Abstract
Base-pairing of U4 and U6 snRNAs during di-snRNP assembly requires large-scale remodeling of RNA structure that is chaperoned by the U6 snRNP protein Prp24. We investigated the mechanism of U4/U6 annealing in vitro using an assay that enables visualization of ribonucleoprotein complexes and faithfully recapitulates known in vivo determinants for the process. We find that annealing, but not U6 RNA binding, is highly dependent on the electropositive character of a 20 Å-wide groove on the surface of Prp24. During annealing, we observe the formation of a stable ternary complex between U4 and U6 RNAs and Prp24, indicating that displacement of Prp24 in vivo requires additional factors. Mutations that stabilize the U6 ‘telestem’ helix increase annealing rates by up to 15-fold, suggesting that telestem formation is rate-limiting for U4/U6 pairing. The Lsm2–8 complex, which binds adjacent to the telestem at the 3′ end of U6, provides a comparable rate enhancement. Collectively, these data identify domains of the U6 snRNP that are critical for one of the first steps in assembly of the megaDalton U4/U6.U5 tri-snRNP complex, and lead to a dynamic model for U4/U6 pairing that involves a striking degree of evolved cooperativity between protein and RNA.
INTRODUCTION
Proteins that stimulate the annealing of RNA to target nucleic acids are ubiquitously important in biology, with examples including the Argonaute family proteins (1,2), the CRISPR-Cas systems (3) and Hfq (4,5). Additionally, RNA remodeling proteins are a broad class of proteins that chaperone the proper folding of RNAs, which have a high propensity to misfold into stable alternative structures (6). A striking example of both protein-mediated RNA remodeling and annealing occurs during spliceosome assembly. Spliceosome assembly requires complex conformational rearrangements among a set of five small nuclear RNAs (U1, U2, U4, U5 and U6 snRNAs) and many proteins (7,8). Once assembled, the spliceosome catalyzes precursor-messenger RNA (pre-mRNA) splicing, an essential process in all eukaryotes.
Structural rearrangements in U6 snRNA are particularly dynamic, involving the unwinding and re-annealing of the U6 internal stem loop (ISL) (9), which coordinates catalytic magnesium ions in the active site of fully-assembled spliceosomes (10). Thus, the spliceosome is an ‘RNPzyme’ with an RNA active site that is assembled and organized by proteins (11,12). Prior to incorporation into the spliceosome, yeast U6 exists in the form of the U6 snRNP, containing U6 snRNA, the tetra-RRM (RNA recognition motif) protein Prp24 and the heteroheptameric Lsm2–8 protein ring (13–21). In addition to being a stable component of the U6 snRNP, Prp24 acts as an RNA chaperone to catalyze unwinding of the U6 ISL and base pairing of the U4/U6 di-snRNA (15,16,22). Despite the stability of the U6 ISL (23), Prp24 accomplishes this task in an adenosine triphosphate (ATP)-independent manner.
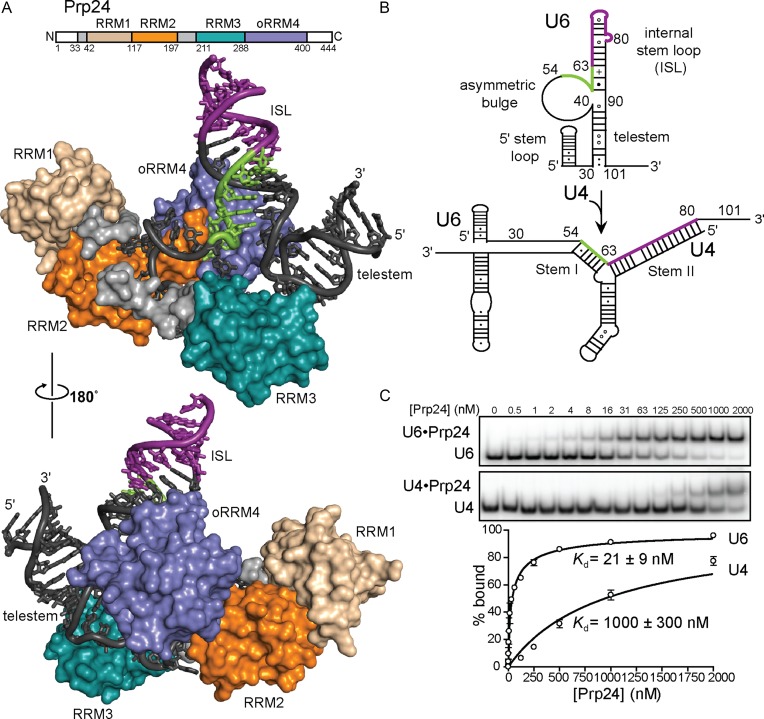
Our recent crystal structure of the U6•Prp24 complex revealed an extensive RNA–protein interface (24). Three of Prp24's four RRMs encircle a large loop in U6 RNA, generating a novel interlocked conformation in which U6 RNA-mediated RRM2–oRRM4 interactions result in topologically interlocked ‘rings’ of RNA and protein (Figure 1A). Thus, U6 RNA and Prp24 must co-fold around each other to form the U6 snRNP core. In the U6 snRNP core, U6 RNA adopts a secondary structure consisting of the ISL (nucleotides 59–88), the large asymmetric bulge (nucleotides 41–58) bound by Prp24 and an additional helix known as the telestem (nucleotides 30–40 and 91–101) (Figure 1B). The structure also revealed a 20 Å-wide electropositive groove within Prp24, composed of RRMs 1, 2 and oRRM4, which does not contact U6 within the complex, but binds duplex RNA from a neighboring complex in the crystal lattice. We previously hypothesized that this groove promotes U4/U6 annealing through stabilization of product duplex in the U4/U6 di-snRNA (24).
Figure 1.
Prp24 binds U6 RNA with high affinity and specificity. (A) Top: primary structure of Prp24. White regions are disordered and were deleted from the crystallization construct as previously described (24). RRM4 is an occluded RRM (oRRM), with terminal α-helices masking its β-sheet face (22,24). Unless indicated otherwise, all assays herein used full-length protein and RNA. Bottom: molecular architecture of the U6 snRNP core. Regions of U6 RNA (black) that form U4/U6 Stem I and Stem II are highlighted in green and purple, respectively. (B) Schematic of U6 RNA annealing to U4 RNA to form U4/U6. (C) Native gel analysis of full-length Prp24 binding to U6 (top) and U4 (bottom) RNAs.
In this study, we analyzed the mechanism of U4/U6 annealing by tracking the formation of all RNA–protein (RNP) species, quantifying their binding affinities, and measuring annealing rates in vitro. These approaches, in combination with extensive mutagenesis of both RNA and protein, provided significant insight into the annealing mechanism. Importantly, our in vitro system faithfully recapitulates a previously observed in vivo defect associated with a mutation in the U6 ISL, as well as its suppression by a second-site mutation (9). We found that Prp24 remained bound to the product U4/U6 in a stable ternary complex, indicating that additional factors are likely required to displace Prp24 from annealed U4/U6 in vivo, and showed that the core regions of U6 and Prp24 present in our crystal structure are optimal for annealing. We verified that the electropositive groove on the surface of Prp24 is indeed critical for annealing, but is not required for binding U6. Stabilization of the U6 telestem helix significantly increases annealing rates, as does binding of the Lsm2–8 complex adjacent to the telestem. Remarkably, a specific mutation in the telestem induces Prp24-independent RNA annealing. Based on these findings, we propose a dynamic model for the U4/U6 annealing pathway.
MATERIALS AND METHODS
Overexpression and purification of proteins
Saccharomyces cerevisiae Prp24 protein was recombinantly overexpressed in Escherichia coli using a pET15b plasmid (EMD Millipore) modified to replace the N-terminal hexahistidine tag with a decahistidine tag and the N-terminal thrombin cleavage site with a TEV cleavage site. Expression plasmids contained Prp24 residues 1–444 (full-length), 34–400 (1234), 1–400 (N1234) or 34–444 (1234C). Mutants of this plasmid were generated using inverse polymerase chain reaction (PCR) with Phusion DNA polymerase (New England Biolabs); PCR products were DpnI treated, self-ligated using T4 DNA ligase and T4 PNK and transformed into E. coli NEB 5-α competent cells (New England Biolabs). Clones were isolated by plasmid minipreps (Qiagen) and the identity of each verified by Sanger sequencing. Resulting clones were transformed into E. coli STAR pLysS cells (Invitrogen) for protein overexpression and purification essentially as described (24), with the exception that dialysis buffer and cation-exchange chromatography buffers did not contain ethylenediaminetetraacetic acid (EDTA), and that 1 mg of TEV protease was added during dialysis into cation-exchange chromatography buffer. All protein samples were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis to assess their purity.
Recombinant S. cerevisiae Lsm2–8 complex was expressed in E. coli from a pQLink vector containing all seven yeast Lsm genes with a C-terminally truncated Lsm4 (residues 1-93), Lsm6 with an N-terminal TEV-cleavable GST tag, and Lsm8 with an N-terminal TEV-cleavable 7XHis tag (a kind gift from Yigong Shi, Tsinghua University) and purified essentially as described (25) but was not subjected to gel filtration.
RNA synthesis
In vitro transcription was used to synthesize RNAs corresponding to S. cerevisiae U6 nucleotides 30–101 with a U100C/U101C double mutation, full-length wild-type and mutant U6 (nucleotides 1–112), full-length U4 (nucleotides 1–160) and a portion of human U1 (nucleotides 1–143). The U6 (30–101) U100C/U101C construct was transcribed from synthetic DNA oligonucleotide templates (Integrated DNA Technologies). Full-length U4 (nucleotides 1–160) was transcribed off a modified pUC118 plasmid template (a kind gift from P. Fabrizio) containing the T7 polymerase promoter sequence, two additional G nucleotides at the beginning of the transcript (for efficient T7 transcription) and a BsaI restriction site at the end of the transcript to allow for run-off transcription. Full-length U6 (nucleotides 1–112) was transcribed off a modified pUC57 plasmid containing the T7 polymerase promoter sequence with an additional G nucleotide at the beginning of the transcript (for efficient T7 transcription) and an HDV ribozyme sequence at the end of the transcript to confer a homogeneous 3' terminus, followed by a BamHI restriction site to allow run-off of the polymerase. A portion of human U1 RNA (nucleotides 1–143) was transcribed off a pUC18 plasmid (a kind gift from A. Hoskins) containing the T7 polymerase promoter sequence, two additional G nucleotides at the beginning of the transcript, an HDV ribozyme sequence and a BamHI restriction site for run-off transcription. All plasmid templates were linearized with either BsaI or BamHI (New England Biolabs) prior to transcription.
All RNAs were transcribed in vitro using recombinant His6-tagged T7 RNA polymerase (26,27) in 40 mM Tris-Cl pH 8.0, 1 mM spermidine, 0.01% Triton X-100, 38 mM MgCl2, 5 mM dithiothreitol (DTT) and 5 mM each of ATP, cytidine triphosphate (CTP), guanosine triphosphate (GTP) and uridine triphosphate (UTP). RNAs were purified from abortive transcripts, linearized plasmid and the HDV ribozyme using an 8% 29:1 acrylamide: bis-acrylamide denaturing gel containing 8 M urea, 89 mM Tris borate, 2 mM EDTA. RNA was visualized using UV shadowing and extracted from the gel by passive diffusion into 0.3 M sodium acetate pH 5.2. RNA was ethanol precipitated and resuspended in water.
RNA labeling
RNAs were dephosphorylated prior to 5' end labeling by incubating 10 pmol of RNA at 37°C with 10 units of CIP (New England Biolabs) in 50 mM potassium acetate, 20 mM Tris-acetate pH 7.9, 10 mM magnesium acetate, 100 μg/ml bovine serum albumin (BSA). This reaction was extracted with phenol:chloroform:isoamyl alcohol 25:24:1 and ethanol precipitated. After precipitation, RNAs were pelleted by centrifugation and resuspended in the 5' end labeling reaction consisting of 10 units of T4 PNK, 70 mM Tris-HCl pH 7.6, 10 mM MgCl2, 5 mM DTT and 0.03 μCi of [γ-32P] ATP (3000 Ci/mmol). The reaction was stopped by the addition of an equal volume of urea loading dye and purification on an 8% denaturing gel. RNAs were extracted from the gel by passive diffusion into 300 mM sodium acetate pH 5.2 and ethanol precipitated.
Fluorescently-labeled U4 and U6 were made via ligation of an in vitro transcribed RNA to a synthesized fluorescently labeled RNA (IDT) using T4 RNA ligase 2. An RNA oligonucleotide consisting of nucleotides 1–12 of U6 with a 5′-Cy3 label was ligated to nucleotides 13–112 of U6 transcribed from a plasmid containing a 5' hammerhead ribozyme (to confer a homogeneous 5′ terminus for ligation) and a 3' HDV ribozyme (modified from a kind gift by Kiyoshi Nagai). Fluorescent U4 was created by ligation of a 5′-Cy5 labeled RNA consisting of U4 nucleotides 1–13 to an in vitro transcribed U4 containing nucleotides 14–160 (modified from the pUC118 plasmid described previously). After in vitro transcription, U4(14–160) was modified as previously described using CIP and T4 PNK (NEB). U6(13–112) was modified using only T4 PNK due to the presence of a 5'-OH group in the original transcript. RNAs were ligated using DNA splints and T4 RNA ligase 2 and purified by urea polyacrylamide gelelectrophoresis as above (28).
Binding and annealing buffer preparation
A stock buffer containing 100 mM KCl, 20% glycerol,10 mM HEPES acid, 10 mM sodium HEPES base, 1 mM EDTA acid, 1 mM TCEP-HCl, 0.01% Triton X-100, pH∼ 7 was prepared. A ‘2× protein dilution buffer’ was made by the addition of BSA to 0.2 mg/ml, and a ‘2× RNA dilution buffer’ was made by the addition of yeast tRNA to 0.2 mg/ml and sodium heparin to 0.02 mg/ml. All buffer stock solutions were passed through a HiTrapSP cation exchange column (GE Healthcare) to remove trace levels of ribonuclease activity. The heparin component of the RNA dilution buffer was added after the buffer had been passed though the column. The final ‘binding buffer’ containing 1× RNA dilution buffer and 1× protein dilution buffer was used for all binding and annealing experiments.
Gel shift assay
Binding of RNAs with Prp24 was performed with trace (<1 nM) [32P]-labeled RNA with variable concentrations of protein. RNAs were heated to 90°C for 2 min in RNA binding buffer, then snap cooled on wet ice. Proteins were prepared as 2× stocks in protein binding buffer. The affinity of Prp24 for RNAs was determined using a 10 μl binding reaction prepared at room temperature containing 5 μl of RNA and 5 μl of 2× protein stock. Samples were incubated at room temperature for 20 min prior to loading onto a 16.5 × 22 cm 6% polyacrylamide gel (29:1 acrylamide:bis-acrylamide, 89 mM Tris borate, 2 mM EDTA pH 8.0). Samples were electrophoresed for 2–3 h at 150V at 4°C. Radioactive gels were dried on BioRad filter paper, exposed to a PhosphorImager screen and imaged on a Typhoon FLA 9000 biomolecular imager. Results were analyzed using ImageJ software and binding curves were fit using nonlinear regression in GraphPad Prism 4 to the Hill equation: % bound = (Bmax * [Prp24]H)/(KdH + [Prp24]H). Bmax was restrained to be between 0 and 100%, and the H (Hill coefficient) and Kd were restrained to be >0. Binding affinities are reported for three technical replicates.
Protein activity was determined as previously described (29) using 5 nM 5′-Cy3 labeled U6 RNA supplemented with 500 nM unlabeled full-length U6. Stoichiometric amounts of protein were added and binding reactions were treated as described above. Fluorescent gels were imaged directly through low fluorescence glass plates (CBS Scientific) on a Typhoon FLA 9000. The percent of labeled U6 in U6/Prp24 was analyzed using ImageJ software and the binding curve of protein concentration vs. percentage bound was plotted (GraphPad Prism 4). The linear region of this curve (protein:RNA stoichiometry <2) was fit to a linear regression. The activity of the protein is expressed as the slope of this line.
U4/U6 annealing assay
Annealing reactions were carried out at 30°C in 10 μl reactions in binding buffer (as described above) containing <1 nM 32P-labeled U4 RNA, 25 nM U6 RNA and 250 nM Prp24 protein. Reactions were stopped by the addition of 2 μl of proteinase K buffer (0.5% sodium dodecyl sulphate, 0.3 mg/ml tRNA, 5 mM CaCl2, 30 mM HEPES pH 7.0, 0.2 mg/ml proteinase K) or by separation on a 6% polyacrylamide gel (29:1 acrylamide:bis-acrylamide, 89 mM Tris borate, 2 mM EDTA pH 8.0) that had been pre-run at 150V for at least 30 min. Samples were electrophoresed for 3 h at 150 V at 4°C, dried, and analyzed as above. Annealing rates were calculated using the ratio of free U4 to U4/U6 in Proteinase K-treated lanes at 0, 15, 30, 60 and 90 min; resulting data were then fit to a one-phase exponential association equation (GraphPad Prism 4). Annealing rates are reported for three technical replicates.
RESULTS
Prp24 binds U6 snRNA with much higher affinity than U4 snRNA
Prp24 protein is capable of binding both U6 and U4 RNAs (Figure 1C), as previously observed (16). Previous studies of U6-Prp24 binding via gel shift showed the appearance of higher order complexes at high Prp24 concentrations, presumably due to non-specific binding events (18). We observed formation of multimeric Prp24 binding is suppressed by the inclusion of tRNA, heparin, and BSA in the buffer conditions (29). Using these conditions, we demonstrated that full-length Prp24 has a Kd of 21 ± 9 nM for full-length U6 RNA, similar to the value of 43 ± 11 nM reported previously (18) (Figure 1C). The tight binding of Prp24 to U6 RNA is in agreement with the large number of protein–RNA contacts present in the crystal structure of the U6•Prp24 complex, as twenty nucleotides in U6 are directly contacted by Prp24 (24). Interestingly, Prp24 binds U6 with a Hill coefficient of 0.50 ± 0.07. This may be due to structural heterogeneity in the RNA population, or interference between multiple binding sites. Prp24 has a ∼50-fold lower affinity of 1000 ± 300 nM for full-length U4 RNA under these conditions, with a Hill coefficient near unity (Figure 1C). Although Prp24 does display relatively weak binding to U4, this binding is not non-specific, as Prp24 displayed tighter binding to U4 than to a control RNA of similar size (human U1 snRNA nucleotides 1–143) (Supplementary Figure S1).
An annealing assay that preserves assembled RNPs
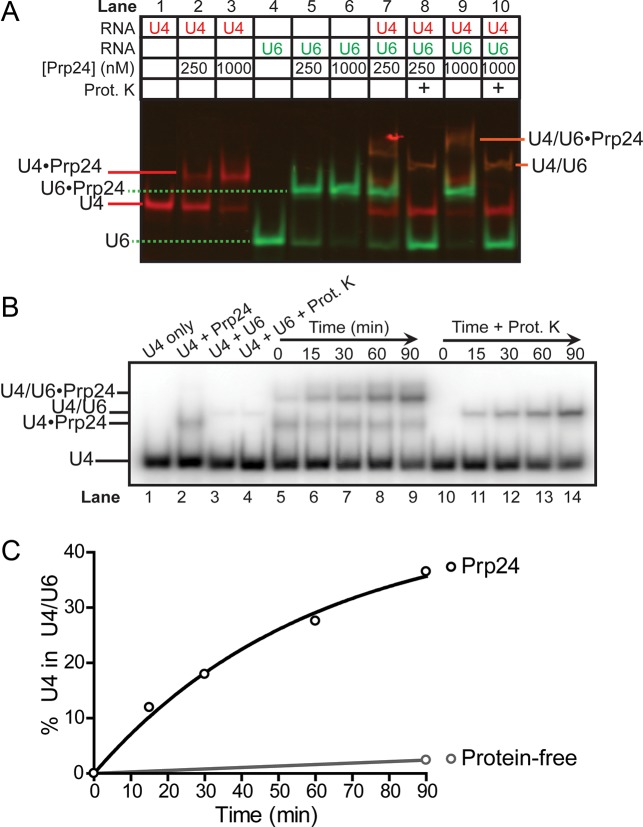
Previous gel-shift based approaches for determining U4/U6 annealing rates used treatment with proteinase K in order for U4/U6 RNA complexes to enter the gel, and so did not directly assess RNP formation (15,16). To define the RNPs formed in the annealing reaction, we employed fluorescently labeled U4 and U6 RNAs and our optimized conditions for specific binding. We observed formation of U6•Prp24 and U4•Prp24 complexes, as well as a U4/U6•Prp24 ternary complex when both RNAs were present (Figure 2A). Upon treatment of the annealing reaction with proteinase K, the U4•Prp24 and U6•Prp24 binary complexes resolve into free U4 and U6 RNA, and the U4/U6•Prp24 complex resolves into U4/U6 di-snRNA, showing that the U4 and U6 RNAs are base-paired in the U4/U6•Prp24 complex.
Figure 2.
Prp24 catalyzes annealing of U4 and U6 RNAs, and remains bound to product di-RNA. (A) Two-color gel demonstrating tight binding of Prp24 to Cy3-U6 (lanes 5 and 6) and weaker binding of Prp24 to Cy5-U4 (lanes 2 and 3). Co-localization of Cy3 and Cy5 fluorescence in the presence of U4 and U6 (lanes 7 and 9) shows that the slowest-migrating species (orange) contains both RNAs, and the increased mobility upon treatment with proteinase K (lanes 8 and 10) shows that the di-snRNA retains bound Prp24. Annealing reactions were incubated at 30°C for 90 min prior to loading. (B) Time-dependent formation of U4/U6, using radiolabeled U4 snRNA and unlabeled U6 and Prp24. Control reactions in lanes 1–4 were incubated for 90 min. (C) Quantification of Prp24-dependent annealing from proteinase K treated lanes (10–14) in (B) compared to protein-independent annealing (lane 4 in B).
Having established the identity of the U4/U6•Prp24 complex, we followed annealing kinetics using radiolabeled U4 RNA and excess unlabeled U6 RNA (Figure 2B). In our assay, U6 was pre-bound by excess Prp24 and the annealing reaction was started by the addition of U4. Under these conditions, we found that Prp24 accelerates the rate of U4/U6 annealing at 30°C at least 20-fold (Figure 2C). In non-deproteinized lanes, we observed formation of a small amount U4•Prp24 binary complex, which is expected based on the observed Kd for U4 and the fact that Prp24 is in excess in the assay. Formation of a Prp24•U4 binary complex is not a prerequisite for annealing in vitro, as titration of Prp24 displayed a K1/2 for annealing of 20 nM (Supplementary Figure S2), almost two orders of magnitude lower than the Kd for U4•Prp24 binding (Figure 1C), and consistent with the Kd for U6. Interestingly, at 10 nM Prp24 and without proteinase K digestion, about half of the product U4/U6 RNA complex is bound to Prp24 (Supplementary Figure S2). Thus, the Kd of Prp24 for U4/U6 under these conditions must be about 10 nM, close to the 18 nM affinity observed by Ghetti et al. (16).
The in vitro annealing assay faithfully recapitulates in vivo phenotypes of U6 ISL mutations
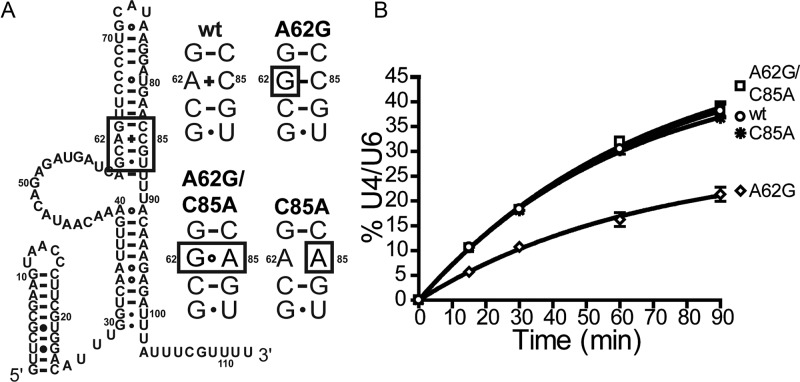
Mutation of U6-A62 to G was previously reported to reduce the levels of U4/U6 RNA complex in yeast cells grown at 30 or 18°C and cause a cold-sensitive growth defect (9). This mutation changes an A-C mismatch at the base of ISL into a stable G-C base pair (Figure 3A). The A62G annealing and growth defects are corrected by cis-acting suppressors that reintroduce a mismatch at this position and restore the stability of the ISL to near wild-type levels (9). We tested U6-A62G and one of its cis-acting suppressors, U6-C85A, which converts the G-C pair formed by U6-A62G into a G-A mismatch and was previously demonstrated to reverse the in vivo U4/U6 annealing defect and cold sensitivity. Both the A62G annealing defect and its suppression by C85A were recapitulated in the in vitro assay (Figure 3B). U6-A62G results in a 2-fold decrease in the rate of annealing, and this annealing defect is corrected by inclusion of U6-C85A. U6-C85A alone did not change the annealing rate in comparison to wild-type U6. Thus, mutations that induce the formation of a stable base pair at the base of the U6-ISL are deleterious to Prp24-catalyzed U4/U6 annealing, and can be rescued by suppressors that disrupt stable pairing, both in vitro and in vivo.
Figure 3.
The in vitro annealing assay recapitulates in vivo phenotypes of U6 RNA substitutions. (A) Secondary structure of U6 RNA bound to Prp24 (24), with the boxed region of the ISL shown with substitutions at right. (B) Time course of U4/U6 annealing for reactions containing the indicated substitutions in U6.
The components of the U6 snRNP core are sufficient for annealing to U4 snRNA
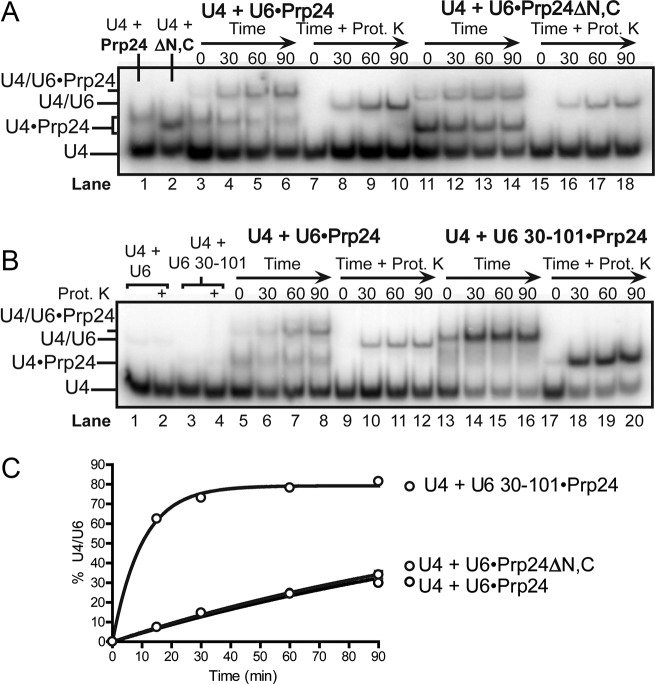
We sought to determine if the truncated forms of U6 and Prp24 present in the crystallized U6 snRNP core structure are sufficient for U4/U6 annealing. The crystallized complex contains Prp24 residues 34–400, including all four RNA recognition motifs but lacking the unstructured N- and C-terminal domains (22,30), and U6 nucleotides 30–101, with stabilizing mutations A62G, U100C and U101C (24). Since the A62G substitution is detrimental to annealing (Figure 3) and not required for crystallization (our unpublished data), this substitution was not included in our analysis. Truncation of Prp24 to remove the N- and C-terminal domains (Prp24ΔN,C) had no effect on U4/U6 annealing (Figure 4A and C). The U4•Prp24 complex is more pronounced in annealing reactions containing the truncated protein because truncation of Prp24 increased its affinity for U4 ∼3-fold (Supplementary Figure S3). Interestingly, truncation and mutation of U6 RNA stimulated annealing (Figure 4B and C).
Figure 4.
The components of the U6 snRNP core efficiently promote annealing. (A) Full-length (residues 1–444) and truncated (residues 34–400) Prp24 catalyze U4/U6 annealing with similar rates. Lane 1 contains U4 RNA with full-length Prp24 and lane 2 contains U4 with truncated Prp24. Lanes 3–18 contain full-length U6 RNA (unlabeled), U4 RNA and the indicated form of Prp24 incubated 0–90 min at 30°C. Samples in lanes 7–10 and 15–18 were treated with proteinase K before electrophoresis. (B) Truncated U6 RNA (30–101) with two stabilizing mutations (U100C/U101C) anneals more rapidly than full length U6 (1–112). Lanes 1 and 2 contain only U4 and full-length U6, while lanes 3 and 4 contain U4 and truncated U6. Lanes 2 and 4 are proteinase K treated. Lanes 5–20 correspond to lanes 3–18 of Panel A, but with full length Prp24 and full length or truncated U6 as indicated. (C) Annealing timecourses for experiments shown in Panels A and B.
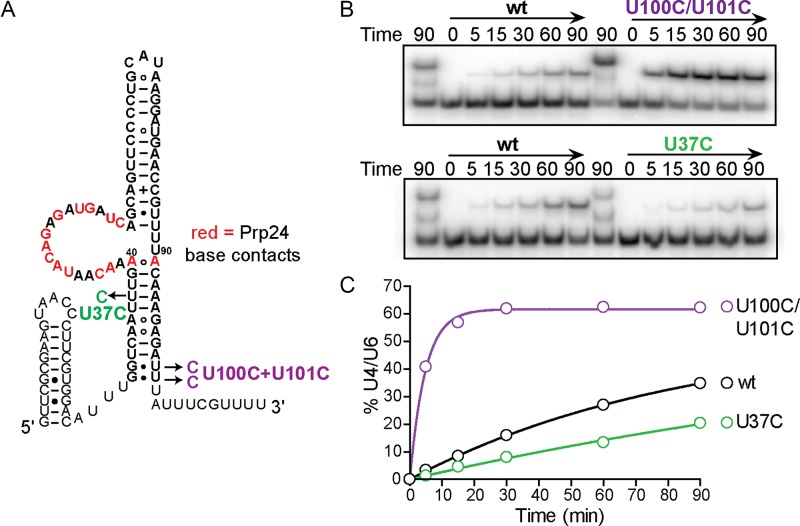
To investigate the basis of the rapid annealing of U6 30–101 (U100C/U101C), we tested the separate contributions of terminal truncation and telestem stabilization to the annealing rate. Truncation of the 3′ end of U6 RNA increased the annealing rate 2- to 3-fold, while 5′ truncations had little or no effect (Supplementary Figure S4). In contrast, stabilization of the telestem within full-length U6 via the U100C/U101C substitutions accelerated annealing ∼10-fold, a rate enhancement similar to that observed in the annealing reaction using the crystal construct (Figure 5). Since the U100C/U101C mutation is expected to increase telestem stability by replacing two terminal G-U wobble pairs with two G-C pairs, we hypothesized that stabilization of the telestem may be responsible for increasing the annealing rate. Consistent with this hypothesis, destabilization of the telestem with mutation U37C, which changes a U-A base-pair into a C-A mismatch, reduced the rate of annealing ∼2-fold (Figure 5).
Figure 5.
Mutations within the telestem affect annealing rate. (A) Secondary structure of U6 showing the position of telestem-stabilizing (U100C/U101C) or destabilizing (U37C) mutations. Base specific contacts with Prp24 are highlighted in red. (B) Annealing gel comparing full-length wild-type U6 to U6-U100C/U101C (top) or U6-U37C (bottom). A control lane containing the annealing reaction at 90 min that was not proteinase K-treated precedes each time course of annealing reactions treated with proteinase K. (C) Annealing timecourses for reactions containing wild-type, U100C/U101C and U37C variants of full-length U6.
Both U6 telestem stability and sequence influence the U4/U6 annealing rate
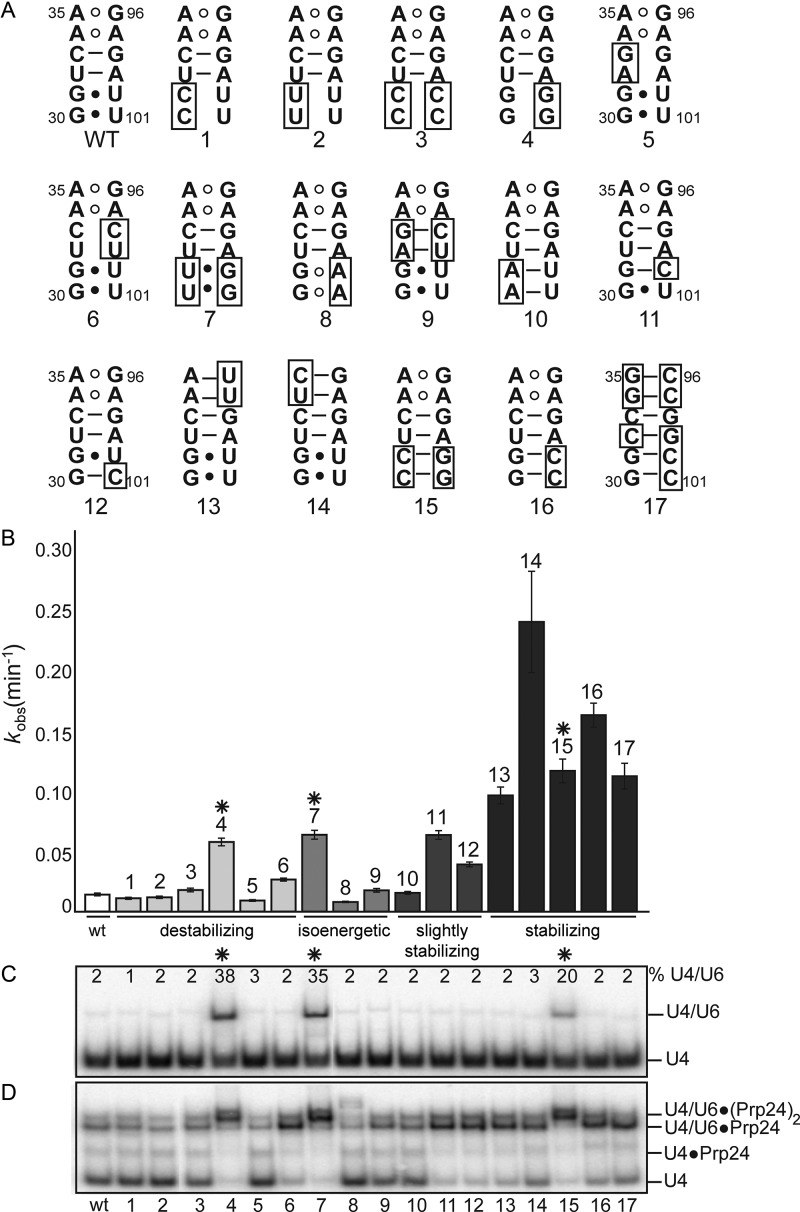
In order to determine if telestem stability generally correlates with annealing rates, we systematically altered the pairing potential of the six base pairs at the base of the telestem (Figure 6A). A clear general trend is that stabilization of the telestem results in a faster annealing rate (Figure 6B).
Figure 6.
Stabilization of the U6 telestem results in significant rate enhancement of U4/U6 annealing. (A) Secondary structure of the lower telestem, with tested mutations boxed. (B) U4/U6 annealing rates of mutant U6 RNAs. Mutations predicted to destabilize the telestem (numbers 1–6) or to be isoenergetic with wild type RNA (nos. 7–9) had little effect on annealing rate, while those predicted to be slightly stabilizing (nos. 10–12) or stabilizing (nos. 13–17) significantly increased the annealing rate. (C) Protein-free annealing of U4 and U6 RNAs at 90 min. The percentage of U4 incorporated into U4/U6 is shown. Mutations that enhance the protein-free annealing rate are marked with an asterisk. (D) Electrophoretic mobility of U4/U6•Prp24 species after 90 min of annealing.
In addition, we found that mutations U100G/U101G in combination with any nucleotides at positions 30 and 31 of U6—either stabilizing or destabilizing—increase the rate of annealing (Figure 6B and C, asterisks). Intriguingly, these mutations substantially increased the rate of annealing even in the absence of Prp24 (Figure 6C), thus accounting for the anomalous kinetic behavior of apparent outlier mutants 4 and 7. RNA secondary structure prediction of the U100G/U101G mutations via MFold analysis (31) suggests that the U100G/U101G mutations induce an alternative fold of U6 RNA that destabilizes the ISL, which could account for the observed increase in protein-free annealing rates (Supplementary Figure S5A and B). In this potential alternate fold, nucleotides 64–68 are single stranded in the loop of a very short helix. These nucleotides form part of U4/U6 Stem II and in wild-type U6 are base-paired in the U6 ISL. Destabilization of the ISL via stabilization of this alternate fold may drive the annealing reaction forward in the absence of Prp24 by exposing nucleotides involved in U4/U6 base pairing. In the presence of Prp24, these mutations decreased the electrophoretic mobility of U4/U6•Prp24 (Figure 6D). This difference in mobility is likely due to multiple copies of Prp24 bound to U4/U6, as increasing the concentration of Prp24 caused the formation of discrete higher bands in U4/U6-U100G/U101G, but not wild-type U4/U6 (Supplementary Figure S5C). Despite the difference in U4/U6-Prp24 binding, the U100G/U101G mutation did not change U6-Prp24 binding affinity and retained a Hill coefficient of about one-half, similar to wild-type RNA (Supplementary Figure S5D and E).
The behavior of the U100G/U101G-containing mutants is in stark contrast to the behavior of a hyperstabilized U6 mutant 17, in which the last 6 base pairs of the telestem are all G-C or C-G (Figure 6A). These mutations accelerated the rate of Prp24-mediated annealing ∼10-fold, but no enhancement of protein-independent annealing was observed and the resulting U4/U6-Prp24 ternary complex retained wild-type electrophoretic mobility (Figure 6D, see lane marked 17). Interestingly, U6 mutant 17 RNA exhibited a ∼10-fold tighter Kd for Prp24 binding than wild-type RNA (Supplementary Figure S6, Table 1) with a Hill coefficient near unity (H = 1.3 ± 0.1 versus H = 0.50 ± 0.07 for wild-type) (Supplementary Figure S6A and B). U6 mutant 17 also displayed ∼5-fold slower off-rate of Prp24 (Supplementary Figure S6C and D, Table 1). The slower off-rate for mutant 17 largely accounts for the lower Kd and is consistent with the observed interlocked topology of the complex and the hypothesis that the telestem must unfold in order for Prp24 to dissociate (24). The mutant 17-Prp24 complex also had a lower electrophoretic mobility than wild-type U6-Prp24 (Supplementary Figure S6A and C). A possible explanation for this behavior is that the wild-type RNA may be in conformational exchange with an alternatively folded, higher mobility species.
Table 1.
| Property | U6 wt | Mutant 17 |
|---|---|---|
| Kd (nM) | 21 ± 9 | 2.2 ± 0.2 |
| Bmax (% bound) | 100 ± 8 | 96 ± 1 |
| Hill coefficient | 0.49 ± 0.07 | 1.3 ± 0.1 |
| koff x 10−5 (s−1) | 20 ± 2 | 4 ± 0.3 |
| Calculated kon x 103 (M−1s−1) | 9.5 | 18 |
From the measured off-rates and dissociation constants, we can calculate that the apparent on-rates are approximately five orders of magnitude slower than diffusion (Table 1). The slow on-rates are also consistent with the interlocked topology of the RNP, which requires the intricate co-folding of RNA and protein (24).
Reduction of net positive charge in the electropositive groove of Prp24 inhibits annealing
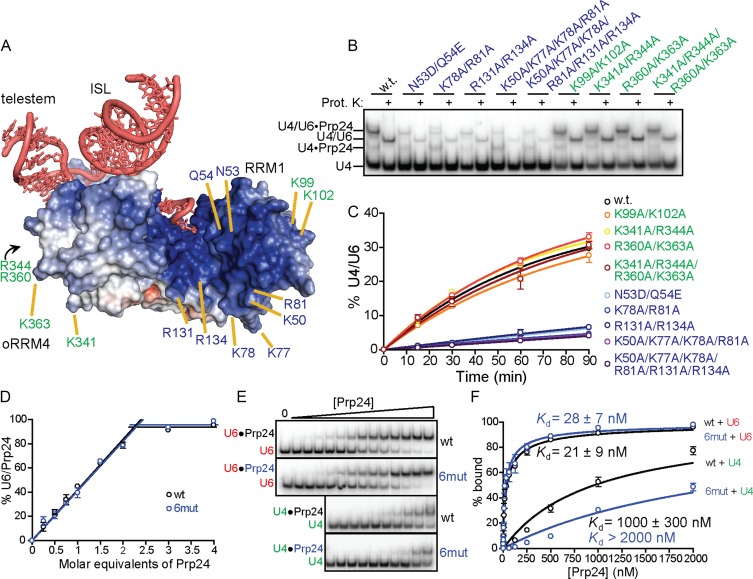
The electropositive groove of Prp24 is comprised of RRMs 1 and 2 and oRRM4 (24). In the crystal structure of U6•Prp24, this region does not bind any nucleotides of U6 within the crystallographic asymmetric unit. However, the ISL of a neighboring U6•Prp24 complex occupies the groove. This groove is well-suited for binding double-stranded RNA, as it has a width of approximately the diameter of double stranded RNA (20 Å) and many positively charged residues for interacting with the negatively charged phosphate backbone of RNA. To test whether this groove may be the active site for U4/U6 annealing, we mutated positively charged residues within the groove and observed the effect on annealing rate. Arginines (R81, R131 and R134) and lysines (K50, K77 and K78) in the electropositive groove were substituted with alanine (Figure 7A). In addition, the amido groups of N53 and Q54 were mutated to carboxylate groups. Five combinations of mutations in these eight residues were tested (Figure 7B). Mutagenesis of the selected residues does not affect protein production or purity (Supplementary Figure S7).
Figure 7.
Reduction of net positive charge in the electropositive groove decreases the rate of U4/U6 annealing without affecting U6 RNA-binding. (A) Electrostatic surface of Prp24, contoured from +8 kT/e (blue) to −8 kT/e (red). Positions of mutations within the electropositive groove (blue text) and outside the groove (green text) are indicated. (B) Annealing gel showing a single time point (90 min) for wild-type Prp24 and each of the nine mutants. Samples were divided into native (odd lanes) and proteinase K-treated (even lanes). Substitutions in the electropositive groove (blue text) reduce the amount of U4/U6 after 90 min, while substitutions outside this region (green text) do not. (C) Rate of U4/U6 formation over time for wild-type (black), constructs with an electropositive groove mutation (blue text) and constructs with mutations outside of this region (green text). (D) Wild-type and ‘6mut’ Prp24 are equally active for U6 binding. Labeled U6 (5 nM Cy5-U6) was supplemented with 500 nM unlabeled U6, and the binding of stoichiometric amounts of Prp24 was monitored. (E) Native gel analysis of U6-Prp24 binding (top) and U4-Prp24 binding (bottom) using wild-type and mutant full-length Prp24. ‘6mut’ refers to the presence of K50A/K77A/K78A/R81A/R131A/R134A mutations. (F) Binding curves (simple one site binding model) of wild-type versus mutant protein with full-length U6 and U4.
When tested for their effect on annealing, all combinations of mutations within the groove significantly decreased the rates of annealing (Figure 7B and C). Both positive to neutral mutations (arginine and lysine to alanine) and neutral to negative mutations (asparagine and glutamine to aspartate and glutamate, respectively) inhibit annealing to a similar extent. The inhibitory effect is correlated with the number of mutations, where the sextuple mutation is most deleterious. In order to exclude the possibility that the inhibitory effect of the electropositive groove mutations is simply due to an overall reduction in charge of the protein, surface arginine and lysines outside of the electropositive groove (Figure 7A) were also mutated and tested in the annealing assay. Similarly to the electropositive groove mutations, these mutations did not affect protein purification or stability (Supplementary Figure S7). All four sets of mutations outside the groove, including a quadruple mutant, did not significantly affect annealing rate (Figure 7A–C).
To verify that the mutations in the electropositive groove were not deficient for annealing due to a reduced binding affinity for U6, the activity and affinity of wild-type and sextuple mutant Prp24 for U6 RNA were determined. The preparations of both of these proteins used for binding and annealing studies displayed the same binding activity (Figure 7D), where both proteins appeared to be ∼45% active for U6 binding. These proteins were then used in a native gel assay to determine the affinity for U6 RNA (Figure 7E). Wild-type protein displayed a Kd of 21 ± 9 nM for U6 RNA, while mutant protein displayed a Kd of 28 ± 7 (Figure 7F). Both proteins bound to U6 with a Hill coefficient of less than unity. In contrast, mutations within the electropositive groove had a measurable effect on the affinity of Prp24 for U4, as the Kd of Prp24 for U4 was at least doubled from 1000 nM to >2000 nM for the mutant protein (Figure 7E and F). These results indicate that the electropositive groove substitutions do not alter Prp24's affinity for U6 RNA, but do diminish its affinity for U4 RNA, consistent with binding of U4 to the groove prior to annealing with U6.
The Lsm2–8 ring enhances Prp24-mediated U4/U6 annealing in vitro
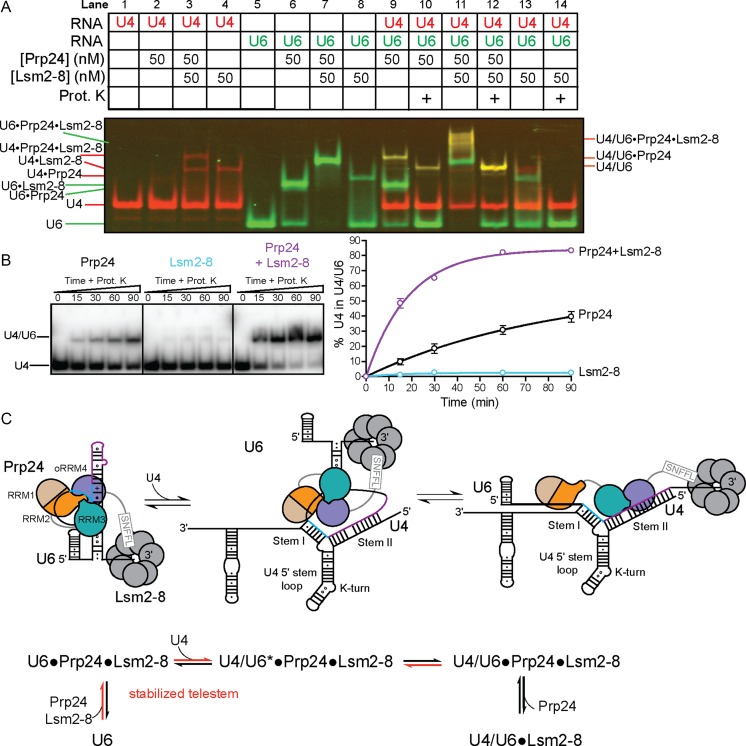
It was previously shown that deletion of the C-terminal decapeptide of Prp24, which binds to the Lsm2-8 complex, decreases the efficiency of Prp24-mediated U4/U6 annealing in extracts (32). Using recombinant Prp24 and Lsm2-8, we directly tested the influence of the Lsm2-8 ring on U4 and U6 annealing in vitro. We again used a two-color gel system to monitor the formation of RNP species that contribute to annealing (Figure 8A). This system reveals that a productive U4/U6•Prp24•Lsm2-8 complex is formed that results in a 4-fold increase in the rate of U4/U6 annealing compared to Prp24 alone (Figure 8B). We also observe that although Lsm2-8 can bind to both U6 and U4 in vitro, it cannot anneal the RNAs in the absence of Prp24 (Figure 8A, lanes 4, 8, 13–14) even under saturating binding conditions (data not shown). The kinetic profile obtained upon addition of the Lsm2-8 proteins is highly similar in terms of rate enhancement to the moderately stabilizing telestem mutations (11, 12 and 13) (Figure 6).
Figure 8.
The Lsm2–8 ring enhances U4/U6 annealing. (A) Two-color native gel with Cy5-U4 (red) and Cy3-U6 (green). The Lsm2–8 ring alone binds U4 and U6 RNAs (lanes 4 and 8), but exhibits strong cooperative binding with Prp24 only on U6 RNA (cf. lanes 3 and 7). Lsm2–8 enhances Prp24-mediated annealing (cf. lanes 10 and 12; yellow shows colocalized RNAs), but does not catalyze annealing alone (lane 14). Annealing reactions were incubated at 30°C for 90 minutes prior to loading. (B) Time-dependent formation of U4/U6, using radiolabeled U4 snRNA and unlabeled U6, Prp24 (50 nM) and Lsm2–8 (50 nM). (C) Model for the rate enhancement conferred by stabilizing the telestem or by inclusion of the Lsm2–8 ring. The C-terminal decapeptide of Prp24 (the SNFFL box) interacts with Lsm2–8.
DISCUSSION
A critical step in spliceosome biogenesis is U4/U6 RNA annealing, which involves large-scale rearrangement of RNA structure. Prp24 accelerates the extensive remodeling of RNA structure in the absence of ATP hydrolysis. ATP-dependent RNA/RNP remodeling is required for later steps in spliceosome assembly, activation and disassembly, controlled in part by the eight DExD/H helicases in the spliceosome (33). Since Prp24 only uses binding free energy to stimulate annealing, the process is likely driven forward by the favorable free energy of U4/U6 base pairing. For example, the predicted standard free energies for the U6 ISL versus U4/U6 stem II are −6.7 and −28.1 kcal/mol, respectively (31). Additionally, ours and other's data (16) indicate that Prp24 binds more tightly to the annealed U4/U6 complex than U6, which is likely to further stabilize the annealed product and help drive the process forward. Therefore, it is likely that additional factors are required to displace Prp24 from U4/U6 upon formation of the U4/U6 di-snRNP, which does not contain Prp24 (14,17). Interestingly, human Prp24 does remain associated with U4/U6 (34), but is displaced upon U4/U6.U5 tri-snRNP assembly (35,36).
In vitro reconstitution of snRNP assembly
Our in vitro reconstitution of U4/U6 annealing faithfully recapitulates known genetic interactions in the U6 ISL and thus shows promise for dissecting the complex series of conformational changes that drive U4/U6 di-snRNP assembly in vivo. Inhibition of U4/U6 annealing by the U6-A62G mutation and correction of this defect by the C85A mutation indicate that the stability of the 62–85 base pair at the base of the ISL is critical for annealing. Since nucleotide 85 is outside of the U4 binding region of U6, we hypothesize that U6 nucleotide A62 must be made available for pairing to U4 at a relatively early stage in the annealing process. Indeed, A62 is at the 5′ terminal end of the U6 ISL, and is adjacent to the single stranded region of U6 that is positioned most closely to the electropositive groove in our crystal structure (24). We did not observe correction of the A62G annealing defect in vitro in the presence of substitutions in Prp24 that suppress the U6-A62G cold-sensitive growth defect, or suppressor substitutions in U6 that lie in the Prp24•U6 interface rather than in the ISL (24) (data not shown). These results suggest that the latter group of suppressors act at a stage of di-snRNP assembly not assayed in our in vitro system, most likely displacement of Prp24 from U4/U6. We predict that, as we add additional splicing factors to our di-snRNP reconstitution assay, we will be able to detect the effect of this latter group of suppressor substitutions.
An active site for U4/U6 annealing
Due to the interlocked topology of the U6 snRNP core, it is unlikely that the electropositive groove is completely formed in the absence of U6 RNA. However, U4 RNA still binds to Prp24 with some specificity, albeit with low (Kd = 1 μM) affinity. It is likely that U4 RNA binding to the pre-formed U6 snRNP core is a much higher affinity interaction, as reflected in the observed 20 nM K1/2 of annealing. Mutation of the electropositive groove impairs U4 RNA binding and annealing, consistent with a model in which U4 RNA binds to the electropositive groove of the pre-formed U6•Prp24 complex to initiate annealing. The U6 nucleotide that is closest to the electropositive groove is A51, which stacks on A53 and is therefore proximal to the U6 nucleotides that participate in formation of U4/U6 Stem I (U6 54–63). Thus, we hypothesize that U4 localization to the electropositive groove positions U4 in close proximity to U6 nucleotides 54–63 to form Stem I (Figure 1A and B). Based on our crystal structure and prior nuclear magnetic resonance data (37), we previously proposed a model in which the single stranded nucleotides 54–60 of U6 dynamically reorient into the electropositive groove to initiate annealing (24). Our proposal that the electropositive groove stabilizes the nascent annealing of U4/U6 is consistent with the observation that mutations that reduce the positive charge within this groove have a strong deleterious effect on annealing efficiency. This proposal is also consistent with the observation that electropositive groove mutations do not affect U6-Prp24 binding, implying that the electropositive groove functions in the annealing process subsequent to U6•Prp24 formation.
The telestem is a prerequisite for efficient U4/U6 annealing
It has previously been demonstrated that truncations to the 5′ and 3′ ends of U6 had only a marginal effect on splicing activity in vitro (38), and our data indicate that these regions are not important for Prp24-mediated U4/U6 annealing. Although hydroxyl radical probing and chemical crosslinking results suggest that Prp24 interacts with and protects the 5′ SL and linker region in U6 (39), we conclude that these interactions are not necessary for efficient annealing in vitro. Truncation of the 3′ end may increase annealing rate by biasing formation of the telestem, or the 3′ end may otherwise occupy the electropositive groove in the absence of the Lsm ring, competing with annealing.
There is a clear correlation between the presence of stabilizing mutations in the telestem and an increased Prp24-mediated annealing rate (Figure 6), leading us to conclude that formation of the telestem is important for the annealing process. Interestingly, there appears to be additional sequence requirements involving the 3′ side of the telestem, as the U100G/U101G mutation increases both the Prp24-catalyzed annealing rate and the protein-free annealing rate. These mutations may stabilize an alternate fold involving the 3′ end of U6 that invades and destabilizes the ISL, while preserving the Prp24 binding site 5′ of the ISL, thereby increasing the annealing rate regardless of the presence of Prp24. The U100G/U101G mutation also stabilizes a second binding site for Prp24, observed as a U4/U6•(Prp24)2 complex with decreased gel mobility (Figure 6 and Supplementary Figure S5). We have no evidence for the biological relevance of this secondary binding site, but hypothesize it is due to sequence similarities between the primary binding site for Prp24 RRM2 (nucleotides 46–54: UACAGAGAU) (24) and the 3′ side of the telestem (nucleotides 90–100: UACAAAGAGAU). Note that the underlined sequences are identical. We hypothesize that formation of the U4/U6 complex disrupts the telestem and exposes this 3′ sequence, allowing for the binding of a second Prp24 molecule.
In the U6•Prp24 complex, the telestem is sandwiched between RRM3 and oRRM4 (Figure 1A). We propose that the telestem acts as a linchpin to stabilize the ‘interlocked ring’ topology of U6•Prp24 required for formation of the electropositive groove (24). In support of this model, we find that the most stable telestem mutant (#17) has a lower Kd and slower off-rate for Prp24 (Supplementary Figure S6).The fact that we observe a general correlation between telestem stability and annealing activity for a large number of mutants (18 total, Figures 5 and 6) with different RNA sequences suggests that the observed rate effect is a function of the thermodynamic stability of the telestem. A model that reconciles our data is shown in Figure 8C. Stabilization of the telestem may be achieved either via mutation of the telestem or by binding of the Lsm2–8 ring to the 3′ end of U6. Thus, we propose that an important function of the Lsm2–8 ring is to ensure proper folding of the U6 telestem region without over-stabilizing the RNA fold. Once U4/U6 annealing has occurred, coaxial stacking of U4/U6 Stems I and II, as observed in the tri-snRNP structure (40), likely splays apart the ends of U6 RNA to destabilize the telestem.
The ability of the components of the crystallized U6 snRNP core (truncated U6 and truncated Prp24) to efficiently anneal, combined with the observation that the U100C/U101C mutations greatly accelerate annealing, strongly suggests that the crystal structure represents an on-pathway conformation in the annealing process. As both Prp24 and the Lsm ring can crosslink to the base of the telestem (39), it is possible that the protein components of the U6 snRNP stabilize the telestem and exert allosteric control over U4/U6 di-snRNP assembly. Future studies will seek to address the mechanism by which Prp24 is displaced from U4/U6 in vivo (14).
Conclusions and prospects
The U4/U6 di-snRNA annealing mechanism illustrates the high level of specificity and complexity required for spliceosome assembly and catalysis. Unlike other nucleic acid annealing proteins, Prp24 has evolved a high degree of specificity for its snRNA substrates. This specificity is illustrated by the interlocked ring topology of U6 and Prp24, which is an extreme (and thus far, apparently unique) example of RNP co-folding. The mechanism of U4/U6 annealing is also intriguing, given that Prp24 must resolve considerable secondary structure in U6 in order to form even more extensive intermolecular base pairing in the U4/U6 di-snRNA. Annealers of short RNAs, such as Argonaute and Hfq, typically do not need to resolve such extensive secondary structure in order to anneal their target RNAs. However, the mechanism of Prp24-mediated annealing is not without analogy to these other annealers, as they all share a common feature in the presence of a positively charged region that is critical for the annealing process. In Hfq, an ‘arginine patch’ along the rim of the protein is essential for annealing sRNAs to their mRNA targets (4). Like the electropositive groove in Prp24, mutation of the arginine patch in Hfq reduces annealing but does not prevent sRNA/mRNA binding to Hfq. Similarly, Argonaute also contains a deep electropositive cleft in which the guide RNA/target RNA duplex is annealed (41).
Like the ribosome, the spliceosome is a key supramolecular enzyme of gene expression. The largest pre-assembled spliceosome particle is the U4/U6.U5 tri-snRNP, which has a maximum dimension of 31 nm, larger than both the post-catalytic Schizosaccharomyces pombe spliceosome (27 nm) (12) and the fully assembled 70S ribosome (24 nm) (42). Assembly pathways for the small ribosomal subunit were initially mapped out in the late 1960's (43–45), and mechanistic descriptions of this process are still being elucidated (46,47). In comparison to the ribosome, relatively little is understood about the assembly mechanism of the spliceosome. The first structures of the U4/U6.U5 tri-snRNP and spliceosome have recently emerged into view (12,40) and the astonishing complexity of these large structures suggests an equally complex assembly pathway. Here we elucidate the molecular requirements for one of the first steps in tri-snRNP biogenesis, formation of the U4/U6 di-snRNA. This work provides a quantitative and structural foundation for understanding the pathway and kinetics of tri-snRNP assembly, starting from the U6 snRNP. Important future directions include understanding the contributions of U4 snRNP proteins to U4/U6 di-snRNP assembly (and Prp24 release), and to understand how the U5 snRNP joins the U4/U6 di-snRNP to form the tri-snRNP.
Supplementary Material
Acknowledgments
We thank Aaron Hoskins, Kiyoshi Nagai and Yigong Shi for plasmids, Aaron Hoskins for critical reading of the manuscript, and members of the Butcher, Brow and Hoskins labs for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM065166 to S.E.B., D.A.B.]. Funding for open access charge: NIH/NIGMS [R01 GM065166].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ameres S.L., Martinez J., Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Herzog V.A., Ameres S.L. Approaching the Golden Fleece a Molecule at a Time: Biophysical Insights into Argonaute-Instructed Nucleic Acid Interactions. Mol. Cell. 2015;59:4–7. doi: 10.1016/j.molcel.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 3.Jiang F., Doudna J.A. The structural biology of CRISPR-Cas systems. Curr. Opin. Struct. Biol. 2015;30:100–111. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panja S., Schu D.J., Woodson S.A. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41:7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajkowitsch L., Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodson S.A. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010;7:677–686. doi: 10.4161/rna.7.6.13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brow D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 8.Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Fortner D.M., Troy R.G., Brow D.A. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev. 1994;8:221–233. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- 10.Fica S.M., Tuttle N., Novak T., Li N.S., Lu J., Koodathingal P., Dai Q., Staley J.P., Piccirilli J.A. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hang J., Wan R., Yan C., Shi Y. Structural basis of pre-mRNA splicing. Science. 2015;349:1191–1198. doi: 10.1126/science.aac8159. [DOI] [PubMed] [Google Scholar]
- 12.Yan C., Hang J., Wan R., Huang M., Wong C.C., Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 13.Mayes A.E., Verdone L., Legrain P., Beggs J.D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon K.W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 15.Raghunathan P.L., Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 16.Ghetti A., Company M., Abelson J. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- 17.Jandrositz A., Guthrie C. Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. EMBO J. 1995;14:820–832. doi: 10.1002/j.1460-2075.1995.tb07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan S.S., Brow D.A. The N- and C-terminal RNA recognition motifs of splicing factor Prp24 have distinct functions in U6 RNA binding. RNA. 2005;11:808–820. doi: 10.1261/rna.2010905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achsel T., Brahms H., Kastner B., Bachi A., Wilm M., Luhrmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pannone B.K., Xue D., Wolin S.L. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salgado-Garrido J., Bragado-Nilsson E., Kandels-Lewis S., Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Tumasz S., Richie A.C., Clos L.J. 2nd, Brow D.A., Butcher S.E. A novel occluded RNA recognition motif in Prp24 unwinds the U6 RNA internal stem loop. Nucleic Acids Res. 2011;39:7837–7847. doi: 10.1093/nar/gkr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sashital D.G., Allmann A.M., Van Doren S.R., Butcher S.E. Structural basis for a lethal mutation in U6 RNA. Biochemistry. 2003;42:1470–1477. doi: 10.1021/bi027137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montemayor E.J., Curran E.C., Liao H.H., Andrews K.L., Treba C.N., Butcher S.E., Brow D.A. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-A resolution. Nat. Struct. Mol. Biol. 2014;21:544–551. doi: 10.1038/nsmb.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou L., Hang J., Zhou Y., Wan R., Lu G., Yin P., Yan C., Shi Y. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature. 2014;506:116–120. doi: 10.1038/nature12803. [DOI] [PubMed] [Google Scholar]
- 26.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 28.Nandakumar J., Ho C.K., Lima C.D., Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- 29.Ryder S.P., Recht M.I., Williamson J.R. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae E., Reiter N.J., Bingman C.A., Kwan S.S., Lee D., Phillips G.N., Jr, Butcher S.E., Brow D.A. Structure and interactions of the first three RNA recognition motifs of splicing factor Prp24. J. Mol. Biol. 2007;367:1447–1458. doi: 10.1016/j.jmb.2007.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rader S.D., Guthrie C. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA. 2002;8:1378–1392. doi: 10.1017/s1355838202020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cordin O., Hahn D., Beggs J.D. Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 2012;24:431–438. doi: 10.1016/j.ceb.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Bell M., Schreiner S., Damianov A., Reddy R., Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabrizio P., Esser S., Kastner B., Luhrmann R. Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science. 1994;264:261–265. doi: 10.1126/science.8146658. [DOI] [PubMed] [Google Scholar]
- 36.Liu S., Rauhut R., Vornlocher H.P., Luhrmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Tumasz S., Reiter N.J., Brow D.A., Butcher S.E. Structure and functional implications of a complex containing a segment of U6 RNA bound by a domain of Prp24. RNA. 2010;16:792–804. doi: 10.1261/rna.1913310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan D.E., Stevens S.W., Abelson J. The 5′ and 3′ domains of yeast U6 snRNA: Lsm proteins facilitate binding of Prp24 protein to the U6 telestem region. RNA. 2002;8:1011–1033. doi: 10.1017/s1355838202026092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karaduman R., Fabrizio P., Hartmuth K., Urlaub H., Luhrmann R. RNA structure and RNA-protein interactions in purified yeast U6 snRNPs. J. Mol. Biol. 2006;356:1248–1262. doi: 10.1016/j.jmb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen T.H., Galej W.P., Bai X.C., Savva C.G., Newman A.J., Scheres S.H., Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schirle N.T., MacRae I.J. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 43.Hosokawa K., Fujimura R.K., Nomura M. Reconstitution of functionally active ribosomes from inactive subparticles and proteins. Proc. Natl. Acad. Sci. U.S.A. 1966;55:198–204. doi: 10.1073/pnas.55.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traub P., Nomura M. Structure and function of Escherichia coli ribosomes. I. Partial fractionation of the functionally active ribosomal proteins and reconstitution of artificial subribosomal particles. J. Mol. Biol. 1968;34:575–593. doi: 10.1016/0022-2836(68)90182-4. [DOI] [PubMed] [Google Scholar]
- 45.Traub P., Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. U.S.A. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H., Abeysirigunawarden S.C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., Ha T., Woodson S.A. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014;506:334–338. doi: 10.1038/nature13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulder A.M., Yoshioka C., Beck A.H., Bunner A.E., Milligan R.A., Potter C.S., Carragher B., Williamson J.R. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.