Abstract
Background:
There are no objective, biological markers that can robustly predict methylphenidate response in attention deficit hyperactivity disorder. This study aimed to examine whether applying machine learning approaches to pretreatment demographic, clinical questionnaire, environmental, neuropsychological, neuroimaging, and genetic information can predict therapeutic response following methylphenidate administration.
Methods:
The present study included 83 attention deficit hyperactivity disorder youth. At baseline, parents completed the ADHD Rating Scale-IV and Disruptive Behavior Disorder rating scale, and participants undertook the continuous performance test, Stroop color word test, and resting-state functional MRI scans. The dopamine transporter gene, dopamine D4 receptor gene, alpha-2A adrenergic receptor gene (ADRA2A) and norepinephrine transporter gene polymorphisms, and blood lead and urine cotinine levels were also measured. The participants were enrolled in an 8-week, open-label trial of methylphenidate. Four different machine learning algorithms were used for data analysis.
Results:
Support vector machine classification accuracy was 84.6% (area under receiver operating characteristic curve 0.84) for predicting methylphenidate response. The age, weight, ADRA2A MspI and DraI polymorphisms, lead level, Stroop color word test performance, and oppositional symptoms of Disruptive Behavior Disorder rating scale were identified as the most differentiating subset of features.
Conclusions:
Our results provide preliminary support to the translational development of support vector machine as an informative method that can assist in predicting treatment response in attention deficit hyperactivity disorder, though further work is required to provide enhanced levels of classification performance.
Keywords: ADHD, machine learning, methylphenidate, prediction, treatment response
Introduction
Attention deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder characterized by symptoms of inattention, hyperactivity, and impulsivity. Methylphenidate (MPH) is the most frequently prescribed first-line therapeutic agent for ADHD, and it is reportedly effective in approximately 70% of children with the disorder (Santosh and Taylor, 2000). Although the response rate is higher relative to other psychiatric medications, the issue of therapeutic response is of great clinical relevance, because responses to treatment varies from one individual to another, as in most psychiatric conditions, and around one-fourth of patients with ADHD show no improvement following the initial treatment with stimulants. However, to date, there are no objective, biological makers that can robustly predict the treatment response of MPH.
ADHD has an estimated heritability of 0.8 and has been theorized to be a complex, polygenic disorder (Biederman and Faraone, 2005). The leading candidate genes based on the catecholamine hypothesis of ADHD include those involved in dopaminergic or noradrenergic neurotransmission, and these genes have also been implicated in modifying the therapeutic response to MPH medication (Froehlich et al., 2010; Arnsten, 2011). The dopamine transporter gene (DAT1) and the dopamine D4 receptor gene (DRD4) are the most extensively studied. The alpha-2A adrenergic receptor gene (ADRA2A) and the norepinephrine transporter gene (SLC6A2) have also been the focus of recent studies (Mick and Faraone, 2008). While genetic factors play an important role in ADHD, it is possible that environmental risk factors (eg, lead, nicotine) and interplay between genetic predisposition and environmental exposure may lead to brain alterations related to action of MPH (Nigg et al., 2010). For instance, recent evidence has indicated that lead exposure might influence ADHD symptoms via the disruption of the dopamine neurocircuits (Eubig et al., 2010; Cho et al., 2013).
The extant literature of functional and structural imaging studies has generally implicated prefrontal and striatal regions as neuroimaging correlates of ADHD and MPH response (Liston et al., 2011). Brain networks involving temporal and limbic regions have also been suggested to contribute to the pathophysiology of ADHD (Cortese, 2012). The presumed therapeutic action of MPH involves the dopaminergic and noradrenergic neurotransmitter systems in prefrontal and striatal regions (Wilens, 2008). Among the imaging measures, resting-state functional magnetic resonance imaging (fMRI) has garnered significant recent interest as a tool for finding clinically relevant biomarkers or measuring response to treatment in psychiatric disorders (Greicius, 2008). As for neuropsychological endophenotypes of ADHD, the continuous performance test (CPT) and the Stroop color word test (SCWT) have often been employed to measure neurocognitive functioning and its changes with MPH treatment in ADHD (Kebir et al., 2009). Thus, these variables may have clinical utility as predictors of treatment response in ADHD.
Machine learning is an area of artificial intelligence concerned with the construction and study of systems that can learn from data (Orru et al., 2012). Recent evidence indicates that the application of machine learning classification techniques to psychiatric data may allow prediction of treatment response at the individual level (Orru et al., 2012). It is hoped that these methods could inform and assist clinicians to make more effective clinical decisions prior to treatment and would lead to fewer unsuccessful trials and higher response rates. ADHD in particular would benefit from more effective predictive analytics because of its high prevalence, clinical heterogeneity, and societal costs (Bohland et al., 2012). There has been a growing interest in the use of machine learning algorithms for analyzing neuroimaging data (Dosenbach et al., 2010). However, limited work has been done integrating genetic information with imaging data, and we are unaware of any study that utilized genetic, environmental, neuroimaging, and neuropsychological information together to predict MPH response in ADHD.
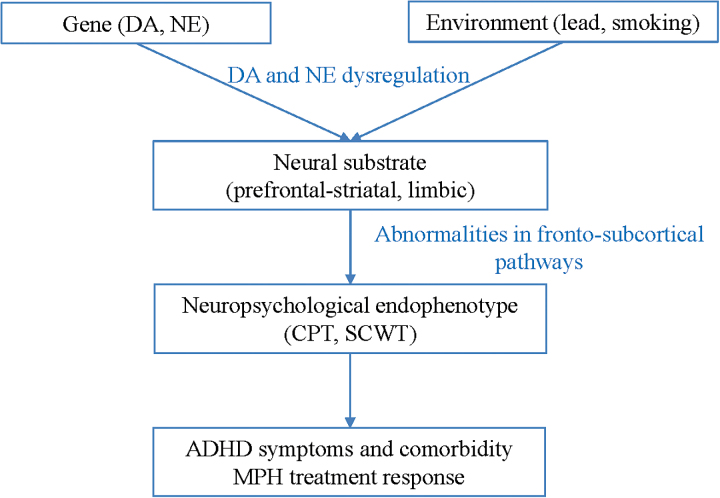
In this study, we applied machine learning approaches using pretreatment demographic, clinical questionnaire, environmental, neuropsychological, neuroimaging, and genetic information to predict the responders or nonresponders to MPH treatment in children and adolescents with ADHD. Based on the extant literature, we hypothesized that the biological/cognitive correlates of dopamine and norepinephrine neurocircuits in ADHD would show significant predictive potential for therapeutic response to MPH administration (Figure 1).
Figure 1.
A research hypothesis for attention deficit hyperactivity disorder (ADHD). CPT, continuous performance test; DA, dopamine; MPH, methylphenidate; NE, norepinephrine; SCWT, Stroop color word test.
Methods
The present study included 83 ADHD subjects (9.5±2.6 years, 65 boys) recruited from the Seoul National University Hospital in Korea. ADHD was diagnosed according to DSM-IV criteria using the Kiddie-Schedule for Affective Disorders and Schizophrenia-Present and Lifetime Version (Kaufman et al., 1997). ADHD patients with an intelligence quotient (IQ) below 70, a past or an ongoing history of either tic disorder, obsessive compulsive disorder, language disorder, learning disorder, convulsive disorder, pervasive developmental disorder, schizophrenia, bipolar disorder, or brain damage, a past history of taking stimulants or atomoxetine longer than 6 months, or a recent history of taking stimulants or atomoxetine during the last 4 weeks were excluded from the study (Hong et al., 2015). The study protocol was approved by the institutional review board for human subjects at the Seoul National University Hospital. Detailed information about the study was given to parents and children, and written informed consent was obtained prior to study entry. At baseline, the parents completed the ADHD Rating Scale-IV (DuPaul, 1998) and Disruptive Behavior Disorder rating scale (DBD) (Silva et al., 2005), and the participants undertook the CPT (Greenberg and Waldman, 1993), SCWT (Golden, 1978), genetic/environmental testing, and resting-state fMRI scans.
The participants were enrolled in an 8-week, open-label trial of MPH. Initial doses of MPH were maintained for 2 weeks, and the doses were adjusted at the second and fourth week of treatment. The doses were titrated upward until sufficient therapeutic effects were achieved, on the basis of the subjects’ and parents’ reports of symptom improvement and adverse effects, and then the doses were maintained for the remainder of the 8 weeks. Global improvement of each patient was assessed on the basis of the investigator-rated Clinical Global Impressions-Improvement scale (Guy, 1976). Subjects with scores of 1 or 2 on the Clinical Global Impressions-Improvement scale after treatment were classified as “good responders,” whereas subjects with scores of 3 to 7 were deemed “poor responders.”
Genomic DNA was extracted from whole blood lymphocytes using a G-DEXTM II Genomic DNA Extraction Kit (Intron). The DRD4 exon III VNTR polymorphism and the 40-base pair VNTR polymorphism located in the 3’-UTR of DAT1 were genotyped, as previously described (Hong et al., 2012). For the ADRA2A and SLC6A2, the detection of a single nucleotide polymorphism was based on analysis of primer extension products generated from previously amplified genomic DNA using a chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform (Sequenom). The ADRA2A (MspI and DraI) and SLC6A2 (G1287A and A-3081T) polymorphisms were genotyped as previously described (Hong et al., 2012). As for environmental factors, we measured blood lead and urine cotinine based on the evidence of our prior ADHD research (Cho et al., 2010). For the lead measurement, a volume of 5mL of venous blood was collected from each child in metal-free tubes, and samples were assayed using previously described methods (Kim et al., 2010). We used urine cotinine as a biomarker for environmental tobacco smoke exposure, and it was measured using cotinine direct ELISA kits (BioQuant, San Diego, CA), as previously described (Cho et al., 2013).
Resting-state fMRI images were acquired on a 3T Siemens scanner (Siemens Magnetom Trio Tim Syngo MR B17) with the following parameters: repetition time 3000ms; echo time 40ms; acquisition matrix 128×128; field of view 240×240mm2; flip angle 90°; voxel size 1.9 mm×1.9 mm×4.0mm; slices 30. The total time of the acquisition was 6 minutes 24 seconds. FMRI analyses were conducted using locally developed NeuroImaging Software (Fissell et al., 2003) and Analysis of Functional Neuroimaging software (Cox, 1996). Functional imaging data were corrected for motion using 3dVolReg implemented in Analysis of Functional Neuroimaging using the first image as a reference. Quadratic trends within runs were removed, and outliers >1.5 interquartile range from the Tukey hinges were Windsorized using niscorrect from NeuroImaging Software. Data were temporally smoothed using a 7 point Gaussian filter and converted to percent change based on the median of the run. Data were coregistered to the Colin-27 Montreal Neurological Institute template using the Automated Image Registration package’s (AIR (Woods et al., 1993) 32 parameter nonlinear automated warping algorithm and spatially smoothed using a 6-mm full-width at half maximum filter. Finally, because even small amounts of motion (<2mm) can affect measures of functional connectivity (Van Dijk et al., 2012), we employed the technique from Power and colleagues (Power et al., 2012) to eliminate contributions of motion.
We conducted region of interest (ROI) analyses on a priori regions, including the right and left amygdala (anatomically defined by hand tracing on the Montreal Neurological Institute Colin 27 brain as in Siegle and colleagues; Siegle et al., 2007) and other regions specified using AFNI’s Talairach atlas, including the area of the dorsolateral prefrontal cortex, defined as the lateral parts of the bilateral middle frontal gyrus (5 < z < 37), bilateral nucleus accumbens, bilateral hippocampus, and ventromedial prefrontal cortex, defined as the anterior part of Brodmann’s area 24 (y > 21) and includes anterior cingulate. For each participant, the representative time series for each seed ROI was extracted by averaging time series across all voxels within ROI. We then calculated the correlation between each seed ROI time series.
We used Waikato Environment for Knowledge Analysis (Hall, 2009), an open source machine learning framework, to apply and evaluate performance of various classification algorithms to differentiate between MPH treatment responders and nonresponders. In the dataset, the above-noted genetic, environmental, neuroimaging, neuropsychological, and clinical attributes were included (Table 1). In addition, demographic and clinical attributes of age, gender, IQ, height and weight at baseline, and initial MPH dose were included for the dataset. We used Wrapper subset evaluation method, which evaluates attribute sets by using a learning scheme and identifies features that optimize the prediction performance (Kohavi and John, 1997). Wrapper subset evaluation method was used for feature evaluation and forward greedy hill-climbing augmented with a backtracking algorithm to search and select the subset of feature space. To avoid the overfitting and increase the generalization of results, Wrapper subset evaluation method was applied with 10-fold cross validation. This gave a list of features along with how many times they were chosen (this varies from 0% to 100% time) out of 10-fold. Features that were chosen in each fold of 10-fold carried a selection weight of 100%, while features that were never selected had a selection weight of 0%. The final feature subset used in this paper was chosen from the feature space generated from each fold of 10-fold cross validation. Generated feature space for this paper mostly included the features that consistently performed well in each fold of 10-fold (30% or more) cross validation stage. Following this method made the results less prone to overfitting and more generalizable to new instances. Features selected have a computational importance with or without clinical relevance or importance; thus, an experienced clinician should decide how to interpret the selected features.
Table 1.
Demographic and Clinical Characteristics, Genotype Frequencies, and Lead and Cotinine Levels of the Good and Poor Responders to MPH in ADHD Participants
| Good Responder (n=48) | Poor Responder (n=30) | P Value | |
|---|---|---|---|
| Age, mean (SD) years | 9.4 (2.3) | 9.8 (2.8) | 0.48 |
| Female, n (%) | 11 (22.9%) | 5 (16.7%) | 0.51 |
| IQ, mean (SD) | 108 (12) | 105 (15) | 0.41 |
| Handedness (right), n (%) | 42 (87.5%) | 28 (93.3%) | 0.34 |
| CPT, mean (SD) | |||
| Omission errors | 64.5 (20.3) | 68.3 (21.8) | 0.45 |
| Commission errors | 63.1 (17.0) | 66.4 (17.9) | 0.43 |
| Response time variability | 64.3 (17.6) | 63.7 (17.3) | 0.88 |
| SCWT | |||
| Word test | 45.1 (10.3) | 45.4 (12.3) | 0.92 |
| Color test | 44.7 (11.0) | 46.0 (10.5) | 0.60 |
| Color-Word test | 44.6 (9.5) | 49.5 (14.9) | 0.12 |
| Interference | 52.0 (11.0) | 55.8 (11.9) | 0.15 |
| ADHD-RS, mean (SD) | |||
| Inattention | 15.5 (6.2) | 14.3 (5.1) | 0.40 |
| Hyperactivity-impulsivity | 10.7 (5.7) | 11.1 (5.9) | 0.77 |
| Total | 26.2 (10.6) | 25.5 (10.5) | 0.76 |
| ADHD subtypes, n (%) | 0.64 | ||
| Combined | 25 (52.1%) | 15 (50.0%) | |
| Inattentive | 19 (39.6%) | 12 (40.0%) | |
| Hyperactive-impulsive | 0 (0.0%) | 1 (3.3%) | |
| Not otherwise specified | 4 (8.3%) | 2 (6.7%) | |
| Comorbid disorders, n (%) | |||
| Oppositional defiant disorder | 4 (8.3%) | 9 (30.0%) | 0.01 |
| Anxiety disorder | 0 (0.0%) | 1 (3.3%) | 0.20 |
| Genotype | |||
| DAT1, n (%) | 0.63 | ||
| With 10/10 | 39 (81.3%) | 23 (76.7%) | |
| Without 10/10 | 9 (18.7%) | 7 (23.3%) | |
| DRD4, n (%) | 0.64 | ||
| With 4/4 | 25 (52.1%) | 14 (46.7%) | |
| Without 4/4 | 23 (47.9%) | 16 (53.3%) | |
| ADRA2A MspI, n (%) | 0.14 | ||
| G/G | 19 (39.6%) | 17 (56.7%) | |
| G/C+C/C | 29 (60.4%) | 13 (43.3%) | |
| ADRA2A DraI, n (%) | 0.87 | ||
| C/C | 12 (25.0%) | 7 (23.3%) | |
| C/T+T/T | 36 (75.0%) | 23 (76.7%) | |
| SLC6A2 G1287A, n (%) | 0.54 | ||
| G/G | 21 (43.8%) | 11 (36.7%) | |
| G/A+A/A | 27 (56.2%) | 19 (63.3%) | |
| SLC6A2 A-3081T, n (%) | 0.68 | ||
| A/A | 17 (35.4%) | 12 (40.0%) | |
| A/T+T/T | 31 (64.6%) | 18 (60.0%) | |
| Environmental measure | |||
| Lead (µg/dL), mean (SD) | 1.5 (0.4) | 1.4 (0.4) | 0.60 |
| Cotinine (µg/g), mean (SD) | 0.6 (0.9) | 0.8 (1.8) | 0.41 |
ADHD, attention deficit hyperactivity disorder; ADHD-RS, ADHD rating scale; ADRA2A, alpha-2A adrenergic receptor gene; CPT, continuous performance test; DAT1, dopamine transporter gene; DRD4, dopamine D4 receptor gene; MPH, methylphenidate; SCWT, Stroop color word test; SLC6A2, norepinephrine transporter gene.
We applied 4 different machine learning algorithms: support vector machines (SVM), a decision tree algorithm (J48), an ensemble learning machine (Random Forest), and Logistic Ridge Regression (a regression-based approach which handles multicollinearity well) with 10-fold cross validation to compare the performance of various algorithms. Researchers recommended that it is better to use 10-fold cross validation for evaluating the performance of classifiers in case of small sample sizes (eg, a sample size <250) (Hawkins et al., 2003). We chose SVM and Random Forest because they are commonly used for modeling complex nonlinear hypothesis spaces when sample sizes are small. J48 decision tree gives an outcome model that is easy to interpret and applicable in clinical practice. We used sequential minimal optimization algorithm for training the SVM classifier (Keerthi et al., 2001). We used SVM with second-order polynomial kernel and tuned the model for optimum cost parameter from 1 to 100 (details are available upon request). Classification accuracy and area under receiver operating characteristic (ROC) curve (AUC) of these algorithms were compared to find the best classifier for treatment response. Based on the cost and burden of obtaining various measures, we set up 4 stages (see below) and examined whether classification accuracy and AUC could be improved by addition of the measures with each stage. We think that examining tests in tranches based on increasing cost provides data to say whether or not different groups of tests are worth the additional cost.
Stage 1: demographics (age, gender, IQ, height/weight)
Stage 2: stage 1 + clinical information (ADHD Rating Scale-IV, DBD, initial MPH dose)
Stage 3: stage 2 + neuropsychological measures (CPT, SCWT)
Stage 4: stage 3 + genetic/environmental/neuroimaging measures (DAT1, DRD4, ADRA2A, SLC6A2, lead, cotinine, resting-state fMRI seed ROI correlations)
Results
Participant Characteristics
Among the 83 ADHD subjects, 5 of them dropped out before completing the 8-week MPH trial and were excluded. Forty-eight ADHD patients were good responders and 30 patients were poor responders. No baseline differences were found between the good and poor responder groups in their age, gender, IQ, ADHD subtypes and symptoms, neuropsychological characteristics, genotype frequencies, and environmental measures (Table 1). The poor responders showed higher comorbidity rate of oppositional defiant disorder relative to the good responders (P=.01).
Prediction of Treatment Response
SVM classification accuracy was 84.6% (AUC 0.84) at stage 4 for predicting MPH response (Table 2). Wrapper subset evaluation method demonstrated the age, weight, ADRA2A MspI and DraI polymorphisms, lead level, SCWT color-word and word performance, and oppositional symptoms of DBD as the most differentiating subset of features. J48, Random Forest, and Logistic Ridge Regression classification accuracies (and AUCs) at stage 4 were 69.2% (0.61), 73.1% (0.79), and 76.9% (0.73), respectively. Figure 2 shows the AUCs of the classifiers on MPH response. Examining SVM, the best performing algorithm, classification accuracy, and AUC continued to improve between all stages and was best at stage 4 (Table 2).
Table 2.
Classification Accuracy and Area Under Receiver Operating Characteristic (ROC) Curve (AUC) Performance of the Classifiers for Predicting MPH Response
| Support Vector Machine | J48 | Random Forest | Logistic Ridge Regression | |||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | AUC | |
| Stage 1 | 64.1% | 0.55 | 61.5% | 0.51 | 61.5% | 0.58 | 66.7% | 0.61 |
| Stage 2 | 70.5% | 0.69 | 62.8% | 0.56 | 66.7% | 0.59 | 65.4% | 0.65 |
| Stage 3 | 74.4% | 0.69 | 68.0% | 0.55 | 66.7% | 0.64 | 73.1% | 0.70 |
| Stage 4 | 84.6% | 0.84 | 69.2% | 0.61 | 73.1% | 0.79 | 76.9% | 0.73 |
Abbreviation: MPH, methylphenidate.
Stage 1: demographics.
Stage 2: stage 1 + clinical information.
Stage 3: stage 2 + neuropsychological measures.
Stage 4: stage 3 + genetic/environmental/neuroimaging measures.
Figure 2.
Comparison of area under the curve (AUC) performance of the classifiers on methylphenidate response. ROC, area under receiver operating characteristic (ROC) curve.
Table 3 shows the confusion matrix of performance of the classifiers on MPH response (stage 4). All of these algorithms provide a decision curve allowing different choices for the tradeoff between sensitivity and false positive rate (1-sensitivity). More sensitivity always comes with a higher false positive rate, and the best tradeoff varies from use to use.
Table 3.
Classification of MPH Response (Stage 4)
| Support Vector Machine | J48 | ||||
|---|---|---|---|---|---|
| Classification (no.) | Classification (no.) | ||||
| Positive | Negative | Positive | Negative | ||
| Response (+) | 41 | 7 | Response (+) | 39 | 9 |
| Response (-) | 5 | 25 | Response (-) | 15 | 15 |
| Random Forest | Logistic Ridge Regression | ||||
| Classification (no.) | Classification (no.) | ||||
| Positive | Negative | Positive | Negative | ||
| Response (+) | 38 | 10 | Response (+) | 39 | 9 |
| Response (-) | 11 | 19 | Response (-) | 9 | 21 |
Abbreviation: MPH, methylphenidate.
Stage 1: demographics.
Stage 2: stage 1 + clinical information.
Stage 3: stage 2 + neuropsychological measures.
Stage 4: stage 3 + genetic/environmental/neuroimaging measures.
Response (+), good responder; Response (-), poor responder
Discussion
To our knowledge, the present study is the first to apply machine learning approaches using genetic, environmental, neuroimaging, and neuropsychological data together to predict MPH response in ADHD. Our findings demonstrate a potentially effective method towards the development of biological/cognitive markers for the prediction of therapeutic response in the disorder.
As noted, the ADRA2A polymorphisms, lead level, and SCWT performance were included in the subset of predictors. First, it is important to note that the ADRA2A MspI and DraI polymorphisms were reported to be associated with treatment response to MPH in ADHD (Hong et al., 2012; Kim et al., 2013). Second, our results suggest that the attributes related to response inhibition or impulsivity contributed to the predictive potential for therapeutic response. As for the lead, recent evidence suggests that lead might influence symptoms of ADHD via disruption of striatal functioning and associated prefrontal-striatal neurocircuits (Eubig et al., 2010). Nigg and colleagues (2008) demonstrated that blood lead was associated with symptoms of hyperactivity-impulsivity in ADHD, and the lead effects on the symptoms were mediated by poor performance on response inhibition task. Interestingly, the predictors that contributed to differentiating between MPH responders and nonresponders in this study included SCWT color-word performance, which is related to inhibitory control functions and impulsivity symptoms of ADHD. The predictor of the oppositional symptoms of DBD might also be related to impulse control functions of individuals with ADHD (King et al., 2003; Herrmann et al., 2010).
The significance of age and weight as predictors of treatment response in ADHD is currently unclear. As for neuroimaging, it is of note that no fMRI measure in this study contributed to predict the treatment response. It is possible that altered functional connectivity patterns throughout the whole brain might contain important information for predicting individual therapeutic outcomes rather than the predetermined regions or networks of interest used for this study.
For prediction of MPH treatment response, the addition of genetic/environmental/neuroimaging measures to demographics, clinical information, and neuropsychological measures improved both the classification accuracy (74.4% to 84.6%) and AUC (0.69 to 0.84). Since ADHD is a clinically heterogeneous disorder, any added ability to point to objective measures might be of high potential significance. Our overall ability to predict treatment response using all variables was substantial. Nevertheless, given the current cost of the imaging and genetic studies and the relatively low risk of treatment with MPH, we do not think that this is of substantial immediate clinical utility.
Neurobiological research may not provide a single magical biomarker for ADHD or other psychiatric disorders. A more feasible opportunity for psychiatry, as with the rest of the medicine, is to utilize and combine the recent evidences in genetics, neuroimaging, and cognitive neuroscience to predict responses/nonresponses or adverse events with respect to specific pharmacological/psychosocial treatments. Machine learning techniques such as SVM may have an invaluable potential to provide adequate classification accuracy and reliability at the individual level through integrating the scientific evidences. We think that our results demonstrate how the use of machine learning approaches with rich multi-domain pretreatment data may soon reach the point to inform clinical practice in psychiatry.
There are several limitations to this study that deserve comment. First, the sample size was small; it provides 80% power to detect a difference in AUC of at least 0.18. Our findings should be replicated in a larger sample (eg, a sample size >250) (Hawkins et al., 2003) with a broader base of biological/cognitive measures. Second, this study was conducted at one university center in Korea and, thus, we are unable to make inference with regard to the generalizability across different research centers or ethnic groups for any of the successful predictors found in our results. Third, while the approach used strongly suggests that a group of variables can predict patient response, inclusion of particular measure in the predictive variables is not evidence that the variable alone or in combination with other variables significantly provides statistically significant prediction. Finally, the pharmacological treatment for the ADHD subjects was with a single medication. The predictive potential for other ADHD medications and nonpharmacological treatments of ADHD requires further investigation. The specificity of the predictive markers of this study should be interpreted cautiously, since there was no placebo treatment arm. Further studies are required that compare patient groups receiving different treatments.
In summary, the results of this study demonstrate that MPH treatment responders in ADHD can be identified at the individual level using a range of biological and cognitive measures, including genetic, environmental, and neuropsychological data. From a clinical perspective, our results provide preliminary support to the translational development of SVM as an informative method that can assist in predicting treatment response in ADHD. The eventual use of this method in clinical practice would require greater levels of classification performance than reported in this study, along with cost-benefit analysis of biological/cognitive measures.
Statement of Interest
None.
Acknowledgments
This research was supported by the Basic Science Program through the National Research Foundation of Korea (2010-0002283 to Dr. Kim). The authors thank Dr. Dae-Yeon Cho at Lab Genomics Clinical Research Institute for genotyping and Drs. Greg J. Siegle and Kyung-Hwa Lee at University of Pittsburgh for processing neuroimaging data for this study.
References
- Arnsten AF. (2011) Catecholamine influences on dorsolateral prefrontal cortical networks. Biol Psychiatry 69:e89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederman J, Faraone SV. (2005) Attention-deficit hyperactivity disorder. Lancet 366:237–248. [DOI] [PubMed] [Google Scholar]
- Bohland JW, Saperstein S, Pereira F, Rapin J, Grady L. (2012) Network, anatomical, and non-imaging measures for the prediction of ADHD diagnosis in individual subjects. Front Syst Neurosci 6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SC, Kim BN, Hong YC, Shin MS, Yoo HJ, Kim JW, Bhang SY, Cho IH, Kim HW. (2010) Effect of environmental exposure to lead and tobacco smoke on inattentive and hyperactive symptoms and neurocognitive performance in children. J Child Psychol Psychiatry 51:1050–1057. [DOI] [PubMed] [Google Scholar]
- Cho SC, Hong YC, Kim JW, Park S, Park MH, Hur J, Park EJ, Hong SB, Lee JH, Shin MS, Kim BN, Yoo HJ, Cho IH, Bhang SY, Hahn S. (2013) Association between urine cotinine levels, continuous performance test variables, and attention deficit hyperactivity disorder and learning disability symptoms in school-aged children. Psychol Med 43:209–219. [DOI] [PubMed] [Google Scholar]
- Cortese S. (2012) The neurobiology and genetics of Attention-Deficit/Hyperactivity Disorder (ADHD): what every clinician should know. Eur J Paediatr Neurol 16:422–433. [DOI] [PubMed] [Google Scholar]
- Cox RW. (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173. [DOI] [PubMed] [Google Scholar]
- Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR, Jr, Barch DM, Petersen SE, Schlaggar BL. (2010) Prediction of individual brain maturity using fMRI. Science 329:1358–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPaul GJ, Power TJ, Anastopoulos AD, Reid R. (1998) The ADHD Rating Scale-IV: checklists, norms, and clinical interpretation. New York: Guildford Press. [Google Scholar]
- Eubig PA, Aguiar A, Schantz SL. (2010) Lead and PCBs as risk factors for attention deficit/hyperactivity disorder. Environ Health Perspect 118:1654–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fissell K, Tseytlin E, Cunningham D, Iyer K, Carter CS, Schneider W, Cohen JD. (2003) Fiswidgets: a graphical computing environment for neuroimaging analysis. Neuroinformatics 1:111–125. [DOI] [PubMed] [Google Scholar]
- Froehlich TE, McGough JJ, Stein MA. (2010) Progress and promise of attention-deficit hyperactivity disorder pharmacogenetics. CNS Drugs 24:99–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden CJ. (1978) The Stroop Color and Word Test. Chicago: Stoelting Company. [Google Scholar]
- Greenberg LM, Waldman ID. (1993) Developmental normative data on the test of variables of attention (T.O.V.A.). J Child Psychol Psychiatry 34:1019–1030. [DOI] [PubMed] [Google Scholar]
- Greicius M. (2008) Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol 21:424–430. [DOI] [PubMed] [Google Scholar]
- Guy W. (1976) ECDEU Assessment manual for psychopharmacology, Revised. Rockville, MD: US Department of Health, Education, and Welfare. [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. (2009) The WEKA data mining software: an update. SIGKDD Explorations 11:10–18. [Google Scholar]
- Hawkins DM, Basak SC, Mills D. (2003) Assessing model fit by cross-validation. J Chem Inf Comput Sci 43:579–586. [DOI] [PubMed] [Google Scholar]
- Herrmann MJ, Biehl SC, Jacob C, Deckert J. (2010) Neurobiological and psychophysiological correlates of emotional dysregulation in ADHD patients. Atten Defic Hyperact Disord 2:233–239. [DOI] [PubMed] [Google Scholar]
- Hong SB, Kim JW, Cho SC, Shin MS, Kim BN, Yoo HJ. (2012) Dopaminergic and noradrenergic gene polymorphisms and response to methylphenidate in Korean children with attention-deficit/hyperactivity disorder: is there an interaction? J Child Adolesc Psychopharmacol 22:343–352. [DOI] [PubMed] [Google Scholar]
- Hong SB, Harrison BJ, Fornito A, Sohn CH, Song IC, Kim JW. (2015) Functional dysconnectivity of corticostriatal circuitry and differential response to methylphenidate in youth with attention-deficit/hyperactivity disorder. J Psychiatry Neurosci 40:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, Williamson D, Ryan N. (1997) Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry 36:980–988. [DOI] [PubMed] [Google Scholar]
- Kebir O, Tabbane K, Sengupta S, Joober R. (2009) Candidate genes and neuropsychological phenotypes in children with ADHD: review of association studies. J Psychiatry Neurosci 34:88–101. [PMC free article] [PubMed] [Google Scholar]
- Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK. (2001) Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Comput 13:637–649. [Google Scholar]
- Kim BN, Kim JW, Cummins TD, Bellgrove MA, Hawi Z, Hong SB, Yang YH, Kim HJ, Shin MS, Cho SC, Kim JH, Son JW, Shin YM, Chung US, Han DH. (2013) Norepinephrine genes predict response time variability and methylphenidate-induced changes in neuropsychological function in attention deficit hyperactivity disorder. J Clin Psychopharmacol 33:356–362. [DOI] [PubMed] [Google Scholar]
- Kim Y, Cho SC, Kim BN, Hong YC, Shin MS, Yoo HJ, Kim JW, Bhang SY. (2010) Association between blood lead levels (<5 mug/dL) and inattention-hyperactivity and neurocognitive profiles in school-aged Korean children. Sci Total Environ 408:5737–5743. [DOI] [PubMed] [Google Scholar]
- King JA, Tenney J, Rossi V, Colamussi L, Burdick S. (2003) Neural substrates underlying impulsivity. Ann N Y Acad Sci 1008:160–169. [DOI] [PubMed] [Google Scholar]
- Kohavi R, John GH. (1997) Wrappers for feature subset selection. Artif Intell 97:273–324. [Google Scholar]
- Liston C, Malter Cohen M, Teslovich T, Levenson D, Casey BJ. (2011) Atypical prefrontal connectivity in attention-deficit/hyperactivity disorder: pathway to disease or pathological end point? Biol Psychiatry 69:1168–1177. [DOI] [PubMed] [Google Scholar]
- Mick E, Faraone SV. (2008) Genetics of attention deficit hyperactivity disorder. Child Adolesc Psychiatr Clin N Am 17:261–284, vii–viii. [DOI] [PubMed] [Google Scholar]
- Nigg J, Nikolas M, Burt SA. (2010) Measured gene-by-environment interaction in relation to attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 49:863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg JT, Knottnerus GM, Martel MM, Nikolas M, Cavanagh K, Karmaus W, Rappley MD. (2008) Low blood lead levels associated with clinically diagnosed attention-deficit/hyperactivity disorder and mediated by weak cognitive control. Biol Psychiatry 63:325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orru G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. (2012) Using Support Vector Machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci Biobehav Rev 36:1140–1152. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. (2012) Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosh PJ, Taylor E. (2000) Stimulant drugs. Eur Child Adolesc Psychiatry 9:I27–43. [DOI] [PubMed] [Google Scholar]
- Siegle GJ, Thompson W, Carter CS, Steinhauer SR, Thase ME. (2007) Increased amygdala and decreased dorsolateral prefrontal BOLD responses in unipolar depression: related and independent features. Biol Psychiatry 61:198–209. [DOI] [PubMed] [Google Scholar]
- Silva RR, Alpert M, Pouget E, Silva V, Trosper S, Reyes K, Dummit S. (2005) A rating scale for disruptive behavior disorders, based on the DSM-IV item pool. Psychiatr Q 76:327–339. [DOI] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. (2012) The influence of head motion on intrinsic functional connectivity MRI. Neuroimage 59:431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilens TE. (2008) Effects of methylphenidate on the catecholaminergic system in attention-deficit/hyperactivity disorder. J Clin Psychopharmacol 28:S46–53. [DOI] [PubMed] [Google Scholar]
- Woods RP, Mazziotta JC, Cherry SR. (1993) MRI-PET registration with automated algorithm. J Comput Assist Tomogr 17:536–546. [DOI] [PubMed] [Google Scholar]