Abstract
Objectives
There is a growing body of evidence suggesting a shared genetic susceptibility between many neuropsychiatric disorders, including schizophrenia, autism, intellectual disability and epilepsy. The sodium channel, voltage-gated type II alpha subunit gene SCN2A has been shown to exhibit loss-of-function mutations in individuals with seizure disorders, intellectual disability, autism and schizophrenia. The role of loss-of-function mutations in schizophrenia is still uncertain with only one such mutation identified to date.
Methods
To seek additional evidence for a role for loss-of-function mutations at SCN2A in schizophrenia we performed mutation screening of the entire coding sequence in 980 schizophrenia cases. Given an absence of LoF mutations in a public exome cohort (ESP6500, N=6503) we did not additionally sequence controls.
Results
We identify a novel, nonsense (i.e. stop-codon) mutation in one case (E169X) that is absent in 4300 European American and 2203 African-American individuals from the NHLBI Exome Sequencing Project. This is the second loss-of-function allele identified in a schizophrenia case to date. We also show a novel, missense variant, V1282F, that occurs in 2-cases and is absent in the control dataset.
Conclusion
We argue that very rare, loss-of-function mutations at SCN2A act in a moderately penetrant manner to increase the risk of developing several neuropsychiatric disorders including seizure disorders, intellectual disability, autism and schizophrenia.
Keywords: Schizophrenia, gene, mutation, nonsense, missense, loss-of-function
Introduction
The application of genome-wide association study (GWAS) technologies to neuropsychiatric disorders has led to the discovery of common risk-variants for schizophrenia and other brain disorders (O'Donovan et al., 2008, Ripke et al., 2013, Purcell et al., 2009, Consortium, 2014). It is now widely accepted that these common, low-penetrance risk-alleles can impart risk of developing related neuropsychiatric disorders (Lee et al., 2013, Purcell et al., 2009), however the underlying pathological mechanisms remain unclear. The application of GWAS and Whole Exome-Sequencing (WES) technologies to the study of rare variations such as Copy Number Variants (CNVs) or loss-of-function (LoF) mutations has implicated a role for rare alleles and mutations in schizophrenia (Purcell et al., 2014, Kirov et al., 2012, Fromer et al., 2014, Li et al., 2015, Kirov et al., 2009), Autism Spectrum Disorder (ASD) (Li et al., 2015, De Rubeis et al., 2014, Guilmatre et al., 2009, Sanders et al., 2011, Sanders et al., 2012, Iossifov et al., 2012), Intellectual Disability (ID) (Cooper et al., 2011, Classen et al., 2013, Allen et al., 2013, Li et al., 2015, Gilissen et al., 2014) and seizure disorders (Carvill and Mefford, 2013, Martin et al., 2014, Allen et al., 2013). The identification of moderate-to-highly penetrant rare alleles and their enrichment in certain sub-sets of genes of known function has implicated glutamatergic post-synaptic protein complexes as one potential common pathological site for schizophrenia, ASD and ID (Fromer et al., 2014, Kirov et al., 2012).
In their WES study of individuals with schizophrenia, Fromer et al (Fromer et al., 2014) showed an enrichment of de novo loss-of-function (LoF) mutations in synapse related genes; LoF mutations are any mutation that prevents formation of a mature protein product and they encompass nonsense, frameshift and splice-site mutations. One of the LoF mutations disrupted a canonical splice-site in the gene SCN2A encoding an alpha subunit of voltage-gated sodium channels, a key protein involved in axon potential propagation (Meisler et al., 2010). Several loss-of-function mutations at this locus have also been shown in ASD (Sanders et al., 2012, Tavassoli et al., 2014, Fromer et al., 2014, Li et al., 2015, Codina-Solà et al., 2015, De Rubeis et al., 2014), ID (Rauch et al., 2012, de Ligt et al., 2013, Li et al., 2015) and a patient with generalised epilepsy, mental decline and autistic behaviour (Kamiya et al., 2004), as well as >20 missense alleles implicated in seizure disorders (Kwong et al., 2015, Boutry-Kryza et al., 2015, Shi et al., 2012).
The data implicating SCN2A in neuropsychiatric disorders is therefore compelling, regardless of the underlying pathological mechanism or specific diagnosis, however the evidence for a role for SCN2A in schizophrenia is weak, relying as it does on one de novo LoF mutation (Fromer et al., 2014, Li et al., 2015). Therefore, we undertook mutation screening of the entire coding-sequence (CDS) of SCN2A in sample of 980 individuals with schizophrenia, aiming to identify LoF alleles to support involvement of this gene in disease pathogenesis.
Methods
Case Samples
We acquired blood-sample genomic DNA from individuals with Treatment-Resistant Schizophrenia (TRS) via samples taken from patients taking clozapine (Clozaril™) from the United Kingdom Clozaril Monitoring Service; these sample have been employed in previous publications and further details can be found there (Hamshere et al., 2013. Strange et al., 2012). The sample consisted of 980 Caucasians with TRS (Male 705, Female 275). Anonymisation of samples from the clozapine monitoring service was maintained and genetic studies on this sample are approved by relevant ethics panels and are in accordance with the UK Human Tissue Act.
Comparison Sample Data
Exome sequencing data were available from the NHLBI Exome Sequencing Project (ESP) via the Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/), release ESP6500 (Exome Variant Server, [Accessed May 2015]). The dataset includes 4300 European Americans and 2203 African American individuals recruited for studies of heart, lung and blood disorders and the sample is frequently used as a comparison dataset (Zaidi et al., 2013, Lim et al., 2013, Li et al., 2015). Genotype calls for insertion and deletion variants (Indels) are less robust than SNP calls having a higher false positive rate and so are considered experimental (Tennessen et al., 2012, Project, 2014). Therefore, insertions and deletions were excluded from comparison analysis.
Polymerase Chain Reaction (PCR), High-Resolution Melting Analysis (HRMA) and Sequencing
PCR was performed using a previously published methodology (Dwyer et al., 2010, Carroll et al., 2011). The oligonucleotide primers for PCR and sequencing are given in Supplementary Table ST1. High Resolution Melting Analysis (HRMA) was performed using LightScanner™ technology (Idaho Technologies) according previously published criteria (Carroll et al., 2011, Dwyer et al., 2010). PCR products from HRMA were cleaned using AMPure™, DNA sequencing reactions used Big-Dye™ terminator chemistry and sequencing reactions were cleaned using cleanSEQ™. Samples were analysed using an Applied Biosystems™ ABI3100 genetic analyzer and then sequencing traces were inspected manually using Sequencher™ software.
Bioinformatics
Genomic positions and sequences were obtained from The UCSC Genome Browser, Feb. 2009, HG19, NCBI Build 37.1 (http://genome.ucsc.edu/) (Kent et al., 2002). Polymorphism details were extracted from dbSNP build 138 (dbSNP 135 was used during study design) (Sherry et al., 1999) and the 1000 Genomes Project (Abecasis et al., 2010). PCR primers were designed using Primer3 (http://frodo.wi.mit.edu/) (Koressaar and Remm, 2007). Identified alleles were annotated using various software programs including SIFT and PolyPhen-2 (Kumar et al., 2009, Artimo et al., 2012, Woolfe et al., 2010, Adzhubei et al., 2013, Punta et al., 2012).
Results
Primary Analysis
The SCN2A locus lies at chromosome 2q24.3 and 3 mRNA transcripts code for 2 protein isoforms according to RefSeq. The 3 mRNA species of SCN2A have 27 exons containing coding sequence. Together with splice sites (≥2bp of adjacent intron), these were covered by 30 PCR amplimers ranging in size from 220-487bp. Oligonucleotide primers did not encompass known genetic variants >1% minor allele frequency (MAF) according to dbSNP build 135 and HapMap CEU samples (Altshuler et al., 2010).
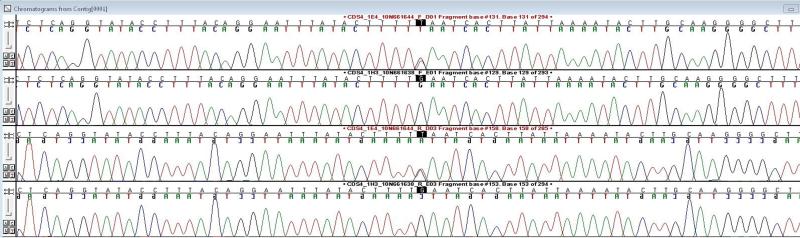
PCR was completed and HRMA performed in all 980 individuals for all 30 PCRs. A total of 19 exonic variants (12 synonymous, 7 non-synonymous, 0 splice-site) (Table 1) were identified in this study, including all variants >1% MAF in dbSNP 138 (HapMap CEU and 1000 Genomes GBR sample (Abecasis et al., 2012, Altshuler et al., 2010)). Five novel variants were found. Of the non-synonymous variants, 6 are missense and one is a novel, nonsense variant introducing a stop-codon at amino acid 169 (169 E>X). No canonical splice-site variants were identified. When compared to the EVS dataset of 6503 sequenced individuals we identify no LoF mutations. An example of HRMA analysis where E169X was identified is shown in Supplementary Figure SF1 and the sequencing traces for the corresponding allele (chr2: 166165204 G/T) are shown in Figure 1.
Table 1.
All 19 exonic variants identified by LightScanner™ HRMA and sequencing analysis of 980 schizophrenia cases. For common alleles (≥0.5%) where HRMA cannot be reliably used for accurate genotyping the HapMap CEU or 1000 Genomes project minor allele frequencies are given. The allele count refers to the number of samples showing the same LightScanner allele profile, which identifies all heterozygotes but may not identify rare homozygotes.
| PCR | Flanking Sequence | rs | Hg19 | Type | MAF (%) | Allele Count |
|---|---|---|---|---|---|---|
| CDS1 | ttctttacca[G/A]ggaatccctt | rs17183814 | 166152389 | 19 R > K | 8.48 | - |
| CDS4 | ttatactttt[G/T]aatcacttat | novel | 166165204 | 169 E > X | Singleton | T=1/G=1959 |
| CDS11 | ctgaatcaag[A/G]gacttcagtg | rs200246820 | 166172013 | Synonymous | 0.10 | G=2/A=1958 |
| CDS12 | actttgctga[T/C]gatgagcaca | rs141815642 | 166179779 | Synonymous | 1.02 | C=20/T=1940 |
| CDS12 | tgttcgtgcc[G/T]cacagacatg | rs114315466 | 166179836 | Synonymous | 0.10 | T=2/G=1958 |
| CDS13 | ctactgaaac[A/G]gaaataagaa | rs147891446 | 166183379 | Synonymous | 1.07 | G=15/A=1945 |
| CDS15 | tcagttctcc[G/C]atcattccgg | novel | 166198966 | 850 R > P | Singleton | C=1/G=1959 |
| CDS16 | aagagctaca[A/G]agaatgtgtc | rs2228980 | 166201225 | 908 K > R | 0.66 | G=13/A=1947 |
| CDS17A | gtttcaggtt[C/T]tgaacctctt | rs375858093 | 166210705 | Synonymous | Singleton | T=1/C=1959 |
| CDS17B | ttgctgttgg[A/C]gaatctgact | rs138143967 | 166211112 | Synonymous | 0.10 | C=2/A=1958 |
| CDS18 | ttgaacctga[G/A]gaatcccttg | novel | 166221739 | Synonymous | Singleton | A=1/G=1959 |
| CDS20 | cttcctgatt[G/T]ttgatgtgag | novel | 166226804 | 1282 V > F | 0.10 | T=2/G=1958 |
| CDS23 | tacaggccac[G/A]tttaagggat | rs138241682 | 166234112 | Synonymous | Singleton | A=1/G=1959 |
| CDS26 | gagtcaagaa[A/G]tgacaaacat | novel | 166243379 | 1559 M > V | Singleton | G=1/A=1959 |
| CDS27A | gaatcctacg[T/A]ctgatcaaag | rs2060198 | 166245230 | Synonymous | 26.83 | - |
| CDS27B | ctgggatgga[T/C]tgctagcacc | rs199698414 | 166245471 | Synonymous | Singleton | T=1/C=1959 |
| CDS27B | agatgttcta[T/C]gaggtttggg | rs200603552 | 166245713 | Synonymous | 0.15 | C=3/T=1957 |
| CDS27C | gattttgcag[A/C]tgccctggat | rs138497939 | 166245784 | 1823 D > A | Singleton | C=1/A=1959 |
| CDS27D | cgtctccacc[C/T]tcgtatgata | rs73025979 | 166246235 | Synonymous | Singleton | T=1/C=1959 |
Figure 1.
Forward and reverse sequencing traces of the E169X heterozygous carrier and a non-carrier. The mutated base is central and highlighted
The EVS database reports an indel that would introduce a frameshift (chr2:166246312, AT/A), however the genotypes for this variant are not in Hardy-Weinberg Equilibrium in both European Americans (AT/AT=1, AT/A=2, A/A=4105, HWE chi-square p<0.0001) and African Americans (AT/AT=1. AT/A=1. A/A=2089, HWE chi-square p<0.0001), in agreement with indel variants being more likely to represent false-positives (Tennessen et al., 2012) and justifying the exclusion of indel variants from analysis.
Secondary Analysis
HRMA analysis precludes the study of rare homozygote genotype calls, as only heterozygote genotypes are detected reliably (Dwyer et al., 2010). Therefore, only alleles with a MAF <0.5% have allele counts shown (Table 1). None of the 980 samples harboured >1 of these 19 rare alleles.
The 6 missense variants identified were analysed using SIFT and Polyphen-2 (Kumar et al., 2009, Adzhubei et al., 2013). Both packages highlighted the same two variants as being possibly of functional significance (damaging with low-confidence or possibly damaging). One is a novel singleton variant (chr2:166198966 G/C R850P) not present in the EVS controls. The other is also a novel variant present in two cases (chr2:166226804, G/T, V1282F) and absent in the EVS dataset and is therefore potentially interesting with the caveat that the observations are rare and the biological significance uncertain (Fisher’s exact test, 2-tailed p-value=0.034). The two samples do not represent a duplicate sample as they have different LightScanner profile genotype calls at rs2121371 and rs2060198.
Mutation screening comparison
Given differences in the mutation detection platforms, we compared the frequency of rare alleles in cases and the EVS dataset. In cases we identified a small but significant deficit of the total number of variants identified when compared to the EVS sample (Pearson chi-square, two-tailed p=0.04, OR 0.61 (0.38-0.98)). There was no significant difference for synonymous variants (p=0.21, OR 0.70 (0.40-1.23)) but there was a trend for an excess of missense variants in the EVS dataset (p=0.07, OR 0.47 (0.20-1.09)). The LightScanner approach employed here may therefore be less-sensitive than WES for the identification of rare alleles, suggesting that our detection of a rare LoF mutation in the sample is not simply due to higher screening efficiency. Alternatively the EVS sample may harbour more missense variants at SCN2A than our case sample.
Discussion
Our understanding of the aetiology of the major psychiatric disorders remains limited, however a large part of the risk of developing these disorders is due to our individual genetic architecture (Owen, 2012). Promising insights into the molecular aetiology of neuropsychiatric diseases have come from the study of rare, moderately penetrant alleles such as inherited or de novo CNVs (Guilmatre et al., 2009, Kirov et al., 2012, Cooper et al., 2011, de Ligt et al., 2013, Girirajan et al., 2013) or more recently smaller LoF mutations (Rauch et al., 2012, Sanders et al., 2012, Fromer et al., 2014, Iossifov et al., 2012, Li et al., 2015, De Rubeis et al., 2014, Purcell et al., 2014). One of the genes implicated by these recent studies of ID, ASD and schizophrenia is SCN2A (Sanders et al., 2012, Fromer et al., 2014, Rauch et al., 2012, de Ligt et al., 2013, Li et al., 2015, Codina-Solà et al., 2015, De Rubeis et al., 2014), encoding the alpha subunit of voltage gated sodium channels. However, amongst these disorders the evidence for SCN2A involvement in schizophrenia is dependent upon one splice-site mutation (Fromer et al., 2014, Li et al., 2015).
We performed mutation screening of SCN2A in 980 individuals with schizophrenia to identify further LoF mutations and support a role for the gene in disease aetiology. Previously, nonsense loss-of-function mutations have been identified in nine unrelated individuals with ASD (Tavassoli et al., 2014, Sanders et al., 2012, Fromer et al., 2014, Codina-Solà et al., 2015, De Rubeis et al., 2014), three unrelated cases with ID (de Ligt et al., 2013, Rauch et al., 2012), an individual with epilepsy and autistic features (Kamiya et al., 2004) and an individual with schizophrenia (Fromer et al., 2014). Therefore, 14 de novo loss-of-function (LoF) mutations were known at SCN2A in individuals that have a neuropsychiatric diagnosis. Genic recurrence of de novo LoF alleles in unrelated individuals with a shared diagnosis unambiguously implicates LoF alleles at SCN2A in the pathogenesis of ASD and ID as the occurrence of ≥2 such alleles is highly-unlikely to occur by chance (Sanders et al., 2012). In 980 individuals with TRS we identify a further LoF mutation to the one identified by Fromer et al (Fromer et al., 2014). Although we cannot establish if the LoF variant identified in this study is inherited or de novo, a caveat of this study, we do not identify any LoF mutations in the EVS comparison dataset of 6503 individuals, which is suggestive of a pathogenic role for the variant. When taken with previous mutations identified in schizophrenia and related disorders our simple study is supportive for the involvement of SCN2A in the pathogenesis of schizophrenia.
Loss-of-function mutations at SCN2A may have a greater or lesser effect on risk of developing disease for each of the neuropsychiatric phenotypes discussed, or their higher prevalence in some samples may reflect screening in larger or smaller samples. Using data from multiple large WES studies of several neuropsychiatric phenotypes (Li et al., 2015), LoF mutations at SCN2A occurred in 7/5893 ASD cases (0.12%, [95% CI 0.03% to 0.21%]), 3/151 cases for ID (1.99%, [95% CI −0.24% to 4.21%]) and 2/1828 cases for schizophrenia (0.11%, 95% CI −0.04% to 0.26%) when the current study is included. These studies however do not include smaller, directed sequencing studies of the locus (Kamiya et al., 2004, Tavassoli et al., 2014). When compared to a mutation rate of 0% in a control sample of 6503 individuals this suggests a moderate-to-high penetrance for these alleles. However, particularly for schizophrenia, ID and seizure disorders much larger scale sequencing studies are warranted to characterise the true frequency and effect-size of LoF mutations at SCN2A in each phenotype. A soon to be published coalition of WES studies using different methodologies, including 60,706 unrelated individuals of varying ancestry, and affection status may help to uncover the true frequency of LoF mutations at SCN2A in the wider population (Exome Aggregation Consortium, ExAC Browser Beta (http://exac.broadinstitute.org) [Accessed: July 2015]). Currently only two LoF mutations are observed in the >120,000 chromosomes included in this dataset confirming a low-frequency of LoF mutations at SCN2A. In addition, the missense variant V1282F is absent in this dataset also. Screening of such large samples will undoubtedly uncover more potentially damaging alleles at SCN2A although the significance of these will be difficult to interpret without accurate phenotypic information and it is likely that large, well-controlled association studies will be required to assess the true effect on disease risk of damaging mutations at this locus.
In further study of schizophrenia samples it may be pertinent to examine a wide phenotypic distribution, given that the present sample of TRS may represent a particularly severe category of schizophrenia. In vitro or in vivo analysis of the handful of loss-of-function mutations at SCN2A may then be justified and necessary to understand the molecular biology of these mutations. Previous studies of the SCN2A protein, NaV1.2 have implicated the protein in redistribution of channels required for neural plasticity, dependent upon scaffolding proteins (ankyrin G) (Garrido et al., 2003) and have also shown a neocortical and hippocampal expression pattern that diminishes throughout neurodevelopment (Liao et al., 2010). Given the key role for SCN2A in neuronal development and the specificity of the molecular lesions identified so far, the gene offers promise to aid understanding of the molecular pathogenesis of a complex neuropsychiatric disorder in some individuals.
Supplementary Material
HRMA using LightScanner™ Call-IT™ software. A 96-well microtitre plate including 4 non-template controls (NTCs) was analysed. The NTCs were identified (burgundy), and 91 individuals were shown to have similar melt curves (grey), i.e. no genetic variation within this group. One sample showed a divergent melt profile (red) and follow-up sequencing identified the E169X mutation.
Sequencing traces of the V1282F non-carriers and heterozygous. The mutated base is highlighted.
Details of the oligonucleotide primers used in the present study for LightScanner™ analysis and Fluorescent Dye-terminator sequencing.
All variants identified in the present study and all variants listed in corresponding regions of the NHLBI Exome Sequencing Project (EVS control dataset). Allele counts are given and association analysis performed where allele counts >1 across both datasets. NS = Non-synonymous, Syn = Synonymous, Int = Intronic.
Acknowledgements
Work funded by grants from the Medical Research Council UK. All authors contributed to the manuscript; LSC prepared the manuscript which was reviewed by MCO and MJO before submission by LSC.
This work was supported by grants from the Wellcome Trust and Medical Research Council
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7, pp. Unit7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40(Web Server issue):W597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry-Kryza N, Labalme A, Ville D, de Bellescize J, Touraine R, Prieur F, et al. Molecular characterization of a cohort of 73 patients with infantile spasms syndrome. Eur J Med Genet. 2015;58(2):51–8. doi: 10.1016/j.ejmg.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Carroll LS, Williams HJ, Walters J, Kirov G, O'Donovan MC, Owen MJ. Mutation screening of the 3q29 microdeletion syndrome candidate genes DLG1 and PAK2 in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(7):844–9. doi: 10.1002/ajmg.b.31231. [DOI] [PubMed] [Google Scholar]
- Carvill GL, Mefford HC. Microdeletion syndromes. Curr Opin Genet Dev. 2013;23(3):232–9. doi: 10.1016/j.gde.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Classen CF, Riehmer V, Landwehr C, Kosfeld A, Heilmann S, Scholz C, et al. Dissecting the genotype in syndromic intellectual disability using whole exome sequencing in addition to genome-wide copy number analysis. Hum Genet. 2013;132(7):825–41. doi: 10.1007/s00439-013-1296-1. [DOI] [PubMed] [Google Scholar]
- Codina-Solà M, Rodríguez-Santiago B, Homs A, Santoyo J, Rigau M, Aznar-Laín G, et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol Autism. 2015;6:21. doi: 10.1186/s13229-015-0017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43(9):838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ligt J, Boone PM, Pfundt R, Vissers LE, Richmond T, Geoghegan J, et al. Detection of clinically relevant copy number variants with whole-exome sequencing. Hum Mutat. 2013;34(10):1439–48. doi: 10.1002/humu.22387. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer S, Carroll L, Mantripragada KK, Owen MJ, O'Donovan MC, Williams NM. Mutation screening of the DTNBP1 exonic sequence in 669 schizophrenics and 710 controls using high-resolution melting analysis. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(3):766–74. doi: 10.1002/ajmg.b.31045. [DOI] [PubMed] [Google Scholar]
- Exome Aggregation Consortium (ExAC) Cambridge, MA: [Accessed: July 2015]. URL: http://exac.broadinstitute.org. [Google Scholar]
- Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: [Accessed: May 2015]. URL: http://evs.gs.washington.edu/EVS/ [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014 doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido JJ, Fernandes F, Moussif A, Fache MP, Giraud P, Dargent B. Dynamic compartmentalization of the voltage-gated sodium channels in axons. Biol Cell. 2003;95(7):437–45. doi: 10.1016/s0248-4900(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511(7509):344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Johnson RL, Tassone F, Balciuniene J, Katiyar N, Fox K, et al. Global increases in both common and rare copy number load associated with autism. Hum Mol Genet. 2013;22(14):2870–80. doi: 10.1093/hmg/ddt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66(9):947–56. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2013;18(6):708–12. doi: 10.1038/mp.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya K, Kaneda M, Sugawara T, Mazaki E, Okamura N, Montal M, et al. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24(11):2690–8. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18(8):1497–503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17(2):142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–91. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Kwong AK, Ho AC, Fung CW, Wong VC. Analysis of mutations in 7 genes associated with neuronal excitability and synaptic transmission in a cohort of children with non-syndromic infantile epileptic encephalopathy. PLoS One. 2015;10(5):e0126446. doi: 10.1371/journal.pone.0126446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Cai T, Jiang Y, Chen H, He X, Chen C, Li X, et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Deprez L, Maljevic S, Pitsch J, Claes L, Hristova D, et al. Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain. 2010;133(Pt 5):1403–14. doi: 10.1093/brain/awq057. [DOI] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77(2):235–42. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HC, Kim GE, Pagnamenta AT, Murakami Y, Carvill GL, Meyer E, Copley RR, et al. Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, O'Brien JE, Sharkey LM. Sodium channel gene family: epilepsy mutations, gene interactions and modifier effects. J Physiol. 2010;588(Pt 11):1841–8. doi: 10.1113/jphysiol.2010.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–5. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Owen MJ. Implications of genetic findings for understanding schizophrenia. Schizophr Bull. 2012;38(5):904–7. doi: 10.1093/schbul/sbs103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;511:421–427. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380(9854):1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45(10):1150–9. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–85. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9(8):677–9. [PubMed] [Google Scholar]
- Shi X, Yasumoto S, Kurahashi H, Nakagawa E, Fukasawa T, Uchiya S, et al. Clinical spectrum of SCN2A mutations. Brain Dev. 2012;34(7):541–5. doi: 10.1016/j.braindev.2011.09.016. [DOI] [PubMed] [Google Scholar]
- Strange A, Riley BP, Spencer CCA, Morris DW, Brinen M, O’Dushlaine CT, et al. Genome-wide association study implicates HLA-C*01:02 as a risk factor at the major histocompatibility complex locus in schizophrenia. Biol Psychiatry. 2012;72(8):620–8. doi: 10.1016/j.biopsych.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli T, Kolevzon A, Wang AT, Curchack-Lichtin J, Halpern D, Schwartz L, et al. De novo SCN2A splice site mutation in a boy with Autism spectrum disorder. BMC Med Genet. 2014;15:35. doi: 10.1186/1471-2350-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Mullikin JC, Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome Biol. 2010;11(2):R20. doi: 10.1186/gb-2010-11-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–3. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HRMA using LightScanner™ Call-IT™ software. A 96-well microtitre plate including 4 non-template controls (NTCs) was analysed. The NTCs were identified (burgundy), and 91 individuals were shown to have similar melt curves (grey), i.e. no genetic variation within this group. One sample showed a divergent melt profile (red) and follow-up sequencing identified the E169X mutation.
Sequencing traces of the V1282F non-carriers and heterozygous. The mutated base is highlighted.
Details of the oligonucleotide primers used in the present study for LightScanner™ analysis and Fluorescent Dye-terminator sequencing.
All variants identified in the present study and all variants listed in corresponding regions of the NHLBI Exome Sequencing Project (EVS control dataset). Allele counts are given and association analysis performed where allele counts >1 across both datasets. NS = Non-synonymous, Syn = Synonymous, Int = Intronic.