Abstract
Circular RNAs (circRNAs) are widespread circles of non-coding RNAs with largely unknown function. Because stimulation of mammary cells with the epidermal growth factor (EGF) leads to dynamic changes in the abundance of coding and non-coding RNA molecules, and culminates in the acquisition of a robust migratory phenotype, this cellular model might disclose functions of circRNAs. Here we show that circRNAs of EGF-stimulated mammary cells are stably expressed, while mRNAs and microRNAs change within minutes. In general, the circRNAs we detected are relatively long-lived and weakly expressed. Interestingly, they are almost ubiquitously co-expressed with the corresponding linear transcripts, and the respective, shared promoter regions are more active compared to genes producing linear isoforms with no detectable circRNAs. These findings imply that altered abundance of circRNAs, unlike changes in the levels of other RNAs, might not play critical roles in signaling cascades and downstream transcriptional networks that rapidly commit cells to specific outcomes.
INTRODUCTION
Growth factors are evolutionarily conserved molecules, which are secreted by specific cells and bind specific receptors on the surface of target cells (1). One family of growth factors comprises 11 epidermal growth factor- (EGF-) like molecules. These ligands regulate proliferation and migration of epithelial and other cell lineages throughout embryonic development and in adulthood, such as in mammary gland development (2). Importantly, growth factors often induce rapid effects on signaling pathways, but their long-term biological effects, such as cell cycle regulation and chemotaxis, require synthesis of new RNAs and proteins (3). Correspondingly, EGF family growth factors induce wave-like bursts of transcription of distinct RNA molecules, starting with a group of immediate early genes (IEGs) and culminating in a large group of fate-determining mRNAs (4).
Like mRNAs, specific microRNAs display dynamic up- and down-regulation in response to growth factors. For example, a group of immediately down-regulated microRNAs (ID-miRs) normally suppresses transcription of the group of IEGs (5). Another type of non-coding RNAs, long non-coding RNAs (lncRNAs), is similarly regulated by growth factors. For example, lncRNA-ATB is activated by the β type transforming growth factors (TGF-β), to promote invasion of hepatocytes (6). Although synthetic circular RNAs have the ability to produce a protein product in vitro (7), in general, natural circRNAs are believed to be non-coding (8,9). Whether or not circRNAs are dynamically regulated following stimulation with growth factors is currently unknown. Circularization of RNAs was recently recognized to broadly expand the transcriptome (8–18). CircRNAs in animals have been discovered more than 30 years ago, but they were largely neglected due to rarity and lack of function. Due to the advent of next generation sequencing, thousands of different circRNAs were recently identified in various organisms, from archaea to human (9,14–16,19,20). The circRNA CDR1as was found to contain an exceptionally high number of binding sites specific to a miRNA and indeed was found to antagonize miRNA activity by a sponge-like mechanism (15,21), which led to the notion that circRNAs may function to sequester miRNAs (22–26). A recent study, however, raised doubts regarding a biological function of most circRNAs (27). Sponge-like recruitment of multiple microRNA would be an attractive mechanism in the context of growth factor activation. Interestingly, analysis of epithelial cells that underwent an epithelial-mesenchymal transition (EMT), after a 21 day-long treatment with TGF-β, revealed that hundreds of circRNAs were regulated during this process (28). Additional reports imply that circRNAs are functional molecules, rather than by-products of mis-splicing. For example, treatment of endothelial cells with tumor necrosis factors revealed that circRNA formation correlates with exon skipping (29), and yet another report has shown that circRNAs regulate transcription by means of interactions with the U1 snRNA (30). Furthermore, in mammalian neural tissues, hundreds of circRNas are highly abundant and change expression during differentiation (12).
To examine the possibility that circRNA expression levels rapidly change in response to growth factors, we selected human mammary epithelial cells, MCF10A (31). In response to EGF treatment, these cells rearrange their actin cytoskeleton and start migrating after induction of the IEG called EGR1 (32). Importantly, this process involves widespread transcript isoform variation, including mRNA alternative splicing and polyadenylation (33), as well as fast downregulation (5) and rapid upregulation of microRNAs (34). Here, by applying deep RNA sequencing, we identified more than 1000 circRNAs in MCF10A cells. Analysis of a randomly selected large group of these circRNAs unraveled their static nature: unlike the rapid (<30 min) changes exhibited by mRNAs and microRNAs, the levels of expression of circRNAs were minimally altered following stimulation. Congruently, we found no statistical evidence supporting a general sponge-like function of MCF10A's circRNAs toward the microRNAs expressed in these cells. Interestingly, the circRNAs of human mammary cells are driven by relatively active promoters, which simultaneously generate also the corresponding linear isoforms. The results we obtained are discussed in terms of the biogenesis of circRNAs and their potential biological functions.
MATERIALS AND METHODS
Cell culture and metabolic labeling
MCF10A cells were cultured as described (32) in DME:F12 medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10 μg ml−1 insulin, 0.1 μg ml−1 cholera toxin, 0.5 μg ml−1 hydrocortisone, 5% heat-inactivated horse serum (Biological Industries, Beit-Haemek, Israel) and 10 ng ml−1 EGF. For time course experiments, cells were starved overnight in medium without additives, and thereafter stimulated with EGF (10 ng ml−1) or dexamethasone (DEX; 100 nM). RNA metabolic labeling was performed in different concentrations of 4-thiouridine (4sU; Sigma), as recommended before (35). The labeling reagent was added to the medium (0.5 mM final concentration) and incubated with cells for 20 min. This was followed by 40 min of labeling (0.3 mM) and further incubations for 1, 2 or 4 h (0.2 mM). When treated with EGF, cells were concomitantly labeled with 4sU.
Sequencing of RNA derived from MCF10A cells
Total RNA was isolated from MCF10A cells using Trizol (Invitrogen). Ribosomal RNA was removed using the Ribominus kit (Invitrogen). A cDNA library was generated and sequenced using an Illumina Hiseq 2500 (1 × 100 bp runs), as instructed by Illumina's RNA-seq protocols.
Computational pipelines for predicting and annotating circRNAs from ribominus sequencing data
We used previously described computational pipelines and classification of circRNAs as Annotated, Intergenic and Antisense (15). Our circRNA analyses conditioned that at least two independent reads support existence of a non-canonical sequence junction.
mRNA and miRNA expression data from microarrays
Expression data of mRNA and miRNA were previously generated in our lab using MCF10A cells and a stimulation protocol identical to the one used in the current study (5,36). We randomly selected 16.1% of mRNAs to obtain the same coverage as we obtained by high-throughput polymerase chain reaction (PCR) for circRNAs identified by sequencing (241 out of 1498; 16.1%). Thereafter, the selected mRNAs were sorted according to their maximal fold change in ascending order for induced genes, and descending order for repressed genes. We applied the same procedure to all microRNAs of the dataset because our selection included only 180 miRNA in total, which did not allow us to apply additional selection steps.
Primer design
For circRNA expression measurements we wrote a program that designs divergent primers on the basis of the Primer3 program. Out of 1498 circRNAs that we identified in MCF10A cells, 47 circRNAs, which are either derived from intergenic regions or they represent antisense transcripts, were excluded. Each of the remaining 1451 circRNAs was intersected with Refseq exons because most circRNAs (>75%) have their introns spliced out (27), and even if introns are retained, this would not usually interfere with primer design. Thereafter, we combined the sequences to obtain the predicted circRNA sequence. In the last step we divided the predicted sequence in half, inverted the two parts and concatenated them to generate the final input for primer design. The Primer3 program was run using default parameters, except that primers were designed to flank non-canonical splicing sites. To design primers for the excluded 47 circRNAs, we used genomic DNA sequences rather than Refseq exons. The output primers from Primer3 flank non-canonical splicing sites and result in a 70–150 nt long amplicons. To exclude off-target amplification, we used the in-silico PCR program of the UCSC server. In parallel, we selected convergent primers that amplify the linear isoform from a pre-built library of qPCR primers covering all possible exon–exon junctions (37).
cDNA preparation for real-time PCR
Total RNA was isolated from cell lysates using Trizol (Invitrogen). RNA was extracted from the aqueous phase with phenol:chloroform (1:1) and precipitated for 30 min at −80°C with isopropanol and Glycoblue (Ambion). One microgram of total RNA was reverse transcribed using random hexamers and the MultiScribe reverse transcriptase from Applied Biosystems.
High-throughput real-time PCR
Pre-amplification of each individual sample was performed using a pool of 96 primer sets. The pre-amplification step facilitates downstream PCR amplification of specific sample-primer set pairs. The product was diluted 1:5 and stored at −80°C. qPCR was carried out in dynamic arrays (Fluidigm Corporation, CA, USA). Reaction mixtures contained forward and reverse primers (20 μM), assay loading reagent (Fluidigm, PN85000746), TE buffer (low EDTA, from VWR) and a mixture consisting of a sample loading reagent (Fluidigm, PN 100–0388), EvaGreen DNA binding dye (Biotium, PN 31 000), along with a pre-amplified cDNA. An IFC controller was used to prime the fluidics array with control line fluid, and thereafter with samples and assay mixtures in the appropriate inlets. After loading, the chip was placed for 10 min in a BioMark Instrument for PCR (at 95°C), followed by 40 cycles at 95°C (each for 15 s) and 60°C (for 1 min). A non-template control (NTC) was included in each chip to detect contaminations or non-specific amplification. Endogenous genes (G6PD, PGK1, B2M and GAPDH) were used to normalize for differences in the amount of total RNA. PCR reactions were performed using the ABI Prism 7700 Sequence Detection System (PE Applied Biosystems). The cycle number (termed cycle threshold, or Ct), at which amplification entered the exponential phase, was determined and this number was used as an indicator of the amount of target RNA, namely: lower Ct values indicated higher quantity of starting RNA. The result obtained for each circRNA or linear RNA was normalized to the mean of the control RNAs from the same sample.
Quantitative PCR
Expression profiles of several circular and linear isoforms obtained using Fluidigm were verified using qPCR. All reactions were performed using Power SYBR Green PCR Master Mix (from Applied Biosystems). The levels of housekeeping genes (G6PD and PGK1) were used as endogenous controls for normalization.
Metabolic labeling of newly transcribed RNA
We isolated total RNA, newly transcribed RNA and preexisting RNA from MCF10A cells that were metabolically pre-labeled using 4sU. The isolated RNA was quantified using Fluidigm and qPCR. For labeling, cells were treated for 20, 40, 60, 120 and 240 min with a combination of 4sU and EGF, or 4sU only. Because a short labeling of 10 min is sufficient for 4sU uptake (35,38), the fraction of unlabeled EGF-responsive genes was negligible in our studies. Differences in the amount of total RNA were corrected by normalizing to the levels of two housekeeping genes (G6PD and PGK1).
Biotinylation and purification of 4sU-labeled RNA
Total cellular RNA was isolated from cells using the Trizol reagent (Invitrogen), and sample quality was tested on a 2100 Bioanalyzer (Agilent). 4sU-labeled RNA (150 μg) was biotinylated using EZ-Link Biotin-HPDP (Pierce), dissolved in dimethylformamide (DMF). Biotinylation was carried out in labeling buffer (10 mM Tris pH 7.4, 1 mM EDTA) containing Biotin-HPDP (0.2 mg/ml; 90 min at room temperature). Unbound Biotin-HPDP was removed using chloroform and MaXtract (high density) tubes (Qiagen). RNA was precipitated at 20 000 g for 20 min with a 1:10 volume of 5 M NaCl and an equal volume of isopropanol. The pellet was washed with an equal volume of 75% ethanol and re-precipitated. The pellet was re-suspended in 100 μl RNase-free water. Biotinylated RNA was captured by incubation with 100 μl Dynabeads MyOne Streptavidin T1 beads (Invitrogen) with rotation for 15 min at 25°C. Beads were magnetically selected and washed with Dynabeads washing buffer. The flow-through was collected for unlabeled pre-existing RNA recovery. RNA-4sU was eluted twice with freshly prepared 100 mM dithiothreitol (DTT; 100 μl). RNA was recovered using RNeasy MinElute Spin columns (Qiagen).
Estimating half-lives of RNA isoforms
A simple linear regression model was employed to normalize the three RNA fractions, newly synthesized (4sU-RNA), total and pre-existing RNA (FT-RNA), to each other, and also determine the median RNA half-life, using the HALO software (39). Newly synthesized to pre-existing RNA ratio was used because this ratio was found to be more precise than the newly synthesized to total RNA ratio for measurements of both short and medium-to-long-lived transcripts (39). The computational methodology has been previously described (35,40). Since newly transcribed and pre-existing RNA should sum up to total RNA, a negative linear correlation should exist between the newly transcribed/total and pre-existing/total RNA ratios. Half-lives were then calculated for each regression-normalized feature using the ratio newly synthesized to pre-existing RNA. All data were corrected for any bias in circRNA and linear RNA half-life calculations caused by shorter length. For this, we used Loess regression and the built-in function of the HALO software (39).
Analysis of basal (unstimulated) gene expression
Out of 1498 circRNAs identified in MCF10A cells we selected circRNAs derived from annotated genomic regions (1451 out of 1498). Next, we excluded circRNAs that were derived from the same host gene. Basal expression levels of the remaining 1081 genes were derived from two different datasets: (i) exon microarrays (41) and (ii) RNA-sequence analysis (42). Minor fractions of these genes, 106 and 30, were not found in the exon array and RNA-Seq expression data, respectively. Our control set consisted of annotated genes (17 126 in the exon array and 22 923 in the RNA-Seq data) minus the genes that have evidence for circRNA production (16 151 in the exon array data and 21 872 in the RNA-Seq data).
Analysis of acetylated lysine 27 of histone 3
Cell fixation, chromatin immunoprecipitation and sequencing were performed as described (43). For immunoprecipitation, we used a rabbit antibody (ab4729 from Abcam) specific to acetylated lysine 27 of histone 3. Chromatin immunoprecipitation (ChIP) sequencing reads were mapped to the hg19 genome assembly using Bowtie2 (44). To avoid biases while comparing H3K27Ac levels in the promoter region of genes hosting both linear and circular RNA isoforms, relative to genes generating linear isoforms only, we filtered out circRNAs whose host genes have more than one potential TSS (according to Refseq). This selected 793 circRNAs, out of a list of 1081 circRNAs derived from known regions. Because 12 genes of this set had no expression data, they were excluded from further analysis. Next, we generated a control set consisting of 4992 genes with single TSS, that have no evidence of circRNA production, and that share exactly the same expression level distribution as single TSS genes that generate circRNAs (altogether 781 genes; see Figure 4A). We generated histograms for both groups of genes (genes with or without evidence for circRNA production) using the function ‘annotatePeaks’ of HOMER (http://biowhat.ucsd.edu/homer/). Libraries were normalized by their size such that each library contains 10 million reads to allow comparison across samples. All tag counts were thus expressed as reads/10 M reads. ChIP-Seq tag read densities were calculated near promoter regions defined as −3 to +3 kb relative to the TSS. Histograms of tag densities within 3 kb of TSS were derived using HOMER (bin = 25 bp) (45).
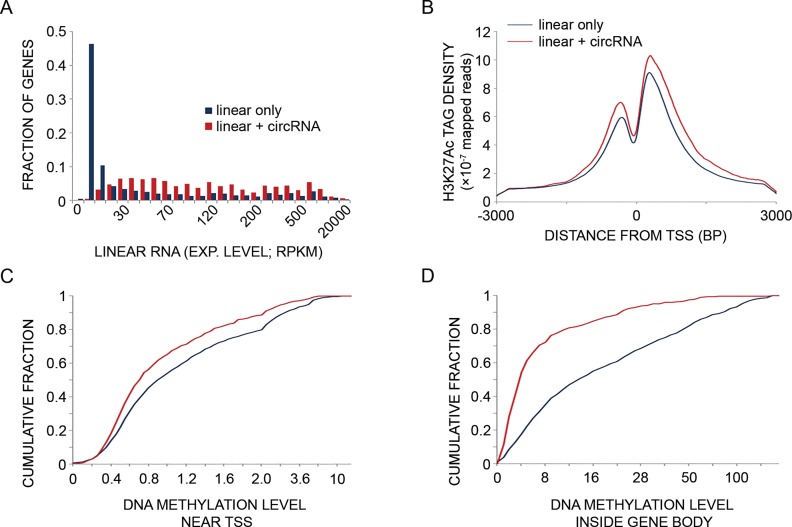
Figure 4.
Genes giving rise to circRNAs frequently express the respective mRNAs, and they are transcriptionally more active than genes transcribed into linear RNA only. (A) Basal (unstimulated) expression levels of mRNAs transcribed from genes that also give rise to circRNAs (red bars; N = 1051) are compared to mRNAs transcribed from genes that give rise only to linear RNAs (no detectable circRNAs; blue bars; N = 21 872). MCF10A RNA-seq data were obtained from a publicly available RNA-sequencing dataset (42). RPKM—reads per kilo basepairs per million reads. P = 6.13e-212, Mann–Whitney-U test. Note that only five circRNAs (leftmost quantiles) displayed no linear counterparts. (B) Basal H3K27 acetylation levels at promoter regions (transcription start site (TSS) ± 3000 bp) were determined in MCF10A cells. The acetylation levels displayed by genes transcribed into both circRNAs and linear RNAs (N = 781; red line) are compared to genes transcribed into linear RNAs with no indication of the respective circRNAs (N = 4992; blue line). The tag counts were normalized to 10 million sequencing tags for each sample. Shown are curves of tag densities of 25 nt bins. Tag density units are presented per basepair (bp) and per gene. Note that H3K27 acetylation levels are higher for genes producing circRNA compared to the control gene set (P = 1.26e-26, Mann–Whitney-U test). (C) Basal DNA methylation levels at the promoter region (TSS ± 1000 bp) of genes transcribed into both circRNAs and linear RNAs (N = 781) are compared to methylation levels displayed by genes that give rise to linear RNA only (N = 4992). DNA methylation data of MCF10A cells was derived from the ENCODE dataset. DNA methylation levels in the promoter region (−1000 to +1000 bp relative to the TSS) of each circRNA-producing host gene and control gene (no evidence for circRNA production) were tallied and a cumulative fraction was calculated and plotted for both groups. Note that DNA methylation levels are lower in genes producing also circRNAs compared to a group of genes giving rise to linear RNAs only (P = 3.15e-12, Mann–Whitney-U test). (D) Basal DNA methylation levels in the region between the TSS and TES (gene body) of genes transcribed into both circRNA and linear RNA (N = 399) are compared to levels of methylation displayed by genes that give rise to linear RNA with no detectable circRNAs (N = 2220). The methylation levels were tallied and normalized to gene length for each circRNA-producing host gene and for control genes (no evidence for circRNA production). A cumulative fraction was calculated and plotted for both groups. Note that DNA methylation levels in the body of genes producing circRNAs are lower compared to a control group of genes (P = 9.46e-57, Mann–Whitney-U test).
DNA methylation analysis
DNA methylation data of MCF10A cells were downloaded from the ENCODE database (450K bead array). We analyzed 781 genes giving rise to circular and linear RNA isoforms, relative to 4992 genes giving rise to linear RNAs only. Methylation of three gene regions was separately analyzed for each control (linear RNA, no detectable circRNA) and circRNA generating gene (linear and circRNAs): (i) promoter regions (−1 to +1 kb relative to the TSS). (ii) Transcription end site (TES) regions (−1 to +1 kb relative to the TES). This was preceded by listing 1081 circRNAs, all derived from known regions, and filtering circRNAs the host gene of which has only one potential TES (according to Refseq; altogether 908 circRNAs). Because 10 genes of this set had no expression data, they were excluded from further analysis. Next, we generated a control set consisting of 5916 genes with a single TES, which have no evidence of circRNA production and they share the same expression level distribution as single TES genes that give rise to circRNAs (total of 898, excluding the aforementioned 10 genes). (iii) Gene body regions. For this, we selected 408 circRNAs with only one possible transcript according to Refseq annotation. A minority of this set, 9 genes, were not found in the expression dataset, hence we excluded these genes from further analysis. For the remaining 399 genes that generate cirRNA we assembled a control set of genes consisting of 2220 single transcript genes that have no evidence of circRNA production and they share exactly the same expression level distribution as single transcript genes that generate circRNAs.
Seed matching analyses
Exonic segments from within each circRNA were concatenated using the Refseq database. circRNAs that do not overlap with Refseq exons were discarded (125 of 1451 circRNAs). Next, we quantified for each circRNA the numbers of canonical miRNA sites as following: 7mer-A1—perfect match to the 6-nt miRNA seed augmented by an A at target position 1, 7mer-m8—perfect match to the 6-nt miRNA seed with an additional match to nucleotide 8 of the miRNA, 8mer sites—comprising the 6 nt seed match flanked by both the match at position 8 and the A at position 1 (46). Three groups of miRNAs were analyzed: (i) the 33 percentile most highly expressed miRNA families in MCF10A (using GSE50064), (ii) the 87 miRNA families conserved across vertebrates and (iii) the 66 miRNA families conserved across mammals. To estimate the distribution of sites expected by chance, the procedure was applied to synthetic miRNA sequences obtained by random permutations preserving the mono-nucleotide composition. Permuted sequences were selected if they preserved the GC dinucleotide and possessed an A at the 3′-most position. For 6-mer matches, windows of 6 nt, starting from the 5′ end of miRNAs, were tested for complementarity to circRNA exons. Windows with the same resulting sequence were merged to avoid duplicate counting of the same circRNA–miRNA pair. To estimate the distribution of sites expected by chance, every window of 6 nt was randomly permuted 1000 times, while preserving its mono-nucleotide and GC dinucleotide composition.
Data deposition
Raw and processed RNA-Seq and ChIP-Seq data were deposited in the Gene Expression Omnibus (GSE71197).
RESULTS
Unlike messenger RNAs and microRNAs, which rapidly change in mammary epithelial cells, circular RNAs display only minor immediate changes in response to EGF treatment
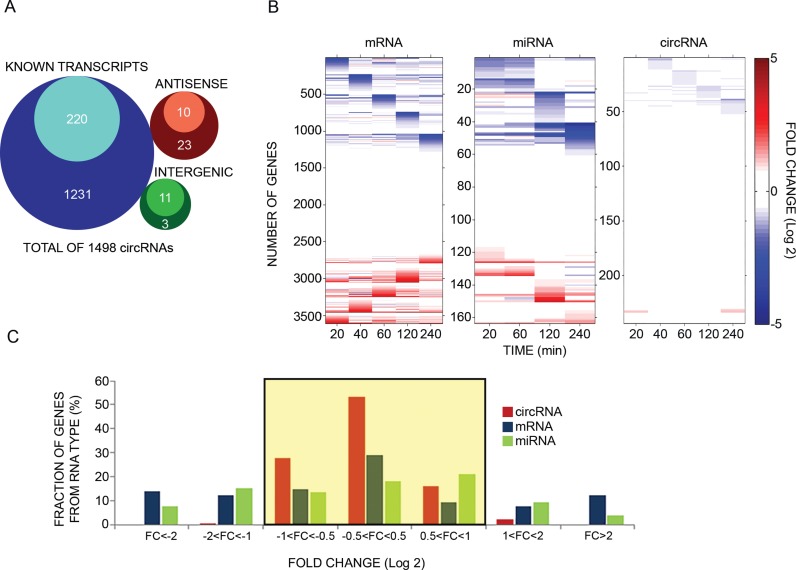
To identify putative EGF-inducible circRNAs, we isolated RNA from MCF10A cells treated for 60 min with EGF and performed three rounds of RNA sequencing using previously described procedures (15). Notably, only part of the circRNAs identified in any of these three rounds were reproducible, meaning that they were shared with at least one more RNA-Seq library. Reads that uniquely aligned to the circRNA-specific head-to-tail junctions were counted, as a measure for circRNA expression. Altogether we identified 1498 distinct circRNA candidates, each presenting at least two distinct head-to-tail spliced reads (average read number per candidate circRNA = 4.25; Supplementary Table S1). As shown in the Venn diagram of Figure 1A, apart from a few antisense and intergenic candidates, most putative circRNAs overlapped known transcripts. In addition, out of the 1498 candidates, 1164 are listed in the circBase database (47). Thirteen of these represent bona fide circRNAs (0.86%) that have been previously validated using PCR and other methods (9,15). PCR that employs divergent primers generally correlates well with RNA circularity (15,20). Hence, in the next step we established a high-throughput RT-PCR assay that employed microfluidic arrays (from Fluidigm) and divergent primers, which validated circularity of a selected group of 241 candidates (represented by the inner circles in Figure 1A). This group included >50% of the most abundant candidates, along with dozens of other circRNAs with varying expression levels (Supplementary Figure S1A). In addition to circularity verification, the high-throughput PCR assay offers precise quantification of circRNA abundance, which we could not derive from the RNA-seq data due to small read numbers. To verify the quantitative feature we confirmed previously reported rapid effects of EGF and dexamethasone (41) on two EGF inducible mRNAs (Supplementary Figure S1B). Note that dexamethasone (DEX) antagonizes many EGF-induced transcription events (41). The results we obtained when stimulating MCF10A cells for up to 4 h are presented in Figure 1B. Also presented are re-analyzed mRNA (36) and microRNA data (5) derived from DNA arrays. Notably, unlike the dynamic patterns exhibited by the other two types of RNAs, no circRNA displayed more than a 2.0 fold-change in two or more consecutive time points. Figure 1C exemplifies the relatively limited inducibility of circRNAs: whereas ∼46 and 40% of the analyzed mRNAs and microRNAs, respectively, showed more than 2.0 fold-change alterations (up- or down-regulation; in at least one time point) in response to EGF, the corresponding fraction of circRNAs was lower than 3%. Notably, the comparison between expression level changes of the different RNA molecules was performed using different techniques (Microarrays versus Fluidigm RT-PCR). For few exemplary genes that we examined (TGFA, PIK3C2G, ITPKC and LDLR), the results were comparable but the Fluidigm methodology presented higher fold changes. Thus, the RT-PCR seems to be more sensitive compared to the microarrays that were used in our study. This lends support to the idea that using different techniques (perhaps also RNA-Seq) for the calculation of fold changes might present slightly different values without affecting the overall conclusion. Importantly, extending the time scale of stimulation with EGF to 8 and 26 h revealed only minimal alterations in circRNAs (N = 89): only 1% of circRNAs changed more than two-fold (Supplementary Figure S1C), in similarity to shorter intervals of cellular activation.
Figure 1.
Unlike mRNAs and microRNAs, circular RNAs of mammary cells display minor changes in response to an extracellular cue. (A) Venn diagrams presenting genomic origins of circRNAs found in MCF10A mammary cells. The analysis comprises 1498 circRNA molecules we identified in MCF10A cells using RNA-sequencing. Of these, 1451 molecules (97%) overlap known transcripts. The remainder 47 circRNAs are either intergenic (14 transcripts) or antisense to known transcripts (N = 33). Inner circles represent Quantified fractions, meaning transcripts we followed also by using PCR; For circRNAs derived from known transcripts, the majority (>90%) of the Quantified fraction refers to circRNAs, the response of which to EGF was assayed using divergent and convergent sets of primers, while for the remainder of the Quantified fraction, including circRNAs derived from either antisense or intergenic regions, measurements were performed using only divergent sets of primers. (B) MCF10A human mammary epithelial cells were starved overnight for serum factors. Thereafter they were treated with EGF (10 ng/ml) for the indicated time intervals. High-throughput PCR and specific primers were applied on isolated RNA samples to amplify 241 of 1498 circRNA species previously identified using RNA sequencing. This group included >50% of the most abundant candidates, along with dozens of other circRNAs with varying expression levels. The presented heatmap (right panel) depicts time-dependent alterations in expression levels of specific circRNAs. These alterations are compared to mRNA and microRNA alterations we previously observed, using microarrays, while stimulating MCF10A under identical conditions (5,36). Note that all previously analyzed miRNAs are represented, but in order to match the size of the circRNA population, only randomly selected, 16.1% of all MCF10A's mRNA molecules, are depicted in the heatmap. Data were normalized to time zero and ordered according to the time point corresponding to the maximal change. Red squares represent an increase and blue squares represent a decrease, as shown in the scale bar on the right. CircRNA results represent biological duplicates performed in technical triplicates. (C) A histogram showing the range of abundance changes of mRNAs, miRNAs and circRNAs (N = 3608, N = 164 and N = 288, respectively) displayed by EGF-stimulated MCF10A cells. To construct the histogram, the maximal change value (induction or repression) along the stimulation interval (240 min) was found for each RNA molecule. Note that circRNAs exhibit narrower dynamic range (highlighted region) than mRNAs and miRNAs (P < 1e-100, F-test, Bonferroni corrected for multiple comparisons). Note that only 3 time points (30, 90 and 240 minutes) were available for 47 of the presented circRNAs.
CircRNAs are significantly less abundant and dynamic than linear isoforms derived from the very same host gene
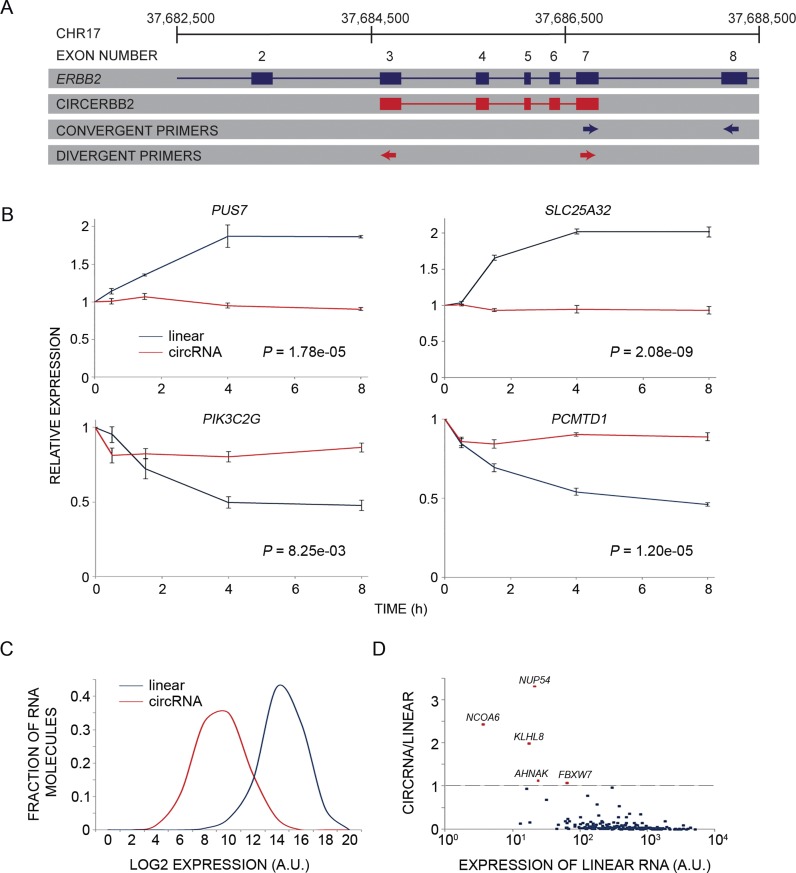
The relatively static patterns of the population of circRNAs we examined motivated us to compare the dynamicity of individual circRNAs and the linear RNAs (usually mRNAs) derived from the very same host gene. For this, we developed a high-throughput, PCR-based quantitative approach that employed combinations of divergent and convergent primers enabling simultaneous measurements of circular isoforms, and the corresponding linear isoform (see an example in Figure 2A). Notably, linear and circRNAs differ in terms of average length, but our estimation of abundance assumed that they are reverse transcribed with similar, length-independent efficiencies. When applied to 203 candidates randomly selected from the 288 shown in Figure 1C, this methodology showed that circRNAs display static abundance also in comparison to the corresponding linear isoforms. Figure 2B presents four examples of mRNAs that changed by 1.8-fold or more (up- or downregulation) in response to EGF, while the respective circular isoforms remained almost unaltered under the same conditions. Further comparative analysis revealed that, in general, circRNAs displayed significantly lower abundance than the corresponding linear transcripts (Figure 2C). Assuming that linear RNAs and the circular forms are amplified, using PCR, at similar efficiencies, we calculated that, on average, circRNAs were 36 times less abundant than their corresponding linear transcripts. Notably, previous studies estimated the transriptome fraction of circRNAs to be even lower than our estimation (20,27). Interestingly, we found weak, but statistically significant, correlation (Pearson coefficient = 0.22) between the mammary cell expression levels of linear and circular RNA isoforms that are derived from the same host gene (Supplementary Figure S2A). Next, we compared the basal expression levels (no EGF stimulation) of individual circRNAs to the expression levels of their linear counterparts. This analysis revealed that only five circRNAs (derived from the following genes: FBXW7, AHNAK, NCOA6, NUP54 and KLHL8) were expressed at higher levels than the corresponding linear isoforms (Figure 2D and Supplementary Figure S2B). In conclusion, circRNAs of growth factor stimulated cells appear less dynamic than mRNAs and miRNAs. This conclusion extends to direct comparisons of circRNAs and their corresponding linear transcripts.
Figure 2.
circRNAs are less dynamic and abundant relative to linear isoforms derived from the same host gene. (A) A linear diagram exemplifying primer pairs (arrows) for parallel measurements of levels of circular and linear RNA isoforms derived from the same host gene. The ERBB2 (chromosome 17) gene is shown as an example. The respective circRNA (red) spans five internal exons. The divergent primers (red arrows; circular transcript) flank the non-canonical splicing (circularization), whereas the convergent primers (blue arrows) were designed for measuring the corresponding linear isoform. (B) PCR and four pairs of primers (designed as in A) were used to follow the response of the indicated linear and circular isoforms of four EGF-regulated genes. Note that mRNAs corresponding to PUS7 and SLC25A32 are induced when MCF10A cells are stimulated with EGF, whereas PCMTD1 and PIK3C2G mRNAs undergo down-regulation under the same conditions. Shown are means ± S.E. of triplicates. P-values were calculated using two-way Anova with time and RNA type as categorical factors. (C) A histogram comparing the abundance of circRNAs and the corresponding linear transcripts (N = 203) of MCF10A mammary cells. Note that circRNAs are, on average, 36 times less abundant than the corresponding linear isoforms (P = 5.45e-78, t-test, two-tail, paired; A.U., arbitrary units). (D) A dot plot presenting the ratio between expression levels of each circRNA of MCF10A cells we characterized and the respective linear transcript, which is derived from the same host gene. The ratios are presented against the abundance of the latter isoform. Note that most circRNAs are expressed at lower levels than the corresponding linear transcripts; only five circRNAs (red dots; identified by names) exceeded the abundance of their respective linear isoforms, but the absolute expression levels of all five circRNAs are relatively low.
Analysis of newly transcribed RNA reveals that circRNAs are more stable than the linear RNA isoforms derived from the same host gene
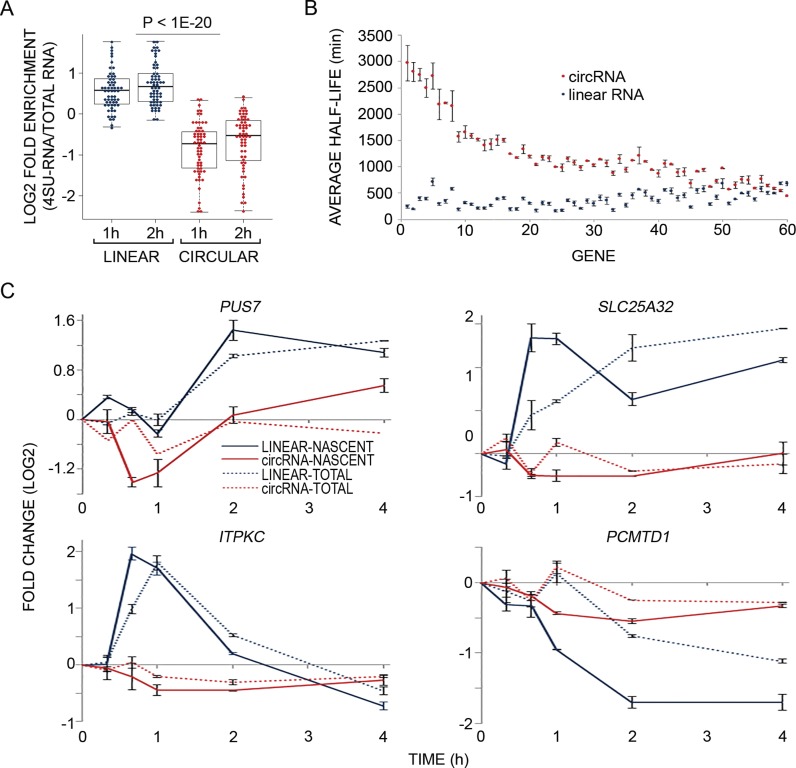
Limited transcript alterations in response to external cues, such as the behavior displayed by circRNAs in response to EGF, might be due to long-lived transcripts (38,48,49). Because circRNAs lack free ends, as well as poly(A) tails and 5′ caps, they normally escape de-adenylation and de-capping. Hence, we set out to determine half-lives of 61 circRNAs in comparison to their linear counterparts. To this end, we metabolically labeled newly transcribed RNA of MCF10A cells using 4sU (35), a method that determines half-lives based on the analysis of three fractions: total cellular RNA, newly transcribed RNA, and unlabeled (pre-existing) RNA. The method is based on the ability of thiol-group containing nucleosides, such as 4sU, to be introduced into nucleoside salvage pathways (35). It allows labeling of newly transcribed RNA, separation from total RNA using thiol-specific biotinylation, and purification on streptavidin-coated magnetic beads. Thus, direct incorporation of 4sU allows determination of the proportion of new RNA, which under steady state conditions is largely specified by the degradation rate. Notably, metabolic labeling with 4sU is suitable for half-life determination of both short—as well as medium-to-long-lived transcripts (39,50), while other methods are inherently imprecise when applied to medium-to-long-lived transcripts (35). Firstly, we confirmed that unlabeled RNA binds only minimally to the avidin beads used for purification, and secondly we determined that EGF-inducible genes are significantly enriched in the labeled RNA fraction (on average, 2.98–4.25-fold depending on normalization; Supplementary Figure S3A and S3B), in line with previously reported similar analyses (38,48,49). Next, we compared the enrichment of circRNAs relative to their linear counterparts, and found that while linear RNAs were enriched, circRNAs were diluted in the newly transcribed fraction compared to the total RNA fraction (Figure 3A). A slightly larger difference was observed when comparing the newly transcribed fraction to pre-existing RNA (Supplementary Figure S3C). This observation implies that circRNAs are longer-lived transcripts, and in light of their overall low abundance, their production rate might be slower, relative to the corresponding linear RNAs.
Figure 3.
Analysis of newly transcribed RNAs reveals that circRNAs are more stable and static than the linear isoforms derived from the same host genes. (A) MCF10A cells were treated with EGF as in Figure 1B and RNA was simultaneously metabolically labeled using 4-thiouridine (4sU), for the indicated time intervals. RNA was extracted (Total-RNA) with Trizol, biotinylated and purified on streptavidin magnetic beads (denoted 4sU-RNA). Flow-through RNA was also collected (denoted FT-RNA). Thereafter, RNA was reverse transcribed and quantified using high-throughput real time PCR (Fluidigm). The boxplot diagram shows enrichment of newly transcribed RNA (4sU labeled) in linear isoforms (blue dots) relative to the respective circRNA isoforms (red dots; P < 1e-20, t-test, two-tailed distribution, unequal variance, Bonferroni corrected for multiple comparisons, N = 61). Shown are the log2 fold enrichments of 4sU-labeled RNA versus total RNA (Y axis). (B) MCF10A cells were metabolically labeled using 4sU, for 1 or 2 h. Thereafter, RNA was extracted, biotinylated and purified on streptavidin magnetic beads. Flow-through RNA was also collected. Next, RNA was reverse transcribed and quantified using high-throughput real time PCR (Fluidigm). Presented are the half-lives of 60 circRNAs and their corresponding linear counterparts. Half-life values were calculated from two samples, which were labeled with 4sU for 1 or 2 h and then averaged. All data were corrected for any bias introduced due to low uridine (short length) of RNA species (see ‘Materials and Methods’ section). The circRNAs (red dots) and their linear counterparts (blue dots) were sorted according to the difference between their half-lives from high to low. Error bars represent standard errors. The calculations of half life were performed using the HALO software (39) and the ratios between newly transcribed (RNA-4sU) and pre-existing RNAs (RNA-FT). (C) PCR and four pairs of primers were used to follow the response of newly transcribed RNA (solid lines) and total RNA (dashed lines) of both linear and circular isoforms of the indicated EGF-regulated genes. Shown are calculated fold changes of expression levels displayed by EGF-treated cells relative to basal (unstimulated) expression levels, measured in cells labeled with 4sU for the indicated intervals (Y axis). Shown are means ± S.E. of duplicates.
Calculating the half-lives of 60 circRNAs and their linear counterparts expressed from the same host gene revealed that the median half-life of circRNAs of mammary cells (18.8–23.7 h) is at least 2.5 times longer than the median half-life of their linear counterparts (4.0–7.4 h; Figure 3B and Supplementary Table S2). As expected, we found no correlation between the half-lives of circRNAs and their linear counterparts after correction for any bias introduced due to shorter RNA length (Supplementary Figure S3D; see ‘Materials and Methods’ section). Because 4sU labeling enables high sensitivity detection of up- and down-regulation events (35,38), we simultaneously treated mammary cells with EGF and 4sU, and exemplified the results with four pairs of linear and circRNAs (Figure 3C). This analysis confirmed differential dynamicity and suggested a general but weak EGF-induced decrease in newly transcribed circRNA abundance, independent of the sign of linear RNA's alteration. In conclusion, although a large fraction of mammary cell expressed genes give rise to both circular and linear RNA isoforms, the circRNAs are, in general, more stable and static compared to the linear species.
Virtually all genes generating circRNAs also generate the respective linear RNAs and their promoters are more active relative to genes hosting only linear RNAs
Although circRNAs of mammary cells are derived mainly from known transcripts, several studies reported antisense configured circRNAs, including the well-characterized ciRS-7/CDR1as, and some circRNAs might overlap unannotated regions (Figure 1A). Thus, circRNA transcription might be independent of linear transcription. To address this issue, using a publically available dataset, we compared in MCF10A cells RNA expression levels and found that the vast majority (>98%) of genes that give rise to circRNAs also express the respective linear transcript (Figure 4A). Moreover, genes hosting both linear and circular RNAs express higher levels of the linear RNAs, as compared to the linear only group (no detectable circRNA; Supplementary Figure S4A). Thus, it is still possible that weakly expressed linear RNAs are also associated with transcription of circRNAs, but the latter are below our detection level. This unexpected coupling of circRNAs expression to linear isoforms led us to re-analyze circRNA data from human mammary epithelial cells (28), which revealed that only 1% of the genes that give rise to circRNAs have RPKM (reads per kilobase of exons per million reads mapped) smaller than one. Consistent with coupled expression, similar analysis of human foreskin fibroblasts concluded that genes that do not express a linear RNA isoform in fibroblasts, also do not express the respective circular isoform. Thus, out of 73 genes we analyzed, three (PIK3C2G, TNS4 and BCL11B) were co-expressed with the cognate circRNAs in MCF10A cells, but neither the linear nor the circular forms were detectable in fibroblasts. Taken together, these observations lend strong support to the possibility that circRNAs of human cells are, in general, co-expressed alongside the cognate, frequently transcribed linear isoforms.
To validate the finding that circRNAs are derived from highly transcribed genes, we examined two markers of promoter activity: histone 3 lysine 27 acetylation (H3K27Ac) and DNA methylation. H3K27Ac levels were determined using DNA sequencing of chromatin immunoprecipitates obtained with an antibody specific to H3K27Ac, whereas DNA methylation data of MCF10A cells was derived from the ENCODE dataset. Next, we constructed a set of 4992 genes with single TSS giving rise to linear RNAs with no detectable circRNAs, and displaying the same expression distribution (matched in quantiles) as genes that also generate circRNAs (altogether 781 genes; Figure 4A). As expected, genes that give rise to circRNAs presented more active promoter regions: significantly higher H3K27Ac and lower DNA methylation were observed in promoter regions of genes that give rise to circRNAs, as compared to genes that generate linear RNA but no detectable circRNA (Figure 4B and C). To substantiate these observations we analyzed DNA methylation within gene bodies, namely the regions lying in between transcription start sites and TES, and also the regions within 1 kb from the TES. Interestingly, both regions displayed less DNA methylation in the group of genes that generates circRNAs compared to the control genes, which generate only linear RNAs (Figure 4D and Supplementary Figure S4B). Overall, our results revealed that genes giving rise to circRNAs are characterized by higher levels of promoter activity. Furthermore, the observed lower intragenic DNA methylation of such genes might relate to the mechanism of circRNA biogenesis.
CircRNAs expressed by mammary cells show no enrichment for binding sites specific for microRNAs co-expressed in these cells
The circRNAs CDR1as and Sry, each containing multiple conserved sites for a specific microRNA, might act as molecular sponges (15,21). According to recent quantitative models, competition between binding sites on different RNAs may lead to regulatory crosstalk between transcripts (51). For example, to enable a sponging function circRNAs must display a greater affinity than the linear RNA for the suspected miRNA. Furthermore, according to recent studies, target repression is released only after adding many competing target sites, independently from miRNA levels (52), and, similarly, thousands of binding sites per cell are required to significantly affect binding of well-expressed miRNAs (51,53). Thus, in order for circRNAs to function as potent miRNA sponges in mammary cells, the product of the formula circRNA molecules × miRNA binding sites should be high relative to other competing molecules. Herein we demonstrated low abundance of circRNAs in MCF10A cells and others have shown this for other non-neuronal cell lines (16). Hence, we examined the other component of the formula, namely enrichment of miRNA seed matches in circRNA sequences in our cellular model. To this end, we concatenated the annotated exons within each circRNA and counted the number of canonical 7- and 8-nt long target sites (54) of the most abundant miRNAs expressed by MCF10A cells (33, 10 or 5% upper percentile). This analysis found no circRNA–miRNA pairs that exceeded the upper limit of results from a negative control (Figure 5A; only the 33 percentile is shown). To substantiate this we extended the analysis to 87 miRNA families conserved across vertebrates, and also to 66 miRNA families conserved across mammals. Once again, no significant enrichment of miRNA binding sites was found in circRNAs expressed in human mammary cells (Supplementary Figure S5A and B).
Figure 5.
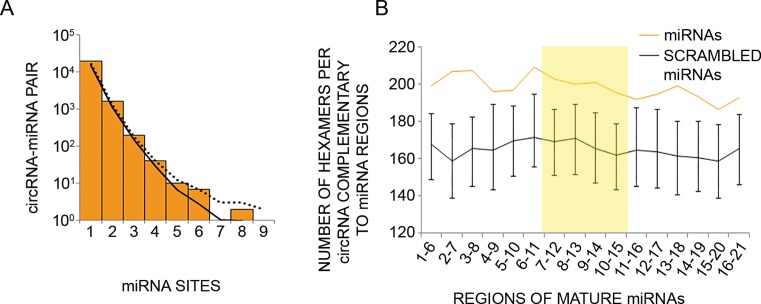
Circular RNAs expressed by human mammary cells show no enrichment for binding sites of the microRNAs found in the same cells. (A) CircRNAs expressed in MCF10A cells were analyzed for the presence of binding sites (7 or 8 nt long) for miRNAs expressed by the same cells (33% uppermost percentile). The number of sites was tallied for each circRNA–miRNA pair, and the distribution of values is plotted. The solid and dotted curves indicate the averaged and 95% upper percentile, respectively, of results when repeating the analysis 1000 times using different permutations of site sequences (see ‘Materials and Methods’ section). (B) Shown are the numbers of hexamers within circRNAs of MCF10A cells that potentially pair to miRNAs expressed in the same cells (orange line), or to scrambled miRNA regions (black line). Error bars represent the distribution of values in 1000 control scrambled miRNA regions. Scrambled miRNA regions preserved mono-nucleotide and GC dinucleotide composition of the original 6 nt window. For each miRNA highly expressed in MCF10A cells (33% upper percentile), starting from the 5′ end, we checked 6-nt-long windows for complementarity to hexamers within the collection of circRNAs expressed in MCF10A cells (N = 1326). The total number of matches was normalized to the number of unique 6 nt windows analyzed. Highlighted is the central region of miRNAs found in a previous study to have statistically significant pairing to coding sequences (55).
Because circRNAs of MCF10A cells are derived mainly from coding exons and because coding sequences were found to be enriched with hexamers matching 6-nt long internal regions of miRNAs (55), we examined complementarity of 6-nt long regions of miRNAs to circRNAs of mammary cells. Surprisingly, we found that the number of hexamers in circRNAs that are complementary to miRNAs’ hexameric regions exceeded the higher limit of complementarity to scrambled hexameric sequences (Figure 5B). However, this higher number corresponded only to three, or lower, matching sites per circRNA–miRNA pair, while higher numbers of circRNA–miRNA pairs did not exceed the higher limit achieved by the control permuted sequence set. We present the matching of nucleotides 7–12 of miRNAs to circRNAs (Supplementary Figure S5C) to demonstrate this finding. In summary, we found that circRNAs expressed by MCF10A human mammary cells do not possess the attributes needed to sponge co-expressed miRNAs, namely: sufficiently high intracellular abundance and matching miRNA binding sites, including sites located within internal regions of miRNAs. Our conclusions, along with the observed long half-life, as well as low abundance and only minor alterations of circRNAs in response to EGF stimulation, imply that circRNAs, unlike mRNAs and miRNAs of mammary cells, might not participate in transcriptional networks that are rapidly stimulated in response to extracellular cues.
DISCUSSION
We detected more than 1400 circRNAs in human mammary epithelial cells. These circRNAs are mainly derived from coding exons, and to a lesser extent from antisense or intergenic transcripts. Thus, our study adds several hundreds of mammary circRNAs to the growing catalog of annotated circRNAs (8,12,13,15,16,28,47,56,57). By directly analyzing a sub-group of the mammary circRNAs (n = 203), we observed much smaller early alterations in response to EGF, not only in comparison to mRNAs and miRNAs, but also in comparison to the linear RNAs expressed from the very same host genes. Interestingly, analysis of newly transcribed circRNAs independently confirmed that the response of these newly synthesized molecules was quite limited, resulting in minimal effects on the cellular pool of circRNAs (Figure 3C). These findings are in agreement with a recent study that analyzed alterations of several intronic circular RNAs in response to treatment of human cells with a peptide toxin, α-amanitin (58). Nevertheless, other studies, which applied much longer time courses, showed that the levels of circRNAs might display relatively large dynamic changes. For example, in differentiating cells, some circRNAs displayed large fold changes in the endpoint compared to the initial levels (28). However, it should be noted that these changes were observed after 21 days of treatment of epithelial cells with TGF-β (28). In general, the high circRNA stability might be one of several parameters leading to slow responses to cellular perturbations: a long half-life translates to a slow approach to steady state. Additionally, the low abundance means that only a few copies of most circRNAs exist in a mammary cell. Therefore, we conclude that, unlike mRNAs and miRNAs, MCF10A's circRNAs are slowly responding components, unlikely to participate in processes that commit cells to a specific, rapidly attainable outcome, such as cell migration or proliferation.
Despite the reported wide diversity of circRNAs (14), we show that the abundance of circRNA molecules in mammary cells is, on average, <3% of the abundance of the corresponding linear isoforms derived from the same host gene. Other studies reported even lower abundance of circRNAs compared to the cognate linear RNAs (16). Remarkably, only five out of the 203 circRNAs we analyzed were expressed at levels higher than their linear counterparts. Still, their absolute expression levels were relatively low (Figure 2C and D). By contrast, several studies showed that hundreds of circRNAs, especially in neuronal organs, are expressed at higher levels than their cognate linear isoforms (10,12,13,28,57). The low abundance of circRNAs found in our cultured mammary cell model and the contrasting high abundance found in neuronal tissues might be due to frequent exon skipping events (57,59) and resulting exon circularization (29) in neuronal tissues. Alternatively, due to their low production rate, circRNAs might slowly accumulate and undergo constant dilution in dividing cells, like MCF10A (56). It should be noted, however, that another study analyzed P19 differentiated non-dividing cells and observed accumulation of circRNAs only after induction of the cells to neurons (12). It is interesting to note that although most circRNAs displayed longer half-lives compared to their linear counterparts, the half-lives of some circRNAs are quite similar to those of the respective linear isoforms. This raises the intriguing possibility of selective degradation, as already reported for the miRNA sponge ciRS-7/CDR1as, which is subject to the slicer activity of Ago2 (60).
Importantly, we found only weak correlation between abundances of circRNAs and levels of expression of cognate linear RNAs, reaffirming previous results (16). For example, it was shown that the levels of circular and linear isoforms of the ZKSCAN1 transcript in different tissues are largely independent of each other (61). These observations suggest that the biogenesis of circular and cognate linear isoforms is regulated independently. Along this vein, we report that not only abundance of circRNAs and linear isoforms are quite independent of each other, but also the degradation rates of circRNAs are independent of the rates of degradation displayed by the corresponding linear RNAs in mammary cells. In conclusion, the remarkably dissimilar levels of cognate circular and linear transcripts in human mammary cells might be ascribed to either differential mechanisms of clearance or to distinct modes of biogenesis. Yet, we cannot rule out the possibility that the depth of our RNA sequencing did not permit detection of a few, very rare circRNAs corresponding to weakly expressed genes.
It is notable that despite dissimilar abundances, we were able to detect linear transcripts of almost all (>98%) circular RNAs of MCF10A cells. This observation raises the possibility that circRNAs are synthesized co-transcriptionally, alongside the linear isoforms. In line with this scenario, it has been reported that circularization and splicing compete against each other (10). Congruent with co-transcriptional generation of circRNAs, our analysis of the promoters of genes that host both circular and linear RNAs revealed higher H3K27Ac and lower DNA methylation relative to promoters hosting only linear RNAs. This epigenetic difference holds even when the expression levels of the respective linear RNAs are equal. Hence, we posit that the promoters of genes hosting both circular and linear transcripts are more permissive, and consequently raise the possibility that back-splicing, which generates circles, is typical to highly active gene promoters. Taken together, the lower abundance of circRNAs of mammary cells, compared to their linear counterparts, as well as their association with both active promoters and lower intragenic DNA methylation, lend support to the possibility that circRNA biogenesis might be secondary to transcription of linear RNA.
The well-established microRNA sponging function of two circular RNAs, namely CDR1as and Sry, spawned models attributing to additional RNA circles the ability to decoy multiple copies of the same microRNA (25,26). However, addressing the decoy model in MCF10A mammary cells yielded no support to this theory: we found no significant enrichment of miRNA seed matches in circRNAs expressed in mammary cells. This observation is in agreement with studies conducted with diverse cell types (8,12,13,15,28), and suggests that circRNAs perform other functions. Indeed, several recent reports propose a plethora of functions, such as promoting transcription of the parent genes (30,58), regulating RNA binding proteins and controlling alternative splicing (10). Although it is presently difficult predicting the major function(s) of circRNAs, the herein reported remarkably high stability of mammary cell circRNAs, along with their relatively static nature, propose a role in long-term processes, such as differentiation and cell lineage acquisition, rather than in highly dynamic processes, such as cell cycle control and growth factor signaling.
Supplementary Material
Acknowledgments
The authors thank Nikolaus Rajewsky, Sebastian Memczak, Marvin Jens and other members of the team for guidance and RNA sequence analysis, Noam Stern-Ginossar and Roni Golan-Lavi for their help. The studies reported herein were conducted in the Marvin Tanner Laboratory for Research on Cancer. Y.Y. is a Research Professor of the Israel Cancer Research Fund and the incumbent of the Harold and Zelda Goldenberg Professorial Chair in Molecular Cell Biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation (ISF) - 280/15; Rising Tide Foundation; European Research Council - AdG 20100317; Dr Miriam and Sheldon G. Adelson Medical Research Foundation. Funding for open access charge: Dr Miriam and Sheldon G. Adelson Medical Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lo H.W., Hung M.C. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br. J. Cancer. 2007;96(Suppl):R16–R20. [PubMed] [Google Scholar]
- 2.Witsch E., Sela M., Yarden Y. Roles for growth factors in cancer progression. Physiology (Bethesda) 2010;25:85–101. doi: 10.1152/physiol.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronnstrand L., Heldin C.H. Mechanisms of platelet-derived growth factor-induced chemotaxis. Int. J. Cancer. 2001;91:757–762. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1136>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 4.Avraham R., Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 5.Avraham R., Sas-Chen A., Manor O., Steinfeld I., Shalgi R., Tarcic G., Bossel N., Zeisel A., Amit I., Zwang Y., et al. EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci. Signal. 2010;3:ra43. doi: 10.1126/scisignal.2000876. [DOI] [PubMed] [Google Scholar]
- 6.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C., et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Chen C.Y., Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 8.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashwal-Fluss R., Meyer M., Nagarjuna R.P., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with Pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 13.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C., et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasda E., Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilusz J.E., Sharp P.A. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danan M., Schwartz S., Edelheit S., Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Ghosal S., Das S., Sen R., Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front. Bioinform. Comput. Biol. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosik K.S. Molecular biology: circles reshape the RNA world. Nature. 2013;495:322–324. doi: 10.1038/nature11956. [DOI] [PubMed] [Google Scholar]
- 24.Li J.-H., Liu S., Zhou H., Qu L.-H., Yang J.-H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taulli R., Loretelli C., Pandolfi P.P. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat. Struct. Mol. Biol. 2013;20:541–543. doi: 10.1038/nsmb.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 31.Tait L., Soule H.D., Russo J. Ultrastructural and immunocytochemical characterization of an immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6087–6094. [PubMed] [Google Scholar]
- 32.Tarcic G., Avraham R., Pines G., Amit I., Shay T., Lu Y., Zwang Y., Katz M., Ben-Chetrit N., Jacob-Hirsch J., et al. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB J. 2012;26:1582–1592. doi: 10.1096/fj.11-194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostler W.J., Zeisel A., Korner C., Tsai J.M., Jacob-Hirsch J., Ben-Chetrit N., Sharma K., Cohen-Dvashi H., Yitzhaky A., Lader E., et al. Epidermal growth-factor - induced transcript isoform variation drives mammary cell migration. PLoS One. 2013;8:e80566. doi: 10.1371/journal.pone.0080566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kedmi M., Ben-Chetrit N., Korner C., Mancini M., Ben-Moshe N.B., Lauriola M., Lavi S., Biagioni F., Carvalho S., Cohen-Dvashi H., et al. EGF induces microRNAs that target suppressors of cell migration: miR-15b targets MTSS1 in breast cancer. Sci. Signal. 2015;8:ra29. doi: 10.1126/scisignal.2005866. [DOI] [PubMed] [Google Scholar]
- 35.Dölken L., Ruzsics Z., Rädle B., Friedel C.C., Zimmer R., Mages J., Hoffmann R., Dickinson P., Forster T., Ghazal P., et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 37.Zeisel A., Yitzhaky A., Bossel Ben-Moshe N., Domany E. An accessible database for mouse and human whole transcriptome qPCR primers. Bioinformatics. 2013;29:1355–1356. doi: 10.1093/bioinformatics/btt145. [DOI] [PubMed] [Google Scholar]
- 38.Rabani M., Levin J.Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N., et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedel C.C., Kaufmann S., Dölken L., Zimmer R. HALO—a Java framework for precise transcript half-life determination. Bioinformatics. 2010;26:1264–1266. doi: 10.1093/bioinformatics/btq117. [DOI] [PubMed] [Google Scholar]
- 40.Lam L.T., Pickeral O.K., Peng A.C., Rosenwald A., Hurt E.M., Giltnane J.M., Averett L.M., Zhao H., Davis R.E., Sathyamoorthy M., et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0041. RESEARCH0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauriola M., Enuka Y., Zeisel A., D'Uva G., Roth L., Sharon-Sevilla M., Lindzen M., Sharma K., Nevo N., Feldman M., et al. Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat. Commun. 2014;5:5073. doi: 10.1038/ncomms6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daemen A., Griffith O.L., Heiser L.M., Wang N.J., Enache O.M., Sanborn Z., Pepin F., Durinck S., Korkola J.E., Griffith M., et al. Modeling precision treatment of breast cancer. Genome Biol. 2013;14:R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 2013:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 44.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glazar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elkon R., Zlotorynski E., Zeller K.I., Agami R. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics. 2010;11:259. doi: 10.1186/1471-2164-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pérez-Ortín J.E., Alepuz P.M., Moreno J. Genomics and gene transcription kinetics in yeast. Trends Genet. 2007;23:250–257. doi: 10.1016/j.tig.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Friedel C.C., Dölken L., Ruzsics Z., Koszinowski U.H., Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jens M., Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2015;16:113–126. doi: 10.1038/nrg3853. [DOI] [PubMed] [Google Scholar]
- 52.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zisoulis D.G., Lovci M.T., Wilbert M.L., Hutt K.R., Liang T.Y., Pasquinelli A.E., Yeo G.W. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bachmayr-Heyda A., Reiner A.T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T.W., Zeillinger R., Pils D. Correlation of circular RNA abundance with proliferation—exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 59.Brown J.B., Boley N., Eisman R., May G.E., Stoiber M.H., Duff M.O., Booth B.W., Wen J., Park S., Suzuki A.M., et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.