Abstract
The epithelial sodium channel (ENaC) is composed of three homologous subunits and allows the flow of Na+ ions across high resistance epithelia, maintaining body salt and water homeostasis. ENaC dependent reabsorption of Na+ in the kidney tubules regulates extracellular fluid (ECF) volume and blood pressure by modulating osmolarity. In multi-ciliated cells, ENaC is located in cilia and plays an essential role in the regulation of epithelial surface liquid volume necessary for cilial transport of mucus and gametes in the respiratory and reproductive tracts respectively.
The subunits that form ENaC (named as alpha, beta, gamma and delta, encoded by genes SCNN1A, SCNN1B, SCNN1G, and SCNN1D) are members of the ENaC/Degenerin superfamily. The earliest appearance of ENaC orthologs is in the genomes of the most ancient vertebrate taxon, Cyclostomata (jawless vertebrates) including lampreys, followed by earliest representatives of Gnathostomata (jawed vertebrates) including cartilaginous sharks. Among Euteleostomi (bony vertebrates), Actinopterygii (ray finned-fishes) branch has lost ENaC genes. Yet, most animals in the Sarcopterygii (lobe-finned fish) branch including Tetrapoda, amphibians and amniotes (lizards, crocodiles, birds, and mammals), have four ENaC paralogs. We compared the sequences of ENaC orthologs from 20 species and established criteria for the identification of ENaC orthologs and paralogs, and their distinction from other members of the ENaC/Degenerin superfamily, especially ASIC family. Differences between ENaCs and ASICs are summarized in view of their physiological functions and tissue distributions. Structural motifs that are conserved throughout vertebrate ENaCs are highlighted. We also present a comparative overview of the genotype-phenotype relationships in inherited diseases associated with ENaC mutations, including multisystem pseudohypoaldosteronism (PHA1B), Liddle syndrome, cystic fibrosis-like disease and essential hypertension.
Keywords: Ion channels, Epithelia, Evolution, Transmembrane proteins, Kidney, Renin-angiotensin-aldosterone system
1. Introduction
As it is well known, 60–70 % of the human body weight is water. About 2/3 of this water is within the cells (intracellular fluid, ICF) and the remaining 1/3 fills the extracellular spaces and the vascular bed in the circulatory system (extracellular fluid, ECF) (Ruth and Wassner, 2006). The cell membrane, as a semi-permeable barrier, is permeable to water molecules. Yet, the net movement of water between ECF and ICF depends on the relative osmolarity of these compartments and the permeability of the membranes (Fischbarg, 2010). In most vertebrates, the osmolarity of both the ECF and ICF is determined mainly by the concentration of electrolytes (dissolved salt ions carrying a net charge, mainly Na+, K+, Ca+2, Mg+2, Cl−, HCO3−, PO43−, SO42−). In the ECF, Na+ is the electrolyte with the highest concentration and thus it is the major determinant of the osmolarity of the ECF (Takei, 2000). Osmolarity-dependent volume changes may lead to shrinking or swelling of cells. To prevent damage from such changes and to protect the nervous system, mammals maintain a common osmotic set-point near 300 mosmol/L (Bourque, 2008). Thus, in vertebrates, the regulation of water and electrolyte homeostasis is highly interdependent (Ruth and Wassner, 2006).
The processes of absorption, secretion and excretion of water and solutes take place in epithelial cell layers that cover the internal and external surfaces of the body. In terms of permeability properties, epithelia are classified into two groups as leaky- and tight-epithelia (Fischbarg, 2010; Reddy and Stutts, 2013). Leaky epithelia are located generally in an isoosmotic environment as in the small intestine and proximal kidney tubules and are highly permeable to water. In contrast to leaky epithelia, the cells in tight epithelia are connected by complex tight junctions that reduce the permeability of the epithelia (Capaldo et al., 2014; Reddy and Stutts, 2013).
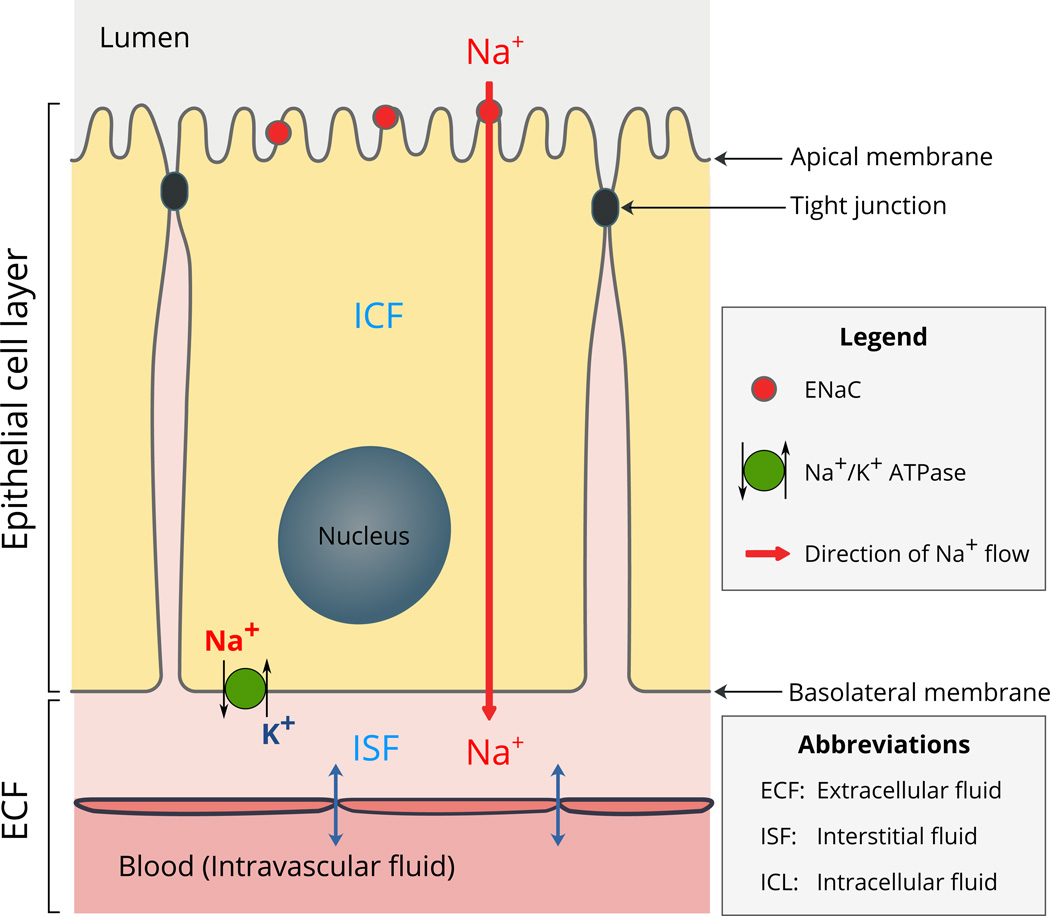
The epithelial sodium channel (ENaC), that is the focus of this review, is located mostly in tight or high-resistance epithelia. As a constitutively active channel, ENaC allows the flow of Na+ ions from the lumen into the epithelial cell, across the apical cell membrane (Garty and Palmer, 1997; Kashlan and Kleyman, 2011; Kellenberger and Schild, 2015) (Fig. 1). The absorbed Na+ ions are then pumped out of the cell into the interstitial fluid by the action of Na+/K+ ATPase located on the basolateral membrane (Fig. 1). As ENaC modulates the amount of Na+ in the ECF, it has a central role in the regulation of ECF volume and blood pressure (Büsst, 2013; Rossier et al., 2015). The activity of ENaC is regulated by the renin-angiotensin-aldosterone system (Asher et al., 1996; Bhalla and Hallows, 2008; Büsst, 2013; Rossier et al., 2015) and a complex variety of extracellular factors including Na+, Cl−, protons, shear stress and proteases (Bhalla and Hallows, 2008; Kashlan and Kleyman, 2012, 2011; Kellenberger and Schild, 2015).
Fig. 1.
Schematic illustration of the location and function of ENaC in epithelia.
The subunits that form ENaC constitute a family within the ENaC/Degenerin superfamily. In addition to ENaC, this superfamily includes acid-sensing ion channels (ASICs) (Deval and Lingueglia, 2015; Kellenberger and Schild, 2015, 2002; Lin et al., 2015; Omerbašić et al., 2014; Waldmann and Lazdunski, 1998), pickpocket genes in the Diptera order including Drosophila and mosquitoes (Zelle et al., 2013), degenerin subunits involved in sensory transduction in nematodes such as Caenorhabditis elegans (Eastwood and Goodman, 2012; Liddle et al., 1963), and peptide-gated Hydra Na+ channels (HyNaC) in hydrozoans (Gründer and Assmann, 2015).
The first sequences of ENaC subunits were based on cDNAs cloned from mRNAs isolated from rat and human tissues (Canessa et al., 1994b; Lingueglia et al., 1993; McDonald et al., 1995, 1994; Voilley et al., 1995, 1994; Waldmann et al., 1995). Later development of rapid genome sequencing techniques has led to the determination of the sequences of ENaC/Degenerin superfamily members in a growing number of species. This review concentrates on the sequences and phylogenetic relationships of ENaC paralogs and orthologs across species and with other homologous proteins that have been mostly revealed by genome sequences of many species.
In biology, the word "homology" is also used to describe functional equivalence and not just sequence and structural similarity. Thus, after inter-species sequence comparisons, we shall also present the physiological implications of the currently available information about ENaC phylogenetic distribution and function.
2. Nomenclature of ENaC homologs
2.1. Definitions: Homolog, paralog, ortholog
In studies of protein evolution, the word "homologous" is used to describe proteins that share significant sequence similarity that is assumed to derive from a common ancestral origin. This concept of homology covers both proteins that are homologous across species as well as proteins that are present in multiple copies in the genome of a single species. To distinguish between these two types of homologous proteins, two separate terms were coined by Walter Fitch (Fitch, 1970): orthologous and paralogous. Within the genome of a single species, there are many genes that represent duplicate copies encoding isoforms of proteins with similar functions. The most common example is the globin family that includes α-globin, β-globin, and myoglobin. Homologous proteins that exist "in parallel" within one species are called "paralogs", a hybrid word combining "parallel" with "homolog". The word "ortholog" is used for homologous proteins that originate from a single ancestral gene in the last common ancestor of the compared species. Continuing the globin example, the ortholog of human α-globin is any of the α-globins in related primates. Further examples of these terms are provided by (Koonin, 2005).
2.2. ENaC paralogs
In the human genome there are nine genes that encode for ENaC paralogs. These paralogs are grouped into two families based on their homology: 1. Non-voltage gated sodium channel family that is composed of four genes encoding ENaC homologs and 2. acid-sensing (proton-gated) ion channels (ASIC) family that is composed of five homologous genes. The four ENaC genes have been assigned abbreviations as SCNN1A, SCNN1B, SCNN1G, and SCNN1D by the Human Genome Organization (HUGO) Gene Nomenclature Committee (http://www.genenames.org/) following the Greek letters assigned to the four ENaC subunits α, β, γ, and δ (Table 1 and Table 2). The second "N" in "SCNN1" was added to distinguish between the NON-voltage gated ENaC and the SCN1 symbol assigned to the "sodium channel, voltage-gated, type I" that is expressed in neurons and muscle. The UniProt protein database (UniProt, 2014) uses an abbreviated code for ENaC subunits (SCNNA, SCNNB, SCNNG and SCNND) to which the abbreviated species name is appended (Table 2). For the mouse genome, the convention for gene nomenclature starts with an uppercase letter, followed by all lowercase letters as shown in Table 1. For mouse, the gene for SCNN1D is not listed as it was not found in the mouse genome (Giraldez et al., 2012). Another common name for ENaC subunits is "amiloride-sensitive sodium channel" as ENaC is inhibited by amiloride (Garty and Palmer, 1997; Kashlan and Kleyman, 2011).
Table 1.
Characteristics of the genes and transcripts encoding for ENaC subunits.*
| Species | Gene | Chro. | CCDS code | Ensembl Transcript ID | Pre-spliced (nt) |
Exons | Coding exons |
|---|---|---|---|---|---|---|---|
| Human | SCNN1A | 12p | 8543.1 | ENST00000228916 | 28,703 | 13 | 12 |
| SCNN1B | 16p | 10609.1 | ENST00000343070 | 79,030 | 13 | 12 | |
| SCNN1G | 16p | 10608.1 | ENST00000300061 | 34,169 | 13 | 12 | |
| SCNN1D | 1p | - | ENST00000400928 | 10,806 | 16 | 13 | |
| Mouse | Scnn1a | 6 | 39641.2 | ENSMUST00000081440 | 23,603 | 12 | 12 |
| Scnn1b | 7 | 21804.1 | ENSMUST00000033161 | 53,691 | 13 | 12 | |
| Scnn1g | 7 | 21803.1 | ENSMUST00000000221 | 33,971 | 13 | 12 | |
| Rat | Scnn1a | 4 | - | ENSRNOT00000067271 | 23,137 | 12 | 12 |
| Scnn1b | 1 | - | ENSRNOT00000067138 | 54,743 | 13 | 12 | |
| Scnn1g | 1 | - | ENSRNOT00000024057 | 33,957 | 13 | 12 |
Based on the NCBI CCDS, and Ensembl databases.
Table 2.
Length and mass of human and mouse ENaC subunits.
| Species | Subunit | Gene | CCDS code | Uniprot name | Length* (aa) |
Mass* (Da) |
|---|---|---|---|---|---|---|
| Human | Alpha | SCNN1A | 8543.1 | SCNNA_HUMAN | 669 | 75,704 |
| Beta | SCNN1B | 10609.1 | SCNNB_HUMAN | 640 | 72,659 | |
| Gamma | SCNN1G | 10608.1 | SCNNG_HUMAN | 649 | 74,270 | |
| Delta | SCNN1D | - | SCNND_HUMAN | 638 | 70,215 | |
| Mouse | Alpha | Scnn1a | 39641.2 | SCNNA_MOUSE | 699 | 78,893 |
| Beta | Scnn1b | 21804.1 | SCNNB_MOUSE | 638 | 72,197 | |
| Gamma | Scnn1g | 21803.1 | SCNNG_MOUSE | 655 | 74,635 | |
| Rat | Alpha | Scnn1a | - | SCNNA_RAT | 698 | 78,888 |
| Beta | Scnn1b | - | SCNNB_RAT | 638 | 71,995 | |
| Gamma | Scnn1g | - | SCNNG_RAT | 650 | 74,066 | |
Based on Uniprot database.
As detailed below, the HUGO nomenclature appears to be sufficient for naming ENaC orthologs in other vertebrate species whose genomes have been sequenced.
2.3. ASICs and other homologs
The five genes that code for the five Acid-Sensing Ion Channel (ASIC) subunits in the human genome have been numbered as ASIC1, ASIC2, ASIC3, ASIC4 and ASIC5 by the HUGO Gene Nomenclature. The same abbreviation is used by the UniProt database (e.g. ASIC1_HUMAN). These channels were previously called as ACCN and BNaC (García-Añoveros et al., 1997). One example of the proliferation of names is ASIC5. The product of this gene was initially named "brain, liver, intestine Na+ channel" (BLINaC) in mouse and rat. The homologous protein in humans was found to be expressed in the intestine. Therefore, it was named "intestine Na+ channel (INaC)" in humans (Schaefer et al., 2000). A more recent study renamed the same protein as "bile acid-sensitive ion channel" (BASIC) (Lefèvre et al., 2014). Although referred to as ASIC5, it is not an acid-activated ion channel. The multiplicity of names for one protein emphasizes the need to adhere to names standardized by international nomenclature.
Many of the ENaC homologs were named based on the protein characteristics such as, sites of expression (e.g. "INaC", "BLINaC"), physiologic consequences of activating mutations (e.g. "degenerin"), ligand interactions (e.g. "FMRFamide-activated", "amiloride-sensitive", "acid-sensing"), organism (e.g. HyNaC for channels in Hydra) and original gene name (e.g. pickpocket in Drosophila). As noted with ASIC5, the use of different terms to name homologous proteins results in unrelated names for proteins that are highly homologous or orthologous. Moreover, homologous proteins may be expressed in different cell types and fulfill multiple functions in different species, as observed with ENaC/Degenerin superfamily members. Thus, assignment of one name for a protein may not be relevant for an orthologous protein in a different species.
As an alternative to naming proteins based on functional characteristics, HUGO has taken the approach of a serial numbering system based on homologous groupings (e.g. ASIC1… ASIC5, SCNN1A…SCNN1D). In our view, this is a better approach for the nomenclature of ENaC/Degenerin superfamily, as it provides identical names to orthologs across species. In the current genomic era, protein sequences are predicted based on genomic sequence analysis that includes comparisons between predicted and known protein sequences. This approach of naming proteins based on sequence homology avoids the problems of names associated with protein characteristics.
In numerous invertebrate Metazoan species there is a multitude of highly divergent proteins that show sequences homologous to ENaC/Degenerin superfamily members, but clearly represent different families based on low sequence similarity. As there is no standardized nomenclature for these proteins, in this review we used the names as in the original database records.
3. Chromosomal location and intron-exon organization of ENaC genes
In the human genome, SCNN1A encoding the α subunit is located on the short arm of chromosome 12 (12p) (Voilley et al., 1994). The genes SCNN1B and SCNN1G encoding the β and the γ subunits are located side by side on the short arm of chromosome 16 (16p) (Shimkets et al., 1994; Voilley et al., 1995). The SCNN1D gene encoding the δ subunit is located in chromosome 1p (Table 1).
In the mouse genome, the gene Scnn1a is located on chromosome 6, and Scnn1b and Scnn1g are juxtaposed at a region of chromosome 7 that shares synteny with the human chromosome 16 (Brooker et al., 1995; Pathak et al., 1996) (Table 1). Mouse genome appears to have lost the gene for the delta subunit (Giraldez et al., 2012). Yet, as detailed in Section 6, most vertebrate genomes have a gene that encodes for the delta subunit.
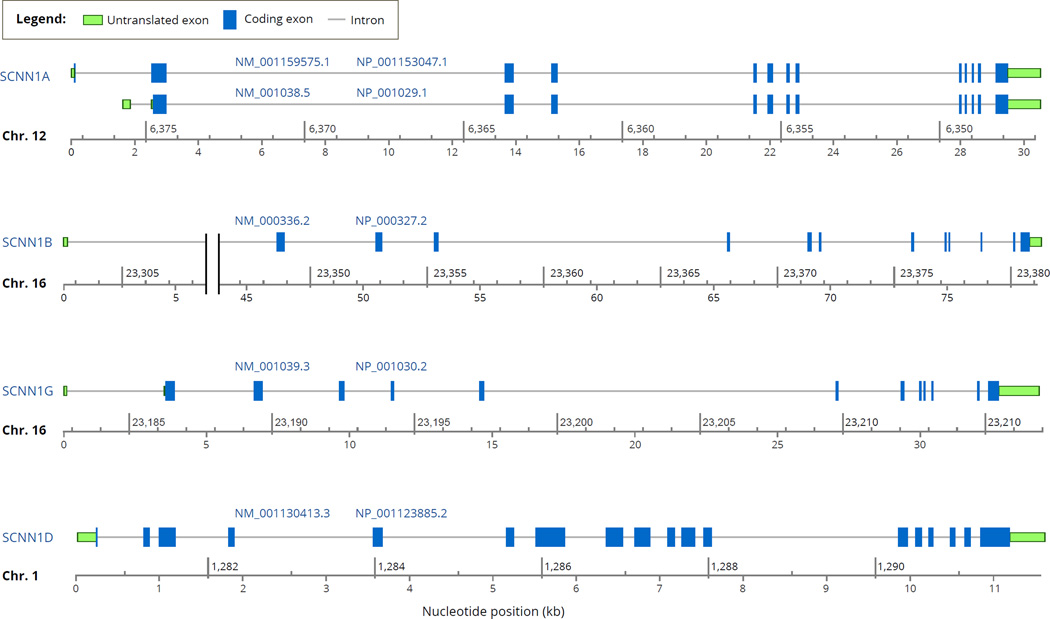
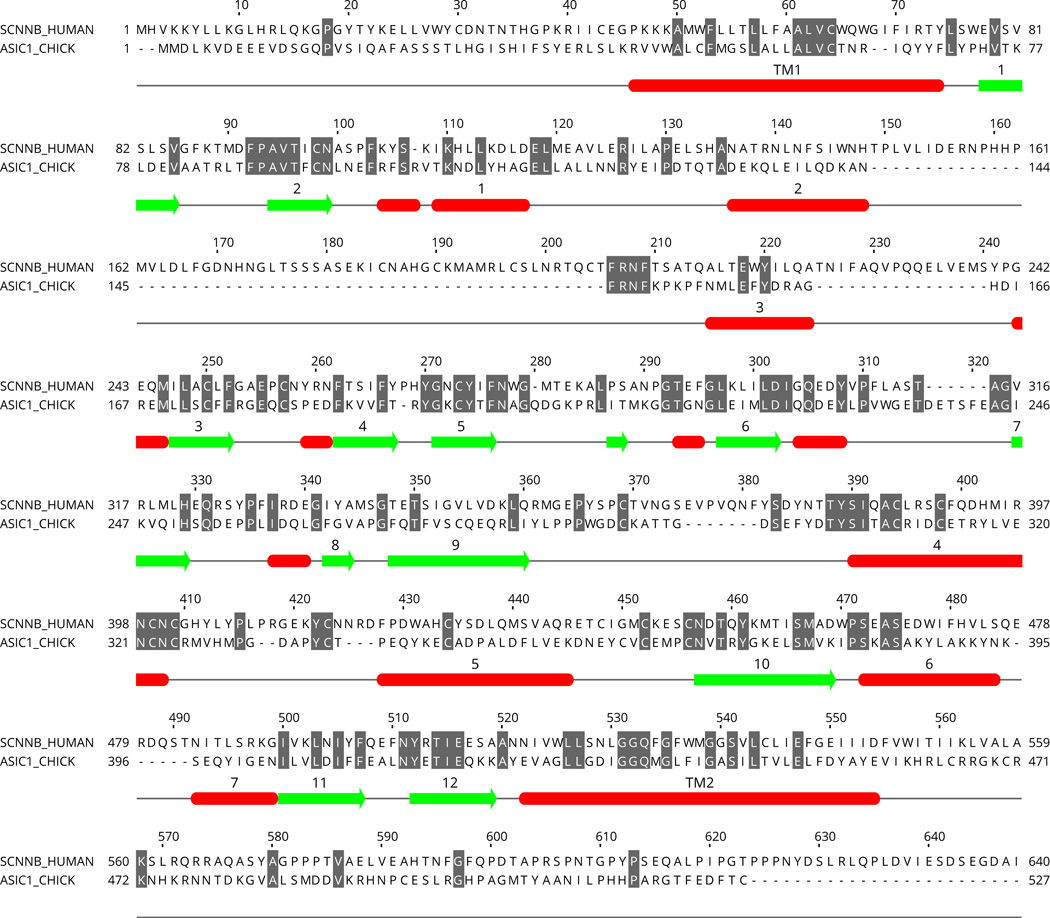
Sequencing of the α, β, and γ genes of the human genome revealed that all three genes include 13 exons but only 12 of these contain translated sequence (Fig. 2) (Table 1) (Ludwig et al., 1998; Saxena et al., 2002, 1998; Thomas et al., 1996). In the human somatic chromosomes, the average number of exons per coding gene ranges from 8.5 to 13.5 (Hubé and Francastel, 2015).
Fig. 2.
Intron-exon organization of the human ENaC genes, SCNN1A, SCNN1B, SCNN1G and SCNN1D and their primary transcripts based on the NCBI Homo sapiens Annotation Release 107 (2015-03-13). The name of each gene and its chromosomal location are noted at the left-edge of the diagrams. Under each exon-intron map, there are two coordinates: the upper one specifies the chromosomal coordinates, and the lower one specifies the position of the nucleotide (in kb) starting at the 5'-end of the RNA transcript (marked as 0). The codes above the diagrams represent the ID numbers of the RNA transcript (starting with NM_) and the encoded protein (starting with NP_) in the NCBI Gene database. For SCNN1A, two transcripts are shown as examples of alternative splicing products. Notes: 1) SCNN1A coordinates are given in a scale that descends from left-to-right because the gene is located in the reverse strand of the chromosome. 2) The x-axis for SCNN1B intron #1 includes a break between 5 kb and 45 kb marks. Display of the full sequence (i.e., without a break) would lead to the visible merger of exons 9 and 10 and hence disappearance of the intron 9 because of the short size of intron 9. Additional information about the genes and their products is provided in Table 1 and Table 2.
In all three genes, SCNN1A, SCNN1B and SCNN1G, the introns are located at identical positions in the coding sequence (Saxena et al., 1998). The SCNN1D gene structure, revealed by the human genome sequencing project, includes at least 16 exons 13 of which are protein coding (Table 1). Despite the conservation of the intron positions within the coding sequence, the sizes of the introns have diverged greatly resulting in significant differences between gene lengths (Fig. 2). The sizes of the primary transcripts prior to splicing range from 10,806 bp (for SCNN1D) to 79,030 (for SCNN1B) (Table 1) (Fig. 2). Among the four genes, the longest intron is intron #1 of SCNN1B (Fig. 2, note that there is a break in the x-axis of nucleotide position). In both SCNN1A and SCNN1B genes, the longest introns are intron #1 or #2 closest to the 5'-end of the transcription initiation site (Fig. 2). This represents a general trend that in genomes the longest introns appear at the 5'-end of the gene (Zhu et al., 2009).
Analyses of the RNA transcripts of the genes encoding ENaC subunits have provided evidence for alternative RNA splicing products and multiple translation initiation sites (see Ensembl records listed in Table 1) (Berman et al., 2015; Bremner et al., 2002; Thomas et al., 2002). Alternative splicing is common in vertebrates and is thought to contribute to a higher level of phenotypic complexity in mammals (Kim et al., 2007). In cases where there was more than one isoform sequence for a gene, we used the UniProt Canonical Sequence or an NCBI Consensus CDS (CCDS) as the representative sequence for the gene in homology analyses for paralogs and orthologs.
4. Assembly of ENaC with paralogs
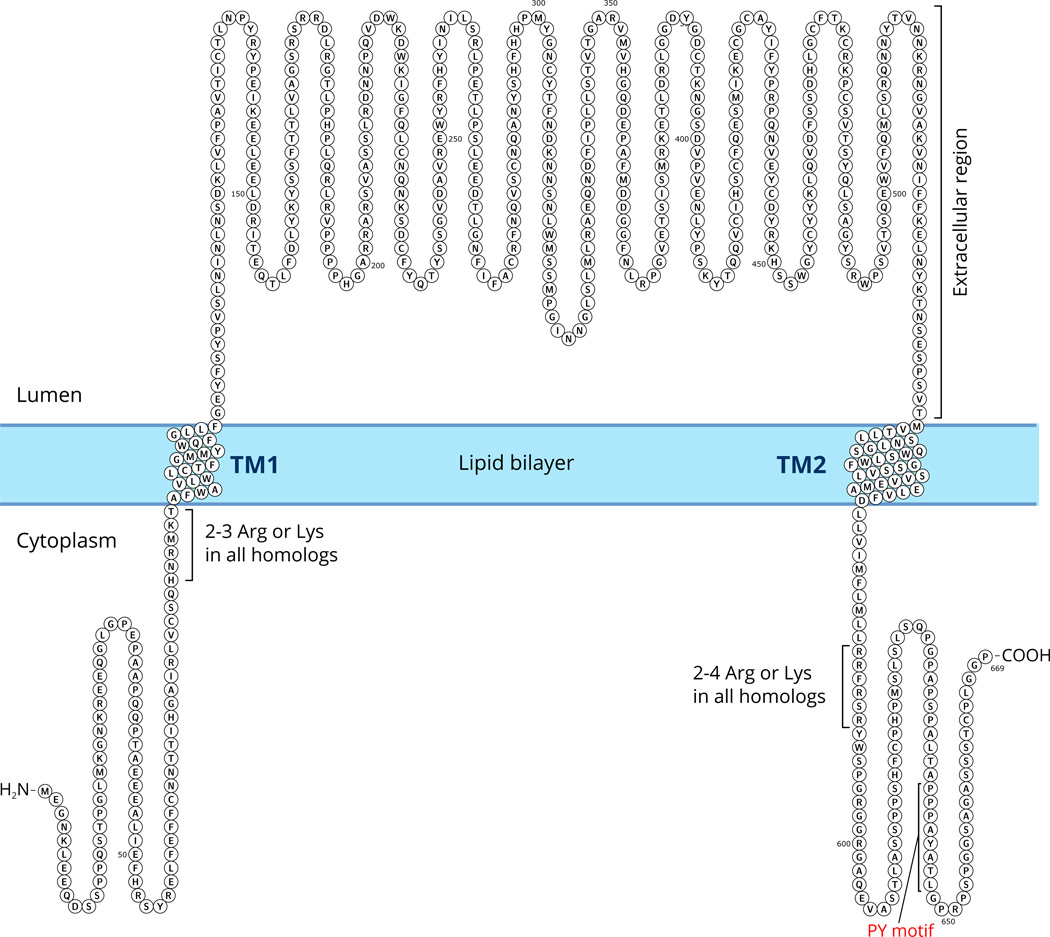
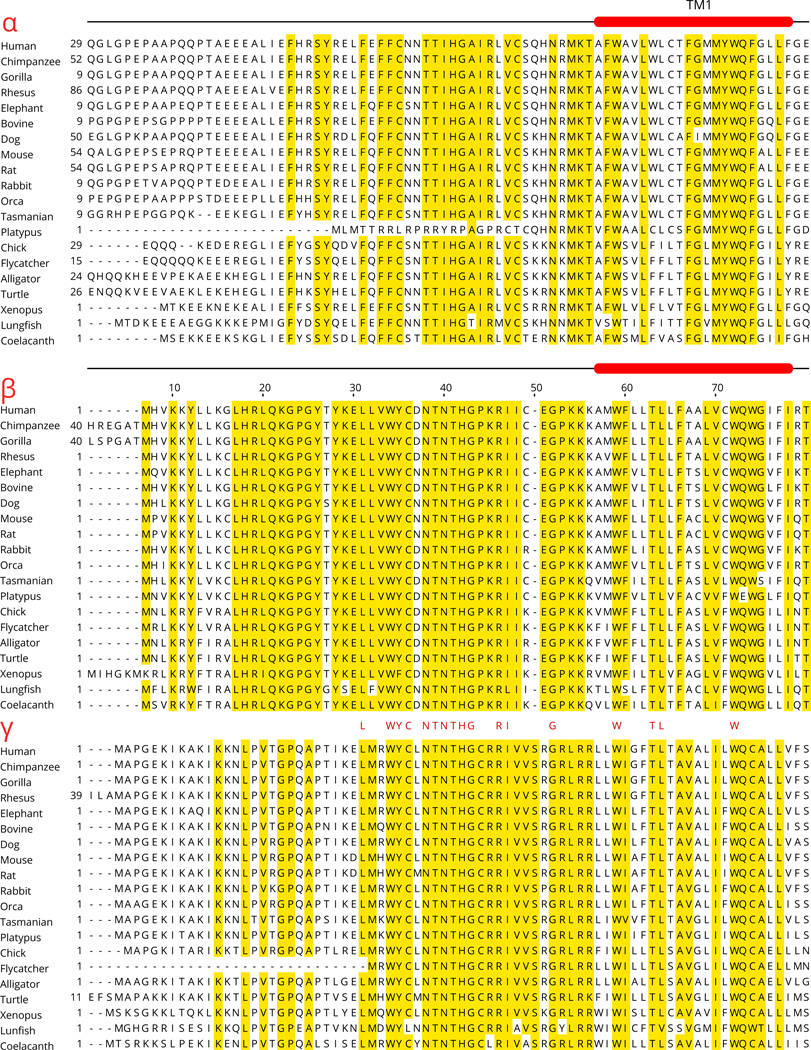
Previous studies have established that ENaC paralogs serve as subunits that form the channel (Canessa et al., 1994b; Kashlan and Kleyman, 2011). The most salient common feature of ENaC paralogs is the presence of two segments that function as two transmembrane (TM) segments embedded in the membrane, referred to as TM1 and TM2 (Fig. 3). In membrane-bound form, the amino (N) and the carboxy (C) termini of ENaC are intracellular, and a large extracellular segment, comprising about 70% of the amino acids of each subunit, connects the TM segments.
Fig. 3.
Schematic illustration of the transmembrane localization of an ENaC subunit. The sequence shown is of human α subunit (see Table 1). All homologous ENaC subunits have two transmembrane segments. The TM segments for this figure was predicted by the Phobius program (see Table 3) and drawn using Protter (Omasits et al., 2014). The extracellular domain includes about 70% of the sequence of amino acids of an ENaC subunit.
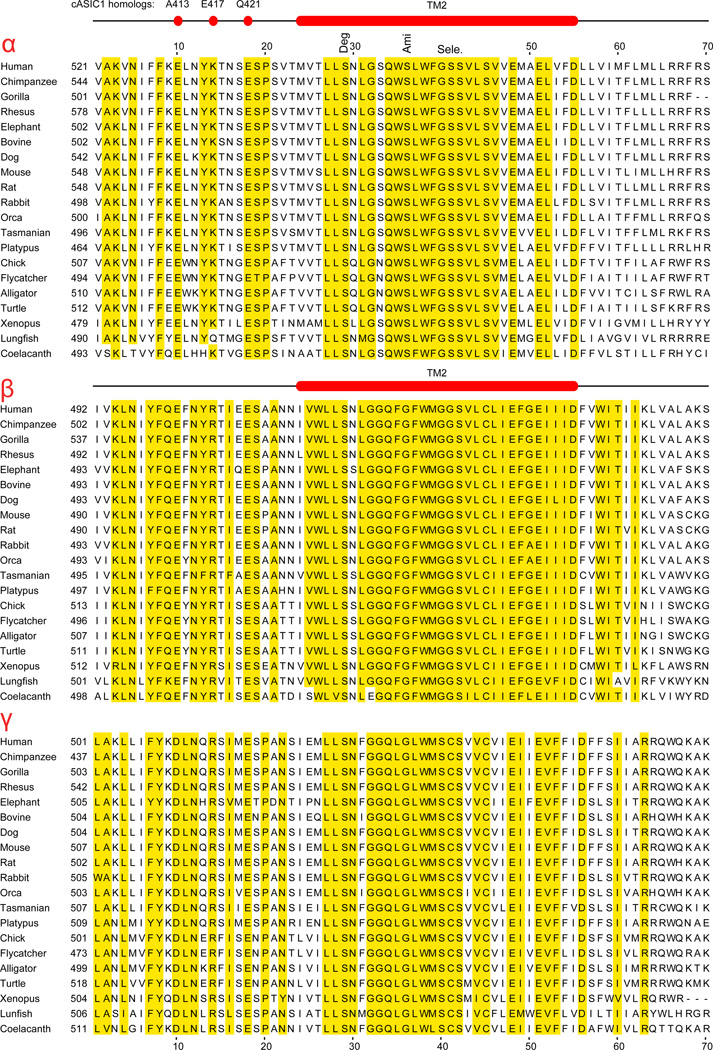
Although the structure of ENaC is not known, the strong hydrophobicity of the TM segments and homology with the resolved ASIC1 structure (Jasti et al., 2007) allows prediction of the TM segments (Table 3) (Fig. 3). In humans, the four ENaC subunits show significant sequence similarity in large segments of the extracellular region (Fig. 4). The most divergent parts of the ENaC paralogs are the N- and C-termini (Fig. 4).
Table 3.
Intracellular, extracellular and transmembrane (TM) segments of human ENaC subunits. The position1 of TM1 was predicted using Phobius software (Käll et al., 2004). The position of TM2 is based on homology with the ASIC1 structure (Jasti et al., 2007).
| Cytoplasmic N-ter | TM1 | Extracellular | TM2 | Cytoplasmic C-ter | |
|---|---|---|---|---|---|
| Alpha | 1–84 | 85–106 | 107–543 | 544–575 | 576–669 |
| Beta | 1–49 | 50–70 | 71–514 | 515–546 | 547–640 |
| Gamma | 1–53 | 54–79 | 80–523 | 524–555 | 556–649 |
| Delta | 1–87 | 88–107 | 108–520 | 521–552 | 553–638 |
Fig. 4.
Aligned sequences of human α, β, γ and δ-ENaC subunits and conserved positions of introns in all four subunits. Residues that are identical in all four subunits are shaded. The numbers (2 to 12) below the sequences mark the position and number of the intron located in or at the end of the codon of the specific residue above the number. In the 5' portion of the gene encoding δ-ENaC subunit there are additional introns that are not shown here. The sequences were aligned using the ClustalW2 program, and the alignment of some residues in the amino and carboxy termini were manually edited to eliminate some gaps without affecting percent identity score. TM1 and TM2 mark the predicted transmembrane segments of the proteins.
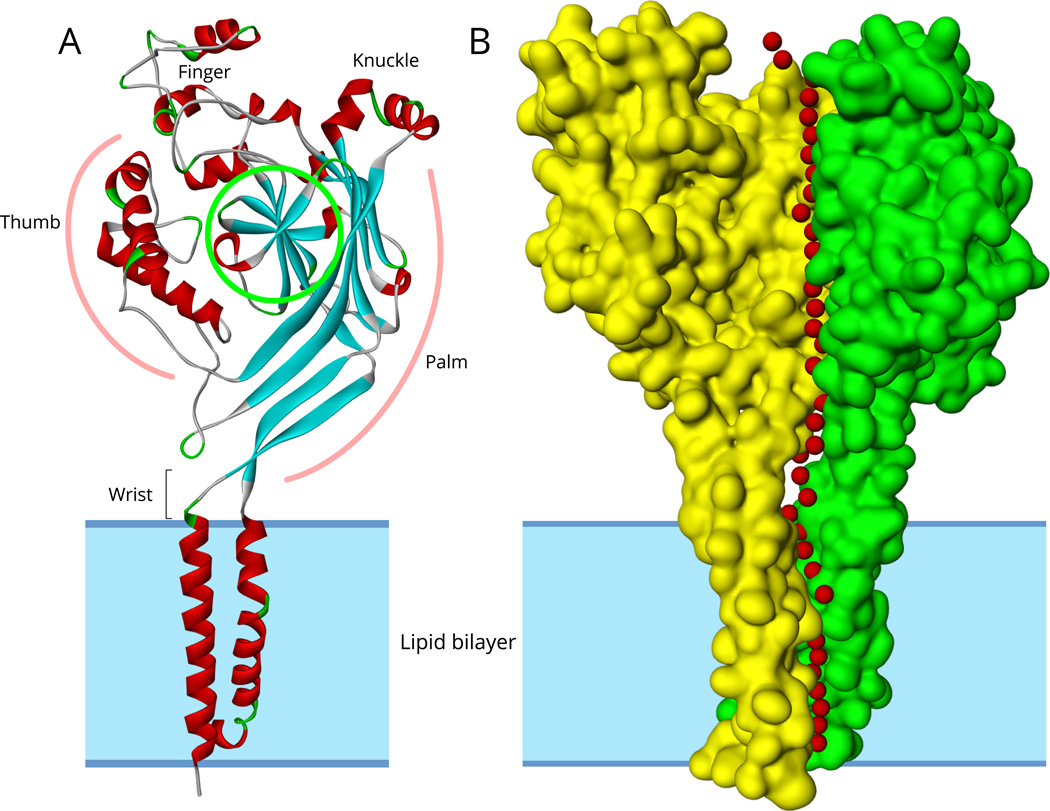
The resolved structures of chicken ASIC1 revealed a homotrimer composed of three identical subunits (Baconguis et al., 2014; Jasti et al., 2007) (Fig. 5). In contrast to ASIC1 structure, independent lines of evidence indicate that ENaC is assembled as a heterotrimer composed of α (or δ), β and γ subunits:
Specific mutations in any one of the three genes coding for the α, β, and γ-ENaC were shown to result in an autosomal recessive disorder termed multi-system pseudohypoaldosteronism type I (PHA) (Chang et al., 1996; Hanukoglu, 1991). The underlying mechanism of multi-system PHA is the unresponsiveness to aldosterone in target organs expressing ENaC including kidney, sweat and salivary glands, reproductive and respiratory tracts (Enuka et al., 2012; Hanukoglu, 1991). In affected patients the disease is characterized by severe hyponatremia, hyperkalemia, dehydration and acidosis that starts in infancy and continues later in life with varying severity (Belot et al., 2008; Chang et al., 1996; Edelheit et al., 2010, 2005; Hanukoglu and Hanukoglu, 2010; Hanukoglu, 1991; Strautnieks et al., 1996). So far, no case of PHA has been identified that is caused by a mutation in the SCNN1D gene encoding δ-ENaC.
Gene knockout studies inactivating the genes coding for the α, β, and γ subunits in mice showed that all three subunits are essential for survival. All gene knockout mice without either α, β, or γ subunits (genotype: −/−) die within < 50 hours after birth, with respiratory insufficiency or kidney dysfunction leading to hyperkalemia, metabolic acidosis and severe dehydration (Barker et al., 1998; Bonny and Hummler, 2000; Hummler et al., 1996).
Robust expression of ENaC activity in Xenopus oocytes requires all three subunits (α, β, and γ) (Canessa et al., 1994b; Edelheit et al., 2014, 2011; Giraldez et al., 2007). Expression of one or two ENaC subunits in Xenopus oocytes yields either minimal or no detectable channel activity (Canessa et al., 1994b; Edelheit et al., 2011; Giraldez et al., 2007).
Assessment of the stoichiometry of ENaC subunits using fluorescently labeled subunits, and imaging of ENaC-antibody complexes by atomic force microscopy indicated that the subunits are assembled as heterotrimers with a ratio of 1:1:1 (Staruschenko et al., 2005; Stewart et al., 2011).
Post-translation processing of the channel, including N-glycan maturation and furin-dependent cleavage, requires expression of all three subunits (Hughey et al., 2004).
Fig 5.
A. Ribbon structure model of subunit A of chicken ASIC1 (PDB ID: 2QTS). Segments in helical conformation are red colored and segments in sheet conformation are blue colored.
B. The surface structure of subunits A and B of ASIC1. The four hydrophobic helices of the A and B subunits are embedded in the lipid bilayer marked by gray shading. The third subunit (C) surface is not shown to allow visibility of the central pore predicted by the Porewalker software. Red colored small spheres represent water molecules placed at the center of the predicted pore and extracellular vestibule in each 3 Angstrom slice of 2QTS calculated by Porewalker.
Studies examining the structure of ENaC by molecular modeling and site-directed mutagenesis of conserved residues support the concept that ENaC structure is homologous to ASIC1 channel. In contrast to ASIC1 that functions as a homotrimer, ENaC is an obligate heterotrimer (Edelheit et al., 2014; Kashlan and Kleyman, 2011; Stockand et al., 2008). A study based on mutagenesis of Cl− inhibitory sites suggests that the clockwise orientation of the subunits is αγβ, when viewed from the top of the channel (Collier and Snyder, 2011).
In summary, the three paralogs encoding the α (or δ), β and γ subunits are essential for the assembly of functional channels. As summarized below, these three paralogs are highly conserved in all vertebrates. The evolutionary conservation of these genes provides further evidence that the subunits encoded by these genes are essential for the assembly of the heterotrimeric channel. The tissue distribution of the δ subunit is different from that of other subunits and its activity has been studied less. Excellent reviews by Giraldez et al. and Ji et al. summarize the characteristics of δ-ENaC (Giraldez et al., 2012; Ji et al., 2012).
4.1. Trimeric structure and channel pore
In the trimeric structure of ASIC1, one of the issues that have been intensively studied is the location of the channel pore through which ions flow across the membrane. ASIC1 has six transmembrane segments - three of each of TM1 and TM2. The structure of ASIC1 revealed that the TM1 and TM2 helices are organized in two separate concentric triads. The central pore is formed by the triad of TM2s. TM1s form a triad around the TM2 triad (Baconguis et al., 2014; Gonzales et al., 2009; Li et al., 2011). Most studies on ENaC suggest a similar organization of the TM segments in ENaC as well (Tolino et al., 2011). Section 11 on conserved motifs presents the properties of these segments in detail.
One of the major unresolved questions in ENaC function is the path(s) of ions into the channel pore in the membrane as described above. On top of the channel pore, the extracellular regions of the three subunits form a tripartite funnel with rotational symmetry (Fig. 5) (Baconguis et al., 2014; Jasti et al., 2007). However, the three subunits are not completely tightly juxtaposed along their entire lengths and there are fenestrations between the subunits above the pore around the region called "extracellular vestibule" (Baconguis et al., 2014). The hollow space along the central axis of rotational symmetry of this channel has been called a "vestibule". This vestibule leads from the top opening in the lumen to the channel pore embedded in the membrane. Under different crystallization conditions, segments of this vestibule may be constricted or expanded (Baconguis et al., 2014). These different states suggest that dynamic vestibule constriction and expansion may regulate ion flow into the channel pore.
The extracellular and central segments of the vestibule are surrounded by beta-strands of the palm domain two of which are connected to the TM helices (β1 to TM1 and β12 to TM2) (Fig. 5). Thus, changes in the angles of TM helices may effect constriction of the vestibule. Conversely, movement of the β1 and β12 strands may effect opening or closing of the channel gate by modulating the position of the TM helices. For ASIC1, there is evidence that the movement of the coiled linker region immediately prior to β1 and β12 strands may effect channel opening and closing (Li et al., 2010; Springauf et al., 2011). The dynamics of these parts are also affected by the interactions between the thumb and finger domains (Gwiazda et al., 2015; Yang et al., 2009). There is a variety of intracellular and extracellular factors that can affect the dynamics of these segments, e.g. cytoplasmic Ca2+ (Gu, 2008), binding to actin and other cytoskeletal proteins (Ilatovskaya et al., 2012; Sasaki et al., 2014), phosphoinositides that serve as second messengers in intracellular signaling cascades (Hille et al., 2015; Pochynyuk et al., 2008), extracellular ions, including Na+ and Cl−, pH and cleavage by extracellular proteases (Kashlan and Kleyman, 2012; Kellenberger and Schild, 2015).
5. Homology between ENaC and ASIC paralogs
To assess the similarity of the ENaC and ASIC sequences, Fasta format of the selected sequences were downloaded from the Uniprot database. Multiple sequence alignments were carried out by the CLUSTALW software (version 2.1) with default parameters (http://www.genome.jp/tools/clustalw/) (Chenna et al., 2003). Percent identity figures were calculated using GeneDoc (Nicholas and Deerfield, 1997). Sequence alignments for the figures were generated using the Jalview program (Waterhouse et al., 2009).
Among the four human ENaC subunits, greatest similarity exists between the α and δ subunits (34% identity) and the β and γ subunits (34% identity) (Table 4). The percent identity between other pairs (e.g. α vs. β or γ) is between 23–27% (Table 4). Since the N- and C-termini of ENaC subunits show divergence, we also determined the sequence identity in the extracellular regions of ENaC subunits. These values indicate a 2–6% higher sequence identity in the extracellular regions (Table 5), as compared to the full-length sequences of ENaC subunits (Table 4).
Table 4.
Percent identity between human ENaC and ASIC subunits along their entire sequences.
| SCNNA | SCNNB | SCNNG | SCNND | ASIC1 | ASIC2 | ASIC3 | ASIC4 | |
|---|---|---|---|---|---|---|---|---|
| SCNNB_HUMAN | 26 | |||||||
| SCNNG_HUMAN | 27 | 34 | ||||||
| SCNND_HUMAN | 34 | 23 | 23 | |||||
| ASIC1_HUMAN | 13 | 16 | 15 | 16 | ||||
| ASIC2_HUMAN | 13 | 15 | 15 | 13 | 64 | |||
| ASIC3_HUMAN | 14 | 14 | 15 | 14 | 46 | 45 | ||
| ASIC4_HUMAN | 12 | 12 | 11 | 14 | 35 | 31 | 32 | |
| ASIC5_HUMAN | 12 | 14 | 13 | 13 | 22 | 22 | 21 | 17 |
The sequences were aligned using ClustalW2 program (version 2.1).
Table 5.
Percent identity between human ENaC and ASIC subunits in the conserved central segment including TM1 + extracellular domain + TM2 (see Fig. 3 and Fig. 4).
| SCNNA | SCNNB | SCNNG | SCNND | ASIC1 | ASIC2 | ASIC3 | ASIC4 | |
|---|---|---|---|---|---|---|---|---|
| SCNNB_HUMAN | 31 | |||||||
| SCNNG_HUMAN | 33 | 36 | ||||||
| SCNND_HUMAN | 37 | 28 | 28 | |||||
| ASIC1_HUMAN | 16 | 19 | 18 | 18 | ||||
| ASIC2_HUMAN | 16 | 18 | 17 | 17 | 74 | |||
| ASIC3_HUMAN | 17 | 17 | 16 | 17 | 52 | 53 | ||
| ASIC4_HUMAN | 15 | 17 | 15 | 17 | 49 | 47 | 46 | |
| ASIC5_HUMAN | 16 | 17 | 16 | 16 | 26 | 27 | 26 | 25 |
In contrast to ENaC subunits, the sequence identities between human ENaC and human ASIC subunits are much lower: in the range of 11 to 16% (Table 4). Thus, clearly ENaC and ASIC paralogs belong to distinct families as marked by the demarcation lines in Table 4. Percent sequence identity between ASIC subunits themselves ranges from 17 to 64% (Table 4). Similar to ENaC, the extracellular segments of ASIC subunits share higher identity than the whole sequences (compare Fig. 7 vs. Fig. 6 for ASIC), reflecting divergence of N- and C- terminal sequences (see Section 11).
Fig. 7.
Secondary structures in the sequence of chicken ASIC1. The positions of the structures were taken from the PDB file of 2QTS. The numbering of the structures is based on (Jasti et al., 2007). Note that some short stretches of helix and β-strand are not numbered. For comparison of sequence conservation, human β-ENaC is globally aligned with the ASIC1 sequence and identical residues were gray color shadowed. Note that most but not all secondary structures are associated with conserved sequences.
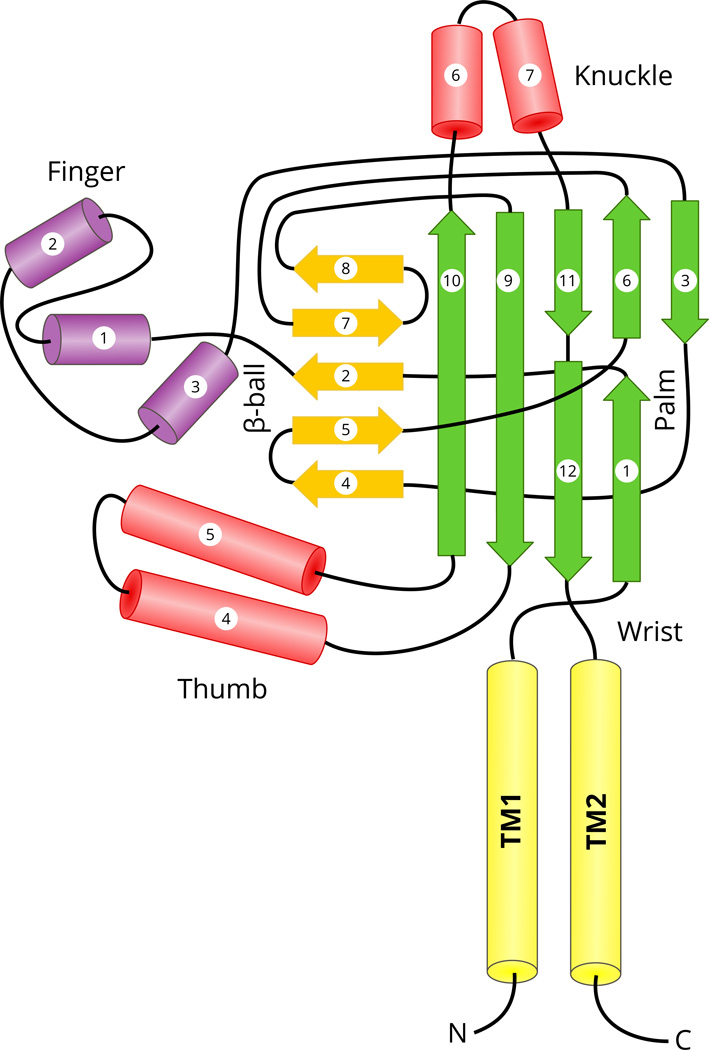
Fig. 6.
Topology diagram of chicken ASIC1 structure. The cylinders represent helical segments, and the arrows represent β-strands. The transmembrane (TM), and secondary structural domains (palm, β-ball, finger, thumb and knuckle) were colored distinctly and named as in (Jasti et al., 2007). Certain features of the diagram were adopted from previous diagrams (Eastwood and Goodman, 2012; Kashlan and Kleyman, 2011).
Comparisons of the sequences of all four ENaC paralogs from six species (in addition to human) indicate that the degree of sequence identity between the four paralogs within each species is quiet similar to that observed in the human genome (compare Table 4 and Table 6).
Table 6.
Percent sequence identity between four paralogous ENaC subunits (α, β, γ and δ) in six species (for comparison of human paralogs see Fig. 4 and Table 4).
| α | β | γ | ||
|---|---|---|---|---|
| Rhesus | β | 24 | ||
| γ | 27 | 31 | ||
| δ | 25 | 16 | 18 | |
| Bovine | β | 28 | ||
| γ | 28 | 34 | ||
| δ | 38 | 23 | 25 | |
| Tasmanian D. | β | 26 | ||
| γ | 27 | 32 | ||
| δ | 37 | 21 | 22 | |
| Xenopus | β | 29 | ||
| γ | 31 | 30 | ||
| δ | 39 | 27 | 28 | |
| Alligator | β | 27 | ||
| γ | 29 | 34 | ||
| δ* | 39 | 25 | 27 | |
| Coelacanth | β | 26 | ||
| γ | 29 | 29 | ||
| δ | 43 | 24 | 27 |
Named by us as the δ-subunit. Named as "alpha like" in the original report.
In the CATH protein structural domain database (Sillitoe et al., 2015), ASIC and ENaC channels are listed as two separate families within the Superfamily number 2.60.470.10 titled "Acid-sensing ion channels like domains". CATH classification system is mostly based on specific local structural domains. The domain selected for the classification is mainly the "palm" domain based on the ASIC1 structure (2QTS). The palm domain is composed of a complex of β-sheets. Therefore within the CATH database, the channel is included under Class 2 for "Mainly beta" type domains. Since the ASIC1 structure is an intricate complex of α-helices and β-sheets this classification does not take into account the full structural view of the channels.
5.1. Sites of divergence among ENaC and ASIC paralogs
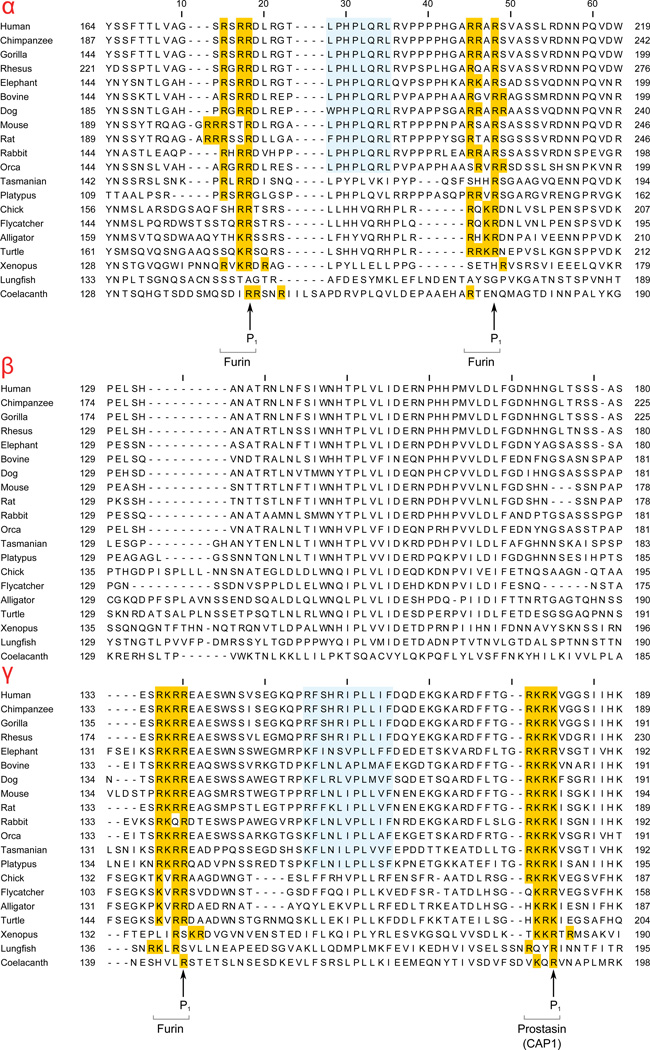
The divergence of N- and C- termini of ENaC/Degenerin superfamily members (noted above) represents a general trend in protein families. Previous studies on other proteins have shown that changes in protein domain architecture are most common in the N- and C-termini of proteins (Björklund et al., 2005; Forslund and Sonnhammer, 2012). In contrast to α- and δ-ENaC, the N- and C-termini of human β- and γ-ENaC are highly conserved. The structures of these terminal segments are currently not known, but there are studies indicating that these cytoplasmic domains interact, either directly or indirectly, with other cytoplasmic and cytoskeletal proteins such as syntaxin (Berdiev et al., 2004; Condliffe et al., 2003), actin (Copeland et al., 2001), ubiquitin ligase Nedd4 and protein kinases (Asher et al., 2001; Bobby et al., 2013; Shi et al., 2002).
Since the structure of the extracellular region of ASIC1 has been resolved and in this region there is a significant homology between ASIC1 and ENaC subunits, we shall present the sites of divergence in this region in terms of the secondary structural segments of ASIC1. The original study on the crystal structure of ASIC1 noted that ASIC1 structure resembles a hand holding a ball (Jasti et al., 2007). Hence, domains within the extracellular regions are referred to as palm, thumb, knuckle, finger and β-ball (Jasti et al., 2007). The palm and β-ball domains are formed by non-contiguous β-strands and loops, and are in close proximity to the membrane (Fig. 6). More peripheral domains (thumb, knuckle and finger) are formed by contiguous α-helices and loops (Fig. 6).
To facilitate location of divergent regions in ENaC relative to the structural domains of ASIC1 in Fig. 7 we provide an alignment of the β-subunit with ASIC1 sequence including marking of the positions of the secondary structural elements according to the PDB ID 2QTS (Fig. 7).
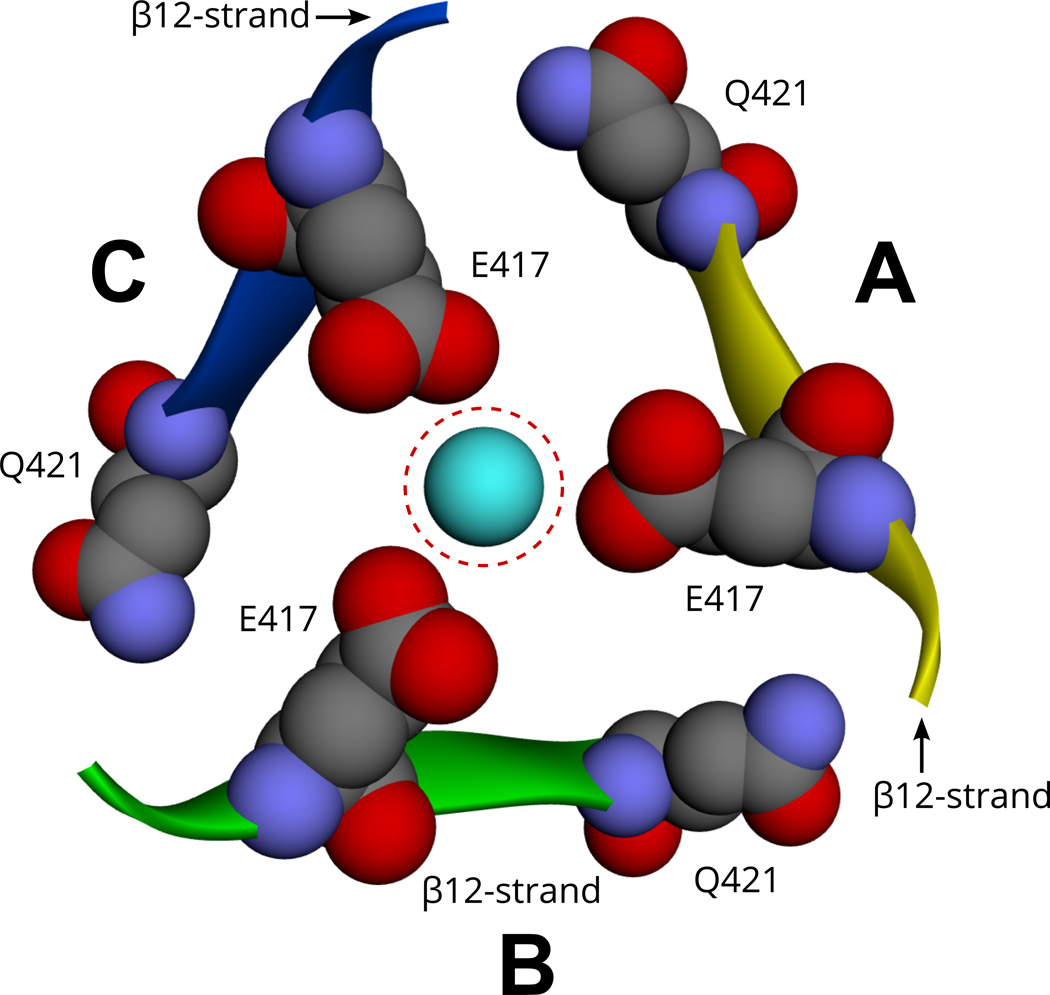
In the extracellular region of ENaC subunits, there are several highly divergent segments where insertions/deletions are found (Fig. 4 and Fig. 7). One divergent area is in the finger domain in between helix #1 and β-strand #3 (Fig. 4, Fig. 6 and Fig. 7). This segment is divergent in four ENaC paralogs and is characterized by poorly aligned sequences including large insertions and deletions (Fig. 4 and Fig. 7). This "finger" domain shows the highest variability among ENaC/Degenerin superfamily members indicating that this region may have an important role in conferring functional specificity (Eastwood and Goodman, 2012; Kashlan and Kleyman, 2011). For example, the α and γ-subunit finger domains have inhibitory tracts that are released following proteolytic processing (Bruns et al., 2007; Carattino et al., 2008a, 2006; Kashlan et al., 2011; Passero et al., 2010).
Another divergent segment in ENaC starts at about residue 376 of the human β-ENaC and includes an insertion of three residues (Fig. 4). In alignment with ASIC1 this region is located in the region between β-9 and α-4 (Fig. 7). This is the region that connects the palm domain of ASIC1 to the thumb domain (Jasti et al., 2007). This region has been proposed to transmit conformational changes in the periphery of the extracellular region to the channel pore and gate (Jasti et al., 2007; Li et al., 2011; Shi et al., 2011). Other divergent areas include the knuckle domain and the loop connecting the β-6 and β-7 strands. Residues in the β-6 - β-7 loop of the α subunit have been proposed to function as an extracellular Na+ binding site that is involved in Na+ self-inhibition (Kashlan et al., 2015).
In conclusion, it appears that areas of divergence that are seen in ENaC and ASIC1 comparisons are located in the connecting segments within the finger and thumb domains. In additions to these, there are a few other sequence differences but the sequence homology predominates especially in the β-strand segments in the palm and β ball domains (Fig. 6, and Fig. 7).
It is interesting that the most divergent areas within members of the ENaC/Degenerin family are in the periphery of the extracellular region. There is growing evidence that these divergent areas have sites of direct interaction with extracellular regulatory factors that modulate channel activity, such as proteases (Bruns et al., 2007; Vallet et al., 1997), inhibitory peptide released by proteases (Carattino et al., 2006; Kashlan et al., 2010), extracellular chloride (Cl−) ions (Collier and Snyder, 2011), extracellular Na+ (Chraibi and Horisberger, 2002; Edelheit et al., 2014; Winarski et al., 2010), protons (Collier et al., 2012; Krauson et al., 2013), and laminar shear stress induced by fluid flow (Shi et al., 2012). As the different ENaC/Degenerin family members are regulated by distinct factors, evolutionary divergence within the peripheral domains may have been a key factor in allowing this family to evolve with different functional properties.
6. Phylogenetic distribution of ENaC orthologs
Determination of genomic sequences of many eukaryotic species has provided ENaC gene sequences from a broad spectrum of vertebrates. Comparison of ENaC gene and protein sequences across species is useful from several perspectives. Knowledge about ENaC orthologs across species can contribute to our understanding of the significance and function of ENaC subunits. Conservation of a gene across species suggests an important physiological function for the organism (see for example (Studer et al., 2011)). Secondly, comparisons of the sequences of the ENaC subunits enhance our understanding of the structural and functional importance of conserved sequence segments. Thirdly, the absence of an ENaC gene in a species is important information as the species may use alternative subunits or channels to fulfill the homeostatic functions of ENaC.
The Ensembl genome database (release 79) of vertebrate and eukaryotic species currently includes 540 genes homologous to ENaC family members, 188 of which encode one of the four ENaC subunits. The remainder represents ASICs or other family members from different species. A BLAST search of the UniProt protein database shows that ENaC subunits are found in vertebrates. BLAST search of UniProt bacteria, fungi and plant protein sequence databases did not reveal orthologs of human ENaC subunits. Here we provide a summary of the appearance of ENaC genes in Metazoan species.
6.1. Cyclostomata and Chondrichthyes (cartilaginous fishes)
In the phylogeny of vertebrates, the most ancient taxon is Cyclostomata, i.e. jawless vertebrates. Lampreys and hagfishes are common extant species that represent this taxon. These fishes have only cartilaginous elements as a primitive skeleton that supports their body parts (Shimeld and Donoghue, 2012). The genome of sea lamprey includes three genes that code for the orthologs of α, β and γ-ENaC, but apparently does not include a gene for the delta subunit (Table 7) (Smith et al., 2013). The sequence of lamprey α subunit is not complete (S4RTA3_PETMA).
Table 7.
Presence (+) or absence (−) of genes encoding SCNN1A, SCNN1B, SCNN1G, and SCNN1D in non-mammalian vertebrates.
| Taxon | Example species | A | B | G | D | Reference |
|---|---|---|---|---|---|---|
| Cyclostomata (jawless vertebrates) | ||||||
| Petromyzontidae (lampreys) | Petromyzon marinus (Sea lamprey) | + | + | + | − | (Smith et al., 2013) |
| Gnathostomata (jawed vertebrates) | ||||||
| Chondrichthyes (cartilaginous fishes) | Callorhinchus milii (Elephant shark) | + | + | + | − | (Venkatesh et al., 2007) |
| Euteleostomi (bony vertebrates) | ||||||
| Actinopterygii (ray-finned fishes) | Danio rerio (Zebrafish) | − | − | − | − | (Venkatesh et al., 2007) |
| Coelacanthiformes (lobe-finned fishes) | Latimeria chalumnae (Coelacanth) | + | + | + | + | (Amemiya et al., 2013) |
| Dipnoi (lungfishes) | Neoceratodus forsteri (Lungfish) | + | + | + | ? | (Uchiyama et al., 2012) |
| Euteleostomi: Tetrapoda: Amphibia: Batrachia: Anura (frogs and toads) | ||||||
| Pipidae (tongueless frogs) | Xenopus tropicalis (Frog) | + | + | + | + | (Hellsten et al., 2010) |
| Euteleostomi: Tetrapoda: Amniota: Sauropsida: Sauria: Archelosauria: Archosauria: Crocodylia | ||||||
| Alligatorinae (alligators) | Alligator mississippiensis (American alligator) | + | + | + | + | (Green et al., 2014) |
| Euteleostomi: Tetrapoda: Amniota: Sauropsida: Sauria: Archelosauria: Archosauria: Dinosauria: Aves (birds) | ||||||
| Galliformes (fowls) | Gallus gallus (Chicken) | + | + | + | + | (Chicken-Genome, 2004) |
| Galliformes | Meleagris gallopavo (Turkey) | + | + | + | + | (Dalloul et al., 2010) |
| Gruiformes | Eurypyga helias (Sunbittern) | + | + | + | + | (Zhang et al., 2014) |
| Passeriformes (perching birds) | Taeniopygia guttata (Zebra finch) | + | + | + | + | (Warren et al., 2010) |
| Passeriformes | Ficedula albicollis (Flycatcher) | + | + | + | + | (Ellegren et al., 2012) |
| Piciformes | Picoides pubescens (Downy woodpecker) | + | + | + | + | (Zhang et al., 2014) |
| Spheniscidae (penguins) | Aptenodytes forsteri (Emperor penguin) | + | + | + | + | (Zhang et al., 2014) |
| Euteleostomi: Tetrapoda: Amniota: Sauropsida: Sauria: Archelosauria: Testudines (turtles) | ||||||
| Trionychidae (soft-shelled turtles) | Pelodiscus sinensis (Soft-shelled turtle) | + | + | + | + | (Z. Wang et al., 2013) |
| Euteleostomi: Tetrapoda: Amniota: Sauropsida: Sauria: Lepidosauria | ||||||
| Squamata (lizards and snakes) | Anolis carolinensis (Green anole lizard) | + | + | + | + | (Alföldi et al., 2011) |
Next steps in the evolution of vertebrates include the development of jaw and skeleton leading to the formation of Gnathostomata (jawed vertebrates) (Donoghue et al., 2006; Kawasaki and Weiss, 2006; Kuratani, 2012). The earliest representatives of this branch include cartilaginous fish species, including rays and sharks. The genome of the cartilaginous elephant shark has been determined and it includes three orthologous ENaC genes (Venkatesh et al., 2007) (Table 7).
6.2. Euteleostomi (bony vertebrates)
In evolution, the development of jaw is followed by the development of bony fishes. The clade of Euteleostomi (bony vertebrates) includes two branches:
-
Actinopterygii (ray-finned fishes): The "ray-finned" description is based on spiny projections in the fins of these fishes.
Comparison of shark, human and teleost ray-finned fish genomes has revealed that 154 genes (including ENaC paralogs) that have orthologs in the shark genome are not present in ray-finned fish genomes (Venkatesh et al., 2007). Thus, the whole clade of Actinopterygii (ray finned-fishes), which includes Zebrafish, do not have ENaC genes. However, they have ASIC genes. During the course of evolution, ENaC genes may have been lost at the onset of the branch of ray-finned fishes for lack of a functional need or were replaced functionally by alternative genes and proteins (Uchiyama et al., 2014; Venkatesh et al., 2007).
The EnsemblCompara GeneTree shows one "SCNN1A" gene for Lepisosteus oculatus (spotted gar) that is a freshwater ray-finned fish. Our comparison of this protein with human ASIC and ENaC paralogs showed that it is more homologous to ASIC than ENaC paralogs. UniProt database includes 9 protein fragments from the spotted-gar genome that show homology to "amiloride-sensitive sodium channel family". Our comparison of 4 partial sequences (with lengths >400 residues) from the UniProt database with human ASIC and ENaC paralogs showed that all four sequences share 49–61% sequence identity with human ASIC1, while they share 13–15% with human ENaC paralogs. Therefore, the naming of the single ray-finned fish spotted gar protein (ENSLOCP00000013400) as "SCNN1A" appears to be in error. Thus with the elimination of this case, so far ray-finned fish genomes do not appear to have ENaC orthologs as noted above.
-
Sarcopterygii (lobe-finned fish): The "lobe-finned" description was given because of their fleshy paired fins which are considered an early form of limb development in tetrapod vertebrates with four limbs. Therefore, this clade also includes all Tetrapoda species.
Sarcopterygii includes two ancient taxa with extant fishes: Coelacanthiformes (lobe finned fishes, coelacanth) and Dipnoi (lungfishes) (Table 7). The three ENaC genes are present in the genomes of these fish (Amemiya et al., 2013; Uchiyama et al., 2014, 2012). Tetrapoda is considered a branch that emerged in parallel to Dipnoi.
6.3. Amphibia
In the evolutionary ladder, the development of bony vertebrates was followed by the emergence of tetrapods with four limbs. Amphibians (frogs, toads and salamanders) represent the first class of tetrapods. Xenopus tropicalis (frog) genome includes genes encoding the four ENaC paralogs (Hellsten et al., 2010) (Table 7).
6.4. Sauropsida
The second group of tetrapods is Amniota (amniotes) characterized by having an egg or embryo covered with an amniotic membrane. Amniotes include two clades: Sauropsida that includes birds and reptiles, and Mammalia (mammals).
The genome sequences of three crocodilians have been recently reported (Green et al., 2014). Currently, NCBI Genome database Genome Assembly and Annotation report (including a list of predicted proteins) is available only for Alligator mississippiensis (American alligator). Search of this database for amiloride-sensitive sodium channel yielded four sequences (XP_006258424.1, XP_006268483.1, XP_006268484.1, and XP_006277862.1). In this report, the first and the fourth sequences were named as "amiloride-sensitive sodium channel subunit alpha-like", while the second and the third sequences were named as "… subunit beta" and "… subunit gamma". Percent identities of these four sequences are shown on Table 6. These results show that the second sequence (XP_006277862.1) that was labeled as "alpha-like", matches other delta-ENaC sequences in terms of its percent identity with the other alligator ENaC subunits (Table 6) and human ENaC subunits (results not shown). Thus, we conclude that this alligator has four ENaC paralogs including one gene coding for the delta subunit.
The Ensembl (release 79) Gene Tree view includes two reptiles: soft-shell turtle and green anole lizard (Table 7). Both of these genome sequences also include four genes coding for the four ENaC paralogs (Table 7).
Bird genomes that are listed in Ensembl (release 79) Gene Tree view, include four genes coding for ENaC subunits. The recently determined genome of sunbittern (Eurypyga helias) (Zhang et al., 2014) is not yet included in the Ensembl database. Similar to the case of alligator genome noted above, NCBI Genome database Genome Assembly and Annotation report includes four amiloride-sensitive sodium channel entries one of which was listed as "…alpha-like". Our sequence identity analysis unequivocally classifies this "alpha-like" as the δ subunit. Therefore, this genome also includes four ENaC heterologs (Table 7). Since birds and crocodilians are considered evolutionary descendants of dinosaurs (Green et al., 2014), it is likely that dinosaurs also had four genes coding for ENaC subunits.
6.5. Mammalia
The class of Mammalia includes three taxa: egg-laying mammals (Monotremata), marsupials (Metatheria) and placental mammals (Eutheria). In nearly all mammals in these three clades, there are four ENaC genes (Table 8). Ensembl genome database (release 79) includes 38 mammalian species, including 34 placental mammals, 3 marsupials (opossum, Tasmanian devil, wallaby) and egg-laying platypus. All of these species have four paralogs of ENaC with the exception of the mouse genome that appears to have lost the gene for the delta subunit (Ensembl Gene Tree for ENaC homologs). The rat genome, that is a very close phylogenetic relative of the mouse, includes four ENaC paralogs, but the δ subunit sequence is presently available only as a fragment (NCBI Accession: NC_005104.4).
Table 8.
Presence (+) or absence (−) of genes encoding SCNN1A, SCNN1B, SCNN1G, and SCNN1D in mammals.
| Taxon | Example species | A | B | G | D | Reference | |
|---|---|---|---|---|---|---|---|
| Monotremata (egg-laying mammals) | Ornithorhynchus anatinus (Platypus) | + | + | + | + | (Warren et al., 2008) | |
| Metatheria (marsupials) | |||||||
| Diprotodontia | Macropus eugenii (tammar wallaby) | + | + | + | + | (Renfree et al., 2011) | |
| Didelphimorphia | Monodelphis domestica (opossum) | + | + | + | + | (Mikkelsen et al., 2007) | |
| Dasyuridae | Sarcophilus harrisii (Tasmanian devil) | + | + | + | + | (Miller et al., 2011) | |
| Eutheria (placental mammals) | |||||||
| Afrotheria | |||||||
| Elephantidae (elephants) | Loxodonta africana (African elephant) | + | + | + | + | Elephant genome project | |
| Tenrecidae (tenrecs) | Echinops telfairi (hedgehog) | + | + | + | + | ||
| Boreoeutheria: Laurasiatheria: Carnivora (carnivores) | |||||||
| Canidae (dog, coyote, wolf, fox) | Canis lupus familiaris (dog) | + | + | + | + | (Lindblad-Toh et al., 2005) | |
| Felidae (cat family) | Felis catus (domestic cat) | + | + | + | + | (Pontius et al., 2007) | |
| Boroeutheria: Laurasiatheria: Cetartiodactyla (whales, hippos, ruminants, pigs, camels etc.) | |||||||
| Cetacea (whales) | Orcinus orca (killer whale) | + | + | + | + | Marine mammal genomics Ensembl | |
| Boreoeutheria: Laurasiatheria: Cetartiodactyla: Ruminantia | |||||||
| Bovinae | Bos taurus (cow) | + | + | + | + | (Elsik et al., 2009) | |
| Caprinae | Ovis aries (sheep) | + | + | + | + | Sheep Genomics Consortium | |
| Boreoeutheria: Laurasiatheria: Perissodactyla (odd-toed ungulates) | |||||||
| Equidae (horses) | Equus caballus (horse) | + | + | + | + | (Wade et al., 2009) | |
| Boreoeutheria: Euarchontoglires: Rodentia | |||||||
| Muridae | Mus musculus (mouse) | + | + | + | − | (Takada et al., 2013) | |
| Muridae | Rattus norvegicus (rat) | + | + | + | + | (Saar et al., 2008) | |
| Boreoeutheria: Euarchontoglires: Primates | |||||||
| Hominidae | Pan troglodytes (chimpanzee) | + | + | + | + | Chimpanzee Sequencing and Analysis Consortium | |
| Hominidae | Gorilla gorilla | + | + | + | + | (Scally et al., 2012) | |
| Hominidae | Pongo abelii (Sumatran orangutan) | + | + | + | + | (Locke et al., 2011) | |
| Cercopithecidae (Old World monkeys) | Macaca mulatta | + | + | + | + | (Zimin et al., 2014) | |
| Platyrrhini (New World monkeys) | Callithrix jacchus (marmoset) | + | + | + | + | (Worley et al., 2014) | |
In the Ensembl (release 79) Gene Tree view, there are only one to three ENaC paralogs for some mammalian species. Our examination of the genome in each of these cases showed that in most cases the genome sequence does include the missing paralog(s); in other cases the genome sequence is incomplete.
6.6. Summary for Tetrapoda
For the genomes of tetrapods where sequence information is available, including amphibians and amniotes (lizards, crocodiles, birds, and mammals) there are four paralogs of ENaC with the exception of mouse that has lost the gene for the delta subunit (Table 7 and Table 8) (Giraldez et al., 2012).
7. Homologs in invertebrates
As noted in the introduction, invertebrate species have many genes encoding polypeptides homologous to ASIC/ENaC such as mec and deg genes in C. elegans, and pickpocket genes in Drosophila (Table 9). In global (end-to-end) sequence alignment, homologous C. elegans (CAEEL) proteins share up to 16% sequence identity with ENaC subunits from 18 vertebrate species (Table 10). In contrast, among ENaC subunits, percent identities are 40–95% depending on the taxonomic distance (Table 11 and Table 12).
Table 9.
Sodium channel families within the DEG/ENaC superfamily.
| Channel/gene name* | Phylum | Genus / species | Reference | |
|---|---|---|---|---|
| Invertebrates | Annelida (annelid worms) | Helobdella (leech) | (Simakov et al., 2013) | |
| Pickpocket (ppk) | Arthropoda | Drosophila Anopheles Tribolium castaneum |
(Zelle et
al., 2013) (Holt et al., 2002) (Kim et al., 2009) |
|
| Hydra Na+ channel (HyNaC) | Cnidaria | Hydra | (Gründer and Assmann, 2015) | |
| Sp-Scnnla Sp-Scnnlg |
Echinodermata | Strongylocentrotus (sea urchin) | Ensembl database | |
| FMRFamide-activated amiloride-sensitive sodium channel (FaNaC) | Mollusca | Aplysia (sea hare) Crassostrea (oyster) Helix aspersa (snail) Planorbella trivolvis |
(Furukawa et
al., 2006) (Zhang et al., 2012) (Lingueglia et al., 2006) |
|
| Degenerin (deg) (mec) (unc) | Nematoda | C. elegans Toxocara canis Trichuris suis |
(Eastwood and
Goodman et al., 2012) (Zhu et al., 2015) (Jex et al., 2014) |
|
| Putative FMRFamide-gated Na+ channel | Platyhelminthes (flatworms) | Schistosoma mansoni Echinococcus |
(Protasio et
al., 2012) (Zheng et al., 2013) |
|
| C3Y149_BRAFL C3ZNH4_BRAFL |
Chordata | Branchiostoma floridae (Florida lancelet) | (Putnam et al., 2008) | |
| Vertebrates | acid-sensing ion channel (ASIC) | Chordata | Wide distribution | (Deval and Lingueglia et al., 2015) |
| Epithelial Na Channel (ENaC) | Chordata | Wide distribution | This review |
Names for the retrieval of sequence records from the Uniprot database.
Table 10.
Percent identity between globally aligned amino acid sequences of selected metazoan ENaC homologs and α ENaC subunit sequences from 18 vertebrate species (see Table 7 and Table 8 for the full names of the species).
| Human | Chimp. | Rhesus | Eleph. | Bovine | Dog | Mouse | Rabbit | Orca | Tasman. | Platypus | Chick | Flycat. | Alligator | Turtle | Xenopus | Lungfish | Coelacanth | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAEEL-deg1 | 12 | 12 | 12 | 13 | 13 | 13 | 12 | 13 | 13 | 13 | 12 | 13 | 12 | 13 | 13 | 12 | 13 | 12 |
| CAEEL-del1 | 16 | 15 | 15 | 15 | 15 | 15 | 16 | 16 | 15 | 16 | 15 | 14 | 14 | 15 | 15 | 15 | 15 | 13 |
| CAEEL-mec4 | 14 | 14 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 14 | 12 | 14 | 14 | 14 | 14 | 13 | 13 | 12 |
| CAEEL-mec10 | 14 | 14 | 13 | 14 | 13 | 14 | 14 | 14 | 14 | 14 | 13 | 13 | 14 | 14 | 13 | 12 | 13 | 12 |
| CAEEL-unc8 | 15 | 15 | 15 | 15 | 14 | 15 | 15 | 14 | 15 | 15 | 14 | 14 | 14 | 14 | 14 | 14 | 13 | 13 |
| CAEEL- unc105 |
14 | 14 | 14 | 13 | 12 | 13 | 13 | 13 | 13 | 12 | 11 | 14 | 13 | 13 | 13 | 12 | 12 | 12 |
| CAEEL-asic1 | 11 | 11 | 10 | 11 | 11 | 10 | 10 | 11 | 11 | 11 | 12 | 11 | 11 | 11 | 11 | 12 | 11 | 11 |
| STRPU-Scnn1a | 18 | 17 | 17 | 17 | 18 | 17 | 17 | 18 | 18 | 16 | 15 | 16 | 17 | 17 | 17 | 17 | 15 | 15 |
| STRPU- Scnn1bL |
15 | 14 | 14 | 14 | 15 | 14 | 15 | 15 | 15 | 16 | 15 | 14 | 14 | 14 | 13 | 15 | 15 | 15 |
| STRPU-Scnn1g | 16 | 15 | 14 | 16 | 16 | 15 | 15 | 15 | 16 | 15 | 16 | 14 | 14 | 14 | 15 | 16 | 15 | 14 |
Sequences were selected from the Uniprot database. Species abbreviation is followed by the gene symbol. Species: Caenorhabditis elegans (CAEEL); S. purpuratus (STRPU, sea urchin).
Table 11.
Percent identity between globally aligned amino acid sequences of α-ENaC orthologs from 18 species of Vertebrata (see Table 7 and Table 8 for the full names of the species).
The first species (Human) listed on the header row is not listed in the first column to avoid including self-comparisons (e.g. Human vs. Human) that obviously equal 100%.
Table 12.
Percent identity between globally aligned amino acid sequences of β- and γ-ENaC orthologs from 18 species.
| SCNN1B | Human | Chimp. | Rhesus | Eleph. | Bovine | Dog | Mouse | Rabbit | Orca | Tasman. | Platypus | Chick | Flycat. | Alligator | Turtle | Xenopus | Lungfish |
| Chimpanzee | 87 | ||||||||||||||||
| Rhesus | 97 | 85 | |||||||||||||||
| Elephant | 82 | 72 | 82 | ||||||||||||||
| Bovine | 85 | 75 | 84 | 81 | |||||||||||||
| Dog | 87 | 77 | 87 | 84 | 86 | ||||||||||||
| Mouse | 83 | 73 | 83 | 80 | 80 | 82 | |||||||||||
| Rabbit | 85 | 75 | 85 | 80 | 83 | 85 | 81 | ||||||||||
| Orca | 83 | 73 | 83 | 81 | 89 | 86 | 80 | 81 | |||||||||
| Tasmanian | 75 | 65 | 75 | 74 | 75 | 75 | 74 | 75 | 73 | ||||||||
| Platypus | 75 | 66 | 74 | 74 | 74 | 75 | 75 | 73 | 75 | 79 | |||||||
| Chick | 61 | 54 | 61 | 59 | 61 | 62 | 60 | 59 | 61 | 63 | 65 | ||||||
| Flycatcher | 64 | 56 | 64 | 62 | 64 | 64 | 62 | 63 | 63 | 66 | 67 | 82 | |||||
| Alligator | 63 | 56 | 63 | 62 | 63 | 64 | 61 | 62 | 63 | 66 | 67 | 78 | 78 | ||||
| Turtle | 61 | 53 | 61 | 58 | 60 | 62 | 59 | 61 | 60 | 61 | 63 | 71 | 70 | 74 | |||
| Xenopus | 57 | 51 | 58 | 57 | 57 | 58 | 56 | 57 | 56 | 59 | 60 | 59 | 58 | 59 | 56 | ||
| Lungfish | 52 | 46 | 52 | 52 | 51 | 53 | 54 | 52 | 52 | 51 | 53 | 52 | 54 | 53 | 49 | 54 | |
| Coelacanth | 48 | 43 | 48 | 47 | 49 | 48 | 49 | 48 | 49 | 49 | 51 | 49 | 49 | 50 | 45 | 47 | 49 |
| SCNN1G | Human | Chimp. | Rhesus | Eleph. | Bovine | Dog | Mouse | Rabbit | Orca | Tasman. | Platypus | Chick | Flycat. | Alligator | Turtle | Xenopus | Lungfish |
| Chimpanzee | 89 | ||||||||||||||||
| Rhesus | 92 | 83 | |||||||||||||||
| Elephant | 83 | 73 | 78 | ||||||||||||||
| Bovine | 85 | 76 | 80 | 81 | |||||||||||||
| Dog | 88 | 79 | 83 | 84 | 85 | ||||||||||||
| Mouse | 85 | 75 | 80 | 79 | 82 | 85 | |||||||||||
| Rabbit | 86 | 77 | 81 | 81 | 82 | 87 | 86 | ||||||||||
| Orca | 78 | 70 | 74 | 77 | 84 | 81 | 77 | 78 | |||||||||
| Tasmanian | 72 | 64 | 68 | 74 | 72 | 73 | 71 | 73 | 68 | ||||||||
| Platypus | 75 | 67 | 71 | 76 | 75 | 75 | 75 | 74 | 70 | 76 | |||||||
| Chick | 63 | 56 | 59 | 63 | 62 | 63 | 62 | 62 | 60 | 61 | 66 | ||||||
| Flycatcher | 59 | 52 | 56 | 60 | 59 | 60 | 59 | 59 | 57 | 58 | 62 | 86 | |||||
| Alligator | 64 | 56 | 60 | 64 | 63 | 63 | 63 | 64 | 59 | 63 | 66 | 81 | 76 | ||||
| Turtle | 62 | 55 | 60 | 62 | 62 | 62 | 62 | 62 | 59 | 62 | 65 | 79 | 74 | 79 | |||
| Xenopus | 55 | 48 | 52 | 55 | 55 | 55 | 55 | 55 | 52 | 54 | 55 | 57 | 54 | 58 | 57 | ||
| Lungfish | 50 | 44 | 48 | 51 | 51 | 50 | 51 | 49 | 48 | 50 | 51 | 51 | 50 | 52 | 52 | 52 | |
| Coelacanth | 55 | 49 | 52 | 53 | 55 | 53 | 54 | 54 | 51 | 54 | 55 | 55 | 53 | 55 | 54 | 54 | 57 |
Table 10 includes only comparisons with the α ENaC subunit. Comparisons with the β and γ sequences from the same 18 species show a highly similar range of identity (data not shown), i.e., there is no significant difference in the similarity of any CAEEL homolog to any of the three ENaC subunits. Similarly, C. elegans homologs share only a low (<15%) sequence identity with the human ASIC isoforms. Thus, these homologs represent a family(s) separate from the ENaC as well as ASIC families. In Table 9 we note only a few references for the Deg family of proteins in nematodes. The UniProt protein database includes many polypeptides that belong to this family in various worms. As these are outside the scope of this review we will not further relate to these sequences.
In addition to the nematode and arthropod species, BLAST search of UniProt protein database shows significant sequence identity in the range of 13–22% between predicted protein sequences from the genome of Strongylocentrotus purpuratus (purple sea urchin) (unpublished yet; available at Ensembl database) (Table 9) and vertebrate ENaC subunit sequences. Sea urchins belong to the phylum Echinodermata (echinoderms). In the records of this genome, some of these homologs have been assigned names as such as "amiloride-sensitive sodium channel subunit alpha", "…beta" and "…gamma". Multi-sequence comparisons of these proteins with vertebrate ENaC sequences show up to 18% partial sequence identity and reveal large areas of sequence insertions. These echinoderm sequences likely constitute an additional family within the ENaC/Degenerin superfamily. The functional characteristics of these proteins have not been determined, and we believe that it is premature yet to call these proteins with names that imply a direct orthologous relationship with vertebrate ENaC subunits. Moreover, sea urchin homologs show greater sequence identity with vertebrate ASIC paralogs than with ENaC. Gene Tree display in Ensembl Metazoa Genome database links between these sea urchin proteins and Deg type proteins from invertebrate species listed in Table 9.
Among invertebrates the taxon that is closest to vertebrates is Cephalochordata that includes lancelets. Cephalochordata and Vertebrata are two of the subphyla of Chordata (Table 9). The genome sequence of Florida lancelet (amphioxus) has been determined (Putnam et al., 2008). BLAST search of the predicted lancelet proteins using ENaC sequences yields many homologous fragments. Most of these lancelet sequences share greater homology with human ASIC (7–24%) than with ENaC paralogs. As many of these sequences are in the status of homology predicted proteins, it is too early to make definitive statements regarding phylogenetic relationships. Nonetheless, the lancelet sequences do not appear to be direct orthologs of human ENaC paralogs.
In summary, among invertebrate species, there are many members of the ENaC/Degenerin superfamily that clearly differ from ENaC. Thus, these homologs do not appear to be direct orthologs or ancestors of ENaC. As we discuss below, the ancestors of ENaC apparently emerged prior to the branching of the first vertebrates but there is not an apparent direct ancestor of ENaC among the invertebrate sequences available at present. The total number of eukaryotic species is estimated as ~8.7 million (Mora et al., 2011) and only a few percent are vertebrate species. Hence, determination of more invertebrate genomes may lead to the findings of new families within the ENaC/Degenerin superfamily.
The multiplicity and divergence of invertebrate sequences that show homology to "amiloride-sensitive sodium channels" require extended efforts to classify these proteins into families based on their homology and phylogenetic distance among other metazoan sequences.
8. Homology between ENaC orthologs
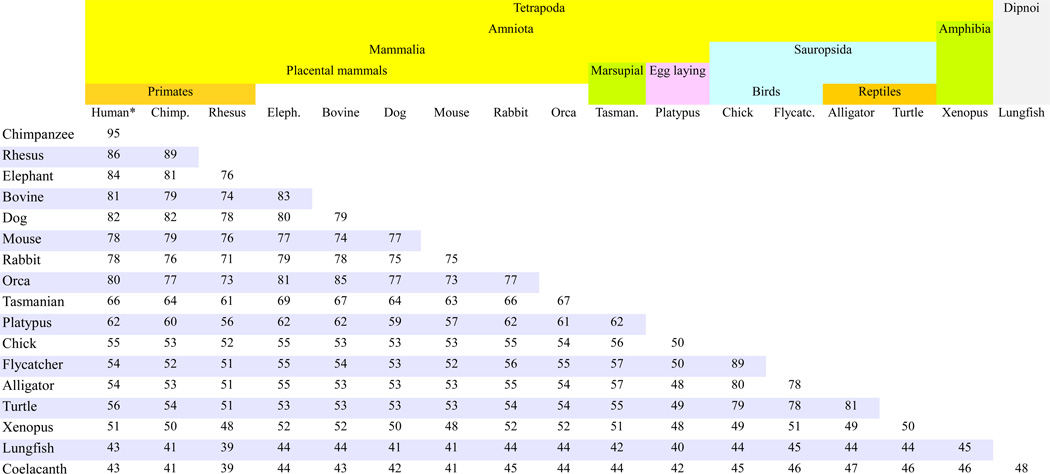
To determine the degree of conservation and sites of divergence of ENaC orthologs, we examined in more detail 20 species for which full sequence of the three ENaC subunits are available. Table 11 and Table 12 show results for 18 species rather than 20 we analyzed (omitted gorilla and rat) because of page and font size limitations. The species selected included representatives of primates (rhesus, chimpanzee, and human), elephant, ruminants (cow), carnivores (dog), rodents (mouse), leporids (rabbit), whales (killer whale Orca), marsupials (Tasmanian devil), egg-laying mammals (platypus), birds (chicken and flycatcher), reptiles (alligator and turtle), amphibians (Xenopus), lobe finned fishes (coelacanth), and lungfish.
Table 11 presents the percent sequence identity for α subunit orthologs from 18 species with headers that mark taxonomic classification. Each cell of the table gives percent identity between two sequences from the species listed in the respective header and the first column. To determine the percent identity in the conserved extracellular domain of α subunit orthologs, we also compared the sequences of the extracellular domain (Fig. 3). On the average across species, sequence identity is ~9% higher in this central segment, than the sequence identity along the entire length of the orthologous proteins (data not shown). Table 12 shows the percent identity between the entire sequences of β- (upper table) and γ subunit orthologs (lower table) in 18 species.
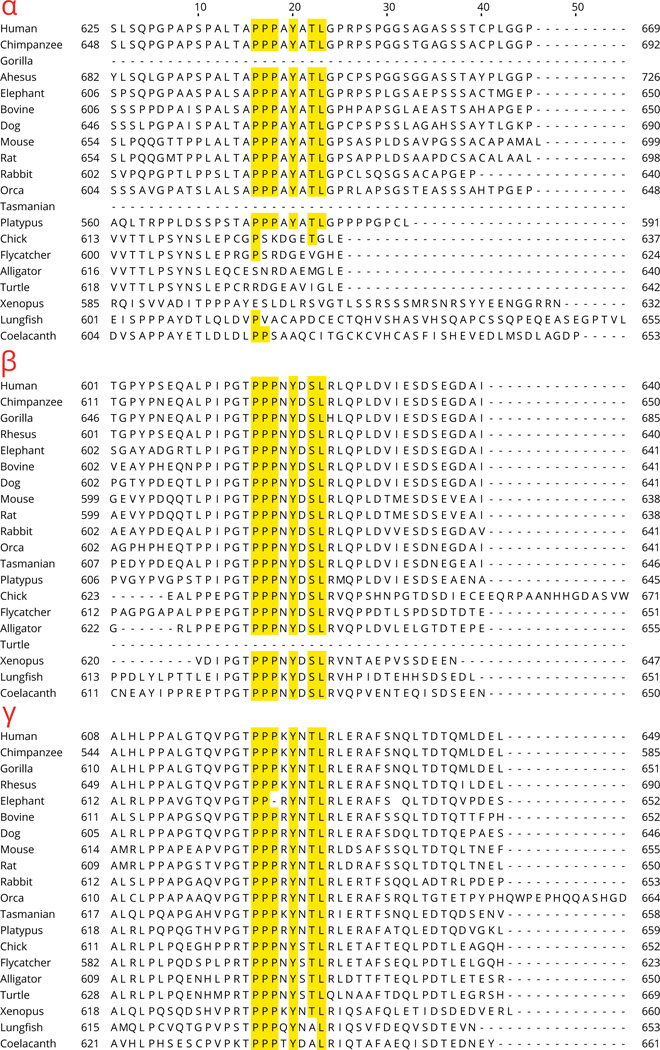
Global alignment of α sequences from 20 species showed that N- and C-termini of orthologous α subunits are divergent across species (see Section 11). Similar to the α subunit, δ subunit orthologs also show high divergence at their N- and C-termini. In contrast, the N- and C-termini of the β and γ subunits are well conserved (see Section 11).
By the comparisons presented here we also wanted to examine if the rate of evolutionary change of ENaC orthologs among different species is similar for the three subunits. Previous studies have indicated that interacting proteins show similar patterns and dynamics of evolution (Lemos et al., 2005). Since the three subunits (α, β and γ) assemble to form a tight complex of a functional channel, we hypothesized that the rate of divergence as measured by the sequence identity would be similar across species for all three subunits.
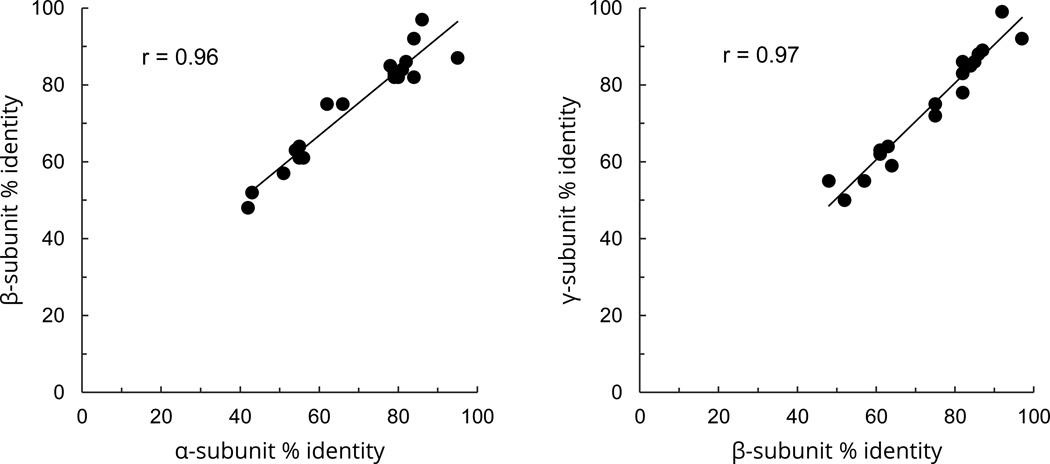
A cursory comparison of the figures in Tables 11 and 12 shows that for each pair of species the percent identity for all three subunits are similar. For example, percent identity between human and turtle α, β and γ sequences is 56, 61 and 62% respectively. The correlation between the sequence identities among α and β subunits and β and γ subunits was r=0.96 and r=0.97 respectively (Fig. 8). Thus, as measured by the percent identity, the divergence of the three subunits has proceeded at similar levels during the species evolution.
Fig. 8.
Correlation between the sequence identities among α, β and γ subunits of ENaC for 20 species relative to human ENaC. A) Correlation of the extent of identity of α and β subunits with their human counterparts. B) Correlation of the extent of identity of β and γ subunits with their human counterparts. The x, y coordinates of each point are percent identities between human sequence and the sequence of another species for the subunit indicated in the x and y axes. The sequences were from human, chimpanzee, gorilla, rhesus, elephant, bovine, dog, mouse, rat, rabbit, orca, Tasmanian devil, platypus, chicken, flycatcher, alligator, turtle, Xenopus, lungfish, and coelacanth.
For all three ENaC subunits (α, β, and γ), sequence identity between orthologs is consistent with the phylogenetic distance between species.
The following list represents some highlights of this phylogeny related homology:
ENaC subunits of extant species within the same taxonomic family share generally >87–96% sequence identity. Example: Human and chimpanzee (family Hominidae) (Table 11 and 12).
All placental mammal sequences, including marine mammal Orcinus orca (killer whale), share >70% identity (Tables 10 and 11).
Birds and reptiles share a common ancestor (Green et al., 2014). Consistent with this phylogenetic relationship, chicken, and flycatcher ENaC sequences share the highest identity (70–81%) with alligator and turtle (Table 11 and 12). In contrast, sequence identity between ENaC sequences from birds versus mammalian species is lower, ranging between 50 to 66% (Tables 11 and 12).
ENaC orthologs in the amphibian Xenopus, share 47–59% identity with the sequences from amniotic animals (Table 11 and 12).
The orthologs in coelacanth that are descendants of the earliest forms of vertebrates share about 39–55% identity with the ENaC sequences from other Vertebrata species (Table 11 and 12).
Lungfish (Table 8), considered a species closest to tetrapods, share 49–57% identity with coelacanth sequence and 39–54% identity with other vertebrates (Table 11 and 12).
8.1. Insertions and deletions in orthologs
In phylogenetic comparisons above, we noted that in some ENaC/Degenerin homologs, in addition to sequence divergence, there are major insertions/deletions (extending for tens to hundreds of residues) relative to ENaC. Thus, we concluded that such proteins belong to different families within the ENaC/Degenerin superfamily. The major differences in the functions of these families of proteins are associated with specific structural features built upon the major common scaffold of these channels. Whereas ENaC is constitutively active and functions in transport of Na+ across epithelia and consequently regulates extracellular fluid volume, ASIC and degenerin type channels fulfill mainly sensory functions (Ben-Shahar, 2011) (see Section 14). The large insertions in the finger domain (Fig. 6) of DEG family of proteins are apparently part of the complex of mechano-sensitivity of these channels (Eastwood and Goodman, 2012).
In the alignment of α subunit sequences from tetrapod species, it can be seen that the N- and C-termini show divergence (see Section 11). However, the extracellular regions do not have major insertions and deletions. Several sequences have deletion/insertion of 2–6 residues relative to the human ortholog. Nearly all of these are located at sites of sequence divergence when compared with ASIC1 (see Section 5.1).
Alignments of β-ENaC sequences also show no major insertions/deletions for 20 species. The anole lizard β-ENaC has a 16-residue insert starting at residue 406. The status of this protein is currently "uncharacterized protein" implying it may have errors.
Alignment of γ subunit sequences from 20 species shows high homology in the extracellular region, with the exception of the chimpanzee sequence that has a ~65 residue deletion. Such a deletion is not found in other mammalian species, and could reflect an error.
Overall, ENaC family orthologs are highly conserved throughout the spectrum of vertebrate species. The degree of their sequence identity is related to their phylogenetic/taxonomic distance. ENaC orthologs do not have major insertions/deletions and can be readily distinguished from members of other families within the ENaC/Degenerin superfamily by their high percent of sequence identity.
9. Identifying ENaC family members within the ENaC/Degenerin superfamily
Members of the ENaC/Degenerin superfamily are readily identified by their common structural features: a large extracellular region connecting two transmembrane domains, and relatively short intracellular N- and C-termini (Fig. 3). Beyond these common structural features, the proteins share sequence homology of varying degrees, depending on their subfamily and the phylogenetic distance between species. Among vertebrates, there are two subfamilies: ASIC and ENaC. Analyses presented above show that ENaC paralogs in vertebrate species can be readily distinguished from ASIC paralogs.
In phylogenetic comparisons, we noted that some homologs are marked as ENaC orthologs in genome analysis. However, our analyses indicate that these are ASIC rather than ENaC orthologs. As more genome sequences are determined, misclassification of orthologs may occur. To avoid this problem, we formulated thresholds of sequence identity that can clearly distinguish ENaC orthologs from other members of ENaC/Degenerin superfamily.
9.1. Threshold for orthologs
The sequences of ENaC orthologs across species show a high degree of conservation with the lowest sequence identity of 39% between tetrapod species and lobe-finned fish coelacanth in global alignment (Tables 10 and 11). The termini of α subunit orthologs are more divergent, while the sequences of the extracellular region have about 10% higher sequence identity. Thus, in a case where the classification of a sequence is unclear, extracellular regions should be compared. Secondly, insertion/deletion of a large segment (>10 residues) should raise concerns regarding subfamily classification (see Section 8.1).
Protein structure database SCOP employed a minimal criteria of 30% sequence identity for assignment of proteins into the same protein family (Murzin et al., 1995). CATH database uses >35% sequence similarity as the criteria for classification as members of a family (Sillitoe et al., 2015). The observation that among ENaC orthologs sequence identity is >38%, matches the requirements of these two databases for the classification of these proteins as members of the same family of ENaC. As sequence identity with ASIC homologs (see Table 4) and other Degenerin type proteins are generally less than 20%, these proteins represent members of families different from ENaC.
9.2. Threshold for paralogs
Multisequence comparisons presented here show a consistent picture. Global alignments within species show that ENaC paralogs generally share >20% sequence identity with one another (Table 4 and Table 6). In contrast, all four ENaC subunits share less than 20% sequence identity with ASIC. This also extends to other homologs, such as Degenerins. Thus within species, 20% sequence identity appears as the cut-off point for the ENaC family as opposed to membership in the ASIC family among vertebrates.
10. Pedigree of ENaC family members
By definition, paralogous proteins emerge as a result of a duplication of a gene in a genome and then diverge as a result of accumulation of mutations in duplicate copies at evolutionary time scale. There are several strong lines of evidence that the four ENaC subunits share a common ancestor:
All four ENaC subunits share the highest homology among themselves as compared to other families.
The genes for all four ENaC subunits have introns in the same locations (Fig. 4) (Saxena et al., 1998) while many introns of other homologs are at different positions.
Within the ENaC family, two pairs appear to have distinct ancestors: 1) the α and δ subunits, and 2) the β and γ subunits. Apparently, an ancestral ENaC sequence underwent a gene duplication that resulted in the formation of two ancestral genes that again underwent independent duplication events. The result is four paralogous genes coding for the four ENaC subunits. The evidence for two duplication events includes the following:
Within each pair of subunits (α and δ; β and γ), there is higher sequence identity than with the other pair of subunits (Table 4 and Table 6).
The genes encoding the β and γ subunits are in adjacent locations on the same chromosome (Brooker et al., 1995), providing evidence that they resulted from a local gene duplication event.
The information provided above on the human genome and other species represents a picture that is true for vertebrates in general. The Ensembl genome database (release 79) of vertebrate and eukaryotic species currently includes 540 homologs of ENaC. A phylogenetic "Gene Tree" constructed for these 540 ENaC homologs using EnsemblCompara GeneTrees paralogy prediction method (Vilella et al., 2009) presents a picture that is consistent with the information provided above.
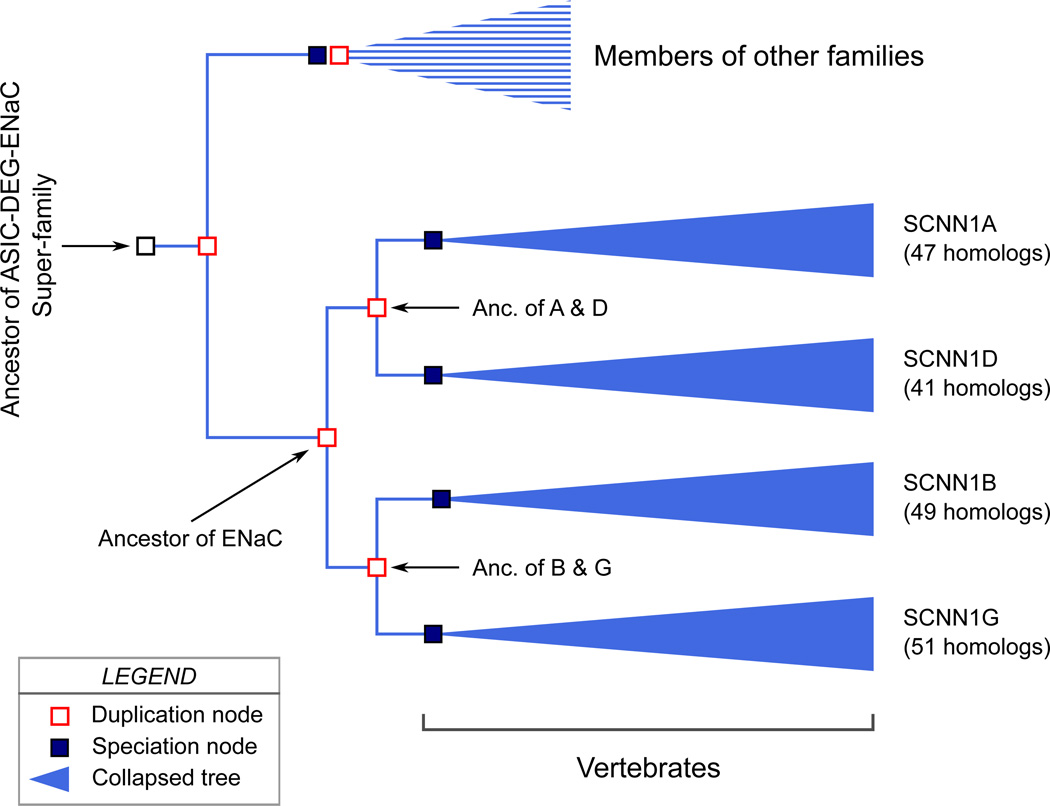
In Fig. 9 we present a hypothetical "pedigree" for the ENaC paralogous genes based on the Ensembl Gene Tree. A phylogenetic tree is analogous to a pedigree. But, phylogeny differs from pedigree in that while in a pedigree ancestor is known, in phylogeny the ancestor is deduced based on homology relationships. The Ensembl Gene Tree predicts a common ancestral gene for all the ENaC homologs that was duplicated. These duplicate genes were once again duplicated to generate the ancestral genes from which the four ENaC genes derive (Fig. 9).
Fig. 9.
A hypothetical phylogenetic tree for paralogs of ENaC. "Anc." is used as an abbreviation for "Ancestor". A "duplication node" represents a gene duplication event that yields two genes within one genome. A "speciation node" represents the formation of a new species that carries the gene of interest. By the convention of Ensembl Gene Tree, collapsed trees for paralogs are shown in blue color. The figure is based on a Gene Tree constructed for 540 ENaC homologs in the Ensembl genome database (release 79) of vertebrate and eukaryotic species using EnsemblCompara GeneTrees paralogy prediction method. The figure includes several modifications from the Gene Tree: The nodes for C. elegans degenerins and one homolog from a fish were omitted from the figure, and the positions of the nodes were modified to show branches in parallel. The number of homologs in each collapsed branch is written on the right side of the collapsed tree marking.
As noted in Section 6, the genes coding for the α, β and γ subunits are present in all vertebrates, except ray-finned fishes, starting with the most ancient jawless vertebrate species such as lamprey (Table 7). SCNN1D gene coding for the δ subunit appears only in Euteleostomi (bony vertebrates) (Table 7).
The widespread phylogenetic spread of the four ENaC subunits provides evidence that the gene duplications that resulted in the formation of these subunits represent an ancient event that preceded the evolution of vertebrates. There is strong evidence for two rounds of whole-genome duplication (2R-WGD) prior to the diversification of the vertebrates (Cañestro et al., 2013; Putnam et al., 2008). 2R-WGD could result in the generation of four copies of duplicated genes. Yet, it is assumed that duplicate copies of many genes were lost after initial duplication. Currently, we do not know whether all paralogs of ENaC are descendants of this 2R-WGD event. Duplicated genes may also originate as a result of local gene duplication events, independent of whole genome duplication (Cañestro et al., 2013). The SCNN1B and SCNN1G genes coding for the β and γ subunits most likely represent products of a local duplication event as they are immediate chromosomal neighbors. A recent review provides a general overview of the evolution of ENaC and other functionally related proteins such as Na+-K+-ATPase and renin-angiotensin-aldosterone system proteins and enzymes (Rossier et al., 2015).
11. Conserved sequence motifs and their functions
Alignments of ENaC orthologs from different species reveal many segments as well as single isolated residues that are conserved in all species. The conservation of these residues and sequence segments suggests that these residues fulfill important functional roles. In this section, we shall summarize conserved sequence motifs and their functions as well as other important functional sites.
11.1. Cytoplasmic amino terminus
As can be seen in the alignments of the human ENaC sequences, the N-termini of α and δ subunits show heterogeneity in both their sequence and length (Fig. 4). A similar pattern of heterogeneity is observed in the alignment of the N-terminal sequences from 20 species (Fig. 10). In contrast to the α and δ subunits, the β and γ subunits from 20 species are highly conserved and most are of similar length (Fig. 10).
Fig. 10.
Comparison of α, β, and γ sequences in the N-terminal, pre-TM1, and TM1 segments from twenty species. For each subunit, residues that are identical in at least 19 out of 20 species (95% identity) are shaded. The location of the predicted TM1 is shown above the sequences. The α subunits have N-termini of highly variable lengths (the numbers at the beginning of each sequence marks the number of additional residues that did not fit into the page), with little or no sequence conservation in this variable region. In contrast, the β and γ subunit N-termini are mostly of similar length and show a high degree of conservation within a ~40 residues-long segment prior to the TM1. The row of red letters, in between the β and γ sequence groups, mark the residues that are identical in both β and γ subunits. In four sequences (β: chicken; γ: gorilla, chicken and coelacanth) 2–5 residues prior to the first methionine were deleted to be consistent with other Uniprot sequences that start with Met as the first translated codon. There may be also sequencing errors in the unusually short platypus α sequence, and flycatcher γ sequence.
Chalfant et al. examined activities of rat ENaC subunits with N-terminal deletions. They found that deletion of residues 2–67 in the N-terminus of the α subunit reduced endocytosis of ENaC and increased the half-life of the channel in the membrane, suggesting that the N-terminus contains an endocytotic motif (Chalfant et al., 1999). Deletion of longer segments of α, β, and γ N-terminus (94, 50, and 94 residues respectively), drastically reduce ENaC activity (Bachhuber et al., 2005).
Yue et al. noted that the N-terminal segments of β and γ subunits contain a stretch of basic residues that is characteristic of Phosphatidylinositol 4,5-bisphosphate (PIP2) binding sites in other proteins (Yue et al., 2002). Mutation of 4 of these basic residues to nonpolar residues in the β but not in the γ subunit drastically reduced ENaC currents in the Xenopus oocyte expression system (Kunzelmann et al., 2005).
In a stretch of about 30 to 40 residues prior to the start of TM1, all four human ENaC paralogs have some strictly conserved residues (Fig. 4 and Fig. 10). These residues are conserved in the four human ENaC paralogs (Fig. 4) as well as in all 20 species examined (Fig. 10). The conservation of these sequences across species suggests that this region has an important functional role. A Gly37Ser missense mutation in this region of the β subunit causes multi-system PHA and this mutation reduces the open probability (Po) of ENaC (Gründer et al., 1997). Mutations of the corresponding residue in the α and γ subunits also reduced channel activity, suggesting that this site has an important role in regulating channel gating (Gründer et al., 1997). Gründer et al. also examined the roles of the residues flanking the key Gly residue by systematic alanine mutagenesis of 28 residues (H77 to H104 in rat α subunit) (Gründer et al., 1999). The expression of ENaC with these mutant subunits in Xenopus oocytes showed that most mutants decreased channel activity, likely due to a reduction in channel Po (Gründer et al., 1999). The stretch of ten residues from T92 to C101 showed the highest sensitivity to alanine mutagenesis, with G95, H94 and R98 mutants showing the strongest reduction (Gründer et al., 1999). These residues are conserved in all 20 species with the exception of the platypus, which has an exceptionally short N-terminus (Uniprot ID: F7F7U2_ORNAN) (Fig. 10). Since the status of this sequence is marked as an "uncharacterized protein" it may have an error.
11.2. TM1
The resolved structure of ASIC1, and sequence similarities between ASICs and ENaCs provide important clues about the stretch of ENaC residues that form TM1. We have also used algorithms to predict the location of the TM1 (Fig. 4 and Table 3). Relative to the alignment with ASIC1 sequence, predicted TM1 segment starts three residues (KKK in human β subunit) after the start of the ASIC1 TM1 (cf. Fig. 4, Fig. 7, and Fig. 10). According to this prediction, TM1 is preceded by 2–3 Arg/Lys residues that are conserved in all ENaC orthologs (Fig. 3, and Fig. 10). Studies on the distributions of charged residues in α-helical TM segments indicated that positively charged residues Arg and Lys are present at much higher proportions on the cytoplasmic side of the TM segment of proteins. This trend was named the "positive-inside" rule (von Heijne, 1992). A recent study has shown that in 191 transmembrane α-helical segments, the residues Arg and Lys are present at highest proportion just before the start of the lipid bilayer (Pogozheva et al., 2014). At this location, the positively charged residues interact with the polar head groups of membrane lipids and contribute to the strength of membrane anchoring of ENaC. The conserved appearance of Arg and Lys just before the predicted TM1 provide support for the predicted location of the TM1 (Table 3, Fig. 3, and Fig. 10).
In the predicted TM1 location, a tryptophan (W) appears as one of the first three residues conserved in all 20 ENaC orthologs (Fig. 10). In ASIC1, a Trp appears as the third residue from the beginning of the first helix (Fig. 7). Analysis of 191 α-helical TM proteins showed that aromatic residues Trp and Tyr are predominantly located at the membrane-water interface (Pogozheva et al., 2014). These aromatic amino acids are known to partition into the interface region of membranes. Hence, it has been suggested that they contribute strongly to the anchoring and precise positioning of TM segments in the lipid bilayer (Hong et al., 2007). The appearance of Trp at the beginning of the TM1 in all ENaC orthologs in 20 species provides further support for the predicted location of the TM1 (Fig. 3, Fig. 4 and Fig. 10). Analyses of TM1 of the α subunit of ENaC by tryptophan-scanning mutagenesis suggested two functionally different regions. N-terminal tryptophan residues altered both channel activity and cation selectivity, with a periodicity consistent with a helical structure. While C-terminal tryptophan residues also affected activity and selectivity, there was no apparent periodicity (Kashlan et al., 2006).
11.3. Extracellular region
The extracellular region, as revealed in the resolved structures of ASIC1, has a complex structure that resembles an outstretched hand holding a ball (Figs. 3 and 4). Hence, domains within the extracellular regions are referred to as palm, thumb, knuckle, finger and β ball (Jasti et al., 2007) (Fig. 7). The palm and β ball domains are formed by non-contiguous β strands and loops, and are in close proximity to the membrane (Figs. 3 and 4). More peripheral domains (thumb, knuckle and finger) are formed by contiguous α helices and loops, and are poorly conserved among ENaC/Degenerin family members, when compared with other parts of the extracellular region. Based on sequence homology and predicted secondary structure, it is likely that the structural features of the extracellular region of ASIC1 is shared among all members of the ENaC/Degenerin superfamily. This is one of the key defining features of this ion channel family. Below we present some of these conserved segments the functions of which have been examined.
Protease cleavage sites
Proteases activate ENaC by cleaving the α and γ subunits at multiple sites within their extracellular finger domains, releasing imbedded inhibitory tracts (Kleyman et al., 2009; Rossier and Stutts, 2009; Vuagniaux et al., 2002). Serine proteases represent one of the largest gene families with 175 predicted genes in the human genome (Szabo and Bugge, 2011). Despite variations in the cleavage sequence specificity of these enzymes, there is a common denominator of one or more Arg or Lys residue immediately preceding the cleavage site (Antalis et al., 2010).
Furin, a member of the proprotein convertase family of serine proteases, cleaves the α subunit twice at sites (RSTR (proximal) and RSAR (distal)) flanking an inhibitory tract (LPHPLQRL) (mouse sequences) (Carattino et al., 2008b, 2006; Hughey et al., 2004; Sheng et al., 2006). Furin cleaves the γ subunit once (RKRR), and cleavage by a second protease at a distal site releases another inhibitory tract (RFLNLIPLLVF) (Bruns et al., 2007; Passero et al., 2010). A polybasic RKRK sequence is one of the distal sites targeted by some of the non-furin proteases that cleave the γ subunit and activate ENaC (Bruns et al., 2007; Passero et al., 2011; Patel et al., 2012). The sequences of the inhibitory tracts in both α and γ subunits are conserved in mammals (marked with blue shading in Fig. 11). While there is a divergence of the sequence of the homologous segment in Sauropsida, Amphibia, lungfish and coelacanth, the fact that amphibian ENaC is activated by proteases (Alli et al., 2012) suggests that this intrinsic inhibitory tract has evolved over time. At present, we are not aware of evidence that other members of the ENaC/Degenerin family are activated by proteases.
Fig. 11.
Serine protease cleavage sites in the extracellular domain of α, β and γ ENaC subunits from 20 species. Key basic amino acids (Arg (R) and Lys (K)) in the putative cleavage site are marked with yellow shading. The sequences of the respective subunits from 20 species were aligned by CLUSTALW. The conserved sequences of the inhibitory tracts located in between the two SP sites are marked light blue background. The residues of the substrate protein that are recognized by proteases are numbered based on their position relative to the cleaved peptide bond. P1 marks the putative residue after which the peptide bond is cleaved by the SP (Antalis et al., 2010).
The α and γ subunit sequences from 20 species indicate that the key protease cleavage sites are strongly conserved in all species with the exception of lungfish and coelacanth (Fig. 11). In Fig. 11, β and γ subunit sequences have been aligned together as these two proteins are products of paralogous duplicated genes. It is noteworthy that in the β sequences the protease cleavage motifs are missing (Fig. 11). In mammals and marsupials, there is a gap instead of the protease cleavage motifs (Fig. 11). Considering that both genes are apparently the descendants of the same gene, either the protease cleavage motifs were deleted from the SCNN1B gene or were later added to the SCNN1G gene.
Disulfide bonds
Within the extracellular regions of ENaC family members, there are 16 highly conserved cysteine residues that likely form eight disulfide bonds (Firsov et al., 1999; Jasti et al., 2007; Sheng et al., 2007). Based on the disulfide bonds in the resolved ASIC1 structure, these include five disulfide bonds in the thumb domain (Sherwood et al., 2012). The 16 extracellular cysteine residues are conserved in all 20 species examined with the exception of the γ subunit of chimpanzee and the β subunit of coelacanth. As noted above, there are two large gaps in the chimpanzee γ subunit sequence. These gaps probably reflect an error in sequence. The structural importance of the conserved cysteines has been demonstrated by site-directed mutagenesis experiments (Firsov et al., 1999; Sheng et al., 2007). There are additional Cys residues in the finger domain of family members in C. elegans, which may form additional disulfide bonds.
Sites of N-linked glycosylation
During the process of translation of proteins that are membrane bound or secreted, oligasacccharides may be attached to the N4 of the asparagine residue at the start of a consensus sequence composed of three amino acids: Asn-Xaa-Ser/Thr. The extracellular region of rat α, β, and γ subunits were shown to have such sites that are glycosylated (Canessa et al., 1994a; Snyder et al., 1994). Alignment of the sequences of subunits from 20 species show that most of the sites identified in the rat sequences are conserved in mammals, but not in birds and lower species. Since the studies on glycosylated residues were carried out using rat subunits, we note here the conserved sites according to the rat subunit residue numbering. Thus, the homologs of the following rat residues are conserved as the first residue in N-glycosylation consensus sequence in most mammals: α subunit: N259, N320, N339, N424, N538; β subunit: N135, N141, N146, N197, N205, N258, N362, N376, N482; γ subunit: N210, N249, N272, N292, N498.
Knuckle domain
In the ASIC1 model, the knuckle domain is composed of two helices (α6 and α7) that are located at the top of each subunit (Figs. 3–5). The sequence of this region is conserved within each subunit, but shows divergence between ENaCs and ASICs (Fig. 7). In all three subunits there is a positively charged residue (Arg or Lys) at a position that corresponds to the end of the α7 helix in ASIC1 (Fig. 7). Mutation of this residue (K498 in the human γ subunit) to alanine was shown significantly to reduce surface expression of ENaC (Edelheit et al., 2014). Recently Chen et al. showed that deletion of the entire knuckle domain (including the conserved Arg/Lys) in mouse β or γ subunit drastically reduced ENaC surface expression and consequently ENaC function (Chen et al., 2015). These consistent findings suggest that the conserved charged residues in this domain may be involved in binding to other proteins that are involved in the transport of ENaC subunits.
In contrast to effects in β or γ subunits, deletion of the knuckle domain in the α subunit resulted in channel activation as a result of a loss of Na+ self-inhibition (Chen et al., 2015).
Palm domain
A stretch of 15 residues prior to TM2 form the two β-strands (β11 and β-12) that are a central component of the palm domain (Fig. 6 and Fig. 7). This region is highly conserved in all three subunits and includes three charged residues that are conserved in all three subunits in all 20 species examined (Fig. 12). These residues are homologous to A413, E417 and Q421 in cASIC1 (Fig. 12). In cASIC1, the R-group of E417 protrudes into the central vestibule (Fig. 13) and has been implicated in proton binding and functional conformational changes in ASIC (Ishikita, 2011). In all three ENaC subunits, the homologous residues are positively charged Lys or Arg (K534, R505 and R514 in human α, β and γ subunits respectively) in all 20 species (Fig. 12). An R514A mutant in human γ subunit significantly leads to a decrease in sodium feedback inhibition consistent with an increase in channel open probability (Edelheit et al., 2014). The other two residues (A413 and Q421) are located at the interface between subunits (Fig. 13) (Jasti et al., 2007). Because of the strict conservation of homologous residues in ENaC subunits, these residues are probably located at symmetrical sites in ENaC subunits as they are in ASIC1 (Fig. 13). Site-directed mutagenesis studies using human ENaC subunits showed that mutation of these residues to alanine also leads to a decrease in sodium feedback inhibition that controls channel open probability (Po) (Edelheit et al., 2014, 2011). A human γ subunit variant in the palm domain (L511Q) is associated with an increase in channel open probability (Chen et al., 2013).
Fig. 12.
Comparison of α, β, and γ sequences in the pre-TM2 and TM2 segment from twenty species. For each subunit, residues that are identical in at least 19 out 20 species (95% identity) are shaded. The location of the TM2 based on homology to ASIC1 is shown above the sequences. In the preTM2 region, only three charged residues are conserved in all three subunits. The positions of these charged residues are marked at the top of the alignments by the corresponding cASIC1 homologs, Ala413, Glu417 and Gln421. Column headers: Deg: degenerin or "Deg" residue. Ami: amiloride binding residues. Sel.: selectivity filter.
Fig. 13.
Location of the cASIC1 E417 and Q421 in ASIC1 structure (PDB ID: 2QTS). The three ribbon structures shown represent the β12-strand region (from L414 to K423) of all three subunits of chicken ASIC1, termed in order A, B, and C (PDB 2QTS). For each subunit, only two residues, E417, and Q421, are shown in CPK style. In 20 species examined, the residue homologous to E417 is an arginine or lysine (K534 in α, R505 in β and R514 in γ subunit of human ENaC). The space in the center of the figure is part of the vestibule along the three-fold axis of symmetry that is thought to be part of the ion pathway.
11.4. TM2
Prior to the report on ASIC1 structure, the location of the TM2 in ENaC subunits was predicted by various software based on hydrophobicity of this region (Canessa et al., 1994a; Saxena et al., 1998). After the publication of the ASIC1 structure in 2007, the helical TM2 segment of ASIC1 has been generally adopted as the location of TM2 in ENaC subunits as well, based on the strong sequence homology in this region and subsequent empirical studies (Kashlan and Kleyman, 2011). While there is overlap between earlier predictions and ASIC1 structure based segment, a segment identified as "pre-M2" (i.e. before the TM2) in earlier work (Kellenberger et al., 1999; Schild et al., 1997), resides within TM2. The terms used in earlier studies should be examined to avoid confusion about regions studied.
Fig. 12 presents alignments of α, β and γ subunits in the TM2 segments in 20 species where the TM2 was marked based on homology with ASIC1. These alignments show that TM2 is highly conserved in all subunits.
A number of key functional sites are within the TM2 region of ENaC subunits, including the channel gate, amiloride binding site and selectivity filter. Fig. 10 illustrates how these sites are conserved through evolution. Functional studies as well as resolved ASIC1 structures suggest that the channel gate is within the outer part of the second membrane spanning domain, within the region encompassing an LLSN motif that is conserved among ENaC subunits across species (Fig. 12). The Ser in this motif is a site where the introduction of large residues has been found to dramatically increase channel open probability (Kellenberger et al., 2002; Sheng et al., 2001a; Snyder et al., 1999). This site has been referred to as the degenerin or "Deg" site, as the introduction of large residues in mechanosensitive ENaC/Degenerin family member in C. elegans results in neurodegeneration, in association with an increase in channel open probability (Goodman et al., 2002; Sherwood et al., 2012).
One of the characteristics of αβγ ENaC is its inhibition by relatively low concentrations of amiloride (Kleyman and Cragoe, 1988). Other family members, including δβγ and ASICs, are inhibited by amiloride or its derivatives at higher concentrations of these drugs (Table 13) (Diochot et al., 2007; Ji et al., 2012). An amiloride binding site has been described at a site in the second membrane-spanning domain, consisting of a Ser in the α subunit, and a Gly in the beta and gamma subunits (Fig. 12). The introduction of specific mutations at these sites led to a profound loss of the efficacy of amiloride (Kashlan et al., 2005; Schild et al., 1997).
Table 13.
Functional characteristics of ASIC and ENaC type channels.
| ENaC (αβγ) | ASIC | |
|---|---|---|
| Channel structure | Hetero-trimer | Homo- / hetero-trimer |
| Channel gating | Constitutively active | H+ activated |
| pH EC50 | Species dependent | 4.8–6.7* |
| Na+/K+ permeability ratio* | >100 | 5–14 |
| Permeable to larger cations | No | Yes |
| Protease activation | Yes | No |
| Amiloride IC50 | 0.1 µM | 10–100 µM |
| Amiloride Ki | 0.35 µM | |
| Extracellular Na+ inhibition | Yes | No |
| Shear stress activation | Yes | No |
| Main functions | Na+ reabsorption across high
resistance epithelia. Maintenance of body salt and water homeostasis. Kidney: Regulation of ECF volume, blood pressure and electrolyte homeostasis. Respiratory airway: Regulation of airway surface liquid (ASL) volume, composition and mucociliary clearance. Reproductive tract: Regulation of epithelial fluid volume necessary for cilial transport of gametes, fertilization and implantation. Skin and exocrine glands: Na+ reabsorption. Taste buds: Salt taste perception. |
Nociception Mechanosensation Synaptic plasticity Fear-related behavior Seizure termination |
Source: (Gründer and Pusch, 2015)
Another defining characteristic of ENaC is its cation selectivity. With regard to its ability to discriminate Na+ and K+, ENaC is the most Na selective mammalian ion channel (Table 13). A three-residue selectivity filter, consisting of a G/S-X-S motif, is present in the TM2 of ENaC subunits (Fig. 12). The introduction of specific mutations in the first or third residue of this motif resulted in channels that allow for modest K+ permeation (Kellenberger et al., 1999; Sheng et al., 2000; Snyder et al., 1999).
At the distal end of TM2 there are three charged residues (within the stretch of EMAELVFD in human α subunit) that are conserved in all 20 species examined (Fig. 12). Mutation of these acidic residues reduced channel conductance (Langloh et al., 2000; Sheridan et al., 2005) and also affect ion selectivity (Sheng et al., 2001b).
11.5. Carboxy terminus
In general, the cytoplasmic C-terminus contains sites of interaction with other proteins, signal transduction molecules, and ions that regulate ENaC function.
Motifs involved in signal transduction
In accordance with the "positive inside" rule noted above (in Subsection 11.2), the region after TM2 is enriched in positively charged Arg and Lys in all three subunits in all 20 species examined (Fig. 12). The proximity of these residues to the cytoplasmic side of the membrane allows interactions of these residues with polar heads of membrane phospholipids and phosphoinositides concentrated at the cytosolic surface of membranes (Di Paolo and De Camilli, 2006). This region in the β and γ subunits has a role in the binding of phosphotidylinositol triphosphate which has an allosteric effect on ENaC open probability (Pochynyuk et al., 2007, 2005). It is noteworthy that positively charged residues at the same analogous region in P2X receptor channels fulfill similar roles (Bernier et al., 2012).
One of the common mechanisms of membrane protein regulation is phosphorylation/dephosphorylation of critical residues by intracellular signal transduction systems. It is likely that the residues in the C-terminus of ENaC subunits may be substrates for such reactions. For example, Volk et al. showed that the activity of ENaC expressed in oocytes could be enhanced by activation of protein kinase C (PKC) by phorbol ester and that this effect was dependent on the presence of an intact C-terminus of α subunit (Volk et al., 2000). The C-terminus also is phosphorylated by the kinases SGK, casein kinase 2, and ERK (Diakov and Korbmacher, 2004; Shi et al., 2002; Yang et al., 2006).
An important regulator of ENaC activity is chloride ions. Truncation of the C-terminus of the β but not of α and γ subunits reduced cytoplasmic Cl− inhibition of ENaC, suggesting that this segment is essential for down-regulation of ENaC by CFTR (Bachhuber et al., 2005; Ji et al., 2000).
Interaction with cytoskeletal elements
Cytoskeletal elements such as microtubules, actin filaments, and associated proteins form an essential part of the complex network of proteins involved in intracellular transport of proteins including endocytosis (Anitei and Hoflack, 2012). As ENaC subunits are transported to the cell membrane via the trans-Golgi network (Butterworth, 2010), cytoplasmic termini of ENaC may interact with cytoskeletal elements during transport and in the cell membrane itself. Indeed, the C-terminus of the α subunit was shown to bind to spectrin that is located in the intracellular side of the membrane. The sequence responsible for binding to the SH3 domain of α-spectrin (PPLALTAPPPA in rat α subunit) (Rotin et al., 1994) starts prior to the PY motif (see below) and partially overlaps with it (Fig. 12). This motif is conserved only in mammals (Fig. 12). In addition to spectrin, there is also evidence for direct interaction of F-actin with the carboxy terminus of the α subunit (Mazzochi et al., 2006; Sasaki et al., 2014).
PY motif
The most conserved motif in the C-terminus of ENaC subunits is the PY motif (Fig. 14). The consensus sequence for the PY motif is PPPXYXXL that is located 65–70 residues after the end of the TM2 in α, β and γ subunits. Delta ENaC orthologs do not have a conserved PY motif. In the 20 species we examined the PY motif is conserved strictly in nearly all species (Fig. 14). Turtle β subunit sequence has a short C-terminus that may be a genome sequencing error. In contrast to the β and γ subunits, in Sauropsida, Amphibia and fishes the PY motif has been lost in the α subunit (Fig. 14). The consistent lack of PY motif in these species makes it very unlikely that the lack of the motif is a sequencing error.
Fig. 14.
Conservation of the PY motif in the C-termini of α, β and γ ENaC subunits from 20 species.
The PY motif is recognized by the WW domains in Nedd4-2 that is an E3 ubiquitin-protein ligase (Rotin and Staub, 2011). WW domains are ~40 residue long segments that are characterized by a conserved sequence that includes two tryptophans (W). Such modules are found in many proteins and bind to proline-rich sequences such as PY motif (Macias et al., 2002). With its WW domains, Nedd4-2 catalyzes ligation of ubiquitin to the ENaC subunit leading to the internalization of ENaC and eventual degradation in a proteasome or lysosome (Rotin and Staub, 2011).
Missense mutations in the β or γ subunit PY motif or truncations that lead to a loss of this motif in these subunits, cause Liddle syndrome (see section 15.2). Mutation or deletion of the PY motif reduces the rate of ENaC ubiquitylation and consequent internalization, leading to accumulation of ENaC in the membrane (Lu et al., 2007). This in turn leads to enhanced absorption of filtered Na+ and consequently increases blood volume and blood-pressure (Rotin, 2008).
12. Tissue distribution of ENaC
Alpha, beta and gamma subunits of ENaC
Most of the studies on tissue localization of ENaC subunits have been carried out by immunohistochemical studies using antibodies generated against small segments of expressed proteins or synthetic peptides that represent short segments of ENaC subunits (Brouard et al., 1999; Coric et al., 2004, 2003; Duc et al., 1994; Hager et al., 2001; Masilamani et al., 1999; Tousson et al., 1989; S. Wang et al., 2013). These studies provided evidence for the localization of ENaC in kidney, lung, salivary glands, skin, placenta and the colon. Expression of ENaC subunits has been also examined by in situ hybridization of tissue sections using cDNA probes (Greig et al., 2003). But, this approach does not provide an image of the intracellular localization of the subunits themselves.
To enhance the immunofluorescence signal, we generated polyclonal antibodies against the complete extracellular region of ENaC subunits (Enuka et al., 2012). These antibodies allowed us to visualize ENaC expression in the bronchial epithelia of human lung and the female reproductive tract extending from the uterus to the fallopian tube at a high resolution by immunofluorescence and 3D confocal microscopy (Enuka et al., 2012).
The expression and sites of localization of ENaC in the kidney nephron tubules have been recently extensively reviewed (Rossier, 2014) and the complexities of this subject are beyond the scope of the present review.
Tissue specificity of expression of ENaC subunits has been also investigated by large-scale microarray and high-throughput RNA sequencing experiments. The results of these studies can be accessed via the EMBL-EBI Expression Atlas database of gene expression (Petryszak et al., 2014). Experiments included in the Expression Atlas (https://www.ebi.ac.uk/gxa/home) report many tissues wherein ENaC subunits are expressed at varying levels. Consistent with the immunohistochemical studies, these studies report highest levels of expression for the α, β and γ subunits in the kidney, lung, and colon. Other tissues reported include the fallopian tube, esophagus, placenta, prostate, skin, stomach, thyroid, tongue and vagina (Expression Atlas).
Some recent studies have reported immunolocalization of ENaC subunits in astrocytes in the brain (Miller and Loewy, 2013), human eye (Krueger et al., 2012), nasal mucosa (Jiang et al., 2015), mouse ear (Morris et al., 2012), rat muscle (Simon et al., 2010), vascular endothelium (Kusche-Vihrog et al., 2014), vascular smooth muscle cells (Drummond et al., 2008), lymphocytes (Ottaviani et al., 2002) and platelets (Cerecedo et al., 2014).
There is also evidence for the expression of ENaC in mammary epithelia (Wang and Schultz, 2014). The functional significance of ENaC in organs, such as kidney, lung and the respiratory tract, sweat and salivary glands and the reproductive tract where ENaC is expressed at relatively high levels, has been established, based on the facts that mutations in ENaC genes result in major dysfunction in these tissues in multi-system PHA (Chang et al., 1996; Enuka et al., 2012; Hanukoglu, 1991; Hanukoglu et al., 2008). However, even in most severe cases of multi-system PHA there does not appear to be an abnormal function in the eye, ear, muscle or neural tissue function. Thus, the physiological significance of the low-level expression of ENaC in other tissues remains to be established.
Expression of ENaC has also been detected in the lingual epithelium and in taste receptor cells (TRCs) (Chandrashekar et al., 2010; Kretz et al., 1999). Most mammals can sense five basic tastes, sweet, sour, bitter, umami and salt by TRCs specific for each taste. In fungiform taste buds of mice there are amiloride-sensitive TRCs that are responsive specifically to NaCl (Shigemura et al., 2008). In genetically engineered mice lacking the α subunit specifically in TRCs, the neural response to low NaCl (<120 mM) in a subpopulation of TRCs was abolished while response to high NaCl remained intact (Chandrashekar et al., 2010). These studies suggest that ENaC in TRCs plays an essential role in the salt-taste receptor system (Chandrashekar et al., 2010; Oka et al., 2013).
In the mammalian order Cetacea that includes whales and dolphins, taste receptor buds have atrophied and appear in degenerate form (Tinker, 1988). Sequencing of the various cetacean genes for the receptors of the five tastes revealed that the receptor genes for four tastes are non-functional pseudogenes because of accumulation of mutations (Zhu et al., 2014). However, in contrast to these, the three ENaC subunits, α, β, and γ, that also serve as salt taste receptor have remained intact and functional. As the authors note, the conservation of ENaC genes in Cetacea is because of the significance of ENaC in osmoregulation and other physiological functions and it is still not known whether Cetacea are capable of sensing salty taste (Zhu et al., 2014).
Delta subunit of ENaC
The first report on the cloning of the human δ subunit also showed that in northern blots the highest levels of its expression are observed in brain, pancreas, testis and ovary with only low levels in the kidney and lung (Waldmann et al., 1995). Consistent with these initial results, the Expression Atlas database results show that the expression of the delta subunit gene (SCNN1D) is relatively much lower in the kidney and lung but highest in neural tissues, (including cerebral cortex, cerebellum, hippocampus, hypothalamus and pituitary gland) and testis (Expression Atlas, 53 GTEx). Expression of the δ subunit has been also detected in the human nasal epithelium (Bangel-Ruland et al., 2010) and eye (Krueger et al., 2012). Overall, the tissue distribution pattern of the δ subunit is distinctly different from that of α, β and γ subunits.
ASICs
ASIC subunits are expressed mainly in the central and peripheral nervous systems and the gastrointestinal tract. ASIC tissue distribution has been recently extensively reviewed (Holzer, 2015; Lin et al., 2015; Zha, 2013).
13. Subcellular and cilial localization
As an ion channel on the apical side of the epithelium, ENaC would be expected to be localized on the apical membrane of cells. Indeed, in human polarized epithelial cells in the female reproductive tract and the respiratory airways, immunohistochemical and immunofluorescence studies show that ENaC is localized at the apical surface of the cells (Enuka et al., 2012). Studies on subcellular localization of α, β and γ subunits in mice and rat kidney nephron and in cultured Madin-Darby Canine Kidney (MDCK) epithelial cell line have yielded varying results probably in part because of hormonal and Na+ treatment conditions (Ackermann et al., 2010; Bao et al., 2014; Chanoux et al., 2013; Hager et al., 2001).
ENaC on cilia
A cilium is a finger like protrusion on the cell surface that has a microtubular skeleton called axoneme. The ciliary membrane that covers the axoneme is continuous with the cell membrane (Satir and Christensen, 2007). Most cells in mammals contain a single cilium (primary cilium) that is built upon the microtubular structure of a centriole. In three major organs, the lung, the reproductive tract, and the central nervous system, the epithelial surface of many cells have multiple cilia (200–300 per cell). These cilia are motile and beat in concert at a predetermined direction generating waves that move the fluid, particles and cells in the lumen of the epithelium (Brooks and Wallingford, 2014).
Using antibodies we generated, we showed that in multi-ciliated cells in the human bronchus and the female reproductive tract (extending from the uterus to the fimbria of fallopian tube), ENaC is specifically located in cilia (Enuka et al., 2012). This was also confirmed by co-localization of ENaC immunofluorescence with that of cilia-specific β-tubulin IV (Enuka et al., 2012).
The discovery that ENaCs are highly expressed in multi-ciliated cells in the lung and the reproductive tract and that they are specifically located over the entire length of cilia has increased our understanding of the function of ENaC in epithelia with motile cilia (Enuka et al., 2012). The depth of the fluid that bathes the cilia in the lumen has to be precisely regulated for normal cilial function (Choi et al., 2015; Tilley et al., 2015). Since Na+ is the major solute in the ECF, regulation of ENaC activity directly affects osmolarity of the periciliary fluid and consequently the flow and volume of the fluid in the lumen. Cilial location allows ENaC to serve as a sensor and regulator of osmolarity of the periciliary fluid along the entire length of the cilia. Thus, ENaC mediated changes in osmolarity would then modulate the fluid volume on the epithelial surface (Enuka et al., 2012). Since all airway epithelia are Na+ absorptive, ENaC plays a key role in pulmonary epithelia, but it should be noted that in the airway epithelium there are additional ion channels and transporters that contribute to the regulation of the composition and volume of the periciliary liquid (Hollenhorst et al., 2011).
The functional importance of ENaC in the multi-ciliated cells of the respiratory airway is illustrated by the contrasting phenotypes of two diseases: 1) In multi-system pseudohypoaldosteronism (see section 15.1), the loss-of-function of ENaC leads to an increase in the volume of airway surface liquid (ASL) (Kerem et al., 1999). 2) In cystic fibrosis (see section 15.3), dysfunction of the chloride transporter CFTR (Cystic Fibrosis Transmembrane Conductance Regulator) leads to reduced inhibition of ENaC by chloride ions. Enhanced activity of ENaC contributes to the drastic reduction of the airway surface liquid (ASL) volume observed in cystic fibrosis. Normally, the undulating movement of the cilia rapidly moves the mucus gel on top of the cilial layer, and together with the mucus, it pushes inhaled particles and microbes in the respiratory airways for clearance via the mouth (Rogers, 2007; Tilley et al., 2015). When ASL volume is reduced, clearance of foreign particles, dust, microbes and viruses is impaired contributing to the chronic infections characteristic of cystic fibrosis.
In epithelia with motile-cilia, the interaction between ENaC and CFTR in the respiratory airways has been extensively studied because of the involvement of these channels in cystic fibrosis (Althaus, 2013). There are conflicting views regarding the mechanism of CFTR and ENaC interactions (Nagel et al., 2005). Some studies suggested that CFTR interacts directly with ENaC (Berdiev et al., 2009). But, while ENaC is located on the cilial surface, CFTR is located on the apical membrane outside of cilial borders (Enuka et al., 2012). Thus, the mechanism of CFTR action in multi-ciliated cells cannot be via direct interaction with ENaC. There is evidence that chloride ions inhibit ENaC (Bachhuber et al., 2005). Thus, CFTR may regulate ENaC activity via its modulation of Cl− levels.
14. Functional differences between ENaC and ASIC
As noted above, ENaCs and ASICs are the only families that are expressed in vertebrates. While ENaCs and ASICs share similar structures, there are major differences in their functional characteristics as observed in heterologous expression systems as well as differences in the physiologic roles in vertebrates (Gründer and Pusch, 2015; Kellenberger and Schild, 2015). The major differences are outlined in Table 13.
ENaCs are constitutively active and facilitate the bulk transport of Na+ across high resistance epithelia in many organs (see Section 12). Transport of sodium is accompanied by a flow of fluid as a result of osmolarity changes. The physiological consequences of these effects depend on the tissue where ENaC is expressed.
ENaC in the distal nephron has an important role in regulating extracellular fluid volume and renal K+ secretion (Rossier et al., 2002). In the respiratory airway and alveoli, ENaC has a major role in regulating the volume of the airway and alveolar fluids (Chambers et al., 2007; Eaton et al., 2009). In the female reproductive tract, ENaC modulates uterine fluid absorption during the reproductive cycles (Ruan et al., 2014; Salleh et al., 2005). Fertilization of the oocyte in the oviduct and fallopian tube requires transport of the oocyte to the ampulla region of the tube. This process is dependent on the ciliary beating along the oviduct in addition to smooth muscle contractions (Coy et al., 2012). Similarly, the transport of embryo to the uterus is also dependent on cilial motion (Lyons et al., 2006). As ENaC is richly expressed on cilia in multi-ciliated cells in the oviduct (see Section 13) and in the uterine glands in the endometrium, reproductive processes of ovum and embryo transport and implantation are dependent on ENaC function (Enuka et al., 2012). In the tongue, ENaC has been identified as the salt taste "receptor" (Chandrashekar et al., 2010; Oka et al., 2013) (see Section 12).
In contrast to ENaC, ASICs are H+-gated ion channels that are are closed in the resting state, and rapidly desensitize following activation. They are expressed in mammalian central and peripheral nervous systems and have roles in nociception, mechanosensation, fear-related behavior and seizure termination (Chen et al., 1998; Deval and Lingueglia, 2015; Price et al., 2001, 2000; Waldmann and Lazdunski, 1998; Ziemann et al., 2009, 2008). ASICs also play an important role in synaptic function regulating neural plasticity in pathological conditions (Zha, 2013). ASICs are also expressed in sensory neurons in the gastrointestinal tract, and play a role in acid-sensing within the gastrointestinal tract and may be involved in human sour taste sensing (Holzer, 2015).
15. Diseases associated with ENaC mutations
In this section, we shall briefly describe hereditary diseases that have been associated with mutations in the genes coding for ENaC subunits. The major characteristics of these diseases are summarized in Table 14.
Table 14.
Characteristics of hereditary disorders that result from mutations or polymorphisms in ENaC subunits.
| Multi system PHA | Liddle syndrome | Cystic fibrosis-like disease | |
|---|---|---|---|
| Inheritance | Autosomal recessive | Autosomal dominant | Polygenetic mechanism. ENaC association in some cases. |
| Mutations in | α, β and γ subunits | PY motif at the C-terminus of β and γ subunits | Heterozygous variants in α, β
or γ subunit genes. ENaC / CFTR genotype (see Table 15). |
| ENaC activity | Loss of function | Gain of function | Increased or decreased activity |
| Water and electrolyte metabolism | Hypovolemia (dehydration), hyponatremia, hyperkalemia | Volume expansion, hypokalemia (usually) | Rarely dehydration |
| Acid-base balance | Metabolic acidosis during recurrent salt wasting episodes | Metabolic alkalosis | Rarely metabolic alkalosis |
| Blood pressure | Hypotension during recurrent salt wasting episodes | Hypertension | Mostly normal |
| Renin-aldosterone system | Hyperreninemia, hyperaldosteronism | Hyporeninemia, Normal-high aldosterone | Mostly normal aldosterone and PRA |
| End organs involved | Kidneys, sweat and salivary glands, respiratory tract, reproductive system, colon | Kidney | Respiratory tract, sweat glands. Less frequent: pancreatic and gastrointestinal tract. |
| Kidneys | Impaired Na+ reabsorption resulting in severe salt loss | Increased Na+ and fluid reabsorption | Normal renal function |
| Respiratory tract | Recurrent lower pulmonary tract infections,
chronic rhinitis. No chronic lung disease |
Normal | Chronic, moderate to severe
bronchitis/bronchiectasis/sinusitis. P. aeruginosa infections |
| Sweat and salivary glands | Increased chloride (>>60 mmol/L) Aggravates renal salt wasting specifically in hot environments. | Normal | Borderline to highly increased sweat chloride
(40–>60 mmol/L), rarely normal. Normal salivary chloride. |
| Reproductive tract | Impaired ciliary function in endometrium and fallopian tube, impaired fertility | Normal | Normal |
| Age of onset/presentation | Infancy | Childhood/young adulthood Rare in infancy | Infancy, childhood, young adulthood |
| Outcome | High mortality in infancy. Decreasing frequency and severity of salt wasting episodes with age. |
Premature death in undiagnosed young adults | Variable. Depends on severity of pulmonary
involvement. No long term data in most studies |
| Therapy | High NaCl supplementation lifelong | Diuretics that block ENaC (amiloride or triamterene) and low salt diet | Variable |
15.1. Multi-system pseudohypoaldosteronism type 1 (PHA1B)
Type I pseudohypoaldosteronism (PHA) is a syndrome of unresponsiveness (also called resistance) to mineralocorticoid hormone aldosterone. This disease was first described by Cheek and Perry as an aldosterone unresponsiveness syndrome resulting in salt wasting in a child (Cheek and Perry, 1958). Subsequently over 50 studies reported many cases of PHA with characteristics of aldosterone resistance with varying degrees of salt wasting. In 1991, Hanukoglu established that PHA includes two independent syndromes (called renal and multi-system forms of PHA) that differ in their pathogenesis, mode of inheritance, the involvement of aldosterone target organs and the severity of salt wasting (Hanukoglu, 1991). Later studies confirmed this distinction and these two forms have been assigned two separate entries in the OMIM database (http://omim.org/): 1) Renal form (PHA1A): OMIM #177735, inherited as an autosomal-dominant disease; and 2) multi-system form (PHA1B): OMIM #264350, inherited as an autosomal-recessive disease.
As commonly observed for other steroid hormone resistance diseases, initially the cause of PHA was suspected to be a mutation(s) in the mineralocorticoid receptor (MR) gene (Armanini et al., 1985). However, analysis of the linkage between PHA and polymorphisms adjacent to the mineralocorticoid receptor gene on chromosome 4 in 10 consanguineous families excluded mutations in the MR gene in multi-system PHA patients (Chung et al., 1995). By further homozygosity mapping, multi-system PHA locus was mapped to two regions coding for SCNN1A and SCNN1B and SCNN1G in chromosomes 12p and 16p respectively (Strautnieks et al., 1996). Indeed, sequencing of the genes coding for the α, β and γ subunits revealed that the multi-system form results from mutations in these three genes (Chang et al., 1996). Two years later Geller et al. reported that the milder renal form (PHA1A) is caused by mutations in the mineralocorticoid receptor gene (Geller et al., 1998).
Clinical presentation
Patients affected by multi-system PHA lose salt from all aldosterone-responsive target organs expressing ENaC including kidney, sweat and salivary glands and respiratory tract (Hanukoglu, 1991). This may necessitate frequent hospitalizations especially during infancy and childhood for severe hyponatremia, hyperkalemia, acidosis and dehydration (Edelheit et al., 2005; Hanukoglu, 1991). These patients also exhibit recurrent pulmonary symptoms such as congestion, wheezing, recurrent lower respiratory tract infections and chronic rhinorrhea due to impaired lung fluid absorption causing excessive airway surface liquid (Hanukoglu et al., 1994; Kerem et al., 1999; Schaedel et al., 1999). They may also fail to conceive due to impaired ENaC expression in cilia lining the fallopian tube and the endometrial mucosa (Enuka et al., 2012). This form carries a high mortality risk especially in infancy, although mortality has been observed even in older patients (Hanukoglu, 1991; Porter et al., 2003; Saxena et al., 2002). Disease manifestations are life long, yet the severity and frequency of salt wasting episodes improve with age (Adachi et al., 2010; Hanukoglu and Hanukoglu, 2010; Hanukoglu et al., 2008).
PHA1B patients require high amounts of sodium chloride (up to 45 g/day) life long, to prevent recurrent salt wasting episodes from multiple organs (Hanukoglu and Hanukoglu, 2010; Hanukoglu et al., 2008; Hogg et al., 1991). The severity of PHA manifestations improves with age, depending on the nature of the mutations, environmental factors such as ambient temperatures and degree of compliance with the therapy (Adachi et al., 2010; Hanukoglu et al., 2008).
Genotype-phenotype relationships
Multi-system PHA patients characterized to date carry homozygous or compound heterozygous mutations in the genes coding for the α, β and γ subunits (Belot et al., 2008; Bonny et al., 2002; Chang et al., 1996; Edelheit et al., 2005; Kellenberger et al., 1999; Saxena et al., 2002; Strautnieks et al., 1996; J. Wang et al., 2013; Welzel et al., 2013). Nonsense, frameshift, and abnormal splicing mutations are associated with a severe phenotype (Edelheit et al., 2005). Generally, missense mutations result in a milder phenotype (Hanukoglu et al., 2008). Functional expression of subunits with frameshift mutations showed these mutations reduce but do not necessarily eliminate ENaC activity (Edelheit et al., 2010).
Transient severe salt loss (severe hyponatremia and hyperkalemia) was reported in a premature baby with a homozygous missense mutation in the SCNN1A gene but not in his brother born at term who carried the same mutation (Dirlewanger et al., 2011). In the neonatal period, the human kidney is characterized by an impaired ability to regulate water and sodium homeostasis and premature babies are even more susceptible to blood volume and electrolyte changes when ENaC activity is partial (Martinerie et al., 2009).
In cases where the mutant subunit has been expressed together with the other two wild-type subunits, a correlation has been observed between the in vitro sodium conductance activity of the mutated channel and the clinical severity of the disease (Hanukoglu et al., 2008).
Subjects carrying the mutations in one allele (e.g. parents) are asymptomatic. Rarely, increased sweat sodium and chloride concentrations have been observed in carriers of a mutation in the α subunit without additional hormonal or clinical phenotypes (Riepe et al., 2009).
15.2. Liddle syndrome
Liddle syndrome (OMIM #177200) is an autosomal dominant disorder characterized by early-onset hypertension associated with hypokalemia, metabolic alkalosis, and low levels of plasma renin activity (PRA) and aldosterone (Bogdanović et al., 2012; Hansson et al., 1995; Liddle et al., 1963) (see Table 14). The degree of phenotypic expression may vary even within the same family (Bogdanović et al., 2012; Findling et al., 1997).
Liddle syndrome is caused by missense mutations in the PY motif of β or γ subunit of ENaC (Fig. 14), and nonsense or frameshift mutations that result in truncation of the C-terminus leading to the loss of the PY motif in these subunits (Furuhashi et al., 2005; Hansson et al., 1995; Ma et al., 2008; Schild et al., 1996; Shimkets et al., 1994). Mutation or elimination of the PY motif disrupts ubiquitin ligase Nedd4-2 binding to the PY motif (Rotin and Staub, 2011). This leads to accumulation of active channels at the cell surface and increased Na+ reabsorption in the kidney, resulting in elevated blood volume and blood pressure. Although in the α subunit there is also a PY-motif (Fig. 14), so far no case of Liddle syndrome has been reported with a mutation in the α subunit.
In addition to mutations in the PY motif region, two mutations in the C-terminus (β subunit, R563Q) and the TM2 segment (γ subunit, N530S) have been reported to be associated with Liddle syndrome phenotype. In the β subunit, an R563Q mutation is associated with low plasma renin activity (PRA), low aldosterone and hypertension in a minority of the individuals carrying the variant (Rayner et al., 2003). In the γ subunit, Hiltunen et al. found an N530S mutation in a patient who developed Liddle syndrome like symptoms at the age of 25 years (Hiltunen et al., 2002). Yet, in the same study the N530S mutation was also found in a healthy person with normal blood pressure (Hiltunen et al., 2002). Thus, the authors raise the possibility that the γ N530S mutation may not be the sole cause of hypertension and that there may be additional factors (such as other genes or environmental factors) responsible for the phenotype observed (Hiltunen et al., 2002). Functional expression of ENaC carrying the N530S mutation showed that the mutation increases ENaC open probability two fold. The increased ENaC activity was observed without a change in the surface expression of ENaC relative to the wild type (Hiltunen et al., 2002). It is probable that more mutations will be found that increase ENaC activity and that are associated with Liddle syndrome like symptoms, similar to the two cases noted above.
Generally Liddle syndrome is a rare disease. However, an extensive study that included a sample of 330 Chinese young hypertensive patients revealed that 1.5% of the patients had mutations associated with a loss of the PY motif (Wang et al., 2015). In a retrospective study in a cohort of 149 hypertensive US veterans, 6% were found to have biochemical abnormalities compatible with Liddle syndrome (Tapolyai et al., 2010). Thus, Liddle syndrome may be a common cause of monogenic hypertension in some populations (Padmanabhan et al., 2015).
Undiagnosed and untreated individuals with Liddle syndrome are at high risk of premature cardiovascular morbidity and mortality. Diuretics that block ENaC activity (amiloride or triamterene) and low salt diet, are usually adequate to control hypertension.
15.3. Cystic fibrosis-like disease
Cystic fibrosis (CF) is an autosomal recessive disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. CFTR dysfunction affects epithelial chloride transport in multiple organs including the digestive system, sweat glands, pancreas, and the reproductive tract, but, progressive lung disease continues to be the major cause of morbidity and mortality.
Respiratory tract infections result from dehydration of airway surfaces that reduces mucociliary clearance and creates an environment conducive to bacterial infections leading to progressive respiratory insufficiency and eventually respiratory failure.
In the lumen of the respiratory tract, ENaC function is essential for normal mucociliary clearance (Althaus, 2013; Hobbs et al., 2013). As noted in section 13, ENaC is expressed in the respiratory tract, located on the surface of cilia and regulates the volume of luminal fluid (Enuka et al., 2012). PHA1B patients with ENaC mutations suffer frequently from lower respiratory tract infections (Hanukoglu et al., 1994; Kerem et al., 1999; Schaedel et al., 1999). Increased airway-specific expression of the β subunit of ENaC results in CF-like symptoms in mice (Mall et al., 2004). This is thought to result in over-expression of channels composed of only α and β subunits, which have a high intrinsic open probability (Mall et al., 2010). Therefore, it has been suggested that mutations in ENaC genes may be involved in some forms of cystic fibrosis. To examine the hypothesis that ENaC mutations may be associated with the degree of severity of CF, a French group screened genomic DNA of 56 CF patients for the presence of variants in SCNN1B and SCNN1G genes (Viel et al., 2008). By using denaturing high-performance liquid chromatography (DHPLC), they found 4 missense mutations in three patients out of 56 (T313M and G589S in β, and L481Q and V546I in γ subunit). However, nasal potential difference measurements did not indicate a functional effect of these variants on Na+ transport, at least in the nasal epithelium of these patients. Thus, the authors concluded that variants in SCNN1B and SCNN1G genes are not associated with CF severity in the cohort examined (Viel et al., 2008).
Cases that present with classical features of cystic fibrosis (such as chronic lung infections with elevated sweat chloride concentration), but without CFTR mutation or a single allele CFTR mutation, have been referred to as cystic fibrosis-like (CF-like) disease (Table 14). These cases lack a genetic diagnosis, and a commonly suspected cause is mutations in ENaC genes (Collawn et al., 2012).
Screening 185 patients with non-classic CF, Sheridan at al. identified 20 patients who had elevated sweat chloride concentrations, and pulmonary disease but without CFTR mutations (Sheridan et al., 2005). Sequencing of the ENaC genes (SCNN1A, B or G) revealed that two of the patients carry compound heterozygous mutations in the SCNN1B gene: a missense mutation (P267L) with a splice site mutation in one patient and two missense mutations (G294S and E539K) in the other. Neither patient had abnormal renin or aldosterone levels. In functional expression studies, P267L and E539K mutants showed decreased activity and G294S mutant showed increased activity (Sheridan et al., 2005). The authors concluded that the compound heterozygous mutations identified in the β ENaC genes that have a mild effect on ENaC activity are associated with CF-like disease without causing severe renal salt loss (Sheridan et al., 2005).
In a multi-center European study, 30 ENaC variants were found in 76 patients with CF-like disease (Azad et al., 2009). Only two (hypoactive F61L and hyperactive V114I in SCNN1A) of the 30 variants were found in patients but not in the control populations. ENaC subunit variants had a significantly higher frequency in the patients as compared to controls. The variant W493R in the α subunit showed the most significant difference, and in functional expression in Xenopus oocytes showed over four-fold higher ENaC activity. Thus, the authors concluded that these variants may be involved in CF-like disease by a polygenetic mechanism (Azad et al., 2009). In a study including 99 Italian patients with CFTR-related diseases, 12 ENaC variants were found, but the allele frequency of these variants was not significantly different from controls (Amato et al., 2012).
Fajac et al. screened a group of 55 patients with diffuse idiopathic bronchiectasis (permanent dilation of the airways as a result of chronic bronchial infection) by sequencing SCNN1B and SCNN1G exons and identified five heterozygous missense variants (S82C, P369T, N288S in β, and G183S, E197K in γ subunit) in eight patients (Fajac et al., 2008). The S82C mutation was found in three unrelated patients who were also heterozygous for a CFTR mutation. The authors thus concluded that trans-heterozygous mutations in ENaC and CFTR may be responsible for the CF-like symptoms.
In a study including 60 Rwandan children with CF-like symptoms, five patients were found to have a heterozygous CFTR mutation. Two of these patients had a missense ENaC variant (V573I in α, V348M and G442V in β subunit); of these only V348M was not found in the control group (Mutesa et al., 2009). Functional expression of the V348M mutant showed that the mutation enhances ENaC activity (Rauh et al., 2013). Since the full ENaC subunit gene sequences were not determined in 55 of the patients, the relationship between ENaC variants and CF-like disease cannot be determined for the whole group.
A recent study on CF-like phenotypes examined the sequences of five genes (CFTR, SCNN1A, SCNN1B, SCNN1G and SERPINA1) in six patients by whole exome sequencing (Ramos et al., 2014). The authors detected three missense variants in SCNN1A (R204W, A357T, C641F) and one missense (R563Q) in SCNN1B, and four additional nucleotide variants. Two of these mutants (C641F and R563Q) appeared also in two CF controls but not in healthy controls. The authors suggest that the variants that appear at a higher frequency in patients with CF-like phenotype than in controls may be responsible for this phenotype. By their family analysis they also stress the importance of genetic /environmental factors in the development of CF-like disease (Ramos et al., 2014).
Brenan et al. examined 33 nonwhite, non-Hispanic patients with a CF-like disease whose CFTR gene analysis was non-diagnostic (19 with no mutations in CFTR gene and 14 with a heterozygous change in the CFTR gene) (Brennan et al., 2015). Sequencing of the exons and introns of SCNN1A, SCNN1B, and SCNN1G in all patients revealed 21 variants. Since the variants found in conjunction with a CFTR mutation were common polymorphisms, the authors concluded that there is no conclusive association of ENaC genetic variants with CF in their cohort.
In Table 15 we listed all the missense mutations identified in the studies reviewed above. Among the three ENaC genes, most of the variants have been observed in SCNN1A and SCNN1B. Proportionately, the highest number of mutants is in SCNN1A. Two of the major studies examined only two genes: SCNN1B and SCNN1G, thus the number of variants in SCNN1A in Table 15 is underrepresented. In our analysis of PHA1B mutants, we observed a similar trend that most of the PHA1B causing mutants appear in SCNN1A and only a few in SCNN1G (Edelheit et al., 2005).
Table 15.
Missense mutations found in patients with a CF-like phenotype.
| Variant | Change in function | Ethnicity | References | |
|---|---|---|---|---|
| SCNN1A | ||||
| V14G | Spanish | (Ramos et al., 2014) | ||
| F61L | ↓ ENaC activity | Caucasian | (Azad et al., 2009) | |
| V114I | ↑ ENaC activity | Caucasian | (Azad et al., 2009) | |
| R181W | ↑ ENaC activity | Caucasian | (Azad et al., 2009; Sheridan et al., 2005) | |
| R204W | Spanish | (Ramos et al., 2014) | ||
| A304P | Spanish | (Ramos et al., 2014) | ||
| A334T | ↓ ENaC activity | Caucasian, nonwhite | (Amato et al., 2012; Azad et al., 2009; Brennan et al., 2015) | |
| A357T | Spanish | (Ramos et al., 2014) | ||
| W493R | ↑ ENaC activity | Caucasian | (Azad et al., 2009) | |
| V573I | African, nonwhite | (Brennan et al., 2015; Mutesa et al., 2009) | ||
| C618F | ↑ ENaC activity | Nonwhite | (Brennan et al., 2015) | |
| C641F | Spanish | (Ramos et al., 2014) | ||
| T663A | ↓ ENaC activity | Caucasian, nonwhite | (Amato et al., 2012; Azad et al., 2009; Brennan et al., 2015) | |
| SCNN1B | ||||
| S82C | None | Caucasian | (Azad et al., 2009; Fajac et al., 2008; Sheridan et al., 2005) | |
| P267L | ↓ ENaC activity | (Sheridan et al., 2005) | ||
| N288S | Caucasian | (Fajac et al., 2008) | ||
| G294S | ↑ ENaC activity | (Sheridan et al., 2005) | ||
| T313M | No change in NPD* | French | (Viel et al., 2008) | |
| V348M | ↑ ENaC activity | African | (Mutesa et al., 2009) | |
| P369T | High basal NPD | Caucasian | (Fajac et al., 2008) | |
| R388C | Nonwhite | (Brennan et al., 2015) | ||
| G442V | African, nonwhite | (Brennan et al., 2015; Mutesa et al., 2009) | ||
| E539K | ↓ ENaC activity | (Sheridan et al., 2005) | ||
| R563Q | Spanish | (Ramos et al., 2014) | ||
| G589S | None, normal NPD | Caucasian, French | (Azad et al., 2009; Viel et al., 2008) | |
| T594M | Nonwhite | (Brennan et al., 2015) | ||
| SCNN1G | ||||
| G183S | African, nonwhite | (Brennan et al., 2015; Fajac et al., 2008) | ||
| E197K | None | Caucasian | (Azad et al., 2009; Fajac et al., 2008) | |
| L481Q | Normal NPD | French | (Viel et al., 2008) | |
| V546I | Normal NPD | French | (Viel et al., 2008) |
NPD: Nasal potential difference.
As detailed in the studies cited, many of the variants are also present in control groups. Yet, some of the mutants that were shown to affect adversely ENaC activity have been reported in independent studies (Table 15). The total number of cases is still too small to reach definitive conclusions about the role of these variants/mutants in causing CF-like disease. In any case, the number of variants that may be associated with CF-like disease represents a small percentage of the total patients in each cohort and differs between ethnic groups.
15.4. Hypertension
In modern industrial societies, hypertension has emerged as one of most widespread health problems (Toka et al., 2013). A minority of the cases of hypertension can be ascribed to monogenic conditions such as Liddle's syndrome or Gordon's syndrome (Padmanabhan et al., 2015). The remainder of the cases is generally grouped as "essential hypertension" with multifactorial etiology, including multiple genetic, humoral, environmental and dietary factors (Su and Menon, 2001). The major systems that are responsible for the regulation of blood pressure include the renin-angiotensin-aldosterone system and Na+ transporters in the kidney (Padmanabhan et al., 2015; Rossier, 2014; Soundararajan et al., 2010; Su and Menon, 2001). Moreover, the Liddle syndrome described above firmly established the importance of ENaC in blood pressure regulation. Therefore, several large-scale studies have examined the association of ENaC variants with essential hypertension. Liu et al. examined 2880 Chinese subjects (GenSalt study) and found an association of blood pressure with SCNN1B and SCNN1G SNPs and variants (Liu et al., 2015). Rayner et al. screened 139 South African black hypertensives for the R563Q variant of β subunit and found that the variant was significantly associated with hypertension (Rayner et al., 2003). In a study in a Finland, the authors sequenced only exon 13 (that codes for TM2 and the C-terminal segment) of β and γ subunits (see Fig. 7) in 27 hypertensive patients. The three identified variants were then screened in 347 hypertensives. The frequency of all variants in the hypertensives was significantly higher (~3 fold) than that in two control groups. Functional expression of one variant (β G589S) showed slightly enhanced activity in Xenopus oocytes (Hannila-Handelberg et al., 2005).
Ambrosius et al. reported a small but significant association between a common αT663A variant and normal blood pressure (Ambrosius et al., 1999). The αA663 variant reduces ENaC activity and functional expression in Xenopus oocytes (Samaha et al., 2004; Tong et al., 2006), but the change in activity was opposite of that predicted by the results of Ambrosius et al. Furthermore, no association was found between αT663A variant and blood pressure in a sample of 247 Japanese hypertensives (Sugiyama et al., 2001). Nonetheless, homozygous A663 allele appears to have a differential effect on lung function (Foxx-Lupo et al., 2011).
Another polymorphism with conflicting reports is β-T594M. In a sample of black hypertensives from London, 8.3% (17 out of 206) were heterozygous for the β T594M variant, but in the control group only 2.1% had the same variant (Baker et al., 1998). However, in a much larger sample, including 1666 Jamaican blacks, no association was found between the β T594M allele and hypertension (Hollier et al., 2006). This variant did not alter ENaC activity in a heterologous expression system (Persu et al., 1998)
Persu et al. identified seven variants in the β subunits of, mostly white, 525 probands of hypertensive families, but could not identify an association between a variant and essential hypertension (Persu et al., 1998).
In summary, currently there does not appear to be a clear association between ENaC variants and essential hypertension. Yet, this area of research is just at its beginnings and requires examination of larger sets of SNPs and variants in selected populations. Some of the past studies have examined only specific segments (such as exon 13) of subunits. This approach skews the results. The SNPs that are accumulating in whole genome sequencing will present larger and more comprehensive databases for future examination. It is likely that such studies will reveal new variants associated with hypertension similar to that found in the Chinese GenSalt study. Since the majority of the variants so far screened do not seem to be associated with hypertension, the proportion of ENaC variants associated with hypertension would be expected to be small.
Highlights.
A comprehensive review of the structure and function of four ENaC subunits from an evolutionary perspective.
Comparison of the sequences of ENaC homologs and identification of structural motifs conserved throughout vertebrates.
Establishing criteria for distinguishing ENaC family members from other families within the ENaC/Degenerin superfamily including ASIC, deg, mec, unc, ppk type gene products.
Review of tissue-specific expression and functions of ENaC paralogs and inherited diseases associated with mutations in ENaC genes.
Acknowledgments
This review and the corresponding Gene Wiki articles are written as part of the Gene Wiki Review series - a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE.
We are grateful to Prof. Thomas Kleyman (University of Pittsburgh) for valuable discussions. This research was funded in part by a grant from the United States-Israel Binational Science Foundation (BSF).
Abbreviations
- ASIC
acid-sensing ion channel
- ASL
airway surface liquid
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- ENaC
epithelial sodium channel
- ECF
extracellular fluid
- ICF
intracellular fluid
- PHA
pseudohypoaldosteronism
- PRA
plasma renin activity
- TM
transmembrane
- TRCs
taste receptor cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann D, Gresko N, Carrel M, Loffing-Cueni D, Habermehl D, Gomez-Sanchez C, Rossier BC, Loffing J. In vivo nuclear translocation of mineralocorticoid and glucocorticoid receptors in rat kidney: differential effect of corticosteroids along the distal tubule. Am J Physiol Renal Physiol. 2010;299:F1473–F1485. doi: 10.1152/ajprenal.00437.2010. [DOI] [PubMed] [Google Scholar]
- Adachi M, Asakura Y, Muroya K, Tajima T, Fujieda K, Kuribayashi E, Uchida S. Increased Na reabsorption via the Na-Cl cotransporter in autosomal recessive pseudohypoaldosteronism. Clin Exp Nephrol. 2010;14:228–232. doi: 10.1007/s10157-010-0277-0. [DOI] [PubMed] [Google Scholar]
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, Ray DA, Boissinot S, Shedlock AM, Botka C, Castoe TA, Colbourne JK, Fujita MK, Moreno RG, ten Hallers BF, Haussler D, Heger A, Heiman D, Janes DE, Johnson J, de Jong PJ, Koriabine MY, Lara M, Novick PA, Organ CL, Peach SE, Poe S, Pollock DD, de Queiroz K, Sanger T, Searle S, Smith JD, Smith Z, Swofford R, Turner-Maier J, Wade J, Young S, Zadissa A, Edwards SV, Glenn TC, Schneider CJ, Losos JB, Lander ES, Breen M, Ponting CP, Lindblad-Toh K. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alli AA, Song JZ, Al-Khalili O, Bao H-F, Ma H-P, Alli AA, Eaton DC. Cathepsin B is secreted apically from Xenopus 2F3 cells and cleaves the epithelial sodium channel (ENaC) to increase its activity. J Biol Chem. 2012;287:30073–30083. doi: 10.1074/jbc.M111.338574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althaus M. ENaC inhibitors and airway re-hydration in cystic fibrosis: state of the art. Curr Mol Pharmacol. 2013;6:3–12. doi: 10.2174/18744672112059990025. [DOI] [PubMed] [Google Scholar]
- Amato F, Bellia C, Cardillo G, Castaldo G, Ciaccio M, Elce A, Lembo F, Tomaiuolo R. Extensive molecular analysis of patients bearing CFTR-related disorders. J. Mol. diagnostics. 2012;14:81–89. doi: 10.1016/j.jmoldx.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Ambrosius WT, Bloem LJ, Zhou L, Rebhun JF, Snyder PM, Wagner MA, Guo C, Pratt JH. Genetic variants in the epithelial sodium channel in relation to aldosterone and potassium excretion and risk for hypertension. Hypertension. 1999;34:631–637. doi: 10.1161/01.hyp.34.4.631. [DOI] [PubMed] [Google Scholar]
- Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels A, De Moro G, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Panji S, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searle SMJ, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, van Heusden P, White S, Williams L, Yandell M, Brinkmann H, Volff J-N, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, Di Palma F, Lander ES, Meyer A, Lindblad-Toh K. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitei M, Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat. Cell Biol. 2012;14:11–19. doi: 10.1038/ncb2409. [DOI] [PubMed] [Google Scholar]
- Antalis TM, Buzza MS, Hodge KM, Hooper JD, Netzel-Arnett S. The cutting edge: membrane-anchored serine protease activities in the pericellular microenvironment. Biochem J. 2010;428:325–346. doi: 10.1042/BJ20100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanini D, Kuhnle U, Strasser T, Dorr H, Butenandt I, Weber PC, Stockigt JR, Pearce P, Funder JW. Aldosterone-receptor deficiency in pseudohypoaldosteronism. N Engl J Med. 1985;313:1178–1181. doi: 10.1056/NEJM198511073131902. [DOI] [PubMed] [Google Scholar]
- Asher C, Chigaev A, Garty H. Characterization of interactions between Nedd4 and beta and gammaENaC using surface plasmon resonance. Biochem Biophys Res Commun. 2001;286:1228–1231. doi: 10.1006/bbrc.2001.5508. [DOI] [PubMed] [Google Scholar]
- Asher C, Wald H, Rossier BC, Garty H. Aldosterone-induced increase in the abundance of Na+ channel subunits. Am. J. Physiol. 1996;271:C605–C611. doi: 10.1152/ajpcell.1996.271.2.C605. [DOI] [PubMed] [Google Scholar]
- Azad AK, Rauh R, Vermeulen F, Jaspers M, Korbmacher J, Boissier B, Bassinet L, Fichou Y, des Georges M, Stanke F, De Boeck K, Dupont L, Balascáková M, Hjelte L, Lebecque P, Radojkovic D, Castellani C, Schwartz M, Stuhrmann M, Schwarz M, Skalicka V, de Monestrol I, Girodon E, Férec C, Claustres M, Tümmler B, Cassiman J-J, Korbmacher C, Cuppens H. Mutations in the amiloride-sensitive epithelial sodium channel in patients with cystic fibrosis-like disease. Hum Mutat. 2009;30:1093–1103. doi: 10.1002/humu.21011. [DOI] [PubMed] [Google Scholar]
- Bachhuber T, König J, Voelcker T, Mürle B, Schreiber R, Kunzelmann K. Cl-interference with the epithelial Na+ channel ENaC. J Biol Chem. 2005;280:31587–31594. doi: 10.1074/jbc.M504347200. [DOI] [PubMed] [Google Scholar]
- Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell. 2014;156:717–729. doi: 10.1016/j.cell.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EH, Dong YB, Sagnella GA, Rothwell M, Onipinla AK, Markandu ND, Cappuccio FP, Cook DG, Persu A, Corvol P, Jeunemaitre X, Carter ND, MacGregor GA. Association of hypertension with T594M mutation in beta subunit of epithelial sodium channels in black people resident in London. Lancet. 1998;351:1388–1392. doi: 10.1016/s0140-6736(97)07306-6. [DOI] [PubMed] [Google Scholar]
- Bangel-Ruland N, Sobczak K, Christmann T, Kentrup D, Langhorst H, Kusche-Vihrog K, Weber W-M. Characterization of the epithelial sodium channel delta-subunit in human nasal epithelium. Am J Respir Cell Mol Biol. 2010;42:498–505. doi: 10.1165/rcmb.2009-0053OC. [DOI] [PubMed] [Google Scholar]
- Bao H-F, Thai TL, Yue Q, Ma H-P, Eaton AF, Cai H, Klein JD, Sands JM, Eaton DC. ENaC activity is increased in isolated, split-open cortical collecting ducts from protein kinase Cα knockout mice. Am J Physiol Renal Physiol. 2014;306:F309–F320. doi: 10.1152/ajprenal.00519.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker PM, Nguyen MS, Gatzy JT, Grubb B, Norman H, Hummler E, Rossier B, Boucher RC, Koller B. Role of gammaENaC subunit in lung liquid clearance and electrolyte balance in newborn mice. Insights into perinatal adaptation and pseudohypoaldosteronism. J Clin Invest. 1998;102:1634–1640. doi: 10.1172/JCI3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belot A, Ranchin B, Fichtner C, Pujo L, Rossier BC, Liutkus A, Morlat C, Nicolino M, Zennaro MC, Cochat P. Pseudohypoaldosteronisms, report on a 10-patient series. Nephrol Dial Transpl. 2008;23:1636–1641. doi: 10.1093/ndt/gfm862. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar Y. Sensory functions for degenerin/epithelial sodium channels (DEG/ENaC) Adv Genet. 2011;76:1–26. doi: 10.1016/B978-0-12-386481-9.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdiev BK, Jovov B, Tucker WC, Naren AP, Fuller CM, Chapman ER, Benos DJ. ENaC subunit-subunit interactions and inhibition by syntaxin 1A. Am J Physiol Renal Physiol. 2004;286:F1100–F1106. doi: 10.1152/ajprenal.00344.2003. [DOI] [PubMed] [Google Scholar]
- Berdiev BK, Qadri YJ, Benos DJ. Assessment of the CFTR and ENaC association. Mol. Biosyst. 2009;5:123–127. doi: 10.1039/b810471a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman JM, Brand C, Awayda MS. A long isoform of the epithelial sodium channel alpha subunit forms a highly active channel. Channels (Austin) 2015;9:30–43. doi: 10.4161/19336950.2014.985478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier L-P, Blais D, Boué-Grabot É, Séguéla P. A dual polybasic motif determines phosphoinositide binding and regulation in the P2X channel family. PLoS One. 2012;7:e40595. doi: 10.1371/journal.pone.0040595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla V, Hallows KR. Mechanisms of ENaC regulation and clinical implications. J Am Soc Nephrol. 2008;19:1845–1854. doi: 10.1681/ASN.2008020225. [DOI] [PubMed] [Google Scholar]
- Björklund AK, Ekman D, Light S, Frey-Skött J, Elofsson A. Domain rearrangements in protein evolution. J Mol Biol. 2005;353:911–923. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- Bobby R, Medini K, Neudecker P, Lee TV, Brimble MA, McDonald FJ, Lott JS, Dingley AJ. Structure and dynamics of human Nedd4-1 WW3 in complex with the αENaC PY motif. Biochim Biophys Acta. 2013;1834:1632–1641. doi: 10.1016/j.bbapap.2013.04.031. [DOI] [PubMed] [Google Scholar]
- Bogdanović R, Kuburović V, Stajić N, Mughal SS, Hilger A, Ninić S, Prijić S, Ludwig M. Liddle syndrome in a Serbian family and literature review of underlying mutations. Eur J Pediatr. 2012;171:471–478. doi: 10.1007/s00431-011-1581-8. [DOI] [PubMed] [Google Scholar]
- Bonny O, Hummler E. Dysfunction of epithelial sodium transport: from human to mouse. Kidney Int. 2000;57:1313–1318. doi: 10.1046/j.1523-1755.2000.00968.x. [DOI] [PubMed] [Google Scholar]
- Bonny O, Knoers N, Monnens L, Rossier BC. A novel mutation of the epithelial Na+ channel causes type 1 pseudohypoaldosteronism. Pediatr Nephrol. 2002;17:804–808. doi: 10.1007/s00467-002-0945-8. [DOI] [PubMed] [Google Scholar]
- Bourque CW. Central mechanisms of osmosensation and systemic osmoregulation. Nat Rev Neurosci. 2008;9:519–531. doi: 10.1038/nrn2400. [DOI] [PubMed] [Google Scholar]
- Bremner HR, Freywald T, O’Brodovich HM, Otulakowski G. Promoter analysis of the gene encoding the beta-subunit of the rat amiloride-sensitive epithelial sodium channel. Am J Physiol Lung Cell Mol Physiol. 2002;282:L124–L134. doi: 10.1152/ajplung.2002.282.1.L124. [DOI] [PubMed] [Google Scholar]
- Brennan M-L, Pique LM, Schrijver I. Assessment of epithelial sodium channel variants in nonwhite cystic fibrosis patients with non-diagnostic CFTR genotypes. J Cyst Fibros. 2015 doi: 10.1016/j.jcf.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Brooker DR, Kozak CA, Kleyman TR. Epithelial Sodium Channel GenesScnn1bandScnn1gAre Closely Linked on Distal Mouse Chromosome 7. Genomics. 1995;29:784–786. doi: 10.1006/geno.1995.9934. [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB. Multiciliated cells. Curr Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard M, Casado M, Djelidi S, Barrandon Y, Farman N. Epithelial sodium channel in human epidermal keratinocytes: expression of its subunits and relation to sodium transport and differentiation. J Cell Sci. 1999;112(Pt 1):3343–3352. doi: 10.1242/jcs.112.19.3343. [DOI] [PubMed] [Google Scholar]
- Bruns JB, Carattino MD, Sheng S, Maarouf AB, Weisz OA, Pilewski JM, Hughey RP, Kleyman TR. Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the gamma-subunit. J Biol Chem, The Journal of biological chemistry. 2007;282:6153–6160. doi: 10.1074/jbc.M610636200. [DOI] [PubMed] [Google Scholar]
- Büsst CJ. Blood pressure regulation via the epithelial sodium channel: from gene to kidney and beyond. Clin Exp Pharmacol Physiol. 2013;40:495–503. doi: 10.1111/1440-1681.12124. [DOI] [PubMed] [Google Scholar]
- Butterworth MB. Regulation of the epithelial sodium channel (ENaC) by membrane trafficking. Biochim Biophys Acta. 2010;1802:1166–1177. doi: 10.1016/j.bbadis.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa CM, Merillat AM, Rossier BC. Membrane topology of the epithelial sodium channel in intact cells. Am J Physiol. 1994a;267:C1682–C1690. doi: 10.1152/ajpcell.1994.267.6.C1682. [DOI] [PubMed] [Google Scholar]
- Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, Rossier BC. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994b;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- Cañestro C, Albalat R, Irimia M, Garcia-Fernàndez J. Impact of gene gains, losses and duplication modes on the origin and diversification of vertebrates. Semin. Cell Dev. Biol. 2013;24:83–94. doi: 10.1016/j.semcdb.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Capaldo CT, Farkas AE, Nusrat A. Epithelial adhesive junctions. F1000Prime Rep. 2014;6:1. doi: 10.12703/P6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattino MD, Hughey RP, Kleyman TR. Proteolytic processing of the epithelial sodium channel gamma subunit has a dominant role in channel activation. J Biol Chem. 2008a;283:25290–25295. doi: 10.1074/jbc.M803931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattino MD, Passero CJ, Steren CA, Maarouf AB, Pilewski JM, Myerburg MM, Hughey RP, Kleyman TR. Defining an inhibitory domain in the alpha-subunit of the epithelial sodium channel. Am J Physiol Renal Physiol. 2008b;294:F47–F52. doi: 10.1152/ajprenal.00399.2007. [DOI] [PubMed] [Google Scholar]
- Carattino MD, Sheng S, Bruns JB, Pilewski JM, Hughey RP, Kleyman TR. The epithelial Na+ channel is inhibited by a peptide derived from proteolytic processing of its alpha subunit. J Biol Chem. 2006;281:18901–18907. doi: 10.1074/jbc.M604109200. [DOI] [PubMed] [Google Scholar]
- Cerecedo D, Martínez-Vieyra I, Alonso-Rangel L, Benítez-Cardoza C, Ortega A. Epithelial sodium channel modulates platelet collagen activation. Eur J Cell Biol. 2014;93:127–136. doi: 10.1016/j.ejcb.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Chalfant ML, Denton JS, Langloh AL, Karlson KH, Loffing J, Benos DJ, Stanton BA. The NH(2) terminus of the epithelial sodium channel contains an endocytic motif. J Biol Chem. 1999;274:32889–32896. doi: 10.1074/jbc.274.46.32889. [DOI] [PubMed] [Google Scholar]
- Chambers LA, Rollins BM, Tarran R. Liquid movement across the surface epithelium of large airways. Respir Physiol Neurobiol. 2007;159:256–270. doi: 10.1016/j.resp.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba NJP, Zuker CS. The cells and peripheral representation of sodium taste in mice. Nature. 2010;464:297–301. doi: 10.1038/nature08783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SS, Grunder S, Hanukoglu A, Rösler A, Mathew PM, Hanukoglu I, Schild L, Lu Y, Shimkets RA, Nelson-Williams C, Rossier BC, Lifton RP. Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1. Nat Genet. 1996;12:248–253. doi: 10.1038/ng0396-248. [DOI] [PubMed] [Google Scholar]
- Chanoux Ra, Shubin CB, Robay A, Suaud L, Rubenstein RC. Hsc70 negatively regulates epithelial sodium channel trafficking at multiple sites in epithelial cells. Am J Physiol Cell Physiol. 2013;305:C776–C787. doi: 10.1152/ajpcell.00059.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheek DB, Perry JW. A salt wasting syndrome in infancy. Arch Dis Child. 1958;33:252–256. doi: 10.1136/adc.33.169.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, England S, Akopian AN, Wood JN. A sensory neuron-specific, protongated ion channel. Proc Natl Acad Sci U S A. 1998;95:10240–10245. doi: 10.1073/pnas.95.17.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kleyman TR, Sheng S. Gain-of-function variant of the human epithelial sodium channel. Am J Physiol Renal Physiol. 2013;304:F207–F213. doi: 10.1152/ajprenal.00563.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ray EC, Yates ME, Buck TM, Brodsky JL, Kinlough CL, Winarski KL, Hughey RP, Kleyman TR, Sheng S. Functional roles of clusters of hydrophobic and polar residues in the epithelial Na+ channel knuckle domain. J Biol Chem. 2015 doi: 10.1074/jbc.M115.665398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicken-Genome. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Choi H-C, Kim CSK, Tarran R. Automated acquisition and analysis of airway surface liquid height by confocal microscopy. Am J Physiol Lung Cell Mol Physiol. 2015;309:L109–L118. doi: 10.1152/ajplung.00027.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chraibi A, Horisberger JD. Na self inhibition of human epithelial Na channel: temperature dependence and effect of extracellular proteases. J Gen Physiol. 2002;120:133–145. doi: 10.1085/jgp.20028612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Hanukoglu A, Rees M, Thompson R, Dillon M, Hanukoglu I, Bistritzer T, Kuhnle U, Seckl J, Gardiner RM. Exclusion of the locus for autosomal recessive pseudohypoaldosteronism type 1 from the mineralocorticoid receptor gene region on human chromosome 4q by linkage analysis. J Clin Endocrinol Metab. 1995;80:3341–3345. doi: 10.1210/jcem.80.11.7593448. [DOI] [PubMed] [Google Scholar]
- Collawn JF, Lazrak A, Bebok Z, Matalon S. The CFTR and ENaC debate: how important is ENaC in CF lung disease? Am J Physiol Lung Cell Mol Physiol. 2012;302:L1141–L1146. doi: 10.1152/ajplung.00036.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier DM, Peterson ZJ, Blokhin IO, Benson CJ, Snyder PM. Identification of extracellular domain residues required for epithelial Na+ channel activation by acidic pH. J Biol Chem. 2012;287:40907–40914. doi: 10.1074/jbc.M112.417519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier DM, Snyder PM. Identification of epithelial Na+ channel (ENaC) intersubunit Cl- inhibitory residues suggests a trimeric alpha gamma beta channel architecture. J Biol Chem. 2011;286:6027–6032. doi: 10.1074/jbc.M110.198127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condliffe SB, Carattino MD, Frizzell RA, Zhang H. Syntaxin 1A regulates ENaC via domain-specific interactions. J Biol Chem. 2003;278:12796–12804. doi: 10.1074/jbc.M210772200. [DOI] [PubMed] [Google Scholar]
- Copeland SJ, Berdiev BK, Ji HL, Lockhart J, Parker S, Fuller CM, Benos DJ. Regions in the carboxy terminus of alpha-bENaC involved in gating and functional effects of actin. Am J Physiol Cell Physiol. 2001;281:C231–C240. doi: 10.1152/ajpcell.2001.281.1.C231. [DOI] [PubMed] [Google Scholar]
- Coric T, Hernandez N, de la R, osa DA, Shao D, Wang T, Canessa CM. Expression of ENaC and serum- and glucocorticoid-induced kinase 1 in the rat intestinal epithelium. Am J Physiol Gastrointest Liver Physiol. 2004;286:G663–G670. doi: 10.1152/ajpgi.00364.2003. [DOI] [PubMed] [Google Scholar]
- Coric T, Zhang P, Todorovic N, Canessa CM. The extracellular domain determines the kinetics of desensitization in acid-sensitive ion channel 1. J Biol Chem. 2003;278:45240–45247. doi: 10.1074/jbc.M304441200. [DOI] [PubMed] [Google Scholar]
- Coy P, García-Vázquez FA, Visconti PE, Avilés M. Roles of the oviduct in mammalian fertilization. Reproduction. 2012;144:649–660. doi: 10.1530/REP-12-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg LA, Bouffard P, Burt DW, Crasta O, Crooijmans RPMA, Cooper K, Coulombe RA, De S, Delany ME, Dodgson JB, Dong JJ, Evans C, Frederickson KM, Flicek P, Florea L, Folkerts O, Groenen MAM, Harkins TT, Herrero J, Hoffmann S, Megens H-J, Jiang A, de Jong P, Kaiser P, Kim H, Kim K-W, Kim S, Langenberger D, Lee M-K, Lee T, Mane S, Marcais G, Marz M, McElroy AP, Modise T, Nefedov M, Notredame C, Paton IR, Payne WS, Pertea G, Prickett D, Puiu D, Qioa D, Raineri E, Ruffier M, Salzberg SL, Schatz MC, Scheuring C, Schmidt CJ, Schroeder S, Searle SMJ, Smith EJ, Smith J, Sonstegard TS, Stadler PF, Tafer H, Tu ZJ, Van Tassell CP, Vilella AJ, Williams KP, Yorke JA, Zhang L, Zhang H-B, Zhang X, Zhang Y, Reed KM. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval E, Lingueglia E. Acid-Sensing Ion Channels and nociception in the peripheral and central nervous systems. Neuropharmacology. 2015 doi: 10.1016/j.neuropharm.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Diakov A, Korbmacher C. A novel pathway of epithelial sodium channel activation involves a serum- and glucocorticoid-inducible kinase consensus motif in the C terminus of the channel’s alpha-subunit. J Biol Chem. 2004;279:38134–38142. doi: 10.1074/jbc.M403260200. [DOI] [PubMed] [Google Scholar]
- Diochot S, Salinas M, Baron A, Escoubas P, Lazdunski M. Peptides inhibitors of acid-sensing ion channels. Toxicon. 2007;49:271–284. doi: 10.1016/j.toxicon.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Dirlewanger M, Huser D, Zennaro MC, Girardin E, Schild L, Schwitzgebel VM. A homozygous missense mutation in SCNN1A is responsible for a transient neonatal form of pseudohypoaldosteronism type 1. Am J Physiol Endocrinol Metab. 2011;301:E467–E73. doi: 10.1152/ajpendo.00066.2011. [DOI] [PubMed] [Google Scholar]
- Donoghue PCJ, Sansom IJ, Downs JP. Early evolution of vertebrate skeletal tissues and cellular interactions, and the canalization of skeletal development. J. Exp. Zool. B. Mol. Dev. Evol. 2006;306:278–294. doi: 10.1002/jez.b.21090. [DOI] [PubMed] [Google Scholar]
- Drummond HA, Grifoni SC, Jernigan NL. A new trick for an old dogma: ENaC proteins as mechanotransducers in vascular smooth muscle. Physiology (Bethesda) 2008;23:23–31. doi: 10.1152/physiol.00034.2007. [DOI] [PubMed] [Google Scholar]
- Duc C, Farman N, Canessa CM, Bonvalet JP, Rossier BC. Cell-specific expression of epithelial sodium channel alpha, beta, and gamma subunits in aldosterone-responsive epithelia from the rat: localization by in situ hybridization and immunocytochemistry. J Cell Biol. 1994;127:1907–1921. doi: 10.1083/jcb.127.6.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood AL, Goodman MB. Insight into DEG/ENaC channel gating from genetics and structure. Physiology (Bethesda) 2012;27:282–290. doi: 10.1152/physiol.00006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DC, Helms MN, Koval M, Bao HF, Jain L. The contribution of epithelial sodium channels to alveolar function in health and disease. Annu Rev Physiol. 2009;71:403–423. doi: 10.1146/annurev.physiol.010908.163250. [DOI] [PubMed] [Google Scholar]
- Edelheit O, Ben-Shahar R, Dascal N, Hanukoglu A, Hanukoglu I. Conserved charged residues at the surface and interface of epithelial sodium channel subunits--roles in cell surface expression and the sodium self-inhibition response. FEBS J. 2014;281:2097–2111. doi: 10.1111/febs.12765. [DOI] [PubMed] [Google Scholar]
- Edelheit O, Hanukoglu I, Dascal N, Hanukoglu A. Identification of the roles of conserved charged residues in the extracellular domain of an epithelial sodium channel (ENaC) subunit by alanine mutagenesis. Am J Physiol Renal Physiol. 2011;300:F887–F897. doi: 10.1152/ajprenal.00648.2010. [DOI] [PubMed] [Google Scholar]
- Edelheit O, Hanukoglu I, Gizewska M, Kandemir N, Tenenbaum-Rakover Y, Yurdakök M, Zajaczek S, Hanukoglu A. Novel mutations in epithelial sodium channel (ENaC) subunit genes and phenotypic expression of multisystem pseudohypoaldosteronism. Clin Endocrinol (Oxf) 2005;62:547–553. doi: 10.1111/j.1365-2265.2005.02255.x. [DOI] [PubMed] [Google Scholar]
- Edelheit O, Hanukoglu I, Shriki Y, Tfilin M, Dascal N, Gillis D, Hanukoglu A. Truncated beta epithelial sodium channel (ENaC) subunits responsible for multi-system pseudohypoaldosteronism support partial activity of ENaC. J Steroid Biochem Mol Biol. 2010;119:84–88. doi: 10.1016/j.jsbmb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T, Künstner A, Mäkinen H, Nadachowska-Brzyska K, Qvarnström A, Uebbing S, Wolf JBW. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, Adelson DL, Eichler EE, Elnitski L, Guigo R, Hamernik DL, Kappes SM, Lewin HA, Lynn DJ, Nicholas FW, Reymond A, Rijnkels M, Skow LC, Zdobnov EM, Schook L, Womack J, Alioto T, Antonarakis SE, Astashyn A, Chapple CE, Chen H-C, Chrast J, Camara F, Ermolaeva O, Henrichsen CN, Hlavina W, Kapustin Y, Kiryutin B, Kitts P, Kokocinski F, Landrum M, Maglott D, Pruitt K, Sapojnikov V, Searle SM, Solovyev V, Souvorov A, Ucla C, Wyss C, Anzola JM, Gerlach D, Elhaik E, Graur D, Reese JT, Edgar RC, McEwan JC, Payne GM, Raison JM, Junier T, Kriventseva EV, Eyras E, Plass M, Donthu R, Larkin DM, Reecy J, Yang MQ, Chen L, Cheng Z, Chitko-McKown CG, Liu GE, Matukumalli LK, Song J, Zhu B, Bradley DG, Brinkman FSL, Lau LPL, Whiteside MD, Walker A, Wheeler TT, Casey T, German JB, Lemay DG, Maqbool NJ, Molenaar AJ, Seo S, Stothard P, Baldwin CL, Baxter R, Brinkmeyer-Langford CL, Brown WC, Childers CP, Connelley T, Ellis SA, Fritz K, Glass EJ, Herzig CTA, Iivanainen A, Lahmers KK, Bennett AK, Dickens CM, Gilbert JGR, Hagen DE, Salih H, Aerts J, Caetano AR, Dalrymple B, Garcia JF, Gill CA, Hiendleder SG, Memili E, Spurlock D, Williams JL, Alexander L, Brownstein MJ, Guan L, Holt RA, Jones SJM, Marra MA, Moore R, Moore SS, Roberts A, Taniguchi M, Waterman RC, Chacko J, Chandrabose MM, Cree A, Dao MD, Dinh HH, Gabisi RA, Hines S, Hume J, Jhangiani SN, Joshi V, Kovar CL, Lewis LR, Liu Y-s, Lopez J, Morgan MB, Nguyen NB, Okwuonu GO, Ruiz SJ, Santibanez J, Wright RA, Buhay C, Ding Y, Dugan-Rocha S, Herdandez J, Holder M, Sabo A, Egan A, Goodell J, Wilczek-Boney K, Fowler GR, Hitchens ME, Lozado RJ, Moen C, Steffen D, Warren JT, Zhang J, Chiu R, Schein JE, Durbin KJ, Havlak P, Jiang H, Liu Y, Qin X, Ren Y, Shen Y, Song H, Bell SN, Davis C, Johnson AJ, Lee S, Nazareth LV, Patel BM, Pu L-L, Vattathil S, Williams RL, Curry S, Hamilton C, Sodergren E, Wheeler DA, Barris W, Bennett GL, Eggen A, Green RD, Harhay GP, Hobbs M, Jann O, Keele JW, Kent MP, Lien S, McKay SD, McWilliam S, Ratnakumar A, Schnabel RD, Smith T, Snelling WM, Sonstegard TS, Stone RT, Sugimoto Y, Takasuga A, Taylor JF, Van Tassell CP, MacNeil MD, Abatepaulo ARR, Abbey CA, Ahola V, Almeida IG, Amadio AF, Anatriello E, Bahadue SM, Biase FH, Boldt CR, Carroll JA, Carvalho WA, Cervelatti EP, Chacko E, Chapin JE, Cheng Y, Choi J, Colley AJ, de Campos TA, De Donato M, Santos IKFdM, de Oliveira CJF, Deobald H, Devinoy E, Donohue KE, Dovc P, Eberlein A, Fitzsimmons CJ, Franzin AM, Garcia GR, Genini S, Gladney CJ, Grant JR, Greaser ML, Green JA, Hadsell DL, Hakimov HA, Halgren R, Harrow JL, Hart EA, Hastings N, Hernandez M, Hu Z-L, Ingham A, Iso-Touru T, Jamis C, Jensen K, Kapetis D, Kerr T, Khalil SS, Khatib H, Kolbehdari D, Kumar CG, Kumar D, Leach R, Lee JC-M, Li C, Logan KM, Malinverni R, Marques E, Martin WF, Martins NF, Maruyama SR, Mazza R, McLean KL, Medrano JF, Moreno BT, More DD, Muntean CT, Nandakumar HP, Nogueira MFG, Olsaker I, Pant SD, Panzitta F, Pastor RCP, Poli MA, Poslusny N, Rachagani S, Ranganathan S, Razpet A, Riggs PK, Rincon G, Rodriguez-Osorio N, Rodriguez-Zas SL, Romero NE, Rosenwald A, Sando L, Schmutz SM, Shen L, Sherman L, Southey BR, Lutzow YS, Sweedler JV, Tammen I, Telugu BPVL, Urbanski JM, Utsunomiya YT, Verschoor CP, Waardenberg AJ, Wang Z, Ward R, Weikard R, Welsh TH, White SN, Wilming LG, Wunderlich KR, Yang J, Zhao F-Q. The Genome Sequence of Taurine Cattle: A Window to Ruminant Biology and Evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y, Hanukoglu I, Edelheit O, Vaknine H, Hanukoglu A. Epithelial sodium channels (ENaC) are uniformly distributed on motile cilia in the oviduct and the respiratory airways. Histochem Cell Biol. 2012;137:339–353. doi: 10.1007/s00418-011-0904-1. [DOI] [PubMed] [Google Scholar]
- Fajac I, Viel M, Sublemontier S, Hubert D, Bienvenu T. Could a defective epithelial sodium channel lead to bronchiectasis. Respir. Res. 2008;9:46. doi: 10.1186/1465-9921-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findling JW, Raff H, Hansson JH, Lifton RP. Liddle’s syndrome: prospective genetic screening and suppressed aldosterone secretion in an extended kindred. J Clin Endocrinol Metab. 1997;82:1071–1074. doi: 10.1210/jcem.82.4.3862. [DOI] [PubMed] [Google Scholar]
- Firsov D, Robert-Nicoud M, Gruender S, Schild L, Rossier BC. Mutational analysis of cysteine-rich domains of the epithelium sodium channel (ENaC). Identification of cysteines essential for channel expression at the cell surface. J Biol Chem. 1999;274:2743–2749. doi: 10.1074/jbc.274.5.2743. [DOI] [PubMed] [Google Scholar]
- Fischbarg J. Fluid transport across leaky epithelia: central role of the tight junction and supporting role of aquaporins. Physiol Rev. 2010;90:1271–1290. doi: 10.1152/physrev.00025.2009. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Forslund K, Sonnhammer ELL. Evolution of protein domain architectures. Methods Mol Biol. 2012;856:187–216. doi: 10.1007/978-1-61779-585-5_8. [DOI] [PubMed] [Google Scholar]
- Foxx-Lupo WT, Wheatley CM, Baker SE, Cassuto NA, Delamere NA, Snyder EM. Genetic variation of the alpha subunit of the epithelial Na+ channel influences exhaled Na+ in healthy humans. Respir Physiol Neurobiol. 2011;179:205–211. doi: 10.1016/j.resp.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M, Kitamura K, Adachi M, Miyoshi T, Wakida N, Ura N, Shikano Y, Shinshi Y, Sakamoto K, Hayashi M, Satoh N, Nishitani T, Tomita K, Shimamoto K. Liddle’s syndrome caused by a novel mutation in the proline-rich PY motif of the epithelial sodium channel beta-subunit. J Clin Endocrinol Metab. 2005;90:340–344. doi: 10.1210/jc.2004-1027. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Miyawaki Y, Abe G. Molecular cloning and functional characterization of the Aplysia FMRFamide-gated Na+ channel. Pflugers Arch. 2006;451:646–656. doi: 10.1007/s00424-005-1498-z. [DOI] [PubMed] [Google Scholar]
- García-Añoveros J, Derfler B, Neville-Golden J, Hyman BT, Corey DP. BNaC1 and BNaC2 constitute a new family of human neuronal sodium channels related to degenerins and epithelial sodium channels. Proc Natl Acad Sci U S A. 1997;94:1459–1464. doi: 10.1073/pnas.94.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garty H, Palmer LG. Epithelial sodium channels: function, structure, and regulation. Physiol Rev. 1997;77:359–396. doi: 10.1152/physrev.1997.77.2.359. [DOI] [PubMed] [Google Scholar]
- Geller DS, Rodriguez-Soriano J, Vallo Boado A, Schifter S, Bayer M, Chang SS, Lifton RP. Mutations in the mineralocorticoid receptor gene cause autosomal dominant pseudohypoaldosteronism type I. Nat Genet. 1998;19:279–281. doi: 10.1038/966. [DOI] [PubMed] [Google Scholar]
- Giraldez T, Afonso-Oramas D, Cruz-Muros I, Garcia-Marin V, Pagel P, González-Hernández T, Alvarez de la Rosa D. Cloning and functional expression of a new epithelial sodium channel delta subunit isoform differentially expressed in neurons of the human and monkey telencephalon. J. Neurochem. 2007;102:1304–1315. doi: 10.1111/j.1471-4159.2007.04622.x. [DOI] [PubMed] [Google Scholar]
- Giraldez T, Rojas P, Jou J, Flores C, Alvarez de la Rosa D. The epithelial sodium channel δ-subunit: new notes for an old song. Am J Physiol Renal Physiol. 2012;303:F328–F338. doi: 10.1152/ajprenal.00116.2012. [DOI] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Ernstrom GG, Chelur DS, O’Hagan R, Yao CA, Chalfie M. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, St. John JA, Capella-Gutierrez S, Castoe TA, Kern C, Fujita MK, Opazo JC, Jurka J, Kojima KK, Caballero J, Hubley RM, Smit AF, Platt RN, Lavoie CA, Ramakodi MP, Finger JW, Suh A, Isberg SR, Miles L, Chong AY, Jaratlerdsiri W, Gongora J, Moran C, Iriarte A, McCormack J, Burgess SC, Edwards SV, Lyons E, Williams C, Breen M, Howard JT, Gresham CR, Peterson DG, Schmitz J, Pollock DD, Haussler D, Triplett EW, Zhang G, Irie N, Jarvis ED, Brochu CA, Schmidt CJ, McCarthy FM, Faircloth BC, Hoffmann FG, Glenn TC, Gabaldon T, Paten B, Ray DA. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science. 2014;346:1254449–1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig ER, Mathialahan T, Boot-Handford RP, Sandle GI. Molecular and functional studies of electrogenic Na(+) transport in the distal colon and rectum of young and elderly subjects. Gut. 2003;52:1607–1615. doi: 10.1136/gut.52.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründer S, Assmann M. Peptide-gated ion channels and the simple nervous system of Hydra. J Exp Biol. 2015;218:551–561. doi: 10.1242/jeb.111666. [DOI] [PubMed] [Google Scholar]
- Gründer S, Firsov D, Chang SS, Jaeger NF, Gautschi I, Schild L, Lifton RP, Rossier BC. A mutation causing pseudohypoaldosteronism type 1 identifies a conserved glycine that is involved in the gating of the epithelial sodium channel. EMBO J. 1997;16:899–907. doi: 10.1093/emboj/16.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründer S, Jaeger NF, Gautschi I, Schild L, Rossier BC. Identification of a highly conserved sequence at the N-terminus of the epithelial Na+ channel alpha subunit involved in gating. Pflugers Arch. 1999;438:709–715. doi: 10.1007/s004249900119. [DOI] [PubMed] [Google Scholar]
- Gründer S, Pusch M. Biophysical properties of acid-sensing ion channels (ASICs) Neuropharmacology. 2015 doi: 10.1016/j.neuropharm.2014.12.016. [DOI] [PubMed] [Google Scholar]
- Gu Y. Effects of [Ca2+]i and pH on epithelial Na+ channel activity of cultured mouse cortical collecting ducts. J Exp Biol. 2008;211:3167–3173. doi: 10.1242/jeb.019646. [DOI] [PubMed] [Google Scholar]
- Gwiazda K, Bonifacio G, Vullo S, Kellenberger S. Extracellular Subunit Interactions Control Transitions Between Functional States of Acid-sensing Ion Channel 1a. J Biol Chem. 2015 doi: 10.1074/jbc.M115.641688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager H, Kwon TH, Vinnikova AK, Masilamani S, Brooks HL, Frøkiaer J, Knepper MA, Nielsen S. Immunocytochemical and immunoelectron microscopic localization of alpha-, beta-, and gamma-ENaC in rat kidney. Am J Physiol Renal Physiol. 2001;280:F1093–F1106. doi: 10.1152/ajprenal.2001.280.6.F1093. [DOI] [PubMed] [Google Scholar]
- Hannila-Handelberg T, Kontula K, Tikkanen I, Tikkanen T, Fyhrquist F, Helin K, Fodstad H, Piippo K, Miettinen HE, Virtamo J, Krusius T, Sarna S, Gautschi I, Schild L, Hiltunen TP. Common variants of the beta and gamma subunits of the epithelial sodium channel and their relation to plasma renin and aldosterone levels in essential hypertension. BMC Med Genet, BMC medical genetics. 2005;6:4. doi: 10.1186/1471-2350-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- Hanukoglu A. Type I pseudohypoaldosteronism includes two clinically and genetically distinct entities with either renal or multiple target organ defects. J Clin Endocrinol Metab. 1991;73:936–944. doi: 10.1210/jcem-73-5-936. [DOI] [PubMed] [Google Scholar]
- Hanukoglu A, Bistritzer T, Rakover Y, Mandelberg A. Pseudohypoaldosteronism with increased sweat and saliva electrolyte values and frequent lower respiratory tract infections mimicking cystic fibrosis. J Pediatr. 1994;125:752–755. doi: 10.1016/s0022-3476(94)70071-0. [DOI] [PubMed] [Google Scholar]
- Hanukoglu A, Edelheit O, Shriki Y, Gizewska M, Dascal N, Hanukoglu I. Renin-aldosterone response, urinary Na/K ratio and growth in pseudohypoaldosteronism patients with mutations in epithelial sodium channel (ENaC) subunit genes. J Steroid Biochem Mol Biol. 2008;111:268–274. doi: 10.1016/j.jsbmb.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Hanukoglu A, Hanukoglu I. Clinical improvement in patients with autosomal recessive pseudohypoaldosteronism and the necessity for salt supplementation. Clin Exp Nephrol. 2010;14:518–519. doi: 10.1007/s10157-010-0326-8. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B, Dickson EJ, Kruse M, Vivas O, Suh B-C. Phosphoinositides regulate ion channels. Biochim Biophys Acta. 2015;1851:844–856. doi: 10.1016/j.bbalip.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen TP, Hannila-Handelberg T, Petäjäniemi N, Kantola I, Tikkanen I, Virtamo J, Gautschi I, Schild L, Kontula K. Liddle’s syndrome associated with a point mutation in the extracellular domain of the epithelial sodium channel gamma subunit. J Hypertens. 2002;20:2383–2390. doi: 10.1097/00004872-200212000-00017. [DOI] [PubMed] [Google Scholar]
- Hobbs CA, Da Tan C, Tarran R. Does epithelial sodium channel hyperactivity contribute to cystic fibrosis lung disease? J Physiol. 2013;591:4377–4387. doi: 10.1113/jphysiol.2012.240861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg RJ, Marks JF, Marver D, Frolich JC. Long term observations in a patient with pseudohypoaldosteronism. Pediatr Nephrol. 1991;5:205–210. doi: 10.1007/BF01095953. [DOI] [PubMed] [Google Scholar]
- Hollenhorst MI, Richter K, Fronius M. Ion transport by pulmonary epithelia. J. Biomed. Biotechnol. 2011;2011:174306. doi: 10.1155/2011/174306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollier JM, Martin DF, Bell DM, Li J-L, Chirachanchai MG, Menon DV, Leonard D, Wu X, Cooper RS, McKenzie C, Victor RG, Auchus RJ. Epithelial sodium channel allele T594M is not associated with blood pressure or blood pressure response to amiloride. Hypertension. 2006;47:428–433. doi: 10.1161/01.HYP.0000200704.45994.ff. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JMC, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin J-J, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O’Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Holzer P. Acid-sensing ion channels in gastrointestinal function. Neuropharmacology. 2015;94:72–79. doi: 10.1016/j.neuropharm.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Park S, Jiménez RHF, Rinehart D, Tamm LK. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J. Am. Chem. Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- Hubé F, Francastel C. Mammalian introns: when the junk generates molecular diversity. Int. J. Mol. Sci. 2015;16:4429–4452. doi: 10.3390/ijms16034429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey RP, Bruns JB, Kinlough CL, Harkleroad KL, Tong Q, Carattino MD, Johnson JP, Stockand JD, Kleyman TR. Epithelial sodium channels are activated by furin-dependent proteolysis. J Biol Chem. 2004;279:18111–18114. doi: 10.1074/jbc.C400080200. [DOI] [PubMed] [Google Scholar]
- Hummler E, Barker P, Gatzy J, Beermann F, Verdumo C, Schmidt A, Boucher R, Rossier BC. Early death due to defective neonatal lung liquid clearance in alpha-ENaC-deficient mice. Nat Genet. 1996;12:325–328. doi: 10.1038/ng0396-325. [DOI] [PubMed] [Google Scholar]
- Ilatovskaya DV, Pavlov TS, Neguliyaev YA, Starushchenko A. Mechanisms of epithelial sodium channel (ENaC) regulation by cortactin: involvement of dynamin. Cell Tissue Biol. 2012;6:52–59. [PubMed] [Google Scholar]
- Ishikita H. Proton-binding sites of acid-sensing ion channel 1. PLoS One. 2011;6:e16920. doi: 10.1371/journal.pone.0016920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- Jex AR, Nejsum P, Schwarz EM, Hu L, Young ND, Hall RS, Korhonen PK, Liao S, Thamsborg S, Xia J, Xu P, Wang S, Scheerlinck J-PY, Hofmann A, Sternberg PW, Wang J, Gasser RB. Genome and transcriptome of the porcine whipworm Trichuris suis. Nat Genet. 2014;46:701–706. doi: 10.1038/ng.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H-L, Zhao R-Z, Chen Z-X, Shetty S, Idell S, Matalon S. δ ENaC: a novel divergent amiloride-inhibitable sodium channel. Am J Physiol Lung Cell Mol Physiol. 2012;303:L1013–L1026. doi: 10.1152/ajplung.00206.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji HL, Chalfant ML, Jovov B, Lockhart JP, Parker SB, Fuller CM, Stanton BA, Benos DJ. The cytosolic termini of the beta- and gamma-ENaC subunits are involved in the functional interactions between cystic fibrosis transmembrane conductance regulator and epithelial sodium channel. J Biol Chem. 2000;275:27947–27956. doi: 10.1074/jbc.M002848200. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xu J, Chen Y, Shi J, Zhang C, Li J. Expression and distribution of epithelial sodium channel in nasal polyp and nasal mucosa. Eur Arch Otorhinolaryngol. 2015 doi: 10.1007/s00405-014-3477-5. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kashlan OB, Adelman JL, Okumura S, Blobner BM, Zuzek Z, Hughey RP, Kleyman TR, Grabe M. Constraint-based, homology model of the extracellular domain of the epithelial Na+ channel alpha subunit reveals a mechanism of channel activation by proteases. J Biol Chem, The Journal of biological chemistry. 2011;286:649–660. doi: 10.1074/jbc.M110.167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Blobner BM, Zuzek Z, Tolino M, Kleyman TR. Na+ inhibits the epithelial Na+ channel by binding to a site in an extracellular acidic cleft. J Biol Chem. 2015;290:568–576. doi: 10.1074/jbc.M114.606152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Boyd CR, Argyropoulos C, Okumura S, Hughey RP, Grabe M, Kleyman TR. Allosteric inhibition of the epithelial Na+ channel through peptide binding at peripheral finger and thumb domains. J Biol Chem. 2010;285:35216–35223. doi: 10.1074/jbc.M110.167064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Kleyman TR. Epithelial Na+ channel regulation by cytoplasmic and extracellular factors. Exp Cell Res. 2012;318:1011–1019. doi: 10.1016/j.yexcr.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Kleyman TR. ENaC structure and function in the wake of a resolved structure of a family member. Am J Physiol Renal Physiol. 2011;301:F684–F696. doi: 10.1152/ajprenal.00259.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashlan OB, Maarouf AB, Kussius C, Denshaw RM, Blumenthal KM, Kleyman TR. Distinct structural elements in the first membrane-spanning segment of the epithelial sodium channel. J Biol Chem. 2006;281:30455–30462. doi: 10.1074/jbc.M604615200. [DOI] [PubMed] [Google Scholar]
- Kashlan OB, Sheng S, Kleyman TR. On the interaction between amiloride and its putative alpha-subunit epithelial Na+ channel binding site. J Biol Chem. 2005;280:26206–26215. doi: 10.1074/jbc.M503500200. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Weiss KM. Evolutionary genetics of vertebrate tissue mineralization: the origin and evolution of the secretory calcium-binding phosphoprotein family. J. Exp. Zool. B. Mol. Dev. Evol. 2006;306:295–316. doi: 10.1002/jez.b.21088. [DOI] [PubMed] [Google Scholar]
- Kellenberger S, Gautschi I, Schild L. An external site controls closing of the epithelial Na+ channel ENaC. J Physiol. 2002;543:413–424. doi: 10.1113/jphysiol.2002.022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S, Gautschi I, Schild L. A single point mutation in the pore region of the epithelial Na+ channel changes ion selectivity by modifying molecular sieving. Proc Natl Acad Sci U S A. 1999;96:4170–4175. doi: 10.1073/pnas.96.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S, Schild L. International union of basic and clinical pharmacology. XCI. structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol. Rev. 2015;67:1–35. doi: 10.1124/pr.114.009225. [DOI] [PubMed] [Google Scholar]
- Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- Kerem E, Bistritzer T, Hanukoglu A, Hofmann T, Zhou Z, Bennett W, MacLaughlin E, Barker P, Nash M, Quittell L, Boucher R, Knowles MR. Pulmonary epithelial sodium-channel dysfunction and excess airway liquid in pseudohypoaldosteronism. N Engl J Med. 1999;341:156–162. doi: 10.1056/NEJM199907153410304. [DOI] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Murphy T, Xia J, Caragea D, Park Y, Beeman RW, Lorenzen MD, Butcher S, Manak JR, Brown SJ. BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 2009;38:D437–D442. doi: 10.1093/nar/gkp807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyman TR, Carattino MD, Hughey RP. ENaC at the cutting edge: regulation of epithelial sodium channels by proteases. J Biol Chem. 2009;284:20447–20451. doi: 10.1074/jbc.R800083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyman TR, Cragoe EJ. Amiloride and its analogs as tools in the study of ion transport. J Membr Biol. 1988;105:1–21. doi: 10.1007/BF01871102. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Krauson AJ, Rued AC, Carattino MD. Independent Contribution of Extracellular Proton Binding Sites to ASIC1a Activation. J Biol Chem. 2013;288:34375–34383. doi: 10.1074/jbc.M113.504324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz O, Barbry P, Bock R, Lindemann B. Differential expression of RNA and protein of the three pore-forming subunits of the amiloride-sensitive epithelial sodium channel in taste buds of the rat. J Histochem Cytochem. 1999;47:51–64. doi: 10.1177/002215549904700106. [DOI] [PubMed] [Google Scholar]
- Krueger B, Schlötzer-Schrehardt U, Haerteis S, Zenkel M, Chankiewitz VE, Amann KU, Kruse FE, Korbmacher C. Four subunits (αβγδ) of the epithelial sodium channel (ENaC) are expressed in the human eye in various locations. Invest. Ophthalmol. Vis. Sci. 2012;53:596–604. doi: 10.1167/iovs.11-8581. [DOI] [PubMed] [Google Scholar]
- Kunzelmann K, Bachhuber T, Regeer R, Markovich D, Sun J, Schreiber R. Purinergic inhibition of the epithelial Na+ transport via hydrolysis of PIP2. FASEB J. 2005;19:142–143. doi: 10.1096/fj.04-2314fje. [DOI] [PubMed] [Google Scholar]
- Kuratani S. Evolution of the vertebrate jaw from developmental perspectives. Evol. Dev. 2012;14:76–92. doi: 10.1111/j.1525-142X.2011.00523.x. [DOI] [PubMed] [Google Scholar]
- Kusche-Vihrog K, Tarjus A, Fels J, Jaisser F. The epithelial Na+ channel: a new player in the vasculature. Curr Opin Nephrol Hypertens. 2014;23:143–148. doi: 10.1097/01.mnh.0000441054.88962.2c. [DOI] [PubMed] [Google Scholar]
- Langloh AL, Berdiev B, Ji HL, Keyser K, Stanton BA, Benos DJ. Charged residues in the M2 region of alpha-hENaC play a role in channel conductance. Am J Physiol Cell Physiol. 2000;278:C277–C291. doi: 10.1152/ajpcell.2000.278.2.C277. [DOI] [PubMed] [Google Scholar]
- Lefèvre CMT, Diakov A, Haerteis S, Korbmacher C, Gründer S, Wiemuth D. Pharmacological and electrophysiological characterization of the human bile acid-sensitive ion channel (hBASIC) Pflugers Arch. 2014;466:253–263. doi: 10.1007/s00424-013-1310-4. [DOI] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. Outlines of the pore in open and closed conformations describe the gating mechanism of ASIC1. Nat. Commun. 2011;2:399. doi: 10.1038/ncomms1409. [DOI] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. Asn415 in the beta11-beta12 linker decreases proton-dependent desensitization of ASIC1. J Biol Chem. 2010;285:31285–31291. doi: 10.1074/jbc.M110.160382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddle GW, Bledsoe T, Coppage WSJ. A familial renal disorder simulating primary aldosteronism but with negligible aldosterone secretion. Trans. Assoc. Am. Phys. 1963;76:199–213. [Google Scholar]
- Lin S-H, Sun W-H, Chen C-C. Genetic exploration of the role of acid-sensing ion channels. Neuropharmacology. 2015 doi: 10.1016/j.neuropharm.2014.12.011. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin C-W, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli K-P, Parker HG, Pollinger JP, Searle SMJ, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger J-P, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O’Neill B, O’Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lingueglia E, Deval E, Lazdunski M. FMRFamide-gated sodium channel and ASIC channels: a new class of ionotropic receptors for FMRFamide and related peptides. Peptides. 2006;27:1138–1152. doi: 10.1016/j.peptides.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Lingueglia E, Voilley N, Waldmann R, Lazdunski M, Barbry P. Expression cloning of an epithelial amiloride-sensitive Na+ channel. A new channel type with homologies to Caenorhabditis elegans degenerins. FEBS Lett. 1993;318:95–99. doi: 10.1016/0014-5793(93)81336-x. [DOI] [PubMed] [Google Scholar]
- Liu F, Yang X, Mo X, Huang J, Chen J, Kelly TN, Hixson JE, Rao DC, Gu CC, Shimmin LC, Rice TK, Li J, Schwander K, He J, Liu D-P, Gu D. Associations of epithelial sodium channel genes with blood pressure: the GenSalt study. J Hum Hypertens. 2015;29:224–228. doi: 10.1038/jhh.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang S-P, Wang Z, Chinwalla AT, Minx P, Mitreva M, Cook L, Delehaunty KD, Fronick C, Schmidt H, Fulton LA, Fulton RS, Nelson JO, Magrini V, Pohl C, Graves TA, Markovic C, Cree A, Dinh HH, Hume J, Kovar CL, Fowler GR, Lunter G, Meader S, Heger A, Ponting CP, Marques-Bonet T, Alkan C, Chen L, Cheng Z, Kidd JM, Eichler EE, White S, Searle S, Vilella AJ, Chen Y, Flicek P, Ma J, Raney B, Suh B, Burhans R, Herrero J, Haussler D, Faria R, Fernando O, Darré F, Farré D, Gazave E, Oliva M, Navarro A, Roberto R, Capozzi O, Archidiacono N, Della Valle G, Purgato S, Rocchi M, Konkel MK, Walker JA, Ullmer B, Batzer MA, Smit AFA, Hubley R, Casola C, Schrider DR, Hahn MW, Quesada V, Puente XS, Ordoñez GR, López-Otín C, Vinar T, Brejova B, Ratan A, Harris RS, Miller W, Kosiol C, Lawson HA, Taliwal V, Martins AL, Siepel A, Roychoudhury A, Ma X, Degenhardt J, Bustamante CD, Gutenkunst RN, Mailund T, Dutheil JY, Hobolth A, Schierup MH, Ryder OA, Yoshinaga Y, de Jong PJ, Weinstock GM, Rogers J, Mardis ER, Gibbs RA, Wilson RK. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Pribanic S, Debonneville A, Jiang C, Rotin D. The PY motif of ENaC, mutated in Liddle syndrome, regulates channel internalization, sorting and mobilization from subapical pool. Traffic. 2007;8:1246–1264. doi: 10.1111/j.1600-0854.2007.00602.x. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Bolkenius U, Wickert L, Marynen P, Bidlingmaier F. Structural organisation of the gene encoding the alpha-subunit of the human amiloride-sensitive epithelial sodium channel. Hum Genet. 1998;102:576–581. doi: 10.1007/s004390050743. [DOI] [PubMed] [Google Scholar]
- Lyons RA, Saridogan E, Djahanbakhch O. The reproductive significance of human Fallopian tube cilia. Hum Reprod Update. 2006;12:363–372. doi: 10.1093/humupd/dml012. [DOI] [PubMed] [Google Scholar]
- Ma B-G, Chen L, Ji H-F, Chen Z-H, Yang F-R, Wang L, Qu G, Jiang Y-Y, Ji C, Zhang H-Y. Characters of very ancient proteins. Biochem Biophys Res Commun. 2008;366:607–611. doi: 10.1016/j.bbrc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- Mall M, Grubb BR, Harkema JR, O’Neal WK, Boucher RC. Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice. Nat Med. 2004;10:487–493. doi: 10.1038/nm1028. [DOI] [PubMed] [Google Scholar]
- Mall MA, Button B, Johannesson B, Zhou Z, Livraghi A, Caldwell RA, Schubert SC, Schultz C, O’Neal WK, Pradervand S, Hummler E, Rossier BC, Grubb BR, Boucher RC. Airway surface liquid volume regulation determines different airway phenotypes in liddle compared with betaENaC-overexpressing mice. J Biol Chem. 2010;285:26945–26955. doi: 10.1074/jbc.M110.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinerie L, Pussard E, Foix-L’Hélias L, Petit F, Cosson C, Boileau P, Lombès M. Physiological partial aldosterone resistance in human newborns. Pediatr Res. 2009;66:323–328. doi: 10.1203/PDR.0b013e3181b1bbec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masilamani S, Kim GH, Mitchell C, Wade JB, Knepper MA. Aldosterone-mediated regulation of ENaC alpha, beta, and gamma subunit proteins in rat kidney. J Clin Invest, The Journal of clinical investigation. 1999;104:R19–R23. doi: 10.1172/JCI7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzochi C, Bubien JK, Smith PR, Benos DJ. The carboxyl terminus of the alpha-subunit of the amiloride-sensitive epithelial sodium channel binds to F-actin. J Biol Chem. 2006;281:6528–6538. doi: 10.1074/jbc.M509386200. [DOI] [PubMed] [Google Scholar]
- McDonald FJ, Price MP, Snyder PM, Welsh MJ. Cloning and expression of the beta- and gamma-subunits of the human epithelial sodium channel. Am J Physiol. 1995;268:C1157–C1163. doi: 10.1152/ajpcell.1995.268.5.C1157. [DOI] [PubMed] [Google Scholar]
- McDonald FJ, Snyder PM, McCray PB, Welsh MJ. Cloning, expression, and tissue distribution of a human amiloride-sensitive Na+ channel. Am J Physiol. 1994;266:L728–L734. doi: 10.1152/ajplung.1994.266.6.L728. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, Jurka J, Kamal M, Mauceli E, Searle SMJ, Sharpe T, Baker ML, Batzer MA, Benos PV, Belov K, Clamp M, Cook A, Cuff J, Das R, Davidow L, Deakin JE, Fazzari MJ, Glass JL, Grabherr M, Greally JM, Gu W, Hore TA, Huttley GA, Kleber M, Jirtle RL, Koina E, Lee JT, Mahony S, Marra MA, Miller RD, Nicholls RD, Oda M, Papenfuss AT, Parra ZE, Pollock DD, Ray DA, Schein JE, Speed TP, Thompson K, VandeBerg JL, Wade CM, Walker JA, Waters PD, Webber C, Weidman JR, Xie X, Zody MC, Graves JAM, Ponting CP, Breen M, Samollow PB, Lander ES, Lindblad-Toh K. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Miller RL, Loewy AD. ENaC γ-expressing astrocytes in the circumventricular organs, white matter, and ventral medullary surface: sites for Na+ regulation by glial cells. J. Chem. Neuroanat. 2013;53:72–80. doi: 10.1016/j.jchemneu.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B, Knight J, Qi J, Zhao F, Wang Q, Bedoya-Reina OC, Katiyar N, Tomsho LP, Kasson LM, Hardie R-A, Woodbridge P, Tindall EA, Bertelsen MF, Dixon D, Pyecroft S, Helgen KM, Lesk AM, Pringle TH, Patterson N, Zhang Y, Kreiss A, Woods GM, Jones ME, Schuster SC. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil) Proc Natl Acad Sci U S A. 2011;108:12348–12353. doi: 10.1073/pnas.1102838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. How many species are there on Earth and in the ocean? PLoS Biol. 2011;9:e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris LM, DeGagne JM, Kempton JB, Hausman F, Trune DR. Mouse middle ear ion homeostasis channels and intercellular junctions. PLoS One. 2012;7:e39004. doi: 10.1371/journal.pone.0039004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Mutesa L, Azad AK, Verhaeghe C, Segers K, Vanbellinghen JF, Ngendahayo L, Rusingiza EK, Mutwa PR, Rulisa S, Koulischer L, Cassiman JJ, Cuppens H, Bours V. Genetic analysis of Rwandan patients with cystic fibrosis-like symptoms: identification of novel cystic fibrosis transmembrane conductance regulator and epithelial sodium channel gene variants. Chest. 2009;135:1233–1242. doi: 10.1378/chest.08-2246. [DOI] [PubMed] [Google Scholar]
- Nagel G, Barbry P, Chabot H, Brochiero E, Hartung K, Grygorczyk R. CFTR fails to inhibit the epithelial sodium channel ENaC expressed in Xenopus laevis oocytes. J Physiol. 2005;564:671–682. doi: 10.1113/jphysiol.2004.079046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Deerfield DWII. GeneDoc: Analysis and visualization of genetic variation. EMBnew.News. 1997;4:14. [Google Scholar]
- Oka Y, Butnaru M, von Buchholtz L, Ryba NJP, Zuker CS. High salt recruits aversive taste pathways. Nature. 2013;494:472–475. doi: 10.1038/nature11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- Omerbašić D, Schuhmacher L-N, Bernal Sierra Y-A, Smith ESJ, Lewin GR. ASICs and mammalian mechanoreceptor function. Neuropharmacology. 2014 doi: 10.1016/j.neuropharm.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Ottaviani E, Franchini A, Mandrioli M, Saxena A, Hanukoglu A, Hanukoglu I. Amiloride-sensitive epithelial sodium channel subunits are expressed in human and mussel immunocytes. Dev. Comp. Immunol. 2002;26:395–402. doi: 10.1016/s0145-305x(01)00097-0. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and Molecular Aspects of Hypertension. Circ Res. 2015;116:937–959. doi: 10.1161/CIRCRESAHA.116.303647. [DOI] [PubMed] [Google Scholar]
- Passero CJ, Carattino MD, Kashlan OB, Myerburg MM, Hughey RP, Kleyman TR. Defining an inhibitory domain in the gamma subunit of the epithelial sodium channel. Am J Physiol Renal Physiol, American journal of physiology. Renal physiology. 2010;299:F854–F861. doi: 10.1152/ajprenal.00316.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passero CJ, Mueller GM, Myerburg MM, Carattino MD, Hughey RP, Kleyman TR. TMPRSS4-dependent activation of the epithelial sodium channel requires cleavage of the gamma subunit distal to the furin cleavage site. Am J Physiol Renal Physiol, American journal of physiology. Renal physiology. 2011 doi: 10.1152/ajprenal.00330.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AB, Chao J, Palmer LG. Tissue kallikrein activation of the epithelial Na channel. Am J Physiol Renal Physiol. 2012;303:F540–F550. doi: 10.1152/ajprenal.00133.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak BG, Shaughnessy JD, Meneton P, Greeb J, Shull GE, Jenkins NA, Copeland NG. Mouse chromosomal location of three epithelial sodium channel subunit genes and an apical sodium chloride cotransporter gene. Genomics. 1996;33:124–127. doi: 10.1006/geno.1996.0168. [DOI] [PubMed] [Google Scholar]
- Persu A, Barbry P, Bassilana F, Houot AM, Mengual R, Lazdunski M, Corvol P, Jeunemaitre X. Genetic analysis of the beta subunit of the epithelial Na+ channel in essential hypertension. Hypertension. 1998;32:129–137. doi: 10.1161/01.hyp.32.1.129. [DOI] [PubMed] [Google Scholar]
- Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, McMurry J, Marioni JC, Malone J, Megy K, Rustici G, Tang AY, Taubert J, Williams E, Mannion O, Parkinson HE, Brazma A. Expression Atlas update--a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochynyuk O, Bugaj V, Stockand JD. Physiologic regulation of the epithelial sodium channel by phosphatidylinositides. Curr Opin Nephrol Hypertens. 2008;17:533–540. doi: 10.1097/MNH.0b013e328308fff3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochynyuk O, Staruschenko A, Tong Q, Medina J, Stockand JD. Identification of a functional phosphatidylinositol 3,4,5-trisphosphate binding site in the epithelial Na+ channel. J Biol Chem. 2005;280:37565–37571. doi: 10.1074/jbc.M509071200. [DOI] [PubMed] [Google Scholar]
- Pochynyuk O, Tong Q, Medina J, Vandewalle A, Staruschenko A, Bugaj V, Stockand JD. Molecular determinants of PI(4,5)P2 and PI(3,4,5)P3 regulation of the epithelial Na+ channel. J Gen Physiol. 2007;130:399–413. doi: 10.1085/jgp.200709800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogozheva ID, Mosberg HI, Lomize AL. Life at the border: adaptation of proteins to anisotropic membrane environment. Protein Sci. 2014;23:1165–1196. doi: 10.1002/pro.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, Schäffer AA, Agarwala R, Narfström K, Murphy WJ, Giger U, Roca AL, Antunes A, Menotti-Raymond M, Yuhki N, Pecon-Slattery J, Johnson WE, Bourque G, Tesler G, O’Brien SJ. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J, Kershaw M, Kirk J, Trevelyan N, Shaw NJ. The use of sodium resonium in pseudohypoaldosteronism. Arch Dis Child. 2003;88:1138–1139. doi: 10.1136/adc.88.12.1138-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MP, Lewin GR, McIlwrath SL, Cheng C, Xie J, Heppenstall PA, Stucky CL, Mannsfeldt AG, Brennan TJ, Drummond HA, Qiao J, Benson CJ, Tarr DE, Hrstka RF, Yang B, Williamson RA, Welsh MJ. The mammalian sodium channel BNC1 is required for normal touch sensation. Nature. 2000;407:1007–1011. doi: 10.1038/35039512. [DOI] [PubMed] [Google Scholar]
- Price MP, McIlwrath SL, Xie J, Cheng C, Qiao J, Tarr DE, Sluka KA, Brennan TJ, Lewin GR, Welsh MJ. The DRASIC cation channel contributes to the detection of cutaneous touch and acid stimuli in mice. Neuron. 2001;32:1071–1083. doi: 10.1016/s0896-6273(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Protasio AV, Tsai IJ, Babbage A, Nichol S, Hunt M, Aslett MA, De Silva N, Velarde GS, Anderson TJC, Clark RC, Davidson C, Dillon GP, Holroyd NE, LoVerde PT, Lloyd C, McQuillan J, Oliveira G, Otto TD, Parker-Manuel SJ, Quail MA, Wilson RA, Zerlotini A, Dunne DW, Berriman M. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl. Trop. Dis. 2012;6:e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PWH, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Ramos MD, Trujillano D, Olivar R, Sotillo F, Ossowski S, Manzanares J, Costa J, Gartner S, Oliva C, Quintana E, Gonzalez MI, Vazquez C, Estivill X, Casals T. Extensive sequence analysis of CFTR, SCNN1A, SCNN1B, SCNN1G and SERPINA1 suggests an oligogenic basis for cystic fibrosis-like phenotypes. Clin. Genet. 2014;86:91–95. doi: 10.1111/cge.12234. [DOI] [PubMed] [Google Scholar]
- Rauh R, Soell D, Haerteis S, Diakov A, Nesterov V, Krueger B, Sticht H, Korbmacher C. A mutation in the β-subunit of ENaC identified in a patient with cystic fibrosis-like symptoms has a gain-of-function effect. Am J Physiol Lung Cell Mol Physiol. 2013;304:L43–L55. doi: 10.1152/ajplung.00093.2012. [DOI] [PubMed] [Google Scholar]
- Rayner BL, Owen EP, King JA, Soule SG, Vreede H, Opie LH, Marais D, Davidson JS. A new mutation, R563Q, of the beta subunit of the epithelial sodium channel associated with low-renin, low-aldosterone hypertension. J Hypertens. 2003;21:921–926. doi: 10.1097/00004872-200305000-00016. [DOI] [PubMed] [Google Scholar]
- Reddy MM, Stutts MJ. Status of fluid and electrolyte absorption in cystic fibrosis. Cold Spring Harb. Perspect. Med. 2013;3:a009555. doi: 10.1101/cshperspect.a009555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Shaw G, Wong ESW, Lefèvre CM, Nicholas KR, Kuroki Y, Wakefield MJ, Zenger KR, Wang C, Ferguson-Smith M, Nicholas FW, Hickford D, Yu H, Short KR, Siddle HV, Frankenberg SR, Chew KY, Menzies BR, Stringer JM, Suzuki S, Hore TA, Delbridge ML, Patel HR, Mohammadi A, Schneider NY, Hu Y, O’Hara W, Al Nadaf S, Wu C, Feng Z-P, Cocks BG, Wang J, Flicek P, Searle SMJ, Fairley S, Beal K, Herrero J, Carone DM, Suzuki Y, Sugano S, Toyoda A, Sakaki Y, Kondo S, Nishida Y, Tatsumoto S, Mandiou I, Hsu A, McColl KA, Lansdell B, Weinstock G, Kuczek E, McGrath A, Wilson P, Men A, Hazar-Rethinam M, Hall A, Davis J, Wood D, Williams S, Sundaravadanam Y, Muzny DM, Jhangiani SN, Lewis LR, Morgan MB, Okwuonu GO, Ruiz SJ, Santibanez J, Nazareth L, Cree A, Fowler G, Kovar CL, Dinh HH, Joshi V, Jing C, Lara F, Thornton R, Chen L, Deng J, Liu Y, Shen JY, Song X-Z, Edson J, Troon C, Thomas D, Stephens A, Yapa L, Levchenko T, Gibbs RA, Cooper DW, Speed TP, Fujiyama A, Graves JAM, O’Neill RJ, Pask AJ, Forrest SM, Worley KC. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riepe FG, van Bemmelen MX, Cachat F, Plendl H, Gautschi I, Krone N, Holterhus PM, Theintz G, Schild L. Revealing a subclinical salt-losing phenotype in heterozygous carriers of the novel S562P mutation in the alpha subunit of the epithelial sodium channel. Clin Endocrinol (Oxf) 2009;70:252–258. doi: 10.1111/j.1365-2265.2008.03314.x. [DOI] [PubMed] [Google Scholar]
- Rogers DF. Physiology of airway mucus secretion and pathophysiology of hypersecretion. Respir. Care. 2007;52:1134–1146. discussion 1146–9. [PubMed] [Google Scholar]
- Rossier BC. Epithelial sodium channel (ENaC) and the control of blood pressure. Curr Opin Pharmacol. 2014;15:33–46. doi: 10.1016/j.coph.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Rossier BC, Baker ME, Studer RA. Epithelial sodium transport and its control by aldosterone: the story of our internal environment revisited. Physiol Rev. 2015;95:297–340. doi: 10.1152/physrev.00011.2014. [DOI] [PubMed] [Google Scholar]
- Rossier BC, Pradervand S, Schild L, Hummler E. Epithelial sodium channel and the control of sodium balance: interaction between genetic and environmental factors. Annu Rev Physiol. 2002;64:877–897. doi: 10.1146/annurev.physiol.64.082101.143243. [DOI] [PubMed] [Google Scholar]
- Rossier BC, Stutts MJ. Activation of the epithelial sodium channel (ENaC) by serine proteases. Annu Rev Physiol. 2009;71:361–379. doi: 10.1146/annurev.physiol.010908.163108. [DOI] [PubMed] [Google Scholar]
- Rotin D. Role of the UPS in Liddle syndrome. BMC Biochem. 2008;9(Suppl 1) doi: 10.1186/1471-2091-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Bar-Sagi D, O’Brodovich H, Merilainen J, Lehto VP, Canessa CM, Rossier BC, Downey GP. An SH3 binding region in the epithelial Na+ channel (alpha rENaC) mediates its localization at the apical membrane. EMBO J. 1994;13:4440–4450. doi: 10.1002/j.1460-2075.1994.tb06766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Staub O. Role of the ubiquitin system in regulating ion transport. Pflugers Arch. 2011;461:1–21. doi: 10.1007/s00424-010-0893-2. [DOI] [PubMed] [Google Scholar]
- Ruan YC, Chen H, Chan HC. Ion channels in the endometrium: regulation of endometrial receptivity and embryo implantation. Hum Reprod Update. 2014;20:517–529. doi: 10.1093/humupd/dmu006. [DOI] [PubMed] [Google Scholar]
- Ruth JL, Wassner SJ. Body composition: salt and water. Pediatr Rev. 2006;27:181–187. doi: 10.1542/pir.27-5-181. [DOI] [PubMed] [Google Scholar]
- Saar K, Beck A, Bihoreau M-T, Birney E, Brocklebank D, Chen Y, Cuppen E, Demonchy S, Dopazo J, Flicek P, Foglio M, Fujiyama A, Gut IG, Gauguier D, Guigo R, Guryev V, Heinig M, Hummel O, Jahn N, Klages S, Kren V, Kube M, Kuhl H, Kuramoto T, Kuroki Y, Lechner D, Lee Y-A, Lopez-Bigas N, Lathrop GM, Mashimo T, Medina I, Mott R, Patone G, Perrier-Cornet J-A, Platzer M, Pravenec M, Reinhardt R, Sakaki Y, Schilhabel M, Schulz H, Serikawa T, Shikhagaie M, Tatsumoto S, Taudien S, Toyoda A, Voigt B, Zelenika D, Zimdahl H, Hubner N. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet. 2008;40:560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salleh N, Baines DL, Naftalin RJ, Milligan SR. The hormonal control of uterine luminal fluid secretion and absorption. J Membr Biol, The Journal of membrane biology. 2005;206:17–28. doi: 10.1007/s00232-005-0770-7. [DOI] [PubMed] [Google Scholar]
- Samaha FFFF, Rubenstein RCRC, Yan W, Ramkumar M, Levy DIDI, Ahn YJYJ, Sheng S, Kleyman TRTR. Functional polymorphism in the carboxyl terminus of the alpha-subunit of the human epithelial sodium channel. J Biol Chem. 2004;279:23900–23907. doi: 10.1074/jbc.M401941200. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Yui N, Noda Y. Actin directly interacts with different membrane channel proteins and influences channel activities: AQP2 as a model. Biochim Biophys Acta. 2014;1838:514–520. doi: 10.1016/j.bbamem.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol, Annual review of physiology. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Saxena A, Hanukoglu I, Saxena D, Thompson RJ, Gardiner RM, Hanukoglu A. Novel mutations responsible for autosomal recessive multisystem pseudohypoaldosteronism and sequence variants in epithelial sodium channel alpha-, beta-, and gamma-subunit genes. J Clin Endocrinol Metab. 2002;87:3344–3350. doi: 10.1210/jcem.87.7.8674. [DOI] [PubMed] [Google Scholar]
- Saxena A, Hanukoglu I, Strautnieks SS, Thompson RJ, Gardiner RM, Hanukoglu A. Gene structure of the human amiloride-sensitive epithelial sodium channel beta subunit. Biochem Biophys Res Commun. 1998;252:208–213. doi: 10.1006/bbrc.1998.9625. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, McCarthy S, Montgomery SH, Schwalie PC, Tang YA, Ward MC, Xue Y, Yngvadottir B, Alkan C, Andersen LN, Ayub Q, Ball EV, Beal K, Bradley BJ, Chen Y, Clee CM, Fitzgerald S, Graves TA, Gu Y, Heath P, Heger A, Karakoc E, Kolb-Kokocinski A, Laird GK, Lunter G, Meader S, Mort M, Mullikin JC, Munch K, O’Connor TD, Phillips AD, Prado-Martinez J, Rogers AS, Sajjadian S, Schmidt D, Shaw K, Simpson JT, Stenson PD, Turner DJ, Vigilant L, Vilella AJ, Whitener W, Zhu B, Cooper DN, de Jong P, Dermitzakis ET, Eichler EE, Flicek P, Goldman N, Mundy NI, Ning Z, Odom DT, Ponting CP, Quail MA, Ryder OA, Searle SM, Warren WC, Wilson RK, Schierup MH, Rogers J, Tyler-Smith C, Durbin R. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedel C, Marthinsen L, Kristoffersson AC, Kornfalt R, Nilsson KO, Orlenius B, Holmberg L. Lung symptoms in pseudohypoaldosteronism type 1 are associated with deficiency of the alpha-subunit of the epithelial sodium channel. J Pediatr. 1999;135:739–745. doi: 10.1016/s0022-3476(99)70094-6. [DOI] [PubMed] [Google Scholar]
- Schaefer L, Sakai H, Mattei M, Lazdunski M, Lingueglia E. Molecular cloning, functional expression and chromosomal localization of an amiloride-sensitive Na(+) channel from human small intestine. FEBS Lett. 2000;471:205–210. doi: 10.1016/s0014-5793(00)01403-4. [DOI] [PubMed] [Google Scholar]
- Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton RP, Rossier BC. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. EMBO J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- Schild L, Schneeberger E, Gautschi I, Firsov D. Identification of amino acid residues in the alpha, beta, and gamma subunits of the epithelial sodium channel (ENaC) involved in amiloride block and ion permeation. J Gen Physiol. 1997;109:15–26. doi: 10.1085/jgp.109.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng S, Carattino MD, Bruns JB, Hughey RP, Kleyman TR. Furin cleavage activates the epithelial Na+ channel by relieving Na+ self-inhibition. Am J Physiol Renal Physiol. 2006;290:F1488–F1496. doi: 10.1152/ajprenal.00439.2005. [DOI] [PubMed] [Google Scholar]
- Sheng S, Li J, McNulty KA, Avery D, Kleyman TR. Characterization of the selectivity filter of the epithelial sodium channel. J Biol Chem. 2000;275:8572–8581. doi: 10.1074/jbc.275.12.8572. [DOI] [PubMed] [Google Scholar]
- Sheng S, Li J, McNulty KA, Kieber-Emmons T, Kleyman TR. Epithelial sodium channel pore region. structure and role in gating. J Biol Chem. 2001a;276:1326–1334. doi: 10.1074/jbc.M008117200. [DOI] [PubMed] [Google Scholar]
- Sheng S, Maarouf AB, Bruns JB, Hughey RP, Kleyman TR. Functional role of extracellular loop cysteine residues of the epithelial Na+ channel in Na+ self-inhibition. J Biol Chem. 2007;282:20180–20190. doi: 10.1074/jbc.M611761200. [DOI] [PubMed] [Google Scholar]
- Sheng S, McNulty KA, Harvey JM, Kleyman TR. Second transmembrane domains of ENaC subunits contribute to ion permeation and selectivity. J Biol Chem. 2001b;276:44091–44098. doi: 10.1074/jbc.M108522200. [DOI] [PubMed] [Google Scholar]
- Sheridan MB, Fong P, Groman JD, Conrad C, Flume P, Diaz R, Harris C, Knowles M, Cutting GR. Mutations in the beta-subunit of the epithelial Na+ channel in patients with a cystic fibrosis-like syndrome. Hum Mol Genet. 2005;14:3493–3498. doi: 10.1093/hmg/ddi374. [DOI] [PubMed] [Google Scholar]
- Sherwood TW, Frey EN, Askwith CC. Structure and activity of the acid-sensing ion channels. Am J Physiol Cell Physiol. 2012;303:C699–C710. doi: 10.1152/ajpcell.00188.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Asher C, Chigaev A, Yung Y, Reuveny E, Seger R, Garty H. Interactions of beta and gamma ENaC with Nedd4 can be facilitated by an ERK-mediated phosphorylation. J Biol Chem. 2002;277:13539–13547. doi: 10.1074/jbc.M111717200. [DOI] [PubMed] [Google Scholar]
- Shi S, Carattino MD, Kleyman TR. Role of the wrist domain in the response of the epithelial sodium channel to external stimuli. J Biol Chem. 2012;287:44027–44035. doi: 10.1074/jbc.M112.421743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Ghosh DD, Okumura S, Carattino MD, Kashlan OB, Sheng S, Kleyman TR. Base of the thumb domain modulates epithelial sodium channel gating. J Biol Chem, The Journal of biological chemistry. 2011;286:14753–14761. doi: 10.1074/jbc.M110.191734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemura N, Ohkuri T, Sadamitsu C, Yasumatsu K, Yoshida R, Beauchamp GK, Bachmanov AA, Ninomiya Y. Amiloride-sensitive NaCl taste responses are associated with genetic variation of ENaC alpha-subunit in mice. Am J Physiol Regul Integr Comp Physiol. 2008;294:R66–R75. doi: 10.1152/ajpregu.00420.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld SM, Donoghue PCJ. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish) Development. 2012;139:2091–2099. doi: 10.1242/dev.074716. [DOI] [PubMed] [Google Scholar]
- Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Ulick S, Milora RV, Findling JW. Liddle’s syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- Sillitoe I, Lewis TE, Cuff A, Das S, Ashford P, Dawson NL, Furnham N, Laskowski RA, Lee D, Lees JG, Lehtinen S, Studer RA, Thornton J, Orengo CA. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015;43:D376–D381. doi: 10.1093/nar/gku947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho S-J, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo D-H, Larsson T, Lv J, Arendt D, Savage R, Osoegawa K, de Jong P, Grimwood J, Chapman JA, Shapiro H, Aerts A, Otillar RP, Terry AY, Boore JL, Grigoriev IV, Lindberg DR, Seaver EC, Weisblat DA, Putnam NH, Rokhsar DS. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493:526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A, Shenton F, Hunter I, Banks RW, Bewick GS. Amiloride-sensitive channels are a major contributor to mechanotransduction in mammalian muscle spindles. J Physiol. 2010;588:171–185. doi: 10.1113/jphysiol.2009.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, Morgan JR, Buxbaum JD, Sachidanandam R, Sims C, Garruss AS, Cook M, Krumlauf R, Wiedemann LM, Sower SA, Decatur WA, Hall JA, Amemiya CT, Saha NR, Buckley KM, Rast JP, Das S, Hirano M, McCurley N, Guo P, Rohner N, Tabin CJ, Piccinelli P, Elgar G, Ruffier M, Aken BL, Searle SMJ, Muffato M, Pignatelli M, Herrero J, Jones M, Brown CT, Chung-Davidson Y-W, Nanlohy KG, Libants SV, Yeh C-Y, McCauley DW, Langeland JA, Pancer Z, Fritzsch B, de Jong PJ, Zhu B, Fulton LL, Theising B, Flicek P, Bronner ME, Warren WC, Clifton SW, Wilson RK, Li W. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–421. 421e1–421e2. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder PM, McDonald FJ, Stokes JB, Welsh MJ. Membrane topology of the amiloride-sensitive epithelial sodium channel. J Biol Chem. 1994;269:24379–24383. [PubMed] [Google Scholar]
- Snyder PM, Olson DR, Bucher DB. A pore segment in DEG/ENaC Na(+) channels. J Biol Chem. 1999;274:28484–28490. doi: 10.1074/jbc.274.40.28484. [DOI] [PubMed] [Google Scholar]
- Soundararajan R, Pearce D, Hughey RP, Kleyman TR. Role of epithelial sodium channels and their regulators in hypertension. J Biol Chem, The Journal of biological chemistry. 2010;285:30363–30369. doi: 10.1074/jbc.R110.155341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springauf A, Bresenitz P, Gründer S. The interaction between two extracellular linker regions controls sustained opening of acid-sensing ion channel 1. J Biol Chem. 2011;286:24374–24384. doi: 10.1074/jbc.M111.230797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staruschenko A, Adams E, Booth RE, Stockand JD. Epithelial Na+ channel subunit stoichiometry. Biophys J. 2005;88:3966–3975. doi: 10.1529/biophysj.104.056804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AP, Haerteis S, Diakov A, Korbmacher C, Edwardson JM. Atomic force microscopy reveals the architecture of the epithelial sodium channel (ENaC) J Biol Chem. 2011;286:31944–31952. doi: 10.1074/jbc.M111.275289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockand JD, Staruschenko A, Pochynyuk O, Booth RE, Silverthorn DU. Insight toward epithelial Na+ channel mechanism revealed by the acid-sensing ion channel 1 structure. IUBMB Life. 2008;60:620–628. doi: 10.1002/iub.89. [DOI] [PubMed] [Google Scholar]
- Strautnieks SS, Thompson RJ, Hanukoglu A, Dillon MJ, Hanukoglu I, Kuhnle U, Seckl J, Gardiner RM, Chung E. Localisation of pseudohypoaldosteronism genes to chromosome 16p12.2–13.11 and 12p13.1-pter by homozygosity mapping. Hum Mol Genet. 1996;5:293–299. doi: 10.1093/hmg/5.2.293. [DOI] [PubMed] [Google Scholar]
- Studer RA, Person E, Robinson-Rechavi M, Rossier BC. Evolution of the epithelial sodium channel and the sodium pump as limiting factors of aldosterone action on sodium transport. Physiol Genomics. 2011;43:844–854. doi: 10.1152/physiolgenomics.00002.2011. [DOI] [PubMed] [Google Scholar]
- Su YR, Menon AG. Epithelial sodium channels and hypertension. Drug Metab Dispos. 2001;29:553–556. [PubMed] [Google Scholar]
- Sugiyama T, Kato N, Ishinaga Y, Yamori Y, Yazaki Y. Evaluation of selected polymorphisms of the Mendelian hypertensive disease genes in the Japanese population. Hypertens. Res. 2001;24:515–521. doi: 10.1291/hypres.24.515. [DOI] [PubMed] [Google Scholar]
- Szabo R, Bugge TH. Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu Rev Cell Dev Biol. 2011;27:213–235. doi: 10.1146/annurev-cellbio-092910-154247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada T, Ebata T, Noguchi H, Keane TM, Adams DJ, Narita T, Shin-I T, Fujisawa H, Toyoda A, Abe K, Obata Y, Sakaki Y, Moriwaki K, Fujiyama A, Kohara Y, Shiroishi T. The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains. Genome Res. 2013;23:1329–1238. doi: 10.1101/gr.156497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y. Comparative physiology of body fluid regulation in vertebrates with special reference to thirst regulation. Jpn J Physiol, The Japanese journal of physiology. 2000;50:171–186. doi: 10.2170/jjphysiol.50.171. [DOI] [PubMed] [Google Scholar]
- Tapolyai M, Uysal A, Dossabhoy NR, Zsom L, Szarvas T, Lengvárszky Z, Fülöp T. High prevalence of liddle syndrome phenotype among hypertensive US Veterans in Northwest Louisiana. J Clin Hypertens (Greenwich) 2010;12:856–860. doi: 10.1111/j.1751-7176.2010.00359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CP, Doggett NA, Fisher R, Stokes JB. Genomic organization and the 5’ flanking region of the gamma subunit of the human amiloride-sensitive epithelial sodium channel. J Biol Chem. 1996;271:26062–26066. doi: 10.1074/jbc.271.42.26062. [DOI] [PubMed] [Google Scholar]
- Thomas CP, Loftus RW, Liu KZ, Itani OA. Genomic organization of the 5’ end of human beta-ENaC and preliminary characterization of its promoter. Am J Physiol Renal Physiol. 2002;282:F898–F909. doi: 10.1152/ajprenal.00268.2001. [DOI] [PubMed] [Google Scholar]
- Tilley AE, Walters MS, Shaykhiev R, Crystal RG. Cilia dysfunction in lung disease. Annu Rev Physiol. 2015;77:379–406. doi: 10.1146/annurev-physiol-021014-071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker SW. Whales of the world. E. J. Brill. 1988 [Google Scholar]
- Toka HR, Koshy JM, Hariri A. The molecular basis of blood pressure variation. Pediatr Nephrol. 2013;28:387–99. doi: 10.1007/s00467-012-2206-9. [DOI] [PubMed] [Google Scholar]
- Tolino LA, Okumura S, Kashlan OB, Carattino MD. Insights into the mechanism of pore opening of acid-sensing ion channel 1a. J Biol Chem. 2011;286:16297–16307. doi: 10.1074/jbc.M110.202366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q, Menon AG, Stockand JD. Functional polymorphisms in the alpha-subunit of the human epithelial Na+ channel increase activity. Am J Physiol Renal Physiol. 2006;290:F821–F827. doi: 10.1152/ajprenal.00312.2005. [DOI] [PubMed] [Google Scholar]
- Tousson A, Alley CD, Sorscher EJ, Brinkley BR, Benos DJ. Immunochemical localization of amiloride-sensitive sodium channels in sodium-transporting epithelia. J Cell Sci, Journal of cell science. 1989;93(Pt 2):349–362. doi: 10.1242/jcs.93.2.349. [DOI] [PubMed] [Google Scholar]
- Uchiyama M, Konno N, Shibuya S, Nogami S. Cloning and expression of the epithelial sodium channel and its role in osmoregulation of aquatic and estivating African lungfish Protopterus annectens. Comp Biochem Physiol A Mol Integr Physiol. 2014;183:1–8. doi: 10.1016/j.cbpa.2014.12.028. [DOI] [PubMed] [Google Scholar]
- Uchiyama M, Maejima S, Yoshie S, Kubo Y, Konno N, Joss JMP. The epithelial sodium channel in the Australian lungfish, Neoceratodus forsteri (Osteichthyes: Dipnoi) Proc. Biol. Sci. 2012;279:4795–4802. doi: 10.1098/rspb.2012.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniprot. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet V, Chraibi A, Gaeggeler HP, Horisberger JD, Rossier BC. An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature. 1997;389:607–610. doi: 10.1038/39329. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh Y-H, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viel M, Leroy C, Hubert D, Fajac I, Bienvenu T. ENaCbeta and gamma genes as modifier genes in cystic fibrosis. J Cyst Fibros. 2008;7:23–29. doi: 10.1016/j.jcf.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voilley N, Bassilana F, Mignon C, Merscher S, Mattéi MG, Carle GF, Lazdunski M, Barbry P. Cloning, chromosomal localization, and physical linkage of the beta and gamma subunits (SCNN1B and SCNN1G) of the human epithelial amiloride-sensitive sodium channel. Genomics. 1995;28:560–565. doi: 10.1006/geno.1995.1188. [DOI] [PubMed] [Google Scholar]
- Voilley N, Lingueglia E, Champigny G, Mattéi MG, Waldmann R, Lazdunski M, Barbry P. The lung amiloride-sensitive Na+ channel: biophysical properties, pharmacology, ontogenesis, and molecular cloning. Proc Natl Acad Sci U S A. 1994;91:247–251. doi: 10.1073/pnas.91.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk KA, Husted RF, Snyder PM, Stokes JB. Kinase regulation of hENaC mediated through a region in the COOH-terminal portion of the alpha-subunit. Am J Physiol Cell Physiol. 2000;278:C1047–C1054. doi: 10.1152/ajpcell.2000.278.5.C1047. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Vuagniaux G, Vallet V, Jaeger NF, Hummler E, Rossier BC. Synergistic activation of ENaC by three membrane-bound channel-activating serine proteases (mCAP1, mCAP2, and mCAP3) and serum- and glucocorticoid-regulated kinase (Sgk1) in Xenopus Oocytes. J Gen Physiol. 2002;120:191–201. doi: 10.1085/jgp.20028598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blöcker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MCT, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guérin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Røed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvänen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, Lander ES, Lindblad-Toh K. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann R, Champigny G, Bassilana F, Voilley N, Lazdunski M. Molecular cloning and functional expression of a novel amiloride-sensitive Na+ channel. J Biol Chem. 1995;270:27411–27414. doi: 10.1074/jbc.270.46.27411. [DOI] [PubMed] [Google Scholar]
- Waldmann R, Lazdunski M. H(+)-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol. 1998;8:418–424. doi: 10.1016/s0959-4388(98)80070-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Yu T, Yin L, Li J, Yu L, Shen Y, Yu Y, Shen Y, Fu Q. Novel mutations in the SCNN1A gene causing Pseudohypoaldosteronism type 1. PLoS One. 2013;8:e65676. doi: 10.1371/journal.pone.0065676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-P, Yang K-Q, Jiang X-J, Wu H-Y, Zhang H-M, Zou Y-B, Song L, Bian J, Hui R-T, Liu Y-X, Zhou X-L. Prevalence of Liddle Syndrome Among Young Hypertension Patients of Undetermined Cause in a Chinese Population. J Clin Hypertens (Greenwich) 2015 doi: 10.1111/jch.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Schultz BD. Cholera toxin enhances Na(+) absorption across MCF10A human mammary epithelia. Am J Physiol Cell Physiol. 2014;306:C471–C484. doi: 10.1152/ajpcell.00181.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, He G, Yang Y, Liu Y, Diao R, Sheng K, Liu X, Xu W. Reduced expression of Enac in Placenta tissues of patients with severe preeclampsia is related to compromised trophoblastic cell migration and invasion during pregnancy. PLoS One. 2013;8:e72153. doi: 10.1371/journal.pone.0072153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pascual-Anaya J, Zadissa A, Li W, Niimura Y, Huang Z, Li C, White S, Xiong Z, Fang D, Wang B, Ming Y, Chen Y, Zheng Y, Kuraku S, Pignatelli M, Herrero J, Beal K, Nozawa M, Li Q, Wang J, Zhang H, Yu L, Shigenobu S, Wang J, Liu J, Flicek P, Searle S, Wang J, Kuratani S, Yin Y, Aken B, Zhang G, Irie N. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 2013;45:701–706. doi: 10.1038/ng.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Künstner A, Searle S, White S, Vilella AJ, Fairley S, Heger A, Kong L, Ponting CP, Jarvis ED, Mello CV, Minx P, Lovell P, Velho TAF, Ferris M, Balakrishnan CN, Sinha S, Blatti C, London SE, Li Y, Lin Y-C, George J, Sweedler J, Southey B, Gunaratne P, Watson M, Nam K, Backström N, Smeds L, Nabholz B, Itoh Y, Whitney O, Pfenning AR, Howard J, Völker M, Skinner BM, Griffin DK, Ye L, McLaren WM, Flicek P, Quesada V, Velasco G, Lopez-Otin C, Puente XS, Olender T, Lancet D, Smit AFA, Hubley R, Konkel MK, Walker JA, Batzer MA, Gu W, Pollock DD, Chen L, Cheng Z, Eichler EE, Stapley J, Slate J, Ekblom R, Birkhead T, Burke T, Burt D, Scharff C, Adam I, Richard H, Sultan M, Soldatov A, Lehrach H, Edwards SV, Yang S-P, Li X, Graves T, Fulton L, Nelson J, Chinwalla A, Hou S, Mardis ER, Wilson RK. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grützner F, Belov K, Miller W, Clarke L, Chinwalla AT, Yang S-P, Heger A, Locke DP, Miethke P, Waters PD, Veyrunes F, Fulton L, Fulton B, Graves T, Wallis J, Puente XS, López-Otín C, Ordóñez GR, Eichler EE, Chen L, Cheng Z, Deakin JE, Alsop A, Thompson K, Kirby P, Papenfuss AT, Wakefield MJ, Olender T, Lancet D, Huttley GA, Smit AFA, Pask A, Temple-Smith P, Batzer MA, Walker JA, Konkel MK, Harris RS, Whittington CM, Wong ESW, Gemmell NJ, Buschiazzo E, Vargas Jentzsch IM, Merkel A, Schmitz J, Zemann A, Churakov G, Kriegs JO, Brosius J, Murchison EP, Sachidanandam R, Smith C, Hannon GJ, Tsend-Ayush E, McMillan D, Attenborough R, Rens W, Ferguson-Smith M, Lefèvre CM, Sharp JA, Nicholas KR, Ray DA, Kube M, Reinhardt R, Pringle TH, Taylor J, Jones RC, Nixon B, Dacheux J-L, Niwa H, Sekita Y, Huang X, Stark A, Kheradpour P, Kellis M, Flicek P, Chen Y, Webber C, Hardison R, Nelson J, Hallsworth-Pepin K, Delehaunty K, Markovic C, Minx P, Feng Y, Kremitzki C, Mitreva M, Glasscock J, Wylie T, Wohldmann P, Thiru P, Nhan MN, Pohl CS, Smith SM, Hou S, Nefedov M, de Jong PJ, Renfree MB, Mardis ER, Wilson RK. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel M, Akin L, Büscher A, Güran TP, Hauffa B, Högler W, Leonards J, Karges B, Kentrup H, Kirel B, Senses EEY, Tekin N, Holterhus P-M, Riepe FG. Five novel mutations in the SCNN1A gene causing autosomal recessive pseudohypoaldosteronism type 1. Eur J Endocrinol. 2013;168:707–715. doi: 10.1530/EJE-12-1000. [DOI] [PubMed] [Google Scholar]
- Winarski KL, Sheng N, Chen J, Kleyman TR, Sheng S. Extracellular allosteric regulatory subdomain within the gamma subunit of the epithelial Na+ channel. J Biol Chem, The Journal of biological chemistry. 2010;285:26088–26096. doi: 10.1074/jbc.M110.149963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley KC, Warren WC, Rogers J, Locke D, Muzny DM, Mardis ER, Weinstock GM, Tardif SD, Aagaard KM, Archidiacono N, Rayan NA, Batzer MA, Beal K, Brejova B, Capozzi O, Capuano SB, Casola C, Chandrabose MM, Cree A, Dao MD, de Jong PJ, del Rosario RC-H, Delehaunty KD, Dinh HH, Eichler EE, Fitzgerald S, Flicek P, Fontenot CC, Fowler RG, Fronick C, Fulton LA, Fulton RS, Gabisi RA, Gerlach D, Graves TA, Gunaratne PH, Hahn MW, Haig D, Han Y, Harris RA, Herrero J, Hillier LW, Hubley R, Hughes JF, Hume J, Jhangiani SN, Jorde LB, Joshi V, Karakor E, Konkel MK, Kosiol C, Kovar CL, Kriventseva EV, Lee SL, Lewis LR, Liu Y, Lopez J, Lopez-Otin C, Lorente-Galdos B, Mansfield KG, Marques-Bonet T, Minx P, Misceo D, Moncrieff JS, Morgan MB, Nazareth LV, Newsham I, Nguyen NB, Okwuonu GO, Prabhakar S, Perales L, Pu L-L, Puente XS, Quesada V, Ranck MC, Raney BJ, Raveendran M, Deiros DR, Rocchi M, Rodriguez D, Ross C, Ruffier M, Ruiz SJ, Sajjadian S, Santibanez J, Schrider DR, Searle S, Skaletsky H, Soibam B, Smit AFA, Tennakoon JB, Tomaska L, Ullmer B, Vejnar CE, Ventura M, Vilella AJ, Vinar T, Vogel J-H, Walker JA, Wang Q, Warner CM, Wildman DE, Witherspoon DJ, Wright RA, Wu Y, Xiao W, Xing J, Zdobnov EM, Zhu B, Gibbs RA, Wilson RK. The common marmoset genome provides insight into primate biology and evolution. Nat Genet. 2014;46:850–857. doi: 10.1038/ng.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yu Y, Li W-G, Yu F, Cao H, Xu T-L, Jiang H. Inherent dynamics of the acid-sensing ion channel 1 correlates with the gating mechanism. PLoS Biol. 2009;7:e1000151. doi: 10.1371/journal.pbio.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L-M, Rinke R, Korbmacher C. Stimulation of the epithelial sodium channel (ENaC) by cAMP involves putative ERK phosphorylation sites in the C termini of the channel’s beta- and gamma-subunit. J Biol Chem. 2006;281:9859–9868. doi: 10.1074/jbc.M512046200. [DOI] [PubMed] [Google Scholar]
- Yue GG, Malik B, Eaton DCDC, Yue GG, Eaton DCDC. Phosphatidylinositol 4,5-bisphosphate (PIP2) stimulates epithelial sodium channel activity in A6 cells. J Biol Chem. 2002;277:11965–11969. doi: 10.1074/jbc.M108951200. [DOI] [PubMed] [Google Scholar]
- Zelle KM, Lu B, Pyfrom SC, Ben-Shahar Y. The genetic architecture of degenerin/epithelial sodium channels in Drosophila. G3 (Bethesda) 2013;3:441–450. doi: 10.1534/g3.112.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha X. Acid-sensing ion channels: trafficking and synaptic function. Mol. Brain. 2013;6:1. doi: 10.1186/1756-6606-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, Xiong Z, Que H, Xie Y, Holland PWH, Paps J, Zhu Y, Wu F, Chen Y, Wang J, Peng C, Meng J, Yang L, Liu J, Wen B, Zhang N, Huang Z, Zhu Q, Feng Y, Mount A, Hedgecock D, Xu Z, Liu Y, Domazet-Lošo T, Du Y, Sun X, Zhang S, Liu B, Cheng P, Jiang X, Li J, Fan D, Wang W, Fu W, Wang T, Wang B, Zhang J, Peng Z, Li Y, Li N, Wang J, Chen M, He Y, Tan F, Song X, Zheng Q, Huang R, Yang H, Du X, Chen L, Yang M, Gaffney PM, Wang S, Luo L, She Z, Ming Y, Huang W, Zhang S, Huang B, Zhang Y, Qu T, Ni P, Miao G, Wang J, Wang Q, Steinberg CEW, Wang H, Li N, Qian L, Zhang G, Li Y, Yang H, Liu X, Wang J, Yin Y, Wang J. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- Zhang G, Li B, Li C, Gilbert MTP, Jarvis ED, Wang J. Comparative genomic data of the Avian Phylogenomics Project. Gigascience. 2014;3:26. doi: 10.1186/2047-217X-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhang W, Zhang L, Zhang Z, Li J, Lu G, Zhu Y, Wang Y, Huang Y, Liu J, Kang H, Chen J, Wang L, Chen A, Yu S, Gao Z, Jin L, Gu W, Wang Z, Zhao L, Shi B, Wen H, Lin R, Jones MK, Brejova B, Vinar T, Zhao G, McManus DP, Chen Z, Zhou Y, Wang S. The genome of the hydatid tapeworm Echinococcus granulosus. Nat Genet. 2013;45:1168–1175. doi: 10.1038/ng.2757. [DOI] [PubMed] [Google Scholar]
- Zhu K, Zhou X, Xu S, Sun D, Zhou K, Yang G. The loss of taste genes in cetaceans. BMC Evol. Biol. 2014;14:218. doi: 10.1186/s12862-014-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Zhang W, Yang S, Chen J-Q, Tian D. Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC Genomics. 2009;10:47. doi: 10.1186/1471-2164-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X-Q, Korhonen PK, Cai H, Young ND, Nejsum P, von Samson-Himmelstjerna G, Boag PR, Tan P, Li Q, Min J, Yang Y, Wang X, Fang X, Hall RS, Hofmann A, Sternberg PW, Jex AR, Gasser RB. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat. Commun. 2015;6:6145. doi: 10.1038/ncomms7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann AE, Allen JE, Dahdaleh NS, Drebot II, Coryell MW, Wunsch AM, Lynch CM, Faraci FM, Howard MA, Welsh MJ, Wemmie JA. The amygdala is a chemosensor that detects carbon dioxide and acidosis to elicit fear behavior. Cell. 2009;139:1012–1021. doi: 10.1016/j.cell.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemann AE, Schnizler MK, Albert GW, Severson MA, Howard MA, Welsh MJ, Wemmie JA. Seizure termination by acidosis depends on ASIC1a. Nat Neurosci. 2008;11:816–822. doi: 10.1038/nn.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, Tharp GK, Marçais G, Roberts M, Ferguson B, Fox HS, Treangen T, Salzberg SL, Yorke JA, Norgren RB. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct. 2014;9:20. doi: 10.1186/1745-6150-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]