Abstract
Prothrombin is activated to thrombin by the prothrombinase complex through sequential cleavage at two distinct sites. This occurs at sites of vascular injury in a highly regulated cascade of serine protease and cofactor activation, where activated platelets provide a suitable surface for protease/cofactor/substrate assembly. The precise structural and conformational changes undergone during the transition from prothrombin to thrombin have been studied for decades, and several structures of prothrombin fragments along the activation pathway have been solved. Here we present a new structure analyzed in context of other recent structures and biochemical studies. What emerges is an unexpected mechanism that involves a change in the mode of binding of the F2 domain (fragment 2) on the catalytic domain after cleavage at Arg320, and a subsequent reorientation of the linker between the F2 and catalytic domain to present the Arg271 site for cleavage.
Keywords: Hemostasis, Protease, Prothrombinase, Regulation, Thrombin
Highlights
-
•
The catalytic domain of thrombin precursors binds to its F2 domain by two distinct modes.
-
•
Cleavage of prothrombin at either Arg271 or Arg320 results in shift from mode 2 to mode 1.
-
•
After cleavage at Arg320, movement of F2 helps to present the second cleavage site at Arg271.
1. Introduction
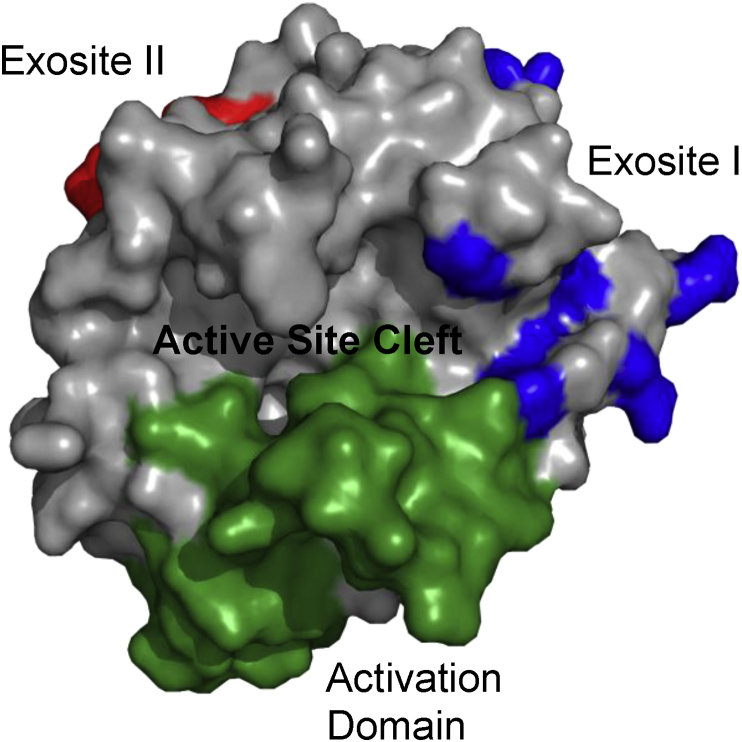
A critical event in hemostasis is the conversion of prothrombin from an inactive zymogen to the active enzyme thrombin. This is the end-point of the blood coagulation cascade and is regulated by many factors [1], [2]. Once formed, thrombin uses two anion-binding exosites (I and II) and its active site to recognize several substrates (Fig. 1) [3], [4]. Maintenance of prothrombin as a zymogen requires the encryption of these three sites to prevent premature interactions. Catalytic inactivity is a general feature of zymogens of serine proteases, but encryption of the two exosites is equally important in prothrombin to prevent the dysregulation of hemostasis. If exosite I were ‘competent’ then prothrombin could saturate thrombomodulin on the vascular endothelium, thereby precluding binding by active thrombin and preventing protein C activation. It could also bind to fibrinogen and potentially block fibrin formation, and similarly bind to circulating fV and fVIII thereby preventing their activation [5], [6], [7], [8], [9]. Exosite II exposure would localize prothrombin to the intact vascular surface via heparan sulfate binding and to platelets via GpIbα binding [10], [11], [12], [13].
Fig. 1.
Structural features of thrombin. Surface representation of thrombin in gray in the standard orientation showing the positions of exosite I (in blue), exosite II (in red), the activation domain (in green), and the active site cleft.
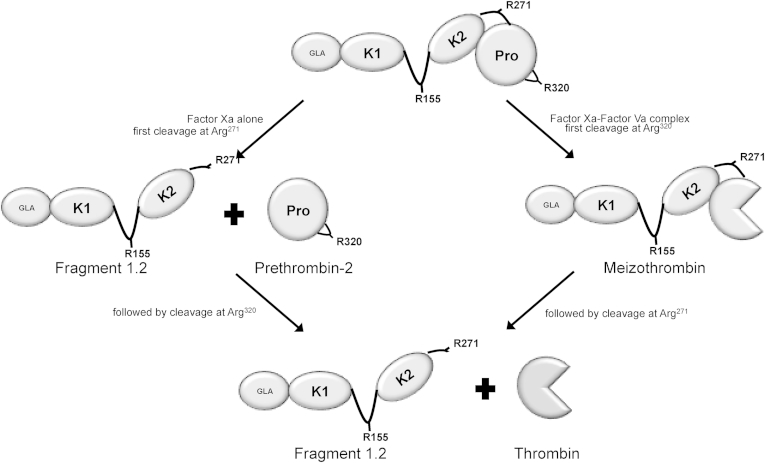
Most serine protease zymogens are converted to active proteases through the cleavage of a single scissile bond between residues 15 and 16 (chymotrypsin numbering). The structural rearrangements that lead to the development of a functional active site cleft is well understood, and involves the ‘insertion’ of the new N-terminus (normally Ile16) into the so-called ‘activation pocket’ [14], [15]. However, it can also be effected in the absence of cleavage by peptides and certain virulence factors such as staphylocoagulase [16], [17], [18]. This results in the formation of the principal substrate-binding pocket (S1) and the oxyanion hole. Prothrombin is an unusual zymogen in the sense that cleavage of the Arg15-Ile16 (Arg320-Ile321 in prothrombin numbering) peptide bond does not result in a fully functional enzyme, but in the disulfide-linked enzyme intermediate meizothrombin (mIIa) [19]. Complete activation to thrombin requires an additional cleavage at residue Arg271 (prothrombin numbering) to release the catalytic domain from the N-terminal fragment 1.2 (F1.2), composed of the Gla domain and two kringle domains (Fig. 2). Two potential pathways for prothrombin activation are available depending on which of these two cleavage events occurs first. With factor Xa alone, cleavage at Arg271 is kinetically favored, and cleavage at Arg320 progressing relatively slowly. Initial cleavage at Arg271 results in formation of the zymogen precursor prethrombin-2 (pre2) and F1.2 [20]. In the presence of factor Va and phospholipid membranes, prothrombinase cleaves first at the Arg320 site, followed by cleavage at Arg271 [21], [22]. The ordered cleavage of Arg320 followed by Arg271 of prothrombin by prothrombinase has been referred to as ‘ratcheting’ because the zymogen to protease conformational change seems to present the second cleavage site to the active site of fXa [23]. A recent study concluded that cleavage by prothrombinase through the meizothrombin intermediate may be attributed to the use of synthetic phospholipids and that activation of prothrombin on platelets proceeds via the pre2 intermediate [24]. However, it is clear that the meizothrombin pathway predominates provided prothrombin is anchored to the same surface as the prothrombinase complex [25], and this must thus be considered the physiological pathway, in spite of the use of synthetic phospholipid vesicles.
Fig. 2.
Prothrombin activation pathways. The proteolytic conversion of the zymogen prothrombin to the enzyme thrombin is the result of proteolytic cleavage at Arg271 and Arg320. In the presence of factor Xa alone, initial cleavage at Arg271 produces F1.2 and pre2 (left pathway), followed by cleavage at Arg320 generating active thrombin. In the presence of factor Va and phospholipids, factor Xa cleaves Arg320 producing the active intermediate meizothrombin, which is further processed at Arg271 to release thrombin from F1.2 (right pathway). The Gla domain (GLA), kringle 1 (K1), kringle 2 (K2), and protease domains (Pro) are shown as ovals showing linkers between each as lines.
Exosite I exists in a flexible, low-affinity state in prothrombin that is ordered upon conversion to thrombin [26]. Exosite II is occupied in prothrombin by the second kringle domain, fragment 2 (F2), blocking exosite II mediated interactions, and effectively encrypting this exosite in prothrombin. F2 binds 2-fold more weakly to thrombin than to the zymogen pre2, and F1.2 binds to thrombin 20-fold more weakly than to pre2 [27]. This suggests that formation of meizothrombin through cleavage at Arg320 may help release the catalytic domain from its interaction with F2. It is unclear how the covalent linkage of the F1 domain affects the binding of F2 to the catalytic domain, but a direct interaction between the F1 and F2 domains is unlikely. The conformational changes in the zymogen activation domain upon cleavage at Arg320 matures exosite I, although meizothrombin is still somewhat deficient in its ability to bind to exosite I-dependent ligands [28]. Thus, cleavage at both Arg320 bond and removal of F1.2 by cleavage at Arg271 is required for the formation of fully competent exosites on thrombin. Here we present a new structure of the pre2/F2 complex in context of other structures of prothrombin fragments containing at least the F2 and catalytic domains, and investigate how conformational change might contribute to prothrombin activation.
2. Materials and methods
2.1. Protein expression and purification
Human prethrombin-2 was expressed in E. Coli using our previously described method [29]. Briefly, wild-type human prethrombin-2 in the pET23(+) vector (Novagen) was converted to the S195A (chymotrypsin numbering) variant using site directed mutagenesis and transformed into the BL21 STAR (DE3) pLysS E. Coli strain (Invitrogen). The protein was expressed into inclusion bodies, washed, and then solubilized in 1 mL 0.1% tri-fluoroacetic acid. The solubilized protein was diluted to 10 mL with the addition of 9 mL of 7 M guanidine hydrochloride and 30 mM l-cysteine, and refolded by drop-wise addition of 2.5 L of 50 mM Tris–HCl, pH7.4, 0.6 M l-arginine, 500 mM NaCl, 1 mM EDTA, 1 mM l-cysteine, 10% glycerol, 0.2% Brij-58. The protein was then loaded onto a 5 mL HiTrap heparin Sepharose column (GE Healthcare) and eluted with a 0.25–1 M NaCl gradient over 5 column volumes. Fractions containing prethrombin-2 were pooled and dialyzed into 20 mM Tris, 100 mM NaCl, pH7.4.
2.2. Digestion of human plasma prothrombin and purification of fragment-2
Human plasma prothrombin was purified as previously described [30]. Prothrombin was digested with 0.5 U recombinant factor Xa per mg prothrombin (Novagen) in 10 mM Tris, 100 mM NaCl, 1 mM CaCl2 overnight at 22 °C. The cleavage products were initially separated using a 5 mL HiTrap heparin Sepharose column where both factor Xa and prethrombin-2 were retained, while fragments 1, 2, and prothrombin were collected in the flow-through. Following 3-fold dilution with 20 mM Tris, pH8.0, the flow-through containing fragments 1 and 2 were loaded onto a 5 mL HiTrap Q Sepharose column and separated with a gradient elution from 0 to 0.6 M NaCl over 10 column volumes. Fractions containing fragment 2 were pooled, concentrated, and dialyzed against 20 mM Tris, pH7.4.
2.3. Crystallization of hF2-preII(S195A) complex
Recombinant prethrombin-2 was combined with a 20% molar excess of plasma derived fragment 2 and concentrated to 8.7 mg/mL. Initial crystallization trials were setup using 200 nL drops on a Screenmaker 96 + 8 Xtal crystallization robot (Innovadyne). Several preliminary crystals were observed after 1–2 days from the Morpheus Screen (Molecular Dimensions). Following optimization, large crystals (200 × 200 × 300 μm) grew in 1–3 days in 10% Alcohols mix, 10% Buffer 1 Mix, 40% EDO_P8K Mix of the Morpheus Screen.
2.4. Data collection, processing, and refinement
Data from a single flash-cooled crystal were collected at Diamond beamline I04 (Didcot, UK) and indexed using Mosflm [31]. The data were processed with the CCP4 program suite using Scala and Truncate [32] followed by molecular replacement using Phaser [33]. Initial refinement was carried out iteratively with successive rounds of model building followed by simulated annealing using CNS [34]. XtalView [35] was used for model building. Data processing and refinement statistics are given in Table 1. Figures were made using Pymol [36], and the interaction area was calculated using AREAIMOL (part of the CCP4 suite). Coordinates and structure factors are deposited in the Protein Data Bank under PDBID code 3K65.
Table 1.
Data processing and refinement statistics for 3K65.
| Crystal | ||
| Space group | P41212 | |
| Cell dimensions (Å) | a = b = 73.53 c = 205.88 α = β = γ = 90 |
|
| Solvent content (%) | 62.4 | |
| Data Processing | ||
| Wavelength (Å) | 0.97 (Diamond I04) | |
| Resolution (Å) | 51.50–1.85 | 1.95–1.85 |
| Total reflections | 369318 | 54430 |
| Unique reflections | 48719 | 7012 |
| Multiplicity | 7.6 | 7.8 |
| <I/σ(I)> | 16.5 | 3.7 |
| Completeness (%) | 99.0 | 99.5 |
| Rmerge | 9.1 | 37.4 |
| Model Details | ||
| # of atoms: | ||
| Protein | 2779 | |
| Water | 231 | |
| Average B-factor (Å2) | 36.4 | |
| Refinement Statistics | 12.00–1.85 | 1.90–1.85 |
| Reflections (working/free) | 46006/2463 | 3286/173 |
| Rfactor/Rfree (%) | 20.8/23.3 | 32.8/31.5 |
| r.m.s. deviation from ideality bonds (Å)/Angles (°) | 0.028/2.19 | |
| Ramachandran plot (%) | ||
| Most favored | 95.6 | |
| Additionally allowed | 4.4 | |
| Outliers | 0.0 | |
3. Structural comparison
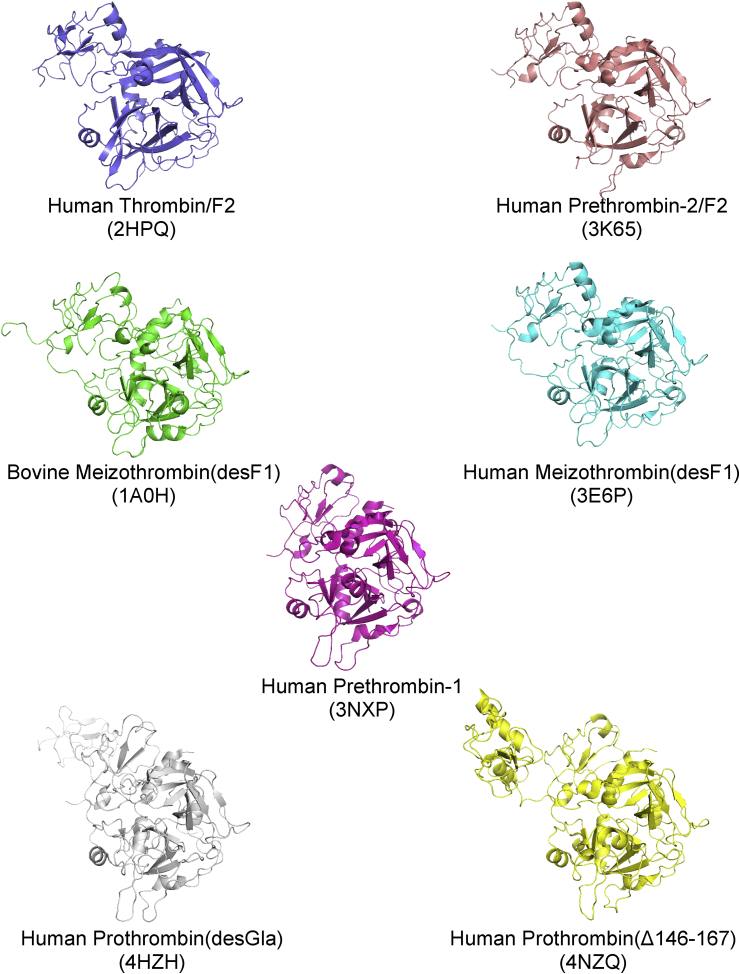
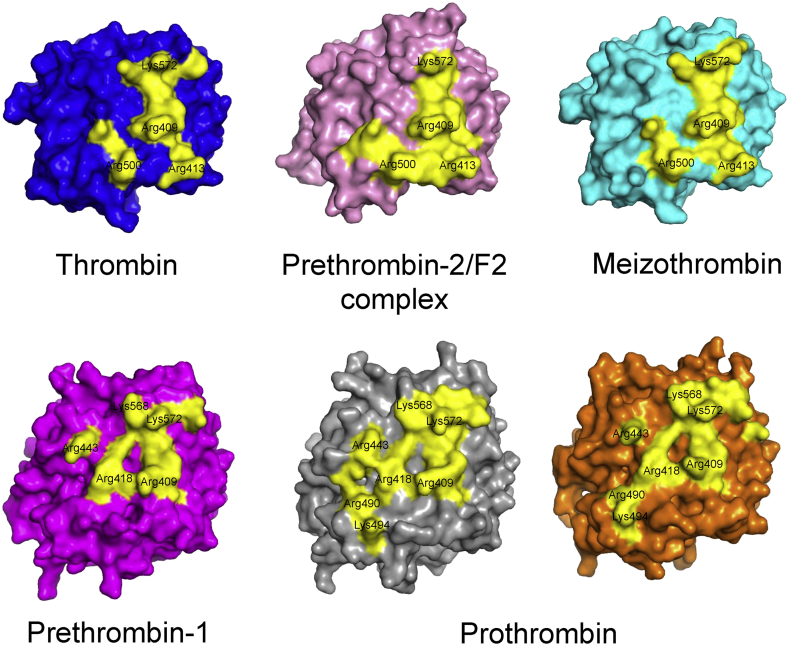
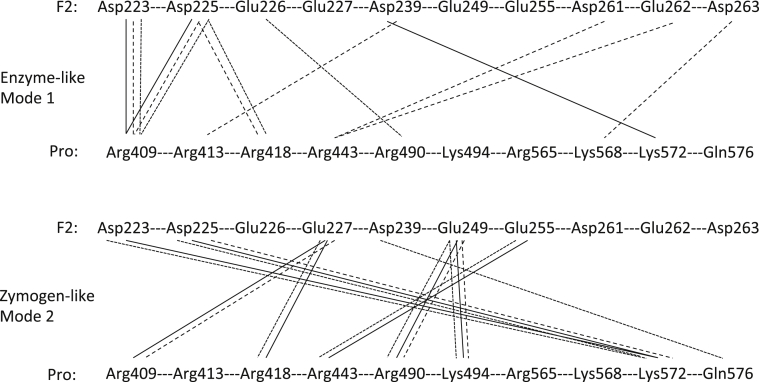
A comparison of all the structures of prothrombin and fragments, both zymogen and enzyme, show several important structural rearrangements that the protein undergoes during activation [37], [38], [39], [40], [41], [42] (Fig. 3). Early structures of the human PPACK-thrombin/F2 complex (PDBID:2HPQ) and bovine PPACK-meizothrombin(desF1) (PDBID:1A0H) show similar binding interfaces for F2 on the catalytic domain, which we have termed ‘mode 1’ (red (in the web version) in Fig. 4). The subsequent structure of human PPACK-meizothrombin(desF1) (PDBID:3E6P) confirmed this binding interface [43]. In all of these structures the catalytic domain has been activated by cleavage at Arg320 and the active site of the enzyme was covalently bound to the inhibitor PPACK (D-phenylalanyl-prolyl-arginyl chloromethylketone). An extensive contact surface between F2 and the catalytic domain is observed in these structures, burying a total of ∼1600 Å2 (Fig. 5), involving several salt-bridges, hydrogen bonds, and hydrophobic contacts (Fig. 6). The interface interposes the basic exosite II of catalytic domain with the highly acidic face of F2. Important ionic interactions include: Asp223 and Asp225 with Arg409 (93 in chymotrypsin numbering) and Arg418(101); Glu226 with Arg490(165); Glu227 with Arg500(175); and Asp239 with Arg413(97). Other basic residues on the C-terminus, Arg565(233), Lys568(236) and Lys572(240), do not form salt-bridges, although they have been shown to be critical for binding other exosite II ligands. In terms of number and types of contacts, the most important residue in exosite II is Arg409(93). It participates in multiple salt-bridges, water-mediated hydrogen bonds, pi-stacking and hydrophobic interactions, and accounts for up to 18% of the buried surface area.
Fig. 3.
Structures of prothrombin and activation fragments. Cartoon ribbon representations of all prothrombin and fragment structures containing the protease domain bound to the F2 domain, oriented with the active site located at the lower right. The corresponding protein data bank identification code (PDBID) is displayed below the structure name in parentheses.
Fig. 4.
Binding modes of fragment-2. Alignment of all structures containing the F2 domain reveal two overlapping binding sites on the protease domain. Stereo view of the protease domain of prethrombin-1 (PDBID:3NXP) is shown as gray surface representation. Fragment 2 in the thrombin/F2 complex (PDBID:2HPQ), pre2/F2 complex (PDBID:3K65), and meizothrombin(desF1) (PDBID:1A0H,3E6P) bind via ‘mode 1’, shown as red cartoon. Fragment 2 in prothrombin (PDBID:4HZH,4NZQ) and prethrombin-1 (PDBID:3NXP) structures binds via ‘mode 2’, shown in green cartoon.
Fig. 5.
Footprint of fragment-2 on the protease domain in mode 1 and 2 structures. Surface representation of various prothrombin/fragment structures showing surfaces within the protease domain that interact with F2. Thrombin is shown in blue; prethrombin-2 is shown in pink; meizothrombin is shown in cyan; prethrombin-1 is shown in magenta; prothrombin from 4NZQ is shown in gray, and from 4HZH is shown in orange. Residues within 3.5 Å of F2 in each structure have been colored yellow with important basic residues of the catalytic domain labeled.
Fig. 6.
Interaction diagram of modes 1 and 2. The interactions between the second kringle domain (F2) and the protease domain (Pro) are shown for the enzyme-like mode 1 and the zymogen-like mode 2. The schematic illustrates that conservation of interactions within modes. Three structures for each mode are represented (Mode 1: 2HPQ solid black lines, 3E6P dashed black lines, 3K65 dotted black lines; Mode 2: 4NZQ solid black lines, 4HZH dashed black lines, 3NXP dotted black lines).
The more recent structures of human prethrombin 1 (PDBID:3NXP), human prothrombin(desGla) (PDBID:4HZH), and human prothrombin(Δ146–167) (PDBID:4NZQ) show an alternate binding mode for F2 on the catalytic domain [37], [38], [42]. This new binding mode (mode 2) shows a large shift in position, with a rotation of approximately 30° accompanied by a translation of ∼6 Å relative to the mode 1 structures (Fig. 4). Mode 2 is slightly more intimate, with a buried surface area of ∼1800 Å2 (Fig. 5). As before, the interface between the F2 and catalytic domain is composed primarily of complimentary ionic interactions (Fig. 6). However, in the mode 2 structures Asp223 and Asp225, which form salt-bridges to Arg409(93) and Arg418(101) in mode 1, now form salt-bridges to Lys572(240) and Lys568(236), and Arg409(93) and Arg418(101) now make contacts with Glu226 and Glu227. Additional differences from binding mode 1 include hydrogen bonding interactions Glu249 with Arg490(165) and Lys494(169), Glu254 with Arg443(126), and Glu255 with Arg443(126).
This suggests that the binding mode of F2 is determined by the conformation of the catalytic domain. In binding mode 1 the catalytic domain is cleaved at Arg320 and locked in its enzyme state through active site inhibition, whereas in binding mode 2, the catalytic domain is still in its zymogen state with an intact Arg320-Ile321 bond. To investigate the role of the conformation of the catalytic domain in the F2 binding mode, we determined a 1.85 Å structure of pre2 in complex with F2 (Table 1). It shows that, even though pre2 is still a zymogen, F2 binds to the catalytic domain using mode 1. The structure also reveals increased disorder within the zymogen activation domain when compared to that of pre2 alone. It appears that cleavage of either Arg271 or Arg320 leads to transition from binding mode 2 to 1.
4. Linker between F2 and protease domains
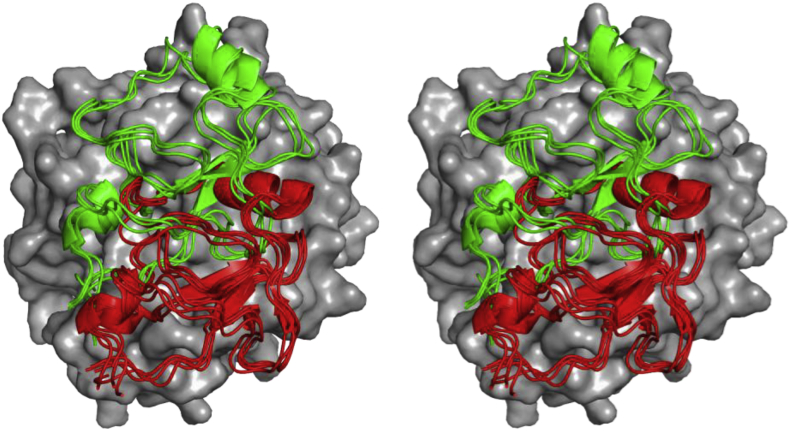
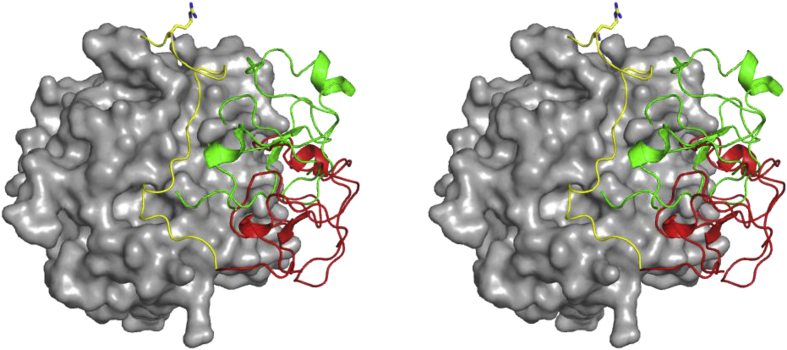
Cleavage at Arg320 causes a shift from mode 2 to mode 1, but it might also result in a rearrangement of the linker containing Arg271 between the F2 and protease domain. The crystal structure of bovine meizothrombin(desF1) (PDBID:1A0H) shows the linker between the 120- and C-terminal helices on the catalytic domain (Fig. 7). This is the only structure displaying electron density for this region. In structures with binding mode 2 the F2 domain occupies the cleft formed by the two helices that bind this linker in meizothrombin. The transition to binding mode 1 shifts the F2 domain away from these helices allowing the linker containing Arg271 to dock. It is possible that this apparent reorientation of the linker between F2 and the catalytic domain is part of the ‘ratcheting’ mechanism of prothrombin activation.
Fig. 7.
Conformational rearrangement of the linker after Arg320 cleavage. The linker between the F2 domain and the protease domain of bovine meizothrombin (PDB:1A0H) is shown as yellow cartoon. Arg271 is shown yellow sticks. The protease domain is displayed as a surface representation and the F2 domain is displayed in red as cartoon (mode 1). The F2 binding in prethrombin-1 is shown as green cartoon (mode 2). The F2 domain bound in mode 1 sterically blocks the linker of in the zymogen conformation (mode 2).
5. The environment of Arg320
Recent structural studies by the Di Cera group have suggested that part of the mechanism of accelerated cleavage of Arg320 by assembled prothrombinase is the ‘exposure’ of the activation loop containing the Arg320-Ile321 bond [39], [44]. This is based on the observation in a crystal structure (3SQE) of a ‘buried’ Arg320 side chain, due to interactions with nearby negatively charged residues, Glu311, Asp318 and Glu323. The authors failed to mention the extensive crystal contact with a symmetry-related molecule in this area intimately involving the side chains of Tyr316, Asp318 and Arg320, and the lack of electron density for the side chain of Glu323 (Indeed, there is strong negative difference density corresponding to where it is placed.). The relevance of the trapping of Arg320 in this ‘anionic cage’ was studied by mutating the three acidic residues, Glu311, Asp318 and Glu323, to alanine. The result was prothrombin capable of autoactivation to thrombin over long incubation periods. The authors concluded that this was evidence of improved flexibility of the Arg320-Ile321 bond due to the disruption of the ionic interactions. However, the authors did not consider the more likely explanation that substitution of acidic residues flanking the scissile bond will have a profound influence on the ability of thrombin to recognize a substrate. Indeed, several reports have concluded that acidic residues at either the P3 or P3′ positions (corresponding to Asp318 and Glu323 in prothrombin) antagonize cleavage by thrombin, and that hydrophobic or basic residues are favored [45], [46], [47], [48], [49]. Thus, altering the activation loop from IDGR-IVE to IAGR-IVA might have an effect on the structure of the region, but it will certainly impact thrombin's ability to cleave it. The argument that it is the sequence that matters, not flexibility, is strengthened by insensitivity of mutation solely at Glu311 that would be predicted to have the most profound effect on the orientation and burying of Arg320 [44]. It would be interesting to know if the neutralizing mutations in the activation loop of prothrombin alters its ability to be cleaved by fXa, alone or in the presence of fVa and membranes, or indeed by other proteases, such as ecarin. The accessibility of the activation loop of prothrombin might be improved upon fVa binding, but it is more likely that it is simply presented to the active site of fXa in the context of the assembled prothrombinase complex.
6. F2 in ratcheting
The exact mechanism of prothrombin activation by prothrombinase, in particular how a single enzyme complex can be responsible for two sequential cleavage events at sites separated >30 Å, has been the matter of some debate over the last 30 years. Some reports have suggested two different interconverting forms of prothrombinase exist, one cleaving at Arg320 and another at Arg271 [50], [51]. However, the overwhelming weight of experimental evidence supports a single enzyme complex that cleaves prothrombin at Arg320 to meizothrombin, and that it is the change in conformation of the substrate that presents the second Arg271 site. A recombinant prothrombin variant in which the sequence immediately following the Arg320 cleavage site was replaced with residues that do not allow transition to an active protease showed normal cleavage at Arg320 by prothrombinase, but subsequent cleavage at Arg271 was impaired [23], resulting in a build-up of meizothrombin. The conclusion is that presentation of the Arg271 cleavage site requires the conversion of the catalytic domain to its active enzyme conformation. To test this, bacterial staphylocoagulase was used to conformationally activate the catalytic domain without cleavage at Arg320. The covalent inhibitor PPACK was then used to lock the catalytic domain in an activated conformation, followed by removal of staphylocoagulase. This prothrombin species showed accelerated cleavage at Arg271 and inhibition of cleavage at Arg320. We now see that the transition from the zymogen to enzyme conformation in the catalytic domain also leads to the reorientation of the F2 domain and possibly the linker between the F2 and catalytic domains. These conformational changes may be important for positioning the Arg271 site for efficient cleavage, thus constituting part of the molecular basis of prothrombin processing.
Data deposition
Coordinates and structure factors are deposited in the Protein Data Bank under PDBID code 3K65.
Acknowledgments
Funding was provided by the British Heart Foundation Grant no. RG/11/3/28732. This work was carried out with the support of the Diamond Light Source.
References
- 1.MacFarlane R.G. An enzyme cascade in the blood clotting mechanism, and its function as a biochemical amplifier. Nature. 1964;202:498–499. doi: 10.1038/202498a0. [DOI] [PubMed] [Google Scholar]
- 2.Davie E.W., Ratnoff O.D. Waterfall sequence for intrinsic blood clotting. Science. 1964;145:1310–1312. doi: 10.1126/science.145.3638.1310. [DOI] [PubMed] [Google Scholar]
- 3.Stubbs M.T., Bode W. A player of many parts: the spotlight falls on thrombin's structure. Thromb. Res. 1993;69(1):1–58. doi: 10.1016/0049-3848(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 4.Huntington J.A. Molecular recognition mechanisms of thrombin. J. Thromb. Haemost. 2005;3(8):1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 5.Esmon C.T., Lollar P. Involvement of thrombin anion-binding exosites 1 and 2 in the activation of factor V and factor VIII. J. Biol. Chem. 1996;271(23):13882–13887. doi: 10.1074/jbc.271.23.13882. [DOI] [PubMed] [Google Scholar]
- 6.Segers K. The role of thrombin exosites I and II in the activation of human coagulation factor V. J. Biol. Chem. 2007;282(47):33915–33924. doi: 10.1074/jbc.M701123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monkovic D.D., Tracy P.B. Activation of human factor V by factor Xa and thrombin. Biochemistry. 1990;29(5):1118–1128. doi: 10.1021/bi00457a004. [DOI] [PubMed] [Google Scholar]
- 8.Myles T. An extensive interaction interface between thrombin and factor V is required for factor V activation. J. Biol. Chem. 2001;276(27):25143–25149. doi: 10.1074/jbc.M011324200. [DOI] [PubMed] [Google Scholar]
- 9.Myles T., Yun T.H., Leung L.L. Structural requirements for the activation of human factor VIII by thrombin. Blood. 2002;100(8):2820–2826. doi: 10.1182/blood-2002-03-0843. [DOI] [PubMed] [Google Scholar]
- 10.Li C.Q. Platelet glycoprotein Ib alpha binds to thrombin anion-binding exosite II inducing allosteric changes in the activity of thrombin. J. Biol. Chem. 2001;276(9):6161–6168. doi: 10.1074/jbc.M004164200. [DOI] [PubMed] [Google Scholar]
- 11.De Cristofaro R. Structural and functional mapping of the thrombin domain involved in the binding to the platelet glycoprotein Ib. Biochemistry. 2001;40(44):13268–13273. doi: 10.1021/bi010491f. [DOI] [PubMed] [Google Scholar]
- 12.Lechtenberg B.C., Freund S.M., Huntington J.A. GpIbalpha interacts exclusively with exosite II of thrombin. J. Mol. Biol. 2014;426(4):881–893. doi: 10.1016/j.jmb.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter W.J., Cama E., Huntington J.A. Crystal structure of thrombin bound to heparin. J. Biol. Chem. 2005;280(4):2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 14.Khan A.R., James M.N. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998;7(4):815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fehlhammer H., Bode W., Huber R. Crystal structure of bovine trypsinogen at 1-8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin. J. Mol. Biol. 1977;111(4):415–438. doi: 10.1016/s0022-2836(77)80062-4. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix H. Activation of human prothrombin by stoichiometric levels of staphylocoagulase. J. Biol. Chem. 1983;258(6):3637–3644. [PubMed] [Google Scholar]
- 17.Hemker H.C., Bas B.M., Muller A.D. Activation of a pro-enzyme by a stoichiometric reaction with another protein. The reaction between prothrombin and staphylocoagulase. Biochim. Biophys. Acta. 1975;379(1):180–188. doi: 10.1016/0005-2795(75)90020-3. [DOI] [PubMed] [Google Scholar]
- 18.Bode W., Huber R. Induction of the bovine trypsinogen-trypsin transition by peptides sequentially similar to the N-terminus of trypsin. FEBS Lett. 1976;68(2):231–236. doi: 10.1016/0014-5793(76)80443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bovill E.G. Evidence that meizothrombin is an intermediate product in the clotting of whole blood. Arterioscler. Thromb. Vasc. Biol. 1995;15(6):754–758. doi: 10.1161/01.atv.15.6.754. [DOI] [PubMed] [Google Scholar]
- 20.Esmon C.T., Jackson C.M. The conversion of prothrombin to thrombin. III. The factor Xa-catalyzed activation of prothrombin. J. Biol. Chem. 1974;249(24):7782–7790. [PubMed] [Google Scholar]
- 21.Krishnaswamy S., Mann K.G., Nesheim M.E. The prothrombinase-catalyzed activation of prothrombin proceeds through the intermediate meizothrombin in an ordered, sequential reaction. J. Biol. Chem. 1986;261(19):8977–8984. [PubMed] [Google Scholar]
- 22.Krishnaswamy S. Activation of human prothrombin by human prothrombinase. Influence of factor Va on the reaction mechanism. J. Biol. Chem. 1987;262(7):3291–3299. [PubMed] [Google Scholar]
- 23.Bianchini E.P. Ratcheting of the substrate from the zymogen to proteinase conformations directs the sequential cleavage of prothrombin by prothrombinase. Proc. Natl. Acad. Sci. U. S. A. 2005;102(29):10099–10104. doi: 10.1073/pnas.0504704102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haynes L.M. Prothrombin activation by platelet-associated prothrombinase proceeds through the prethrombin-2 pathway via a concerted mechanism. J. Biol. Chem. 2012;287(46):38647–38655. doi: 10.1074/jbc.M112.407791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford H.N., Orcutt S.J., Krishnaswamy S. Membrane binding by prothrombin mediates its constrained presentation to prothrombinase for cleavage. J. Biol. Chem. 2013;288(39):27789–27800. doi: 10.1074/jbc.M113.502005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q. Activation-induced exposure of the thrombin anion-binding exosite. Interactions of recombinant mutant prothrombins with thrombomodulin and a thrombin exosite-specific antibody. J. Biol. Chem. 1994;269(5):3725–3730. [PubMed] [Google Scholar]
- 27.Kamath P., Krishnaswamy S. Fate of membrane-bound reactants and products during the activation of human prothrombin by prothrombinase. J. Biol. Chem. 2008;283(44):30164–30173. doi: 10.1074/jbc.M806158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q. Activation-induced exposure of the thrombin anion-binding exosite. Interactions of recombinant mutant prothrombins with thrombomodulin and a thrombin exosite-specific antibody. J. Biol. Chem. 1994;269(5):3725–3730. [PubMed] [Google Scholar]
- 29.Lechtenberg B.C. NMR resonance assignments of thrombin reveal the conformational and dynamic effects of ligation. Proc. Natl. Acad. Sci. U. S. A. 2010;107(32):14087–14092. doi: 10.1073/pnas.1005255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orcutt S.J., Pietropaolo C., Krishnaswamy S. Extended interactions with prothrombinase enforce affinity and specificity for its macromolecular substrate. J. Biol. Chem. 2002;277(48):46191–46196. doi: 10.1074/jbc.M208677200. [DOI] [PubMed] [Google Scholar]
- 31.Leslie A.W., Powell H. Processing diffraction data with mosflm. In: Read R., Sussman J., editors. Evolving Methods for Macromolecular Crystallography. Springer; Netherlands: 2007. pp. 41–51. [Google Scholar]
- 32.Leslie A.G.W. 1992. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography; p. 26. [Google Scholar]
- 33.McCoy A.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D. Biol. Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 34.Brunger A.T. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.McRee D.E. A visual protein crystallographic software system for X11/XView. J. Mol. Graph. 1992;10:3. [Google Scholar]
- 36.DeLano W.L. The PyMOL molecular graphics system. 2002. http://www.pymol.org On world wide web.
- 37.Pozzi N. Crystal structure of prothrombin reveals conformational flexibility and mechanism of activation. J. Biol. Chem. 2013;288(31):22734–22744. doi: 10.1074/jbc.M113.466946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pozzi N. The linker connecting the two kringles plays a key role in prothrombin activation. Proc. Natl. Acad. Sci. U. S. A. 2014;111(21):7630–7635. doi: 10.1073/pnas.1403779111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pozzi N. Crystal structures of prethrombin-2 reveal alternative conformations under identical solution conditions and the mechanism of zymogen activation. Biochemistry. 2011;50(47):10195–10202. doi: 10.1021/bi2015019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arni R.K. Structures of the noncovalent complexes of human and bovine prothrombin fragment 2 with human PPACK-thrombin. Biochemistry. 1993;32(18):4727–4737. doi: 10.1021/bi00069a006. [DOI] [PubMed] [Google Scholar]
- 41.Martin P.D. New insights into the regulation of the blood clotting cascade derived from the X-ray crystal structure of bovine meizothrombin des F1 in complex with PPACK. Structure. 1997;5(12):1681–1693. doi: 10.1016/s0969-2126(97)00314-6. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z., Pelc L.A., Di C.E. Crystal structure of prethrombin-1. Proc. Natl. Acad. Sci. U. S. A. 2010;107(45):19278–19283. doi: 10.1073/pnas.1010262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papaconstantinou M.E. Na+ binding to meizothrombin desF1. Cell Mol. Life Sci. 2008;65(22):3688–3697. doi: 10.1007/s00018-008-8502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pozzi N. Autoactivation of thrombin precursors. J. Biol. Chem. 2013;288(16):11601–11610. doi: 10.1074/jbc.M113.451542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Bonniec B.F. Characterization of the P2′ and P3′ specificities of thrombin using fluorescence-quenched substrates and mapping of the subsites by mutagenesis. Biochemistry. 1996;35(22):7114–7122. doi: 10.1021/bi952701s. [DOI] [PubMed] [Google Scholar]
- 46.Le Bonniec B.F., MacGillivray R.T., Esmon C.T. Thrombin Glu-39 restricts the P′3 specificity to nonacidic residues. J. Biol. Chem. 1991;266(21):13796–13803. [PubMed] [Google Scholar]
- 47.Le Bonniec B.F., Esmon C.T. Glu-192----Gln substitution in thrombin mimics the catalytic switch induced by thrombomodulin. Proc. Natl. Acad. Sci. U. S. A. 1991;88(16):7371–7375. doi: 10.1073/pnas.88.16.7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishii K. Determinants of thrombin receptor cleavage. Receptor domains involved, specificity, and role of the P3 aspartate. J. Biol. Chem. 1995;270(27):16435–16440. doi: 10.1074/jbc.270.27.16435. [DOI] [PubMed] [Google Scholar]
- 49.Harris J.L. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. U. S. A. 2000;97(14):7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brufatto N., Nesheim M.E. Analysis of the kinetics of prothrombin activation and evidence that two equilibrating forms of prothrombinase are involved in the process. J. Biol. Chem. 2003;278(9):6755–6764. doi: 10.1074/jbc.M206413200. [DOI] [PubMed] [Google Scholar]
- 51.Kim P.Y., Nesheim M.E. A revisit of the two-form kinetic model of prothrombinase: A rebuttal. Biophys. Chem. 2012;160(1):75–76. doi: 10.1016/j.bpc.2011.09.004. author reply 77–8. [DOI] [PubMed] [Google Scholar]