Abstract
Toxin-antitoxin (TA) systems play key roles in bacterial persistence, biofilm formation and stress responses. The MazF toxin from the Escherichia coli mazEF TA system is a sequence- and single-strand-specific endoribonuclease, and many studies have led to the proposal that MazF family members exclusively target mRNA. However, recent data indicate some MazF toxins can cleave specific sites within rRNA in concert with mRNA. In this report, we identified the repertoire of RNAs cleaved by Mycobacterium tuberculosis toxin MazF-mt9 using an RNA-seq-based approach. This analysis revealed that two tRNAs were the principal targets of MazF-mt9, and each was cleaved at a single site in either the tRNAPro14 D-loop or within the tRNALys43 anticodon. This highly selective target discrimination occurs through recognition of not only sequence but also structural determinants. Thus, MazF-mt9 represents the only MazF family member known to target tRNA and to require RNA structure for recognition and cleavage. Interestingly, the tRNase activity of MazF-mt9 mirrors basic features of eukaryotic tRNases that also generate stable tRNA-derived fragments that can inhibit translation in response to stress. Our data also suggest a role for tRNA distinct from its canonical adapter function in translation, as cleavage of tRNAs by MazF-mt9 downregulates bacterial growth.
INTRODUCTION
Tuberculosis is an ancient and resilient disease caused by Mycobacterium tuberculosis, a bacterial pathogen characterized by a slow growth rate and the ability to cause a long-term asymptomatic infection, known as latent tuberculosis infection (LTBI), in which the bacteria are thought to persist in a dormant state. The molecular events that underlie entry into and exit from this persistent state remain unclear. However, toxin-antitoxin (TA) systems have been suggested to play a role in LTBI because toxins can regulate growth and in Escherichia coli can induce a ‘quasi-dormant’ state (1) bearing a striking resemblance to the nonreplicating persistent state of M. tuberculosis. TA systems also play key roles in biofilm formation and stress responses (2–4), both of which are critical for M. tuberculosis pathogenesis. Curiously, the M. tuberculosis genome harbors an unusually high number of potential TA systems (>80), considerably more than any other pathogen (5,6). Several of these TA transcripts or proteins in M. tuberculosis are either induced or downregulated under stresses that the pathogen might encounter during infection, such as hypoxia (6–8), DNA damage (9), nutrient limitation (8,10,11), macrophage infection (6,12–14), heat shock (15) and treatment with antibiotics (8,16–18). Collectively, these observations are consistent with a role for TA systems in M. tuberculosis persistence, dormancy and stress responses (19–22).
The well-studied E. coli mazEF TA system serves as a useful foundation to examine the role of TA systems in M. tuberculosis. This bicistronic operon encodes the intracellular toxin MazF and its inhibitor, antitoxin MazE. The two proteins interact to form a stable complex in which the toxin is neutralized in the absence of stress (23). However, cellular proteases degrade MazE upon certain stresses (23,24) and release MazF. This MazF toxin is an endoribonuclease that cleaves single-stranded ACA sequences to inhibit translation and bacterial growth (25,26). However, sustained toxin expression can lead to cell death (27–29). Unlike endoribonuclease toxins in the RelE family, MazF toxins do not require association with ribosomes for RNA cleavage activity (25).
The M. tuberculosis genome harbors nine TA systems that are members of the mazEF family, whereas most prokaryotes only have one or two (5,6). Although the roles of so many seemingly redundant genes are still unknown, each M. tuberculosis MazF toxin (MazF-mt1 through MazF-mt9) appears to recognize and cleave a unique RNA sequence. Using primer extension studies, MazF-mt1 (locus Rv2801c) was reported to specifically recognize and cleave U↓AC sequences of RNA (30), where ‘↓’ indicates the exact location of cleavage. Using the same method, MazF-mt7 (Rv1495) was reported to cleave U↓CGCU sequences (31), and MazF-mt6 (Rv1102c) was reported to cleave at UU↓CCU sequences (32,33). In prior work, we developed an RNA-seq approach termed MORE RNA-seq (mapping by overexpression of an RNase in E. coli) to identify the sequence specificities of endoribonuclease toxins (34). Using this method, we determined that MazF-mt3 (Rv1991c) actually recognizes U↓CCUU (34), not UU↓CCU or CU↓CCU as originally reported by primer extension (31). Although MazF orthologs were originally proposed to exclusively target mRNA (25,26,30,31,35,36), we and others have demonstrated that at least three MazF toxins—E. coli MazF, MazF-mt3 and MazF-mt6—also target 23S or 16S rRNA at specific sites (32–34,37).
Because the conditions that activate MazF toxins in M. tuberculosis are unknown and since the characterized MazF family members in M. tuberculosis each cleave a unique RNA sequence and have the potential to target mRNA or rRNA, each MazF in this pathogen may have a distinct physiological trigger, effect and role. In fact, individual MazF toxins from M. tuberculosis are induced either during nutrient starvation (8,10,11), upon overexpression of a cell cycle regulation protein (38), during hypoxia or antibiotic treatment (8) or during conversion to a ‘non-culturable’ state (39). In addition, expression of a MazF toxin from M. tuberculosis increases the level of persisters (40), simultaneous deletion of three mazF genes impairs formation of M. tuberculosis persister cells (8) and expression or deletion of individual mazF genes alters persister recovery in a drug-specific manner in M. tuberculosis (18). Finally, a triple mazF deletion strain of M. tuberculosis hinders mycobacterial growth in guinea pigs and elicits reduced pathology in the lungs and liver relative to the wild-type strain, suggesting some MazF toxins may work together as virulence factors (8).
To better understand the physiological role of the nine M. tuberculosis mazEF TA systems, we characterized the features of the MazF-mt9 toxin (locus Rv2063A) by employing our specialized RNA-seq approach to pinpoint its RNA targets. Our biochemically validated RNA-seq results demonstrate that recognition of tRNA and the targeted selection of only a few tRNAs from among the 45 total in M. tuberculosis occurs in a manner that is unprecedented for a MazF toxin because it involves recognition of both sequence and structural determinants. Thus, all MazF toxins do not primarily target mRNAs, and protein sequence and structure alone does not always predict precise functional relationships between members within toxin families.
MATERIALS AND METHODS
Strains, plasmids and reagents
The E. coli strains BW25113Δ6 [lacIq rrnBT14 Δlac-ZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 ΔchpBIK ΔdinJ-yafQ ΔhipBA ΔmazEF ΔrelBE ΔyefM-yoeB] (41) and BL21(DE3) [F− ompT hsdSβ(rβ−, mβ−) dcm gal λ(DE3); Novagen] were used for all RNA cleavage and protein expression studies, respectively. E. coli Mach1-T1 [F− ΔrecA1398 endA1 tonA φ80(lacZ)ΔM15 ΔlacX74 hsdR(rκ−, mκ+); Invitrogen] cells were used for all cloning experiments. M. tuberculosis strain H37Rv (ATCC) was used for all growth and toxicity experiments. Plasmids used were pET-21c and pET-28a (Novagen), pBAD33 (42), pColdTF-FT (43) and pMC1s (ATCC). The mazF-mt9 (Rv2063A locus) and mazE-mt9 (Rv2063 locus) genes were PCR-amplified from M. tuberculosis strain H37Rv genomic DNA with 5′-NdeI/BamHI-3′ ends to create pET-21c-mazF-mt9 and pET-28a-mazE-mt9, respectively. To create pBAD33-mazF-mt9, the pET-21c-mazF-mt9 plasmid was digested with XbaI and HindIII to include the highly efficient T7 phage ribosome binding site, and the resulting fragment was cloned into pBAD33. To create pColdTF-FT-mazF-mt9 and pMC1s-mazF-mt9, pET-21c-mazF-mt9 was digested with NdeI and BamHI, and the resulting fragment was cloned into pColdTF-FT and pMC1s, respectively. To generate sequencing ladders for primer extension analysis in E. coli, the entire ompF transcript or the lpp and ompA CDSs were PCR-amplified from E. coli strain BW25113Δ6 cultures and ligated into Strataclone PCR cloning vectors (Agilent) to create pSC-A-ompF, pSC-A-lpp or pSC-A-ompA, respectively. M. tuberculosis tRNA genes were PCR-amplified as described in more detail later and ligated into a Strataclone PCR cloning vector to create pSC-A-tRNAPro14 or pSC-A-tRNALys43. Clones were confirmed by DNA sequence analysis. All E. coli cells were grown at 37°C in M9 minimal medium supplemented with a final concentration of either 0.2% glucose or 0.1% glycerol. All M. tuberculosis cells were grown in 7H9 Middlebrook medium supplemented with a final concentration of 0.05% Tween 80, 0.5% bovine albumin, 0.2% glucose and 0.085% NaCl (7H9-TW80-ADN). The working concentrations of ampicillin and chloramphenicol in E. coli were 100 and 25 μg/ml, respectively, while the final concentration of kanamycin in M. tuberculosis was 20 μg/ml.
Growth and toxicity assays
MazF-mt9 was expressed in E. coli strain BW25113Δ6 from an arabinose-inducible promoter in pBAD33-mazF-mt9 using a final concentration of 0.2% arabinose and in M. tuberculosis strain H37Rv from a tetracycline-inducible promoter in pMC1s-mazF-mt9 using a final concentration of 200 ng/ml anhydrotetracycline (ATC). For assessing toxicity of MazF-mt9 expression, E. coli transformants harboring either pBAD33 or pBAD33-mazF-mt9 were plated on solid M9 medium with 0.1% glycerol and either with or without a final concentration of 0.2% arabinose and were incubated overnight at 37°C. For growth profiles, E. coli transformants harboring pBAD33-mazF-mt9 were grown overnight at 37°C in M9 liquid medium with 0.2% glucose and diluted to an OD600nm of ≈0.15 in M9 liquid medium with 0.1% glycerol. Cultures were grown at 37°C to an OD600nm of 0.28 (1.5 h post-dilution), the cultures were split into two flasks and arabinose was added to one culture to a final concentration of 0.2% to induce MazF-mt9. For assessing toxicity of MazF-mt9 expression in M. tuberculosis, transformants harboring either pMC1s or pMC1s-mazF-mt9 were serially diluted, plated on solid 7H9-TW80-ADN medium supplemented with or without a final concentration of 200 ng/ml ATC, and incubated overnight at 37°C. For growth profiles in M. tuberculosis, transformants harboring either pMC1s or pMC1s-mazF-mt9 were grown in 7H9-TW80-ADN liquid medium at 37°C with a final concentration of 200 ng/ml ATC to induce MazF-mt9 expression. ATC was added every 48 h, and spent culture supernatant was used as specified in Sharp et al. (44).
Assessment of protein synthesis in vivo
E. coli BW25113Δ6 cells harboring pBAD33-mazF-mt9 were grown in M9 minimal medium with a final concentration of 0.1% glycerol and 1 mM of all amino acids except Cys and Met at 37°C until reaching an OD600nm between 0.2 and 0.3. The culture was then split into equal portions. Arabinose was added to one portion at a final concentration of 0.2%. Aliquots were removed at the indicated times and incubated with 30 μCi of [35S]Met at 37°C. After 1 min of incorporation, the samples were chased with 0.3 mg of cold methionine at 37°C for 5 min. A portion of the culture was applied to a cellulose filter disc (Whatman). The filters were boiled for 30 min in 10% trichloroacetic acid (TCA) and washed three times with 10% TCA and once with acetone. The amount of incorporated radioactivity was determined using a liquid scintillation counter. Cells from each time point were centrifuged, and pellets were resuspended in equivalent volumes of Tris-EDTA buffer (pH 8.0) and 2× Laemmli buffer corresponding to the measured OD600nm. Incorporation was visualized by autoradiography following 17.5% SDS-PAGE.
RNA isolation
Total RNA was isolated from E. coli strain BW25113Δ6 harboring either pBAD33 or pBAD33-mazF-mt9 or M. tuberculosis strain H37Rv grown to mid-logarithmic phase. When E. coli cultures reached an OD600nm of 0.4, the cultures were split into two flasks, arabinose was added to one culture to a final concentration of 0.2%, and growth continued for an additional 15-, 30-, 60-, 90- or 120-min post-induction. Cells were pelleted by centrifugation at 2000 × g for 10 min, and supernatants were removed. E. coli cell pellets were resuspended in acid phenol and lysed for 10 min at 60°C. Lysates were extracted with chloroform and precipitated with ethanol. RNA pellets were dissolved in nuclease-free water, treated with TURBO DNase (Invitrogen) for 45 min at 37°C, extracted with acid phenol chloroform and precipitated with ethanol. M. tuberculosis cell pellets were resuspended in TRIzol Reagent (Invitrogen) and lysed by bead-beating. Lysates were extracted with chloroform and precipitated with isopropanol. RNA pellets were dissolved in nuclease-free water and treated with TURBO DNase (Invitrogen) for 60 min at 37°C.
Northern analysis of steady-state mRNA and tRNA levels
To detect mRNA, 25 μg of total RNA from E. coli was loaded onto a 1.2% (wt/vol) agarose, 2% (vol/vol) formaldehyde gel and visualized by adding ethidium bromide and exposing to UV light. To detect tRNA, 2 μg of total RNA from E. coli was separated on a 9% polyacrylamide, 7 M urea gel and visualized by SYBR Gold (Invitrogen) staining. RNA was transferred to a nylon Hybond-N+ membrane (GE Healthcare) and hybridized with radiolabeled DNA overnight at 42°C. To detect E. coli tRNALys, a DNA oligonucleotide specific for that tRNA species (NWO2358, 5′- AGTCAACTGCTCTACCAACTGAGC -3′) was radiolabeled at the 5′ end by treating with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (PerkinElmer) for 1 h at 37°C. To detect mRNA, the radiolabeled DNA for northern analysis was generated with a random-primed DNA labeling kit (Roche) using PCR products amplified from the CDSs of E. coli genes lpp, ompA, ompC, ompF, ompX, rplB, rpsA and tsf, or from a portion of two transcripts containing tufA. E. coli contains two genes, tufA and tufB, that encode elongation factor EF-Tu and whose nucleotide sequences are 99% identical. To detect tufA but not tufB transcripts, a tufA-specific fragment was created using primers that overlap a portion of the upstream gene fusA and end just before the start of the tufA CDS. This fragment hybridizes to four mRNAs, including a transcript containing fusA and tufA, two transcripts of roughly equal size containing only tufA, and one containing rpsL, rpsG, fusA and tufA.
In vivo primer extension analysis
Total RNA (25 μg) from E. coli expressing MazF-mt9 was used in primer extension reactions, and sequencing ladders were generated by using 5 μg of plasmids carrying E. coli genes (pSC-A-lpp, pSC-A-ompA or pSC-A-ompF), [α-32P]dCTP for radiolabeling and a Sequenase version 2.0 DNA sequencing kit (Affymetrix) according to the Sequenase kit protocol, essentially as described in Sharp et al. (44) and Schifano et al. (34). DNA oligonucleotides were radiolabeled at the 5′ end by treating with T4 polynucleotide kinase and [γ-32P]ATP for 1 h at 37°C. The oligonucleotide NWO1171 (5′-TTACTTGCGGTATTTAGTAGCC-3′) was used to detect cleavage in the lpp CDS, the primer NWO1582 (5′-CACTGGTATTCCAGACGGG-3′) was used for the ompA CDS, and primer NWO1614 (5′-CGCTTCATCATTATTTATTACCCTCATGG-3′) was used for the ompF 5′ UTR. Cleavage products were detected by extending 1.4 pmol of gene-specific 5′-end-radiolabeled oligonucleotides with 5 U of avian myeloblastosis virus reverse transcriptase (AMV RT; New England Biolabs) and 20 U of RNase inhibitor (Roche) for 1 h at 42°C. All reactions were electrophoresed on a 6% (wt/vol) polyacrylamide 7 M urea gel and visualized by autoradiography.
Preparation of recombinant MazE-mt9 and MazF-mt9
pET-28a-mazE-mt9 or pColdTF-FT-mazF-mt9 BL21(DE3) transformants were used to inoculate one or four liters of M9 liquid medium and grown to an OD600nm of 0.50 or 0.35, respectively. For pET-28a-mazE-mt9 BL21(DE3) transformants, cultures were induced with a final concentration of 1 mM IPTG and expressed at 37°C for 4 h. For pColdTF-FT-mazF-mt9, the method of purifying a toxin protein was similar to our group's published work (32,43). Cultures were transferred to a 15°C water bath and incubated for 30 min before the protein was induced with a final concentration of 1 mM IPTG and expressed for 20 h. Cells were disrupted by sonication, and extracts were purified by nickel-nitrilotriacetic acid affinity chromatography (Qiagen). To produce sufficient amounts of toxic protein, MazF-mt9 was expressed as a fusion to the 55-kDa molecular chaperone trigger factor (TF). Presumably the relatively large size of the chaperone fusion precludes the cytotoxic effects of overexpressing a toxin in culture. We suspect that TF either blocks dimerization of toxin monomers, precludes proper folding of the toxin or inhibits interaction between the toxin and its RNA target. After cell lysis, His6-TF-FLAG-MazF-mt9 was cleaved with Factor Xa (New England Biolabs) to excise TF from the target protein. FLAG-MazF-mt9 was further purified over an anti-FLAG resin to remove His6-TF, which resulted in approximately 8 nmol of protein. Purity of the toxin was confirmed by SDS-PAGE analysis and silver staining.
In vitro primer extension analysis of MS2 RNA
Cleavage products were detected and DNA sequencing ladders were generated using the following oligonucleotide primers: NWO1724, 5′-CGGCAGTGTACGCCTTCACGAGCG-3′; NWO1726, 5′-GCTGGATACGACAGACGGCCATC-3′; B3, 5′-AGCACACCCACCCCGTTTAC-3′; D, 5′-CCGCTCTCAGAGCGCGGGGG-3′; NWO1463, 5′-GCTTTGTGAGCAATTCGTCCCTTAAG-3′; and NWO1727, 5′-CGCGAAAGAGCCCGGACACGAACG-3′. Oligonucleotides were radiolabeled as above. For primer extension, RNA cleavage and antitoxin rescue was performed with or without a final concentration of 5.0 μM MazF-mt9 or 10.0 μM MazE-mt9 in 10 mM Tris (pH 7.8). For antitoxin inhibition of RNA cleavage, 50 pmol MazF-mt9 was pre-incubated with 100 pmol MazE-mt9 for 20 min at 4°C and 5 min at room temperature before the RNA substrate was added. Cleavage reactions using 0.8 μg (0.77 pmol) of MS2 RNA (Roche) as a substrate were incubated with or without 50 pmol of MazF-mt9 for 45 min at 37°C. Primer extension reactions were then performed at 50°C for 1 h with 1 pmol of 5′-end-radiolabeled primer, 10 U of RNase inhibitor and 2.5 U AMV RT in the supplied enzyme buffer. For DNA sequencing reactions, 1 pmol of 5′-end-radiolabeled primer was annealed to 0.8 μg (0.77 pmol) of MS2 RNA for 30 s at 80°C and allowed to cool to room temperature. The sequencing reactions were then performed at 50°C for 1 h using 10 U of RNase inhibitor and 1 U of AMV RT, as well as dNTPs and ddNTPs from the Sequenase version 2.0 DNA sequencing kit at the concentration recommended by the manufacturer. Samples were electrophoresed and visualized as above.
Identification of MazF-mt9 cleavage sites by RNA-seq
Preparation of RNA for high-throughput sequencing
The procedure to prepare RNA for high-throughput sequencing was essentially as described in Schifano et al. (34), with the major alteration of only isolating RNA with a 5′-OH end and the inclusion of an additional set performed with M. tuberculosis RNA incubated with MazF-mt9 toxin in vitro. Total RNA was harvested from either E. coli cells harboring a pBAD33 plasmid without MazF-mt9, E. coli cells expressing MazF-mt9 from pBAD33-mazF-mt9 or plasmid-free M. tuberculosis cells. For in vivo experiments, total RNA was harvested 90-min post-induction from two E. coli biological replicates to coincide with the start of growth arrest in MazF-mt9-expressing cells (Figure 1B). RNA from each of these biological replicates was also split into two technical replicates, one treated with a MICROBExpress kit (Ambion) to deplete rRNAs and one in which rRNAs were not depleted. For in vitro experiments, 4.48 μg of M. tuberculosis total RNA was incubated with or without 692 pmol of recombinant MazF-mt9 protein for 45 min at 37°C in a final concentration of 10 mM Tris (pH 7.8), and rRNAs were not depleted. With a goal of retaining RNA cleavage fragments <200 nt, total RNA was not passed through an RNeasy Mini Kit (Qiagen). To isolate RNAs with a 5′-OH, 3.25 μg of E. coli RNA or 3.90 μg of M. tuberculosis RNA was treated with 1 U of Terminator 5′-Phosphate-Dependent Exonuclease (Epicentre) to remove RNAs with a 5′-monophosphate (5′-P), followed by phosphorylation by 50 U of OptiKinase (Affymetrix) to convert 5′-OH to 5′-P suitable for ligation. The resultant RNAs were then ligated to the 5′ SOLiD (Sequencing by Oligonucleotide Ligation and Detection) RNA adaptor (5′-CCACUACGCCUCCGCUUUCCUCUCUAUGGGCAGUCGGUGAU-3′). Ligation reactions contained 5′-P RNA, 75 pmol 5′ SOLiD adaptor and 20 U T4 RNA Ligase I (New England Biolabs). Ligations were incubated for 20 h at 16°C. Ligation reactions were electrophoresed on a 6% (wt/vol) polyacrylamide 7 M urea gel in TBE buffer, and RNAs that migrated above the free 5′ adaptor were isolated by gel excision. After ligation, cDNAs were generated by reverse transcription using a primer that contained nine degenerate nucleotides at the 3′ end and a common ‘3’ SOLiD adaptor’ sequence (5′-CTGCTGTACGGCCAAGGCGNNNNNNNNN-3′). For E. coli RNA, the annealing step was performed by mixing 350 to 600 ng of 5′ adaptor-ligated RNA with 60 pmol of the RT primer, while for M. tuberculosis RNA, 250 to 360 ng of 5′ adaptor-ligated RNA was mixed with 50 pmol of the RT primer. These mixtures were annealed by incubating for 3 min at 85°C to allow unfolding of secondary structures in rRNA and tRNA followed by 5 min at 4°C. Reverse transcription was performed by adding a cocktail containing 300 U SuperScript III reverse transcriptase (Invitrogen), reaction buffer, dNTPs and RNase inhibitor to the RT primer-RNA mixture and incubating for 5 min at 25°C, then 60 min at 55°C. The reverse transcriptase was then inactivated by incubating for 15 min at 70°C. Next, to remove the RNA strand from RNA-DNA hybrids, 20 U of RNase H (Ambion) was added, and reactions were incubated for 20 min at 37°C. Samples were electrophoresed on a 10% (wt/vol) polyacrylamide 7 M urea gel, and cDNAs that migrated between ≈125 nt and ≈500 nt were isolated by gel excision. PCR of cDNA was performed with an initial denaturation step of 30 s at 98°C, amplification for 11–14 cycles (denaturation for 10 s at 98°C, annealing for 20 s at 62°C and extension for 10 s at 72°C) and a final extension for 7 min at 72°C using reagents from a SOLiD total RNA-seq kit and primers from a SOLiD RNA barcoding kit (Applied Biosystems). After electrophoresis on a non-denaturing 10% (wt/vol) polyacrylamide gel, amplified DNA that migrated between the 200 bp and 400 bp DNA standards was isolated by gel excision and sequenced using an Applied Biosystems SOLiD system, version 4.0.
Figure 1.
Expression of MazF-mt9 arrests translation and growth. (A) E. coli BW25113Δ6 cells transformed with arabinose-inducible pBAD33 or pBAD33-mazF-mt9 were streaked onto M9 minimal medium plates with (right, +Arab) or without 0.2% arabinose (left, -Arab) and incubated overnight at 37°C. (B) Growth profile with (+F9, grey diamonds) or without (-F9, black squares) addition of arabinose to E. coli cells harboring pBAD33-mazF-mt9. (C) [35S]methionine incorporation in E. coli cells with or without MazF-mt9 expression. Equivalent amounts of cell lysate relative to OD600nm were subjected to SDS-PAGE followed by autoradiography. Numbers indicate the time (min) after MazF-mt9 induction. Data shown are representative of two independent experiments. (D) Quantification of [35S]methionine incorporation relative to OD600nm in E. coli cells with (+F9, grey diamonds) or without (-F9, black squares) MazF-mt9 expression. The initial rate was set to 100%. (E) Effect on colony formation with (grey) or without (black) addition of anhydrotetracycline (ATC) to M. tuberculosis H37Rv cells harboring ATC-inducible pMC1s or pMC1s-mazF-mt9. (F) Growth profile of M. tuberculosis cells harboring pMC1s (black squares) or pMC1s-mazF-mt9 (grey diamonds) upon addition of ATC. (B, D, E, F) Data points are the mean of two (D), three (B, F) or four (E) independent experiments and error bars represent the S.D.
RNA-seq data analysis
We obtained between 4 and 10 million sequencing reads that aligned to the E. coli MG1655 genome (Supplementary Table S1) and between 6 and 7 million sequencing reads that aligned to the M. tuberculosis strain H37Rv genome (Supplementary Table S2). Sequencing reads for which the first 15 or 20 bases mapped with zero mismatches to the E. coli or M. tuberculosis genomes, respectively, were identified using Bowtie version 1.0.0 (45). The number of sequencing reads whose first base aligned to each position in the genome (#5′-ends) was calculated. For E. coli libraries, replicates were averaged. Next, we added a pseudocount to the genomic positions for which the #5′-ends was zero. We then divided the #5′-ends from the analysis of RNA isolated from cells containing MazF-mt9 with the #5′-ends from cells that did not contain MazF-mt9. We identified genomic positions for which this ratio was ≥50 either in the analysis of both biological replicates in E. coli or in the single M. tuberculosis cDNA library. In addition, we required that the position of enrichment represented local maxima within a 30-base window spanning 15 bases up- and downstream. In the analysis of E. coli RNAs carrying a 5′-OH, we identified 163 positions that met these criteria, while in the analysis of M. tuberculosis RNAs carrying a 5′-OH, we identified 48 positions that met these criteria (Supplementary Data 1). For cleavage sites that mapped to more than one position in the E. coli genome due to redundant sequences, the sequence surrounding each cleavage site was only counted once to determine the consensus sequence in Figure 2E. Values for fold-change in Figure 2A, C and F were calculated from the average of the two biological replicates whose rRNAs were not depleted. The RNA sequencing data were deposited in the NCBI Sequence Read Archive under accession code PRJNA289381.
Figure 2.
MORE RNA-seq uncovers MazF-mt9 sequence requirements and tRNA as a new target. (A, C, F) Histograms representing the ratio of sequencing reads identified using MORE RNA-seq at each nt position within the ompA (A) or ompF (C) transcripts or within tRNALys (F) in E. coli cells that did (+F9) or did not express (-F9) MazF-mt9. The cleavage site is labeled with an asterisk (*). (C, D) Negative nucleotide positions correspond to the 5′ UTR (shaded in green), while positive values correspond to the ompF CDS. (B, D, H) The RNA sequences surrounding the cleavage sites (indicated by a black arrow) identified using MORE RNA-seq (A, C, F) and validated with primer extension (Supplementary Figure S2B and C) or northern analysis (Figure 2G), with flanking position numbers from the corresponding E. coli CDS, 5′ UTR, or mature tRNA, respectively. (E) Sequence logo (75) generated by aligning the RNA sequences surrounding the 163 cleavage sites identified using MORE RNA-seq on E. coli cells expressing MazF-mt9. The numbering reflects the nt position relative to the exact point of cleavage, indicated by the yellow arrow. (G) Northern analysis of E. coli tRNALys with expression of toxin MazF-mt9 for the times shown (in min), relative to empty vector (EV) after adding inducer. The red arrow indicates the position of a cleavage product. Tick marks denote a DNA marker and numbers indicate length in nt. (I) Secondary structure (above) of E. coli tRNALys, showing the location of MazF-mt9 cleavage (inset below). The acceptor stem at the 5′ end is highlighted in red, the D-arm in orange, the anticodon arm in light green, the anticodon in dark green, the T-arm in blue and the CCA tail at the 3′ end in purple. Yellow arrows indicate a cleavage site. Numbering in the anticodon indicates mature tRNA nucleotide position. (B, D, H, I) Nt identical to the MazF-mt9 motif UU↓U are in red text.
In vitro synthesis of tRNA
M. tuberculosis tRNAs were synthesized in vitro following the methods described by Sisido et al. (46) with minor modifications. A synthetic DNA oligonucleotide containing the T7 RNA polymerase promoter and the 5′-end of the tRNA gene was annealed to a second oligonucleotide corresponding to the 3′-end of the tRNA. The following oligonucleotide primers were used (T7 RNA polymerase promoter is underlined): tRNAPro14 gene proT forward (Fwd), NWO2098, 5′-GCAGCGAAATTAATACGACTCACTATAGCGGGCTGTGGCGCAGTTTGGTAGCGCACTTGACT-3′; tRNAPro14 gene proT reverse (Rev), NWO2143, 5′-TCGGGCTGACAGGATTTGAACCTGCGACCACTTGACCCCCAGTCAAGTGCGCTACCAAACTGCG-3′; tRNALys43 gene lysT Fwd, NWO2078, 5′-GCAGCGAAATTAATACGACTCACTATAGGCCCCTATAGCTCAGTTGGTAGAGCTACGGAC-3′; tRNALys43 gene lysT Rev, NWO2079, 5′-TTGTGCCCCCACCAGGACTCGAACCTGGGACCTGCGGATTAAAAGTCCGTAGCTCTACCAACTGAGCTATAGGG-3′. The annealed oligonucleotides were then extended using Taq DNA polymerase to create dsDNA containing the entire tRNA gene preceded by the T7 promoter. The product was electrophoresed on a 2% (wt/vol) agarose gel to confirm its size and purified using the QIAquick gel extraction kit (Qiagen). The sequence of the product was confirmed by DNA sequence analysis. 200 ng of tRNA gene dsDNA was then transcribed in vitro for 4 to 6 h at 37°C using the RiboMAX Large Scale RNA Production System (Promega) as recommended by the manufacturer. The transcription reaction was then electrophoresed on a 9% (wt/vol) polyacrylamide 7 M urea gel and visualized by staining with ethidium bromide to confirm the size and purity of the transcribed tRNA. The tRNA transcript was excised from the gel and incubated for 18 h at 37°C in elution buffer (1 mM EDTA, 0.5 M ammonium acetate, 10 mM magnesium acetate, 0.1% SDS). The eluate was collected, and the tRNA was precipitated by ethanol.
In vitro primer extension analysis of tRNAs
Cleavage of tRNAs produced via T7 transcription was performed in vitro following the assay described by Winther and Gerdes (47) with slight modifications. Cleavage reactions using 3 pmol of tRNA were incubated with 30 pmol of MazF-mt9 at 37°C for 5.5 h in tRNA cleavage buffer (10 mM HEPES (pH 7.5), 15 mM potassium chloride, 3 mM magnesium chloride and 10% glycerol). For antitoxin inhibition of RNA cleavage, 30 pmol MazF-mt9 was pre-incubated with 20 pmol MazE-mt9 for 15 min at room temperature before the RNA substrate was added. Cleavage products were detected and DNA sequencing ladders were generated using the following oligonucleotide primers: tRNAPro14 gene proT, NWO2144, 5′-TCGGGCTGACAGGATTTGAACC-3′; tRNALys43 gene lysT NWO2080, 5′-TTGTGCCCCCACCAGGAC-3′. Oligonucleotides were radiolabeled exactly as above, while primer extension reactions were the same as in the ‘In vitro primer extension analysis of MS2 RNA’ section except they were incubated at 55°C. DNA sequencing reactions using 3.5 μg plasmid DNA (pSC-A-tRNAPro14 or pSC-A-tRNALys43) as a template and [α-32P]dATP for radiolabeling were performed with a Sequenase version 2.0 DNA sequencing kit as recommended by the manufacturer. The samples were incubated at 95°C for 5 min prior to electrophoresis on a 15% (wt/vol) polyacrylamide, 7 M urea gel followed by autoradiography.
Statistical analysis of proline, lysine and asparagine codon frequency in M. tuberculosis CDS
The M. tuberculosis strain H37Rv genome (accession number NC_000962) was retrieved from NCBI RefSeq, and all pseudogenes were removed. The counts of each of the 64 possible codons in all CDSs were tallied. These individual counts were divided by the sum of all counts to obtain the probability of each codon appearing in any M. tuberculosis CDS. We defined tRNAPro14-recognized codons as CCC, the perfect reverse complement of the tRNAPro14 anticodon, and CCU, an ‘orphan’ proline codon in M. tuberculosis that does not have a natural anticodon match but can be recognized by tRNAPro14 due to wobble base-pair rules (48). Thus, the probabilities of proline (CCC, CCU), lysine (AAA, AAG) and asparagine (AAC, AAU) codons appearing anywhere in an M. tuberculosis CDS are 0.02047, 0.02041 and 0.02532, respectively. Let p denote their combined probability of 0.0662. Let L be 30 to indicate the first 30 codons in the CDS. The expected number, E, of the described codons among the first 30 codons is:
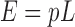 |
Let K be the actual number of these three sets of codons. Then the probability, P, of having K or more of these codons among the first L codons of the CDS is:
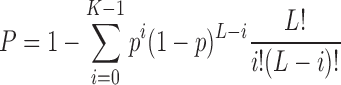 |
To calculate the P-value for the entire CDS from a given mRNA, the value for L is replaced with the number of codons in the CDS. A CDS with a very small P-value suggests that its translation may be sensitive to MazF-mt9 cleavage of tRNAPro14, tRNALys and tRNAAsn isoforms.
In vitro cleavage of tRNALys43 and several short RNAs based on the tRNALys43 ASL
Cleavage of tRNALys43 produced via T7 transcription and of 15-nt RNA oligonucleotides based on the wild-type or mutant tRNALys43 ASL was performed in vitro in a manner similar to that in the tRNA primer extension analysis above. We designed RNA oligonucleotides that either match the wild-type tRNALys43 ASL or contain mutations that disrupt RNA sequence or secondary structure as illustrated in Figure 4. The RNA oligonucleotides based on the tRNALys43 ASL are as follows, with stem-forming bases underlined and mutated bases in bold: NWO2233, wild-type, 5′-CGGACUUUUAAUCCG-3′, Figure 4A, lanes 4–6, and Figure 4B, lanes 1–3; NWO2234, ‘-UUU,’ 5′-CGGACUAAAAAUCCG-3′, Figure 4A, lanes 7–9; NWO2238, ‘shifted UUU,’ 5′-CGGACUAAUUUUCCG-3′, Figure 4A, lanes 10–12; NWO2235, ‘-stem, 5' side,’ 5′-GCCUCUUUUAAUCCG-3′, Figure 4A, lanes 13–15; NWO2236, ‘-stem, -UUU,’ 5′-GCCUCUAAAAAUCCG-3′, Figure 4A, lanes 16–18; NWO2237, ‘restore stem,’ 5′-GCCUCUUUUAAAGGC-3′, Figure 4B, lanes 4–6; and NWO2387, ‘-stem, 3′ side,’ 5′-CGGACUUUUAAAUAU-3′, Figure 4B, lanes 7–9. We used the program RNAstructure (49) to confirm the presence or absence of a stem as desired for each RNA oligonucleotide. To radiolabel RNA oligonucleotides, RNA was first treated with calf intestinal alkaline phosphatase (New England Biolabs) for 60 min at 50°C to remove the 5′-P. Treated RNA was extracted with 25:24:1 phenol:chloroform:isoamyl alcohol and precipitated with ethanol. For size markers, a DNA oligonucleotide and two RNA oligonucleotides were radiolabeled at the 5′ end by treating with T4 polynucleotide kinase and [γ-32P]ATP for 1 h at 37°C. The DNA oligonucleotide NWO1693 (5′-GGAATTCCATATGTCTACATCCACGACGATTAGGG-3′) is 35-nt, while the RNA oligonucleotides are 10-nt (5′-AUCCGGAAUC-3′) and 5-nt (5′-CGCCU-3′). Treated RNA was also radiolabeled at the 5′ end as with the oligonucleotides but with the following exceptions: (i) RNA was initially incubated without enzyme for 10 min at 70°C and immediately placed on ice, (ii) 2.5× more T4 polynucleotide kinase was used, (iii) polyethylene glycol 8000 was added to a final concentration of 5% (wt/vol) and (iv) the labeling reaction was incubated for 1 h at 51°C. Cleavage reactions using 0.1 pmol of radiolabeled RNA were incubated with 1.0 pmol of MazF-mt9 at 37°C for 60 min in tRNA cleavage buffer. For antitoxin inhibition of RNA cleavage, 1.0 pmol MazF-mt9 was pre-incubated with 0.67 pmol MazE-mt9 for 45 min at room temperature before the RNA substrate was added. Radiolabeled or non-radiolabeled samples were electrophoresed on a 15% (wt/vol) polyacrylamide, 7 M urea gel followed by autoradiography or staining with SYBR Gold.
Figure 4.
MazF-mt9 requires both sequence and structural determinants for tRNA cleavage. (A, B) tRNALys43 and mutated ASL derivatives illustrated as in Figure 3. RNA incubated with or without MazF-mt9 (F9) or MazE-mt9 (E9) as indicated. Numbering in RNA oligonucleotides corresponds to the nt position of anticodon residues in the full-length tRNALys43. Dark green highlights the anticodon position. Nt identical to the MazF-mt9 UUU motif are in red text, while nt mutated from the wild-type tRNALys43 ASL are in blue text. Yellow arrows indicate cleavage sites. Black lines connecting complementary nt indicate base-pairing as part of the anticodon stem. (A) RNA was radiolabeled at the 5′ end prior to addition of toxin. The red arrows correspond to a ≈35-nt or a ≈8-nt RNA fragment generated by MazF-mt9 cleavage. Tick marks indicate either a 35-nt DNA oligonucleotide or 10-nt and 5-nt RNA oligonucleotides, each containing a 5′-end-radiolabel. (B) RNA cleavage was visualized by staining with SYBR Gold. We empirically determined that a 10-nt but not a 5-nt RNA oligonucleotide can be visualized by SYBR Gold staining and that RNA oligonucleotides predicted to form a stem stain with more intensity than linear oligonucleotides. Thus, the ≈8-nt RNA fragments expected in lanes 3 and 6 cannot be seen here, and RNA in lanes 7–9 appears less intense than RNA in lanes 1, 2, 4 and 5 despite the fact that equal molar amounts of all oligonucleotides were loaded in all lanes. Tick marks indicate a DNA marker.
Northern analysis of M. tuberculosis tRNA cleaved in vitro
To assess MazF-mt9 cleavage activity, 2 μg of M. tuberculosis total RNA was incubated with 30 pmol of MazF-mt9 in a 10 μl reaction containing tRNA cleavage buffer at 37°C for 0, 6 or 24 h. To assess for cleavage inhibition by the cognate antitoxin and control from contaminating ribonucleases, 20 pmol MazE-mt9 was pre-incubated with MazF-mt9 for 15 min at room temperature before the RNA substrate was added and incubated at 37°C for 24 h. The products of these reactions were separated on a 9% (wt/vol) polyacrylamide, 7 M urea gel and visualized by SYBR Gold staining. RNA was transferred to nitrocellulose, and RNA was then hybridized with a radiolabeled oligonucleotide specific for M. tuberculosis tRNALys43, NWO2267, 5′-GCAGTGAGCCCCATTCG-3′.
RESULTS
Expression of MazF-mt9 arrests translation and growth
Expression of the MazF-mt9 toxin leads to growth arrest in E. coli (50) and M. smegmatis (6). To establish whether or not the growth inhibition observed upon MazF-mt9 expression (Figure 1A and B) is the result of translation inhibition, we metabolically labeled E. coli cells with [35S]Met in the presence or absence of MazF-mt9 at intervals corresponding to those in the growth profile from 0 to 180 min (Figure 1C and D). To limit potential trans-activation of endogenous E. coli toxins (51–53), we performed all E. coli experiments in a BW25113Δ6 strain carrying deletions of six TA systems—chpBIK, dinJ-yafQ, hipBA, mazEF, relBE and yefM-yoeB (41). The results of this analysis indicated that inhibition of translation was the underlying cause of the growth arrest by MazF-mt9 since translation levels declined significantly before growth arrest was detected. In particular, there was no difference between the growth rate of MazF-mt9-induced and control cultures until 60 min post-induction (Figure 1B), yet there was a steep decline in de novo protein synthesis between 20 and 30 min post-induction (Figure 1C and D). In addition, we demonstrated that MazF-mt9 expression in M. tuberculosis also led to a reduction in the recovery of colony forming units and to growth arrest (Figure 1E and F). Expression of MazF-mt9 inhibited colony formation by nearly 200-fold relative to the uninduced control (10 versus 1914 CFU/ml; Figure 1E) and arrested growth in M. tuberculosis for up to 12 d post-induction (Figure 1F). Therefore, our data revealed that MazF-mt9 expression inhibits growth in E. coli and M. tuberculosis, consistent with published reports in E. coli (50) and M. smegmatis (6).
Determinants for MazF-mt9 cleavage recognition are more complex than for other MazF toxins
To define the RNA sequence determinants for MazF-mt9 cleavage, we first used a conventional approach involving MazF-mt9 expression in E. coli followed by a combination of northern and primer extension analyses on naturally abundant mRNAs. Upon northern analysis of the lpp, ompA and ompF transcripts, we detected a gradual decrease in the levels of full length RNA upon MazF-mt9 expression (Supplementary Figure S1). We then performed primer extension on these three RNAs and detected only a single cleavage site in each (Supplementary Figure S2). Although the three cleavage sites were all flanked by uridine nucleotides on either side (U↓U) and two of the cleavage sites contained a UU↓U motif, there was not enough similarity in the RNA sequences surrounding the cleavage sites to determine a definitive consensus sequence. Therefore, we next surveyed six additional mRNAs—ompC, ompX, rplB, rpsA, tsf and tufA—by northern analysis but were unable to detect any evidence of MazF-mt9 cleavage (Supplementary Figure S3). Finally, we incubated the ≈3.5-kb bacteriophage MS2 RNA with or without purified MazF-mt9 toxin and performed primer extension analysis to obtain more MazF-mt9 cleavage sites (Supplementary Figure S4). In total, we identified 11 MazF-mt9 cleavage sites, including five ‘major’ sites that exhibited robust cleavage and six ‘minor’ sites with relatively weak signals (Supplementary Table S3). Alignment of the RNA sequences flanking the MazF-mt9 cleavage sites only revealed a U↓U sequence in common between these 11 sites (Supplementary Table S3). This 2-nt UU sequence is predicted to occur once every 42 or 16 nucleotides, or ≈1,400 times collectively in the 10 RNAs we tested, assuming an equal base content, equal representation of all potential cleavage motifs and no secondary structure. Because we observed cleavage at <1% of the UU sequences, MazF-mt9 appeared to require more than a UU motif for substrate recognition and cleavage.
MORE RNA-seq uncovers MazF-mt9 sequence requirements and tRNA as a new target
To further define the sequence determinants for MazF-mt9 cleavage, we used MORE RNA-seq (34) to survey the transcriptome of E. coli. MORE RNA-seq is designed to isolate a single class of RNA depending on the nature of the chemical moiety (a monophosphate or a hydroxyl group) at the 5′ end of a transcript. Since cleavage by MazF toxins produces an RNA fragment with a 5′-hydroxyl (5′-OH) group (25,34,36,54,55), MORE RNA-seq enables identification of cleavage sites by isolating RNA with a 5′-OH and searching for genomic positions enriched when the MazF toxin is induced compared to when the toxin is absent. Having previously demonstrated that MORE RNA-seq readily determines a cleavage recognition sequence and precisely maps the position of cleavage (34), we applied this approach to MazF-mt9. To this end, we harvested total RNA from E. coli cells harboring either a vector that directs the synthesis of MazF-mt9 or an empty plasmid, selected for transcripts bearing a 5'-OH, prepared cDNA for high-throughput sequencing, and identified 163 unique cleavage sites. As validation of our MORE-RNA-seq approach, two of the 163 sites—one within the ompA coding sequence (CDS) (Figure 2A and B) and another within the ompF 5′ UTR (Figure 2C and D)—matched cleavage sites we previously identified by northern (Supplementary Figure S1) and primer extension analyses (Supplementary Figure S2B and C). Alignment of the RNA sequences 20 bases up- and downstream of the 163 cleavage sites revealed a 3-base UUU consensus sequence from –2 to +1 (where the position of cleavage is immediately 5′ of the +1 position; Figure 2E).
Surprisingly, six of the 163 cleavage sites were within tRNA, widely presumed to be refractory to MazF toxin recognition because of its extensive secondary structure (1,26,35,56,57). However, MazF-mt9 cleaved these tRNAs predominantly within their single-stranded loop regions. In fact, a cleavage site within the UU↓U anticodon of tRNALys (identical genes lysT, lysW, lysY, lysZ, lysQ and lysV; Figure 2F–I) was enriched 620-fold relative to the control, ranking third among all MazF-mt9 target RNAs identified by MORE RNA-seq. To validate the tRNA cleavage identified by MORE RNA-seq, we performed northern analysis of tRNALys upon expression of MazF-mt9 (Figure 2G) and observed the appearance of an RNA fragment whose size was consistent with cleavage within the UUU anticodon (Figure 2H and I). In addition, tRNALys was cleaved between 0 and 15 min after MazF-mt9 induction (Figure 2G), preceding translation inhibition at 30 min (Figure 1C and D) and arrest of bacterial growth at 60 min (Figure 1B). However, cleavage of three mRNAs was not observed until 60 min (Supplementary Figure S1), the same time that growth arrest started. Therefore, MORE RNA-seq and northern analyses led to three important conclusions: (i) that the UUU sequence is essential for MazF-mt9 cleavage in vivo, (ii) that tRNAs represent a new class of target for this MazF toxin family member and (iii) that tRNA cleavage—not mRNA cleavage—appears to be the underlying cause of MazF-mt9-triggered inhibition of protein synthesis and bacterial growth.
MazF-mt9 preferentially targets a subset of M. tuberculosis tRNAs
Having identified cleavage sites of MazF-mt9 in E. coli using MORE RNA-seq, we then sought to identify RNA targets in the native context of the M. tuberculosis transcriptome. We adapted our RNA-seq approach to identify cleavage sites generated upon incubation of M. tuberculosis total RNA with recombinant MazF-mt9. We identified 48 cleavage sites that were enriched in the presence of MazF-mt9 (Supplementary Data 1). Alignment of the RNA sequences 20 bases up- and downstream of the 48 enriched sites revealed a very strong 3-base UUU consensus sequence from –2 to +1 (Figure 3A). This cleavage consensus sequence is in agreement with that derived from MORE RNA-seq (Figure 2E), but is much stronger, as 90% (43 of 48) of the cleavage sites are identical to the consensus and 96% (46 of 48) match at two out of three positions. Taken together, our RNA-seq data established that the preferred MazF-mt9 recognition motif is UU↓U. However, this short 3-nt motif alone does not account for MazF-mt9 target recognition as evidenced by the relatively few cleavage sites detected. As with E. coli, the identity of RNAs targeted provided insight into the determinants required for MazF-mt9 cleavage.
Figure 3.
MazF-mt9 preferentially targets a subset of M. tuberculosis tRNAs. (A) Sequence logo (75) generated by aligning the RNA sequences surrounding the 48 cleavage sites identified by applying our RNA-seq method to M. tuberculosis total RNA incubated with MazF-mt9. (B, F) Histograms representing the ratio of sequencing reads at each nt position within tRNAPro14 (B) or tRNALys43 (F) in M. tuberculosis total RNA incubated with (+F9) or without (-F9) MazF-mt9. The cleavage site is labeled with an asterisk (*). (C, G) Primer extension analysis of M. tuberculosis tRNAPro14 (C) or tRNALys43 (G) incubated with or without toxin MazF-mt9 (F9) and antitoxin MazE-mt9 (E9). ‘G, A, T, C,’ sequencing ladder. The red arrow indicates the position of a cleavage product. (D, H) The RNA sequences surrounding the cleavage sites (indicated by a black arrow) identified by RNA-seq (B, F) and primer extension analysis (C, G), with flanking position numbers from the corresponding M. tuberculosis tRNA. (E, I) Secondary structure (above) of M. tuberculosis tRNAPro14 (E) or tRNALys43 (I), showing the location of MazF-mt9 cleavage (inset below). The sequence of the region of tRNALys43 shown is identical to E. coli tRNALys in Figure 2I. Labeling of tRNA secondary structures is the same as in Figure 2.
Remarkably, tRNAs comprise two of the highest ranking MazF-mt9-mediated cleavage sites detected by RNA-seq (Supplementary Data 1). A cleavage site in tRNAPro14 (gene proT; numbering based on the Genomic tRNA Database (58); Figure 3B–E) exhibited the overall highest fold increase and was enriched 1,617-fold relative to the control (Figure 3B). Cleavage of tRNALys43 (gene lysT; Figure 3F–I) was enriched 511-fold relative to the control (Figure 3F), and occurred at an analogous position to its highly conserved tRNALys counterpart cleaved in vivo in E. coli (Figure 2I).
To validate the tRNA cleavage identified by RNA-seq, we performed primer extension analysis of tRNAPro14 and tRNALys43 incubated with MazF-mt9 (Figure 3C and G) and confirmed that MazF-mt9 cleaved these tRNAs within the UUU motif (Figure 3D and H), each at the same location (Figure 3E and I) as detected by RNA-seq. Additionally, four other tRNAs—tRNALys19, tRNAVal22, tRNALeu13 and tRNAAsn36—were among the 48 targets, although they were enriched an order of magnitude less than tRNALys43. Overall, MazF-mt9 cleaved six of the 45 M. tuberculosis tRNAs, confirming tRNA as a new target for a MazF toxin.
MazF-mt9 cleavage of tRNAPro14, tRNALys and tRNAAsn is predicted to reduce the abundance of many PE/PPE proteins and essential components of the translation machinery
Given that MazF-mt9 only cleaves a small subset of tRNAs—including all isoacceptor tRNAs for tRNALys (two) and tRNAAsn (one), as well as one of the three isoacceptor tRNAs for proline, tRNAPro14—this toxin might not inhibit the synthesis of all proteins in M. tuberculosis equally. Thus, we explored the potential effects of MazF-mt9 activity on translation by performing a series of statistical analyses on M. tuberculosis CDSs to identify protein-coding transcripts that contain more or fewer than the expected number of these proline, lysine or asparagine codons. Transcripts containing a large number of codons recognized by tRNAPro14, tRNALys or tRNAAsn should be more sensitive to translation inhibition by MazF-mt9, while transcripts with few or none of these codons should be resistant to translation inhibition. Antitoxin MazE-mt9 is among only five CDSs that do not contain any of the codons targeted by MazF-mt9. Curiously, the class of proteins predicted to be the most susceptible to MazF-mt9 is different depending on the parameters used to sort the data.
We first analyzed all M. tuberculosis CDSs for the abundance of proline, lysine or asparagine codons. Among the top 22 proteins predicted to be susceptible to MazF-mt9 based on our statistical analysis described in the Materials and Methods (Supplementary Table S4), 86% (19 of the top 22) belong to the proline-glutamate (PE)/proline-proline-glutamate (PPE) family, so named because they contain a conserved, single PE or PPE motif at their N terminus. Likewise, of the top 25 CDSs with the highest actual number of MazF-mt9-targeted codons (Supplementary Table S5), 16 (64%) are members of the PE/PPE protein family. Members of this large PE/PPE protein family (comprising nearly 10% of the M. tuberculosis CDS) have intriguing connections to pathogenesis, virulence, antigenic variation and the host immune response (59,60). The proposed high sensitivity of PE/PPE proteins to MazF-mt9 cleavage of tRNAs is likely due to the overrepresentation of asparagine codons in (GGAGGN)n or GN-rich amino acid repeats in approximately half of the PE/PPE family members.
In our third analysis, we prioritized the data based on the abundance of proline, lysine or asparagine codons at the beginning of all M. tuberculosis CDSs. We chose the first 30 codons since there is experimental evidence—ribosome profile experiments (61) and translation efficiency studies with early rare codons (62–64)—demonstrating that translation proceeds at a slower rate and has a greater chance of stalling within the first 25 to 40 codons. Of the top 28 proteins whose synthesis was predicted to be the most sensitive to MazF-mt9 using this analysis, 36% (10 of 28) are directly involved in translation (ribosomal proteins; Supplementary Table S6). This analysis suggests that by targeting tRNAPro14, tRNALys and tRNAAsn, MazF-mt9 broadly inhibits translation by blocking synthesis of essential components of the M. tuberculosis translation machinery.
MazF-mt9 recognizes RNA sequence and structure within the tRNALys43 anticodon stem-loop
To ascertain which sequence or structural elements in RNA are required for MazF-mt9 target selection and cleavage, we first sought to find a minimal tRNA-based MazF-mt9 substrate. We selected the anticodon stem-loop (ASL) of M. tuberculosis tRNALys43 for three reasons. First, although MazF-mt9 cleaves each of the two tRNALys isoacceptors present in M. tuberculosis, only tRNALys43 contains a complete UUU motif. Second, we confirmed MazF-mt9 cleavage of M. tuberculosis tRNALys43 upon northern analysis using an isoacceptor-specific oligonucleotide complementary to the ASL (Supplementary Figure S5). Third, an E. coli tRNALys isoacceptor has a high degree of sequence and structural identity to M. tuberculosis tRNALys43 and is also cleaved within this conserved UUU motif in the anticodon, indicating that this tRNA is cleaved in vitro and in vivo.
We next developed an in vitro cleavage assay to test whether secondary structure contributes to MazF-mt9 recognition of tRNA. Briefly, short RNA oligonucleotides were designed that either match the wild-type tRNALys43 ASL or contain mutations that disrupt RNA sequence or secondary structure as illustrated at the top of Figure 4A and B. We then incubated each of these 5′-end-radiolabeled RNAs with or without the MazF-mt9 toxin or MazE-mt9 antitoxin, separated the resulting RNA fragments by gel electrophoresis and visualized the radiolabeled RNA by autoradiography. As a control, we first demonstrated that full-length tRNALys43 was cleaved by MazF-mt9 in vitro (Figure 4A, lane 3), consistent with our RNA-seq (Figure 3F) and primer extension (Figure 3G) data. Addition of MazF-mt9 to tRNALys43 produced an RNA fragment ≈35 nt in length, consistent with cleavage at 34UU↓U36 within the anticodon. This cleavage resulted from MazF-mt9 and not a contaminating RNase, since incubation of the MazE-mt9 antitoxin with MazF-mt9 prior to addition of RNA substrate abolished cleavage (Figure 4A, lane 2).
We then tested whether MazF-mt9 was also able to cleave a 15-nt RNA oligonucleotide consisting of the tRNALys43 ASL (Figure 4, lanes 4–6, and Figure 4B, lanes 1–3). As with the entire tRNALys43, MazF-mt9 produced an RNA cleavage fragment (Figure 4A, lane 6, and Figure 4B, lane 3) that was prevented by preincubation with MazE-mt9 (Figure 4A, lane 5, and Figure 4B, lane 2). The RNA fragment resulting from MazF-mt9-mediated cleavage was ≈8 nt in length, consistent with cleavage between the residues that correspond to U35 and U36 within the anticodon.
Having established a minimal RNA substrate for MazF-mt9 cleavage, we then tested which sequence or structural elements are required for MazF-mt9 recognition. First, we replaced the three U nucleotides comprising the anticodon with A residues (Figure 4A, lanes 7–9), which resulted in no detectable MazF-mt9-mediated cleavage (lane 9). This result indicates that the UUU motif is required for MazF-mt9 recognition. Second, we shifted the UUU motif from a position in the anticodon loop directly opposite the stem to the side of the loop immediately adjacent to the stem (Figure 4A, lanes 10–12). This also resulted in no detectable cleavage (Figure 4A, lane 12), suggesting that the UUU motif must be positioned at a specific location within a loop. Third, we mutated the 5′ half of the stem to be identical to the 3′ half instead of complementary to it, thereby destroying the stem and forming a linear RNA with a UUU motif (Figure 4A, lanes 13–15). Despite the presence of the UUU motif in this linear oligonucleotide, this RNA was not cleaved (Figure 4A, lane 15), suggesting that the stem is necessary for MazF-mt9 recognition. Finally, an RNA oligonucleotide lacking both the stem and the UUU motif was not cleaved (Figure 4A, lane 18), which was expected by the lack of cleavage when either the sequence or structural determinant alone was removed (Figure 4A, lanes 9 and 15).
To confirm whether MazF-mt9 requires the stem in the ASL to recognize and cleave tRNALys43, we reconstituted the stem that we disrupted in Figure 4A, lanes 13–15. We mutated the 3′ half of the stem to be complementary to the prior mutations in the 5′ half of the stem, thereby restoring the structure but not the sequence of the stem from the wild-type tRNALys43 ASL (Figure 4B, lanes 4–6). MazF-mt9 cleaved this RNA oligonucleotide alone (Figure 4B, lane 6) but not when preincubated with MazE-mt9 (lane 5), confirming that the stem is necessary for MazF-mt9 recognition. To rule out the possibility that the sequence in the 5′ half of the stem is recognized by MazF-mt9, we mutated the 3′ half of the stem to form a linear oligonucleotide with a UUU motif in the anticodon (Figure 4B, lanes 7–9). This RNA was not cleaved (Figure 4B, lane 9), just as with the linear oligonucleotide mutated at the 5′ half of the stem (Figure 4A, lane 15), confirming that the structure and not the sequence of the stem is crucial. Thus, the results of our in vitro cleavage assays establish that MazF-mt9 recognizes its RNA targets on the basis of both the presence of a UUU sequence and its structural context.
DISCUSSION
MazF-mt9 exhibits two properties distinct from all other MazF family members. First, it appears to preferentially cleave tRNA within single-stranded loops, and second, it exhibits exquisite selectivity for this novel target by requiring both sequence and structural determinants. By comparison, recognition and cleavage by all other characterized MazF toxins require only (i) the presence of the cleavage recognition motif for to that particular toxin, (ii) that this motif resides within a single-stranded region of RNA and (iii) that the motif is free of interacting proteins or a tertiary fold that can sterically block the toxin. In addition, our findings also contradict earlier assumptions that all MazF toxins only cleave mRNAs and cannot cleave tRNA (1,26,35,56,57) due to its extensive double-stranded regions and conserved tertiary fold. Finally, our results also expose unforeseen functional parallels between MazF-mt9 and some VapC toxins that also primarily target tRNA (47,65). Relationships between toxin families have been forged based on compelling sequence and structural similarities (5,66–68). However, unlike the clear relationships within the MazF toxin family (including ChpBK and PemK/Kid) and the RelE family (including HigB and YoeB), the MazF and VapC families have been kept functionally distinct because they do not exhibit significant sequence or structural similarities. However, our results reveal that protein sequence and structure alone cannot always predict functional relationships between individual toxins.
Collectively, our data indicate that mRNA is not a preferred target of MazF-mt9. We only observed 23 CDSs cleaved by MazF-mt9 in our RNA-seq results, a small fraction of the 3803 M. tuberculosis protein-coding genes. By comparison, 99% of E. coli CDSs are susceptible to targeting by the ACA-cleaving MazF. This dearth of sites within CDSs in our RNA-seq data set is consistent with the relative lack of MazF-mt9-mediated cleavage detected by northern and primer extension analyses despite interrogation of a large number of mRNA substrates (Supplementary Figures S1–S3 and Table S3). In fact, MazF-mt9 cleaved UUU motifs within these mRNA transcripts at a rate of only 2%, suggesting that most mRNAs do not contain the required structural determinant(s) for MazF-mt9 cleavage. In contrast, the relatively few mRNAs that are cleaved appear to be targeted because they possess the sequence and structure required by MazF-mt9. For example, when we predicted the secondary structure of the top 10 MORE RNA-seq cleavage sites in mRNA using RNAstructure (49), all 10 mRNA structures cleaved by MazF-mt9 resemble tRNA arms (Supplementary Figure S6), with the cleavage sites located in a short (≤11-nt) single-stranded loop directly opposite a stem as with the MazF-mt9-mediated tRNA cleavage sites we observed (Figures 2I and 3E and I, Figure 4, and Supplementary Figure S5). Likewise, nine of the top 10 mRNAs in M. tuberculosis cleaved by MazF-mt9 and detected using our RNA-seq approach resemble an arm of tRNA (Supplementary Figure S7). Finally, MazF-mt9 cleaved tRNALys (Figure 2G) at a faster rate than three mRNAs (Supplementary Figure S1), suggesting that tRNA cleavage and not mRNA cleavage was the underlying cause of MazF-mt9-mediated translation inhibition and subsequent growth arrest. There was a sharp decline in tRNALys between 0 and 15 min after induction of MazF-mt9 (Figure 2G) that preceded translation inhibition around 30 min (Figure 1C and D) and growth inhibition at 60 min (Figure 1B). However, cleavage of the three mRNAs was not detected until 60 min (Supplementary Figure S1), the same time as commencement of growth inhibition, suggesting that at least these three mRNAs are secondary targets of MazF-mt9 when compared with tRNALys.
In terms of sequence requirements, MazF-mt9 prefers to cleave RNA within a UU↓U motif. Yet while the UUU sequence is necessary for cleavage, it is not sufficient. The UUU must be in the preferred structural context. Our initial experiments revealed that MazF-mt9 only cleaved 2% (6 of 251) of the UUU motifs in all the RNA substrates we tested using primer extension analysis of abundant E. coli mRNAs and MS2 RNA (Supplementary Table S3). By contrast, other MazF toxins typically cut at all or nearly all cleavage sequences. For example, MazF-mt6 cleaves all UUCCU cleavage motifs present in the three E. coli mRNAs (ompA, ompF and tufA) (32). Also, the MazF-mt9 recognition motif UUU is represented 18,701 times in the M. tuberculosis transcriptome (either as a solitary UUU sequence or as a 4- to 8-nt string of U residues), yet we detected a very limited number of individual RNAs preferentially cut by MazF-mt9 in our RNA-seq analysis (i.e. 48, or 0.3% of the UUU sequences in the transcriptome). Then, once we identified certain tRNAs as a primary targets we noted that several UUU motifs in single-stranded regions of these tRNAs were not cleaved, suggesting MazF-mt9 also requires another determinant for target recognition. We also determined that the MazF-mt9-mediated cleavage sites in mRNA (Supplementary Figures S5 and S6) were largely in the same context as the cleavage sites within tRNAs (i.e. in a stem directly opposite a loop), while the majority of uncut UUU motifs in mRNAs were not. Finally, mutagenesis of the tRNALys43 ASL to create variants in the sequence or position of the UUU motif or in the structure of the stem revealed that cleavage by MazF-mt9 requires that the UUU be positioned in the proper structural context.
Interestingly, at least two major positions in the tRNA architecture were cleaved, both occurring in the loop portion of a stem-loop. MazF-mt9 cleaved tRNAPro14 within a UU↓U sequence in the D-loop (Figure 3E) and cleaved tRNALys43 within the UU↓U anticodon in the ASL (Figure 3I). Importantly, M. tuberculosis tRNALys43 (Figure 3I) and E. coli tRNALys (Figure 2I) are identical in nucleotide length and structure, and the anticodon loop of tRNALys43 is 100% identical to its counterpart in E. coli tRNALys, which was also cleaved by MazF-mt9 in vivo (Figure 2F and G). Therefore, tRNALys isoacceptors from both bacteria are cleaved at the same position within the UU↓U anticodon (Figures 2I and 3I). However, no counterpart of MazF-mt9 substrate tRNAPro14 from M. tuberculosis is cleaved in E. coli, most likely because tRNAPro14 is the only tRNA of all 45 M. tuberculosis tRNAs and all 88 E. coli tRNAs that contains a complete UUU motif in the D-loop. The ability of MazF-mt9 to cleave tRNA at these two positions means this toxin not only generates tRNA halves but also smaller tRNA-derived fragments, which together comprise an intriguing class of small RNAs recently found in RNA-seq data sets from all domains of life and whose functions are just beginning to be determined experimentally (69,70). Overall, MazF-mt9 cleaved the majority of its tRNA targets within the anticodon arm, similar to bacterial tRNases including colicins D and E5 (71), anticodon nuclease PrrC, which exclusively targets E. coli tRNALys (72), and three recently characterized VapC toxins (47,65).
Cleavage of tRNA is presumably sufficient to impair its function as a translation adaptor molecule and inhibit protein synthesis. Therefore, we performed statistical analyses to identify M. tuberculosis proteins whose translation is predicted to be impaired upon reduction of the major tRNAs cleaved by MazF-mt9 (Supplementary Tables S4–S6). Depending on the parameters selected for sorting, we identified two predominant classes of susceptible proteins: PE/PPE proteins or ribosomal proteins. It is not clear which of these predicted scenarios most closely reflects in vivo events. If the goal is to arrest bacterial growth or initiate cell death, however, it is unclear why these toxins and other bacterial tRNases only target specific, and sometimes low-usage, tRNAs. As such, it is intriguing to speculate that MazF-mt9-generated tRNA halves or fragments may have additional functional roles or be able to inhibit translation by interacting directly with the translation machinery as in eukaryotes or archaea (69,70).
In addition to having a preference for cleaving tRNA, MazF-mt9 also targeted other M. tuberculosis noncoding regions. The 12 cleavage sites in our RNA-seq data set that mapped to 5′ untranslated regions (5′ UTRs) or intergenic regions (IGRs) should affect translation of multiple proteins, as all the downstream CDSs in a polycistronic operon normally depend on translation of the upstream genes. Four of the cleavage sites within IGRs are upstream of the three M. tuberculosis rRNA genes, which are located in a single operon. Since maturation of rRNAs requires distinct secondary structures in the precursor transcript to be recognized in a series of stepwise cleavage events by several endoribonucleases, it is expected that MazF-mt9-mediated cleavage at any of these four IGRs would inhibit rRNA maturation and ribosome biogenesis. These predictions are complementary to the results of our statistical analyses in Supplementary Table S6, where ribosomal proteins comprise >35% of the top 28 M. tuberculosis proteins whose synthesis are predicted to be lessened as a result of the downstream effects of MazF-mt9 cleavage of its tRNA targets.
Our RNA-seq data also showed that M. tuberculosis rRNAs were cleaved at six different locations by MazF-mt9, including a single site in 16S rRNA and five others in 23S rRNA (Supplementary Data 1). However, these sites appear to be an artifact of cleaving M. tuberculosis rRNA in the absence of ribosomal proteins, because the corresponding rRNA sites were not cleaved when MazF-mt9 was expressed in E. coli, despite the overall high degree of sequence and structural conservation between rRNAs from these two bacteria. It is likely that these putative rRNA cleavage sites, which arose from an in vitro cleavage assay using M. tuberculosis total RNA, would normally be protected by ribosomal proteins under physiological conditions. This argument is strengthened by the fact that specific ribosomal proteins bind immediately adjacent to or within all six of the potential cleavage sites in M. tuberculosis rRNA (73,74). Conversely, other RNA species from E. coli and M. tuberculosis were cleaved by MazF-mt9 at identical sites, such as several UUU or UU motifs conserved between tRNAs in both bacteria. This suggests that although the putative rRNA cleavage sites in M. tuberculosis would not be cleaved by MazF-mt9 in the context of the native ribosome, most other M. tuberculosis target RNAs (Supplementary Data 1) likely represent bona fide MazF-mt9 targets since none of them are known to form stable interactions with protein complexes.
In total, our data suggest that MazF-mt9-mediated cleavage of specific tRNAs controls M. tuberculosis growth by directly and indirectly targeting essential components of the translation machinery. This mechanism has intriguing parallels to the eukaryotic stress response, where the accumulation of tRNA halves and other species analogous to those created by MazF-mt9 are implicated in regulation of not only translation, but also apoptosis and RNA interference (69,70). Since MazF-mt9 cleaves all lysine tRNAs, the only asparagine tRNA and the second most abundant tRNAPro (codon usage 29%) in M. tuberculosis, MazF-mt9 is predicted to inhibit or reduce the rate of protein synthesis from any transcript coding for an abundance of these three amino acids. One of the three computational approaches we performed to predict proteins sensitive to MazF-mt9 revealed a large percentage involved in translation (Supplementary Table S6). In addition, MazF-mt9 cleavage within the rRNA precursor transcript at regions upstream of all three rRNAs likely inhibits their maturation and assembly into the ribosome, which would also negatively impact translation. This targeting of a handful of tRNAs, along with cleavage of other RNAs, suggests that MazF-mt9 launches a multipronged attack on the translation machinery to downregulate protein synthesis as a tool to regulate cell growth.
Supplementary Material
Acknowledgments
We thank Jared Sharp for determining the ideal conditions for expression of toxins in M. tuberculosis, Jared Knoblauch for bioinformatics analyses and Christine Dunham for helpful discussions.
Author contributions: J.M.S. designed and performed experiments, analyzed data, and wrote the paper; J.W.C. designed and performed experiments and analyzed data; R.E. performed experiments; I.O.V. designed experiments and analyzed data; M.O. developed analytic tools and analyzed data; and R.N.H., B.E.N., and N.A.W. designed experiments, analyzed data and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R21 AI072399 to N. A. W. and R. N. H., R01 GM095693 to N. A. W., R56 AI119055 to N. A. W., R01 GM088343 and GM096454 to B.E.N., and T32 AI007403 to J. M. S. and J.W.C., awarded to G. Brewer].
Conflict of interest statement. None declared.
REFERENCES
- 1.Suzuki M., Zhang J., Liu M., Woychik N.A., Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell. 2005;18:253–261. doi: 10.1016/j.molcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 3.Maisonneuve E., Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 4.Wang X., Wood T.K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey D.P., Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramage H.R., Connolly L.E., Cox J.S. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rustad T.R., Harrell M.I., Liao R., Sherman D.R. The enduring hypoxic response of Mycobacterium tuberculosis. PloS One. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiwari P., Arora G., Singh M., Kidwai S., Narayan O.P., Singh R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 2015;6:6059. doi: 10.1038/ncomms7059. [DOI] [PubMed] [Google Scholar]
- 9.Rand L., Hinds J., Springer B., Sander P., Buxton R.S., Davis E.O. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 2003;50:1031–1042. doi: 10.1046/j.1365-2958.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 10.Albrethsen J., Agner J., Piersma S.R., Hojrup P., Pham T.V., Weldingh K., Jimenez C.R., Andersen P., Rosenkrands I. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol. Cell. Proteomic. 2013;12:1180–1191. doi: 10.1074/mcp.M112.018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betts J.C., Lukey P.T., Robb L.C., McAdam R.A., Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 12.Cappelli G., Volpe E., Grassi M., Liseo B., Colizzi V., Mariani F. Profiling of Mycobacterium tuberculosis gene expression during human macrophage infection: upregulation of the alternative sigma factor G, a group of transcriptional regulators, and proteins with unknown function. Res. Microbiol. 2006;157:445–455. doi: 10.1016/j.resmic.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Fontan P., Aris V., Ghanny S., Soteropoulos P., Smith I. Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect. Immun. 2008;76:717–725. doi: 10.1128/IAI.00974-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korch S.B., Contreras H., Clark-Curtiss J.E. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 2009;191:1618–1630. doi: 10.1128/JB.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart G.R., Wernisch L., Stabler R., Mangan J.A., Hinds J., Laing K.G., Young D.B., Butcher P.D. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 16.Denkin S., Byrne S., Jie C., Zhang Y. Gene expression profiling analysis of Mycobacterium tuberculosis genes in response to salicylate. Arch. Microbiol. 2005;184:152–157. doi: 10.1007/s00203-005-0037-9. [DOI] [PubMed] [Google Scholar]
- 17.Provvedi R., Boldrin F., Falciani F., Palu G., Manganelli R. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology. 2009;155:1093–1102. doi: 10.1099/mic.0.024802-0. [DOI] [PubMed] [Google Scholar]
- 18.Singh R., Barry C.E. 3rd, Boshoff H.I. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 2010;192:1279–1291. doi: 10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao M.C., Rubin E.J. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu. Rev. Microbiol. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 20.Gengenbacher M., Kaufmann S.H. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 2012;36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerdes K., Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 22.Keren I., Minami S., Rubin E., Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio. 2011;2:e00100–e00111. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aizenman E., Engelberg-Kulka H., Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen S.K., Pedersen K., Hansen F.G., Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Zhang J., Hara H., Kato I., Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Zhang J., Hoeflich K.P., Ikura M., Qing G., Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 27.Amitai S., Yassin Y., Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 2004;186:8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazan R., Sat B., Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 2004;186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erental A., Sharon I., Engelberg-Kulka H. Two programmed cell death systems in Escherichia coli: an apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012;10:e1001281. doi: 10.1371/journal.pbio.1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu L., Zhang Y., Teh J.S., Zhang J., Connell N., Rubin H., Inouye M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- 31.Zhu L., Phadtare S., Nariya H., Ouyang M., Husson R.N., Inouye M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 2008;69:559–569. doi: 10.1111/j.1365-2958.2008.06284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schifano J.M., Edifor R., Sharp J.D., Ouyang M., Konkimalla A., Husson R.N., Woychik N.A. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 2013;110:8501–8506. doi: 10.1073/pnas.1222031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schifano J.M., Woychik N.A. 23S rRNA as an a-Maz-ing new bacterial toxin target. RNA Biol. 2014;11:101–105. doi: 10.4161/rna.27949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schifano J.M., Vvedenskaya I.O., Knoblauch J.G., Ouyang M., Nickels B.E., Woychik N.A. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014;5:3538. doi: 10.1038/ncomms4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Zhang Y., Zhu L., Suzuki M., Inouye M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 2004;279:20678–20684. doi: 10.1074/jbc.M314284200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Zhu L., Zhang J., Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 2005;280:26080–26088. doi: 10.1074/jbc.M502050200. [DOI] [PubMed] [Google Scholar]
- 37.Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A.C., Engelberg-Kulka H., Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez M.V., Dawson C.C., Crew R., England K., Slayden R.A. MazF6 toxin of Mycobacterium tuberculosis demonstrates antitoxin specificity and is coupled to regulation of cell growth by a Soj-like protein. BMC Microbiol. 2013;13:240. doi: 10.1186/1471-2180-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salina E.G., Mollenkopf H.J., Kaufmann S.H., Kaprelyants A.S. M. tuberculosis gene expression during transition to the ‘non-culturable’ state. Acta Nat. 2009;1:73–77. [PMC free article] [PubMed] [Google Scholar]
- 40.Han J.S., Lee J.J., Anandan T., Zeng M., Sripathi S., Jahng W.J., Lee S.H., Suh J.W., Kang C.M. Characterization of a chromosomal toxin-antitoxin, Rv1102c-Rv1103c system in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2010;400:293–298. doi: 10.1016/j.bbrc.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 41.Prysak M.H., Mozdzierz C.J., Cook A.M., Zhu L., Zhang Y., Inouye M., Woychik N.A. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009;71:1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- 42.Guzman L.M., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothenbacher F.P., Suzuki M., Hurley J.M., Montville T.J., Kirn T.J., Ouyang M., Woychik N.A. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 2012;194:3464–3474. doi: 10.1128/JB.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp J.D., Cruz J.W., Raman S., Inouye M., Husson R.N., Woychik N.A. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 2012;287:12835–12847. doi: 10.1074/jbc.M112.340109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sisido M., Ninomiya K., Ohtsuki T., Hohsaka T. Four-base codon/anticodon strategy and non-enzymatic aminoacylation for protein engineering with non-natural amino acids. Methods. 2005;36:270–278. doi: 10.1016/j.ymeth.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 47.Winther K.S., Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy F.V.t., Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 49.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta A. Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2009;290:45–53. doi: 10.1111/j.1574-6968.2008.01400.x. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Pino A., Christensen-Dalsgaard M., Wyns L., Yarmolinsky M., Magnuson R.D., Gerdes K., Loris R. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J. Biol. Chem. 2008;283:30821–30827. doi: 10.1074/jbc.M805654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kasari V., Mets T., Tenson T., Kaldalu N. Transcriptional cross-activation between toxin-antitoxin systems of Escherichia coli. BMC Microbiol. 2013;13:45. doi: 10.1186/1471-2180-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winther K.S., Gerdes K. Ectopic production of VapCs from Enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol. Microbiol. 2009;72:918–930. doi: 10.1111/j.1365-2958.2009.06694.x. [DOI] [PubMed] [Google Scholar]
- 54.Kamphuis M.B., Bonvin A.M., Monti M.C., Lemonnier M., Munoz-Gomez A., van den Heuvel R.H., Diaz-Orejas R., Boelens R. Model for RNA binding and the catalytic site of the RNase Kid of the bacterial parD toxin-antitoxin system. J. Mol. Biol. 2006;357:115–126. doi: 10.1016/j.jmb.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 55.Pellegrini O., Mathy N., Gogos A., Shapiro L., Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol. Microbiol. 2005;56:1139–1148. doi: 10.1111/j.1365-2958.2005.04606.x. [DOI] [PubMed] [Google Scholar]
- 56.Baik S., Inoue K., Ouyang M., Inouye M. Significant bias against the ACA triplet in the tmRNA sequence of Escherichia coli K-12. J. Bacteriol. 2009;191:6157–6166. doi: 10.1128/JB.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu Z., Donegan N.P., Memmi G., Cheung A.L. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J Bacteriol. 2007;189:8871–8879. doi: 10.1128/JB.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan P.P., Lowe T.M. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohli S., Singh Y., Sharma K., Mittal A., Ehtesham N.Z., Hasnain S.E. Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H37Rv and H37Ra reveal novel and interesting differences with implications in virulence. Nucleic Acids Res. 2012;40:7113–7122. doi: 10.1093/nar/gks465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sampson S.L. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin. Dev. Immunol. 2011;2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen G.F., Inouye M. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 1990;18:1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldman E., Rosenberg A.H., Zubay G., Studier F.W. Consecutive low-usage leucine codons block translation only when near the 5′ end of a message in Escherichia coli. J. Mol. Biol. 1995;245:467–473. doi: 10.1006/jmbi.1994.0038. [DOI] [PubMed] [Google Scholar]
- 64.Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 65.Cruz J.W., Sharp J.D., Hoffer E.D., Maehigashi T., Vvedenskaya I.O., Konkimalla A., Husson R.N., Nickels B.E., Dunham C.M., Woychik N.A. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nature Commun. 2015;6:7480. doi: 10.1038/ncomms8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anantharaman V., Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leplae R., Geeraerts D., Hallez R., Guglielmini J., Dreze P., Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makarova K.S., Wolf Y.I., Koonin E.V. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson P., Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gebetsberger J., Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Masaki H., Ogawa T. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie. 2002;84:433–438. doi: 10.1016/s0300-9084(02)01425-6. [DOI] [PubMed] [Google Scholar]
- 72.Kaufmann G. Anticodon nucleases. Trends Biochem. Sci. 2000;25:70–74. doi: 10.1016/s0968-0004(99)01525-x. [DOI] [PubMed] [Google Scholar]
- 73.Brodersen D.E., Clemons W.M., Jr, Carter A.P., Wimberly B.T., Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 74.Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 75.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






