Summary
Interspecies protein-protein interactions are essential mediators of infection. While bacterial proteins required for host cell invasion and infection can be identified through bacterial mutant library screens, information about host target proteins and interspecies complex structures has been more difficult to acquire. Using an unbiased chemical cross-linking/mass spectrometry approach, we identified interspecies protein-protein interactions in human lung epithelial cells infected with Acinetobacter baumannii. These efforts resulted in identification of 3076 total cross-linked peptide pairs and 46 interspecies protein-protein interactions. Most notably, the key A. baumannii virulence factor, OmpA, was identified cross-linked to host proteins involved in desmosomes, specialized structures that mediate host cell-to-cell adhesion. Co-immunoprecipitation and transposon mutant experiments were used to verify these interactions and demonstrate relevance for host cell invasion and acute murine lung infection. These results shed new light on A. baumannii-host protein interactions and their structural features and the presented approach is generally applicable to other systems.
Introduction
Interspecies protein interactions and the underlying structural interfaces are essential for bacterial infection
The molecular-level arms race between hosts and pathogens is carried out on multiple fronts, but predominantly takes place through evolutionary adaptation of protein structural landscapes[1-5]. Bacteria commandeer host resources through evolutionarily optimized bacterial protein structures that bind with high specificity to host protein cognates. Pathogen proteins target diverse host proteins involved in metabolite acquisition [4], molecular trafficking to the cell membrane [2], cytoskeletal rearrangement [6], and cell adherence complexes [7]. As an example, iron is necessary for biochemical processes in both bacteria and hosts and can be sequestered by the vertebrate membrane protein transferrin to defend against bacterial infection[4, 8]. In response, bacteria such as Neisseria gonorrhoeae and Haemophilus influenzae have evolved transferrin-binding proteins (TbpA) capable of binding and scavenging iron directly from transferrin to overcome sequestration[8]. Barber and Elde[4] showed that single point mutations in transferrin alter TbpA affinity at the interface of the two proteins and are responsible for establishing the host range of the bacteria and modulating host nutritional immunity. Therefore knowledge of not only the proteins involved in host-pathogen protein interactions but also the manner of their interaction, i.e. structural insight into interfacial regions, can profoundly advance understanding of bacterial infection and provide insight for developing new antimicrobial therapies[4].
Technologies have evolved to allow large-scale protein interaction identification, but relevant information on host-pathogen interspecies interactions and structures is still limited
Two-hybrid[9], affinity purification mass spectrometry[10], and protein compliment[11] methods have made the large-scale study of protein-protein interactions (PPIs) possible. Although recent efforts with these techniques have demonstrated the ability to identify PPIs relevant to host-pathogen interactions, including the virus-human protein interactions of HIV[12] and H1N1[13], host pathogen PPIs remain a general challenge to identify. Furthermore, structural details pertaining to host-pathogen protein interactions are exceedingly sparse. Many aspects of host-pathogen interactions are mediated by membrane proteins, as exemplified by the transferrin case above. With roles in quorum sensing, secretion, adhesion and invasion, membrane proteins play pivotal roles in bacterial pathogenesis, yet they often require significant dedicated efforts for interaction studies, are less suitable for many large-scale methods, and are equally challenging for conventional structural characterization[14].
Alternative technologies have the potential to shed light on interspecies PPIs and their structural interfaces
Chemical crosslinking mass spectrometry (XL-MS) approaches are beginning to have a greater impact on protein interaction studies[15-20]. Because of the finite crosslinker length, covalent linkage of two amino acid sidechains indicates their proximity during the crosslinking reaction period. Identification of crosslinked peptide pairs provides useful distance constraints for development and assessment of structural models, as illustrated for the interactions of protein in purified complexes from the protein phosphatase 2A network[16]. Chemical crosslinking can be carried out with mixtures of proteins in cell lysates[19, 21, 22] or on living cells[23-25], whereby interaction identification and structural details on complexes can be performed in an unbiased manner[26-29]. This approach holds great potential for the determination of transient or long-lived interactions that have been chemically stabilized[22], particularly for the identification of protein interactors and structural details of membrane proteins[27]. For example, the outer membrane protein OmpA in Escherichia coli is important for adhesion to host cells, catheters and implants among its other roles[30]. OmpA has been among the most heavily studied bacterial membrane proteins over the past 30 or more years. However, the protein was only recently shown to exist as a multimer through in vivo crosslinked sites within its C-terminal domain[31]. This finding was recently verified in vitro by site-directed mutation based on our reported crosslinked sites and native mass spectrometry measurements[32]. Our recent efforts have further shown that in vivo crosslinking can yield large scale interactions and structural details on complexes in pathogenic bacterial cells, such as Pseudomonas aeruginosa[27]. These results demonstrated that OprF, an OmpA-homolog in P aeruginosa, exists as a multimer in vivo. Additionally, these data showed the potential to gain information from living cells on the interactions and structures of membrane and soluble protein complexes in systems where molecular biology-based strategies are less developed or even unsuitable.
Given the unbiased capabilities for membrane protein interactions and structural characterization, chemical crosslinking technologies offer the potential for identification of interspecies interactions that can help increase our understanding of bacterial infection
The results presented here illustrate the initial crosslinking application of Protein Interaction Reporter technologies to the study of interspecies PPIs in human lung epithelial cells infected with the nosocomial pathogen, Acinetobacter baumannii. These efforts produced the first large-scale interspecies crosslink dataset, including 46 host-pathogen PPIs, several of which involve the key A. baumannii virulence factor OmpA.
Results
Determination of interspecies protein interactions
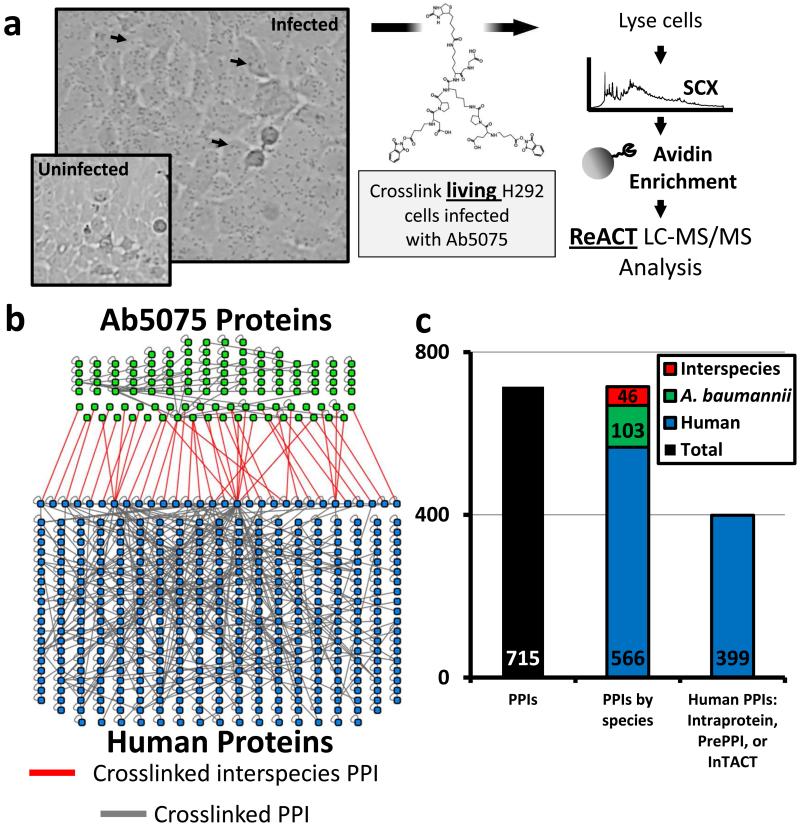
In vivo proteomic XL-MS analysis of proteins from infected lung epithelial cells generated 16,758 crosslinked peptide-peptide relationships (Figure 1A). Of these we identified 3,076 non-redundant peptide-peptide relationships across three biological replicates at a relationship false discovery rate of 0.24% (Table S1). Crosslinked peptide-peptide relationships were mapped to a network of 715 PPIs attributed to 488 human proteins and 113 bacterial proteins (Figure 1B, Figure 1C, Figure S1). Identified human proteins covered an abundance range of greater than five orders of magnitude (Figure S1C)[33]. Relationships between two peptides from the same protein constituted the majority of identifications (intraprotein PPIs). Two residues within a single protein have a high likelihood of being in close physical contact within a cell, are therefore more frequently cross-linked, and make up a higher proportion of interactions in crosslinked datasets[22, 34]. Importantly, intraprotein interactions define proximal residues within a protein, and thereby provide valuable structural coordinates for identified proteins even when no known structure exists for this protein (Figure 2). Alternatively, interprotein PPIs were derived from crosslinked relationships between peptides from two different proteins. Interprotein PPIs were used to generate interaction networks (Figure 1B, Figure 3) and yielded structural data pertaining to the interaction interfaces of protein complexes. Within the interprotein relationships, we determined that 3.7% of the total peptide-peptide crosslinks, and 6.4% of all PPIs, were interspecies interactions Figure 1B, Figure 1C). No bacterial proteins or interspecies interactions were detected in uninfected, crosslinked H292 cells (Table S2). Based on these cross-linked relationships we constructed a protein interaction network that included: host-host, pathogen-pathogen, and host-pathogen PPIs (Figure 1B).
Figure 1. Identification of protein interactions from infected lung epithelial cells.
(a) H292 cells were infected with Ab5075, crosslinked using BDP-NHP. Digested peptides were enriched and analyzed by LC-MS/MS. (b) PPI map of human (blue) and bacterial (green) proteins. Interspecies crosslinks are highlighted in red. (c) Total number of PPIs within the dataset, their breakdown by species (Human-human, Ab5075-Ab5075, or Ab5075-Human), and the relative matched interactions from orthogonal data (i.e. previously observed in PrePPI[35]/IntAct[36] or intraprotein PPIs within a single protein).
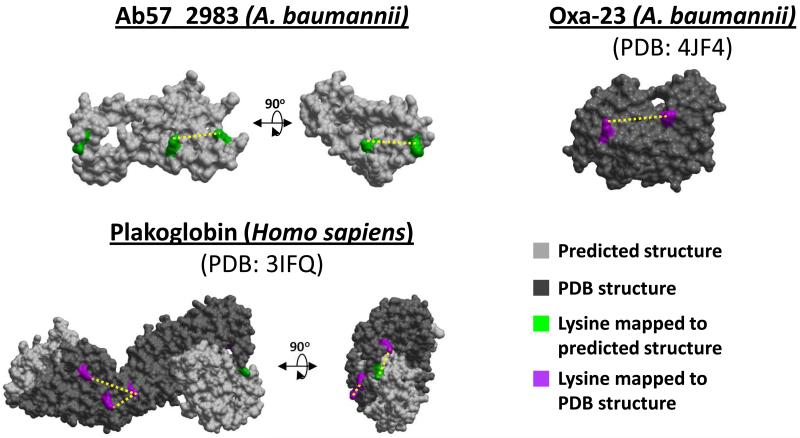
Figure 2. Intraprotein interactions mapped to known and predicted protein structures.
Crosslinked sites identified in the large-scale proteomic infection experiment were mapped to Phyre2 predicted structures [83] (green lysine sites, light grey model) or known crystal structures (magenta lysine sites, dark grey model) for the bacterial proteins Ab57_2983 and Oxa23 and the human protein plakoglobin. For plakoglobin, while the central domain has been crystalized (PDB: 3IFQ), the N- and C- terminal portions have not been crystalized, yet a site within the C-terminus was identified in an intraprotein crosslinked relationship and shown within the predicted structural model of this region.
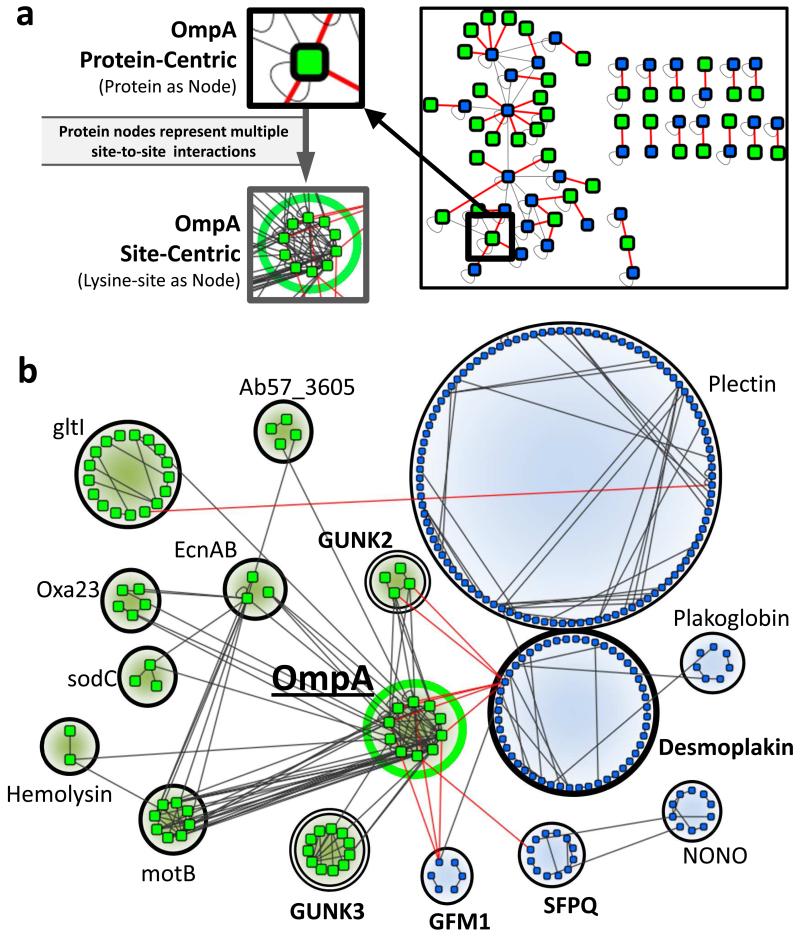
Figure 3. Depth of interspecies and OmpA specific intercellular interactions.
(a) Force-directed network of the interspecies protein interactions identified between A. baumannii and human proteins. Insets depict how multiple site-to-site crosslink interactions underlie each PPI. Interspecies crosslinks are red. (b) Site-to-site interactions for all proteins (bacterial and human) interacting with OmpA in the cell infection model. Nodes are individual lysine sites identified in crosslinked relationships between human (blue nodes) and bacterial (green nodes) proteins. Interspecies links are red.
To verify that PIR technology could detect physiological protein complexes and interactions, we quantified the number of human PPIs that were (1) intraprotein interactions or (2) were previously annotated in a PPI databases (i.e. PrePPI[35] and IntAct[36]). Out of 566 unique human PPIs 70% were attributed to either intraprotein interactions or identified in a PPI database (Figure 1C). Subnetworks of PPIs from known complexes included: host cytoskeleton, heterogeneous ribonucleoproteins, integrins, histones, cohesin, and ATP synthase (Figure S2) [37]. Identified A. baumannii PPIs included interactions with virulence factors, membrane integrity proteins, metabolic complexes, transcription and translation machinery, and many Genes of UNKnown function (GUNKs) (Table S1). The last group represents a large, and potentially important, subset of the A. baumannii proteome. Though they make up approximately 30% of the all A. baumannii proteins (Figure S1D), functional characterization of GUNK proteins is a challenge [38]. Owing to structural and network information derived from identified crosslinked peptide relationships this work provides insight on functional roles for several GUNK proteins.
A. baumannii interactions with host proteins
Bacterial adherence and internalization require host-microbe protein interactions involving bacterial virulence factors binding host signaling complexes, host membrane adherence proteins, and host cytoskeletal proteins [7, 39, 40]. Bacterial membrane proteins and proteins secreted via OMVs were previously suggested to be a major mechanism by which A. baumannii adhere to host cells[41] and deliver effector molecules [42]. Interestingly, the bacterial proteins observed in interspecies PPIs were enriched for the presence of predicted signal peptides (p-value = 0.019)[43] and proteins observed in outer membrane vesicles (OMVs) (p-value = 0.026) [42] (Figure S1E). Furthermore, we observed crosslinked relationships involving bacterial proteins previously determined to be A. baumannii virulence factors, including: OmpA, Lon protease, Oxa-23, hemolysin, TolB, TonB, and several lipoproteins (Figure 3, Table S1) [44]. We found crosslinked relationships between several of these virulence factors (lipoproteins, OmpA and Lon protease) and human proteins from the cytoskeleton (keratin-7, keratin-8, keratin-18, actin B, plectin, Arp 2/3-1B) and junctional adherence proteins (desmoplakin [DSP], plakoglobin, plectin) (Figure 3B)[40, 41, 45].
Verification of OmpA as an essential virulence factor in Ab5075
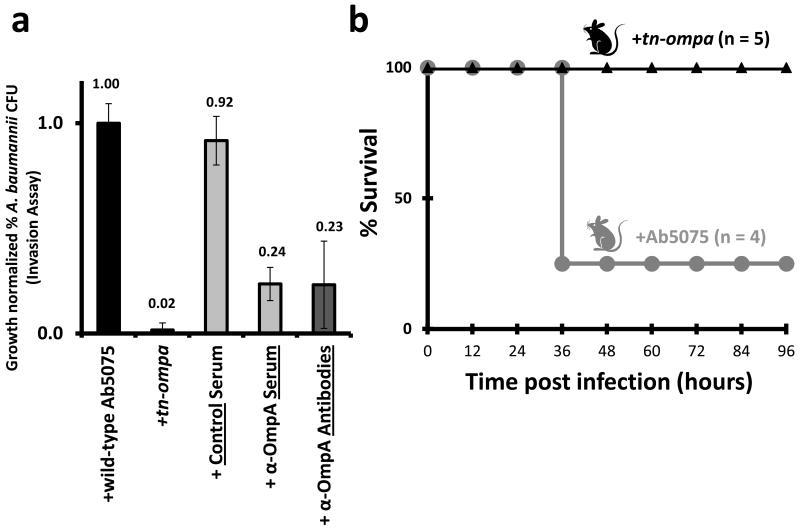
Interactions identified between OmpA and host or bacterial proteins were of special interest as OmpA orthologs in other bacteria and A. baumannii strains are necessary for bacterial invasion [30, 41]. Verification of this critical role for OmpA in Ab5075 was necessary to better understand the significance of the in vivo cross-linking results and establish the pathogenic characteristics of this clinical isolate [46]. To investigate the role of OmpA in the A. baumannii strain Ab5075, we first tested bacterial invasion of a gene inactivation mutant of OmpA (transposon insertion, tn-ompa) in the Ab5075 genetic background. Invasion of host cells was severely attenuated for the tn-ompa mutant compared to wild type Ab5075 (WT, Figure 4A, Figure S3A). Second, pre-treatment with α-OmpA serum prior to infection significantly attenuated Ab5075’s ability to invade host cells (Figure 4A). Third, we quantified bacterial invasion after pre-treatment of bacterial cells with novel, purified, and targeted OmpA antibodies (Figure 4A, Figure S3B). Three antibodies, targeting two extracellular loops and a C-terminal portion of OmpA, were generated and pooled (Figure S3B). We observed that these targeted antibodies were able to attenuate bacterial invasion to the same degree as α-OmpA serum (Figure 4A). Finally in an acute murine lung infection model, all 5 mice treated with tn-ompa survived bacterial challenge, while 3 of the 4 mice treated with WT Ab5075 succumbed to infection within 36 hours (Figure 4B). Taken together, these findings establish the significance of OmpA in Ab5075 virulence and highlight new tools for studying Ab5075 virulence (i.e. tn-ompa strain and targeted antibodies)
Figure 4. OmpA-related virulence in Ab5075.
(a) Ab5075 invasion assays comparing WT Ab5075, Ab5075 with transposon disruption of OmpA (tn-ompA), and WT Ab5075 treated with control IgG serum, α-OmpA serum, or purified α-OmpA antibodies. Mean +/− SEM. (b) Murine intratracheal infection with WT Ab5075 (grey) or tn-ompa (black, log-rank p-value = 0.025).
Identification of interactions between a host desmosome and A. baumannii proteins
Bacterial outer membrane proteins have been proposed to be involved in host cell adherence and evasion of host immunity[47] and the outer membrane protein OmpA appears critical in several bacterial species[30, 41]. Therefore, identification of OmpA cross-linked to several host proteins in infected cells, including an obligate component of desmosomes and hemi-desmosomes (Figure 3B) offers new insight on OmpA function during cell invasion. Peptide sequences from DSP and OmpA were unique to the human and A. baumannii proteomes, respectively (Table S1).
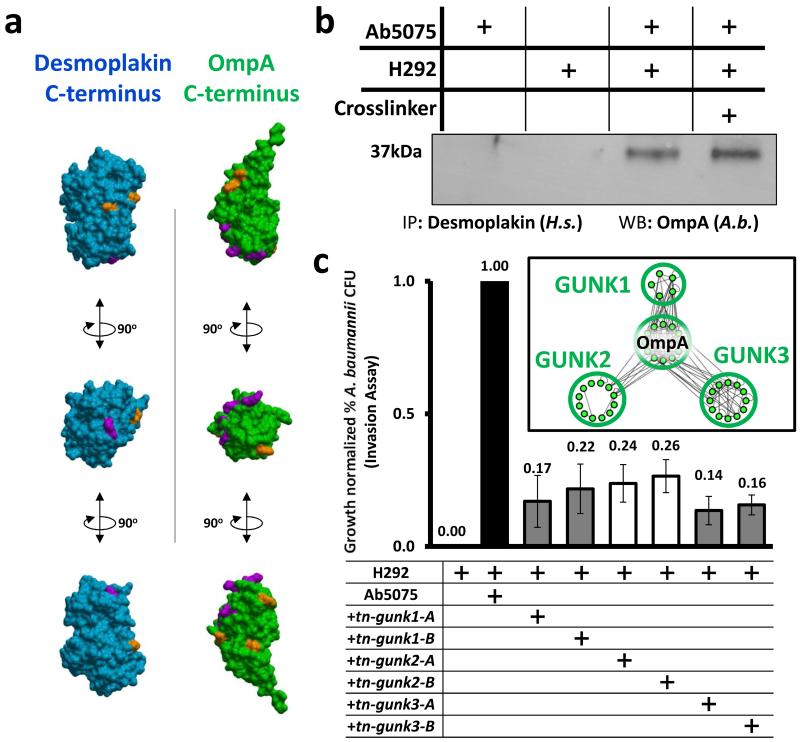
Bacterial infiltration of host epithelial layers has been demonstrated to involve close proximal relationships between desmosomes and bacterial cells[48]. Desmosomes are cell adhesion complexes that mediate cell-to-cell contact in host epithelia [45, 49, 50], and – along with tight junctions, gap junctions, and adherens junctions – create a physical barrier to prevent bacterial intrusion through the epithelium[51]. DSP-OmpA crosslinked peptides that we identified correspond to sites within the C-termini of both proteins. Disruption of the DSP C-terminus has been reported to interfere with epithelial integrity [50], and therefore virulence factor binding the DSP C-terminal could serve to destabilize host cell-to-cell interactions to bypass the barrier function of host epithelia[52]. Sites of crosslinking within both proteins were mapped to C-terminal crystal structures of OmpA and DSP (Protein Databank: 4G4Y [OmpA] and 1LM5 [DSP] [53, 54]) (Figure 5A) [55]. Consistent with our crosslinking results, when we modeled the protein docking of the C-termini of DSP and OmpA, the linked lysine residues between the two proteins in the predicted model were within the crosslinker distance constraint for the BDP-NHP crosslinker (Figure S4B).
Figure 5. Functional significance of uncharacterized interspecies interactors and OmpA interactors.
(a) Space filling models for crystal structures of OmpA (green, PDB: 4G4Y) and DSP (blue, PDB: 1LM5). Identified sites of crosslinking are highlighted in orange. Sites crosslinked between DSP and OmpA are highlighted in magenta. (b) Immunoprecipitation of DSP from Ab5075 cells, H292 cells alone or Ab5075-infected H292 cells (with and without crosslinker). (c) Ab5075 invasion assays comparing WT Ab5075 to Ab5075 with transposon disruption mutants of GUNK1, GUNK2, GUNK3 (Ab57_1108, Ab57_2521, and Ab57_2983). Mean +/− SEM. Inset: crosslinked site interactions between GUNK proteins and OmpA from Ab5075 crosslinked alone (nodes: non-redundant crosslinked lysines; edges: crosslinked relationships).
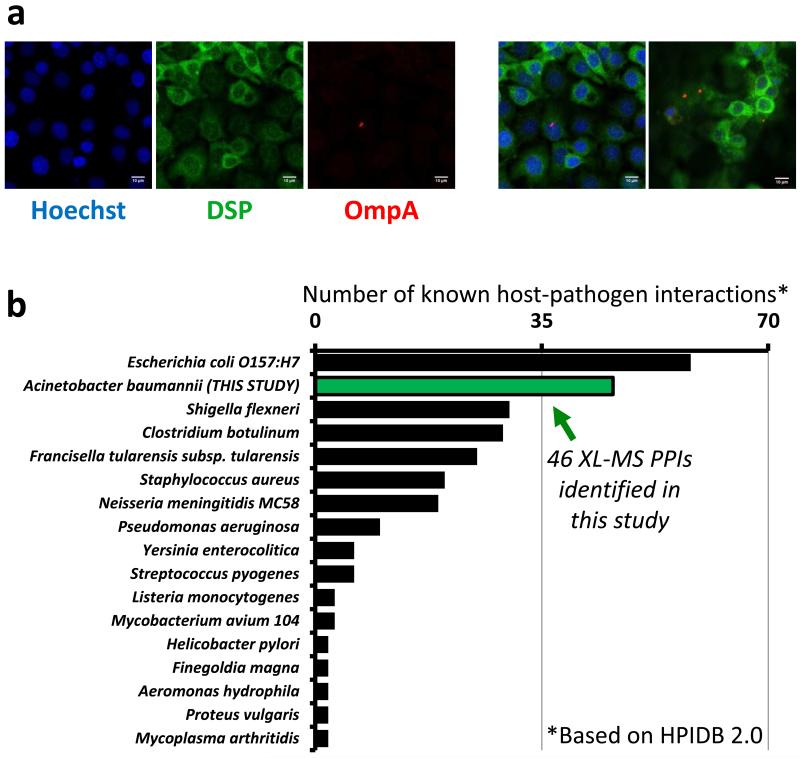
To validate this interaction, we immunoprecipitated DSP from Ab5075-infected H292 cells and uninfected H292 cells and blotted for the presence of OmpA. OmpA precipitated with DSP from infected H292 cells in the presence and absence of crosslinker (Figure 5B, Figure S4A), confirming the interaction observed through large-scale, in vivo crosslinking. Notably, two high-mass bands, both greater than 250,000 Da in size, were present only in the crosslinked sample (Figure S4A). These high molecular weight bands were presumed to be the crosslinked protein complex formed by DSP (>260 kDa) and OmpA (38 kDa). By confocal immunofluorescence, DSP and OmpA proteins colocalized during Ab5075 infection of H292 cells (Figure 6A). In addition, we observed an A. baumannii GUNK protein crosslinked with DSP. The GUNK identified in multiple peptide-peptide interactions with DSP was Ab57_2521 (later referred to as GUNK2), a signal-peptide containing protein that also physically interacts with OmpA in infected host cells. The random chance of matching multiple interactors across species in this way was miniscule (ρ = 2.35×10−9, Figure 3B, Figure S5A).
Figure 6. Ab5075 host-pathogen interactions.
(a) Confocal micrographs of Ab5075-infected H292 cells stained with primary antibodies against desmoplakin (green) and OmpA (red). DNA was stained with Hoechst 33342 (blue). Confocal images and overlaid images from replicate analyses at a minimal thickness of 1.1μm, scale bars represent 10μm. (b) The 46 host-pathogen interactions identified in this study (green bar) compared to the number of interspecies interactions identified for other bacterial species (HPIDB 2.0 database) [60]. If substrains were present in the database, the strain with the highest number of interactions was shown. Due to scale, HPIDB 2.0 host pathogen interactions for Yersinia pestis (n=4018), Bacillus anthracis (n=3061), and Francisella tularensis subsp. tularensis SCHU S4 (n=1346) were not shown.
Guilt-by-association: Functional significance of genes of unknown function
Defining the function and biological significance of GUNK proteins remains a challenge [38]. The unbiased determination of interactions between GUNK proteins and annotated gene products from a bacterial genome offers clues for putative functional characterization of GUNKs[56]. In addition to cross-linking infected host cells, we harvested Ab5075 alone and crosslinked the bacterial cells to improve network and structural coverage of bacterial interprotein and intraprotein crosslinks within Ab5075 (Table S3). Of particular interest were GUNK proteins we observed linked to OmpA, since OmpA appears necessary for bacterial infection (Figure 4). In both infected host cells and A. baumannii cells alone we observed crosslinks between OmpA and several GUNK proteins (Table S1, Table S3). We quantified bacterial invasion in host cells using transposon insertion mutants of three unannotated GUNKs most frequently identified cross-linked to OmpA – hereafter GUNK1, GUNK2, and GUNK3 (Figure 5C, Table S1). Transposon insertion mutants of GUNK1, GUNK2, and GUNK3 in the AB-5075 background showed reduced host cell invasion as compared to WT Ab5075 (Figure 5C). Two independent transposon mutants (A and B) for each GUNK gene were tested and observed to have consistent phenotypes (Figure 5C). While the attenuation of mutant bacterial invasion was not as great for these GUNK proteins as for OmpA, the reduction was still statistically significant compared to WT Ab5075 (p-values < 2×10−4, < 7×10−5, and < 9×10−7 for GUNK1, 2 and 3, respectively) (Figure 5C).
The intersection of PPIs and bacterial genes essential for persistence in host lungs
We compared our dataset of interspecies crosslinked proteins to a recently established set of A. baumannii genes determined to be essential for bacterial persistence in the murine lung [57]. Of the bacterial proteins we identified in crosslinked interspecies interactions (n=31), we observed a significant enrichment of essential persistence genes (7/31, p-value = 0.000664) (Figure S5B). Discrepancies between these two datasets can, in part be explained by three factors. First, a different, less virulent strain of A. baumannii (Ab17978) was used to test persistence in the murine lung[46, 57]. Second, not all proteins essential for invasion may interact with host proteins (proteins identified in persistence study but not in the proteomic interactome). Third, not all protein interactions between host and pathogen proteins may be essential (proteins identified in proteomic interactome but not in persistence study). Nonetheless, these results from in vivo host-pathogen cross-linking provide the first identification of host partners for seven A. baumannii proteins previously found to be essential for bacterial persistence in the host lung.
Discussion
Interspecies protein interactions are critical determinants in bacterial infection and pathogenesis[2]. Improved knowledge on which interspecies PPIs exist and how the involved proteins interact can greatly advance understanding of molecular mechanisms involved in host invasion and provide new opportunities for antibacterial therapies. As the growing threat of antibiotic resistant pathogens appears likely to surpass all currently known antibiotics [58, 59], new knowledge and strategies that can help treat multidrug resistant bacterial infections are of critical importance. Many strains of Acinetobacter baumannii, such as the clinical strain Ab5075 used in these studies, exhibit increased resistance to all antibiotics, including carbapenems [46, 58]. Using unbiased in vivo protein crosslinking we identified physical interspecies PPIs during microbial infection to enable the first visualization of the A. baumannii-host interaction network. In total, more than 3000 cross-linked peptide pairs were identified defining the existence and structural features of more than 700 PPIs and 46 interspecies PPIs representing one of the largest host-bacteria protein interaction datasets and the first large-scale PPI analysis of host-pathogen interactions for A. baumannii (based on HPIDB2.0, Figure 6B) [60].
Previous studies have shown the outer membrane protein OmpA to be critical to attachment and invasion in many pathogenic bacteria [30], including A. baumannii [41, 61]. OmpA was also identified as a required gene for microbial persistence in murine lung infection (ρ=7.6×10−79)[57]. Though our experimental design did not specifically target OmpA, the membrane porin was identified in cross-linked relationships to several host proteins in infected epithelial cells. During infection OmpA migrates to at least three subcellular locations within host cells: mitochondria, nuclei and cell-surfaces [62, 63]. Consistent with these findings, we identified OmpA interspecies interactions with a mitochondrial protein that affects reactive oxygen species generation (GFM-1) [64, 65], a nuclear protein involved in the DNA damage response[66] and circadian rhythm regulation[67] (SFPQ), and the obligate desmosomal adhesion protein DSP (Figure 3B).
Bacterial targeting of adhesion complexes is a well-established mechanism for microbial intrusion and invasion [68, 69]. Gram-negative bacteria, such as Staphylococcus aureus[70] and Campylobacter jejuni[71], have previously been observed to bind and disrupt desmosomes during infection. Van Schilfgaarde et al. showed that bacterial intrusion into human lung epithelial cells placed infiltrating bacteria in close contact with desmosomal plaques[48]. The in vivo cross-linking results presented here linking OmpA and DSP are consistent with these observations and is corroborated by the fact that OmpA co-localizes with DSP in infected human cells (Figure 6A). OmpA has previously been shown to be a component of secreted A. baumannii OMVs[61]. OmpA and DSP could therefore colocalize after bacterial internalization [72] (Figure 4A, Figure 6A) or through the local release of bacterial OMVs [61] and the subsequent integration of these vesicular membranes into the host membrane [73]. For the latter case, altering desmosomal integrity could aid infiltration through host epithelia. Finally, because desmosomes integrate signals between the host cell surface and cytoskeleton [74], and a functional host cytoskeleton is required for A. baumannii pathogenesis [75], binding and targeted disruption of the desmosome by bacterial proteins could be a means for A. baumannii to modulate the requisite host cytoskeleton and facilitate epithelial intrusion and host-cell invasion. This speculation requires further investigation of course, but knowledge of the OmpA-DSP interaction will enable these future studies.
With extensive evidence linking OmpA and orthologous proteins to pathogenicity, the discovery of GUNK proteins linked to OmpA established a preliminary association between these GUNKs and bacterial invasion. While two GUNK proteins were A. baumannii-specific (GUNK2 and GUNK3), GUNK1 shares sequence identity with hypothetical proteins from Bacillus (52%) and Pseudomonas (43%) species and META-domain/HslJ proteins from Bordetella (38%) and E. coli (32%). The HslJ protein has previously been related to antibiotic resistance to novobiocin [76], and overexpression of META proteins in Leishmania resulted in increased virulence[77]; META-domain proteins, often observed in hypothetical bacterial proteins [78], have also been implicated in bacterial motility. The determination of multiple crosslinked relationships with OmpA provided interaction-based evidence (‘guilt-by-association’ [56]) leading to further characterization of these GUNK proteins. We believe that sequence similarity potentiating the involvement of GUNK1 in motility/resistance and the attenuated host invasion by all three GUNK transposon insertion mutants (Figure 5C) further support these GUNK proteins as putative A. baumannii virulence factors. These findings established the utility of an unbiased approach to identify novel interactions and disseminate functional characterization from PPIs between known virulence factors and GUNK proteins.
The techniques underlying our study of in vivo protein crosslinking in bacterially infected human cells provide a unique opportunity to view networks of intercellular PPIs. Recent studies have suggested that interactomes, even incomplete ones, can enable discovery of molecular commonalities between phenotypically-related disease pathologies that may not share primary disease genes [79]. When extrapolated to the study of bacterial pathogenesis, overlapping interspecies interactomes could reveal common paradigms of infection exploited by microbes. In vivo cross-linking of host-pathogen systems is a generally applicable technology and future applications with other pathogens will help to explore the possibility of common strategies used among multiple pathogens.
Significance
Protein interactions are essential mediators of bacterial pathogenesis. While methods to elucidate bacterial proteins required for infection have been employed extensively, information regarding the host proteins targeted by bacteria and interspecies complex structures has been more difficult to determine. In this proof-of-principle study, we used chemical cross-linking of proteins in combination with large-scale mass spectrometry to identify interspecies interactions between proteins in cultured human cells and Gram-negative bacterial proteins during bacterial infection. These efforts resulted in identification of interspecies protein-protein interactions between human proteins and known bacterial virulence factors. Most notably we identified bacterial virulence factors interacting with host structural proteins that mediate host cell-to-cell adhesion. Our study shows the potential of chemical cross-linking of proteins in combination with large-scale mass spectrometry to shed new light on host-pathogen protein interactions and their structural features. We demonstrate the ability of said methods to interrogate protein interactions underlying complex pathogenic systems and the presented methodologies are generally applicable to other pathogen systems. The broad application of these methods could aid the rapid expansion of understanding of how diverse bacterial species target and manipulate host proteins during pathogenesis.
Experimental Procedures
Cell culture
Ab5075 cells were grown to stationary phase in nutrient broth (BD). H292 cells were grown in RPMI-1640 (Thermo) with 10% fetal bovine serum and 1% penicillin/streptomycin (37°C, 5% CO2). Transposon mutants in Ab5075 were attained from Dr. Colin Manoil [80].
Crosslinker synthesis, infection, Stage-1 database creation, sample preparation
Biotin-Aspartate Proline-PIR n-hydroxyphthalimide (BDP-NHP) was synthesized as previously described [28, 81]. Confluent H292 cells were washed, released and pelleted before resuspended in crosslinking (XL)-buffer (0.17M potassium phosphate) containing Ab5075 cells at an MOI of 500 for 2 hours. Five experiments were run: three biological replicates of Ab5075-infected H292 cells (Figure S6), and two biological replicates of uninfected H292 cells (Table S2). BDP-NHP (8mM) was added to the cell suspension for 1 hour. H292 cells infected with A. baumannii (i.e. HA-1, HA-2, HA-3) and uninfected cells were washed, pelleted, and frozen at −80°C.
Infected H292 cell pellets were resuspended in lysis buffer: 8M urea, 100mM Tris-Cl pH 8.0, 150mM NaCl, and protease inhibitor tablets (Roche); then lysed by cryo-grinding and sonication. Proteins were reduced and alkylated. Protein lysates were diluted with 100mM Tris-Cl, pH 8.0. 1mg of protein was removed to enrich full length crosslinked proteins with monomeric avidin beads. Enriched, crosslinked proteins were digested with sequencing grade trypsin (Promega), and injected on a C-8 column eluting into an LTQ-XL or LTQ-Velos-FT-ICR mass spectrometer to create a search database of proteins that had been crosslinked (Stage-1 database)[28]. Digested Stage-1 peptide samples were shot in quadruplicate on a 4 hour reverse phase data dependent Top5 method. Spectra were searched using SEQUEST. 30880 peptide-spectral matches (10304 unique peptides) were identified at a false discovery rate (FDR) of less than 1%, based on a concatenated, target-decoy database of all human and A. baumannii (AB0057) proteins [28]. The remaining protein lysates were digested with sequencing grade trypsin. Digested peptides were desalted and fractionated by strong cation exchange (SCX). Eluted peptides were incubated with monomeric avidin beads to enrich crosslinked peptides.
LC-MS/MS/MS, ReACT, database searching, and data analysis
Crosslinked peptides were resuspended in 5% ACN/2% formic acid and injected onto an in-house pulled C-8 column (Magic, 200A, 5um) run on 4 hour gradients as with the Stage-1 mass spectral analysis; eluted peptides were analyzed on an LTQ-Velos-FT-ICR. Crosslinked peptides were fragmented in a data-dependent ReACT mode (Real-time Analysis for Crosslinked peptide Technology)[28]. Briefly, high-charge state precursor ions (MS1, z > 4+) were isolated and fragmented at low energy (Q = 0.20) to release crosslinked peptides and a reporter ion (m/z = 752.41). Data-dependent selection of fragmented MS2-ions that sum to the precursor mass minus the reporter ion were further fragmented for MS3 spectra and peptide sequencing.
All parameters and filtering was done as previously described to identify unique PPIs and site-site interactions[28]. The final relationship FDR was calculated to be 0.24% [(2 * 20 decoy relationships with at least 1 reverse hit)/16758 total relationships]. The final PPI FDR was 1% [(2 * 4 unique decoy PPIs)/719 total unique PPI’s]. All interaction data, including pep.xml files, can be found at http://brucelab.gs.washington.edu/xlinkdb/. Protein interaction networks were created using Cytoscape 3.0. Protein structures for OmpA and desmoplakin were downloaded from the Protein Databank (4G4Y and 1LM5, respectively). Protein structure interactions were modeled using PatchDock[55]. KEGG pathway enrichment p-values were determined using STRING v9.1 [37].
Gentamicin protection assays
H292 cells were plated in 24-well plates and allowed to attach for 16 hours. H292’s were incubated with Ab5075 or Ab5075-tn-ompa for 3 hrs in serum-free, antibiotic-free RPMI-1640 media (RPMI). The supernatant was saved to normalize AB5075 growth during infection. H292 cells were incubated for 1 hour with 200μg/ml gentamicin in RPMI at 37°C. Lysis buffer (PBS with 0.1% Trition-X100) was added to each well, and the plate was shaken at 200rpm for 10 min. Lysates (50ul) were cultured on NB-agar plates (16 hours, 37°C). Colonies were counted and normalized to WT AB5075 growth. Values are the average of technical duplicates from at least three experiments performed on three different days. For antibody blocking of OmpA-based invasion, Ab5075 cells were pre-incubated with α-OmpA or PBS-control serum (Dr. Michael McConnell) or purified Genscript synthesized antibodies. Pre-treated Ab5075 cells were then incubated with H292 cells as above.
Confocal immunofluorescence and brightfield microscopy
Confluent H292 cells were infected with Ab5075 (MOI = 100) on 3.5cm plates with glass coverslips (No. 1.5, Mattek). For confocal immunofluorescence, cells were fixed (formalin), blocked (3% milk in PBST) and incubated overnight at 4°C with α-DSP antibody (rabbit, Abcam) and primary mouse α-OmpA serum (Dr. Michael McConnell) in blocking buffer. Microscopy was performed with a Nikon A1 confocal mounted on a Nikon TiE inverted microscope (Garvey Cell Imaging Lab) at 20× magnification, n=1 (air), NA=0.75. Depth of field measured based on λ = 595nm. For brightfield microscopy, H292 cells were grown to confluency on 24 well plates, incubated in crosslinking buffer +/− Ab5075 cells (MOI=100), washed with PBS and imaged at 20× magnification.
Murine acute lung infection
WT Ab5075 or tn-ompA were streaked on LB agar or LB agar with 5ug/ml tetracycline (LBtet) from frozen stocks. PBS (50ul) containing ~2×108 CFU/ml bacteria were administered intratracheally to anesthetized mice as described before[82]. Animals that became moribund, distressed or were unable to eat/drink were euthanized using a CO2 chamber. Experiments were approved by the University of Washington Institutional Animal Care and Use Committee (protocol number 4113-01).
Co-immunoprecipitation (Co-IP) and western blotting
H292 cells were grown to confluency and washed prior to co-incubation with Ab5075 cells (MOI=500) for 2 hours at RT. Subsequently, either DMSO or BDP-NHP in DMSO was added to the samples followed by 1 hour incubation. Cells were pelleted and resuspended in IP buffer (10mM Tris-Cl pH 8.0, 100mM NaCl, 1% Triton-X100, 1mM EDTA, protease inhibitors, 100μg/ml lysozyme) and incubated on ice for 30min to lyse bacterial cells; samples were syringe pumped (27.5 gauge needle) and sonicated to further lyse cells and fragment DNA.
Lysates were pre-cleared with 50μl of Protein-G-agarose (Thermo) mixed at 4°C for 1 hour. α-Desmoplakin antibody was added for 16.5 hours at 4°C. For immunoprecipitation, 50μl of Protein-G-agarose was added for 3 hours at 4°C. Beads were pelleted and thoroughly washed. Co-IP proteins were eluted at 95°C for 10 min with a 3:1 ratio of 1% SDS, 15% glycerol, 50mM Tris-Cl pH 8.0, 150mM NaCl: XT Sample Buffer (Biorad). Finally, proteins were detected by SDS-PAGE and western blot (primary: α-OmpA serum; secondary: α-mouse antibodies [IRDye, Li-Cor]).
Supplementary Material
Highlights.
3076 cross-linked peptide pairs identified from infected cells.
Forty-six interspecies protein-protein interactions were identified from cells.
Interspecies interactions involving unannotated A. baumannii proteins were identified.
Structural features of OmpA-desmoplakin interaction revealed.
Acknowledgements
This work was supported by National Institutes of Health grants: U19-AI107775-02 (JEB and CM), R01-AI101307-03 (PKS, JEB and CM), R01-GM086688-06 (JEB) and R01-HL110879-04 (PKS and JEB).The authors thank: Chunxiang Zheng and Jimmy Eng, and Drs. Arti Navare, and Priska von Haller for advice concerning experimentation, technology and programming; Dr. Nicholas Foti for help with statistics; Drs. Rustin Lovewell and Brent Berwin for advice concerning bacterial invasion assays; Dr. Michael McConnell for control and α-OmpA serum; and Dr. Ron Seifert (UW Garvey Cell Imaging Lab) for microscopy assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Further description of Experimental Procedures can be found in the Supplemental Experimental Procedures.
References
- 1.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nature reviews. Microbiology. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- 3.Demogines A, Abraham J, Choe H, Farzan M, Sawyer SL. Dual host-virus arms races shape an essential housekeeping protein. PLoS biology. 2013;11:e1001571. doi: 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber MF, Elde NC. Nutritional immunity. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346:1362–1366. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel MR, Loo YM, Horner SM, Gale M, Jr., Malik HS. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS biology. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cossart P, Lecuit M. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. The EMBO journal. 1998;17:3797–3806. doi: 10.1093/emboj/17.14.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda J, Hayashi N, Okamoto M, Sawada S, Minagawa S, Yano Y, Gotoh N. Translocation of Pseudomonas aeruginosa from the intestinal tract is mediated by the binding of ExoS to an Na,K-ATPase regulator, FXYD3. Infection and immunity. 2010;78:4511–4522. doi: 10.1128/IAI.00428-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarantonelli ML, Szatanik M, Giorgini D, Hong E, Huerre M, Guillou F, Alonso JM, Taha MK. Transgenic mice expressing human transferrin as a model for meningococcal infection. Infection and immunity. 2007;75:5609–5614. doi: 10.1128/IAI.00781-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 12.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter EP, Beis K, Cameron AD, Iwata S. Overcoming the challenges of membrane protein crystallography. Current opinion in structural biology. 2008;18:581–586. doi: 10.1016/j.sbi.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang XT, Munske GR, Siems WF, Bruce JE. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 16.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 17.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nature reviews. Molecular cell biology. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 18.Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass spectrometry reviews. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 19.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, et al. Identification of cross-linked peptides from complex samples. Nature methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 20.Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, et al. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Identification of cross-linked peptides from large sequence databases. Nature methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subbotin RI, Chait BT. A pipeline for determining protein-protein interactions and proximities in the cellular milieu. Molecular & cellular proteomics: MCP. 2014;13:2824–2835. doi: 10.1074/mcp.M114.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Tang X, Munske GR, Tolic N, Anderson GA, Bruce JE. Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Molecular & cellular proteomics: MCP. 2009;8:409–420. doi: 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X, Yi W, Munske GR, Adhikari DP, Zakharova NL, Bruce JE. Profiling the membrane proteome of Shewanella oneidensis MR-1 with new affinity labeling probes. Journal of proteome research. 2007;6:724–734. doi: 10.1021/pr060480e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaake RM, Wang X, Burke A, Yu C, Kandur W, Yang Y, Novtisky EJ, Second T, Duan J, Kao A, et al. A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Molecular & cellular proteomics: MCP. 2014;13:3533–3543. doi: 10.1074/mcp.M114.042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavez JD, Hoopmann MR, Weisbrod CR, Takara K, Bruce JE. Quantitative proteomic and interaction network analysis of cisplatin resistance in HeLa cells. PloS one. 2011;6:e19892. doi: 10.1371/journal.pone.0019892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navare AT, Chavez JD, Zheng C, Weisbrod CR, Eng JK, Siehnel R, Singh PK, Manoil C, Bruce JE. Probing the Protein Interaction Network of Pseudomonas aeruginosa Cells by Chemical Cross-Linking Mass Spectrometry. Structure. 2015;23:762–773. doi: 10.1016/j.str.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisbrod CR, Chavez JD, Eng JK, Yang L, Zheng C, Bruce JE. In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J Proteome Res. 2013;12:1569–1579. doi: 10.1021/pr3011638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng C, Weisbrod CR, Chavez JD, Eng JK, Sharma V, Wu X, Bruce JE. XLink-DB: database and software tools for storing and visualizing protein interaction topology data. J Proteome Res. 2013;12:1989–1995. doi: 10.1021/pr301162j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Confer AW, Ayalew S. The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Veterinary microbiology. 2013;163:207–222. doi: 10.1016/j.vetmic.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Zheng C, Yang L, Hoopmann MR, Eng JK, Tang X, Weisbrod CR, Bruce JE. Cross-linking measurements of in vivo protein complex topologies. Molecular & cellular proteomics: MCP. 2011;10 doi: 10.1074/mcp.M110.006841. M110 006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcoux J, Politis A, Rinehart D, Marshall DP, Wallace MI, Tamm LK, Robinson CV. Mass spectrometry defines the C-terminal dimerization domain and enables modeling of the structure of full-length OmpA. Structure. 2014;22:781–790. doi: 10.1016/j.str.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Molecular systems biology. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zybailov BL, Glazko GV, Jaiswal M, Raney KD. Large Scale Chemical Cross-linking Mass Spectrometry Perspectives. Journal of proteomics & bioinformatics. 2013;6:001. doi: 10.4172/jpb.S2-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang QC, Petrey D, Garzon JI, Deng L, Honig B. PrePPI: a structure-informed database of protein-protein interactions. Nucleic acids research. 2013;41:D828–833. doi: 10.1093/nar/gks1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic acids research. 2014;42:D358–363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier M, Sit RV, Quake SR. Proteome-wide protein interaction measurements of bacterial proteins of unknown function. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:477–482. doi: 10.1073/pnas.1210634110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 40.Swanson JA, Baer SC. Phagocytosis by zippers and triggers. Trends in cell biology. 1995;5:89–93. doi: 10.1016/s0962-8924(00)88956-4. [DOI] [PubMed] [Google Scholar]
- 41.Choi CH, Lee JS, Lee YC, Park TI, Lee JC. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC microbiology. 2008;8:216. doi: 10.1186/1471-2180-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon SO, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS microbiology letters. 2009;297:150–156. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 43.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 44.McConnell MJ, Actis L, Pachon J. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS microbiology reviews. 2013;37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 45.Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harbor perspectives in biology. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio. 2014;5:e01076–01014. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galdiero S, Falanga A, Cantisani M, Tarallo R, Della Pepa ME, D’Oriano V, Galdiero M. Microbe-host interactions: structure and role of Gram-negative bacterial porins. Current protein & peptide science. 2012;13:843–854. doi: 10.2174/138920312804871120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Schilfgaarde M, van Alphen L, Eijk P, Everts V, Dankert J. Paracytosis of Haemophilus influenzae through cell layers of NCI-H292 lung epithelial cells. Infection and immunity. 1995;63:4729–4737. doi: 10.1128/iai.63.12.4729-4737.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nekrasova O, Green KJ. Desmosome assembly and dynamics. Trends in cell biology. 2013;23:537–546. doi: 10.1016/j.tcb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrod D, Chidgey M. Desmosome structure, composition and function. Biochimica et biophysica acta. 2008;1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 51.Lievin-Le Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clinical microbiology reviews. 2006;19:315–337. doi: 10.1128/CMR.19.2.315-337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nature reviews. Immunology. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 53.Choi HJ, Park-Snyder S, Pascoe LT, Green KJ, Weis WI. Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure. Nature structural biology. 2002;9:612–620. doi: 10.1038/nsb818. [DOI] [PubMed] [Google Scholar]
- 54.Park JS, Lee WC, Yeo KJ, Ryu KS, Kumarasiri M, Hesek D, Lee M, Mobashery S, Song JH, Kim SI, et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2012;26:219–228. doi: 10.1096/fj.11-188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic acids research. 2005;33:W363–367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang PI, Marcotte EM. It’s the machine that matters: Predicting gene function and phenotype from protein networks. Journal of proteomics. 2010;73:2277–2289. doi: 10.1016/j.jprot.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang N, Ozer EA, Mandel MJ, Hauser AR. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio. 2014;5:e01163–01114. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiology and molecular biology reviews: MMBR. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nature reviews. Microbiology. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 60.Kumar R, Nanduri B. HPIDB--a unified resource for host-pathogen interactions. BMC bioinformatics. 2010;11(Suppl 6):S16. doi: 10.1186/1471-2105-11-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PloS one. 2011;6:e17027. doi: 10.1371/journal.pone.0017027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JS, Choi CH, Kim JW, Lee JC. Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting. Journal of microbiology. 2010;48:387–392. doi: 10.1007/s12275-010-0155-1. [DOI] [PubMed] [Google Scholar]
- 63.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, Chae JP, Yoo SM, Lee JC. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cellular microbiology. 2008;10:309–319. doi: 10.1111/j.1462-5822.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 64.Prasad T, Hameed S, Manoharlal R, Biswas S, Mukhopadhyay CK, Goswami SK, Prasad R. Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS yeast research. 2010;10:587–596. doi: 10.1111/j.1567-1364.2010.00639.x. [DOI] [PubMed] [Google Scholar]
- 65.Soiferman D, Ayalon O, Weissman S, Saada A. The effect of small molecules on nuclear-encoded translation diseases. Biochimie. 2014;100:184–191. doi: 10.1016/j.biochi.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 66.Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of Matrin and SFPQ/NONO in the DNA damage response. Cell cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- 67.Guillaumond F, Boyer B, Becquet D, Guillen S, Kuhn L, Garin J, Belghazi M, Bosler O, Franc JL, Francois-Bellan AM. Chromatin remodeling as a mechanism for circadian prolactin transcription: rhythmic NONO and SFPQ recruitment to HLTF. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25:2740–2756. doi: 10.1096/fj.10-178616. [DOI] [PubMed] [Google Scholar]
- 68.Guttman JA, Li Y, Wickham ME, Deng W, Vogl AW, Finlay BB. Attaching and effacing pathogen-induced tight junction disruption in vivo. Cellular microbiology. 2006;8:634–645. doi: 10.1111/j.1462-5822.2005.00656.x. [DOI] [PubMed] [Google Scholar]
- 69.Guttman JA, Kazemi P, Lin AE, Vogl AW, Finlay BB. Desmosomes are unaltered during infections by attaching and effacing pathogens. Anatomical record. 2007;290:199–205. doi: 10.1002/ar.20414. [DOI] [PubMed] [Google Scholar]
- 70.Stanley JR, Amagai M. Pemphigus, bullous impetigo, and the staphylococcal scalded-skin syndrome. The New England journal of medicine. 2006;355:1800–1810. doi: 10.1056/NEJMra061111. [DOI] [PubMed] [Google Scholar]
- 71.Wine E, Chan VL, Sherman PM. Campylobacter jejuni mediated disruption of polarized epithelial monolayers is cell-type specific, time dependent, and correlates with bacterial invasion. Pediatric research. 2008;64:599–604. doi: 10.1203/PDR.0b013e31818702b9. [DOI] [PubMed] [Google Scholar]
- 72.Rumbo C, Tomas M, Fernandez Moreira E, Soares NC, Carvajal M, Santillana E, Beceiro A, Romero A, Bou G. The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infection and immunity. 2014;82:4666–4680. doi: 10.1128/IAI.02034-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS pathogens. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. The Journal of investigative dermatology. 2007;127:2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 75.Rottner K, Stradal TE, Wehland J. Bacteria-host-cell interactions at the plasma membrane: stories on actin cytoskeleton subversion. Developmental cell. 2005;9:3–17. doi: 10.1016/j.devcel.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 76.Lilic M, Jovanovic M, Jovanovic G, Savic DJ. Identification of the CysB-regulated gene, hslJ, related to the Escherichia coli novobiocin resistance phenotype. FEMS microbiology letters. 2003;224:239–246. doi: 10.1016/S0378-1097(03)00441-5. [DOI] [PubMed] [Google Scholar]
- 77.Uliana SR, Goyal N, Freymuller E, Smith DF. Leishmania: overexpression and comparative structural analysis of the stage-regulated meta 1 gene. Experimental parasitology. 1999;92:183–191. doi: 10.1006/expr.1999.4410. [DOI] [PubMed] [Google Scholar]
- 78.Ramos CS, Franco FA, Smith DF, Uliana SR. Characterisation of a new Leishmania META gene and genomic analysis of the META cluster. FEMS microbiology letters. 2004;238:213–219. doi: 10.1016/j.femsle.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 79.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabasi AL, Disease networks Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347 doi: 10.1126/science.1257601. 1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. Journal of bacteriology. 2015;197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang X, Bruce JE. A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Molecular bioSystems. 2010;6:939–947. doi: 10.1039/b920876c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. The Journal of clinical investigation. 2007;117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.