Abstract
Single gene mutations that primarily affect pancreatic β-cell function account for approximately 1–2% of all cases of diabetes. Overlapping clinical features with common forms of diabetes makes diagnosis of monogenic diabetes challenging. A genetic diagnosis often leads to significant alterations in treatment, allows better prediction of disease prognosis and progression, and has implications for family members. Currently, genetic testing for monogenic diabetes relies on selection of appropriate individual genes for analysis based on the availability of often-limited phenotypic information, decreasing the likelihood of making a genetic diagnosis. We thus developed a targeted next-generation sequencing (NGS) assay for the detection of mutations in 36 genes known to cause monogenic forms of diabetes, including transient or permanent neonatal diabetes mellitus (TNDM or PNDM), maturity-onset diabetes of the young (MODY) and rare syndromic forms of diabetes. A total of 95 patient samples were analyzed: 19 with known causal mutations and 76 with a clinically suggestive phenotype but lacking a genetic diagnosis. All previously identified mutations were detected, validating our assay. Pathogenic sequence changes were identified in 19 out of 76 (25%) patients: 7 of 32 (22%) NDM cases, and 12 of 44 (27%) MODY cases. In 2 NDM patients the causal mutation was not expected as consanguinity was not reported and there were no clinical features aside from diabetes. A 3 year old patient with NDM diagnosed at 3 months of age, who previously tested negative for INS, KCNJ11 and ABCC8 mutations, was found to carry a novel homozygous mutation in EIF2AK3 (associated with Wolcott–Rallison syndrome), a gene not previously suspected because consanguinity, delayed growth, abnormal bone development and hepatic complications had not been reported. Similarly, another infant without a history of consanguinity was found to have a homo-zygous GCK mutation causing PNDM at birth. This study demonstrates the effectiveness of multi-gene panel anal ysis in uncovering molecular diagnoses in patients with monogenic forms of diabetes.
Keywords: Next-generation sequencing, Monogenic diabetes, Diagnostic evaluation, Targeted sequencing
1. Introduction
Monogenic diabetes mellitus includes a heterogeneous group of diabetes types that are caused by mutations in one of an expanding list of genes [1]. It can be familial or sporadic and if familial, the inheritance can be dominant, recessive or X-linked. It is estimated that the monogenic forms of diabetes together could represent as much as 1–2% of all cases of diabetes mellitus [2]. The main phenotypes suggestive of an underlying monogenic cause include transient or permanent neonatal diabetes mellitus (TNDM or PNDM), maturity-onset diabetes of the young (MODY) and rare diabetes-associated syndromes. More than twenty genes highly expressed in the pancreatic beta-cell have been identified in these monogenic subtypes, and many other genes have been implicated in syndromes that often include diabetes. Several etiological mechanisms of dysfunction are involved including impairment of pancreatic beta-cell development and/or gene expression, failure of glucose sensing, disruption of insulin synthesis, disorders of ion channels and increased endoplasmic reticulum stress leading to destruction of the beta-cell [3–5].
It is likely that the majority of patients with monogenic diabetes go unrecognized [6] and continue to be misdiagnosed as type 1 or type 2 diabetes [7–9]. In addition to elucidating the etiology of the patient's diabetes and explaining other associated clinical features, establishing the underlying monogenic cause can provide important prognostic and therapeutic information. Attention has recently focused on the most common forms of PNDM caused by heterozygous activating mutations in the KCNJ11 and ABCC8 genes, which encode the protein subunits (Kir6.2 and SUR1) of the ATP-sensitive potassium (KATP) channel [10–12]. The majority of patients with KATP channel mutations may be treated successfully with oral sulfonylureas alone in lieu of multiple daily insulin injections. This transition results in improved glycemic control and supports a crucial role for genetic testing in all neonatal diabetes patients [13–15]. In addition, HNF1A and HNF4A mutations cause forms of MODY that are often sensitive to low-dose sulfonylurea therapy [16,17], while heterozygous mutations in the GCK gene generally lead to a mild fasting hyperglycemia that seldom needs treatment and is not associated with significant complications [18,19]. Thus, uncovering a genetic basis by making an accurate molecular diagnosis is extremely important for optimal treatment of these patients and may lead to dramatic improvement in their quality of life. Moreover, once a mutation is established, at-risk family members can be screened and predictive genetic testing can be offered to relatives after appropriate genetic counseling. As monogenic diabetes is a genetically heterogeneous group of disorders, the ability to use next-generation sequencing (NGS) technology to sequence several genes simultaneously is a potentially cost-effective means of increasing the rate of molecular diagnosis in affected probands [20–22]. In this study, we describe the development of the first panel-based NGS assay for monogenic diabetes available in the United States.
2. Methods
2.1. Study subjects
Subjects with neonatal diabetes or with a clinical and/or family history suggestive of MODY or syndromic forms of diabetes were consented for participation through the University of Chicago Monogenic Diabetes Registry (http://monogenicdiabetes.uchicago.edu/registry/) through which longitudinal information regarding the diagnosis and treatment of diabetes, other medical problems or complications, family history and genetic testing results, was collected through surveys and medical records [23]. A neonatal diabetes phenotype was defined as persistent hyperglycemia requiring treatment diagnosed under 1 year of age. A MODY phenotype was defined as diabetes diagnosed under 35 years of age in a non-obese individual with either a linear family history of diabetes or the absence of diabetes-related autoantibodies. All subjects were consented for participation through protocols approved by the Institutional Review Board at the University of Chicago. Genomic DNA was isolated from patients’ saliva or blood samples using the Oragene OG-300 non-invasive saliva sampling kit (DNA Genotek Inc., Ottawa, ON, Canada) or the PureGene DNA isolation kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions. DNA integrity was verified using a Qubit Fluorometer (Life technologies, Darmstadt, Germany). A total of 95 families were chosen to examine the utility of a gene panel-based next-generation sequencing approach: 19 with known mutations and 76 with a clinically suggestive phenotype but no known genetic etiology.
2.1.1. Gene selection
The monogenic diabetes panel included the most common genes causing NDM/MODY (ABCC8, GCK, HNF1A, HNF4A, HNF1B, INS, KCNJ11), genes involved in less common known causes of NDM/MODY and congenital hyperinsulinism (AKT2, BLK, CEL, CISD2, CP, EIF2AK3, FOXP3, GATA6, GLIS3, GLUD1, HADH, KLF11, IER3IP1, INSR, NEUROD1, NEUROG3, PAX4, PDX1, PTF1A, RFX6, SLC2A2, TBC1D4, WFS1, ZFP57) and genes involved in extremely rare syndromic forms of diabetes mellitus (ALMS1, DCAF17, SLC19A2, SLC29A3, PAX6) (Supplementary Table 1). Only genes that had been proven to cause disease were included in the panel.
2.1.2. Next-generation sequencing
The targeted NGS approach was based on HaloPlex enrichment (Agilent Technologies, Santa Clara, CA, USA) followed by MiSeq Illumina NGS. HaloPlex probes were designed following Agilent's recommendations [24,25] to enrich all exons, plus 10-bp at each end, of the 36 selected genes previously associated with NDM, MODY and very rare syndromic forms of diabetes mellitus (Table 1). HaloPlex enriched Illumina libraries were obtained following Agilent's recommendations with the exception that the gDNA-probe hybrids were amplified using the KAPA HiFi HotStart PCR kit (KAPA Biosystem) (1X KAPA HiFi Fidelity GC buffer, 0.8 mM dNTPs, 1 μM PCR primers, 20 μM Acetic Acid and 1U of KAPA HiFi HotStart polymerase), and the following program in the Applied Biosystems 2720 thermocycler: 98 °C for 2 min; 21 cycles of 98 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min; followed by 72 °C for 10 min then hold on 8 °C. HaloPlex enriched Illumina libraries were quantified using the Agilent 2100 Bioanalyzer, multiplexed (8 samples) and 150PE sequenced on the Illumina MiSeq system following Agilent's recommendations. Data from each sequence run was de-multiplexed and reads aligned to the reference human genome (hg18) using the Burrows–Wheeler Aligner (version 0.6.2). Reads in the regions of the genome that are susceptible to alignment artifacts due to the presence of repetitive sequences were locally realigned using GATK (version 1.0.5506). The NGS data analysis and variant calling were performed using an in-house custom-developed bioinformatics pipeline and a commercial software package (NextGENe, Softgenetics, State College, PA). Raw variant calls were filtered based on various quality metrics such as depth, quality by depth score and directional bias. Variants were then annotated in regard to their positions in transcripts of interest, position relative to the coding sequence, consequence for the protein or mRNA and a collection of direct and indirect evidentiary tools and databases including NCBI dbSNP, 1000 Genomes Project, Exome Sequencing Project (ESP), GERP, Conseq, PolyPhen-2, SIFT and the Human Gene Mutation Database (HGMD).
Table 1.
Subjects with known mutations used to validate the gene panel.
| Patient | Gender | Gene | Nucleotide | Amino Acid | Zygosity |
|---|---|---|---|---|---|
| 1 | M | ABCC8 | c.3440T>G, c.4135C>T | p.Leu1147Arg, p.Arg1379Cys | Comp HET |
| 2 | M | EIF2AK3 | c.1267dup | p.Ile423Asnfs*26 | HOMO |
| 3 | M | GATA6 | c.1088_1098del | p.Gln363Argfs*96 | HET |
| 4 | F | KCNJ11 | c.602G>A | p.Arg201His | HET |
| 5 | F | KCNJ11 | c.5T>C | p.Leu2Pro | HET |
| 6 | M | RFX6 | c.779A>C | p.Lys260Thr | HOMO |
| 7 | F | FOXP3 | c.1044+5G>A | p.? | HET |
| 8 | M | FOXP3 | c.340C>T | p.Arg114Trp | HEMI |
| 9 | M | FOXP3 | c.1227_1235del | p.Asp409_Leu411del | HOMO |
| 10 | M | GCK | c.449T>A | p.Phe150Tyr | HET |
| 11 | F | GCK | c.617C>T | p.Thr206Met | HET |
| 12 | M | GCK | c.766G>A | p.Glu256Lys | HET |
| 13 | M | HNF1A | c.476G>A | p.Arg159Gln | HET |
| 14 | M | HNF1A | c.872dup | p.Gly292Argfs*25 | HET |
| 15 | F | HNF1A | c.862G>T | p.Gly288Trp | HET |
| 16 | F | HNF1A | c.872del | p.Pro291Glnfs*51 | HET |
| 17 | M | HNF4A | c.416C>T | p.Arg134Gln | HET |
| 18 | F | INSR | c.1268+2T>C | p.? | HOM |
| 19 | M | PDX1 | c.188del | p.Pro63Argfs*60 | HET |
2.1.3. Sanger sequencing
All sequence variants with putatively deleterious effects were confirmed by conventional Sanger sequencing in both forward and reverse directions on a 3730xl DNA Analyzer (Life Technologies, Grand Island, NY). Sequence analysis was performed using Mutation Surveyor software version 3.01 (SoftGenetics, State College, PA).
3. Results
A total of 372 exons in 36 genes were enriched using the HaloPlex technology and sequenced with Illumina NGS. A mean depth of sequencing coverage >1000× was observed and 99.3 ± 0.16% of the targeted regions were successfully sequenced with a depth of coverage >15×. All 20 pathogenic sequence changes were identified in the 19 positive control samples, validating the sensitivity and the accuracy of our platform. These included 14 base substitutions, two-base duplications and four deletions ranging from 1 to 10 bases (Table 1). 352 of the 372 exons were fully sequenced in all samples, 11 exons in more than half of the samples and 9 exons (in the ALMS1, CEL, CISD2, EIF2AK3, HNF4A and PTF1A genes) were poorly sequenced in all positive control samples (Supplementary Table 2).
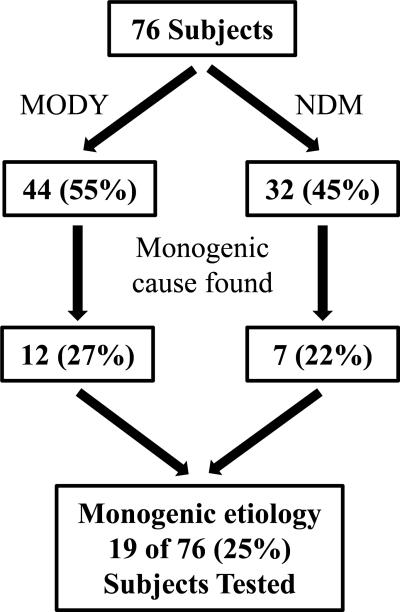
Among the 76 patients with either neonatal diabetes or a MODY phenotype but without an established genetic etiology, a molecular genetic diagnosis was obtained for 19 (25%) patients (Fig. 1). 26 (46%) of 57 patients whose sequencing did not reveal a mutation had been previously analyzed for some of the more commonly seen forms of monogenic diabetes (Supplementary Table 3). Among 32 patients with NDM, a causative mutation was identified in 7 cases (22%) (Table 2, patients 20–26). The majority of the mutations identified in this study had previously been reported. Examples include: the most common mutation in the KCNJ11 gene c.602G>A (p.Arg201His), which causes oral sulfonylurea-responsive NDM [26–28], in a 41 year old female (Table 2, patient 20) diagnosed with NDM at 6 months of age; the INS c.94G>A (p.Gly32Ser) mutation [29] in a 60 year old female (Table 2, patient 21) diagnosed at 9 months of age; and the c.188-31G>A sequence change in the INS gene [30] previously reported to affect proper splicing of exon 2 in the INS gene identified in a patient with NDM diagnosed at 10 months of age (Table 2, patient 22). A novel, likely pathogenic c.204G>C (p.Trp68Cys) change in the KCNJ11 gene was identified in a patient with NDM (Table 2, patient 23) in the de-novo state. This sequence change affects a highly conserved amino acid residue located in a functional domain of the Kir6.2 protein and it is predicted to be damaging by in-silico prediction tools (Supplementary Table 4). A different nucleotide change, c.202T>C, affecting the same codon but resulting in a different amino acid substitution, p.Trp68Arg, has been previously reported and functional studies demonstrated that like other NDM mutations, it prevents channel closure and insulin secretion [31].
Fig. 1.
Summary of unknown samples analyzed and results obtained in this study.
Table 2.
Summary of the 19 pathogenic sequences identified in this study.
| Patient | Gender | Race- ethnicity |
Gene | Reference Sequence |
Nucleotide Change |
Amino acid | Zygosity | Age at Diagnosis |
Pharmacotherapy | Generations with Hyperglycemia |
|---|---|---|---|---|---|---|---|---|---|---|
| 20 | F | H | KCNJ11 | NM_000525.3 | c.602G>A | p.Arg201His | HET | 6 months | Insulina | 1 |
| 21 | F | W | INS | NM_000207.2 | c.94G>A | p.Gly32Ser | HET | 9 months | Insulin | 1 |
| 22 | M | W | INS | NM_000207.2 | c.188-31G>A | p.? | HET | 10 months | Insulin | 1 |
| 23 | F | AS | KCNJ11 | NM_000525.3 | c.204G>C^ | p.Trp68Cys | HET | 1.5 months | Insulina | 1 |
| 24 | M | W | ABCC8 | NM_000352.3 | c.3989-9G>A; c.4174T>G | p.?; p.Phe1392Val | HET; HET | 6 months | Insulina | 1 |
| 25 | M | H | EIF2AK3 | NM_004836 | c.2758C>T^ | p.Gln920* | HOMO | 3 months | Insulin | 1 |
| 26 | M | H | GCK | NM_000162.3 | c.706G>A | p.Glu236Lys | HOMO | Birth | Insulin | 1 |
| 27 | M | W | HNF1A | NM_000545.5 | c.391C>T | p.Arg131Trp | HET | 10 years | Insulina | 1 |
| 28 | M | W | HNF1A | NM_000545.5 | c.586A>G | p.Thr196Ala | HET | 3 years | None | 3 |
| 29 | F | W | GCK | NM_000162.3 | c.863+1G>A^ | p.? | HET | 5 years | None | 3 |
| 30 | F | W | GCK | NM_000162.3 | c.148del^ | p.His50Metfs*6 | HET | 11 years | Metformina | 3 |
| 31 | M | W | GCK | NM_000162.3 | c.544G>A | p.Val182Met | HET | 4 years | Insulina | 1 |
| 32 | M | W | GCK | NM_000162.3 | c.616A>C | p.Thr260Pro | HET | 17 years | None | 3 |
| 33 | M | W | GCK | NM_000162.3 | c.626C>T | p.Thr209Met | HET | 11 years | None | 3 |
| 34 | F | W | GCK | NM_000162.3 | c.676G>A | p.Val226Met | HET | 39 years | Metformin/Sitagliptina | 2 |
| 35 | F | W | GCK | NM_000162.3 | c.689G>A^ | p.Cys230Tyr | HET | 25 years | Metformin/Sitagliptin/Sulfonylureaa | 3 |
| 36 | F | W | GCK | NM_000162.3 | c.787T>C | p.Ser263Pro | HET | 23 years | Metformina | 3 |
| 37 | M | H | GCK | NM_000162.3 | c.1003delA | p.Val335Cysfs*18 | HET | 6 years | Metformina | 4 |
| 38 | M | AA | GCK | NM_000162.3 | c.1112G>T | p.Cys371Phe | HET | 8 years | None | 4 |
Genetic results may allow transition to more appropriate drug therapy or discontinuation of drug therapy
Novel to this study
AA- African American, AS-Asian, H- Hispanic of any race, W- White
Several atypical NDM cases were also identified. A 27 year old white male presented with diabetic ketoacidosis at 6 months of age, with no family history of diabetes, was found to be a carrier of two ABCC8 sequence changes (c.3989-9G>A and c.4174T>G) (Table 2, patient 24), which have been reported as recessively inherited in patients with congenital hyperinsulinism [32,33]. His birth weight was 2722 g at term and he is currently treated with 0.55 units/kg/day of insulin. Parents were not available to determine whether these sequence changes were in cis or in trans in the patient. A novel truncating mutation in homozygous state, c.2758C>T (p.Gln920*), in the EIF2AK3 gene was identified in a 3 year old male with PNDM since 3 months of age (Table 2, patient 25). Mutations in the EIF2AK3 gene have been associated with Wolcott–Rallison syndrome (WRS). This patient had previously been extensively tested only for the more common monogenic causes of NDM as there were no reported features suggestive of WRS such as consanguinity, delayed growth, abnormal bone development, and hepatic complications. Similarly, a rare case of a homozygous inactivating GCK mutation, c.706G>A (p.Glu236Lys), was identified in a patient (Table 2, patient 26) with NDM diagnosed at birth. This mutation had been previously reported in the heterozygous state in two independent MODY2 families [34]. The patient's parents were not known to be related or have diabetes. Both parents are heterozygous for the mutation and are reported to have mild fasting hyperglycemia while the paternal grandfather was diagnosed with diabetes at 40 years of age. The proband's older sibling had marked hyperglycemia and died from presumed diabetic ketoacidosis in infancy.
Pathogenic sequence changes were found in 12 out of 44 patients with a MODY phenotype (27%) (Table 2, patients 27–38). For all these patients the mutated gene had not been analyzed previously. Two patients harbored missense mutations in the HNF1A gene that were previously reported in patients with MODY3 [35,36]; the c.586A>G (p.Thr196Ala) identified in a 3 year old male patient with “prediabetes” (Table 2, patient 28) was also found in his 12 year old brother with a similar glucose pattern. Mutations in GCK were identified in 10/44 cases with a MODY phenotype (23%) (Table 2, patients 29–38); 4 of them are novel to this study and predicted to be damaging by in-silico predictions (Supplementary Table 4). The c.689G>A (p.Cys230Tyr) variant affects a conserved amino acid residue located in a functional domain of the glucokinase protein where other pathogenic missense sequence changes have been described and it is also predicted to be damaging (Supplementary Table 4). Of 10 probands identified with heterozygous GCK mutations, 8 had at least a three-generation linear history of hyperglycemia. Comprehensive family screening has begun and, to date, 5 relatives have been confirmed to carry a heterozygous GCK mutation.
4. Discussion
Because monogenic diabetes is a genetically heterogeneous group of disorders, selection of appropriate gene(s) to test is challenging when based solely on clinical grounds. Pathogenic variants within several genes can present with similar phenotypic characteristics, while other features that are more gene specific may not yet have manifested at the time of initial presentation with hyperglycemia. Despite these challenges, confirming a molecular diagnosis can be greatly beneficial, especially for patients who can have their medical management significantly changed based on an understanding of the underlying molecular defect causing their diabetes (Table 2). Even in cases where drug therapy remains unchanged, patients and families can benefit from having more specific information about their diagnosis, such as the predicted clinical course and familial risk, and receive genetic counseling.
Variable awareness of monogenic diabetes and unequal access to genetic testing are potential reasons that the majority of those with monogenic diabetes go unrecognized [6]. Currently available genetic testing for the most common forms of MODY and NDM is primarily based on Sanger sequencing of multiple genes in a sequential manner, which is time consuming and potentially costly, and limits the diagnosis to a few selected genes. Targeted single gene Sanger sequencing remains the least costly option in cases with gene specific clinical features. If the first round of single gene sequencing is negative, the cost of sequencing the other genes may preclude further testing. Furthermore, the choice of genes to be tested using this approach depends on the availability of reliable and comprehensive phenotypic information and a detailed family history. We identified a mutation in the EIF2AK3 gene in a neonatal patient who did not yet exhibit epiphyseal dysplasia and other characteristic features associated with Wolcott–Rallison syndrome at the time of diabetes diagnosis. We also found a rare GCK homozygous mutation in a patient with diabetes diagnosed at birth for whom consanguinity was not reported. These two examples demonstrate some limitations of the candidate gene approach in that the diagnosis of some forms of monogenic diabetes is not always clear-cut and may be complicated by incomplete or absent clinical and/or family history information.
The exponential growth in the fields of high-throughput capture and NGS has made this approach among the most promising and economic techniques to identify causative mutations in genetically heterogeneous diseases [37–39]. In this study, we describe a targeted NGS assay for the molecular diagnosis of monogenic forms of diabetes. In total, 36 genes were simultaneously sequenced with deep coverage. A genetic diagnosis was obtained for 19/76 (25%) patients in whom testing had been limited to a subset of genes or had not yet been performed. Among 32 patients with neonatal diabetes, a causative mutation was identified in 7 cases (22%). For the 3 patients with KATP channel mutations, the test results are likely to allow them to be effectively managed with sulfonylurea therapy instead of insulin injections.
A causative mutation was identified in 12 of 44 (27%) patients with a MODY phenotype. Heterozygous inactivating GCK mutations result in mild elevation in serum glucose levels with fasting serum glucose typically ranging from 100 to 144 mg/dL [34]. There is minimal deterioration of glycemic control over time and diabetes-related complications are rare, making drug therapy unnecessary for most individuals [40,41]. Those with HNF1A-MODY often exhibit a marked sensitivity to oral sulfonylurea agents, which is the established first-line therapy that results in equal or improved control as compared to insulin therapy [16]. Our data demonstrates that diagnosing an individual with MODY often allows multiple members across many generations within a family to be screened.
In 75% of patients selected for testing, a genetic cause was not identified (Supplementary Table 3). There is significant clinical overlap between monogenic diabetes and the more common forms of diabetes but there are several other possible explanations for this finding. Patients may harbor mutations in intronic or regulatory regions, or large copy number alterations that are not covered by our method. The cause of diabetes in some patients may be a mutation in a more recently described gene not included in this current panel design. Another possibility is that these patients have a hitherto unknown gene causing diabetes. Indeed, an alternative approach would be to perform whole exome sequencing (WES) in lieu of testing specific genes in patients with suspected forms of monogenic diabetes, as recently proposed [42,43]. Johansson et al. performed WES in nine patients with suspected MODY. They analyzed a pre-defined set of 111 genes implicated in glucose metabolism and identified three pathogenic variants leading to a genetic diagnosis in three patients in the ABCC8, HNF4A and PPARG genes [42]. Bonnefond et al. carried out WES in a patient with NDM and identified a novel de-novo mutation in the ABCC8 gene [43]. While the use of WES and whole-genome sequencing (WGS) and their implementation as a diagnostic tool is exciting, those pathogenic sequence changes would have been identified with a gene-based NGS panel approach at a lower cost and at a higher sensitivity and specificity due to more adequate sequence coverage of the genes of interest. Indeed, coverage of the genes that are known to be associated with the pheno-type or disease of interest in both WES and WGS is currently not comparable to the coverage achieved with the gene-based NGS panel approach. WES and WGS also target a much wider genomic area than panel-based targeted NGS and produce larger amounts of sequence data to reach comparable per base coverage values. Furthermore, due to their broader hypothesis-free approach, WES and WGS provide a significantly larger number of variants of unknown clinical significance (VOUS), so more detailed phenotypic information is needed to assist the laboratory in analyzing and interpreting the results of testing. In addition to a larger number of VOUS, WES and WGS may uncover genetic predisposition to previously undiagnosed medical or psychiatric conditions that are unrelated to the condition for which the patient is being assessed. Thus, one needs to carefully consider the impact of incidental findings in genes known to cause diseases other than the disease that prompted the initial genetic test. Consenting patients for potential incidental findings with uncertain implications raises a number of ethical and regulatory issues [44,45]. A NGS gene-panel approach can significantly reduce these concerns reserving WES for cases with a high clinical suspicion for a monogenic cause of diabetes but in which disease-targeted panel NGS testing has not demonstrated evidence for a genetic etiology.
The flexibility of our targeted strategy coupled with NGS allows for continued refinement. We have recently changed the enrichment method to SureSelect (Agilent Technologies) and obtained 100% coverage of the targeted regions with a depth coverage >15×. The m.3243 region of the mitochondrial genome (where the m.3243A>G mutation causes Maternally Inherited Diabetes and Deafness or MIDD) [46] was not tested due to the complexity of simultaneously capturing/sequencing nuclear as well as mitochondrial loci. Because the mitochondrial copy number is many times greater than the nuclear copy number (and the rates of capture and sequencing are proportional), true mitochondrial genome reads will grossly outnumber those observed for nuclear loci thus significantly reducing their coverage. This is a propriety of the design and can be altered easily—for example by reducing the density and the replication of RNA baits targeting the mitochondrial genome [47]. We are currently in the process of optimizing the assay to include the m.3243A>G mutation in the next iteration of this panel. In addition, recently described genes will be added to the panel — the UCP2 and SLC16A1 genes associated with congenital hyperinsulinism [48–50] and exercise-induced hyperinsulinism [51] respectively, as well as the NKX2-2 and MNX1 genes recently associated with NDM [52]. Future directions will include the evaluation of algorithms for the detection of copy number alterations (large intragenic deletions or duplications) using NGS data.
In summary, we developed the most comprehensive monogenic diabetes panel now available in the United States and demonstrated the effectiveness of multi-gene panel analysis in identifying an accurate genetic diagnosis in patients with monogenic forms of diabetes and ultimately guiding appropriate pharmacotherapy. Moreover, this approach allowed rapid diagnosis in those atypical cases that may have been missed using a candidate gene base strategy in addition to expanding established gene and mutation specific phenotypes.
5. Conclusions
Our focused approach utilizing a targeted gene panel for monogenic diabetes exploits the improved testing speed and falling cost of NGS technology without many of the difficulties associated with WES and WGS (e.g. informed consent, cost, lower sensitivity and specificity, incidental findings, reporting obligations). This demonstrates how NGS technology can be leveraged to improve genetic testing for monogenic diabetes, which because of frequent resulting alterations in treatment has been shown to be cost effective or even cost saving [53,54].
Supplementary Material
Acknowledgments
This work was supported by funding from the Institute for Translational Medicine/CTSA (NIH UL1 TR000430) at the University of Chicago and USPHS grants P30DK020595 and K23DK094866. We also thank all of the patients and families who participated in these studies.
Abbreviations
- NGS
Next-Generation Sequencing
- TNDM
Transient NeonatalDiabetes
- PNDM
Permanent Neonatal Diabetes
- MODY
Maturity-Onset Diabetes of the Young
- NDM
Neonatal Diabetes
- GATK
Genome Analysis Tool Kit
- ESP
Exome Sequencing Project
- GERP
Genomic Evolutionary Rate Profiling
- PolyPhen-2
Polymorphism Phenotyping v2
- SIFT
Sorting Intolerant from Tolerant
- HGMD
Human Gene Mutation Database
- VOUS
Variant of Unknown Clinical Significance
- WES
Whole Exome Sequencing
- WGS
Whole Genome Sequencing
Footnotes
Contributions
A.A.G. performed the genetic analyses, analyzed the results, wrote the manuscript, and made the figures and tables; C.D. provided clinical information, and wrote the manuscript; C.Y.W. implemented Haloplex NGS technology and performed the genetic analysis; N.V. performed the bioinformatic analysis of NGS data; M.L. performed Sanger Sequencing, D.J.T. collected patient samples; D.S. performed critical revision of the manuscript; S.W.G. provided insights on study concept and design; and D.G.D. conceived the study and revised the manuscript. All authors critically revised the manuscript and approved of the final version.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ymgme.2014.09.007.
Conflict of interest disclosure
The authors report no financial conflict of interest.
References
- 1.Greeley SA, Naylor RN, Philipson LH, Bell GI. Neonatal diabetes: an expanding list of genes allows for improved diagnosis and treatment. Curr. Diab. Rep. 2011;11:519–532. doi: 10.1007/s11892-011-0234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eide SA, et al. Prevalence of HNF1A (MODY3) mutations in a Norwegian population (the HUNT2 Study) Diabet. Med. 2008;25:775–781. doi: 10.1111/j.1464-5491.2008.02459.x. [DOI] [PubMed] [Google Scholar]
- 3.Philipson LH, Murphy R, Ellard S, Hattersley AT, Stoy J, Greeley SA, Bell GI, Polonsky KS. Genetic diagnosis of endocrine disorders. In: Weiss RE, Refetoff S, editors. Genetic Diagnosis of Endocrine Disorders. Elsevier; London: 2010. [Google Scholar]
- 4.Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat. Clin. Pract. Endocrinol. Metab. 2008;4:200–213. doi: 10.1038/ncpendmet0778. [DOI] [PubMed] [Google Scholar]
- 5.Porter JR, Barrett TG. Monogenic syndromes of abnormal glucose homeostasis: clinical review and relevance to the understanding of the pathology of insulin resistance and beta cell failure. J. Med. Genet. 2005;42:893–902. doi: 10.1136/jmg.2005.030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shields BM, et al. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia. 2010;53:2504–2508. doi: 10.1007/s00125-010-1799-4. [DOI] [PubMed] [Google Scholar]
- 7.Slingerland AS, Hattersley AT. Mutations in the Kir6.2 subunit of the KATP channel and permanent neonatal diabetes: new insights and new treatment. Ann. Med. 2005;37:186–195. doi: 10.1080/07853890510007287. [DOI] [PubMed] [Google Scholar]
- 8.Lambert AP, et al. Identifying hepatic nuclear factor 1alpha mutations in children and young adults with a clinical diagnosis of type 1 diabetes. Diabetes Care. 2003;26:333–337. doi: 10.2337/diacare.26.2.333. [DOI] [PubMed] [Google Scholar]
- 9.Møller AM, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in Caucasian families originally classified as having Type I diabetes. Diabetologia. 1998;41:1528–1531. doi: 10.1007/s001250051101. [DOI] [PubMed] [Google Scholar]
- 10.Gloyn AL, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 11.Pearson ER, et al. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N. Engl. J. Med. 2006;355:467–477. doi: 10.1056/NEJMoa061759. [DOI] [PubMed] [Google Scholar]
- 12.Babenko AP, et al. Activating mutations in the ABCC8 gene in neonatal diabetes mellitus. N. Engl. J. Med. 2006;355:456–466. doi: 10.1056/NEJMoa055068. [DOI] [PubMed] [Google Scholar]
- 13.Ellard S, Bellanné-Chantelot C, Hattersley AT, E. M. G. Q. N. E. M. group Best practice guidelines for the molecular genetic diagnosis of maturity-onset diabetes of the young. Diabetologia. 2008;51:546–553. doi: 10.1007/s00125-008-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Støy J, et al. Diagnosis and treatment of neonatal diabetes: a United States experience. Pediatr. Diabetes. 2008;9:450–459. doi: 10.1111/j.1399-5448.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattersley A, Bruining J, Shield J, Njolstad P, Donaghue KC. The diagnosis and management of monogenic diabetes in children and adolescents. Pediatr. Diabetes. 2009;10(Suppl. 12):33–42. doi: 10.1111/j.1399-5448.2009.00571.x. [DOI] [PubMed] [Google Scholar]
- 16.Pearson ER, et al. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362:1275–1281. doi: 10.1016/S0140-6736(03)14571-0. [DOI] [PubMed] [Google Scholar]
- 17.Pearson ER, et al. Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4alpha mutations in a large European collection. Diabetologia. 2005;48:878–885. doi: 10.1007/s00125-005-1738-y. [DOI] [PubMed] [Google Scholar]
- 18.Stride A, et al. The genetic abnormality in the beta cell determines the response to an oral glucose load. Diabetologia. 2002;45:427–435. doi: 10.1007/s00125-001-0770-9. [DOI] [PubMed] [Google Scholar]
- 19.Stride A, et al. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia. 2014;57:54–56. doi: 10.1007/s00125-013-3075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellard S, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013;56:1958–1963. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnefond A, et al. Highly sensitive diagnosis of 43 monogenic forms of diabetes or obesity through one-step PCR-based enrichment in combination with next-generation sequencing. Diabetes Care. 2014;37:460–467. doi: 10.2337/dc13-0698. [DOI] [PubMed] [Google Scholar]
- 22.Gao R, et al. Evaluation of a target region capture sequencing platform using monogenic diabetes as a study-model. BMC Genet. 2014;15:13. doi: 10.1186/1471-2156-15-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greeley SA, et al. Creation of the Web-based University of Chicago Monogenic Diabetes Registry: using technology to facilitate longitudinal study of rare subtypes of diabetes. J. Diabetes Sci. Technol. 2011;5:879–886. doi: 10.1177/193229681100500409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berglund EC, et al. Accurate detection of subclonal single nucleotide variants in whole genome amplified and pooled cancer samples using HaloPlex target enrichment. BMC Genomics. 2013;14:856. doi: 10.1186/1471-2164-14-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagen JV, et al. Permanent neonatal diabetes due to mutations in KCNJ11 encoding Kir6.2: patient characteristics and initial response to sulfonylurea therapy. Diabetes. 2004;53:2713–2718. doi: 10.2337/diabetes.53.10.2713. [DOI] [PubMed] [Google Scholar]
- 27.Klupa T, et al. The identification of a R201H mutation in KCNJ11, which encodes Kir6.2, and successful transfer to sustained-release sulphonylurea therapy in a subject with neonatal diabetes: evidence for heterogeneity of beta cell function among carriers of the R201H mutation. Diabetologia. 2005;48:1029–1031. doi: 10.1007/s00125-005-1731-5. [DOI] [PubMed] [Google Scholar]
- 28.Proks P, Girard C, Ashcroft FM. Functional effects of KCNJ11 mutations causing neonatal diabetes: enhanced activation by MgATP. Hum. Mol. Genet. 2005;14:2717–2726. doi: 10.1093/hmg/ddi305. [DOI] [PubMed] [Google Scholar]
- 29.Støy J, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garin I, et al. Permanent neonatal diabetes caused by creation of an ectopic splice site within the INS gene. PLoS One. 2012;7:e29205. doi: 10.1371/journal.pone.0029205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Männikkö R, et al. A conserved tryptophan at the membrane–water interface acts as a gatekeeper for Kir6.2/SUR1 channels and causes neonatal diabetes when mutated. J. Physiol. 2011;589:3071–3083. doi: 10.1113/jphysiol.2011.209700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaser B, et al. ABCC8 mutation allele frequency in the Ashkenazi Jewish population and risk of focal hyperinsulinemic hypoglycemia. Genet Med. 2011;13:891–894. doi: 10.1097/GIM.0b013e31821fea33. [DOI] [PubMed] [Google Scholar]
- 33.Bellanné-Chantelot C, et al. ABCC8 and KCNJ11 molecular spectrum of 109 patients with diazoxide-unresponsive congenital hyperinsulinism. J. Med. Genet. 2010;47:752–759. doi: 10.1136/jmg.2009.075416. [DOI] [PubMed] [Google Scholar]
- 34.Osbak KK, et al. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum. Mutat. 2009;30:1512–1526. doi: 10.1002/humu.21110. [DOI] [PubMed] [Google Scholar]
- 35.Glucksmann MA, et al. Novel mutations and a mutational hotspot in the MODY3 gene. Diabetes. 1997;46:1081–1086. doi: 10.2337/diab.46.6.1081. [DOI] [PubMed] [Google Scholar]
- 36.Bellanné-Chantelot C, et al. The type and the position of HNF1A mutation modulate age at diagnosis of diabetes in patients with maturity-onset diabetes of the young (MODY)-3. Diabetes. 2008;57:503–508. doi: 10.2337/db07-0859. [DOI] [PubMed] [Google Scholar]
- 37.Ku CS, et al. Exome sequencing: dual role as a discovery and diagnostic tool. Ann. Neurol. 2012;71:5–14. doi: 10.1002/ana.22647. [DOI] [PubMed] [Google Scholar]
- 38.Sule G, et al. Next-generation sequencing for disorders of low and high bone mineral density. Osteoporos. Int. 2013;24:2253–2259. doi: 10.1007/s00198-013-2290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glöckle N, et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur. J. Hum. Genet. 2014;22:99–104. doi: 10.1038/ejhg.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin D, et al. Long-term follow-up of oral glucose tolerance test-derived glucose tolerance and insulin secretion and insulin sensitivity indexes in subjects with glucokinase mutations (MODY2) Diabetes Care. 2008;31:1321–1323. doi: 10.2337/dc07-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steele AM, et al. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA. 2014;311:279–286. doi: 10.1001/jama.2013.283980. [DOI] [PubMed] [Google Scholar]
- 42.Johansson S, et al. Exome sequencing and genetic testing for MODY. PLoS One. 2012;7:e38050. doi: 10.1371/journal.pone.0038050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnefond A, et al. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010;5:e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGuire AL, Caulfield T, Cho MK. Research ethics and the challenge of whole-genome sequencing. Nat. Rev. Genet. 2008;9:152–156. doi: 10.1038/nrg2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabor HK, Berkman BE, Hull SC, Bamshad MJ. Genomics really gets personal: how exome and whole genome sequencing challenge the ethical framework of human genetics research. Am. J. Med. Genet. A. 2011;155A:2916–2924. doi: 10.1002/ajmg.a.34357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy R, Turnbull DM, Walker M, Hattersley AT. Clinical features, diagnosis and management of maternally inherited diabetes and deafness (MIDD) associated with the 3243ANG mitochondrial point mutation. Diabet. Med. 2008;25:383–399. doi: 10.1111/j.1464-5491.2008.02359.x. [DOI] [PubMed] [Google Scholar]
- 47.Perry GH, Marioni JC, Melsted P, Gilad Y. Genomic-scale capture and sequencing of endogenous DNA from feces. Mol. Ecol. 2010;19:5332–5344. doi: 10.1111/j.1365-294X.2010.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Astrup A, et al. Impact of the v/v 55 polymorphism of the uncoupling protein 2 gene on 24-h energy expenditure and substrate oxidation. Int. J. Obes. Relat. Metab. Disord. 1999;23:1030–1034. doi: 10.1038/sj.ijo.0801040. [DOI] [PubMed] [Google Scholar]
- 49.Snider KE, et al. Genotype and phenotype correlations in 417 children with congenital hyperinsulinism. J. Clin. Endocrinol. Metab. 2013;98:E355–E363. doi: 10.1210/jc.2012-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.González-Barroso MM, et al. Mutations in UCP2 in congenital hyperinsulinism reveal a role for regulation of insulin secretion. PLoS One. 2008;3:e3850. doi: 10.1371/journal.pone.0003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otonkoski T, et al. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic beta cells. Am. J. Hum. Genet. 2007;81:467–474. doi: 10.1086/520960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flanagan SE, et al. Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab. 2014;19:146–154. doi: 10.1016/j.cmet.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naylor RN, et al. Cost-effectiveness of MODY genetic testing: translating genomic advances into practical health applications. Diabetes Care. 2014;37:202–209. doi: 10.2337/dc13-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greeley SA, et al. The cost-effectiveness of personalized genetic medicine: the case of genetic testing in neonatal diabetes. Diabetes Care. 2011;34:622–627. doi: 10.2337/dc10-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.