Abstract
Microdeletions of chromosomal region 2q23.1 that disrupt MBD5 contribute to a spectrum of neurodevelopmental phenotypes, however the impact of this locus in human psychopathology has not been described. To characterize the structural variation landscape of MBD5 disruptions and the associated psychopathology, 22 individuals with genomic disruption of MBD5 (translocation, point mutation, and deletion) were identified through whole-genome sequencing or cytogenomic microarray at 11 molecular diagnostic centers. The genomic impact ranged from a single base pair to 5.4 Mb. Parents were available for 11 cases, all of which confirmed the rearrangement arose de novo. Phenotypes were largely indistinguishable between patients with full-segment 2q23.1 deletions and those with intragenic MBD5 rearrangements, including alterations confined entirely to the 5′UTR, confirming the critical impact of non-coding sequence at this locus. We found heterogeneous, multi-system pathogenic effects of MBD5 disruption and characterized the associated spectrum of psychopathology, which includes sensory integration disorder, anxiety, self-hugging, bipolar disorder and others. Importantly, unique features of the oldest assessed patient were early-onset dementia and behavioral regression. Analyses also revealed phenotypes that distinguish MBD5 disruptions from seven well-established syndromes with significant diagnostic overlap. This study indicates that haploinsufficiency of MBD5 causes diverse phenotypes, yields insight into the spectrum of resulting neurodevelopmental and behavioral psychopathology, and provides clinical context for interpretation of MBD5 structural variations. Empirical evidence also suggests that disruption of non-coding MBD5 regulatory regions is sufficient for clinical manifestation, highlighting the limitations of exon-focused assessments. These results suggest an ongoing perturbation of neurological function throughout the lifespan, including risks for neurobehavioral regression and early-onset dementia.
Keywords: MBD5, 2q23.1 microdeletion syndrome, chromosomal microarray, next generation sequencing, regression
Introduction
The increasing resolution of genomic technologies, including cytogenomic microarrays and next-generation sequencing, has led to remarkable advances in our understanding of the highly heterogeneous genetic etiology of neurodevelopmental conditions such as autism spectrum disorders (ASD). Among the largest known genetic risk factors contributing to ASD are recurrent structural variations such as chromosomal microdeletions and microduplications. However, interpretation of the associated clinical outcomes is complicated by the varying sizes of these dosage imbalances which typically disrupt many genes. For example, the 16p11.2 microdeletion and microduplication syndromes are associated with a spectrum of features including ASD, schizophrenia, obesity, dysmorphic characteristics, and numerous other neuropsychiatric and behavioral disorders.1, 2 Only a small number of recurrent chromosomal microdeletion syndromes have a single known contributing gene, such as SATB2 in 2q33.13, 4 and EHMT1 in 9q34.34, 5. The localization of these contributing loci provides a route to understanding pathogenic mechanisms as well as deeply characterizing associated phenotypic features of these typically heterogeneous and complex syndromes. In the current study, we perform extensive phenotypic analyses of the diverse psychopathology associated with a single necessary and sufficient causal locus in the chromosome 2q23.1 microdeletion syndrome and describe the clinical features that overlap with multiple known syndromes.
The pathogenesis of 2q23.1 microdeletion syndrome was recently defined by Talkowski et al6 and others7–15 to be due to haploinsufficiency of a single gene, MBD5 (methyl-CpG-binding domain protein 5), through characterization of the minimal region of overlap from non-recurrent genomic alterations in a cohort of 65 cases with chromosomal deletions or translocations. The mRNA isoform 1 of MBD5 comprises 15 exons with exons 1–5 forming the non-translated 5′UTR. Monoallelic expression of this locus was found to be highly penetrant as both partial and complete deletions of MBD5 involving coding and/or non-translated exons resulted in diverse neurodevelopmental phenotypes including ASD, intellectual disability, and seizures. In addition, no alterations of MBD5 exons were observed in 7,878 controls or in the Database of Genomic Variants (DGV).6
The association of MBD5 disruption with ASD and other neurobehavioral features is supported by its expression in the brain and the suggested function of its methyl-CpG-binding domain in heterochromatin and epigenetic regulation.16, 17 In addition, sequencing of the MBD5 coding region identified a missense mutation (p.79Gly>Glu) in this functionally critical domain that was significantly overrepresented in 747 ASD patients compared to 2,043 individuals from an exome sequencing study of patients with disorders of the heart, lungs and blood (NHLBI Exome Sequencing Project; initial p = 0.012, OR = 5.5).6 Notably, the recent completion and revised variant calling in the final exome sequencing analyses of this mutation provides a more robust estimate of its association and effect size in ASD (p = 0.0039, OR = 5.2).18 The methyl-CpG-binding domain is also shared with MECP2, a causal locus for Rett syndrome. Clinical features associated with MBD5 deletions have considerable overlap with Rett syndrome and can also masquerade as Smith-Magenis (SMS), Cornelia de Lange (CdLS), Angelman (AS), Prader-Willi (PWS), Kleefstra, and Pitt-Hopkins (PTHS) syndromes.
We report here phenotypic characterization of 22 individuals with MBD5 structural alterations collected through a multi-institution collaboration, revealing a complex clinical syndrome that involves a diverse range of neurodevelopmental features with variable severity. A significant inclusion in this cohort is the oldest assessed MBD5 deletion patient who provides important confirmatory evidence that behavioral regression is a potential outcome. Interestingly, a subset of these patients with disruptions confined to the non-coding region of MBD5 manifest a similar spectrum of clinical features as those with coding region rearrangements, confirming a pathogenic role for non-coding regulatory domains. We further identify considerable phenotypic overlap between MBD5 disruptions and other known diagnoses, and provide clinical context for syndrome differentiation with an emphasis on neurobehavioral considerations.
Methods
Subjects
Diagnostic screening of postnatal peripheral blood specimens from 17,477 consecutive patients tested in the Mayo Clinic Cytogenetics Laboratory from 2008–2012 using either the 4×44K or 4×180K oligonucleotide-based whole-genome cytogenomic microarray (Agilent Technologies, Santa Clara, CA) identified eight deletions spanning genomic segments including MBD5 (Cases 1–8), six of which were of sufficient size for detection (>100 kb) and had cells available for confirmation by fluorescence in situ hybridization (FISH). Clinical phenotypic information was obtained from medical records following a Mayo Clinic IRB-approved protocol. A multi-institution collaboration of clinical diagnostic laboratories resulted in inclusion of additional patients not previously published from the Greenwood Genetic Center (Cases 9–12), Pathology Associates Medical Laboratories (Cases 13–14), Virginia Commonwealth University (Case 15), Fullerton Genetics Center (Case 16), and Boston Children’s Hospital (Case 17) that were identified with different cytogenomic microarray platforms. Phenotypes were further considered in the analysis of two MBD5 deletion cases reported by Motobayashi et al19 (Motobayashi/Case 18) and Noh et al20 (Noh/Case 19) as well an intragenic frameshift mutation c.150del or p.Thr52Hisfs*31 described by Kleefstra et al5 (Kleefstra/Case 20) and an intragenic indel of -TC at position 148,942,435 in hg18 of exon 9 described by O’Roak et al21 (O’Roak/Case 21), all of which were published after the Talkowski et al study6. The final patient, DGAP235 (Case 22), was enrolled in the Developmental Genome Anatomy Project (DGAP; www.dgap/harvard.edu) based on karyotypic detection of a de novo, balanced translocation 46,XY,t(2;5)(q22;q22), which was found to disrupt MBD5 by massively parallel paired-end sequencing; this case was independent of two previously published translocations targeting MBD5.6
Genetic analysis of translocation case
Whole genome sequencing was performed using large-insert fragments in a mate-pair method that was previously optimized and applied to cases with abnormal karyotypes.4, 22, 23 Sequencing paired-ends of fragments separated by 2,000 bases in genomic distance enabled high coverage of mapped inserts in a genome-wide manner with minimal individual nucleotide coverage required. In brief, genomic DNA was randomly sheared and size selected for 2 kb fragments. A cap adaptor containing an EcoP15I restriction site was ligated to the fragment ends and circularized with an internal adaptor containing a subject specific bar code and a single biotinylated thymine. The circularized fragments were restriction digested, junction fragments were isolated by binding the biotinylated adaptor to streptavidin beads, and an Illumina library was prepared directly on the beads. Multiplexed paired-end 25 cycle sequencing was performed on an Illumina HiSeq 2000 and 45.7% of all reads corresponded to DGAP235 (87,079,316 reads pairs). Quality control was assessed using FASTQC (Babraham Bioinformatics) followed by distributed parallel alignment of FASTQ read data to the human genome reference hg19 using BWA 0.5.924, merging of aligned BAM files with SAMtools 0.1.1824, and coordinate and name-sorting of aligned read-pairs using Picard Tools 1.5.8 (http://picard.sourceforge.net). A single-linkage clustering of anomalous read-pairs was competed with subsequent filtering of clusters based on size, mapping quality, uniqueness, and presence in a previous database of whole genome sequencing samples from a neurodevelopmental cohort4 using a custom pipeline developed in C++ (BamStat) and MATLAB (Mathworks). The average mapped insert was 1,862 bp (standard deviation = 332 bp) and 94.1% of all reads aligned, yielding an average coverage of mappable inserts of approximately 39.2X (Supplementary Figures S1). See Supplementary Table S1 and Supplementary Results for library metrics, alignment information and bioinformatics analysis.
Phenotypic analysis
The developmental history and neurological, behavioral, and physical characteristics of the new patients were assembled. In addition, the original phenotypic data from the 65 cases reported by Talkowski et al6 were examined. Age-dependent characteristics were considered relative to developmental stage. Individuals were considered to have “autistic-like symptoms” if autistic-like behaviors were specifically reported or a diagnosis of pervasive developmental disorder (PDD-NOS) was given. Phenotypic features were classified as present, absent, or not evaluated, and were reported in Table 3 only if present in more than one individual.
Table 3.
Phenotypes in Intragenic MBD5 Disruptions and 2q23.1 Microdeletions Versus Syndromes in the Differential Diagnosis
| Features | SMS1 | CdLS1 | AS1 | PWS1 | Rett1 | Kleefstra1 | PTHS1 | MBD5 | 2q23.1 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Behavioral | |||||||||||
| Aggression/Temper | +2 | + | − | + | − | + | + | + | + | ||
| Anxiety | + | + | − | − | − | − | + | +/−3 | +/− | ||
| Apathy/Catatonia after puberty | − | − | − | − | − | + | − | − | − | ||
| Autistic-like behaviors (stereotypic repetitive) | + | + | + | + | + | + | + | + | + | ||
| Bruxism (teeth grinding) | + | − | − | − | + | − | + | − | + | ||
| Distractibility/Short attention span | + | + | + | − | + | + | − | + | + | ||
| Eye contact (intense) and/or eye pointing | − | − | − | − | + | − | − | − | − | ||
| Hand-flapping | + | − | + | − | − | − | + | + | + | ||
| Hyperactivity | + | + | + | + | + | − | − | − | + | ||
| Hyperphagia | + | − | − | + | − | − | − | + | + | ||
| Inappropriate happy demeanor | − | − | + | − | + | − | + | − | + | ||
| Inconsolable crying | − | − | − | − | + | − | − | − | − | ||
| Insertion of hand in mouth or handbiting | + | − | − | − | − | − | − | +/− | + | ||
| Obsessive-compulsive or routine-bound behaviors | + | + | − | + | − | − | + | +/− | + | ||
| Panic-like attacks | − | − | − | − | + | − | − | − | − | ||
| Self-injurious behaviors including skin and eye picking | + | + | − | + | − | + | + | + | + | ||
| Sensory integration disorder | + | − | − | − | − | + | − | +/− | + | ||
| Sleep disturbances | + | + | + | + | + | + | + | + | + | ||
| Social withdrawal | − | + | − | − | − | − | − | +/− | +/− | ||
| Temperature intolerance | − | + | + | + | − | − | − | − | − | ||
| Central Nervous System | |||||||||||
| Ataxia/Unusual gait | + | + | + | − | + | − | + | − | + | ||
| Breathing abnormalities | − | − | − | − | + | − | + | − | − | ||
| Developmental delay/Intellectual disability | + | + | + | + | + | + | + | + | + | ||
| Feeding difficulties | + | + | + | + | + | − | − | + | + | ||
| Generalized lethargy (infancy) | + | − | − | + | − | − | − | − | − | ||
| Hyporeflexia | + | − | − | + | − | − | − | − | − | ||
| Hypotonia/Abnormal muscle tone | + | + | + | + | + | + | + | + | + | ||
| Low-pitched cry in infancy | − | + | − | − | + | − | − | − | − | ||
| Motor delay | + | + | + | + | + | + | + | + | + | ||
| Oropharyngeal and gastroesophageal incoordination | − | − | − | − | + | − | − | − | − | ||
| Seizures | + | + | + | + | + | + | + | + | + | ||
| Speech delay | + | + | + | + | + | + | + | + | + | ||
| Tremulous movement of limbs | − | − | + | − | + | − | − | − | − | ||
| Velopharyngeal insufficiency/Vocal-fold abnormalities | + | − | − | − | − | − | − | − | − | ||
| Growth/Endocrine | |||||||||||
| Delayed sexual maturation | +** | − | − | + | − | − | − | − | − | ||
| Failure to thrive | + | + | − | + | − | − | − | − | − | ||
| Growth retardation | + | + | − | − | + | − | + | + | + | ||
| Hirsutism | − | + | − | − | − | − | − | − | + | ||
| Hypercholesterolemia | + | − | − | − | − | − | − | − | − | ||
| Low thyroxine/Hypothyroidism | + | − | − | + | − | − | − | − | − | ||
| Obesity | + | + | + | + | − | + | − | − | + | ||
| Precocious puberty | + | − | − | − | − | − | − | − | − | ||
| Short stature | + | + | − | + | + | + | + | + | + | ||
| Craniofacial | |||||||||||
| Cranium | |||||||||||
| Brachycephaly | + | + | − | − | − | + | − | + | + | ||
| Flat occiput | − | − | + | − | − | − | − | − | − | ||
| Metopic ridging | − | − | − | − | − | − | − | − |
|
||
| Microcephaly | − | + | + | − | + | + | + | +/−*** | + | ||
| Plagiocephaly | − | − | − | − | − | − | − | − |
|
||
| Broad forehead | + | − | − | − | − | − | − | + | + | ||
| Ears/Nose | |||||||||||
| Nasal abnormalities | + | + | − | − | − | + | + | − | + | ||
| Outer ear abnormalities | + | + | − | − | − | + | + | + | + | ||
| Eyes | |||||||||||
| Almond-shaped eyes | − | − | − | + | − | − | − | +/− | +/− | ||
| Astigmatism | − | − | + | + | − | − | + | +/− | + | ||
| Coloboma of optic nerve | − | + | − | − | − | − | − | − | − | ||
| Deep-set eyes | + | − | − | − | − | − | + | − | − | ||
| Glaucoma | − | + | − | − | − | − | − | − | − | ||
| Hypotelorism | − | − | − | − | − | − | + | − | + | ||
| Iris anomalies | + | − | − | − | − | − | − | − | − | ||
| Long eyelashes | − | + | − | − | − | − | − | − | − | ||
| Microcornea | + | + | − | − | − | − | − | − | − | ||
| Myopia/Hypermetropia/Corrective Lenses | + | + | − | + | − | − | + | + | + | ||
| Nasolacrimal duct stenosis | − | + | − | − | − | − | − | − | − | ||
| Optic atrophy | − | + | − | − | − | − | − | − | − | ||
| Optic nerve hypoplasia | + | − | − | − | − | − | − | − | + | ||
| Palpebral fissures - downslanting | − | + | − | − | − | − | − | +/− | + | ||
| Palpebral fissures - long | − | − | − | − | − | − | − | − |
|
||
| Palpebral fissures - upslanting | + | + | − | − | − | + | + | +/− | + | ||
| Strabismus | + | + | + | + | + | − | + | − | + | ||
| Synophrys | + | + | − | − | − | + | − | + | + | ||
| Thick/Arched eyebrows | + | + | − | − | − | + | + | +/− | + | ||
| Face | |||||||||||
| Coarse facies | + | − | − | − | − | + | + | − | + | ||
| Midfacial hypoplasia | + | − | − | − | − | + | − | +/− | + | ||
| Mouth/Chin | |||||||||||
| Cleft palate | + | + | − | − | − | − | − | − | + | ||
| Dental abnormalities | + | + | − | − | − | + | + | + | + | ||
| Downturned mouth corners or everted lower lip | − | + | − | + | − | + | − | + | + | ||
| Long, smooth philtrum | − | + | − | − | − | − | − | − | − | ||
| Macroglossia or protruding tongue | − | − | + | − | − | + | − | − | + | ||
| Micrognathia | + | + | − | − | − | − | − | + | + | ||
| Open mouth | + | − | − | − | − | − | − | + | + | ||
| Prognathia | + | − | + | − | − | + | + | − | − | ||
| Thin upper lip | − | + | − | + | − | − | − | + | + | ||
| Wide mouth | − | − | + | − | − | − | + | − | + | ||
| Neck/Thorax/Vertebrae | |||||||||||
| Hypoplastic nipples | − | + | − | − | − | − | − | − | − | ||
| Short neck | − | + | − | − | − | − | − | − | − | ||
| Skeletal | |||||||||||
| Extremities | |||||||||||
| Brachydactyly | + | − | − | − | − | + | − | − | + | ||
| Clubbing of digits | − | − | − | − | − | − | + | − | − | ||
| Fifth finger clinodactyly | + | + | − | − | − | + | + | +/− | + | ||
| Flat feet | + | − | − | + | − | − | + | + | + | ||
| Lower limb amyotrophy | − | − | − | − | + | − | − | − | − | ||
| Sandal gap | − | − | − | − | − | − | − |
|
|
||
| Short fifth digit | − | − | − | − | − | − | − |
|
|
||
| Small hands or feet | + | + | − | + | + | − | + | +/− | + | ||
| Toe syndactyly | + | + | − | − | − | + | − | − | − | ||
| Upper limb reduction defects | − | + | − | − | − | − | − | − | − | ||
| Other | − | − | |||||||||
| Hip dysplasia | − | + | − | + | − | − | − | − | + | ||
| Joint laxity | − | − | − | − | − | + | − | + | − | ||
| Scoliosis | + | + | + | + | + | − | + | + | + | ||
| Gastrointestinal | |||||||||||
| Abdominal bloating from excessive air swallowing | − | − | − | − | + | − | − | − | − | ||
| Bowel malrotation and obstruction | − | + | − | − | − | − | − | − | − | ||
| Constipation | + | − | + | + | + | − | + | + | + | ||
| Gastroesophageal reflux | − | + | + | − | + | + | + | − | + | ||
| Gallbladder dysfunction | − | − | − | − | + | − | − | − | − | ||
| Heart | |||||||||||
| Atrial septal defect | + | + | − | − | − | + | − | − | − | ||
| Ventricular septal defect | + | + | − | − | − | + | − | +/− | +/− | ||
| Pulmonary stenosis | + | + | − | − | − | + | − | − | − | ||
| Tetrology of Fallot | + | + | − | − | − | + | − | − | − | ||
| Unspecified congenital heart defect | + | + | − | − | − | + | − | − | − | ||
| Immune Function | |||||||||||
| Immunoglobulin deficiency | + | − | − | − | − | − | − | − | − | ||
| Skin | |||||||||||
| Cutis marmorata | − | + | − | − | − | − | − | − | + | ||
| Hypopigmentation | − | − | + | + | − | − | − | − | − | ||
| Urogenital | |||||||||||
| Hypoplastic genitalia | − | + | − | + | − | + | + | − | + | ||
| Other urogenital anomalies | + | + | − | − | − | + | − | − | − | ||
Denoted as positive (+) when the feature was present in at least two cases.
Denoted as potentially positive (+/−) when the feature was present in one case of MBD5 plus one or more cases of 2q23.1 and vice versa.
Horizontal hash marks indicate features present in MBD5 and/or 2q23.1 but not in the other syndromes.
The clinical features of the new cohort (n = 22) were assessed for both overlap with and divergence from those cases in the published original cohort that had phenotypic data available (n = 48) to enable a more comprehensive phenotypic profile associated with MBD5 disruption. A list of the 48 cases analyzed from the original cohort of Talkowski et al6 is provided in the Supplementary Results. The phenotypic spectrum of the combined original and new MBD5 disruptions (n = 70) was then compared and contrasted to well-established syndromes with diagnostic overlap (SMS, CdLS, AS, PWS, Rett, Kleefstra and PTHS). In each of these evaluations, cases were grouped by genotype, i.e. intragenic MBD5 disruptions versus overlapping 2q23.1 deletions.
Results
Identification and molecular characterization of MBD5 disruption cases
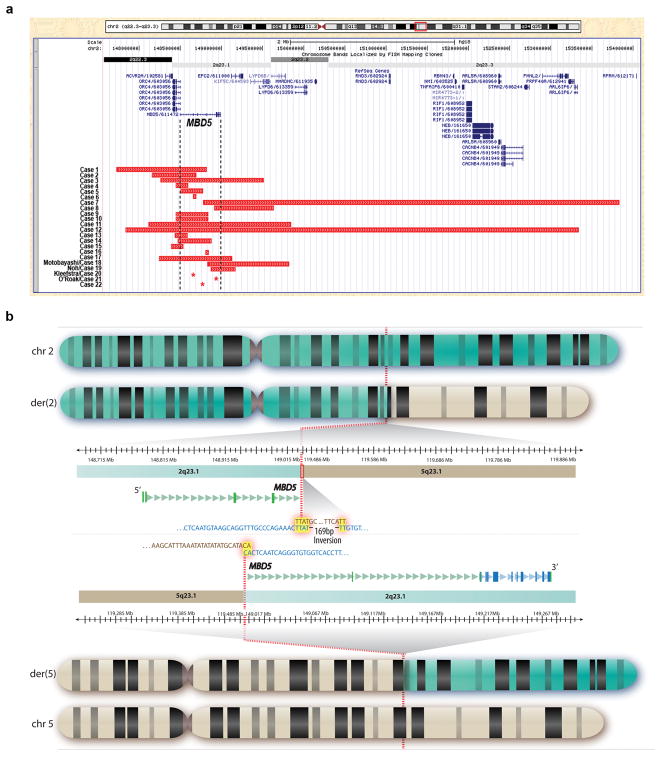
The frequency of MBD5 deletion was estimated to be 0.05% based on the identification of 8 cases out of 17 477 consecutive Mayo Clinic peripheral blood samples submitted for clinical testing by cytogenomic microarray. These 8 deletions, along with 10 cases from collaborators and 4 cases identified recently in the literature, define a new cohort that is independent of the subjects in Talkowski et al6. The new cohort (n = 22) includes one frameshift mutation, one indel mutation, one inverted translocation directly disrupting MBD5, three intragenic MBD5 deletions, and 16 larger deletions. The rearrangements arose de novo in all cases for which parental testing was available (n = 11). However, two cases without parental genetic or phenotypic information involve brothers with similar deletions, suggesting likely parental inheritance or gonadal mosaicism. No bias in gender or age of diagnosis was observed in the 13 males and nine females ranging in age from two weeks to 69 years. The disrupted region varied from a single base pair point mutation to a 5.4 Mb microdeletion, with 17 of the alterations being no more than 1.1 Mb (Figure 1a). Five of these alterations are within MBD5, all but one of which is confined entirely to the non-coding 5′UTR. In addition, Case 2 is a high percentage mosaic with the deletion present in 28/30 metaphases and 77.5% of interphase nuclei from a cultured specimen as well as 81.0% of interphase nuclei from a direct preparation.
Figure 1.
Structural alterations disrupting MBD5. (a) UCSC genome browser (genome build hg18) demonstrating 22 cases of intragenic MBD5 and larger 2q23.1 disruptions (red bars) identified by cytogenomic microarray. MBD5 is at position 148,495,050–148,987,514 and contains 15 exons of which exons 1–5 are the non-coding 5′UTR and exons 6–15 are protein coding. Cases 5, 6, 16, 20 and 21 involve disruptions confined to the non-coding region of MBD5 and Case 22 is confined to the coding region of MBD5, while all other cases overlap at least one other gene. Asterisks (*) denote a single base pair change resulting in a frameshift mutation in Case 20, a two base pair deletion in Case 21, and the translocation breakpoint in Case 22. (b) Whole genome sequencing of Case 22 delineated that an apparently balanced translocation between chromosomes 2 and 5 directly disrupted MBD5 at 2q23.1 and did not affect any annotated functional sequence on chromosome 5. A local microinversion of 169 bases was also detected at the breakpoint of the chromosome 5 material on the der(2) chromosome. The GTG-banded idiograms depict the normal chromosomes 2 and 5 as well as the derivative chromosomes from the translocation. The breakpoint regions on the derivative chromosomes are expanded in the middle, providing genomic coordinates, cytogenetic bands, precise breakpoints (dotted red line), and surrounding nucleotide sequence of the junctions including microhomology (highlighted in yellow) at the translocation and inversion breakpoints. The 5′ and 3′ non-coding UTR regions of the disrupted MBD5 transcript are highlighted in green, the translated region is highlighted in blue, and exons are denoted by rectangles.
Case 22 harbored a de novo translocation between chromosomes 2 and 5 that was balanced at karyotypic resolution. Whole-genome sequencing by large-insert jumping libraries revealed direct disruption of MBD5 at the breakpoint on chromosome 2. The orientation of a small subset of reads relative to the reference sequence also supported the presence of a small local inversion at the chromosome 5 breakpoint on the der(2) chromosome, suggesting a translocation with an accompanying inversion (Figure 1b). We previously discovered that such microinversions are a pervasive feature of karyotypically balanced translocations.22 Capillary sequencing confirmed the translocation breakpoints on both derivative chromosomes and the presence of a 169 bp microinversion of chromosome 5 on the der(2). The chromosome 2 breakpoints (der(2):148,732,432; der(5):148,733,228 in human genome build hg18) directly disrupt the 5′ non-coding region of MBD5 that was previously shown to be interrupted in two independent translocations sequenced in Talkowski et al6. In sum, a total genomic imbalance of 795 bp was observed at the der(5) breakpoint (including both deleted sequence and duplication of microhomology), and the der(2) translocation and inversion events resulted in 41 bp of deleted sequence. The chromosome 5 breakpoint occurred in an intergenic region without annotated coding sequence within a 500 kb window of the breakpoint. Sequencing thus revised the interpretation of the karyotype from 46,XY,t(2;5)(q22;q22) to 46,XY,der(2)t(2;5)(q23.1;q23.1)inv(5)(q23.1q23.1),der(5)t(2;5)(q23.1;q23.1)dn. See Supplementary Table S2 for complete breakpoint information.
In the original cohort described by Talkowski et al6 there were 12 intragenic MBD5 deletions, two other translocations disrupting the same untranslated region of MBD5 as the present case, and 41 larger 2q23.1 microdeletions encompassing MBD5 and additional genes ranging in size from 38 kb to 19.3 Mb. After removal of the Talkowski et al cases without phenotypic information, the remaining 48 individuals were added to the 22 new cases for analysis. This combined cohort (n = 70) involved three translocations, one point mutation, one indel, and 65 deletions; 16 are intragenic MBD5 alterations, all but one of which are confined to the 5′UTR. The following analyses are derived from the phenotypic features of these 70 individuals.
MBD5 disruption is associated with neurodevelopmental features and clinical variation including behavioral regression
Comparison of the original raw data from the MBD5 disruption cases in Talkowski et al6 to a new cohort of 22 cases demonstrated an independent confirmation of substantial genetic and phenotype overlap between the two groups. Combined assessment of these cohorts showed that MBD5 disruption has a multi-system effect, leading to behavioral, growth/endocrine, craniofacial, skeletal, gastrointestinal, heart, urogenital and central nervous system anomalies.
The phenotypes are largely indistinguishable between intragenic MBD5 and larger deletions, whether considering neurodevelopmental and behavioral abnormalities specifically or all other features (Table 1). However, a small subset of the characteristics may have resulted from loss of neighboring genes. This is suggested by features that were confined to larger deletion cases in both the new cohort and separately in the original cohort6 that were assessed in at least 4 patients, specifically bruxism, hyperphagia, ataxia, obesity, brachycephaly, metopic ridging, optic nerve hypoplasia, strabismus, coarse facies, wide mouth, brachydactyly and hypoplastic genitalia. It is of note that only 2 of these twelve features are neurodevelopmental or psychiatric in origin, which further implicates MBD5 as the causative gene for the majority of such phenotypes. Additional features potentially arising from genes near MBD5 are demonstrated in the combined cohort analysis (Table 1). However, definitive genotype/phenotype correlations are challenging due to the relative rarity of intragenic MBD5 disruptions (n = 16) compared to 2q23.1 microdeletions (n = 54), warranting further consideration in even larger cohorts.
Table 1.
Phenotypes in Intragenic MBD5 Disruptions and 2q23.1 Microdeletions in the Combined Cohort1
| Features | MBD5 | 2q23.1 | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Number Reported2 | Total Cohort3 | Number Reported2 | Total Cohort3 | |||
| Behavioral | ||||||
| Aggression/Temper | 3/3 (100%) | 3/16 (19%) | 10/11 (90%) | 10/54 (19%) | ||
| Anxiety | 1/1 (100%) | 1/16 (6%) | 1/2 (50%) | 1/54 (2%) | ||
| Autistic-like behaviors | 9/9 (100%) | 9/16 (56%) | 43/44 (97%) | 43/54 (80%) | ||
| Bruxism (teeth grinding) | -- | 0/16 (0%) | 5/5 (100%) | 5/54 (9%) | ||
| Distractibility/Short attention span | 5/5 (100%) | 5/16 (31%) | 15/15 (100%) | 15/54 (29%) | ||
| Hand flapping | 2/2 (100%) | 2/16 (13%) | 2/3 (66%) | 2/54 (4%) | ||
| Hyperactivity | -- | 0/16 (0%) | 2/4 (50%) | 2/54 (4%) | ||
| Hyperphagia | 0/4 (0%) | 0/16 (0%) | 7/11 (64%) | 7/54 (13%) | ||
| Inappropriate happy demeanor | -- | 0/16 (0%) | 5/5 (100%) | 5/54 (9%) | ||
| Insertion of hand in mouth or hand biting | 1/1 (100%) | 1/16 (6%) | 6/6 (100%) | 6/54 (11%) | ||
| Obsessive-compulsive or routine-bound behaviors | -- | 0/16 (0%) | 4/5 (80%) | 4/54 (7%) | ||
| Self-injurious behaviors including skin and eye picking | 2/4 (50%) | 2/16 (13%) | 18/27 (66%) | 17/54 (31%) | ||
| Sleep disturbances | 5/8 (63%) | 5/16 (31%) | 17/22 (77%) | 17/54 (32%) | ||
| Social withdrawl | 1/1 (100%) | 1/16 (6%) | 1/2 (50%) | 1/54 (2%) | ||
| Sensory processing disorder | -- | 0/16 (0%) | 2/3 (66%) | 2/54 (4%) | ||
| Central Nervous System | ||||||
| Ataxia/Unusual gait | 0/4 (0%) | 0/16 (0%) | 18/26 (72%) | 18/54 (33%) | ||
| Developmental delay/Intellectual disability | 15/15 (100%) | 15/16 (94%) | 52/52 (100%) | 52/54 (96%) | ||
| Feeding difficulties | 2/3 (67%) | 2/16 (13%) | 15/16 (94%) | 15/54 (29%) | ||
| Hypotonia/Abnormal muscle tone | 3/4 (75%) | 3/16 (19%) | 22/23 (96%) | 22/54 (41%) | ||
| Motor delay | 6/6 (100%) | 6/16 (38%) | 32/32 (100%) | 32/54 (60%) | ||
| Seizures | 11/12 (92%) | 10/16 (63%) | 31/38 (82%) | 31/54 (57%) | ||
| Speech delay | 7/10 (70%) | 7/16 (44%) | 37/37 (100%) | 37/54 (69%) | ||
| Growth/Endocrine | ||||||
| Growth retardation | 2/5 (40%) | 2/16 (13%) | 23/40 (56%) | 23/54 (43%) | ||
| Hirsutism | 0/5 (0%) | 0/16 (0%) | 4/20 (20%) | 4/54 (7%) | ||
| Obesity | 0/1 (0%) | 0/16 (0%) | 5/9 (56%) | 5/54 (9%) | ||
| Short stature | 3/6 (50%) | 3/16 (19%) | 25/33 (70%) | 25/54 (41%) | ||
| Craniofacial | ||||||
| Cranium | ||||||
| Brachycephaly | 0/6 (0%) | 0/16 (0%) | 12/28 (43%) | 12/54 (22%) | ||
| Metopic ridging | -- | 0/16 (0%) | 2/4 (50%) | 2/54 4%) | ||
| Microcephaly | 1/8 (13%) | 1/16 (6%) | 26/32 (81%) | 26/54(48%) | ||
| Plagiocephaly | -- | 0/16 (0%) | 2/4 (50%) | 2/54 (4%) | ||
| Broad forehead | 2/2 (100%) | 2/16 (13%) | 3/5 (60%) | 3/54 (6%) | ||
| Ears/Nose | ||||||
| Nasal abnormalities | 6/6 (100%) | 6/16 (38%) | 34/35 (97%) | 34/54 (63%) | ||
| Outer ear abnormalities | 2/6 (33%) | 2/16 (13%) | 18/21 (86%) | 18/54 (33%) | ||
| Eyes | ||||||
| Almond shaped eyes | 1/1 (100%) | 1/16 (6%) | 2/2 (100%) | 2/54 (4%) | ||
| Astigmatism | 1/1 (100%) | 1/16 (6%) | 2/4 (50%) | 2/54 (4%) | ||
| Hypotelorism | 0/5 (0%) | 0/16 (0%) | 7/22 (36%) | 7/54 (13%) | ||
| Myopia/Hypermetropia/Corrective Lenses | 2/2 (100%) | 2/16 (13%) | 6/9 (67%) | 6/54 (11%) | ||
| Optic nerve hypoplasia | -- | 0/16 (0%) | 2/4(50%) | 2/54 (4%) | ||
| Palpebral fissures - downslanting | 1/1 (100%) | 1/16 (6%) | 2/4 (50%) | 2/54 (4%) | ||
| Palpebral fissures - long | -- | 0/16 (0%) | 3/4 (75%) | 3/54 (6%) | ||
| Palpebral fissures - upslanting | 1/1 (100%) | 1/16 (6%) | 1/3 (33%) | 1/54(2%) | ||
| Strabismus | -- | 0/16 (0%) | 2/5 (40%) | 2/54 (4%) | ||
| Synophrys | 2/6 (33%) | 2/16 (13%) | 10/20 (50%) | 10/54(19%) | ||
| Thick/Arched eyebrows | 1/1 (100%) | 1/16 (6%) | 17/22 (77%) | 17/54 (31%) | ||
| Face | ||||||
| Coarse facies | -- | 0/16 (0%) | 3/4 (75%) | 3/54 (6%) | ||
| Midfacial hypoplasia | 1/6 (17%) | 1/16 (6%) | 14/26 (54%) | 14/54 (27%) | ||
| Mouth/Chin | ||||||
| Dental abnormalities | 4/7 (57%) | 4/16 (25%) | 13/26 (50%) | 13/54 (25%) | ||
| Downturned mouth corners or everted lower lip | 2/4 (50%) | 2/16 (13%) | 16/22 (73%) | 16/54 (29%) | ||
| Macroglossia or protruding tongue | 2/7 (29%) | 2/16 (13%) | 6/24 (25%) | 6/54 (11%) | ||
| Micrognathia | 3/5 (60%) | 3/16 (19%) | 13/24 (54%) | 13/54 (25%) | ||
| Open mouth | 2/7 (29%) | 2/16 (13%) | 23/25 (92%) | 23/54 (43%) | ||
| Tented upper lip | 1/5 (20%) | 1/16 (6%) | 17/24 (71%) | 17/54 (31%) | ||
| Thin upper lip | 2/2 (100%) | 2/16 (13%) | 19/25 (76%) | 19/54(36%) | ||
| Wide mouth | 0/6 (0%) | 1/16 (6%) | 14/18 (77%) | 14/54 (27%) | ||
| Skeletal | ||||||
| Extremities | Brachydactyly | 0/4 (0%) | 0/16 (0%) | 12/27 (47%) | 12/54(23%) | |
| Fifth finger clinodactyly | 1/5 (20%) | 1/16 (6%) | 22/31 (72%) | 22/54 (41%) | ||
| Flat feet | 3/3 (100%) | 3/16 (19%) | 3/4 (75%) | 3/54 (6%) | ||
| Sandal gap | 3/6 (50%) | 3/16 (19%) | 8/24 (33%) | 8/54 (16%) | ||
| Short fifth digit | 1/5 (20%) | 1/16 (6%) | 13/25 (52%) | 13/54 (25%) | ||
| Small hands or feet | 1/5 (20%) | 1/16 (6%) | 22/30 (73%) | 22/54 (41%) | ||
| Other | Hip dysplasia | -- | 0/16 (0%) | 2/3 (67%) | 2/54 (4%) | |
| Joint laxity | 2/3 (67%) | 2/16 (13%) | 2/5 (40%) | 2/54 (4%) | ||
| Scoliosis | 2/2 (100%) | 2/16 (13%) | 2/6 (33%) | 2/54 (4%) | ||
| Gastrointestinal | ||||||
| Constipation | 6/7 (86%) | 6/16 (38%) | 13/15 (87%) | 13/54(25%) | ||
| Gastroesophageal reflux | -- | 0/16 (0%) | 3/3 (100%) | 3/54 (6%) | ||
| Heart | ||||||
| Atrial septal defect | 0/7 (0%) | 0/16 (0%) | 1/24 (4%) | 1/54(2%) | ||
| Ventricular septal defect | 1/7 (14%) | 1/16 (6%) | 1/24 (4%) | 1/54(2%) | ||
| Pulmonary stenosis | 0/7 (0%) | 0/16 (0%) | 1/24 (4%) | 1/54(2%) | ||
| Unspecified heart defect | 0/7 (0%) | 0/16 (0%) | 1/24 (4%) | 1/54(2%) | ||
| Urogenital | ||||||
| Hypoplastic genitalia | 0/1 (0%) | 0/16 (0%) | 5/8 (63%) | 5/54 (9%) | ||
The combined cohort is the new cases plus those cases from Talkowski et al with phenotype data (n = 70).
Each denominator reflects the total number of cases with this feature specifically noted to be present or absent.
Each denominator reflects the total number of cases in the cohort which may or may not represent assessment for the specific feature.
Potential novel behavioral features linked with MBD5 disruption are suggested in the new cohort including sensory integration disorder, anxiety, bipolar disorder and self-hugging, as well as a number of physical anomalies (Table 2). The associations of MBD5 disruption with optic nerve hypoplasia and with social withdrawal were also potentially confirmed through observation of a second affected patient (Case 3 and Case 21, respectively). The new cohort further includes the oldest assessed MBD5 disruption patient, a 44-year old man who had the novel features of early-onset dementia and cataracts (Case 3). Importantly, he demonstrated behavioral regression with worsening skin picking and obsessive-compulsive tendencies. Regression of motor and verbal skills was also noted in a second case in the new cohort (Case 19).20
Table 2.
New Features Potentially Associated withMBD5 Region Disruptions
| Case Number | Age (years) | Gender | Genes Affected1 | Inheritance | New Features |
|---|---|---|---|---|---|
|
| |||||
| 1 | 19 | F | 3 | De novo | --2 |
| 2 | 2 | M | 3 | Unknown | -- |
| 3 | 44 | M | 5 | De novo | Cataracts; Behavioral regression (worsening skin picking and obsessive-compulsive actions); Anxiety; Bipolar Disorder |
| 4 | 5 | F | 2 | De novo | Polydactyly; Skin tags; Ventricular septal defect |
| 5 | 12 | F | MBD5 only | Unknown | -- |
| 6 | <1 | M | MBD5 only | Unknown | Diaphragmatic eventration; Ventricular septal defect |
| 7 | 25 | F | 20 | Unknown | Spina bifida |
| 8 | 2 | M | 4 | De novo | -- |
| 9 | 14 | M | 2 | Likely inherited 3 | Cutis mamorata; Sensory integration disorder; Anxiety |
| 10 | 12 | M | 2 | Likely inherited 3 | -- |
| 11 | 20 | F | 8 | De novo | Flat occiput |
| 12 | 3 | F | 22 | De novo | -- |
| 13 | 5 | F | 2 | Unknown | -- |
| 14 | 7 | F | 2 | Unknown | -- |
| 15 | 7 | M | 3 | Unknown | -- |
| 16 | 7 | M | MDB5 only | Unknown | Bipolar disorder |
| 17 | 6 | M | 4 | De novo | -- |
| Motobayashi/18 | 5 | M | 4 | De novo | -- |
| Noh/19 | 2 | F | 2 | De novo | Short neck |
| Kleefstra/20 | >13 | M | MBD5 only | De novo | Self-hugging; Anxiety |
| O’Roak/21 | 13 | M | MBD5 only | De novo | -- |
| 22 | 2 | M | MBD5 only | Unknown | -- |
Minimum number of affected RefSeq genes.
Indicates no new phenotype observed in this case.
Although no parental testing has been performed, Cases 9 and 10 are siblings indicating the deletion is likely inherited.
Phenotypic presentation of MBD5 disruption overlaps with known syndromes
We analyzed extensive phenotypic data in a large cohort of subjects with MBD5 disruptions to understand the consequences of multiple mutational mechanisms impacting transcription of this gene and to provide an assessment algorithm for clinicians to identify affected patients. However, devising an appropriate testing strategy presents a significant challenge due to substantial heterogeneity in both the type and severity of MBD5-related phenotypes which span cognitive, behavioral and physical domains. Moreover, MBD5 disruption is a potent masquerader, resulting in a number of clinical features previously thought to be unique to other syndromes. Based on our analyses, it is clear that a first-tier diagnostic cytogenomic microarray test will aid in the partitioning of subjects into gene-specific disorders characterized by dosage imbalance. Such a recommendation would be consistent with recent guidelines.25, 26 Absence of detectable dosage imbalance will then require clinicians to decide if karyotype or targeted gene-specific testing, such as sequencing for intragenic point mutations, small deletions or duplications, is appropriate based on the phenotypic gestalt. Of note, although whole exome sequencing is moving towards replacing cytogenomic arrays and gene or gene panel sequencing, targeted gene analysis could still have an important role in the future as UTR variants, including those in multiple cases in the current study, might not otherwise be detected.
To aid in determining the next diagnostic step in the absence of a detectable dosage imbalance, we present expansion of the clinical heterogeneity associated with MBD5 disruption, as well as the distinguishing phenotypes or constellations of anomalies which would suggest additional diagnostic testing for MBD5 versus targeting a classic syndrome with overlapping features including SMS27–32, PWS33–38, AS39, 40, CdLS41–44, Rett45–47, Kleefstra48–50, or PTHS51–53. Thus, the phenotypes in the combined cohort separated by intragenic MBD5 versus larger 2q23.1 deletions that occurred in two or more cases were compared to those of other syndromes considered in the differential diagnosis (Table 3).
Neurodevelopmental and behavioral characteristics common to all of these syndromes include autistic-like actions, intellectual disability, developmental delay, speech delay, motor delay, hypotonia, sleep disturbances and seizures. There are also features confined solely to each of the syndromes in the differential diagnosis as detailed in the Supplementary Results. Although single characteristics can be helpful, the phenotypic diversity noted with MBD5 disruptions remains a diagnostic challenge, as illustrated in the divergence of facial dysmorphism in MBD5 deletion cases, ranging from isolated ear anomalies to multiple atypical characteristics (Figure 2). A clear understanding of how the breadth of phenotypes compares and contrasts for these multiple diagnoses is required. The differentiating and overlapping neurodevelopmental, behavioral, and other features presented in Table 3 are further described in detail for each syndrome in the Supplementary Results.
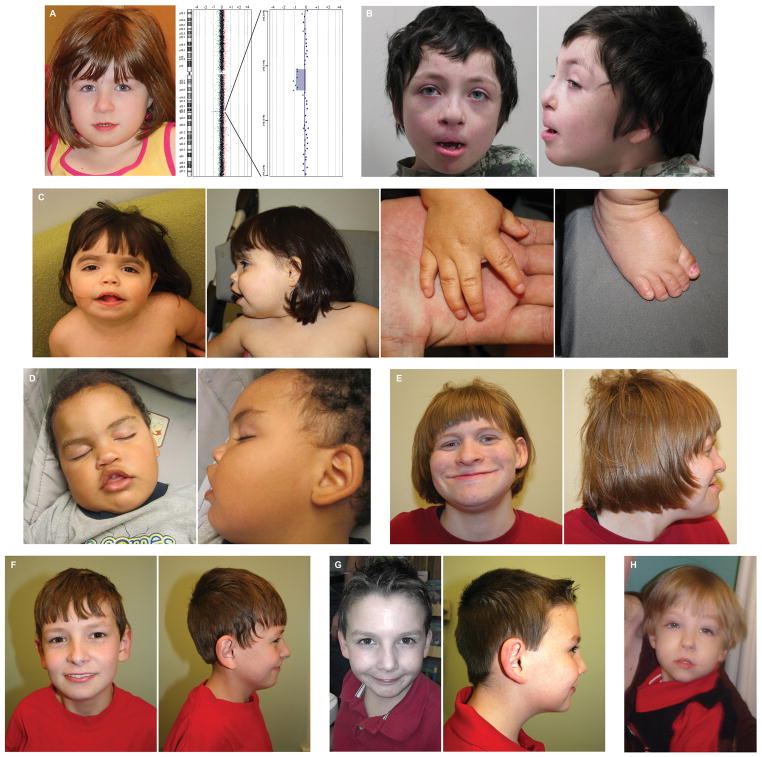
Figure 2.
Clinical features associated with MBD5 disruption.
Patients with MBD5 deletions have a broad range of physical features both in type and severity. A, Case 4 (6 year 9 month old female) has a round face, midface hypoplasia, flat nose and thin upper lip, which along with ID/DD, hearing loss, and unusual behavior of self-injury and altered sleep cycle, were highly suggestive of SMS. Cytogenomic microarray demonstrated a de novo deletion (153 kb) of the MBD5 5′ UTR while subsequent sequencing of the RAI1 gene associated with SMS was negative. B, Case 15 (7 year 9 month old male) has brachycephaly, midface hypoplasia, depressed nasal bridge, a thin and tented upper lip, open mouth and dental crowding. C, Case 12 (3 year old female) has dysmorphic facies with synophrys, slightly downslanting palpebral fissures, tented upper lip, depressed nasal bridge with an upturned nose and somewhat prominent ears with attached lobes as well as a low posterior hairline, right supernumerary nipple, short neck, short thumbs and fifth fingers, and mildly dysplastic fifth toenails. D, Case 8 (2 year old male) has an open mouth with downturned corners, a depressed nasal bridge, and an overfolded helix. E, Case 11 (20 year old female) has a thin upper lip, a slightly smooth philtrum, mild epicanthal folds with slightly upslanting palpebral fissures and a prominent columella with a thickened nasal tip. F, Case 8 (14 year old male) has mildly prominent and overfolded ears with thickened helices. G, Case 9 (12 year old male) is the brother of Case 8 and has similar atypical ears. H, Case 16 (7 year old male) has a thin upper lip, long philtrum, prominent nasal bridge, arched eyebrows, epicanthal folds and almond-shaped eyes.
Discussion
Deletions encompassing MBD5 are highly penetrant and result in a broad phenotypic spectrum that includes many neurodevelopmental and psychiatric traits. Our analyses of 22 cases with extensive clinical information confirm the phenotypes previously described in an independent cohort of 65 subjects described in Talkowski et al6 and define the significant psychopathology associated with MBD5 disruptions. When combined, these data demonstrate similar phenotypic traits between individuals with intragenic MBD5 alterations and larger 2q23.1 microdeletions, consistent with MBD5 being the necessary and sufficient pathogenic target in 2q21.3 microdeletion syndrome. The new cohort further expands the potential phenotypic spectrum to include the behavioral characteristics of sensory integration disorder, anxiety, bipolar disorder, and self-hugging, as well as the physical anomalies polydactyly, skin tags, diaphragmatic eventration, ventricular septal defect (VSD), spina bifida, cutis marmorata, flat occiput and short neck. The associations with optic nerve hypoplasia and with social withdrawal, each observed in a single patient in the original cohort6, were also demonstrated by independent cases in this new cohort.
The potential for MBD5 to perturb neurological function throughout life was demonstrated by the oldest assessed MBD5 deletion patient at 44 years of age (Case 3). Such a delayed diagnosis is consistent with the difficulty in clinically identifying MBD5 deletions and suggests potential longevity for affected individuals; a normal lifespan is further implied by a 69 year old male (O’Roak/Case 21), although his associated features are only known through 4 years of age. The 44 year old patient had early-onset dementia, cataracts and behavioral regression with a significant increase in skin picking and obsessive-compulsive activities. He thus represents the third case for which regression has been documented and the first case with early-onset dementia. Regression also occurred in a second patient included within the new cohort (Noh/Case 19), a 2 year old girl with a 300 kb deletion overlapping two genes at 2q23.1. This child walked independently at 28 months of age but an examination at four years showed that she had lost a subset of her vocabulary and the ability to walk, standing only with support.20 Another case of developmental regression was previously noted to occur suddenly at six years of age in a girl with a de novo approximately 4 MB deletion involving 15 genes at 2q23.1-q23.3. This child had progressive difficulties with fine motor skills and balance, worsening behavior, and loss of the ability to draw lines and circles.9 It is of note that this child was originally tested for Rett and Angelman syndromes with normal results before the MBD5 deletion was identified. These results merit careful evaluation of future cases for developmental and behavioral regression and emphasize the recommendation of cytogenomic microrarray as a first-tier test for all such cases.
Many features found in MBD5 deletions such as intellectual disability, motor and speech delays, sleep disturbances, microcephaly, and seizures are non-specific and commonly found in multiple genetic syndromes. A more specific spectrum of behavioral and other phenotypes in individuals with MBD5 disruptions may be diagnostically useful, although it remains challenging to differentiate from the profiles in other syndromes including SMS, CdLS, AS, PWS, Rett, Kleefstra, and PTHS. In fact, many patients with MBD5 deletions previously reported in the literature were referred for genetic testing for one or more of these disorders, all of which were negative, prior to cytogenomic microarray providing an accurate diagnosis; this is illustrated well in a study of 2q23.1 microdeletion syndrome patients (n = 11) who had normal test results for AS (n = 8), SMS (n = 5) and Rett (n = 4) syndrome.8 In the current cohort, the known additional testing included karyotype (n = 6), fragile X analysis (n = 6), RAI1 sequencing for SMS (n = 2), MECP2 and CDKL5 sequencing for Rett syndrome (n = 1), methylation analysis and UBE3A sequencing for PWS/AS (n = 1), and TCF4 sequencing for PTHS (n = 1). It is of note that guidelines indicate fragile X testing should be performed for any patient with intellectual disability54, a feature also found in MBD5 disruption, although there is no overlap of more specific phenotypes between these two syndromes which suggests that they should be clinically distinguishable.
The ability of MBD5 deletions to manifest significant clinical heterogeneity that overlaps with previously defined syndromes in a highly penetrant manner confirms the advantage of using unbiased, high resolution genome-wide surveys such as cytogenomic microarrays when any of the above mentioned syndromes is suspected. However, cytogenomic microarrays do not detect balanced abnormalities or small pathogenic mutations, as occurred in cases 20 through 22, suggesting that implementation of whole-genome sequencing will also have a significant diagnostic impact and karyotyping may reveal additional balanced rearrangements. Our results further raise important issues about the current interpretation of variants of unknown significance and coding region-focused analyses such as exome sequencing as we found highly penetrant pathogenic rearrangements confined to the putative 5′UTR of MBD5 (as in cases 5, 6, 16, 20 and 22) to be contributory towards diverse phenotypes. Therefore, a focused phenotypic annotation was undertaken in the current study to complement the recent genetic characterization6 of MBD5 disruptions and to identify features that contrast with syndromes for which MBD5 can be mistaken. This may help determine an appropriate testing strategy and avoid unnecessary medical expenses.
In summary, careful phenotyping of patients with MBD5 disruptions confirms that abnormalities at this locus are highly penetrant risk factors for neurodevelopmental and other abnormalities. We also extend the associated clinical heterogeneity to a broad range of neuropsychiatric features including early-onset dementia, provide a differential diagnostic context, and suggest that this locus is potentially associated with risk for neurobehavioral regression.
Supplementary Material
Acknowledgments
This work was funded by grants from the NIH GM061354 (CCM, JFG) and MH09586 (MET), and from the Fondation Jerome Lejeune (SHE). MET is also supported by the Simons Foundation for Autism Research and the Nancy Lurie Marks Family Foundation. We thank the patients and their families and physicians for participating in this study.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11. 2 and autism. N Engl J Med. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 2.Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11. 2 locus. Nature. 2011;478(7367):97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenfeld JA, Ballif BC, Lucas A, Spence EJ, Powell C, Aylsworth AS, et al. Small deletions of SATB2 cause some of the clinical features of the 2q33. 1 microdeletion syndrome. PLoS One. 2009;4(8):e6568. doi: 10.1371/journal.pone.0006568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, Vissers LE, et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet. 2012;91(1):1–10. doi: 10.1016/j.ajhg.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talkowski ME, Mullegama SV, Rosenfeld JA, van Bon BW, Shen Y, Repnikova EA, et al. Assessment of 2q23. 1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet. 2011;89(4):551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams SR, Mullegama SV, Rosenfeld JA, Dagli AI, Hatchwell E, Allen WP, et al. Haploinsufficiency of MBD5 associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. Eur J Hum Genet. 2010;18(4):436–441. doi: 10.1038/ejhg.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Bon BW, Koolen DA, Brueton L, McMullan D, Lichtenbelt KD, Ades LC, et al. The 2q23. 1 microdeletion syndrome: clinical and behavioural phenotype. Eur J Hum Genet. 2010;18(2):163–170. doi: 10.1038/ejhg.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung BH, Stavropoulos J, Marshall CR, Weksberg R, Scherer SW, Yoon G. 2q23 de novo microdeletion involving the MBD5 gene in a patient with developmental delay, postnatal microcephaly and distinct facial features. Am J Med Genet Part A. 2011;155A(2):424–429. doi: 10.1002/ajmg.a.33821. [DOI] [PubMed] [Google Scholar]
- 10.Jaillard S, Dubourg C, Gerard-Blanluet M, Delahaye A, Pasquier L, Dupont C, et al. 2q23.1 microdeletion identified by array comparative genomic hybridisation: an emerging phenotype with Angelman-like features? J Med Genet. 2009;46(12):847–855. doi: 10.1136/jmg.2008.058156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, Messa J, et al. Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet. 2007;44(12):750–762. doi: 10.1136/jmg.2007.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003;73(6):1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagenstaller J, Spranger S, Lorenz-Depiereux B, Kazmierczak B, Nathrath M, Wahl D, et al. Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. Am J Hum Genet. 2007;81(4):768–779. doi: 10.1086/521274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koolen DA, Vissers LE, Nillesen W, Smeets D, van Ravenswaaij CM, Sistermans EA, et al. A novel microdeletion, del(2)(q22.3q23.3) in a mentally retarded patient, detected by array-based comparative genomic hybridization. Clin Genet. 2004;65(5):429–432. doi: 10.1111/j.0009-9163.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- 15.de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77(4):606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laget S, Joulie M, Le Masson F, Sasai N, Christians E, Pradhan S, et al. The human proteins MBD5 and MBD6 associate with heterochromatin but they do not bind methylated DNA. PLoS One. 2010;5(8):e11982. doi: 10.1371/journal.pone.0011982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motobayashi M, Nishimura-Tadaki A, Inaba Y, Kosho T, Miyatake S, Niimi T, et al. Neurodevelopmental features in 2q23.1 microdeletion syndrome: Report of a new patient with intractable seizures and review of literature. Am J Med Genet Part A. 2012;158A(4):861–868. doi: 10.1002/ajmg.a.35235. [DOI] [PubMed] [Google Scholar]
- 20.Noh GJ, Graham JM., Jr 2q23.1 microdeletion of the MBD5 gene in a female with seizures, developmental delay and distinct dysmorphic features. Eur J Med Genet. 2012;55(1):59–62. doi: 10.1016/j.ejmg.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 21.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012;44(4):390–397. S391. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talkowski ME, Ernst C, Heilbut A, Chiang C, Hanscom C, Lindgren A, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88(4):469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning M, Hudgins L. Use of array-based technology in the practice of medical genetics. Genet Med. 2007;9(9):650–653. doi: 10.1097/gim.0b013e31814cec3a. [DOI] [PubMed] [Google Scholar]
- 27.Chen RM, Lupski JR, Greenberg F, Lewis RA. Ophthalmic manifestations of Smith-Magenis syndrome. Ophthalmology. 1996;103(7):1084–1091. doi: 10.1016/s0161-6420(96)30563-0. [DOI] [PubMed] [Google Scholar]
- 28.Greenberg F, Lewis RA, Potocki L, Glaze D, Parke J, Killian J, et al. Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2) Am J Med Genet. 1996;62(3):247–254. doi: 10.1002/(SICI)1096-8628(19960329)62:3<247::AID-AJMG9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 29.Gropman AL, Duncan WC, Smith AC. Neurologic and developmental features of the Smith-Magenis syndrome (del 17p11.2) Pediatr Neurol. 2006;34(5):337–350. doi: 10.1016/j.pediatrneurol.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Edelman EA, Girirajan S, Finucane B, Patel PI, Lupski JR, Smith AC, et al. Gender, genotype, and phenotype differences in Smith-Magenis syndrome: a meta-analysis of 105 cases. Clin Genet. 2007;71(6):540–550. doi: 10.1111/j.1399-0004.2007.00815.x. [DOI] [PubMed] [Google Scholar]
- 31.Smith AC, Dykens E, Greenberg F. Sleep disturbance in Smith-Magenis syndrome (del 17 p11.2) Am J Med Genet. 1998;81(2):186–191. [PubMed] [Google Scholar]
- 32.Martin SC, Wolters PL, Smith AC. Adaptive and maladaptive behavior in children with Smith-Magenis Syndrome. J Autism Dev Disord. 2006;36(4):541–552. doi: 10.1007/s10803-006-0093-2. [DOI] [PubMed] [Google Scholar]
- 33.Gunay-Aygun M, Schwartz S, Heeger S, O’Riordan MA, Cassidy SB. The changing purpose of Prader-Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics. 2001;108(5):E92. doi: 10.1542/peds.108.5.e92. [DOI] [PubMed] [Google Scholar]
- 34.Jin DK. Systematic review of the clinical and genetic aspects of Prader-Willi syndrome. Korean J Pediatr. 2011;54(2):55–63. doi: 10.3345/kjp.2011.54.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinnema M, Maaskant MA, van Schrojenstein Lantman-de Valk HM, van Nieuwpoort IC, Drent ML, Curfs LM, et al. Physical health problems in adults with Prader-Willi syndrome. Am J Med Genet A. 2011;155A(9):2112–2124. doi: 10.1002/ajmg.a.34171. [DOI] [PubMed] [Google Scholar]
- 36.Vaiani E, Herzovich V, Chaler E, Chertkoff L, Rivarola MA, Torrado M, et al. Thyroid axis dysfunction in patients with Prader-Willi syndrome during the first 2 years of life. Clin Endocrinol (Oxf) 2010;73(4):546–550. doi: 10.1111/j.1365-2265.2010.03840.x. [DOI] [PubMed] [Google Scholar]
- 37.West LA, Ballock RT. High incidence of hip dysplasia but not slipped capital femoral epiphysis in patients with Prader-Willi syndrome. J Pediatr Orthop. 2004;24(5):565–567. doi: 10.1097/00004694-200409000-00018. [DOI] [PubMed] [Google Scholar]
- 38.Hered RW, Rogers S, Zang YF, Biglan AW. Ophthalmologic features of Prader-Willi syndrome. J Pediatr Ophthalmol Strabismus. 1988;25(3):145–150. doi: 10.3928/0191-3913-19880501-10. [DOI] [PubMed] [Google Scholar]
- 39.Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genet Med. 2012;12(7):385–395. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- 40.Michieletto P, Bonanni P, Pensiero S. Ophthalmic findings in Angelman syndrome. J Aapos. 2011;15(2):158–161. doi: 10.1016/j.jaapos.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Jackson L, Kline AD, Barr MA, Koch S. de Lange syndrome: a clinical review of 310 individuals. Am J Med Genet. 1993;47(7):940–946. doi: 10.1002/ajmg.1320470703. [DOI] [PubMed] [Google Scholar]
- 42.Kline AD, Krantz ID, Sommer A, Kliewer M, Jackson LG, FitzPatrick DR, et al. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A. 2007;143A(12):1287–1296. doi: 10.1002/ajmg.a.31757. [DOI] [PubMed] [Google Scholar]
- 43.Levin AV, Seidman DJ, Nelson LB, Jackson LG. Ophthalmologic findings in the Cornelia de Lange syndrome. J Pediatr Ophthalmol Strabismus. 1990;27(2):94–102. doi: 10.3928/0191-3913-19900301-11. [DOI] [PubMed] [Google Scholar]
- 44.Wygnanski-Jaffe T, Shin J, Perruzza E, Abdolell M, Jackson LG, Levin AV. Ophthalmologic findings in the Cornelia de Lange Syndrome. J Aapos. 2005;9(5):407–415. doi: 10.1016/j.jaapos.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Neul JL, Kaufmann WE, Glaze DG, Christodoulou J, Clarke AJ, Bahi-Buisson N, et al. Rett syndrome: revised diagnostic criteria and nomenclature. Ann Neurol. 2010;68(6):944–950. doi: 10.1002/ana.22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Percy AK, Lane JB. Rett syndrome: model of neurodevelopmental disorders. J Child Neurol. 2005;20(9):718–721. doi: 10.1177/08830738050200090301. [DOI] [PubMed] [Google Scholar]
- 47.Smeets EE, Pelc K, Dan B. Rett Syndrome. Mol Syndromol. 2012;2(3–5):113–127. doi: 10.1159/000337637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleefstra T, van Zelst-Stams WA, Nillesen WM, Cormier-Daire V, Houge G, Foulds N, et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J Med Genet. 2009;46(9):598–606. doi: 10.1136/jmg.2008.062950. [DOI] [PubMed] [Google Scholar]
- 49.Stewart DR, Kleefstra T. The chromosome 9q subtelomere deletion syndrome. Am J Med Genet C Semin Med Genet. 2007;145C(4):383–392. doi: 10.1002/ajmg.c.30148. [DOI] [PubMed] [Google Scholar]
- 50.Willemsen MH, Vulto-van Silfhout AT, Nillesen WM, Wissink-Lindhout WM, van Bokhoven H, Philip N, et al. Update on Kleefstra Syndrome. Mol Syndromol. 2012;2(3–5):202–212. doi: 10.1159/000335648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marangi G, Ricciardi S, Orteschi D, Lattante S, Murdolo M, Dallapiccola B, et al. The Pitt-Hopkins syndrome: report of 16 new patients and clinical diagnostic criteria. Am J Med Genet A. 2011;155A(7):1536–1545. doi: 10.1002/ajmg.a.34070. [DOI] [PubMed] [Google Scholar]
- 52.Peippo M, Ignatius J. Pitt-Hopkins Syndrome. Mol Syndromol. 2012;2(3–5):171–180. doi: 10.1159/000335287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Balkom IDC, Vuijk PJ, Franssens M, Hoek HW, Hennekam RC. Development, cognition, and behaviour in Pitt-Hopkins syndrome. Dev Med Child Neurol. 2012 doi: 10.1111/j.1469-8749.2012.04339.x. In Press. [DOI] [PubMed] [Google Scholar]
- 54.Curry CJ, Stevenson RE, Aughton D, Byrne J, Carey JC, Cassidy S, et al. Evaluation of mental retardation: recommendations of a Consensus Conference: American College of Medical Genetics. Am J Med Genet. 1997;72(4):468–477. doi: 10.1002/(sici)1096-8628(19971112)72:4<468::aid-ajmg18>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.