Abstract
Human Mps1 (hMps1) is a protein kinase essential for mitotic checkpoints and the DNA damage response. Here, we present new evidence that hMps1 also participates in the repair of oxidative DNA lesions and cell survival through the MDM2-H2B axis. In response to oxidative stress, hMps1 phosphorylates MDM2, which in turn promotes histone H2B ubiquitination and chromatin decompaction. These events facilitate oxidative DNA damage repair and ATR-CHK1, but not ATM-CHK2 signaling. Depletion of hMps1 or MDM2 compromised H2B ubiquitination, DNA repair and cell survival. The impairment could be rescued by re-expression of WT but not the phospho-deficient MDM2 mutant, supporting the involvement of hMps1-dependent MDM2 phosphorylation in the oxidative stress response. In line with these findings, localization of RPA and base excision repair proteins to damage foci also requires MDM2 and hMps1. Significantly, like MDM2, hMps1 is upregulated in human sarcoma, suggesting high hMps1 and MDM2 expression may be beneficial for tumors constantly challenged by an oxidative micro-environment. Our study therefore identified an hMps1-MDM2-H2B signaling axis that likely plays a relevant role in tumor progression.
INTRODUCTION
Human Mps1 (hMps1) or TTK is a protein kinase with dual specificity (1,2). The kinase has been shown to be required for safeguarding spindle assembly and centresome duplication in eukaryotes from yeasts to mammals (3–7). Furthermore, it colocalizes with mitotic checkpoint proteins on unattached kinetochores (3). In addition to spindle checkpoint regulation, our previous studies demonstrated that hMps1 can be activated by DNA damage and phosphorylates CHK2 at Thr68, resulting in CHK2 activation and arrest of the cell cycle at G2/M. Reciprocally, hMps1 can be phosphorylated at Thr288 and stabilized by CHK2 after DNA damage (8,9). The tumor suppressor protein p53 is another hMps1 substrate in the tetraploid checkpoint where phosphorylation at Thr18 by hMps1 disrupts p53-MDM2 interaction and causes stabilization and activation of p53 (10). hMps1 has also been reported to phosphorylate c-Abl and controls its nuclear targeting under oxidative stress (11). Collectively, these studies indicate that, in addition to regulation of the spindle assembly checkpoint (SAC), hMps1 may also participate in other stress responses.
MDM2 is an E3 ubiquitin ligase which functions as an important negative regulator of p53 by targeting the protein for proteasomal degradation. In addition to p53, other substrates of MDM2, for example, APE1 (12), Mdmx (13) and histone H2B (14), have been identified. Modification of MDM2 has been reported to regulate either its enzymatic activity or protein stability. Acetylation of the RING domain diminishes its ability to promote p53 ubiquitination (15). Phosphorylation by AKT at the S166 and S188 stabilizes MDM2 protein and promotes its nuclear translocation (16). Furthermore, phosphorylation by Ataxia telangiectasia mutated (ATM) inhibits MDM2 RING domain oligomerization and E3 processivity (17). Although MDM2 has been considered as an oncogene due to its overexpression in many human cancers and its ability to ubiquitinate p53, accumulating evidence suggests that MDM2 might also act as a tumor suppressor by inhibiting the G0/G1–S phase transition in normal human diploid cells; in support of this, the growth repressor domains of MDM2 have been identified (18–20). Moreover, MDM2 has been reported to ubiquitinate histone H2B at Lys120 and Lys125 in human cells to repress transcription (14), and more recently, MDM2 has been implicated in H2B ubiquitination in response to oxidative DNA damage to control chromatin relaxation for repair, though no direct evidence was provided (21).
Histone H2B ubiqutination is known to be involved in the regulation of various cellular pathways such as transcription elongation, chromatin reorganization and DNA replication (22–25). H2B ubiquitination has also been shown to be associated with DNA damage responses (DDR) in human cells (26,27) and in budding yeast (28–30). Human E3 ubiquitin ligase, RNF20 and RNF40 are the orthologs of Bre1 that monoubiquitinates histone H2B at Lys123 in budding yeast (31–34). Like Bre1, RNF20/40 monoubiquitinates H2B at Lys120 in mammals (23,24,35). Histone H2B ubiquitination also plays important role in trans-tail H3 histone methylation (36,37). The underlying mechanism that renders H2B ubiquitination so versatile can be attributed to reduced chromatin compaction as a result of this modification (38,39).
We observed previously that coexpression with hMps1 increases a slower migrating form of MDM2, suggesting that hMps1 might have an impact on MDM2 (10). Here, we explore the possible interplay between the two proteins and show that hMps1 can interact with and phosphorylate MDM2, and that the functional interaction contributes to oxidative DDR and repair through the regulation of H2B ubiquitination.
MATERIALS AND METHODS
Cell lines
293T, MCF-7 and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; HyClone), and HCT116 and H1299 cells were kept in RPMI-1640 medium (Gibco) with 10% fetal bovine serum (HyClone) and antibiotics (Gibco). HeLa cells inducibly expressing Myc-MDM2 WT, the 3A mutant and the vector control (10–3) were cultured in DMEM medium with G418 (400 μg/ml), puromycin (0.4 μg/ml) and doxycycline (1 μg/ml).
Plasmids and siRNAs
The MDM2 constructs were generated by cloning the cDNA amplified from pCMV-MDM2 (kind gift of B. Vogelstein, Johns Hopkins) in between the BamH I and Xho I sites of the pXJ-HA or the pXJ-myc vector (40) for mammalian expression, or the pRSET-A (Invitrogen) and pGEX4T-1 (Amersham Biosciences) vectors for the expression of His-or GST-tagged proteins in Escherichia coli. All tags are at the N-terminus of MDM2. To generate the constructs for inducible expression of siRNA-resistant myc-MDM2 WT or the 3A mutant, primers with Pvu II and Nhe I cutting sites were used to amplify the corresponding sequences, and the polymerase chain reaction (PCR) products were cloned into the vector, pTRE2pur (Clontech). The DsRed-MDM2 expression plasmid was generated by cloning the DsRed coding sequence in between the EcoR I and BamH I of pXJ-MDM2. Plasmids for the expression of hMps1 were described previously (10). GST-hMps1-His was generated by insertion of the His-tag into the C-terminus of GST-hMps1. The DNA sequences encoding H2B were amplified from LNCaP cells and cloned in between the BamH I and Xho I sites of the pXJ vector or pCMV-Tag 2B (Stratagene) with an N-terminal Flag-tag. The H2B 2KR mutant was generated by PCR-based site-directed mutagenesis. Plasmids were transfected using TurboFect (Thermo Scientific) for HeLa cells and Lipofectamine 2000 (Invitrogen) for other cells.
siRNAs were synthesized by Sigma and transfected using Oligofectamine (Invitrogen). The targeted sequences were (5′ to 3′): MDM2, GTCTGTTGGTGCACAAAAA (siMDM2) and CCTACTGATGGTGCTGTAA (siMDM2 #2); and hMps1, TGAACAAAGTGAGAGACAT (sihMps1) and TGGTTGAGTTTGTTGCTCA (sihMps1 #3).
Antibodies
Antibodies used for western blotting were as follows: anti-Myc (sc-40), anti-hMps1 (sc-540), anti-MDM2 (sc-965), anti-GST (sc-138), anti-His (sc-803), anti-CHK1 (sc-7898), anti-ATRIP (sc-365383) and anti-RPA2 (sc-56770) antibodies from Santa Cruz; anti-H2B-Ub (05–1312) and anti-histone H3 (07–690) antibodies from Millipore; anti-CHK1 pS345 (#2348), anti-CHK2 pT68 (#2661) and anti-ATM pS1981 (#2355) antibodies from Cell Signaling; anti-Flag M2 (F3165) and anti-actin (A2066) from Sigma; anti-H2B from Epitomics (1847–1) and GeneTex (GTX115955); anti-γH2Ax (#27505) from Upstate; anti-RPA2 pS33 (NB100–544) from Novus; and anti-HA (MMS-101p) from Covance. Anti-Myc and anti-HA used for immunoprecipitation were from LTK Biolaboratories (Touyuan, Taiwan). Anti-XRCC1 (sc-56254, sc-11429), anti-CRM1 (A300–469A), anti-8-oxoG (MAB3560) and anti-PARP1 (11835238001) used for immunofluorescence were from Santa Cruz, Bethyl, Millipore and Roche, respectively.
Phospho-specific antibodies
Rabbit polyclonal antibodies against phospho-Thr4 or Thr306 of MDM2 were raised, respectively, against phosphopeptides H-CNpTNMSVPT-OH or H-YWKCpTpSCNEM-OH synthesized by Kelowna International Scientific Inc. (Taipei, Taiwan). Rabbit inoculation and crude serum production were performed by LTK Biolaboratories (Touyuan, Taiwan). Antibodies were affinity-purified by binding to a phosphopeptide column. The eluted antibodies were further purified by passing through the unphosphorylated peptide column to remove antibodies that cross-react with unphosphorylated epitopes.
In vivo ubiquitination assay
293T cells transfected with His-tagged ubiquitin, HA-hMps1, Flag-H2B and pCMV-MDM2 WT or its mutants were collected and sonicated in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole, pH 8.0). When indicated, MG132 (Sigma-Aldrich) at a final concentration of 25 μM was added 4 h before harvest. Cell lysates were incubated with Ni-NTA agarose beads (Qiagen) to pulldown the His-tagged ubiquitinated proteins. The beads were washed once with buffer A, once with Ti-A mixed buffer (3:1) and three times with Ti buffer (20 mM imidazole, 0.2% Triton X-100, 25 mM Tris–HCl, pH 6.8). The beads were boiled in sample buffer and analyzed by western blotting.
Preparation of recombinant proteins
His-tagged and GST fusion proteins were expressed in E. coli strain BL21 (DE3) LysE and purified as described previously (8).
GST pulldown assay
Assays were performed essentially as previously described (8) using recombinant GST-hMps1 and His-tagged MDM2.
Coimmunoprecipitation
HeLa or transfected 293T cells were lysed in buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES) containing 0.1% NP-40, protease inhibitor cocktail (Roche), 10 mM NaF, 10 mM β-glycerophosphate and 1 mM dithiothreitol (DTT). Cell lysates were sonicated and incubated with antibodies and protein G beads (Thermo Scientific) for 2 h at 4°C. The beads were washed three times in cytoskeleton (CSK) buffer and the bound proteins were analyzed by immunoblotting.
In vitro kinase assay
Purified recombinant hMps1 was incubated with recombinant MDM2 in kinase buffer (20 mM Tris, pH 7.5, 10 mM MgCl2, and 1 mM DTT) in the presence of 50 μM ATP and 3 μCi [γ-32P] ATP for 15 min at 30°C and analyzed by autoradiography and immunoblotting.
Comet assay
The alkaline comet assay was performed using the comet assay kit (Trevigen). Briefly, cells were treated with 0.1 mM H2O2 for 15 min and incubated in drug-free medium for the indicated time before collection. Cells were lysed and electrophoresis was conducted under alkaline conditions. DNA was visualized by staining with yoyo-1. Comet tails were analyzed and quantified using CometScore.
HPRT mutation assay
The mutation frequency at the HPRT locus was determined as previously described (41) with the following modification. In brief, cells were first grown in HAT medium (DMEM with 100 μM hypoxanthine, 0.4 μM aminopterin and 16 μM thymidine) for 3 days to eliminate those with a pre-existing HPRT mutation. Cells were then grown in HT medium for 1 day and in fresh DMEM medium for 3 days. Following the HAT procedure, 5 × 105 cells were seeded and either untreated or treated with 0.1 mM H2O2 for 10 min and then grown in fresh medium for 10 d to allow the mutation to stabilize. Next, 106 cells/100 mm dish were plated and cultured in medium containing 2.5 μg/ml 6-TG for the first 4 days then in normal medium until colonies appeared. For the plating control, 2500 cells were grown in parallel in the absence 6-TG. Colonies formed after 3 weeks of incubation were counted after crystal violet staining.
Immunofluorescence assay
Cells grown on coverslips were treated with 0.5 or 1 mM H2O2 for 15 min and then recovered in drug-free medium for the indicated time. Cells were extracted with CSK buffer containing 0.5% Triton X-100, a protease inhibitor cocktail, 10 mM NaF, 10 mM β-glycerophophate and 1 mM DTT for 2 min on ice followed by methanol/acetone fixation at 4°C for 20 min. After blocking in 5% bovine serum albumin (BSA) in phosphate buffered saline (PBS) for 30 min at RT, cells were incubated with primary antibodies diluted in 1% BSA/PBS overnight at 4°C and the next day with secondary DyLight™488-conjugated goat anti-mouse IgG, TRITC-conjugated goat anti-rabbit or mouse IgG antibodies (Jackson ImmunoResearch). Images were obtained using a Carl Zeiss LSM510 confocal microscope.
Preparation of chromatin fraction
Cells were lysed in CSK containing 0.1% NP-40, protease inhibitor cocktail, 10 mM NaF, 10 mM β-glycerophophate and 1 mM DTT and centrifuged at 13 000 rpm for 10 min. The supernatant was reserved and designated as the soluble fraction. The pellet was washed twice with CSK and extracted with 0.2 N HCl for 20 min at 4°C followed by neutralization with 1/4 volume of 1.5 M Tris (pH 8.8). The supernatant after centrifugation was collected and designated as the chromatin fraction.
Clonogenic survival assay
HeLa cells transfected with siRNA for 24 h were replated in a 35 mm dish at a density of 104 cells per dish. Cells were treated with the indicated concentration of H2O2 for 15 min and incubated in drug-free medium for 14–21 days. Colonies were stained with 0.5% crystal violet before counting.
Statistical analysis
All statistical analyses except that in Figure 9A were conducted with Student's t-test. For Figure 9A, Fisher's exact test was used. * and ** denote P < 0.05 and P < 0.01, respectively.
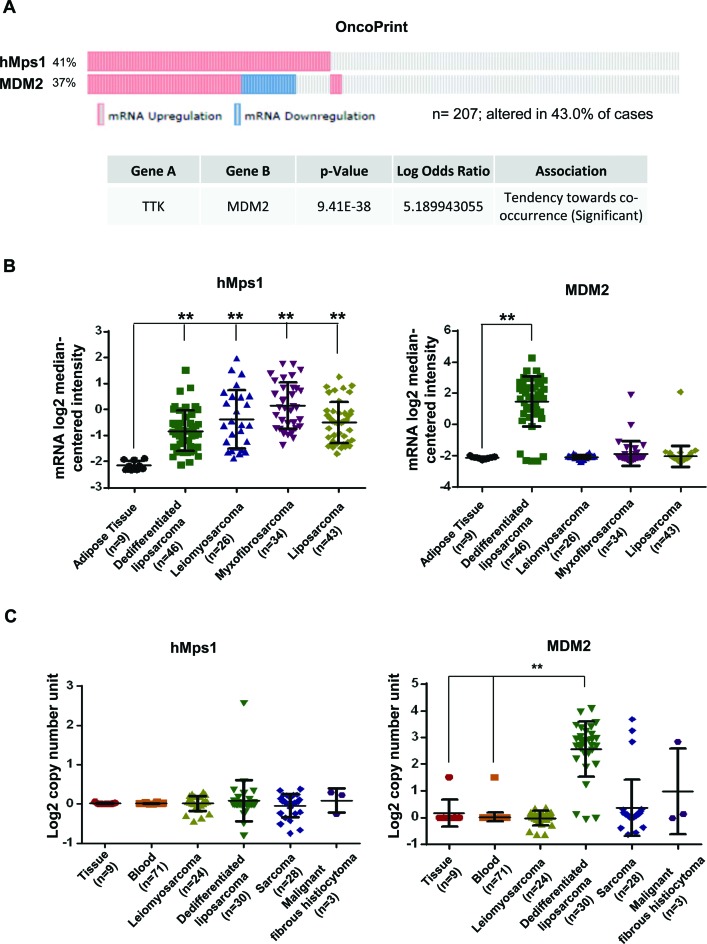
Figure 9.
Upregulation of hMps1 in human sarcoma. (A) Co-occurrence of the hMps1 and MDM2 alteration in gene expression. Shown are OncoPrints for 207 high-grade adult soft tissue sarcomas across seven subtypes of disease collected from MSKCC, the Broad Institute and Barretina et al. (47). Data were downloaded and analyzed on the cBioPortal for Cancer Genomics platform (http://www.cbioportal.org). (B) mRNA expression of hMps1 and MDM2 in 158 sarcoma specimens. Data source: Barretina sarcoma (47). (C) Copy number variation of the hMps1 and MDM2 genes in 165 human sarcoma samples. Data source: TCGA sarcoma. Datasets in (B) and (C) were downloaded from Oncomine® (https://www.oncomine.com) and analyzed by Prism. **P < 0.01 by Student's t-test.
RESULTS
hMps1 interacts with MDM2 in vivo and in vitro
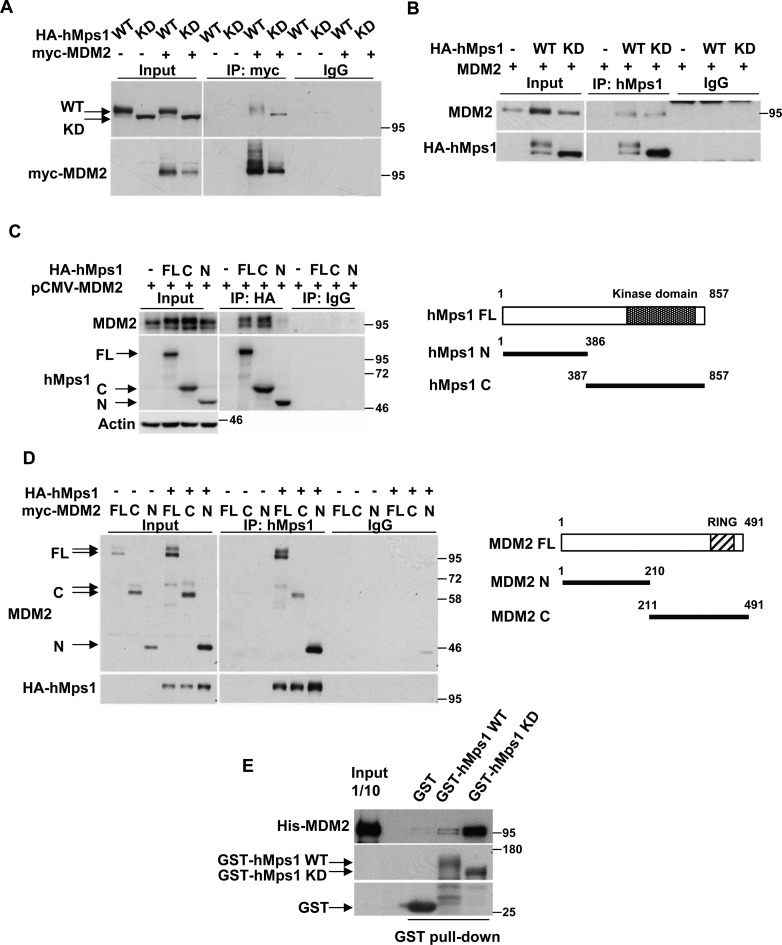
To decipher the functional relationship between human Mps1 (hMps1) and MDM2, we first investigated whether hMps1 interacts with MDM2. We coexpressed hMps1 wild-type (WT) or the kinase-dead mutant (KD) with MDM2 in 293T cells and immunoprecipitated hMps1 or MDM2. Both WT and KD hMps1 can be coprecipitated with MDM2 (Figure 1A) and, reciprocally, MDM2 was coprecipitated with either WT or KD hMps1 (Figure 1B). The interaction was mediated through the C-terminal domain of hMps1 (Figure 1C) binding to either the N- or the C-terminal domain of MDM2 (Figure 1D). The fact that both N- and C-terminal domains of MDM2 interact with hMps1 suggests there are more than one Mps1 binding modules in MDM2. Furthermore, the recombinant GST-fused WT and KD hMps1 also pulled down the His-tagged full-length MDM2 in vitro (Figure 1E), suggesting that the interaction can be direct.
Figure 1.
hMps1 interacts with MDM2. (A and B) MDM2 interacts with hMps1 in cells. 293T cells were transfected with MDM2 together with either wild-type (WT) or kinase-dead (KD) hMps1 and cell lysates were collected and analyzed by immunoprecipitation (IP) using the anti-myc antibody (A) or the anti-hMps1 antibody (B). Anti-His antibody was used as IgG control. (C) MDM2 binds the C-terminal domain of hMps1. 293T cells were transfected as in (A) but with the full-length (FL), the N- (amino acids 1–386) or the C-terminal (amino acids 387–857) domain of HA-hMps1. Immunoprecipitation was performed using anti-HA antibody. (D) hMps1-interacting domains in MDM2. 293T cells were transfected as in (A) but with FL, the N- (amino acids 1–210) or the C-terminal (amino acids 211–491) domain of myc-MDM2. (E) Direct interaction of MDM2 with hMps1 in vitro. GST pulldown assays were performed with His-tagged recombinant FL MDM2 and the recombinant GST-hMps1 WT or KD. Results were analyzed by immunoblotting using the indicated antibodies.
MDM2 is phosphorylated by hMps1
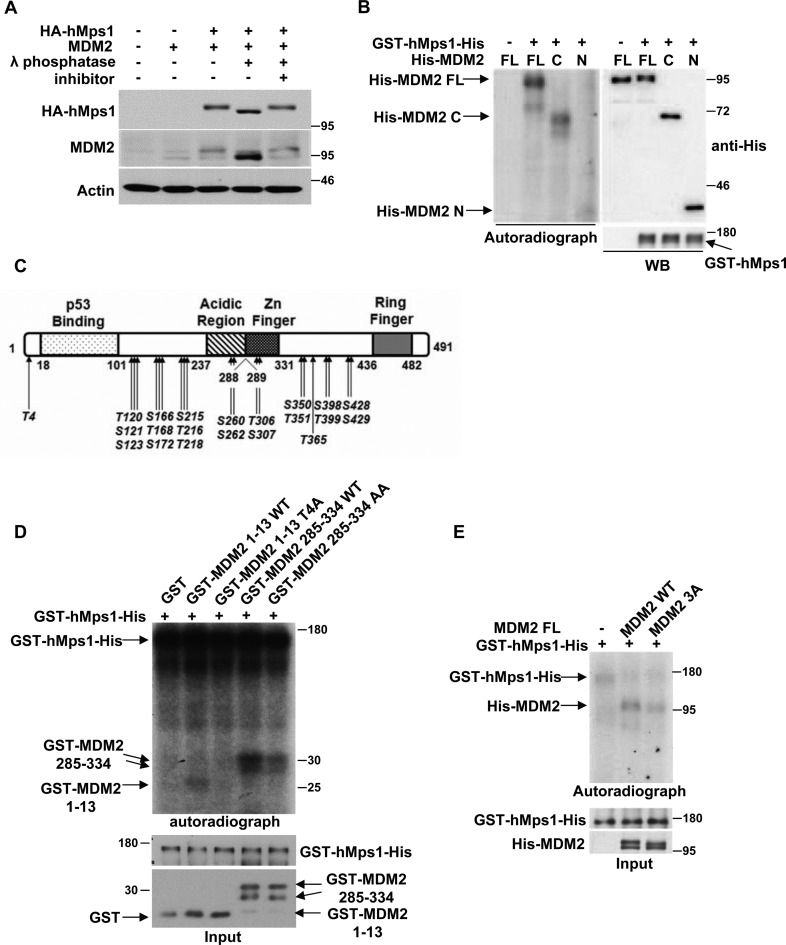
The fact that coexpression of hMps1 and MDM2 increased the slower migrating form of MDM2 (10) suggested to us the possibility of MDM2 phosphorylation by hMps1. Indeed, when the lysates were treated with λ phophatase, the slower migrating form of MDM2 was reduced, whereas the faster migrating form was increased (Figure 2A). Furthermore, the recombinant hMps1 phosphorylated the His-tagged MDM2 at both the N-terminal and C-terminal domains in vitro (Figure 2B). These results suggest that MDM2 is a substrate of hMps1.
Figure 2.
MDM2 is phosphorylated by hMps1. (A) MDM2 was phosphorylated upon coexpression of hMps1 in 293T cells. Lysates were treated with λ phosphatase at room temperature for 30 min in the absence or presence of the inhibitor (10 mM Na3VO4) prior to gel electrophoresis and western blot analysis. (B) MDM2 was phosphorylated by hMps1 in vitro. Kinase assays were conducted in vitro with recombinant GST-hMps1-His and recombinant FL, N- or C- region of His-tagged MDM2 in the presence of [γ-32P]ATP. Results were analyzed by autoradiography and immunoblotting. (C) Summary of hMps1 phosphorylation sites in MDM2 identified by LC-MS-MS after reaction in vitro. (D and E) MDM2 was phosphorylated by hMps1 at Thr4, Thr306 and Ser307 in vitro. Kinase assays were performed as in (B) but using truncated MDM2 carrying WT sequences or Ala substitution at Thr4 (T4A) or at Thr306/Ser307 (AA) (D). A mutant carrying all three residues mutated to Ala (3A) in the context of FL was also assayed in (E).
Using the full-length MDM2 as the substrate in vitro, the hMps1 phosphorylation sites were subsequently analyzed by mass spectrometry. Altogether, 22 phosphorylation sites were identified (Figure 2C). Interrogation of these sites by Thr/Ser to Ala substitution revealed that Thr4, Thr306 and Ser307 are the major hMps1 target sites (Supplementary Figure S1A and B), which are conserved among mammals (Supplementary Figure S1C). Mutations of Thr4 to Ala (T4A) in the truncated MDM2 (amino acids 1–13) and Thr306/Ser307 to Ala (AA) in MDM2 (amino acids 285–334) compromised the phosphorylation by hMps1 (Figure 2D). In the context of the MDM2 full-length protein, mutation of Thr4/Thr306/Ser307 to Ala (3A) also resulted in reduced phosphorylation by hMps1 (Figure 2E). This was not because the mutation had disrupted the interaction, as the 3A mutant and WT MDM2 were comparable in interaction with hMps1 (Supplementary Figure S1D). Taken together, these results demonstrate that MDM2 can be phosphorylated by hMps1 at Thr4, Thr306 and Ser307 in vitro.
hMps1 phosphorylation promotes MDM2-mediated histone H2B ubiquitination
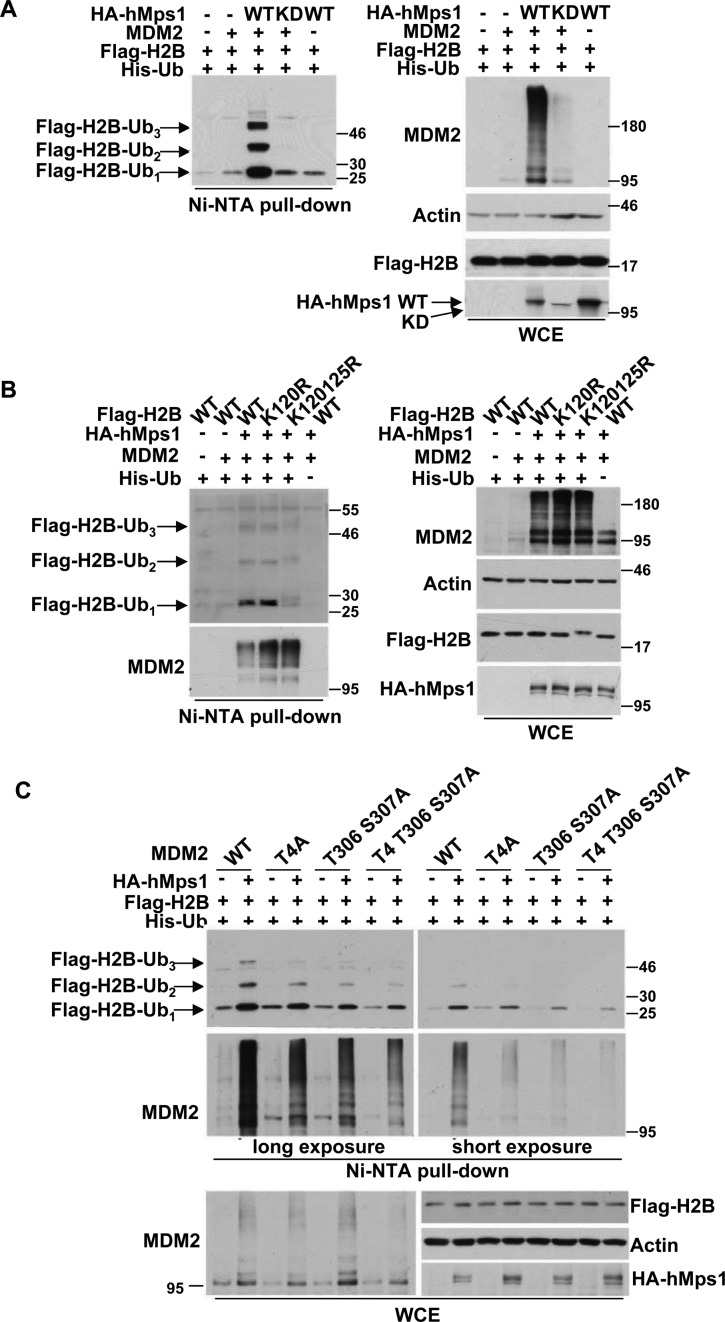
MDM2 has been reported to ubiquitinate histone H2B at Lys120/125 (14), and H2B ubiquitination has been implicated in the DDR upon oxidative stress (21). Based on the results described above, we hypothesized that hMps1 may act upstream of the MDM2-H2B axis in response to oxidative stress. To address this possibility, we first determined whether hMps1 phosphorylation regulates MDM2-mediated H2B ubiquitination. By coexpression of WT or KD hMps1with MDM2 and His-tagged ubiquitin in 293T cells, we found that hMps1 phosphorylation increased H2B ubiquitination (Figure 3A). Such an increase was not observed when the ligase-dead (LD) MDM2 mutant was used, indicating that the enhanced H2B ubiquitination was indeed mediated by the E3 ligase activity of MDM2 (Supplementary Figure S2A). In addition to the enhanced activity, we also observed a marked increase in MDM2 protein levels. As coexpression of hMps1 WT but not the KD mutant increased the protein half-life of MDM2 (Supplementary Figure S2B), we concluded that the effect on the activity was at least in part attributable to the increased stability of the MDM2 protein. In support, the MDM2 3A mutant was less stable than the WT protein when coexpressed with hMps1 (Supplementary Figure S2B and C).
Figure 3.
hMps1 promotes MDM2-mediated H2B ubiquitination. (A) WT but not KD hMps1 promotes MDM2-mediated H2B ubiquitination. 293T cells were transfected with the indicated plasmids and lysates were prepared for the Ni-NTA bead pulldown assay. Ubiquitinated proteins were analyzed by western blotting using the indicated antibodies. (B) MDM2-mediated H2B ubiquitination was abolished in the H2B K120/125R mutant. In vivo ubiquitination was performed as in (A) but with the H2B WT, K120R or K120/125R double mutant. (C) MDM2-mediated H2B ubiquitination was diminished by mutation of the hMps1 phosphorylation sites. Ubiquitination was carried out as in (A) but with MDM2 mutated at hMps1 phosphorylation sites singularly or in combination.
The ubiquitination site on H2B, in this event, was also investigated. We compared ubiquitination of WT H2B with the H2B mutants with Arg substitution at Lys120 (K120R) or at Lys120 and Lys125 (2KR). As a result, while ubiquitination of K120R was similar to WT, ubiquitination of 2KR was markedly reduced, suggesting that ubiquitination was mainly at Lys125 (Figure 3B).
Next, we compared H2B ubiquitination by WT MDM2 and the Ala mutants. The results show that Ala substitution at MDM2 Thr4, Thr306 and Ser307, singularly or in combination dampened the hMps1-promoted MDM2-mediated H2B ubiquitination (Figure 3C). The difference appears to be an in vivo event as the recombinant MDM2 WT and 3A proteins prepared from bacteria are equally competent in ubiquitination of p53 in vitro (Supplementary Figure S1E). Collectively, these results indicate that MDM2 phosphorylation at Thr4, Thr306 and Ser307 by hMps1 plays a decisive role in MDM2-mediated H2B ubiquitination.
To exclude the possibility that hMps1 may inhibit the proteasome to render the effect, we performed the H2B ubiquitination assay in the presence or absence of MG132, a proteasome inhibitor. The results showed that although MG132 generally increased the observed ubiquitination, hMps1 stills markedly enhanced WT MDM2-mediated H2B ubiquitination compared to the 3A mutant (Supplementary Figure S2D), suggesting that the impact of hMps1 on MDM2 are most likely to be direct, through MDM2 phosphorylation.
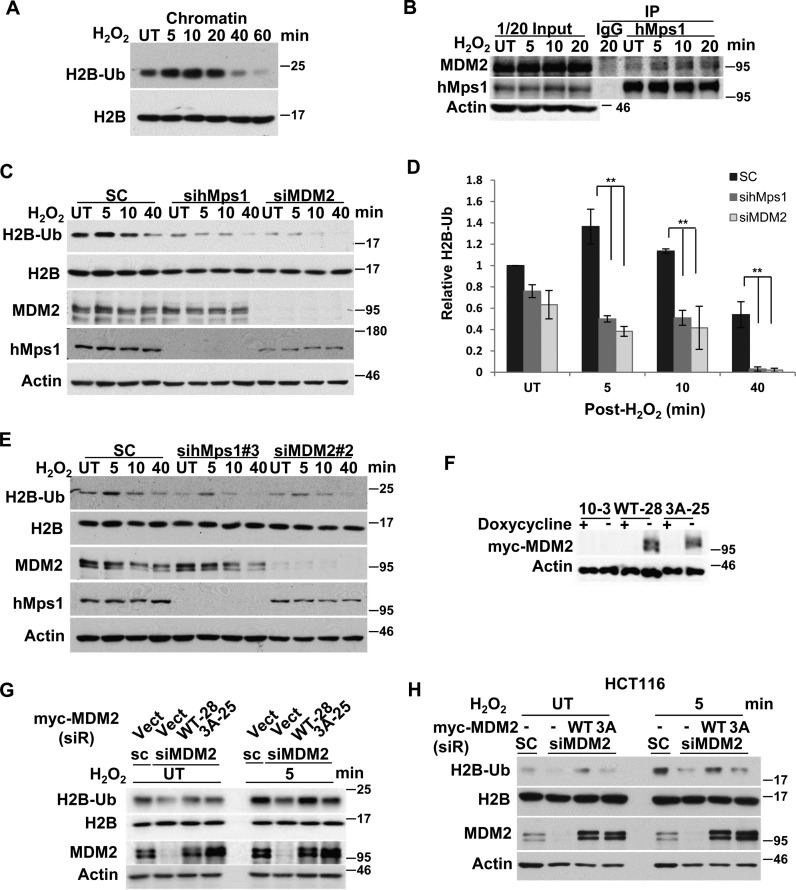
hMps1 and MDM2 are required for H2B ubiquitination after H2O2 treatment
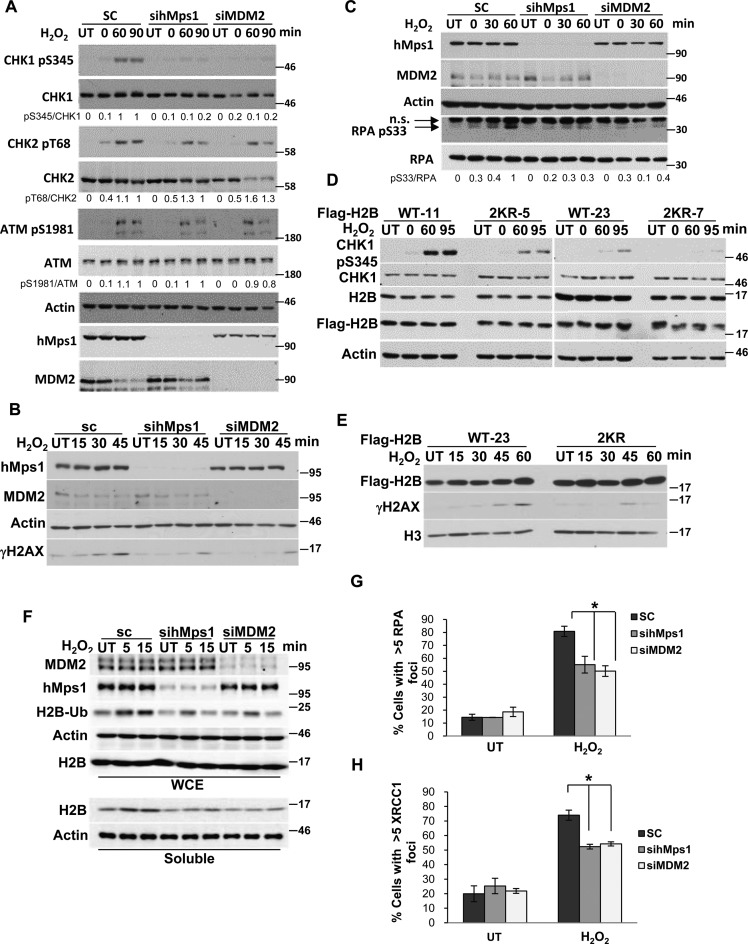
Human Mps1 has been reported to be activated by oxidative stress to control c-Abl cellular localization (11). Therefore, we explored the possibility that hMps1, upon activation by oxidative stress, may act on MDM2 to affect H2B ubiquitination. First, we investigated whether H2B ubiquitination is altered under oxidative stress. Extraction of the chromatin fraction after H2O2 treatment in HeLa cells demonstrated that ubiquitination of H2B was increased transiently within minutes after treatment (Figure 4A). Concomitantly, the interaction between endogenous MDM2 and hMps1 were detected (Figure 4B). Moreover, knockdown of hMps1 or MDM2 by small interfering RNA (siRNA) markedly reduced both basal and induced H2B ubiquitination (Figure 4C and D). Similar effects were observed in MCF-7 and HCT116 cells (Supplementary Figure S3A and B), or by using a second set of siRNAs (Figure 4E), thus excluding off-target effects. Notably, the knockdown effect was also observed in p53-null H1299 cells (Supplementary Figure S3C), suggesting that the hMps1-MDM2-H2B signaling axis can be independent of p53.
Figure 4.
hMps1 and MDM2 regulate ubiquitination of endogenous histone H2B upon oxidative stress. (A) H2B ubiquitination was increased by H2O2 treatment. HeLa cells were untreated (UT) or treated with 0.5 mM H2O2 and collected at the indicated time points. Chromatin fractions were isolated and analyzed by immunoblotting. (B) H2O2 treatment promotes interaction between endogenous hMps1 and MDM2 in HeLa cells. Co-immunoprecipitation was performed using the anti-hMps1 antibody. (C) H2B ubiquitination was diminished in cells depleted of hMps1 and MDM2. HeLa cells transfected with scramble control (SC), sihMps1 or siMDM2 siRNA were incubated for 2 days before treatment with 0.5 mM H2O2 for the indicated time. (D) Quantification of results shown in (C) after normalization to total H2B. Mean ± SD from three independent experiments is shown. **P < 0.01. (E) Effects of hMps1 and MDM2 knockdown on H2B ubiquitination verified using a second set of siRNAs. (F) Inducible expression of siRNA resistant WT or 3A mutant of myc-MDM2 in Tet-off HeLa cells. (G) MDM2 WT but not the 3A mutant restores H2B ubiquitination in MDM2-depleted HeLa cells. Cells as in (F) were transfected with MDM2 siRNA then treated or not with 0.5 mM H2O2. (H) Complementation with WT but not 3A myc-MDM2 restored H2B ubiquitination in MDM2 knockdown HCT116 cells. Cells were first transfected with control or MDM2 siRNA then the following day with plasmids expressing siRNA-resistant WT or 3A MDM2. Treatment and analysis were as in (G).
Next, rescue experiments were performed to further confirm the involvement of MDM2 and hMps1 in H2B ubiquitination. To that end, we established HeLa cell lines with Tet-off inducible expression of the siRNA-resistant MDM2 WT or MDM2 3A (Figure 4F). In these cells, induced WT MDM2 was better than the 3A mutant at restoring H2B ubiquitination upon depletion of endogenous MDM2 (Figure 4G). Similar results were observed using a second MDM2 WT and 3A clones (Supplementary Figure S4), indicating that the activity was not a clonal effect. Likewise, transient expression of MDM2 WT but not the 3A mutant also restored H2B ubiquitination in MDM2-depleted HCT116 cells (Figure 4H). Cumulatively, our results suggest that hMps1-mediated MDM2 phosphorylation is involved in H2B ubiquitination in H2O2-treated cells and possibly also in undisturbed cells under a basal level of oxidative stress.
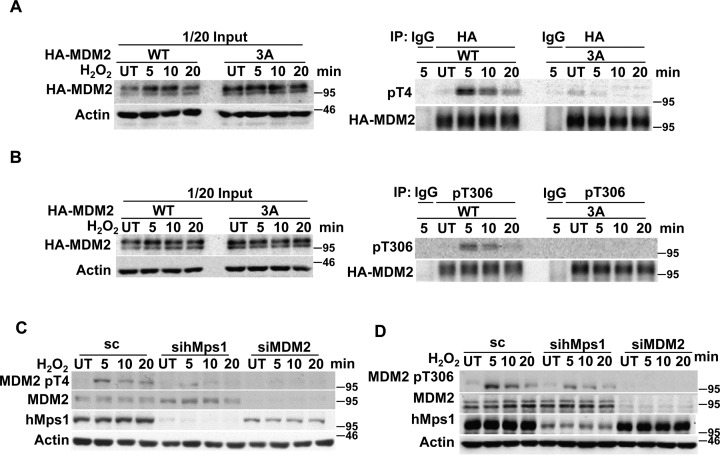
MDM2 Thr4 and Thr306 are phosphorylated by hMps1 upon oxidative stress
To determine whether MDM2 Thr4, Thr306 and Ser307 can be phosphorylated upon oxidative stress in cells, we generated phospho-specific antibodies against MDM2 phospho-Thr4 (pT4) and Thr306 (pT306). The antibodies preferentially recognized the T4 and T306 phosphorylated peptides compared with the unphosphorylated peptides (Supplementary Figure S5A and B), and reacted with the in vitro phosphorylated full-length (Supplementary Figure S5C and D) or truncated (Supplementary Figure S5E and F) WT MDM2, but not the corresponding Ala mutants. These results demonstrate the specificity of these antibodies.
Whether MDM2 Thr4 and Thr306 are phosphorylated upon oxidative damage was studied using the pT4 and pT306 antibodies. Our results show that the pT4 and pT306 antibodies preferentially reacted with WT but not the 3A mutant immunoprecipitated from transfected 293T cells, and both peaked at 5 min after H2O2 treatment (Figure 5A and B). Furthermore, the pT4 and pT306 antibodies detected endogenous phosphorylated MDM2 in HeLa cells after H2O2 treatment, and the detection was reduced in cells depleted of hMps1 or MDM2 (Figure 5C and D). These results together demonstrated that MDM2 can be phosphorylated by hMps1 at Thr4 and Thr306 in cells under oxidative stress. The attempt to generated pSer307-specific antibody was not successful. Whether Ser307 is also phosphorylated in cells remains unclear.
Figure 5.
MDM2 Thr4 and Thr306 phosphorylation is induced by oxidative stress. (A and B) The ectopically expressed WT MDM2 but not the 3A mutant reacted with anti-MDM2 pT4 and pT306 antibodies after H2O2 treatment. HA-tagged MDM2 was first immunoprecipitated from the transfected 293T lysates, then analyzed by western blot using the anti-MDM2 pT4 (A) or pT306 (B) antibody. (C and D) Endogenous MDM2 is phosphorylated at Thr4 (C) and Thr306 (D) after H2O2 treatment in an hMps1-dependent manner. HeLa cells transfected with control, hMps1 or MDM2 siRNA were untreated (UT) or treated with 0.5 mM H2O2 for the indicated time and collected for western blot analysis using the indicated antibodies.
Next, Thr4, Thr306 and Ser307 were mutated to Asp to mimic constitutive phosphorylation, and their H2B E3 activity was examined in 293T cells. As shown in Supplementary Figure S5G and quantified in Supplementary Figure S5H, T4/T306D and T4/S307D double substitution mutants but not the T4/T306/S307D triple mutant exhibited modest increase in the activity to promote H2B ubiquitination. These results suggest that phosphorylation at at least two of the sites, T4 and T306 or T4 and S307, is involved in regulating the H2B E3 activity of MDM2.
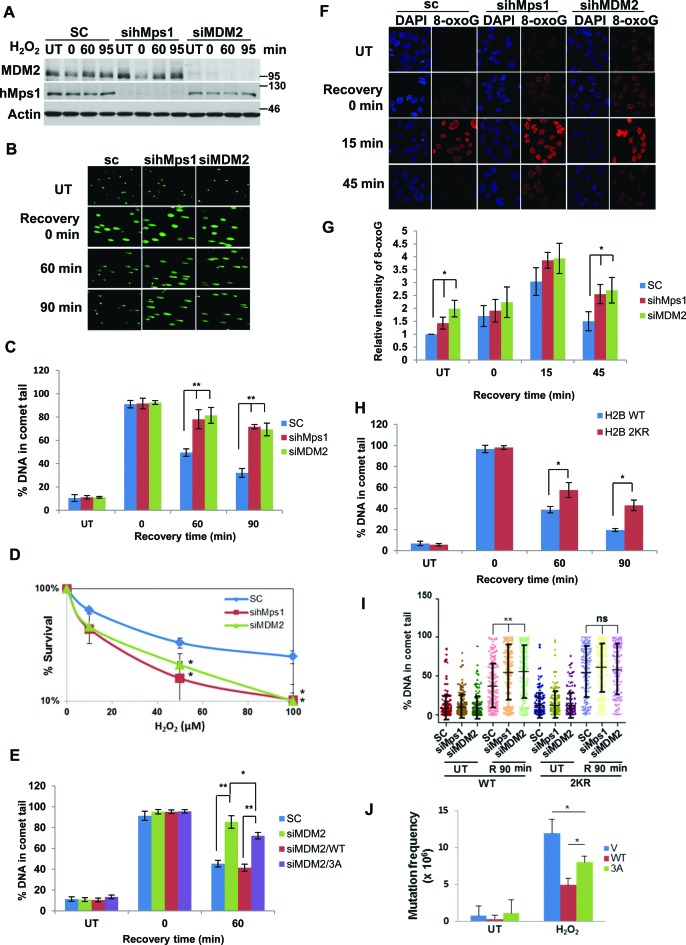
Oxidative DNA damage repair is impaired in hMps1 and MDM2-depleted cells
H2B ubiquitination has been shown to regulate DNA double strand break repair (26) and control the recruitment of the chromatin-remodeling factor SNF2h (27). Moreover, H2B ubiquitination by itself could lead to chromatin relaxation in vitro (38,39). Based on the data presented above, we wondered whether hMps1 and MDM2 are involved in repair of oxidative DNA damage through regulation of H2B ubiquitination. We first downregulated hMps1 or MDM2 in HeLa cells with siRNA (Figure 6A), then assessed the oxidative damage repair using the comet assay (Figure 6B). The results indicate that depletion of either hMps1 or MDM2 significantly hampered DNA repair, evidenced by sustained tail DNA (Figure 6C) and consequently reduced clonogenic cell survival (Figure 6D). Complementation with siRNA-resistant MDM2 WT but not the 3A mutant in MDM2 knockdown cells completely restored the repair (Figure 6E, Supplementary Figure S6A and B), suggesting the three hMps1 phosphorylation sites in MDM2 are required for oxidative DNA damage repair.
Figure 6.
Depletion of hMps1 or MDM2 dampens DNA repair and cell survival in cells encountering oxidative stress. (A) Western blots showing downregulation of hMps1 and MDM2 by siRNA in HeLa cells. sc, scramble control. (B) Comet assays demonstrating delayed repair of oxidative DNA damage in hMps1 and MDM2 knockdown cells. HeLa cells transfected with sc, hMps1 or MDM siRNA were treated or not with 0.1 mM H2O2 for 15 min and then recovered in drug-free fresh medium for 0, 60 or 90 min. UT, untreated. (C) Quantification of comet tails shown in (B). Mean ± SD from three independent experiments is shown. (D) Downregulation of hMps1 or MDM2 sensitized HeLa cells to H2O2 treatment. Colony survival assay was performed using HeLa cells transfected with control (sc), hMps1 or MDM2 siRNA. Cells were untreated or treated with various concentrations of H2O2 for 15 min before plating. Shown are results from three independent experiments. (E) Re-expression of WT MDM2 but not the 3A mutant restored DNA repair in MDM2 knockdown cells. Comet assays were performed as in (B) but using the Tet-off HeLa cell lines induced to express siRNA-resistant WT or 3A MDM2 (Supplementary Figure S6A and B). Mean ± SD from three independent experiments is shown. (F and G) Delayed repair of 8-oxoguanine (8-oxoG) lesions in hMps1 and MDM2-downregulated cells. HeLa cells treated as in (A) were fixed and stained with an anti-8-oxoG antibody for immunofluorescence microscopy (F). Quantification of the 8-oxoG fluorescence intensity from three experiments is shown in (G). (H) Expression of the H2B 2KR mutant hampered oxidative DNA damage repair. Repair in HeLa cells stably expressing WT or the 2KR (K120/125R) mutant of Flag-H2B was assessed by the comet assay (Supplementary Figure S6E). Quantification of comet tails from three experiments is shown. Expression of WT and 2KR H2B is shown in Supplementary Figure S6F. (I) H2B ubiquitination acts downstream of hMps1 and MDM2 in oxidative DNA damage repair. hMps1 or MDM2 was first depleted with siRNA in H2B WT and 2KR cells, and comet assays were performed after 0.1 mM H2O2 treatment. For each treatment, 150 cells were quantified. The results were analyzed by Prism. (J) The MDM2 3A mutation compromises the ability of MDM2 to suppress mutation caused by oxidative stress. The HPRT mutation assay was conducted using HeLa Tet-off cells expressing the empty vector (V), MDM2 WT, or 3A. * and **P < 0.05 and 0.01, respectively.
In addition to the comet assay, we also sought supporting evidence by directly assessing the repair of the oxidative damage lesion 8-oxoguanine (8-oxoG.) using immunofluorescence staining. The 8-oxoG positive cells appeared minutes after the removal of H2O2 whereas the unrelated protein CRM1 stained uniformly before and after the treatment, thus validating the specificity of the assay (Supplementary Figure S6C). Similar to the observations with the comet assay, elimination of 8-oxoG was compromised in hMps1 or MDM2 downregulated cells (Figure 6F and G).
To determine if the impact of MDM2 and hMps1 in repair is mediated through H2B ubiquitination, we first established HeLa cells stably expressing either Flag-H2B WT or 2KR (Supplementary Figure S6D). The results of comet assays with H2O2 indicate that the repair was less efficient in the ubiquitination-deficient H2B 2KR—than in H2B WT—expressing cells (Figure 6H, Supplementary Figure S6E and F). Moreover, knockdown of MDM2 or hMps1 increased comet tails in WT but not in 2KR cells (Figure 6I and Supplementary Figure S6G), suggesting that H2B ubiquitination functions downstream of hMps1 and MDM2 in oxidative DNA damage repair. Taken together, these data provide further support for the involvement of the hMps1-MDM2-H2B signaling axis in oxidative DNA damage repair.
Defect in hMps1-MDM2 signaling promotes accumulation of mutation
If indeed hMps1-mediated MDM2 phosphorylation is required for oxidative DNA damage repair, one would expect that the damage would accumulate in phosphorylation-deficient cells, which would then lead to increased mutation. To address this, the HPRT mutation assay was employed. The Tet-off HeLa cells inducibly expressing either MDM2 WT or 3A were either untreated or treated with H2O2 and then selected with 6-TG for mutation in HPRT. Our data show that while minimal mutation was observed in untreated cells, elevated mutation was seen after H2O2 treatment in cells expressing empty vector (Figure 6J). The mutation frequency was effectively downregulated by WT but less so by the 3A mutant (Figure 6J). This result further supports a role for hMps1-dependent MDM2 phosphorylation in oxidative DNA damage repair.
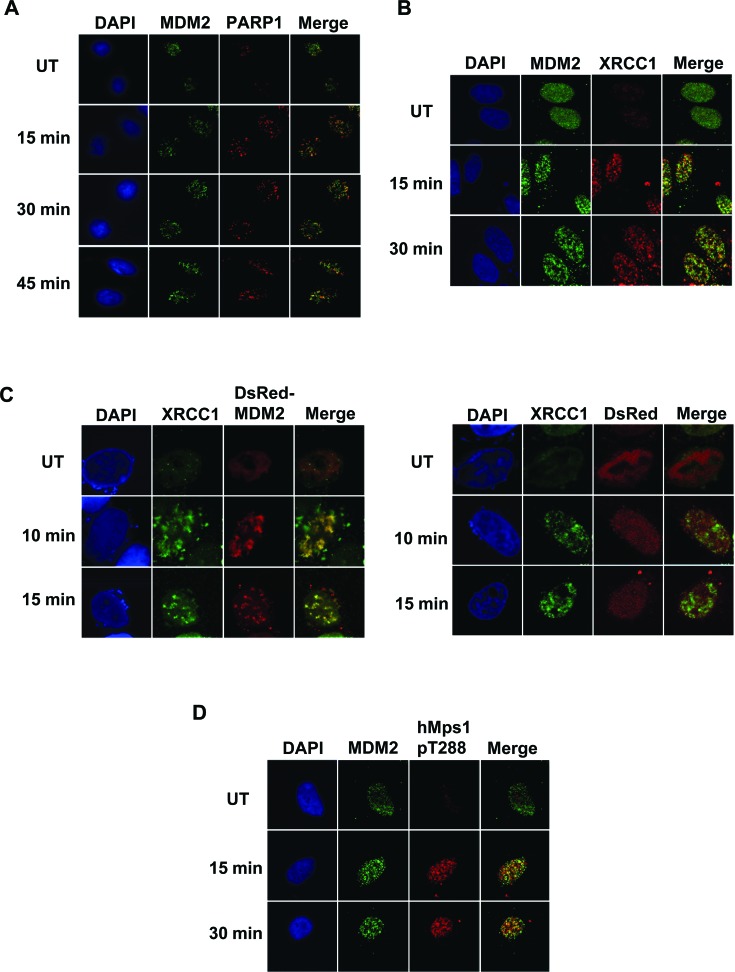
Colocalization of MDM2 with base excision repair proteins after oxidative damage
To further confirm the involvement in oxidative DNA damage repair, we next investigated the localization of MDM2, hMps1 and the proteins known to be involved in base excision repair after oxidative damage. Following H2O2 treatment, MDM2 relocalized to form foci in the nuclei, partially overlapped with those formed by PARP-1 and XRCC1, two key effectors participating in repair of oxidative DNA lesions (Figure 7A and B). In addition, DsRed-MDM2 but not DsRed formed foci that colocalized with XRCC1 after H2O2 treatment (Figure 7C). Moreover, we observed partial colocalization of MDM2 foci with those of hMps1 phosphorylated at Thr288 (Figure 7D), which has been reported to colocalize with γH2AX after ionizing irradiation and might function as a damage response marker (9). Our data suggest that at least some of the MDM2 foci represent damage sites and that MDM2 can colocalize with activated hMps1 and base excision repair machinery at these sites after oxidative damage.
Figure 7.
Colocalization of MDM2 with base excision repair proteins. (A) Colocalization of MDM2 with PARP1 in cells treated with H2O2. HeLa cells untreated (UT) or treated with 0.5 mM H2O2 were fixed at the indicated time after H2O2 removal for immunofluorescence staining using anti-MDM2 (SMP14) and anti-PARP1 antibodies. (B) Colocalization of MDM2 and XRCC1 foci. Cells were treated as in (A) but co-stained with anti-MDM2 and anti-XRCC1 antibodies. (C) Colocalization of DsRed-MDM2 with XRCC1 foci. HeLa cells transfected with DsRed or DsRed-MDM2 were treated with 1 mM H2O2 and fixed at the indicated time after removal of H2O2. (D) Colocalization of MDM2 with hMps1 phosphorylated at Thr288. Cells were co-stained with anti-MDM2 and anti-hMps1 pT288 antibodies.
Oxidative stress-induced ATR-CHK1 signaling is obliterated in hMps1 and MDM2-depleted cells
As depletion of hMps1 and MDM2 impaired oxidative DNA damage repair (Figure 6C and G), we wondered whether the DNA damage signaling was also affected. Immunoblot analysis of cell lysates using phospho-specific antibodies indicated that depletion of hMps1 or MDM2 dampened the activation of the checkpoint kinase CHK1, but not CHK2 or ATM after H2O2 treatment (Figure 8A). Concurrently, we also observed reduced γH2AX in these cells (Figure 8B). These data indicate that the ATR-CHK1 pathway but not the ATM-CHK2 pathway was specifically affected by hMps1 and MDM2 downregulation. In support, phosphorylation of Replication protein A (RPA) Ser33, a target site of Ataxia telangiectasia mutated and Rad3 related (ATR) , was also reduced in hMps1 and MDM2-knockdown cells (Figure 8C).
Figure 8.
The hMps1-MDM2-H2B pathway is required for ATR but not ATM signaling in response to oxidative stress. (A) Phosphorylation of CHK1 but not CHK2 or ATM was compromised in hMps1 or MDM2-downregulated cells. HeLa cells as treated in Figure 6A were analyzed by western blotting using the indicated antibodies. (B) Impaired induction of γH2AX in hMps1 and MDM2-depleted cells. (C) RPA phosphorylation was reduced in hMps1 and MDM2-depleted cells. HeLa cells treated as in (A) were analyzed by immunoblotting using the indicated antibodies. n.s., non-specific. (D and E) Expression of the H2B 2KR mutant interferes with phosphorylation of CHK1 (D) and H2AX (E) after H2O2 treatment. HeLa cells stably expressing WT or the 2KR mutant of Flag-H2B were treated and analyzed as in (A). (F) Eviction of histones, represented by soluble H2B, following H2O2 treatment was compromized in hMps1 and MDM2-knockdown HeLa cells. (G and H) RPA (G) and XRCC1 (H) foci were reduced in hMps1 and MDM2-downregulated HeLa cells. Shown are data from untreated cells (UT) and cells in recovery for 60 min after H2O2 treatment. Representative confocal images are shown in Supplementary Figure S7A and B, respectively. Mean ± SD from three independent experiments is shown. *P < 0.05.
Whether the signaling defect is related to H2B ubiquitination was examined using cells stably expressing the H2B 2KR mutant. Similar to hMps1 and MDM2-depleted cells, these cells showed reduced CHK1 activation and γH2AX when compared to the WT H2B-expressing cells (Figure 8D and E).
H2B ubiquitination has been reported to regulate chromatin compaction (38). DNA damage including oxidative stress was shown to cause chromatin remodeling, histone exchange and increased soluble histones (42–44). Consistent with these studies, we found that soluble H2B was increased after H2O2 treatment. More important, this increase was diminished in hMps1 and MDM2-downregulated cells (Figure 8F), suggesting that in these cells chromatin is more compact and less accessible. In support, we found reduced RPA and XRCC1 foci in hMps1 and MDM2-depleted cells (Figure 8G and H, Supplementary Figure S7), which at least in part explains the defective repair in these cells.
hMps1, like MDM2, is highly expressed in human soft tissue sarcoma
To explore the physiological relevance of the hMps1-MDM2 relationship, we analyzed expression of hMps1 and MDM2 in human cancers using data from available public databases. Data were downloaded and analyzed using the cBioPortal for Cancer Genomics platform (http://cbioportal.org) (45,46). Of the several human cancer types analyzed, soft tissue sarcoma stood out with the highest proportion of tumors showing hMps1 mRNA upregulation. Forty-one percent of the 207 samples from the sarcoma studies by Barretina et al. (47) exhibited hMps1 upregulation, compared to ∼3–9% in breast, colorectal and lung cancers from other studies (data not shown) (Figure 9A). Interestingly, the alteration was found to associate significantly with that of MDM2 expression (mostly upregulation and some downregulation), with the co-occurrence log odds ratio reaching 5.190 (P = 9.41E-38). Of the 207 samples analyzed, 54 (26%) showed upregulation in both MDM2 and Mps1 mRNA, and these samples were mainly from the subtype of dedifferentiated liposarcoma (Figure 9B). This, together with the prevalence of hMps1 upregulation, highlights the importance of hMps1-MDM2 signaling in the development of sarcoma.
To determine whether this upregulation of hMps1 is at the DNA level, we analyzed available TCGA sarcoma data and found that the copy number variation of the hMps1 gene among the 165 samples appeared to be negligible (Figure 9C), whereas consistent with previous findings (48,49), the MDM2 gene was amplified in dedifferentiated liposarcoma. These analyses show that the hMps1 upregulation occurs mostly at the transcriptional or post-transcriptional level, and affects all subtypes analyzed (Figure 9B). In comparison, MDM2 mRNA upregulation was observed mostly in dedifferentiated liposarcoma (Figure 9B) and appeared to be correlated with the MDM2 gene amplification (Figure 9C). The results of these analyses together with the experimental data from our study suggest the possibility that, in sarcoma that does not have MDM2 amplification, upregulation of hMps1 may, among other things, promote MDM2 function and cancer cell survival in an oxidative tumor micro-environment.
DISCUSSION
In this study, we identified an hMps1-MDM2-ubH2B signaling axis that plays an important role in oxidative DNA damage repair and signaling. We demonstrated that hMps1-mediated MDM2 phosphorylation is required for ATR-CHK1 signaling and the recruitment of repair proteins after oxidative DNA damage. This is likely mediated through chromatin remodeling as a result of MDM2-mediated H2B ubiquitination.
Compared to the response to DNA double strand break, the signaling in response to oxidative DNA damage is less well characterized. It has been reported that hMps1 may be activated under oxidative stress (11). hMps1 and H2B ubiquitination have each been shown to play a role in regulating the response to DNA damage not limited to oxidative damage (8,9,11,26,27). MDM2 has also been implicated to modulate base excision repair through ubiquitination of APE1 (12). In contrast to these previous studies, our data presented here provide a new perspective and a mechanistic explanation linking signaling (by hMps1) to chromatin remodeling (by MDM2 and histone H2B) and further to the oxidative DDR and repair.
H2B ubiquitination and the Bre1 mammalian orthologs, RNF20 and RNF40, have been demonstrated to regulate the DDR in human cells upon DNA double-strand break (26,27). It was shown that H2B ubiquitination is required for the recruitment of the chromatin-remodeling factor SNF2h, and that the requirement for RNF20 in HR repair could be partially bypassed by forced chromatin relaxation (27). As chromatin structure can be influenced by H2B ubiquitination by directly disrupting the contact between histone and DNA (38,39) or by the recruitment of remodeling factors such as SNF2h or FACT (24,27), we propose here that hMps1 regulates H2B ubiquitination and chromatin compaction by phosphorylating MDM2 to promote DNA damage signaling and repair under oxidative stress. The signaling axis is reminiscent of the ATM-RNF20/RNF40-H2B signaling characterized by Moyal et al. (26) upon DNA double-strand break; however, whether hMps1 and ATM are interchangeable for these two signaling pathways remains to be determined. Our data appear to disfavor the involvement of ATM in oxidative repair since, despite normal activation of ATM, cells were unable to repair oxidative damage efficiently upon hMps1 depletion.
MDM2 has been shown to inhibit DNA double-strand break repair and induce chromosome/chromatid breaks (50,51), which is in conflict with the role of MDM2 we identified here in oxidative DNA damage repair. Several possibilities could account for the differences. Apart from the usage of different cell lines and DNA damage agents, different approaches were employed. In those previous studies, MDM2 was overexpressed in cells, which according to our proposed model and a report from Minsky and Oren (14), may promote H2B ubiquitination and likely reduce chromatin compaction. This could possibly explain why increased chromosome/chromatid breaks were observed in those studies, as it has also been demonstrated that loosening of chromatin increases its susceptibility to DNA damage agents (52–55). In contrast to their studies, we mainly utilized an siRNA-mediated knockdown approach to assess the role of MDM2 and hMps1 in DNA damage signaling and repair. In addition, our data suggest that MDM2 may have a housekeeping surveillance function in unstressed cells as its depletion in cells reduced H2B ubiquitination under not only the stressed but also the basal condition (Figure 4C, Supplementary Figure S3) and its knockdown increased 8-oxoG staining in untreated cell (Figure 6G). It should be noted that although our data implicate a role of H2B ubiquitination, there exists the possibility that additional MDM2 and hMps1-regulated players may also be involved. Our data do not support a role of the p53 response in this signaling axis, as comparable results were observed with HeLa cells (p53-compromised) (Figure 4C and E), H1299 cells (p53-null) (Supplementary Figure S3C), and MCF7 and HCT116 cells (p53 competent) (Supplementary Figure S3A and B).
A growing body of evidence supports the involvement of the chromatin structure in the regulation of DNA-associated processes such as transcription, replication, recombination and the DDR (56–58). Compacted chromatin limits the access of protein regulators and must be relaxed to allow efficient progression of DNA-associated processes, such as the access of effectors to detect and repair the DNA lesions and also to deliver damage signals. Our findings here demonstrate that the hMps1-MDM2 pathway may join in to affect the DDR and repair, at least in cells under oxidative stress. It was, however, somewhat perplexing that depletion of hMps1 or MDM2 only affected the ATR-CHK1 but not the ATM-CHK2 pathway. The mechanism by which a global impact on chromatin would preferentially elicit one signaling pathway but not the other remains unclear. One possible explanation for this phenomenon might be that there are different needs for chromatin decompaction in different response pathways. Chromatin compaction might compromise the initial cleavage and thus the formation of ssDNA, which is required for RPA binding and activation of the ATR-CHK1 pathway. For example, activation of the ATR-CHK1 pathway after UV or base damage requires initial cleavage, which is impinged on by the nucleotide or base excision repair machinery (59–62). On the other hand, the ATM-CHK2 pathway could be activated by the cellular redox state and therefore may be independent of DNA lesions (63). Our results suggest that, upon oxidative damage, hMps1 and MDM2 may work together to loosen the chromatin, therefore facilitating initial DNA cleavage by the repair machinery. We believe that the model can explain the reduced RPA and XRCC1 foci and impaired ATR-CHK1 signaling in hMps1 andMDM2 depleted cells.
As hMps1 is also required for the SAC, a relevant question is that whether the observations we made in hMps1 knockdown cells were somehow linked to an inefficient SAC. Several lines of evidence suggest that this may not be the case. Our earlier studies indicated that hMps1 knockdown (10) or expression of a kinase-deficient hMps1 mutant (8) did not grossly alter cell cycle unless cells were treated with DNA damage agent or spindle poison. In addition, there was no apparent comet tail (Figure 6B) or increased ATM-CHK2 signaling (Figure 8A) before H2O2 treatment, arguing that DNA break was minimal. Furthermore, knockdown of MDM2, which has not been linked to SAC, yielded similar results. Collectively, our data are more consistent with an oxidative damage response independent of its role in SAC. The staining of 8-oxoG (Figure 6F and G) lends further support.
Because of the unique metabolic activity and the chronic inflammatory tumor micro-environment, cancer cells are constantly challenged by oxidative stress (64). Our analysis of clinical sarcoma samples implicates a role of the hMps1-MDM2 pathway in ensuring the survival of cancer cells, perhaps also the surrounding supporting stroma cells, at least through facilitating oxidative DNA damage repair. It would be interesting to see if inhibition of this pathway, for example, by using an hMps1 inhibitor, would synergize with chemotherapeutic agents in treating human sarcoma.
Supplementary Material
Acknowledgments
We thank the Proteomics Core at IBMS, Academia Sinica for their expert analysis of phosphopeptides.
Footnotes
Present Address: Yi-Fu Huang, Institute of Molecular and Cell Biology, 61 Biopolis Drive, Proteos 138673, Singapore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [NSC102-2628-B-001-006-MY3]; Academia Sinica, Taiwan (to S.-Y. S.). Funding for open access charge: Ministry of Science and Technology [NSC102-2628-B-001-006-MY3]; Academia Sinica, Taiwan (to S.-Y. S.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindberg R.A., Fischer W.H., Hunter T. Characterization of a human protein threonine kinase isolated by screening an expression library with antibodies to phosphotyrosine. Oncogene. 1993;8:351–359. [PubMed] [Google Scholar]
- 2.Mills G.B., Schmandt R., McGill M., Amendola A., Hill M., Jacobs K., May C., Rodricks A.M., Campbell S., Hogg D. Expression of TTK, a novel human protein kinase, is associated with cell proliferation. J. Biol. Chem. 1992;267:16000–16006. [PubMed] [Google Scholar]
- 3.Abrieu A., Magnaghi-Jaulin L., Kahana J.A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D.W., Labbe J.C. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 4.Fisk H.A., Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 5.Stucke V.M., Sillje H.H., Arnaud L., Nigg E.A. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss E., Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winey M., Huneycutt B.J. Centrosomes and checkpoints: the MPS1 family of kinases. Oncogene. 2002;21:6161–6169. doi: 10.1038/sj.onc.1205712. [DOI] [PubMed] [Google Scholar]
- 8.Wei J.-H., Chou Y.-F., Ou Y.-H., Yeh Y.-H., Tyan S.-W., Sun T.-P., Shen C.-Y., Shieh S.-Y. TTK/hMps1 participates in the regulation of DNA damage checkpoint response by phosphorylating CHK2 on threonine 68. J. Biol. Chem. 2005;280:7748–7757. doi: 10.1074/jbc.M410152200. [DOI] [PubMed] [Google Scholar]
- 9.Yeh Y.-H., Huang Y.-F., Lin T.-Y., Shieh S.-Y. The cell cycle checkpoint kinase CHK2 mediates DNA damage-induced stabilization of TTK/hMps1. Oncogene. 2009;28:1366–1378. doi: 10.1038/onc.2008.477. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y.-F., Chang M.D., Shieh S.-Y. TTK/hMps1 mediates the p53-dependent postmitotic checkpoint by phosphorylating p53 at Thr18. Mol. Cell. Biol. 2009;29:2935–2944. doi: 10.1128/MCB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nihira K., Taira N., Miki Y., Yoshida K. TTK/Mps1 controls nuclear targeting of c-Abl by 14–3–3-coupled phosphorylation in response to oxidative stress. Oncogene. 2008;27:7285–7295. doi: 10.1038/onc.2008.334. [DOI] [PubMed] [Google Scholar]
- 12.Busso C.S., Iwakuma T., Izumi T. Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene. 2009;28:1616–1625. doi: 10.1038/onc.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y., Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minsky N., Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Wang X., Taplick J., Geva N., Oren M. Inhibition of p53 degradation by Mdm2 acetylation. FEBS Lett. 2004;561:195–201. doi: 10.1016/S0014-5793(04)00168-1. [DOI] [PubMed] [Google Scholar]
- 16.Feng J., Tamaskovic R., Yang Z., Brazil D.P., Merlo A., Hess D., Hemmings B.A. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J. Biol. Chem. 2004;279:35510–35517. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Q., Chen L., Li Z., Lane W.S., Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28:3857–3867. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown D.R., Thomas C.A., Deb S.P. The human oncoprotein MDM2 arrests the cell cycle: elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 1998;17:2513–2525. doi: 10.1093/emboj/17.9.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang J., Kuo M.L., Eischen C.M., Stepanova L., Sherr C.J., Roussel M.F. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002;62:1222–1230. [PubMed] [Google Scholar]
- 20.Zhou R., Frum R., Deb S., Deb S.P. The growth arrest function of the human oncoprotein mouse double minute-2 is disabled by downstream mutation in cancer cells. Cancer Res. 2005;65:1839–1848. doi: 10.1158/0008-5472.CAN-03-3755. [DOI] [PubMed] [Google Scholar]
- 21.Khoronenkova S.V., Dianova I.I., Parsons J.L., Dianov G.L. USP7/HAUSP stimulates repair of oxidative DNA lesions. Nucleic Acids Res. 2011;39:2604–2609. doi: 10.1093/nar/gkq1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleming A.B., Kao C.F., Hillyer C., Pikaart M., Osley M.A. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Kim J., Guermah M., McGinty R.K., Lee J.S., Tang Z., Milne T.A., Shilatifard A., Muir T.W., Roeder R.G. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 25.Trujillo K.M., Osley M.A. A role for H2B ubiquitylation in DNA replication. Mol. Cell. 2012;48:734–746. doi: 10.1016/j.molcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyal L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.Y., Eppink B., Chung Y.M., Shalev G., Shema E., et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D.V., Shimada M., Tauchi H., Suzuki H., Tashiro S., et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Game J.C., Chernikova S.B. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair (Amst) 2009;8:470–482. doi: 10.1016/j.dnarep.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Game J.C., Williamson M.S., Spicakova T., Brown J.M. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics. 2006;173:1951–1968. doi: 10.1534/genetics.106.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 31.Hwang W.W., Venkatasubrahmanyam S., Ianculescu A.G., Tong A., Boone C., Madhani H.D. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 32.Kao C.F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M.A. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robzyk K., Recht J., Osley M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 34.Wood A., Krogan N.J., Dover J., Schneider J., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 35.Zhu B., Zheng Y., Pham A.D., Mandal S.S., Erdjument-Bromage H., Tempst P., Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 36.Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 37.Weake V.M., Workman J.L. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 38.Fierz B., Chatterjee C., McGinty R.K., Bar-Dagan M., Raleigh D.P., Muir T.W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee C., McGinty R.K., Fierz B., Muir T.W. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 40.Xiao J.H., Davidson I., Matthes H., Garnier J.-M., Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 41.Glaab W.E., Risinger J.I., Umar A., Barrett J.C., Kunkel T.A., Tindall K.R. Resistance to 6-thioguanine in mismatch repair-deficient human cancer cell lines correlates with an increase in induced mutations at the HPRT locus. Carcinogenesis. 1998;19:1931–1937. doi: 10.1093/carcin/19.11.1931. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y., Parry J.A., Chin A., Duensing S., Duensing A. Soluble histone H2AX is induced by DNA replication stress and sensitizes cells to undergo apoptosis. Mol. Cancer. 2008;7:61–70. doi: 10.1186/1476-4598-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y., Ayrapetov M.K., Xu C., Gursoy-Yuzugullu O., Hu Y., Price B.D. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muthurajan U.M., Hepler M.R.D., Hieb A.R., Clark N.J., Kramer M., Yao T., Luger K. Automodification switches PARP-1 function from chromtin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12752–12757. doi: 10.1073/pnas.1405005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barretina J., Taylor B.S., Banerji S., Ramos A.H., Lagos-Quintana M., Decarolis P.L., Shah K., Socci N.D., Weir B.A., Ho A., et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliner J.D., Kinzler K.W., Meltzer P.S., George D.L., Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 49.Cordon-Cardo C., Latres E., Drobnjak M., Oliva M.R., Pollack D., Woodruff J.M., Marechal V., Chen J., Brennan M.R., Levine A.J. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- 50.Alt J.R., Bouska A., Fernandez M.R., Cerny R.L., Xiao H., Eischen C.M. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J. Biol. Chem. 2005;280:18771–18781. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- 51.Bouska A., Lushnikova T., Plaza S., Eischen C.M. Mdm2 promotes genetic instability and transformation independent of p53. Mol. Cell. Biol. 2008;28:4862–4874. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douki T., Bretonniere Y., Cadet J. Protection against radiation-induced degradation of DNA bases by polyamines. Radiat. Res. 2000;153:29–35. doi: 10.1667/0033-7587(2000)153[0029:parido]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 53.Spotheim-Maurizot M., Ruiz S., Sabattier R., Charlier M. Radioprotection of DNA by polyamines. Int. J. Radiat. Biol. 1995;68:571–577. doi: 10.1080/09553009514551561. [DOI] [PubMed] [Google Scholar]
- 54.Takata H., Hanafusa T., Mori T., Shimura M., Iida Y., Ishikawa K., Yoshikawa K., Yoshikawa Y., Maeshima K. Chromatin compaction protects genomic DNA from radiation damage. PLoS One. 2013;8:e75622. doi: 10.1371/journal.pone.0075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warters R.L., Newton G.L., Olive P.L., Fahey R.C. Radioprotection of human cell nuclear DNA by polyamines: radiosensitivity of chromatin is influenced by tightly bound spermine. Radiat. Res. 1999;151:354–362. [PubMed] [Google Scholar]
- 56.Happel N., Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Lans H., Marteijn J.A., Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet. Chromatin. 2012;5:4–17. doi: 10.1186/1756-8935-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soria G., Polo S.E., Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Marini F., Nardo T., Giannattasio M., Minuzzo M., Stefanini M., Plevani P., Falconi M.M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.SenGupta T., Torgersen M.L., Kassahun H., Vellai T., Simonsen A., Nilsen H. Base excision repair AP endonucleases and mismatch repair act together to induce checkpoint-mediated autophagy. Nat. Commun. 2013;4:2674–2686. doi: 10.1038/ncomms3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sertic S., Pizzi S., Cloney R., Lehmann A.R., Marini F., Plevani P., Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13647–13652. doi: 10.1073/pnas.1108547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willis J., Patel Y., Lentz B.L., Yan S. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2013;110:10592–10597. doi: 10.1073/pnas.1301445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo Z., Koziov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 64.Costa A., Scholer-Dahirel A., Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Semin. Cancer Biol. 2014;25:23–32. doi: 10.1016/j.semcancer.2013.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.