Abstract
Growth of a microorganism in a host is essential for infection, and bacterial pathogens have evolved to utilize specific metabolites to enhance replication in vivo. Now, emerging data demonstrate that pathogens rely on microbiota-derived metabolites as a form of bacterial-bacterial communication to gain information about location within a host and modify virulence gene expression accordingly. Thus, metabolite-sensing is critical for pathogens to establish infection. Here, we highlight recent examples of how the foodborne pathogen enterohemorrhagic Escherichia coli O157:H7 (EHEC) exploits microbiota-derived metabolites to recognize the host intestinal environment and control gene expression that results in controlled expression of virulence traits.
Graphical Abstract
Introduction
The gastrointestinal (GI) tract is inhabited by trillions of commensal bacteria collectively referred to as the resident microbiota that aid in the development of the immune and digestive systems as well as in vitamin and nutrient production [1]. Additionally, the microbiota act as a barrier against infection by invading pathogens through efficiently utilizing nutrients, and thereby limiting the growth of pathogens within the host. This process is called colonization resistance [1]. However, bacterial pathogens have also evolved means of overcoming colonization resistance by utilizing non-competitive substrates for growth and/or exploiting dysbiosis, which is a disturbance in the microbiota that commonly results from antibiotic use [2, 3]. In addition to selectively using metabolites for in vivo replication, emerging data have revealed that microbiota-derived metabolites form the basis for communicating spatiotemporal information to bacterial pathogens. This is used to adapt to distinct host niches and precisely control the expression of virulence traits.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 causes major outbreaks of hemorrhagic colitis and hemolytic uremic syndrome (HUS) worldwide. EHEC colonizes the large intestine and forms attaching and effacing (AE) lesions on the intestinal surface. AE lesions are characterized by intimate attachment of EHEC to enterocytes and effacement of the microvilli [4–6]. The locus of effacement (LEE) pathogenicity island encodes a type three secretion system and most of the effectors required for AE lesions [7], as well as the transcription factor Ler that is the master regulator of the LEE [8]. EHEC also produces a Shiga toxin that causes HUS and can lead to fatal outcomes associated with EHEC infections [9]. Significantly, EHEC has a very low infectious dose (as low as 50 colony forming units) [10], which is a major factor contributing to outbreaks, and indicates that EHEC has evolved mechanisms to outcompete commensal bacteria for nutrients and rapidly respond to environmental cues to coordinate expression of virulence traits. In this review, we highlight recent, notable examples of commensal-derived metabolites that EHEC senses as signals of the intestinal environment and the corresponding regulatory cascades that modulate expression of the LEE and Shiga toxin, as well as other virulence factors important for host colonization and infection.
Ethanolamine
Ethanolamine is a component of phosphatidylethanolamine, an abundant lipid in eukaryotic and bacterial cell membranes. As a component of phosphatidylethanolamine and other modified lipid molecules, ethanolamine is an important signaling molecule and influences immunomodulation, cell division, nutritional intake, and energy balance [11–14]. The exfoliation of enterocytes as well as the turnover of bacterial cells releases an abundant and replenished supply of ethanolamine in the GI tract. Although ethanolamine can serve as a carbon and/or nitrogen source for bacteria, the resident microbiota do not readily metabolize ethanolamine [15]. Thus, intestinal pathogens, including EHEC, utilize ethanolamine to sidestep nutritional competition and enhance growth during infection [3, 15–17].
Significantly, bacterial pathogens respond to ethanolamine as a signaling molecule to activate virulence gene expression [18–20••] (Fig. 1). In EHEC, ethanolamine activates the expression of genes critical for colonization of the GI tract, including fimbrial adhesins and the LEE, as well as genes encoding Shiga toxin [19••, 20]. Fimbrial adhesins are extracellular proteinaceous structures that mediate binding of bacteria to surfaces, including host epithelial cells. EHEC encodes 16 distinct fimbrial loci [21, 22], and these fimbriae may be important for initial adherence to enterocytes, that precedes intimate, LEE-dependent adherence [23]. However, the contribution of many fimbrial loci to EHEC pathogenesis has been elusive due to the difficulties of expressing fimbrial genes in vitro [24]. Thus, the finding that the biologically relevant molecule ethanolamine promotes expression of EHEC fimbriae suggests that these fimbriae play a role in the ability of EHEC to establish infection. Additionally, ethanolamine activates expression of global regulators in EHEC [20], suggesting that ethanolamine plays a central role in integrating multiple cues, to optimize timing of virulence gene expression.
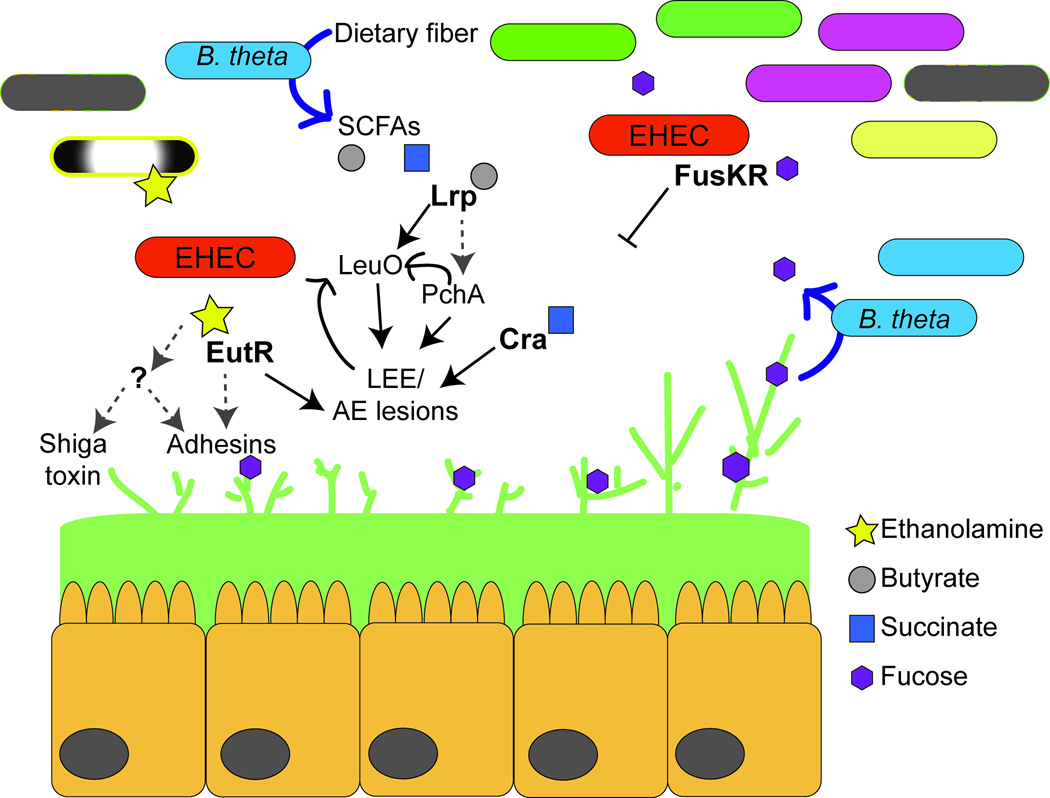
Figure 1.
EHEC responds to microbiota-derived metabolites to activate complex regulatory cascades. EHEC senses ethanolamine, butyrate, and succinate to activate virulence gene expression, whereas the two-component system FusKR represses LEE expression in response to fucose. Ethanolamine is sensed through EutR as well as an unidentified ethanolamine sensor; butyrate is sensed through the transcription factor Lrp; succinate is sensed by Cra. Black lines indicate direct interaction, and grey hashed arrows indicate indirect regulation. For details, refer to main text.
EHEC and other members of the Enterobacteriaceae carry the ethanolamine utilization operon, which contains 17 genes that encodes for the transport and breakdown of ethanolamine [25, 26]. This operon also encodes EutR, which belongs to the AraC/XylS family of transcriptional regulators. EutR directly senses ethanolamine to promote eut transcription [20, 27]. Biochemical studies revealed that EutR directly regulates LEE expression by binding to the ler promoter [28•], and genetic data suggest that EutR also regulates a subset of EHEC fimbriae [19]. Importantly, EutR-dependent virulence gene regulation is independent of ethanolamine metabolism, as a deletion of the ethanolamine catalytic enzymes did not affect the ability of EHEC to sense ethanolamine and activate expression of the LEE and Shiga toxin [20]. Genetic data also indicate that EHEC encodes a second ethanolamine sensor, as a eutR deletion strain of EHEC is able to promote gene expression in response to ethanolamine [20]. These findings suggest that ethanolamine is a critical signal for EHEC to recognize the host intestinal environment.
Microbiota-liberated sugars
The intestinal epithelium is covered by stratified layers of mucus. In addition to regulating water, vitamin, mineral, and electrolyte absorption, the mucus serves as a physical and chemical barrier separating the resident microbiota and epithelium [29]. Mucus is composed of glycosylated proteins, called mucins, antimicrobial peptides, enzymes, as well as monosaccharides, including fucose [30, 31]. Some members of the resident microbiota, such as Bacteroides thetaiotamicron (B. theta), contain fucosidases that cleave fucose from host mucin, which leads to fucose availability in the GI tract [30, 32, 33].
EHEC uses a two-component system FusKR to sense luminal fucose and repress LEE expression [34••] (Fig. 1). Two-component systems are composed of a histidine kinase and a response regulator. The histidine kinase phosphorylates in response to a particular environmental stimulus and then transfers the phosphate to a response regulator that typically binds DNA to promote or repress gene transcription [35]. FusK is the histidine kinase that senses fucose and relays this information to the response regulator FusR [34]. FusR directly represses LEE expression, which results in a corresponding decrease in AE lesion formation [34]. In vivo studies demonstrated that FusKR enhances EHEC colonization of the GI tract [34] because it functions to decrease unnecessary energy expenditure by repressing the expression of the LEE virulence genes in the lumen where fucose is abundant and expression of the LEE-encoded type three secretion system and effectors would be unproductive. When EHEC reaches the epithelium FusKR is no longer active due to lower fucose concentrations as well as transcriptional repression of fusK and fusR by the QseBC two-component system that senses epinephrine and norepinephrine [34, 36].
Short chain fatty acids
Short chain fatty acids (SCFAs), which include acetate, butyrate, propionate, and succinate [37–41], are a subset of fatty acids that are produced by the gut microbiota during the fermentation of partially digestible and nondigestible polysaccharides [42]. The diet also provides a source of SFCAs; however, the anaerobic members of the microbiota are largely responsible for the generation of SFCAs, as significantly lower amounts of SCFAs are detected in the intestines of germ-free mice compared to conventionally colonized mice [43]. SCFAs play important roles in host physiology by serving as a major energy source for enterocytes, regulating diverse cellular processes, and influencing inflammatory signaling and the immune responses [44–47].
The largest concentrations of SCFAs are measured in the proximal colon [42]; therefore, it is not surprising that bacterial pathogens rely on sensing SCFAs as a signal to indicate arrival at this site. EHEC responds generally to SCFAs to activate expression of genes encoding flagella and motility [48]; however, butyrate, specifically, enhances LEE gene expression and adherence to epithelial cells [49]. In response to butyrate, the leucine-responsive regulatory (Lrp) protein initiates a signaling cascade that promotes expression of pchA [49], which encodes a direct activator (PchA) of the LEE [50, 51]. Recent work by Takao et al. revealed that butyrate-dependent activation of the LEE is even more complex and also includes the transcription factor LeuO [52•]. Lrp directly promotes expression of leuO, which in turn also binds the ler promoter to activate expression of the LEE and microcolony formation. Interestingly, LeuO activation of the LEE genes required PchA and both PchA and Ler activated leuO expression. This positive feedback mechanism is hypothesized to function in prolonging expression of the LEE [52] (Fig. 1).
EHEC also senses succinate to activate virulence gene expression [53••] (summarized in Fig. 1). Succinate is a major by-product of fermentation by Bacteroides species [54], which are a significant component of the resident anaerobic microbiota. EHEC senses succinate through the transcription factor Cra to enhance virulence gene expression and AE lesion formation [53].
The important role of succinate in bacterial virulence was corroborated using a mouse model of infection with Citrobacter rodentium. C. rodentium is a murine pathogen that carries the LEE and recapitulates EHEC colonization during infection (recently reviewed in [55]). During C. rodentium infection, mice reconstituted with B. theta presented with more severe disease manifestations compared to mice that were depleted of the normal microbiota. This was due to increased concentrations of succinate in mice with B. theta compared to mice in which B. theta was absent [53]. C. rodentium carries the cra gene (also annotated as fruR) [56], which shares 99% homology to EHEC Cra [21, 22] (based on amino acid sequences); therefore, succinate sensing through Cra may be a conserved mechanism that AE pathogens use to gauge gluconeogenic versus glycolytic conditions (in conjunction with fucose sensing) within the intestine [57].
Host-derived metabolites
It should be noted that host metabolites also influence EHEC virulence gene expression and host colonization. For example, host-generated bicarbonate influences EHEC and C. rodentium virulence [58–60]. Gastric and duodenal mucosae secrete bicarbonate to the lumen to regulate intestinal pH and maintain homeostasis [61, 62]. In C. rodentium, bicarbonate is sensed through the global regulator RegA. RegA belongs to the AraC/XylS family of transcriptional regulators and shares homology to important virulence regulators, including Rns in enterotoxigenic E. coli, AggR in enteroaggregative E. coli, and PerA in enteropathogenic E. coli [59]. Upon sensing bicarbonate, RegA expression and activity is stimulated, which results in activation of the LEE genes, through direct regulation of the LEE-encoded regulator grlA [63]. RegA also controls expression of fimbrial and afimbrial adhesins that impact C. rodentium adherence to epithelial cells [59, 60]. In addition to bicarbonate, the vitamin biotin and the amino acid D-serine influence LEE expression [64•, 65•]. The small intestine is the main site of biotin absorption, and therefore, biotin concentrations are likely to higher at this site compared to the large intestine [65, 66]. Moreover, D-serine is present in the large intestine, but is most abundant at extraintestinal sites, including in the urinary tract [67, 68]. These high concentrations of biotin and D-serine repress LEE expression, suggesting a model in which biotin and D-serine confer niche specificity by preventing EHEC colonization of sites outside of the large intestine [64, 65]. Altogether, these studies highlight complex regulatory pathways based on host- and bacterial-derived cues to ensure precise and coordinated expression of virulence genes.
Conclusions
Growth within a host is a requisite for bacterial infection, and bacterial pathogens take advantage of non-competitive metabolites and/or host dysbiosis to survive and replicate within a host. However, it is becoming increasingly clear that metabolites contribute to pathogenesis beyond promoting growth: metabolites are important signals that pathogens exploit to recognize specific host niches and appropriately modulate virulence gene expression. Here, we described how EHEC relies on sensing microbiota-derived metabolites to integrate several cues to precisely regulate expression of virulence traits. In addition to uncovering novel and complex signaling pathways, these studies have revealed the physiological conditions that promote expression of uncharacterized virulence factors. Importantly, as research continues to focus on the impact of the microbiota-derived metabolites on host physiology, including resistance and susceptibility to infectious diseases, and for their potential use as probiotic therapies [44, 69–71], it is exceedingly important to better elucidate how microbiota-derived metabolites impact bacterial virulence. A greater understanding of the interaction between bacterial virulence and microbiota-derived metabolites will provide a more comprehensive understanding of the impact of current therapies on host physiology as well as assist in the development of novel treatments for infectious diseases.
Highlights.
The commensal microbiota communicate to pathogens through the release of nutrients
Pathogens sense microbiota-derived metabolites as cues to recognize host niches and control gene expression
Better understanding of metabolite-based signaling pathways is necessary to study bacterial virulence factors and develop novel therapies to treat infectious diseases
Acknowledgments
This work was supported by NIH NIAID grant R01AI118732 to MMK. DHL received support through NIH Training Grant 5 T32 AI055432. The authors apologize to colleagues whose work could not be cited due to the length limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2012;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–101. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. 1995;92(17):7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jerse AE, Yu J, Tall BD, Kaper JB. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;14:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 7.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. 1995;92(5):1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol. Microbiol. 1999;33(2):296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 9.Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. Escherichia coli cytotoxin, haemolyticuraemic syndrome, and haemorrhagic colitis. Lancet. 1983;2:1299–1300. doi: 10.1016/s0140-6736(83)91167-4. [DOI] [PubMed] [Google Scholar]
- 10.Tilden JJ, Young W, McNamara AM, Custer C, Boesel B, Lambert-Fair MA, Majkowski J, Vugia D, Werner SB, Hollingsworth J, et al. A new route of transmission for Escherichia coli : infection from dry fermented salami. Am. J. Public Health. 1996;86(8):1142–1145. doi: 10.2105/ajph.86.8_pt_1.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakovic M, Fullerton MD, Michel V. Metabolic and moleclar aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyl-transferase (Pcyt2) Biochem. Cell Biol. 2007;85:283–300. doi: 10.1139/o07-006. [DOI] [PubMed] [Google Scholar]
- 12.Gibellini F, Smith TK. The Kennedy pathway- de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. Life. 2010;62(6):414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 13.Meijerink J, Plastina P, Vincken JP, Poland M, Attya M, Balvers M, Gruppen H, Gabriele B, Witkamp RF. The ethanolamide metabolite of DHA, docosahexaenoylethanolamine, shows immunomodulating effects in mouse peritoneal and RAW264.7 macrophages: evidence for a new link between fish oil and inflammation. Br. J. Nutr. 2011;105:1798–1807. doi: 10.1017/S0007114510005635. [DOI] [PubMed] [Google Scholar]
- 14.Sugiura T, Kobayashi Y, Oka S, Waku K. Biosynthesis and degradation of anandamide and 2-arachidonoylglycerol and their possible physiological significance. Prostaglandins Leukot Essent Fatty Acids. 2002;66:173–192. doi: 10.1054/plef.2001.0356. [DOI] [PubMed] [Google Scholar]
- 15.Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 2011;13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 16.Maadani A, Fox KA, Mylonakis E, Garsin DA. Enterococcus faecalis mutations affecting virulence in Caenorhabditis elegans model host. Infect. Immun. 2007;75:2634–2637. doi: 10.1128/IAI.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellin JR, Koutero M, Dar D, Nahori MA, Sorek R, Cossart P. Riboswitches. Sequestration of a twocomponent response regulator by a riboswitch-regulated noncoding RNA. Science. 2014;345:940–943. doi: 10.1126/science.1255083. [DOI] [PubMed] [Google Scholar]
- 18.Anderson CJ, Clark DE, Adli M, Kendall MM. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog. 2015;11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••. Gonyar LA, Kendall MM. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2014;82:193–201. doi: 10.1128/IAI.00980-13. This paper reported expression of several fimbrial loci for the first time.
- 20••. Kendall MM, Gruber CC, Parker CT, Sperandio V. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio. 2012;3:e00050-12. doi: 10.1128/mBio.00050-12. The first description of ethanolamine as a signaling molecule.
- 21.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 22.Perna NT, Plunkett Gr, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 23.Farfan MJ, Torres AG. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 2012:903–913. doi: 10.1128/IAI.05907-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Low AS, Holden N, Rosser T, Roe AJ, Constantinidou C, Hobman JL, Smith DGE, Low JC, Gally DL. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 2006;8(6):1033–1047. doi: 10.1111/j.1462-2920.2006.00995.x. [DOI] [PubMed] [Google Scholar]
- 25.Roof DM, Roth JR. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 1988;170(9):3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsoy O, Ravcheev D, Mushegian A. Comparative genomics of ethanolamine utilization. J. Bacteriol. 2009;191(23):7157–7164. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roof DM, Roth JR. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 1992;174:6634–6643. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•. Luzader DH, Clark DE, Gonyar LA, Kendall MM. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2013;195:4947–4953. doi: 10.1128/JB.00937-13. First biochemical and mechanistic description of EutR binding ethanolamine and target promoters.
- 29.Sellers RS, Morton D. The colon: from banal to brillant. Toxicol. Path. 2014;42:67–81. doi: 10.1177/0192623313505930. [DOI] [PubMed] [Google Scholar]
- 30.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of Oglycans in normal human mucins along the intestinal tract. Biochem J. 2004;384(Pt 2):307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13(7):41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 32.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10(4):336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaswal VM, Babbar HS, Mahmood A. Changes in sialic acid and fucose contents of enterocytes across the crypt-villus axis in developing rat intestine. Biochem Med Metab Biol. 1988;39(1):105–110. doi: 10.1016/0885-4505(88)90064-3. [DOI] [PubMed] [Google Scholar]
- 34••. Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. First description of the TCS FusKR and demonstrated a mechanism for how EHEC distinguishes luminal from epithelial niches in the host.
- 35.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 36.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc. Natl. Acad. Sci. 2006;103(27):10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.denBesten G, vanEunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62(1):67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 40.Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70(6):443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 41.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81(3):1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 42.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 43.Hoverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J. Nutr. 1986;116:1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 44.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 45.Hamer HM, Jonkers D, Venema K, Vanhoutvin ST FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol. Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 46.Millard AL, Mertes PM, Ittelet D, Villard F, Jeannesson P, Bernard J. Butyrate affects differentiation, maturation and function of human monocyte-derived dendritic cells and macrophages. Clin. Exp. Immunol. 2002;130:245–255. doi: 10.1046/j.0009-9104.2002.01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mills E, O'Neill LAJ. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 2014;24:313–320. doi: 10.1016/j.tcb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Tobe T, Nakanishi N, Sugimoto N. Activation of motility by sensing short-chain fatty acids via two steps in a flagellar gene regulatory cascade in enterohemorrhagic Escherichia coli. Infect. Immun. 2011;79:1016–1024. doi: 10.1128/IAI.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology. 2009;155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 50.Abe A, Miyahara A, Oshima T, Tashiro K, Ogura Y, Kuhara S, Ogasawara N, Hayashi T, Tobe T. Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 2008;15:25–38. doi: 10.1093/dnares/dsm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyoda S, Watanabe H. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology. 2004;150(7):2357–2571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- 52•. Takao M, Yen H, Tobe T. LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 2014;93:1302–1313. doi: 10.1111/mmi.12737. This report further investigates the mechanistic aspects of butyrate signaling in EHEC virulence regulation and identified a new regulator in this cascade.
- 53••. Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric enfection through modification of the metabolic landscape. Cell Host Microbe. 2014;16(6):759–769. doi: 10.1016/j.chom.2014.11.005. This paper shows in vitro and in vivo data that microbiota-derived succinated promotes EHEC virulence.
- 54.Macy JM, Ljungdahl LG, Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J. Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins JW, Keeney KM, Crepin VF, Rathinam VAK, Fitzgerald KA, Finlay BB, Frankel G. Citrobacter rodentium : infection, inflammation, and the microbiota. Nat. Rev. Microbiol. 2014;12:612–623. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- 56.Petty NK, Bulgin R, Crepin VF, Cerdeno-Tarraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron AM, Clark L, et al. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 2010;192(2):525–538. doi: 10.1128/JB.01144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio. 2012;3:e00280-00212. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abe H, Tatsuno I, Tobe T, Okutani A, Sasakawa C. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2002;70:3500–3509. doi: 10.1128/IAI.70.7.3500-3509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hart E, Yang J, Tauschek M, Kelly M, Wakefield MJ, Frankel G, Hartland EL, Robins-Browne RM. RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect. Immun. 2008;76:5347–5256. doi: 10.1128/IAI.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J, Hart E, Tauschek M, Price GD, Hartland EL, Strugnell RA, Robins-Browne RM. Bicarbonatemediated transcriptional activation of divergent operons by the virulence regulatory protein, RegA, from Citrobacter rodentium. Mol. Microbiol. 2008;68:314–327. doi: 10.1111/j.1365-2958.2008.06171.x. [DOI] [PubMed] [Google Scholar]
- 61.Knutson L, Flemstrom G. Duodenal mucosal bicarbonate secretion in man. Stimulation by acid and inhibition by the alpha 2-adrenoceptor agonist clonidine. Gut. 1989;30(12):1708–1715. doi: 10.1136/gut.30.12.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer JH, Way LW, Grossman MI. Pancreatic bicarbonate response to various acids in duodenum of the dog. Am. J. Physiol. 1970;219:964–970. doi: 10.1152/ajplegacy.1970.219.4.964. [DOI] [PubMed] [Google Scholar]
- 63.Tauschek M, Yang J, Hocking D, Azzopardi K, Tan A, Hart E, Praszkier J, Robins-Browne RM. Transcriptional analysis of the grlRA virulence operon from Citrobacter rodentium. J. Bacteriol. 2010;192:3722–3734. doi: 10.1128/JB.01540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•. Connolly JPR, Goldstone RJ, Burgess K, Cogdell RJ, Beatson SA, Vollmer W, Smith DGE, Roe AJ. The host metabolite D-serine contributes to bacterial niche specificity through gene selection. ISME J. 2015;9:1052. doi: 10.1038/ismej.2015.17. An interesting example of how negative regulation is important to confer colonization specificity within a host.
- 65•. Yang B, Feng L, Wang F, Wang L. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat. Commun. 2015;6:6592. doi: 10.1038/ncomms7592. An interesting example of how negative regulation is important to confer colonization specificity within a host.
- 66.Said HM. Cellular uptake of biotin: mechanisms and regulation. J. Nutr. 1999;129(2S Suppl):490S–493S. doi: 10.1093/jn/129.2.490S. [DOI] [PubMed] [Google Scholar]
- 67.Anfora AT, Haugen BJ, Roesch P, Redford P, RA W. Roles of serine accumulation and catabolism in the colonization of the murine urinary tract by Escherichia coli CFT073. Infect. Immun. 2007;75:5298–5304. doi: 10.1128/IAI.00652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Y, Nishikawa T, Satoh K, Iwata T, Fukushima T, Santa T, Homma H, Imai K. Urinary excretion of D-serine in human: comparison of different ages and species. Biol. Pharm. Bull. 1998;21:156–162. doi: 10.1248/bpb.21.156. [DOI] [PubMed] [Google Scholar]
- 69.Frankel W, Lew J, Su B, Bain A, Klurfeld D, Einhorn E, MacDermott RP, Rombeau J. Butyrate increases colonocyte protein synthesis in ulcerative colitis. J Surg Res. 1994;57:210–214. doi: 10.1006/jsre.1994.1133. [DOI] [PubMed] [Google Scholar]
- 70.Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N. Engl. J. Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 71.Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DC, O'Brien AD. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc. Natl. Acad. Sci. 2013;110:E2126–E2133. doi: 10.1073/pnas.1222014110. [DOI] [PMC free article] [PubMed] [Google Scholar]