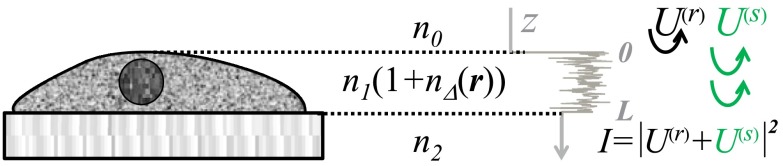Abstract.
The spectrum registered by a reflected-light bright-field spectroscopic microscope (SM) can quantify the microscopically indiscernible, deeply subdiffractional length scales within samples such as biological cells and tissues. Nevertheless, quantification of biological specimens via any optical measures most often reveals ambiguous information about the specific structural properties within the studied samples. Thus, optical quantification remains nonintuitive to users from the diverse fields of technique application. In this work, we demonstrate that the SM signal can be analyzed to reconstruct explicit physical measures of internal structure within label-free, weakly scattering samples: characteristic length scale and the amplitude of spatial refractive-index (RI) fluctuations. We present and validate the reconstruction algorithm via finite-difference time-domain solutions of Maxwell’s equations on an example of exponential spatial correlation of RI. We apply the validated algorithm to experimentally measure structural properties within isolated cells from two genetic variants of HT29 colon cancer cell line as well as within a prostate tissue biopsy section. The presented methodology can lead to the development of novel biophotonics techniques that create two-dimensional maps of explicit structural properties within biomaterials: the characteristic size of macromolecular complexes and the variance of local mass density.
Keywords: microscopy, biomedical optics, backscattering, spectroscopy, biophotonics, signal processing
1. Introduction
Visible-light microscopy and spectroscopy techniques remain the principal tools of biological cell and tissue examination in fields from basic science to medical diagnostics. Accordingly, the limitations of these techniques also remain an open problem. Specifically, due to their weakly scattering transparent nature, biomaterials are notoriously difficult to analyze without the use of exogenous labels. In addition, microscopy-based techniques cannot image structures smaller than the diffraction limit of light (, depending on optics setup) and spectroscopy-based techniques only quantify sample bulk properties, lacking the spatial resolution of obtained information.
It has been recently demonstrated that a tandem application of spectroscopy and microscopy enhances the advantages and mitigates the disadvantages of each technique, showing great promise in a variety of application fields.1 Thus, a spectroscopic microscope (SM), configured to detect interference spectra of backscattered light in the far zone, can quantify the statistics of nanometer scale refractive-index (RI) distribution via the spectral variance () of the acquired bright-field image. Further, it has been determined that can sense RI fluctuations at any spatial frequency whatsoever and its lengthscale sensitivity range is limited only by the signal-to-noise ratio (SNR) of the instrument.2
Despite the remarkable ability to sense subtle, microscopically indiscernibe structural alterations within weakly scattering label-free media, the quantification of sample’s internal structure via is also associated with a degree of ambiguity. As with most light-scattering markers of structure, it is not always clear which of the two structural properties, the characteristic lengthscale or the amplitude of RI fluctuations, cause a change in during any particular experiment. In addition, the value of is also affected by the sample thickness in a nonlinear manner.2 In this work, we establish that the spectrum registered by an epi-illumination bright-field wavelength-resolved microscope can be analyzed to accurately and explicitly measure sample’s internal structure in terms of physical rather than optical parameters: the standard deviation and characteristic lengthscale of the spatial RI distribution.
2. Theoretical Background
Consider a spatially varying RI object sandwiched between two semi-infinite homogeneous media (Fig. 1). The RIs of the three media are from top to bottom: , (as a function of location ), and . We assume to approximate the typical case of fixed biological media on a glass slide.3,4 The unit-amplitude plane wave incident normally onto the sample has two distinct sources of reflection: the first is caused by the RI mismatch on one side of the sample (top, air-sample interface in Fig. 1), which is further referred to as reference arm reflectance, and the second is composed of the light scattered from weak RI fluctuations within the sample of interest, which comprises the sample arm. The reference and sample arms are combined to form the wavelength-resolved far-field microscope image. The optical interference of these two components results in spectral fluctuations of registered intensity, and the variance of these fluctuations is the nanoscale-sensing marker .1
Fig. 1.
Sample geometry represented by three layers: RI of the middle layer is random (the sample of interest), RIs of the top and bottom layers are constant and spatially uniform; RI fluctuations as a function of depth are depicted in gray. Registered signal is a coherent sum of and .
We emphasize that no assumptions of one-dimensional light propagation are made in the underlying optics theory, and is derived from full three-dimensional (3-D) consideration of light scattering and propagation, as well as 3-D specification of RI distribution within the sample.1
Thus, the instrument specifics of the SM technique are summarized as white-light epi-illumination, bright-field microscope with spectrally resolved image acquisition, small numerical aperture (NA) of illumination (), moderate NA of collection [], and with a pixel size of microscope image corresponding to an area in sample space that is smaller than the diffraction limit of light. For simplicity and SNR enhancement, the collection NA used in this work is constant, . In turn, the sample geometry includes: (i) a weakly scattering sample of interest, (ii) sample thickness not greater than the microscope’s depth of focus (for most setups, 5 to ), (iii) in the axial dimension, the sample should be RI matched on one side (substrate in Fig. 1) and have a strong RI mismatch on the other (air in Fig. 1) to ensure reference and sample arm light reflections.
Since SM measures interference between a fixed reference-arm reflection and the waves scattered from within the sample, the variance of registered spectral oscillations can be decomposed into two components:
| (1) |
where is fully described by the RI contrast at the bottom surface, and is defined by the scattering events occurring within the sample.
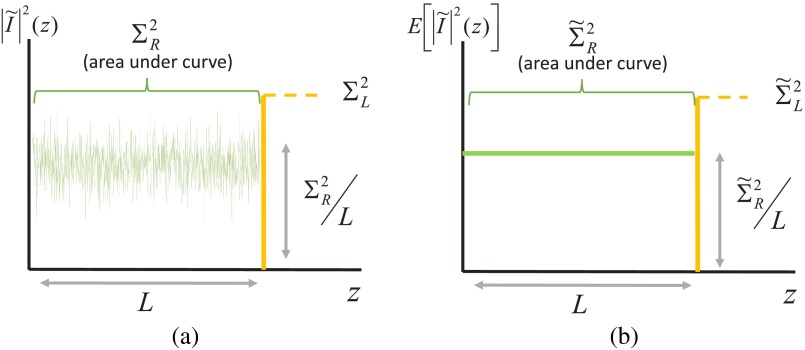
In Eq. (1), the optical path difference (OPD) between the interfering waves contributing to is within 0 and , and the OPD of interfering waves contributing to is always . It follows that it should be possible to independently measure the two components and from the spectral frequency composition of the SM spectrum, i.e., its Fourier transform . Essentially, the Fourier transform of an SM spectrum shows the amount of scattering that has occurred at depth inside the sample. For illustration purposes, frequency-space spectrum as a function of depth corresponding to an infinite spectral bandwidth is shown in Fig. 2.
Fig. 2.
of spectroscopic microscopy signal in an idealized case of an infinite bandwidth, pictured for (a) single realization of a random medium and (b) denoting the ensemble mean of . According to Wiener–Khinchine theorem, the area under the square of the frequency-space spectrum equals the spectral variance.
According to Parseval’s theorem, the spectral variance is related to the Fourier transform of the spectrum as . Therefore, and , where denotes the expected value of a random variable.
However, in practice, the spectral bandwidth is naturally limited to the visible-light wave number range , and the experimental is the infinite-bandwidth convolved with a sinc function . Hence, a closed-form analytical equation allowing to independently measure and from does not exist. Nevertheless, this relation can be obtained empirically.
In this work, we develop empirical signal processing algorithm for calculating spectral markers and independently. Further, we demonstrate that two optical measures and yield two physical measures of sample structure: the standard deviation and the characteristic lengthscale of RI distribution. Using SM data synthesized via finite-difference time-domain (FDTD) solutions of Maxwell’s equations, we validate the developed algorithm on samples with a wide range of structural properties within the example of exponential spatial correlation of RI. We then apply the validated algorithm to experimental data from biological cells and tissues, measuring the explicit physical characteristics of their internal organization.
3. Materials and Methods
3.1. Finite-Difference Time-Domain Simulations
In order to develop the inverse algorithm for and determination from the spectral-frequency composition of SM signal, we simulate a physical experiment using rigorous FDTD method, which calculates the light-scattering response of arbitrary inhomogeneous materials via numerical solution of Maxwell’s equations of electromagnetics.5–8 It is based on the discretization of a 3-D volume into a Cartesian grid consisting of small compared to the wavelength cubic voxels, and the solution of Maxwell’s equations for the evolution of the electric and magnetic field at discrete positions on this Cartesian grid. The core algorithm of the FDTD method was proposed by Yee9 in 1966, and popularized by Taflove in the ’80s and ’90s, who also coined the term FDTD. Compared to other electromagnetic approximation methods, such as the finite-element method or the method-of-moments, the FDTD method is more intuitive and simpler to implement. The ease with which inhomogeneous materials are handled in FDTD has made it very attractive for biological applications.10–12
We have an in-house software implementation of the FDTD method, called Angora.6,8 It can accurately calculate microscope images of arbitrary inhomogeneous samples under various imaging parameters, incorporating RI fluctuations as fine as 10 nm.7 We have used Angora to synthesize all reported herein bright-field plane-wave epi-illumination microscope images at 30 different wavelengths between 500 and 700 nm, equally spaced in wave number space.
Sample RI geometry was set to resemble that of fixed biomaterials on glass microscopy slides. RI of dehydrated cells and tissues is reported to be between 1.50 and 1.55,3,4 with the exact values being poorly investigated. In this study, we evaluated the average RI using Gladstone–Dale relation , where is the RI of water, is the specific refractive increment (), and is the cell dry density, which was approximated here as that of stratum mucosum ().13,14 Thus, we set 3,4 and .15 The spatial RI correlation was set to be exponential, and the RIs of the top and bottom media were and . To cover the biologically relevant range of structural properties, samples with 6 different thicknesses between 0.5 and , 4 RI standard deviation values between 0.02 and 0.05, and 20 RI correlation lengths between 20 and 250 nm were considered; spectrally resolved pixel microscope images of 20 different samples per statistical condition were synthesized (1 image pixel corresponded to area in sample plane, with the diffraction limit for the described setup being ). Following the conventional use of discrete Fourier transforms of limited-bandwidth signals, the synthesized reflectance spectra of every pixel were multiplied by a discrete Hann window (to minimize aliasing), zero-padded to total points (to increase frequency-space sampling frequency and thus reduce the minimal error in to 28 nm), after which fast Fourier transform was performed using built-in MATLAB function fft, yielding the spectral-frequency spectrum for each microscope image pixel. Then, squared absolute values of the frequency-space spectra are averaged across all pixels per statistical condition, and the ensemble average is obtained.
3.2. Prediction Rule Derivation
3.2.1. Calculation of and
In general, for a sample with RI fluctuation spatial correlation function and power spectral density of RI fluctuations (these two entities are related through the Wiener–Khinchine theorem), and are analytically expressed as1
| (2) |
| (3) |
where is a combination of Fresnel reflectance and transmission coefficients, , , . Here, both and are isotropic, as they are defined for an unbounded medium , with a physical size much larger than the characteristic lengthscale of RI fluctuations. The sample, in turn, is defined as a horizontal slice of with thickness , where can be comparable in magnitude to lengthscales of sample’s RI fluctuations. We also note that is a linear function of . Thus, in this work, we specifically aim to compute the internal-property marker , which is independent of sample thickness .
In an example of exponential functional form of with RI fluctuation variance , spatial correlation length , , and is1
| (4) |
| (5) |
where is the unitless parameter of size with respect to wavelength.
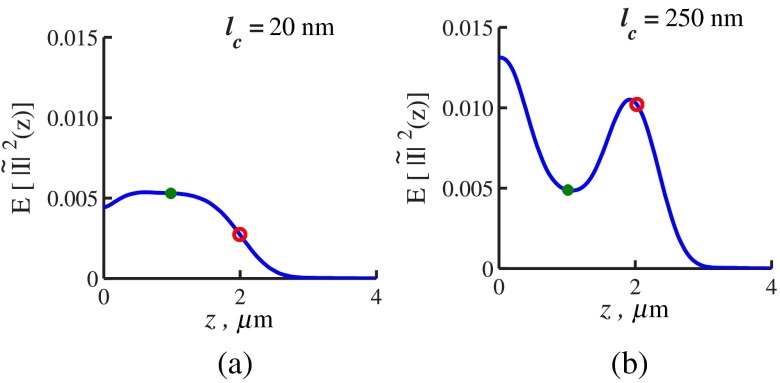
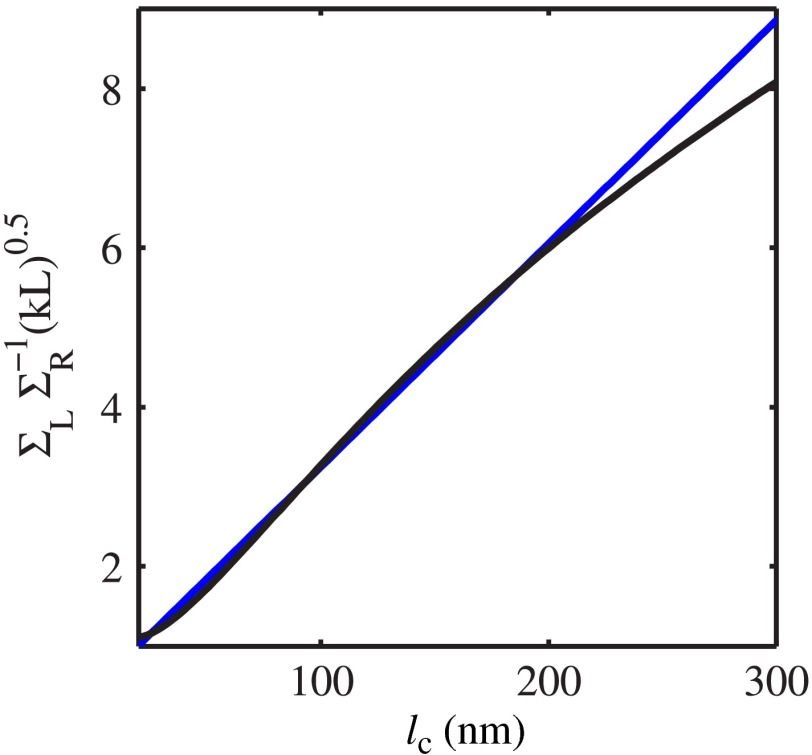
From the FDTD-generated library of SM images of samples with thickness and known internal properties ( standard deviation , and RI correlation lengths from 20 to 250 nm), we empirically obtained equations relating and to (examples of in Fig. 3):
| (6) |
| (7) |
where , , and .
Fig. 3.
is obtained by FDTD. Sample thickness , exponential functional form of RI spatial distribution with correlation length (a) (exemplifying a case when ) and (b) (exemplifying a case when ). On each curve, the depth at which is evaluated shown by red circles, and depth is used for evaluation by green dots. The shape of changes with due to the difference in the functional forms of and .
Since in this simulation, the sample thickness is known a priori, was readily evaluated at and , after which and were found according to Eqs. (6) and (7). Figure 5(a) illustrates the match between and obtained from the FDTD data according to the derived algorithm with those calculated by analytical Eqs. (4) and (5) for the known sample parameters.
Fig. 5.
Accuracy of sample parameters measured from the FDTD-generated SM signal from a sample with a known thickness: (a) and as a function of the correlation length obtained for a sample with thickness according to analytical equations (solid lines) and calculated from FDTD simulation (circles with error bars corresponding to the standard deviation between nine ensembles per statistical condition). (b) Corresponding true and (solid lines) and reconstructed from FDTD-synthesized SM signal (circles with error bars corresponding to the standard deviation between nine ensembles per statistical condition).
Most importantly, as seen from Eqs. (4) and (5), it is possible to reconstruct the statistics of the internal structure and once and are recovered. As both and are linear functions of , the relation between and is only dependent on system parameters controlled by the user ( and NA):
| (8) |
While a solution for can be found numerically, for simplicity and computational speed, we exploit the fact that is well approximated as a linear function for a wide range of correlation lengths above 15 nm and NA: for any NA within 0 to 0.6, the of linear regressions for for between 15 and 600 nm is above 0.98. In the case of considered throughout this work, can be found for any sample thickness simply as
| (9) |
with the fit shown in Fig. 4. The obtained value of can be substituted into Eq. (4) to find as
| (10) |
Fig. 4.
as a function of correlation length and the linear fit optimized for the subdiffractional range (, ).
Thus, our computations (results depicted in Fig. 5) postulate that and can be independently calculated from SM signal. The condition necessary for using the corresponding empirical Eqs. (6) and (7) followed by Eqs. (9) and (10) is the knowledge of sample’s thickness .
3.2.2. Reconstruction of sample thickness from spectroscopic microscope data
We next develop signal processing algorithm for accurate measurement of sample thickness from SM data. Finding sample thickness is complicated in part by the fact that the sample-substrate interface does not always reflect light due to the low RI contrast at the bottom interface typical to fixed biomaterials on glass. Specifically, this is the case when is smaller than the diffraction-limited spot [as in Fig. 3(a)] and the frequency-space spectrum does not necessarily contain an evident peak at .
Nevertheless, since no light scattering events occur at , always decays at . The shape of decay “tail” at has no closed-form analytical expression and depends on the sample’s internal structure as well as the spectral bandwidth of light.
Based on the FDTD-synthesized SM data used to develop Eqs. (6) and (7), a fourth order polynomial was fitted to all calculated frequency-space spectra at . Since the shape of in general, and its decay at in particular, depends on , this was done for SM images of samples with all 20 values of (between 20 and 250 nm). As a result, a matrix of corresponding polynomial coefficients was stored. Then, a MATLAB function was created to find a best fit between an arbitrary experimentally measured decay and the created library of decays for various values of . Finally, the sample thickness is determined from the location of the best fit of the experimental and FDTD-obtained decay tails.
To summarize, determining sample thickness from an arbitrary experimentally obtained spectrum allows the evaluation of at and , yielding and [Eqs. (6) and (7)], as well as and [Eqs. (8) and (10)], which completes the developed inverse algorithm here.
3.2.3. Considerations of sample roughness
It is important to consider that in true experimental conditions the surface roughness properties are most often unknown. Thus, an optimal spectral processing algorithm must universally apply to samples with rough as well as smooth surfaces. In cases when the sample top surface is rough, the registered spectroscopic microscopy signal has an additional, low spectral frequency component.16 Thus, to remove this spectral feature related to a property of sample surface, a second order polynomial is fitted to the registered spectrum and subtracted from it16 prior to converting the spectrum into spectral frequency space. We validate this step by applying our inverse algorithm to FDTD-synthesized SM images of weakly scattering samples with a rough surface profile that is characteristic of biological cells and tissues16 (average thickness of , standard deviation of nanoscale height variations within a diffraction-limited area 22 nm, correlation length of height variations 170 nm, internal RI , and ).
3.3. Spectroscopic Microscopy Instrument
For SM experimental measurements, we used our in-house instrument previously built for high-throughput partial wave spectroscopy measurements:17 epi-illumination bright-field microscope with small illumination , moderate collection , and magnification (objective lens from LUCPlanFL N, Olympus, Center Valley, Pennsylvania). Koehler illumination scheme was implemented for uniformity of incident light intensity throughout the image. Wavelength-resolved image acquisition was accomplished by using Xenon whitelight lamp illumination and spectral filtration of the light incident onto the sample via acousto-optical tunable filter (AOTF, HSI-300, Gooch & Housego, Orlando, Florida; filter bandwidth of 3 nm). As a result, each measurement recorded a 3-D data cube consisting of sample bright-field microscope images obtained at 200 1 nm-spaced wavelengths within the spectral range of 500 to 700 nm.
After the data were collected, spectral noise was removed from spectra corresponding to each pixel of the acquired wavelength-resolved microscope image by a low-pass spectral filter using sixth order butterworth filter with a 0.2 frequency cutoff. Then, data postprocessing was performed in exact accordance with algorithm developed and applied to FDTD-synthesized spectra, which included second-order polynomial fitting and subtraction, Fourier transformation of spectra with the use of Hann window and zero-padding, followed by the analysis of the resultant spectrum.
3.4. Colon Cancer Cell Lines
The performance of the proposed analysis was first tested on HT29 human colonic adenocarcinoma cell line models. The experiment included two groups, control vector HT29 (CV) cells and epidermal growth factor receptor (EGFR) knockdown HT29 cells, a less aggressive genetic variant that is histologically indistinguishable from the CV.18–20
HT29 CV and EGFR knockdown cells were collected in centrifuge tubes and centrifuged for 5 min at 1000 rpm. The supernatant was removed, after which the cells were plated on a glass chamber slide: 2 mL of fresh cell culture medium was added to each chamber slide, which was then incubated at 37°C for 6 h. After incubation, the medium was completely removed, the slides were washed and then immediately fixed using 70% ethanol, which completed sample preparation. Slides were stored at 4°C until the spectroscopic microscopy data from 18 CV cells and 17 EGFR knockdown variant cells were acquired. Topography of the same cells was later obtained via an atomic force microscope (AFM) to validate the cell thickness predictions obtained from SM data.
3.5. Tissue Section
The second biological model for analysis algorithm application and testing included human prostate tissue biopsy section. Collection of the human sample was approved by the Institutional Review Board at NorthShore University HealthSystem. Sample was obtained from the NorthShore University active surveillance trial initiated in November 2008. Informed written consent was obtained from the participant.
Transrectal biopsy was obtained with 3-D ultrasound guidance, fixed in ethanol and embedded in paraffin. Then, the sample was sectioned, and two sections were applied to a glass slide, after which they were deparaffinized following standard histological procedures. Using one section, both SM and AFM data were collected from the same region of the sample for structural property determination. The other section was stained with hematoxylin and eosin (H&E) to aid in organelle visualization, which was used only for illustration purposes.
3.6. Atomic Force Microscope
Height map of the biological samples was determined at room temperature by peak force tapping mode using a Bruker Dimension Icon AFM system with silicon OTESPA-R AFM probes (Bruker AXS).
For cell lines, image was obtained with pixel size of 46.9 nm and for tissue section, image was obtained with pixel size of 176 nm. Image magnification (and, therefore, pixel resolution) for tissue section image was chosen so that the AFM-imaged area of sample surface captures the same area as within the SM field of view. At the same time, larger magnification of cell line images was chosen due to their surface area being much smaller than the microscope’s field of view.
4. Results
4.1. Algorithm Validation on Finite-Difference Time-Domain Generated Data
We validate the proposed inverse algorithm by applying it to a validation set of Angora-synthesized SM data for samples with a wide range of internal and surface properties.
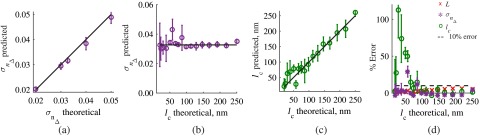
First, we confirm that the developed algorithm is accurate for experimentally realistic samples with uneven surface. To accomplish this, we apply the analysis algorithm to FDTD-synthesized SM images of 20 rough samples with RI distribution , , and average . The roughness of these samples was set to resemble that of biological cells and tissues.16 Our analysis results reconstructed the internal structure characteristics of the imaged samples with excellent accuracy: predicted (6.9% error from true value), (3.7% error) and predicted (2.3% error).
This validates that subtraction of low-frequency components from the registered spectra via a second-order polynomial efficiently removes surface roughness contributions to the SM signal and allows accurate calculation of the internal properties. Note that the lowest frequencies removed by the polynomial correspond to small values of in profile, which are never sampled when and are evaluated. As a result, our reconstruction algorithm remains accurate despite this additional spectral processing step.
In order to keep the spectral processing algorithm independent of the sample surface features, we subtract the low-frequency components from spectra obtained from all samples, including the validation set below as well as the biological cells and tissue.
Next, as a validation set for the inverse algorithm, we analyzed SM images of samples with various combinations of all three measured parameters—, , and —covering the biologically relevant range of structural properties. Figure 6(a) illustrates the excellent accuracy in the prediction of varying obtained by the inverse algorithm from the FDTD-generated spectrally resolved microscope images. In this set, and were fixed and were predicted within 15-nm accuracy for and 6% accuracy for . Figures 6(b)–6(d) present prediction accuracy in a more complex case where RI correlation length was varied within the subdiffractional range, and a new thickness value was tested. Note that at where the percentage of error in prediction is relatively high, the predicted values of are still within tens of nanometers from the corresponding true value [Fig. 6(b)].
Fig. 6.
Accuracy of structural properties predicted from the FDTD-generated SM signal (a) predictions from wavelength-resolved reflectance measurements simulated by FDTD for samples with varying , , and . (b) and (c) predictions from samples with varying RI correlation lengths , , and , and (d) the corresponding percentage of error in calculation of , , and from the generated reflectance spectra.
4.1.1. Minimum thicknesses for analysis validity
Importantly, the finite spectral bandwidth of light naturally imposes limitations to the closest spectral frequencies of the SM data that can be resolved. In turn, since OPDs of all interfering waves are within the range of 0 to , the sample thickness confines the range of spectral frequencies present in the SM signal. As a consequence, in the limit of very small , values of at cannot be resolved from those at , and the key Eqs. (6) and (7) cannot be used. Thus, there must be a lower limit to the sample thickness in order for the developed herein analysis of spectral-frequency profile to apply.
We determine the lower limit of for our algorithm accuracy using sets of SM images generated for samples with various thicknesses ( as low as 500 nm was tested). For every thickness, SM images of samples with 20 subdiffractional within 20 to 250 nm were obtained in order to ensure that the thickness limitations will be determined for a general case, independent of the internal RI distribution.
We applied the inverse algorithm to calculate all three parameters: , , and and subsequently evaluated the error between true and predicted parameters (as in Fig. 6). Results of the error evaluation are summarized in Table 1.
Table 1.
Range of sample thicknesses for which quantification of the internal ( and ) as well as external () properties of the sample is accurate.
| Property | Applicable samples | Error |
|---|---|---|
| , | ||
| , |
Our analysis shows that accuracy in measuring is a greater challenge than that in for the inverse algorithm. Moreover, predictions remain extremely robust even in cases when the error in the predicted is large [see Figs. 5(b), 6(a), and 6(c)]. We believe this to be explained by the fact that while the shape of SM spectral-frequency profile depends strongly on [due to the difference in and ], both and scale linearly with . Thus, subtle errors in the quantification of the shape of have a stronger effect on the accuracy of rather than on that of .
To summarize, the inverse algorithm was developed on the testing set of samples with and a subdiffractional range of RI correlation lengths 20 to 250 nm. Then, we confirmed that the algorithm is accurate even in the cases when samples have a rough surface. Finally, we applied the algorithm to accurately reconstruct the internal structure from SM data generated for a large set of inhomogeneous weakly-scattering samples, where, in addition to different values, we varied thickness from 0.5 to Fig. 7, and from 0.02 to 0.05. Results of the above testing and validation procedures specify the accuracy as well as the applicability range of the proposed methodology (Table 1), positioning us for data processing from complex experimental samples such as biological cells and tissues.
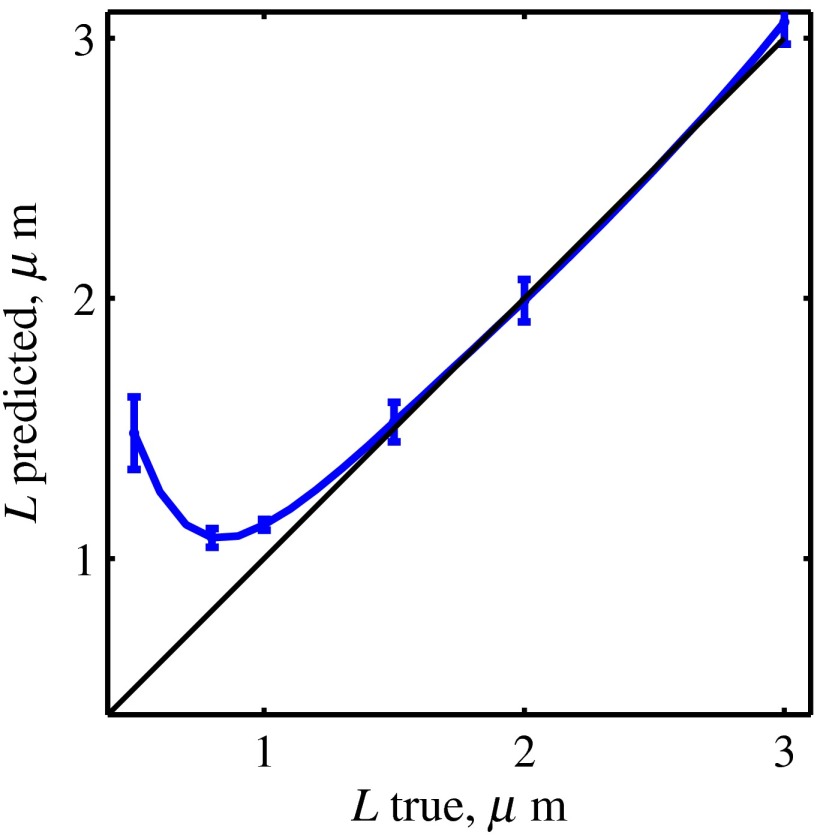
Fig. 7.
Thickness prediction from SM frequency spectra, errors are standard deviations across 20 samples with different RI correlations lengths per thickness.
4.2. Cell Lines
After validating our methodology on FDTD solutions of Maxwell’s equations, we proceeded with experiments on isolated biological cells. As a model, we chose HT29 human colonic adenocarcinoma cell line. We have also used a genetic variant of HT29 cells with EGFR knockdown, which partially suppresses the proliferation aggressiveness of the cell line without changing its microscopically visible morphological qualities.18–20
Our theoretical derivations as well as FDTD simulation have shown that the single necessary condition for obtaining the and values from SM data is accurate determination of sample thickness. Thus, we specifically ensure that the predictions of obtained from experimental SM data are accurate by comparing them to the values measured by AFM from the same cells.
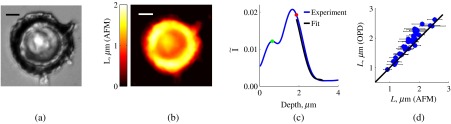
Results of this comparison are shown in Fig. 8. White-light epi-illumination microscope image of an isolated HT29 cell [Fig. 8(a)] and the corresponding AFM image of the same cell [Fig. 8(b)] illustrate the spatial height variations and roughness of cell surface that emphasize the complexity of biological cell measurements. Note that to obtain the ensemble average , Fourier transforms of independent spectra are averaged. In the case of biological cells, this entails averaging data from areas with different thicknesses. Despite this, our predictions of 35 cell-averaged thickness values with the assumption of average RI to be 1.53 (RI of fixed biological cells and tissues)3,4 show an excellent match with the physical thickness of cells measured with AFM [Fig. 8(d)].
Fig. 8.
Experimental results from biological cells: (a) representative bright-field reflectance white-light image of a cell; (b) representative height map obtained by AFM from the same cell (EGFR-knockdown variant shown here), scale bar ; (c) tail-fitting of experimentally obtained Fourier transform of reflectance intensity averaged throughout the cell area of interest; red dot corresponds to the predicted thickness, where is estimated and the green dot corresponds to the midsample depth, where is estimated for this particular spectrum. (d) Match between the cell thickness evaluated from the frequency content of its reflectance intensity compared to that obtained with AFM; horizontal error bars indicate the range (standard deviation) of thicknesses within the sample as measured by AFM.
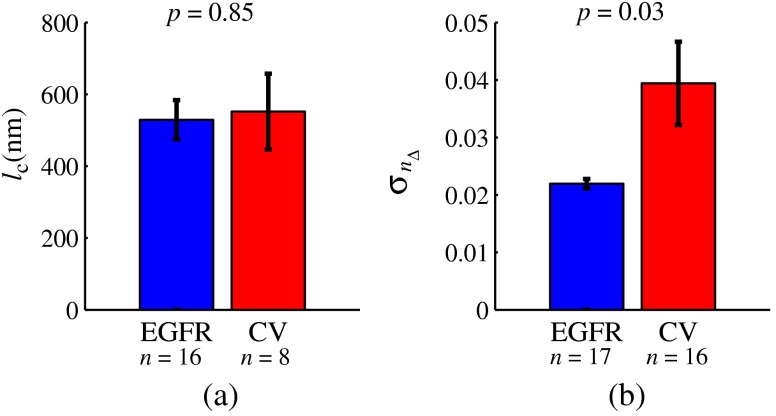
Next, we obtained and predictions for the same biological cells. Our results showed that the characteristic length scale of RI distribution within the two genetic variants is approximately the same (552 nm inside the CV and 529 nm in EGFR-knockdown variant). However, the standard deviation of RI within those two genetic variants was drastically different: 0.04 inside the CV and 0.02 inside the less aggressive EGFR-knockdown variant (Fig. 9). This difference was statistically significant with a -value of 0.03.
Fig. 9.
Length scale and standard deviation of RI distribution: (a) RI correlation lengths within CV and EGFR cells as measured from SM data; error bars are standard errors; (b) a drastic difference between the RI standard deviation within CV and EGFR cells measured from SM data following the reconstruction algorithm; error bars are standard errors.
4.3. Tissue Section
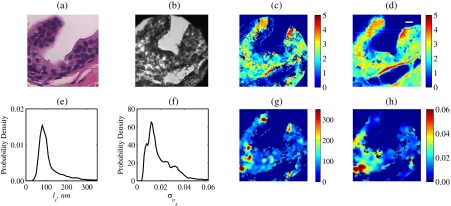
Last, we demonstrate that the developed methodology can be used for spatially resolved quantification of the internal structure of biological tissue specimens on an example of a sectioned human prostate tissue biopsy. Transmission bright-field microscopy image of the neighboring hematoxylin- and eosin-stained section is shown in Fig. 10(a) and bright-field reflectance microscope image of the unstained sample of interest is shown in Fig. 10(b). Following the above-developed procedure, we first confirm the accuracy of the reconstruction algorithm by comparing AFM-measured sample thickness with that predicted using SM data [Figs. 10(c) and 10(d)]. Here, the large (compared to single isolated biological cells) sample area allowed data analysis in a spatially resolved manner, and the obtained thickness map of the sample within microscope’s field of view showed an excellent match with that measured with AFM (pixels with were excluded from the SM data analysis). Accurate evaluation of the observed match between the AFM- and the SM-calculated sample topology is complicated by the differences in pixel sizes, spatial resolutions, as well as sample orientation in the respective images acquired by the two techniques. Thus, after applying a Gaussian blur to the AFM image to approximate a microscope’s diffraction-limited resolution, rotating and extrapolating images (to match the pixel size), we have estimated that 74% of pixels used for internal property reconstruction had an SM-measured thickness within 20% from that measured by AFM.
Fig. 10.
Experimental results from a tissue section: (a) Transmission microscopy image of a neighboring H&E stained section. (b) Bright-field white-light reflectance microscope image of the analyzed (unstained) tissue section, with its topography. (c) Reconstructed from SM data analysis and (d) directly measured by AFM. Probability density functions of the reconstructed spatial distributions of (e) RI correlation lengths and (f) RI standard deviations . (g) Spatially resolved map of and (h) spatially resolved map of .
Spatially resolved values of and were then obtained, as depicted in Figs. 10(g) and 10(h). As per algorithm limitations determined and reported above based on FDTD data analysis (Table 1), we have calculated only for areas with thickness (which comprised 48% of sample area) and only for areas with thickness (70% of sample area).
We also note that the relative blurriness of and distributions is due to the fact that each value is a cumulative statistical characteristic of sample structure within a moving window of , which corresponds to . For the studied tissue sample, we found the mean and most common values to be 118 and 80 nm correspondingly. In turn, the mean and most common values were found to be 0.020 and 0.012. Based on this very limited dataset, the overall shape of both and value distributions appeared to follow a lognormal functional form.
5. Discussion and Conclusions
In this work, we demonstrate that the spectral-frequency composition of a wavelength-resolved image registered by a reflected-light, bright-field microscope can be analyzed to independently obtain two explicit physical measures of the RI distribution within weakly scattering samples such as biological cells and tissues: the standard deviation and the spatial correlation length. Since the local mass density is a linear function of RI within biomaterials (Gladstone–Dale relation),13 these measures of RI distribution directly translate into statistics of mass density distribution inside biological cells and tissues: the correlation length of mass density is exactly the same as that of RI, and the standard deviation of mass is that of RI divided by the RI-mass proportionality coefficient . In biological terms, variance of local mass density quantifies the compaction degree of macromolecular complexes (folded proteins, chromatin aggregates, etc.) contained within the volume underneath a diffraction-limited area surrounding each pixel .21 In turn, is the characteristic size of macromolecular complexes within that same volume. Hence, measurement of and is an important tool in studies of structure–functional relationship in crucial biological processes including cancer initiation and progression (epigenetic changes observed in fixed-cell nucleus,22,23 cytoplasm,24 extracellular matrix,25,26 etc.), cell proliferation,20,27 as well as genome dysregulation and potential therapy.28,29
Conceptually, the developed algorithm utilizes the physical meaning behind the spectral-frequency profile , which allows to obtain several independent parameters of sample’s organization by evaluating at different . First, no scattering events occur at , which we use to measure . Second, at , is predominantly defined by the amount of light reflected at the sample–substrate interface, which we use to measure . Third, at , represents the amount of scattering from within the sample, which determines . Essentially, the reflection at is defined by the two-dimensional (2-D) statistics of RI distribution, , and the scattering at is defined by the 3-D statistics of RI, which is why and probe the sample structure in a truly independent manner. We also note that due to the statistical homogeneity considered here, could be computed from the value of for virtually any between 0 and . However, we have chosen to calculate at the midpoint of in order to minimize the inevitable (due the finite spectral bandwidth) contributions from surface roughness at low and at . Finally, at low , also contains information about the sample surface roughness profile in addition to its internal inhomogeneity.16 Since the present work does not aim to measure surface statistics, the surface-related contributions are simply removed from the signal.
Technically, the underlying algorithm is composed of (i) numerical curve fitting to measure , (ii) an empirical step to obtain the exact values of and from , and (iii) reconstruction of structural properties and based on analytical closed-form equations.
We have tested and validated the inverse algorithm using FDTD solutions of Maxwell’s equations. The two important advantages of FDTD for algorithm validation are (1) sample exact structure known a priori and thus the technique precision can be readily evaluated and (2) experimental noise and other sources of error are absent. The testing set included SM images synthesized for samples with thickness of , and RI correlation lengths of 20 to 250 nm. Then, the algorithm accuracy has been extensively studied and validated on a larger set of samples within biologically relevant structural properties, including 6 different thicknesses, 5 different RI standard deviation values, and 20 correlation lengths for each value of , as well as samples with surface roughness. Our results demonstrated an excellent accuracy in measuring the correlation length within samples with , standard deviation of RI for those with , and thickness for , indicating the applicability of the proposed technique to dehydrated squamous epithelial cell nuclei (), columnar epithelial cells (), and tissue sections (thickness chosen by the user). These results also show that the accuracy in measuring is a greater challenge for the inverse algorithm than that in measuring (Table 1). In fact, predictions remain extremely robust even when the error in the predicted is relatively large [see Figs. 5(b), 6(a), and 6(c)]. Accordingly, our validation studies were specifically focused on testing a wide range of values along with a smaller set of values.
Next, the validated algorithm was applied to quantify the structure within fixed, label-free biological cells. After confirming with AFM that the necessary condition for our algorithm accuracy—precise knowledge of cell thickness—has been satisfied, we measured the intracellular and of two genetic variants of human adenocarcinoma HT29 cell lines. Due to their columnar epithelial cell type, 85% to 90% of the measured cell volume30 was occupied by the cell nucleus, and thus the measured structure was predominantly determined by the nuclear organization of these cells. Results of our analysis showed that while both variants of HT29 cells have a similar RI spatial correlation length , the standard deviation of RI within the CV exceeded that in the EGFR-knockdown variant by a factor of 2. Thus, the cancer cell line with a more aggressive proliferating behavior was found to have a similar characteristic lengthscale but a much higher amplitude of the intracellular macromolecular mass density variations. This observation is in agreement with the previously published reports on an increased “degree of inhomogeneity” within the CV compared to its EGFR-knockdown variant.20 Importantly, these two genetic variants are microscopically indistinguishable,18,19 which may be in part reflected in the similarity between the measured microscale RI correlation lengths.
Finally, on an example of experimental data from a biological tissue section, we show that our algorithm can reconstruct the internal structure of weakly scattering biomaterials in a spatially resolved manner. This spatially resolved quantification of structure is possible when the lateral size of the sample (here ) is much larger than the size of a diffraction-limited spot and therefore, superpixel averaging of the frequency-space SM signal yields enough statistics to measure the local and , corresponding to a neighboring area. The measured values of RI standard deviation within the isolated cells were only slightly higher than those in tissue (0.02 to 0.04 in cancer cells and 0.012 to 0.02 in tissue). At the same time, the RI correlation lengths measured in the two experiments were very different (120 nm in tissue and 500 nm in cells). Importantly, the two experiments were performed on entirely different models from the biological perspective: isolated cells of a colon cancer cell line and a continuous section of microscopically normal tissue from patient prostate biopsy and hence, their structural properties are also expected to differ. In addition, there are slight sample-geometry differences in the two sample types, which may have contributed to the difference in . First, intact isolated cells always have cytoplasm at the sample–substrate interface and hence, is mostly affected by the cytoplasmic structure. In turn, the sectioned tissue can have an arbitrary organelle touching the substrate and thus, measured the sample structure in a more statistically accurate manner. Second, only the nuclear area with characteristically large, microscale chromatin aggregates was analyzed in cancer cell lines, while whole cells with no microscopically discernible marcomolecular aggregates were included in the tissue section analysis. We believe that all of the above factors must have contributed to the observed fourfold difference in the RI correlation lengths within cancer cell lines and histologically normal tissue. Lastly, based on previous studies focused on the quantification of the internal organization of biomaterials via light or electron microscopy,20,23 we believe that data acquired from 10 to 30 fields of view (30 to 150 biological cells depending on cell type) will be sufficient to account for biological variability and determine and values typical for a given biological sample. In the future, automated image acquisition can be implemented to acquire and analyze whole-slide images of biological samples (up to 1500 images per slide as has already been implemented elsewhere).31
The presented algorithm has been developed with the specific application focus of measuring internal structure of fixed biomaterials. As a result, the sample mean RI was accordingly set to 1.53 throughout the algorithm testing and validation procedures. The observed match between sample thickness determined by AFM (which measures physical thickness) and SM (which measures optical thickness and recovers using the assumption of mean RI) confirms the accuracy of assumed average RI value for fixed cells and tissue. In addition, the choice of glass slides as sample substrates has also eliminated deterministic light reflection at the bottom interface due to the match between glass and sample average RIs. While these average RI choices do not change the nanoscale structure sensitivities of SM1 and do not affect our algorithm derivations from the physics perspective, they define the scope of the sample/substrate microscale properties tested in the presented work. Following the outlined framework, the algorithms presented herein can be easily extended to other sample/substrate properties as well as other applications of biophotonics. For example, one could introduce an RI mismatch at the sample–substrate interface to accentuate the peak at and subsequently remove this deterministic contribution to when is calculated.1
From the perspective of structure parametrization, the internal properties and measured by the inverse algorithm correspond to the height and width of the spatial RI correlation function. Thus, the value of is independent of the sample’s lengthscale composition and, therefore, of the shape of spatial correlation function. At the same time, our definition of as the correlation length of RI presented here involves an assumption of exponential RI correlation. We note that previous calculations based on electron microscopy images of biological cell nuclei have shown that the SM signal predicted based on their experimentally measured RI distribution matches that predicted based on an value that assumes an exponential RI correlation.2 Thus, even if under certain experimental conditions, the exact value of can be prone to error, we still firmly believe that it provides a valuable measure of sample’s lengthscale composition regardless of the functional form of its RI correlation function.
The technique presented herein is unique in its nanoscale sensitivity, versatility, and ease of application: by an automated analysis of a wavelength-resolved far-field microscope image, it can explicitly measure physical properties native to weakly scattering samples. Moreover, it requires no external labels or labor-intensive sample fixation/processing procedures. However, this great advantage also defines the main limitation of the present work, as the exact values of and measured from biological samples cannot be corroborated by another independent technique. Thus, corroboration of the and values predicted by SM would require imaging of the 3-D native mass distribution within the same biological cells with nanometer resolution, which is prohibitive with current state-of-the-art technology. Nevertheless, the measured spatial standard deviation of RI from both sets of experimental data is within the interval of (0.02, 0.06), which agrees with the estimates based on a discrete-particle model of soft tissue.15 Analogous estimates for correlation length of RI fluctuations inside label-free ethanol-fixed biological cells and tissues are not reported in previous literature, owing by large to the abovementioned technical limitations. Future work will focus on validation of the presented algorithm on experimentally measured data from samples of known internal structure (controlled phantoms or biological samples quantified via emerging nanoscale-imaging methodologies such as correlative light-electron microscopy).32
In summary, we establish that the spectrum registered by a reflected-light microscope can be analyzed to independently reconstruct two physical measures of internal structure within samples such as biological cells and tissues. Applying this approach can lead to the development of novel biophotonics techniques capable of creating 2-D images of intracellular mass-distribution properties such as characteristic size of macromolecular complexes and variance of local mass-density. The ease of utilization as well as the most intuitive physical meaning of measured parameters (as opposed to optical markers of structure) will make this approach widely applicable for users in fields from basic biology, material science to medical diagnostics.
Acknowledgments
This study was supported by the National Institutes of Health under Grant Nos. R01CA200064, R01CA155284, R01CA165309, and R01EB016983, and by the Lungevity Foundation. The FDTD simulations in this paper were made possible by a computational allocation from the Quest high-performance computing facility at Northwestern University. H.S. and V.B. are cofounders and/or shareholders in Nanocytomics LLC.
Biographies
Lusik Cherkezyan is a postdoctoral fellow in the Department of Biomedical Engineering at Northwestern University, Illinois, where she has also received her PhD. Her work focuses on the use of physics and engineering principles to the study of biological systems. In particular, she is interested in the technology development for the spectroscopic quantification of biomaterials at nanometer scales. Her work has been published in multiple high-profile journals, including Physical Review Letters, BMC Cancer, Optics Letters, Endoscopy, etc.
Di Zhang received his BS degree in electrical engineering from Beijing Jiaotong University, China, in 2009. He is currently working toward his PhD degree in biomedical engineering at Northwestern University, Evanston, Illinois, United States. His research interests include computational electromagnetics for modeling light interaction with biological tissue and optical imaging techniques for early stage cancer detection.
Hariharan Subramanian received his PhD in biomedical engineering from Northwestern University and is currently a research professor of biomedical engineering at Northwestern University. He is the cofounder and chief technology officer of NanoCytomics (www.nano-cytomics.com), an in vitro medical diagnostic company developing screening strategies for different types of cancers (e.g., lung, colon, prostate, etc.). He has considerable experience in biomedical optics, cancer biology, and clinical research with numerous publications appearing in leading peer-reviewed journals.
Allen Taflove is a professor in the Department of Electrical Engineering and Computer Science of Northwestern University, Evanston, Illinois. Since 1972, he has pioneered finite-difference time-domain (FDTD) computational solutions of Maxwell’s equations, for which he received the 2014 IEEE Electromagnetics Award. His major publication—Computational Electrodynamics: The Finite-Difference Time-Domain Method—is the seventh most-cited book in physics.
Vadim Backman is the Walter Dill Scott professor of biomedical engineering at Northwestern University and a program leader at the Robert H. Lurie Comprehensive Cancer Center in Chicago, Illinois. An internationally renowned expert in biomedical optics, he develops revolutionary nanoscale imaging technologies that allow researchers to explore previously intractable questions in biology, disease diagnosis and progression, with a focus on detecting cancer at its earliest and most treatable stages.
References
- 1.Cherkezyan L., et al. , “Interferometric spectroscopy of scattered light can quantify the statistics of subdiffractional refractive-index fluctuations,” Phys. Rev. Lett. 111, 033903 (2013). 10.1103/PhysRevLett.111.033903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherkezyan L., Subramanian H., Backman V., “What structural length scales can be detected by the spectral variance of a microscope image?,” Opt. Lett. 39(15), 4290–4293 (2014). 10.1364/OL.39.004290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook D., Cellular Pathology: An Introduction to Techniques and Applications, pp. 67–104, Scion, Bloxham: (2006). [Google Scholar]
- 4.Crossmon G. C., “Mounting media for phase microscope specimens,” Stain Technol. 24, 241–247 (1949). 10.3109/10520294909139615 [DOI] [PubMed] [Google Scholar]
- 5.Taflove A., Hagness S. C., Computational Electrodynamics: The Finite-Difference Time-Domain Method, 3rd ed., Artech House, Norwood, Massachusetts: (2005). [Google Scholar]
- 6.Capoglu I. R., “Angora: a free software package for finite-difference time-domain (FDTD) electromagnetic simulation,” 2012, http://www.angorafdtd.org/ (accessed April 2012).
- 7.Capoglu I. R., et al. , “Chapter 1 - the microscope in a computer: Image synthesis from three-dimensional full-vector solutions of Maxwell’s equations at the nanometer scale,” in Progress in Optics, Wolf E., Ed., Vol. 57, pp. 1–91, Elsevier, Philadelphia, Pennsylvania: (2012). [Google Scholar]
- 8.Capoglu I., Taflove A., Backman V., “Angora: a free software package for finite-difference time-domain electromagnetic simulation,” IEEE Antennas Propag. Mag. 55(4), 80–93 (2013). 10.1109/MAP.2013.6645144 [DOI] [Google Scholar]
- 9.Yee K., “Numerical solution of initial boundary value problems involving Maxwell’s equations in isotropic media,” IEEE Trans. Antennas Propag. 14, 302–307 (1966). 10.1109/TAP.1966.1138693 [DOI] [Google Scholar]
- 10.Hollmann J. L., Dunn A. K., DiMarzio C. A., “Computational microscopy in embryo imaging,” Opt. Lett. 29, 2267–2269 (2004). 10.1364/OL.29.002267 [DOI] [PubMed] [Google Scholar]
- 11.Winters D., et al. , “Three-dimensional microwave breast imaging: dispersive dielectric properties estimation using patient-specific basis functions,” IEEE Trans. Med. Imaging 28, 969–981 (2009). 10.1109/TMI.2008.2008959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starosta M. S., Dunn A. K., “Three-dimensional computation of focused beam propagation through multiple biological cells,” Opt. Express 17, 12455–12469 (2009). 10.1364/OE.17.012455 [DOI] [PubMed] [Google Scholar]
- 13.Davies H. G., et al. , “The use of the interference microscope to determine dry mass in living cells and as a quantitative cytochemical method,” Q. J. Microsc. Sci. s3–95(31), 271–304 (1954). [Google Scholar]
- 14.Barer R., Joseph S., “Refractometry of living cells part i. Basic principles,” Q. J. Microsc. Sci. 3(32), 399–423 (1954). [Google Scholar]
- 15.Schmitt J. M., Kumar G., “Optical scattering properties of soft tissue: a discrete particle model,” Appl. Opt. 37, 2788–2797 (1998). 10.1364/AO.37.002788 [DOI] [PubMed] [Google Scholar]
- 16.Zhang D., et al. , “Spectroscopic microscopy can quantify the statistics of subdiffractional refractive-index fluctuations in media with random rough surfaces,” Opt. Lett. 40, 4931–4934 (2015). 10.1364/OL.40.004931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandler J. E., et al. , “High-speed spectral nanocytology for early cancer screening,” J. Biomed. Opt. 18(11), 117002 (2013). 10.1117/1.JBO.18.11.117002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts R. B., et al. , “Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis,” Proc. Natl. Acad. Sci. 99(3), 1521–1526 (2002). 10.1073/pnas.032678499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Slyke P., et al. , “Dok-R mediates attenuation of epidermal growth factor-dependent mitogen-activated protein kinase and Akt activation through processive recruitment of c-Src and Csk,” Mol. Cell. Biol. 25(9), 3831–3841 (2005). 10.1128/MCB.25.9.3831-3841.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian H., et al. , “Optical methodology for detecting histologically unapparent nanoscale consequences of genetic alterations in biological cells,” Proc. Natl. Acad. Sci. 105(51), 20118–20123 (2008). 10.1073/pnas.0804723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J. S., et al. , “The influence of chromosome density variations on the increase in nuclear disorder strength in carcinogenesis,” Phys. Biol. 8(1), 015004 (2011). 10.1088/1478-3975/8/1/015004 [DOI] [PubMed] [Google Scholar]
- 22.Stypula-Cyrus Y., et al. , “HDAC up-regulation in early colon field carcinogenesis is involved in cell tumorigenicity through regulation of chromatin structure,” PLoS One 8(5), e64600 (2013). 10.1371/journal.pone.0064600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherkezyan L., et al. , “Nanoscale changes in chromatin organization represent the initial steps of tumorigenesis: a transmission electron microscopy study,” BMC Cancer 14(1), 189 (2014). 10.1186/1471-2407-14-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stypula-Cyrus Y., et al. , “End-binding protein 1 (EB1) up-regulation is an early event in colorectal carcinogenesis,” FEBS Lett. 588(5), 829–835 (2014). 10.1016/j.febslet.2014.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadiarnykh O., et al. , “Alterations of the extracellular matrix in ovarian cancer studied by second harmonic generation imaging microscopy,” BMC Cancer 10, 94 (2010). 10.1186/1471-2407-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backman V., Roy H. K., “Advances in biophotonics detection of field carcinogenesis for colon cancer risk stratification,” J. Cancer 4(3), 251 (2013). 10.7150/jca.5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papantonis A., Cook P. R., “Genome architecture and the role of transcription,” Curr. Opin. Cell Biol. 22(3), 271–276 (2010). 10.1016/j.ceb.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misteli T., “Higher-order genome organization in human disease,” Cold Spring Harbor Perspect. Biol. 2(8), a000794 (2010). 10.1101/cshperspect.a000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egger G., et al. , “Epigenetics in human disease and prospects for epigenetic therapy,” Nature 429(6990), 457–463 (2004). 10.1038/nature02625 [DOI] [PubMed] [Google Scholar]
- 30.Damania D., et al. , “Role of cytoskeleton in controlling the disorder strength of cellular nanoscale architecture,” Biophys. J. 99, 989–996 (2010). 10.1016/j.bpj.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palcic B., “Malignancy associated changes in bronchial epithelial cells and clinical application as a biomarker,” Lung Cancer 3(19), 161–166 (1998). [DOI] [PubMed] [Google Scholar]
- 32.van Rijnsoever C., Oorschot V., Klumperman J., “Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections,” Nat. Methods 5(11), 973–980 (2008). 10.1038/nmeth.1263 [DOI] [PubMed] [Google Scholar]