Abstract
Aggregation of amyloid β-peptide (Aβ) is implicated in the pathology of Alzheimer’s disease (AD), with the soluble, Aβ oligomeric species thought to be the critical pathological species. Identification and characterization of intermediate species formed during the aggregation process is crucial to the understanding of the mechanisms by which oligomeric species mediate neuronal toxicity and following disease progression. Probing these species proved to be extremely challenging, as evident by the lack of reliable sensors, due to their heterogeneous and transient nature. We describe here an oligomer-specific fluorescent chemical probe, BoDipy-Oligomer (BD-Oligo), developed through the use of the diversity-oriented fluorescent library approach (DOFLA) and high-content, imaging-based screening. This probe enables dynamic oligomer monitoring during fibrillogenesis in vitro and shows in vivo Aβ oligomers staining possibility in the AD mice model.
Graphical Abstract
INTRODUCTION
Protein misfolding diseases represent a group of disorders that have tissue deposition of β-sheet-rich, filamentous protein aggregates, known as amyloid fibrils in common.1 Alzheimer’s disease (AD) is one of the most studied protein misfolding diseases in which amyloid β-peptide (Aβ) aggregates, forming extracellular neuritic plaques in the brain. AD affects well over 35 million worldwide, and this number is expected to grow dramatically as the population ages.2 Amyloidogenic proteins and peptides can adopt a number of distinct assembly states, and a key issue is which of these assembly states are more closely associated with pathogenesis. Fibrillization of Aβ resulting in plaque deposition has long been regarded as the cause of neurodegeneration in AD. However, recent data suggest that oligomeric soluble Aβ is principally responsible for the pathogenesis of AD, and its levels are more important in disease progression.3–6 The concept of Aβ intermediate involvement in the development of AD has been used to explain why amyloid pathology, defined by Aβ plaque load, is only poorly correlated with clinical AD presentation, effectively suggesting that amyloid plaque is a relatively nontoxic aggregated form of Aβ. Hence, there is an urgent need for the development of detection methods that are able to identify a variety of morphologically distinct Aβ peptides.
Aβ plaques have been detected using a number of fibril-specific dyes, such as Congo Red (CR) or Thioflavin T (ThT),7 which preferably bind to mature amyloid fibrils. Neither CR nor ThT was suitable for in vivo use; nonetheless, they serve as the basis for development of improved imaging agents to detect amyloid accumulation, which gave rise to compounds such as PiB.8 Despite extensive research for many decades, it was only until recently that a brain imaging agent, Florbetapir, was approved by the Food and Drug Administration (FDA) to evaluate AD.9 In recent years, however, there has been a paradigm shift with numerous reported efforts involved in the development of effective methods for Aβ oligomers detection, including oligomer-specific antibody,10 oligomer-specific peptide-FlAsh system,11,12 peptide-based fluorescent protein,13 as well as the ELISA method.14 Yet, these detection methods often involve laborious construction methods, complicated instrumentation, or a long testing time, which make them inconvenient to use. In addition, their inability to cross the blood–brain barrier (BBB) makes them inappropriate for in vivo application. Small fluorescent molecular probes, which yield high sensitivity and easy visibility, would offer a convenient and straightforward approach for the detection of Aβ oligomers. One of the reported oligomer-specific fluorescence sensors showed the capability of distinguishing soluble Aβ from Aβ of ordered conformation but fell short of discriminating oligomers from fibrils and lack demonstration of biological application capabilities.15,16
Here, we describe BD-Oligo, a novel fluorescent chemical probe that preferentially recognizes Aβ oligomeric assemblies over monomers or fibrils, by using diversity-oriented fluorescence library (DOFL) screening and computational techniques. DOFL was generated in house through combinatorial synthesis by the modification of side chains of different fluorescent dye backbones and has proven its versatility in sensor development.17–21 BD-Oligo demonstrates a dynamic oligomer-monitoring ability during Aβ peptide fibrillogenesis, as Aβ was induced to form oligomers and eventually fibrils over time. More importantly, BD-Oligo also shows BBB penetration with capabilities of staining Aβ oligomers in vivo.
RESULTS
Oligomer-Specific Sensor Discovery (BD-Oligo) and Characterization
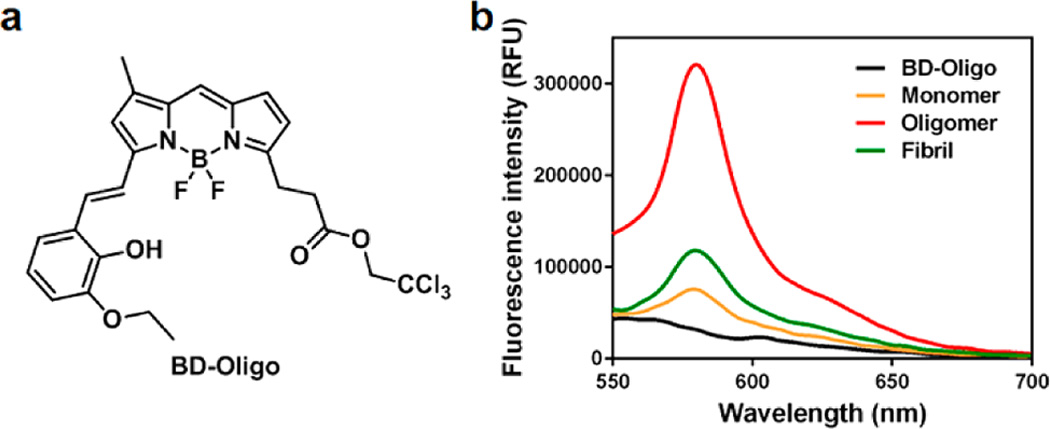
Since the proposed role of Aβ oligomers in the pathophysiology of AD, synthetic Aβ oligomers have been used as tools for the development of therapeutics and biomarkers. To develop an Aβ oligomer-selective probe in a living system, we incubated 7PA2 cells, which were reported to be enriched in Aβ oligomers,5 with 3500 DOFL compounds.22–24 When in the absence of mechanistic cues to rationally design probes for Aβ oligomers, we envisioned high-throughput screening to be crucial in helping us identify promising leads. By expanding this strategy, 5 candidate compounds were selected based on their higher fluorescence intensity in 7PA2 cells than in CHO cells, from which the 7PA2 cells were propagated. We then sought to further narrow these candidates by a more direct approach using a synthetically stabilized oligomer of Aβ in comparison to monomer and fibrils. While Aβ monomers and fibrils have been used widely, Aβ oligomer is challenging to form or maintain due to its dynamic nature. In this study, Aβ 1–40 peptide was solubilized in borate-buffered saline (50 mM BBS/PBS) and reacted with 5 mM glutaraldehyde overnight at 37 °C to produce covalently stabilized Aβ oligomers, as previously described.25,26 The most selective oligomer fluorescence turn-on probe was dubbed BoDipy-Oligomer or BD-Oligo for short. With BD-Oligo, the highest fluorescence enhancement is observed upon incubation with Aβ oligomers, indicating a preference for these intermediary conformations of Aβ aggregation over monomers or fibrils (Figure 1).
Figure 1.
Conformational specificity of BD-Oligo. (a) Chemical structure of BD-Oligo. (b) Emission spectra of BD-Oligo alone and when incubated with monomers, oligomers, and fibrils of Aβ (λex = 530 nm, dye 5 µM, Aβ 20 µM).
We confirmed the conformations of different Aβ peptide preparation by dot blot assays, and the results showed that the oligomer responded most strongly to the antioligomer antibody (A11), which has been reported to specifically recognize a generic epitope common to prefibrillar oligomers but not monomers or fibrils27 (Figure S1a). By blotting a replicate membrane with anti-Aβ 1–16 (6E10) antibody, which does not discriminate different conformations of Aβ, a similar amount of protein was shown in all 3 Aβ preparations. Amyloid fibrils probe ThT showed fluorescence response in the increasing order of freshly prepared Aβ monomers, followed by oligomer and fibrils as expected (Figure S1b).
The photophysical properties of BD-Oligo are characterized and summarized in Figure S2. To quantify the affinity of BD-Oligo for Aβ oligomers, we measured the apparent binding constant (Kd) of BD-Oligo by conducting a saturation assay. Transformation of the saturation binding data to a Scatchard plot indicated the affinity of BD-Oligo for oligomers with a Kd value of 0.48 µM (Figure S3).
BD-Oligo Detects Oligomers on Fibril Formation Pathway
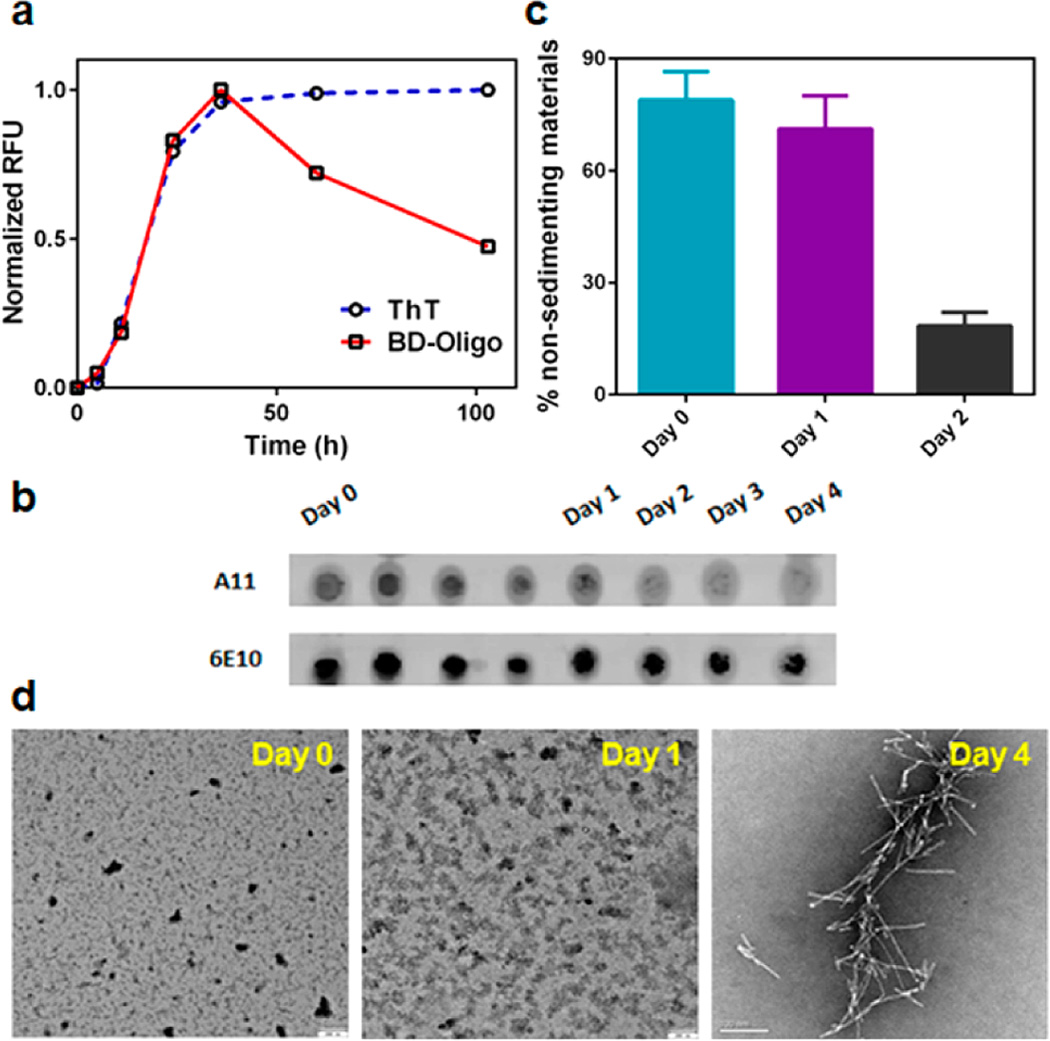
Next, we investigated the oligomer-sensing ability of BD-Oligo over the course of Aβ fibril formation using the same peptide preparation instead of 3 different preprepared conformations as described earlier. To do this we subjected Aβ peptide to fibril-forming conditions, and at each selected time point, a small aliquot was sampled and added to BD-Oligo for fluorescence measurement. Concurrently, Aβ fibril formation samples were monitored with ThT, which reaches a maximum fluorescence after about 1 day and plateaus for the remaining incubation period. Measurements with BD-Oligo observed a gradual increase in fluorescence, which reaches the maximum fluorescence intensity at about day 1 incubation, followed by a decrease in signal over the remaining incubation period (Figure 2a, Figure S4). Fluorescence measurement of BD-Oligo alone in the same manner reveals no change in its signal intensity (data not shown). We postulate that the observed change in fluorescence signal is an indication of BD-Oligo detecting Aβ oligomeric species on-fibril pathway, whereby the signal increases as monomers aggregate into oligomers but decreases as more Aβ assemble into fibrils.
Figure 2.
Biophysical characterization of oligomer-specific response. (a) Time-dependent fibril formation of Aβ was monitored by ThT, whereas BD-Oligo detects on-fibril pathway oligomers (dye 5 µM, Aβ 20 µM). (b) Kinetics of oligomer-specific immunoreactivity during fibrillogenesis, as probed by oligomer-specific A11 antibody and 6E10 antibody against Aβ. (c) Pelleting assay for Aβ at various time points after fibril formation time course has been initiated. (d) Transmission electron microscopy (TEM) images of Aβ at day 0, day 1, and day 4 of fibrillogenesis.
To elucidate the aggregated species or the changes in protein conformations that BD-Oligo may be recognizing, we performed biophysical characterization of the sample during Aβ fibril formation. Particular attention was paid toward the day 1 species, where the probe has been observed to yield maximum fluorescence enhancement. Dot blots over the course of fibril formation showed that A11 recognizes earlier species in the incubation, most intense at 3–5 h, as compared to BD-Oligo, which recognizes the later (day 1) species (Figure 2b). Pelleting assay showed that at day 1, quite similar to day 0, the majority of Aβ are still in solution and have not aggregated into large sedimenting materials. This implies that the aggregated species which enhanced the fluorescence of BD-Oligo are soluble, which is in stark contrast to the decrease in the fraction of soluble protein after 2 days incubation (Figure 2c). At the same time, transmission electron microscopic (TEM) images taken at the end of the 4 day incubation confirmed the presence of Aβ fibrils. In contrast, TEM images captured either immediately after fibril formation has been initiated (day 0) or after 1 day incubation did not yet show any signs of fibrils (Figure 2d). The secondary structure of Aβ analyzed by circular dichroism (CD) spectroscopy at selected time points indicated that Aβ is a random coil when freshly initiated to form fibrils (day 0), consistent with reports in the literature,28 while day 1 species is observed to possess β-sheet content, similar to fibrils formed at day 4 (Figure S5). Taken together, the presence of β-sheet structure alone does not suffice to explain the binding specificity of our probe.
Structural Characteristics of Aβ Oligomer Complex with BD-Oligo
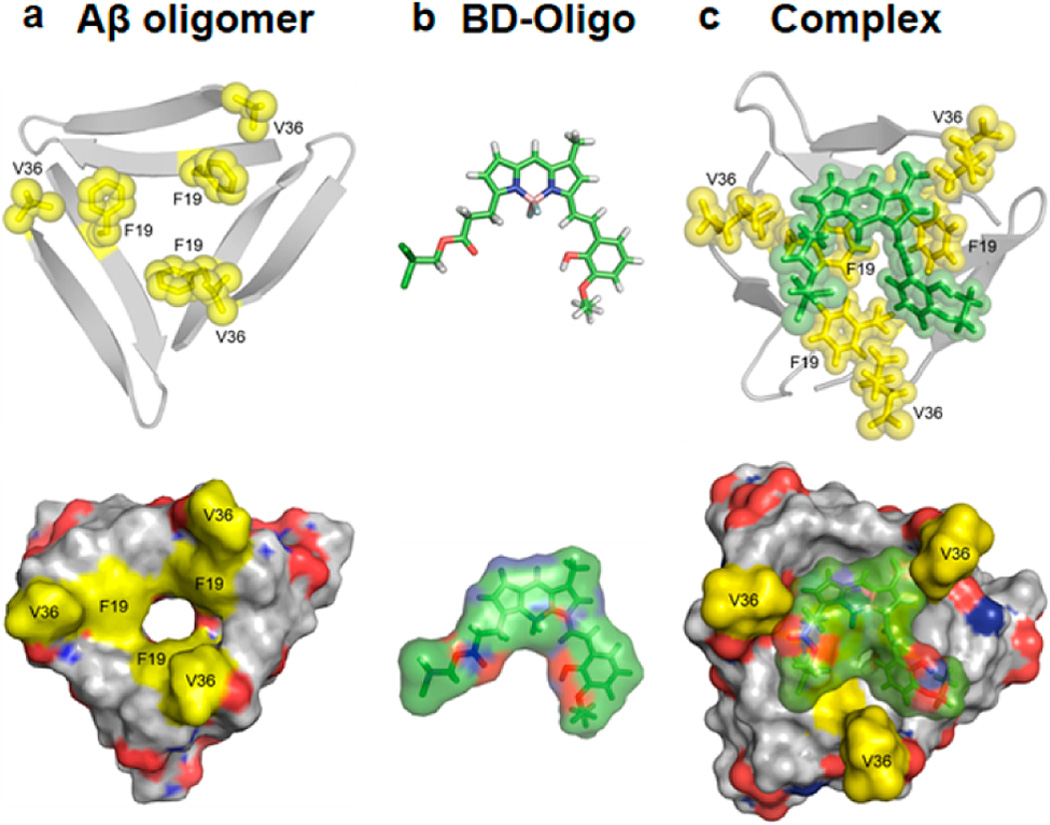
To understand the structural features and the binding specificity of BD-Oligo for Aβ oligomer over Aβ monomer and fibrils, we performed quantum mechanical calculations for BD-Oligo followed by a molecular docking search and molecular dynamics (MD) simulations for the complex of BD-Oligo and Aβ oligomer. To construct Aβ oligomer structure, we used X-ray-determined Aβ trimers derived from the β-amyloid peptide as a working model for toxic Aβ oligomer associated with neurodegeneration in AD (Figure 3a).29 Though not a true depiction of the structure, the described computation methods offer a possible approximation as a starting point. BD-Oligo is most stable as a planar form in the gas phase as well as in an aqueous environment based on quantum mechanical calculations at the B3LYP/6-31G* level (Figure 3b). To search for the stable complex structure of BD-Oligo with Aβ oligomer, we performed a molecular docking search followed by all-atom, explicit water MD simulations (see Supporting Information for detailed computational methods). Upon complexation, BD-Oligo adopts a conformational transition from planar to twisted geometry in order to maximize the interaction with Aβ oligomer (Figure 3c). The main binding mode is π–π stacking interactions between the aromatic rings of BD-Oligo and the exposed hydrophobic patches of Aβ oligomer. More specifically, the BODIPY ring and the phenyl ring of BD-Oligo are recognized by hydrophobic F19/V36 residues in Aβ oligomer. Moreover, the carbonyl group of BD-Oligo forms CH---O bonding with V36 side chain. These binding modes between BD-Oligo and F19/V36 residues of Aβ oligomer are oligomer specific, since F19/V36 residues are exposed to solvent only in Aβ oligomer but not in Aβ fibrils.30 In addition, the F19/V36 residues are also less exposed to solvents in Aβ monomer, which displays intrinsic disorder in aqueous environments.31 The exposed F19/V36 residues which are only present in Aβ oligomer and not (or much less) in Aβ fibril (Aβ monomer) are quite suitable for BD-Oligo recognition by executing π–π stacking interactions as well as H bonding between them. This structural analysis offers the molecular motif on why BD-Oligo is an Aβ oligomer-specific detector.
Figure 3.
BD-Oligo complex with Aβ oligomers. (a) Aβ oligomer from X-ray (4NTR) from ref 29. F19 and V36 residues are shown in yellow. (b) Optimized BD-Oligo structure at the B3LYP/6-31G* level. (c) Simulated complex structure of BD-Oligo and Aβ oligomer.
Thermodynamic Calculations for BD-Oligo Complex with Aβ Oligomer
To further characterize the molecular origin and binding affinity upon complexation of BD-Oligo with Aβ oligomer, we computed the changes in total internal energy (ΔEu), solvation free energy (ΔGsolv), and free energy (Δf) upon its complexation. The internal energy was directly computed from the force field used for the simulations, whereas the solvation free energy was calculated using the integralequation theory of liquids.32 By combining the internal energy and solvation free energy, we obtain the free energy (f = Eu + Gsolv). The binding free energy upon BD-Oligo complexation with Aβ oligomer is computed to be −27.2 kcal/mol in aqueous environments. On the basis of the site-directed thermodynamics analysis33 of the binding free energy, it is evident that the hydrophobic residues of F19/V36 in Aβ oligomer contribute most distinctively to the binding free energy upon complexation (Figure S6). Thermodynamic analysis based on the simulated complex structure confirms that the hydrophobic patches of F19/V36 in Aβ oligomer are the main contributors to recognize BD-Oligo in aqueous environments.
Aβ Oligomer Staining with BD-Oligo in Live AD Brain
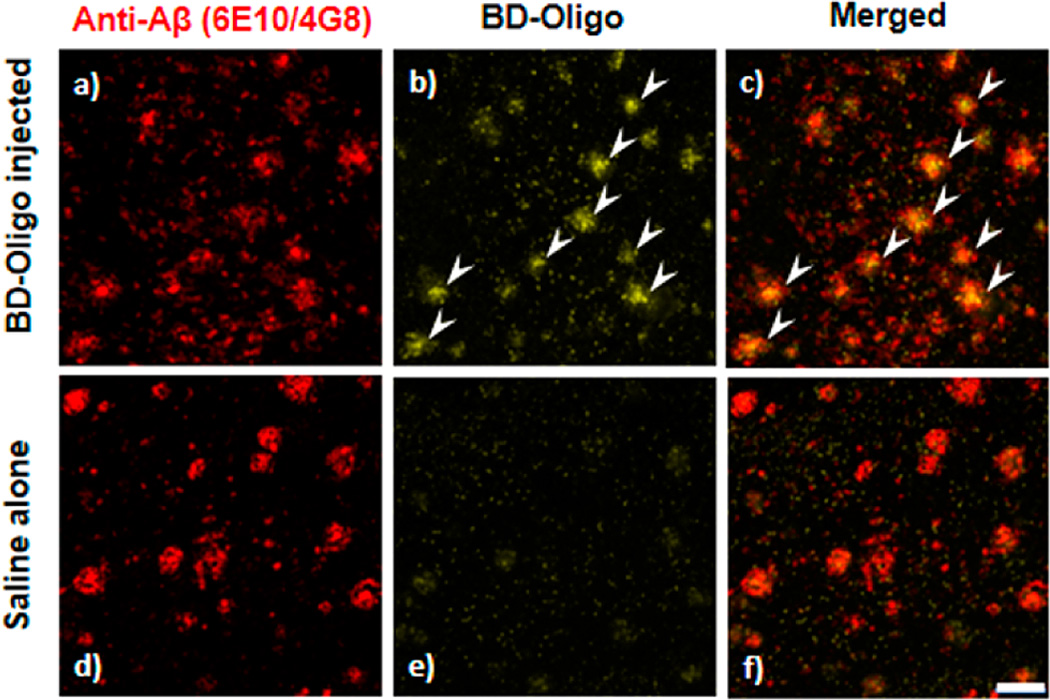
Encouraged by the in vitro findings, we further investigate the oligomer detection ability of BD-Oligo in biological sample using a set of brain tissue fluorescence imaging experiments. Immunofluorescence analysis of 18 month old APP/PS1 transgenic (Tg) mouse brain with anti-Aβ (6E10/4G8) antibody showed that extracellular Aβ deposition is evident. In addition, 6E10/4G8 also identified sites of Aβ intracellular accumulation (Figure 4a). Intraperitoneal (ip) injection of BD-Oligo resulted in fluorescent labeling of AD brain tissue of APP/PS1 Tg mice, which indicates that BD-Oligo is able to cross the BBB and that there is no apparent toxicity associated with in vivo injection. Interestingly, BD-Oligo labeling not only appeared in the central core of the plaques but is also present in the less compacted periphery of plaques indicating oligomer staining (Figure 4b). In addition, there appeared to be some punctate, possibly intraneuronal staining with BD-Oligo surrounding plaques, which had brighter intensity than the endogenous autofluorescence present in the control APP/ PS1Tg mouse brains. Figure 4c indicated the labeling of BD-Oligo, which colocalized with the labeling using anti-Aβ antibodies 4G8/6E10. Fluorescent staining was not present in the APP/PS1 mouse injected with saline alone (Figure 4e). Taken together, BD-Oligo successfully penetrates the BBB to show Aβ oligomers detection capabilities in the brains of the AD transgenic mice model without toxicity.
Figure 4.
Ex vivo binding of BD-Oligo in 18 month old AD mouse brains. (a, b, and c) Fluorescence in the APP/PS1 mouse brain injected with BD-Oligo using the channel for 6E10/4G8 labeling, BD-Oligo labeling, and the merged image, respectively. BD-Oligo fluorescence was present in the brain 24 h after an ip injection of BD-Oligo (see b), which colocalized with the Aβ labeling (see c). Arrows indicate plaques with colocalization. (d, e, and f) Fluorescence in the APP/PS1 mouse brain injected saline alone using the channel for 6E10/4G8 labeling, BD-Oligo labeling, and the merged image, respectively. There are no plaques seen in the BD-Oligo channel in the control saline-injected mice, indicating the specificity of the BD-Oligo oligomer labeling. Scale bar 100 µm.
DISCUSSION
Studies over the past decade have suggested that oligomers of Aβ are now thought to play a central role in neurodegeneration in Alzheimer’s disease. Despite the great personal and economic toll associated with the disease, progress in developing effective treatments remains slow. A significant factor is the lack of powerful diagnostic methods, especially for the earliest stage of Alzheimer’s disease, which are needed for effective disease intervention and management.
BD-Oligo was found through a systematic screening of 3500 fluorescent compounds selected from our in-house diversity-oriented fluorescence libraries. DOFL has shed light on sensor development in the past decade.17–21 The rationale for adopting such a tedious approach is due to the lack of mechanistic cues to rationally design a probe for Aβ oligomers. While the structures of Aβ fibrils are relatively well understood, knowledge regarding the structures of oligomers is still limited, largely due to their heterogeneous and transient nature. Our results show that BD-Oligo is capable of differentiating Aβ oligomers-containing samples from controls as well as the versatility of detecting Aβ oligomeric species on-fibril pathway during Aβ fibril formation.
The hydrophobic central and C-terminal regions of Aβ are known to participate in aggregation to form fibrils and are likely involved in the aggregation of oligomers.34 Although many molecular details of the aggregation processes are yet to be elucidated, the formation of β-sheets appears to be involved. In the current study, biophysical characterization of Aβ peptide sample during fibrillogenesis renders the presence of β-sheet structure alone insufficient to explain the binding specificity of BD-Oligo. Whatever assembly state or conformational change of Aβ BD-Oligo may recognize exists in soluble, prefibrillar Aβ aggregates. It is believed that aggregated Aβ peptides which have not attained the final mature form of an amyloid fibril display exposed hydrophobic patches. In fact, 4,4-bis-1-phenylamino-8-naphthalenesulfonate (bis-ANS) was shown to bind oligomeric intermediates,35,36 which has been widely used in the protein folding field for many decades as a marker for surface-exposed hydrophobic patches and molten-globule-like characteristics.37 Moreover, MD simulations for the complex of BD-Oligo and Aβ oligomers revealed the main binding mode to be π–π-stacking interactions in addition to H bonding between BD-Oligo and the exposed hydrophobic patches of Aβ oligomers. The proposed interactions are deemed oligomer specific, since the hydrophobic patches are exposed to solvent only in Aβ oligomers but not in Aβ fibrils or Aβ monomer. As most BODIPY dyes tend to form aggregates in polar solutions due to their relatively hydrophobic nature,38 we postulate that the interaction of BD-Oligo and Aβ oligomers is strong enough to disassemble BD-Oligo aggregates, which subsequently manifests as an enhancement in fluorescence signal.
It has been suggested that insoluble amyloid plaques may represent a reservoir that releases toxic soluble oligomers.3 We postulate that the tissue staining pattern is a reflection of this phenomenon, where BD-Oligo-labeled-soluble Aβ intermediates are associated with plaque cores, as well as with the periphery of plaques. Further support for this hypothesis is provided by the observation of a halo of enlarged, abnormal neuronal processes surrounding amyloid plaques, suggesting that the source of synaptoxicity resides within the plaque and can diffuse to distant locations.39,40 Moreover, considering the fact that the kinetic data (Figure 2) shows BD-Oligo to be labeling later assembly states of Aβ while A11 recognizes earlier prefibrillar, Aβ oligomers, it may explain why our probe labeling is associated with plaques and the periphery of such areas. On the other hand, it is also possible that BD-Oligo labels the transient, unstable oligomer species on transition to elongating fibrils, which may be present in the amyloid plaques and its periphery.
CONCLUSION
In summary, through high-content DOFL screening, we discovered BD-Oligo as a promising fluorescence sensor for the detection of Aβ oligomers. BD-Oligo demonstrated dynamic oligomer monitoring during Aβ fibrillogenesis, as Aβ peptide was induced to form fibrils over time. The sensing process is based on π–π-stacking interactions in addition to H bonding between BD-Oligo and the exposed hydrophobic patches of Aβ oligomers, as determined by computational techniques. BD-Oligo is able to cross the BBB to give rise to oligomers detection in the brains of AD transgenic mice model without toxicity. Imaging agents than can detect Aβ oligomers in vivo are believed to be essential for disease diagnosis, progress, and medical treatment monitoring and are therefore greatly needed. As such, BD-Oligo provides a good starting point for further probe development applicable in the studies and to assist the research of AD associated with oligomer sensing.
MATERIALS AND METHODS
Diversity-Oriented Fluorescence Library (DOFL) High-Throughput/Content Screening
DOFL compounds were diluted from 1 mM DMSO stock solutions with the culture medium to make a final concentration of 1 µM. Chinese Hamster Ovary (CHO) cells and 7PA2 cells, which were both kindly donated by Dr. Edward H. Koo (University of California, San Diego), were plated side by side in 384 well plates and incubated with DOFL compounds for 2 h at 37 °C. 7PA2 cells were stably transfected with plasmid encoding APP751 with V717F mutation and reported to produce low MW Aβ oligomers (up to 4-mer) in intracellular vesicles prior to secretion into the cell culture medium.41 Detailed characterization of 7PA2 cells has been reported in the literature.42,43 The fluorescence cell images of two regions per well were acquired using an ImageXpress Microcellular imaging system (Molecular Device, Sunnyvale, CA) with 10× objective lens, and the intensity was analyzed by MetaXpress image processing software (Molecular Devices, Sunnyvale, CA) and manual observation. The compounds which stained 7PA2 cells with brighter appearance than CHO cells were selected as candidates.
Peptide Preparation
Synthetic Aβ1–40 was purchased from American Peptide Co. (Sunnyvale, CA) in lyophilized form. Dry peptide was dissolved in 1,1,1,3,3,3-hexafluoro-2-isopropanol (HFIP) and incubated at 25 °C for 1 h to remove any preformed aggregates. It was aliquoted into small aliquots and dried using a speed-vac. The dry peptide was stored at −20 °C until required, where each aliquot was then dissolved in 5 M GuHCl 10 mM Tris·Cl pH 8 to 1 mM peptide solution. After sonication in a sonicating water bath for 15 min, the solution was diluted with phosphate-buffered saline (PBS), pH 7.4, and stored on ice until use. This freshly prepared sample is referred to as monomer.28 To form fibrils, 100 µM sample is incubated for 24 h at 37 °C with 5 s shaking at a 7 min interval. Preformed oligomers were prepared by Aβ1–40 peptide solubilized in borate-buffered saline (50 mM BBS/PBS) and reacted with 5 mM glutaraldehyde overnight at 37 °C to produce stable oligomers by controlled polymerization, as previously described.25,26 The solution was neutralized with Tris buffer and then dialyzed against deionized distilled water overnight and lyophilized. Prior to fluorescence assays, it is resolubilized in deionized distilled water and diluted in PBS. Western blot performed on the sample with anti-Aβ 4G8/6E10 as primary antibody revealed major band of about 80 kDa and higher without monomers. By electron microscopy, the sample makes spheres of 10–20 nm.
Time-Dependent Fibril Formation
For monitoring of fibril formation over time, 40 µM peptide solution of Aβ1–40 was prepared as above and incubated at 37 °C with 5 s shaking at every 7 min interval. Fluorescence readings were taken at various time point intervals by mixing a 30 µL aliquot of peptide solution to 10 µM dye. ThT signal was monitored at 480 nm by 444 nm excitation, whereas BD-Oligo was excited at 530 nm and its emission detected at 585 nm. Fluorescence was measured using a SpectraMax M2 spectrophotometer (Molecular Devices, Sunnyvale, CA). Aβ1–40 was also coincubated with dye to study any effects the dye may have on fibril formation.
Dot Blot Analysis
A 3 µL amount of 40 µM Aβ1–40 sample was spotted onto nitrocellulose membrane (Bio-Rad) at selected time points. The membranes were blocked by 10% (w/v) fat-free milk in 50 mM Tris 150 mM NaCl, pH 7.4, and 0.05% (v/v) Tween-20 (TBST buffer) for 1 h at room temperature, followed by incubation with antioligomer polyclonal A11 antibody (1:1000 dilution; Invitrogen) or Aβ1–16 (6E10) monoclonal antibody (1:1000 dilution; Covance) in 5% (w/v) fat-free milk and TBST buffer overnight at 4 °C. The membranes were washed 3 times in TBST before incubation with antirabbit or antimouse antibody (1:5000 dilution) in 5% (w/v) fat-free milk and TBST buffer at room temperature for 1 h.
Pelleting Assay
Aβ1–40 samples were incubated at 37 °C. At selected time points, aliquots of 150 µL were removed and subjected to centrifugation at 100 000 rpm (TL-100 rotor, Beckman) for 20 min at 4 °C. Under these centrifugation conditions, monomeric Aβ does not sediment significantly. The concentration of monomeric Aβ in the supernatant after centrifugation was monitored using fluorescence measurements based on the reaction of fluorescamine with primary amine groups. The supernatants (45 µL) were added to a microtiter plate along with 15 µL of 1 mg/mL fluorescamine in DMSO. Samples were incubated at room temperature for 5 min, and fluorescence intensities were measured using a SpectraMax M2 spectrophotometer (Molecular Devices, Sunnyvale, CA) with excitation and emission filters of 355 and 460 nm, respectively.
Transmission Electron Microscopy
At selected time points, Aβ1–40 sample incubated at 37 °C was removed and applied to freshly glow-discharged carbon-coated copper grids. The grids were then stained with several drops of 2% potassium phosphotungstate, pH 6.8, and examined using an FEI Tecnai 12 transmission electron microscope operating at 120 kV. Images were obtained using an Olympus SiS MegaViewIII charge-coupled device camera.
Ex Vivo Imaging of Brains
For ex vivo imaging, a stock solution of BD-Oligo was made at 10 mM in 100% DMSO. Eighteen month old APP/PS1 transgenic (Tg) AD model mice were given intraperitoneal (ip) injections with either 1.25 µL of BD-Oligo diluted in 500 µL of saline (n = 2) or 500 µL of saline alone (n = 2). APP/PS1 Tg mice develop amyloid plaques from 4 months of age.44 Mice were anesthetized with an overdose of sodium pentobarbital and perfused 0.1 M PBS, pH 7.4. Brains were removed 24 h after the ip injection and fixed by immersion in periodate-lysine-paraformaldehyde for 24 h, cryo-protected in 30% sucrose for 3 days, and sectioned into 40 µm coronal sections using a cryostat. Brain sections from the BD-Oligo-injected mouse and the control APP/PS1 mouse that received a saline alone injection were then stained for Aβ using fluorescent immunohistochemistry. Briefly, free floating sections were incubated with MOM blocking reagent (Vector) followed by an overnight incubation at 4 °C with anti-Aβ antibodies 4G8 and 6E10 diluted in MOM protein concentrate (Vector), as we previously published.26,45 Sections were then incubated with a 488 conjugated secondary antibody (Jackson Immunoresearch) for 2 h at room temperature, mounted onto slides, and cover slipped. Staining was visualized using a LMD6500 fluorescent microscope (Leica); 6E10/4G8 staining was imaged using in the green (488) channel, and BD-Oligo was imaged in the red (561) channel.
Computational Details
The geometry of BD-Oligo was quantum mechanically optimized in the gas phase as well as in the aqueous phase. The stable complex structure of BD-Oligo with Aβ oligomer was executed by molecular docking search followed by all-atom, explicit water molecular dynamics simulations. Thermodynamic analysis was then performed by applying the liquid integral-equation theory to simulated complex conformations. Further details are provided in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the A*STAR Joint Council Office, Singapore (JCO DP Grant 1230400020), a Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2010-T2-2-030), NIH grants NS073502 and AG20245, and Samsung Science and Technology Foundation under Project Number SSTF-BA1401-13. Chinese Hamster Ovary (CHO) cells and 7PA2 cells were both kindly donated by Dr. Edward H. Koo (University of California, San Diego). We thank Dr. Tim Ryan (Florey Department of Neuroscience and Mental Health, Melbourne) for discussion, Dr. Sung-Jin Park (Singapore Bioimaging Consortium) for assistance with animal-related work, and Joint IMB-IMCB Electron Microscopy Suite and SBIC-Nikon Imaging Centre for microscopy facilities.
Y.T.C., D.S. C.L.T., and S.S. are the inventors of BD-Oligo for which a patent has been applied.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Synthesis and characterization data and additional spectroscopic information (PDF)
The authors declare no competing financial interest.
Contributor Information
Sihyun Ham, Email: sihyun@sookmyung.ac.kr.
Thomas Wisniewski, Email: thomas.wisniewski@nyumc.org.
Young-Tae Chang, Email: chmcyt@nus.edu.sg.
REFERENCES
- 1.Pepys MB. Annu. Rev. Med. 2006;57:223. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 2.Brookmeyer R, Evans DA, Hebert L, Langa KM, Heeringa SG, Plassman BL, Kukull WA. Alzheimer's Dementia. 2011;7:61. doi: 10.1016/j.jalz.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haass C, Selkoe DJ. Nat. Rev. Mol. Cell Biol. 2007;8:101. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 4.Caughey B, Lansbury PT. Annu. Rev. Neurosci. 2003;26:267. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 5.Walsh DM, Selkoe DJ. J. Neurochem. 2007;101:1172. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 6.Rijal Upadhaya A, Kosterin I, Kumar S, von Arnim CA, Yamaguchi H, Fandrich M, Walter J, Thal DR. Brain. 2014;137:887. doi: 10.1093/brain/awt362. [DOI] [PubMed] [Google Scholar]
- 7.Westermark GT, Johnson KH, Westermark P. Methods Enzymol. 1999;309:3. doi: 10.1016/s0076-6879(99)09003-5. [DOI] [PubMed] [Google Scholar]
- 8.Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergstrom M, Savitcheva I, Huang GF, Estrada S, Ausen B, Debnath ML, Barletta J, Price JC, Sandell J, Lopresti BJ, Wall A, Koivisto P, Antoni G, Mathis CA, Langstrom B. Ann. Neurol. 2004;55:306. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Rieves D, Ganley CN. Engl. J. Med. 2012;367:885. doi: 10.1056/NEJMp1208061. [DOI] [PubMed] [Google Scholar]
- 10.Morgado I, Wieligmann K, Bereza M, Ronicke R, Meinhardt K, Annamalai K, Baumann M, Wacker J, Hortschansky P, Malesevic M, Parthier C, Mawrin C, Schiene-Fischer C, Reymann KG, Stubbs MT, Balbach J, Gorlach M, Horn U, Fandrich M. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12503. doi: 10.1073/pnas.1206433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, Su B, Kim CS, Hernandez M, Rostagno A, Ghiso J, Kim JR. Chem Bio Chem. 2010;11:2409. doi: 10.1002/cbic.201000435. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Su B, Zheng H, Kim JR. Mol. BioSyst. 2012;8:2741. doi: 10.1039/c2mb25148e. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi T, Mihara H. Chem. Commun. 2012;48:1568. doi: 10.1039/c1cc14552e. [DOI] [PubMed] [Google Scholar]
- 14.Bruggink KA, Jongbloed W, Biemans EALM, Veerhuis R, Claassen JAHR, Kuiperij HB, Verbeek MM. Anal. Biochem. 2013;433:112. doi: 10.1016/j.ab.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Jameson LP, Dzyuba SV. Bioorg. Med. Chem. Lett. 2013;23:1732. doi: 10.1016/j.bmcl.2013.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith NW, Alonso A, Brown CM, Dzyuba SV. Biochem. Biophys. Res. Commun. 2010;391:1455. doi: 10.1016/j.bbrc.2009.12.091. [DOI] [PubMed] [Google Scholar]
- 17.Kang NY, Ha HH, Yun SW, Yu YH, Chang YT. Chem. Soc. Rev. 2011;40:3613. doi: 10.1039/c0cs00172d. [DOI] [PubMed] [Google Scholar]
- 18.Vendrell M, Zhai D, Er JC, Chang YT. Chem. Rev. 2012;112:4391. doi: 10.1021/cr200355j. [DOI] [PubMed] [Google Scholar]
- 19.Lee JS, Vendrell M, Chang YT. Curr. Opin. Chem. Biol. 2011;15:760. doi: 10.1016/j.cbpa.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Vendrell M, Lee JS, Chang YT. Curr. Opin. Chem. Biol. 2010;14:383. doi: 10.1016/j.cbpa.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Lee JS, Kim YK, Vendrell M, Chang YT. Mol. BioSyst. 2009;5:411. doi: 10.1039/b821766c. [DOI] [PubMed] [Google Scholar]
- 22.Im CN, Kang NY, Ha HH, Bi X, Lee JJ, Park SJ, Lee SY, Vendrell M, Kim YK, Lee JS, Li J, Ahn YH, Feng B, Ng HH, Yun SW, Chang YT. Angew. Chem., Int. Ed. 2010;49:7497. doi: 10.1002/anie.201002463. [DOI] [PubMed] [Google Scholar]
- 23.Kang NY, Lee SC, Park SJ, Ha HH, Yun SW, Kostromina E, Gustavsson N, Ali Y, Chandran Y, Chun HS, Bae M, Ahn JH, Han W, Radda GK, Chang YT. Angew. Chem., Int. Ed. 2013;52:8557. doi: 10.1002/anie.201302149. [DOI] [PubMed] [Google Scholar]
- 24.Yun SW, Kang NY, Park SJ, Ha HH, Kim YK, Lee JS, Chang YT. Acc. Chem. Res. 2014;47:1277. doi: 10.1021/ar400285f. [DOI] [PubMed] [Google Scholar]
- 25.Goni F, Prelli F, Ji Y, Scholtzova H, Yang J, Sun YJ, Liang FX, Kascsak R, Kascsak R, Mehta P, Wisniewski T. PLoS One. 2010;5:e13391. doi: 10.1371/journal.pone.0013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goni F, Herline K, Peyser D, Wong K, Ji Y, Sun Y, Mehta P, Wisniewski T. J. Neuroinflammation. 2013;10:150. doi: 10.1186/1742-2094-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 28.Ryan TM, Friedhuber A, Lind M, Howlett GJ, Masters C, Roberts BR. J. Biol. Chem. 2012;287:16947. doi: 10.1074/jbc.M111.321778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spencer RK, Li H, Nowick JS. J. Am. Chem. Soc. 2014;136:5595. doi: 10.1021/ja5017409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, Schubert D, Riek RP. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17342. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C, Ham S. J. Comput. Chem. 2011;32:349. doi: 10.1002/jcc.21628. [DOI] [PubMed] [Google Scholar]
- 32.Imai T, Harano Y, Kinoshita M, Kovalenko A, Hirata F. J. Chem. Phys. 2006;125:024911. doi: 10.1063/1.2213980. [DOI] [PubMed] [Google Scholar]
- 33.Chong SH, Ham S. J. Chem. Phys. 2011;135:034506. doi: 10.1063/1.3610550. [DOI] [PubMed] [Google Scholar]
- 34.Sandberg A, Luheshi LM, Sollvander S, Pereira de Barros T, Macao B, Knowles TP, Biverstal H, Lendel C, Ekholm-Petterson F, Dubnovitsky A, Lannfelt L, Dobson CM, Hard T. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15595. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frare E, Mossuto MF, de Laureto PP, Tolin S, Menzer L, Dumoulin M, Dobson CM, Fontana A. J. Mol. Biol. 2009;387:17. doi: 10.1016/j.jmb.2009.01.049. [DOI] [PubMed] [Google Scholar]
- 36.Bolognesi B, Kumita JR, Barros TP, Esbjorner EK, Luheshi LM, Crowther DC, Wilson MR, Dobson CM, Favrin G, Yerbury J. J. ACS Chem. Biol. 2010;5:735. doi: 10.1021/cb1001203. [DOI] [PubMed] [Google Scholar]
- 37.Ptitsyn OB. Adv. Protein. Chem. 1995;47:83. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 38.Zhai D, Xu W, Zhang L, Chang YT. Chem. Soc. Rev. 2014;43:2402. doi: 10.1039/c3cs60368g. [DOI] [PubMed] [Google Scholar]
- 39.Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Nature. 2008;451:720. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koffie RM, Meyer-Luehmann M, Hashimoto T, Adams KW, Mielke ML, Garcia-Alloza M, Micheva KD, Smith SJ, Kim ML, Lee VM, Hyman BT, Spires-Jones TL. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4012. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 42.Podlisny MB, Ostaszewski BL, Squazzo SL, Koo EH, Rydell RE, Teplow DB, Selkoe DJ. J. Biol. Chem. 1995;270:9564. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- 43.Podlisny MB, Walsh DM, Amarante P, Ostaszewski BL, Stimson ER, Maggio JE, Teplow DB, Selkoe DJ. Biochemistry. 1998;37:3602. doi: 10.1021/bi972029u. [DOI] [PubMed] [Google Scholar]
- 44.Holcomb L, Gordon MN, McGowan E, Yu X, Benkovic S, Jantzen P, Wright K, Saad I, Mueller R, Morgan D, Sanders S, Zehr C, O’Campo K, Hardy J, Prada CM, Eckman C, Younkin S, Hsiao K, Duff K. Nat. Med. 1998;4:97. doi: 10.1038/nm0198-097. [DOI] [PubMed] [Google Scholar]
- 45.Scholtzova H, Chianchiano P, Pan J, Sun Y, Goni F, Mehta PD, Wisniewski T. Acta Neuropathol. Commun. 2014;2:101. doi: 10.1186/s40478-014-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.