Abstract
The conformational dynamics of a macromolecule can be modulated by a number of factors, including changes in environment, ligand binding and interactions with other macromolecules, among others. We present a method that quantifies the differences in macromolecular conformational dynamics and automatically extracts the structural features responsible for these changes. Given a set of molecular dynamics (MD) simulations of a macromolecule, the norms of the differences in covariance matrices are calculated for each pair of trajectories. A matrix of these norms thus quantifies the differences in conformational dynamics across the set of simulations. For each pair of trajectories, covariance difference matrices are parsed to extract structural elements that undergo changes in conformational properties. As a demonstration of its applicability to biomacromolecular systems, the method, referred to as DIRECT-ID, was used to identify relevant ligand-modulated structural variations in the β2-adrenergic (β2AR) G-protein coupled receptor. Micro-second MD simulations of the β2AR in an explicit lipid bilayer were run in the apo state and complexed with the ligands: BI-167107 (agonist), epinephrine (agonist), salbutamol (long-acting partial agonist) or carazolol (inverse agonist). Each ligand modulated the conformational dynamics of β2AR differently and DIRECT-ID analysis of the inverse-agonist vs. agonist-modulated β2AR identified residues known through previous studies to selectively propagate deactivation/activation information, along with some previously unidentified ligand-specific microswitches across the GPCR. This study demonstrates the utility of DIRECT-ID to rapidly extract functionally relevant conformational dynamics information from extended MD simulations of large and complex macromolecular systems.
Keywords: conformational dynamics, covariance analysis, norm of matrix, ligand efficacy, microswitches, β2-adrenergic G-protein coupled receptor
Introduction
Knowledge about macromolecular conformational dynamics is essential for understanding biological function.1 Molecular dynamics (MD) simulations are extremely powerful towards this understanding in that they provide high-resolution details of spatial arrangement of particles in a system over time. Atomistic MD simulations spanning a wide range of time-scales have been employed extensively in the past to understand different aspects of dynamics and functions of biomacromolecules of different sizes.2-5 However, the overwhelming amount of information in MD trajectories detailing the time-evolution of the system is often under-utilized.6 Thus, it is beneficial to refine analysis methods to optimize the extraction of functionally relevant conformational information.
Covariance matrices quantify coupling between atomic displacements and provide a detailed means of extracting comprehensive information from MD trajectories. Information from the covariance matrices has been useful for quantifying configurational entropy,7,8 identifying quasi-harmonic modes,9 and extracting functionally relevant global motions.10 In Cartesian space, for an N-atom system, a covariance matrix contains 3N × 3N elements, making extraction of relevant information challenging for large systems, given the high dimensionality. In Principal Component Analysis (PCA),11 the covariance matrices are diagonalized to yield collective variables (eigenvectors) sorted according to their contribution to the total mean-squared fluctuation.6 In other words, the MD trajectory is projected onto principal components (collective variables), and it has been found that these collective coordinates constitute a low-dimensional subspace that represents a significant part of the macromolecular motion.12 Collective coordinates thereby drastically reduce the number of descriptors that are required to characterize the dynamics of the macromolecule.13
In certain cases, large-amplitude functionally relevant deformations are harmonic, aligning with the macromolecular intrinsic normal modes.14,15 Consequently, several studies have attempted to project macromolecular dynamics using elastic network models (ENM),16,17 where the macromolecule is defined as a set of nodes connected by harmonic springs.18 Normal modes/collective coordinates are then derived from this simplistic harmonic approximation of the potential energy function near equilibrium. Covariance-based methods delineating modes from ENM have been used to identify domains within a macromolecule that cluster structural features with concerted structural fluctuations.19-21 While this is functionally relevant information,22,23 ENM-based methods suffer from two major limitations. First, by either treating the macromolecule in isolation, or using implicit solvent schemes to screen inter-atomic interactions, the methods likely fail to account for configurational entropy of the surrounding environment. Second, given the ruggedness of the potential energy surface and anharmonicity of the energy barriers for conformational transitions, harmonic approximations tend to lose critical information relevant for characterizing macromolecular conformational behavior.14,24
A number of factors modulate the conformational dynamics of a macromolecule. These include changes in ion25,26 or osmolyte27,28 concentration, ligand binding or dissociation,29-31 and interactions with other macromolecules,32-34 to list a few. While some of these factors modulate large domain motions,35,36 others are more subtle.37,38 Often there is a large overlap in the low-frequency global motions across these different conformational equilibria, with only small changes occurring in the high frequency motions. All of these effects are likely to be missed when characterizing the dynamics through ENM or normal mode analysis.
In the present work, we describe a method called DIRECT-ID (Dimensionality Reduction through Covariance matrix Transformation for Identification of Differences in dynamics) that quantifies changes in the macromolecular conformational dynamics using the norm of the differences in the covariance matrices calculated from multiple MD trajectories. The MD simulations can be independent runs under different conditions to gain insight into the modulation of macromolecular conformational dynamics. For each pair of trajectories, structural features with the largest changes in atomic motions are identified. The covariance matrices are constructed from non-hydrogen atoms of the macromolecule, thus the method is sensitive to both global motion changes as well as smaller higher frequency mode changes. Given that the bonds involving hydrogen atoms are generally constrained in most biomolecular MD simulations, their dynamics are dominated by the associated non-hydrogen atoms and thus not included in the current implementation of DIRECT-ID.
The ability of DIRECT-ID to identify functionally relevant features is evaluated by probing the modulation of conformational dynamics of the β2-adrenergic G-protein coupled receptor (β2AR) by ligands of different functions. Studies have shown that instead of a previously hypothesized two state active-inactive model, GPCRs have multiple, distinct active-inactive conformations.39 Ligands of different efficacies modulate the conformations of GPCRs differently,40 ultimately affecting the downstream functional signaling profiles. Consequently, a number of computational41,42 and experimental31 studies have probed the effect of ligand binding on β2AR. Thus, the β2AR is a good system to validate the ability of DIRECT-ID to quantify changes in the conformational dynamics and identify structural features that are sensitive to these changes.
Method
1) Multiple MD simulations of the macromolecule of interest are initially performed. These simulations may vary in any manner dictated by the user. Each of these simulations is recommended to span similar time-scales and should be tested for convergence with any of the conventionally used metrics (e.g., RMSD of the backbone atoms) or using our convergence analysis (see below). From the trajectories, the Cartesian coordinates of the atoms of interest are used for the calculation of the covariance matrices and the analysis as follows.
2) In the Cartesian space, the covariance between two atoms, along an axis x, is calculated as ⟨r1x · Δr2x⟩, where Δr1x = r1x – r10x, and Δr2x = r2x – r20x, r1x and r10x, r2x and r20x are the positions at a time t and in a reference conformation along an axis x for atoms 1 and 2, respectively. Angle brackets denote ensemble average. For a system containing N atoms being analyzed, the covariance matrix is symmetric with 3N × 3N elements,
3) Given a set of MD simulations, i,j,…, M of the N-atom system, the covariance matrices Ci, Cj,…,CM are calculated with respect to a single reference configuration, r0.
4) Differences between two covariance matrices Ci and Cj are calculated as
It is important that the same reference configuration r0 be used for the calculation of the covariance matrices from the different simulations, instead of the mean structures from within the simulations. This is because, when the same r0 is used in calculation of Ci, and Cj across two trajectories i and j, then elements of the Di–j matrix, , (where p and q refer to a row and column, respectively) characterize the differences in the atomic displacements between the two trajectories. On the other hand, when the mean structures from the simulations are used in Ci, then , characterize the differences in the variance of the motion.
5) Differences in conformational dynamics across two trajectories are quantified using the norm of the difference matrix. Norm, ∥Di–j∥1, is the maximum absolute column sum of the difference matrix and can be considered as the magnitude of the matrix. A higher norm signifies a greater difference between the two corresponding covariance matrices. Given the symmetric nature of the covariance and difference matrices, calculations are invariant between ∥Di–j∥1 and ∥Di–j∥∞, the maximum absolute row sum. Thus, for M trajectories, we have a matrix of norms,
that is a triangular matrix with diagonal elements equal to 0.
6) Thus, differences across M covariance matrices of 3N × 3N elements are transformed into a single norm matrix containing M × M elements. NM×M can be parsed to identify trajectories with the biggest differences in their conformational dynamics. Difference matrices, Di–j, between each pair of trajectories, i and j, can be parsed to identify elements with the highest magnitude. The elements correspond to pairs of non-hydrogen atoms in the macromolecule with largest changes in the correlated motions between the two trajectories. These atom pairs are then used to establish a list of residues/nucleotides (depending on the nature of the macromolecule) associated with the corresponding non-hydrogen atoms; an example is shown in Table 1. Thus, the final output is a table that lists macromolecular residues with the most prominent changes in their dynamics between the two trajectories.
Table 1.
β2AR residues with the biggest changes in conformational motions between two trajectories. Ballesteros-Weinstein numbering of the residues is marked in the superscript.
| Trajectories | ∥ D i–j ∥ 1 | Residues with biggest change in conformational motions |
Published mutations and their effect on the receptor |
|---|---|---|---|
| APO-CARA | 194.4 | R1514.43, F1664.58, I1694.61 Q1704.62, Q1975.37, L2756.37, C3277.54 |
|
| APO-BIA | 403.1 | E1223.41, R1514.43-W1584.50, T1644.56, Y3267.53 | E122W: inactivation,79 W158F: reduce agonist affinity,80 T164I: reduce basal activity,81 Y326A: inactivation82 |
| APO-EPNE | 233.6 | G351.34-M361.35, I381.37, L421.41, F2085.47, F2175.56, Y2195.58, L2756.37, I2776.39, F2826.44, N3187.45, Y3267.53-R3287.55 |
|
| APO-SALB | 140.2 | G351.34, F491.48, F1664.58, Q1704.62, Q1975.37, V2065.46, F2175.56, I2786.40, H2966.58, I2986.60, E3067.32, L3107.36, I3257.52-R3287.55 |
|
| CARA-BIA | 415.1 | E1223.41, R1514.43-V1604.52, L1634.55, T1644.56, Y3267.53, C3277.54 |
T164V: reduce agonist-driven activation81 |
| CARA-EPNE | 252.4 | R1514.43, F1664.58, I1694.61, Q1704.62, Q1975.37, F2175.56, L2756.37, E3067.32, N3187.45, C3277.54 |
L275A: reduce agonist activation83 |
| CARA-SALB | 249.4 | R1514.43, F1664.58, I1694.61 Q1704.62, Q1975.37, F2175.56, L2756.37, E3067.32, C3277.54 |
|
| EPNE-BIA | 414.8 | E1223.41, R1514.43, I1534.45-I1594.51, L1634.55, T1644.56, F2175.56, N3187.45 Y3267.53 |
F217A: reduce activity,84 N318A: inactivation82 |
| EPNE-SALB | 209.4 | G351.34-M361.35, I381.37, L421.41, F2085.47, F2175.56, Y2195.58, L2756.37, I2776.39, T2816.43, N3187.45, Y3267.53-R3287.55 |
|
| SALB-BIA | 425.0 | E1223.41, R1514.43-L1554.47, V1574.49-I1594.51, T1644.56, F2175.56, Y3267.53, C3277.54 |
DIRECT-ID is written as a C++ program that can handle covariance matrices outputted from CHARMM43 or the GROMACS44 utility “gmx covar.” The code extracts structural features using these matrices. Note that in general any molecular simulation software can be used to generate the trajectories and calculate the covariance matrices, as the C++ code only requires as ASCII covariance matrix of the form shown above in step 2.
Simulation Details
One micro-second MD simulations of the β2AR were run in both the apo form and in complex with ligands: BI-167107 (BIA),35 epinephrine (EPNE),45 salbutamol (SALB),46 and carazolol (CARA).47 The active conformation of β2AR from PDB 3P0G35 was used to initiate the MD simulations. The region of the structure corresponding to T4 lysozyme, which was inserted between TM5 and TM6 to facilitate crystallization, was deleted and the third intracellular loop (ICL3) was built in its place using MODELLER48. In total, 100 models were generated and ranked using the Discrete Optimized Protein Energy (DOPE) method49 and the highest ranking model was used as a starting structure. Binding poses of ligands EPNE and CARA were obtained from PDB 4LDO and 2RH1, respectively. SALB was docked using Autodock-Vina,50 and the top scoring binding pose used in the simulation had an orientation similar to that of EPNE. Disulphide bridges between residues 106-191 and 184-190 were maintained. All simulations were initiated from the same starting conformation which was also used as the reference in the DIRECT-ID analysis.
Empirical force field parameters for β2AR and lipids were taken from CHARMM36,51-53 ligand topologies and parameters were derived from CGenFF54 and water was treated using the TIP3P model.55,56 β2AR was inserted into a rectangular palmitoyl oleoyl phosphatidyl choline (POPC) membrane containing ~252 lipids, including 10% cholesterol using CHARMM-GUI;57 the remainder of the unit cell volume was filled with ~0.15 M NaCl aqueous solution including sufficient counterions to neutralize the system. All systems were energy minimized in two steps, first by 1500 steps of steepest descent (SD)58 and the next 1500 steps with the adopted basis Newton-Raphson (ABNR) algorithm.59 During minimization, the non-hydrogen atoms of the protein backbone and side chains were harmonically restrained using force constants of 10 kcal/mol/Å2 and 5 kcal/mol/Å2, respectively. Influx of water molecules into the hydrophobic core of the TM region was prevented using a harmonic restraining potential of 2.5 kcal/mol/Å2 along the x-y plane at positions of ±11 Å along the z-axis from the center of the receptor. The same force was also used to keep the heads and tails of the lipids in place, and configurations of the lipids were maintained with harmonic dihedral restraints with a force constant of 250 kcal/mol/rad2. The same restraints were used during the 375 ps of equilibration with a 1-fs time step, and the restraint forces were gradually reduced as described in our previous work.60 The first 50 ps of equilibration was performed using Langevin dynamics with a friction coefficient of 3 ps−1 on all non-hydrogen atoms. The remaining 325 ps of the equilibration used constant pressure-temperature (NPT) dynamics using the Langevin Piston integrator.61 Covalent bonds involving hydrogens were fixed at the equilibrium length by the SHAKE algorithm.62 Lennard-Jones (LJ) interactions were switched to zero from 10 to 12 Å and the non-bonded pair list was generated out to 14 Å and updated heuristically.63 Electrostatic interactions were calculated by the particle-mesh Ewald64 summation method with a real space cutoff of 12 Å. These minimization and equilibration simulations were performed using CHARMM.59
Following equilibration, production runs were performed using GROMACS44 and with the leapfrog integrator (GROMACS integrator “md”) with a time step of 2 fs. The Nosé–Hoover method65,66 was used to maintain the temperature at 300 K with the protein and the remainder of the system separately coupled to heat baths. Pressure was maintained at 1 bar using the Parrinello–Rahman67 barostat. The time constant used for temperature and pressure coupling were both set to 1 ps. The LINCS68,69 algorithm was used to constrain all covalent bonds involving hydrogen atoms and water geometries were kept rigid with SETTLE.70 LJ interactions were switched off smoothly in the range of 10–12 Å, and the particle mesh Ewald (PME) method64,71 was used to treat long-range electrostatics with cubic B-spline interpolation and a maximum grid spacing of 1.2 Å. Long-range dispersion correction was applied to the energy and pressure terms. No restraints were applied during the production.
Results
Across the five 1-μs MD simulations, the overall structure of the β2AR was stable, with the root mean square deviation (RMSD) of the backbone atoms across the helices H1-H8 (excluding the loops), plateauing at around 0.4 μs (SI, Fig. S1). Ligands were also stable in the β2AR binding pocket, maintaining interactions with the residues in helices H3, H5, H6 and H7 (SI, Fig. S2). EPNE was slightly more flexible, with a deviation from the crystallographic conformation modulated by a break in its hydrogen bonding with N3127.38 (superscript indicates Ballesteros-Weinstein numbering), while preserving interactions with the rest of the binding pocket (SI, Fig. S2C). Despite the similarity in the backbone RMSD, important differences were noted in the conformational dynamics across the five simulations, as detailed below.
Ligand Modulated G-protein Binding Cavity in β2AR
Structural studies have identified two distinct conformations of β2AR, in which the opening and closing of this cavity are correlated with the activation and inactivation of the GPCR, respectively.35,72 Helix α5 in Gαs of the G-protein heterotrimer enters into this cavity which is located at the intracellular side of the GPCR, and interactions between α5 and the GPCR are known to initiate the subsequent intracellular signaling downstream.73,74 To probe the heterogeneity in the conformational dynamics of β2AR in the apo state and with the different ligands, this G-protein binding cavity was traced using the distances between the center of mass of intracellular side of helices H2-H6 and H3-H7. Residues considered for the center of mass calculation were: V67-Y70(H2), S120-V129(H3), C265-H269(H6) and G320-S329(H7). Using these two distances as reaction coordinates, approximate free energy surfaces were calculated as F(n) = −kBT ln(Nn), where kB is the Boltzmann constant (kcal/mol/K), T is the temperature (300 K), and Nn is the number of times the grid point is visited during the length of a simulation. These reaction coordinates were based on previous conformational dynamics studies that probed GPCR-G-protein coupling specificity75 and passage of water through a GPCR channel.76
As shown in Fig. 1, conformational dynamics of β2AR are clearly different between its apo state and when in complex with different ligands. Apo and CARA-bound β2AR transitioned to distinct “inactive-like” conformations, in which the cavity that accommodates α5 of Gαs is not formed (Fig. 1B and C). Agonists, on the other hand, retained the G-protein cavity, but the size of this cavity was smaller than that found in the active crystal structures (Fig. 1D-F), in agreement with recent studies.75,76 Conformational sampling was also distinct between the agonists, such that BIA-bound β2AR more frequently sampled “active-like” conformations with bigger cavities, than when bound to EPNE or SALB. SALB-bound β2AR also visited more inactive-like conformations, indicating a need for a second ligand or a G-protein to stabilize the active conformation in the presence of this partial agonist. Consistent with previous studies,42 the opening of the cavity is found to be correlated with a break in the bond between R131 of H3 and E268 of H7 SI, Fig. S3). This ionic-bond formed most predominantly in the APO state of β2AR and not observed in the agonist-bound simulations. Notably, although R131-E268 can characterize the transitions between the end-states of β2AR, at least two reaction coordinates are needed to delineate heterogeneity in ligand modulated changes to conformational dynamics of β2AR. For instance, cavity opening can also be traced using distances between H3-H6 and H3-H7 (SI, Fig. S4). These surfaces also depict the conformational heterogeneity similar to that shown in Fig. 1.
Figure 1.
A) View of the transmembrane helices (H1-H7) from the intracellular side. Structural studies indicate the largest conformational differences are between the inactive (green, PDB:2RH1) and active (orange, PDB: 3P0G) states across helices H5 and H6, which consequently modulate the cavity between the helices H2-H6 and H3-H7. Approximate free energy surfaces (in kcal/mol) trace the change in the cavity in the B) absence of ligand (APO), presence of C) inverse agonist, carazolol (CARA), agonists (D) BI-167107 (BIA), E) epinephrine (EPNE) and F) salbutamol (SALB). H2-H6 and H3-H7 distances in the crystal structures are marked with black dots. Residues used for the calculation were: V67-Y70(H2), S120-V129(H3), C265-H269(H6) and G320-S329(H7).
DIRECT-ID Analysis
It is important to note that the choice of the reaction coordinates for the surfaces in Fig. 1 was based on knowledge derived from previous structural studies. These surfaces describe the more global domain-like motions in the receptor without any information about conformational changes of individual residues. Recent studies have shown that conformational changes in a subset of residues, often called “microswitches”, manifest into macroscopic conformational changes indicative of activation/inactivation of the receptor.77,78 To characterize the changes in the conformational equilibria without prior knowledge of reaction coordinates, and to identify structural features such as the microswitches that are responsive to these changes, DIRECT-ID analysis was performed using the trajectories from the five MD simulations. For each simulation, covariance of the atomic displacements was calculated over the 1-μs trajectory for all the non-hydrogen atoms in the seven transmembrane helices (excluding loops), yielding five covariance matrices that each contained 3879 × 3879 elements. Differences between these covariance matrices contain all the variations in conformational dynamics, but given the size of these systems, representation of these matrices becomes intractable (see Fig. 2A). Thus the differences are quantified using ∥Di–j∥1 as shown in Fig. 2B. The largest differences all involve the agonist BIA with the all other states, with the smallest difference occurring between the apo state and the inverse-agonist CARA, and the partial agonist SALB. The values of ∥Di–j∥1 are included in Table 1.
Figure 2.
A) Differences in the covariance matrices of β2AR bound to CARA vs. BIA ( DCARA–BIA ). B) DIRECT-ID analysis quantifies conformational differences across 1-μs MD trajectories of β2AR in the apo state and when bound to CARA, BIA, EPNE and SALB. C) β2AR structural elements identified as having biggest changes in conformational dynamics, retrieved from DCARA–BIA.
Ligand Efficacy using DIRECT-ID
As agonists maximally activate the receptor, and activation is coupled with conformational transition from the apo state, efficacy can be estimated from ∥Dagonist–apo∥1. As shown in Fig.2B, ∥DBIA–APO∥1 (403.1) > ∥DEPNE–APO∥1 (233.6) > ∥DSALB–APO∥1 (140.2). This outcome is consistent with previous observations that BIA has a greater efficacy than the natural ligand, EPNE, and the partial agonist induced submaximal activity even at saturating concentrations.35 Thus, DIRECT-ID is able to quantify the effect of changes to the global motion of the protein, which is critical to the understanding of effects of different ligands.
Atomic Details of Conformational Variations
Once the systems with the largest differences are identified using the NM×M matrix, the corresponding Di–j matrices may be parsed to identify structural features (residues) with the most prominent changes between the two trajectories. Table 1 lists the residues thus obtained across all the pairs of the trajectories. Notably, residues that respond to changes in ligand properties are not limited to the binding pocket or helices H5 and H6, rather they are distributed all across the receptor as described in the following sections.
Agonists Mediated Differences
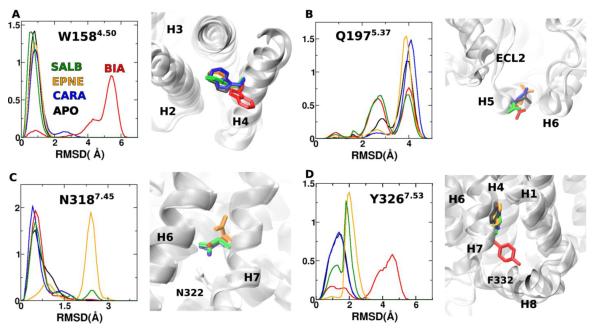
Distinct differences were found between conformational dynamics of β2AR when in complex with agonists versus the inverse-agonist, CARA. This is particularly significant when comparing dynamics between BIA- and CARA-bound β2AR (Fig. 2A,B). Structural features with the biggest changes in their motion between these two simulations span helices H3, H4, H6 and H7 (Fig. 2C, Table 1). Several of these residues (E1223.41, T1644.56, Y3267.53) have been identified through previous network information analysis methods78,85 and site-directed mutational studies (see Table 1) to behave as microswitches and participate in communication pathways that relay activation/inactivation information from the ligand binding pocket to the intracellular G-protein binding regions. In addition, DIRECT-ID identified residues that selectively responded to changes in conformational equilibria of the receptor mediated by only some of the ligands and not others. For instance, W1584.50 underwent a conformational transition that involved a χ2-rotation, pushing it out of the largely hydrophobic cavity between the helices H2 and H3, only when β2AR was bound to BIA (Fig. 3A). Similarly, displacement of Q1975.37 that broke its interaction with H6 was also observed only in the BIA-bound β2AR (Fig. 3B). Y3267.53, is a known microswitch, whose conformational transition allows passage of water through another family A GPCR, the adenosine receptor, was modulated only by its agonist.76 Consistent with this mechanism, only in the BIA-bound β2AR did Y326 undergo a χ1-rotation that disrupted its hydrogen-bond network with H1, H2, and H6 and moved into an alternate rotameric conformation that involved an aromatic stacking interaction with the conserved F3328.50 in TM8 (Fig. 3D).
Figure 3.
RMSD distributions of A) W158, B) Q197, C) N318 and D) Y326. These residues were identified using DIRECT-ID to have significant differences in their conformational motions between β2AR bound to agonists (BIA or EPNE) vs. CARA. Snapshot at 400 ns (where the RMSD values are near the maximum of the probability distributions) reveal the differences in conformations for each of these residues of β2AR in the apo state (black), bound to CARA (blue), BIA (red), EPNE (orange) and SALB (green).
In contrast, the presence of EPNE in the β2AR binding pocket modulated conformational transitions in N3187.45 that were not seen in BIA-bound β2AR (Fig. 3C). Specifically, interactions between N318 and helices H2, H3 and H6, and the N3187.45-N3227.49 (another known microswitch) hydrogen bond were broken. Although occurring at different locations along the receptor, each of these transitions allow for greater flexibility within the transmembrane helices, resulting in a larger opening of G-protein binding cavities (Fig. 1D-F) that correlate with the activation of the receptor.
Partial-Agonist Mediated Differences
SALB is a long-acting partial agonist of β2AR. Functional studies have determined that SALB-mediated signaling is different from other agonists such as EPNE.86 These changes were attributed to conformational transitions that significantly reduced GRK-modulated phosphorylation at the ICL3 and C-terminal tail of β2AR.87,88 Both ICL3 and the C-terminal tail were missing in the crystal structures and not included in the current analysis. Nevertheless, DIRECT-ID identified residues along the receptor with changes in their conformational motions selectively mediated by SALB-bound β2AR (Table 1). For example, F1664.58 (Fig. 4A) and Q1975.37 (Fig. 3B) had transitions similar to those seen in β2AR bound to agonists but different from those in the apo state or the CARA-bound β2AR. Others, such as the F2175.56 (Fig. 4B) underwent conformational transitions that were different from the agonists. χ2-rotation of F217 in SALB-bound β2AR disrupted its interaction with Y1323.51 moving into a hydrophobic cluster with L2125.51, V2135.52 and V2165.55. This transition likely creates a strain in H5 that could propagate downstream and modulate the phosphorylation of ICL3, although additional studies are needed to confirm this hypothesis. Notably, DIRECT-ID correctly identified these changes in side-chain conformations in SALB-bound β2AR, despite similarity in the more “global” conformational motions of H6 and H7 to inactive-like conformations seen in the apo state and CARA-bound β2AR (Fig. 1B,C vs. F).
Figure 4.
RMSD distributions for A) F166 and B) F217, identified to have large differences in their conformational motion between the SALB-bound β2AR and apo state. Snapshots at 400 ns (where the RMSD values are near the maximum of the probability distributions) reveal the differences in conformations for these residues across the different simulations. Coloring scheme is the same as in Fig. 3.
DIRECT-ID analysis with the different ligands revealed the existence of multiple allosteric communication pathways/microswitches that selectively propagate information depending upon the state of the receptor. Some residues such as the R1514.43, I1534.45, and T1644.56 were consistently found to have differences in their conformational motion when comparing the apo state versus agonists bound β2AR. T164 is known to be polymorphic, with mutational changes modulating the receptor functional profile differently. T164V reduces basal activity and agonist-induced activation, while T164I was found to alter Salmeterol’s (agonist) exosite binding and conventional agonist-driven coupling between GPCR and Gαs of G-protein.81 Thus, changing the nature of the residue at a conserved position could modulate the communication pathway adopted and the consequent receptor functioning.
As a more thorough understanding of the effect of these residues on the signaling pathways emerges through future site-directed mutational studies, they (along with the others listed in Table 1) could be targeted using new allosteric modulators to bias the signaling profile of the receptor.
Convergence Assessment using DIRECT-ID
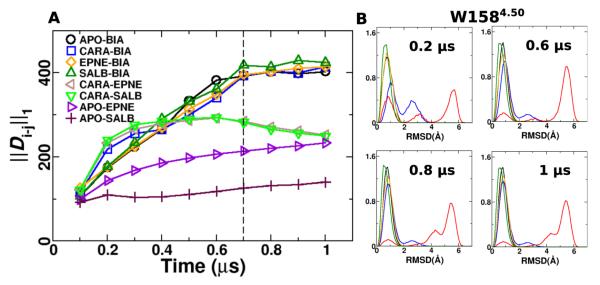
Resetting a fully activated β2AR into the inactivated state requires at least 100 ns,41,76 leading to recent MD simulations studies on the order of several microseconds in an effort to observe fully reversible transitions.76,89 To determine the time scales required to quantify the differences in conformational dynamics using DIRECT-ID, ∥Di–j∥1 was calculated over 0.1-μs increments through the 1-μs trajectories. As shown in Fig. 5A, differences in ∥Di–j∥1 converged at around 0.7 μs for all the trajectories. Differences in local conformational transitions for residues identified using DIRECT-ID, such as the W1584.50 also converged at these time-scales (Fig. 5B). Thus, while longer MD simulations may be useful for fully characterizing reversible transitions across different states of the receptor, DIRECT-ID analysis demonstrates that the present simulations are sufficient for quantifying differences in conformational dynamics.
Figure 5.
A) Convergence of ∥Di–j∥1 through 1-μs simulations. Values were calculated in 0.1- μs increments. B) RMSD distributions of W158 in β2AR from apo simulations (black) and when bound to CARA (blue), BIA (red), EPNE (orange) and SALB (green) calculated through 0.2, 0.6, 0.8 and 1 μs of the MD simulations.
Discussion
Biased agonism is a novel and emerging concept, particularly in GPCR research.90 Recent drug-design efforts have focused on developing ligands that stabilize a particular active conformation of the receptor and subsequently bias a beneficial signaling profile downstream. As the mechanistic link between macromolecular conformations and downstream signaling emerges, allosteric modulators could be developed to target residues identified using DIRECT-ID that selectively modulate a beneficial signaling profile. For instance, by targeting residues identified using DIRECT-ID to selectively respond to SALB, ligands could potentially reduce GRK-modulated phosphorylation of the ICL3 and the C-terminal tail, and thereby reduce β2AR desensitization, a known problem with the current drugs targeting β2AR.91 Additionally, DIRECT-ID will be useful towards both quantification of efficacies of the new ligands and identification of unique structural features that are responsive to the changes in conformational dynamics associated with these ligands.
The present results with the β2AR demonstrate the utility of DIRECT-ID in linking the heterogeneity of free energy surfaces delineating “global” changes (such the G-protein binding cavity in Fig. 1) with the more specific changes in the “local” residues associated with the conformational transitions (Fig. 3,4) without any loss of information. Specifically, this procedure allows for rapid identification of residues that contribute to the selective conformational changes of the receptor. For the 1-μs trajectories, generation of covariance matrices using the gmx covar utility in GROMACS took ~6 min on a 12 core Xeon X5675 3.07 GHz CPU workstation. Extraction of structural features from a pair of these covariance matrices each containing 3879 × 3879 elements took ~1 min using DIRECT-ID. As the method operates with covariance matrices from MD simulations, anharmonicity of the conformational differences are implicitly taken into account. Dynamics across simulations can be restricted to a single global minimum or can span multiple macrostates. As the covariance matrices are defined based on the non-hydrogen atoms, even small conformational transitions such as the side-chain displacements will be identified. For instance, akin to the SALB-bound vs apo state β2AR, conformational landscapes between CARA-bound β2AR and the apo state are also similar (Fig. 1B,C) with the β2AR visiting inactive-like conformations through both the simulations. Consequently, ∥DCARA–APO∥1 (194.1) is smaller than ∥Dagonists–APO∥1, thus quantifying the differences between the CARA-bound and the apo state β2AR. Despite these similarities in conformational landscapes, DIRECT-ID was still able to extract structural features with more subtle changes in conformational transitions (Table 1), demonstrating its power in connecting microscopic and macroscopic dynamics that might otherwise go unnoticed.
A caveat of the DIRECT-ID method is that, if there is a structural shift, such as a change in the tertiary structure, without a change in the local conformational motions of elements of the secondary structure, then these elements may rank low in Di–j. However, it is important to note that such large-scale structural changes are unlikely to be independent of local coupled changes such as breakage of an ionic or hydrogen bond, disruption of hydrophobic packing, etc. Often the structural shift allows for greater conformational variability of the elements in the secondary structure. Each of these events, on the other hand, will rank high in Di–j, and hence will be extracted. Thus, DIRECT-ID will invariably identify a subset of structural features with significant changes in their conformational variability, and as demonstrated in the case of β2AR, these structural features are likely to have critical roles in the modulating the function of the macromolecule.
Importantly, no prior knowledge of the reaction coordinates of the macromolecule is needed to study the modulation of its dynamics with DIRECT-ID. As the changes in the conformational motions across the non-hydrogen atoms of a macromolecule are reported, the method identifies structural features best suited for projecting the conformational dynamics of the macromolecule. It is important to note that the utility of DIRECT-ID is not restricted to GPCRs or even ligand-modulated changes in dynamics. The method is readily applicable to studying the modulation of conformational dynamics by a variety of factors and, as the results with the GPCR highlight, even very large systems can be analyzed. In its current implementation, from any set of M covariance matrices and a reference coordinate file in PDB format, the code outputs an NM×M matrix with ∥Di–j∥1 as its elements as shown in Fig. 2B, along with the list of residues as shown in Table 1. Given the overarching importance of characterizing the modulation of conformational properties of a macromolecule and the consequent implications on its functional profile, a wide applicability of this method is anticipated. Scripts and the code are made available at http://mackerell.umaryland.edu.
Supplementary Material
Acknowledgements
This work was supported by NIH grants GM051501, GM070855, and GM072558. J.A.L. is supported by NIH fellowship F32GM109632. The authors acknowledge computer time and resources from the Computer Aided Drug Design (CADD) Center at the University of Maryland, Baltimore, and the XSEDE supercomputing grid.
References
- 1.Karplus M. Angew. Chem. Int. Ed. 2014;53:9992–10005. doi: 10.1002/anie.201403924. [DOI] [PubMed] [Google Scholar]
- 2.McCammon JA, et al. Nature. 1977;267:585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 3.Perilla JR, et al. Curr. Opin. Struct. Biol. 2015;31:64–74. doi: 10.1016/j.sbi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw DE, et al. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 5.Lakkaraju SK, et al. J. Chem. Info. Model. 2013;53:1337–1349. doi: 10.1021/ci400160x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berendsen HJ, Hayward S. Curr. Opin. Struct. Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 7.Karplus M, Kushick JN. Macromolecules. 1981;14:325–332. [Google Scholar]
- 8.Frappier V, Najmanovich R. Protein Sci. 2015;24:474–483. doi: 10.1002/pro.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy RM, et al. Macromolecules. 1984;17:1370–1374. [Google Scholar]
- 10.Balsera MA, et al. J. Phys. Chem. 1996;100:2567–2572. [Google Scholar]
- 11.Jolliffe I. Principal component analysis. Wiley Online Library; 2002. [Google Scholar]
- 12.Amadei A, et al. Proteins Structure Function and Bioinformatics. 1994:412–425. [Google Scholar]
- 13.Kitao A, Go N. Curr. Opin. Struct. Biol. 1999;9:164–169. doi: 10.1016/S0959-440X(99)80023-2. [DOI] [PubMed] [Google Scholar]
- 14.Ma J. Structure. 2005;13:373–380. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Lakkaraju SK, Hwang W. Phys. Rev. Lett. 2009;102:118102. doi: 10.1103/PhysRevLett.102.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keskin O, et al. Biochemistry. 2002;41:491–501. doi: 10.1021/bi011393x. [DOI] [PubMed] [Google Scholar]
- 17.Bahar I, et al. Curr. Opin. Struct. Biol. 2007;17:633–640. doi: 10.1016/j.sbi.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lezon TR, et al. Handbook on Biological Networks. 2009:129–158. [Google Scholar]
- 19.Wriggers W, Schulten K. Proteins: Struct., Func., Genetics. 1997;29:1–14. [PubMed] [Google Scholar]
- 20.Koike R, et al. J. Mol. Biol. 2014;426:752–762. doi: 10.1016/j.jmb.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 21.Wieninger SA, Ullmann GM. J. Chem. Theory. Comput. 2015 doi: 10.1021/acs.jctc.5b00150. [DOI] [PubMed] [Google Scholar]
- 22.Wieninger SA, et al. J. Mol. Biol. 2011;409:450–465. doi: 10.1016/j.jmb.2011.03.061. [DOI] [PubMed] [Google Scholar]
- 23.Hayward S, Berendsen HJ. Proteins: Struct., Func., Genetics. 1998;30:144–154. [PubMed] [Google Scholar]
- 24.Cui Q, et al. J. Mol. Biol. 2004;340:345–372. doi: 10.1016/j.jmb.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 25.Montange RK, Batey RT. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 26.Draper DE, et al. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 27.Denning EJ, et al. Biophys. Chem. 2013;184:8–16. doi: 10.1016/j.bpc.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priyakumar UD, et al. J. Am. Chem. Soc. 2009;131:17759–17761. doi: 10.1021/ja905795v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichhorn CD, et al. Nucleic Acids Res. 2011:gkr833. [Google Scholar]
- 30.Gong Z, et al. PloS one. 2014;9:e92247. doi: 10.1371/journal.pone.0092247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nygaard R, et al. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huxley H. Cold Spring Harbor Symposia on Quantitative Biology. 1973:361–376. doi: 10.1101/sqb.1952.017.01.007. [DOI] [PubMed] [Google Scholar]
- 33.McGough A, et al. J. Cell. Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sindelar CV, Downing KH. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen SG, et al. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bueren-Calabuig JA, Michel J. P.L.O.S. Comput. Biol. 2015;11:e1004282. doi: 10.1371/journal.pcbi.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kidd BA, et al. P.L.O.S. Comput. Biol. 2009;5:e1000484. doi: 10.1371/journal.pcbi.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badireddy S, et al. Mol. Cell. Proteom. 2011;10:M110.004390. doi: 10.1074/mcp.M110.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobilka BK, Deupi X. Trends Pharmacol. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Vaidehi N, Kenakin T. Curr. Opin. Pharmacol. 2010;10:775–781. doi: 10.1016/j.coph.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Dror RO, et al. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dror RO, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks BR, et al. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hess B, et al. J. Chem. Theory. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 45.U'Prichard D, et al. J. Biol. Chem. 1978;253:5090–5102. [PubMed] [Google Scholar]
- 46.Larsson I. J. Vet. Pharmacol. Therapeut. 1998;21:388–392. doi: 10.1046/j.1365-2885.1998.00156.x. [DOI] [PubMed] [Google Scholar]
- 47.Innis RB, et al. Life Sci. 1979;24:2255–2264. doi: 10.1016/0024-3205(79)90102-4. [DOI] [PubMed] [Google Scholar]
- 48.Fiser A, et al. Protein Sci. 2008;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen M. y., Sali A. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Best RB, et al. J. Chem. Theory. Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacKerell AD, et al. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 53.Klauda JB, et al. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanommeslaeghe K, et al. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen WL, et al. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 56.Durell SR, et al. J. Phys. Chem. 1994;98:2198–2202. [Google Scholar]
- 57.Jo S, et al. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levitt M, Lifson S. J. Mol. Biol. 1969;46:269–279. doi: 10.1016/0022-2836(69)90421-5. [DOI] [PubMed] [Google Scholar]
- 59.Brooks BR, et al. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 60.Lakkaraju SK, et al. J. Chem. Info. Model. 2015;55:700–708. doi: 10.1021/ci500729k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feller SE, et al. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 62.Ryckaert JP, et al. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 63.Steinbach PJ, Brooks BR. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 64.Darden T, et al. J Chem Phys. 1993;98:10089. [Google Scholar]
- 65.Nosé S. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 66.Hoover WG. Phys. Rev. A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 67.Parrinello M, Rahman A. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 68.Hess B. J. Chem. Theory. Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 69.Hess B, et al. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 70.Miyamoto S, Kollman PA. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 71.Perera L, et al. J Chem Phys. 1995;102:450–456. [Google Scholar]
- 72.Cherezov V, et al. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rasmussen SG, et al. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dror RO, et al. Science. 2015;348:1361–1365. doi: 10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rose AS, et al. J. Am. Chem. Soc. 2014;136:11244–11247. doi: 10.1021/ja5055109. [DOI] [PubMed] [Google Scholar]
- 76.Yuan S, et al. Nat. Commn. 2014;5 [Google Scholar]
- 77.Nygaard R, et al. Trends Pharmacol. Sci. 2009;30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 78.Lee Y, et al. P.L.O.S. Comput. Biol. 2015;11:e1004044–e1004044. doi: 10.1371/journal.pcbi.1004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roth CB, et al. J. Mol. Biol. 2008;376:1305–1319. doi: 10.1016/j.jmb.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wess J, et al. EMBO J. 1993;12:331. doi: 10.1002/j.1460-2075.1993.tb05661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Green SA, et al. Eur J Pharmacol. 2001;421:141–147. doi: 10.1016/s0014-2999(01)01049-4. [DOI] [PubMed] [Google Scholar]
- 82.Barak LS, et al. J. Biol. Chem. 1994;269:2790–2795. [PubMed] [Google Scholar]
- 83.Greasley PJ, et al. Mol Pharmacol. 2002;61:1025–1032. doi: 10.1124/mol.61.5.1025. [DOI] [PubMed] [Google Scholar]
- 84.van Rhee AM, Jacobson KA. Drug Devel. Res. 1996;37:1. doi: 10.1002/(SICI)1098-2299(199601)37:1<1::AID-DDR1>3.0.CO;2-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhattacharya S, Vaidehi N. Biophys. J. 2014;107:422–434. doi: 10.1016/j.bpj.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Green SA, et al. Am. J. Resp. Cell. Mol. Biol. 1995;13:25–33. doi: 10.1165/ajrcmb.13.1.7598936. [DOI] [PubMed] [Google Scholar]
- 87.Granier S, et al. J. Biol. Chem. 2007;282:13895–13905. doi: 10.1074/jbc.M611904200. [DOI] [PubMed] [Google Scholar]
- 88.Seibold A, et al. Mol Pharmacol. 2000;58:1162–1173. doi: 10.1124/mol.58.5.1162. [DOI] [PubMed] [Google Scholar]
- 89.Kohlhoff KJ, et al. Nat. Chem. 2014;6:15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whalen EJ, et al. Trends Mol. Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lefkowitz RJ, Caron MG. Curr. Topic Cell. Regul. 2014;28:209–231. doi: 10.1016/b978-0-12-152828-7.50007-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.