Abstract
The P1 adhesin (aka Antigen I/II or PAc) of the cariogenic bacterium Streptococcus mutans is a cell surface-localized protein involved in sucrose-independent adhesion and colonization of the tooth surface. The immunoreactive and adhesive properties of S. mutans suggest an unusual functional quaternary ultrastructure comprised of intact P1 covalently attached to the cell wall and interacting with non-covalently associated proteolytic fragments thereof, particularly the ~57-kDa C-terminal fragment C123 previously identified as Antigen II. S. mutans is capable of amyloid formation when grown in a biofilm and P1 is among its amyloidogenic proteins. The C123 fragment of P1 readily forms amyloid fibers in vitro suggesting it may play a role in the formation of functional amyloid during biofilm development. Using wild-type and P1-deficient strains of S. mutans, we demonstrate that solid state NMR (ssNMR) spectroscopy can be used to 1) globally characterize cell walls isolated from a Gram-positive bacterium and 2) characterize the specific binding of heterologously expressed, isotopically-enriched C123 to cell wall-anchored P1. Our results lay the groundwork for future high-resolution characterization of the C123/P1 ultrastructure and subsequent steps in biofilm formation via ssNMR spectroscopy, and they support an emerging model of S. mutans colonization whereby quaternary P1-C123 interactions confer adhesive properties important to binding to immobilized human salivary agglutinin.
Keywords: Gram-positive bacteria, Streptococcus mutans, dental caries, adhesin, amyloid, cell surface, solid-state NMR
Introduction
Streptococcus mutans is an established etiologic agent of human dental caries, commonly known as cavities, one of the most common infectious diseases in the world (reviewed in (Forssten et al. 2010)). This organism is particularly effective at colonizing hard tissues of the human oral cavity, in part by adhering to the high molecular weight salivary agglutinin glycoprotein complex (SAG) contained within the salivary pellicle immobilized on tooth surfaces (reviewed in (Brady et al. 2010)). The highly resilient acidogenic and acid tolerant Gram-positive bacterium ferments a wide variety of dietary carbohydrates causing a rapid drop in environmental pH resulting in demineralization of the tooth and caries formation and progression. It was also recently shown that S. mutans is among those microorganisms capable of producing amyloid during growth in biofilm cultures and that known inhibitors of amyloid fibrillization inhibit biofilm formation by S. mutans (Oli et al. 2012).
S. mutans adherence is mediated by both sucrose-dependent and independent mechanisms. In the presence of dietary sugars such as sucrose, S. mutans produces sticky glucan polymers to facilitate attachment and colonization (reviewed in (Banas 2004)). In addition, the Mr~185 kDa extracellular cell wall-associated adhesin P1 (Forester et al. 1983), also known as Antigen I/II (Russell et al. 1980a) or PAc (Okahashi et al. 1989a), facilitates attachment to the acquired salivary pellicle on teeth in the absence of sucrose. Antigen I/II family molecules are encoded in the genomes of most oral streptococci, as well as certain strains of Streptococcus pyogenes (Zhang et al. 2006) and Streptococcus agalactiae (Chuzeville et al. 2015). P1 was originally identified as a dual antigen dubbed Antigen I/II (Russell et al. 1980a) and shown in numerous studies to represent a target of protective immunity, hence its study as a potential vaccine candidate (reviewed in (Brady et al. 2010; Russell et al. 2004; Yan 2013)). When the gene encoding P1 was cloned and sequenced by two independent groups (Kelly et al. 1989; Kelly et al. 1990; Lee et al. 1988; Okahashi et al. 1989a; Okahashi et al. 1989b), it was recognized that a single gene encoded it and that Antigen II (AgII) represented a C-terminal protease resistant fragment of the full-length Antigen I/II (AgI/II) protein. When the entire spaP gene encoding P1 was deleted from S. mutans, the mutant strain was shown to be less virulent in a rat dental caries model (Crowley et al. 1999) as well as altered in its cell surface properties (Lee et al. 1989) and biofilm-formation characteristics (Ahn et al. 2008). AgI/II family molecules mediate interactions with other oral bacteria and extracellular matrix proteins such as laminin, collagen, and fibronectin (reviewed in (Brady et al. 2010)). The most widely studied substrate for S. mutans P1 is SAG, which consists predominantly of the scavenger receptor gp340 (Loimaranta et al. 2005), as well as trace amounts of sIgA and amylase. There are two distinct and independent binding sites for SAG within P1 (Hajishengallis et al. 1994). The nature of cell surface P1’s interaction with SAG varies depending on whether SAG is immobilized on a surface or is present in fluid-phase (Brady et al. 1992; Loimaranta et al. 2005). This is evidenced by differential inhibition by anti-P1 monoclonal antibodies (MAbs) of different specificities of S. mutans aggregation in the presence of fluid-phase SAG compared to S. mutans adherence to SAG immobilized on a surface such as hydroxyapatite (Brady et al. 1992; Oli et al. 2006). Because fluid-phase gp340 represents an innate host defense mechanism whereby microbes can be agglutininated and cleared (Reichhardt et al. 2014), the ability of S. mutans to adhere to immobilized SAG represents an important physiological adaptation that benefits the bacterium rather the host.
P1 is a large molecule consisting of ~1565 amino acids, depending on the strain. Crystal structures of various fragments of P1, i.e. A3VP1 (Larson et al. 2010), C123 (Larson et al. 2011), and NA1 in complex with P3C (Heim et al. 2014), combined with ultracentrifugation studies of intact P1 (Larson et al. 2010), have now enabled construction of a complete tertiary structure model (Heim et al. 2014) of this highly unusual protein. Within the primary sequence are a series of 3 tandem 82 residue alanine-rich repeats (A-region) and a series of 3 tandem 39 residue proline-rich repeats (P-region) intervened by a beta sheet-rich globular domain (Troffer-Charlier et al. 2002) that contains the so-called variable (V-) region where the majority of sequence differences between strains are clustered (Brady et al. 1991a) (Figure 1A). A highly extended stalk (~ 50 nm) is formed by the high-affinity association of the alpha and polyproline type II helices of the discontinuous A- and P-regions (Larson et al. 2010) that is thought to project the V-region containing globular domain away from the cell wall (Figure 1B). At the other end of the hybrid helix lays another beta sheet-rich globular region. C123 represents a series of three contiguous domains, each of which adopts a DE-variant IgG fold (Larson et al. 2011). At the far C-terminus of P1 is an LPXTG consensus motif that represents the cleavage site for Sortase A, the transpeptidase that cleaves cell surface-localized substrates such as P1 and covalently links them to the cell wall peptidoglycan (Schneewind and Missiakas 2012). It was believed for some time that the C-terminal segment of P1 existed exclusively at the cell wall end of the helical stalk and that the reason for low of reactivity of MAbs mapping within C123 was their lack of accessibility to this segment. More recently, we have identified quaternary interactions between cell-surface anchored P1 and released fragments of P1 (Heim et al. 2015). These studies were prompted by our observation that S. mutans cells subjected to mechanical extraction are significantly diminished in adherence to immobilized SAG but remain readily aggregated by fluid-phase SAG and also remain immunoreactive with anti-P1 MAbs known to react with S. mutans whole cells. Additionally, we found that bacterial adherence to immobilized SAG was restored by incubation of post-extracted cells with P1 fragments including C123, which corresponds to the naturally occurring P1 derivative AgII. In contrast to untreated cells, glutaraldehyde-fixed bacteria gained reactivity with anti-C-terminal MAbs, whereas reactivity of MAbs against other portions of the molecule was diminished suggesting masking of their epitopes (Heim et al. 2015). This and other studies (Heim et al. 2015; Homonylo-McGavin et al. 1999; Lee 1995) have revealed a mechanism by which binding of certain adherence inhibiting antibodies triggers release of non-covalently attached P1 to disrupt the adhesive layer.
Figure 1.
A schematic representation of the primary structure (A) and a tertiary model (B) of full-length P1. Approximate binding sites of anti-P1 MAbs used in this study are indicated on the tertiary model. Gram stain images of whole and lysed NG8 cells (C) show complete cellular lysis of the cell wall samples. Western dot blots (D) of NG8 and PC3370 whole cells, as well as cell wall samples before and after the SDS/trypsin treatment, indicate that P1 is only present on NG8 cell walls and is fully removed by SDS/trypsin treatment.
As stated above, we have found that S. mutans produces amyloid when grown as a biofilm on a surface, but not during growth in planktonic cultures (Oli et al. 2012). There is a growing recognition of functional amyloid formation by microorganisms growing within biofilms (reviewed in (Blanco et al. 2012; DePas and Chapman 2012; Hobley et al. 2015; Syed and Boles 2014)), which may serve to stabilize the biofilm and that can also interact with other components of the biofilm matrix including highly repetitive polysaccharide polymers such as cellulose (de Jong et al. 2009; Saldana et al. 2009). Amyloid material is present in human dental plaque, and amyloid formation is a common property of both laboratory strains and clinical isolates of S. mutans. P1 is among the amyloid forming proteins produced by S. mutans and self-aggregates to form fibrils with common biophysical properties ascribed to amyloids including uptake of Thioflavin T and Congo Red (CR), CR-induced birefringence, and visualization of fibrillar aggregates by transmission electron microscopy (Oli et al. 2012). Its globular domains in particular are associated with uptake of amyloidophilic dyes during self-aggregation, and can also interact with one another.
Surface plasmon resonance, ELISA, and atomic force microscopy experiments now support a model of P1/C123 interactions in which the AgII (C123) fragment binds to the apical region of cell surface-anchored intact P1 (Heim et al. 2015). However, specific structural details regarding this interaction are unknown. Of particular interest in understanding mechanisms of P1-mediated adherence and subsequent biofilm development is whether the C123 domain retains its native monomeric structure, as seen in the crystal structures (Heim et al. 2013; Larson et al. 2011), on binding to P1 or whether a structural change may occur upon intermolecular binding which could predispose or seed the formation of functional amyloid. Here we describe the results of initial fundamental experiments to characterize the interaction between recombinant isotopically-labeled C123 and P1 contained in S. mutans cell walls using Magic Angle Spinning (MAS) solid-state nuclear magnetic resonance spectroscopy (ssNMR). Cell walls derived from a P1-deficient mutant and cell walls treated with SDS and trypsin to eliminate surface exposed P1 were used as negative controls in these experiments. Our results verify the specificity of C123 binding to cell wall-localized P1, demonstrate that binding is at sufficiently high levels to enable further structural characterizations of quaternary protein interactions important to biofilm formation via ssNMR, and lay the groundwork for future characterization of the high-resolution structure of C123 when it is bound to wall-anchored P1 and/or contained within amyloid fibrils.
Materials and methods
Cell wall preparation
S. mutans strain NG8 (serotype c) (Knox et al. 1986) was used in this study, with the isogenic ΔP1 mutant PC3370 (Crowley et al. 1999) as the negative control. Each strain was grown for 12–15 h in 20 mL Terleckyj defined medium (TDM) (Terleckyj and Shockman 1975; Terleckyj et al. 1975) at 37 °C and then used to inoculate a 2 L culture and grown for 48 h. Cells were harvested by centrifugation at 5400 g, 4 °C for 30 min. The cell pellets were resuspended in 20 mL of phosphate buffered saline (PBS), pH 7.0, containing 200 μL DNase I solution at 1 mg/mL and 100 μL cOmplete® mini EDTA-free protease inhibitor cocktail (Roche) solution; incubated at room temperature for 15 min; and transferred into 5 mL Eppendorf tubes with a 1 mL suspension of 0.1 mm zirconia/silica beads. The cell suspensions were subjected to ten 1 min on/off cycles in a mini Beadbeater-8 (BioSpec Products) alternated with cooling on ice. Lysed cells were pipetted into clean centrifuge tubes and centrifuged at 3100 g, 4 °C for 10 min to remove residual whole cells. Fragmented cell walls were subsequently harvested by centrifugation at 25,000 g for 1 h, washed three times with PBS, pH 7.0, and stored as a suspension at 4 °C until use.
As a basis for comparison with untreated cell walls that display exposed P1, half of each cell wall sample was treated with SDS and trypsin to remove surface accessible proteins. In this case, the cell wall pellets were resuspended in 4% SDS solution in water, boiled for 30 min, and left to cool overnight at room temperature. SDS-treated cell walls were collected by centrifugation at 25,000 g for 30 min at room temperature, washed five times with PBS, and resuspended in 20 mL of PBS containing 8 mg of trypsin from bovine pancreas (Sigma). Following incubation for 24 h at 37° C on an orbital shaker, the protease treated cell walls were collected by centrifugation at 25,000 g for 30 min, and washed three times with PBS. The SDS/trypsin treated cell wall preparations were stored as a suspension at 4 °C until use.
Gram staining of S. mutans cells and cell wall preparations
Intact cells and cell wall samples, before and after the SDS/trypsin treatment, were smeared onto microscope slides using inoculation loops and air dried to form thin films. Crystal violet was added and allowed to stain for 60 seconds. The slides were then gently rinsed with water followed by the addition of iodine. After another 60 seconds, the slides were rinsed again with water before a brief wash with acetone/isopropanol. Finally, the slides were counterstained with safranin and allowed to dry before microscopic examination.
Reactivity of S. mutans cells and cell wall preparations with anti-P1 monoclonal antibodies
S. mutans cells and cell wall preparations were each resuspended in PBS to reflect original cell culture volumes. For each sample, 200 μL of serial two-fold dilutions in PBS, beginning at 1:8, were applied to a PBS-soaked nitrocellulose membrane under vacuum using a 96-well manifold dot blot apparatus (Minifold I; Whatman). Membranes were blocked for 1 h at room temperature with PBS containing 0.2% Tween-20 (PBS-Tw), then reacted with P1 specific monoclonal antibodies (MAbs) obtained from previously established hybridomas (Ayakawa et al. 1987). MAbs included 6–8C, which reacts with P1’s C2 and C3 domains; 4–10A, which reacts with the hybrid helical stalk; and 1–6F, which reacts with the globular head domain (Heim et al. 2015). Anti-P1 MAbs were used as ascites fluids diluted 1:1000 in PBS-Tw. Following overnight incubation at room temperature, the membranes were washed four times for 15 min with PBS-Tw, reacted for 2 h with horseradish peroxidase-labeled goat-anti-mouse secondary antibody (ICN/Cappell Biomedicals) diluted 1:1000 in PBS-Tw, washed twice with PBS-Tw and twice with PBS, then developed with 4-chloro-1-naphthol solution (7 mL PBS, 1 mL 4-chloro-1-naphthol [Sigma, 3 mg/ml in ice cold methanol], and 8 μL of 30% hydrogen peroxide).
15N, 13C-enrichment and purification of the P1 C123 fragment
Cloning of DNA encoding the C-terminal C123 domains of P1 (amino acids 1000–1486) was described previously (Heim et al. 2015). Recombinant E. coli were grown overnight in 20 mL M9 media and inoculated into 2 L M9 media containing 15N-enriched ammonium chloride and 13C-enriched glucose (Cambridge Isotope Laboratories) as the nitrogen and carbon sources, respectively. Expression was induced at OD600 1.0 with 0.2% L-arabinose. After incubation overnight at 22 °C, cells were harvested by centrifugation at 5400 g for 30 min at 4° C, lysed with an EmulsiFlex-C5 cell disruptor (Avestin), and the cell lysate loaded onto a HisTRAP HP column (GE Healthcare) with 30 mM Tris/100 mM NaCl/10 mM imidazole, pH 7.4, as binding buffer. The rC123 polypeptide was eluted with 30 mM Tris/100 mM NaCl/300 mM imidazole, pH 7.4, and further purified using a HiTrap Q FF column with 15 mM Tris, 10 mM NaCl, pH 8.4, as the binding buffer and 15 mM Tris, 1M NaCl, pH 8.4, as the elution buffer. The eluted product was then polished and exchanged into storage buffer using a HiLoad 16/600 Superdex 200 pg column with 30 mM Tris/100mM NaCl, pH 6.0.
Binding of 15N, 13C-enriched C123 to cell walls
Approximately 50 mg of cell walls (wet weight) isolated from S. mutans strains NG8 and PC3370 were each gently resuspended in 1 ml of 30 mM Tris/100mM NaCl, pH 6.0, containing 3 mg/mL 15N, 13C-enriched C123. After incubation for 30 min at room temperature, cell walls were pelleted by centrifugation at 16,000 g for 5 min at room temperature, washed with 30 mM Tris/100mM NaCl, pH 6.0, and packed into zirconia rotors for NMR experiments. Separate samples were prepared using either cell walls isolated from the wild type NG8 strain or the corresponding P1-deficient mutant PC3370 strain and using either untreated cell wall fragments or those stripped of protein by treatment with SDS and trypsin. To stabilize the bound 15N, 13C-enriched C123 for longer NMR experiments, additional samples were made as above and fixed with glutaraldehyde added to a final concentration of 0.25% for 30 min prior to the centrifugation and washing steps.
NMR experiments
Initial chemical shift spectra were collected using a Bruker Avance III 500 MHz system and a 3.2 mm E-free® Probe operating at 4 °C (based on independent temperature calibration) with samples spun at 12 kHz to remove spinning sidebands. 13C spectra were collected using both cross-polarization and direct detection methods. Cross-polarization magic angle spinning (CPMAS) (Schaefer and Stejskal 1976) experiments employed a ramped (30–60 kHz) 1H field and a 60 kHz 13C field during 1.5 ms of cross polarization. For excitation and decoupling, a 100 kHz 1H field was utilized. Spectra were acquired using a recycle delay of 3 s and 10k scans for each spectrum. Direct detection experiments employed a 110 kHz 13C field for excitation, 100 kHz 1H decoupling during detection, a recycle delay of 3 s and signal averaging for 10k scans.
Rotational-Echo DOuble-Resonance (REDOR) experiments (Gullion and Schaefer 1989) and further CPMAS experiments were carried out on a Bruker Avance III HD 750 MHz system with a 3.2 mm Low-E HXY MAS probe designed and constructed at the National High Magnetic Field Lab (NHMFL) operating at −40 °C (based on independent temperature calibration) with samples spun at 10 kHz; this temperature was utilized to minimize protein motions which would attenuate dipolar couplings. 13C-1H cross polarization employed a ramped pulse (50–100 kHz) on the 1H channel and a 60 kHz 13C field during the 1.5 ms CP process. A 100 kHz 1H field was utilized for both excitation and detection. For REDOR experiments, 13C 180° pulses at 114 kHz and 15N 180° pulses at 62 kHz were used. Again, 3 s was used as the recycle delay and each spectrum was collected for 1–4k scans.
All one dimensional CPMAS, DPMAS, and REDOR experiments were identically processed followed by Fourier transformation and third order baseline correction. To correct for varying levels of cell wall present in the samples, spectra were scaled based on the intensities of the anomeric carbons (100–105 ppm) and sugar ring carbons (65–75 ppm) present in the peptidoglycan. Various regions of the spectra were then defined for integration purposes, specifically: carbonyl carbons (170–180 ppm), anomeric carbons (100–105 ppm), sugar ring carbons (65–75 ppm) and α-carbons (50–60 ppm).
Results
Characterization of cell wall preparations
To ensure that the bacterial cells were lysed successfully, samples were Gram-stained and inspected by microscopy. During Gram staining, the crystal violet dye penetrates bacterial cell walls. Upon addition of iodine, the iodine-dye complex is not removed by destaining and is retained by the thick peptidoglycan layer of intact Gram-positive cells. In contrast, Gram-negative cells or lysed Gram-positive cells are destained by acetone-isopropanol washes that remove the iodine-dye precipitate, and thus appear pink upon counterstaining with safranin dye. As expected, both the NG8 and the PC3370 strains appeared purple after Gram-staining; however, the lysed cells with disrupted cell walls appeared pink (Figure 1C). No intact cells were observed in the cell wall samples after centrifugation steps following cell lysis.
To verify the presence of P1 on the isolated cell walls, and its absence following their treatment with SDS and trypsin, whole cells and the treated and untreated cell wall preparations were characterized by dot blot assay (Figure 1D). Three anti-P1 MAbs, namely 6–8C, 4–10A and 1–6F, which recognize epitopes spanning the entire molecule and contained within the C-terminal AgII moiety, stalk and globular head regions of P1, respectively, were used as primary antibodies followed by detection with a horseradish peroxidase-labeled goat anti-mouse secondary reagent. These assays confirmed the presence of P1 on the surface of NG8 whole cells and showed that P1 remained associated with the cell walls after cell lysis. We found increasing the number of bead beater cycles increased the yield of cell wall fragments, but did not significantly affect P1 levels on the NG8 cell wall surface (Figure S2). Exposed P1 was successfully removed from the NG8 cell walls by treatment with SDS and trypsin. There was no reactivity of the anti-P1 MAbs with cell walls or whole cells of the P1-deficient mutant PC3370.
Characterization of cell walls with magic angle spinning solid state NMR spectroscopy
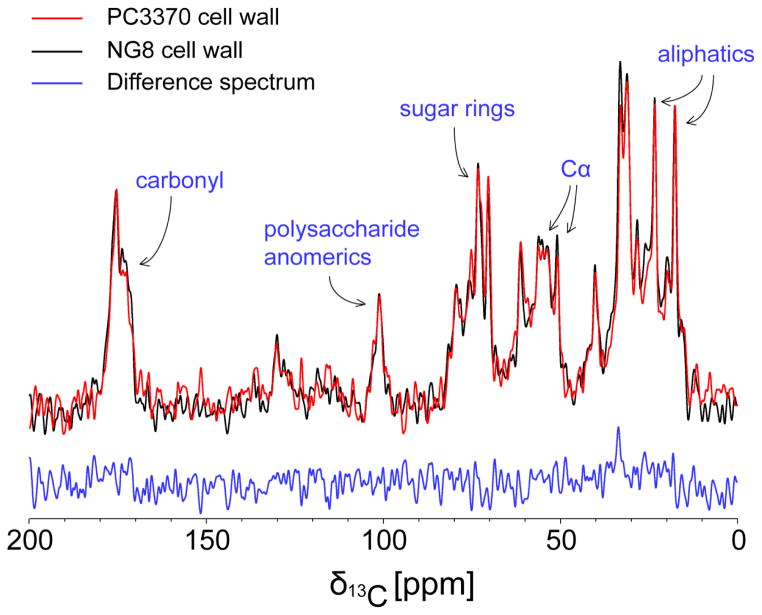
We used NMR spectroscopy to examine cell walls prepared from both the P1-producing and P1-deficient strains NG8 and PC3370, respectively, via one-dimensional 13C cross-polarization (CPMAS) and direct-polarization (DPMAS) magic angle spinning experiments. For these spectra we observed 13C signals at natural abundance (1.1%) since cells were not grown in isotopically enriched media. The CPMAS spectra are presented in Figure 2 with a comparison of CPMAS and DPMAS spectra for NG8 provided in Figure S2. Both NG8 (black) and PC3370 (red) cell wall spectra exhibit resonances for protein and peptidoglycan species with expected relative intensities (Reichhardt et al. 2015). Specifically, our observation of protein chemical shifts (resonances for carbonyl carbons at 170–180 ppm, α-carbons at 50–60 ppm, aliphatic carbons at 10–40 ppm) as well as polysaccharide chemical shifts (acetyl carbons at 175–180ppm, anomeric carbons at 95–100 ppm, sugar-ring carbons at 70–80 ppm, methyl acetylation carbons at 10–20 ppm) agree well with previously published studies of Gram-positive cell walls (Nygaard et al. 2015). A difference spectrum (NG8-PC3370, in blue) shows no substantial differences between the two cell wall preps. This is consistent with the PC3370 isogenic mutant producing the remaining complement of cell surface proteins and intact peptidoglycan in the absence of P1. We note a small, but difficult to quantitate, increase in signal at 170–175 ppm and 20–60 ppm within the NG8 sample spectrum which may be attributable to P1. A noticeable absence of resonances at 140–160 ppm indicates the successful removal of DNA, RNA and other intracellular nucleic acid components. Comparison of isolated cell wall and whole cell spectra (Figure S3) show significant attenuation of signals at 30 ppm, consistent with removal of lipids, as well as at 50–80 ppm and 170–175, consistent with removal of intracellular proteins. Thus, both Gram-staining and NMR spectroscopy confirm lysis of the cells and removal of intracellular components from both cell wall preparations. CPMAS vs. DPMAS spectra indicate limited motion of the peptidoglycan and cell surface attached proteins
Figure 2.
One-dimensional 13C CPMAS spectra of NG8 (black) and PC3370 (red) cell walls, as well as their difference spectrum (blue). 13C chemical shift regions are assigned to specific moieties within the samples based on previous literature.
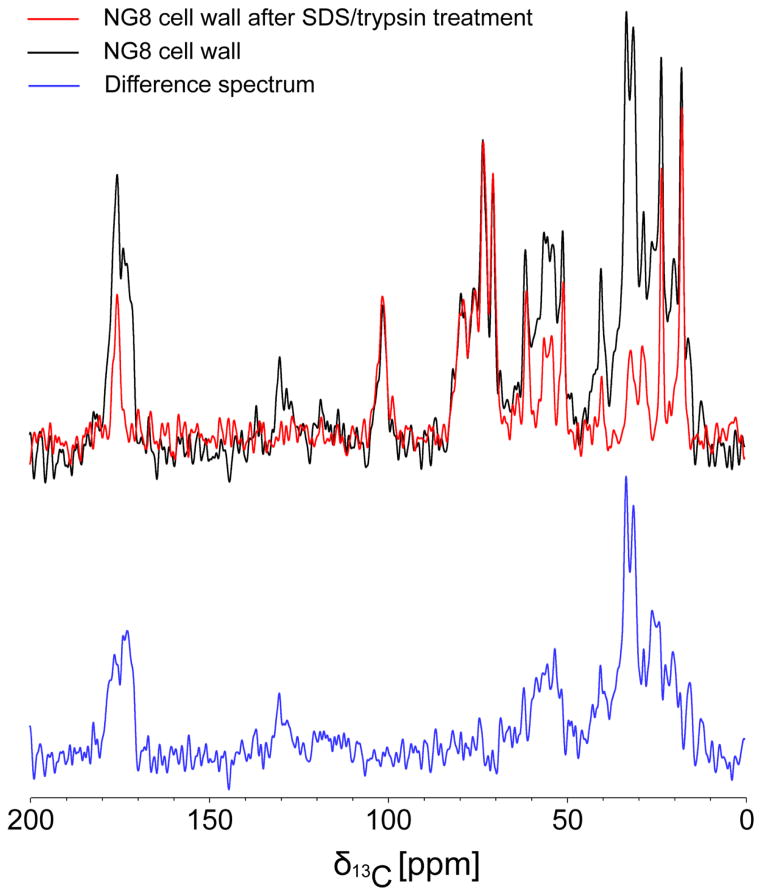
Next, we characterized NG8 and PC3370 cell walls which had been treated by boiling in 4% SDS followed by digestion with trypsin to remove exposed proteins from the peptidoglycan matrix. A comparison of CPMAS spectra for NG8 cell walls before (black) and after (red) the SDS/trypsin treatment (Figure 3) demonstrates successful removal of protein, with significant loss of signals at 20–40 ppm, 50–60 ppm and 170–180 ppm. In contrast, signals for the peptidoglycan at 175–180 ppm (acetyl carbons), 95–100 ppm (anomeric carbons), 70–80 ppm (sugar-ring carbons) and 10–20 ppm (methyl acetylation carbons) remain strong. Thus, the majority of protein was shown to have been successfully removed from the cell walls, including those prepared from NG8, by the SDS/trypsin treatment. This is consistent with the dot blot verification that P1 had been removed from the NG8 cell walls by treatment with SDS/trypsin (Figure 1). CPMAS spectra of PC3370 cell walls (Figure S4) also demonstrated changes in resonant intensities after SDS/trypsin treatment that would stem from elimination of all non-P1 proteins. Comparison of the treated and untreated NG8 and PC3370 cell wall spectra enabled us to estimate signal contributions from peptidoglycan (PG), which is resistant to treatment with SDS and trypsin. PG carbonyl/acetyl carbons at 170–180 ppm contribute ~20–25% of signals observed for NG8 cell walls and ~30% of signals observed for PC3370 prior to SDS/treatment. Signal contributions from N-modified sugar carbons at 40–60 ppm account for ~40% of the signals observed in this region.
Figure 3.
One-dimensional 13C CPMAS spectra of NG8 cell walls before (black) and after (red) SDS/trypsin treatment as well as the difference spectrum (blue). The peptidoglycan signals remain after treatment while protein signals are lost.
Binding of 15N, 13C-enriched C123 to NG8 cell walls observed by NMR spectroscopy
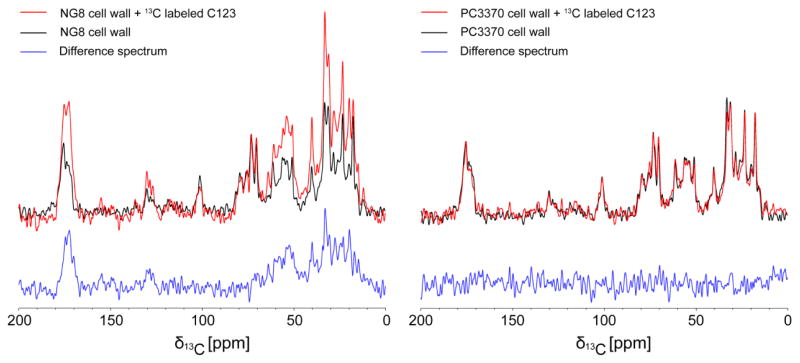
In the next experiment, we added purified 15N, 13C-enriched C123 to both untreated and SDS/trypsin treated cell walls derived from both NG8 and PC3370 strains followed by a washing step to remove unbound or weakly bound protein. Because P1 is only produced by NG8 cells, and SDS/trypsin removes P1 as well as other surface-localized proteins, specific binding of C123 to P1 should only occur for the NG8 cell walls that were not treated with SDS/trypsin. Any nonspecific binding of C123 would be observed for both NG8 and PC3370 cell walls, hence inclusion of the PC3370 negative control. CPMAS spectra (Figure 4a) indicate significant binding of 15N, 13C-enriched C123 to the NG8 cell walls. DPMAS spectra (Figure S5) are in agreement with CPMAS results. Difference spectra (cell walls after C123 binding–cell walls before C123 binding, in blue) clearly indicate the addition of resonant intensities consistent with protein at 170–175 ppm (carbonyl carbons), 125–130 ppm (aromatic carbons), 50–60 ppm (α-carbons) and 10–40 ppm (aliphatic carbons). Very little C123 protein binding to PC3370 cell walls was observed (Figure 4b). DPMAS spectra (Figure S5) gave similar results. No binding of C123 or change in signal was observed for SDS/trypsin treated cell walls prepared from either strain (Figure S6). Because the PC3370 mutant is derived from the NG8 parent strain and lacks only the spaP gene encoding P1, these results indicate that the C123 polypeptide, which corresponds to the naturally occurring AgII C-terminal fragment of P1, interacts specifically with P1, and not with other proteins attached to the cell wall. Of particular note, the spread in resonances seen for C123 upon its interaction with NG8 cell walls is sufficiently broad to suggest that this polypeptide fragment likely retains a folded structure when bound to P1.
Figure 4.
NMR measurements of 15N, 13C-enriched C123 binding to NG8 (left panel) and PC3370 (right panel) cell walls. One-dimensional 13C CPMAS spectra of cell walls before (black) and after (red) binding along with the difference spectra (blue) indicate specific binding of C123 to NG8 cell walls.
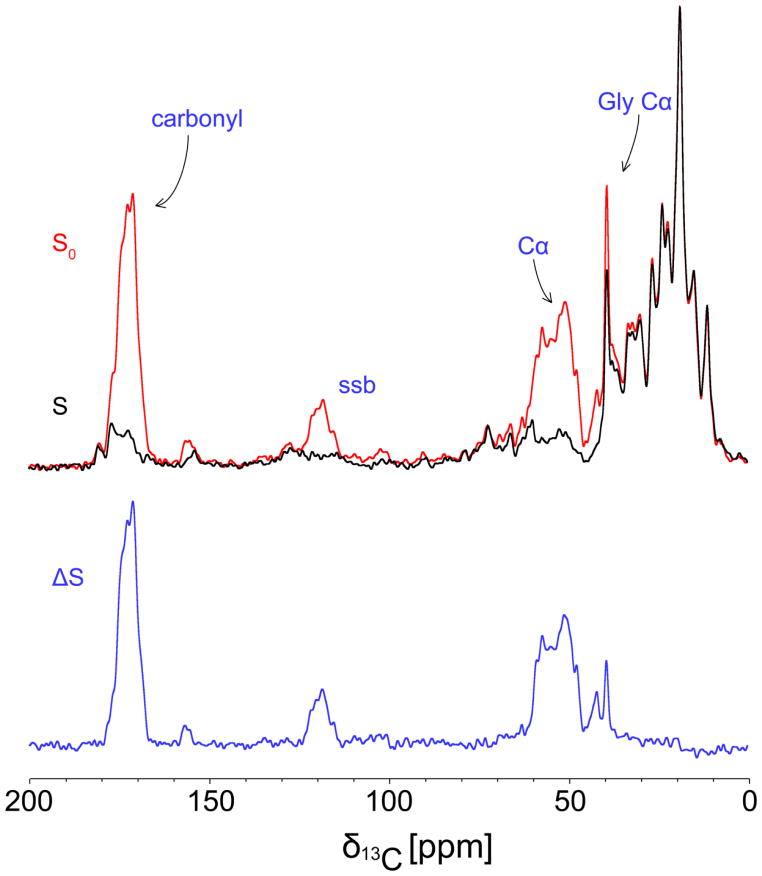
To independently characterize the signals arising directly from 15N, 13C-enriched C123 bound to NG8 cell walls, in contrast to the natural abundance protein and PG signals, we employed REDOR difference spectroscopy. REDOR spectra were recorded in pairs for each sample. An initial experiment (Figure 5, black) was collected with rotor-synchronized dephasing pulses (S), with a dephasing time selected to affect only 13C sites that are directly bonded to 15N sites (carbonyl carbons and α-carbons in C123). A second control experiment (Figure 5, red) was also collected without such pulses (S0). Spectral subtraction provides a REDOR difference spectrum (ΔS) in which signals arising solely from the added, 15N, 13C-enriched C123 are visible (Figure 5, blue). The difference spectrum shown in Figure 5 matches the carbonyl carbon (170–175 ppm) and α-carbon (40–60 ppm) regions of the difference spectrum shown in Figure 4a. A difference is observed between the two experiments in the aliphatic region (10–40 ppm) due to the fact that the REDOR experiment only dephases signals from 13C nuclei directly bonded to 15N nuclei. As a result, aliphatic 13C signals are not dephased and consequently do not appear in the REDOR difference spectrum. The data in Figure 5 were collected at −40 °C in order to eliminate residual protein motions within the fully hydrated sample. REDOR experiments collected at −5 °C (Figure S7a) showed dephasing of C123 bound to the NG8 cell walls, but with incomplete dephasing due to protein motion attenuating the dipolar couplings. Again, a negative control experiment utilizing PC3370 cell walls (Figure S7b) verified that no isotopically enriched protein was bound in the absence of P1. This indicates, therefore, that the additional signals arising in the spectrum of the NG8 cell wall sample upon addition of 15N, 13C-enriched C123 arises from specific binding of C123 to the cell wall-localized P1.
Figure 5.
13C/15 N REDOR spectra of NG8 cell walls after binding of 15N, 13C-enriched C123 and glutaraldehyde fixation. The dephased S spectrum (black) is subtracted from the S0 control spectrum (red) to yield a difference spectrum (blue) thus enabling quantitation of 13C signals arising from 13C nuclei directly bonded to 15N nuclei in isotopically-enriched C123. A spinning sideband arising from the carbonyl 13C signals is indicated by ‘ssb’.
Based on signal integrations and comparison of CPMAS spectra obtained at 500 MHz and 4 °C (Figures 2–4), we estimate from our initial experiments that binding of C123 to NG8 cell walls accounts for ~53% of the protein signals in Figure 4a. When fresh samples were made for REDOR spectra collected at 750 MHz and −40 °C (Figure 5), we observed that C123 binding could account for >80% of the protein signals. Based on systematic checks regarding the integrity of P1 on the NG8 cell wall between sample preps (Figure S1) and the reproducibility of NMR spectra at a given field and temperature with different cell wall preps (Figure S8), these differences in NMR intensities for C123 are due in part to variations in cross polarization conditions, magnetic fields and temperature between the experiments. However, there is likely also some variability in the level of C123 binding between sample preps. Nonetheless, we consistently observed specific binding of C123 to NG8 cell walls (and not PC3370 cell walls) with sufficient S/N for NMR experiments at both 4 °C and −40 °C. Given the level of P1 present is not sufficient to distinguish NG8 cell wall spectra from PC3370 cell wall spectra, the observed levels of 13C/15N enriched C123 binding to the NG8 cell walls is remarkable and suggests a strong interaction between C123 and P1 with the possibility of multiple copies of C123 binding per copy of P1. Using the PRISM webserver (Baspinar et al. 2014; Tuncbag et al. 2011) and crystal structures of different regions of P1 (PDB id: 3QE5, 3IPK) to simulate protein-protein interactions, several energetically favorable interactions are predicted for the native form of C123 with the V-region of P1 (Figure S9). These predictions give us testable hypotheses regarding the binding of C123 which can be addressed in future work.
Discussion
While it has long been known that S. mutans adhesin P1 contains two separate and distinct binding sites for SAG (Hajishengallis et al. 1994), that the protein is processed such that a protease resistant C-terminal fragment known as AgII is recoverable from cell extracts and spent culture supernatants (Russell et al. 1980a; Russell et al. 1980b), and that the P1-mediated interaction of S. mutans with SAG differs depending on whether SAG is immobilized on a surface or present in fluid-phase (Brady et al. 1992; Loimaranta et al. 2005), the limited structural information regarding P1 precluded the development of a comprehensive model to integrate all of these findings. The field has advanced rapidly in recent years with the development of a complete P1 tertiary model based on electron microscopy (Larson et al. 2011) and ultracentrifugation studies (Larson et al. 2010) in conjunction with crystal structures of numerous recombinant fragments (Heim et al. 2014; Larson et al. 2011; Larson et al. 2010; Troffer-Charlier et al. 2002). In addition, curious findings in which anti-C-terminal MAbs that did not appear to react with the cell surface could in some way inhibit adhesion of S. mutans to immobilized SAG (Brady et al. 1991b; Brady et al. 1992; McArthur et al. 2007) were explained by the recognition that P1 does not exist in a uniform single layer of covalently attached protein. Rather, an additional hierarchical ultrastructure exists in which P1 fragments, C123 in particular, associate with covalently linked P1 to form the functional adhesive layer (Heim et al. 2015). When cells are stripped of these fragments, they are readily agglutinated by fluid-phase SAG (gp340), but are impaired in adherence to immobilized SAG. Certain anti-P1 MAbs, particularly those mapping to the C-terminal domains, trigger the release of these adhesive fragments and thereby disrupt the quaternary architecture such that the ability of the MAbs to bind to intact bacterial cells went unrecognized until glutaraldehyde fixation experiments were performed.
Our goal in the current study was to utilize ssNMR to observe and characterize the specific binding of the C-terminal segment of P1 to covalently attached P1 contained within isolated S. mutans cell walls to lay the necessary groundwork for future structural studies. Previously, surface plasmon resonance measurements demonstrated that recombinant C123 could interact with another recombinant polypeptide known as A3VP1, which contains the globular head and one-third of the extended helical stalk of P1 (Heim et al. 2015). ELISA measurements also indicated that the globular head of P1 alone was capable of interaction with the isolated C-terminus. However, these binding experiments were performed with purified polypeptides outside of the native context of the cell wall. The NMR results we present here indicate that C123, a recombinant polypeptide that corresponds to the originally identified biological moiety known as AgII, indeed binds specifically to P1 contained within S. mutans cell walls. More importantly, comparison of NMR spectra enables us to directly quantify the amount of C123 specifically binding to the cell walls. Early work characterizing the cell surface localized proteins of S. mutans found P1 to be a major component (Hardy et al. 1986; Knox et al. 1986). In this study, we were not able to reproducibly quantify P1 signals within the S/N of the NMR spectra but observed sufficient signals from isotopically-enriched C123 to suggest a significant level of quaternary C123/P1 assembly and to establish that further ssNMR experiments will enable us to explore the structure-function relationship of this interaction. Our data are consistent with the assertion, based on previous studies, that a quaternary C123/P1 assembly plays an integral functional role in mediating bacterial adhesion to SAG.
We have previously found that P1can form fibrils with hallmark characteristics of amyloid. Amyloid material is present when S. mutans is grown on a surface as a biofilm, but not when it is grown in planktonic liquid cultures. In addition, a Sortase A mutant which is incapable of covalently linking P1 and several other surface-localized proteins to the cell wall does not make amyloid during biofilm growth, although amyloid forming proteins can be found in spent culture supernatants (Oli et al. 2012). This suggests that a cell surface nucleation event is critical to the process of amyloidogenesis in S. mutans. Recently, we systematically studied the ability of naturally occurring fragments from the cell surface localized proteins of S. mutans to form amyloids; C123 in particular was found to readily form fibrils with all the hallmarks of amyloid formation (manuscript under review). Interestingly, other bacteria that make functional amyloid require a nucleation event at the cell surface (Wang et al. 2008), and several of the chaplin proteins of the soil microorganism Streptomyces coelicolor which self-assemble into amyloid fibrils are sortase substrates (Claessen et al. 2003). A question remaining therefore is whether the two observations, namely that C123 bound to cell wall-attached P1 promotes S. mutans adherence to SAG and that C123 can readily form amyloid, are linked or whether they represent separate functions of C123 in the formation of quaternary structures important to bacterial adherence and/or biofilm formation, respectively. This raises the intriguing question of whether C123 on binding to P1 retains a native globular structure, previously solved by crystallography, with adhesive properties, or whether it templates fibrillization of additional copies to form functional amyloid within the context of the biofilm matrix. In our current study, the NMR signals we observe for C123 bound to cell wall-associated P1 was sufficient to be consistent with C123 binding P1 in a 1:1 stoichiometry or greater, suggesting multiple copies of C123 may bind a single P1 copy.. We also note that the similarities in the CPMAS and DPMAS spectra suggest that C123 bound to P1 is relatively immobilized with little flexibility in the protein backbone. In the current study, the level of signal-to-noise for C123 bound to cell walls was insufficient to perform conventional multi-dimensional NMR experiments to characterize more fully the structure and assembly of the bound protein. However, an order or two magnitude increase in signal via DNP enhancement would certainly make this feasible. Alternatively, isotopic enrichment of amino acid residues that are most likely to change conformation on transition of C123 from a globular to a fibrillar structure could help distinguish which form(s) of C123 can interact with P1 on the cell surface. These approaches are the subject of ongoing work. The PRISM predicted interactions of the native form of C123 with the V-region of P1 agree with our previously published observation that C123 can bind the A3VP1 polypeptide that represents the globular head region and top third of the helical stalk contained within intact P1 (Heim et al. 2015). However, this interaction is reversible and we note that C123 signal intensities decrease quickly as we apply additional washing steps after binding C123 to intact NG8 cell walls. The prediction of a specific binding interface for C123 to A3VP1 also provides guidance on potential isotopic enrichment schemes for delineating specific protein-protein interactions.
Taken as a whole, our results show that C123 specifically binds to cell wall-localized P1 of S. mutans at a sufficiently high enough level to form functional quaternary structures. The intricate details of how P1 and its fragments are assembled on the cell surface to confer its full range of capabilities can potentially be elucidated by further ssNMR studies if sufficient signal intensities can be measured via either dynamic nuclear polarization or 1H-detection methodologies at high field. Determining the quaternary organization of C123 and P1 will enable a deeper understanding of the adhesive behavior of P1 including how protective immunity against it might be achieved and/or how the functional ultrastructure might be disrupted with appropriate inhibitors. Structural measurements might also elucidate any role of C123 functional amyloid fibril formation in stabilizing S. mutans biofilms.
Supplementary Material
Acknowledgments
We would like to thank Drs. Kyle Heim and Richard Besingi for helpful discussions. This work was supported in part by National Institutes of Health Grants R01DE08007 and R01DE21789 from the NIDCR. A portion of this work was performed in the McKnight Brain Institute at the National High Magnetic Field Laboratory’s AMRIS Facility, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490, the State of Florida, and an NIH award, S10RR031637, for magnetic resonance instrumentation.
Abbreviations
- MAb
Monoclonal Antibody
- MAS
Magic Angle Spinning
- NMR
Nuclear Magnetic Resonance
- ssNMR
solid-state NMR
- CR
Congo Red
- SAG
salivary agglutinin glycoprotein complex
References
- Ahn SJ, Ahn SJ, Wen ZT, Brady LJ, Burne RA. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect Immun. 2008;76:4259–4268. doi: 10.1128/IAI.00422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayakawa GY, Boushell LW, Crowley PJ, Erdos GW, McArthur WP, Bleiweis AS. Isolation and characterization of monoclonal antibodies specific for antigen P1, a major surface protein of mutans streptococci. Infect Immun. 1987;55:2759–2767. doi: 10.1128/iai.55.11.2759-2767.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA. Virulence properties of Streptococcus mutans. Front Biosci. 2004;9:1267–1277. doi: 10.2741/1305. [DOI] [PubMed] [Google Scholar]
- Baspinar A, Cukuroglu E, Nussinov R, Keskin O, Gursoy A. PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42:W285–289. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Crowley PJ, Ma JK, Kelly C, Lee SF, Lehner T, Bleiweis AS. Restriction fragment length polymorphisms and sequence variation within the spaP gene of Streptococcus mutans serotype c isolates. Infect Immun. 1991a;59:1803–1810. doi: 10.1128/iai.59.5.1803-1810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol. 2010;77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Piacentini DA, Crowley PJ, Bleiweis AS. Identification of monoclonal antibody-binding domains within antigen P1 of Streptococcus mutans and cross-reactivity with related surface antigens of oral streptococci. Infect Immun. 1991b;59:4425–4435. doi: 10.1128/iai.59.12.4425-4435.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Piacentini DA, Crowley PJ, Oyston PC, Bleiweis AS. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuzeville S, Dramsi S, Madec JY, Haenni M, Payot S. Antigen I/II encoded by integrative and conjugative elements of Streptococcus agalactiae and role in biofilm formation. Microb Pathog. 2015;88:1–9. doi: 10.1016/j.micpath.2015.07.018. [DOI] [PubMed] [Google Scholar]
- Claessen D, et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun. 1999;67:1201–1206. doi: 10.1128/iai.67.3.1201-1206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong W, Wosten HA, Dijkhuizen L, Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol. 2009;73:1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- DePas WH, Chapman MR. Microbial manipulation of the amyloid fold. Res Microbiol. 2012;163:592–606. doi: 10.1016/j.resmic.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester H, Hunter N, Knox KW. Characteristics of a high molecular weight extracellular protein of Streptococcus mutans. J Gen Microbiol. 1983;129:2779–2788. doi: 10.1099/00221287-129-9-2779. [DOI] [PubMed] [Google Scholar]
- Forssten SD, Bjorklund M, Ouwehand AC. Streptococcus mutans, caries and simulation models. Nutrients. 2010;2:290–298. doi: 10.3390/nu2030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullion T, Schaefer J. Rotational-Echo Double-Resonance. NMR Journal of Magnetic Resonance. 1989;81:196–200. doi: 10.1016/0022-2364(89)90280-1. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Koga T, Russell MW. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73:1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- Hardy LN, Knox KW, Brown RA, Wicken AJ, Fitzgerald RJ. Comparison of extracellular protein profiles of seven serotypes of mutans streptococci grown under controlled conditions. J Gen Microbiol. 1986;132:1389–1400. doi: 10.1099/00221287-132-5-1389. [DOI] [PubMed] [Google Scholar]
- Heim KP, Crowley PJ, Brady LJ. An intramolecular interaction involving the N terminus of a streptococcal adhesin affects its conformation and adhesive function. J Biol Chem. 2013;288:13762–13774. doi: 10.1074/jbc.M113.459974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim KP, Crowley PJ, Long JR, Kailasan S, McKenna R, Brady LJ. An intramolecular lock facilitates folding and stabilizes the tertiary structure of Streptococcus mutans adhesin P1. Proc Natl Acad Sci U S A. 2014;111:15746–15751. doi: 10.1073/pnas.1413018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim KP, et al. Identification of a supramolecular functional architecture of Streptococcus mutans adhesin P1 on the bacterial cell surface. J Biol Chem. 2015;290:9002–9019. doi: 10.1074/jbc.M114.626663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015 doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homonylo-McGavin MK, Lee SF, Bowden GH. Subcellular localization of the Streptococcus mutans P1 protein C terminus. Can J Microbiol. 1999;45:536–539. [PubMed] [Google Scholar]
- Kelly C, et al. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 1989;258:127–132. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- Kelly C, et al. Sequencing and characterization of the 185 kDa cell surface antigen of Streptococcus mutans. Arch Oral Biol. 1990;35(Suppl):33S–38S. doi: 10.1016/0003-9969(90)90128-w. [DOI] [PubMed] [Google Scholar]
- Knox KW, Hardy LN, Wicken AJ. Comparative studies on the protein profiles and hydrophobicity of strains of Streptococcus mutans serotype c. J Gen Microbiol. 1986;132:2541–2548. doi: 10.1099/00221287-132-9-2541. [DOI] [PubMed] [Google Scholar]
- Larson MR, Rajashankar KR, Crowley PJ, Kelly C, Mitchell TJ, Brady LJ, Deivanayagam C. Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J Biol Chem. 2011;286:21657–21666. doi: 10.1074/jbc.M111.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MR, et al. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices. Proc Natl Acad Sci U S A. 2010;107:5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF. Active release of bound antibody by Streptococcus mutans. Infect Immun. 1995;63:1940–1946. doi: 10.1128/iai.63.5.1940-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Progulske-Fox A, Bleiweis AS. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect Immun. 1988;56:2114–2119. doi: 10.1128/iai.56.8.2114-2119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Progulske-Fox A, Erdos GW, Piacentini DA, Ayakawa GY, Crowley PJ, Bleiweis AS. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II) Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loimaranta V, Jakubovics NS, Hytonen J, Finne J, Jenkinson HF, Stromberg N. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect Immun. 2005;73:2245–2252. doi: 10.1128/IAI.73.4.2245-2252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur WP, Rhodin NR, Seifert TB, Oli MW, Robinette RA, Demuth DR, Brady LJ. Characterization of epitopes recognized by anti-Streptococcus mutans P1 monoclonal antibodies. FEMS Immunol Med Microbiol. 2007;50:342–353. doi: 10.1111/j.1574-695X.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- Nygaard R, Romaniuk JAH, Rice DM, Cegelski L. Spectral Snapshots of Bacterial Cell-Wall Composition and the Influence of Antibiotics by Whole-Cell. NMR Biophys J. 2015;108:1380–1389. doi: 10.1016/j.bpj.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Cloning of a surface protein antigen gene from serotype c Streptococcus mutans. Mol Microbiol. 1989a;3:221–228. doi: 10.1111/j.1365-2958.1989.tb01811.x. [DOI] [PubMed] [Google Scholar]
- Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol Microbiol. 1989b;3:673–678. doi: 10.1111/j.1365-2958.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Oli MW, McArthur WP, Brady LJ. A whole cell BIAcore assay to evaluate P1-mediated adherence of Streptococcus mutans to human salivary agglutinin and inhibition by specific antibodies. J Microbiol Methods. 2006;65:503–511. doi: 10.1016/j.mimet.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Oli MW, et al. Functional amyloid formation by Streptococcus mutans. Microbiology. 2012;158:2903–2916. doi: 10.1099/mic.0.060855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhardt C, Fong JC, Yildiz F, Cegelski L. Characterization of the Vibrio cholerae extracellular matrix: a top-down solid-state NMR approach. Biochim Biophys Acta. 2015;1848:378–383. doi: 10.1016/j.bbamem.2014.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhardt MP, et al. The salivary scavenger and agglutinin in early life: diverse roles in amniotic fluid and in the infant intestine. J Immunol. 2014;193:5240–5248. doi: 10.4049/jimmunol.1401631. [DOI] [PubMed] [Google Scholar]
- Russell MW, Bergmeier LA, Zanders ED, Lehner T. Protein antigens of Streptococcus mutans: purification and properties of a double antigen and its protease-resistant component. Infect Immun. 1980a;28:486–493. doi: 10.1128/iai.28.2.486-493.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MW, Childers NK, Michalek SM, Smith DJ, Taubman MA. A Caries Vaccine? The state of the science of immunization against dental caries. Caries Res. 2004;38:230–235. doi: 10.1159/000077759. [DOI] [PubMed] [Google Scholar]
- Russell MW, Zanders ED, Bergmeier LA, Lehner T. Affinity purification and characterization of protease-susceptible antigen I of Streptococcus mutans. Infect Immun. 1980b;29:999–1006. doi: 10.1128/iai.29.3.999-1006.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldana Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Giron JA. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol. 2009;11:992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J, Stejskal EO. Carbon-13 nuclear magnetic resonance of polymers spinning at the magic angle. Journal of the American Chemical Society. 1976;98:1031–1032. [Google Scholar]
- Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AK, Boles BR. Fold modulating function: bacterial toxins to functional amyloids. Front Microbiol. 2014;5:401. doi: 10.3389/fmicb.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleckyj B, Shockman GD. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect Immun. 1975;11:656–664. doi: 10.1128/iai.11.4.656-664.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleckyj B, Willett NP, Shockman GD. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun. 1975;11:649–655. doi: 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troffer-Charlier N, Ogier J, Moras D, Cavarelli J. Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 A resolution suggests a sugar preformed binding site. J Mol Biol. 2002;318:179–188. doi: 10.1016/S0022-2836(02)00025-6. [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using. PRISM Nat Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. Salivary IgA enhancement strategy for development of a nasal-spray anti-caries mucosal vaccine. Sci China Life Sci. 2013;56:406–413. doi: 10.1007/s11427-013-4473-5. [DOI] [PubMed] [Google Scholar]
- Zhang S, Green NM, Sitkiewicz I, Lefebvre RB, Musser JM. Identification and characterization of an antigen I/II family protein produced by group A Streptococcus. Infect Immun. 2006;74:4200–4213. doi: 10.1128/IAI.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.