 3 | 3 | 48603.7 | |
| 104 | Q13283 | Ras GTPase-activating protein-binding protein 1 | 9 | 3 | 4 | 52162.8 | |
| 105 | Q1KMD3 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 | 9 | 6 | 7 | 85105.2 | |
| 106 | P05556 | Integrin beta-1 | 6 | 4 | 4 | 88415.1 | |
| 107 | O14786 | Neuropilin-1 | 19 | 12 | 13 | 103136.6 | |
| 108 | P08865 | 40S ribosomal protein SA | 19 | 3 | 3 | Cell Structure/Architecture | 32854.1 |
| 109 | P60709 | Actin, cytoplasmic 1 | 19 | 5 | 6 | 41737.8 | |
| 110 | P68363 | Tubulin alpha-1B chain | 6 | 2 | 5 | 50151.7 | |
| 112 | P55081 | Microfibrillar-associated protein 1 | 17 | 5 | 6 | 51958.7 | |
| 113 | P17661 | Desmin | 4 | 2 | 8 | 53536.6 | |
| 114 | P41219 | Peripherin | 4 | 2 | 10 | 53652.0 | |
| 115 | P08670 | Vimentin | 67 | 30 | 142 | 53652.7 | |
| 116 | Q16352 | Alpha-internexin | 6 | 3 | 4 | 55392.3 | |
| 117 | Q07065 | Cytoskeleton-associated protein 4 | 48 | 23 | 27 | 66022.2 | |
| 118 | Q16643 | Drebrin | 6 | 2 | 2 | 71428.6 | |
| 119 | P23284 | Peptidyl-prolyl cis-trans isomerase B | 21 | 4 | 4 | Protein Folding | 23743.2 |
| 120 | Q9UKD2 | mRNA turnover protein homolog 4 | 16 | 3 | 3 | Ribosome Assembly | 27561.3 |
| 121 | Q99848 | Probable rRNA-processing protein EBP2 | 27 | 6 | 8 | 34852.9 | |
| 122 | Q15050 | Ribosome biogenesis regulatory protein homolog | 24 | 7 | 9 | 41194.4 | |
| 123 | O00541 | Pescadillo homolog | 6 | 4 | 4 | 68004.9 | |
| 124 | Q14692 | Ribosome biogenesis protein BMS1 | 4 | 3 | 3 | 145812.2 | |
| 125 | P54652 | Heat shock-related 70 kDa protein 2 | 5 | 3 | 3 | Cell Cycle | 70023.0 |
| 126 | Q5TAP6 | U3 small nucleolar RNA-associated protein 14 homolog A | 12 | 7 | 7 | 87979.4 | |
| 127 | P46087 | Putative ribosomal RNA methyltransferase NOP2 | 9 | 5 | 5 | 89303.6 | |
| 128 | Q8WTT2 | Nucleolar complex protein 3 homolog | 3 | 2 | 2 | 92552.2 | |
| 129 | A0LTM5 | Cell division protein 11B | 3 | 2 | 2 | 92709.0 | |
| 130 | O75400 | Pre-mRNA-processing factor 40 homolog A | 5 | 5 | 5 | 108807.3 | |
| 131 | Q14980 | Nuclear mitotic apparatus protein 1 | 5 | 9 | 10 | 238259.7 | |
| 132 | Q9BXY0 | Protein MAK16 homolog | 15 | 3 | 4 | Unknown | 35370.0 |
| 133 | Q14257 | Reticulocalbin-2 | 32 | 9 | 12 | 36877.6 | |
| 134 | Q9NZM5 | Glioma tumor suppressor candidate region gene 2 protein | 14 | 6 | 6 | 54390.7 |
Proteins found to form covalent cross-links to chromosomal DNA following exposure to PM (Table 1) were categorized based on their cellular distribution, molecular function, and biological process using the GO database available from the European Bioinformatics Institute (http://www.ebi.ac.uk/GOA).25 A large portion of the cross-linked proteins are classified as nuclear (67 total, 50%), including histones, high mobility group proteins, protein MAK16 homolog, and U1 small nuclear ribonucleoprotein 70 kDa (Figure 4A, Table 1). This is not unexpected as nuclear proteins are most likely to form DPCs due to their proximity to nuclear DNA. The remaining proteins were classified as cytoplasmic (38 total, 28.4%), membrane bound/cytoskeleton (18 total, 13.4%), endoplasmic reticulum (7 total, 5.2%), mitochondrial (5 total, 3.7%), golgi apparatus (2, 1.5%) and unknown (2 total, 1.5%) (Figure 4A). It is important to note that many of the proteins may be present in multiple cellular compartments due to their participation in many diverse biological processes. For example, S100 A9 is classified as a cytoplasmic protein, but it has been shown to enter the nucleus and bind the promoter region of complement component 4 (C3), upregulating C3 expression.26
Figure 4.

GO annotations for the proteins involved in PM-induced DPC formation in human HT1080 cells according to cellular distribution, molecular function, and biological processes. The numbers of proteins falling into each category are included.
In respect to their molecular function, the majority of the cross-linked proteins are known to bind DNA and RNA (69 total, 37.7%). In addition, 10 of the proteins are known to participate in transcription factor binding, while 7 are involved in translation initiation factor binding (Figure 4).
In regard to biological processes, many of the cross-linked proteins are known to be involved in transcriptional regulation (20 total, 12.0%), mRNA/RNA processing (17 total, 10.2%), translation (40 total, 23.9%), DNA damage response (8 total, 4.8%), and chromatin organization (7, 3.8%) (Figure 4). Included in this group are the nucleolar complex protein 2 (inhibitor of histone deacetyltransferase and p53/TP53-regulated target promoters),27 Myb-binding protein 1A (binds sequence-specific DNA-binding proteins),28,29 double-strand-break repair protein rad21homolog,30 and chromodomain-helicase-DNA-binding protein 4 (regulation of homologous recombination DNA repair).31 Some of the proteins were identified as structural constituents of the ribosome (27 total, 14.8%) (Figure 4). These include the 40S ribosomal protein S6, 60S ribosomal protein L7, 40S ribosomal protein S2, and 40S ribosomal S3 (Table 1). This is not a result of RNA contamination in our experiments, because HPLC analysis of enzymatic digests has confirmed that DNA isolated by our methodology was free of RNA (Supplemental Figure S-3). An alternative explanation is that many of known RNA binding proteins may also have an affinity for DNA. We also observed proteins involved in cell signaling (15 total, 8.9%) such as the heterogeneous nuclear ribonucleoprotein U-like protein 2, integrin β-1,32 and neurophilin 1,33 cell cycle/homeostasis (21 total, 11.5%) such as the Lon protease homolog34 and dehydrogenase/reductase SDR family member 2,35 and cell structure/architecture (11 total, 6.6%) such as actin,36 tubulin alpha-1B chain, and microfibrillar associated protein 1 (Table 1).
Many of the gene products identified in our study were counted into multiple GO categories due to their diverse cellular functions and their involvement in multiple biological processes. For example, the FACT complex subunit SPT16 is involved in regulating transcription from RNA polymerase II promoters,37–39 cellular response to DNA damage stimulus,40 and DNA replication.41 As a result, it is listed under three GO annotation categories: DNA repair, DNA replication, and transcriptional regulation. It is also possible that the GO annotation does not take into account every protein’s secondary cellular distributions and molecular functions. The identities of a subset of proteins detected by mass spectrometry-based proteomics were further confirmed by western blot analysis using commercially available antibodies (Supplemental Figure S-4).
HPLC-ESI+-MS/MS Detection of Cys-N7G-NOR Conjugates
To quantify DPC formation in NOR-treated fibroblasts, HT1080 cells (in triplicate) were treated with 50, 100, 250, or 500 μM NOR, and DPC-containing chromosomal DNA was extracted as described above. Following thermal hydrolysis to release DNA and proteolytic digestion of proteins to amino acids, isotope dilution HPLC-ESI+-MS/MS was used to quantify N-[2-[cysteinyl]ethyl]-N-[2-(guan-7-yl)ethyl]methylamine (Cys-NOR-N7G) conjugates, which correspond to cross-linking between the N7-position of guanine in DNA and cysteine thiols of proteins (Scheme 2 and Figure 5A). Cys-NOR-N7G adduct levels increased linearly with increasing concentrations of NOR, ranging between 3 and 14 adducts per 108 nucleotides (Figure 5B). These results confirm that PM and NOR treatment induces DPC formation between the N7-position of guanine and the cysteine thiols of proteins, as was previously reported for other nitrogen mustard drugs.5,6
Figure 5.

Representative HPLC-ESI+-MS/MS trace (A) and quantitative results for Cys-NOR-N7G conjugates in HT1080 cells treated with 0–500 μM NOR for 3 h. Following treatment, genomic DNA containing DPCs was extracted, hydrolyzed to release DNA-protein conjugates, and digested by proteinase K to amino acids. NOR-induced Cys-NOR-N7G conjugates were analyzed by isotope dilution HPLC-ESI+-MS/MS using the corresponding 15N5-labeled internal standard.
Adduct formation in repair deficient cells
To gain insight into potential mechanisms of repair of NOR-induced DPCs in human cells, we examined PM toxicity and DPC formation in repair proficient and repair deficient cells. HT1080 (wild type), XPA (NER deficient), PD20 (FANCD2 deficient), and PD20 corrected cells (FANCD2 proficient) were employed in these experiments. Both NER-deficient Xeroderma pigmentosum cells (XPA) cells and Fanconi Anemia pathway-deficient cells (PD20) exhibited an increased sensitivity towards PM than the corresponding wild type cells (EC50 = 146 ± 31 and 168 ± 36 respectively, Figures 6A and S-5A). PD20-corrected cells had similar sensitivity as HT1080 cells (EC50 = 313 ± 65 vs 317 ± 43, Figures 6A and S-5). These results suggest that both NER and FA repair pathways help protect cells against PM-mediated toxicity.
Figure 6.
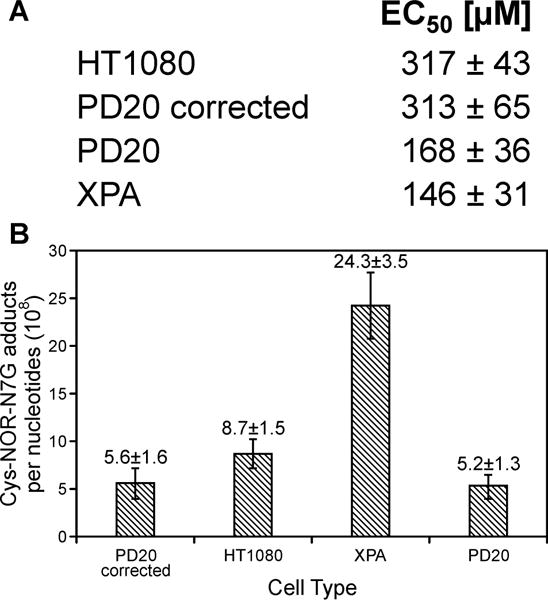
Cellular viability (A) and DPC formation (B) in HT1080, XPA, PD20, and PD20- corrected cells treated with 250 μM NOR for 3 h. Following treatment, NOR-containing media was removed, and the cells were allowed to recover in fresh media for 4 h. Cys-NOR-N7G conjugates were quantified by isotope dilution HPLC-ESI+-MS/MS.
To determine whether the observed differences in sensitivity to PM can be attributed to impaired removal of DPC lesions, HT0180, XPA, PD20, and PD-corrected cells were treated with NOR (250 μM) and allowed to recover in drug-free media for 4h to allow for repair of NOR-induced DPCs. HPLC-ESI+-MS/MS analyses have revealed that NER-deficient XPA cells had the highest numbers of Cys-NOR-N7G adducts 24.3 ± 3.5 adducts/108 nucleotides, Figure 6B as compared to HT1080 (8.7 ± 1.5 adducts/108 nucleotides), PD20 (5.2 ± 1.3 adducts/108 nucleotides) and PD20-corrected cells (5.6 ± 1.6 adducts/108 nucleotides) (Figure 6B). To measure NOR-induced DPC repair over time, HT1080, XPA, PD20, and PD-corrected cells were treated with NOR (250 μM) and allowed to recover in drug-free media for 0, 4, 12, or 24 h. HPLC-ESI+-MS/MS analysis revealed that XPA cells had the highest Cys-NOR-N7G adduct levels at each time point measured (14.7 ± 6.2, 19.2 ± 5.1, 10.2 ± 6.1, and 7.2 ± 3.3 at 0, 4, 12, and 24 h respectively, Figure S-6). After 24 hours of repair, Cys-NOR-N7G adduct levels in all four cell types decreased to below 10 adducts/108 nucleotides (Figure S-6). Taken together, the results shown in Figures 6, S-5A, S-5B, and S-6 are consistent with an involvement of nucleotide excision repair in the removal of cyclophosphamide-mediated DPCs. Fanconi Anemia pathway deficient cells (PD20) were more sensitive to PM (Figure S-5), but contained similar Cys-NOR-N7G adducts levels as compared to corrected cells (Figure 6B). This could be rationalized by FA-mediated removal of other toxic DNA lesions induced by PM, such as DNA-DNA cross-links.42–44
Discussion
DNA-protein cross-links are bulky DNA adducts that can form upon exposure to bis-electrophiles, reactive oxygen species, and transition metals.1,13 DPC formation is thought to contribute to the biological activity of bis-electrophiles. For example, the cytotoxicity and mutagenicity of dibromoethane and diepoxybutane is enhanced in bacteria overexpressing O6-alkylguanine DNA transferase (AGT) protein due to the formation of covalent AGT-DNA conjugates.45 Furthermore, proteins functionalized with DNA-reactive 2-hydroxy-3,4-epoxybutyl groups induced cell death and mutations in human cells.46
Identification of the proteins involved in DPC formation in living cells and determining the mechanisms of DPC repair are important for our understanding of the etiology of cardiovascular disease, age-related neurodegeneration, and cancer. DPCs accumulate in the heart and brain tissues of mice with age47 and are generated as a result of ischemia-reperfusion injury (Groehler et al., unpublished observations). DPC formation is likely to contribute to both target and off-target toxicity of DNA bis-alkylating agents commonly used as antitumor drugs, and single nucleotide polymorphisms in DPC repair genes may contribute to inter-individual differences in response to antitumor therapy.
In the first part of this study, a mass spectrometry-based approach was used to characterize DNA-protein cross-linking following exposure of human cells to two biologically active metabolites of CPA (PM and NOR, Scheme 1). We found that PM and NOR induced covalent cross-links between DNA and over 130 cellular proteins, including those participating in chromatin remodeling, translation, DNA replication, DNA repair, RNA metabolism, transcriptional regulation, and apoptosis (Table 1). These proteins are involved in a variety of cellular functions including transcriptional regulation (e.g. prohibitin-2, protein FAM50A, and transcription activator BRG1), RNA splicing/processing (e.g. splicing factor, arginine/serine-rich 3, splicing factor 3B subunit 3, and 116 kDa U5 small nuclear ribonucleoprotein component), chromatin organization (e.g. core histone macro-H2A.2, SWI/SNF-related matrix associated actin dependent regulator of chromatin subfamily E member 1, chromodomain-helicase-DNA-binding protein 4), protein transport (e.g. charged multivesicular body protein 6, AP-3 complex subunit beta-1, and clathrin heavy chain1), cellular signaling (14-3-3 protein zeta/delta, DnaJ homolog subfamily B member 11, and neuropilin-1), and cell structure/architecture (actin, cytoplasmic-1, desmin, and vimentin).48–50 When the list of proteins participating in cross-linking to DNA in the presence of PM was compared to an analogous lists proteins cross-linked to DNA in the presence of DEB,22 a total of 47 proteins (30.9%) were found in common (Figure 7A). When compared to a list of cisplatin cross-linked proteins, 106 (41.4%) proteins were found in common (Figure 7B). These results suggest that nitrogen mustards and platinum drugs may target the same group of proteins for cross-linking to DNA, while a distinct group of proteins is cross-linked by DEB. Further studies will establish whether these differences in specificity could contribute to different biological outcomes of DEB (known carcinogen)51–53 and cyclophosphamide (useful antitumor agent).14–16
Figure 7.

Venn diagrams showing the overlaps between the lists of proteins that form DPCs in human cells following exposure to PM, DEB, and cisplatin.
HPLC-ESI+-MS/MS analysis have revealed that similarly to other nitrogen mustards, PM and NOR cross-link cysteine thiols of proteins to the N-7 position of guanine in DNA. Cys-NOR-N7G conjugates were formed in a concentration-dependent manner in NOR-treated cells (Figure 5). Although N7-guanine cross links are expected to have limited hydrolytic stability, they may persist long enough to affect cellular replication and transcription, potentially leading to toxicity and mutations.
The biological outcomes of DNA-protein cross-linking are currently under active investigation. Nakano et al. have shown that covalent DPCs larger than 16.0 kDa blocked the progression of DNA helicases, thus preventing the unwinding of double stranded DNA ahead of the replication fork.54 In our experiments with recombinant human DNA polymerases, we found that proteins and large peptides (> 20 amino acids) conjugated to the C-5 position of dT blocked DNA synthesis in the presence of lesion bypass polymerases η and κ, while smaller DNA-peptide conjugates (< 10-mer) were bypassed by in an error-prone manner, giving rise to deletions and point mutations.55,56 Recent in vivo studies of model peptide cross-links in human embryonic kidney cells have provided preliminary evidence for the ability of these conjugates to cause both targeted and off-target mutations (Pande et al., manuscript in preparation).
The processes by which covalent protein-DNA complexes are recognized as DNA damage and removed from genomic DNA are incompletely understood. Recently, Duxin et al used a plasmid containing a site-specific DPC to demonstrate that in Xenopus egg extracts, DPC repair is coupled to DNA replication.57 These authors proposed that the collision of the replisome/CMG helicase with a DPC located on the DNA leading strand triggers proteolytic degradation of the protein constituent of DPC.57 In the yeast, the metalloprotease Wss1 has been identified as the DPC-specific protease.58 The resulting DNA-peptide cross-links may serve as substrates for nucleotide excision repair (NER).59–61 Alternatively, Nakano et al proposed that large DPCs do not undergo proteolysis, but rather are directly repaired by the HR pathway.62,63
Our quantitative isotope dilution HPLC-ESI+-MS/MS results are consistent with a role for NER in removal of NOR-induced DPCs (Figure 6B). Cys-NOR-N7G numbers were significantly higher in cells deficient in nucleotide excision repair as compared with repair proficient cells. In contrast, the presence of functional FA pathway (PD 20 vs PD20 corrected) did not influence the numbers of Cys-NOR-N7G adducts observed in NOR-treated human cells (Figure 6B). These results are consistent with a model that in replicating human cells, NOR-induced DPCs are proteolytically digested to smaller DNA-NOR-peptide conjugates, which are subject to repair by NER. Further investigations are needed to elucidate the relative contributions of NER and HR in protecting human cells form toxic effects of DPCs and to establish the role of DNA-specific proteases such as Dvc1/Spartan58,64 in initiating DPC repair.
Supplementary Material
Acknowledgments
We thank Erin Michaelson-Richie, Teshome B. Gherezghiher, and Xun Ming for their contributions to the development of proteomics methodologies for DNA-protein cross-links, Prof. Alexandra Sobeck (University of Minnesota) for providing FA-D2 and the corresponding corrected cells, and Robert Carlson (University of Minnesota) for creating graphics for this manuscript.
Funding Sources:
This study was supported by a grant from the National Institutes of Health (ES023350, CA100670).
Abbreviations
- PM
phosphoramide mustard
- NM
nornitrogen mustard
- DPC
DNA-Protein cross-link
- Cys-NOR-N7G
N-[2-[cysteinyl]ethyl]-N-[2-(guan-7-yl)ethyl]amine
- XPA
Xeroderma Pigmentosum
- CP
cyclophosphamide
- AGT
O6-alkylguanine DNA alkyltransferase
- NER
nucleotide excision repair
- PMSF
phenylmethanesulfonyl fluoride
- Boc-Cys-OH
Boc-L-cysteine
- TFA
triflouroacetic acid
- DTT
dithiothreitol
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- PBS
phosphate-buffered saline
- EDTA
ethylenediaminetetraacetic acid
- UV
ultraviolet light
- kDa
kilodaltons
- TIC
total ion current
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PARP
poly-(ADP-ribose) polymerase 1
- Ar
argon
- DEB
diepoxybutane
- MEC
mechlorethamine
- GO
gene ontology
Footnotes
Notes
The authors declare no competing financial interest.
Supporting information
Supplementary information is provided including comparative cytotoxicity of PM, mechlorethamine, and DEB in HT1080 cells, representative MS/MS spectra of a tryptic peptide, representative HPLC-UV traces of nucleosides, western blots, and toxicity curves for PM in HT1080, XPA, PD20 and PD20-corrected cells. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Tretyakova NY, Groehler A, Ji S. DNA-Protein cross-links: formation, structural identities, and biological outcomes. Acc Chem Res. 2015;48:1631–1644. doi: 10.1021/acs.accounts.5b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhitkovich A, Voitkun V, Kluz T, Costa M. Utilization of DNA-protein cross-links as a biomarker of chromium exposure. Environ Health Perspect. 1998;106(Suppl 4):969–974. doi: 10.1289/ehp.98106s4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarti SK, Bai C, Subramanian KS. DNA-Protein crosslinks induced by nickel compounds in isolated rat renal cortical cells and its antagonism by specific amino acids and magnesium ion. Toxicol Appl Pharmacol. 1999;154:245–255. doi: 10.1006/taap.1998.8584. [DOI] [PubMed] [Google Scholar]
- 4.Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J Biol Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 5.Michaelson-Richie ED, Ming X, Codreanu SG, Loeber RL, Liebler DC, Campbell C, Tretyakova NY. Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2011;10:2785–2796. doi: 10.1021/pr200042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker JM, Parish JH, Curtis JP. DNA-DNA and DNA-protein crosslinking and repair in Neurospora crassa following exposure to nitrogen mustard. Mutat Res. 1984;132:171–179. doi: 10.1016/0167-8817(84)90035-x. [DOI] [PubMed] [Google Scholar]
- 7.Ewig RA, Kohn KW. DNA damage and repair in mouse leukemia L1210 cells treated with nitrogen mustard, 1,3-bis(2-chloroethyl)-1-nitrosourea, and other nitrosoureas. Cancer Res. 1977;37:2114–2122. [PubMed] [Google Scholar]
- 8.Zwelling LA, Anderson T, Kohn KW. DNA-protein and DNA interstrand cross-linking by cis- and trans-platinum(II) diamminedichloride in L1210 mouse leukemia cells and relation to cytotoxicity. Cancer Res. 1979;39:365–369. [PubMed] [Google Scholar]
- 9.Kloster M, Kostrhunova H, Zaludova R, Malina J, Kasparkova J, Brabec V, Farrell N. Trifunctional dinuclear platinum complexes as DNA-protein cross-linking agents. Biochemistry. 2004;43:7776–7786. doi: 10.1021/bi030243e. [DOI] [PubMed] [Google Scholar]
- 10.Miller CA, III, Cohen MD, Costa M. Complexing of actin and other nuclear proteins to DNA by cis-diamminedichloroplatinum(II) and chromium compounds. Carcinogenesis. 1991;12:269–276. doi: 10.1093/carcin/12.2.269. [DOI] [PubMed] [Google Scholar]
- 11.Chvalova K, Brabec V, Kasparkova J. Mechanism of the formation of DNA-protein cross-links by antitumor cisplatin. Nucleic Acids Res. 2007;35:1812–1821. doi: 10.1093/nar/gkm032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewig RA, Kohn KW. DNA-protein cross-linking and DNA interstrand cross-linking by haloethylnitrosoureas in L1210 cells. Cancer Res. 1978;38:3197–3203. [PubMed] [Google Scholar]
- 13.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Emadi A, Jones RJ, Brodsky RA. Cyclophosphamide and cancer: golden anniversary. Nat Rev Clin Oncol. 2009;6:638–647. doi: 10.1038/nrclinonc.2009.146. [DOI] [PubMed] [Google Scholar]
- 15.Yule SM, Foreman NK, Mitchell C, Gouldon N, May P, McDowell HP. High-dose cyclophosphamide for poor-prognosis and recurrent pediatric brain tumors: a dose-escalation study. J Clin Oncol. 1997;15:3258–3265. doi: 10.1200/JCO.1997.15.10.3258. [DOI] [PubMed] [Google Scholar]
- 16.Brodsky RA. High dose cyclophosphamide treatment for autoimmune disorders. ScientificWorldJournal. 2002;2:1808–1815. doi: 10.1100/tsw.2002.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemminki K. Binding of metabolites of cyclophosphamide to DNA in a rat liver microsomal system and in vivo in mice. Cancer Res. 1985;45:4237–4243. [PubMed] [Google Scholar]
- 18.Hemminki K. DNA-binding products of nornitrogen mustard, a metabolite of cyclophosphamide. Chem Biol Interact. 1987;61:75–88. doi: 10.1016/0009-2797(87)90020-2. [DOI] [PubMed] [Google Scholar]
- 19.Benson AJ, Martin CN, Garner RC. N-(2-hydroxyethyl)-N-[2-(7-guaninyl)ethyl]amine, the putative major DNA adduct of cyclophosphamide in vitro and in vivo in the rat. Biochem Pharmacol. 1988;37:2979–2985. doi: 10.1016/0006-2952(88)90285-7. [DOI] [PubMed] [Google Scholar]
- 20.Hansson J, Lewensohn R, Ringborg U, Nilsson B. Formation and removal of DNA cross-links induced by melphalan and nitrogen mustard in relation to drug-induced cytotoxicity in human melanoma cells. Cancer Res. 1987;47:2631–2637. [PubMed] [Google Scholar]
- 21.Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. Cross-linking of the DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem Res Toxicol. 2008;21:787–795. doi: 10.1021/tx7004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gherezghiher TB, Ming X, Villalta PW, Campbell C, Tretyakova NY. 1,2,3,4-Diepoxybutane-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J Proteome Res. 2013;12:2151–2164. doi: 10.1021/pr3011974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien J, Wilson I, Orton T, Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 24.Yates JR, III, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 25.Camon E, Barrell D, Brooksbank C, Magrane M, Apweiler R. The Gene Ontology Annotation (GOA) Project--Application of GO in SWISS-PROT, TrEMBL and InterPro. Comp Funct Genomics. 2003;4:71–74. doi: 10.1002/cfg.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schonthaler HB, Guinea-Viniegra J, Wculek SK, Ruppen I, Ximenez-Embun P, Guio-Carrion A, Navarro R, Hogg N, Ashman K, Wagner EF. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity. 2013;39:1171–1181. doi: 10.1016/j.immuni.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Hublitz P, Kunowska N, Mayer UP, Muller JM, Heyne K, Yin N, Fritzsche C, Poli C, Miguet L, Schupp IW, van Grunsven LA, Potiers N, van Dorsselaer A, Metzger E, Roemer K, Schule R. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev. 2005;19:2912–2924. doi: 10.1101/gad.351205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz VM, Mori S, Longobardi E, Menendez G, Ferrai C, Keough RA, Bachi A, Blasi F. p160 Myb-binding protein interacts with Prep1 and inhibits its transcriptional activity. Mol Cell Biol. 2007;27:7981–7990. doi: 10.1128/MCB.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori S, Bernardi R, Laurent A, Resnati M, Crippa A, Gabrieli A, Keough R, Gonda TJ, Blasi F. Myb-binding protein 1A (MYBBP1A) is essential for early embryonic development, controls cell cycle and mitosis, and acts as a tumor suppressor. PLoS One. 2012;7:e39723. doi: 10.1371/journal.pone.0039723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKay MJ, Troelstra C, van der SP, Kanaar R, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers JH. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- 31.Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem. 2012;287:6764–6772. doi: 10.1074/jbc.M111.287037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Argraves WS, Suzuki S, Arai H, Thompson K, Pierschbacher MD, Ruoslahti E. Amino acid sequence of the human fibronectin receptor. J Cell Biol. 1987;105:1183–1190. doi: 10.1083/jcb.105.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 34.Fu GK, Markovitz DM. The human LON protease binds to mitochondrial promoters in a single-stranded, site-specific, strand-specific manner. Biochemistry. 1998;37:1905–1909. doi: 10.1021/bi970928c. [DOI] [PubMed] [Google Scholar]
- 35.Gabrielli F, Donadel G, Bensi G, Heguy A, Melli M. A nuclear protein, synthesized in growth-arrested human hepatoblastoma cells, is a novel member of the short-chain alcohol dehydrogenase family. Eur J Biochem. 1995;232:473–477. doi: 10.1111/j.1432-1033.1995.473zz.x. [DOI] [PubMed] [Google Scholar]
- 36.Vandekerckhove J, Weber K. Mammalian cytoplasmic actins are the products of at least two genes and differ in primary structure in at least 25 identified positions from skeletal muscle actins. Proc Natl Acad Sci U S A. 1978;75:1106–1110. doi: 10.1073/pnas.75.3.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 38.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 39.Wada T, Orphanides G, Hasegawa J, Kim DK, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, Handa H. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell. 2000;5:1067–1072. doi: 10.1016/s1097-2765(00)80272-5. [DOI] [PubMed] [Google Scholar]
- 40.Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H, Goodman R, Lozano G, Zhao Y, Lu H. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell. 2001;7:283–292. doi: 10.1016/s1097-2765(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 41.Belotserkovskaya R, Reinberg D. Facts about FACT and transcript elongation through chromatin. Curr Opin Genet Dev. 2004;14:139–146. doi: 10.1016/j.gde.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Clauson C, Scharer OD, Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guainazzi A, Scharer OD. Using synthetic DNA interstrand crosslinks to elucidate repair pathways and identify new therapeutic targets for cancer chemotherapy. Cell Mol Life Sci. 2010;67:3683–3697. doi: 10.1007/s00018-010-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knipscheer P, Raschle M, Scharer OD, Walter JC. Replication-coupled DNA interstrand cross-link repair in Xenopus egg extracts. Methods Mol Biol. 2012;920:221–243. doi: 10.1007/978-1-61779-998-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valadez JG, Liu L, Loktionova NA, Pegg AE, Guengerich FP. Activation of bis-electrophiles to mutagenic conjugates by human O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- 46.Tretyakova NY, Michaelson-Richie ED, Gherezghiher TB, Kurtz J, Ming X, Wickramaratne S, Campion M, Kanugula S, Pegg AE, Campbell C. DNA-reactive protein monoepoxides induce cell death and mutagenesis in mammalian cells. Biochemistry. 2013;52:3171–3181. doi: 10.1021/bi400273m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izzotti A, Cartiglia C, Taningher M, De Flora S, Balansky R. Age-related increases of 8-hydroxy-2′-deoxyguanosine and DNA-protein crosslinks in mouse organs. Mutat Res. 1999;446:215–223. doi: 10.1016/s1383-5718(99)00189-8. [DOI] [PubMed] [Google Scholar]
- 48.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 49.Paulin D, Li Z. Desmin: a major intermediate filament protein essential for the structural integrity and function of muscle. Exp Cell Res. 2004;301:1–7. doi: 10.1016/j.yexcr.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Dave JM, Bayless KJ. Vimentin as an integral regulator of cell adhesion and endothelial sprouting. Microcirculation. 2014;21:333–344. doi: 10.1111/micc.12111. [DOI] [PubMed] [Google Scholar]
- 51.Sasiadek M, Norppa H, Sorsa M. 1,3-Butadiene and its epoxides induce sister-chromatid exchanges in human lymphocytes in vitro. Mutat Res. 1991;261:117–121. doi: 10.1016/0165-1218(91)90057-s. [DOI] [PubMed] [Google Scholar]
- 52.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: I. Mutagenic potential of 1,2-epoxybutene, 1,2,3,4-diepoxybutane and 3,4-epoxy-1,2-butanediol in cultured human lymphoblasts. Carcinogenesis. 1994;15:713–717. doi: 10.1093/carcin/15.4.713. [DOI] [PubMed] [Google Scholar]
- 53.Himmelstein MW, Turner MJ, Asgharian B, Bond JA. Metabolism of 1,3-butadiene: inhalation pharmacokinetics and tissue dosimetry of butadiene epoxides in rats and mice. Toxicology. 1996;113:306–309. doi: 10.1016/0300-483x(96)03462-2. [DOI] [PubMed] [Google Scholar]
- 54.Nakano T, Miyamoto-Matsubara M, Shoulkamy MI, Salem AM, Pack SP, Ishimi Y, Ide H. Translocation and stability of replicative DNA helicases upon encountering DNA-protein cross-links. J Biol Chem. 2013;288:4649–4658. doi: 10.1074/jbc.M112.419358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickramaratne S, Boldry EJ, Buehler C, Wang YC, Distefano MD, Tretyakova NY. Error-prone translesion synthesis past DNA-peptide cross-links conjugated to the major groove of DNA via C5 of thymidine. J Biol Chem. 2015;290:775–787. doi: 10.1074/jbc.M114.613638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeo JE, Wickramaratne S, Khatwani S, Wang YC, Vervacke J, Distefano MD, Tretyakova NY. Synthesis of site-specific DNA-protein conjugates and their effects on DNA replication. ACS Chem Biol. 2014;9:1860–1868. doi: 10.1021/cb5001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duxin JP, Dewar JM, Yardimci H, Walter JC. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014;159:346–357. doi: 10.1016/j.cell.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014;158:327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 59.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 60.Reardon JT, Cheng Y, Sancar A. Repair of DNA-protein cross-links in mammalian cells. Cell Cycle. 2006;5:1366–1370. doi: 10.4161/cc.5.13.2892. [DOI] [PubMed] [Google Scholar]
- 61.Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O’Connor TR. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J Biol Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 62.Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AM, Izumi S, Pack SP, Makino K, Ide H. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 63.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, Van Houten B, Iijima K, Tauchi H, Ide H. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maskey RS, Kim MS, Baker DJ, Childs B, Malureanu LA, Jeganathan KB, Machida Y, van Deursen JM, Machida YJ. Spartan deficiency causes genomic instability and progeroid phenotypes. Nat Commun. 2014;5:5744. doi: 10.1038/ncomms6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


