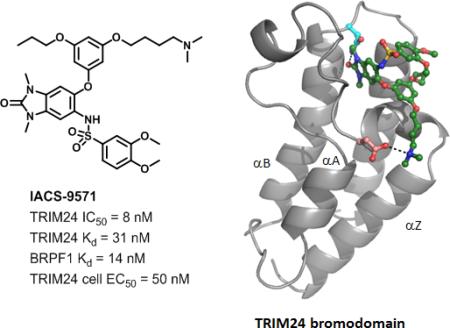
Abstract
The bromodomain containing proteins TRIM24 (Tripartite motif containing protein 24) and BRPF1 (bromodomain and PHD finger containing protein 1) are involved in the epigenetic regulation of gene expression and have been implicated in human cancer. Overexpression of TRIM24 correlates with poor patient prognosis and BRPF1 is a scaffolding protein required for the assembly of histone acetyltransferase complexes, where the gene of MOZ (monocytic leukemia zinc finger protein) was first identified as a recurrent fusion partner in leukemia patients (8p11 chromosomal rearrangements). Here, we present the structure guided development of a series of N,N-dimethyl benzimidazolone bromodomain inhibitors through the iterative use of X-ray cocrystal structures. A unique binding mode enabled the design of a potent and selective inhibitor, 8i (IACS-9571) with low nanomolar affinities for TRIM24 and BRPF1 (ITC Kd = 31 nM and 14 nM, respectively). With its excellent cellular potency (EC50 = 50 nM) and favorable pharmacokinetic properties (F = 29%), 8i is a high-quality chemical probe for the evaluation of TRIM24 and/or BRPF1 bromodomain function in vitro and in vivo.
Keywords: Bromodomain, TRIM24, BRPF1, structure-guided, chemical probe, epigenetics
INTRODUCTION
Bromodomains are protein interaction modules found within a diverse set of chromatin-regulator proteins and specifically recognize, or “read”, acetylated lysine (KAc) residues on the histone tails of chromatin as well as KAc residues on other proteins.1, 2 Inhibitors of bromodomains, and in particular the BET (bromodomain and extra terminal) subfamily, have gained much attention with the development of selective chemical probes3-6 and the advancement of several inhibitors into the clinic for various oncology indications.7-10 Additionally, targeting bromodomains is being explored for the treatment of inflammatory, neurodegenerative and cardiovascular diseases.11, 12 The development of this class of inhibitors has demonstrated the potential utility of treating diseases by the disruption of the protein-protein interaction network of chromatin reader modules, and is enabling pharmacological studies into how bromodomain-containing proteins (BCPs) regulate gene transcription and cell signaling.1, 13
The functions of BCPs, other than BET bromodomains, are much less well understood; however, selective chemical probes6, 14-17 are now beginning to emerge, which should help unravel their biological role in normal development and human disease. Equally encouraging are inhibitors that have now been identified for more difficult to drug bromodomains,18 such as BAZ2B19, 20 and ATAD2A.21-23 Unlike the BET sub-family, which contain two bromodomains that primarily drive their binding and therefore function, a large proportion of the other BCPs contain auxiliary domains, many with diverse and sometimes unknown functions. These multi-domain containing BCPs can also be part of multi-protein complexes. Accordingly, the relative contribution of the bromodomain within these complexes has yet to be elucidated. For example, the bromodomain-PHD finger protein (BRPF) family acts as scaffolding proteins to assemble complexes of MYST-family histone acetyltransferases (HATs).24 BRPF1 is a subunit of the monocytic leukemic zinc finger (MOZ) complex whose translocations are associated with an aggressive form of acute myeloid leukemia.25 The BRPF1 protein contains multiple conserved domains involved in gene transcription, chromatin binding and remodeling. The BRPF1 bromodomain has been demonstrated to bind to several histone peptides, including H2AK5ac, H4K12ac, H4K8ac, H4K5ac, and H3K14ac; knowledge of the importance of these observations and the biological function of the BRPF1 bromodomain is largely unknown.26
The tripartite motif containing proteins (TRIMs) are a large family of proteins whose structures are characterized by a highly conserved sequence of domains in the N-terminal region; RING domain, B-box zinc-fingers, and a coiled-coil region (also collectively known as an RBCC domain).27 Many functions have been ascribed to TRIM proteins, including their ability to act as E3 ligases for ubiquitin and small-ubiquitin like modifier (SUMO) activities.28, 29 A sub-group of TRIM proteins (TRIM24, 28 and 33) also contain a dual C-terminal plant homeodomain and bromodomain motif (PHD-bromo) which makes these interesting targets for exploring epigenetic regulation through bromodomain inhibition.30 TRIM24 was originally identified as a transcriptional intermediary factor (TIF1α) which acts as a co-repressor of the retinoic acid receptor that interacts with multiple nuclear receptors via an LXXLL motif.31 The C-terminal PHD-bromo domain of TRIM24 acts as a dual “reader” of unmodified H3K4 and acetylated H3K23 marks within the same histone tail,32 additionally, TRIM24 can also bind to non-histone KAc marks on other proteins, including p53.33 Genetic knock-down of TRIM24 in cancer cells results in antiproliferative phenotypes, and overexpression of TRIM24 has also been linked to poor prognosis for several cancers, including breast,32, 34 head and neck,35 non-small cell lung,36 hepatocellular,37 and glioblastoma.38 Because of the interest in TRIM24 here at MD Anderson and the druggability of BCPs,18 we set out to develop a potent (cellular EC50 < 100 nM) and selective TRIM24 chemical probe in order to interrogate the role of the bromodomain in human disease.
RESULTS AND DISCUSSION
Identification of scaffolds
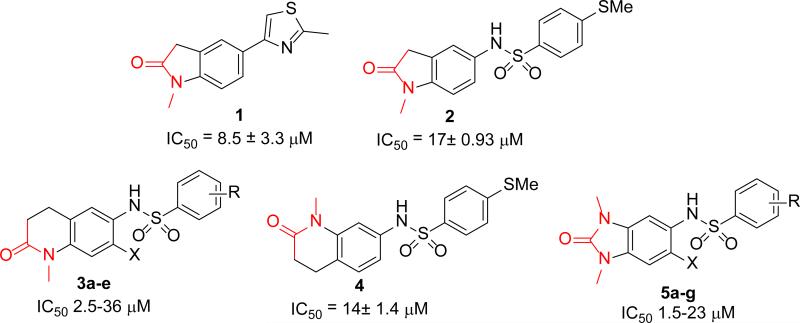
Since there were no known small-molecule TRIM24 inhibitors39 at the start of our study, we explored three different hit identification approaches for finding novel scaffolds: virtual in silico high-throughput screening (HTS), construction of a focused KAc mimetic library, as well as a traditional small-molecule library HTS. Three different chemotypes, as shown in Fig 1, were identified from this diverse approach: an N-methyl indolinone (1 and 2), the N-methyl tetrahydro quinolin-2-one (3 and 4), and the N,N-dimethylbenzimidazolone(5).14
Figure 1.
Chemotypes for TRIM24 identified by three different hit-finding approaches: in silico virtual screening (1), acetyl-lysine mimetic (highlighted in red) library building and SAR exploration (2–5), as well as HTS screening (5f and 5g). TRIM24 IC50 ± SEM of at least 3 determinations.
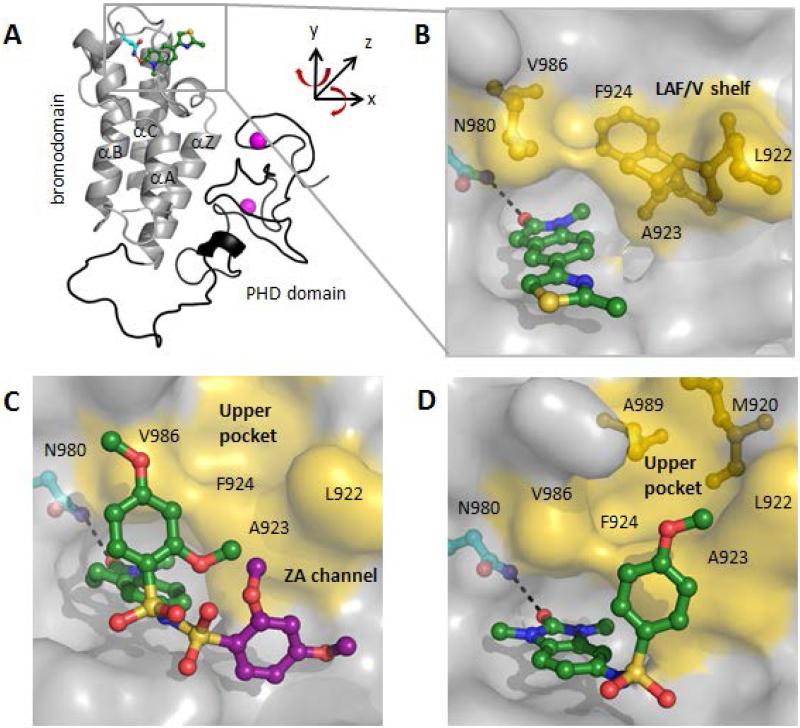
The in silico virtual screening was performed using the ‘Virtual Screening Workflow’ implemented within Schrodinger's Maestro modeling software suite and the reported TRIM24 X-ray crystal structures (PDB ID 3O33 and 3O34, apo and peptide substrate-bound complexes, respectively). Commercially available compounds within the ZINC library40 that bound to the KAc binding pocket, with a constraint of making at least one hydrogen bond (H-bond) interaction with N980 sidechain, were docked, scored, and ranked. After visual inspection of the top ranked poses and applying appropriate physicochemical property filters, the highest scoring hits were evaluated for binding to the bromodomain of TRIM24 using a biochemical bromodomain-H3K23(Ac) peptide displacement assay (AlphaScreen™).41 Fragment 1 (Fig 1) was identified as a TRIM24 inhibitor with an IC50 = 8.5 μM and subsequent X-ray cocrystal structure determination of 1 with TRIM24(PHD-bromo) (Fig 2A) validated the initially proposed docking hypothesis. Compound 1 was verified to be a KAc mimetic, whereby the carbonyl of the methyl-oxindole moiety interacts with the conserved N980 side-chain through an H-bond (Fig 2B). Although 1 was ligand efficient (LE = 0.41), the linked-thiazole group is co-planar with the oxindole template and is primarily solvent exposed, which made further modifications unlikely to interact with the protein. We sought to improve potency by interacting with the lipophilic sequence of residues 922-LAF-924 on the ZA-loop and V986 on the B-loop. This LAF/Vsequence highlighted in yellow (Fig 2B) forms a lipophilic shelf, or “LAF/V shelf”, akin to the “WPF” shelf of the BET-family of bromodomains (BRD2-4 and BRDT) whereby ligand interactions within this region have been demonstrated to be a way to improve potency for a wide variety of inhibitors.42, 43 In order to vector off orthogonally from the template and interact with the LAF/V-shelf we reasoned that a sulfonamide linkage to an aromatic substituent, as in PFI-1,5 would be a way to achieve this.
Figure 2.
TRIM24 (PHD-Bromo) X-ray complexes of initial virtual screening hit and lead templates. (A) Compound 1 bound to TRIM24 (1.9 Å resolution) bromodomain (grey ribbon), PHD domain (black), N980 (cyan), and Zinc (magenta) are depicted (PDB 4YAB). (B) Acetyllysine binding site with 1, lipophilic residues L922, A923, and F924 on the ZA-loop and V986 on the BC α-helix form an “LAF/V-shelf” (yellow sticks) (PDB 4YAB). (C) Complex of 3b (1.7 Å resolution) shown in two poses based upon partial occupancy, second pose (purple) in ZA-channel (PDB 4YAD). (D) Complex of 5b (2.2 Å resolution) with arylsulfonamide interacting with LAF/V-shelf (PDB 4YAT).
We explored a variety of template replacements for the indolinone and constructed a KAc mimetic library of 5,6- and 6,6-fused heterocycles containing a sulfonamide linkage to an aryl-group. In contrast to the indolinone sulfonamide 2 which lost potency (IC50 = 17 μM), both the tetrahydro isoquinolin-2-one 3 and the N,N-dimethyl-benzimidazol-2-one 5 scaffolds were identified as viable chemotypes with micromolar inhibition for TRIM24 (Fig 1). A summary of structure-activity relationships (SAR) for the scaffolds is shown in Table 1. Both 3a and 5a have comparable potencies of 4.8 and 4.9 μM, respectively, and were reasonable starting points from which to optimize. Interestingly, the regioisomeric derivative 4, which has the same sulfonamide attachment point relative to the KAc mimetic (red) compared to 5a, was less potent with an IC50 = 14 μM. Both the template, as well as the positioning of the sulfonamide-linker appeared to be important for obtaining potency, therefore evaluation of the relative binding-modes between the two templates would be important for selecting a series to progress.
Table 1.
Structure-Activity Relationship for Sulfonamides 3a–e and 5a–g
| Cmpd | R | X | TRIM-24 IC50 (μM)a |
|---|---|---|---|
| 3a | 4-SMe | H | 4.8 ± 2.9 (5) |
| 3b | 2,4-di-OMe | H | 2.5 ± 0.56 (3) |
| 3c | 4-O-iso-butyl | H | 3.7 ± 3.2 (3) |
| 3d | 4-cyclohexyl | H | 2.8 ± 1.3 (6) |
| 3e | H | OMe | 36 ± 24 (9) |
| 5a | 4-SMe | H | 4.9 ± 0.82 (5) |
| 5b | 4-OMe | H | 9.3 ± 5.1 (3) |
| 5c | H | H | 23 ± 2.6 (3) |
| 5d | 4-O-iso-butyl | H | 4.0 ± 1.3 (5) |
| 5e | 4-cyclohexyl | H | 1.7 ± 0.69 (5) |
| 5f | 3-CN | OMe | 10 ± 2.7 (6) |
| 5g | H | O(4-MeO-Ph) | 1.5 ± 0.79 (11) |
Mean ± SEM (number of measurements)
The TRIM24 cocrystal structure complexes of 3b (Fig 2C) and 5b (Fig 2D) informed on the importance of the relative regiochemistry of the sulfonamide-linker attachment and the role in defining how the aryl-group interacts with the lipophilic LAF/V-shelf. Both templates interact with N980 and bind similarly within the KAc binding site; however, the positioning of the aryl-group was remarkably different between the two compounds. The complex with 3b also revealed the conformational mobility of the aryl-sulfonamide; two poses of the aryl-group were observed within the same chain of the complex based upon partial occupancy. The relative atomic densities suggested an approximately 60:40 ratio between the two conformations (Fig 2C) with the aryl-group (green) only partially interacting with V986 of the BC-loop and in the less populated conformation (purple) directing the aryl-group towards the ZA-channel. In contrast, the complex of 5b showed a single conformation of the aryl-sulfonamide which makes lipophilic contacts with both A923 and F924 of the LAF/V-shelf (Fig 2D). Another attractive feature of scaffold 5 relative to 3, is the symmetrical nature of the two N-methyl groups, which allows the scaffold to always place a methyl-group deep into the pocket. Scaffold 5 was also attractive synthetically as it allowed for a more rapid development of this series relative to the regiochemical control necessary for the prosecution of 3.
The positioning of the aryl-group in the complex of 5b looked ideal for targeting the lipophilic region just beyond the LAF/V-shelf or “upper pocket” (depicted as a yellow surface in Fig 2D). The “upper pocket” is defined by lipophilic residues A989 and M920, which we thought we could improve potency by exploiting this region, therefore we evaluated a series of para-substituted aryl derivatives for both templates (Table 1). The SAR with substituents in the para-position showed very little improvement going from the thiomethyl group of 3a and 5a (IC50 = 4.8 and 4.9 μM, respectively) to the much larger cyclohexyl group of 3d and 5e (IC50 = 2.8 and 1.7 μM, respectively). The poor ligand binding efficiency of adding the larger cyclohexyl group (LE = 0.28 for 5e) was discouraging. After evaluating a library of aryl sulfonamides, there did not appear to be a likelihood of achieving the desired nanomolar TRIM24 potency through this approach.
Concurrent with the lead optimization of this series, an HTS screen was performed at the Texas Screening Alliance for Cancer Therapeutics with a 61,000 compound library using the same AlphaScreen conditions we employed for our in-vitro assay. Derivative 5f, containing a 5-methoxy subsituent, was identified as a hit and verified as a modest TRIM24 inhibitor (IC50 = 10 μM). This is in contrast to 3e with an IC50 = 36 μM. Knowledge of the binding mode of the un-substituted series, as previously discussed, made substitution at the 5-position of the dimethylbenzimidazolone a non-obvious vector from which to explore further changes. We subsequently performed a hit expansion of 4-arylsulfonamide-5-substituted compounds and identified 5g as a promising new lead with an IC50 = 1.5 μM.
New Binding Mode Allowed for Significant Improvements in Potency
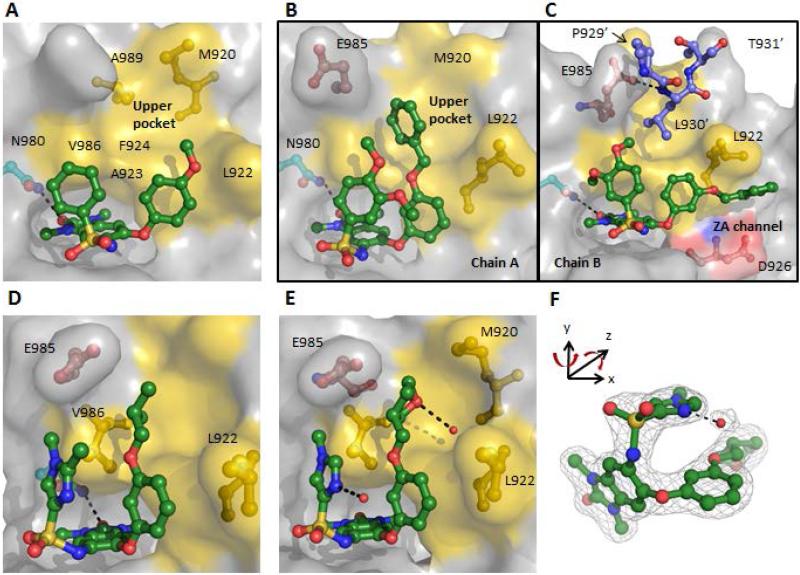
The cocrystal structure of 5g with TRIM24 quite surprisingly revealed a new “flipped’ binding-mode in which the aryl-ether group now interacted with the LAF/V-shelf and the aryl-sulfonamide group was oriented towards the conserved asparagine (Fig 3A). The aryl-ether ring of 5g makes lipophilic contacts with A923 (3.75Å between the Cα and the center of the aryl ring), while the phenyl-sulfonamide makes lipophilic contact with V986. The lone-pairs of the oxygen of the methoxy-group directed towards L922 would appear to be an unfavorable interaction and may influence the positioning of this residue. The meta-position appeared to be more favorable as it allows more room for additional derivatizations, directing groups towards the “upper pocket”. Although the ligand efficiency of 5g was lower (LE = 0.26), we recognized that this unexpected binding mode offered a new opportunity to optimize interactions with the LAF/V-shelf and target the “upper pocket” in order to improve potencies.
Figure 3. New unexpected binding mode enabled the design of interactions with the “upper pocket”.
(A) Cocrystal structure of 5g (2.3 Å resolution) depicting a “flipped” binding mode with the aryl-ether group now interacting with the LAF/V-shelf (PDB 4YAX). (B and C) Cocrystal structure of 7b (1.5 Å resolution) depicted in two binding conformations in the asymmetric unit (PDB 4YBM): (B) Chain A, shows the benzyl-group filling the “upper pocket”; (C) Chain B, with the benzyl-group occupying the ZA-channel and depicted in blue are three residues of Chain A TRIM24 protein with L983’ occupying the “upper pocket”. (D) Cocrystal structure of 7g (1.8 Å resolution) with the iso-butyl ether group occupying the “upper pocket” (PDB 4YBS). (E) Cocrystal stucture of 7l (1.8 Å resolution) with the 3-tetrahydropyranyl group H-bonded to a conserved water in the “upper pocket” and the imidazole H-bonded to a water that is π-stacked with the aryl-ether group (PDB 4YBT). (F) 2Fo-Fc map of 7l and the π-stacked water molecule contoured at 1.0σ (PDB 4YBT).
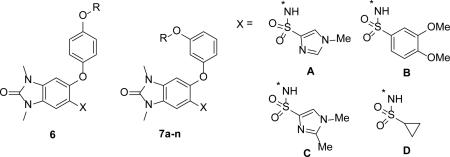
We evaluated the SAR of a series of aryl-ether substitutions to improve the potency and assess the role of the sulfonamide functionality on enhancing the solubility of the molecule (Table 2). In order to develop a potent cellular molecular probe, compounds were evaluated in an AlphaLisa cellular target engagement assay, which measured the displacement of ectopically expressed TRIM24(PHD-bromo) from endogenous histone H3 in HeLa cells. The effect of substituents on the kinetic solubility of the compound at pH 7.0 was also monitored. The larger para-benzyl substituent of 6 (IC50 = 2.0 μM) did not improve upon the potency of 5g and had no cellular activity; however, the meta-regioisomer 7a was 10-fold more potent than 6 with an IC50 = 0.22 μM and cellular EC50 = 6.2 μM, thereby confirming the meta-position to be the best vector for further exploration.
Table 2.
Structure-Activity Relationships for Improved In-vitro, Cellular Potencies and Solubility
| |||||
|---|---|---|---|---|---|
| Cmpd | R | X | TRIM-24 IC50 (μM)a | Cell EC50b (μM)a | Solubilityc (μM) |
| 6 | Benzyl | A | 2.0 ± 0.93 (6) | >33 (4) | nt |
| 7a | Benzyl | A | 0.22 ± 0.041 (5) | 6.2 ± 2.0 (4) | 59 |
| 7b | Benzyl | B | 0.14 ± 0.056 (29) | 3.9 ± 0.87 (13) | 0.6 |
| 7c | Benzyl | C | 0.27 ± 0.094 (6) | 4.9 ± 1.3 (4) | nt |
| 7d | Et | C | 0.18 ± 0.045 (5) | 2.3 ± 1.1 (4) | 74 |
| 7e | n-propyl | B | 0.043 ± 0.0037 (6) | 0.83 ± 0.17 (7) | 1.1 |
| 7f | n-propyl | C | 0.053 ± 0.015 (9) | 0.95 ± 0.31 (9) | 62 |
| 7g | CH2CH(Me)2 | C | 0.057 ± 0.016 (65) | 1.3 ± 0.27 (53) | 87 |
| 7h | n-Butyl | C | 0.16 ± 0.055 (4) | 3.0 ± 0.81 (5) | nt |
| 7i | CH2CH(Me)2 | D | 0.11 ± 0.023 (7) | 5.0 ± 0.53 (7) | 0.78 |
| 7j | CH2CH(Me)2 | H | 2.4 ± 1.6 (11) | >36 (5) | 2.6 |
| 7k | (CH2)3OMe | C | 0.13 ± 0.035 (5) | 2.7 ± 0.36 (6) | 100 |
| 71 | CH2(tetrahydro-furan-3-yl) | A | 0.10 ± 0.038 (5) | 1.9 ± 0.41 (5) | 77 |
| 7m | (CH2)4NMe2 | B | 0.27 ± 0.12 (4) | 3.2 ± 0.44 (2) | 66 |
Mean ± SEM (number of measurements)
TRIM24 AlphaLisa assay in HeLa cells
Kinetic solubility in phosphate buffer pH = 7.0.
nt = not tested
We found that a wide variety of aryl- and hetero-aromatic sulfonamides were tolerated and had minor effects on the intrinsic potency of the molecules. The sulfonamide appears to play a structural role in positioning the aryl-ether group, since few contacts with the protein are made by this functional group. In fact, this moiety could be used to modify physical properties, such as solubility and permeability, without adversely affecting the binding affinity of the molecule. For example, imidazole 7a had an improved aqueous solubility of 59 μM compared with aryl-sulfonamide 7b (0.6 μM) while maintaining similar in-vitro potencies of 0.22 versus 0.14 μM, respectively. In this example, the improved solubility was accompanied by a reduction in permeability, as determined by a CACO-2 permeability assay; compound flux from the apical to basolateral side was determined to be 4.0 Papp×10−6 cm·s−1 for 7a versus 21 for 7b. The cellular EC50 of 6.3 and 3.9 μM for 7a versus 7b, respectively, resulted in identical 28-fold cell-shifts for both compounds. For further development, we focused on improving both the potency as well as the cell-shift, since this would best reflect improvements in overall physical properties of the molecule.
In the presence of 7b, TRIM24 cocrystalized with two molecules in the asymmetric unit. In the first TRIM24 protein (Chain A), the 1.5Å resolution structure confirmed that the benzyl-ether targeted the “upper pocket” as we initially envisioned (Fig 3B) and that the conformation of the arylsulfonamide now undergoes an off-set π-π interaction with the aryl-ether ring, positioning the other aryl-group to interact with the LAF/V-shelf (3.8Å from the methoxy-group of the arylsulfonamide and the center of aryl-ether ring). Conversely, in the second TRIM24 protein (Chain B), the complex revealed a second conformation with 7b bound, such that the benzyl-ether occupies the ZA-channel (Fig 3C). Interestingly, although the ligand was not occupying the “upper pocket” in this alternate binding conformation, residue L930’ from Chain A involved in the crystal packing, does occupy the “upper pocket”. The excellent density and high resolution of the leucine showed an ideal and tight fit with the lipophilic residues of the “upper pocket”. This observation suggested that alkyl-ether substituents could be more ligand efficient replacements for the benzyl-group. Therefore, a series of alkyl-ether substituents were evaluated and we found that a three-carbon atom length appeared to be the optimal for potency, exemplified by the n-propyl (7e, f) and iso-butyl (7g) analogues, which improved intrinsic potencies by 3- to 5-fold with respect to 7a. Improved ligand efficiency was also achieved comparing 7e (IC50 = 0.43 μM, LE = 0.27) with 7b (IC50 = 0.14 μM, LE = 0.23).
The TRIM24 cocrystal complex of 7g (Fig 3D) nicely recapitulated the leucine interactions previously observed in Fig 3C. Since the aryl-ether ring appeared to be driving the binding affinity and the interactions with the “upper pocket” appeared to be optimized, we sought to minimize or completely remove the sulfonamide group. However, replacement of the aryl-sulfonamide with a smaller cyclopropyl-group as in 7i (IC50 = 0.11 μM) resulted in a 3-fold loss in cellular potency (EC50 = 5.0 μM) relative to 7g and complete removal of the sulfonamide (7j, IC50 = 2.4 μM) resulted in a 44-fold loss in intrinsic potency and no cellular activity. Other polar functional-groups on the alkyl-ether chain were also well tolerated, such as the methyl ether (7k, IC50 = 0.13 μM), tetrahydrofuran (7l, IC50 = 0.10 μM), and dimethyl amine (7m, IC50 = 0.27 μM) providing good solubilities; however, cellular potencies were weaker with respect to 7e.
Although the sulfonamide group makes minimal contact with the protein, the SAR and structural data demonstrate that the aryl-sulfonamide is an important functional group that helps fill the much more open and wider binding pocket of TRIM24 compared to the smaller more defined pocket of the BET family bromodomains. This is also reflected by the lower druggability scores (Dscore) calculated for TRIM24 (0.67) compared to those for the BET family (0.86–0.94).18 Furthermore, the cocrystal complex of 7l with TRIM24 (Fig 3E) also supports the notion that the sulfonamide plays an important structural role in positioning the aryl-ether ring in order to direct substituents into the “upper pocket”. The basic nitrogen of the imidazole is H-bonded to a structural water molecule which is positioned directly above the aryl-ether group. A different orientation of 7l (Fig 3F) shows the folded U-shape conformation of the molecule and the ligand density map clearly demonstrates this solvent exposed water undergoing a π-stacking interaction with the aryl-ether. The importance of the sulfonamide stacking interaction with the aryl-ether may also explain the observed loss of potency for 7j which has no sulfonamide-group. Interesting to note, that a similar intramolecular aromatic stacking interaction was also an important feature in the design of a potent BAZ2A/B molecular probe.20
Targeting the ZA-channel for Improved Cellular Potencies
Both 7f and 7g displayed the best balance between potency and solubility reflected by their sub-μM cell potencies (EC50 = 0.95 and 1.3 μM, respectively); however, they exhibited large cell-shifts of 18 and 23-fold, respectively. We sought to further improve cell potencies by improving the intrinsic potencies and/or lowering the cell-shift. Interactions of the n-propyl ether group with the “upper pocket” already appeared optimal, so we sought another region of the protein to interact with, in order to further improve potencies. The ZA-channel of TRIM24 offered a second site to pick up further interactions. The two different bound conformations of the benzyl-group in the 7b cocrystal (Fig 3B and 3C) inspired the strategic design of a bi-directional aryl-ether, whereby one vector was used to interact with D926 in the ZA-channel, while the other vector maintained the interaction with the “upper pocket”.
A series of bi-directional aryl-ethers were then evaluated and their SAR are shown in Table 3. The substantial improvement in potency (16-fold) going from the Boc-protected amine 8a (IC50 = 160 nM) to the basic amine analogue 8b (IC50 = 10 nM), as well as the excellent cellular potency of 8b (EC50 = 170 nM), suggested that the bi-directional design principles were realized. The importance of the basic amine is also further demonstrated by the alcohol analogue 8c which is 11-fold less potent in cells compared to 8b. Shortening of the chain-length to 4-carbons, as in 8f, improved both the in-vitro and cellular potencies (IC50 = 7.9 and 120 nM, respectively) as well as the observed cell-shift (15-fold). We also found that with the use of the basic amine on the bi-directional molecules, the imidazole-group was no longer needed for solubility. In fact, the dimethoxy-phenyl sulfonamide 8g, with a kinetic solubility of 42 μM (at pH 7.0), displayed a lower cell-shift compared to the imidazole 8f (7 versus 15-fold, respectively).
Table 3.
Structure-Activity Relationship of Di-substituted Aryl-Ethers
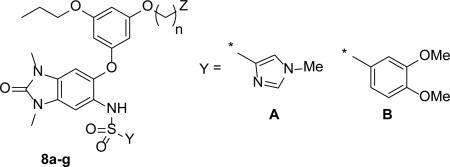
| ||||||
|---|---|---|---|---|---|---|
| Cmpd | Y | n | Z | TRIM-24 IC50 (μM)a | Cell EC50b (μM)a | Cell-shift |
| 8a | A | 6 | NHBoc | 0.16 ± 0.026 (6) | 5.5 ± 0.32 (5) | 34 |
| 8b | A | 6 | NH2 | 0.010 ± 0.0025 (14) | 0.17 ± 0.069 (11) | 17 |
| 8c | A | 6 | OH | 0.060 ± 0.017 (8) | 1.8 ± 0.40 (8) | 30 |
| 8d | B | 6 | NH2 | 0.013 ± 0.0020 (15) | 0.17 ± 0.059 (18) | 13 |
| 8e | A | 5 | NH2 | 0.011 ± 0.0021 (6) | 0.11 ± 0.023 (5) | 10 |
| 8f | A | 4 | NH2 | 0.0079 ± 0.0029 (8) | 0.12 ± 0.0032 (8) | 15 |
| 8g | B | 4 | NH2 | 0.0083 ± 0.0026 (4) | 0.059 ± 0.011 (4) | 7 |
| 8h | B | 3 | NH2 | 0.013 ± 0.0048 (4) | 0.12 ± 0.0066 (4) | 9 |
| 8i | B | 4 | NMe2 | 0.0076 ± 0.0029 (16) | 0.050 ± 0.013 (14) | 7 |
Mean ± SEM (number of measurements)
TRIM24 AlphaLisa assay in HeLa cells
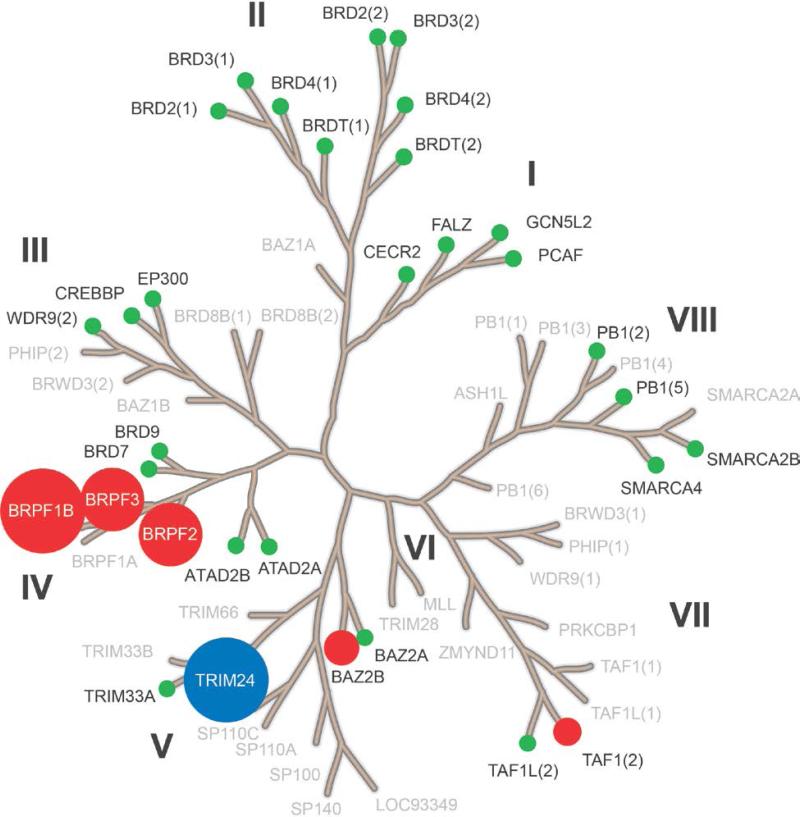
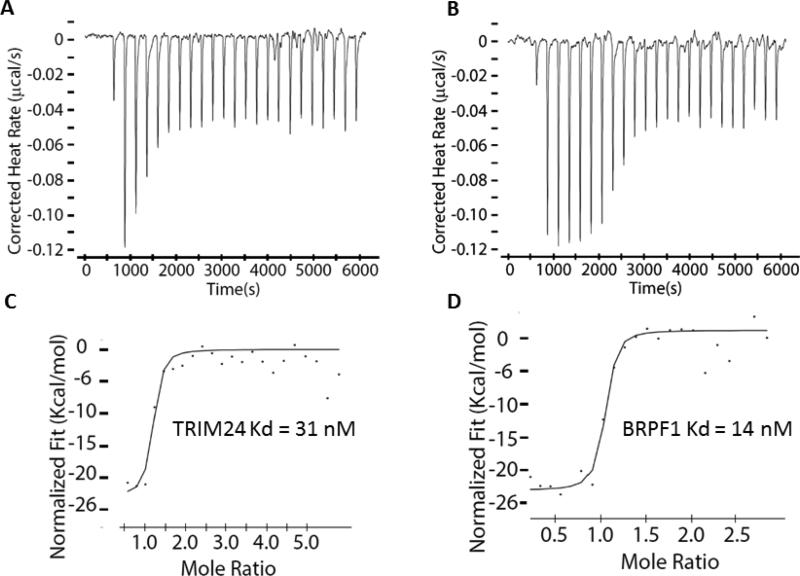
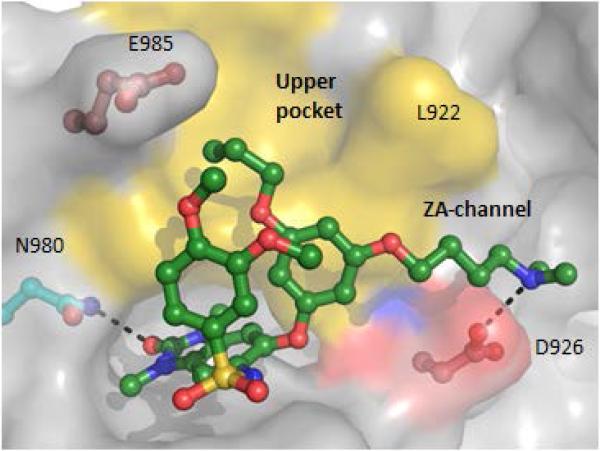
The dimethylamine analogue, 8i (IACS-9571) is one of the most potent TRIM24 inhibitors within this series, with an IC50 of 7.6 nM in the biochemical AlphaScreen and a cellular EC50 of 50 nM in the AlphaLisa cellular target engagement assay. 8i has excellent solubility (76 μM) and a low-cell shift (7-fold). Selectivity profiling of 8i against a panel of 32 bromodomains (DiscoveRx) at 1 μM confirmed the TRIM24 interaction specificity and also revealed strong interactions with family IV bromodomains, BRPF1-3 (Fig 4 and Table S2, Supporting Information).43 Subsequent dose-response determinations demonstrated 8i to be a selective dual TRIM24/BRPF1 inhibitor (Kd = 1.3/2.1 nM) with 9- and 21-fold selectivity against BRPF2 and BRPF3, respectively (Table 4). Much weaker affinities with BAZ2B (Kd = 400 nM) and the second domain of TAF1 (Kd = 1,800 nM) were also observed. Importantly, 8i does not interact with the BET sub-family of bromodomains, displaying greater than 7,700-fold selectivity versus BRD4(1, 2) relative to TRIM24. No interactions were detected against 26 other bromodomains (green circles) at 1 μM concentration of 8i. The dual TRIM/BRPF1 inhibitor profile of 8i was also confirmed by ITC resulting in Kd values of 31 nM and 14 nM, respectively (Fig 5). The cocrystal structure of 8i with TRIM24(PHD-Bromo) ultimately confirmed the bi-directional design principle we envisioned, with the n-propyl ether occupying the “upper pocket” as well as the salt-bridge interaction of the dimethylamine group with D926 in the ZA-channel (Fig 6). The dual TRIM24/BRPF1 potency of 8i is not surprising since potent BRPF1 dimethyl benzimidazolone inhibitors have been described,14 and BRPF1 also contains an acidic residue (E655) in the ZA-channel which the dimethylamine could potentially interact with.
Figure 4.
Selectivity profile of 8i against a panel of 32 bromodomains (DiscoveRx) at 1 μM test concentration. The larger the circle size corresponds to the greater the inhibition: TRIM24(PHD-Bromo) in blue; BRPF1-3, BAZ2B, and TAF1(2) in red; 26 bromodomains with percent of control >30 in green. Bromodomains in gray not tested. See also Table S2, Supporting Information.
Table 4.
Binding Affinity Data for 8i
| Bromodomain | Kd (nM)a |
|---|---|
| TRIM24(PHD-bromo) | 1.3 |
| BRPF1 | 2.1 |
| BRPF2b | 12 |
| BRPF3 | 27 |
| BAZ2B | 400 |
| TAF1(domain 2) | 1,800 |
| BRD4(domains 1, 2) | >10,000 |
Mean of 4 determinations in bromoELECTSM recombinant protein binding assays performed at DiscoveRx (http://www.discoverx.com).
Also known as BRD1.
Figure 5.
ITC data for 8i (A) Release of energy for the titration with TRIM24(PHD-Bromo). (B) Release of energy for the titration with BRPF1B. (C) Integrated binding heats for (PHD-Bromo), Kd = 31 nM. (D) Integrated binding heats for BRPF1B, Kd = 14 nM.
Figure 6.
Cocrystal structure of 8i with TRIM24(PHD-bromo)(1.8 Å resolution) showing the n-propyl ether group occupying the “upper pocket” and the dimethylamino group forming a salt-bridge to D926 in the ZA-channel (PDB 4YC9).
The pharmacokinetics of 8i was studied in female CD1 mice and after iv administration of a 1 mg/kg dose, a moderate clearance of 43 mL·min−1·kg−1 was observed, representing approximately half the rate of hepatic blood flow. The terminal iv half-life was 0.7 h and after oral dosing of a 10 mg/kg dose, the bioavailability of 8i was 29%, making the inhibitor useful for in vivo studies.
CONCLUSIONS
Iterative use of structural biology and medicinal chemistry optimization allowed us to develop a series of highly potent and selective dual inhibitors of TRIM24 and BRPF1 starting from a diverse hit-finding approach that utilized in silico virtual and small-molecule HTS screens as well as KAc mimetic library design. The unique binding mode observed while investigating new substitution patterns on the benzimidazolone template enabled the design of potent and selective dual inhibitors. Also, utilization of a TRIM24 cellular target engagement assay and targeting low cell-shifts allowed for the optimization of compounds that displayed the best physicochemical properties. The excellent cellular potency, bromodomain selectivity and desirable physical properties displayed by 8i provides an ideal chemical probe to investigate the biological and pharmacological role of TRIM24 and/or BRPF1 bromodomain inhibition in vitro and in vivo. The biological activity of 8i and related compounds will be reported in a subsequent manuscript.
CHEMISTRY
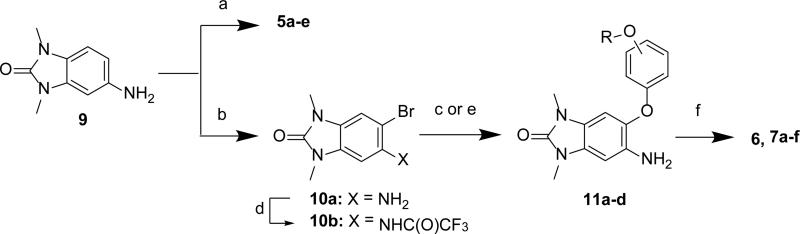
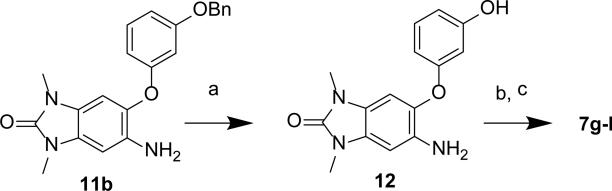
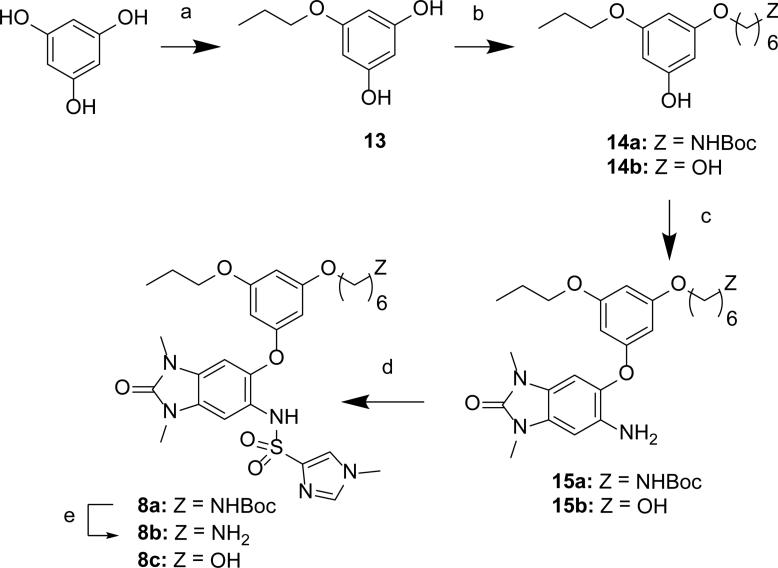
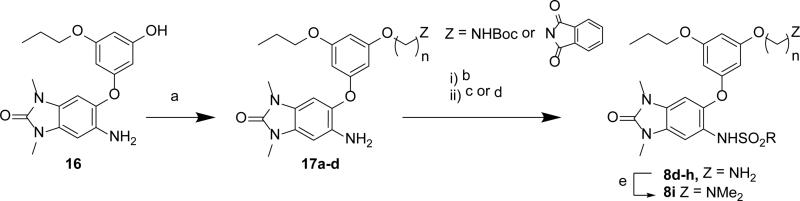
The sulfonamides 5a-e were synthesized from aniline 9 using the corresponding sulfonyl chlorides (Scheme 1). The 5-phenylether derivatives 6, 7a-f were synthesized from either bromide 10a or 10b, and a phenol, through either one of two different Ullmann coupling methods45 to give intermediates 11a-d. Use of the more electrophilic trifluoroacetamide 10b and a milder Ullmann coupling conditions46 with copper iodide, N,N-dimethyl-glycine, and cesium carbonate in dioxane at 80 °C was the prefered method for generating intermediates 11a-d. An alternative and more convergent route was utilized for the synthesis of 7g-l via alkylation of the intermediate phenol 12 (Scheme 2). The bi-directional derivatives 8a-c were synthesized through iterative alkylation of phloroglucinol to give the bi-functionalized phenols 14a and 14b, then Ullmann coupling with 10b to give intermediates 15a and 15b, followed by sulfonamide formation to give 8a and 8c (Scheme 3). Alternatively, the advanced phenol intermediate 16 was alkylated and transformed in a similar manner to give amines 8d-h (Scheme 4). Reductive alkylation of 8h using formaldehyde and sodium cyanoborohydride gave the dimethyl amine 8i.
Scheme 1. Synthesis of Sulfonamides 5a–e, 6, 7a–f.
Reagents and conditions: (a) sulfonyl chloride, pyr, DCM; (b) Br2, AcOH, CHCl2, 0 °C; (c) phenol intermediate, CuCl, quinolin-8-ol, potassium phosphate, diglyme, 120-130 °C; (d) TFA2O, DMAP, Et3N, DCM; (e) phenol intermediate, CuI, N,N-dimethyl-glycine, Cs2CO3, dioxane, 80 °C; (f) sulfonyl chloride, pyr, DCM.
Scheme 2. Synthesis of Sulfonamides 7g–l.
Reagents and conditions: (a) BBr3, DCM, −78 °C; (b) R-Br, K2CO3, DMF; (c) sulfonyl chloride, pyr, DCM
Scheme 3. Synthesis of Derivatives 8a–c.
Reagents and conditions: (a) 1-bromopropane, K2CO3, DMF; (b) Z(CH2)6-Br, K2CO3, DMF; (c) 10b, CuI, N,N-dimethyl-glycine, Cs2CO3, dioxane, 80 °C; (d) 1-methyl-1H-imidazole-4-sulfonyl chloride, pyr, DCM; (e) TFA, DCM.
Scheme 4. Synthesis of Amines 8d–i.
Reagents and conditions: (a) Z(CH2)nBr, K2CO3, DMF; (b) sulfonyl chloride, pyr, DCM; (c) TFA, DCM; (d) hydrazine hydrate, MeOH, 80 °C; (e) formaldehyde, AcOH, NaBH(OAc)3, Et3N, MeOH.
Synthetic Methods
Column chromatography was performed on a Biotage system using Biotage SNAP columns with Biotage KP-Sil silica or Biotage Zip Si columns with Biotage KP-Sil silica, or a Teledyne ISCO system with RediSep Rf normal phase silica cartridges (unless otherwise stated). All NMR spectra were recorded on Bruker instruments operating at 300, 500, or 600 MHz. NMR spectra were obtained as CDCl3, CD3OD, D2O, (CD3)2SO, (CD3)2CO, C6D6, or CD3CN solutions (reported in ppm), using tetramethylsilane (0.00 ppm) or residual solvent (CDCl3: 7.26 ppm; CD3OD: 3.31 ppm; D2O: 4.79 ppm; (CD3)2SO: 2.50 ppm; (CD3)2CO: 2.05 ppm; C6D6: 7.16 ppm; CD3CN: 1.94 ppm) as the reference standard. When peak multiplicities are reported, the following abbreviations are used: s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), br-s (broadened singlet), dd (doublet of doublets), dt (doublet of triplets). Coupling constants, when reported, are reported in Hertz (Hz). All temperatures are reported in degrees Celsius. The purity of all final compounds was >95% and was confirmed by LC/MS analysis. Low-resolution mass spectral (MS) data were obtained on either a Waters H class UPLC with a Waters Acquity UPLC® BEH C18 1.7 μm 2.1×50 mm column, UV detection between 200 and 400 nm, evaporating light scattering detection, and a SQ Detector mass spectrometer with ESI ionization or a Water I class UPLC with a Waters Acquity UPLC® CSH™ C18 1.7 μm 2.1×50 mm column, UV detection at 254 and 290 nm, evaporating light scattering detection, and a SQ Detector 2 mass spectrometer with ESI ionization. Preparative HPLC was performed using a Waters Autopurify system with a Waters Xbridge™ Prep C18 5 μm OBD™ 19×150 mm or 50×100 mm column and SQ Detector mass spectrometer with ESI ionization. Reagents were purchased from commercial suppliers such as Sigma-Aldrich, Alfa Aesar, TCI, or Acros and were used without further purification unless otherwise indicated. Anhydrous solvents (e.g. THF, DMF, DMA, DMSO, MeOH, DCM, toluene) were purchased from Sigma-Aldrich and used directly. The following compounds were obtained from commercial vendors: 1 Enamine cat# Z118580532, 3b Life Chemicals cat# F2278-0147, 3c Life Chemicals cat# F2278-0177, 5f ChemDiv cat# G433-0348, 5g ChemDiv cat# G433-0914.
General procedure for the synthesis of 2, 3a, 3d and 5a–e (Method A – sulfonamide coupling)
The following commercially available intermediates were used: 5-amino-1-methyl-1,3-dihydro-2H-indol-2-one (Combi-Blocks cat# ST-1422 for 2); 6-amino-1-methyl-3,4-dihydroquinolin-2(1H)-one (Enamine cat# EN300-69848 for 3a and 3d); 5-amino-1,3-dimethyl-1H-benzo[d]imidazol-2(3H)-one (Enamine cat# EN300-59698 for 5a–e) (0.1 mmol) in DCM (1 ml) was treated with the sulfonyl chloride (0.1–0.20 mmol) and pyridine (20 μl, 0.2 mmol). The reaction mixture was stirred for 2–24 hours at ambient temp, then concentrated and purified by mass-directed preparative HPLC to give sulfonamides 2, 3a, 3d and 5a–e.
N-(1-methyl-2-oxo-2,3-dihydro-1H-indol-5-yl)-4-(methylsulfanyl)benzene-1-sulfonamide (2)
1H NMR (600 MHz, DMSO-d6) δ 9.95 (br-s, 1H), 7.58 (dt, J = 8.6, 2.0 Hz, 2H), 7.35 (dt, J = 8.6, 2.0 Hz, 2H), 7.01 (s, 1H), 6.93 (dd, J = 8.3, 2.1 Hz, 1H), 6.82 (d, J = 8.3 Hz, 1H), 3.48 (s, 2H), 3.03 (s, 3H), 2.99 (s, 3H). MS (ESI) m/z 349 [M+H]+.
N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)-4-(methylsulfanyl)benzene-1-sulfonamide (3a)
23% yield. 1H-NMR (500 MHz, DMSO-d6) δ 10.01 (s, 1H), 7.95 (q, J = 7 Hz, 4H), 6.88 (s, 1H), 6.80 (q, J = 5 Hz, 1H), 6.67 (d, J = 6 Hz, 1H), 3.33 (s, 3H), 3.15 (s, 3H), 2.76 (t, J = 6 Hz, 2H), 2.37 (t, J = 8 Hz, 2H). MS (ESI) m/z 363 [M+H]+.
4-cyclohexyl-N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzene-1-sulfonamide (3d)
81% yield. 1H NMR (600 MHz, DMSO-d6) δ 10.10 (s, 1H), 7.65 (d, J = 8.3 Hz, 2H), 7.39 (d, J = 8.3 Hz, 2H), 6.99–6.91 (m, 3H), 3.15 (s, 3H), 2.75 (t, J = 6.9 Hz, 2H), 2.59–2.53 (m, 1H), 2.46 (t, J = 6.9 Hz, 2H), 1.81–1.72 (m, 4H), 1.73-1.64 (m, 1H), 1.43–1.29 (m, 4H), 1.27–1.15 (m, 1H). MS (ESI) m/z 399 [M+H]+.
Synthesis of N-(7-methoxy-1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)benzenesulfonamide (3e)
Step 1: To a solution of 7-hydroxy-3,4-dihydroquinolin-2(1H)-one (1.64 g, 10 mmol) in 30 mL of H2SO4 was added water (7.6 mL) dropwise with stirring. The reaction mixture was cooled to 0 °C and HNO3 (0.76 mL) added dropwise while stirring. The reaction mixture was stirred for 15 min at 0 °C then quenched by adding reaction mixture to a slurry of 100 mL of H2O and ice. The resulting solution was extracted with EtOAc (3 × 10 mL) and the combined organic layers were concentrated under reduced pressure to afford 7-hydroxy-6-nitro-3,4-dihydroquinolin-2(1H)-one (1.3 g, 63%) which was used without further purification. MS (ESI) m/z 209 [M+H]+.
Step 2: To a solution of 7-hydroxy-6-nitro-3,4-dihydroquinolin-2(1H)-one (1.1 g, 5 mmol) in 10 mL of DMF was added iodomethane (2.5 mL, 20 mmol) and K2CO3 (5.47 g, 20 mmol). The suspension was stirred at RT overnight. The reaction mixture was then diluted with water (20 mL) and extracted with EtOAc (3 × 10 mL). The combined organic layers were washed with water (10 mL), brine (10 mL) then dried over anhydrous sodium sulfate, concentrated under reduced pressure, then the residue was purified by flash chromatography on silica (EtOAc/hexanes = 1:2) to afford 7-methoxy-1-methyl-6-nitro-3,4-dihydroquinolin-2(1H)-one as a yellow solid (950 mg, 81%). MS (ESI) m/z 237 [M+H]+.
Step 3: To a solution of 7-methoxy-1-methyl-6-nitro-3,4-dihydroquinolin-2(1H)-one (900 mg, 3.8 mmol) in 30 mL of EtOH and 15 mL of H2O was added iron power (640 mg, 11.4 mmol) and ammonium chloride (2.1 g, 38 mmol). The mixture was heated at reflux for 2 h and then cooled to RT. The resulting suspension was filtered through celite and the filtrate was diluted with 50 mL of water. The filtrate was extracted with EtOAc (3 × 10 mL) and washed with water (10 mL) and brine (10 mL), dried over anhydrous sodium sulfate, then the combined organic layers were concentrated under reduced pressure to afford a crude product. The residue was purified by flash chromatography (hexanes/EtOAc= 1:1) on silica to afford 6-amino-7-methoxy-1-methyl-3,4-dihydroquinolin-2(1H)-one as a white solid (700 mg, 81%). 1H NMR (500 MHz, CDCl3) δ: 6.54 (s, 1 H), 6.48 (s, 1 H), 3.87 (s, 3 H), 3.33 (s, 3 H), 2.77–2.74 (m, 2H), 2.61–2.58 (m, 2H). MS (ESI) m/z 207 [M+H]+.
Step 4: Sulfonamide formation with 6-amino-7-methoxy-1-methyl-3,4-dihydroquinolin-2(1H)-one using method A afforded 3e as a white solid (100 mg, 58%). 1H NMR (500 MHz, DMSO) δ: 9.39 (s, 1H), 7.68 (d, J = 7.0 Hz, 2H), 7.59 (m, 1H), 7.53 (m, 2H), 6.99 (s, 1H), 6.79(s, 1H), 3.31 (s, 3H), 3.27 (s, 3H), 3.26 (s, 3H). MS (ESI) m/z 347 [M+H]+.
N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)-4-(methylsulfanyl)benzene-1-sulfonamide (4)
Step 1: A mixture of 7-amino-3,4-dihydroquinolin-2(1H)-one (100 mg, 0.62 mmol), phthalic anhydride (110 mg, 0.74 mmol) in AcOH (1 mL) was stirred at 100°C overnight, cooled to RT and poured into water, and the resulting suspension was filtered and collected solids dried to give 2-(2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)isoindoline-1,3-dione as a white solid (120 mg, 67%). MS (ESI) m/z 293 [M+H]+.
Step 2: To a mixture of 2-(2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)isoindoline-1,3-dione (120 mg, 0.41 mmol) and iodomethane (300 mg, 2.05 mmol) in DMF (2 mL) was added with K2CO3 (113 mg, 0.82 mmol), the mixture was stirred at RT for 48 h. The reaction mixture was diluted with water and the resulting suspension was filtered and collected solids dried to give 2-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)isoindoline-1,3-dione as a white solid (120 mg, 100%). MS (ESI) m/z 307 [M+H]+.
Step 3: To a solution of 2-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-7-yl) isoindoline-1,3-dione (120 mg, 0.39 mmol) in MeOH (5 ml) was added hydrazine hydrate (122 mg, 1.95 mmol), the mixture was stirred at RT for 1h then concentrated and purified by reverse-phase HPLC to give 7-amino-1-methyl-3,4-dihydroquinolin-2(1H)-one (50 mg, 72%). MS (ESI) m/z 177 [M+H]+.
Step 4: Synthesized 4 by Method A (28 mg, 45%). 1H-NMR (500 MHz, CDCl3) δ 7.67 (d, J = 8.5 Hz, 2H), 7.23 (d, J = 9 Hz, 2H), 6.99 (d, J = 7.5 Hz, 1H), 6.99 (br-s, 1H), 6.80 (d, J = 2.5 Hz, 1H), 6.60 (dd, J = 7.5, 2.5 Hz, 1H), 3.26 (s, 3H), 2.80 (t, J = 7.5 Hz, 2H), 2.61 (t, J = 7.5 Hz, 2H), 2.49 (s, 3H). MS (ESI) m/z 363[M+H]+.
N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-(methylsulfanyl) benzene-1-sulfonamide (5a)
1H-NMR (500 MHz, MeOD-d4) δ 7.56 (d, J = 8.5 Hz, 2H), 7.26 (d, J = 8.5 Hz, 2H), 6.93–6.97 (m, 2H), 6.76 (dd, J = 8.0, 1.5 Hz, 1H), 3.35 (s, 3H), 3.34 (s, 3H), 2.47 (s, 3H). MS (ESI) m/z 364 [M+H]+.
N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-methoxybenzene-1-sulfonamide (5b)
39% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.87 (s, 1H), 7.62 (d, J = 8.9 Hz, 2H), 7.02 (d, J = 8.9 Hz, 2H), 6.96 (d, J = 8.2 Hz, 1H), 6.86 (d, J = 1.9 Hz , 1H), 6.68 (dd, J = 8.3, 1.9 Hz, 1H), 3.77 (s, 3H), 3.24 (s, 3H), 3.23 (s, 3H). MS (ESI) m/z 348 [M+H]+.
N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)benzenesulfonamide (5c)
53% yield; 1H NMR (600 MHz, DMSO-d6) δ 10.02 (s, 1H), 7.71–7.68 (m, 2H), 7.60-7.57 (m, 1H), 7.54–7.49 (m, 2H), 6.96 (d, J = 8.3 Hz, 1H), 6.86 (d, J = 1.9 Hz , 1H), 6.68 (dd, J = 8.3, 1.9 Hz, 1H), 3.24 (s, 3H), 3.23 (s, 3H). MS (ESI) m/z 318 [M+H]+.
N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-4-(2-methylpropoxy)benzene-1-sulfonamide (5d)
29% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.86 (s, 1H), 7.60 (d, J = 8.9 Hz, 2H), 7.01 (d, J = 8.9 Hz, 2H), 6.96 (d, J = 8.2 Hz, 1H), 6.86 (d, J = 1.9 Hz, 1H), 6.68 (dd, J = 8.3, 1.9 Hz, 1H), 3.76 (d, J = 6.5 Hz, 2H), 3.24 (s, 3H), 3.23 (s, 3H), 2.03–1.94 (m, 1H), 0.95 (d, J = 6.7 Hz, 6H). MS (ESI) m/z 390 [M+H]+.
4-cyclohexyl-N-(1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)benzene-1-sulfonamide (5e)
40% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.97 (s, 1H), 7.61 (d, J = 8.3 Hz, 2H), 7.37 (d, J = 8.3 Hz, 2H), 6.97 (d, J = 8.3 Hz , 1H), 6.84 (d, J = 1.9 Hz, 1H), 6.71 (dd, J = 8.3, 1.9 Hz, 1H), 3.24 (s, 3H), 3.22 (s, 3H), 2.57–2.50 (m, 1H), 1.81–1.65 (m, 5H), 1.41–1.28 (m, 4H), 1.25–1.15 (m, 1H). MS (ESI) m/z 400 [M+H]+.
5-amino-6-bromo-1,3-dimethyl-1H-benzo[d]imidazol-2(3H)-one (10a)
To a 0 °C solution of 5-amino-1,3-dimethyl-1H-benzo[d]imidazol-2(3H)-one (4.0 g, 23 mmol) in 25 mL of CHCl3 and 25 mL of AcOH was carefully added bromine (3.5 g, 23 mmol) dropwise. The mixture was stirred at RT for 30 min, then concentrated and purified by silica gel chromatography (pet ether/EtOAc 1:1) to afford 10a as a yellow solid (3.2 g, 69%). 1H NMR (500 MHz, DMSO-d6) δ 7.18 (s, 1H), 6.59 (s, 1H), 4.96 (s, 2H), 3.22 (s, 3H), 3.21 (s, 3H). MS (ESI) m/z 257 [M+H]+.
N-(6-bromo-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)-2,2,2-trifluoroacetamide (10b)
To a 0 °C solution of 10a (1.50 g, 5.9 mmol) in DCM (45 ml) was added DMAP (72 mg, 0.59 mmol), triethylamine (1.63 ml, 11.7 mmol) and trifluoroacetic anhydride (0.91 ml, 6.4 mmol). The reaction mixture was stirred for 2 h and warmed to ambient temperature. The reaction mixture was then quenched with water and the organic phase was washed with brine, dried over sodium sulfate, filtered and evaporated to give 10b as a yellow solid (2.20 g, 100%). 1H NMR (600 MHz, DMSO-d6) δ 11.23 (br-s, 1H), 7.57 (s, 1H), 7.31 (s, 1H), 3.34 (s, 3H), 3.32 (s, 3H). MS (ESI) m/z 353 [M+H] +.
Representative procedure for the synthesis of 6 and 7a–c (Method B – Ullmann coupling)
Step 1: A mixture of 3-(benzyloxy)phenol (156 mg, 0.78 mmol), quinolin-8-ol (30 mg, 0.21 mmol), copper(I) chloride (10 mg, 0.10 mmol), potassium phosphate (200 mg, 0.94 mmol) and 10a (200 mg, 0.78 mmol) in diglyme (10 mL) was degassed under a nitrogen atmosphere, then the reaction mixture was heated to 130 °C for 72 hours. The cooled reaction mixture was filtered through a pad of silica gel. The collected filtrate was then concentrated and purified by column chromatography (0-100% EtOAc in hexanes and then 0–40% methanol in EtOAc) to give 5-amino-6-(3-(benzyloxy)phenoxy)-1,3-dimethyl-1H-benzo[d]imidazol-2(3H)-one (11b) as a solid (213 mg, 73%). MS (ESI) m/z 376 [M+H] +. 1H NMR (600 MHz, CDCl3) δ 7.34 (m, 5H), 7.18 (t, J = 7.8 Hz, 1H), 6.65 (d, J = 2.8 Hz, 1H), 6.58 (s, 1H), 6.55 (m, 2H), 6.48 (s, 1H), 4.98 (s, 2H), 3.66 (br-s, 2H), 3.34 (s, 3H), 3.27 (s, 3H). Step 2: Sulfonamide formation using Method A.
Representative procedure for the synthesis of 7d–f (Method C – Ullmann coupling)46
Step 1: A mixture of 10b (50 mg, 0.14 mmol), 2-(dimethylamino)acetic acid (15 mg, 0.14 mmol), 3-ethoxyphenol (29 mg, 0.21 mmol), copper(I) iodide (8 mg, 0.04 mmol) and cesium carbonate (139 mg, 0.43 mmol) were charged in a flask with dioxane (1 ml). The reaction mixture was heated to 80 °C and stirred for 16 h. The reaction was monitored for complete de-protection of trifluoroacetamide (addition of methanol was used to facilitate this step). The cooled reaction mixture was diluted in methanol, filtered and the filtrate was evaporated and purified by column chromatography (hexanes/EtOAc 1:1) to give 5-amino-6-(3-ethoxyphenoxy)-1,3-dimethyl-1H-benzo[d]imidazol-2(3H)-one (11c) as an orange solid (13 mg, 29%). MS (ESI) m/z 314 [M+H]+. Step 2: Sulfonamide formation using Method A.
N-{6-[4-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (6)
6% yield. 1H NMR (600 MHz, DMSO-d6) δ 9.32 (s, 1H), 7.57 (d, J = 3.3 Hz, 2H), 7.46 (d, J = 7.5 Hz, 2H), 7.38 (t, J = 7.1 Hz, 2H), 7.30 (t, J = 6.7 Hz, 1H), 7.08 (s, 1H), 6.95 (d, J = 9.5 Hz, 2H), 6.82 (d, J = 9.5 Hz, 2H), 6.67 (s, 1H), 5.06 (s, 2H), 3.57 (s, 3H), 3.28 (s, 3H), 3.17 (s, 3H). MS (ESI) m/z 520 [M+H]+.
N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7a)
21% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.35 (s, 1H), 7.56 (d, J = 3.5 Hz, 2H), 7.41 (d, J = 7.5 Hz, 2H), 7.36 (t, J = 7.1 Hz, 2H), 7.31 (t, J = 6.7 Hz, 1H), 7.17 (t, J = 8.7 Hz, 1H), 7.09 (s, 1H), 6.75 (s, 1H), 6.70 (dd, J = 2.4, 8.3 Hz, 1H), 6.29–6.31 (m, 2H), 5.05 (s, 2H), 3.55 (s, 3H), 3.29 (s, 3H), 3.19 (s, 3H). MS (ESI) m/z 520 [M+H]+.
N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide (7b)
13% yield; 1H NMR (600 MHz, CDCl3) δ 7.30–7.40 (m, 5H), 7.24 (dd, J = 8.3, 2.0 Hz, 1H), 7.06 (m, 2H), 6.82 (s, 1H), 6.70 (d, J = 8.6 Hz, 1H), 6.65 (dd, J = 8.3, 2.0 Hz, 1H), 6.42 (s, 1H), 6.17 (t, J = 2.6 Hz, 1H), 6.05 (dd, J = 8.3, 2.0 Hz, 1H), 4.97 (s, 2H), 3.79 (s, 3H), 3.60 (s, 3H), 3.45 (s, 3H), 3.25 (s, 3H). MS (ESI) m/z 576 [M+H]+.
N-{6-[3-(benzyloxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide (7c)
47% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.31 (s, 1H), 7.48 (s, 1H), 7.42–7.34 (m, 4H), 7.33–7.29 (m, 1H), 7.18 (t, J = 8.3 Hz, 1H), 7.15 (s, 1H), 6.73 (s, 1H), 6.70 (dd, J = 8.3, 2.2 Hz, 1H), 6.31–6.26 (m, 2H), 5.04 (s, 2H), 3.42 (s, 3H), 3.29 (s, 3H), 3.19 (s, 3H), 2.07 (s, 3H). MS (ESI) m/z 534[M+H]+.
N-[6-(3-ethoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]-1,2-dimethyl-1H-imidazole-4-sulfonamide (7d)
36% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.34 (s, 1H), 7.50 (s, 1H), 7.18–7.14 (m, 2H), 6.76 (s, 1H), 6.61 (dd, J = 7.7, 2.2 Hz, 1H), 6.26 (dd, J = 8.0, 2.2 Hz, 1H), 6.19 (t, J = 2.2 Hz, 1H), 3.96 (q, J = 6.9 Hz, 2H), 3.45 (s, 3H), 3.29 (s, 3H), 3.20 (s, 3H), 2.09 (s, 3H), 1.30 (t, J = 7.0 Hz, 3H). MS (ESI) m/z 472 [M+H]+.
N-[1,3-dimethyl-2-oxo-6-(3-propoxyphenoxy)-2,3-dihydro-1H-1,3-benzodiazol-5-yl]-3,4-dimethoxybenzene-1-sulfonamide (7e)
66% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.51 (s, 1H), 7.19–7.14 (m, 2H), 7.11 (s, 1H), 7.08–7.02 (m, 1H), 6.89 (d, J = 9.0 Hz, 1H), 6.74 (s, 1H), 6.55 (dd, J = 8.2, 2.2 Hz, 1H), 6.08 (dd, J = 8.1, 2.0 Hz, 1H), 6.05–6.03 (m, 1H), 3.79 (t, J = 6.5 Hz, 2H), 3.77 (s, 3H), 3.57 (s, 3H), 3.30 (s, 3H), 3.19 (s, 3H), 1.73–1.64 (m, 2H), 0.95 (t, J = 7.4 Hz, 3H). MS (ESI) m/z 528 [M+H]+.
N-[1,3-dimethyl-2-oxo-6-(3-propoxyphenoxy)-2,3-dihydro-1H-1,3-benzodiazol-5-yl]-1,2-dimethyl-1H-imidazole-4-sulfonamide (7f)
93% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.32 (s, 1H), 7.49 (s, 1H), 7.17–7.13 (m, 2H), 6.76 (s, 1H), 6.61 (dd, J = 7.7, 2.2 Hz, 1H), 6.25 (dd, J = 8.0, 2.2 Hz, 1H), 6.23 (t, J = 2.2 Hz, 1H), 3.86 (t, J = 6.5 Hz, 2H), 3.45 (s, 3H), 3.29 (s, 3H), 3.20 (s, 3H), 2.08 (s, 3H), 1.73–1.66 (m, 2H), 0.96 (t, J = 7.5 Hz, 3H). MS (ESI) m/z 486 [M+H]+.
5-amino-6-(3-hydroxyphenoxy)-1,3-dimethyl-1H-benzo[d]imidazol-2(3H)-one (12)
To a −78 °C solution of 11b (400 mg, 1.07 mmol) in DCM (20 mL) was added tribromoborane (5.3 mL, 5.3 mmol). The reaction mixture was allowed to gradually warm to RT, then quenched by the dropwise addition of methanol, concentrated and purified by column chromatography (20–100% EtOAc/hexanes and then 0–40% methanol/EtOAc) to give 12 as a solid (240 mg, 79%). 1H NMR (600 MHz, DMSO-d6) 9.38 (s, 1H), 7.06 (t, J = 8.1, 1H), 6.78 (s, 1H), 6.58 (s, 1H) 6.39 (dd, J =8.0, 2.2, 1H), 6.33 (dd, J = 8.1, 2.3, 1H), 6.23 (t, J = 2.3, 1H), 4.57 (br-s, 2H), 3.24 (s, 3H), 3.20 (s, 3H). MS (ESI) m/z 286 [M+H]+.
General Procedure for the Synthesis of Sulfonamides 7g–i, 7k, 7l
To a solution of 12 (50 mg, 0.18 mmol) in anhydrous DMF (1 ml) was added potassium carbonate (0.2–0.4 mmol) and alkyl bromide (0.18–0.25 mmol). The reaction mixture was stirred at ambient temperature for 1 day, then the reaction mixture was diluted with water and extracted with EtOAc. The seperated organic layer was dried over sodium sulfate, filtered and concentrated to give the intermediate, 5-amino-6-(3-(alkyloxy)phenoxy)-1,3-dimethyl-1H-benzo[d]imidazol-2(3H)-one, as a crude residue which was used without further purification in the sulfonamide formation step using Method A to give 7g–i, 7k, 7l.
N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide (7g)
3% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.27 (s, 1H), 7.47 (s, 1H), 7.17–7.14 (m, 2H), 6.77 (s, 1H), 6.63 (d, J = 8.9 Hz, 1H), 6.27 (s, 1H), 6.24 (d, J = 8.2 Hz, 1H), 3.69 (d, J = 6.5 Hz, 1H), 3.40 (s, 1H), 3.29 (s, 3H), 3.29 (s, 3H), 3.20 (s, 3H), 2.09 (s, 3H), 2.01–1.97 (m, 1H), 0.96 (d, J = 6.7, 6H). MS (ESI) m/z 501 [M+H]+.
N-[6-(3-butoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl]-1,2-dimethyl-1H-imidazole-4-sulfonamide (7h)
13% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.33 (s, 1H), 7.50 (s, 1H), 7.16 (t, J = 7.3 Hz, 2H), 6.77 (s, 1H), 6.62 (dd, J = 8.3, 2.5 Hz , 1H), 6.25 (m, 2H), 3.91 (t, J = 6.4 Hz, 3H), 3.29 (s, 3H), 3.20 (s, 1H), 2.09 (s, 3H), 1.68–1.65 (m, 2H), 1.43–1.39 (m, 2H), 0.93 (t, J = 7.5 Hz, 3H). MS (ESI) m/z 501 [M+H]+.
N-{1,3-dimethyl-6-[3-(2-methylpropoxy)phenoxy]-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}cyclopropanesulfonamide (7i)
28% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.11 (s, 1H), 7.21 (t, J = 8.2 Hz, 1H), 7.17 (s, 1H), 6.92 (s, 1H), 6.66–6.63 (m, 1H), 6.53–6.47 (m, 2H), 3.69 (d, J = 6.4 Hz, 2H), 3.33 (s, 3H), 3.25 (s, 3H), 1.97 (m, 1H), 0.95 (d, J = 6.7 Hz, 6H), 0.85–0.80 (m, 4H). MS (ESI) m/z 446 [M+H]+.
1,3-dimethyl-5-[3-(2-methylpropoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-2-one (7j)
Synthesized from 5-bromo-1,3-dimethyl-1,3-dihydro-2H-benzo[d]imidazol-2-one using Method C, 32% yield. 1H NMR (500 MHz, DMSO-d6) δ 7.21 (t, J = 8.5 Hz, 1H), 7.14 (d, J = 8.5 Hz, 1H), 6.98 (d, J = 2.2 Hz, 1H), 6.77 (dd, J = 8.4, 2.2 Hz, 1H), 6.65–6.60 (m, 1H), 6.48–6.44 (m, 2H), 3.69 (d, J = 6.5, 2H), 3.33 (s, 3H), 3.29 (s, 3H), 1.97 (m, 1H), 0.94 (d, J = 6.7, 6H). MS (ESI) m/z 327 [M+H]+.
N-{6-[3-(3-methoxypropoxy)phenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1,2-dimethyl-1H-imidazole-4-sulfonamide (7k)
13% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.35 (s, 1H), 7.50 (s, 1H), 7.18–7.15 (m, 2H), 6.78 (s, 1H), 6.63 (d, J = 8.3 Hz, 1H), 6.25 (dd, J = 8.4 Hz, 2H), 3.96 (t, J = 6.4 Hz, 2H), 3.46–3.43 (m, 5H), 3.29 (s, 3H), 3.23 (s, 3H), 3.20 (s, 3H), 2.09 (s, 3H), 1.92 (m, 2H). MS (ESI) m/z 516 [M+H]+.
N-{1,3-dimethyl-2-oxo-6-[3-(oxolan-3-ylmethoxy)phenoxy]-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (7l)
5% yield; 1H NMR (600 MHz, DMSO-d6) δ 9.32 (s, 1H), 7.56 (d, J = 3.5 Hz, 2H), 7.17 (t, J = 8.7 Hz, 1H), 7.09 (s, 1H), 6.78 (s, 1H), 6.63 (dd, J = 8.3, 2.4 Hz, 1H), 6.28 (m, 2H), 3.88 (dd, J = 9.3, 6.4 Hz, 1H), 3.83 (t, J = 7.7 Hz, 1H), 3.73 (m, 1H), 3.65 (dd, J = 15.3, 7.4 Hz, 1H), 3.03 (s, 3H), 3.52 (dd, J = 8.3, 5.4 Hz, 1H), 3.27 (s, 3H), 3.20 (s, 3H), 2.63 (m, 1H), 1.99 (m, 1H), 1.64 (m, 1H). MS (ESI) m/z 515 [M+H]+.
Synthesis of N-(6-{3-[4-(dimethylamino)butoxy]phenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-3,4-dimethoxybenzene-1-sulfonamide (7m)
Step 1: To a solution of resorcinol (585 mg, 5.32 mmol) in DMF (15 ml) was added K2CO3 (735 mg, 5.32 mmol) and 2-(4-bromobutyl)isoindoline-1,3-dione (500 mg, 1.772 mmol). The mixture was stirred at 50 °C overnight and then diluted with water (50 mL) then adjusted to acidic pH with 1N HCl. The product was extracted with EtOAc (2×50 mL), washed with brine (20 mL), dried over Na2SO4, and concentrated. Purification by silica gel chromatography (0% EtOAc/hexane to 100% EtOAc/hexane). Purification by prep-HPLC using a gradient of 40-80% ACN/water containing 0.1% TFA to afford 2-(4-(3-hydroxyphenoxy)butyl)isoindoline-1,3-dione (263 mg, 48 % yield) as a white solid. 1H NMR (600 MHz, DMSO-d6) δ 9.32 (s, 1H), 7.90–7.80 (m, 4H), 7.05–6.97 (m, 1H), 6.34–6.29 (m, 2H), 6.30–6.25 (m, 1H), 3.90 (t, J = 6.1 Hz, 2H), 3.63 (t, J = 6.6 Hz, 2H), 1.79–1.65 (m, 4H). MS (ESI) m/z 312 [M+H]+.
Step 2: 2-(4-(3-((6-amino-1,3-dimethyl-2-methylene-2,3-dihydro-1H-benzo[d]imidazol-5-yl)oxy)phenoxy)butyl)isoindoline-1,3-dione was prepared from 10b using Method C to give the product as as an orange liquid (118 mg, 35 % yield). 1H NMR (600 MHz, DMSO-d6) δ 7.89-7.79 (m, 4H), 7.19-7.12 (m, 1H), 6.79 (s, 1H), 6.58 (s, 1H), 6.55 (dd, J = 2.2, 8.3 Hz, 1H), 6.42-6.37 (m, 2H), 4.57 (br-s, 2H), 3.92 (t, J = 5.9 Hz, 2H), 3.61 (t, J = 6.5 Hz, 2H), 3.24 (s, 3H), 3.20 (s, 3H), 1.76-1.64 (m, 4H). MS (ESI) m/z 487 [M+H]+.
Step 3: N-(6-(3-(4-(1,3-dioxoisoindolin-2-yl)butoxy)phenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)-3,4-dimethoxybenzenesulfonamide was synthesized using Method A to give the product as a light-orange liquid (160 mg, 99 % yield). 1H NMR (600 MHz, DMSO-d6) δ 9.53 (s, 1H), 7.90–7.80 (m, 4H), 7.18–7.13 (m, 2H), 7.12 (s, 1H), 7.06–7.00 (m, 1H), 6.87 (d, J = 9.0 Hz, 1H), 6.73 (s, 1H), 6.53 (dd, J = 8.2, 2.1 Hz, 1H), 6.06 (dd, J = 7.9, 2.0 Hz, 1H), 6.06–6.02 (m, 1H), 3.86 (t, J = 5.9 Hz, 2H), 3.75 (s, 3H), 3.63 (t, J = 6.5 Hz, 2H), 3.56 (s, 3H), 3.30 (s, 3H), 3.18 (s, 3H), 1.78–1.65 (m, 4H). MS (ESI) m/z 687 [M+H]+.
Step 4: To a solution of N-(6-(3-(4-(1,3-dioxoisoindolin-2-yl)butoxy)phenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)-3,4-dimethoxybenzenesulfonamide (160 mg, 0.233 mmol) in MeOH (4 mL) was added hydrazine hydrate (29 μl, 0.47 mmol) and the reaction mixture was heated to 80 °C for 90 min. The cooled reaction mixture was concentrated then purified by prep-HPLC using a gradient of 20-60% ACN/water containing 0.1% TFA to afford the TFA salt of N-(6-(3-(4-aminobutoxy)phenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)-3,4-dimethoxybenzenesulfonamide (103 mg, 66 % yield) as a white solid. 1H NMR (600 MHz, DMSO-d6) δ 9.52 (s, 1H), 7.68 (br-s, 3H), 7.20–7.15 (m, 2H), 7.09 (s, 1H), 7.09–7.04 (m, 1H), 6.89 (d, J = 9.0 Hz, 1H), 6.73 (s, 1H), 6.55 (dd, J = 8.2, 2.1 Hz, 1H), 6.12 (dd, J = 8.2, 2.1 Hz, 1H), 6.08–6.04 (m, 1H), 3.85 (t, J = 5.9 Hz, 2H), 3.77 (s, 3H), 3.59 (s, 3H), 3.30 (s, 3H), 3.19 (s, 3H), 2.89–2.81 (m, 2H), 1.78–1.62 (m, 4H). MS (ESI) m/z 557 [M+H]+.
Step 5: To a solution of N-(6-(3-(4-aminobutoxy)phenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)-3,4-dimethoxybenzenesulfonamide TFA salt (50 mg, 0.075 mmol) in MeOH (3 ml) was added triethylamine (10 μl, 0.08 mmol), acetic acid (9 μl, 0.15 mmol), formaldehyde (16 μl, 0.60 mmol), and sodium triacetoxyborohydride (40 mg, 0.19 mmol). The reaction mixture was stirred at RT for 3 then was quenched with a few drops of TFA and concentrated. Purification by prep-HPLC using a gradient of 20-60% acetonitrile/water containing 0.1% TFA to afford the TFA salt of 7m (14 mg, 27 % yield) as a yellow liquid. 1H NMR (600 MHz, DMSO-d6) δ 9.50 (s, 1H), 9.34 (br-s, 1H), 7.21–7.15 (m, 2H), 7.12–7.05 (m, 2H), 6.89 (d, J = 9.0 Hz, 1H), 6.73 (s, 1H), 6.56 (dd, J = 8.2, 2.0 Hz, 1H), 6.14 (dd, J = 8.1, 1.9 Hz, 1H), 6.09–6.04 (m, 1H), 3.86 (t, J = 5.9 Hz, 2H), 3.77 (s, 3H), 3.59 (s, 3H), 3.29 (s, 3H), 3.19 (s, 3H), 3.14–3.06 (m, 2H), 2.77 (d, J = 4.9 Hz, 6H), 1.79–1.64 (m, 4H). MS (ESI) m/z 585 [M+H]+.
Synthesis of tert-butyl (6-(3-((1,3-dimethyl-6-(1-methyl-1H-imidazole-4-sulfonamido)-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)oxy)-5-propoxyphenoxy)hexyl)carbamate (8a)
Step 1: 5-propoxybenzene-1,3-diol (13). A solution of benzene-1,3,5-triol (1.0 g, 7.9 mmol) in DMF (10 ml) was treated with potassium carbonate powder (1.2 g, 8.7 mmol) and 1-bromopropane (0.80 ml, 8.7 mmol), the mixture was stirred at 50 °C for 16 h. The cooled reaction mixture was diluted with water, carefully quenched with 1N HCl until acidic, then extracted with EtOAc. The organic layer was washed with brine and dried over sodium sulfate, concentrated and purified by column chromatography(EtOAc/hexanes 3:7) to give 5-propoxybenzene-1,3-diol (13) as a yellow liquid (0.49 g, 39%). 1H NMR (600 MHz, CDCl3) δ 6.00 (d, J = 2.1 Hz, 2H), 5.96 (t, J = 2.1 Hz, 1H), 5.48 (br-s, 2H), 3.84 (t, J = 6.5 Hz, 2H), 1.80-1.72 (m, 2H), 1.00 (t, J = 7.4 Hz, 3H). MS (ESI) m/z 169 [M+H]+.
Step 2: To a solution of 13 (344 mg, 2.05 mmol) and tert-butyl (6-bromohexyl)carbamate (268 mg, 0.956 mmol) in 3 mL of DMF was added potassium carbonate (326 mg, 2.36 mmol) and the resulting mixture was stirred at 50°C for 16 h. The cooled reaction mixture was neutralized with 1M HCl, diluted with EtOAc, and the seperated organic layer was washed with sat. aq. NaCl solution, dried over sodium sulfate, filtered and concentrated under reduced pressure. The residue was purified via silica gel chromatography (1:9 to 1:1 EtOAc/hexanes) to give 14a (174 mg, 23%) as a light-brown viscous liquid. 1H NMR (600 MHz, CDCl3) δ 6.05 (m, 1H), 6.01 (m, 2H), 5.15 (br-s, 1H), 4.53 (br-s, 1H), 3.90 (m, 2H), 3.86 (t, J = 6.5 Hz, 2H), 3.12 (m, 2H), 1.81–1.72 (m, 4H), 1.54–1.33 (m, 6H), 1.44 (s, 9H), 1.01 (t, J = 7.5 Hz, 3H). MS (ESI) m/z 368 [M+H]+.
Step 3: A solution of 10b (100 mg, 0.284 mmol), 14a (147 mg, 0.400 mmol) and 2-(dimethylamino)acetic acid (51 mg, 0.50 mmol) in diethylene glycol dimethyl ether (6 ml) was degassed with nitrogen. Copper(I) iodide (34 mg, 0.18 mmol) and Cs2CO3 (410 mg, 1.26 mmol) were added and the mixture was degassed with nitrogen for an additional 2 minutes. The reaction mixture was heated to 80°C for 1.5 days. The cooled reaction mixture was filtered through a pad of celite, concentrated then purified by flash chromatography (EtOAc/hexanes 1:9 to 1:1) to give 15a as a solid (54 mg, 35%). 1H NMR (600 MHz, CDCl3) δ: 6.63 (s, 1H), 6.47 (s, 1H), 6.15 (t, J = 2.1 Hz, 1H), 6.08 (t, J = 2.1 Hz, 1H), 6.06 (t, J = 2.1 Hz, 1H), 4.50 (br-s, 1H), 3.87 (t, J = 6.4 Hz, 2H), 3.84 (t, J = 6.6 Hz, 2H), 3.64 (br-s, 2H), 3.37 (s, 3H), 3.31 (s, 3H), 3.11 (m, 2H), 1.80–1.70 (m, 4H), 1.54–1.34 (m, 6H), 1.44 (s, 9H), 1.00 (t, J = 7.4 Hz, 3H). MS (ESI) m/z 543 [M+H]+.
Step 4: Method A gave 8a as an amorphous solid (68% yield). 1H NMR (600 MHz, CDCl3) δ: 7.97 (br-s, 1H), 7.60 (s, 1H, overlapped with CHCl3), 7.35 (s, 1H), 7.31 (s, 1H), 7.20 (s, 1H), 6.57 (s, 1H), 6.14 (s, 1H), 5.77 (br-s, 1H), 5.72 (br-s, 1H), 3.86 (t, J = 6.4 Hz, 2H), 3.83 (t, J = 6.5 Hz, 2H), 3.62 (s, 3H), 3.45 (s, 3H), 3.31 (s, 3H), 3.12 (br-t, J = 6.3 Hz, 2H), 1.76 (m, 4H), 1.51 (m, 2H), 1.46 (m, 2H), 1.44 (s, 9H), 1.37 (m, 2H), 1.01 (t, J = 7.3 Hz, 3H). MS (ESI) m/z 687 [M+H]+.
N-(6-{3-[(6-aminohexyl)oxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide (8b)
To a solution of 8a (18 mg, 0.026 mmol) in DCM (4 ml) was added TFA (500 μl, 6.49 mmol) and the resulting mixture was stirred at 23 °C for 16 h. The reaction mixture was concentrated to give the TFA salt of 8b as a brown-colored viscous liquid (17 mg, 93%). 1H NMR (DMSO-d6) δ 8.52 (br-s, 1H), 7.94 (br-s, 3H), 7.48 (br-s, 1H), 7.24 (s, 1H), 7.18 (s, 1H), 6.56 (s, 1H), 6.11 (s, 1H), 5.85 (s, 1H), 5.56 (s, 1H), 3.89 (t, J = 6.1 Hz, 2H), 3.79 (t, J = 6.6 Hz, 2H), 3.61 (s, 3H), 3.41 (s, 3H), 3.28 (s, 3H), 2.97 (br-s, 2H), 1.77–1.65 (m, 6H), 1.46 (m, 2H), 1.42 (m, 2H), 0.98 (t, J = 7.3 Hz, 3H). MS (ESI) m/z 587 [M+H]+.
Synthesis of N-(6-{3-[(6-hydroxyhexyl)oxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide (8c)
Step 1: Prepared in a similar manner to 14a from 13 using 6-bromohexan-1-ol to give 14b as a viscous brown liquid (39% yield). 1H NMR (600 MHz, CDCl3) δ 6.06 (m, 1H), 6.03–5.98 (m, 2H), 5.25 (br-s, 1H), 3.90 (t, J = 6.5 Hz, 2H), 3.86 (t, J = 6.7 Hz, 2H), 3.67 (t, J = 6.5 Hz, 2H), 1.82–1.73 (m, 4H), 1.64–1.57 (m, 2H), 1.52–1.36 (m, 5H), 1.01 (t, J = 7.4 Hz, 3H). MS (ESI) m/z 269 [M+H]+.
Steps 2 and 3: Prepared using Method C to give 15b and used without further purification. MS (ESI) m/z 444 [M+H]+. 15b was converted to 8c using Method A (38% yield). 1H NMR (600 MHz, DMSO-d6) δ 9.30 (s, 1H), 7.60 (m, 1H), 7.58 (m, 1H), 7.07 (s, 1H), 6.80 (s, 1H), 6.17 (s, 1H), 5.83 (m, 2H), 4.38 (t, J = 6.5 Hz, 1H), 3.86 (t, J = 6.5 Hz, 2H), 3.83 (t, J = 6.5 Hz, 2H), 3.37 (t, J = 6.5 Hz, 2H), 3.59 (s, 3H), 3.27 (s, 3H), 3.21 (s, 3H), 1.67 (m, 4H), 1.45–1.28 (m, 6H), 0.94 (t, J = 7.5 Hz, 3H). MS (ESI) m/z 588 [M+H]+.
Synthesis of N-(6-(3-((6-aminohexyl)oxy)-5-propoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)-3,4-dimethoxybenzenesulfonamide (8d)
Step 1: Prepared from 13 using Method C to give 16 as a brown solid (43% yield). 1H NMR (600 MHz, DMSO-d6) δ 9.40 (s, 1H), 6.81 (s, 1H), 6.66 (br-s, 1H), 5.99 (s, 1H), 5.93 (s, 1H), 5.85 (s, 1H), 3.80 (t, J = 6.6 Hz, 2H), 3.26 (s, 3H), 3.21 (s, 3H) 1.66 (m, 2H), 0.93 (t, J = 7.5, 3H). MS (ESI) m/z 344 [M+H]+.
Step 2: To a solution of 16 (800 mg, 2.33 mmol) in anhydrous DMF (60 ml) was added potassium carbonate (1.29 g, 9.32 mmol) and 2-(6-bromohexyl)isoindoline-1,3-dione (2.89 g, 9.32 mmol) in a vial. The reaction mixture was stirred at 80 °C for 1 hr. To the cooled reaction mixture was added water (50 mL) and the aqueous phase was extracted with EtOAc (3 × 50 mL), the combined organic layers were concentrated under reduced pressure and the residue was purified by silica gel chromatography (EtOAc/hexanes 1:4 to 100% EtOAc) to give 2-(6-(3-((6-amino-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)oxy)-5-propoxyphenoxy)hexyl)isoindoline-1,3-dione 17a as a viscous liquid (800 mg, 60%). 1H NMR (600 MHz, DMSO-d6) δ 7.87–7.82 (m, 4H), 6.83 (s, 1H), 6.69 (br-s, 1H), 6.17 (s, 1H), 6.00 (s, 2H), 3.86 (t, J = 6.2 Hz, 2H), 3.84 (t, J = 6.5 Hz, 2H), 3.56 (t, J = 7.1 Hz, 2H), 3.26 (s, 3H), 3.21 (s, 3H), 1.70–1.56 (m, 6H), 1.39 (m, 2H), 1.31 (m, 2H), 0.93 (t, J = 7.4 Hz, 3H). MS (ESI) m/z 573 [M+H]+.
Step 3: To a solution of 17a (150 mg, 0.262 mmol) in anhydrous DCM (1 ml) was added pyridine (0.042 ml, 0.524 mmol) and 3,4-dimethoxybenzene-1-sulfonyl chloride (74.4 mg, 0.314 mmol). The mixture was stirred for 1 h before quenching with MeOH (0.5 mL). The resulting mixture was treated with hydrazine hydrate (8.22 μl, 0.262 mmol) then concentrated and purified by mass-triggered preparative HPLC (Mobile phase: A = 0.1% TFA/H2O, B = 0.1% TFA/ACN; Gradient: B = 20 - 60%; 12 min; Column: C18) to give 8d as a white solid. 30% yield (over 2-steps); 1H NMR (600 MHz, DMSO-d6) δ 9.44 (s, 1H), 7.61 (s, 2H), 7.19–7.18 (m, 2H), 7.07 (s, 1H), 6.90 (d, J = 8.9 Hz, 1H) 6.75 (s, 1H), 6.11 (s, 1H), 5.69 (s, 1H), 5.66 (s, 1H), 3.81 (t, J = 6.4 Hz, 2H), 3.78–3.75 (m, 5H), 3.35 (s, 3H), 3.29 (s, 3H), 3.20 (s, 3H), 2.80–2.76 (m, 2H), 1.69-1.64 (m, 4H), 1.55–1.52 (m, 2H), 1.40–1.33 (m, 4H), 0.95 (t, J = 7.5 Hz, 3H). MS (ESI) m/z 643 [M+H]+.
N-(6-{3-[(5-aminopentyl)oxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-1-methyl-1H-imidazole-4-sulfonamide (8e)
Was prepared in a similar manner to 8d except using a BOC-protected intermediate. 27% yield (3 steps). 1H NMR (600 MHz, DMSO-d6) δ 9.28 (s, 1H), 7.60 (s, 4H), 7.63 (s, 1H), 7.05 (s, 1H), 6.80 (s, 1H) 6.17 (s, 1H), 5.87 (t, J = 2.1 Hz, 2H), 3.91 (t, J = 6.1 Hz, 2H), 3.84 (t, J = 6.5 Hz, 2H), 3.61 (s, 3H), 3.26 (s, 3H), 3.22 (s, 3H), 2.85–2.82 (m, 2H), 1.68 (m, 4H), 1.56 (m, 2H), 1.42 (m, 2H), 0.95 (t, J = 7.3 Hz, 3H). MS (ESI) m/z 573 [M+H]+.
N-{6-[3-(4-aminobutoxy)-5-propoxyphenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-1-methyl-1H-imidazole-4-sulfonamide (8f)
Was prepared in a similar manner to 8d. 50% yield (3 steps). 1H NMR (600 MHz, DMSO-d6) δ 9.27 (s, 1H), 7.64 (s, 3H), 7.61 (s, 1H), 7.07 (s, 1H), 6.80 (s, 1H) 6.18 (s, 1H), 5.89–5.87 (m, 2H), 3.91 (t, J = 6.1 Hz, 2H), 3.84 (t, J = 6.5 Hz, 2H), 3.61 (s, 3H), 3.27 (s, 3H), 3.22 (s, 3H), 2.85–2.82 (m, 2H), 1.74–1.64 (m, 7H), 0.95 (t, J = 7.3 Hz, 3H). MS (ESI) m/z 559 [M+H]+.
N-{6-[3-(4-aminobutoxy)-5-propoxyphenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide (8g)
Was prepared in a similar manner to 8d. 26% yield (3 steps). 1H NMR (600 MHz, DMSO-d6) δ 9.47 (s, 1H), 7.59 (s, 3H), 7.25 (d, J = Hz, 1H), 7.09–7.06 (m, 2H), 7.00 (s, 1H), 6.81 (s, 1H) 6.00 (s, 1H), 5.71 (s, 1H), 5.62 (s, 1H), 3.84 (s, 3H), 3.75 (t, J = 6.5 Hz, 2H), 3.70 (s, 3H), 3.31 (s, 3H), 3.25 (s, 3H), 1.69–1.65 (m, 3H), 1.38–1.35 (m, 2H), 0.95 (t, J = 7.5 Hz, 4H). MS (ESI) m/z 615 [M+H]+.
N-{6-[3-(3-aminopropoxy)-5-propoxyphenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide (8h)
Was prepared in a similar manner to 8d except using a BOC-protected intermediate. 13% yield (3 steps). 1H NMR (600 MHz, DMSO-d6) δ p9.44 (s, 1H), 7.68 (br-s, 3H), 7.20–7.17 (m, 2H), 7.06 (s, 1H), 6.90 (d, J = 9.0 Hz, 1H) 6.73 (s,1H), 6.13 (t, J = 2.1 Hz, 1H), 5.72 (t, J = 2.1 Hz, 1H), 5.67 (t, J = 2.1 Hz, 1H), 3.89 (t, J = 6.1 Hz, 2H), 3.775 (t, J = 6.5 Hz, 2H), 3.770 (s, 3H), 3.61 (s, 3H), 3.29 (s, 3H), 3.20 (s, 3H), 2.95–2.89 (m, 2H), 1.98–1.90 (m, 2H), 1.71–1.63 (m, 2H), 0.94 (t, J = 7.5 Hz, 3H). MS (ESI) m/z 601 [M+H]+.
N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-3,4-dimethoxybenzene-1-sulfonamide (8i)
To a solution of 8h TFA salt (180 mg, 0.25 mmol) in methanol (3 ml) was added triethylamine (34 ul, 0.25 mmol), acetic acid (28 uL, 0.49 mmol), formaldehyde (0.054 ml, 1.98 mmol), and sodium triacetoxyborohydride (131 mg, 0.618 mmol). The reaction mixture was stirred at RT for 3 h then concentrated under reduced pressure. The residue was purified by prep-HPLC using a gradient of 20-60% ACN/water containing 0.1% TFA to afford the TFA salt of 8i (106 mg, 57%) as a white solid. 1H NMR (600 MHz, DMSO-d6) δ 9.46 (s, 1H), 9.30 (br-s, 1H), 7.19 (m, 2H), 7.07 (s, 1H), 6.90 (d, J = 9.0 Hz, 1H), 6.75 (s, 1H), 6.13 (t, J = 2.2 Hz, 1H), 5.71 (t, J = 2.0 Hz, 1H), 5.67 (t, J = 2.0 Hz, 1H), 3.84 (t, J = 5.9 Hz, 2H), 3.77 (m, 5H), 3.62 (s, 3H), 3.29 (s, 3H), 3.20 (s, 3H), 3.12–3.05 (m, 2H), 2.78 (d, J = 4.7 Hz, 6H), 1.77–1.63 (m, 6H), 0.95 (t, J = 7.3 Hz, 3H). 13C NMR (600 MHz, DMSO-d6) δ 160.3, 160.0, 159.3, 154.1, 152.0, 148.4, 143.9, 131.8, 128.2, 126.0, 121.9, 120.5, 110.4, 109.4, 106.4, 100.6, 95.9, 95.8, 95.2, 68.9, 66.7, 56.3, 55.6, 55.4, 42.1, 27.1, 27.0, 25.6, 21.9, 20.7, 10.4. MS (ESI) m/z 644 [M+H]+.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Michelle Barton for providing TRIM24 plasmids and Gilbert Lee IV at the CBSF (Core for Biomolecular Structure and Function) for providing BRPF1 purified protein. The authors would also like to thank James Holton and George Meigs for technical help at the Advance Light Source synchrotron facility at the Lawrence Berkeley National Laboratory. We thank Eun Jeong Cho and Kevin N. Dalby at the University of Texas at Austin and the TxSACT (Texas Screening Alliance for Cancer Therapeutics) program for performing the TRIM24 HTS screen. The TxSACT-New Drug Discovery program is supported by CPRIT (Cancer Prevention Research Institute of Texas) grant RP110532. We thank Stefan Knapp at the Structural Genomics Consortium for providing the template for Fig 4, and Richard Lewis for feedback and proofreading of the manuscript.
ABBREVIATIONS
- ACN
acetonitrile
- AcOH
acetic acid
- Alpha
amplified luminescence proximity homogeneous assay
- BET
bromodomain and extra-terminal
- BCP
bromodomain-containing protein
- BOC
tert-butyl carbonate
- BRD4
bromodomain containing protein 4
- BRPF1
bromodomain and PHD finger containing 1
- DCM
dichloromethane
- DMF
dimethyl formamide
- EtOAc
ethyl acetate
- H-bond
hydrogen bond
- HTS
high throughput screening
- ITC
isothermal calorimetry
- KAc
acetylated lysine
- PHD
plant homeodomain
- RT
room temperature
- SAR
structure activity relationship
- SEM
standard error of the mean
- SUMO
small-ubiquitin like modifier
- TBAF
tetrabutylammonium fluoride
- TFA
trifluoroacetic acid
- TRIM24
Tripartite motif containing protein 24
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information. Assay conditions, materials and methods, crystallographic data collection and refinement statistics (Table S1), bromodomain profiling data for 8i (Table S2), and 1H-NMR spectral data for 8i. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes. The models and structure factors have been deposited with PDB accession codes: 1 4YAB, 3b 4YAD, 5b 4YAT, 5g 4YAX, 7b 4YBM, 7g 4YBS, 7l 4YBT, 8i 4YC9.
Accession Codes:
PDB accession codes of TRIM24 in complex ligands: 1 4YAB, 3b 4YAD, 5b 4YAT, 5g 4YAX, 7b 4YBM, 7g 4YBS, 7l 4YBT, 8i 4YC9
REFERENCES
- 1.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 2.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picaud S, Da Costa D, Thanasopoulou A, Filippakopoulos P, Fish PV, Philpott M, Fedorov O, Brennan P, Bunnage ME, Owen DR, Bradner JE, Taniere P, O'Sullivan B, Muller S, Schwaller J, Stankovic T, Knapp S. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res. 2013;73:3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fish PV, Filippakopoulos P, Bish G, Brennan PE, Bunnage ME, Cook AS, Federov O, Gerstenberger BS, Jones H, Knapp S, Marsden B, Nocka K, Owen DR, Philpott M, Picaud S, Primiano MJ, Ralph MJ, Sciammetta N, Trzupek JD. Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit. J. Med. Chem. 2012;55:9831–9837. doi: 10.1021/jm3010515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. http://www.thesgc.org/chemical-probes/epigenetics.
- 7.Mirguet O, Gosmini R, Toum J, Clement CA, Barnathan M, Brusq JM, Mordaunt JE, Grimes RM, Crowe M, Pineau O, Ajakane M, Daugan A, Jeffrey P, Cutler L, Haynes AC, Smithers NN, Chung CW, Bamborough P, Uings IJ, Lewis A, Witherington J, Parr N, Prinjha RK, Nicodeme E. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J. Med. Chem. 2013;56:750, 1–7515. doi: 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- 8.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nature Rev. Drug Discovery. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 9.Garnier JM, Sharp PP, Burns CJ. BET bromodomain inhibitors: a patent review. Expert Opin. Ther. Pat. 2014;24:185–199. doi: 10.1517/13543776.2014.859244. [DOI] [PubMed] [Google Scholar]
- 10.Gallenkamp D, Gelato KA, Haendler B, Weinmann H. Bromodomains and their pharmacological inhibitors. ChemMedChem. 2014;9:438–464. doi: 10.1002/cmdc.201300434. [DOI] [PubMed] [Google Scholar]
- 11.Mirguet O, Lamotte Y, Donche F, Toum J, Gellibert F, Bouillot A, Gosmini R, Nguyen VL, Delannee D, Seal J, Blandel F, Boullay AB, Boursier E, Martin S, Brusq JM, Krysa G, Riou A, Tellier R, Costaz A, Huet P, Dudit Y, Trottet L, Kirilovsky J, Nicodeme E. From ApoA1 upregulation to BET family bromodomain inhibition: discovery of I-BET151. Bioorg. Med. Chem. Lett. 2012;22:2963–2967. doi: 10.1016/j.bmcl.2012.01.125. [DOI] [PubMed] [Google Scholar]
- 12.McLure KG, Gesner EM, Tsujikawa L, Kharenko OA, Attwell S, Campeau E, Wasiak S, Stein A, White A, Fontano E, Suto RK, Wong NC, Wagner GS, Hansen HC, Young PR. RVX-208, an inducer of ApoA-I in humans, is a BET bromodomain antagonist. PloS One. 2013;8:e83190. doi: 10.1371/journal.pone.0083190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Josling GA, Selvarajah SA, Petter M, Duffy MF. The role of bromodomain proteins in regulating gene expression. Genes. 2012;3:320–343. doi: 10.3390/genes3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demont EH, Bamborough P, Chung CW, Craggs PD, Fallon D, Gordon LJ, Grandi P, Hobbs CI, Hussain J, Jones EJ, Le Gall A, Michon AM, Mitchell DJ, Prinjha RK, Roberts AD, Sheppard RJ, Watson RJ. 1,3-Dimethyl Benzimidazolones Are Potent, Selective Inhibitors of the BRPF1 Bromodomain. ACS Med. Chem. Lett. 2014;5:1190–1195. doi: 10.1021/ml5002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay DA, Fedorov O, Martin S, Singleton DC, Tallant C, Wells C, Picaud S, Philpott M, Monteiro OP, Rogers CM, Conway SJ, Rooney TP, Tumber A, Yapp C, Filippakopoulos P, Bunnage ME, Muller S, Knapp S, Schofield CJ, Brennan PE. Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomains. J. Am. Chem. Soc. 2014;136:9308–9319. doi: 10.1021/ja412434f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picaud S, Strocchia M, Terracciano S, Lauro G, Mendez J, Daniels DL, Riccio R, Bifulco G, Bruno I, Filippakopoulos P. The 9H-purine scaffold reveals induced-fit pocket plasticity of the BRD9 bromodomain. J. Med. Chem. 2015;58:2718–2736. doi: 10.1021/jm501893k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vangamudi B, Paul TA, Shah PK, Kost-Alimova M, Nottebaum L, Shi X, Zhan Y, Leo E, Mahadeshwar HS, Protopopov A, Futreal A, Tieu TN, Peoples M, Heffernan TP, Marszalek JR, Toniatti C, Petrocchi A, Verhelle D, Owen DR, Draetta G, Jones P, Palmer WS, Sharma S, Andersen JN. The catalytic SMARCA2/4 ATPase domain, but not the bromodomain, is a drug target in SWI/SNF mutant cancers: Insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-3798. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidler LR, Brown N, Knapp S, Hoelder S. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J. Med. Chem. 2012;55:7346–7359. doi: 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson FM, Fedorov O, Chaikuad A, Philpott M, Muniz JRC, Felletar I, von Delft F, Heightman T, Knapp S, Abell C, Ciulli A. Targeting low druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain. J. Med. Chem. 2013;56:10183–10187. doi: 10.1021/jm401582c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drouin L, McGrath S, Vidler LR, Chaikuad A, Monteiro O, Tallant C, Philpott M, Rogers C, Fedorov O, Liu M, Akhtar W, Hayes A, Raynaud F, Müller S, Knapp S, Hoelder S. Structure enabled design of BAZ2-ICR, a chemical probe targeting the bromodomains of BAZ2A and BAZ2B. J. Med. Chem. 2015;58:2553–2559. doi: 10.1021/jm501963e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harner MJ, Chauder BA, Phan J, Fesik SW. Fragment-based screening of the bromodomain of ATAD2. J. Med. Chem. 2014;57:9687–9692. doi: 10.1021/jm501035j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaikuad A, Petros AM, Fedorov O, Xu J, Knapp S. Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain. Med. Chem. Comm. 2014;5:1843–1848. [Google Scholar]
- 23.Poncet-Montange G, Zhan Y, Bardenhagen JP, Petrocchi A, Leo E, Shi X, Lee GR, Leonard PG, Geck Do MK, Cardozo MG, Andersen JN, Palmer WS, Jones P, Ladbury JE. Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2. Biochem. J. 2015;466:337–346. doi: 10.1042/BJ20140933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson S, Glass KC. The MOZ histone acetyltransferase in epigenetic signaling and disease. J. Cell. Physiol. 2014;229:1571–1574. doi: 10.1002/jcp.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown T, Swansbury J, Taj MM. Prognosis of patients with t(8;16)(p11;p13) acute myeloid leukemia. Leuk. Lymphoma. 2012;53:338–341. doi: 10.3109/10428194.2011.614703. [DOI] [PubMed] [Google Scholar]
- 26.Lubula MY, Eckenroth BE, Carlson S, Poplawski A, Chruszcz M, Glass KC. Structural insights into recognition of acetylated histone ligands by the BRPF1 bromodomain. FEBS lett. 2014;588:3844–3854. doi: 10.1016/j.febslet.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatakeyama S. TRIM proteins and cancer. Nature Rev. Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 28.Marin I. Origin and diversification of TRIM ubiquitin ligases. PloS One. 2012;7:e50030. doi: 10.1371/journal.pone.0050030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herquel B, Ouararhni K, Davidson I. The TIF1α-related TRIM cofactors couple chromatin modifications to transcriptional regulation, signaling and tumor suppression. Transcription. 2011;2:231–236. doi: 10.4161/trns.2.5.17725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Douarin B, Zechel C, Garnier JM, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai W, W., Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, Aronow B, Gozani O, Fischle W, Hung MC, Patel DJ, Barton MC. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, Bergmann A, Johnson RL, Barton MC. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA. 2009;106:11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambon M, Orsetti B, Berthe ML, Bascoul-Mollevi C, Rodriguez C, Duong V, Gleizes M, Thénot S, Bibeau F, Theillet C, Cavaillès V. Prognostic significance of TRIM24/TIF-1α gene expression in breast cancer. Am. J. Pathol. 2011;178:1461–1469. doi: 10.1016/j.ajpath.2010.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui Z, Cao W, Li J, Song X, Mao L, Chen W. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PloS One. 2013;8:e63887. doi: 10.1371/journal.pone.0063887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Sun L, Tang Z, Fu L, Xu Y, Li Z, Luo W, Qiu X, Wang E. Overexpression of TRIM24 correlates with tumor progression in non-small cell lung cancer. PloS One. 2012;7:e37657. doi: 10.1371/journal.pone.0037657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Huang Y, Yang D, Li X, Liang J, Lin L, Zhang M, Zhong K, Liang B, Li J. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PloS One. 2014;9:e85462. doi: 10.1371/journal.pone.0085462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang LH, Yin AA, Cheng JX, Huang HY, Li XM, Zhang YQ, Han N, Zhang X. TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K/Akt signaling pathway. Oncogene. 2014;34:1–11. doi: 10.1038/onc.2013.593. [DOI] [PubMed] [Google Scholar]
- 39.Bennett JM, Federov O, Tallant C, Monteiro OP, Meier J, Gamble V, Savitski P, Nunez-Alonso GA, Haendler B, Rogers C, Brennan PE, Müller S, Knapp S. Discovery of a Chemical Tool Inhibitor Targeting the Bromodomains of TRIM24 and BRPF. J. Med. Chem. 2015 doi: 10.1021/acs.jmedchem.5b00458. DOI: 10.1021/acs.jmedchem.5b00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irwin JJ, Shoichet BK. ZINC – a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philpott M, Yang J, Tumber T, Federov O, Uttarkar S, Filippakopoulos P, Picaud S, Keates T, Felletar I, Ciulli A, Knapp S, Heightman TD. Bromodomain-peptide displacement assays for interactome mapping and inhibitor discovery. Mol. Biosyst. 2011;7:2899–2908. doi: 10.1039/c1mb05099k. [DOI] [PubMed] [Google Scholar]
- 42.Hewings DS, Fedorov O, Filippakopoulos P, Martin S, Picaud S, Tumber A, Wells C, Olcina MM, Freeman K, Gill A, Ritchie AJ, Sheppard DW, Russell AJ, Hammond EM, Knapp S, Brennan PE, Conway SJ. Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands. J. Med. Chem. 2013;56:3217–3227. doi: 10.1021/jm301588r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hewings DS, Rooney TP, Jennings LE, Hay DA, Schofield CJ, Brennan PE, Knapp S, Conway SJ. Progress in the development and application of small molecule inhibitors of bromodomain-acetyl-lysine interactions. J. Med. Chem. 2012;55:9393–9413. doi: 10.1021/jm300915b. [DOI] [PubMed] [Google Scholar]
- 44.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nature Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 45.Monnier F, Taillefer M. Catalytic C-C, C-N, and C-O Ullmann-type coupling reactions. Angew. Chem. Int. Ed. 2009;48:6954–6971. doi: 10.1002/anie.200804497. [DOI] [PubMed] [Google Scholar]
- 46.Cai Q, Zou B, Ma D. Mild Ullmann-type biaryl ether formation reaction by combination of ortho-substituent and ligand effects. Angew. Chem. Int. Ed. 2006;45:1276–1279. doi: 10.1002/anie.200503538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.