Abstract
Post-translational modifications (PTMs) regulate many aspects of protein function and are indispensable for the spatio-temporal regulation of cellular processes. The proteome-wide identification of PTM targets has made significant progress in recent years, as has the characterization of their writers, readers, modifiers and erasers. One of the most elusive PTMs is poly(ADP-ribosyl)ation (PARylation), a nucleic acid-like PTM involved in chromatin dynamics, genome stability maintenance, transcription, cell metabolism and development. In this article, we provide an overview on our current understanding of the writers of this modification and their targets, as well as the enzymes that degrade and thereby modify and erase poly(ADP-ribose) (PAR). Since many cellular functions of PARylation are exerted through dynamic interactions of PAR-binding proteins with PAR, we discuss the readers of this modification and provide a synthesis of recent findings, which suggest that multiple structurally highly diverse reader modules, ranging from completely folded PAR-binding domains to intrinsically disordered sequence stretches, evolved as PAR effectors to carry out specific cellular functions.
INTRODUCTION
Much of the complexity observed at the proteome level is due to post-translational modifications (PTMs) of proteins. PTMs can regulate all major aspects of protein function, including protein localization, interactions, protein stability and enzymatic activities. When considering proteins as the workhorses of a cell, PTMs could be seen as the equestrians that guide all efforts into the right direction. This direction might change over time, in particular when cells have to respond to internal and external cues, and most PTMs therefore do not constitute stable protein changes, but instead provide a means to dynamically regulate protein functions. This is due to the reversibility of most PTMs, and specific enzymes have evolved to antagonistically regulate PTMs by removing modifications from their target proteins. Thus, the interplay between the enzymes that covalently attach PTMs onto proteins, i.e. the writers, and the enzymes that revert these reactions, i.e. the erasers, determines the extent of protein modifications at any given point in time. Adding to this complexity, several PTMs can be modified themselves, and we are only beginning to understand how such modifications of PTMs contribute to the regulation of protein function. An important feature of many PTMs is that they can be recognized by specific protein domains, which thereby act as readers of PTMs, and the identification and characterization of such readers has become as pressing as the identification of PTM targets themselves. Moreover, in many cases reader proteins interact only transiently with their targets, and capturing these dynamics is important if we want to understand how PTMs and their binding partners regulate cellular functions.
Poly(ADP-ribosyl)ation (PARylation) is a PTM that has attracted considerable attention over the last decades due to its manifold cellular functions and the recently uncovered promises associated with its inhibition in cancer therapy. PARylation is defined by the successive conjugation of ADP-ribosyl units derived from NAD+ to generate polymeric ADP-ribose chains (1–3). Consequently, PARylation is significantly different from other typical PTMs in that it is neither a small moiety modification, such as phosphorylation, acetylation or methylation, nor does it represent a polypeptide PTM such as ubiquitylation or sumoylation. Rather, PARylation is characterized by the extensive conjugation of identical molecular building blocks, i.e. ADP-ribosyl units, which together form long and highly negatively charged linear or branched polymers. Despite these differences, PARylation shares many features with other PTMs: its formation relies on writers, i.e. enzymes capable of synthesizing ADP-ribose chains, and it is reversible, with modifiers and erasers working together to degrade poly(ADP-ribose) (PAR) chains (4). Moreover, several readers of PAR chains have been identified in recent years, and the structural diversity within this growing family of PAR-binding domains suggest that PAR can function as a versatile scaffold that dynamically regulates intracellular protein assembly.
In this article, we discuss recent developments that shed new light onto the multiple cellular functions of PAR and the enzymes involved in its generation and turnover. We then focus on PAR-binding modules, the readers of poly(ADP-ribose), and highlight how their structural and functional diversity makes them fit for purpose. Specifically, we discuss how the structural complexity of PAR itself is matched by the high degree of structural diversity found in its readers, ranging from completely folded PAR-binding domains to intrinsically disordered sequence stretches that make multivalent interactions with PAR and can phase separate to dynamically compartmentalize the intracellular space. As a unifying theme, we propose that the different modes of interaction are tightly linked to the functional consequences of PAR binding, and we discuss the implications for cellular PAR functions and their relevance for human disease.
Poly(ADP-ribosyl)ation
The first discovery of poly(ADP-ribosyl)ation was made by Chambon and colleagues more than 50 years ago (5). Interestingly, the polymer identified by Chambon and coworkers was initially assumed to resemble polyadenylic acid and was only later found to constitute a PTM rather than the product of an RNA polymerase. Nevertheless, the similarities between PAR and nucleic acids remain striking, and consequently the enzymes responsible for PAR formation were later called PAR polymerases (PARPs). PARP enzymes use nicotinamide adenine dinucleotide (NAD+) as their substrate, which they cleave into ADP-ribose and nicotinamide (NAM) (6). While the ADP-ribosyl moiety is covalently attached onto target proteins, NAM gets released. For PAR chains to be generated, additional NAD+ molecules have to be cleaved and the resulting ADP-ribosyl units have to be attached onto already existing ones. Thus, sequential reaction cycles are needed to transform mono-ADP-ribose into oligo(ADP-ribose) and eventually into poly(ADP-ribose) (7). The resulting polymer, which can be both linear and branched, is highly anionic due to two negative charges associated with each ADP-ribosyl moiety. The negative charge is one of the features shared between PAR chains and nucleic acids, which contain one (single stranded RNA) or two (double-stranded DNA) negative charges per unit. PAR synthesis is tightly controlled in mammalian cells with steady-state levels being kept relatively low. Certain stimuli such as genotoxic stress or transcriptional activity can activate PARP enzymes, resulting in a rapid increase in PAR levels (7). PAR synthesis is transient, however, and PAR chains are efficiently removed by PAR degrading enzymes (8), exemplifying how the regulated interplay between writers and erasers dynamically controls the extent and duration of this PTM.
PAR polymerases and their cellular functions
At least eighteen proteins exist in human cells with a catalytic domain similar to the one present in PARP1, the first enzyme identified to synthesize PAR. However, not all of these proteins seem to be active or capable of generating PAR chains (8,9). Consequently, it has been proposed to rename the PARP protein family and refer to this class of proteins collectively as ADP-ribosyltransferases of the diphtheria toxin-like type (ARTDs) (Table 1) (10). Within this class, only four proteins have well documented PARylation activity: PARP1, PARP2 and the two tankyrases (8–10). The other family members include mono-ADP-ribosyltransferases as well as ARTD-like proteins without measurable enzymatic activity, and their emerging cellular functions have recently been covered elsewhere (7–11).
Table 1. The PARP/ARTD protein family.
| PARP | ARTD | Alternative names | PAR formation |
|---|---|---|---|
| PARP1 | ARTD1 | yes | |
| PARP2 | ARTD2 | yes | |
| PARP3 | ARTD3 | (?) | |
| PARP4 | ARTD4 | vPARP | |
| PARP5A | ARTD5 | Tankyrase 1 | yes |
| PARP5B | ARTD6 | Tankyrase 2 | yes |
| PARP6 | ARTD17 | ||
| PARP7 | ARTD14 | tiPARP, RM1 | |
| PARP8 | ARTD16 | ||
| PARP9 | ARTD9 | BAL1 | |
| PARP10 | ARTD10 | ||
| PARP11 | ARTD11 | ||
| PARP12 | ARTD12 | ZC3HDC1 | |
| PARP13 | ARTD13 | ZC3HAV1, ZAP1 | |
| PARP14 | ARTD8 | BAL2, CoaSt6 | |
| PARP15 | ARTD7 | BAL3 | |
| PARP16 | ARTD15 | ||
| TPT1 | ARTD18 |
Overview of the 18 PARP/ARTD family members, their alternative names and their confirmed activity as PAR polymerases.
PAR synthesis can be observed under a variety of cellular conditions, most prominently under conditions of cell stress. Genotoxic stress in particular causes a rapid and pronounced increase in PAR levels, and PARylation has thus been studied mostly in the context of DNA damage signaling and repair (12). However, more recently also other functions of PARylation have emerged, including roles in DNA replication, transcription and RNA processing (13,14). Deregulated PAR homeostasis can thus impact cellular differentiation, embryonic development, inflammation, cellular metabolism, cancer development and aging (6–9). While PARP1 is responsible for most of the PAR synthesis observed in mammalian cells, PARP2 is also involved in several PAR-dependent functions and may have at least in part overlapping functions with PARP1, a notion that is supported by the embryonic lethality of mice lacking both enzymes and the elevated genomic instability observed in double-deficient embryonic fibroblasts (15). Thus, both PARP1 and PARP2 have important functions in the maintenance of genome integrity (16). Besides PARP1 and PARP2, also PARP3 plays important roles in the repair of DNA damage, although its capability to generate PAR has remained controversial (16–21).
The tankyrases, first identified as telomere-associated proteins (22), generate linear PAR chains of up to 20 ADP-ribosyl units (23). They function in spindle formation during mitosis (24), control centrosome function (25,26), regulate proteasome assembly and protein turnover to tune Wnt/β-catenin, PTEN-AKT and Hippo signaling, (27–31), and their regulation of protein stability has been implicated in the inflammatory bone destruction observed in Cherubism (32,33).
While all writers of PAR generate the same anionic nucleic acid-like molecule, the cellular context in which the different PAR writers are activated can determine the extent of PARylation (chain length, branching complexity) and which proteins are targeted, providing a means for specific regulation of various cellular functions by PARylation.
Cellular targets of poly(ADP-ribosyl)ation
Due to the polydisperse and labile nature of PAR chains, its structural complexity and rapid turnover in cells, it has been difficult to unambiguously identify cellular targets of this modification by mass spectrometry-based proteomics. However, in recent years significant technological progress has been made and various PAR enrichment strategies have been established to identify PAR-associated proteins and map PAR acceptor sites (34–45). This has led to the discovery that a plethora of nuclear proteins associates with PAR, including many DNA repair factors, chromatin remodelers, cell cycle regulators, RNA-binding proteins and transcription factors (42–45). Many of these proteins are likely to both interact non-covalently with PAR and to be targeted by covalent PARylation. The ever increasing number of PARylation targets is remarkable, as is the emerging diversity in PAR acceptor sites: lysine, arginine, glutamate, aspartate, cysteine, diphthamide, serine, threonine, phosphoserine and asparagine have all been identified as residues targeted by ADP-ribosylation, with the charged amino acids lysine, arginine, glutamate and aspartate being the ones most commonly targeted by covalent PARylation (9,40). This raises questions about the specificity of PAR attachment onto acceptor proteins and about the target amino acid preference. Given that the rate-limiting step for PAR formation is the enzymatic cleavage of NAD+ during the initiation reaction, one possibility could be that the subsequent attachment of ADP-ribose onto target proteins is favored by the availability of suitable amino acids on the exposed surface areas of proteins associated with or in the vicinity of activated PARP enzymes. In such a scenario, PARylation would comprise the context-dependent activation of PARP enzymes, the ensuing nucleophilic attack of the substrate NAD+, as well as the attachment of ADP-ribose moieties onto available acceptor amino acids or onto growing PAR chains. While clearly distinct from non-enzymatic ADP-ribosylation this mechanism might explain the multitude of target proteins and acceptor amino acids identified to date, including the large number of acceptor sites found on PARP1 itself (18,37–39,45). Similar to the synergistic protein group modification employed by the SUMO system in response to DNA damage (46), the specificity of the system would be derived from locally confined activation of the enzymes rather than from the site-specific modification of target proteins. In analogy to the SUMO concept (46), PAR might thus act as a molecular glue to enhance protein residence times and interactions. Of note, and consistent with such a model, PARP1 is the major acceptor of its own reaction product, and it could be that PARP1, but also other non-diffusible, chromatin-associated PAR acceptors such as the histones, primarily serve as a matrix for the PAR scaffold, which can then exert many of its functions by non-covalent interactions with PAR-binding proteins. Such a model is not mutually exclusive with specific functions of certain PAR acceptor sites, and further work is needed to dissect target site specificity and the functional consequences of site-specific PARylation.
Readers of poly(ADP-ribose) and their functions
The recent advancements in proteome-wide analyses of the cellular PARylome provide a hitherto unsurpassed opportunity to globally assess the relative contribution of PARP enzymes for PARylation in different cellular conditions and to identify their targets. This avenue will be particularly fruitful if combined with cellular assays to determine the functional role of PARylation sites in acceptor proteins. But already now, PAR proteomics has tremendously broadened our knowledge on PAR-associated proteins and thus provides a basis to better understand how PAR can exert its many biological functions. Hundreds of proteins interact directly or indirectly with PAR (42,43), and localized PAR formation can thus have profound effects on subcellular protein re-distribution and thereby influence many different signaling pathways, including the DNA damage response, transcriptional regulation, protein stability, cell fate decisions and cell death (47–50). Over the last years, several PAR-binding modules have been described (Figure 1), and their structural properties range from completely folded PAR-binding domains to disordered sequence stretches that can make multivalent interactions with PAR (Table 2). In light of this growing complexity, unifying models can carve out the essence of how proteins read the PAR signal and how the different PAR readers evolved to appear designed to be fit for purpose.
Figure 1.
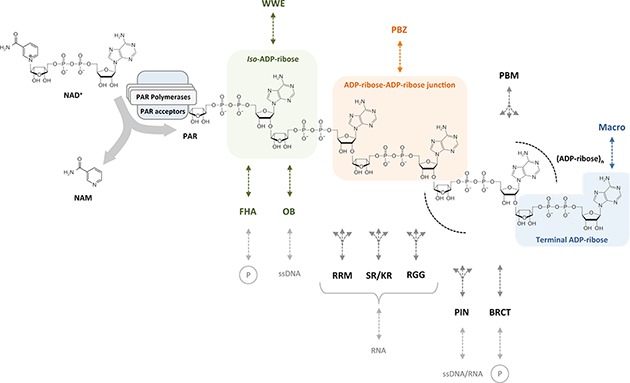
Readers of poly(ADP-ribose). PAR polymerases use NAD+ to generate highly anionic linear and branched (not shown) PAR chains of different size and branching complexity. Besides the classical, well-characterized PAR reader modules WWE, PBZ, PBM, and macrodomains (top) also newly emerging PAR reader modules such as FHA, OB-fold, PIN domain, RRM, SR and KR repeats, RGG repeats and BRCT (bottom) appear as PAR readers and effectors. Multi-branched arrows indicate that the exact binding sites have not been defined.
Table 2. Readers of poly(ADP-ribose).
| Reader Module | Module size | Defined protein fold | Interaction mode | Main functions | Examples | Key references |
|---|---|---|---|---|---|---|
| PBM | ≈20 residues | no | unknown, potentially electrostatic interactions | DNA replication and repair, cell cycle regulation, chromatin architecture, RNA metabolism | H1, H2A, H2B, H3, H4, p21, p53, XRCC1, XPA, MSH6, ERCC6, ATM, MRE11, DNA-PKcs, KU70, WRN, DNA Ligase 3, Polymerase epsilon, TERT, DEK, CAD, CENP-A, CENP-B, Lamin A/C, BUB3, hCAP-D2, HK1, HKDC1, G3BP1, hnRNPA1, hnRNPK, hnRNPH, hnRNPG, hnRNPM, hnRNPA2B1, hnRNPC1C2, AURKAIP1, NF-kappaB, iNOS | Pleschke et al. (2000), Gagné et al. (2003), Fahrer et al. (2007), Gagné et al. (2008), Haince et al. (2008), Fahrer et al. (2010), Malanga and Althaus (2011), Gagné et al. (2012), Kalisch et al. (2012), Krietsch et al. (2013), Popp et al. (2013) |
| PBZ | ≈30 residues | yes | C2-H2-type zinc finger binds to two consecutive ADP-ribose moieties | DNA damage signaling and repair | APLF, CHFR | Ahel et al. (2008), Rulten et al. (2008), Isogai et al. (2010), Oberoi et al. (2010), Eustermann et al. (2010), Li et al. (2010) |
| Macrodomains | ≈130–190 residues | yes | Recognizes the terminal ADP-ribose unit | Chromatin remodeling | macroH2A, ALC1/CHD1L, C6orf130/TARG | Ahel et al. (2009), Gottschalk et al. (2009), Timinszky et al. (2009) |
| WWE | ≈80–100 residues | yes | Binds to iso-ADP-ribose | Protein turnover | RNF146/Iduna | Zhang et al. (2011), Kang et al. (2011), Wang et al. (2012), DaRosa et al. (2015) |
| FHA/BRCT | ≈80–100 residues | yes | Phosphate-binding pockets interact with ADP-ribose or iso-ADP-ribose | DNA damage signaling and repair | APTX, PNKP, XRCC1, NBS1, BARD1, DNA Ligase 4 | Li et al. (2013), Li and Yu (2013), Breslin et al. (2015) |
| RRM | ≈60–80 residues | yes | unknown, potentially electrostatic interactions | DNA damage signaling and repair, RNA metabolism | ASF/SF2, NONO, RBMX, TAF15 | Malanga et al. (2008), Krietsch et al. (2012), Adamson et al. (2012), Izhar et al. (2015) |
| SR repeats and KR-rich motifs | variable | no | unknown, potentially electrostatic interactions | Gene expression, RNA metabolism | ASF/SF2, dMi-2 | Malanga et al. (2008), Murawska et al. (2011) |
| OB-fold | ≈70–150 residues | yes | Binds to iso-ADP-ribose | DNA damage signaling and repair | SSB1, BRCA2 | Zhang et al. (2014), Zhang et al. (2015) |
| PIN domains | ≈130–150 residues | yes | unknown, potentially electrostatic interactions | DNA damage signaling and repair | EXO1 | Zhang et al. (2015) |
| RG/RGG repeats | variable | no | unknown, potentially electrostatic interactions | Stress granule assembly, liquid demixing, DNA repair | MRE11, G3BP1, SAFB1, FUS/TLS, EWS/EWSR1, TAF15 | Haince et al. (2008), Isabelle et al. (2012), Altmeyer et al. (2013), Mastrocola et al. (2013), Rulten et al. (2014), Altmeyer et al. (2015) |
Currently known PAR reader modules are listed together with their structural features, interaction mode with PAR, and main functions associated with PAR-binding. Examples and key references are provided. Notably, several proteins contain multiple PAR-binding modules, i.e. have a modular organization that combines multiple PAR reader domains within the same polypeptide chain (e.g. RRM, RG/RGG and PBM), which may cooperate for PAR binding.
The basic PAR-binding motif (PBM)
The first reader module of PAR was described already 15 years ago as a loosely defined 20 amino acid cluster of hydrophobic amino acids spaced by basic residues (51). Following the original discovery of the PBM, a more recent in silico analysis revealed that more than 800 proteins contain this motif (42). Even a refined search using a more stringent motif based on experimentally derived interaction data still yielded over 500 predicted hits, suggesting that a sizable fraction of the proteome, including several DNA replication and repair proteins, cell cycle regulators, chromatin modifiers and RNA binding proteins, are able to directly interact with PAR (42). Structural data on the PBM and its interaction with PAR are not available and its affinity to PAR is likely due to electrostatic interactions between the positively charged amino acids present in the PBM consensus ([HKR]-X-X-[AIQVY]-[KR]-[KR]-[AILV]-[FILPV]) and the negatively charged PAR chains. Nevertheless, these interactions can reach relatively high affinities with KD values in the sub-micromolar (10−7) to nanomolar (10−9) range (47), in particular when multiple PBMs within the same protein cooperate to bind long PAR chains. Given the large number of proteins containing the PBM consensus, the function of this PAR reading module might go well beyond the recruitment of just a few specific factors to sites of PAR formation, for instance at genomic lesions, and it could be that the collective or protein group assembly at PARylation sites fosters mutual interactions within multi-protein complexes that make specific reactions more efficient. Multivalent interactions with long chains of PAR may both increase affinity and specificity (52–55), and several nuclear proteins contain not just one but multiple PBMs (42,51), which may cooperate in their binding to PAR chains. Thus, the PBM seems to have evolved as a versatile PAR-reading module capable of dynamically interacting with PAR chains in a manner that takes polymer length and branching complexity into account to drive multi-protein assemblies at PARylation sites.
The PAR-binding zinc finger (PBZ)
In 2008, a Cys2-His2 type zinc finger motif with PAR-binding properties was identified in the two DNA damage responsive proteins APLF (aprataxin and PNK-like factor) and CHFR (checkpoint with forkhead and ring finger domains) (56). The structure of this relatively small motif of less than 30 amino acids with the consensus sequence [K/R]-X-X-C-X-[F/Y]-G-X-X-C-X-[K/R]-[K/R]-X-X-X-X-H-X-X-X-[F/Y]-X-H was solved two years later (57–60). The biochemical and structural evidence suggests that two consecutive ADP-ribose moieties within the PAR chain are bound by the PBZ (59,60). Interestingly, and in contrast to the single PBZ present in CHFR, APLF contains a tandem PBZ, and the two zinc finger motifs cooperate to achieve high affinity PAR binding (59,60). Besides APLF and CHFR a third human protein, SNM1A/DCLRE1A (DNA cross-link repair 1A protein), contains a PBZ (56). However, compared to APLF and CHFR, the PBZ of SNM1A/DCLRE1 lacks critical PAR-binding residues, suggesting that it may not bind to PAR (58). Thus, only two proteins in the human proteome seem to have acquired high affinity PAR-binding zinc fingers, and their cellular functions are closely linked to PAR induction: the recruitment of APLF to sites of DNA damage to promote repair by non-homologous end-joining (NHEJ) is critically dependent on PAR formation and on the tandem PBZ (20,61), while similarly the CHFR-dependent antephase checkpoint is dependent on PAR synthesis and an intact PBZ (56,58). The PBZ thus represents a highly specialized PAR reader module, which is present in only very few proteins and specifically regulates their functions in the DNA damage response and cell cycle regulation. In addition to the canonical PBZ, a putative variation of the PBZ motif was recently identified in the checkpoint kinase CHK1, and shown to confer affinity to PAR (62). Potentially, other hitherto unknown variations of the PBZ might exist and their PAR-binding potential remains to be tested.
Macrodomains
In contrast to PBM and PBZ, macrodomains are sizable, globular reader modules containing between 130 and 190 amino acids. Macrodomains bind to various molecules that comprise ADP-ribose as component, including O-acetyl-ADP-ribose, mono- and also poly(ADP-ribose) (63–65). It should be noted, however, that based on structural data macrodomains interact only with the terminal ADP-ribose moiety of the PAR chain (65). This suggests that macrodomains for which PAR binding has been observed could be considered mono-ADP-ribose readers that acquired the property to also recognize the terminal ADP-ribose unit in PAR. The experimentally derived affinities to ADP-ribose reach KD values in the sub-micromolar range (10−7) (47). A comprehensive understanding of the potential interplay between different ADP-ribose-containing molecules and macrodomain-containing ADP-ribose readers in vivo is still missing. Based on amino acid sequence comparison, 11 human proteins contain a macrodomain, including the PARP/ARTD family members PARP9, PARP14 and PARP15, the histone variants macroH2A1.1, macroH2A1.2 and macroH2A2, as well as macroD1/MDO1, macroD2/MDO2, C6orf130/TARG, ALC1/CHD1L and GDAP2. Interestingly, the macrodomain-containing PARP/ARTD family members contain two (PARP9, PARP15) or even three consecutive macrodomains (PARP14) (66). Not all macrodomains identified seem to be able to bind to PAR, but some of the ones that are able to recognize PAR have even acquired catalytic activity and participate in the degradation of PAR (8). Thus, relatively subtle sequence variations within the macrodomain fold can transform this module from a non-PAR-binder to a high affinity PAR reader, and even to an ADP-ribose eraser—a unique feature among the growing number of PAR readers (see below).
WWE domains
The WWE domain, named after its most conserved amino acids tryptophan (W) and glutamate (E), is found in 12 human proteins. Interestingly, these PAR readers are present in only two classes of proteins: PARP/ARTD family members themselves, and ubiquitin ligases (67). The presence of both WWE domains and macrodomains in several PARP/ARTD family members suggests an intriguing and largely unexplored interplay between writing and reading the PAR signal. In this regard, it is noteworthy that within the PARP/ARTD family a clear separation into PAR writers and readers seems to exist, because macro- and WWE domains are present only in family members that do not synthesize PAR, and they are absent from the PAR generating enzymes PARP1, PARP2, PARP5A and PARP5B. Unlike macrodomains, the WWE domains interact with iso-ADP-ribose, a structure comprising parts of two consecutive ADP-ribosyl units within the PAR chain (67). Consistent with a binding preference for iso-ADP-ribose, the experimentally derived KD values are relatively low for ADP-ribose (in the millimolar range), but reach sub-micromolar values (10−7) for iso-ADP-ribose (47). Four residues of the WWE domain are crucial for the interaction with iso-ADP-ribose and these are conserved in PARP11, PARP13 and the first WWE of PARP12, but neither in the second WWE of PARP12, nor in PARP7, or PARP14 (67). Thus, similar to the macrodomains, not all WWE domains seem to possess PAR-binding potential. The four residues crucial for PAR binding are also present in the WWE domains of several ubiquitin E3 ligases, including RNF146/Iduna, HUWE1/Mule, TRIP12/ULF, and Deltex1, Deltex2 and Deltex4. In contrast to the poorly understood role of WWE-mediated PAR reading by members of the PARP/ARTD family, a clear functional link has been established between PAR binding and ubiquitin ligase activity. The best-studied example is the E3 ligase RNF146/Iduna, which catalyzes PAR-dependent ubiquitylation of target proteins to mediate their proteasomal degradation (67,68). In this case, PAR-recognition on target proteins by the WWE domain is a prerequisite for their ubiquitylation. PAR-dependent ubiquitylation is thus reminiscent of the SUMO-dependent ubiquitylation that is carried out by SUMO-targeted ubiquitin ligases (STUbLs) (69). Interestingly, PAR-dependent ubiquitylation is employed both by the cellular response to DNA damage, where PARP1 mediates the PARylation required for RNF146/Iduna targeting (68), and in Wnt/β-catenin signaling, where the tankyrases PARylate the protein axin to allow for its recognition by RNF146/Iduna (28,29). PAR-dependent ubiquitylation by RNF146/Iduna depends on the allosteric activation of its RING domain via a major intramolecular conformational rearrangement upon PAR-binding (70). Thus, reading the PAR signal can provide a switch that turns an inactive catalytic domain into an active one. The list of substrates for PAR-dependent ubiquitylation is growing, and it includes the regulator of Wnt/β-catenin signaling axin (28,29), adaptor protein 3BP2 (32,33), and the DNA repair factors Ku70, XRCC1, DNA ligase III, PARP1 and PARP2 (68). Whether the ubiquitin ligase activities of Deltex1, Deltex2 and Deltex4, involved in notch signaling, or of the HECT domain-containing ubiquitin E3 ligases HUWE1/Mule and TRIP12/ULF, both involved in genome integrity maintenance (71,72), are regulated by WWE domain-mediated PAR recognition remains to be investigated. Nevertheless, PAR-dependent ubiquitylation by PAR-targeted ubiquitin ligases (which, by analogy to STUbLs, could be dubbed PTUbLs) has emerged as one of the most exciting cellular functions of reading the PAR signal.
FHA and BRCT domains
FHA (Forkhead-associated) and BRCT (BRCA1 C-terminal) domains have been studied mainly as readers of phosphorylation marks and modulators of protein-protein interactions (73). Recently, however, it was shown that FHA and BRCT domains also confer affinity to PAR (74–76). Similar to phosphorylation, the PAR signal is negatively charged, which could mediate electrostatic interactions with the phosphor-binding pockets of FHA and BRCT domains. Interestingly, the FHA domains of APTX (Aprataxin) and PNKP (Polynucleotide kinase-3′-phosphatase) interact with iso-ADP-ribose, the structure also recognized by WWE domains, whereas the BRCT domains of DNA Ligase IV, XRCC1 and NBS1 recognize the ADP-ribosyl moiety within PAR (74,75). Also the BRCT domains of BARD1 (BRCA1-associated RING domain protein 1) have affinity for PAR and contribute to the recruitment of the BRCA1/BARD1 complex to DNA double-strand breaks (76). The wave of DNA damage-induced PARylation might thus serve as a molecular trap to catch proteins required during subsequent stages of repair, and cooperate with additional modifications, such as the phosphorylation of H2AX by DNA damage response (DDR) kinases and the ensuing chromatin ubiquitylation, to ensure their sustained retention on the damaged chromatin.
RNA and DNA binding motifs
One of the most intriguing recent discoveries is that PAR can be recognized by protein motifs that are known to bind to RNA or DNA. This development gives rise to the notion that important cellular functions of PAR could reside in its early recognized similarity to other nucleic acids and its potential to outcompete the binding of RNA- and DNA-associated proteins to their target nucleic acids under conditions when PAR levels peak.
The RRM
The RNA recognition motif (RRM) is one of the most abundant RNA binding domains with a broad spectrum of target RNAs and capable of recognizing multiple RNA sequences and structures. It is therefore not too surprising that also PAR as a nucleic acid-like molecule lacking sequence information can be recognized by RRMs. In the cellular context, where not only affinities matter but also the dynamic changes in local concentrations of available molecules to interact with, even relatively unspecific interactions can dynamically alter macromolecular assemblies and can therefore be relevant for cell function. Several examples of RRM-containing proteins, which assemble at sites of PAR formation to promote genome stability, exist, including proteins of the heterogeneous nuclear ribonucleoprotein (hnRNP) family (77,78), the RNA processing factors NONO and RBMX (79,80), and a growing number of transcription factors (81). Since RNA and PAR can compete for the same binding proteins (79), peaks in PAR levels may dynamically outcompete RNA binding and assemble various RNA-binding proteins at PARylation sites, including sites of genomic lesions.
SR repeats and KR-rich motifs
Serine/Arginine repeats (SR repeats) are found in a second group of RNA-binding proteins, in particular in regulators of splicing, and several splicing factors were shown to interact with PAR, including ASF/SF2, SF3A1, SF3B1 and SF3B2 (43,82). The affinity for both RNA and PAR is likely to be mediated by the cluster of positive charges associated with the arginine-rich repeats, and electrostatic interaction with PAR therefore is a recurrent theme in reading the PAR signal. In line, PAR-binding was also observed for lysine- and arginine-rich (KR-rich) motifs present in the nucleosome remodeler dMi-2 (83).
The OB-fold
The oligonucleotiode/oligosaccharide-binding fold (OB-fold) is a 70–150 amino acid containing five-stranded beta-barrel with a terminating alpha-helix found in proteins that bind to single-stranded nucleic acids or oligosaccharides. Recent evidence suggests that, similar to the RNA-binding motifs discussed above, also the OB-fold is a reader of PAR (84,85). The OB-fold recognizes the PAR-specific iso-ADP-ribose and this binding is required to bring the single-stranded DNA-binding protein SSB1 to sites of DNA damage (84), as well as for the early recruitment of BRCA2 to DNA break sites (85). Just like RNA and PAR, single-stranded DNA is a flexible polymer with free rotation around the phosphodiester backbone that links adjacent nucleotides, and its binding to OB-folds relies at least in part on electrostatic interactions, allowing for a dynamic competition between single-stranded DNA and PAR for OB-fold interactions.
PIN domains
PIN domains, named after the amino-terminus of the PilT protein (PilT N-terminus), are evolutionarily conserved single-stranded DNA- and RNA-recognition domains with nucleolytic function, and represent the latest addition to the growing spectrum of PAR-binding domains (86). The PIN domains of EXO1, GEN1 and SMG5 recognize PAR with relatively high affinity (a dissociation constant of 200–300 nM was determined for the EXO1 PIN domain), and the PIN domain mediated interaction with PAR is required for the early recruitment of EXO1 to DNA break sites for efficient DNA end resection (86). Together with the recent finding that the DNA-binding domains of many transcription factors contribute to their PAR-dependent recruitment to DNA damage sites (81) these data suggest that DNA- and PAR-binding are in fact overlapping features of a considerable number of proteins.
RG/RGG motifs
The RG/RGG motifs, regions rich in arginines (R) and glycines (G), also termed glycine-arginine-rich (GAR) domains, are found in more than 1000 human proteins involved in numerous cellular functions, including DNA damage signaling, transcription and RNA processing (87). The accumulation of positive charges not only confers affinity to RNA, but has recently also been shown to mediate interactions with PAR. This was first suggested by a study showing that the rapid recruitment of the nuclease MRE11 to genomic lesions was dependent on its GAR domain as well as on the formation of PAR (88). Also the stress granule component G3BP1 binds PAR via its GAR domain, and the G3BP1-mediated stress granule assembly in response to genotoxic stress is impaired by PARP inhibition (43). More recently, RGG-rich regions in the RNA-binding proteins FUS/TLS, EWS/EWSR1, TAF15, SAFB1, SAF-A and hnRNPUL1/2 were shown to be involved in the PAR-dependent assemble of these proteins at sites of DNA damage (81,89–94). Extended RGG repeats, as observed in several RNA-binding proteins, can be regarded as low complexity domains, and, similar to the PBM, their structural flexibility combined with their positive charge may allow them to make multivalent electrostatic interactions with polydisperse PAR chains leading to PAR-nucleated multi-protein assemblies.
PAR-seeded liquid demixing
Interestingly, in the case of FUS/TLS, EWS/EWSR1 and TAF15, the 18–22 RGG repeats within the carboxyl-terminal half of the proteins are coupled to extended prion-like, aggregation-prone low complexity domains at the amino-termini of these proteins (95). The high degree of low complexity sequences allows these proteins to undergo intracellular phase transitions and thereby demix from the soluble environment (96,97). Such liquid demixing by phase separation can dynamically re-organize the soluble intracellular space by generating membrane-less compartments (98,99). Interestingly, the nucleic acid-like polymer PAR can function as a molecular seed for the dynamic liquid demixing of these proteins at sites of DNA damage (95,100). While genomic lesions could be a prime trigger for PAR-seeded liquid demixing, we envision that dynamic, PAR-seeded compartmentalization might be a more common theme in PAR biology. Indeed, PAR-seeded phase separations may occur not only in response to DNA damage, but in a variety of cellular contexts in which PAR has been reported to initiate protein assemblies into membrane-less compartments. These include the formation of stress granules (43,101), nucleoli (102,103), Cajal bodies (104), spliceosomes (105), viral stress granules (11) and transcriptosomes (106). In the case of transcriptional regulation, phase separation of the low complexity domain-containing proteins FUS/TLS, EWS/EWSR1 and TAF15 at gene promoters was recently proposed to provide a landing platform for the low complexity carboxyl-terminal domain (CTD) of RNA Polymerase II (107), and it is tempting to speculate that PAR formation could be the nucleation event that initiates this process at certain promoters. Thus, PAR reading not only involves the recruitment of enzymes and protein scaffolds to promote biochemical reactions, but also the assembly of proteins poised to undergo phase separation when reaching local threshold concentrations to alter the physico-chemical environment and dynamically re-organize the subcellular space by PAR-seeded membrane-less compartmentalization (Figure 2).
Figure 2.
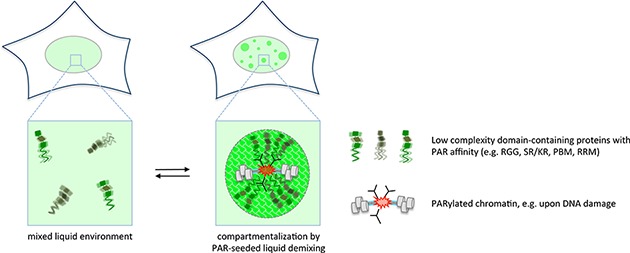
PAR-seeded liquid demixing. PAR chains assemble hundreds of proteins, including many intrinsically disordered, low complexity domain-containing proteins, at sites of PAR formation, which collectively re-shape the local environment. This can lead to dynamic compartmentalization by liquid demixing, indicating that PAR can function as a general organizer of the soluble intracellular space.
Modifiers and erasers of poly(ADP-ribose)
The assembly of numerous cellular proteins at sites of PAR formation bears inherent risks. First, several PAR reader modules possess dual affinity for PAR and RNA/DNA, and—while cells may want to transiently assemble these proteins at sites of PARylation—it would be detrimental to stably sequester them away from their RNA or DNA targets. Second, while the PAR-seeded assembly of intrinsically disordered proteins with extended stretches of low complexity sequences can sub-divide the intracellular space through phase separation, it would be fatal if these delicate compartments turned into irreversible protein aggregates. Third, the generation of PAR is associated with the consumption of significant amounts of NAD+ and has the potential to cause a cellular energy crisis by inhibiting glycolysis (108,109). For all these reasons, it is at least as important to terminate PAR signaling in a timely manner, as it is to trigger PAR formation in the first place. Several enzymes have been identified that can cleave PAR and remove ADP-ribose from modified proteins (8). The most important enzyme for the degradation of chromatin-associated PAR is poly(ADP-ribose) glycohydrolase (PARG). PARG exhibits both exo- and endo-glycohydrolase activity, with the latter being capable of generating protein-free PAR fragments, which can potentially serve as binding partners for various PAR readers (110,111). However, its predominant exo-glycohydrolase activity suggests that the primary mode how PARG degrades PAR is through sequential removal of ADP-ribose moieties from the distal end of the polymer (112). Interestingly, although unrelated in amino acid sequence, the PARG catalytic domain has structural homology to PAR-binding macrodomains (113), linking, from a structure-function point of view, reading of PARylation to modifying the PAR chain and erasing the polymer. PAR degradation requires accessibility of PAR chains, but whether certain PAR readers might constitute obstacles for PARG is largely unknown. Of note, PARG was described to be unable to hydrolize the protein-proximal ADP-ribose unit, potentially due to steric hindrance (113–115). However, Nudix hydrolases (see below) and certain macrodomain-containing proteins, which have acquired catalytic activity, are capable of cleaving the protein-ADP-ribose bond. This group of macrodomain-containing erasers includes macrodomain-containing protein 1 (macroD1/MDO1), macrodomain-containing protein 2 (macroD2/MDO2) and terminal ADP-ribose glycohydrolase (TARG/C6orf130) (116–118). While macroD1/MDO1 and macroD2/MDO2 act specifically only on mono-ADP-ribosylated substrates (116,117), TARG/C6orf130 was reported to remove PAR chains en bloc from target proteins and thereby release free PAR, which may be further processed or serve as a scaffold for PAR-binding proteins (118). Interestingly, all three proteins show a preference for ADP-ribose attached to glutamic acid (116–118). This, together with the notion that ADP-ribose attachment may occur on both acidic and basic residues, suggests that writers and erasers might have evolved distinct substrate specificities, and that therefore different classes of enzymes might exist to counteract the reaction product of a single class of PAR polymerases. Room remains for putative enzymes to be discovered that can remove ADP-ribose from basic residues. In addition to PARG and the macrodomain-containing enzymes, ADP-ribosylhydrolase 3 (ARH3), a member of a structurally distinct class of ADP-ribosylhydrolases, can also degrade PAR (8). The major fraction of ARH3 is found in the cytoplasm, and consistent with its inability to cleave the last, protein-bound ADP-ribose, its physiological function could be the hydrolysis of free PAR chains released during apoptotic cell death (119). More recently, Nudix hydrolases (Nucleoside diphosphate-linked moiety X hydrolases) emerged as new PAR processing enzymes, which can remove PAR and convert protein-bound ADP-ribose into ribose-5′-phosphate (R5P) marks, releasing phosphoribosyl-AMP (PRAMP) as byproduct (120). Nudix hydrolases might thus counteract PARP activity both directly by PAR degradation and indirectly by generating products that may interfere with PARP-dependent ADP-ribosylation and PAR chain elongation. The physiological relevance of the R5P modification and whether enzymes exist to revert it is currently not known.
From recent biochemical, structural and in vivo work it has become clear that the importance of processing and erasing PAR chains is at least equal to the importance of generating PAR, and that multiple enzymes, which combine PAR-recognition and PAR-degradation within their structures, are involved in limiting PAR production and ensuring its timely removal.
Targeting PARylation in human diseases
Inhibitors against the writers of PAR have been available for many years, and these compounds gained widespread attention when it was found that PARP inhibition specifically kills cancer cells with defects in the homologous recombination (HR) DNA repair pathway (121,122). The synthetic lethality conferred by PARP inhibition in HR-defective cancer cells has sparked a variety of pre-clinical and clinical trials, and in 2014 the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) approved the PARP inhibitor Olaparib as a therapy against HR-compromised cancers. While the last decade has seen promising attempts to extend the spectrum of cancers that may respond to PARP inhibition beyond the originally characterized BRCA1- and BRCA2-deficient cancer cells, also the first resistance mechanisms appeared. These include loss of PARP protein expression, up-regulation of the P-glycoprotein multidrug efflux pump, and restoration of HR function—either by re-activating mutations in the BRCA1/2 genes, or by loss of components of the competing non-homologous end-joining (NHEJ) pathway (123). Nevertheless, PARP inhibition continues to be a promising avenue in targeted cancer therapies based on the concept of synthetic lethality, and we may expect the spectrum of cancers that respond well to PARP inhibition to grow further, but also anticipate the appearance of additional resistance mechanisms that have to be considered when designing treatment regimens.
While the use of PARP inhibitors in cancer has advanced quickly during the last decade, the limited toxicity and good tolerability together with the broad spectrum of cellular processes regulated by PARylation make PARP inhibitors candidate drugs to treat also non-oncologic human diseases. Indeed, clinical potential has been described in disease settings related to cardiovascular dysfunction and inflammation (124–126). Moreover, efforts to further improve PARP inhibitors and make these compounds more specific toward single PARP family members have led to the notion that besides PARP1 and PARP2 also the tankyrases and potentially other ARTD family members might be promising targets in certain medical conditions including cancer (127–129). In addition to the inhibition of PAR writers, the development of potent, cell-permeable inhibitors of PAR erasers, in particular of PARG, has become an active field of research (130,131). Given the delicate and highly dynamic balance between PAR formation and its degradation, PARG inhibitors might have therapeutic potential in settings in which PAR metabolism is deregulated, or in which impaired PAR degradation and the ensuing adverse effects on genome maintenance and survival would be clinically beneficial. Indeed, PARG is required to prevent detrimental accumulation of PAR upon prolonged replicative stress, and cells lacking PARG activity are hypersensitive to the replication inhibitor hydroxyurea (132), suggesting that PARG inhibition might synergize with chemotherapeutic approaches aimed at deliberately enhancing replication stress in cancer cells.
While targeting the writers and erasers of PARylation continues to be a promising avenue, little attention has been given to the possibility of targeting the readers of PARylation, despite the fact that many PAR functions are closely linked to non-covalent interactions between PAR-binding proteins and the anionic polymer. Although it might be challenging to design compounds that target the interface between PAR and PAR-binding proteins, in particular where structural information is still missing, promising examples exist in other areas of chromatin biology and provide a paradigm that PTM readers can be efficiently targeted, for instance in the case of epigenetic readers of the bromodomain and extra-terminal (BET) protein family (133). The disruption of interactions between PTMs and their readers could have significant, largely unexploited therapeutic potential. In particular with regard to PAR signaling, it might allow for the inhibition of sub-groups of PAR readers from binding to the polymer scaffold and thereby confer a higher level of specificity as compared to blocking PARP enzymes. Given that different PAR reader modules have evolved to perform specific cellular functions, targeting them individually or in sub-groups rather than abolishing PAR formation altogether may have a clear therapeutic potential. It may also help dissect different PAR functions and how they relate to different classes of PAR binders. For instance, specifically blocking genome caretakers from interacting with PAR at sites of DNA damage may compromise repair efficiency and result in synthetic lethal conditions, whereas blocking intrinsically disordered proteins from PAR-seeded self-assembly might decelerate their aggregation in certain pathological conditions. Mutations in these PAR-responsive proteins can enhance their aggregation propensity, thereby increasing the risk of turning dynamic, PAR-seeded phase transitions into irreversible aggregates that can compromise cell function (95,100). Likewise, deregulated PAR metabolism can result in pathologic PAR accumulation and cause fatal neurodegeneration (118,134,135). PARP activation and PAR formation have also been implicated in the onset of Alzheimer′s disease (136). Such neurodegenerative disease-associated changes in writing, reading and erasing PARylation provide a rationale to test whether targeting PARylation by enzyme inhibitors or by yet to be developed competitive inhibitors of PAR-binding may have a clinical potential in neurodegenerative disorders.
CONCLUDING REMARKS
Due to the many facets of PAR biology and its relevance for human health and disease, significant attention has been attributed to the identification and characterization of writers, modifiers and erasers of this unusual PTM. Moreover, it was realized that reading the PAR signal by PAR-binding proteins constitutes a major aspect of PAR biology. Indeed, non-covalent interactions with PAR can drive the dynamic re-localization and site-specific assembly of hundreds of proteins, including many DNA repair factors, chromatin remodelers, histone variants, transcription factors and RNA processing factors. In light of the large number and functional diversity of proteins responsive to PAR signaling, unifying concepts may help us grasp this ever-increasing complexity. While certain PAR reader modules seem to possess highly specific PAR-binding folds, which recognize defined biochemical structures present in PAR, e.g. PBZ, WWE, and macrodomains, other reader modules seem to comprise sequence elements, which confer affinity to PAR, but which may also allow for interactions with nucleic acids or other PTMs. Examples include the PBM, FHA and BRCT domains, RRM, RG/RGG, SR and KR repeats, as well as OB-folds and PIN domains. Their structural and chemical properties enable them to undergo electrostatic, often multivalent interactions with PAR chains, which can cooperate to reach nanomolar affinities. In silico analyses of the human proteome as performed previously to identify PBMs (42) and RG/RGG motifs (87), combined with mass-spectrometric identification of PAR-associated complexes and their dynamics as well as in vitro affinity measurements will help to quantitatively define PAR-mediated protein assemblies under various cellular conditions. The tight control of PAR levels through context-dependent regulation of PARP enzymes as well as the rapid PAR turnover by erasers make this nucleic-acid like modification ideally suited to transiently outcompete other interactions in order to dynamically re-organize the soluble intracellular space. Consistent with PAR being able to outcompete protein-nucleic acid and protein-PTM interactions, non-consensus binding sites in highly expressed RNAs can compete efficiently with high-affinity consensus sequences in RNAs expressed at lower levels (137). Thus, rather than viewing interactions as either specific or nonspecific, it is necessary to realize that proteins possess affinity to a range of potential interaction partners and that dynamic changes in their relative availability determines which types of interactions manifest at any given time. Similar to highly expressed RNAs with low-affinity non-consensus binding sites, PAR could be regarded as an inducible nucleic acid-mimicking scaffold that lacks sequence information but is nevertheless able to transiently trap and sequester dozens to hundreds of proteins at sites of PARylation. This dynamic re-organization of multi-protein assemblies involves the phase separation of low complexity domain-containing proteins, and it is tempting to speculate that various aspects of cellular PAR functions are closely linked to PAR-seeded compartmentalization and the accompanying changes in the physicochemical environment. In this regard, PAR would share many features with RNAs as structural elements that control the biogenesis of nuclear bodies (138), closing the circle to the first description of PAR in 1963 as a polyadenylic acid (5).
Acknowledgments
We apologize to authors whose work could not be cited due to space limitations and are thankful to members of our institute for helpful discussions and comments on the manuscript. Research in the lab of M.A. is supported by the Swiss National Science Foundation (Grant PP00P3_150690/1) and by the University of Zurich Association Research Talent Development Fund.
FUNDING
Funding for open access charge: University of Zurich, Switzerland.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schreiber V., Dantzer F., Ame J.C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Burkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 3.Hassa P.O., Hottiger M.O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 4.Barkauskaite E., Jankevicius G., Ladurner A.G., Ahel I., Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013;280:3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- 5.Chambon P., Weill J.D., Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Bioch. Biophys. Res. Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 6.Kraus W.L. PARPs and ADP-Ribosylation: 50 Years … and Counting. Mol. Cell. 2015;58:902–910. doi: 10.1016/j.molcel.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai P. Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance. Mol. Cell. 2015;58:947–958. doi: 10.1016/j.molcel.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 8.Barkauskaite E., Jankevicius G., Ahel I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol. Cell. 2015;58:935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Hottiger M.O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem. 2015;84:227–263. doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- 10.Hottiger M.O., Hassa P.O., Luscher B., Schuler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Bock F.J., Todorova T.T., Chang P. RNA Regulation by Poly(ADP-Ribose) Polymerases. Mol. Cell. 2015;58:959–969. doi: 10.1016/j.molcel.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smeenk G., van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu. Rev. Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- 13.Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 14.Luo X., Kraus W.L. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Ame J.C., Dierich A., LeMeur M., et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck C., Robert I., Reina-San-Martin B., Schreiber V., Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014;329:18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Boehler C., Gauthier L.R., Mortusewicz O., Biard D.S., Saliou J.M., Bresson A., Sanglier-Cianferani S., Smith S., Schreiber V., Boussin F., et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altmeyer M., Messner S., Hassa P.O., Fey M., Hottiger M.O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loseva O., Jemth A.S., Bryant H.E., Schuler H., Lehtio L., Karlberg T., Helleday T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 2010;285:8054–8060. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rulten S.L., Fisher A.E., Robert I., Zuma M.C., Rouleau M., Ju L., Poirier G., Reina-San-Martin B., Caldecott K.W. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R.T., Ahel I., Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith S., Giriat I., Schmitt A., de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 23.Cook B.D., Dynek J.N., Chang W., Shostak G., Smith S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang P., Coughlin M., Mitchison T.J. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 25.Kim M.K., Dudognon C., Smith S. Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. 2012;13:724–732. doi: 10.1038/embor.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozaki Y., Matsui H., Asou H., Nagamachi A., Aki D., Honda H., Yasunaga S., Takihara Y., Yamamoto T., Izumi S., et al. Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Mol. Cell. 2012;47:694–706. doi: 10.1016/j.molcel.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 27.Cho-Park P.F., Steller H. Proteasome regulation by ADP-ribosylation. Cell. 2013;153:614–627. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S., et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G.A., Schirle M., Shi X., Hild M., Bauer A., et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 30.Li N., Zhang Y., Han X., Liang K., Wang J., Feng L., Wang W., Songyang Z., Lin C., Yang L., et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 2015;29:157–170. doi: 10.1101/gad.251785.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W., Li N., Li X., Tran M.K., Han X., Chen J. Tankyrase Inhibitors Target YAP by Stabilizing Angiomotin Family Proteins. Cell Rep. 2015;13:524–532. doi: 10.1016/j.celrep.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levaot N., Voytyuk O., Dimitriou I., Sircoulomb F., Chandrakumar A., Deckert M., Krzyzanowski P.M., Scotter A., Gu S., Janmohamed S., et al. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guettler S., LaRose J., Petsalaki E., Gish G., Scotter A., Pawson T., Rottapel R., Sicheri F. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 34.Pic E., Gagne J.P., Poirier G.G. Mass spectrometry-based functional proteomics of poly(ADP-ribose) polymerase-1. Exp. Rev. Proteomic. 2011;8:759–774. doi: 10.1586/epr.11.63. [DOI] [PubMed] [Google Scholar]
- 35.Simbulan-Rosenthal C.M., Rosenthal D.S., Smulson M.E. Purification and characterization of poly(ADP-ribosyl)ated DNA replication/repair complexes. Methods Mol. Biol. 2011;780:165–190. doi: 10.1007/978-1-61779-270-0_11. [DOI] [PubMed] [Google Scholar]
- 36.Matic I., Ahel I., Hay R.T. Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat. Methods. 2012;9:771–772. doi: 10.1038/nmeth.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman J.D., Gagne J.P., Poirier G.G., Goodlett D.R. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2013;12:1868–1880. doi: 10.1021/pr301219h. [DOI] [PubMed] [Google Scholar]
- 38.Gagne J.P., Ethier C., Defoy D., Bourassa S., Langelier M.F., Riccio A.A., Pascal J.M., Moon K.M., Foster L.J., Ning Z., et al. Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair. 2015;30:68–79. doi: 10.1016/j.dnarep.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Daniels C.M., Ong S.E., Leung A.K. Phosphoproteomic Approach to Characterize Protein Mono- and Poly(ADP-ribosyl)ation Sites from Cells. J. Proteome Res. 2014;13:3510–3522. doi: 10.1021/pr401032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniels C.M., Ong S.E., Leung A.K. The Promise of Proteomics for the Study of ADP-Ribosylation. Mol. Cell. 2015;58:911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenthal F., Hottiger M.O. Identification of ADP-ribosylated peptides and ADP-ribose acceptor sites. Front. Biosci. 2014;19:1041–1056. doi: 10.2741/4266. [DOI] [PubMed] [Google Scholar]
- 42.Gagne J.P., Isabelle M., Lo K.S., Bourassa S., Hendzel M.J., Dawson V.L., Dawson T.M., Poirier G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isabelle M., Gagne J.P., Gallouzi I.E., Poirier G.G. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 2012;125:4555–4566. doi: 10.1242/jcs.106963. [DOI] [PubMed] [Google Scholar]
- 44.Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M.O., Nielsen M.L. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol. Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Wang J., Ding M., Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 46.Psakhye I., Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 47.Krietsch J., Rouleau M., Pic E., Ethier C., Dawson T.M., Dawson V.L., Masson J.Y., Poirier G.G., Gagne J.P. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 2013;34:1066–1087. doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gagne J.P., Haince J.F., Pic E., Poirier G.G. Affinity-based assays for the identification and quantitative evaluation of noncovalent poly(ADP-ribose)-binding proteins. Methods Mol. Biol. 2011;780:93–115. doi: 10.1007/978-1-61779-270-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malanga M., Althaus F.R. Noncovalent protein interaction with poly(ADP-ribose) Methods Mol. Biol. 2011;780:67–82. doi: 10.1007/978-1-61779-270-0_5. [DOI] [PubMed] [Google Scholar]
- 50.Kalisch T., Ame J.C., Dantzer F., Schreiber V. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 2012;37:381–390. doi: 10.1016/j.tibs.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 52.Fahrer J., Kranaster R., Altmeyer M., Marx A., Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fahrer J., Popp O., Malanga M., Beneke S., Markovitz D.M., Ferrando-May E., Burkle A., Kappes F. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49:7119–7130. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popp O., Veith S., Fahrer J., Bohr V.A., Burkle A., Mangerich A. Site-specific noncovalent interaction of the biopolymer poly(ADP-ribose) with the Werner syndrome protein regulates protein functions. ACS Chem. Biol. 2013;8:179–188. doi: 10.1021/cb300363g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fischer J.M., Popp O., Gebhard D., Veith S., Fischbach A., Beneke S., Leitenstorfer A., Bergemann J., Scheffner M., Ferrando-May E. Poly(ADP-ribose)-mediated interplay of XPA and PARP1 leads to reciprocal regulation of protein function. FEBS J. 2014;281:3625–3641. doi: 10.1111/febs.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 57.Isogai S., Kanno S., Ariyoshi M., Tochio H., Ito Y., Yasui A., Shirakawa M. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- 58.Oberoi J., Richards M.W., Crumpler S., Brown N., Blagg J., Bayliss R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR) J. Biol. Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eustermann S., Brockmann C., Mehrotra P.V., Yang J.C., Loakes D., West S.C., Ahel I., Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat. Struct. Mol. Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li G.Y., McCulloch R.D., Fenton A.L., Cheung M., Meng L., Ikura M., Koch C.A. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9129–9134. doi: 10.1073/pnas.1000556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rulten S.L., Cortes-Ledesma F., Guo L., Iles N.J., Caldecott K.W. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell. Biol. 2008;28:4620–4628. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min W., Bruhn C., Grigaravicius P., Zhou Z.W., Li F., Kruger A., Siddeek B., Greulich K.O., Popp O., Meisezahl C., et al. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat. Commun. 2013;4:2993. doi: 10.1038/ncomms3993. [DOI] [PubMed] [Google Scholar]
- 63.Ahel D., Horejsi Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P., et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gottschalk A.J., Timinszky G., Kong S.E., Jin J., Cai Y., Swanson S.K., Washburn M.P., Florens L., Ladurner A.G., Conaway J.W., et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Timinszky G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E.H., Scheffzek K., et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 66.Aguiar R.C., Takeyama K., He C., Kreinbrink K., Shipp M.A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagne J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G., et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perry J.J., Tainer J.A., Boddy M.N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 70.DaRosa P.A., Wang Z., Jiang X., Pruneda J.N., Cong F., Klevit R.E., Xu W. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature. 2015;517:223–226. doi: 10.1038/nature13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khoronenkova S.V., Dianov G.L. The emerging role of Mule and ARF in the regulation of base excision repair. FEBS Lett. 2011;585:2831–2835. doi: 10.1016/j.febslet.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 72.Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grofte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S., et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 73.Reinhardt H.C., Yaffe M.B. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 2013;14:563–580. doi: 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- 74.Breslin C., Hornyak P., Ridley A., Rulten S.L., Hanzlikova H., Oliver A.W., Caldecott K.W. The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res. 2015;43:6934–6944. doi: 10.1093/nar/gkv623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li M., Lu L.Y., Yang C.Y., Wang S., Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013;27:1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li M., Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gagne J.P., Hunter J.M., Labrecque B., Chabot B., Poirier G.G. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ji Y., Tulin A.V. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40:10287–10301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Izhar L., Adamson B., Ciccia A., Lewis J., Pontano-Vaites L., Leng Y., Liang A.C., Westbrook T.F., Harper J.W., Elledge S.J. A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Rep. 2015;11:1486–1500. doi: 10.1016/j.celrep.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malanga M., Czubaty A., Girstun A., Staron K., Althaus F.R. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008;283:19991–19998. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- 83.Murawska M., Hassler M., Renkawitz-Pohl R., Ladurner A., Brehm A. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet. 2011;7:e1002206. doi: 10.1371/journal.pgen.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang F., Chen Y., Li M., Yu X. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7278–7283. doi: 10.1073/pnas.1318367111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang F., Shi J., Bian C., Yu X. Poly(ADP-Ribose) Mediates the BRCA2-Dependent Early DNA Damage Response. Cell Rep. 2015;13:678–689. doi: 10.1016/j.celrep.2015.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F., Shi J., Chen S.H., Bian C., Yu X. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv939. doi:10.1093/nar/gkv939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thandapani P., O'Connor T.R., Bailey T.L., Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 88.Haince J.F., McDonald D., Rodrigue A., Dery U., Masson J.Y., Hendzel M.J., Poirier G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 89.Altmeyer M., Toledo L., Gudjonsson T., Grofte M., Rask M.B., Lukas C., Akimov V., Blagoev B., Bartek J., Lukas J. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol. Cell. 2013;52:206–220. doi: 10.1016/j.molcel.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 90.Mastrocola A.S., Kim S.H., Trinh A.T., Rodenkirch L.A., Tibbetts R.S. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rulten S.L., Rotheray A., Green R.L., Grundy G.J., Moore D.A., Gomez-Herreros F., Hafezparast M., Caldecott K.W. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Polo S.E., Blackford A.N., Chapman J.R., Baskcomb L., Gravel S., Rusch A., Thomas A., Blundred R., Smith P., Kzhyshkowska J., et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hong Z., Jiang J., Ma J., Dai S., Xu T., Li H., Yasui A. The role of hnRPUL1 involved in DNA damage response is related to PARP1. PLoS One. 2013;8:e60208. doi: 10.1371/journal.pone.0060208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Britton S., Dernoncourt E., Delteil C., Froment C., Schiltz O., Salles B., Frit P., Calsou P. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grofte M., Rask M.B., Streicher W., Jungmichel S., Nielsen M.L., et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat. Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 98.Hyman A.A., Simons K. Cell biology. Beyond oil and water–phase transitions in cells. Science. 2012;337:1047–1049. doi: 10.1126/science.1223728. [DOI] [PubMed] [Google Scholar]
- 99.Hyman A.A., Weber C.A., Julicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 100.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 101.Leung A.K., Vyas S., Rood J.E., Bhutkar A., Sharp P.A., Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boamah E.K., Kotova E., Garabedian M., Jarnik M., Tulin A.V. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 2012;8:e1002442. doi: 10.1371/journal.pgen.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guetg C., Scheifele F., Rosenthal F., Hottiger M.O., Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 104.Kotova E., Jarnik M., Tulin A.V. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet. 2009;5:e1000387. doi: 10.1371/journal.pgen.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ji Y., Tulin A.V. Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins. Int. J. Mol. Sci. 2013;14:16168–16183. doi: 10.3390/ijms140816168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kraus W.L., Hottiger M.O. PARP-1 and gene regulation: progress and puzzles. Mol. Aaspects Med. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 107.Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J.L., McKnight S.L. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andrabi S.A., Umanah G.K., Chang C., Stevens D.A., Karuppagounder S.S., Gagne J.P., Poirier G.G., Dawson V.L., Dawson T.M. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10209–10214. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fouquerel E., Goellner E.M., Yu Z., Gagne J.P., Barbi de Moura M., Feinstein T., Wheeler D., Redpath P., Li J., Romero G., et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014;8:1819–1831. doi: 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Z., Gagne J.P., Poirier G.G., Xu W. Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase. PLoS One. 2014;9:e86010. doi: 10.1371/journal.pone.0086010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brochu G., Duchaine C., Thibeault L., Lagueux J., Shah G.M., Poirier G.G. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim. Biophys. Acta. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 112.Barkauskaite E., Brassington A., Tan E.S., Warwicker J., Dunstan M.S., Banos B., Lafite P., Ahel M., Mitchison T.J., Ahel I., et al. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat. Commun. 2013;4:2164. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dunstan M.S., Barkauskaite E., Lafite P., Knezevic C.E., Brassington A., Ahel M., Hergenrother P.J., Leys D., Ahel I. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat. Commun. 2012;3:878. doi: 10.1038/ncomms1889. [DOI] [PubMed] [Google Scholar]
- 115.Lambrecht M.J., Brichacek M., Barkauskaite E., Ariza A., Ahel I., Hergenrother P.J. Synthesis of dimeric ADP-ribose and its structure with human poly(ADP-ribose) glycohydrolase. J. Am. Chem. Soc. 2015;137:3558–3564. doi: 10.1021/ja512528p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jankevicius G., Hassler M., Golia B., Rybin V., Zacharias M., Timinszky G., Ladurner A.G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosenthal F., Feijs K.L., Frugier E., Bonalli M., Forst A.H., Imhof R., Winkler H.C., Fischer D., Caflisch A., Hassa P.O., et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 118.Sharifi R., Morra R., Appel C.D., Tallis M., Chioza B., Jankevicius G., Simpson M.A., Matic I., Ozkan E., Golia B., et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mashimo M., Kato J., Moss J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2013;110:18964–18969. doi: 10.1073/pnas.1312783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Palazzo L., Thomas B., Jemth A.S., Colby T., Leidecker O., Feijs K.L., Zaja R., Loseva O., Puigvert J.C., Matic I., et al. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem. J. 2015;468:293–301. doi: 10.1042/BJ20141554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 122.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 123.Lord C.J., Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 2013;19:1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 124.Feng F.Y., de Bono J.S., Rubin M.A., Knudsen K.E. Chromatin to Clinic: The Molecular Rationale for PARP1 Inhibitor Function. Mol. Cell. 2015;58:925–934. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Curtin N.J., Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol. Aspects Med. 2013;34:1217–1256. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hottiger M.O. Poly(ADP-ribose) polymerase inhibitor therapeutic effect: are we just scratching the surface. Expert Opin. Ther. Targets. 2015;19:1149–1152. doi: 10.1517/14728222.2015.1073262. [DOI] [PubMed] [Google Scholar]
- 127.Haikarainen T., Krauss S., Lehtio L. Tankyrases: structure, function and therapeutic implications in cancer. Curr. Pharm. Des. 2014;20:6472–6488. doi: 10.2174/1381612820666140630101525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Riffell J.L., Lord C.J., Ashworth A. Tankyrase-targeted therapeutics: expanding opportunities in the PARP family. Nat. Rev. Drug Disc. 2012;11:923–936. doi: 10.1038/nrd3868. [DOI] [PubMed] [Google Scholar]
- 129.Vyas S., Chang P. New PARP targets for cancer therapy. Nat. Rev. Cancer. 2014;14:502–509. doi: 10.1038/nrc3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Steffen J.D., Coyle D.L., Damodaran K., Beroza P., Jacobson M.K. Discovery and structure-activity relationships of modified salicylanilides as cell permeable inhibitors of poly(ADP-ribose) glycohydrolase (PARG) J. Med. Chem. 2011;54:5403–5413. doi: 10.1021/jm200325s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Finch K.E., Knezevic C.E., Nottbohm A.C., Partlow K.C., Hergenrother P.J. Selective small molecule inhibition of poly(ADP-ribose) glycohydrolase (PARG) ACS Chem. Biol. 2012;7:563–570. doi: 10.1021/cb200506t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Illuzzi G., Fouquerel E., Ame J.C., Noll A., Rehmet K., Nasheuer H.P., Dantzer F., Schreiber V. PARG is dispensable for recovery from transient replicative stress but required to prevent detrimental accumulation of poly(ADP-ribose) upon prolonged replicative stress. Nucleic Acids Res. 2014;42:7776–7792. doi: 10.1093/nar/gku505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cozzi A., Cipriani G., Fossati S., Faraco G., Formentini L., Min W., Cortes U., Wang Z.Q., Moroni F., Chiarugi A. Poly(ADP-ribose) accumulation and enhancement of postischemic brain damage in 110-kDa poly(ADP-ribose) glycohydrolase null mice. J. Cereb. Blood Flow Metab. 2006;26:684–695. doi: 10.1038/sj.jcbfm.9600222. [DOI] [PubMed] [Google Scholar]
- 135.Hanai S., Kanai M., Ohashi S., Okamoto K., Yamada M., Takahashi H., Miwa M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Abeti R., Duchen M.R. Activation of PARP by oxidative stress induced by beta-amyloid: implications for Alzheimer's disease. Neurochem. Res. 2012;37:2589–2596. doi: 10.1007/s11064-012-0895-x. [DOI] [PubMed] [Google Scholar]
- 137.Jankowsky E., Harris M.E. Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2015;16:533–544. doi: 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dundr M., Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]


