Abstract
In the environment, most bacteria form surface-attached cell communities called biofilms. The attachment of single cells to surfaces involves an initial reversible stage typically mediated by surface structures such as flagella and pili, followed by a permanent adhesion stage usually mediated by polysaccharide adhesives. Here, we determine the absolute and relative timescales and frequencies of reversible and irreversible adhesion of single cells of the bacterium Caulobacter crescentus to a glass surface in a microfluidic device. We used fluorescence microscopy of C. crescentus expressing green fluorescent protein to track the swimming behavior of individual cells prior to adhesion, monitor the cell at the surface, and determine whether the cell reversibly or irreversibly adhered to the surface. A fluorescently labeled lectin that binds specifically to polar polysaccharides, termed holdfast, discriminated irreversible adhesion events from reversible adhesion events where no holdfast formed. In wild-type cells, the holdfast production time for irreversible adhesion events initiated by surface contact (23 s) was 30-times faster than the holdfast production time that occurs through developmental regulation (13 min). Irreversible adhesion events in wild-type cells (3.3 events/min) are 15-times more frequent than in pilus-minus mutant cells (0.2 events/min), indicating the pili are critical structures in the transition from reversible to irreversible surface-stimulated adhesion. In reversible adhesion events, the dwell time of cells at the surface before departing was the same for wild-type cells (12 s) and pilus-minus mutant cells (13 s), suggesting the pili do not play a significant role in reversible adhesion. Moreover, reversible adhesion events in wild-type cells (6.8 events/min) occur twice as frequently as irreversible adhesion events (3.3 events/min), demonstrating that most cells contact the surface multiple times before transitioning from reversible to irreversible adhesion.
In the environment, bacteria are most often found adhered to surfaces on which they form multicellular structures called biofilms. Biofilms provide protection from environmental stress and antibacterial agents such as antibiotics and other bacteriocides.1 Biofilms, therefore, provide a reservoir of infectious bacteria that is particularly difficult to eradicate. Furthermore, biofilm formation underlies the establishment and persistence of many bacterial infections in humans.2 Recently, important research efforts have sought to identify methods to inhibit biofilm maturation or to stimulate biofilm dispersal.3–5 Relatively little attention has been paid to the first stage of biofilm formation, the attachment of single bacterial cells to surfaces,1 although pioneering work with parallel-plate flow chambers and flow cells has shown the value of single cell analysis.6–9
Single bacterial cells exhibit both reversible and irreversible adhesion events.10–13 Reversible adhesion is typically mediated by thin proteinaceous structures such as fimbriae, pili, and flagella. Flagellum-driven motility brings a cell to the surface, where low-affinity reversible interactions between the cell and surface are mediated by the flagellum itself and by fimbriae, pili, or both. Irreversible adhesion results from secretion of an adhesin, typically composed of polysaccharides, proteins, or both, which tethers the bacterium to the surface. Understanding the detailed mechanisms by which bacteria attach to surfaces and the biosynthesis and properties of the adhesives that mediate their attachment is essential in order to elucidate the mechanisms of biofilm formation, bacterial infection, and bacterial persistence.
We are studying adhesion events of the fresh water bacterium Caulobacter crescentus. During the dimorphic life cycle of C. crescentus, a newborn motile swarmer cell is released from a stalked, non-motile cell.14 This morphological distinction simplifies the process of isolating and tracking motile cells that have not yet synthesized their permanent adhesin. C. crescentus adheres to surfaces with a polar adhesive, called holdfast, which resides at the pole of the cell. Moreover, this bacterium is genetically well-characterized, enabling experimental manipulation of cellular components, e.g., flagella and pili, and other mechanisms that contribute to adhesion.15–17 Because of the interest in bacterial adhesives, the biosynthetic pathway of holdfast formation18 and the protein and polysaccharide moieties,19–20 elasticity,21 and strength22 of the holdfast have been characterized.
Initially, adhesion of C. crescentus was believed to be regulated by developmentally programmed formation of holdfast during the differentiation of motile swarmer cells into the sessile, stalked stage of the life cycle.23 However, more recent work has identified the presence of holdfast in swarmer cells and the contributions of the flagellum and pili to initial surface attachment. Two distinct stages of adhesion result: transient surface interactions and irreversible adhesion with holdfast.17,24–25 In a study of C. crescentus adhesion,26 surface contact by the flagellum and pili was found to stimulate rapid production of holdfast (< 2 min), which is significantly faster than development-regulated formation of holdfast (13 min).
To study reversible and irreversible adhesion in more detail, we are using microfluidic devices, which have many similarities to conventional flow cells,8,27 including control of the local environment in which the cells reside, precise flow-rate management, single-cell tracking with microscopy, and higher cell throughput. In contrast to conventional flow cells, microfluidic devices can be designed in any two-dimensional architecture, fabricated with more precise dimensions, and manufactured inexpensively to be disposable. Consequently, microfluidic technologies are increasingly being applied to cell-based assays28–29 and have been used to generate chemical gradients for chemotaxis,30–31 to synchronize cells,32 and to analyze the behavior of single bacteria or ensembles of bacteria.33 Bacterial adhesion,34–36 biofilm formation,36–37 and bacterial growth38–40 have been studied in both flow cells and microfluidic devices.
In this paper, we determine the absolute and relative timescales and frequencies of reversible and irreversible adhesion events. Fluorescence microscopy of green fluorescent protein (GFP) and fluorescently labeled lectin (wheat germ agglutinin-Alexa Fluor 555) was used to track the position of C. crescentus cells and detect the presence of holdfast, respectively. With the microfluidic devices, we track the swimming behavior of individual cells prior to adhesion events (reversible or irreversible), monitor the dwell time of the cell at the surface, and determine whether the cell reversibly or irreversibly adhered to the surface. We confirm the role of pili in the transition from reversible to irreversible adhesion is critical. During reversible adhesion events, the length of time that pili-minus mutant cells dwell at the surface before departing (13 s) is identical to the dwell time of the wild-type cells (12 s). However, the pilus-minus mutant cells exhibited irreversible adhesion (0.2 events/min) 15-times less frequently than wild-type cells (3.3 events/min), suggesting that pili have an important function during the transition from reversible to irreversible adhesion. Our results also show that cells sample the surface multiple times before transitioning from reversible to irreversible adhesion. In wild-type cells, reversible adhesion events (6.8 events/min) were 2-times more frequent than irreversible adhesion events (3.3 events/min). For pili-minus mutant cells where surface-initiated holdfast development is less frequent, the ratio of reversible adhesion events to irreversible adhesion events is even higher at 20. Lastly, in wild-type cells, the time needed to produce holdfast is 30-times faster for surface-initiated adhesion (23 s) compared to development-regulated holdfast production (13 min). Our results highlight the complexity of adhesion, with cells sampling the surface multiple times before transiting to permanent adhesion.
Experimental Section
Materials
We purchased wheat germ agglutinin lectin labeled with Alexa Fluor 555 (WGA-AF555) or Alexa Fluor 594 (WGA-AF594) from Life Technologies Corp.; Sylgard 184 Silicone Elastomer Kit (poly(dimethylsiloxane) (PDMS)) from Dow Corning Corp.; SU-8 2010 and NanoPG Developer from MicroChem Corp.; Goldseal No. 1 coverslips (25 mm × 50 mm) from Thermo Fisher Scientific, Inc.; and glass slides (50 mm × 50 mm × 1 mm) from Corning, Inc. All other chemicals were purchased from Sigma Aldrich. Complex peptone yeast extract (PYE) medium14 was made from 2 g bactopeptone, 1 g yeast extract, 0.3 g MgSO4·7H2O, and 0.0735 g CaCl2·2H2O in 1 L of ultrapure water (18 MΩ·cm, Millipore Corp.) and autoclaved before use.
Device Fabrication
Microfluidic devices with single microchannels were fabricated in PDMS and sealed with glass coverslips. Briefly, the master was fabricated in SU-8 2010 on a glass substrate (50 mm × 50 mm × 1 mm) that was cleaned with methanol and dried in a stream of nitrogen. Adhesion promoter (2% titanium diisopropoxide bis(acetylacetonate) in propanol) was spin-coated onto the master prior to spin coating a 10 µm-thick layer of SU-8. This SU-8 base layer on the master was soft-baked on a hotplate (732P, PMC Industries) for 2 min at 60 °C (100 °C/h ramp), followed by 3 min at 90 °C (100 °C/h ramp). The SU-8 was then exposed to 200 mJ/cm2 of UV light (205S, Optical Associates, Inc.) and post-baked similar to the soft-bake to crosslink the SU-8 base layer. Next, a 20-µm thick layer of SU-8 2010 was spin-coated onto the master, soft-baked, and exposed to 200 mJ/cm2 of UV light through a transparency photomask (Photoplot Store) to transfer the microchannel pattern onto the master. The master was post-baked, developed for 2 min in NanoPG Developer, rinsed with isopropanol, and dried in a stream of nitrogen. Dimensions of the raised single-channel feature were measured with a stylus profiler (Dektak 6M, Veeco Instruments, Inc.) and were 110 ± 1 µm wide by 20 ± 0.1 µm high.
PDMS replicas were cast on the SU-8 master from 1 mL of a 10:1 mixture of PDMS prepolymer:curing agent. The PDMS was dispensed onto the master, degassed under vacuum for 20 min, and cured in an oven (DKN-600; Yamato Scientific America, Inc.) at 70 °C for 15 min. The replica was removed from the master, fluid access holes were punched 1 cm apart, and the replica was reversibly sealed to a No. 1 glass coverslip. Glass reservoirs (6-mm tall and 4-mm i.d.) were affixed over the access holes with uncured PDMS, and the assembled device was cured for an additional 15 min at 70 °C in the oven.
The devices above used the PDMS substrates without further treatment, and the PDMS surface was hydrophobic with a contact angle of 98°. To evaluate the influence of hydrophobicity on bacterial adhesion, devices were fabricated where the PDMS surfaces were plasma-treated in an air plasma (PDC-32G, Harrick Plasma) prior to assembly. The plasma-treated PDMS substrates and glass coverslips were hydrophilic with contact angles < 30°. We used the plasma-treated PDMS substrates for the adhesion experiments within 1–2 h after plasma treatment to ensure surfaces were hydrophilic.
Cell Culture and Synchronization
Two strains of C. crescentus were used in these studies: wild-type cells expressing green fluorescent protein (CB15 miniTn7gfp3 (YB4789)) and pilus-minus mutant cells expressing green fluorescent protein (CB15 ΔpilA miniTn7gfp3 (YB4795)). We used a modified plate-release technique to culture and collect ~80% swarmer enriched populations.41 For each strain, 3 mL of PYE medium was inoculated with a single colony and was shaken for 5–6 h at 30 °C. A 1-mL aliquot of the grown culture was diluted in 50 mL of PYE medium, placed in a 150-mm diameter sterile polystyrene petri dish, and shaken overnight (16–18 h) at room temperature on a reciprocal shaker at 70 rpm. Four hours before swarmer cell collection, the plate was rinsed with 100 mL of sterile ultrapure water, and an additional 50 mL of PYE medium was added to restore exponential growth. Just prior to swarmer cell collection, the plate was rinsed with 100 mL of PYE medium, a 1-mL aliquot of PYE medium was added to the plate, and the plate was shaken for 5 min on the reciprocal shaker at 70 rpm. The 1-mL aliquot of the swarmer enriched medium was collected from the plate, centrifuged at 5000 × g for 5 min at 4 °C, and concentrated to a final volume of 100 µL by removal of the supernatant. A 1-µL aliquot of 50 µg/mL of WGA-AF555, which binds specifically to holdfast, was added to the concentrated cell suspension.
Adhesion Assays
The microfluidic device was filled initially with media by application of vacuum to the waste reservoir. To introduce swarmer cells into the microchannel, hydrostatic flow was generated by addition of 100 and 10 µL of media to the cell and waste reservoirs, respectively. The swarmer cell enrichment (~10 µL) were added to the cell reservoir and mixed thoroughly. After swarmer cells were observed in the analysis area, the fluid levels were adjusted to 50 µL in the cell reservoir and 45 µL in the waste reservoir to reduce the hydrostatic velocity to 30–35 µm/s. With 110-µm wide and 20-µm deep channels and a flow velocity of 30–35 µm/s, less than 50 nL of buffer (i.e., less than 0.1% of the volume in the reservoir) is consumed over the course of a 10-min experiment.
To monitor single-cell adhesion events, we used an inverted optical microscope (TE2000-U, Nikon Instruments, Inc.) configured for epifluorescence and equipped with a CCD camera (C9100-13, Hamamatsu Corp.). In addition, a two-channel imaging system (DV2 Dual View, Photometrics) was placed in front of the CCD camera and had GFP/DsRed filters (Chroma Technology) to monitor the fluorescence signal from the cell body (GFP) and the lectin-labeled holdfast (WGA-AF555) simultaneously. Video data were collected with MetaMorph imaging software (Molecular Devices, LLC.). To minimize photobleaching of the GFP and WGA-AF555 during the experiments, we acquired full-frame videos at 4 fps for 5 s, then delayed acquisition for 5 s during which a mechanical shutter (Lambda SC, Sutter Instruments) was closed. Acquisition at this 10-s duty cycle (5-s on and 5-s off) continued until the experiment was completed. For longer duration experiments, photobleaching of the GFP signal from the cell body limited the length of time an adhered cell was observed, but in most cases, the necessary information was obtained because either the holdfast formed or the cell detached from the surface before photobleaching occurred.
Analysis of Adhesion Events
We analyzed all video data with the MetaMorph software and open-source ImageJ software (http://rsbweb.nih.gov/ij/). First, in MetaMorph, each full-frame video was parsed into two 512-pixel × 256-pixel frames that corresponded to the GFP signal and WGA-AF555 signal. For each 5-s data segment, 20 frames of GFP data were left in video format, and the corresponding 20 frames of the WGA-AF555 data were averaged in a single frame to improve the signal-to-noise ratio. Then, the video was reassembled with the GFP video data first, followed by the averaged WGA-AF555 frame. This sequence was selected to ensure that any cells that arrived with a holdfast (motile or non-motile) were not considered in the adhesion analysis.
The compiled video was then processed with ImageJ to identify irreversible adhesion with holdfast formation and reversible adhesion with and without holdfast formation. To determine irreversible adhesion with holdfast formation, the averaged WGA-AF555 frame was analyzed with a modified cell counter plug-in that allowed the operator to identify individual holdfast signals with a signal-to-noise ratio (SNR) ≥ 10. The operator recorded the time and location (i.e., x- and y-coordinates) when and where the holdfast appeared, reviewed the GFP video data in reverse chronological order, and recorded the arrival time of a cell at the surface. From this information, the holdfast production time is calculated as the time between initial adhesion at the surface and when holdfast is detected at SNR ≥ 10.
The operator used the same video data to track reversible adhesion events with and without holdfast formation. The only difference is that reversibly adhered cells left the surface either before or after holdfast formation. For the reversible events without holdfast formation, the dwell time is the difference between arrival and departure times of a cell at the surface. These reversible adhesion events did not have a corresponding WGA-AF555 signal indicating holdfast formation.
Agarose Pad Assays
PYE medium containing 1% agarose and 0.5 µg/ml of WGA-AF594 was applied to a 25-mm by 75-mm glass slide to form an agarose pad (22 mm × 22 mm × 0.5 mm) capable of supporting the growth of bacteria. Exponential phase cell culture was diluted to an OD600 of 0.05–0.2, and 0.8 µL of the cell culture was spotted on the agarose pad. The agarose pad was covered with a coverslip and sealed with a 1:1:1 mixture of Vaseline, lanolin, and paraffin. An upright optical microscope (Eclipse 90i, Nikon Instruments, Inc.) equipped with a 100× DIC Plan Apo VC oil objective was used for differential interference contrast (DIC) and epifluorescence microscopy. Images were captured at 1 min intervals with a CCD camera (Cascade 1K, Photometrics), triple filter cube (83700, Chroma Technology Corp.), and MetaMorph imaging software. Series of images were used to determine the time of cell division and subsequent time of holdfast detection at the pole of the newborn swarmer cell. For the agarose pad assays, the time for holdfast production is the time between cell division and initial holdfast detection.
Measurement of Young’s Modulus
We measured the Young’s modulus (or modulus of elasticity) of the glass, PDMS, and agarose substrates with an atomic force microscope (AFM; MFP-3D, Asylum Research, Inc.) and a 160-µm long cantilever (0.01–0.2 ohm-cm silicon; Bruker OTESPA-R3). The spring constants of each substrate were determined by fitting the slope of the approach (or force) curve (nN) versus nanoindentation (nm) with the JKR model. Prior to each measurement, the spring constant of the cantilever was calibrated on a mica surface. The measured Young’s moduli were 87 ± 3 GPa for glass, 4.9 ± 0.03 MPa for PDMS, and 140 ± 60 kPa for 1% agarose for n = 3 on each substrate.
Results and Discussion
Cell Adhesion Events in Microchannels
With our single microchannel device, we were able to determine the holdfast production times for irreversible adhesion events and dwell times at the surface for reversible events. The microchannel environment offered several advantages over similar adhesion experiments conducted on microscope slides with a coverslip.26 The microchannel effectively confined the lateral movement of bacteria and enhanced our ability to track individual motile cells and subsequent adhesion events. To observe more adhesion events in each experiment, we opted to use a larger field of view (20× objective) and maintained sufficient signal-to-noise ratios for cell body- and holdfast-associated fluorescence to identify each type of adhesion event. With the 20× objective, the entire 100-µm wide microchannel was imaged with the Dual View system. Moreover, a microchannel depth of 20-µm helped to promote swarmer cell contact with the surface and to keep motile cells in focus longer during cell tracking. The gentle hydrostatic flow velocity (30–35 µm/s) continuously delivered new swarmer cells to the analysis region and fresh WGA-AF555 to cells producing holdfast. As seen in Video 1 in the Supporting Information, the swarmer cells easily swim axially and laterally in the microchannel, but had a net displacement in the axial (vertical) direction of the flow over the course of the experiment. The swim velocity of the swarmer cells was 42 µm/s without hydrostatic flow and 45 µm/s in a hydrostatic flow with a velocity of 30 µm/s. Comparable swim velocities are not unexpected in hydrostatic flow with a really low Reynolds number (e.g., Re < 0.001 for a 110-µm wide and 20-µm deep microchannel and a linear flow velocity of 30–35 µm/s). As a point of reference, fluorescent probes with molecular weights of 400–600 Da diffuse at rates of 20–30 µm/s.42
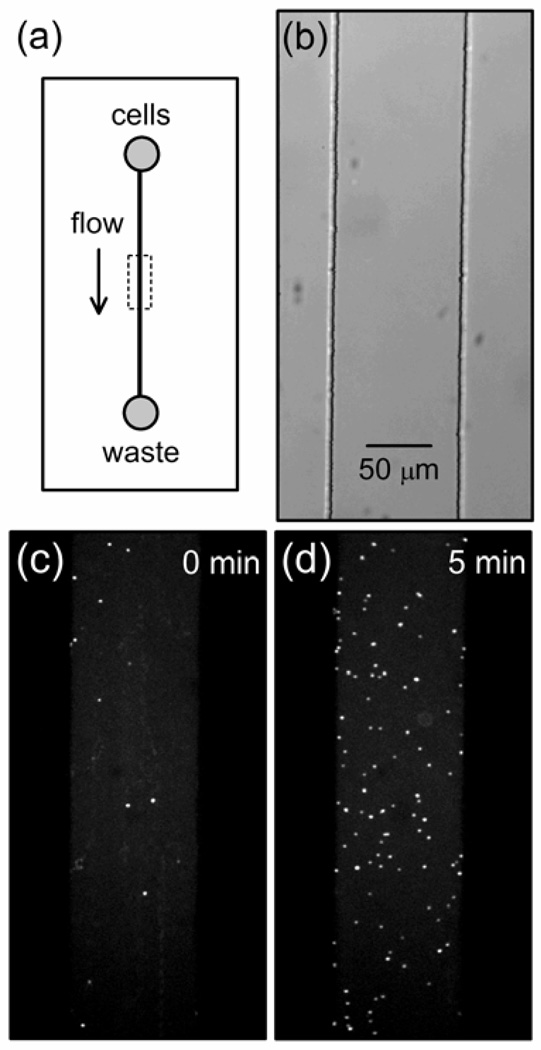
Figure 1b shows a bright-field image of the microchannel cast in PDMS in which adhesion events were monitored. Figures 1c–d are GFP fluorescence images of cells adhered in the channel at the start of the experiment (0 min) and after 5 min. As seen in Figure 1d, ~90 cells have accumulated on the glass surface over the 5-min experiment. Video 1 shows the cells being drawn into the analysis region by hydrostatic flow, swimming in the microchannel, and accumulating on the glass surface over the same 5-min experiment shown in Figure 1c–d. The video is sped up 17-fold.
Figure 1.
(a) Schematic of a microfluidic device with a single poly(dimethylsiloxane) (PDMS) microchannel sealed with a glass cover plate. To deliver bacteria to the analysis region (dashed box), hydrostatic flow is generated by a higher fluid level in the cell reservoir relative to the waste reservoir. (b) Bright-field image of a 100-µm wide microchannel cast in PDMS with a glass cover plate. (c)–(d) Fluorescence images of the GFP signal from wild-type C. crescentus cells adhered to the glass cover plate at (c) 0 and (d) 5 min after the start of the experiment. The images were averaged for 5 s (or 20 frames) to minimize signal from the motile swarmer cells.
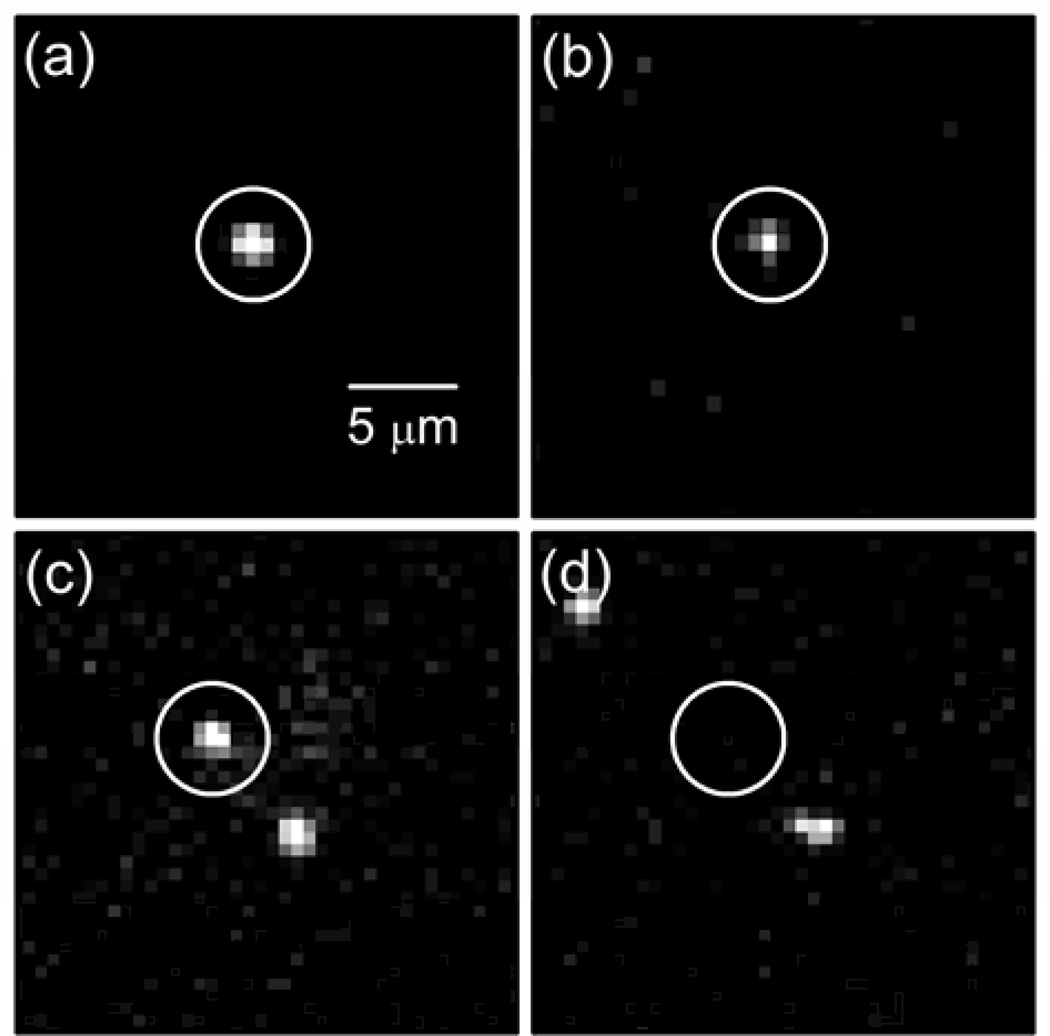
Figures 2a–d are fluorescence images that demonstrate two types of adhesion events. Figures 2a and 2b show fluorescence images of wild-type C. crescentus (GFP) and WGA-AF555 bound to holdfast, respectively. In these two images, one cell irreversibly adhered to the surface with lectin-bound holdfast (circled) and had a holdfast production time of 19 s. Figures 2c and 2d highlight a reversible adhesion event of wild-type C. crescentus. The cell arrived at the surface but did not develop holdfast (cell circled in Figure 2c), and subsequently, the cell swam away from the surface and had a dwell time of 12 s (Figure 2d). To determine the holdfast production time, a constant concentration of the WGA-AF555 (0.5 µg/mL) was maintained in the cell suspension to overcome issues associated with the relatively weak binding affinity of WGA to various oligosaccharides.43 As the WGA-AF555 accumulated at the holdfast, the fluorescence signal at the holdfast increased in intensity relative to the background concentration of the WGA-AF555. Most experiments, especially with wild-type cells, were limited by the absolute number of adhesion events that could be resolved in the field of view. Therefore, only the first 5–6 min of data were typically analyzed before the field of view became too crowded.
Figure 2.
(a) GFP fluorescence image of an irreversible (permanent) adhesion event of a wild-type cell at the glass surface and (b) fluorescence image of labeled lectin (WGA-AF555) bound to the holdfast of the cell in panel (a). The holdfast production time for the cell was 19 s. GFP fluorescence images of (c) a reversible adhesion event of wild-type cell at the glass surface and (d) the same location as panel (c) after the cell detached from the surface. The dwell time for the cell was 12 s. Images in panels (a)–(d) were acquired at 20× magnification and have the same scale, and the white circles highlight the events described.
The distribution of reversible and irreversible adhesion events, their frequencies, and timescales that were observed in typical experiments with wild-type cells and pilus minus mutant cells are summarized in Table 1. For wild-type cells, the breakdown of these events is as follows: (1) 24% were irreversible adhesion events where the cells produced holdfast to tether the cell permanently to the surface, (2) 50% were reversible adhesion events during which cells dwelled at the surface for a brief period of time then departed, and (3) 26% were reversible adhesion events where the cells arrived at the surface with holdfast then departed or reversible adhesion events for which the cells produced holdfast then departed. For pilus-minus mutant cells, (1) 4% were irreversible adhesion events, (2) 86% reversible adhesion events, and (3) 10% were cells arriving at the surface with holdfast or departing from the surface after secreting holdfast. One possible explanation for the cells that depart from the surface after holdfast formation is that the holdfast had not yet made surface contact. Pilus retraction may properly position the cell pole and holdfast in order to facilitate irreversible binding.26 Adhesion events in which cells arrived at the surface with a holdfast or departed after secreting a holdfast were not analyzed further because their history prior to contacting the surface or after departing the surface could not be accounted for accurately. Below, we focus our analysis and discussion on irreversible adhesion with holdfast formation and reversible adhesion without holdfast formation.
Table 1.
Distribution, frequencies, and timescales of reversible and irreversible adhesion events for wild-type and pilus-minus mutant cells.
| wild-type cells |
pilus-minus mutant cells |
|
|---|---|---|
| distribution of total events | ||
| irreversible adhesion (%) | 24 | 4 |
| reversible adhesion (%) | 50 | 86 |
| othera (%) | 26 | 10 |
| irreversible adhesionb | ||
| frequency (events/min) | 3.3 | 0.2 |
| holdfast formation time (s) | 23 | 55 |
| reversible adhesionc | ||
| frequency (events/min) | 6.8 | 4.4 |
| dwell time (s) | 12 | 13 |
The “other” category includes cells that enter with holdfast and cells that develop holdfast then depart the surface.
For wild-type cells, 631 events were counted in 192 min, and for pilus-minus mutant cells, 10 events were counted in 45 min.
For wild-type cells, 171 events were counted in 25 min, and for pilus-minus mutant cells, 159 events were counted in 36 min.
Irreversible Adhesion Events
Image analysis of collected video data permitted accurate tracking of irreversible adhesion events and the time needed for the cell to secrete (or synthesize) holdfast after initially adhering to the surface (holdfast production time). We used PYE medium in these experiments because the frequency of permanent adhesion events of C. crescentus to glass surfaces is typically higher in PYE than in other media such as minimal growth medium with 0.2% glucose (M2G)41 where a low proportion of cells synthesize holdfasts.44 For wild-type C. crescentus cells, a histogram of the holdfast production times for irreversible adhesion events is shown in Figure 3. Of the 631 total events, 73% of holdfasts were produced in fewer than 50 s with a time constant for holdfast production of 23.1 ± 1.1 s, determined from a single exponential decay fitted to the data. The histogram indicates just how rapidly a cell produces holdfast after making contact with a surface. Strikingly, ~30% of the cells synthesized a holdfast within 10 s of surface contact. This observation is consistent with previous reports in which TIRF microscopy revealed that holdfast can be detected less than 2 min after surface contact.26 However, the ability to monitor holdfast production following surface contact within a microchannel increases both the number of cells monitored and the temporal resolution of holdfast detection. The rapidity of holdfast synthesis after surface contact supports an intriguing possibility that secretion of an already synthesized holdfast is triggered.26
Figure 3.
Histogram of holdfast production time after wild-type cells adhere to the glass surface. The holdfast production time is the time needed for the cell to secrete (or synthesize) holdfast after initially adhering to the surface. The histogram includes only cells that were motile prior to irreversible adhesion. Total number of irreversible adhesion events is 631, and the bin size is 10 s. The fitted line is a single exponential decay with a time constant for the holdfast production time of 23.1 ± 1.1 s.
Pili are important for initial attachment to surfaces17,24–25 and are critical for irreversible attachment events following surface contact.26 When similar experiments were conducted for pilus-minus mutant cells, the number of observed irreversible adhesion events was significantly reduced. For the pilus-minus mutant cells, we observed only 10 irreversible adhesion events and calculated an average of 0.2 events/min from these 10 events over six experiments with an average duration of 7.5 min (45 min of total time) (Table 1). Under similar conditions for the wild-type cells, an average of 3.3 events/min of irreversible adhesion events were estimated from 631 permanent adhesion events from 30 experiments with average duration of 6.4 min (192 min of total time). For the 10 irreversible adhesion events observed for the pilus-minus mutant cells, the holdfast production times ranged from 7 to 139 s with an average time of 55 s. The low frequency of events prevented the data from being fitted with a single-exponential decay. These results confirmed the critical importance of the pili for the stimulation of holdfast synthesis following surface contact.
To evaluate the effects of the surface hydrophobicity on bacterial adhesion, the number of cells with holdfasts attached to either the glass bottom or PDMS top of devices without and with plasma treatment of the PDMS substrate were quantitated. The contact angle of the untreated PDMS surface was 98°, and the contact angles of the plasma-treated PDMS and glass coverslips were below 30° and could not be accurately measured with our instrument. After plasma treatment, the PDMS substrates were used within 1–2 h for adhesion experiments to ensure the surfaces remained hydrophilic. In three 30-min experiments, 1820 cells with holdfast were counted, and 60% of the cells were adhered to the untreated (hydrophobic) PDMS surface, and 40% of the cells were adhered to the glass coverslip. In another three 30-min experiments, 1600 cells with holdfast were counted, and 21% of the cells were adhered to the plasma-treated (hydrophilic) PDMS substrate to the 79% of the cells were adhered to the glass surface. These results demonstrate that the cells prefer the hydrophobic surface of the untreated PDMS compared to the more hydrophilic surfaces of the plasma-treated PDMS and glass coverslip. The reason for the preference to adhere to the glass coverslip relative to the plasma-treated PDMS is unclear and may have to do with the surface chemistry or the modulus of elasticity of the material (see below). In complementary experiments, the holdfast adheres with greater force to hydrophobic surfaces (e.g., graphite) than to hydrophilic surfaces (e.g., mica and glass).45
Developmental Holdfast Production on Agarose Pads
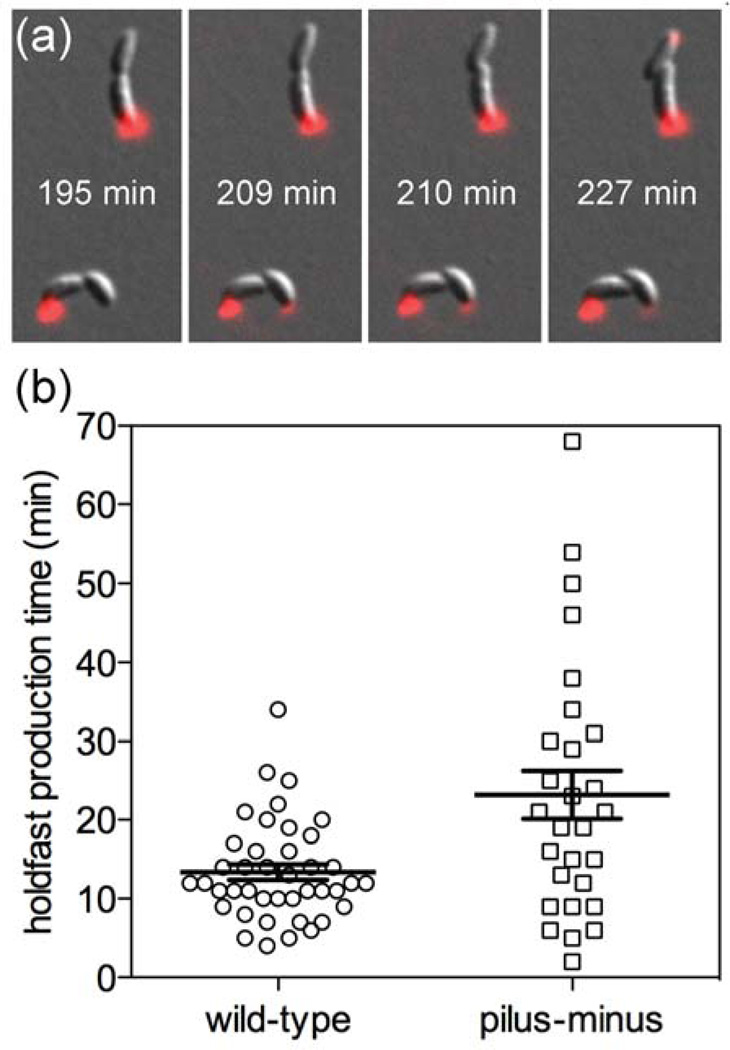
We determined the times for holdfast production to occur developmentally, not by surface contact stimulation, for both wild-type and pilus-minus mutant cells growing on agarose pads. We used agarose pads covered with glass coverslips, not microfluidic devices, for these assays because agarose pads have a low Young’s modulus (see below). Consequently, alignment of the agarose along the cell body prevents polar contact with the glass, and surface contact stimulation of the holdfast is minimized. DIC and fluorescence microscopy were used to identify the time of cell division and subsequent developmental holdfast production in wild-type and pilus-minus C. crescentus cells. Figure 4a shows composite images of two wild-type cells that have undergone division at 195 min (bottom image) and 210 min (top image). Because motility of the swarmer cell is hindered by confinement in the agarose pad, the swarmer cell does not move away from the stalked cell, although the two cells have divided. Monitoring of the swarmer cells continued, and holdfast formation (red spot at the cell pole) was detected at 209 min and 227 min, respectively, for the cells in the bottom and top images. By subtracting the division time from the holdfast formation time, elapsed times for holdfast formation were determined. The average times for holdfast production were 13.4 ± 1.0 min for 41 wild-type cells and 23.2 ± 3.1 min for 28 pilus-minus mutant cells. We hypothesize that the difference between the two strains is due to the fact that the soft agarose still permits some surface contact stimulation for wild-type cells, albeit less efficiently than a hard surface. From these data, we can conclude that surface contact initiates holdfast production at least 30-times faster than cells would produce holdfast through a normal developmental cycle.
Figure 4.
Holdfast production time on agarose pads. (a) Composite images from DIC and fluorescence microscopy of wild-type cells used to determine the time of cell division (195 min for the cell on the bottom; 210 min for the cell on the top) and the time of holdfast detection (209 min for the cell on the bottom; 227 min for the cell on the top). The holdfast production time is the time of holdfast detection minus the time of cell division. (b) Holdfast production times for wild-type cells and pilus-minus mutant cells on agarose pads. The average times and standard deviations for holdfast production, indicated by the bars, are 13.4 ± 1.0 min for 41 wild-type cells and 23.2 ± 3.1 min for 28 pilus-minus mutant cells. For these experiments, agarose pads, instead of microfluidic devices, were used to minimize polar contact and surface contact stimulation.
As noted above, the 1% agarose pads do not stimulate holdfast production through surface contact, whereas the glass and PDMS substrates stimulate holdfast production. The Young’s moduli were measured to be 87 ± 3 GPa for glass, 4.9 ± 0.03 MPa for PDMS, and 140 ± 60 kPa for 1% agarose and are comparable to reported values for D263 glass,46 PDMS,47 and agarose48 substrates. The Young’s modulus of 1% agarose pads is too low to stimulate holdfast production in C. crescentus, whereas the Young’s moduli of PDMS and glass are sufficient to stimulate holdfast production. As observed in these experiments, both the modulus of elasticity and surface chemistry (e.g., hydrophilic and hydrophobic) play important roles in stimulating holdfast production and require further investigation.
Dwell Time Analysis
The developmental program of C. crescentus triggered holdfast production in both wild-type and pilus-minus mutant cells, suggesting that the absence of pili does not prevent holdfast production. In order to determine if the pilus-minus mutants are impaired in reversible binding, we compared the dwell times of reversible adhesion events of these cells to wild-type cells. As noted above, the pilus-minus mutants seldom exhibited irreversible adhesion with holdfast production; however, the pilus-minus mutant cells would contact the surface, reside for a period of time, and then detach without having formed holdfast (Table 1). We define this time that the cell resides at the surface during a reversible event as the dwell time.
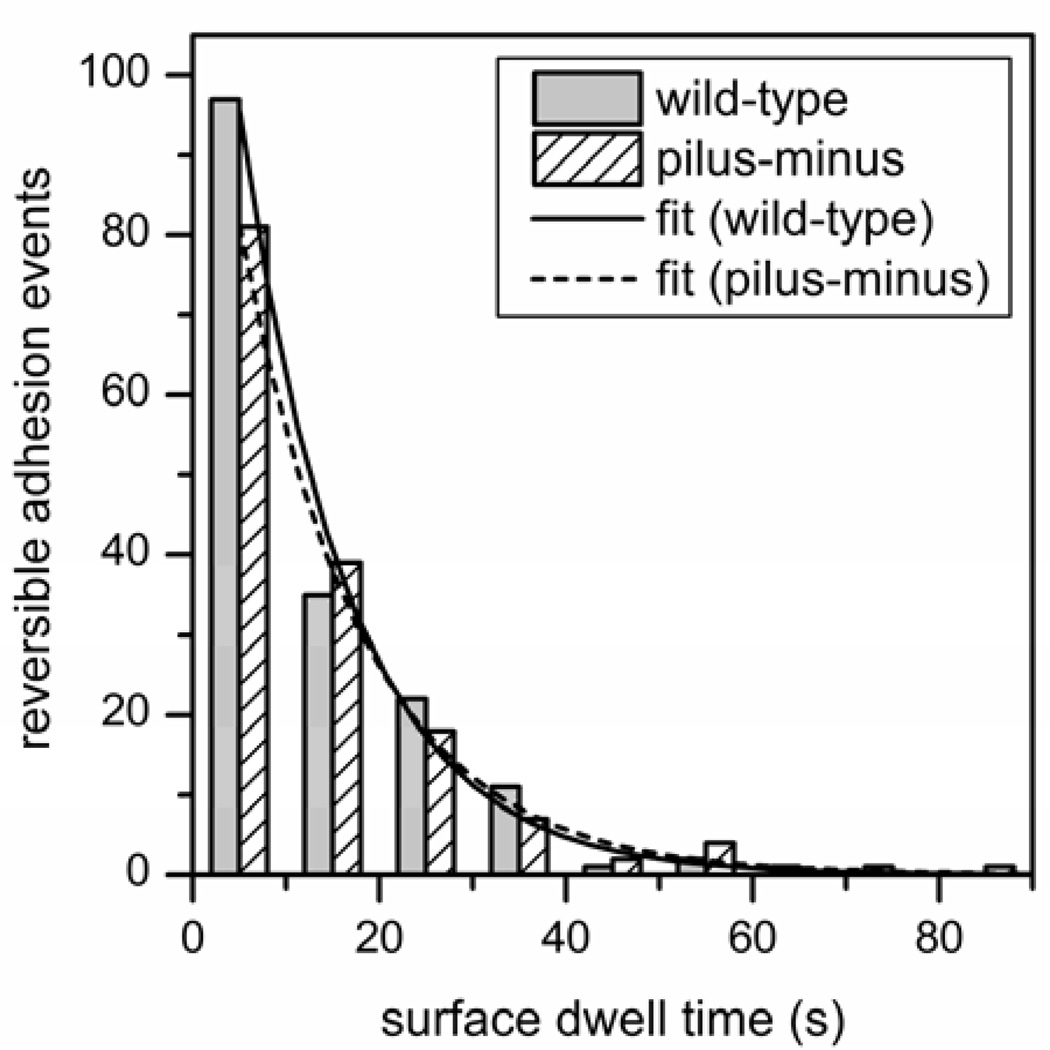
Figure 5 compares the histograms of dwell times for wild-type cells and pilus-minus mutant cells. Total counts for wild-type cells and pilus-minus mutants are 171 and 159, respectively. For the wild-type cells, the 171 reversible events were collected in four experiments with an average duration of 6.3 min (25 min of total time), corresponding to a frequency of 6.8 events/min. For the pilus-minus mutants, the 159 reversible events were collected in six experiments with an average duration of 6.0 min (36 min of total time), corresponding to a frequency of 4.4 events/min. Each histogram was fitted with a single exponential decay, and the time constants for the dwell times were 11.7 ± 0.5 s for the wild-type cells and 13.2 ± 0.3 s for pilus-minus mutants. Both the dwell times and frequency of events are similar for the wild-type and pilus-minus mutant cells, indicating the pili do not influence reversible adhesion.
Figure 5.
Histograms of dwell times at the surface for wild-type cells and the pilus-minus mutant cells. Dwell time is the time of a reversible adhesion event during which a cell adheres to the surface, resides without forming a holdfast, and then detaches. Total numbers of reversible adhesion events are 171 for the wild-type cells and 159 for the pilus-minus mutants, and the bin size is 10 s. Fitted lines are single exponential decays with time constants of 11.7 ± 0.5 s for the wild-type cells and 13.2 ± 0.3 s for the pilus-minus mutant cells.
Surface contact by wild-type cells begins by cell tethering followed by rapid arrest of flagellum motor rotation. Conversely, pili-minus mutant cells cannot arrest flagellum rotation after tethering or stimulate holdfast synthesis, and they eventually leave the surface.26 These previous results indicated that pili, which are located at the same cell pole as the flagellum, play an important role in the transition from reversible to irreversible attachment, but whether pili also played a role in the initial cell tethering to the surface remained unclear. The results reported here clearly indicate the pilus-minus mutants are not defective in surface tethering and reversible attachment; therefore, initial cell tethering is mediated by the flagellum.
Conclusion
We have identified the timescales and frequencies for holdfast formation in irreversible adhesion events and cell residence at a surface during reversible adhesion events. Holdfast formation after polar surface contact stimulation was more than 30 times faster than holdfast formed developmentally on an agarose pad. Moreover, reversible adhesion events with no holdfast formation were comparable for wild-type cells and pilus-minus mutants. These results indicate that pili do not play a major role in initial tethering to surfaces, but instead mediate the transition from reversible to irreversible adhesion by stopping flagellum rotation and the resulting activation of the holdfast synthesis or export machinery. Indeed, newborn swarmer cells already have an assembled holdfast synthesis-anchoring machinery.49 Because swarmer cells harbor between one and seven pili,50 we hypothesize that the time necessary for wild-type cells to stimulate holdfast synthesis upon surface contact may be a function of the number of pili they harbor.
In these experiments, the microchannel system afforded several advantages over traditional biology assays because we can precisely control the local environment in which the cells reside and promote surface contact. These advantages led to observation of hundreds of unique adhesion events and thus a more precise determination of holdfast formation time compared to previous studies.26 Furthermore, using a microfluidic device enabled quantification of the dwell time at the surface for reversible events and the relative frequencies of reversible and irreversible adhesion events. Future work on behavioral or environmental cues will further elucidate the roles of the flagellum and pili in these adhesion processes.
Supplementary Material
Acknowledgments
This work was supported in part by NIH R01 GM113172 for S.C.J. and Y.V.B, NIH R01 GM102841 for Y.V.B., and the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. Support was also provided by NIH National Research Service Awards for P.J.B.B. (F32AI072992 from NIAID) and D.T.K. (F32GM083581 from NIGMS). The authors thank Andrew S. Wilkens for assistance with cell velocity measurements and image and movie preparation and Courtney K. Ellison and Dr. Zach D. Harms for the measurements of the Young’s modulus.
Footnotes
Supporting Information Available. Video of cells swimming in the microchannel and adhering to the glass cover plate over 5 min. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Karatan E, Watnick P. Microbiol. Mol. Biol. Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potera C. Science. 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- 3.Sintim HO, Smith JAI, Wang J, Nakayama S, Yan L. Future Med. Chem. 2010;2:1005–1035. doi: 10.4155/fmc.10.185. [DOI] [PubMed] [Google Scholar]
- 4.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. Nat. Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasko DA, Sperandio V. Nat. Rev. Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 6.Lecuyer S, Rusconi R, Shen Y, Forsyth A, Vlamakis H, Kolter R, Stone HA. Biophys. J. 2011;100:341–350. doi: 10.1016/j.bpj.2010.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busscher HJ, Norde W, Van der Mei HC. Appl. Environ. Microbiol. 2008;74:2559–2564. doi: 10.1128/AEM.02839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busscher HJ, van der Mei HC. Clin. Microbiol. Rev. 2006;19:127–141. doi: 10.1128/CMR.19.1.127-141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Utada AS, Bennett RR, Fong JCN, Gibiansky ML, Yildiz FH, Golestanian R, Wong GCL. Nat. Commun. 2014;5:4913. doi: 10.1038/ncomms5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigeant MAS, Ford RM, Wagner M, Tamm LK. Appl. Environ. Microbiol. 2002;68:2794–2801. doi: 10.1128/AEM.68.6.2794-2801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 12.Agladze K, Wang X, Romeo T. J. Bacteriol. 2005;187:8237–8246. doi: 10.1128/JB.187.24.8237-8246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Dellen KL, Houot L, Watnick PI. J. Bacteriol. 2008;190:8185–8196. doi: 10.1128/JB.00948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poindexter JS. Bacteriol. Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nierman WC, et al. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown PJB, Hardy GG, Trimble MJ, Brun YV. In: Advances in Microbial Physiology. Poole RK, editor. Vol. 54. London: Academic Press Ltd-Elsevier Science Ltd; 2009. pp. 1–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodenmiller D, Toh E, Brun YV. J. Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toh E, Kurtz HD, Brun YV. J. Bacteriol. 2008;190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merker RI, Smit J. Appl. Environ. Microbiol. 1988;54:2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li GL, Smith CS, Brun YV, Tang JX. J. Bacteriol. 2005;187:257–265. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alipour-Assiabi E, Li GL, Powers TR, Tang JX. Biophys. J. 2006;90:2206–2212. doi: 10.1529/biophysj.105.064592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsang PH, Li GL, Brun YV, Ben Freund L, Tang JX. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janakiraman RS, Brun YV. J. Bacteriol. 1999;181:1118–1125. doi: 10.1128/jb.181.4.1118-1125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levi A, Jenal U. J. Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Entcheva-Dimitrov P, Spormann AM. J. Bacteriol. 2004;186:8254–8266. doi: 10.1128/JB.186.24.8254-8266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li GL, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Mol. Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busscher HJ, Vandermei HC. Methods Enzymol. 1995;253:455–477. doi: 10.1016/s0076-6879(95)53039-8. [DOI] [PubMed] [Google Scholar]
- 28.Salieb-Beugelaar GB, Simone G, Arora A, Philippi A, Manz A. Anal. Chem. 2010;82:4848–4864. doi: 10.1021/ac1009707. [DOI] [PubMed] [Google Scholar]
- 29.Kovarik ML, Gach PC, Ornoff DM, Wang YL, Balowski J, Farrag L, Allbritton NL. Anal. Chem. 2012;84:516–540. doi: 10.1021/ac202611x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovarik ML, Brown PJB, Kysela DT, Berne C, Kinsella AC, Brun YV, Jacobson SC. Anal. Chem. 2010;82:9357–9364. doi: 10.1021/ac101977f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KS, Boccazzi P, Sinskey AJ, Ram RJ. Lab Chip. 2011;11:1730–1739. doi: 10.1039/c1lc20019d. [DOI] [PubMed] [Google Scholar]
- 32.Madren SM, Hoffman MD, Brown PJB, Kysela DT, Brun YV, Jacobson SC. Anal. Chem. 2012;84:8571–8578. doi: 10.1021/ac301565g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Chen ZZ, Li QL. Microchim. Acta. 2010;168:177–195. [Google Scholar]
- 34.Boks NP, Norde W, van der Meil HC, Busscher HJ. Microbiology. 2008;154:3122–3133. doi: 10.1099/mic.0.2008/018622-0. [DOI] [PubMed] [Google Scholar]
- 35.Kucik DF, Wu C. Methods Mol. Biol. 2005;294:43–54. doi: 10.1385/1-59259-860-9:043. [DOI] [PubMed] [Google Scholar]
- 36.Kim HN, Hong Y, Lee I, Bradford SA, Walker SL. Biomacromolecules. 2009;10:2556–2564. doi: 10.1021/bm900516y. [DOI] [PubMed] [Google Scholar]
- 37.Meyer MT, Roy V, Bentley WE, Ghodssi R. J. Micromech. Microeng. 2011;21:054023. [Google Scholar]
- 38.Lambert G, Liao D, Vyawahare S, Austin RH. J. Bacteriol. 2011;193:1878–1883. doi: 10.1128/JB.01430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottschamel J, Richter L, Mak A, Jungreuthmayer C, Birnbaumer G, Milnera M, Bruckl H, Ertl P. Anal. Chem. 2009;81:8503–8512. doi: 10.1021/ac901420u. [DOI] [PubMed] [Google Scholar]
- 40.Volfson D, Cookson S, Hasty J, Tsimring LS. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15346–15351. doi: 10.1073/pnas.0706805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Degnen ST, Newton A. J. Mol. Biol. 1972;64:671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- 42.Culbertson CT, Jacobson SC, Ramsey JM. Talanta. 2002;56:365–373. doi: 10.1016/s0039-9140(01)00602-6. [DOI] [PubMed] [Google Scholar]
- 43.Lienemann M, Paananen A, Boer H, de la Fuente JM, Garcia I, Penades S, Koivula A. Glycobiology. 2009;19:633–643. doi: 10.1093/glycob/cwp030. [DOI] [PubMed] [Google Scholar]
- 44.Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. PLoS Genet. 2014;10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berne C, Ma X, Licata NA, Neves BRA, Setayeshgar S, Brun YV, Dragnea B. J. Phys. Chem. B. 2013;117:10492–10503. doi: 10.1021/jp405802e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Product Data Sheet: D263 T Borosilicate Thin Glass. Schott North America, Inc.; [Google Scholar]
- 47.Product Data Sheet: Sylgard 184 Elastomer Kit. Dow Corning Corp.; [Google Scholar]
- 48.Normand V, Lootens DL, Amici E, Plucknett KP, Aymard P. Biomacromolecules. 2000;1:730–738. doi: 10.1021/bm005583j. [DOI] [PubMed] [Google Scholar]
- 49.Hardy GG, Allen RC, Toh E, Long M, Brown PJB, Cole-Tobian JL, Brun YV. Mol. Microbiol. 2010;76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skerker JM, Shapiro L. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.