Abstract
This review covers conductivity detection in fabricated nanochannels and nanopores. Improvements in nanoscale sensing are a direct result of advances in fabrication techniques, which produce devices with channels and pores with reproducible dimensions and in a variety of materials. Analytes of interest are detected by measuring changes in conductance as the analyte accumulates in the channel or passes transiently through the pore. These detection methods take advantage of phenomena enhanced at the nanoscale, such as ion current rectification, surface conductance, and dimensions comparable to the analytes of interest. The end result is the development of sensing technologies for a broad range of analytes, e.g., ions, small molecules, proteins, nucleic acids, and particles.
Introduction
We review the use of nanofluidic devices for chemical analysis with a special focus on conductivity-based detection. Conductivity-based detection methods have the benefit of being label-free and are able to sense a wide range of analytes from ions to proteins to nucleic acids to particles when coupled to a nanoscale channel. We limit this review to experiments that use synthetic and solid state nanofluidic pores and channels for chemical analysis. Reviews that cover biological nanopores1,2 and the theory of nanoscale transport3,4 can be found elsewhere.
Nanofluidic channels and pores can have one, two, or three dimensions with nanometre length scales. For this review, we refer to nanochannels as conduits having at least one dimension confined to the nanoscale, usually channel depth or width. Nanopores are defined simply as having at least their lateral dimensions (i.e., diameter or width and depth) confined to the nanoscale We survey a range of device formats from conventional nanopore sensors, in which a three-dimensional pore is used to sense changes in conductivity, to in-plane nanopore devices, in which the pore is integrated directly into a micro- or nanofluidic channel.
To accomplish label-free conductivity detection, some element must be incorporated into the nanofluidic device that is sensitive to the presence of the analyte of interest. Nanopores have constrictions through which particles pass transiently and typically cause an increase in the pore resistance that is proportional to particle volume. In-plane nanopores are fabricated directly into micro- and nanofluidic devices and can be used to sense particles multiple times. An alternative strategy to axial current measurement is lateral current measurement with electrodes positioned perpendicularly to the nanofeature and measure current as the analyte passes the electrodes. Electrodes for transverse measurements are placed on both sides of a nanochannel or nanopore and are composed of metal electrodes or additional nanofluidic channels. Ion current rectification is exploited to build detectors based on modification of nanopore surface charge, and the on/off behaviour is used to build diodes and transistors for enhanced device functionality. Time-delayed electrical responses of nanofluidic devices provide impedance measurements for the determination of device dimensions and reaction kinetics. Lastly, conductivity measurements with these devices are complemented by optical visualization.
Resistive-Pulse Sensing with Out-of-Plane Nanopores
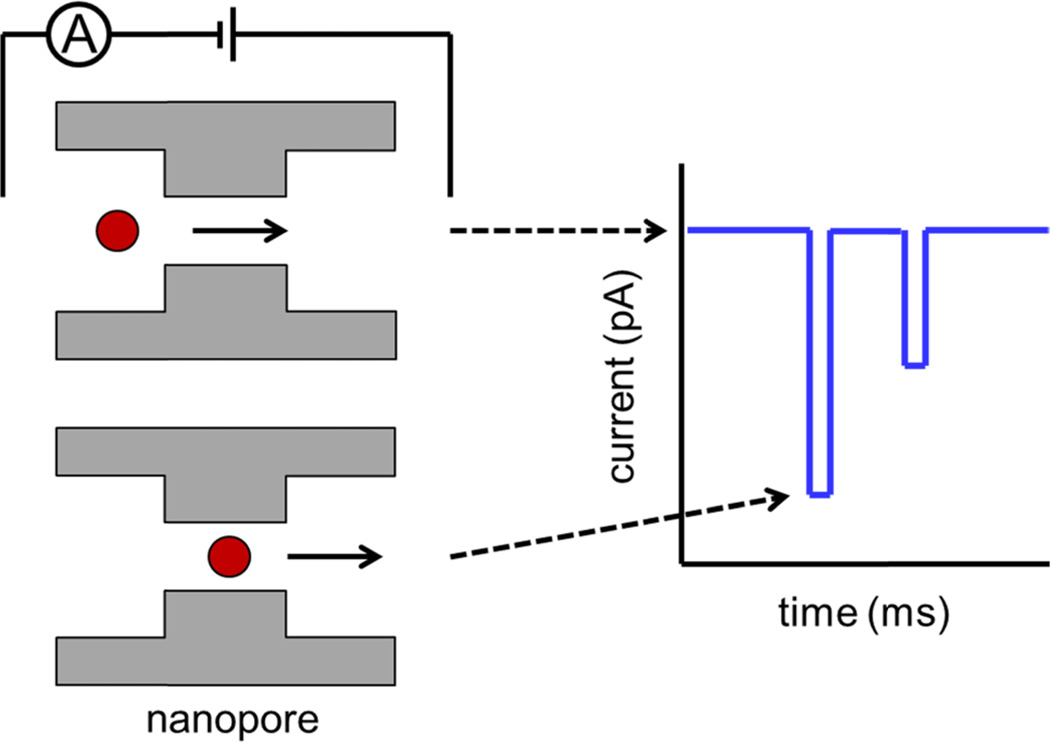
Resistive-pulse sensing with nanopores is used in a wide variety of research areas including virology, bacteriology, and DNA studies. Resistive-pulse sensing uses a constriction (pore) with dimensions comparable to the analyte of interest, and the pore separates two conductive electrolyte solutions. As analyte is driven through the pore by an electrical potential or pressure difference, changes in conductivity from transient blockages in ion current are measured, as shown in Fig. 1. Each transient decrease in current or ‘pulse’ corresponds to the transit of a single particle through the pore. Change in current is proportional to the change in nanopore resistance during translocation: thus, the amplitude of the current pulse increases with particle volume and decreases with pore volume.5 Pore resistance is the sum of the geometrical and access resistance terms, and both must be taken into account for accurate particle sizing.6 However, pulse amplitude is also affected by particle charge, and the counter ions moving through the pore. At low salt concentration, the conductivity of counter ions solvating DNA strands is greater than the solution conductivity, resulting in an increase in baseline current.7 Charged polystyrene particles in low conductivity solutions show an increase in baseline current at the exit of the nanopore.8 Pulse shape is a convolution of pore and particle geometry, and pulse shape reflects pore topography.9 Pulse width is the residence time within the pore, decreases with particle velocity,10 and increases with particle-pore interactions, such as adsorption.11 Particle trajectory through the pore also plays an important role in both pulse width and amplitude.12
Fig. 1.
Principle of resistive-pulse sensing. A potential is applied across a nanopore to electrokinetically drive the particle through the pore. As a particle transits the pore, the resistance typically increases, and current decreases. Transient decreases in current are observed as a series of ‘pulses’ that are proportional to particle size. The width of each pulse is proportional to the velocity of the particle through the pore.
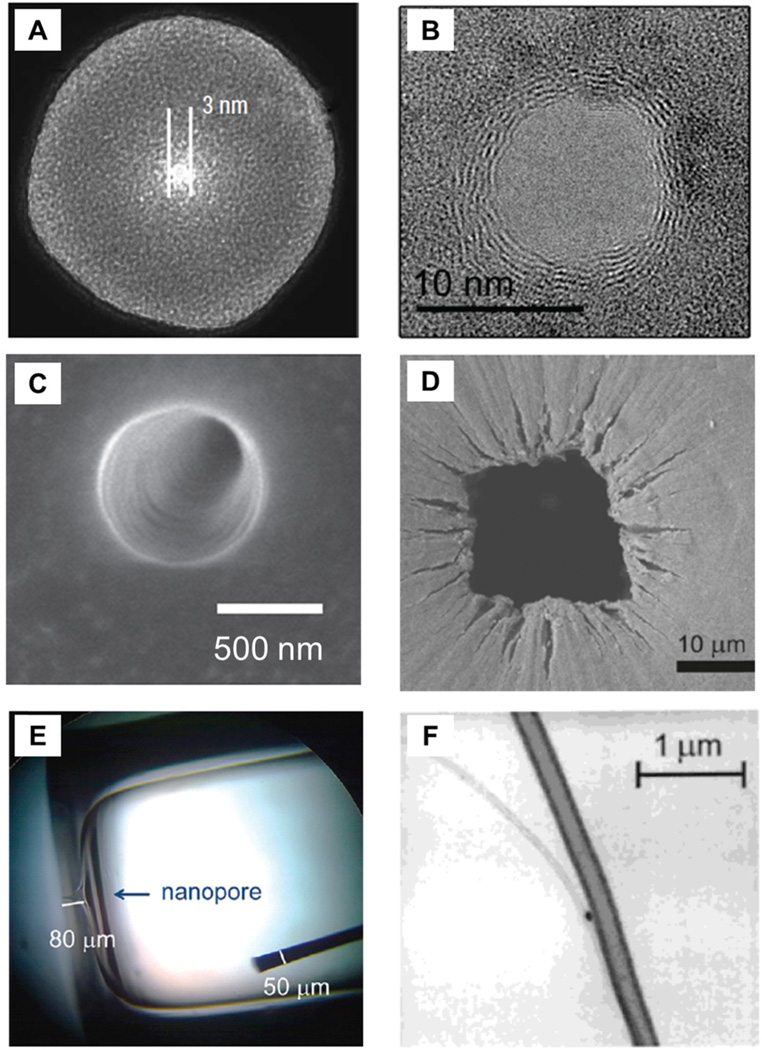
Over the last 60 years, a number of different types of sensors have been developed, and today, nanopores are fabricated in a variety of materials, e.g., polymers, SiN and SiO2 membranes, Si and SiO2 substrates, glass nanopipettes, carbon nanotubes, and graphene sheets (see Fig. 2). The original resistive-pulse sensor is described in a patent by Coulter in 1953 and consists of a glass test tube with a pinhole in the bottom.13 The “Coulter Counting” principle was originally developed to count blood cells,14 and within two years, these devices had sufficient sensitivity to detect bacteria such as E. coli and B. megaterium.15,16 The ability of early Coulter counting devices to study dynamic processes is reported for the determination of PbCO3 and CaC2O4 crystal growth rates.17
Fig. 2.
Nanopores for resistive-pulse sensing. The variety of fabrication techniques and available substrates permit the pore and its dimensions to be tailored to suit the application of interest. Shown are nanopores in (A) SiN membrane, (B) graphene sheet, (C) poly(ethylene terephthalate) membrane, (D) tunable polyurethane membrane, (E) glass substrate, and (F) carbon nanotube. Panel A adapted with permission from Li et al.38 Copyright 2003 Macmillan Publishers Ltd. Panel B adapted with permission from Merchant et al.39 Copyright 2010 American Chemical Society. Panel D adapted with permission from Vogel et al.40 Copyright 2011 American Chemical Society. Panel E adapted with permission from Lan et al.28 Copyright 2012 American Chemical Society. Panel F adapted with permission from Sun et al.34 Copyright 2000 American Chemical Society.
One of the first examples of nanoscale resistive-pulse sensing is detection of virus capsids passing through a nanopore in a polymer membrane. The Nucleopore method was developed to create filters for the screening of biological materials.18 Damage tracks are created by bombarding a membrane with high-energy fission products and are wet chemically etched to create pores with diameters ranging from nm to um. In initial experiments, sub 100-nm particles are detected with a polycarbonate pore.19 Electrokinetic mobility measurements are made on latex nanoparticles and viruses, and a 6-nm difference in particle diameter is resolved from the difference in pulse amplitudes.5 Additionally, polycarbonate pores are used to study several types of bacteriophages and type C Oncornavirus, demonstrating the usefulness of resistive-pulse sensing to accurately count and size biologically relevant species in the 100-nm diameter range.20.
Track-etched polymer membranes continue to offer a low-cost, high-performance option for nanopore fabrication. Commonly used polymers include poly(ethylene terephthalate) (PET) and polyimide (PI). These commercially available membranes have a single heavy-ion damage track and are chemically etched with a two-step process to produce pores with reproducible base and tip diameters.21 A polymer membrane with a single pore is used for resistive-pulse sensing of individual DNA fragments22 and porphyrin molecules.10 PET pores are coated with gold, functionalized with poly(ethylene glycol), and used to study binding of bovine serum albumin to antibodies.23,24 PET pores contain carboxyl groups on the surface, and the pore surface is covalently modified with triethylene glycol to minimize electroosmotic flow and particle adsorption. These modified pores are used to resolve T = 3 and T = 4 Hepatitis B Virus capsids.25
Nanopores fabricated in glass offer low noise26 and are more resistant to collapse than pores formed in polymers, and the surfaces of glass pores are easily modified by various silane chemistries. Recently, glass nanopores are used to characterize the transport of polystyrene nanoparticles27,28 and to examine the deformation of microgels as they pass through the nanopore.29 Laser-pulled glass nanopipettes are easy and inexpensive to fabricate as resistive-pulse sensors and are capable of analysing single DNA strands labelled with nanoparticles.30 Femtosecond-pulsed lasers are used to fabricate conical nanopores in glass for studying immune complexes.31 Antibody binding in complex media is accomplished at protein concentrations down to 30 nM. Laser pulling combined with wet chemical etching provides excellent control over pore size, and pipettes formed with this method are used to distinguish between enantiomers by immobilization of β-cyclodextrin within the pore.32 Glass pipettes are also functionalized to study biotin-streptavidin and antibody-antigen interactions.33
Resistive-pulse sensors are also made from carbon nanotubes embedded in non-conducting polymer blocks. The pore length is defined by the thickness of the polymer block, and the nanotube diameter is tuned from a few nanometres to hundreds of nanometres by the synthesis process. Carbon nanotube sensors accurately measure size and electrokinetic mobility of individual polystyrene nanoparticles.34,35 Because carbon nanotubes are smooth-walled cylinders, calculations of particle size and mobility are accurate and precise and compare favourably to traditional methods such as dynamic light scattering, transmission electron microscopy, and phase analysis light scattering.36 Carbon nanotubes are grown on Si wafers and isolated in polymer blocks for the analysis of single-stranded (ss) DNA.37
Solid state nanopores are typically fabricated in Si or SiN membranes with the advantages of ease of handling, well-characterized material properties, and well-established fabrication protocols. Electron and ion beams provide fine control over nanopore diameter. SiN pores with single nanometre resolution are fabricated with a focused ion beam (FIB) instrument, and pore size is reduced by ion beam exposure due to a thermal annealing process.41 The size of ion-beam milled pores can also be tuned by electron-beam (e-beam) assisted deposition of SiO2 in which the electron beam catalyses a reaction between a precursor gas and a SiN substrate containing a nanopore.42 This fabrication method can be easily scaled up. An array of nanopores drilled in a SiN membrane with a TEM beam record current measurements in parallel.43
SiN pores are used to analyse DNA, and DNA conformation is determined from the magnitude of current displacement in multi-level events.38 SiN pores are also fabricated by TEM drilling for rapid analysis of pM concentrations of DNA, assisted by a salt gradient across the pore.44 Additionally, TEM-drilled pores permit control over pore thickness for discrimination between small nucleic acids.45 Of particular interest is a SiN pore coated with a lipid bi-layer. Ligands incorporated into the bi-layer interact with targeted proteins to slow their transport and to increase the resolution between individual events.46 Additional studies include antibiotic/RNA interactions,47 protein unfolding studies,48 and sizing of polystyrene nanoparticles.49
Solid state nanopores are also created in SiO2 membranes. The diameter of SiO2 pores is tuned by imaging with a TEM to promote oxide growth and create 10-nm diameter pores for analysis of ss- DNA and double-stranded (ds) DNA.50,51 The magnitude of current displacement for DNA translocations depends on salt concentration, and an increase in current is observed at low salt concentration, whereas a decrease in current occurs at high salt concentration.7 E-beam lithography and wet chemical etching are used to create a pyramidal pore in SiO2 for simultaneous measurement of electric charge and zeta potential for nanoparticles of varying composition.52
Recently, nanopores are fabricated in graphene membranes. These sensors are being developed to discriminate individual DNA nucleotides, because pores in atomically thin graphene sheets provides higher axial resolution than pores formed in conventional membranes.39,53 A stack of alternating graphene and aluminium oxide sheets enables each graphene layer to be biased individually, where local potentials retard the transport of DNA through these pores, thus increasing resolution of individual base pairs.54
One limitation of conventional sensors is that pore size must be tuned to the size of the analyte. Polyurethane films are used to create pores with diameters that can be tuned in real time to match analyte size. These membranes are punctured with tungsten needles to create the pore, and then pressure is applied to the membrane which causes the pore to stretch or relax as a function of the pressure. Pore diameters range from hundreds of nanometres to tens of microns. Accurate sizing of polymer nanoparticles and Adenovirus particles is accomplished, as confirmed by light scattering and TEM measurements.40 Mobility of polystyrene nanoparticles is measured as a function of nanopore size and pH for analysis of pulse shape.55 Tuneable pores in commercially available nanopore sensors are shown to detect a wide size range of polystyrene beads56 and have also monitored the binding of DNA to streptavidin-functionalized beads.57
Resistive-Pulse Sensing with In-Plane Nanopores
Integration of in-plane nanopores into microfluidic devices enhances device portability and ease of use, improves mass transfer of the analyte to the pore, lowers sample consumption, and allows integration of enhanced functions. Furthermore, pore geometry is directly characterized, and experiments can also be monitored optically. Planar pores are fabricated in glass, silicon, or polymers with fabrication processes tuned to suit the application of interest.
FIB techniques provide precise fabrication of sub-50 nm features.58 Nanofluidic channels are patterned directly into silicon59 or quartz60 substrates to study the transport dynamics of λ-DNA. Low surface roughness and uniformity of these channels permit a thorough investigation of DNA mobility as a function of potential, measuring the energy barrier to elongate DNA strands as they enter confined space. FIB-milled channels are also replicated in soft polymers, e.g., poly(dimethylsiloxane) (PDMS), such that a single master may yield multiple replicas that are used to track DNA strands with a high degree of device-to-device reproducibility.61 In addition, the deformability of the polymer channels permits the channel cross-section to be dynamically controlled during the experiment by the application of mechanical force to the substrate.62 Compression of the channel cross-section slows DNA translocation by an order of magnitude with implications for increasing measurement sensitivity towards discrimination of DNA nucleotides.
Traditional lithographic processes pattern in-plane features that can be hundreds to thousands of nanometres in width. E-beam lithography is used to write metal lines, which are subsequently replicated in PDMS.63 These devices provide hours of measurement time, and the effect of driving potential on DNA conformation during translocation is studied. Multi-level events are correlated to the folded shapes of the DNA strands. PDMS-replicated nanopores are also used to perform immunoassays in series and parallel. A protein, such as streptavidin, is attached to a latex colloid.64 Binding of anti-streptavidin increases the diameter of the particle, and this size change is easily detected. Furthermore, channels can be patterned in parallel and allows for several immunoassays to be performed simultaneously.65 A microfluidic device in PDMS combines a nanopore with a fluidic balancing arm and is used for high-throughput analysis of nanoparticles and bacteriophage T7.66 Accurate sub-100 nm particle sizing is accomplished at a rate of 500,000 particles per second.
Three-dimensional pores are also incorporated into planar devices. A glass nanopipette is inserted into a PDMS device for detection of λ-DNA folding inside the pore and probes the effects of voltage and salt concentration on the translocation pathways.67,68 Glass devices exhibit a high tolerance to pH and electrical potential, and the close proximity of the pipette to a glass cover slip enables simultaneous optical imaging. Nanopipette sensors have also been used to detect latex69 and polystyrene nanoparticles.70
In-plane sensors may be scaled up to micrometre dimensions, such that cells or bacteria can be studied. A simple PDMS micropore device tracks E. coli, allows accurate measurement of the number and size of bacteria, and may hold promise for early detection of food and waterborne diseases.71 In addition, the lithographic process used to create these pores permits integration of sensing electrodes. Microchannels are filled with a liquid gallium-indium alloy that creates an electrode surface when cured. These devices are used for experiments at low frequencies, as the enhanced surface area alleviates polarization effects.72 Recently, a planar PDMS micropore is used to detect microparticles functionalized with antibodies.73
Multiple Resistive-Pulse Measurements on Single Particles
A particle can be driven back and forth through a single pore by reversal of the applied pressure or potential to allow multiple measurements to be made on a single particle. Measurement precision is increased, and the time between events yields additional information. This principle is first demonstrated by passing 27-µm diameter polystyrene beads back and forth through a glass micropipette.74 The same device is used to measure the dissolution rate of air bubbles and the effect of bovine serum albumin on surface tension of the bubble.75 Multiple measurements made on ss-DNA with a PDMS nanopore increases measurement precision.76 Recapture of ss-DNA is accomplished with a solid state nanopore for dynamic information of strand conformation.77 Nanoparticle dynamics are measured by multiple passes through a single glass nanopipette78 and a single track-etched pore in a PET membrane.79
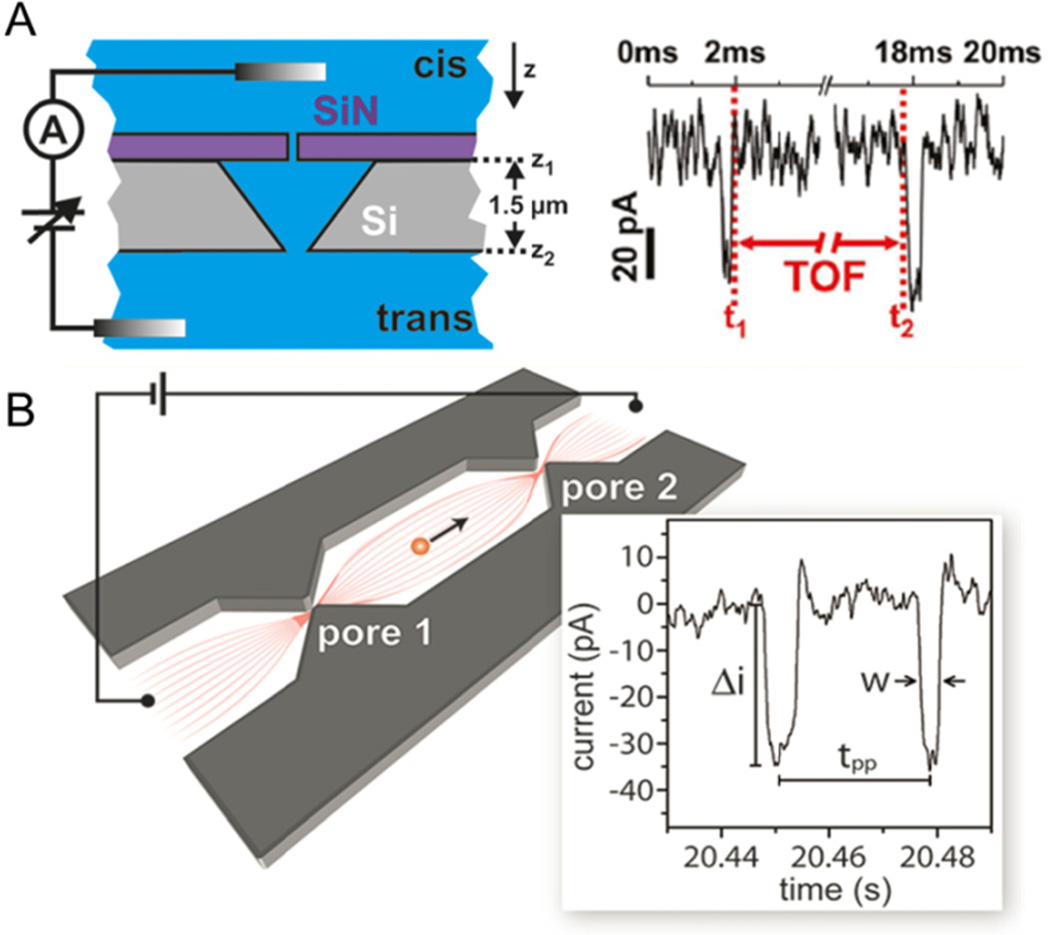
When multiple pores are arranged in series, a single particle is measured multiple times without the need to pass the particle back and forth through a single pore. Two different types of multi-pore devices are shown in Fig. 3, one fabricated with out-of-plane pores and one fabricated with in-plane pores. In Fig. 3A, traditional, out-of-plane pores are used. A SiN/Si bilayer is fabricated by e-beam lithography and wet chemical etching, such that two pores of comparable diameter are stacked in series.80 DNA is trapped in the area between the pores, and diffusion out of the device can be precisely monitored. The same device also tracks DNA time-of-flight from the first pore to the second pore.81 DNA conformation can be tracked by comparing changes in pulse amplitudes at both pores, indicating conformational changes that take place during translocation.
Fig 3.
Resistive-pulse sensing with two nanopores in series. (A) Two solid state pores are stacked in a pore-cavity-pore configuration for measuring DNA time-of-flight (TOF). A single event is shown on the right, where two pulses are observed for the transit of a single DNA strand, and the time between pulses is the transit time between pores. (B) Schematic of an in-plane nanochannel device with two pores in series fabricated in SiO2. Two pores measure the transit of single Hepatitis B Virus capsids, where each capsid is measured twice, as shown on the current trace on the right. Pulse amplitude (Δi) is proportional to capsid size, and the time between pulses (tpp) is used to calculate the electrophoretic mobility of the capsid. Panel A adapted with permission from Langecker et al.81 Copyright 2011 American Chemical Society. Panel B adapted with permission from Harms et al.82 Copyright 2011 American Chemical Society.
One key advantage of in-plane nanopore devices is the freedom to control the number and arrangement of sensing nanopores within a device. Two nanopores in series are fabricated by e-beam lithography and reactive-ion etching in SiO2 to measure the electrophoretic mobility of Hepatitis B Virus (HBV) capsids (Fig. 3B).82 Capsid velocity is shown to increase linearly with field strength and is used to calculate the electrokinetic mobility of the capsids. Nanochannels with two pores in series are milled with an FIB instrument to form channels and pores with three-dimensional topography.83 These devices track both the size and mobility of individual T = 3 and T = 4 HBV capsids and are well-suited to study virus capsid assembly in which the assembly process can be captured with single particle resolution and in real time.
Another serial sensing design uses a long nanochannel that features an arbitrary pattern of expansions (or nodes) that is encoded on each current pulse.84 With this ‘node-pore’ technique, the current trace exhibits a local minimum for each constriction encountered and a local maximum for each expansion. Unique current signatures permit identification of events with a low signal-to-noise ratio, expanding the useful dynamic range of the experiment.
As with single-pore sensors, multi-pore techniques can also be scaled up to interrogate cells. A series of 5 to 10 vertical electrode pairs are patterned along the length of a PDMS microchannel, and multiple impedance measurements are made on latex microparticles and fibroblast cells.85,86 A passive mixing region and dielectrophoretic focusing of the analyte prior to detection highlight the capability of microfluidic devices that incorporate additional on-device functions.
Transverse Conductivity Sensing
Efforts to sequence the human genome more rapidly and cost effectively have inspired new electrical sensing strategies that couple transverse conductance measurements with nanopore devices. Such devices incorporate electrodes perpendicular to the translocation pathway that measure the transverse current as a function of time. Two nanoelectrodes embedded in a nanopore and functioning as conducting probes are predicted to differentiate individual nucleotides.87 Similar theories have suggested this method could significantly reduce sequencing times88 and have high sensitivity.89,90
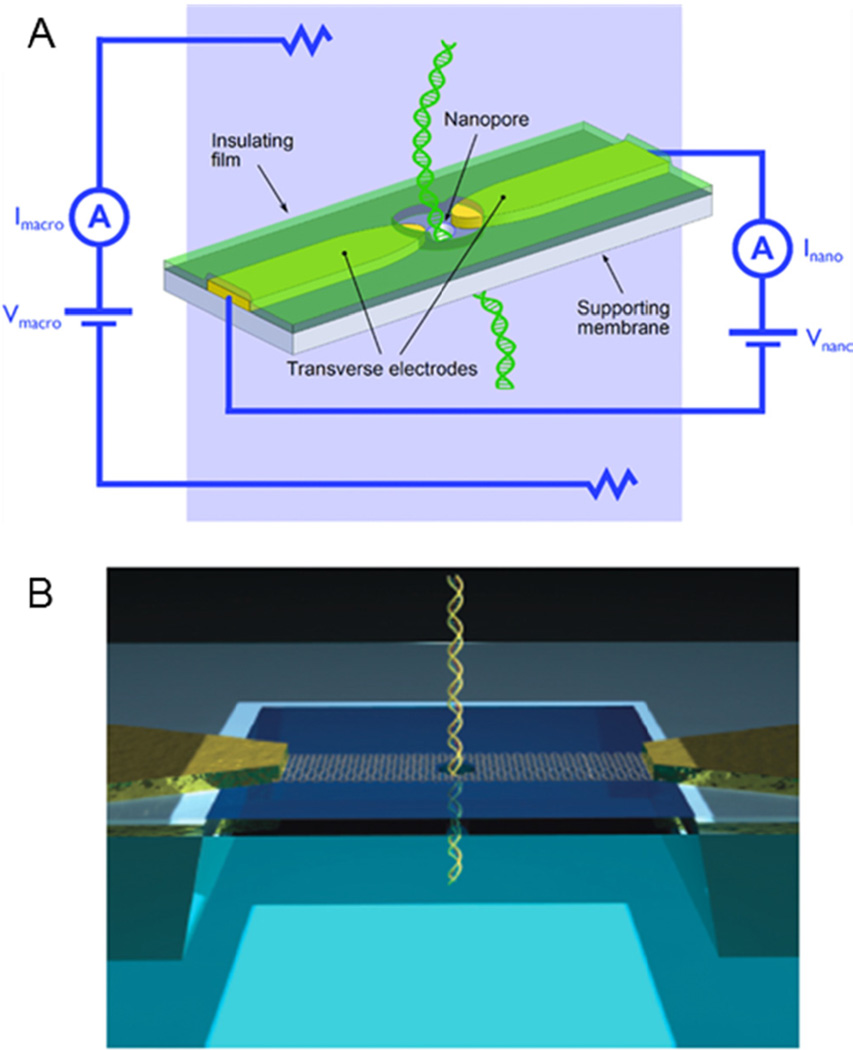
Transverse sensing devices consist of exposed nanogap electrodes separated by 1–2 nm within a nanopore (Fig. 4). Several fabrication methods are used to create pores: FIB milling through metal91 and carbon nanotubes,92 nanofabricated mechanically controllable break junctions,93 and TEM drilling.94–96 Experimentally, analytes are detected as they translocate through the pore by monitoring the tunnelling current across the nanogap electrodes as a function of time. Similar to sensing ionic current, translocation events are observed as pulses in a current trace. Tunnelling current through a 0.55 nm diameter gold nanojunction has been used to experimentally differentiate between single amino acids.97
Fig. 4.
Principle of transverse conductivity sensing. (A) As DNA is driven through the nanopore, the base pairs modulate the electric field between the transverse electrodes. Pores are often fabricated in SiN, and the metal electrodes are typically gold. (B) Recently, nanopore devices are formed in graphene which acts as both the nanopore substrate and electrode. Panel A adapted with permission from Healy et al.95 Copyright 2012 John Wiley and Sons, Inc. Panel B adapted with permission from Radenovic et al104. Copyright 2013 Macmillan Publishers Ltd.
Sensing platforms are also designed to alleviate the need for a nanopore size-matched to the analyte of interest, easing fabrication constraints and increasing the detectable range of particle sizes.98 Similar to the transverse conductivity sensing with nanoelectrodes, straight nanofluidic channels through which molecules transit are bisected by a conductivity sensor. Nanogap electrodes within a nanofluidic channel are capable of sensing ds-DNA when the electrode gap is < 13 nm.99 Another device consists of a nanochannel milled by an FIB instrument through a metal nanoelectrode.100 Recently, two FIB-milled nanochannels are designed to intersect and sense DNA translocation.101 Unlike the other sensors, this particular device monitors the ionic conductivity across a short nanochannel that intersects a longer nanochannel through which the DNA strands pass. Nanogap electrodes adjacent to TEM-drilled nanopores also detect single DNA molecules.102 Simulations suggest that the current signature from each base pair is different, and a bisected nanochannel geometry may be useful for future DNA sequencing.103
Established sensing methods have reported unique current signatures for specific bases.93,105 However, due to the finite size of metal electrodes and ionic channels, single-base resolution of DNA strands has eluded transverse conductivity sensors. Recent simulations suggest that graphene, because of its single atom thickness and high conductance, is capable of single-base resolution by transverse conductivity measurements during DNA translocation through a nanogap in the graphene sheet.106–108 These initial models conclude that the differing structures of each nucleotide is sufficient for base differentiation,107 irrespective of the orientation of the nucleotide within the pore.106 Several studies verify that atomically thin graphene is capable of sensing DNA by axial conductance39,53,109 as well as by lateral conductance.104 These results suggest next-generation devices with graphene sheets may be capable of both longitudinal sensing of DNA translocation and single-base discrimination by transverse conductivity measurement.
Ion Current Rectification
Ion current rectification (ICR) results from an uneven flux of cations and anions through nanofluidic structures with an asymmetry in surface charge or geometry110,111,112 (Fig. 5) or through physical blockages.113,114 Rectification is manifested as high and low conductance states that approximate the ‘on’ and ‘off behaviour of electronic diodes, respectively.115,116,117 The magnitude of ICR is reported as the rectification ratio (RR), which is the absolute value of the ion current at one potential (typically −1 V) divided by the ion current at an equal but opposite potential (+1 V), i.e., RR = | I− / I+ |. A RR > 1 is typically observed when the surface charge is negative and, conversely, RR < 1 is commonly observed when the surface charge is positive. The degree of rectification also depends on the identity of the cations in solution and is shown experimentally to increase with increasing cation diameter.118 ICR is highly dependent on local surface charge,119 which can be controlled by pH and surface functionalization to build nanofluidic sensors with a high degree of sensitivity and selectivity. Furthermore, diode-like behaviour permits use of nanofluidics in simple logic circuits for enhanced device functionality.120,121 Additional terminals, e.g., electrodes or fluidic channels, can further modulate ion current for transistor-like behaviour.
Fig. 5.
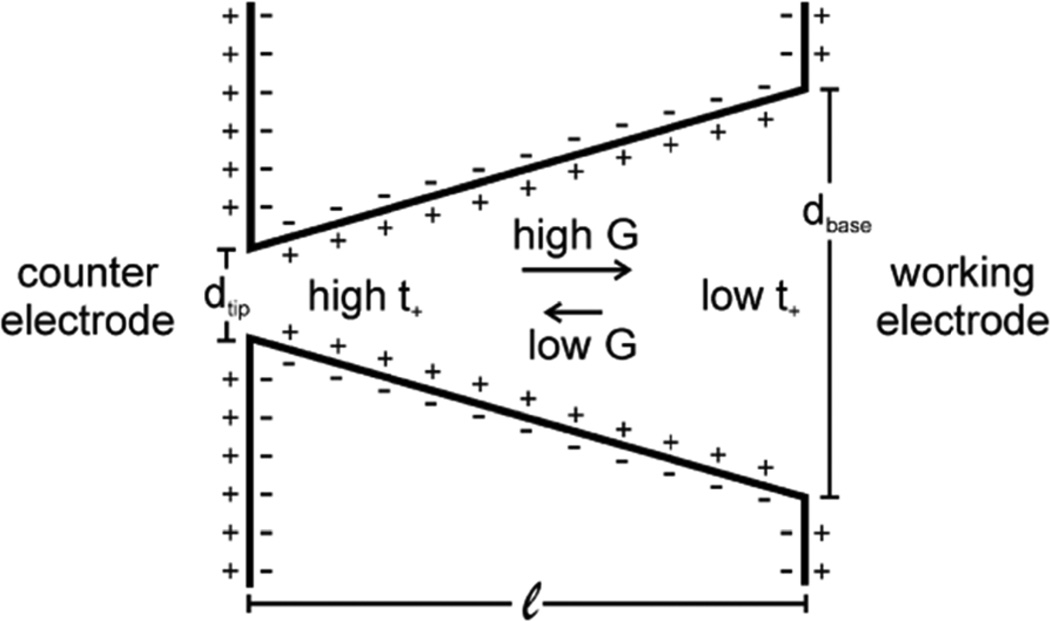
Schematic of an asymmetric nanopore with negative surface charge. Arrows show direction of cation flux in high and low conductance states (G), which, when present, leads to ion current rectification. Figure adapted with permission from Kovarik et al.112 Copyright 2009 American Chemical Society.
To regulate the rectification of ion current, conical polymer nanopores are functionalized with groups that respond to light or heat. Temperature-responsive polymers attached to the surface of a polyimide pore shrink or swell to control ion current.122 Current decreases as temperature increases due to collapse of the polymer brushes within the pore. Nanopores are modified with copolymer brushes that are responsive to both temperature and pH for independent control of ion flux and rectification ratio.123 Photolabile protecting groups that act to neutralize surface charge are removed by UV radiation to return the nanopore to a charged state by an ‘off to ‘on’ activation.124 UV radiation is also used to control pore size in a membrane that is modified with C4-DNA motors, due to a pH triggered conformational change initiated by photo-induced OH− ions.125 Light activation is combined with pH-modulation in a pore modified with spiropyran, where irradiation creates a negatively charged surface that is tuned by solution pH.126
The magnitude and direction of rectification depends strongly on surface charge density and polarity, which are conveniently manipulated by solution pH. Unmodified polymer nanopores have a carboxyl groups on the surface with an isoelectric point around pH 3, where no rectification is observed. At a pH below the isoelectric point, RR < 1, whereas at a pH above the isoelectric point, RR > 1, due to the resulting net positive or negative surface charge, respectively.127 Surface modification of polymer pores with L-lysine,128,129 L-histidine,129 and phosphate brushes130 shifts the isoelectric point and, consequently, the pH-dependent behaviour. Conical glass nanopores are also modified with an aminated silane131,132 and amphoteric chitosan133 for pH-dependent rectification.
Rectification-Based Sensing
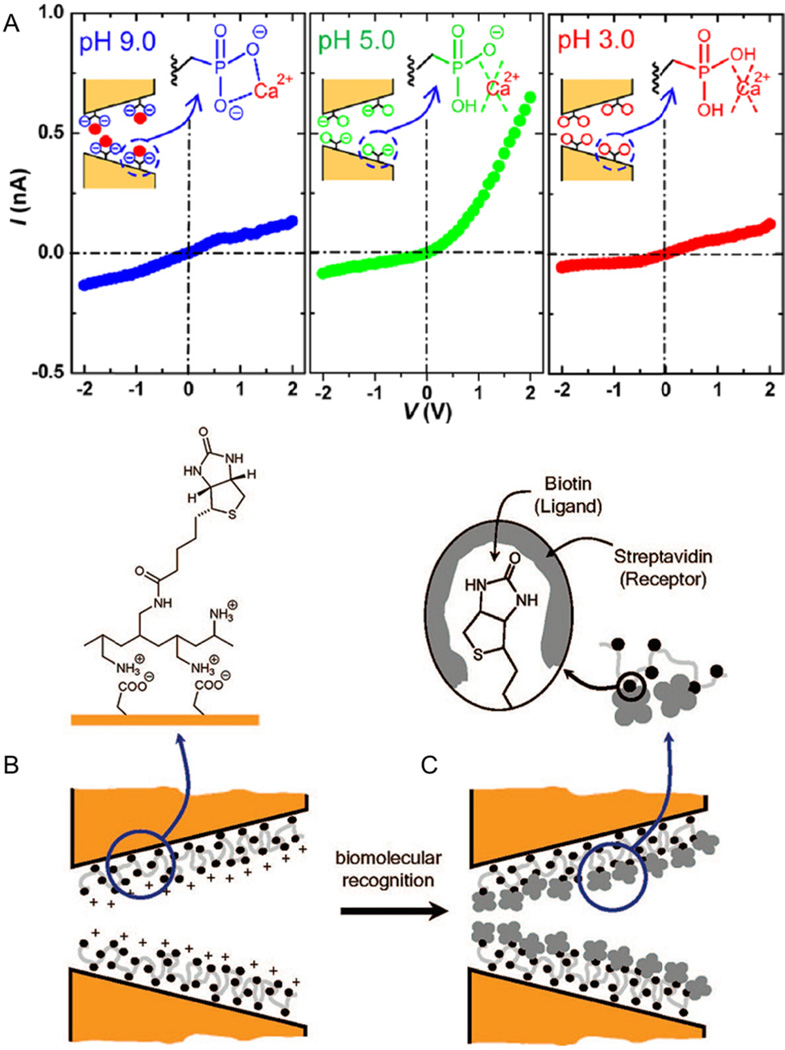
The surfaces of nanopores can be functionalized with groups that bind ions to disrupt surface charge and, in turn, the degree of rectification. Pores fabricated in polymer membranes, such as poly(ethylene) terephthalate (PET), contain carboxyl groups on their surface, that may be functionalized. These systems provide a biomimetic platform for studying ion channel regulation. Polymer pores functionalized with G4 DNA respond to K+ by undergoing a conformational change.134 With K+ in the pore, the DNA folds into a densely packed structure that blocks ion current. Ca2+ and cobalt sepulchrate invert rectification on a negatively charged pore,135 and Ca2+ binds to a phosphonic acid-modified pore for pH dependent rectification as shown in Fig. 6A.136 pH-dependent rectification is similarly achieved by modifying PET nanopores with ss-DNA to create electrostatic meshes.137 Nanopore surfaces functionalized with zinc finger peptides bind Zn2+ to induce a conformation change and cause a fluctuation in ion current.138 Glass nanopipettes are modified with imidazole-terminated silanes139 to bind Co2+ and calcium binding protein (calmodulin)140 to bind Ca2+ and Mg2+. Ca2+ is also detected with calcein-modified PET pores that exhibit a lower RR in the presence of Ca2+ at high pH.141
Fig. 6.
Two strategies for rectification-based sensing. (A) A polymer-based nanopore is modified with phosphonic polyacid chains. These pores exhibit pH-dependent ion current rectification (ICR) and are used to detect calcium binding. (B–C) A nanopore modified with a carboxylated polymer electrostatically binds biotin ligand. Subsequently, the ligand binds streptavidin, which minimizes the excess surface charge and results in decreased ICR as well as decreased total ion current. Panel A adapted with permission from Ali et al.136 Copyright 2012 American Chemical Society. Panels B-C adapted with permission from Ali et al.146 Copyright 2008 American Chemical Society.
Pores with asymmetric geometries that exhibit ICR can also be used to study small molecules. ICR increases cation concentration within a polymer nanopore above the bulk solubility product to precipitate CoHPO4 and CaHPO4, demonstrating the ability of nanopores to probe kinetics and early stages of crystallization.142,143 As the precipitation occurs within the nanopore, millisecond timescales are accessed, and the reversibility of the reaction is highly repeatable to obtain good counting statistics. Quartz nanopores containing poly(vinylpyridine) modified with benzylboronic acid groups respond to monosaccharide polymers by collapsing to a zwitterionic state, and ICR reverses.144 A track-etched polymer pore modified with boronic acid reversibly binds saccharides and glycoproteins.145 Sensors that can accurately determine carbohydrate and glycoprotein concentrations have applications in clinical settings.
Pores with modified surfaces are well suited to study binding of biomolecules. Polymer pores coated with Au are readily modified with substituted thiol groups and are used to study the binding of biotin-streptavidin, protein-G-immunoglobulin, and antibody-ricin.119 PET pores are also directly functionalized with biotin to study streptavidin binding.146,147,48 An example of rectification-based detection is shown in Fig. 6B, where biotin-streptavidin binding eliminates surface charge on the pore, and binding events are detected as a decrease in ICR.146 Pores are modified with horseradish peroxidase enzymes that feature sugar groups for monitoring the binding of the lectin protein, concanavalin A.149 Functionalization of glass and polymer pores with dendrimers150 and peptide nucleic acid probes,151 respectively, enables detection of complementary DNA strands with high selectivity. Peptide nucleic acids are DNA analogues containing repeating glycine units bearing nucleotide bases and can be used to discriminate between complementary DNA strands and those containing a single base mismatch.
Addition of an extra terminal perpendicular to a nanofluidic channel is used to modulate ion current for transistor-like behaviour. The nanochannel functions as the source and drain, and the extra metal contact acts as the gate. These devices have been realized with arrays of silica nanochannels and with isolated silica nanotubes.152,153,154 Field-effect control of proteins is promising for preconcentration and separation applications. A silicon nanochannel in a transistor circuit is used to concentrate avidin.155 Nanofluidic transistors are also fabricated in 3D solid state pores156 for more efficient capture of DNA by reducing electroosmotic flow through application of a gate voltage.157 These pores are fabricated in SiN membranes that sandwich a metal film and are coated with an oxide layer. Alternatively, an all-silicon device, where the membrane itself is used as the gate electrode, demonstrates that voltage gating enhances the rectification ratio by > 100-fold for high on-off ratios.158 Manipulation of bovine serum albumin demonstrates the potential of this system to control protein transport.
Conductance-Based Detection
Conductance measurements provide additional means to probe nanofluidic phenomena. Measurements of nanochannel and nanopore conductance are used to characterize device geometry and surface chemistry and to probe surface binding reactions. Furthermore, if conductance is measured as a function of time when driven with AC waveforms, dynamic processes such as reaction kinetics can be tracked.
Nanochannel conductance depends on both channel geometry and surface charge. As the channel depth (or width) decreases and the surface-to-volume ratio increases, channel conductance deviates from bulk conductance due to surface conductance.159,160 Device conductance is proportional to number of nanochannels and is found to reach a plateau at low ionic strength due to conductance of surface charge.161 Geometric parameters of solid state nanopores, such as pore diameter, are estimated by conductance measurements.162
Electrochemical impedance spectroscopy (EIS) measures the impedance response of nanochannel devices as a function of frequency. EIS spectra taken as a function of solution conductivity are used to model arrays of glass nanocapillaries as equivalent circuit elements for characterization of pore dimensions and the electrical double layer. Equivalent circuit models are useful tools for understanding the behaviour of nanofluidic devices and provide accurate estimates of pore shape and diameter.163
EIS is used to investigate the role of surface charge on ion transport through 50-nm deep amorphous-Si/glass channels. Surface charge as a function of salt concentration is measured in a 50-nm wide quartz nanochannel with an embedded transverse Ti/Au electrode, where double layer overlap is observed at low ion concentration.164 Mass transport through an amine-modified glass nanopore is affected by the surface charge as well as the geometry of the pore itself165
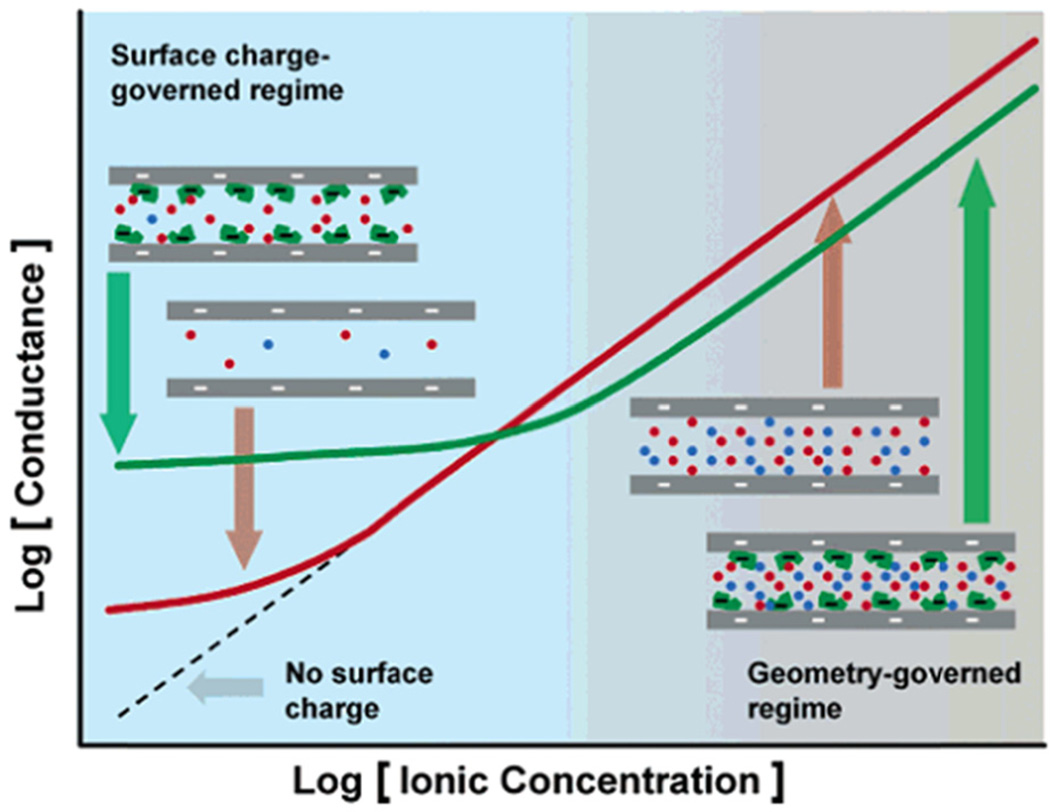
At low ionic strength, nanochannel conductance is dominated by surface charge, and as BSA adsorbs to the channel surface, the channel conductance decreases.167 An array of 30-nm deep silica nanochannels modified with streptavidin is used to measure avidin binding, in which nanochannel conductance increases with biomolecule charge and decreases with biomolecule volume, as shown in Fig. 7.166 Binding of streptavidin to a PEG-biotin in a 50-nm deep Pyrex nanochannel is measured under convective flow and shows the ability of nanofluidics to probe diffusion-limited reactions with a significantly reduced response time.168 An array of nanochannels in a PET membrane is modified with streptavidin for monitoring T4 polynucleotide kinase activity through a cleavage reaction with a biotin-labelled ds-DNA probe.169
Fig. 7.
Conductance-based detection. Nanochannels are biotinylated and bind streptavidin, which leads to a decrease in conductance at high ion concentration and an increase in conductance at low ion concentration. Figure adapted with permission from Karnik et al.166 Copyright 2005 American Chemical Society
Simultaneous Optical and Electrical Measurements
Despite the high sensitivity of conductivity-based sensing methods, detection of analytes of interest is often performed blindly. To overcome this limitation, optical measurements made simultaneously with electrical measurements of particles provide additional and complementary information that electrical measurements alone cannot provide. Tracking fluorescently tagged particles permits unambiguous correlation of a deviation in baseline current to a fluorescent signal, verifying the passing of a particle through the sensing region. Optical detection also provides information about analyte movement within the sensing region, which may explain the variation between individual current pulses. Particle trajectory is also monitored to improve device design for optimized sensing.
Simultaneous sensing of ds-DNA molecules transiting a 4-nm SiN pore is performed in a home-built cell by total internal reflectance fluorescence microscopy.170 Translocation events are monitored by electrical measurements made across the pore showing typical current deviations at the same time fluorescence spikes are detected. Similar measurements are made by confocal fluorescence microscopy in three dimensions to visualize the motion of DNA through a SiN pore embedded into a microfluidic device171 while current is simultaneously measured to confirm the passing of the particle. Fluorescence detection of DNA is also performed with in-plane FIB-milled nanochannels101 to verify that deviations in current are caused by transiting DNA molecules.
Carbon nanotubes positioned between two reservoirs are used to detect single dye molecules by measuring the ionic current passing through the tube as a function of time.172 Electrical detection of a molecule is again measured as a deviation from the baseline current, and optical detection is made possible by a laser focused just past the end of the tube that excites the dye exiting the tube. Simultaneous fluorescence and conductivity measurements of small, fluorescently tagged polystyrene beads transiting through a SiO2-PDMS pore correlate the dependence of the electrical peak shape to the characteristic shape of the pore.173
Conclusions and Outlook
Advances in fabrication techniques have driven research in nanofluidics over the past decade. Conductivity measurements offer a simple, yet powerful means to investigate nanoscale systems. These measurements have the added benefit of being label-free and can detect a wide variety of analyte types and sizes. Furthermore, nanofluidic phenomenona are often exploited to enhance sensing by selective transport and pre-concentration.
The ability to fabricate nanopores with dimensions similar to an analyte of interest makes them appealing for resistive-pulse sensing. One or more nanopores may be used to sense the transport of single particles by the change in resistance upon nanopore translocation. Nanopores can also be used to measure single particles multiple times, and multiple nanopores can be arranged in series to obtain more information and increase measurement precision. Electrodes are added to nanochannels or nanopores for transverse conductivity measurements, providing extra detection functionality. Metal or fluidic electrodes are used to monitor changes in current as analyte passes through a nanogap. Enhanced spatial resolution over traditional resistive-pulse sensing measurements is obtained with the potential application of DNA sequencing. A simple bulk conductance measurement in nanochannels is also used to probe analyte binding to nanochannel walls, and time-dependant signals are used to characterize device geometry.
Nanoscale conduits with an asymmetry in geometry or surface charge may be exploited for ion current rectification-based sensing. Surface charge is modulated by pH and adsorption for control over rectification, and functional groups are used to decorate the surface for specific interactions with target analytes. Finally, optical measurements are used to complement electrical measurements.
Conductivity-based sensing methods are poised to become cost-effective bioanalytical tools and will continue to develop as nanofabrication techniques improve. Future generations of conductivity sensors will build upon the work described above in order to improve detection efficiency and address a broad range of analytical applications. A number of challenges currently face electrical sensing technologies and include increasing device-to-device reproducibility, decreasing fabrication costs, and improving detection sensitivity. As these challenges are being met, conductivity-based detection becomes a practical and cost-effective alternative for a number of applications. Most notably, the development of reliable lab-on-a-chip devices capable of quickly and inexpensively analysing nanoparticles and DNA are an exciting breakthrough in the field. Commercial nanopore systems have been realized for DNA analysis (www.nanoporetech.com and www.nabsys.com), high throughput nanoparticle counting (www.spectradynellc.com), and extended dynamic range particle sizing (hwww.izon.com).
Acknowledgments
This work was supported in part by NIH R01 GM100071 and NSF CHE-1308484.
Notes and References
- 1.Howorka S, Siwy Z. Chem. Soc. Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 2.Oukhaled A, Bacri L, Pastoriza-Gallego M, Betton J-M, Pelta J. ACS Chem. Bio. 2012;7:1935–1949. doi: 10.1021/cb300449t. [DOI] [PubMed] [Google Scholar]
- 3.Sparreboom W, van den Berg A, Eijkel JCT. Nat. Nanotech. 2009;4:713–720. doi: 10.1038/nnano.2009.332. [DOI] [PubMed] [Google Scholar]
- 4.Schoch RB, Han J, Renaud P. Rev. Mod. Phys. 2008;80:839–883. [Google Scholar]
- 5.Deblois RW, Bean CP, Wesley RKA. J. Colloid Interface Sci. 1977;61:323–335. [Google Scholar]
- 6.Davenport M, Healy K, Pevarnik M, Teslich N, Cabrini S, Morrison AP, Siwy ZS, Letant SE. ACS Nano. 2012;6:8366–8380. doi: 10.1021/nn303126n. [DOI] [PubMed] [Google Scholar]
- 7.Smeets RMM, Keyser UF, Krapf D, Wu MY, Dekker NH, Dekker C. Nano Lett. 2006;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 8.Menestrina J, Yang C, Schiel M, Vlassiouk I, Siwy ZS. J. Phys. Chem. C. 2014;118:2391–2398. [Google Scholar]
- 9.Pevarnik M, Healy K, Toimil-Molares ME, Morrison A, Letant SE, Siwy ZS. ACS Nano. 2012;6:7295–7302. doi: 10.1021/nn302413u. [DOI] [PubMed] [Google Scholar]
- 10.Heins EA, Siwy ZS, Baker LA, Martin CR. Nano Lett. 2005;5:1824–1829. doi: 10.1021/nl050925i. [DOI] [PubMed] [Google Scholar]
- 11.Sexton LT, Mukaibo H, Katira P, Hess H, Sherrill SA, Horne LP, Martin CR. J. Am. Chem. Soc. 2010;132:6755–6763. doi: 10.1021/ja100693x. [DOI] [PubMed] [Google Scholar]
- 12.Saleh OA, Sohn LL. Rev. Sci. Instrum. 2002;73:4396–4398. [Google Scholar]
- 13.U.S.A. Pat. 2656508. U.S. patent. 1953
- 14.Coulter WH. Proceedings of the National Electronics Conference; Chicago. 1957. [Google Scholar]
- 15.Kubitschek HE. Nature. 1958;182:234–235. doi: 10.1038/182234a0. [DOI] [PubMed] [Google Scholar]
- 16.Smither R. J. Appl. Bacteriol. 1975;39:157–165. doi: 10.1111/j.1365-2672.1975.tb00557.x. [DOI] [PubMed] [Google Scholar]
- 17.Marković M, Komunjer L. J. Cryst. Growth. 1979;46:701–705. [Google Scholar]
- 18.Fleischer RL, Price PB, Symes EM. Science. 1964;143:249–250. doi: 10.1126/science.143.3603.249. [DOI] [PubMed] [Google Scholar]
- 19.Deblois RW, Bean CP. Rev. Sci. Instrum. 1970;41:909. [Google Scholar]
- 20.DeBlois RW, Wesley RKA. J. Virol. 1977;23:227–233. doi: 10.1128/jvi.23.2.227-233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apel PY, Korchev YE, Siwy Z, Spohr R, Yoshida M. Nucl. Instrum. Methods Phys. Res. B. 2001;184:337–346. [Google Scholar]
- 22.Mara A, Siwy Z, Trautmann C, Wan J, Kamme F. Nano Lett. 2004;4:497–501. [Google Scholar]
- 23.Wharton JE, Jin P, Sexton LT, Horne LP, Sherrill SA, Mino WK, Martin CR. Small. 2007;3:1424–1430. doi: 10.1002/smll.200700106. [DOI] [PubMed] [Google Scholar]
- 24.Sexton LT, Horne LP, Sherrill SA, Bishop GW, Baker LA, Martin CR. J. Am. Chem. Soc. 2007;129:13144–13152. doi: 10.1021/ja0739943. [DOI] [PubMed] [Google Scholar]
- 25.Zhou KM, Li LC, Tan ZN, Zlotnick A, Jacobson SC. J. Am. Chem. Soc. 2011;133:1618–1621. doi: 10.1021/ja108228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uram JD, Ke K, Mayer M. ACS Nano. 2008;2:857–872. doi: 10.1021/nn700322m. [DOI] [PubMed] [Google Scholar]
- 27.Lan W-J, Holden DA, Liu J, White HS. J. Phys. Chem. C. 2011;115:18445–18452. [Google Scholar]
- 28.Lan W-J, White HS. ACS Nano. 2012;6:1757–1765. doi: 10.1021/nn2047636. [DOI] [PubMed] [Google Scholar]
- 29.Holden DA, Hendrickson G, Lyon LA, White HS. J. Phys. Chem. C. 2011;115:2999–3004. doi: 10.1021/jp111244v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karhanek M, Kemp JT, Pourmand N, Davis RW, Webb CD. Nano Lett. 2005;5:403–407. doi: 10.1021/nl0480464. [DOI] [PubMed] [Google Scholar]
- 31.Uram JD, Ke K, Hunt AJ, Mayer M. Angew. Chem. Int. Ed. 2006;45:2281–2285. doi: 10.1002/anie.200502862. [DOI] [PubMed] [Google Scholar]
- 32.Gao C, Ding S, Tan Q, Gu L-Q. Anal. Chem. 2009;81:80–86. doi: 10.1021/ac802348r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umehara S, Karhanek M, Davis RW, Pourmand N. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4611–4616. doi: 10.1073/pnas.0900306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, Crooks RM. J. Am. Chem. Soc. 2000;122:12340–12345. [Google Scholar]
- 35.Ito T, Sun L, Henriquez RR, Crooks RM. Acc. Chem. Res. 2004;37:937–945. doi: 10.1021/ar040108+. [DOI] [PubMed] [Google Scholar]
- 36.Ito T, Sun L, Bevan MA, Crooks RM. Langmuir. 2004;20:6940–6945. doi: 10.1021/la049524t. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, He J, Tang J, Liu H, Pang P, Cao D, Krstic P, Joseph S, Lindsay S, Nuckolls C. Science. 2010;327:64–67. doi: 10.1126/science.1181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. Nat. Mater. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 39.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo ZT, Johnson ATC, Drndić M. Nano Lett. 2010;10:2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 40.Vogel R, Willmott G, Kozak D, Roberts GS, Anderson W, Groenewegen L, Glossop B, Barnett A, Turner A, Trau M. Anal. Chem. 2011;83:3499–3506. doi: 10.1021/ac200195n. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 42.Danelon C, Santschi C, Brugger J, Vogel H. Langmuir. 2006;22:10711–10715. doi: 10.1021/la061321c. [DOI] [PubMed] [Google Scholar]
- 43.Kim MJ, Wanunu M, Bell DC, Meller A. Adv. Mater. 2006;18:3149–3153. [Google Scholar]
- 44.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Nat. Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndić M. Nat. Nanotechnol. 2010;5:807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 46.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. Nat. Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wanunu M, Bhattacharya S, Xie Y, Tor Y, Aksimentiev A, Drndić M. ACS Nano. 2011;5:9345–9353. doi: 10.1021/nn203764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freedman KJ, Jurgens M, Prabhu A, Ahn CW, Jemth P, Edel JB, Kim MJ. Anal. Chem. 2011;83:5137–5144. doi: 10.1021/ac2001725. [DOI] [PubMed] [Google Scholar]
- 49.Tsutsui M, Hongo S, He Y, Taniguchi M, Gemma N, Kawai T. ACS Nano. 2012;6:3499–3505. doi: 10.1021/nn300530b. [DOI] [PubMed] [Google Scholar]
- 50.Storm AJ, Storm C, Chen JH, Zandbergen H, Joanny JF, Dekker C. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 51.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Phys. Rev. E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 52.Arjmandi N, Van Roy W, Lagae L, Borghs G. Anal. Chem. 2012;84:8490–8496. doi: 10.1021/ac300705z. [DOI] [PubMed] [Google Scholar]
- 53.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LMK, Dekker C. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 54.Venkatesan BM, Estrada D, Banerjee S, Jin X, Dorgan VE, Bae M-H, Aluru NR, Pop E, Bashir R. ACS Nano. 2012;6:441–450. doi: 10.1021/nn203769e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willmott GR, Parry BET. J. Appl. Phys. 2011;109:094307. [Google Scholar]
- 56.Eldridge JA, Willmott GR, Anderson W, Vogel R. J. Colloid Interface Sci. 2014;429:45–52. doi: 10.1016/j.jcis.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 57.Billinge ER, Broom M, Platt M. Anal. Chem. 2014;86:1030–1037. doi: 10.1021/ac401764c. [DOI] [PubMed] [Google Scholar]
- 58.Menard LD, Ramsey JM. Nano Lett. 2011;11:512–517. doi: 10.1021/nl103369g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campbell LC, Wilkinson MJ, Manz A, Camilleri P, Humphreys CJ. Lab Chip. 2004;4:225–229. doi: 10.1039/b312592k. [DOI] [PubMed] [Google Scholar]
- 60.Menard LD, Ramsey JM. Anal. Chem. 2013;85:1146–1153. doi: 10.1021/ac303074f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fanzio P, Mussi V, Manneschi C, Angeli E, Firpo G, Repetto L, Valbusa U. Lab Chip. 2011;11:2961–2966. doi: 10.1039/c1lc20243j. [DOI] [PubMed] [Google Scholar]
- 62.Fanzio P, Manneschi C, Angeli E, Mussi V, Firpo G, Ceseracciu L, Repetto L, Valbusa U. Sci. Rep. 2012;2:791. doi: 10.1038/srep00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sen Y-H, Karnik R. Anal. Bioanal. Chem. 2009;394:437–446. doi: 10.1007/s00216-008-2529-3. [DOI] [PubMed] [Google Scholar]
- 64.Saleh OA, Sohn LL. Proc. Natl. Acad. Sci. U. S. A. 2003;100:820–824. doi: 10.1073/pnas.0337563100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carbonaro A, Sohn LL. Lab Chip. 2005;5:1155–1160. doi: 10.1039/b504827c. [DOI] [PubMed] [Google Scholar]
- 66.Fraikin J-L, Teesalu T, McKenney CM, Ruoslahti E, Cleland AN. Nat. Nanotech. 2011;6:308–313. doi: 10.1038/nnano.2011.24. [DOI] [PubMed] [Google Scholar]
- 67.Steinbock LJ, Otto O, Chimerel C, Gornall J, Keyser UF. Nano Lett. 2010;10:2493–2497. doi: 10.1021/nl100997s. [DOI] [PubMed] [Google Scholar]
- 68.Steinbock LJ, Lucas A, Otto O, Keyser UF. Electrophoresis. 2012;33:3480–3487. doi: 10.1002/elps.201100663. [DOI] [PubMed] [Google Scholar]
- 69.Terejanszky P, Makra I, Fuerjes P, Gyurcsanyi RE. Anal. Chem. 2014;86:4688–4697. doi: 10.1021/ac500184z. [DOI] [PubMed] [Google Scholar]
- 70.Lan W-J, Kubeil C, Xiong J-W, Bund A, White HS. J. Phys. Chem. C. 2014;118:2726–2734. [Google Scholar]
- 71.Sun J, Kang Y, Boczko EM, Jiang X. Electroanalysis. 2013;25:1023–1028. [Google Scholar]
- 72.Richards AL, Dickey MD, Kennedy AS, Buckner GD. J. Micromech. Microeng. 2012;22:115012. [Google Scholar]
- 73.Han Y, Wu H, Liu F, Cheng G, Zhe J. Anal. Chem. 2014;86:9717–9722. doi: 10.1021/ac502270n. [DOI] [PubMed] [Google Scholar]
- 74.Berge LI, Feder J, Jøssang T. Rev. Sci. Instrum. 1989;60:2756–2763. [Google Scholar]
- 75.Berge LI. J. Colloid Interface Sci. 1990;134:548–562. [Google Scholar]
- 76.Sen Y-H, Jain T, Aguilar CA, Karnik R. Lab Chip. 2012;12:1094–1101. doi: 10.1039/c2lc20771k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gershow M, Golovchenko JA. Nat. Nanotechnol. 2007;2:775–779. doi: 10.1038/nnano.2007.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.German SR, Luo L, White HS, Mega TL. J. Phys. Chem. C. 2013;117:703–711. [Google Scholar]
- 79.Schiel M, Siwy ZS. J. Phys. Chem. C. 2014;118:19214–19223. [Google Scholar]
- 80.Pedone D, Langecker M, Abstreiter G, Rant U. Nano Lett. 2011;11:1561–1567. doi: 10.1021/nl104359c. [DOI] [PubMed] [Google Scholar]
- 81.Langecker M, Pedone D, Simmel FC, Rant U. Nano Lett. 2011;11:5002–5007. doi: 10.1021/nl2030079. [DOI] [PubMed] [Google Scholar]
- 82.Harms ZD, Mogensen KB, Nunes PS, Zhou K, Hildenbrand BW, Mitra I, Tan Z, Zlotnick A, Kutter JP, Jacobson SC. Anal. Chem. 2011;83:9573–9578. doi: 10.1021/ac202358t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harms ZD, Haywood DG, Kneller AR, Selzer L, Zlotnick A, Jacobson SC. Anal. Chem. 2015;87:699–705. doi: 10.1021/ac503527d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balakrishnan KR, Anwar G, Chapman MR, Trongtuong N, Kesavaraju A, Sohn LL. Lab Chip. 2013;13:1302–1307. doi: 10.1039/c3lc41286e. [DOI] [PubMed] [Google Scholar]
- 85.Wu Y, Benson JD, Almasri M. Biomed. Microdevices. 2012;14:739–750. doi: 10.1007/s10544-012-9655-6. [DOI] [PubMed] [Google Scholar]
- 86.Wu Y, Benson JD, Critser JK, Almasri M. J. Micromech. Microeng. 2010;20:085035. [Google Scholar]
- 87.Zwolak M, Di Ventra M. Nano Lett. 2005;5:421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]
- 88.Lagerqvist J, Zwolak M, Di Ventra M. Nano Lett. 2006;6:779–782. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krems M, Zwolak M, Pershin YV, Di Ventra M. Biophys. J. 2009;97:1990–1996. doi: 10.1016/j.bpj.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sigalov G, Comer J, Timp G, Aksimentiev A. Nano Lett. 2008;8:56–63. doi: 10.1021/nl071890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ivanov AP, Instuli E, McGilvery CM, Baldwin G, McComb DW, Albrecht T, Edel JB. Nano Lett. 2011;11:279–285. doi: 10.1021/nl103873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang Z, Mihovilovic M, Chan J, Stein D. J. Phys. Condens. Matter. 2010;22:454114. doi: 10.1088/0953-8984/22/45/454114. [DOI] [PubMed] [Google Scholar]
- 93.Tsutsui M, Rahong S, Iizumi Y, Okazaki T, Taniguchi M, Kawai T. Sci. Rep. 2011;1:46. doi: 10.1038/srep00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gierhart BC, Flowitt DG, Chen SJ, Zhu Z, Kotecki DE, Smith RL, Collins SD. Sensors Actuat. B-Chem. 2008;132:593–600. doi: 10.1016/j.snb.2007.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Healy K, Ray V, Willis LJ, Peterman N, Bartel J, Drndić M. Electrophoresis. 2012;33:3488–3496. doi: 10.1002/elps.201200350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie P, Xiong QH, Fang Y, Qing Q, Lieber CM. Nat. Nanotechnol. 2012;7:119–125. doi: 10.1038/nnano.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ohshiro T, Tsutsui M, Yokota K, Furuhashi M, Taniguchi M, Kawai T. Nat. Nanotechol. 2014;9:835–840. doi: 10.1038/nnano.2014.193. [DOI] [PubMed] [Google Scholar]
- 98.Di Ventra M, Krems M, Wilson J, Pershin YV. Methods Mol. Biol. 2012;870:149–163. doi: 10.1007/978-1-61779-773-6_9. [DOI] [PubMed] [Google Scholar]
- 99.Liang XG, Chou SY. Nano Lett. 2008;8:1472–1476. doi: 10.1021/nl080473k. [DOI] [PubMed] [Google Scholar]
- 100.Maleki T, Mohammadi S, Ziaie B. Nanotechnology. 2009;20:105302. doi: 10.1088/0957-4484/20/10/105302. [DOI] [PubMed] [Google Scholar]
- 101.Menard LD, Mair CE, Woodson ME, Alarie JP, Ramsey JM. ACS Nano. 2012;6:9087–9094. doi: 10.1021/nn303322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fanget A, Traversi F, Khlybov S, Granjon P, Magrez A, Forró L, Radenovic A. Nano Letters. 2013;14:244–249. doi: 10.1021/nl403849g. [DOI] [PubMed] [Google Scholar]
- 103.Di Ventra M. Nanotechnology. 2013;24:342501. doi: 10.1088/0957-4484/24/34/342501. [DOI] [PubMed] [Google Scholar]
- 104.Traversi F, Raillon C, Benameur SM, Liu K, Khlybov S, Tosun M, Krasnozhon D, Kis A, Radenovic A. Nat. Nanotechnol. 2013;8:939–945. doi: 10.1038/nnano.2013.240. [DOI] [PubMed] [Google Scholar]
- 105.Tsutsui M, Taniguchi M, Yokota K, Kawai T. Nat. Nanotechnol. 2010;5:286–290. doi: 10.1038/nnano.2010.42. [DOI] [PubMed] [Google Scholar]
- 106.Nelson T, Zhang B, Prezhdo OV. Nano Lett. 2010;10:3237–3242. doi: 10.1021/nl9035934. [DOI] [PubMed] [Google Scholar]
- 107.Postma HWC. Nano Lett. 2010;10:420–425. doi: 10.1021/nl9029237. [DOI] [PubMed] [Google Scholar]
- 108.Prasongkit J, Grigoriev A, Pathak B, Ahuja R, Scheicher RH. J. Phys. Chem. C. 2013;117:15421–15428. [Google Scholar]
- 109.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Nature. 2010;467:190–193. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wei C, Bard AJ, Feldberg SW. Anal. Chem. 1997;69:4627–4633. [Google Scholar]
- 111.Cervera J, Schiedt B, Neumann R, Mafé S, Ramírez P. J. Chem. Phys. 2006;124:104706. doi: 10.1063/1.2179797. [DOI] [PubMed] [Google Scholar]
- 112.Kovarik L, Zhou KM, Jacobson SC. J. Phys. Chem. B. 2009;113:15960–15966. doi: 10.1021/jp9076189. [DOI] [PubMed] [Google Scholar]
- 113.Harrell CC, Kohli P, Siwy Z, Martin CR. J. Am. Chem. Soc. 2004;126:15646–15647. doi: 10.1021/ja044948v. [DOI] [PubMed] [Google Scholar]
- 114.Ali M, Ramirez P, Nasir S, Nguyen Q-H, Ensinger W, Mafe S. Appl. Phys. Lett. 2014;104:043703. [Google Scholar]
- 115.Karnik R, Duan C, Castelino K, Daiguji H, Majumdar A. Nano Lett. 2007;7:547–551. doi: 10.1021/nl062806o. [DOI] [PubMed] [Google Scholar]
- 116.Vlassiouk I, Siwy ZS. Nano Lett. 2007;7:552–556. doi: 10.1021/nl062924b. [DOI] [PubMed] [Google Scholar]
- 117.Yan R, Liang W, Fan R, Yang P. Nano Lett. 2009;9:3820–3825. doi: 10.1021/nl9020123. [DOI] [PubMed] [Google Scholar]
- 118.Gamble T, Decker K, Plett TS, Pevarnik M, Pietschmann J-F, Vlassiouk I, Aksimentiev A, Siwy ZS. J. Phys. Chem. C. 2014;118:9809–9819. doi: 10.1021/jp501492g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Siwy Z, Heins E, Harrell CC, Kohli P, Martin CR. J. Am. Chem. Soc. 2004;126:10850–10851. doi: 10.1021/ja047675c. [DOI] [PubMed] [Google Scholar]
- 120.Ali M, Mafé S, Ramírez P, Neumann R, Ensinger W. Langmuir. 2009;25:11993–11997. doi: 10.1021/la902792f. [DOI] [PubMed] [Google Scholar]
- 121.Ali M, Ramírez P, Hung Quoc N, Nasir S, Cervera J, Mafé S, Ensinger W. ACS Nano. 2012;6:3631–3640. doi: 10.1021/nn3010119. [DOI] [PubMed] [Google Scholar]
- 122.Yameen B, Ali M, Neumann R, Ensinger W, Knoll W, Azzaroni O. Small. 2009;5:1287–1291. doi: 10.1002/smll.200801318. [DOI] [PubMed] [Google Scholar]
- 123.Guo W, Xia H, Cao L, Xia F, Wang S, Zhang G, Song Y, Wang Y, Jiang L, Zhu D. Adv. Funct. Mater. 2010;20:3561–3567. [Google Scholar]
- 124.Ali M, Nasir S, Ramírez P, Ahmed I, Quoc Hung N, Fruk L, Mafé S, Ensinger W. Adv. Funct. Mater. 2012;22:390–396. [Google Scholar]
- 125.Wen L, Ma J, Tian Y, Zhai J, Jiang L. Small. 2012;8:838–842. doi: 10.1002/smll.201101661. [DOI] [PubMed] [Google Scholar]
- 126.Zhang M, Hou X, Wang J, Tian Y, Fan X, Zhai J, Jiang L. Adv. Mater. 2012;24:2424–2428. doi: 10.1002/adma.201104536. [DOI] [PubMed] [Google Scholar]
- 127.Guo Z, Wang J, Ren J, Wang E. Nanoscale. 2011;3:3767–3773. doi: 10.1039/c1nr10434a. [DOI] [PubMed] [Google Scholar]
- 128.Yameen B, Ali M, Neumann R, Ensinger W, Knoll W, Azzaroni O. J. Am. Chem. Soc. 2009;131:2070–2071. doi: 10.1021/ja8086104. [DOI] [PubMed] [Google Scholar]
- 129.Ali M, Ramírez P, Mafé S, Neumann R, Ensinger W. ACS Nano. 2009;3:603–608. doi: 10.1021/nn900039f. [DOI] [PubMed] [Google Scholar]
- 130.Yameen B, Ali M, Neumann R, Ensinger W, Knoll W, Azzaroni O. Chem. Comm. 2010;46:1908–1910. doi: 10.1039/b920870d. [DOI] [PubMed] [Google Scholar]
- 131.Liu J, Wang D, Kvetny M, Brown W, Li Y, Wang G. Anal. Chem. 2012;84:6926–6929. doi: 10.1021/ac301791e. [DOI] [PubMed] [Google Scholar]
- 132.Liu J, Kvetny M, Feng J, Wang D, Wu B, Brown W, Wang G. Langmuir. 2012;28:1588–1595. doi: 10.1021/la203106w. [DOI] [PubMed] [Google Scholar]
- 133.Zhang L-X, Cao X-H, Zheng Y-B, Li Y-Q. Electrochem. Commun. 2010;12:1249–1252. [Google Scholar]
- 134.Hou X, Guo W, Xia F, Nie F-Q, Dong H, Tian Y, Wen L, Wang L, Cao L, Yang Y, Xue J, Song Y, Wang Y, Liu D, Jiang L. J. Am. Chem. Soc. 2009;131:7800–7805. doi: 10.1021/ja901574c. [DOI] [PubMed] [Google Scholar]
- 135.He Y, Gillespie D, Boda D, Vlassiouk I, Eisenberg RS, Siwy ZS. J. Am. Chem. Soc. 2009;131:5194–5202. doi: 10.1021/ja808717u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ali M, Nasir S, Ramírez P, Cervera J, Mafé S, Ensinger W. ACS Nano. 2012;6:9247–9257. doi: 10.1021/nn303669g. [DOI] [PubMed] [Google Scholar]
- 137.Buchsbaum SF, Nguyen G, Howorka S, Siwy ZS. J. Am. Chem. Soc. 2014;136:9902–9905. doi: 10.1021/ja505302q. [DOI] [PubMed] [Google Scholar]
- 138.Tian Y, Hou X, Wen L, Guo W, Song Y, Sun H, Wang Y, Jiang L, Zhu D. Chem. Comm. 2010;46:1682–1684. doi: 10.1039/b918006k. [DOI] [PubMed] [Google Scholar]
- 139.Sa N, Fu Y, Baker LA. Anal. Chem. 2010;82:9963–9966. doi: 10.1021/ac102619j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vilozny B, Actis P, Seger RA, Vallmajo-Martin Q, Pourmand N. Anal. Chem. 2011;83:6121–6126. doi: 10.1021/ac201322v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meng Z, Jiang C, Li X, Zhai J. ACS Appl. Mater. Interfaces. 2014;6:3794–3798. doi: 10.1021/am5002822. [DOI] [PubMed] [Google Scholar]
- 142.Powell MR, Sullivan M, Vlassiouk I, Constantin D, Sudre O, Martens CC, Eisenberg RS, Siwy ZS. Nat. Nanotech. 2008;3:51–57. doi: 10.1038/nnano.2007.420. [DOI] [PubMed] [Google Scholar]
- 143.Innes L, Powell MR, Vlassiouk I, Martens C, Siwy ZS. J. Phys. Chem. C. 2010;114:8126–8134. [Google Scholar]
- 144.Vilozny B, Wollenberg AL, Actis P, Hwang D, Singaram B, Pourmand N. Nanoscale. 2013;5:9214–9221. doi: 10.1039/c3nr02105j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Quoc Hung N, Ali M, Neumann R, Ensinger W. Sensors Actuat. B-Chem. 2012;162:216–222. [Google Scholar]
- 146.Ali M, Yameen B, Neumann R, Ensinger W, Knoll W, Azzaroni O. J. Am. Chem. Soc. 2008;130:16351–16357. doi: 10.1021/ja8071258. [DOI] [PubMed] [Google Scholar]
- 147.Vlassiouk I, Kozel TR, Siwy ZS. J. Am. Chem. Soc. 2009;131:8211–8220. doi: 10.1021/ja901120f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ali M, Schiedt B, Neumann R, Ensinger W. Macromol. Biosci. 2010;10:28–32. doi: 10.1002/mabi.200900198. [DOI] [PubMed] [Google Scholar]
- 149.Ali M, Ramírez P, Tahir MN, Mafé S, Siwy Z, Neumann R, Tremel W, Ensinger W. Nanoscale. 2011;3:1894–1903. doi: 10.1039/c1nr00003a. [DOI] [PubMed] [Google Scholar]
- 150.Fu Y, Tokuhisa H, Baker LA. Chem. Comm. 2009:4877–4879. doi: 10.1039/b910511e. [DOI] [PubMed] [Google Scholar]
- 151.Ali M, Neumann R, Ensinger W. ACS Nano. 2010;4:7267–7274. doi: 10.1021/nn102119q. [DOI] [PubMed] [Google Scholar]
- 152.Karnik R, Fan R, Yue M, Li DY, Yang PD, Majumdar A. Nano Lett. 2005;5:943–948. doi: 10.1021/nl050493b. [DOI] [PubMed] [Google Scholar]
- 153.Fan R, Yue M, Karnik R, Majumdar A, Yang PD. Phys. Rev. Lett. 2005;95:086607. doi: 10.1103/PhysRevLett.95.086607. [DOI] [PubMed] [Google Scholar]
- 154.Guan W, Fan R, Reed MA. Nat. Commun. 2011;2:506. doi: 10.1038/ncomms1514. [DOI] [PubMed] [Google Scholar]
- 155.Karnik R, Castelino K, Majumdar A. Appl. Phys. Lett. 2006;88:123114. [Google Scholar]
- 156.Nam S-W, Rooks MJ, Kim K-B, Rossnagel SM. Nano Lett. 2009;9:2044–2048. doi: 10.1021/nl900309s. [DOI] [PubMed] [Google Scholar]
- 157.Paik K-H, Liu Y, Tabard-Cossa V, Waugh MJ, Huber DE, Provine J, Howe RT, Dutton RW, Davis RW. ACS Nano. 2012;6:6767–6775. doi: 10.1021/nn3014917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.James T, Kalinin YV, Chan C-C, Randhawa JS, Gaevski M, Gracias DH. Nano Lett. 2012;12:3437–3442. doi: 10.1021/nl300673r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stein D, Kruithof M, Dekker C. Phys. Rev. Lett. 2004;93 doi: 10.1103/PhysRevLett.93.035901. [DOI] [PubMed] [Google Scholar]
- 160.Haywood DG, Harms ZD, Jacobson SC. Analytical chemistry. 2014;86:11174–11180. doi: 10.1021/ac502596m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schoch RB, van Lintel H, Renaud P. Phys. Fluids. 2005;17:100604. [Google Scholar]
- 162.Frament CM, Dwyer JR. J. Phys. Chem. C. 2012;116:23315–23321. doi: 10.1021/jp305381j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vitarelli MJ, Jr, Prakash S, Talaga DS. Anal. Chem. 2011;83:533–541. doi: 10.1021/ac102236k. [DOI] [PubMed] [Google Scholar]
- 164.Hatsuki R, Yujiro F, Yamamoto T. Microfluid. Nanofluidics. 2013;14:983–988. [Google Scholar]
- 165.Feng J, Liu J, Wu B, Wang G. Anal. Chem. 2010;82:4520–4528. doi: 10.1021/ac100440z. [DOI] [PubMed] [Google Scholar]
- 166.Karnik R, Castelino K, Fan R, Yang P, Majumdar A. Nano Lett. 2005;5:1638–1642. doi: 10.1021/nl050966e. [DOI] [PubMed] [Google Scholar]
- 167.Durand NFY, Renaud P. Lab Chip. 2009;9:319–324. doi: 10.1039/b811006a. [DOI] [PubMed] [Google Scholar]
- 168.Schoch RB, Cheow LF, Han J. Nano Lett. 2007;7:3895–3900. doi: 10.1021/nl0724788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lin L, Liu Y, Yan J, Wang X, Li J. Anal. Chem. 2013;85:334–340. doi: 10.1021/ac302875p. [DOI] [PubMed] [Google Scholar]
- 170.Soni GV, Singer A, Yu Z, Sun Y, McNally B, Meller A. Rev. Sci. Instrum. 2010;81:014301. doi: 10.1063/1.3277116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kurz V, Nelson EM, Shim J, Timp G. ACS Nano. 2013;7:4057–4069. doi: 10.1021/nn400182s. [DOI] [PubMed] [Google Scholar]
- 172.Song W, Pang P, He J, Lindsay S. ACS Nano. 2013;7:689–694. doi: 10.1021/nn3050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yukimoto N, Tsutsui M, He Y, Shintaku H, Tanaka S, Kawano S, Kawai T, Taniguchi M. Sci. Rep. 2013;3:1–7. doi: 10.1038/srep01855. [DOI] [PMC free article] [PubMed] [Google Scholar]