The DNA-binding protein PRDM9 directs positioning of the double strand breaks (DSBs) initiating meiotic recombination, in mice and humans. Prdm9 is the only mammalian speciation gene yet identified and is responsible for sterility phenotypes in male hybrids of certain mouse subspecies. To investigate PRDM9 binding and its role in fertility and meiotic recombination, we humanized PRDM9’s DNA-binding domain in C57BL/6 mice. This change repositions DSB hotspots and completely restores fertility in male hybrids. We show that alteration of one Prdm9 allele impacts the behaviour of DSBs controlled by the other allele at chromosome-wide scales. These effects correlate strongly with the degree to which each PRDM9 variant binds both homologues at the DSB sites it controls. Furthermore, higher genome-wide levels of such “symmetric” PRDM9 binding associate with increasing fertility measures and comparisons of individual hotspots suggest binding symmetry plays a downstream role in the recombination process. These findings reveal that subspecies-specific degradation of PRDM9 binding sites by meiotic drive, which steadily increases asymmetric PRDM9 binding, has impacts beyond simply changing hotspot positions, and strongly support a direct involvement in hybrid infertility. Because such meiotic drive occurs across mammals, PRDM9 may play a wider, yet transient, role in early stages of speciation.
In spite of its central role in evolution, the molecular mechanisms underlying speciation are not well understood. Only a small number of genes involved in speciation have been documented1, with only one such gene, Prdm9, known in mammals2,3. Prdm9 contributes to hybrid sterility in male (PWD×B6)F1 mice from crosses between male Mus musculus domesticus C57BL/6 (hereafter B6) and female Mus musculus musculus PWD/Ph (hereafter PWD)4. Although its genetic basis is only partially understood5,6, this hybrid sterility is characterised by failure of pairing (synapsis) of homologous chromosomes and an arrested meiotic prophase due to lack of repair of recombination intermediates2. Homologous recombination, and synapsis, are interdependent, essential meiotic processes7, and evidence suggests synapsis often nucleates at recombination sites8. Aside from the PWDxB6 cross, Prdm9 allele and dosage have been associated with variation in measures of fertility and successful meiosis in many additional mouse crosses9.
PRDM9 has several functional domains, including a DNA-binding zinc finger (ZF) array, and a PR/SET domain responsible for histone H3 lysine 4 trimethylation (H3K4me3)10 (Fig. 1a). By binding to specific DNA sequence targets, PRDM9 directs the positions of the double-strand break (DSB) events that initiate meiotic recombination11. This results in DSBs, and downstream recombination events, clustering into small discrete regions called hotspots12,13. The PRDM9 ZF-array, encoded by a minisatellite repeat, is highly polymorphic within and across mammalian species3,14-16 and is among the fastest evolving regions in the genome, with strong evidence of natural selection influencing this evolution3. It is unknown whether PRDM9 ZF-array polymorphism has additional impacts, aside from direct alterations of DSB hotspot positions.
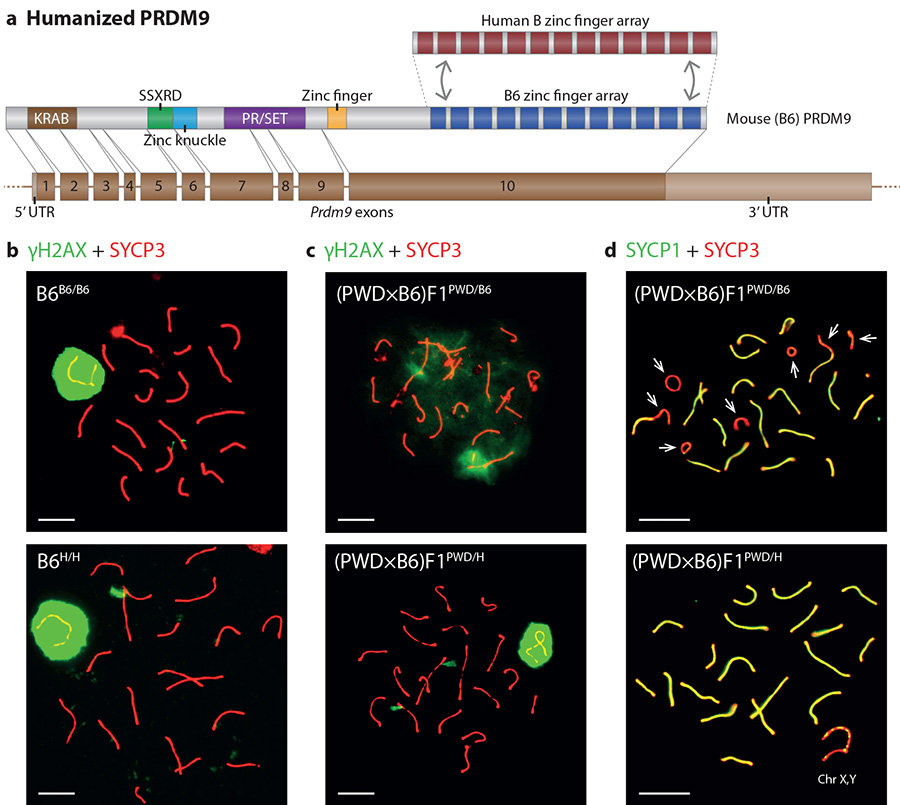
Figure 1. Humanizing the ZF domain of PRDM9 does not impact fertility.
a, Domain structure of the re-engineered PRDM9 protein b, γH2AX staining of the sex body (green), SYCP3 staining of the chromosome axis (red) in late pachytene in B6B6/B6 (top) and B6H/H (bottom). c, As b, but for (PWD×B6)F1PWD/B6 and (PWD×B6)F1PWD/H. d, SYCP1 staining of the synaptonemal-complex transverse filament (green), and SYCP3 staining of the chromosome axis (red) in pachytene for (PWD×B6)F1PWD/B6 and (PWD×B6)F1PWD/H. Arrows: unsynapsed autosomes. Scale bars: 10 μm.
Humanizing Prdm9 restores hybrid fertility
To explore the DNA-binding characteristics of PRDM9, we generated a line of humanized B6 mice, by replacing the portion of mouse Prdm9 exon 10 encoding the ZF-array with the orthologous sequence from the human reference PRDM9 allele (the “B” allele) (Fig. 1a, Extended Data Fig. 1). A feature of PRDM9 (explored further below) is the co-evolution of its ZF-array with the genomic background in which it sits13,17. Minisatellite mutational processes at PRDM9 can produce new alleles with duplications, deletions or rearrangements within the ZF-array, yielding an almost complete change in PRDM9 binding sites, and thus hotspot locations14. Because the human PRDM9 ZF-array evolved on a lineage separated from mice for ~150M years, our experimental approach allows assessment of functional properties of a PRDM9 ZF-array unaffected by changes it has induced in the background genome, similar to new alleles randomly arising in the population.
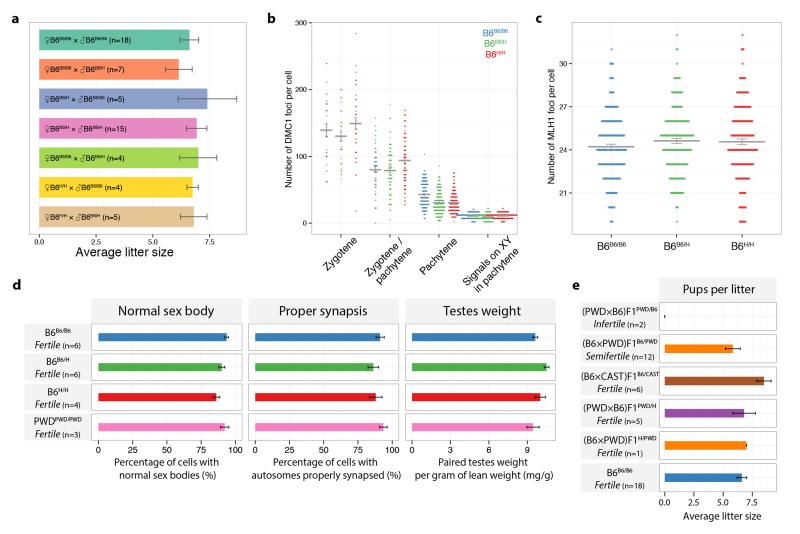
Humanization of the Prdm9 ZF-array in B6 inbred mice had no effect on fertility (Extended Data Fig. 2) and cytogenetic comparisons revealed no significant impact on zygotene DSB counts (DMC1 immunoreactivity, Extended Data Fig. 2b), crossover counts (MLH1 foci, Extended Data Fig. 2c), normal sex body formation (γH2AX immunostaining, Fig. 1b) or quantitative measures of fertility and successful synapsis (see later). The full fertility of humanized mice implies there are unlikely to be any specific essential PRDM9 binding sites. One mechanism underlying speciation in many settings involves Dobzhansky-Muller incompatibilities: hybrid dysfunction arising from incorrect epistatic interactions1. Based on the above, it seems likely that if such interactionsPrdm9 involving PRDM9 occur, they do not reflect constrained co-evolution of Prdm9 with specific genes.
To directly explore PRDM9’s role in fertility, we crossed PWD females with B6B6/H males. As expected18, male (PWD×B6)F1PWD/B6 hybrids (we use superscripts to indicate Prdm9 genotypes and write the female strain first in crosses) exhibited hybrid sterility as evidenced by failures in siring pups (Extended Data Fig. 2e), sex body formation (Fig. 1c) and synapsis (Fig. 1d). In contrast, all these defects were completely rescued in (PWD×B6)F1PWD/H hybrids inheriting the engineered humanized ZF-array (Fig. 1c,d, Extended Data Fig. 2e). Thus the ZF domain of PRDM9, and hence likely the DNA-binding properties of this protein, underlies Prdm9’s role in hybrid sterility.
Although (PWD×B6)F1PWD/B6 male mice are completely sterile, the male progeny of the reciprocal cross (B6×PWD)F1B6/PWD are semi-fertile9. A particular 4.7Mb locus (Hstx2), on the PWD X-chromosome, influences these fertility differences6. We also tested the impact of humanization in this reciprocal cross, and full fertility (from semi-fertility) was again restored (see below and Supplementary Information). Thus humanization of PRDM9 acts at least partially independently of Hstx2.
Our reprogramming of the PRDM9 binding sites mimics the consequences of mutational changes in its ZF-array. The restoration of hybrid fertility suggests that the same rescue is likely to occur for newly arising alleles that also reset PRDM9 binding sites, and hence hybrid sterility between subspecies driven by Prdm9 will be evolutionarily transient. This raises the question, which we return to below, of what properties are possessed by Prdm9 alleles that are associated with reduced fertility.
Humanizing the recombination landscape
To characterize the consequences of re-engineering the ZF domain on recombination, we generated high-resolution DSB maps for mice with different Prdm9 alleles and genomic backgrounds, using ChIP-seq single-stranded DNA sequencing19 on adult testes. This approach identifies single-stranded 3′ sequence ends decorated with DMC1, which arise as intermediates following creation of DSBs by SPO11. In addition to mapping DSB hotspots, our hotspot-calling algorithm estimates a hotspot “heat”, proportional to the fraction of cells marked by DMC1 at that locus (Supplementary Information). This DMC1 heat depends on both the relative frequency of DSB formation and on how long DMC1 marks persist20. We also obtained complementary information by performing ChIP-seq to measure H3K4me3, a histone modification directly introduced in cis by PRDM9 binding11.
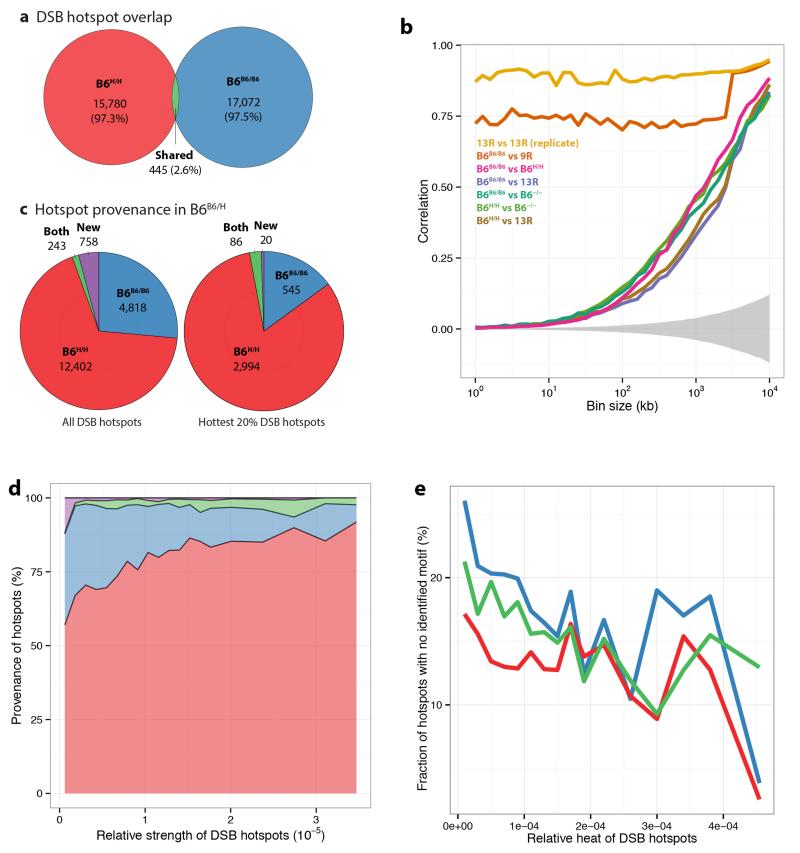
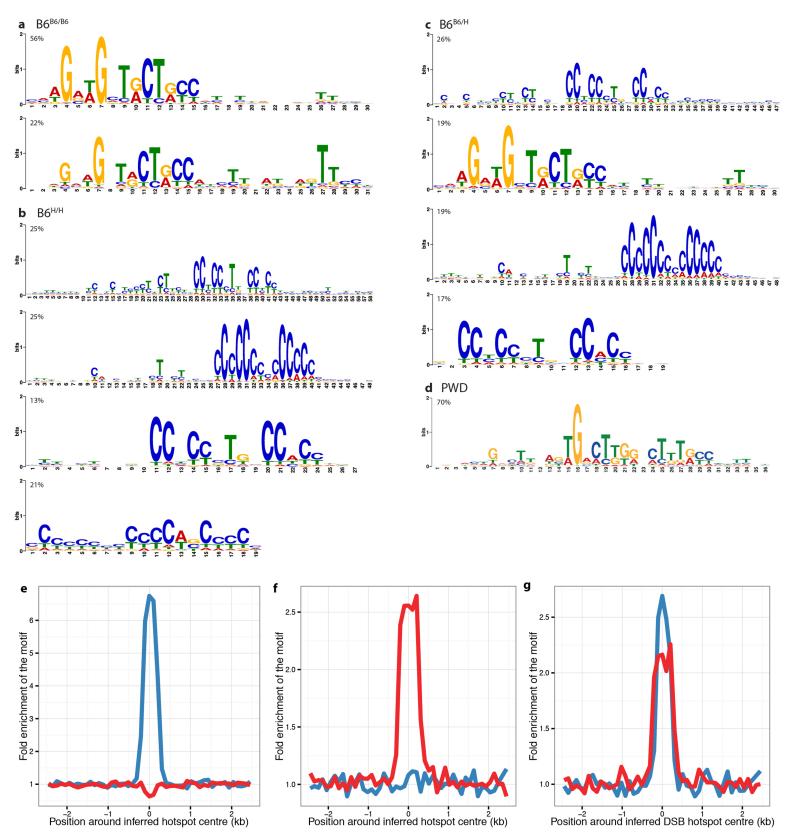
Relative to wild-type B6 mice21, B6H/H mice showed completely changed hotspot landscapes (2.6% overlap; Extended Data Fig. 3), with hotspots in the humanized mouse showing strong enrichment for a motif matching the previously reported human PRDM9 binding motif13 (Extended Data Fig. 4). Most DSB hotspots overlapped H3K4me3 peaks (89%, p<0.05). Correlation between the wild-type and humanized mice in total DSB heats increased over larger genomic scales (Extended Data Fig. 3b), consistent with earlier studies showing large scale crossover rates depend on factors other than PRDM916,20,22.
In the heterozygous mouse, despite the presence of two different Prdm9 alleles, we found a similar number of hotspots to homozygous mice (Supplementary Table 1). Furthermore, almost all B6B6/H hotspots (95.8%) were found in either the B6B6/B6 or B6H/H mice (Extended Data Fig. 3c, 4c, Supplementary Table 2). The human allele exhibited 2.7-fold dominance over the wild-type allele (Supplementary Table 2), with even stronger dominance for hotter hotspots (Extended Data Fig. 3c). Comparison of homozygous and heterozygous hotspot heats (Extended Data Fig. 3d, 3e) implies B6 hotspots operate similarly, but are proportionally less active, in the heterozygote. For additional DSB hotspot analyses, see Supplementary Information and Extended Data Fig. 5.
Humanization restores symmetric binding
Next we examined DSB hotspot maps for hybrid males: infertile (PWD×B6)F1PWD/B6, reciprocal semi-fertile (B6×PWD)F1B6/PWD, humanized rescue (PWD×B6)F1PWD/H, reciprocal humanized rescue (B6×PWD)F1H/PWD, with wild-type PWD for comparison. Sequence differences between the PWD and B6 genomes allowed us to determine whether individual hotspots in these hybrids were “symmetric”, with DSBs occurring equally on both chromosomes, or “asymmetric”, with a preference towards either the PWD or B6 chromosome (Supplementary Information Section 5).
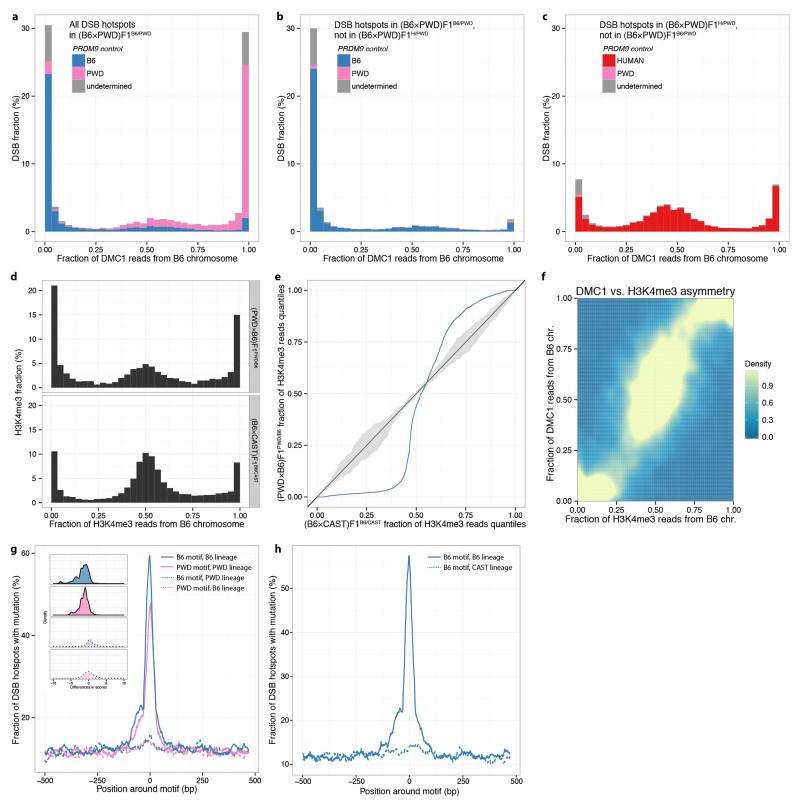
We found that most DMC1 signal (71.8%) in (PWD×B6)F1PWD/B6 or (B6×PWD)F1B6/PWD hybrids occurs within asymmetric DSB hotspots (Fig. 2a, Extended Data Fig. 6). Further, DSBs associated with the PWD allele occur largely on the B6 chromosome and those associated with the B6 allele occur largely on the PWD chromosome. We also measured asymmetry of the H3K4me3 mark at each hotspot and found the same pattern, confirming that DSB asymmetry largely reflects underlying differences in PRDM9 binding and methylation between the two homologues. This H3K4me3 asymmetry resembles that previously described for (B6×CAST)F1B6/CAST hybrids17, but is considerably more extreme. Sequence differences directly disrupting PRDM9 binding motifs explain almost all cases of binding asymmetry (83.4% of PWD hotspots; 91.3% of B6 hotspots), and result from rapid mutational accumulation along the separate lineages from the common ancestor of B6 and PWD (Extended Data Fig. 6g).
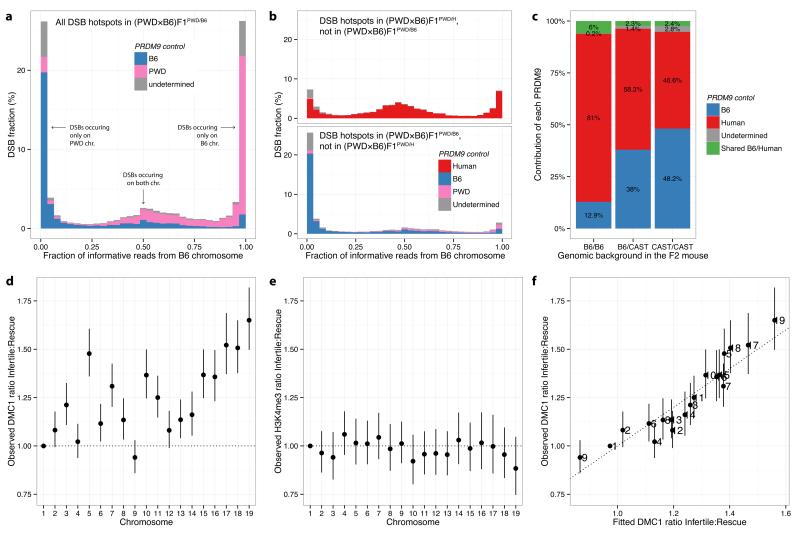
Figure 2. DSB hotspot asymmetry in hybrids.
a, Distribution of the fraction of reads originating from the B6 chromosome in the infertile (PWD×B6)F1PWD/B6. PRDM9 control at each hotspot is attributed to B6 (blue), PWD (pink) or undetermined (grey). b, As a, but for non-shared hotspots, unique to either the rescue (PWD×B6)F1PWD/H (top) or the infertile (PWD×B6)F1PWD/B6 (bottom). c, Relative contributions of B6 and humanized PRDM9 to DMC1 signal in (B6/CAST)F2B6/H. Bars represent the three possible genomic backgrounds. d, Individual chromosome effects (relative to Chromosome 1) when comparing DMC1 signals in (PWD×B6)F1PWD/B6 relative to (PWD×B6)F1PWD/H, for shared DSB hotspots. Bars: 1 SE. e, As d, but for H3K4me3. f, Comparison of DMC1 chromosome effects (as in d) with the fitted chromosome effects, using a model including the symmetric hotspot measures for the three Prdm9 alleles. Bars: 3 SE (95% simultaneous confidence level for 19 chromosomes).
Such asymmetry can arise through meiotic drive to favour mutations disrupting PRDM9 binding motifs, within populations where these motifs are active. Any new mutation disrupting PRDM9 binding at a hotspot is preferentially transmitted to offspring: in individuals heterozygous for the mutation, DSBs occur preferentially on the non-mutant chromosome and are then repaired by copying from the mutant chromosome23. This phenomenon has been observed at PRDM9 binding motifs in human13 and mouse17 and causes a rapid accumulation of mutations disrupting PRDM9 binding. B6 and PWD Prdm9 alleles are largely subspecies-specific15, so only the B6 lineage has experienced strong erosion of the B6 binding motif, and only the PWD lineage has experienced strong erosion of the PWD binding motif. This asymmetric erosion explains the highly asymmetric PRDM9 binding sites in F1 hybrids.
Because the human allele has not been present in mice, its binding sites have not experienced erosion in the mouse genome. As a consequence, DSBs at hotspots attributable to the human allele occur mostly (57%) in symmetric hotspots, with the remaining, asymmetric hotspots mainly (84.2%) explained by the presence of mutations that coincidentally fall within the human PRDM9 binding motif (Fig. 2b). Conversely, only 30% of DSBs at hotspots attributable to the B6 allele occur in symmetric hotspots. An identical pattern is seen in the reciprocal crosses. Thus, a genome-wide effect of humanizing the mouse is to reprogram hotspot positions with the consequence that hotspot asymmetry is reduced in the hybrids.
Meiotic drive might also explain dominance, as seen for the human Prdm9 allele over the B6 allele in the B6B6/H mouse, because B6 motifs are heavily eroded on the B6 background. To test this, we created F2 mice to analyse the behaviour of the B6 and humanized alleles on a neutral Mus musculus castaneous (CAST/EiJ) background which has been unaffected by B6 motif erosion (Extended Data Fig. 6h). Dominance of the human allele disappeared in regions of the genome with two copies of the CAST genome – removing the effect of motif erosion removes the dominance (Fig. 2c). This result excludes some factors which might influence dominance (Supplementary Information), and also suggests that recently arisen Prdm9 alleles might be dominant over older alleles, for which meiotic drive will have had more time to degrade binding motifs.
Chromosome-specific trans effects of humanization
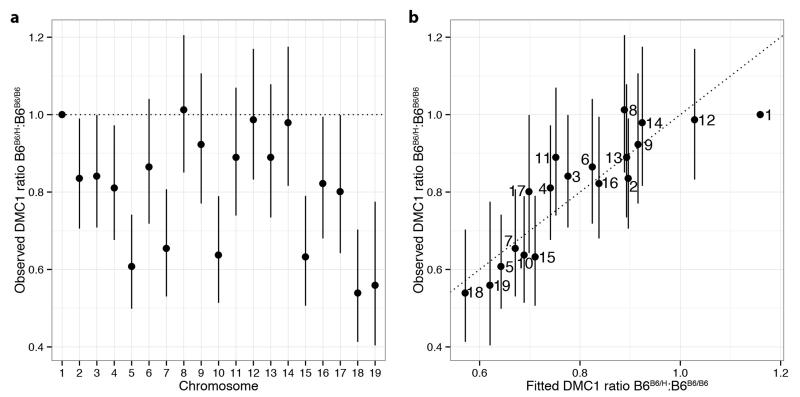
The infertile and humanized rescue mice share some hotspots, controlled by the PWD allele. These shared hotspots show strong correlation (r2=0.63) in DMC1 heat, but nevertheless far weaker than that between hotspots in the infertile and reciprocal mice (0.95). To explore this weaker correlation, we compared DMC1 heats in the two mice for each shared hotspot and calculated their ratio. We observed substantial differences in these ratios across different chromosomes (Fig. 2d). Thus, substituting the B6 allele for the human allele impacts hotspots that neither allele binds directly, in trans, and this impact is observed at broad genomic scales. This trans effect might reflect differences in either the formation, or downstream processing, of DSBs. In contrast to DMC1, the H3K4me3 heat showed no significant chromosomal ratio differences (Fig. 2e), implying the trans effect likely operates downstream of PRDM9 binding. Furthermore, comparison of DMC1 heats between B6B6/B6 and B6B6/H mice also revealed chromosome effects (Extended Data Fig. 7). This implies that such trans effects do not depend on SNP presence (the B6 background is fully homozygous), and cannot simply be a consequence of asynapsis (observed only in the infertile mouse).
Next, we sought to understand the drivers of these chromosome-specific differences in DMC1 heat by testing various potential predictors of these differences between the infertile and humanized rescue mice (Supplementary Information). After exhaustive search over possible models, given the predictors considered, the best-fitting model was highly predictive (r2=0.84; Fig. 2f) and included only symmetric hotspot measures – the total H3K4me3 signal from PRDM9 binding on both homologues (i.e. symmetrically) at the same hotspots, summed over the entire chromosome – for each of the three Prdm9 alleles (p<0.01 in each case). The trans effect is thus explained by knowledge of only the direct differences in PRDM9 binding targets across mice – without any additional information regarding other features such as SNP diversity – consistent with the sole difference between the infertile and rescue mice being the ZF-array of Prdm9. Moreover, only symmetric hotspots (in the infertile mouse, a minority) provide predictive power.
The fitted model implies that lower overall symmetric binding results in increased DMC1 heat, at a chromosome level. The same properties (p<0.0002; Supplementary Information) hold true in the comparison between B6B6/B6 and B6B6/H mice. Although the B6 background is completely homozygous, so PRDM9 is predicted to mark H3K4me3 equally on both homologues, different total levels of H3K4me3 marking across chromosomes still occur and these correlate with observed differences in DMC1 heat between the two genotypes. This excludes sequence differences at or near hotspots, or asynapsis itself, as a cause, and suggests that the total amount of symmetric binding on each chromosome, as opposed to a simple lack of asymmetric binding, plays an important role in predicting DMC1 heat. The direction of causality is reasonably clear (binding predates DSB formation, and the H3K4me3 mark lacks similar chromosome effects), while confounding influences should always be shared between the mice being compared and thus cannot alone explain the observed inter-chromosomal differences. It therefore appears differences in the level of overall symmetric binding by PRDM9 drive downstream trans effects at chromosomal scales, with lower symmetric binding somehow increasing the number, or repair time, of DSBs even at distant hotspots.
PRDM9 binding symmetry and synapsis
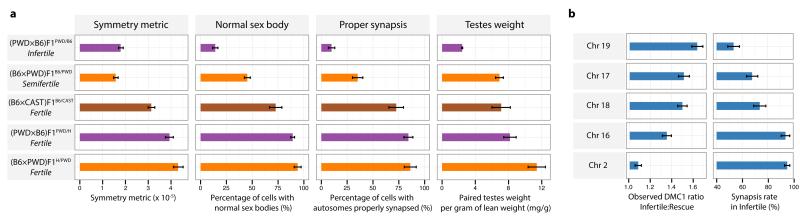
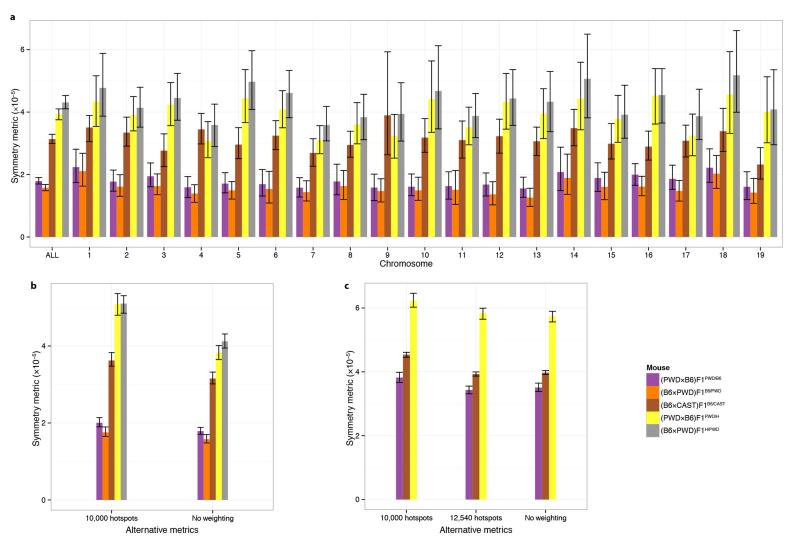
Sterile (PWD×B6)F1PWD/B6 hybrids show very high rates of asynapsis, particularly at specific chromosomes5, and failure to form the sex body during early meiosis5,9. In contrast, these phenotypes are completely rescued in (PWD×B6)F1PWD/H hybrids harbouring the humanized Prdm9 allele (Fig. 1c,d). Having seen a relationship between PRDM9 binding symmetry and the recombination process, we examined binding symmetry in relation to fertility. For different male mice, we measured three quantitative fertility phenotypes24 (Fig. 3a), and calculated several genome-wide measures of hotspot symmetry (Extended Data Fig. 8; Supplementary Information). We observed a significant correlation (p = 0.0083; rank correlation permutation test) between the DMC1 symmetry measures and the rate of proper synapsis among all nine mice studied. In humanized hybrid mice, the observed increase in symmetry was accompanied by improved fertility. Strikingly, this improvement effect is stronger than the Hstx2 modifier, responsible for the difference in asynapsis and fertility observed between the sterile and reciprocal hybrids5 (Fig. 3a). An additional mouse hybrid, (B6×CAST)F1B6/CAST, showed intermediate PRDM9 binding symmetry17 and also an intermediate asynapsis level. Symmetry measures in homozygous mice (PWD, B6B6/B6, B6B6/H, B6H/H) are as expected much higher than hybrids, and these mice show the highest synapsis rates and fertility measures.
Figure 3. Humanizing PRDM9 restores proper synapsis and rescues fertility in hybrids.
a, Fertility metrics in hybrid mice. Bars: bootstrap 95% CI (symmetry metric), or 1 SE. b, Chromosome effects in DMC1 signals (as Fig. 2d) versus previously reported5 asynapsis rates for five chromosomes in infertile (PWD×B6)F1PWD/B6. Bars: 1 SE.
Previous work5 showed that in the infertile (PWD×B6)F1 mouse, synapsis failure occurs at different rates among five chromosomes tested. We compared the reported asynapsis rates for these five chromosomes with the chromosome-specific DMC1 heat effects described above and found an identical ranking (p=0.017 by rank correlation permutation test; Fig. 3b). Because these DMC1 heat effects are strongly predicted by symmetric H3K4me3 levels in the infertile mouse, this result implies that chromosomes with lower symmetric PRDM9 binding experience higher asynapsis rates. This may explain why lower symmetric PRDM9 binding genome-wide accompanies higher overall asynapsis rates among different mice.
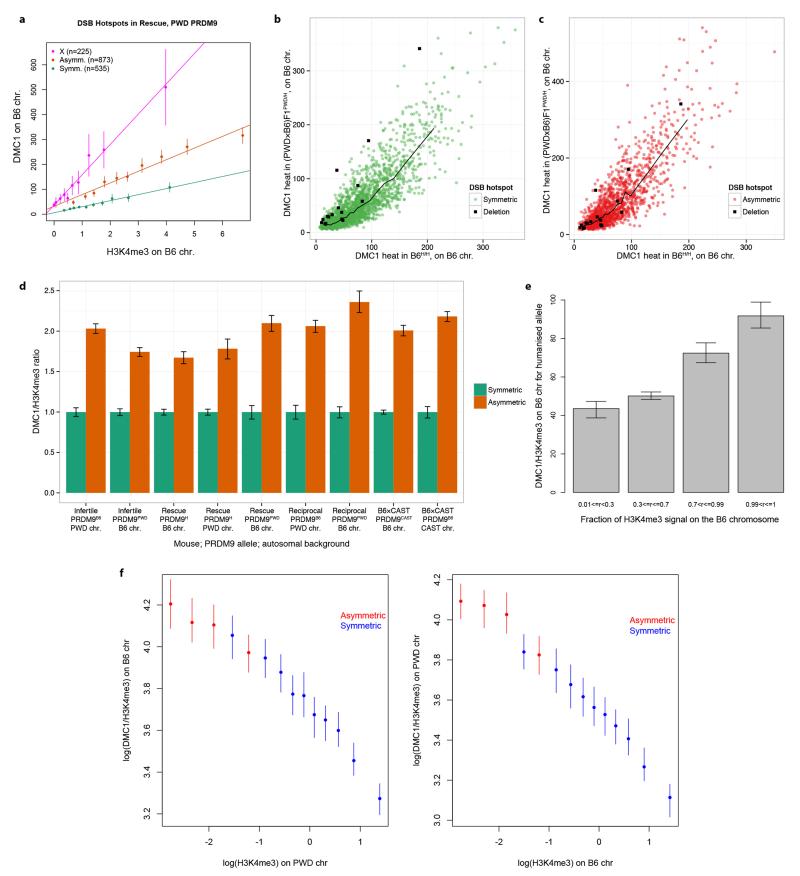
Having found elevated DMC1 heat on chromosomes influenced by asynapsis (where homologous pairing fails), we examined DMC1 and H3K4me3 heats in two additional settings, where no homologue exists at all and thus homologous chromosome pairing cannot occur: the X-chromosome in male mice, and separately in humanized hybrid mice at autosomal hotspots where the human PRDM9 binding motif lies within a region deleted in the PWD genome. In both these settings, we observed an elevation of DMC1 heat relative to autosomal hotspots bound symmetrically by PRDM9 (Extended Data Fig. 9). Elevation of DMC1 heat might, therefore, be a consistent signature of non-pairing of homologous chromosomes during meiosis. DMC1 elevation might be explained by an increased probability of a DSB occurring at that site, or by the DMC1 coating at breaks persisting for longer (delayed repair). However, the total number of RAD51-marked DSBs initiated per cell is tightly regulated25, remaining unchanged even in Prdm9 knock-outs26, while in both knock-outs and infertile hybrids, DSB marks indeed persist late into pachytene suggesting a failure of repair5,9,26. Therefore, the elevated DMC1 signals we observe may be explained by persistence of DMC1 where homologous repair is compromised or delayed.
PRDM9-dependent homologue interactions
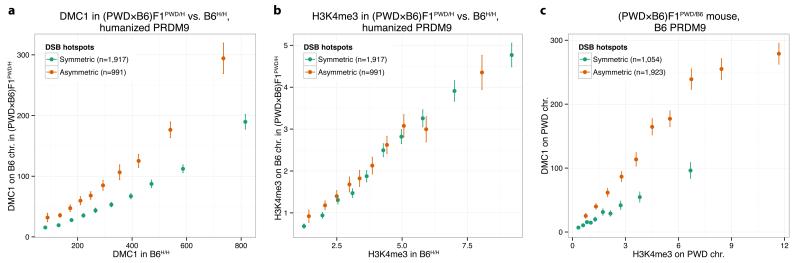
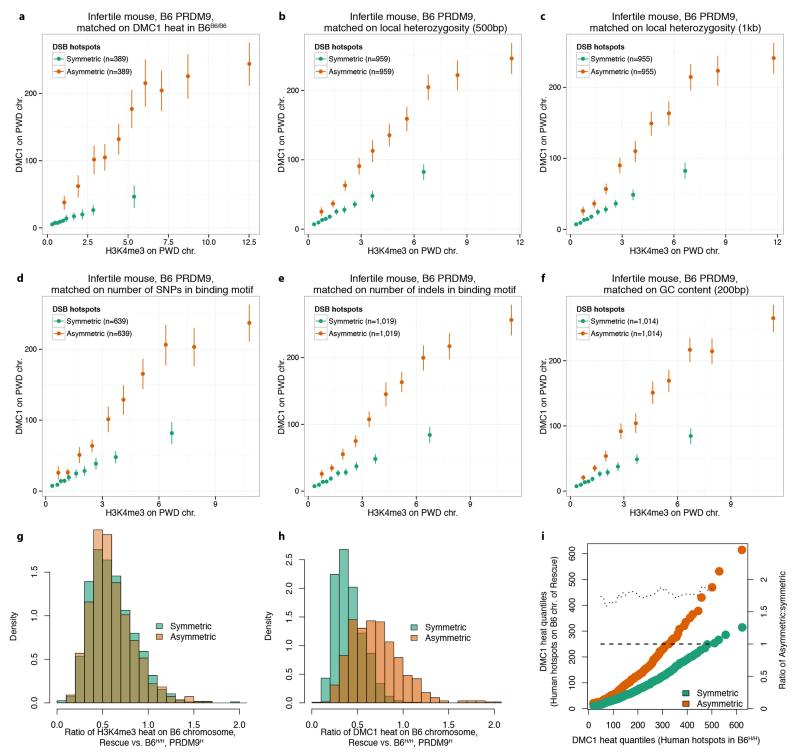
Given our chromosome-scale observations, we next asked whether symmetric binding at individual hotspots might also influence DMC1 heat. At each human-controlled hotspot in the humanized rescue, we measured the component of total DMC1 heat contributed by the B6 chromosome only, and compared this to the DMC1 heat for the same hotspot in B6H/H. The comparison revealed (Fig. 4a) a remarkably strong, and clear, elevation in DMC1 heat in the hybrid mouse for the asymmetric hotspots (>90% asymmetry, towards binding of only the B6 chromosome), relative to the symmetric hotspots (those within 10% of complete symmetry). However, similar to the chromosomal analysis, H3K4me3 enrichment showed no difference whatsoever between symmetric and asymmetric sites in these mice (Fig. 4b). Indeed a comparison of H3K4me3 and DMC1 heat revealed a far higher (Fig. 4c) ratio of average DMC1 heat to H3K4me3 enrichment for asymmetric relative to symmetric hotspots, across all hybrid mice, backgrounds, and Prdm9 alleles tested (Extended Data Fig. 9d). This effect reflects a consistent elevation of DMC1 heat at DSB sites on individual chromosomes when the homologue is not bound strongly (Extended Data Fig. 9e,f). This phenomenon cannot easily be explained by factors including local heterozygosity within or outside the PRDM9 motif, the type of mutation(s) disrupting PRDM9 binding, or outlier effects (Extended Data Fig. 9,10; Supplementary Information Section 13).
Figure 4. Asymmetric DSB hotspots show elevated DMC1 signals but no H3K4me3 elevation.
a, Comparison of B6H/H and (PWD×B6)F1PWD/H DMC1 signals (medians shown). Signals are compared for symmetric and asymmetric hotspots in (PWD×B6)F1PWD/H, on the shared B6 chromosomes. Bars: 95% CIs. b, As a, but for H3K4me3. c, Comparison of DMC1 signals in (PWD×B6)F1PWD/B6, at symmetric and asymmetric hotspots, binned by H3K4me3 enrichment (medians shown). H3K4me3 and DMC1 signals are estimated on the PWD chromosome only, for hotspots associated with B6 PRDM9.
Thus, elevation of DMC1 heat on the bound chromosome appears to be a universal feature of hotspots where PRDM9 binds asymmetrically, relative to symmetrically bound hotspots. In contrast, the results for H3K4me3 suggest the mark is deposited in an independent manner on each homologue. This implies the DMC1 heat elevation depends on a process involving symmetric PRDM9 binding, downstream of H3K4me3 deposition, involving both homologues. While we cannot exclude the possibility that somehow more DSBs occur at asymmetric hotspots, this would require early, precise, pairing of homologues, at least at hotspots, prior to DSB formation, to determine which hotspots are symmetrically bound. Although there is some evidence of pre-meiotic homologue association27, current data do not suggest the existence of precise pairing prior to DSB formation28. The alternative and more plausible explanation is that sites where PRDM9 binds asymmetrically simply experience a delay in DSB processing, delaying DMC1 removal compared to symmetric DSB hotspots. Whilst our data represent the collective behaviour of populations of cells, this model suggests a mechanism of PRDM9-dependent interaction between homologues influencing downstream DSB processing operating within individual cells, which we discuss below (also Supplementary Information Section 14).
Discussion
Only one mammalian speciation gene, Prdm9, has so far been identified. Humanizing the ZF-array of Prdm9 redirects binding, thereby entirely reprogramming recombination hotspots and in doing so reverses the hybrid infertility between musculus and domesticus subspecies. This modification mimics the consequences of a newly arising allele and thus suggests that Prdm9 evolution (e.g. rapid fixation of particular existing variants3,15 or novel alleles arising by mutation) in either or both subspecies would also restore hybrid fertility.
Multiple lines of evidence in our data, at chromosomal, whole organism, and individual hotspot scales, strongly suggest novel roles for PRDM9 in the formation or processing of DSBs downstream of H3K4me3 deposition, dependent upon symmetric binding. Several aspects of our, and published, data (comparison between B6B6/B6 and B6B6/H mice, see also Supplementary Information) also mean that our results cannot be fully explained simply by sequence differences within or around hotspots, which do not specifically impact binding symmetry.
Pervasive asynapsis is proposed to be the underlying cause of infertility in hybrid mice5. We observed a positive relationship between symmetric PRDM9 binding and correct synapsis of homologous chromosomes later in meiosis. Replacing the B6 allele with the humanized allele in hybrids greatly increases symmetric binding, restoring proper synapsis and fertility. Many apparently complex relationships have previously been reported between naturally occurring mouse Prdm9 alleles, allelic dosage, and quantitative fertility measures in hybrids9. Each of ten manipulations shown or predicted to increase PRDM9 binding symmetry also increases meiotic success and fertility (Supplementary Information), supporting the idea that the link between binding symmetry and fertility might be very general, and causal.
The erosion of PRDM9 binding sites through meiotic drive17 also occurs at human hotspots13 and likely across many mammals. In two populations separated for sufficient time, differential PRDM9 binding site erosion will decrease symmetry in hybrids, which is likely to decrease fertility levels (though not necessarily to the extreme of sterility). Therefore, PRDM9 may affect hybrid fertility levels across many mammalian species and so might repeatedly act in driving early speciation steps, although the rapid evolution of PRDM9’s ZF-array implies an unexpected transience of this direct role. However, even subtle or transient PRDM9-driven reductions in fertility might still provide a selective advantage to additional mutations contributing towards speciation. This mechanism is different from the previously characterized causes of intrinsic hybrid incompatibilities, such as differences in ploidy, chromosomal rearrangements, or incompatibilities between genes. The extent to which it has been responsible for speciation in the natural world appears an interesting question for further research.
One plausible mechanism for the impacts of (a)symmetry involves a role for PRDM9 binding in aiding homology search - a process thought to involve invasion of the homologous chromosome to probe for homology by single-stranded DNA formed around DSBs29. It has been suggested that synaptonemal complex proteins are loaded at some DSB sites and synapsis begins to spread7,8. Extending this model, to incorporate the property that asymmetrically bound sites are less favourable for homology search, would parsimoniously predict each symmetry-related phenomenon we observed: DSBs at asymmetric hotspots would repair more slowly, elevating their DMC1 signal, and chromosomes with fewer symmetric hotspots overall would show delayed DSB repair and higher asynapsis rates, ultimately causing subfertility or sterility in animals with low symmetric binding. It is not known how homology search occurs efficiently in the nuclear environment, given the enormous potential search-space of the genome30, or why hotspots exist at all. Both phenomena could be explained by the above model in which homology search is focussed at least partly on hotspot positions. Indeed hotspots might massively increase search efficiency by directing homology search to PRDM9 binding sites.
Methods
Gene targeting in embryonic stem cells
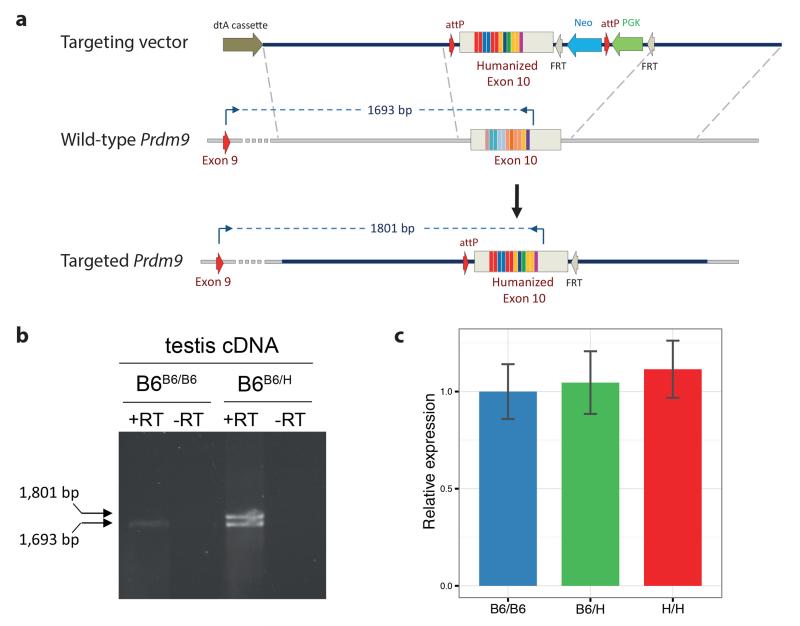
A C57BL/6J (B6) mouse genomic BAC clone (RP23-159N6) encompassing the Prdm9 gene was used for subcloning of homology regions. A 7 kb XmaI / SpeI fragment upstream of exon 10 and a 2.5 kb BamHI / SpeI fragment downstream of exon 10 were used as 5′ and 3′ homology regions, respectively. The intervening 4 kb SpeI / BamHI encoding exon 10 and flanking intronic regions were subcloned and an internal 1.4 kb BglII-NheI fragment, containing the coding region of the zinc finger array, was replaced with a synthesized fragment (Life Technologies) encoding the ZF-array from the human B allele. All coding sequence 5′ of the first zinc finger and all 3′ untranslated regions (UTR) downstream of the stop codon were left as mouse. This humanized fragment was then assembled between the two homology arms, upstream of a neomycin selection cassette. PhiC31 attP sites were incorporated immediately downstream of the 5′ homology arm and between the PGK promoter and the neomycin phosphotransferase open reading frame to equip the locus with PhiC31 integrase cassette exchange machinery for subsequent manipulations31.
The completed targeting vector was linearised with ApaI and electroporated into mycoplasma free C57BL/6N JM8F6 embryonic stem cells (Extended Data Fig. 1a). JM8F6 cells were a gift from Dr. Bill Skarnes, Wellcome Trust Sanger Institute. Following selection in 210 μg/ml G418, recombinant clones were screened by PCR to detect homologous recombination over the 3′ arm. A forward primer (5′-TACCGGTGGATGTGGAATGTG-3′) binding within the PGK promoter was used together with a reverse primer (5′-TGACAGCAAAAACCACCTCTA-3′) binding downstream of the 3′ homology arm to amplify a 2.7 kb fragment from correctly recombined clones. Positive clones were examined for correct recombination at the 5′ end by long range PCR using a forward primer (5′-CAGAGGACCTTTAGTCTGTGAGGG-3′) binding upstream of the 5′ homology arm and a reverse primer (5′-AGCAGAGGCTTGACCTATCGCTAA-3′) binding within the humanized region. Correctly targeted clones yielded a 10.4 kb amplicon. Sanger sequence analysis of the 10.4 kb amplicon encompassing the 5′ homology arm with primer 5′-CCTTTCTCAATGATCCACAAAT-3′ confirmed the correct integration of the 5′ attP sequence, necessary for future manipulations of the locus. Southern blotting using a probe against neomycin was used to confirm that only a single integration event had occurred.
Mouse production and matings
Mice were housed in individually ventilated cages and received food and water ad libitum. All studies received local ethical review approval and were performed in accordance with UK Home Office Animals (Scientific Procedures) Act 1986. Experimental groups were determined by genotype and were therefore not randomized, with no animals excluded from the analysis. Sample size for fertility studies and cytogenetics (see below) were selected on the basis of previously published studies5,9,32. All phenotypic characterization was performed blind to experimental group.
ES cells from correctly targeted clones were injected into albino C57BL/6J blastocysts and the resulting chimeras were mated with albino C57BL/6J females. Successful germline transmission yielded black pups and F1 mice harbouring the humanized Prdm9 allele were identified using the above attP screening PCR. F1 heterozygous male mice were bred with C57BL/6J Flp recombinase deleter mice (Tg(ACTB-Flpe)9205Dym (Jax stock 005703)) and offspring were screened for the deletion of the selection cassette using a forward primer (5′-TTCTGCCATCACTTCCTTCGGTGA-3′) binding immediately upstream of the cassette and a reverse primer (5′- TCTGAAGCCCAACTATTTCATTAATACCCC-3′) binding immediately downstream of the cassette. A 677 bp amplicon was obtained from the Flp deleted humanized allele and a 491 bp amplicon was obtained from the wild-type allele. Heterozygous humanized mice without the selection cassette were then backcrossed with C57BL/6J to remove the Flp transgene prior to intercrossing to obtain experimental cohorts of heterozygous, homozygous and wild-type mice which were genotyped with the above PCR. PWD/PhJ mice were a kind gift of Prof. Jiri Forejt, Institute of Molecular Genetics, Prague, Czech Republic and CAST/EiJ were sourced from MRC Harwell.
Fertility was assessed in male mice between the ages of 2 and 4 months by recording the average number of pups obtained when bred with 7-week-old wild-type C57BL/6J female mice. Paired testes weight was recorded and normalized against lean body weight, as assessed using EchoMRI-100 Small Animal Body Composition Analyzer.
Immunohistochemistry analyses
Spermatocytes from mice at approximately 9 weeks of age were prepared for immunohistochemistry by surface spreading33,34. Briefly, the testis tunica was removed, the tubules cut with a razor blade and disassembled by pipetting, in PBS, containing protease inhibitors (Complete, Roche). Following centrifugation at 5800g for 5 minutes, the cells were resuspended in 0.1M sucrose, and spread onto the surface of slides in a drop of 1% paraformaldehyde in PBS. The slides were left to dry for 3 hours at room temperature, in a humidified box, then washed in 0.4% Photo-Flo 200 (Kodak), and either used immediately, or stored at −80°C. For immunohistochemistry the following antibodies were used: mouse anti-MLH1 (BD, 51-1327GR); mouse anti-phospho-H2A.X (Millipore 05-636, clone JBW301); rabbit anti-SYCP1 (Novus Biological, NB300-229); rabbit anti-DMC1 (Santa Cruz Biotechnology sc-22768, H-100); mouse anti-SYCP3 (Santa Cruz Biotechnology sc-74569, D-1); rabbit anti-SYCP3 (Abcam ab15093). Non-specific binding sites were blocked by incubating the cells with 0.2% BSA, 0.2% gelatin, 0.05% Tween-20 in PBS (B/ABD buffer). Cells were incubated with the primary antibodies overnight at 4°C. Following washes in B/ABD buffer and detection with secondary antibodies, the slides were mounted in DAPI/Vectashield (Oncor) and analysed with an Olympus BX60 microscope for epifluorescence, equipped with a Sensys CCD camera (Photometrics, USA), using Genus Cytovision software (Leica).
Spermatocytes were staged based on SYCP3 staining. For MLH1 analysis, only pachytenes with 19 or more foci, colocalising with SYCP3, were considered, according to criteria defined by ref. 35. For DMC1 analysis, randomly selected cells, from any stage, were scored. The number of DMC1 foci per cell was counted using the PointPicker macro in ImageJ64. For SYCP1 analysis, only cells in pachytene were considered. Cells with 19 fully synapsed autosomes, with colocalising SYCP1 and SYCP3 signals, and one XY body, were considered normal. For characterisation of gamma-H2AX, cells in pachytene or diplotene were scored, and we considered normal those where only a clearly identifiable XY body was covered by gamma-H2AX signal.
Prdm9 expression via RT-PCR analysis
To verify the correct expression of the humanized Prdm9, we performed exon-spanning endpoint RT-PCR on whole testis cDNA prepared using Tetro reverse transcriptase (Bioline) using a forward primer binding to exon 9 (5′-CATTAAGTGGGGAAGCAAGA-3′) and a reverse primer binding within the 3′ UTR, immediately downstream of the humanized zinc finger domain encoded by exon 10 (5′-GGGATTTAATTCCCTTTTCTAGTCA-3′) (Extended Data Fig. 1b). Q-PCR analysis of Prdm9 transcripts was performed using two primer pairs (5′-GAATGAGAAAGCCAACAGCA-3′ and 5′-GGACAACCAGACTGCACAGA-3; 5′-AGCCAACAGCAATAAAACCA-3′ and 5′-GGGATTTAATTCCCTTTTCTAGTCA-3′), amplifying regions within the 3′ UTR, normalizing against a housekeeping gene (Hprt; 5′-AGCTACTGTAATGATCAGTCAACG-3′ and 5′-AGAGGTCCTTTTCACCAGCA-3′) using the Power SYBR Green PCR Master mix (Applied Biosystems) and a BioRad CFX96 cycler as per manufacturer's instructions. Relative expression was calculated using the Livak method. Expression of the humanized Prdm9 allele was unaffected by the genetic manipulation (Extended Data Fig. 1b,c).
Single-stranded DNA sequencing and double-strand break (DSB) detection
Testis cells from B6H/H, B6B6/H, wild-type PWD, the infertile (PWD×B6)F1PWD/B6, the reciprocal semi-fertile (B6×PWD)F1PWD/B6, the humanized rescue (PWD×B6)F1PWD/H, (B6×CAST)F1B6/CAST, (B6/CAST)F2B6/H males were subjected to single-stranded DNA sequencing (SSDS) as previously described19. In addition, we used the sample C57BL/6 (sample 1) from ref. 21 aligned to mm9/NCBI37. This sample was also re-mapped to mm10/NCBI38 with a modified BWA mapper19. Other samples from ref. 21, 9R (sample 2), 13R (samples 1 and 2) and Prdm9 knockout (B6−/−) (sample 1)10, were also used in the comparative analysis of DSB maps (Extended Data Fig. 3e). B6H/H and B6B6/H libraries were prepared in Daniel Camerini-Otero's lab (NIH) and sequenced on a HiSeq 2000 platform, using paired-end reads (read 1: 36bp; read 2: 40bp). These samples were aligned to the mouse mm9/NCBI37 reference genome. Wild-type PWD, the infertile (PWD×B6)F1PWD/B6, the reciprocal semi-fertile (B6×PWD)F1PWD/B6, the reciprocal rescue (PWD×B6)F1PWD/H, (B6×CAST)F1B6/CAST, (B6/CAST)F2B6/H samples were prepared in The Wellcome Trust Centre for Human Genetics and sequenced on HiSeq 2000 and HiSeq 2500 platforms, using paired-end reads (50bp for each read). These samples were aligned to the mouse mm10/NCBI38 reference genome with a modified BWA mapper19. Variation in the number of sequenced fragments results from the difficulty to precisely assess the DNA concentration before sequencing. Only fragments with high mapping quality (at least 20) were retained for DSB hotspot calling, and only one copy of each duplicate fragment was conserved (here, a fragment is duplicated if there exists at least one other fragment mapping to the same genomic position). Supplementary Table 1 gives details about the samples considered in this study.
H3K4me3 ChIP-seq
ChIP-seq was performed as previously described36 with several modifications (noted here). Briefly, the testis tunica was removed, the tubules disassociated with tweezers and fixed in 1% formaldehyde in PBS for 5 minutes followed by glycine quenching (125 mM final conc.) for 5 minutes at room temperature. Following washing steps, pellets were resuspended in 900 μl cold RIPA lysis buffer, dounced 20 times and sonicated in 300 ul aliquots in a Bioruptor Twin sonication bath at 4°C for three 10-minute periods of 30s on, 30s off at high power, then cell debris was pelleted and removed and aliquots were pooled. For each sample, 50 μl of equilibrated magnetic beads were resuspended in 100 μl PBS/BSA and added to the chromatin samples for pre-clearing for 2h at 4°C with rotation. Beads were removed, and 100 μl of pre-cleared chromatin was set aside for the input control. 5 μl rabbit polyclonal anti-H3K4me3 antibody (Abcam ab8580) was added to the remaining pre-cleared chromatin and incubated overnight at 4°C with rotation. 50 μl beads were washed and resuspended as before, then incubated with the chromatin samples for 2h at 4°C with rotation. Beads were then washed and decrosslinked at 65°C as described36, and for input controls, 50 μl of pre-cleared chromatin was used. After descrosslinking, samples were further incubated with 80 μg RNAse A at 37°C for 60 minutes and then with 80 μg Proteinase K at 55°C for 90 minutes. DNA was purified using a Qiagen MinElute reaction cleanup kit.
ChIP and total chromatin DNA samples were sequenced in multiplexed paired-end Illumina libraries, yielding 51bp reads. We prepared two biological replicates plus one genomic input control each for the infertile (PWD×B6)F1PWD/B6, reciprocal (B6×PWD)F1B6/PWD, and rescue (PWD×B6)F1PWD/H mice, yielding roughly 40-50 million usable read pairs per replicate. For the B6B6/B6 and B6H/H mice, we prepared one biological replicate each (yielding 70-80 million usable read pairs per sample) and later split read pairs into pseudoreplicates. Sequencing reads were aligned to mm10 using BWA aln37 (v. 0.7.0) followed by Stampy38 (v. 1.0.23, option bamkeepgoodreads), and reads not mapped in a proper pair with insert size smaller than 10kb were removed. Read pairs representing likely PCR duplicates were also removed by samtools rmdup. Pairs for which neither read had a mapping quality score greater than 0 were removed. Fragment coverage was computed at each position in the genome and in 100bp non-overlapping bins using in-house code and the samtools39 and bedtools40 packages.
DSB hotspot detection and map comparison
To analyse DMC1 data, we developed a novel ChIP-seq peak caller, specific to DSB hotspots, which takes advantage of the shift in the mapping of single stranded DNA (ssDNA) reads between the 5′ and the 3′ DNA strands to call hotspots. These ssDNA segments are a consequence of the resection of DNA ends that accompanies a DSB and are isolated by DMC1 ChIP19. For each hotspot, the caller estimates in particular the centre of the hotspot, and its heat, loosely defined as the number of reads mapping to this DSB hotspot and predicted to represent real signal. The caller handles sample replicates and is able to call hotspots using several samples jointly. Details are given in Supplementary Information. DSB hotspots from two different samples are considered to overlap if their centres are at most 600bp apart. DMC1 hotspot heats have been normalised so that the sum of hotspot heats is identical in each sample (and equals the sum of hotspot heats in B6B6/B6 (sample 1)).
H3K4me3 enrichments have been computed at DSB hotspots identified by DMC1 ChIP-seq, using our previously published method36 (Supplementary Information Subsection 7.1). H3K4me3 hotspots have also been called de novo, without using DSB hotspots, using the same approach36. The de novo calls were used to generate a list of regions likely to be trimethylated independently of PRDM9, by intersecting calls in mice with different Prdm9 alleles. In comparisons involving both DMC1 and H3K4me3 data, we excluded DSB hotspots contained in any of the PRDM9-independent trimethylated regions, and we used H3K4me3 enrichments computed at DSB hotspots (Supplementary Information). We only used de novo calls for analysis in Extended Data Fig. 6d, 6e.
DNA binding motif analyses
We developed a new, Bayesian, approach to identify DNA motifs enriched at DSB hotspots (Supplementary Information). We used FIMO (MEME Suite version 4.9.1) to find the locations of those motifs genome-wide. Using Mus famulus and Mus caroli as outgroups, we reconstructed an ancestral reference genome for B6 and PWD. We could therefore identify on which lineage (B6 or PWD) mutations between these two mouse strains occurred. See Supplementary Information for details.
DSB hotspot assignment in hybrids
Using SNPs between the B6 and PWD genomes, each read pair from a hybrid DSB library (DMC1 ChIP-seq) is assigned to one of the categories “B6”, “PWD”, “unclassified” or “uninformative” using criteria detailed in Supplementary Information. For each DSB hotspot, the ratio of informative reads from the B6 chromosome was then computed as the fraction of “B6” reads mapped within 1kb of the hotspot centre, over the sum of “B6” and “PWD” reads in that region. We followed a similar approach for H3K4me3 ChIP-seq, but we further corrected for background signal.
Chromosome effects
To test for statistically significant differential elevation of DMC1 (or H3K4me3) heats between chromosomes following Prdm9 humanization of the infertile (PWD×B6)F1PWD/B6 mice, we fitted a quasi-Poisson model to these heats, including predictors for each chromosome. Specifically, we fitted , where dinfertile and drescue are the DMC1 heats of a particular hotspot which is shared between the infertile (PWD×B6)F1PWD/B6 and rescue (PWD×B6)F1PWD/H mice and c is a categorical variable which represents the chromosome on which the DSB hotspot occurs. Furthermore, for one of the hybrid mice we considered, for a given autosome, we defined the “total H3K4me3 signal from PRDM9 binding on both homologues (i.e. symmetrically) at the same hotspots, summed over the entire chromosome”, also referred to as “the sum of ‘symmetric’ heats”, as , where ri is the fraction of DMC1 reads coming from the B6 chromosome for hotspot i, hi is the H3K4me3 heat of that hotspot, and the sum is taken over all the hotspots on that chromosome which are under the control of a specific (PWD, B6, or humanized) PRDM9. (Our analyses always refer to this sum of symmetric heats for a specific allele.) When we considered the B6 mouse (which of course has two B6 chromosomes), we defined this sum of symmetric heats to be (which is the special case of the formula above with ri = 1/2, corresponding to all hotspots being fully symmetric). Under the assumptions we describe in the Supplementary Information, this can also be interpreted as being proportional to the expected number of hotspots with PRDM9 bound on both homologues. Details and motivations for defining this quantity are given in Supplementary Information Section 8, together with a slight adjustment we used in practice to provide robustness against outliers in the value of .
We proceeded similarly in the B6B6/B6–B6B6/H comparison. The observed effects reported in Fig. 2d-f and Extended Data Fig. 7 are normalised to the effect for Chromosome 1. Precise definitions for the model, and for the 14 chromosome effect predictors tested, are given in Supplementary Information.
Analysis code availability and source data
Analysis code used for analysis in this study is available at https://github.com/anjali-hinch/hybrid-rescue. The source data generated in this publication has been deposited in NCBI's Gene Expression Omnibus (Accession number GSE73833).
Extended Data
Extended Data Figure 1. Humanization of the zinc finger domain of Prdm9.
a, Top panel: the targeting vector used for the humanization of the ZF-array encoded by a portion of exon 10. Middle panel: wild-type Prdm9 allele. Lower panel: the targeted humanized allele, following the action of Flp recombinase which removes the FRT flanked neomycin selection cassette. The positions of primers used for the exon spanning RT-PCR are shown along with the sizes of the predicted amplification products from cDNA. b, RT-PCR analysis using the exon spanning primers shown in a from testis cDNA prepared from wild-type (B6B6/B6) and heterozygous humanized (B6B6/H) mice. For gel source data, see Supplementary Figure 1. c, Relative expression of the Prdm9 transcript from testis cDNA prepared from wild-type (B6B6/B6), heterozygous (B6B6/H) and humanized (B6H/H) testis cDNA, normalised to Hprt (n=2 for each genotype). Bars: 1 SE.
Extended Data Figure 2. Effects of the humanization of the Prdm9 zinc finger domain on fertility parameters.
a, The average litter size is shown for all combinations of genotype matings. Bars: 1 SE. b, Numbers of DMC1 foci colocalising with SYCP3 immunoreactivity per cell, grouped according to meiotic stage (wild-type (B6B6/B6): n=5 mice; heterozygous (B6B6/H): n=7 mice; homozygous (B6H/H): n=6 mice; cell numbers counted: zygotene: 32, 38, 37; zygotene/pachytene: 55, 96, 90; pachytene: 188, 210, 176; signals on XY in pachytene: 188, 210, 175 for B6B6/B6, B6B6/H and B6H/H, respectively). Mean values are shown c, Number of MLH1 foci per cell in pachytene stage meiotic spreads. (B6B6/B6: n=6 mice, 180 cells; B6B6/H: n=6 mice, 185 cells; B6H/H: n=6 mice, 183 cells). Mean values are shown. d, Comparison of fertility metrics in four mice with homozygous genetic background (B6 or PWD). Across all four mice, there is no statistically significant evidence of differences in these fertility parameters (ANOVA, Bonferroni corrected p-values > 0.08). Bars: 1 SE. e, Average litter sizes in F1 crosses. Bars: 1 SE.
Extended Data Figure 3. Further features revealed by DMC1 signal analysis in mice with homozygous genetic background.
a, Effect of humanization of the Prdm9 zinc finger domain on DSB hotspots. A total of 16,225 and 17,517 DSB hotspots were localized in the homozygous humanized and wild-type mice, respectively. Only 2.6% of these hotspots overlap. b, Correlations between DSB hotspot maps at different scales. Autosomes are divided into bins of given length, and correlations between the sums of the heats of the hotspots falling into each bin are reported, for different bin sizes. Grey region: empirical 95% confidence envelope for the correlation under the null hypothesis of no association between the B6B6/B6 and B6H/H DSB maps. DSB maps for B6B6/B6, 13R, 9R and Prdm9 knockout (B6−/−) mice come from ref. 21. B6B6/B6 and 9R have the same Prdm9 allele, but different genomic backgrounds. c, Breakdown of hotspot provenance (defined by overlap) in the heterozygous humanized mouse for all DSB hotspots (left panel) and for the hottest 20% of hotspots (right panel). d, Distributions of hotspot provenance in the heterozygous humanized mice as a function of the estimated hotspot heats (blue: wild-type B6 mouse, red: humanized homozygous mouse, green: humanized heterozygous mouse, purple: undetermined). The human allele dominates over the mouse allele in terms of heat, as the proportion of DSB hotspots found in the heterozygous mouse that are shared with the homozygous humanized mouse increases with estimated heat. The relative heat/strength of a hotspot is the ratio of this hotspot’s estimated heat to the sum of all the estimated heats (on autosomes). e, Hotter hotspots present a PRDM9 binding motif more often than weaker hotspots in all samples (same colour legend).
Extended Data Figure 4. Inferred PRDM9 binding motifs are enriched at DSB hotspot centres.
a-d, Refined PRDM9 binding motifs detected in the wild-type B6 mouse (a), in the homozygous humanized mouse (b), in the heterozygous humanized mouse (c) and in wild-type PWD (d). Percentages above each motif indicate the fraction of DSB hotspots that are found to harbour this motif, with each DSB hotspot assigned at most to one motif. In logo plots, letter height in bits of information determines degree of base specificity. e-g, Enrichment of the most prevalent 15bp wild-type (blue) and humanized homozygous (red) motifs within 100bp bins across a 5kb window centred on the DSB hotspot centres. Enrichments were computed for the wild-type (e), humanized (f) and heterozygous humanized (g) mice DSB hotspots.
Extended Data Figure 5. Differential epigenetic mark distributions at PRDM9 binding motifs.
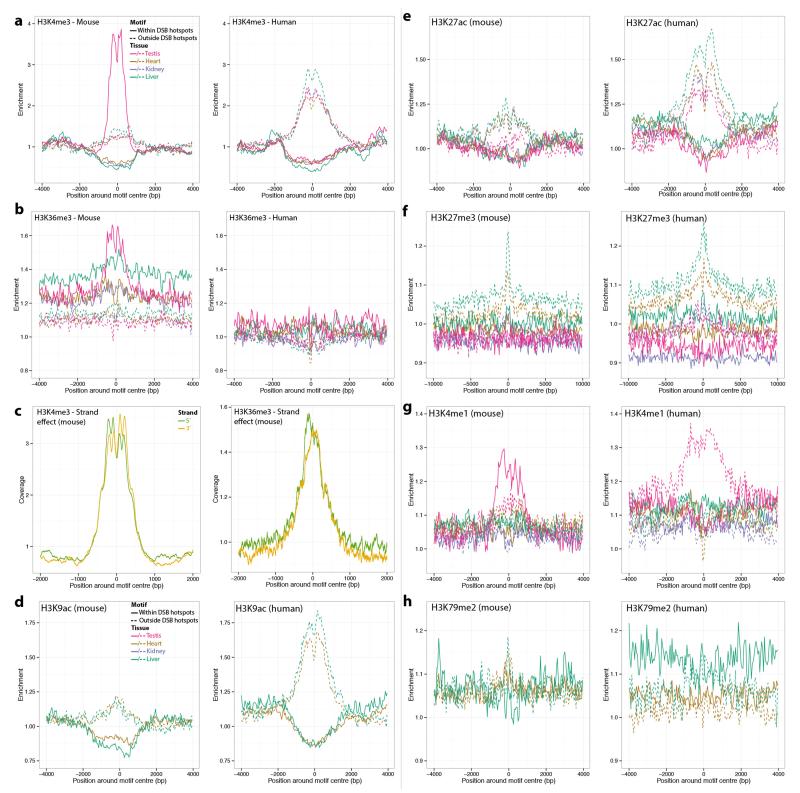
a, Enrichment of H3K4me3 marks at mouse motifs that are either within a B6 (left) or human (right) PRDM9 allele controlled DSB hotspot, or outside such a hotspot. The enrichment is relative to a control genomic track. Given the spread of the distributions, the interaction range between the histones and the DSB hotspot seems to be ~1.5 kb on each side of the motif. b, As a, for H3K36me3 marks. c, Mean coverage of H3K4me3 (left) or H3K36me3 (right) signal around the mouse motif nearest to each B6 DSB hotspot, split according to the strand on which the motif lies. d-h, As a, for H3K9ac (d), H3K27ac (e), H3K27me3 (f), H3K4me1 (g) and H3K79me2 (h) marks. All ChIP-seq data for histone modifications used in this analysis were obtained from the Mouse Encode Project.
Extended Data Figure 6. Further features of DSB hotspot asymmetry.
a-c, DSB hotspot asymmetry in (B6×PWD)F1B6/PWD and in (B6×PWD)F1H/PWD. a, Distribution of the fraction of (DMC1) informative reads originating from the B6 chromosome in the reciprocal (B6×PWD)F1B6/PWD mouse. PRDM9 control at each DSB is attributed either to the B6 allele (blue) or the PWD allele (pink) or is undeterminable (grey). b-c, As a, but showing fractions only for non-shared hotspots, unique to either the reciprocal (B6×PWD)F1B6/PWD (b) or the reciprocal rescue (B6×PWD)F1H/PWD (c) mice. d-e, Comparison of the levels of asymmetric binding in the (PWD×B6)F1PWD/B6 and (B6×CAST)F1B6/CAST mice, using H3K4me3 signal. d, Distributions of the fraction of H3K4me3 reads from the B6 chromosome in the two mice. We used raw data from ref. 17 for the (B6×CAST)F1B6/CAST mouse, and processed both data sets in the same way. H3K4me3 heats were capped at the 95th percentile in each case, and only H3K4me3 binding peaks not inferred to be independent of PRDM9 binding (Supplementary Information Section 7), and overlapping with a DMC1 hotspot in the same mouse, were considered. e, Quantile-quantile plot for the distributions shown in d (blue). Dark grey: y=x line; light grey: 95% confidence band. f, Density plot comparing, for each hotspot in the (PWD×B6)F1PWD/B6 mouse, its DMC1 and H3K4me3 asymmetries. The correlation between the two measures is 0.93. g, Mutations within 1kb regions around B6 and PWD PRDM9 motifs, on the B6 and PWD genomes. Main plot: For each combination of motif and lineage (PWD or B6), we plot the fraction of 30bp windows, along the 1kb regions surrounding motif occurrences within DSB hotspots, where at least one SNP or indel mutation occurred along the respective lineage. Inset plot: Distribution of motif score differences (derived-ancestral) for motif changes shown in the main plot. Motif score was defined as the logarithm of the probability that a motif was drawn from the motif’s position weight matrix, in the ancestral sequence and in the current-day mouse. A negative difference indicates the motif match worsened along the corresponding lineage. This panel is based on the (PWD×B6)F1PWD/B6 DMC1 map. h, Mutations within 1kb regions around B6 PRDM9 motifs, on the B6 and CAST genomes, as in g, using the (B6×CAST)F1B6/CAST DMC1 map. We see no evidence of erosion of B6 PRDM9 motifs on the CAST genome.
Extended Data Figure 7. Chromosome effects following Prdm9 humanization in B6B6/B6.
a, Individual chromosome effects (relative to chromosome 1) when comparing DMC1 signals in the B6B6/H mouse relative to the B6B6/B6 mouse, for the DSB hotspots that are shared between these two mice. b, Comparison of the observed chromosome effects for DMC1 signals with the fitted chromosome effects, using the 2-predictor model including the sum of symmetric H3K4me3 heats in B6B6/B6 and in B6H/H. Bars conservatively show 3 SEs in both plots.
Extended Data Figure 8. Value by chromosome and sensitivity analysis for the symmetry metric.
a, Symmetry metric, as defined in the main text, for each sample (ALL), and for each autosome amongst those samples. Error bars represent bootstrap 95% CIs in all panels. b, Alternative symmetric metrics (to the ones reported in the main text), using only 10,000 hotspots per sample, or without weighting each chromosome specific metric, to compute the average metric genome-wide. Both metrics are computed using the DMC1 maps. c, Alternative symmetric metrics using H3K4me3 maps, similarly to b. The threshold of 12,540 hotspots per sample corresponds to the number of hotspots with ratio estimates in the (PWDxB6)F1PWD/H mouse, which was the lowest amongst the three samples shown here.
Extended Data Figure 9. Asymmetric hotspots, hotspots on the X chromosome and hotspots opposite deletions show systematic increase of DMC1 heat, relative to symmetric hotspots.
a, For the PWD allele in the humanized rescue (PWD×B6)F1PWD/H mouse, mean DMC1 signal is plotted in decile bins of H3K4me3 enrichment on the B6 chromosome (or the PWD X-chromosome), with error bars showing 95% CIs and lines of best fit (as in Fig. 4c). The slope of the line for asymmetric hotspots is 2.5-fold greater than that of the symmetric hotspots, and the slope for hotspots on the X-chromosome is 5.2-fold greater, illustrating that the DMC1 signal at asymmetric sites is elevated in a similar fashion to hotspots on the X-chromosome, which do not repair until late in meiosis. We found similar results in all cases tested. b, Comparison of DMC1 heats on B6 chromosome for hotspots shared between the humanized B6H/H and the humanized rescue (PWD×B6)F1PWD/H mice, under humanized PRDM9 control. We show symmetric hotspots (fraction of DMC1 informative reads between 0.4 and 0.6, green), and hotspots opposite a deletion on the PWD chromosome (deletion of at least 200bp, encompassing a human PRDM9 binding motif, black). The black line represents the median DMC1 heat for symmetric hotspots. c, As b, but showing the asymmetric hotspots (fraction of DMC1 informative reads above 0.9, red), with the corresponding median line. Hotspots opposite PWD deletion show a significant elevation in DMC1 heat relative to symmetric hotspots (14/16 hotspots above the symmetric median line, p=0.004). This elevation is similar to the one showed by asymmetric hotspots (9/16 hotspots above the asymmetric median line, p=0.80). d, Barplot showing the genome-wide ratio of mean DMC1 heat to mean H3K4me3 enrichment for asymmetric hotspots relative to symmetric hotspots in 9 scenarios studied, each for a different combination of mouse, Prdm9 allele, and haplotype, with error bars representing 95% bootstrap CIs for the ratio of means. In all cases, asymmetric hotspots show an elevation in DMC1 signal for a given H3K4me3 signal. e, Ratio of mean DMC1 and H3K4me3 signals on the B6 chromosome for the humanized allele in the humanized rescue mouse. Hotspots are clustered according to the fractions of their H3K4me3 signal that is on the B6 chromosome (r), and the ratio of the mean DMC1 and H3K4me3 signals in each class is shown here. The whiskers show 95% CIs for the mean, estimated using bootstrapping. When r>0.5, the B6 chromosome has greater H3K4me3 than the PWD chromosome, and vice versa. The ratio could not be estimated for r<=0.01 due to H3K4me3 levels being zero or nearly zero in those cases. f, (Left) Ratio of mean DMC1 and H3K4me3 signals on the B6 chromosome compared with the H3K4me3 signal on the PWD chromosome (log scale) in the infertile mouse. Asymmetric hotspots were defined as those with H3K4me3 fraction on the B6 chromosome > 0.9, and symmetric hotspots were those with the fraction between 0.1 and 0.9. Hotspots that we estimated to be completely asymmetric (H3K4me3 fraction=0 on either chromosome) or those with H3K4me3 enrichment on either chromosome close to zero (enrichment < 0.05) were excluded to avoid singularities on either axis. Asymmetric hotspots were binned into 4 bins of equal size and symmetric hotspots were binned into 10 bins of equal size. Different numbers of bins were used for asymmetric and symmetric hotspots to get approximately similar confidence intervals (error bars represent 95% CIs) to enable comparison. We did not observe many weak symmetric hotspots as we have limited power to detect such hotspots, which is why there are no symmetric bins with very low H3K4me3 levels on the homologue (Right). As (Left), but with the ratio determined for the PWD chromosome relative to H3K4me3 on the B6 chromosome. Accordingly, asymmetric hotspots are defined as those with H3K4me3 fraction on the PWD chromosome > 0.9.
Extended Data Figure 10. Elevation of DMC1 asymmetric heat is not explained by GC content, local heterozygosity, differences in binding motif-disrupting mutations or by outliers.
a-f, Comparison of DMC1 signals in the infertile (PWD×B6)F1PWD/B6 mouse, at symmetric and asymmetric hotspots respectively, binned by H3K4me3 enrichment, after matching symmetric and asymmetric hotspots on various features: a, DMC1 heat in B6B6/B6; b, local heterozygosity outside the PRDM9 binding motif, in a 500bp window; c, as b, but for a 1kb window; d, number of SNPs in binding motif; e, number of indels in binding motif; f, local GC content, computed in a 200bp window around hotspot centre. g, Distributions of the ratios of H3K4me3 heats on the B6 chromosome, in the rescue (PWD×B6)F1PWD/H vs humanized B6H/H mice, for the symmetric (fraction of informative DMC1 reads in the range 0.4-0.6, green) and asymmetric (fraction 0.9-1, orange) hotspots under humanized PRDM9 control shared between the two mice. The distributions are very close, suggesting similar trimethylation by PRDM9 on the B6 chromosome in both mice. h, As g, but for the DMC1 heats. Despite similar trimethylation marking by PRDM9 in both mice, we observed striking changes in the distribution of DMC1 ratios. This could be due to either more breaks occurring at the asymmetric sites, or a longer time taken to repair them. i, Quantile-quantile plots of DMC1 heats for hotspots under the control of the human allele on the B6 chromosome in the rescue (PWD×B6)F1PWD/H (y-axis, left) vs the humanized B6H/H (x-axis) mice, for symmetric (green) and asymmetric (orange) hotspots. Dotted line represents the ratios of asymmetric to symmetric quantiles (excluding distribution tails; y-axis, right). Dashed line represents expected ratio if there were no differences between symmetric and asymmetric hotspots. The observed ratio of DMC1 quantiles is constant across DMC1 heats, emphasizing that the increase in DMC1 heat at asymmetric sites is very similar across the whole range of DMC1 heats, and does not simply result from a few outlying hotspots.
Supplementary Material
Acknowledgments
We thank Nicole Hortin, Sheny Chen and Robert Davies for technical assistance, the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics for the generation of the sequencing data and Robert Esnouf and Jon Diprose for assistance with computing facilities. PWD/PhJ mice were a kind gift of Prof. Jiri Forejt. This work was supported by the Wellcome Trust Core Award Grant 090532/Z/09/Z, Senior Investigator Award 095552/Z/11/Z (to P.D.), Investigator Award 098387/Z/12/Z (to S.R.M.) and the NIDDK Intramural Research Program (R.D.C.O.). E.H. is funded by a Nuffield Department of Medicine Prize Studentship. J.G.H. is an EPAC/Linacre Junior Research Fellow funded by the Human Frontiers Postdoctoral Program (LT-001017/2013-L).
Footnotes
Supplementary Information is available in the online version of the paper.
The authors declare no competing financial interests.
References
- 1.Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11:175–180. doi: 10.1038/nrg2718. 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 2.Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 3.Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS genetics. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregorova S, Forejt J. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies--a valuable resource of phenotypic variations and genomic polymorphisms. Folia biologica. 2000;46:31–41. [PubMed] [Google Scholar]
- 5.Bhattacharyya T, et al. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E468–477. doi: 10.1073/pnas.1219126110. 10.1073/pnas.1219126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharyya T, et al. X chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS genetics. 2014;10:e1004088. doi: 10.1371/journal.pgen.1004088. 10.1371/journal.pgen.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao H, et al. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 2012;8:e1002790. doi: 10.1371/journal.pgen.1002790. 10.1371/journal.pgen.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson KA, Keeney S. Synaptonemal complex formation: where does it start? Bioessays. 2005;27:995–998. doi: 10.1002/bies.20310. 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- 9.Flachs P, et al. Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. PLoS genetics. 2012;8:e1003044. doi: 10.1371/journal.pgen.1003044. 10.1371/journal.pgen.1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 11.Grey C, et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS biology. 2011;9:e1001176. doi: 10.1371/journal.pbio.1001176. 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. 10.1038/ng1001-217. ng1001-217 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nature genetics. 2010;42:859–863. doi: 10.1038/ng.658. 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kono H, et al. Prdm9 Polymorphism Unveils Mouse Evolutionary Tracks. DNA research : an international journal for rapid publication of reports on genes and genomes. 2014 doi: 10.1093/dnares/dst059. 10.1093/dnares/dst059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auton A, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker CL, et al. PRDM9 Drives Evolutionary Erosion of Hotspots in Mus musculus through Haplotype-Specific Initiation of Meiotic Recombination. PLoS Genet. 2015;11:e1004916. doi: 10.1371/journal.pgen.1004916. 10.1371/journal.pgen.1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forejt J, Ivanyi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet Res. 1974;24:189–206. doi: 10.1017/s0016672300015214. [DOI] [PubMed] [Google Scholar]
- 19.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012;22:957–965. doi: 10.1101/gr.130583.111. 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. doi:nature11089 [pii] 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paigen K, et al. The recombinational anatomy of a mouse chromosome. PLoS Genet. 2008;4:e1000119. doi: 10.1371/journal.pgen.1000119. 10.1371/journal.pgen.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31:267–271. doi: 10.1038/ng910. 10.1038/ng910. ng910 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Flachs P, et al. Prdm9 incompatibility controls oligospermia and delayed fertility but no selfish transmission in mouse intersubspecific hybrids. PLoS One. 2014;9:e95806. doi: 10.1371/journal.pone.0095806. 10.1371/journal.pone.0095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange J, et al. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun F, et al. Nuclear localization of PRDM9 and its role in meiotic chromatin modifications and homologous synapsis. Chromosoma. 2015 doi: 10.1007/s00412-015-0511-3. 10.1007/s00412-015-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell. 2013;24:196–205. doi: 10.1016/j.devcel.2012.12.002. 10.1016/j.devcel.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishiguro K, et al. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014;28:594–607. doi: 10.1101/gad.237313.113. 10.1101/gad.237313.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renkawitz J, Lademann CA, Jentsch S. Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol. 2014;15:369–383. doi: 10.1038/nrm3805. 10.1038/nrm3805. [DOI] [PubMed] [Google Scholar]
- 30.Weiner A, Zauberman N, Minsky A. Recombinational DNA repair in a cellular context: a search for the homology search. Nat Rev Microbiol. 2009;7:748–755. doi: 10.1038/nrmicro2206. 10.1038/nrmicro2206. [DOI] [PubMed] [Google Scholar]
Additional references for methods
- 31.Chen CM, Krohn J, Bhattacharya S, Davies B. A comparison of exogenous promoter activity at the ROSA26 locus using a PhiiC31 integrase mediated cassette exchange approach in mouse ES cells. PloS one. 2011;6:e23376. doi: 10.1371/journal.pone.0023376. 10.1371/journal.pone.0023376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniel K, et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nature cell biology. 2011;13:599–610. doi: 10.1038/ncb2213. 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barchi M, et al. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 2008;4:e1000076. doi: 10.1371/journal.pgen.1000076. 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 35.Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinch AG, Altemose N, Noor N, Donnelly P, Myers SR. Recombination in the human Pseudoautosomal region PAR1. PLoS Genet. 2014;10:e1004503. doi: 10.1371/journal.pgen.1004503. 10.1371/journal.pgen.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome research. 2011;21:936–939. doi: 10.1101/gr.111120.110. 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.