Abstract
The identification of somatic genetic alterations that confer sensitivity to pharmacologic inhibitors has led to new cancer therapies. To identify mutations that confer an exceptional dependency, shRNA-based loss-of-function data were analyzed from a dataset of numerous cell lines to reveal genes that are essential in a small subset of cancer cell lines. Once these cell lines were determined, detailed genomic characterization from these cell lines was utilized to ascertain the genomic aberrations that led to this extreme dependency. This method, in a large subset of lung cancer cell lines, yielded a single lung adenocarcinoma cell line, NCI-H1437, which is sensitive to RNA interference of MAP2K1 expression. Notably, NCI-H1437 is the only lung line included in the dataset with a known activating mutation in MAP2K1 (Q56P). Subsequent validation using shRNA and CRISPR-Cas9 confirmed MAP2K1 dependency. In vitro and in vivo inhibitor studies established that NCI-H1437 cells are sensitive to MEK1 inhibitors, including the FDA-approved drug trametinib. Like NCI-H1437 cells, the MAP2K1 mutant cell lines SNU-C1 (colon) and OCUM-1 (gastric) showed decreased viability after MAP2K1 depletion via Cas9-mediated gene editing. Similarly, these cell lines were particularly sensitive to trametinib treatment compared to control cell lines. Based on these data cancers that harbor driver mutations in MAP2K1 should benefit from treatment with MEK1 inhibitors. Furthermore, this functional data mining approach provides a general method to experimentally test genomic features that confer dependence in tumors.
Keywords: MAP2K1, MEK, Targeted Therapy, NSCLC, Lung Cancer
Introduction
Targeted therapies have revolutionized cancer gene discovery and drug development. Unlike conventional cancer treatments such as radiation and general chemotherapy, targeted therapies seek to inhibit somatically mutated or dysregulated genes found preferentially in cancer cells. For example, the drug imatinib inhibits the essential BCR-ABL1 gene fusion found in most chronic myelogenous leukemia patients, and estrogen receptor (ER) inhibitors benefit patients with ER positive luminal breast cancer (1,2). These discoveries have influenced cancer researchers to identify essential driver oncogenes that could lead to targeted drug development efforts in specific tumor types.
Retrospective data from clinical trials allow a converse strategy that couples effective targeted therapies to specific cancer mutations by sequencing tumors of the few cancer patients that show a positive response to a drug that fails to have an effect on the majority of patients in the trial (3). Identifying a common mutant gene in these patients may reveal the cancer specific target that is affected by the drug. Such “exceptional responder” patient studies have identified sensitivity to the drug everolimus in bladder cancers harboring TSC1 mutations, and revealed mutant ARAF as a potential target in a patient with an outlier response to the drug sorafenib (3–5). While relatively rare, these N-of-1 studies are a powerful resource for identifying specific genetic events that allow certain drugs to be effective in actual cancer patients, information that is elusive for many cancer drugs.
The advent of next generation sequencing has facilitated the genomic characterization of patients’ tumors and enabled large-scale functional genomic analyses in vitro. The Cancer Cell Line Encyclopedia (CCLE, http://www.broadinstitute.org/ccle) and Sanger Cell Lines Project (cancer.sanger.ac.uk) have created publicly accessible databases documenting genomic alterations in hundreds of cancer cell lines (6,7). These data can potentially be paired with genome-scale functional genomics screens of essentiality to match genetic dependencies with mutations or gene expression patterns. One such genome-scale shRNA screen performed in 216 cancer cell lines across a variety of lineages is publicly available (www.broadinstitute.org/achilles) (8). The uniquely large number and variety of cell lines screened allows for comparison of survival differences in different genetic contexts after knockdown of individual genes, leading to hypotheses generation and subsequent preclinical validation.
To mirror “exceptional responder” studies in vitro, we mined cell line RNAi and genomic data sets to identify genes upon which a small number of cell lines are uniquely dependent (6–8). We focused on the 21 lung cancer lines tested by shRNA analysis. Knockdown and genomic data for these cell lines were then mined to link extreme dependencies to genetic features.
Materials and Methods
Data mining and analysis
Cancer cell line sequencing data were acquired from the public CCLE (http://www.broadinstitute.org/ccle) and Sanger Cell Line (cancer.sanger.ac.uk) data portals. The shRNA-level data for Project Achilles version 2.4.3 are available at http://www.broadinstitute.org/achilles/datasets/all (8). shRNA level dependency values are expressed as the log2 fold change in shRNA abundance after 16 population doublings or 40 days in culture compared to the initial DNA plasmid pool. To calculate gene-level scores for all genes included in this data set we used the ATARiS module on GenePattern (http://www.broadinstitute.org/cancer/ataris) using a null significance value of 1 (9). The gene-level score is a normalized value indicating the magnitude of dependence (negative score) or relative enhancement (positive score) of survival after shRNA knockdown.
Cancer cell lines
Cancer cell lines were grown and maintained in RPMI media supplemented with 10% fetal bovine serum (FBS), penicillin, streptomycin, and L-glutamine. The following lung cancer cell lines were originally provided by the laboratory of Dr. John Minna: A549, NCI-H460, NCI-H1299, NCI-H1650, HCC366, and NCI-H2126. The following cancer cell lines were provided by the CCLE: SW1573, NCI-H1355, NCI-H196, NCI-H226 (lung); SNU-C1, RKO, SW48, HCT116, NCI-H508 (colorectal); OCUM-1, AGS, MKN7, NCI-N87 (stomach).
Immunoblots
Cells were lysed in RIPA lysis buffer (Thermo Scientific) supplemented with 1× protease and phosphatase inhibitor cocktails (Roche). Protein extracts were analyzed by standard immunoblotting with the following antibodies: total MEK1/2 (9122), phospho-ERK Y202/Y204 D13.14.4E (4370), total ERK 3A7 (9107), phosphor-AKT S473 193H12 (4058), total AKT 2H10 (2967) from Cell Signaling; β-actin C4 (sc-47778) from Santa Cruz.
Inhibitor treatment analysis
For drug treatment assays, cells were plated at a density of 2500 cells per well in a 96-well assay plate. The following day, cells were treated with the corresponding MEK1 inhibitors: selumetinib (AZD6244) S1008, trametinib (GSK1120212) S2673, and PD0325901 S1036 from Selleck Chemicals and refametinib (RDEA119) from Chemie Tek. Control wells were treated with dimethylsulfoxide (DMSO). Cell viability was assayed 6 days after drug treatment with CellTiter-Glo Luminescent Cell Viability Assay kit (Promega). Viability was normalized to DMSO controls. IC50 estimates were obtained using least-squares nonlinear regression on a standard 4-parameter logistic model.
Lentiviral-delivered shRNA
The following sequences targeting GFP, LacZ, and MAP2K1 were obtained from The RNAi Consortium (TRC) portal (http://www.broadinstitute.org/rnai/public) and cloned into the lentiviral vector pLKO.1:
GFP shRNA ACAACAGCCACAACGTCTATA
LacZ shRNA CGACCACGCAAATCAGCGATT
MAP2K1 shRNA-1 GCTTCTATGGTGCGTTCTACA
MAP2K1 shRNA-2 GAGGGAGAAGCACAAGATCAT
For virus production, each pLKO.1 vector and packaging vectors were introduced into 293T cells via calcium phosphate transfection (Clontech). Lentivirus was harvested in RPMI media supplemented with 10% FBS and filtered before addition to each cancer cell line. Infected cells were selected in 2 µg/ml puromycin for 5 days and harvested for protein lysates or for proliferation assays in 96 well plates and grown in the presence of RPMI containing 10% FBS and 1 µg/ml puromycin. Cell viability was assayed after 6 days using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega).
CRISPR-Cas9 lentiviral deletions
The following guide RNA sequences targeting MAP2K1 were designed using the sgRNA Designer tool on The RNAi Consortium (TRC) portal (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design):
MAP2K1 sgRNA-1 ACATCCTAGTCAACTCCCGT (sgRNA score = 0.70)
MAP2K1 sgRNA-2 GCTGATGTTTGGGTGCCAGG (sgRNA score = 0.70)
The sgRNA sequences targeting GFP were described previously (10):
GFP sgRNA-1 GGAGCGCACCATCTTCTTCA
GFP sgRNA-2 GAAGTTCGAGGGCGACACCC
These sequences were cloned into the Cas9 expressing lentiviral vector CRISPRv2 (11). Virus production, infection, and selection procedures were as described for shRNA experiments. Immunoblotting confirmed decreased MEK1 protein levels across the pool of infected cells. Cell viability was assayed 3 and 6 days later with the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega).
Xenografts
Xenografts were performed as previously described (12). Two hundred microliters of each cell line suspended in matrigel were injected into each of two injection sites in the flank of 4-week-old female nu/nu mice (Jackson Laboratory). Tumor volume was measured with a digital caliper twice weekly and calculated using the formula 0.5 X L X W2 where L is the longest diameter and W is the diameter perpendicular to L. Drug treatments began once established tumors grew to 200 mm3 total volume. Trametinib was suspended in 5% DMSO and a dose of 0.3 mg/kg was administered daily by oral gavage. Control mice were treated with 5% DMSO. All animal experiments were carried out in accordance with the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee guidelines.
Results
To explore associations between potential drug targets and specific genomic features, we mined genome-scale shRNA-mediated loss of function data from Project Achilles. This large-scale project targeted 11,194 genes with 54,020 shRNAs in 216 human cancer cell lines and the depletion or enrichment of specific shRNAs was measured after cells were grown in vitro for several weeks (8). The use of five shRNAs per gene and the large number of cell lines permits one to identify shRNAs that show similar phenotypic patterns to reduce the contribution of off-target effects. ATARiS is a computational method that uses this information to compute a gene-level dependency score (9).
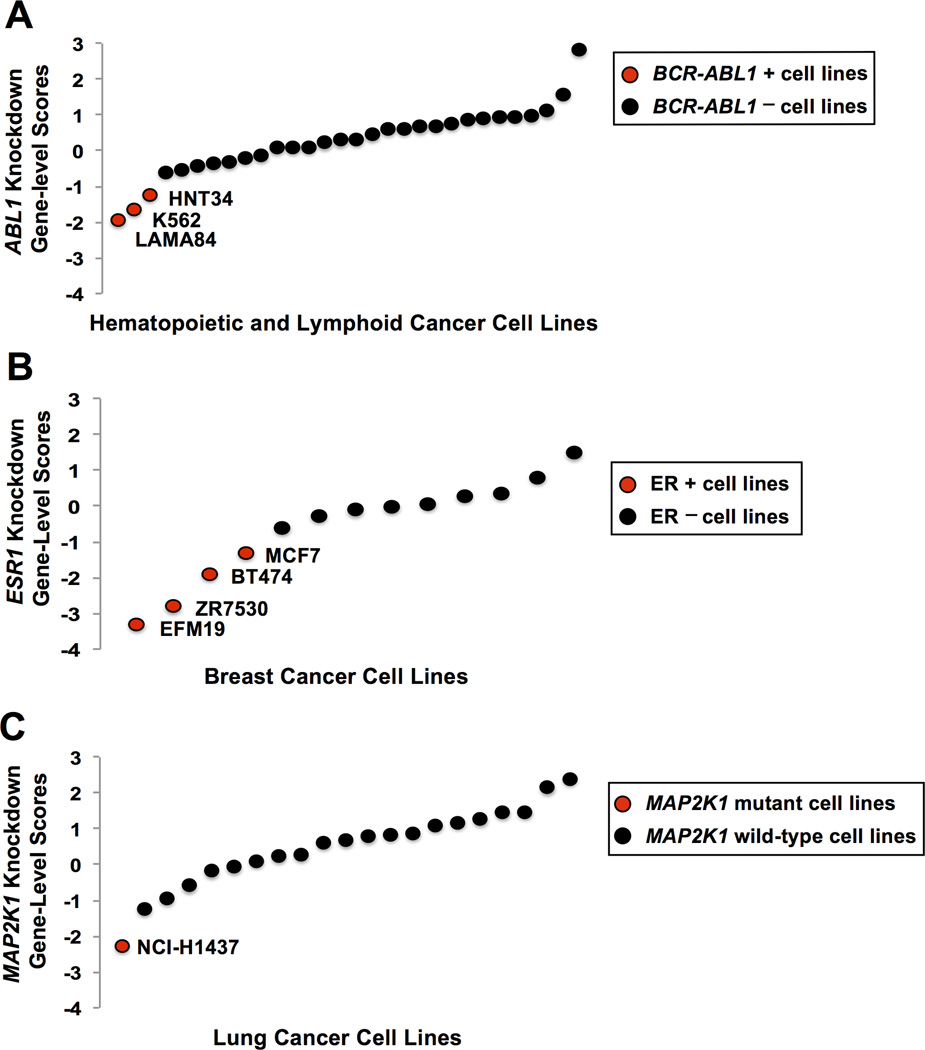
We first explored known relationships between the dependencies and genomic features seen in current targeted therapies. One example discussed above is the inhibition of cells harboring the BCR-ABL1 translocation by the tyrosine kinase inhibitor imatinib. To evaluate genetic dependency in such cells, we assessed the impact of ABL1 knockdown in the 30 hematopoietic and lymphoid cell lines included in the data set. Of these lines, the three known to harbor the BCR-ABL1 translocation are the most sensitive to ABL1 knockdown (Figure 1A). The large difference in the scores between these three cell lines compared to the BCR-ABL1-negative cell line with the lowest score (LAMA84 = 1.4; K562 = 1.0; HNT34 = 0.63) indicates an outlier dependency pattern. This was seen using both ATARiS gene solutions generated from the screen (Supplemental Figure 1) (9). We found a similar pattern across the 13 breast cancer cell lines profiled, as the four ER-positive lines were the most sensitive to ESR1 knockdown, which parallels anti-estrogen treatments in patients with ER-positive breast tumors (Figure 1B). These observations demonstrate that relationships between strong dependencies and genomic features could be detected in this RNAi data set.
Figure 1.
(A) Each data point represents the normalized gene-level dependency scores for ABL1 calculated using ATARiS (gene solution 1) in the 30 hematopoietic and lymphoid cell lines tested (8). The three most sensitive lines (indicated by red circles) are the only lines within this data set known to harbor the BCR-ABL1 translocation.
(B) Each data point represents the normalized gene-level dependency scores for ESR1 in the 13 breast cancer cell lines tested (8). The four most sensitive lines (indicated by red circles) are the only breast lines within this data set known to be positive for the estrogen receptor.
(C) Each data point represents the normalized gene-level dependency scores for MAP2K1 in the 21 lung cancer cell lines tested (8). The most sensitive line (indicated by the red circle) is the only lung line within this data set known to harbor a MAP2K1 mutation.
To perform an unbiased search for outlier dependencies in lung cancer cell lines, we ordered all genes in the 21 lung cancer cell lines included in the data set by dependency score. We then calculated an outlier score: the difference in gene-level scores between the cell line with the lowest score and the cell line with the gene-level score at the 90th percentile. Once we generated a ranked list of outlier scores across the lung cancer cell lines, we searched for genes with the highest outlier scores that are also significantly mutated in lung cancers to identify genes whose exceptional dependency may be due to known oncogenic drivers in lung cancers. Of the known oncogenic drivers in lung cancers, MAP2K1 showed the highest outlier score (1.7, top 1% of outlier scores) (Figure 1C). The MAP2K1 gene encodes the protein kinase MEK1, known to be part of the oncogenic ras/mitogen-activated protein kinase (RAS/MAPK) pathway. This pro-survival and proliferative pathway is commonly activated by oncogenic mutations in many tumor types including lung adenocarcinoma (13–16). The importance of this pathway is highlighted by the many established and on-going drug discovery efforts to inhibit different nodes in this signaling pathway such as mutant EGFR, mutant KRAS, mutant BRAF, and downstream wild-type MEK1.
MEK1 acts downstream from more commonly mutated RAS/MAPK pathway members including KRAS, EGFR, BRAF, RIT1, MET, and ERBB2 (16). MAP2K1 mutations occur at a significant but low frequency in lung adenocarcinoma (<1%) (13–15,17), and MAP2K1 mutations have been observed and characterized in melanoma (18), gastric cancer (19), Langerhans cell histiocytosis (20,21), and hairy-cell leukemia (22). Notably, a recent comprehensive study of MAP2K1 somatic mutations in lung adenocarcinoma reveals that about 86% of the MAP2K1 mutations clustered between amino acid residues 53–57 in exon 2, which encodes a critical alpha-helix domain (13). The recurrence and concentration of these mutations in this region indicate functional significance, and mutations in this region are predicted to affect MEK1 kinase activity. This study also demonstrated that overexpression of MEK1 proteins with mutations in this region, including F53L, Q56P, and K57N, leads to downstream ERK phosphorylation and increased colony formation that is inhibited with the MEK1/2 inhibitor selumetinib (AZD6244) (13). In addition, large-scale genomic characterizations of lung adenocarcinomas found that MAP2K1 mutations in this region do not co-occur with oncogenic mutations in receptor tyrosine kinases or RAS/MAPK pathway members, suggesting that oncogenic mutant MAP2K1 drives RAS/MAPK pathway activation in these cases (14,15). Interestingly, two lung adenocarcinomas that harbor atypical MAP2K1 mutations, M146I and S331R, also bear activating KRAS mutations, suggesting that these uncharacterized MAP2K1 mutations may not be potent oncogenic driver mutations (13,14).
Plotting the gene-level dependency scores for MAP2K1 shows the most sensitive cell line, NCI-H1437, as a potential outlier compared to the rest of the lung cancer cell lines tested, with a relatively high outlier score (1.7) in our analysis (Figure 1C). Interestingly, NCI-H1437 is the only lung line included in the data set that has a MAP2K1 mutation (Q56P). Consistent with the mutant MAP2K1 lung adenocarcinoma cases in the study described above, sequencing analyses show no other oncogenic driver mutations of the RAS/MAPK pathway in the NCI-H1437 line (13).
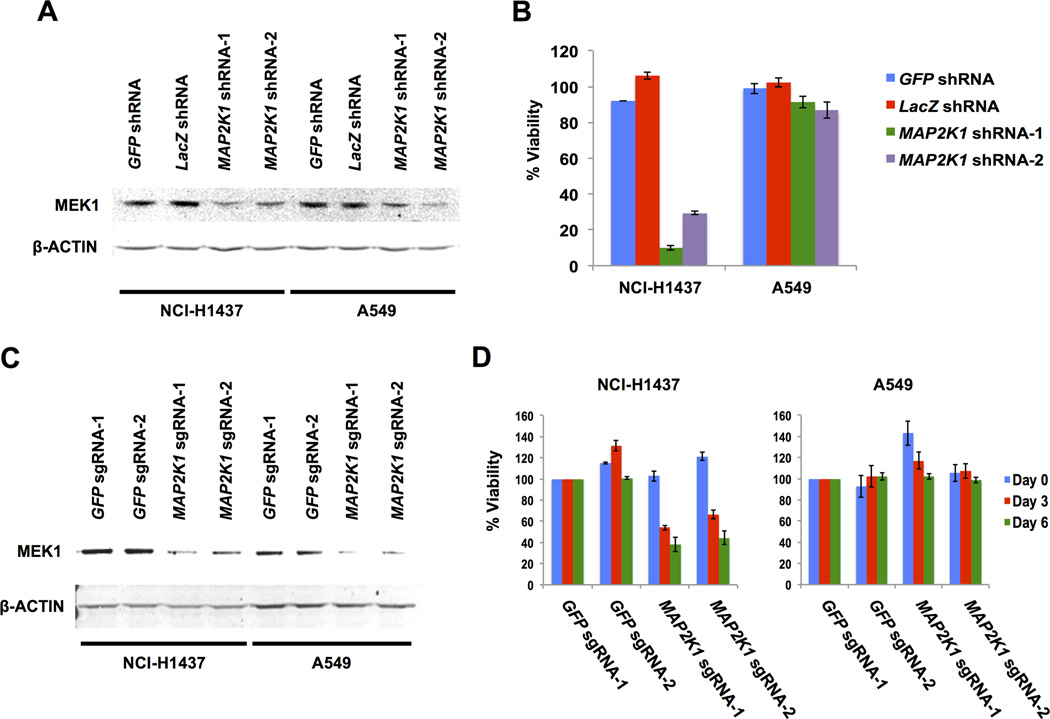
To confirm MAP2K1 dependence in NCI-H1437 cells, we used two shRNAs targeting MAP2K1 and two shRNA controls. The MAP2K1 shRNAs effectively decreased MEK1 protein levels (Figure 2A). MAP2K1 knockdown resulted in decreased viability in the NCI-H1437 cells but not in A549 cells, which harbor an activating KRAS mutation (G12S) (Figure 2B). Consistent with our results, the MAP2K1 dependency score for A549 cells indicates no effect (gene-level score = −0.2),
Figure 2.
(A) Immunoblot of MEK1 and β-ACTIN (loading control) protein levels 5 days after lentiviral infection of control shRNAs (GFP and LacZ) and two independent MAP2K1 shRNAs in NCI-H1437 and A549 cells.
(B) Cell viability beginning 7 days after lentiviral infections. Viability is normalized to mock infected controls.
(C) Immunoblot of MEK1 and β-ACTIN (loading control) protein levels from pooled population of cells 5 days after lentiviral infection of Cas9 with two independent control sgRNAs each targeting either GFP or MAP2K1 in NCI-H1437 and A549 cells.
(D) Cell viability beginning 7 days after Cas9/sgRNA infections. Viability is normalized to control GFP sgRNA-1 for each cell line on each day.
To confirm these observations, we used CRISPR-Cas9 gene-editing technology to deplete MAP2K1 (10). Pooled populations of cells were assayed after lentiviral infection with Cas9 and a small guide RNA (sgRNA) targeting either GFP or MAP2K1. Introduction of both MAP2K1-targeting sgRNAs resulted in decreased MEK1 protein expression in NCI-H1437 and A549 cells (Figure 2C). Similar to the shRNA knockdown experiments, MAP2K1 knockout resulted in decreased cell viability in NCI-H1437 cells, whereas A549 cells were unaffected (Figure 2D). These results confirmed the strong dependency on MAP2K1 expression in NCI-H1437 cells.
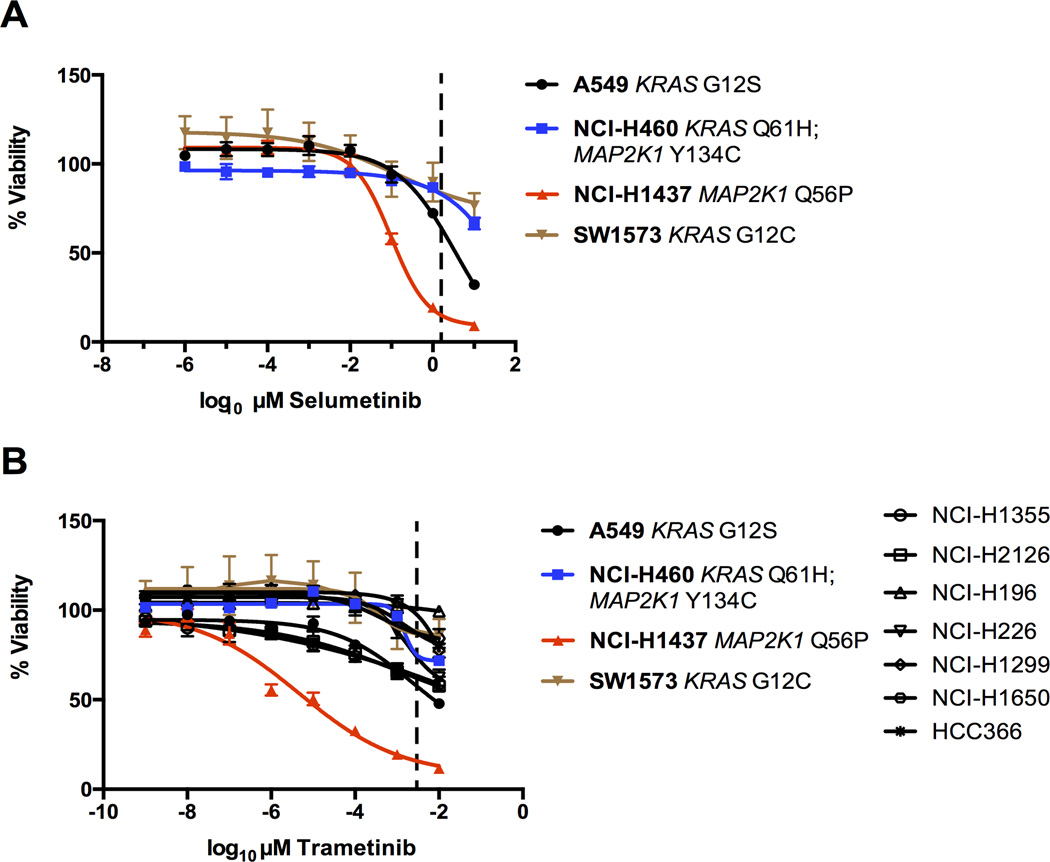
Our analysis and validation experiments suggest that NCI-H1437 cells would also show increased sensitivity to MEK1 inhibition using a small molecule. To test this, we performed dose response curves in a variety of lung cancer cell lines treated with MEK1/MEK2 inhibitors. We included the only other lung cancer cell line that harbors a MAP2K1 mutation: NCI-H460 cells have an atypical MAP2K1 Y134C substitution and also have an activating KRAS mutation (Q61H). This cell line was not included in the 216-cell line gene knockdown data set.
A previous study demonstrated that NCI-H1437 cells are sensitive to selumetinib, a MEK1/MEK2 inhibitor (23). In agreement with this result and our genetic knockdown and gene-knockout data, we found that NCI-H1437 cells were sensitive to selumetinib treatment, with an IC50 value in the nanomolar range, approximately ten-fold less than the maximal serum concentration achieved in patients with the recommended dose from Phase I trials (Figure 3A, dashed line) (24). Strikingly, NCI-H1437 cells showed sensitivity in the picomolar range upon treatment of the FDA-approved MEK1/MEK2-targeting drug, trametinib (GSK1120212), with an estimated IC50 value 1000-fold lower than the IC50 values from a panel of similarly-treated lung cancer cell lines (Figure 3B, Supplemental Figure 2A). This dose was also over 100-fold less than the maximal serum concentration achieved in patients treated with the recommended dose of trametinib from Phase I trials (Figure 3B, dashed line) (25). We also tested additional MEK1 inhibitors, refametinib (RDEA119) and PD0325901, and again found that NCI-H1437 cells were the most sensitive to both drugs, although the difference in sensitivity compared to other cell lines was less pronounced than with trametinib treatment (Supplemental Figure 2B). These results further underscore the acute dependence of NCI-H1437 cells on functional MEK1.
Figure 3.
(A) Selumetinib dose response curve after 6 days. Cell viability was normalized to DMSO treated controls. The dashed line approximately represents the maximal serum concentration achieved in Phase I clinical trials (24).
(B) Trametinib dose response curve after 6 days. Cell viability normalized to DMSO treated controls. The dashed line approximately represents the serum concentration achieved from the recommended dose determined from Phase I clinical trials (25).
While MEK1 inhibition strongly suppressed growth of NCI-H1437 cells, the only other lung line with a MAP2K1 mutation, NCI-H460, was not as affected. The MAP2K1 mutation in NCI-H460 cells (Y134C) has not been reported in any cancer study or additional cell line to date, and is not functionally characterized. In addition, NCI-H460 cells have an activating KRAS mutation (Q61H). Thus, the KRAS mutation or other activated oncogenic pathways in NCI-H460 cells may favor cell survival even in the presence of MEK1 inhibitors. We treated lung cancer cell lines with trametinib and assayed the effect on the KRAS signaling pathway. Trametinib resulted in a clear decrease in phosphorylated ERK levels in all cells tested; however, only the NCI-H1437 cells lacked detectable phosphorylated AKT and trametinib treatment had no effect on the elevated phosphorylated AKT in the KRAS mutant lines A549, NCI-H460, and SW1573 (Supplementary Figure 2C). The lack of compensatory signaling by phosphorylated AKT in NCI-H1437 cells may help to explain the exquisite sensitivity of this cell line to MEK1 inhibition.
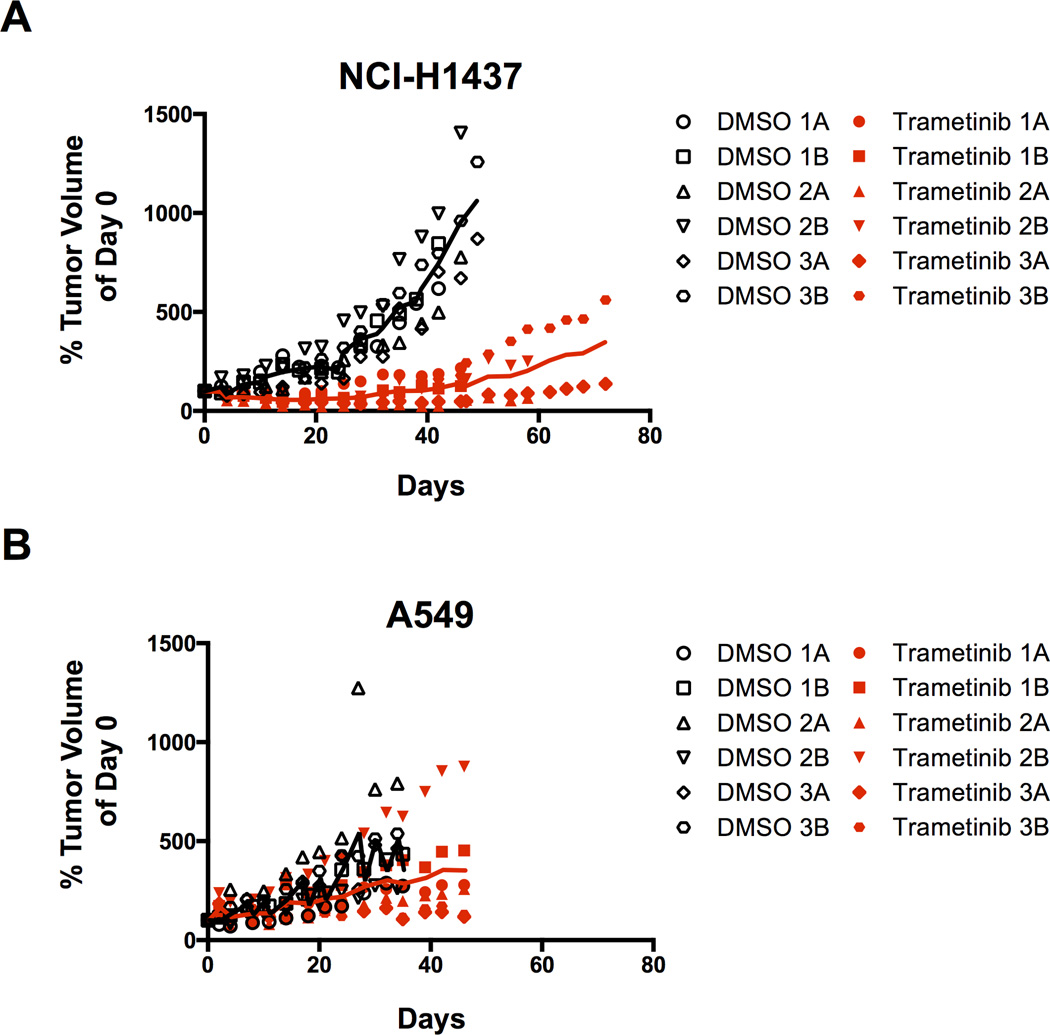
The drug trametinib displayed potent effects preferentially in the NCI-H1437 cells compared to all the other lung cell lines assayed. To validate this in vivo, we performed trametinib drug treatments in mice bearing NCI-H1437 and A549-derived xenografts. Once tumors were established, we treated the mice daily with 0.3 mg/kg trametinib or vehicle. While control (DMSO-treated) NCI-H1437 tumors grew readily, tumors of mice treated with trametinib showed little to no growth for the duration of the experiment (Figure 4A). This was repeated in an independent experiment, which gave similar results (Supplementary Figure 3). In comparison, A549 tumors grew regardless of drug treatment (Figure 4B). Of note, all A549 mice and control NCI-H1437 mice were sacrificed due to excessive tumor burden.
Figure 4.
(A) Xenograft tumor growth after NCI-H1437 subcutaneous injection in nude mice. Mice were treated daily with either vehicle (5% DMSO) or 0.3mg/kg trametinib. Each number represents a single mouse and the letters A and B represent each tumor. Tumor volume was normalized to its size at the start of drug treatment. Solid lines indicate the average of the six tumors for each treatment. The differences between trametinib and control treatments for all days after day 8 were statistically significant (p<0.007). Control mice were all sacrificed during the experiment due to excessive tumor burden. All trametinib treated mice were sacrificed at the conclusion of the experiment.
(B) Xenograft tumor growth after A549 subcutaneous injection in nude mice. Mice were treated daily with either vehicle (5% DMSO) or 0.3mg/kg trametinib. For each tumor, the volume was normalized to the size at the start of drug treatment. Control and trametinib treated mice were all sacrificed during the experiment due to excessive tumor burden.
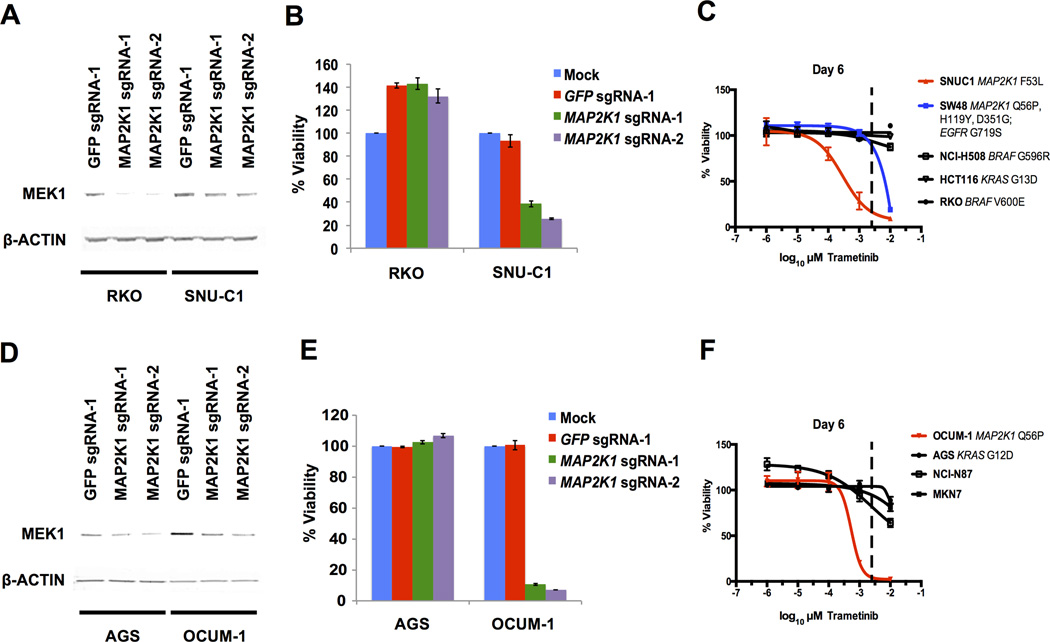
Our analysis of MAP2K1 dependence and activating MAP2K1 driver mutations specifically in lung cancer cell lines is limited to NCI-H1437 cells, as this is the only lung cancer cell line with an activating MAP2K1 mutation to our knowledge. Therefore, we tested additional mutant MAP2K1 cell lines from different lineages to expand our study and determine whether the relationship between mutant MAP2K1 and sensitivity to MEK1 inhibition is observed across different cancer types. This is particularly important as many emerging umbrella trials, such as the NCI-MATCH trials, pair targeted therapies to patients with tumors harboring specific oncogenic mutations regardless of tumor type (26). Our search produced no cancer cell lines that harbor a MAP2K1 K57N mutation, the most frequently observed MAP2K1 mutation in lung adenocarcinoma that lies within the inhibitory alpha helical domain of MEK1 (13). However, we did find two cancer cell lines with activating mutations in MAP2K1 within this region as the sole oncogenic driver of the RAS/MAPK pathway. The first of these is SNU-C1, a colorectal cancer cell line that has an F53L MAP2K1 substitution. This line is also included in the Project Achilles knockdown data set and displayed the lowest dependence score to MAP2K1 knockdown (−2.9) of any of the 216 cell lines assayed. To validate this dependency, we used CRISPR-Cas9 gene editing to decrease MEK1 levels in a control colorectal cell line, RKO (MAP2K1 knockdown dependence score = −0.2), and SNU-C1 cells (Figure 5A). While the RKO cells were insensitive to MAP2K1 knockout, SNU-C1 viability was diminished, confirming what was observed in the original pooled screen (8). Treatment of different colorectal cancer cell lines with trametinib revealed strong sensitivity in the SNU-C1 cells, similar to what we observed in trametinib treated NCI-H1437 cells (Figure 5C).
Figure 5.
(A) Immunoblot of MEK1 and β-ACTIN (loading control) protein levels from pooled population of cells 5 days after lentiviral infection of Cas9 with control sgRNA targeting GFP or two independent sgRNAs targeting MAP2K1 in RKO and SNU-C1 cells.
(B) Cell viability beginning 7 days after Cas9/sgRNA infections. Viability is normalized to mock infected controls for each cell line.
(C) Trametinib dose response curve after 6 days. Cell viability normalized to DMSO treated controls. The dashed line approximately represents the serum concentration achieved from the recommended dose determined from Phase I clinical trials (25).
(D) Immunoblot of MEK1 and β-ACTIN (loading control) protein levels from pooled population of cells 5 days after lentiviral infection of Cas9 with control sgRNA targeting GFP or two independent sgRNAs targeting MAP2K1 in AGS and OCUM-1 cells.
(E) Cell viability beginning 7 days after Cas9/sgRNA infections. Viability is normalized to mock infected controls for each cell line.
(F) Trametinib dose response curve after 6 days. Cell viability normalized to DMSO treated controls. The dashed line approximately represents the serum concentration achieved from the recommended dose determined from Phase I clinical trials (25).
In addition to the SNU-C1 and control colorectal cell lines with BRAF or KRAS mutations, we also tested the sensitivity of SW48 cells to trametinib treatment. SW48 is a colorectal cancer cell line with three detected mutations in MAP2K1 (Q56P, H119Y, and D351G), as well as an activating mutation in EGFR (G719S). Our laboratory previously characterized the dependency of this cell line on EGFR signaling using the EGFR inhibitor cetuximab both in vitro and in xenografts (27). SW48 cells were also included in Project Achilles, resulting in a moderate dependence score to MAP2K1 knockdown (−0.3). SW48 cells displayed increased sensitivity to trametinib compared to control cell lines, but not to the same degree of SNU-C1 cells (Figure 5C). As activating MAP2K1 mutations in cancer patient samples are almost always mutually exclusive with other mutations of the RAS/MAPK pathway, the SW48 cells present an atypical case where activated EGFR signaling may compensate for MEK1 inhibition.
The only other cell line identified to date with a mutation within the inhibitory alpha helix of MAP2K1 is the gastric cancer cell line OCUM-1 (Q56P). Like NCI-H1437 and SNU-C1 cells, mutant MAP2K1 was the only RAS/MAPK pathway driver mutation in OCUM-1 cells. A previous report describing MAP2K1 mutations in epithelial tumors determined that OCUM-1 cells were sensitive to selumetinib treatment compared to two wild-type MAP2K1 gastric cancer cell lines (19). To further confirm this we used CRISPR-Cas9 mediated gene editing to deplete MEK1 levels in pools of infected OCUM-1 and the control gastric cancer cell line AGS (Figure 5D). This resulted in greatly decreased viability in the OCUM-1 cells but not in the AGS cells (Figure 5E). Importantly, we tested the sensitivity of OCUM-1 and three other gastric cancer cell lines with wild-type MAP2K1 to trametinib treatment (Figure 5F). OCUM-1 cells were sensitive to picomolar concentrations of trametinib, similar to what we observed in the NCI-H1437 and SNU-C1 cells. These results further support outlier dependencies in cell lines with mutant MAP2K1 driven RAS/MAPK pathway activation to MEK1 inhibition.
Discussion
To reveal potential associations between cancer cell line features and dependencies, we mined existing large-scale functional genomic data sets. This approach allowed us to search for potential “exceptional responder” lung cancer cell lines in order to discover specific drug targets. We believe this approach is a simple, unbiased method that can identify robust dependencies in a subset of cell lines from large cell line cohorts tested in these knockdown experiments. Furthermore, while inherent, powerful off-target effects of shRNAs can lead to false results, our analysis should reliably identify very strong cancer dependencies that can be further validated.
Our analysis revealed that knockdown of MAP2K1, significantly mutated in lung adenocarcinoma, resulted in an outlier phenotype in a single cell line, NCI-H1437, in the original data set, (8, 13–15,17). Importantly, this is the only lung cell line in this data set with a MAP2K1 mutation, the previously established activating Q56P substitution (13). As different MEK1 inhibitors such as trametinib, which was recently approved to treat mutant BRAF-driven tumors in melanoma patients, and selumetinib are being tested in clinical trials for lung cancer patients, our study indicates that these drug treatments may be the most beneficial in the small but significant percentage of patients whose tumors harbor activating MAP2K1 mutations.
A previous study characterizing the first reported MAP2K1 mutations in lung cancer demonstrated that NCI-H1437 cells were sensitive to MEK1 inhibition by selumetinib (23). This study stated that NCI-H1437 cells displayed sensitivity to selumetinib in the nanomolar range. Our results support this work and provide independent evidence of the exceptional dependency of NCI-H1437 cells on MEK1-driven signaling in vitro. In addition, we observed a dramatic reduction in growth of NCI-H1437 tumor xenografts upon treatment with specific MEK1 inhibitors. Our initial data analysis proposing this dependence was also in the context of 21 different lung cancer cell lines, further corroborating the hypothesis that MAP2K1 mutant cells and cancers may be extremely sensitive to MEK1 inhibitors, particularly trametinib (28).
MEK1 inhibitors were developed to block aberrant RAS/MAPK signaling in cancer. Because MEK1 lies downstream in this pathway, MEK1 inhibitors are thought to indirectly suppress activating oncogenes that lie upstream in this signaling cascade, such as mutant KRAS (28). While shRNA knockdown screen data do not always correlate with drug efficacy in patients, it is interesting to note that sensitivity to MAP2K1 knockdown did not correlate with KRAS mutation or any other oncogenic driver mutations across the lung cell line cohort except mutant MAP2K1 (Supplemental Table 1). However, it is important to note that many small molecules that target MEK1 also target MEK2, while MAP2K1-targeting shRNAs should not affect MAP2K2 expression.
Our work further highlights the need to identify the oncogenic drivers that are present in each cancer patient. Efforts for systematic hybrid capture sequencing can identify patients with MAP2K1 activating mutations that do not occur with other known driver oncogene mutations (29, 30). Based on our results, these patients may benefit the most from trametinib treatment. Additionally, an activating MAP2K1 mutation was recently described as the potential basis for resistance to treatment with ceritinib in an ALK-positive tumor (31). Therefore, sequence knowledge at different stages of tumor evolution in response to drug treatment might suggest combination treatments with MEK1 inhibitors should MAP2K1 mutations be detected. Lung adenocarcinoma is one of the deadliest cancer types, and overall survival of MAP2K1-mutant lung adenocarcinoma patients has been shown to be decreased compared to patients with other oncogene-driven tumors (13). Therefore, matching MAP2K1-mutant lung cancer patients to MEK1 inhibitor treatments will be important and is clinically achievable.
Identifying specific oncogenic driver mutations in patients will also be critical in the design and retrospective analysis of clinical trials assessing MEK1 inhibition in different tumor types. Due to the low frequency of MAP2K1 mutations, those few patients with these oncogenic alterations might be neglected in trials comparing other factors, e.g. KRAS wild-type and KRAS mutant tumors. Based on our results and the current literature it will be important to identify and include patients with mutant MAP2K1 in clinical trials for MEK1 inhibition. Multi-institutional clinical trials that include a larger number of patients with MAP2K1 mutant cancers might be needed to truly assess MEK1 inhibitor efficacy in this population. Likewise, previously conducted trials that included appropriate sequencing data could uncover responses in mutant MAP2K1 patients. Excitingly, a very recent study of an ovarian cancer patient with a long-term, complete response to selumetinib treatment revealed a novel somatic 15 base-pair deletion of the inhibitory alpha helix domain in MAP2K1 (32). This independent evidence further supports the findings of this work and the need for further clinical testing of MEK1 inhibitors in patients harboring activating MAP2K1 mutations.
As functional genomic screens continue to evolve, greater numbers of hypotheses regarding putative drug targets can be generated and tested. Emerging technologies using CRISPR-mediated gene editing allow specific and efficient gene knockout screens to be added as an important orthogonal method to knockdown and chemical screens. Importantly, all of these screens are being performed in increasingly larger numbers of established and newly developed cancer cell lines, thus increasing the power to find genuine associations between gene dependencies and genomic alterations. As new resources and analysis methods emerge, it will be important to further validate findings such as acute MAP2K1 dependence in MAP2K1 mutant cells. Our data suggest that patients with MAP2K1 mutant tumors could benefit from treatment with MEK1 inhibitors, such as the approved drug trametinib, and we hope that this strategy is evaluated in a clinical setting.
Supplementary Material
Acknowledgments
We would like to thank A. Ramachandran, S. Bullman, D. Cai, P. Choi, W. Lin, A. Taylor, H. Watanabe, and X. Zhang for helpful discussion during the course of this work and in manuscript preparation.
Financial Support: NIH F32 CA196141, U01 CA176058, NCI 1R35 CA197568, DOD W81XWH-12-1-0269, and the American Cancer Society Research Professorship
Footnotes
Conflict of Interest: MM receives research support from Bayer Pharmaceuticals. WCH is a consultant for Novartis and Blueprint Medicines.
Authors’ contributions:
Conception and design: H.S. Gannon and M. Meyerson
Development of methodology: H.S. Gannon, A. Tsherniak, F. Vazquez, and M. Meyerson
Acquisition of data: H.S. Gannon and N. Kaplan
Analysis and interpretation of data: H.S. Gannon, N. Kaplan, and M. Meyerson
Writing, review and/or revision of the manuscript: H.S. Gannon, F. Vazquez, B. Weir, W. Hahn, and M. Meyerson
Administration, technical, or material support: H.S. Gannon, F. Vazquez, B. Weir, A. Tsherniak, and W. Hahn
Study supervision: M. Meyerson
Implications: Cancers with an activated RAS/MAPK pathway driven by oncogenic MAP2K1 mutations may be particularly sensitive to MEK1 inhibitor treatments.
References
- 1.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. New England Journal of Medicine. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 2.Early Breast Trialists Collaborative Group (EBTCG) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. The Lancet. 2011;378(9793):771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338(6104):221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagle N, Grabiner B, Van Allen EM, Hodis E, Jacobus S, Supko JG, et al. Activating mTOR Mutations in a Patient with an Extraordinary Response on a Phase I Trial of Everolimus and Pazopanib. Cancer Discovery. 2014;4(5):546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imielinski M, Greulich H, Kaplan B, Araujo L, Amann J, Horn L, et al. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. Journal of Clinical Investigation. 2014;12(6):4–8. doi: 10.1172/JCI72763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Wilson CJ, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Research. 2014;43(D1):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Scientific Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao DD, Tsherniak A, Gopal S, Weir BA, Tamayo P, Stransky N, et al. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Research. 2013;23:665–678. doi: 10.1101/gr.143586.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalem O, Sanjana NE, Hartenian E, Shi S, Scott DA, Mikkelson T, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2013;343(6166):84–87. doi: 10.1126/science.1247005. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark GJ, Cox AD, Graham SM, Der CJ. Biological assays for Ras transformation. Methods in Enzymology. 1995;255:395–412. doi: 10.1016/s0076-6879(95)55042-9. [DOI] [PubMed] [Google Scholar]
- 13.Arcila ME, Drilon A, Sylvester BE, Lovly CM, Borsu L, Reva B, et al. MAP2K1 (MEK1) Mutations Define a Distinct Subset of Lung Adenocarcinoma Associated with Smoking. Clinical Cancer Research. 2015;21(8):1935–1943. doi: 10.1158/1078-0432.CCR-14-2124. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Cancer Genome Atlas. 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downward J. Targeting RAS Signalling Pathways in Cancer Therapy. Nature Reviews Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nature Genetics. 2011;44(2):133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 19.Choi YL, Soda M, Ueno T, Hamada T, Haruta H, Yamato A, et al. Oncogenic MAP2K1 mutations in human epithelial tumors. Carcinogenesis. 2012;33(5):956–961. doi: 10.1093/carcin/bgs099. [DOI] [PubMed] [Google Scholar]
- 20.Nelson DS, Van Halteren A, Quispel WT, Van Den Bos C, Bovée JVMG, Patel B, et al. MAP2K1 and MAP3K1 Mutations in Langerhans Cell Histiocytosis. Genes, Chromosomes & Cancer. 2015;49(1):9–16. doi: 10.1002/gcc.22247. [DOI] [PubMed] [Google Scholar]
- 21.Chakraborty R, Hampton OA, Shen X, Simko SJ, Shih A, Abhyankar H, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2015;124(19):3007–3015. doi: 10.1182/blood-2014-05-577825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterfall JJ, Arons E, Walker RL, Pineda M, Roth L, Killian JK, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34 expressing hairy-cell leukemia. Nature Genetics. 2014;46(1):8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marks JL, Gong Y, Chitale D, Golas B, Mclellan MD, Kasai Y, et al. Novel MEK1 Mutation Identified by Mutational Analysis of Epidermal Growth Factor Receptor Signaling Pathway Genes in Lung Adenocarcinoma. Cancer Research. 2008;(14):5524–5529. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. Journal of Clinical Oncology. 2008;26(13):2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Infante JR, Fecher LA, Falchook GS, Nallapareddy S, Gordon MS, Becerra C, et al. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: A phase 1 dose-escalation trial. The Lancet Oncology. 2012;13(8):773–781. doi: 10.1016/S1470-2045(12)70270-X. [DOI] [PubMed] [Google Scholar]
- 26.Mullard A. NCI-MATCH trial pushes cancer umbrella trial paradigm. Nature Reviews Drug Discovery. 2015;14(8):513–515. doi: 10.1038/nrd4694. [DOI] [PubMed] [Google Scholar]
- 27.Cho J, Bass AJ, Lawrence MS, Cibulskis K, Cho A, Lee S-N, et al. Colon cancer-derived oncogenic EGFR G724S mutant identified by whole genome sequence analysis is dependent on asymmetric dimerization and sensitive to cetuximab. Molecular Cancer. 2014;13(1):141. doi: 10.1186/1476-4598-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nature Reviews Cancer. 2015;15(10):577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 29.Wagle N, Berger MF, Davis MJ, Blumenstiel B, De Felice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discovery. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) The Journal of Molecular Diagnostics. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grisham RN, Sylvester BE, Won H, McDermott G, DeLair D, Ramirez R, et al. Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer. Journal of Clinical Oncology. 2015 Aug 31; doi: 10.1200/JCO.2015.62.4726. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.