Abstract
Significant advances in our knowledge of cancer genomes are rapidly changing the way we think about tumor biology and the heterogeneity of cancer. Recent successes in genomically-guided treatment approaches accompanied by more sophisticated sequencing techniques have paved the way for deeper investigation into the landscape of genomic rearrangements in cancer. While considerable research on solid tumors has focused on point mutations that directly alter the coding sequence of key genes, far less is known about the role of somatic rearrangements. With many recurring alterations observed across tumor types, there is an obvious need for functional characterization of these genomic biomarkers in order to understand their relevance to tumor biology, therapy, and prognosis. As personalized therapy approaches are turning toward genomic alterations for answers, these biomarkers will become increasingly relevant to the practice of precision medicine. This review discusses the emerging role of genomic rearrangements in breast cancer, with a particular focus on fusion genes. In addition, it raises several key questions on the therapeutic value of such rearrangements and provides a framework to evaluate their significance as predictive and prognostic biomarkers.
Keywords: rearrangements, genomic instability, fusion genes, biomarkers, breast cancer, personalized therapy
Introduction
The role of chromosomal rearrangements in tumorigenesis and cancer progression has received substantial attention in recent years.1–3 Chromosomal rearrangements were initially described in hematologic malignancies such as chronic myelogenous leukemia (CML)4,5 and Burkitt’s lymphoma,6 where they have been used both for diagnosis and to direct targeted therapies. Subsequently, recurrent translocations were also found in rare classes of soft tissue tumors such as Ewing’s sarcoma7,8 and synovial sarcoma.9 Recurrent translocations were not initially identified in many of the common solid, epithelial tumors, in part, because of the limitations of standard cytogenetic analyses and the underlying biological diversity. However, with the emergence of new technologies that allow more comprehensive genomic analysis of solid cancers, genomic rearrangements have been identified in many solid tumors, including breast cancer. This has enabled the identification of subsets of common solid tumors that harbor novel fusions or rearrangements that were not previously appreciated, eg, ALK fusions in non-small cell lung cancer (NSCLC), and has provided novel therapeutic approaches.
Tumor genomic profiling encompasses a variety of sequencing techniques that use next-generation sequencing methods, eg, DNA and RNA sequencing (RNA-seq). Genomically-guided therapy or targeted therapy refers to the selection of a treatment strategy based on the results of tumor genomic profiling, where a clinical response is more likely to occur in the presence of the relevant genomic target. Association between the presence of a genomic alteration and drug response defines a genomic alteration as a predictive biomarker. Such biomarkers have been critical to personalizing the approach to cancer treatment and improving patient outcomes. In contrast, prognostic biomarkers define disease trajectory in the untreated individual. Although some biomarkers can be both predictive and prognostic, biomarkers that are only prognostic can be useful in defining subsets of patients at risk for poor outcomes. Such knowledge allows the treating physicians to determine whether more aggressive or alternative approaches should be undertaken for those patients. Many genomic alterations, including point mutations, deletions/insertions, amplifications, and rearrangements, serve as predictive biomarkers, prognostic biomarkers, or both.
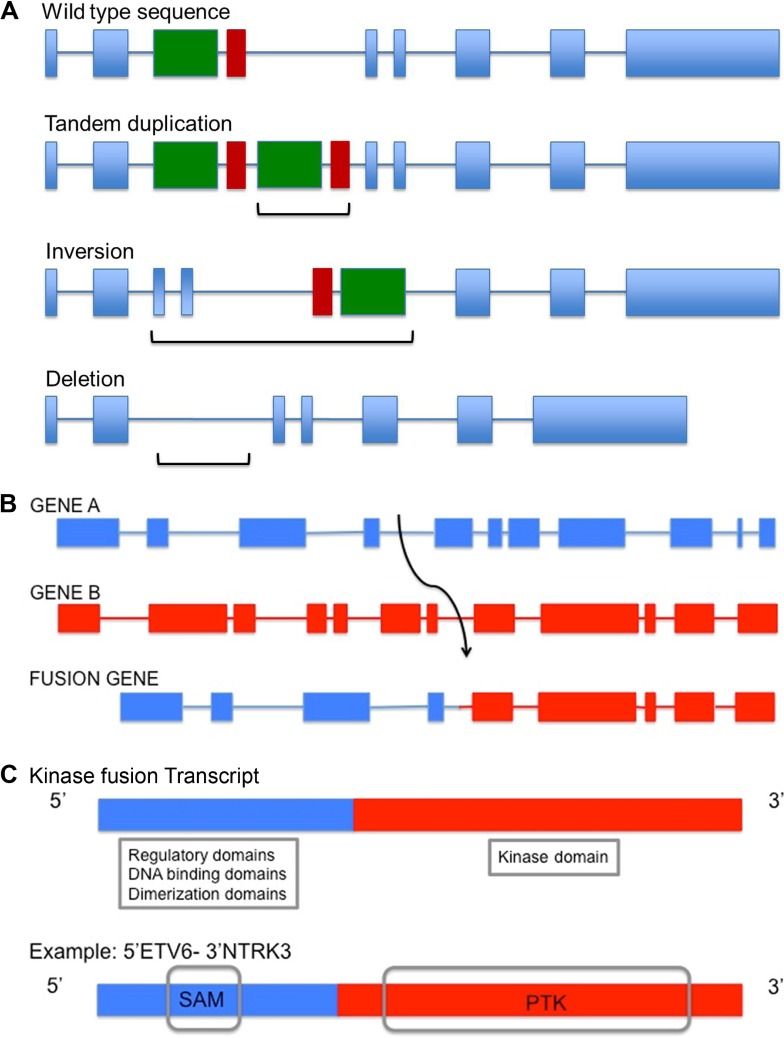
Genomic rearrangements refer to structural changes in the genome that are caused by breakage of DNA followed by erroneous rejoining and replication. These include events that alter copy number, such as deletion, tandem duplication, and amplification, as well as others that maintain copy number, such as reciprocal translocations and inversions (Fig. 1A–C). Rearrangements encompass gross alterations of the whole chromosome or part of a chromosome and do not include the more commonly studied single base mutations or small deletions and insertions of a few base pairs in length. A special class of rearrangements known as interchomosomal or intrachromosomal rearrangements is the result of interactions between distant regions of the genome or even within the same chromosome, respectively. This type of rearrangement can lead to fusion of two disrupted genes, resulting in an altered transcript and a fusion protein (Fig. 1B). These fusions can potentially activate, reduce, or eliminate the original function of the gene product(s) or generate a chimeric protein. Neomorphic functions may also result and have been described, eg, gain of function TP53 mutations and specific PIK3R1, MYOD1, and IDH1 mutations, and are implicated as driver mutations in cancer.10–13
Figure 1.
Illustration of genomic rearrangements and gene fusions. (A) Representation of genomic rearrangements involving tandem duplication, inversion, or deletion involving two exons (green and red boxes) within a single gene. Brackets indicate region of alteration for each mechanism. (B) Representation of a larger genomic rearrangement involving intrachromosomal or interchromosomal translocation leading to the fusion of two independent genes: Gene A (blue), B (red). This event may also involve a change in gene orientation. (C) Receptor tyrosine kinases (RTKs) are often involved in fusion events. A portion of Gene A is fused to that of Gene B, an RTK. Gene A may contribute putative regulatory, coiled-coiled, dimerization or DNA binding domains which may result in the transcription and activation of the kinase portion of the fusion gene. An example of such a fusion is ETV6-NTRK3 in secretory breast carcinoma, where the rearrangement encodes the sterile alpha motif (SAM) dimerization domain of transcription factor ETV6 and the protein tyrosine kinase (PTK) domain of the neurotrophin 3-receptor kinase, NTRK3.68
Detection of recurrent genomic alterations provides new prognostic biomarkers, enables selection of patient groups that may most benefit from specific targeted agents, predicts their response to targeted therapy, and affords the opportunity to elucidate both intrinsic, tissue-specific and acquired resistance mechanisms. With the advent of personalized medicine in cancer, the need for comprehensive genomic profiling of difficult-to-treat tumors is becoming more apparent. While a wealth of information is being generated in the process, characterizing biomarkers for patient classification, prognosis, predicting drug response, and resistance to treatment is crucial.
Defining prognostic and predictive biomarkers in breast cancer is more complicated than in other tumor types. This is primarily because breast cancer represents a heterogeneous set of diseases with distinct molecular features, natural course of disease, and response to treatment. Recognition of this heterogeneity in more recent studies has allowed more precise understanding of molecular characteristics that influence drug response and patient outcomes. Breast cancers are clinically subtyped based on three biomarkers: expression of estrogen receptor (ER) and progesterone receptor (PR) as assayed by immunohistochemistry (IHC) and expression of human epidermal growth factor receptor 2 (HER2) or amplification of erb b2 receptor tyrosine kinase 2 (ERBB2) as assayed by IHC or fluorescence in situ hybridization (FISH), respectively.14,15 In addition to patient stratification, these biomarkers are useful for both prognostication and for predicting response to targeted therapy, eg, ER expression for endocrine therapy;16 ERBB2 amplification for trastuzumab.17 The breast cancer subtype that lacks the expression of ER, PR, and HER2 or ERBB2 amplification, commonly referred to as triple-negative breast cancer (TNBC), is associated with poor prognosis, and standard cytotoxic chemotherapy remains the mainstay of treatment. Gene expression profiling assays (such as OncotypeDx®, Mammaprint®) can also further stratify ER+ cancers to identify those cancers that may benefit from addition of chemotherapy to standard hormonal therapy. ERBB2 amplification is currently the only genomic alteration that is routinely assayed as part of clinical care of breast cancer.
Genomic profiling using high-throughput, next-generation sequencing technologies (ie, whole-exome, whole-genome, and RNA-seq) has identified recurrent point mutations in breast cancer subtypes: PIK3CA mutations in ER+ breast cancers as opposed to TP53, PTEN, and BRCA1 mutations in TNBCs.18–20 Tumors associated with germline BRCA1 mutations can be specifically targeted, either with PARP inhibitors21 or with platinum agents.22 Multiple PIK3CA inhibitors are also in development, currently in both early and late phase clinical trials. Unlike other genomic alterations, the relevance of fusions and rearrangements in breast cancer and its treatment has been less well described.
In this review, the current knowledge of chromosomal instability (CIN) in breast cancer and implications of prognosis for the different molecular subtypes is explored. The patterns and frequency of genomic rearrangements in breast cancer are also discussed. Since more recent knowledge on genomic rearrangements relies heavily on the technique used to study them, the most relevant roles of different technologies and the information acquired are described. Given the therapeutic potential of fusions in cancer, the benefits of identifying and characterizing such rearrangements in breast cancer is outlined. Finally, the current knowledge of breast cancer-related fusions as predictive biomarkers, the future of this evolving field, and the clinical potential for improving therapeutic options for patients with breast cancer is discussed.
Chromosomal Instability and Breast Cancer Prognosis
Genomic rearrangements are closely associated with CIN, which is defined as a dynamic state in which gains or losses of whole or parts of chromosomes occur. Such instability can alter the number of chromosomes, a phenomenon known as aneuploidy. Some cancers such as breast and colorectal cancers harbor more CIN as compared to others.23–25 Though the mechanism of CIN is poorly understood, the implications of its extent have been investigated in relation to clinical outcomes in breast cancer subtypes.
Several groups have analyzed the overall patterns of CIN of breast cancer subtypes and the relevance to clinical outcomes (Table 1). In these studies, CIN was measured based on DNA copy number changes and losses or gains of chromosomal regions or chromosomal number changes in tumor nuclei.26–28 To evaluate the prognosis in some cases, data were retrospectively compared to the clinicopathological details of treatment-naïve patients. CIN is generally considered to be associated with poor prognosis in solid tumors.29,30 While analysis between ER+ and ER− subtypes confirmed that this is true for ER+ cancers, extreme CIN did not show a clear association with survival outcome or prognosis in ER− cancers.26,31 Another study evaluating CIN on the basis of copy number changes found a correlation between increased CIN and poor survival outcomes in ER+ and HER2+ subtypes.27 Similar to the previous reports, CIN score was higher in ER− and TNBC samples. However, there was no correlation with survival outcome in these tumors. Major differences between the studies, including small sample size within CIN study cohorts, different methods for evaluating CIN, presence of confounders, absence of detailed patient and treatment profiles, and other parameters, make interpretation and comparison of such data difficult. These studies highlight the complexity of breast cancer genomes and point to the fact that while instability might be used to assess risk in ER+ cases, additional distinct biological markers for predicting clinical outcome in ER− cases are needed.
Table 1.
Association between chromosomal instability and outcomes in breast cancer subtypes.
| TOOLS USED TO MEASURE CHROMOSOMAL INSTABILITY | BREAST CANCER SUBTYPEa | ASSOCIATION WITH CLINICAL OUTCOME |
|---|---|---|
| Dual centromeric FISH for evaluating modal changes of chromosomes in tumor nuclei26 | ER+ | Higher CIN, poor prognosis |
| ER− | Inconclusive | |
| Microarray to assess copy number gain or loss27 | ER+, HER2+ | Higher CIN, poor metastasis-free survival |
| ER−/PR−/HER2− | Inconclusive | |
| Dual centromeric FISH for evaluating modal changes of chromosomes in tumor nuclei31 | ER− | Higher CIN, improved disease-free survival |
| ER−/HER2− | ||
| Gene expression by microarray of 25 genes to infer aneuploidy29 | Tumor grade 1 | Higher CIN25, poor recurrence-free survival |
| Tumor grade 2 | Higher CIN25, poor recurrence-free survival, metastasis-free survival | |
| Tumor grade 3 | No association | |
| Gene expression by qPCR of 4 genes and ploidy by flow cytometry33 | Tumor grade 2 | Higher CIN4, poor recurrence-free survival |
Notes:
ER−/HER2− means both markers are not expressed; ER−/PR−/HER2− indicates that all three markers are not expressed; ER+, HER2+ indicates either ER+ or HER2+.
Abbreviations: CIN, chromosomal instability; FISH, fluorescence in situ hybridization; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; qPCR, quantitative polymerase chain reaction.
Other groups have developed methods to identify gene expression signatures that reflect CIN and analyze associations with relapse,32 prognosis,33 and survival outcomes29 (Table 1). Tumor grade, which reflects the differentiation and proliferation potential of tumor cells, is a routinely used histological measure. Based on the expression of the top 25 genes in a 25-gene expression assay known as CIN25, grade 1 and grade 2 breast tumors from three data sets were stratified to high or low scores.29 The genes within the panel were selected based on strong associations between altered gene expression and tumor aneuploidy. A higher CIN25 score was associated with a worse clinical outcome for patients with either grade 1 or grade 2 tumors. Similarly, another study using only four genes (CIN4) found a significant association with the proliferation marker Ki67 in grade 2 tumors. CIN4 was used to further stratify patients with grade 2 tumors into good and poor prognosis groups.33 Higher CIN4 was associated with worse recurrence-free survival. Data from these studies, summarized in Table 1, highlight the differences in CIN and the outcomes observed between breast cancer subtypes.
Patterns and Frequency of Genomic Rearrangements in Breast Cancer
An increase in genomic instability has been linked to a concomitant increase in the frequency of gene rearrangements or fusions.2,34 Next-generation sequencing strategies have led to genomic and transcriptomic analysis of large cohorts across cancer types as well as detailed analysis of the patterns of genomic rearrangements in breast cancer.34–36 Some breast cancers showed genome-wide rearrangements, whereas others were reported as clusters in regions of amplification. Array-based comparative genomic hybridization (array CGH) studies of copy number alterations have previously defined structural changes in the genome in terms of gains or losses of specific chromosomal regions such as 1q/16 for low-grade ER+ tumors, commonly amplified sites such as 8p11–12 (FGFR1), 8q24 (MYC), 11q13 (CCND1), and 17q12 (ERBB2) for high-grade ER+ cancers, and numerous low amplitude gains and losses for TNBCs.28,37–40 The presence of 8p11, 8q24, 17q12, and 17q24 amplicons in high-grade ER+ breast cancer has been demonstrated to be associated with poor outcome in multiple studies.41,42
Using next-generation technologies, quantification of the number of rearrangements occurring within chromosomes (intrachromosomal rearrangements such as duplications, inversions, amplifications, and deletions) and also those occurring between different chromosomes (interchromosomal rearrangements) has been depicted by Kwei et al39 using circos plots for breast cancer. Circos plots are circular illustrations for visualizing the structural relationships between regions of chromosomes. These comparisons clearly show distinct patterns related to breast cancer subtypes.39 Low-grade ER+ breast tumors generally display few rearrangements and amplifications, whereas high-grade ER+ breast cancers and TNBCs display a large amount of both large- and small-scale rearrangements, especially increased frequency of intrachromosomal rearrangements such as tandem duplications in TNBCs. This implies that subtype-specific differences in genetic instability may mechanistically contribute to different gene expression patterns observed in breast cancer subtypes.36,39,43
By analyzing 24 breast cancer genomes by paired-end sequencing, Stephens et al36 showed that intrachromosomal alterations are much more prevalent than anticipated across a broad spectrum of molecular subtypes based on ER/PR/ERBB2 status. Tandem duplications were the most commonly observed rearrangement. This feature was largely underappreciated using prior array CGH profiles. Strikingly, there was a large variation between genomes, with some harboring almost no duplications and others having hundreds, ranging from 3 kb to >1 Mb of duplicated segments.36 When subtype analysis was performed, unfavorable ER− cancers harbored comparatively more tandem duplications than ER+ subtypes. In this study, tandem duplications were not found to be associated with breast cancers arising in the setting of germline mutations in the DNA repair genes BRCA1/2. Notably, another group that analyzed tandem duplications in ovarian cancers also reported a lack of correlation of duplications with BRCA1/2 mutations.44 This emphasizes the likelihood of other unknown defects in DNA repair/maintenance in TNBCs, perhaps contributing to the increase in tandem duplications and the relatively poor prognosis of this breast cancer subtype.
Although it has been shown that tandem duplications are the most commonly observed rearrangement in breast cancer genomes, the frequency of specific tandem duplications is currently unknown as these alterations are not easily detected with standard techniques such as array CGH or FISH. Next-generation sequencing techniques will achieve improved clarification of the genes and gene regions that are frequently involved in such duplications.
Role of Technological Advances in Identifying Genomic Rearrangements
The advantages of newer methods to study breast cancer genome have resulted in a greater understanding of the patterns of genomic instability and the underlying gene rearrangements observed within breast cancer (Table 2). Some of these methods include FISH, break-apart FISH, array CGH, polymerase chain reaction (PCR)-based techniques, whole-exome sequencing, whole-genome sequencing (WGS), RNA-seq, single primer enrichment technology (SPET), and anchored multiplex PCR (AMP).
Table 2.
Comparison of methods used to identify rearrangements.
| METHOD | ADVANTAGES | DISADVANTAGES |
|---|---|---|
| Cytogenetics/karyotyping | • Whole genome coverage • Can detect large rearrangements |
• Cannot resolve rearrangements that occur within the same chromosomal band |
| Fluorescence in situ hybridization (FISH) | • Common fusion partners can be identified if partners known a priori | • Fusions with at least one unknown partner result in false negatives • Cannot resolve small intrachromosomal rearrangements less than 3 Mb |
| Break-apart FISH | Common gene involved in fusions can be identified if gene is known a priori | • Does not identify gene fusion partner • may result in false negatives • Cannot identify exact breakpoints if non-canonical |
| Array-based comparative genomic hybridization (array CGH) | • Detects large inter- and intrachromosomal rearrangements | • Does not detect intragenic rearrangements |
| PCR-based techniques | • Can detect known partners in fusion if partners known a priori • Can be used to detect such “intra-band” and intron events only when breakpoints are known |
• May miss small insertions/deletions if allele-specific primers are not used |
| Whole-genome sequencing (WGS) | • Detects small and large rearrangements | • Higher false-negative rate due to lower depth of coverage |
| Exome only sequencing | • Improved false-negative rate as compared to WGS | • Does not detect intronic rearrangements |
| Exome sequencing with targeted intron capture | • Improved false-negative rate as compared to WGS • Detects rearrangements with introns or other regions of genome, e.g. promoter |
• Requires additional coverage of introns known to be involved in fusion breakpoints • Introns have to be “well behaved” (not too big; not too many internal repeats) • May miss less common breakpoints involving other introns • Cannot infer frame of read for all intronic fusions |
| Transcriptome sequencing | • Detects presence and abundance of fusion events (but will require high sequencing depth) | • Higher fail rate with FFPE* tissue • Will miss rearrangements that only affect promoter elements (ie IGH-CCND1 rearrangement) |
| Hybrid capture enriched RNA sequencing | • High sensitivity for fusions involving targeted genes | • Difficult to generate good cDNA libraries from FFPE* • Only detects events involving target genes • Will miss rearrangements involving only promoter regions |
| Single primer enrichment technology (SPET) | • Detects low abundance gene fusion transcripts from fresh or FFPE* tissues • Requires only one partner of the fusion to detect novel fusions |
• Only provides targeted enrichment for desired genes |
Abbreviation:
FFPE, formalin-fixed, paraffin-embedded.
FISH is a powerful tool for detecting specific genomic alterations and has gained popularity due to its clinical application in identifying ERBB2 amplifications in breast cancer,17,45 BCR-ABL fusions in leukemia,46 and EML4-ALK fusions in lung cancer.47 However, since it uses predesigned gene probes, a major drawback is the inability to detect novel alterations. Further, even with known, locus-specific, high-resolution probes, cytogenetic techniques are currently limited in the ability to differentiate between duplications, inversions, or deletions occurring in small regions of a few hundred kilo-bases or less.48 Therefore, standard FISH methods for clinical evaluation may significantly underestimate the prevalence of pathogenic gene rearrangements in solid tumors (Table 2). For example, FISH relies on use of gene-specific probes. Common receptor tyrosine kinase (RTK) fusions with known, recurrent gene partners can be identified using this method. However, fusions with an unknown partner can lead to false negative results. Break-apart FISH describes a technique where two fluorescent probes are designed to recognize one gene. Break-apart FISH can detect the presence of novel fusions with the known gene. In the presence of a rearrangement involving a second gene, the two probes are no longer found in proximity, ie, they break apart. While this technique can establish the presence of a fusion affecting the designated gene, it will not identify novel gene partners, the location of breakpoints in the gene partner, or the functionality of the fusion. Therefore, other methods such as reverse transcriptase-PCR (RT-PCR) and RNA-seq are needed to confirm the presence of such gene fusions. Fusions that occur within the same chromosomal band or with intronic breakpoints are beyond the resolution of cytogenetic techniques. PCR-based assays can be used to detect such intra-band and intron events only when breakpoints are known.
The development of more efficient high-throughput sequencing methods and data analysis pipelines has made the identification of pathogenic rearrangement events across all malignancies more affordable and more accessible. WGS and RNA-seq provide an unbiased view of the genome and expressed transcripts, respectively (Table 2). A subset of these next-generation sequencing methods is designed to capture rearrangement events in particular, eg, intron capture and RNA-Seq. Using WGS and RNA-seq, Stephens et al36 showed that multiple rearrangements are present in many breast cancers, with >50% of them occurring within coding regions.36 These rearrangements can lead to deregulation of gene expression and/or the formation of fusion transcripts, resulting in novel fusion proteins.49
To detect rearrangements, read pairs with unexpected separation distances or orientations that discordantly map to two distinct genes are identified. In order to determine the exact position of the breakpoint, reads that partially map to both genes are then reviewed. Carrara et al50 have comprehensively reviewed the RNA-seq fusion detection tools, and Lin et al51 have detailed the WGS statistical algorithms that detect such variations (structural variation callers). High-throughput sequencing technologies suffer from greater error and shorter reads than traditional sequencing methods, and the bioinformatic pipelines often result in a long list of fusion candidates that require extensive experimental validation.52 Advanced computational analyses are, therefore, required to decipher true rearrangements from false positives that are due to reverse transcriptase template switching, incorrect mapping, read-through transcripts from the splicing of two adjacent genes, and other systematic errors. Further analysis can also help in determining the pathogenic significance of these candidates.53,54 These methodologies have led to the discovery of important fusion events in hematologic and solid tumors.55–57
Whole-exome sequencing can identify rearrangement events whose fusion junctions also occur in the coding region.56,58 More recently, deep sequencing platforms have been developed that capture introns and can detect fusions with intronic breakpoints, previously undetected by exome sequencing only.59 Further, rearrangements occurring in other noncoding regions of the genome can place a gene (or part of it) under the regulation of a different gene promoter, eg, IGH-CCND1 in mantle cell lymphoma.60 Such events are not detectable by transcriptome or hybrid capture RNA-seq (Table 2).
Despite the increased sensitivity in detecting rare events, massively parallel sequencing is prone to error at many levels such as library preparation, analysis, and referencing due to the vastness of the genome and similarity between genes (Table 2).61,62 Due to the reduced quality of formalin-fixed, paraffin-embedded samples, tumor tissue contamination, and low frequency representation of chimeric transcripts, the reduced number and depth of reads can lead to false-negative results in RNA-seq. Newer methodologies are being developed to combine RNA-seq and WGS data to improve sensitivity and specificity.63 For a more specific and reliable detection of fusions, SPET and AMP are currently being explored.64,65
Clinical Significance of Fusions in Cancer
Gene fusions observed repeatedly in certain tumor types are referred to as recurrent gene fusions. Though previously underappreciated in comparison to hematologic malignancies, more recently discovered recurrent gene fusions have been described in solid cancers, including TMPRSS2-Ets family in prostate cancer,66 EML4-ALK fusions in NSCLC,67 and ETV6-NTRK3 in secretory breast carcinoma.68 These recurrent gene fusions have been recognized as tumor-specific oncogenic drivers and have challenged the previously held view that gene fusions are rare in solid tumors and may not play an important role in the development of epithelial tumors.
A recent study using RNA-seq, DNA copy number analysis, and gene mutation analysis of ~4000 primary tumors showed that tumors harboring transcript fusions had significantly fewer driver mutations, suggesting a tumorigenic role for gene fusions.34 Tumors with recurrent, in-frame fusion transcripts were found to have reduced number of gene mutations in comparison to those without recurrent in-frame fusions. This was a common finding in many cancers types, including breast cancer, suggesting that fusions can drive cancer growth and progression in epithelial tumors.
It is important to note that rearrangements not only create novel driver oncogenes but can also disable critical tumor suppressors. In a recently published study, it was shown that ~16% of the osteosarcomas, which lacked commonly known hotspot TP53 mutations, were instead reported to have recurrent rearrangements in intron 1 of TP53.69 Some of these rearrangements resulted in fusions that led to the loss of p53 tumor suppressor function. There are many other reports of out-of-frame fusions with functional consequences.70–72 Such fusions involving tumor suppressor genes, transcriptional repressors, phosphatases, or other regulatory processes can also have a role not only in tumorigenesis but also in drug resistance and malignant phenotype.
Recurrent gene fusions represent a unique class of rearrangements that can serve as predictive or prognostic biomarkers or both. The most commonly observed gene classes involved in fusions are kinases and transcription factors, together representing >50% of all genes found in fusions.73 Recurrent fusions that result in the activation of tyrosine kinases such as BCR-ABL in CML and RET fusions in thyroid cancers have been studied extensively as these cancers are sensitive to kinase inhibitors. This has led to FDA-approved RTK inhibitor (RTKi) therapies for these tumors types, with the fusions serving as predictive biomarkers. Since kinase activation drives cancer-relevant pathways such as growth and proliferation, it is not surprising that oncogenic fusions often retain an intact kinase catalytic domain. Further, many of these kinases are often fused with one of a variety of partners, each with the potential for a unique mechanism of activation such as constitutive dimerization or ligand-independent autophosphorylation, depending on the domains involved in the fusion.74 Table 3 lists a few examples of recurrent kinase fusions that have been reported with more than one fusion partner.
Table 3.
Examples for kinase fusions with multiple 5′ partners, relevant inhibitors, and reported tumor tissue types.
| 3′ KINASE IN FUSION | CURRENT OR POTENTIAL INHIBITORS FOR TARGETED THERAPY | 5′ PARTNERS | REPORTED PRIMARY TISSUES |
|---|---|---|---|
| ALK | 1st and 2nd generation ALK inhibitors, EGFR inhibitors and antibodies, HSP90 inhibitors | EML4 | Lung, Breast, Colorectal |
| NPM1 | Hematopoietic, Lymphoid | ||
| TPM3, TPM4 | Hematopoietic, Lymphoid, Soft tissue | ||
| CLTC | Hematopoietic, Lymphoid, Soft tissue | ||
| RANBP2 | Hematopoietic, Lymphoid, Soft tissue | ||
| BRAF | BRAF and MEK inhibitors | KIAA1549 | Central Nervous System |
| SND1 | Pancreas | ||
| AKAP9 | Thyroid | ||
| PAPSS1 | Skin | ||
| TRIM24 | Skin | ||
| FAM131B | Brain | ||
| MET | MET inhibitors | CAPZA2 | Breast, Lung |
| TFG | Thyroid | ||
| KIF5B | Lung | ||
| NTRK1 | NTRK1/TRKA selective inhibitor, tyrosine kinase inhibitors (TKIs) | TPM3 | Thyroid, Intestine |
| TFG | Thyroid | ||
| LMNA | Skin | ||
| NTRK3 | Pan NTRK inhibitors | ETV6 | Breast, Kidney, Thyroid, Salivary gland, Hematopoietic, Lymphoid, Soft tissue |
| RBPS | Thyroid | ||
| LYN | Head and neck | ||
| RET | RET and pan TKI | CCDC6 | Thyroid, Lung |
| NCOA4 | Thyroid, Lung, Soft tissue | ||
| KIF5B | Lung | ||
| PRKAR1a | Thyroid | ||
| TRIM33 | Lung | ||
| ERC1 | Breast | ||
| ROS1 | ALK/ROS1 inhibitors | CD74 | Lung |
| SLC34A | Lung, Stomach | ||
| SDC4 | Lung | ||
| EZR | Lung | ||
| GOPC | Central Nervous System, Ovary, Lung |
While some fusions serve only as predictive biomarkers for RTKi therapy, others may play a role both as predictive and prognostic biomarkers. Exemplifying this concept is the recurrent fusion between KIAA1549 and BRAF found in >50% of pilocytic astrocytomas. Presence of this fusion was associated with an improved five-year progression-free survival in patients receiving chemotherapy for this childhood brain tumor.75,76 As survival was not correlated to a specific chemotherapy, the KIAA1549-BRAF fusion represents a prognostic marker in this setting. Moreover, this fusion has relevance for targeted therapy with RTKi, which could also classify it as a predictive biomarker. However, absolute associations between the presence of a fusion and either a beneficial or detrimental drug response may not be apparent without validation. Such is the case with KIAA1549-BRAF, where BRAF inhibitors have been evaluated for drug sensitivity and pathway inhibition in fusion-positive cell lines.77 Strikingly, paradoxical activation of the MAPK pathway, as measured by phospho-ERK, and in vivo tumor growth occurred with a first-generation BRAF inhibitor, but not with a second-generation inhibitor. This suggests that the presence of fusions can elicit differential response as compared to kinase-activating mutations and even unpredicted drug responses, which need to be further explored in order to appropriately stratify patients for targeted agents.77,78
In contrast to kinase fusions that most often result in kinase activation, transcription factor fusions can lead to gene activation or to altered gene expression affecting integral mediators of cell function. This can have a dominant negative effect on the cell. In follicular thyroid cancers with PAX8-PPARγ fusions, wild-type PPARγ inhibition is regarded as the mechanism of cellular transformation.79 The relevance of PPARγ agonists in treating fusion-positive, thyroid cancer patients with advanced disease stage is now being tested in phase II trials (clinicaltrials.gov). Intriguingly, a clinical study measuring PPARγ as a surrogate marker for the overexpressed fusion protein showed an association with favorable prognosis and improved clinical outcome.80
Approximately 50% of prostate cancer cases are known to harbor fusions in the Ets transcription factor family with TMPRSS2, a serine protease.66,73,81 Almost 80% of these fusions are TMPRSS2-ERG, which in some studies correlate with poorer prognosis and increased disease recurrence and is thus implicated as a prognostic biomarker.82,83 A genomic deletion event that fuses the 5′ region of the androgen-responsive serine protease TMPRSS2 with the 3′ region of the transcription factor ERG results in an androgen-dependent overexpression of ERG leading to an altered, oncogenic transcriptome. Due to the high frequency and specificity of TMPRSS2-ERG fusions seen in prostate cancer, its clinical applicability as a biomarker for diagnosis84–86 and treatment87,88 is also being explored.
Due to their role, recurrence, and functional implications, fusions thus serve as powerful, predictive biomarkers for targeted therapy, have prognostic implications, and display significant translational relevance.
Current Knowledge of Fusions in Breast Cancer
Large-scale studies involving transcriptome and genomic sequencing have revealed the presence of several gene fusions in the more common types of breast cancer.3,34,36 These include fusions that have been noted in other cancers (Tables 3 and 4). Though several in-frame fusions were observed, a recurrent gene fusion is yet to be identified.3,36 With a few exceptions, the majority of fusions reported in breast cancer are uncommon and present only in a limited number of samples. Whether these are truly single events or are not observed in more samples due to limited sample size, difference in specific breast cancer subtypes included in study cohorts, and/or the appropriate technologies for detection is yet to be determined.
Table 4.
Therapeutic implications for fusions reported in breast cancer.
| FUSION | ONCOGENIC FUNCTION | PREVALENCE IN OTHER CANCERS | POTENTIAL DRUGS/THERAPIES | BREAST CANCER SUBTYPE |
|---|---|---|---|---|
| ETV6-NTRK368,104–108 | Promotes transformation and tumor formation | Mesoblastic nephroma, congenital fibrosarcoma, acute myeloid leukemia, salivary gland secretory carcinoma | Small molecule broad-spectrum kinase inhibitors, NTRK inhibitors, IGF1R/INSR inhibitors | Secretory breast cancer |
| ESR1-CCDC17095 | Increases cell motility, anchorage-independent growth, reduced endocrine sensitivity and xenograft tumor formation | ND | Precise oncogenic pathways involved under investigation | ER+ (luminal B) |
| MAGI3-AKT396 | Constitutive activation of the AKT3 kinase domain and downstream signaling, loss of contact inhibition | ND | ATP-competitive Akt inhibitors, mTOR inhibitors | TNBC |
| ERBB2-fusions89,109 | ND | ND | HER2-targeting agents | Relapsed invasive lobular breast cancer, invasive carcinoma |
| SCNN1A-TNFRSF1A93 | ND | ND | Unknown | ER+, TNBC, breast cancer cell lines |
| CTSD-IFITM1093 | siRNA knockdown decreases live cells | ND | Unknown | ER+, TNBC, breast cancer cell lines |
| EML4-ALK112,113 | Growth inhibition in some cell lines upon EML4 or ALK siRNA knockdown | Colorectal cancer, NSCLC | ALK inhibitors | Basal, luminal, and HER2+ breast cancers, inflammatory breast cancer, breast cancer cell lines |
| ERC1–RET3,34 | ND | Thyroid cancer | RET inhibitors | Invasive carcinoma |
| TBL1XR1–PIK3CA3,98 | ND | Prostate adenocarcinoma | PI3K, AKT or mTOR inhibitors | Invasive carcinoma, metaplastic breast cancer |
Abbreviations: ND, not determined; ER+, estrogen receptor positive; TNBC, triple negative breast cancer; siRNA, small interfering RNA; NSCLC, non-small cell lung cancer.
Thompson et al89 recently reported fusion transcript analysis by five different groups from 813 breast tumors from The Cancer Genome Atlas (TCGA). Although the presence of specific recurrent fusion transcripts was low, most tumors were reported to have at least one or more fusions. ESR1-CCDC170, C20orf3-ACSS1, and USP22-MYH10 fusions were detected in ≥10 tumors in this cohort. The authors also found ERBB2 to be one among the most common 5′ and 3′ fusion partners and was seen in >10 tumors. Several other groups have focused on smaller cohorts of breast cancer cell lines and/or tumor samples and have recently reported the recurrence of specific types of fusions.90–94 Varley et al93 used RNA-seq in 28 breast cancer cell lines and identified six candidate fusion transcripts. Five of these fusion transcripts were confirmed to be present in primary human breast tumors. Comparison of matched adjacent normal tissue with fusion-positive tumor revealed that two of the fusion transcripts, SCNN1A-TNFRSF1A and CTSD-IFITM10, were highly tumor specific (Table 4). Another group investigated 14 breast cancer cell lines for the association of gene fusions to well-known chromosomal loci and reported that 60% of the gene fusions were localized to recurrent amplicons, suggesting that these fusions are a by-product of amplification.91 Regarding data from cancer cell lines, it is important to note that alterations may represent those found in the original tumor or be de novo mutations arising during culture.
Several groups have identified recurrent fusions with potential clinical implications. Robinson et al92 identified the presence of several recurrent gene rearrangements involving MAST and NOTCH family genes using paired-end RNA-seq in 89 cell lines and 69 breast cancer samples. They reported that these genes were repeatedly fused in ~4% and 6% of cases for MAST and NOTCH gene families, respectively. Functional studies suggesting the oncogenic potential of these fusions and of gamma secretase inhibitor in xenograft models showed reduced tumor growth for NOTCH family fusion-positive cell lines. Clinical trials with gamma secretase inhibitors are in progress for breast cancer (clinicaltrials.gov). Yet, most studies include unselected populations and are agnostic to NOTCH genotype, eg, mutations, fusions. Therefore, evaluation of genotype-phenotype associations may define whether these fusions are even relevant as markers for response to gamma secretase inhibitors.
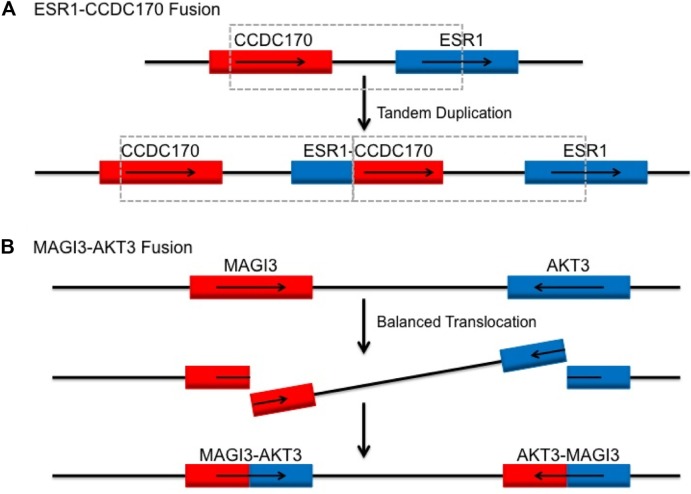
Enrichment of some fusions may also reflect specific molecular subsets of breast cancer. For instance, an integrative pipeline to probe TCGA revealed ESR1-CCDC170 fusions as a top candidate. The frequency of the rearrangement between ESR1 and its neighboring gene CCDC170 occurred in 4% of ER+ breast tumors (n = 200).95 Based on the genomic locations of these two genes and orientation of fusions observed, the authors suggest tandem duplications as the potential mechanism behind the generation of such fusions, as illustrated in Figure 2A. The role of this fusion in oncogenesis and in potentially mediating endocrine resistance is under further investigation.
Figure 2.
Genetic mechanism for reported recurrent fusions ESR1-CCDC170 and MAGI3-AKT3 in breast cancer subtypes, ER+ and TNBC, respectively. (A) Tandem duplication as potential mechanism of generation of ESR1-CCDC170 gene fusion in breast cancers.95 (B) MAGI3-AKT3 fusion as a result of balanced translocation from intron 9 in MAGI3 to intron 1 in AKT3.96
Other breast cancer subtypes with distinct biological behavior have been genomically evaluated using next-generation sequencing strategies. One study that sequenced TNBC samples in Mexican and Vietnamese populations reported ~7% recurrence of MAGI3-AKT3 fusion (Fig. 2B).96 Expression of the fusion in a breast cancer cell line resulted in constitutive activation of the AKT3 kinase domain and downstream pathway signaling. More functional assays are required to clarify the clinical relevance of the MAGI3-AKT3 fusion and the therapeutic benefit of Akt inhibitors in fusion-positive TNBC (Table 4). Another follow-up study aimed at verifying and determining the frequency of these fusions in a larger cohort of TNBCs from Caucasian women failed to detect any MAGI3-AKT3 fusions.97 The first study performed RT-PCR of cDNA from samples followed by Sanger sequencing. The second study analyzed samples using dual color FISH probes for MAGI3 and AKT3 and did not find any signals either by fusion or break-apart probes. They also used RT-PCR for a fraction of samples and analyzed the RNA-seq data of another cohort from TCGA. While there are major differences in the methods used for detection and in cohort populations, these studies underline the importance and challenges of validation and characterization of recurrent fusions in tumors.
Metaplastic breast cancer is a particularly aggressive form of TNBC affecting mesenchymal breast cells. Because it does not respond to standard therapies, the prognosis is worse for patients with this breast cancer subtype as compared to other TNBCs. Characterization of a small cohort (n = 17) revealed the presence of nine in-frame fusions, eg, TBL1XR1-PIK3CA, although none were reported as recurrent in this cohort.98 The therapeutic actionability of TBL1XR1-PIK3CA with PI3K inhibitors, AKT inhibitors, or mTOR inhibitors is yet to be evaluated.
ETV6-NTRK3 is the first recurrent fusion to be identified in a rare breast cancer subtype known as secretory breast carcinoma. These cancers have a good prognosis and rarely metastasize.68 With >90% occurrence in this subtype, the fusion was reported to transform mammary epithelial cells and promote tumor formation in mice, strongly suggesting its role as the dominant acting oncogene in this tumor type.68 NTRK3 is a protein kinase that is normally activated by ligand binding, dimerization, and autophosphorylation. The ETV6-NTRK3 fusion transcripts encode the oligomerization domain of the transcription factor, ETV6, fused to the C-terminal protein kinase domain of NTRK3 (Fig. 1C), leading to ligand-independent dimerization and constitutive autophosphorylation of NTRK3.99,100 Therapeutic targeting of this fusion has been reported with broad-spectrum kinase inhibitors101,102 as well as more specific and mechanism-based IGF1R/INSR inhibitors.103 A phase II clinical trial investigating a selective NTRK1/2/3 inhibitor, LOXO-101, for patients with advanced solid tumors harboring NTRK fusions is ongoing (clinicaltrials.gov). Interestingly, ETV6-NTRK3 was also the first fusion reported to occur in tumors from multiple tissue origins, including epithelial, mesenchymal,104–106 and hematopoietic origins.107,108 With massively parallel sequencing, it has become evident that many such recurrent fusions are prevalent across tumor and tissue types, indicating that different cell types may share similar mechanisms of tumorigenesis.
An analysis of relapsed invasive lobular breast cancers reported the presence of a novel ERBB2 fusion, ERBB2-GRB7 (Table 4).109 Presence of ERBB2 amplifications in breast cancer is well known and routinely evaluated in diagnostic specimens. However, fusions will not be detected by routine IHC and FISH techniques that are currently used for HER2 or ERBB2 evaluation. Considering the impact of HER2-directed therapy in breast cancer, detection and functional testing of such rearrangements offers the potential for treating such tumors with clinically available HER2-targeting agents.
As this is a rapidly evolving field and new information is cataloged daily, some useful websites and data portals for gene fusions in breast cancer include those through the National Cancer Institute,110 Wellcome Trust Sanger Institute,111 and TCGA fusion gene data portal.34
Therapeutic Implications of Fusions in Breast Cancer
Adaptation of trial design has become more important, given the growing knowledge of genotype–drug response associations. Tumor-specific trials still have relevance; however, many recurrent genomic alterations, including rearrangements, identified in one cancer subtype are also seen in other cancers. This underscores the need for expanded eligibility criteria for enrolling patients in clinical trials. ALK and RET fusions are examples of kinase fusions that were initially found in lung and thyroid cancers, respectively, but have also been reported in other cancers, including breast cancers.3,112,113
As more pan-cancer studies are revealing recurrent fusions across tumor types, the concept of basket clinical trials is now being evaluated where patients are matched based on their genomic alteration rather than solely on the basis of tumor type. Basket trials that expand eligibility of cancers with novel genomic alterations would allow investigation of efficacy of targeted agents for previously unreported alterations. Singh et al56 reported the discovery of a highly oncogenic fusion protein, FGFR3-TACC3, in 3% of glioblastoma multiforme patients and demonstrated a high efficacy of FGFR inhibitors against this fusion protein. This fusion was later identified and found to be active in bladder,57 lung,55 and cervical cancers.114 Similarly, fusions involving other FGFR tyrosine kinases, eg, FGFR1/2, present at either the 5′ or 3′ end and retaining the tyrosine kinase domains have also been reported in breast cancer and other cancers.3,34,115 As a result, these fusions provide a therapeutic opportunity for FGFR inhibitors in multiple cancer patient subpopulations.
A caveat to basket trials is that rearrangements may not exhibit the predicted response to drugs. Such an example is the BRAF fusion in comparison to the BRAF V600E point mutation. It is established that melanomas, pilocytic astrocytomas, and lung cancers with BRAF V600E mutations are sensitive to BRAF inhibitors, eg, vemurafenib. However, paradoxical activation can occur in tumors with BRAF fusions. In patients with these fusions, alternative inhibitors, eg, sorafenib, which inhibits feedback signaling through CRAF, or trametinib, which inhibits downstream MEK, may serve as better options in inhibiting tumor growth.77,116,117 Thus, fusions may confer differential functionality as compared to activating point mutations in terms of the mechanism of signaling and sensitivity to inhibition.
Although the prevalence of fusions in breast cancer has been reported in many studies,3,34,36 much less is known about the role of fusions in breast cancer. Functional evaluation of such genes is critical to understand the scope of therapeutic relevance. Validation of top candidates found in cancer cell lines and tumor tissue cohorts suggest potential oncogenic mechanisms and therapeutic opportunities. Table 4 summarizes some of the fusions reported to be recurrent in breast cancer cell lines and tumor subtypes, their oncogenic characterization, prevalence in other cancers, and potential therapeutic opportunities for each. In the latest precision medicine approach for difficult-to-treat cancers, regardless of the recurrence of fusion genes, even the presence of one or more actionable kinase fusions is applicable for targeted therapy.
Small molecule kinase inhibitors are the most commonly used targeted approach (Table 3). However, kinase fusion partners may give rise to their own functional consequences and have also been exploited for the development of targeted treatment strategies. Use of ABL kinase inhibitors for BCR-ABL fusion-positive CML is a successful strategy, but drug resistance may be encountered due to point mutations in the kinase domain. Thus, an alternative is targeting the coil–coil dimerization domain located within the N terminus fusion partner BCR. It is believed that this dimerization domain drives kinase activation. Its targeting has been attempted in cell lines.118–121 The specificity and success of the strategy are yet to be evaluated in vivo. Since many kinase fusion partners are found to have coiled coil domains,3,74,122 eg, EML4 of EML4-ALK; ERC1 of ERC1-RET seen in breast cancer,3 this is a potentially important strategy, given the development of resistance to kinase inhibitors during tumor evolution.
Similar to kinase fusions, transcription factor fusions have also been explored for targeted therapy. For the acute promyelocytic leukemia-associated PML-RARA fusion, arsenic trioxide and all-trans retinoic acid are currently used to target PML, the 5′ moiety, and RARA, the 3′ moiety of the fusion protein, respectively. The precise molecular mechanism of action in both cases involves degradation of the PML-RARA oncoprotein, albeit by different pathways.123,124 Interestingly, the mechanism was elucidated only long after the identification of their therapeutic efficacy. Several groups have reported many transcription factor fusions in breast cancer cell lines90,91,93 and tumors, which suggest their widespread involvement in fusion genes with potential functional implications in breast cancer.
Future Perspectives
Detection of relevant and actionable genomic alterations is at the forefront of personalized therapeutic practice in cancer. Apart from therapy selection and prognosis, there have been clinical reports indicating the usefulness of rearrangements in providing diagnostic clarity,125,126 investigating mechanisms of acquired drug resistance,127 and exploring novel combinatorial therapy. High-throughput genomic sequencing studies, such as exome sequencing of human cancer by TCGA and use of limited hotspot panels, have focused on identifying point mutations and rearrangements specifically involving exons. However, intronic rearrangements are also common in many solid tumors, including breast cancer, and may represent a large class of actionable genomic alterations that are missed by standard short-read sequencing approaches. Inexpensive, rapid turnaround, and clinically implementable sequencing approaches that readily identify potentially actionable genomic rearrangements are clearly needed in conjunction with continued characterization of novel fusion genes.
While our understanding of functional rearrangements in breast cancer is emerging, other overarching challenges remain. These challenges include how to ensure the quality and depth of sequencing reads, standardized reporting and validation across studies, tumor sample purity, clonal heterogeneity, and multifocality. Less well-understood transposable elements such as LINE1 and Alu, which have not been addressed in this review, are also being investigated to define their role in genomic instability and cancer.128,129 Newer techniques of genome editing such as CRISPR, which allow precise manipulation of the genome at a desired location, are under study to model genomic alterations more efficiently and also to develop gene therapy.130–132 Nevertheless, as our understanding grows with affordable, but sophisticated, sequencing strategies and metagenomic approaches, rearrangement-based biomarkers will be pivotal for the practice of precision medicine in breast cancer.
Acknowledgments
We would like to acknowledge Ms. Jacqueline Harris for her support and assistance.
Footnotes
ACADEMIC EDITOR: Barbara Guinn, Editor in Chief
PEER REVIEW: Seven peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1324 words, excluding any confidential comments to the academic editor.
FUNDING: This research was supported by a generous gift to the Genetics Diagnostics to Cancer Treatment Program of the Rutgers Cancer Institute of New Jersey and RUCDR Infinite Biologics, The Val Skinner Foundation, NIH/NCI P30CA072720, AHEPA Foundation, and The Ruth Estrin Goldberg Memorial for Cancer Research. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the first draft of the manuscript: BSP and KMH. Contributed to the writing of the manuscript: BSP, SCD, HK, SG, and KMH. Jointly developed the structure and arguments for the paper: BSP and KMH. Made critical revisions and approved final version: BSP, SCD, HK, LRR, SG, and KMH. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Drier Y, Lawrence MS, Carter SL, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23(2):228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36(4):331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 3.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 5.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36(1):93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 6.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79(24):7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene. 2005;363:1–14. doi: 10.1016/j.gene.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 9.Dal Cin P, Rao U, Jani-Sait S, Karakousis C, Sandberg AA. Chromosomes in the diagnosis of soft tissue tumors. I. Synovial sarcoma. Mod Pathol. 1992;5(4):357–362. [PubMed] [Google Scholar]
- 10.Cheung Lydia WT, Yu S, Zhang D, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell. 2014;26(4):479–494. doi: 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohsaka S, Shukla N, Ameur N, et al. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet. 2014;46(6):595–600. doi: 10.1038/ng.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzymatic activity that converts α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(S2):S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 15.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 16.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17(5):1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 17.Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20(3):719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 18.Balko JM, Giltnane JM, Wang K, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4(2):232–245. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.André F, Bachelot T, Campone M, Arnedos Ballester M, Commol F, Gonçalves A. Array CGH and DNA sequencing to personalize therapy for metastatic breast cancer: a prospective national trial (UNICANCER SAFIR-01) Ann Oncol. 2012;23(suppl):9. [Google Scholar]
- 20.Pierga J, Asselain B, Alsafadi S. A prospective randomized trial evaluating gene expression arrays to select neoadjuvant chemotherapy regimen for operable breast cancer: first report of the REMAGUS04 trial; Paper presented at: 2012 ESMO Congress; 2012. Abstract 2012. [Google Scholar]
- 21.Livraghi L, Garber JE. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med. 2015;13(1):188. doi: 10.1186/s12916-015-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 23.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138(6):2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 25.Yoon D-S, Wersto RP, Zhou W, et al. Variable levels of chromosomal instability and mitotic spindle checkpoint defects in breast cancer. Am J Pathol. 2002;161(2):391–397. doi: 10.1016/S0002-9440(10)64194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roylance R, Endesfelder D, Gorman P, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomark Prev. 2011;20(10):2183–2194. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smid M, Hoes M, Sieuwerts AM, et al. Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Res Treat. 2011;128(1):23–30. doi: 10.1007/s10549-010-1026-5. [DOI] [PubMed] [Google Scholar]
- 28.Loo LW, Grove DI, Williams EM, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64(23):8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 29.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 30.Kronenwett U, Huwendiek S, Östring C, et al. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64(3):904–909. doi: 10.1158/0008-5472.can-03-2451. [DOI] [PubMed] [Google Scholar]
- 31.Jamal-Hanjani M, A’Hern R, Birkbak NJ, et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: a prospective validation cohort study from the TACT trial. Ann Oncol. 2015;26(7):1340–1346. doi: 10.1093/annonc/mdv178. [DOI] [PubMed] [Google Scholar]
- 32.McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14(3):R72. doi: 10.1186/bcr3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szasz AM, Li Q, Eklund AC, et al. The CIN4 chromosomal instability qPCR classifier defines tumor aneuploidy and stratifies outcome in grade 2 breast cancer. PLoS One. 2013;8(2):e56707. doi: 10.1371/journal.pone.0056707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshihara K, Wang Q, Torres-Garcia W, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34(37):4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens PJ, McBride DJ, Lin M-L, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergamaschi A, Kim YH, Wang P, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45(11):1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 38.Haverty PM, Fridlyand J, Li L, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47(6):530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 39.Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol. 2010;4(3):255–266. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nordgard SH, Johansen FE, Alnaes GI, et al. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer. 2008;47(8):680–696. doi: 10.1002/gcc.20569. [DOI] [PubMed] [Google Scholar]
- 41.Bilal E, Vassallo K, Toppmeyer D, et al. Amplified loci on chromosomes 8 and 17 predict early relapse in ER-positive breast cancers. PLoS One. 2012;7(6):e38575. doi: 10.1371/journal.pone.0038575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russnes HG, Vollan HK, Lingjaerde OC, et al. Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci Transl Med. 2010;2(38):38ra47. doi: 10.1126/scitranslmed.3000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride DJ, Etemadmoghadam D, Cooke SL, et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J Pathol. 2012;227(4):446–455. doi: 10.1002/path.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Press MF, Bernstein L, Thomas PA, et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol. 1997;15(8):2894–2904. doi: 10.1200/JCO.1997.15.8.2894. [DOI] [PubMed] [Google Scholar]
- 46.Tkachuk D, Westbrook C, Andreeff M, et al. Detection of bcr-abl fusion in chronic myelogeneous leukemia by in situ hybridization. Science. 1990;250(4980):559–562. doi: 10.1126/science.2237408. [DOI] [PubMed] [Google Scholar]
- 47.McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thoracic Oncol. 2012;7(2):348–354. doi: 10.1097/JTO.0b013e3182381535. [DOI] [PubMed] [Google Scholar]
- 48.Bishop R. Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. Biosci Horizons. 2010;3(1):85–95. [Google Scholar]
- 49.Inaki K, Hillmer AM, Ukil L, et al. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res. 2011;21(5):676–687. doi: 10.1101/gr.113225.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrara M, Beccuti M, Lazzarato F, et al. State-of-the-art fusion-finder algorithms sensitivity and specificity. Biomed Res Int. 2013;2013:340620. doi: 10.1155/2013/340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin K, Smit S, Bonnema G, Sanchez-Perez G, de Ridder D. Making the difference: integrating structural variation detection tools. Brief Bioinform. 2015;16(5):852–864. doi: 10.1093/bib/bbu047. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q, Xia J, Jia P, Pao W, Zhao Z. Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief Bioinform. 2013;14(4):506–519. doi: 10.1093/bib/bbs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shugay M, Ortiz de Mendibil I, Vizmanos JL, Novo FJ. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29(20):2539–2546. doi: 10.1093/bioinformatics/btt445. [DOI] [PubMed] [Google Scholar]
- 54.Abate F, Zairis S, Ficarra E, et al. Pegasus: a comprehensive annotation and prediction tool for detection of driver gene fusions in cancer. BMC Syst Biol. 2014;8:97. doi: 10.1186/s12918-014-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230(3):270–276. doi: 10.1002/path.4209. [DOI] [PubMed] [Google Scholar]
- 56.Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22(4):795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chmielecki J, Crago AM, Rosenberg M, et al. Whole exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet. 2013;45(2):131–132. doi: 10.1038/ng.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirts BH, Salipante SJ, Casadei S, et al. Deep sequencing with intronic capture enables identification of an APC exon 10 inversion in a patient with polyposis. Genet Med. 2014;16(10):783–786. doi: 10.1038/gim.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li JY, Gaillard F, Moreau A, et al. Detection of translocation t(11;14)(q13;q32) in mantle cell lymphoma by fluorescence in situ hybridization. Am J Pathol. 1999;154(5):1449–1452. doi: 10.1016/S0002-9440(10)65399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lou DI, Hussmann JA, McBee RM, et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A. 2013;110(49):19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sleep JA, Schreiber AW, Baumann U. Sequencing error correction without a reference genome. BMC Bioinformatics. 2013;14:367. doi: 10.1186/1471-2105-14-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, White NM, Schmidt HK, et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res. 2015;26(1):108–118. doi: 10.1101/gr.186114.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scolnick JA, Dimon M, Wang IC, Huelga SC, Amorese DA. An efficient method for identifying gene fusions by targeted RNA sequencing from fresh frozen and FFPE samples. PLoS One. 2015;10(7):e0128916. doi: 10.1371/journal.pone.0128916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 66.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 67.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 68.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 69.Ribi S, Baumhoer D, Lee K, et al. TP53 intron 1 hotspot rearrangements are specific to sporadic osteosarcoma and can cause Li-Fraumeni syndrome. Oncotarget. 2015;6(10):7727–7740. doi: 10.18632/oncotarget.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. New Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36(8):945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 72.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 73.Prensner JR, Chinnaiyan AM. Oncogenic gene fusions in epithelial carcinomas. Curr Opin Genet Dev. 2009;19(1):82–91. doi: 10.1016/j.gde.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18(3):378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 75.Jones DT, Gronych J, Lichter P, Witt O, Pfister SM. MAPK pathway activation in pilocytic astrocytoma. Cell Mol Life Sci. 2012;69(11):1799–1811. doi: 10.1007/s00018-011-0898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hawkins C, Walker E, Mohamed N, et al. BRAF-KIAA1549 fusion predicts better clinical outcome in pediatric low-grade astrocytoma. Clin Cancer Res. 2011;17(14):4790–4798. doi: 10.1158/1078-0432.CCR-11-0034. [DOI] [PubMed] [Google Scholar]
- 77.Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A. 2013;110(15):5957–5962. doi: 10.1073/pnas.1219232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haynes HR, Camelo-Piragua S, Kurian KM. Prognostic and predictive biomarkers in adult and pediatric gliomas: toward personalized treatment. Front Oncol. 2014;4:47. doi: 10.3389/fonc.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gregory Powell J, Wang X, Allard BL, et al. The PAX8/PPAR[gamma] fusion oncoprotein transforms immortalized human thyrocytes through a mechanism probably involving wild-type PPAR[gamma] inhibition. Oncogene. 2004;23(20):3634–3641. doi: 10.1038/sj.onc.1207399. [DOI] [PubMed] [Google Scholar]
- 80.Sahin M, Allard BL, Yates M, et al. PPARγ staining as a surrogate for PAX8/PPARγ fusion oncogene expression in follicular neoplasms: clinicopathological correlation and histopathological diagnostic value. J Clin Endocrinol Metabol. 2005;90(1):463–468. doi: 10.1210/jc.2004-1203. [DOI] [PubMed] [Google Scholar]
- 81.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45(7):717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 82.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60(11):1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshimoto M, Joshua AM, Cunha IW, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21(12):1451–1460. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 84.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13(17):5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 85.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20(5):538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 86.Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2015;68(5):e108. doi: 10.1016/j.eururo.2015.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Graff RE, Pettersson A, Lis RT, et al. The TMPRSS2:ERG fusion and response to androgen deprivation therapy for prostate cancer. Prostate. 2015;75(9):897–906. doi: 10.1002/pros.22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang S, Kollipara RK, Srivastava N, et al. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proc Natl Acad Sci U S A. 2014;111(11):4251–4256. doi: 10.1073/pnas.1322198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thompson EA AY, Su X, et al. A comprehensive analysis of fusion transcripts in breast cancer reveals associations between number of fusion transcripts, copy number events, gene expression profiles, and potentially clinical outcome. San Antonio Breast Cancer Symposium. 2016;16(suppl):S4–02. [Google Scholar]
- 90.Edgren H, Murumagi A, Kangaspeska S, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12(1):R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kalyana-Sundaram S, Shankar S, DeRoo S, et al. Gene fusions associated with recurrent amplicons represent a class of passenger aberrations in breast cancer. Neoplasia. 2012;14(8):702–708. doi: 10.1593/neo.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson DR, Kalyana-Sundaram S, Wu Y-M, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17(12):1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Varley KE, Gertz J, Roberts BS, et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat. 2014;146(2):287–297. doi: 10.1007/s10549-014-3019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weirather JL, Afshar PT, Clark TA, et al. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 2015;43(18):e116. doi: 10.1093/nar/gkv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Veeraraghavan J, Tan Y, Cao X-X, et al. Recurrent ESR1 CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mosquera J-M, Varma S, Pauli C, et al. MAGI3-AKT3 fusion in breast cancer amended. Nature. 2015;520(7547):E11–E12. doi: 10.1038/nature14265. [DOI] [PubMed] [Google Scholar]
- 98.Piscuoglio S, Ng CKY, Cowell CF, et al. Genomic and transcriptomic heterogeneity in metaplastic breast carcinomas. San Antonio Breast Cancer Symposium. 2016;16(suppl):P6-03–10. [Google Scholar]
- 99.Liu Q, Schwaller J, Kutok J, et al. Signal transduction and transforming properties of the TEL-TRKC fusions associated with t(12;15)(p13;q25) in congenital fibrosarcoma and acute myelogenous leukemia. EMBO J. 2000;19(8):1827–1838. doi: 10.1093/emboj/19.8.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wai DH, Knezevich SR, Lucas T, Jansen B, Kay RJ, Sorensen PH. The ETV6-NTRK3 gene fusion encodes a chimeric protein tyrosine kinase that transforms NIH3T3 cells. Oncogene. 2000;19(7):906–915. doi: 10.1038/sj.onc.1203396. [DOI] [PubMed] [Google Scholar]
- 101.Chi HT, Ly BT, Kano Y, Tojo A, Watanabe T, Sato Y. ETV6-NTRK3 as a therapeutic target of small molecule inhibitor PKC412. Biochem Biophys Res Commun. 2012;429(1–2):87–92. doi: 10.1016/j.bbrc.2012.10.087. [DOI] [PubMed] [Google Scholar]
- 102.Taipale M, Krykbaeva I, Whitesell L, et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotech. 2013;31(7):630–637. doi: 10.1038/nbt.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tognon CE, Somasiri AM, Evdokimova VE, et al. ETV6-NTRK3-mediated breast epithelial cell transformation is blocked by targeting the IGF1R signaling pathway. Cancer Res. 2011;71(3):1060–1070. doi: 10.1158/0008-5472.CAN-10-3096. [DOI] [PubMed] [Google Scholar]
- 104.Knezevich SR, Garnett MJ, Pysher TJ, Beckwith JB, Grundy PE, Sorensen PHB. ETV6-NTRK3 gene fusions and trisomy 11 establish a histogenetic link between mesoblastic nephroma and congenital fibrosarcoma. Cancer Res. 1998;58(22):5046–5048. [PubMed] [Google Scholar]
- 105.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18(2):184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- 106.Rubin BP, Chen C-J, Morgan TW, et al. Congenital mesoblastic nephroma t(12; 15) is associated withETV6-NTRK3 gene fusion: cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol. 1998;153(5):1451–1458. doi: 10.1016/S0002-9440(10)65732-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kralik JM, Kranewitter W, Boesmueller H, et al. Characterization of a newly identified ETV6-NTRK3 fusion transcript in acute myeloid leukemia. Diagn Pathol. 2011;6:19. doi: 10.1186/1746-1596-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eguchi M, Eguchi-Ishimae M, Tojo A, et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25) Blood. 1999;93(4):1355–1363. [PubMed] [Google Scholar]
- 109.Ross JS, Wang K, Sheehan CE, et al. Relapsed classic E-cadherin (CDH1)-mutated invasive lobular breast cancer shows a high frequency of HER2 (ERBB2) gene mutations. Clin Cancer Res. 2013;19(10):2668–2676. doi: 10.1158/1078-0432.CCR-13-0295. [DOI] [PubMed] [Google Scholar]
- 110.Balko J, Mayer I, Levy M, et al. Molecular profiling of breast cancer. My Cancer Genome. 2015. http://www.mycancergenome.org/content/disease/breast-cancer/2015.
- 111.Bamford S. The COSMIC (catalogue of somatic mutations in cancer) Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. http://cancer.sanger.ac.uk/cosmic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7(9):1466–1476. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]
- 113.Robertson FM, Petricoin Iii, EF Van, Laere SJ, et al. Presence of anaplastic lymphoma kinase in inflammatory breast cancer. SpringerPlus. 2013;2:497. doi: 10.1186/2193-1801-2-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carneiro BA, Elvin JA, Kamath SD, et al. FGFR3-TACC3: a novel gene fusion in cervical cancer. Gynecol Oncol Rep. 2015;13:53–56. doi: 10.1016/j.gore.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Medves S, Demoulin JB. Tyrosine kinase gene fusions in cancer: translating mechanisms into targeted therapies. J Cell Mol Med. 2012;16(2):237–248. doi: 10.1111/j.1582-4934.2011.01415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14(7):455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hutchinson KE, Lipson D, Stephens PJ, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res. 2013;19(24):6696–6702. doi: 10.1158/1078-0432.CCR-13-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beissert T, Hundertmark A, Kaburova V, et al. Targeting of the N-terminal coiled coil oligomerization interface by a helix-2 peptide inhibits unmutated and imatinib-resistant BCR/ABL. Int J Cancer. 2008;122(12):2744–2752. doi: 10.1002/ijc.23467. [DOI] [PubMed] [Google Scholar]
- 119.Beissert T, Puccetti E, Bianchini A, et al. Targeting of the N-terminal coiled coil oligomerization interface of BCR interferes with the transformation potential of BCR-ABL and increases sensitivity to STI571. Blood. 2003;102(8):2985–2993. doi: 10.1182/blood-2003-03-0811. [DOI] [PubMed] [Google Scholar]
- 120.Mian AA, Oancea C, Zhao Z, Ottmann O, Ruthardt M. Oligomerization inhibition, combined with allosteric inhibition, abrogates the transformation potential of T315I-positive BCR/ABL. Leukemia. 2009;23(12):2242–2247. doi: 10.1038/leu.2009.194. [DOI] [PubMed] [Google Scholar]
- 121.Woessner DW, Eiring AM, Bruno BJ, et al. A coiled-coil mimetic intercepts BCR-ABL1 dimerization in native and kinase-mutant chronic myeloid leukemia. Leukemia. 2015;29(8):1668–1675. doi: 10.1038/leu.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Monaco C, Visconti R, Barone MV, et al. The RFG oligomerization domain mediates kinase activation and re-localization of the RET/PTC3 oncoprotein to the plasma membrane. Oncogene. 2001;20(5):599–608. doi: 10.1038/sj.onc.1204127. [DOI] [PubMed] [Google Scholar]
- 123.Lallemand-Breitenbach V, Zhu J, Puvion F, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor α degradation. J Exp Med. 2001;193(12):1361–1372. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu J, Gianni M, Kopf E, et al. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor α (RARα) and oncogenic RARα fusion proteins. Proc Nat Acad Sci. 1999;96(26):14807–14812. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wagner BP, Epperla N, Medina-Flores R. Diagnostic dilemma: late presentation of amelanotic BRAF-negative metastatic malignant melanoma resembling clear cell sarcoma: a case report. Diagn Pathol. 2013;8:192. doi: 10.1186/1746-1596-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Patel RM, Downs-Kelly E, Weiss SW, et al. Dual-color, break-apart fluorescence in situ hybridization for EWS gene rearrangement distinguishes clear cell sarcoma of soft tissue from malignant melanoma. Mod Pathol. 2005;18(12):1585–1590. doi: 10.1038/modpathol.3800503. [DOI] [PubMed] [Google Scholar]
- 127.Dillon R, Nilsson CL, Shi SDH, Lee NV, Krastins B, Greig MJ. Discovery of a novel B-Raf fusion protein related to c-met drug resistance. J Proteome Res. 2011;10(11):5084–5094. doi: 10.1021/pr200498v. [DOI] [PubMed] [Google Scholar]
- 128.Ayarpadikannan S, Kim HS. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Informat. 2014;12(3):98–104. doi: 10.5808/GI.2014.12.3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee E, Iskow R, Yang L, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li Y, Park AI, Mou H, et al. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol. 2015;16:111. doi: 10.1186/s13059-015-0680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sachdeva M, Sachdeva N, Pal M, et al. CRISPR/Cas9: molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Ther. 2015;22(11):509–517. doi: 10.1038/cgt.2015.54. [DOI] [PubMed] [Google Scholar]
- 132.Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]