Abstract
Mechanisms for transcription factor recognition of specific DNA base sequences are well characterized and recent studies demonstrate that the shape of these cognate binding sites is also important. Here, we uncover a new mechanism where the transcription factor GabR simultaneously recognizes two cognate binding sites and the shape of a 29 bp DNA sequence that bridges these sites. Small-angle X-ray scattering and multi-angle laser light scattering are consistent with a model where the DNA undergoes a conformational change to bend around GabR during binding. In silico predictions suggest that the bridging DNA sequence is likely to be bendable in one direction and kinetic analysis of mutant DNA sequences with biolayer interferometry, allowed the independent quantification of the relative contribution of DNA base and shape recognition in the GabR–DNA interaction. These indicate that the two cognate binding sites as well as the bendability of the DNA sequence in between these sites are required to form a stable complex. The mechanism of GabR–DNA interaction provides an example where the correct shape of DNA, at a clearly distinct location from the cognate binding site, is required for transcription factor binding and has implications for bioinformatics searches for novel binding sites.
INTRODUCTION
Transcription factor recognition of specific DNA binding sites is fundamentally important for mediating gene expression and repression in different cellular contexts. These proteins recognize target nucleotide sequences via hydrogen bonds and hydrophobic contacts between amino acid side chains and DNA bases. The structural details of this ‘base readout’ mechanism have been well established for multiple DNA binding motifs including the zinc-finger (1), helix-turn-helix (HTH) (2), leucine zipper (3) and transcription activator-like effector (TALE) domains (4,5). However, transcription factor site selection involves additional levels of complexity. For example, transcription factors can distinguish between several similar binding sequences within the same cell in a context-dependent manner (6). To achieve this specificity, transcription factors also recognize the local three-dimensional (3D) shape of the DNA at the protein binding site (7) including sequence-dependent narrowing of the DNA minor groove (8). These ‘shape readout’ mechanisms can operate independently of base readout (9) and are well described in eukaryotic (8) and prokaryotic organisms (10,11)
The transcription factor GabR from Bacillus subtilis is a member of the GntR family of metabolite-responsive regulators that have evolved by fusion of an N-terminal HTH DNA-binding domain with a C-terminal domain that is homologous to type I aminotransferases and associated with a pyridoxal phosphate (PLP) cofactor (12). GabR regulates the expression of enzymes in the gabTD operon that are directly involved in glutamate production from γ-aminobutyric acid (GABA), a nitrogen source in many bacteria. GabR binds to DNA with high affinity (Kd = ∼1 nM), via two direct repeat sequences (ATACCA) within a 47 bp region that overlaps with the promoter regions of the gabTD operon and its own divergently expressed gabR gene (Figure 1A) (13,14). Binding of GABA to the aminotransferase domain switches the regulator from being a repressor to an activator of the gabTD operon (14). The crystal structure of GabR revealed a head-to-tail domain swapped dimer: the C-terminal aminotransferase-like domains form the dimeric core of the structure, and are connected via a long linking peptide to the N-terminal winged helix-turn helix (wHTH) domains whereby each wHTH domain binds to the aminotransferase domain of its dimeric partner (12). The binding of GabR to DNA is thought to occur via an interaction between the wHTH domain and the repeated ATACCA sequence. How two wHTH domains, which are located on opposing ends of the GabR dimer can simultaneously contact both ATACCA sequences in the promoter region is an unresolved question. Two binding models have been proposed on the basis of the crystal structure (12): in the first, two GabR dimers bind at the promoter, whereby only one wHTH domain of each dimer contacts one of the two ATACCA repeats. In the second model a single GabR dimer binds to its recognition site, but at least one wHTH domain dissociates from the aminotransferase core allowing one dimer to simultaneously occupy both repeated ATACCA binding sites (Supplementary Figure S1).
Figure 1.
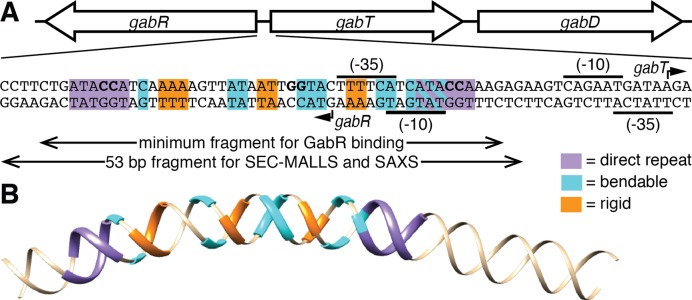
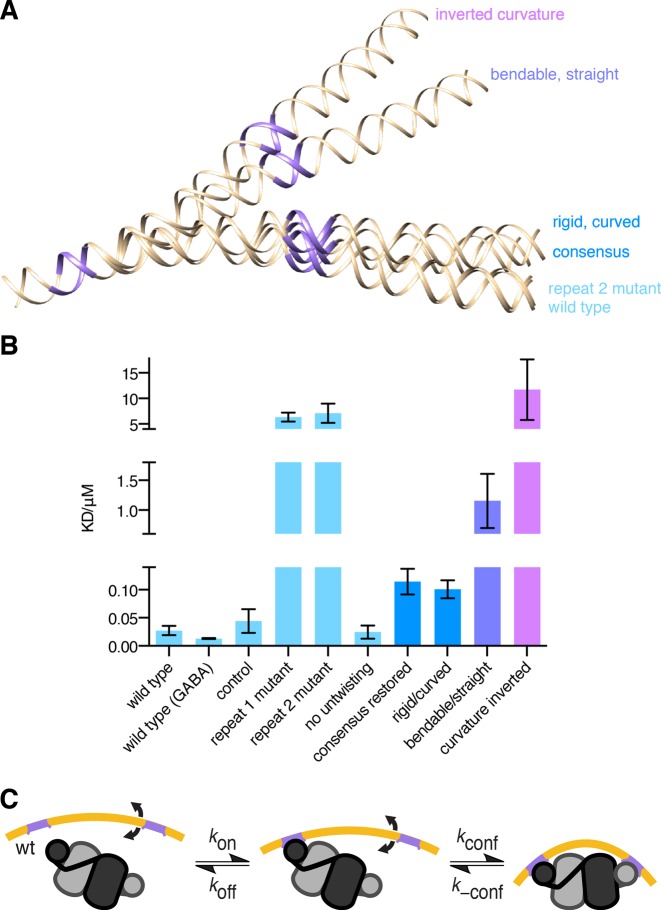
The DNA recognition sequence that binds to GabR. (A) Genetic map of the gabRTD region (top) and sequence of the regulatory/promoter regions between the divergently transcribed gabR gene and gabTD operon (bottom). The transcription start sites (arrows) and −10 and −35 promoter regions (black lines) of the gabR and gabT genes are indicated. (B) 3D model of the DNA sequence shown in A generated using the ‘consensus’ scale for an anisotropic bendability model in an online DNA curvature analysis tool (www.lfd.uci.edu/∼gohlke/dnacurve/). The direct repeat sequences (purple) and regions of high (cyan) and low (orange) bendability are highlighted.
Here we probe the shape of GabR and its complex with DNA in solution with small-angle X-ray scattering (SAXS) to investigate the DNA binding mechanism. These data suggest that the DNA bends and wraps around a positive electrostatic ridge on the dimeric core of GabR. This allows both wHTH domains to interact with their cognate DNA binding sites simultaneously without requiring a conformational rearrangement of the protein. Analysis of the DNA sequence of the 29 bp bridging sequence that separates the two repeat cognate binding sites, revealed a propensity of the DNA to bend in the direction required for complex formation. The physical separation between base pairs dictating DNA shape from the cognate DNA binding sites allowed us to independently probe the importance of base and shape recognition for the GabR–DNA interaction. DNA mutations in the cognate binding site as well as those designed to disrupt the intrinsic curvature of the bridging sequence reduce or abrogate the affinity between GabR and DNA. GabR therefore provides an example of a mechanism where both DNA base sequence and shape recognition are required for the protein–DNA interaction to occur. Moreover, our data reinforce the importance of considering shape information for predicting DNA–protein interactions and suggest that sequences in between or surrounding putative cognate binding sites may also play an important role in DNA site-specific recognition.
MATERIALS AND METHODS
Protein expression and purification
GabR with C-terminal His6-tag was expressed using a pETite vector in Escherichia coli strain BL21 (DE3) Hi-Control cells (Lucigen). Transformed cells were grown in 1 L LB medium containing kanamycin (50 μg = /ml) at 37°C in a shaking incubator (250 rpm) until the cell density reached OD600 = 0.5–0.6. The temperature was then reduced to 25°C and protein expression induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside. Cells were harvested by centrifugation and resuspended in 10–15 ml of wash buffer pH 7.5 consisting of 2× phosphate buffered saline (PBS), 5 mM imidazole, 5% glycerol and Complete protease inhibitor EDTA-free cocktail (Roche). The cells were lysed by sonication and the cell debris was removed by centrifugation. The His6-tagged GabR protein was purified using immobilized metal affinity chromatography on a 5 ml Ni-NTA column (GE Healthcare). Peak fractions were pooled and concentrated using a centrifugal filtration device (Millipore) and the protein was further purified by gel filtration using a Superdex 200 Tricorn 10/300GL column equilibrated in 2× PBS, 375 mM imidazole, 5% glycerol, 0.1 mM PLP.
DNA duplex formation
Complementary DNA oligonucleotides (Sigma) (100 μM in 10 mM Tris pH 7.5, 100 mM NaCl) were mixed at equimolar concentration and hybridized by heating to 95°C followed by cooling to 25°C.
Analytical size exclusion chromatography (SEC) and multi-angle laser light scattering (MALLS)
Analytical size exclusion chromatography (SEC)-MALLS was performed with a Superdex 200 Tricorn 10/300GL column (GE Healthcare). For SEC-MALLS experiments, the column was connected upstream of the flow cell of a multi angle laser light scattering instrument (Viscotek SEC-MALS, Malvern, UK). The system was equilibrated in 2× PBS, 375 mM imidazole, 5% glycerol, 0.1 mM PLP, before injection of 50 μl of sample containing GabR, DNA or mixtures of GabR and DNA (1:1, 2:1 or 4:1 molar ratio) incubated at room temperature for 10–15 min before injection onto the SEC column.
Small-angle X-ray scattering experiments
X-ray scattering data were collected immediately after elution from an SEC column at the Australian Synchrotron. GabR, DNA or GabR + DNA samples at 10 mg/ml were injected into a 23 ml sephacryl S-200 SEC column (GE Healthcare) at a flow rate of 0.5 ml min−1. The outflow was piped directly into a temperature controlled 1.5 mm quartz capillary at 20°C through which monochromatic X-rays were passed at a flux of 4 × 1012 photons per second. SAXS data was collected with exposure times of five seconds on a Pilatus 1M photon counting detector (Dectris, Baden Switzerland), which was set at a distance of 1.48 m from the sample capillary.
Data reduction was performed using a beamline specific software package known as scatterbrain (Australian Synchrotron, Clayton Australia, https://www.synchrotron.org.au/aussyncbeamlines/saxswaxs/software-saxswaxs). Software for data processing were from the ATSAS suite of programs for SAXS data processing (22) including DATOP, PRIMUS, DATAVER, DAMMIN, DAMAVER, DATPOROD and CRYSOL. Custom software was written to generate plots of average intensity of binned wavelength-independent scattering angles (Q) across all q-ranges. This was used to visualize the elution profile from SEC and select appropriate frames (20–50 frames) containing buffer only to subtract from scattering data. Averaged buffer scattering profiles were then subtracted from each acquisition frame using the program DATOP. The radius of gyration was then calculated using AUTORG for each buffer-subtracted frame and plotted over the average scattering at low q, which allowed the identification of frames consisting of monodispersed protein (Supplementary Figure S2). Subtracted data from monodispersed protein was then scaled and averaged Initial Gunier plots and probability atom distance distribution functions (P(r)) were plotted using the program PRIMUS. These data were used to generate P(r) plots, which were subsequently used to generate ab initio shape restorations using the program DAMMIN and. To allow a direct comparison all P(r) plots were scaled to have an area under the curve of 1. At least 20 ab initio dummy-atom shape restorations were performed for each dataset, which were aligned with the software DAMMIN.
Sequence analysis and modeling of DNA
Bendability profiles for wild-type and mutant sequences were calculated using the sequence-dependent anisotropic bendability model with the consensus scale (26). 3D structural models were generated using the DNA curvature analysis tool DNAcurve (C. Gohlke, http://www.lfd.uci.edu/∼gohlke/dnacurve/).
Biolayer interferometry
The kinetics of GabR association and dissociation with DNA was monitored using biolayer interferometry (BLItz, fortèBIO Inc.). Super streptavidin biosensors (fortèBIO Inc.) were hydrated in 2× PBS, 375 mM imidazole, 5% glycerol, 0.1 mM PLP containing 1% BSA at 25°C for at least 15 min. After recording an initial baseline, the sensors were immersed in a solution of biotinylated DNA duplexes (60 nM in 2× PBS, 375 mM imidazole, 5% glycerol, 0.1 mM PLP) formed from oligonucleotides (Integrated DNA Technologies, see Supplementary Figure S8 for sequences) for 120 s. Sensors were then washed before monitoring protein association (1.1 nM–10 μM GabR) followed by dissociation. The on-rate constant was determined from local fits of a Langmuir model to the association phase of biolayer interferometry traces measured at a range of GabR concentrations. The off-rate was determined from local fits of a single exponential decay with a y-offset to the dissociation phase of biolayer interferometry traces measured at a range of GabR concentrations.
Structural modeling
Bent DNA models were generated using the webserver 3D DART (15) (http://haddock.science.uu.nl/dna/dna.php). Periodicities were specified at 10.5 bp per turn and DNA was bent ‘globally’ over the DNA bases that bridge the ATACCA repeat sequence. This resulted in an even curvature across this bridging sequence. To compare different DNA models, these were structurally superimposed using the program Coot (16). Electrostatic potentials were calculated using PDB2PQR (17) and the Adaptive Poisson Boltzmann Solver software package (18). All structures and surfaces were rendered using The PyMOL Molecular Graphics System, (Schrödinger, LLC).
RESULTS
GabR binds to DNA as a dimer
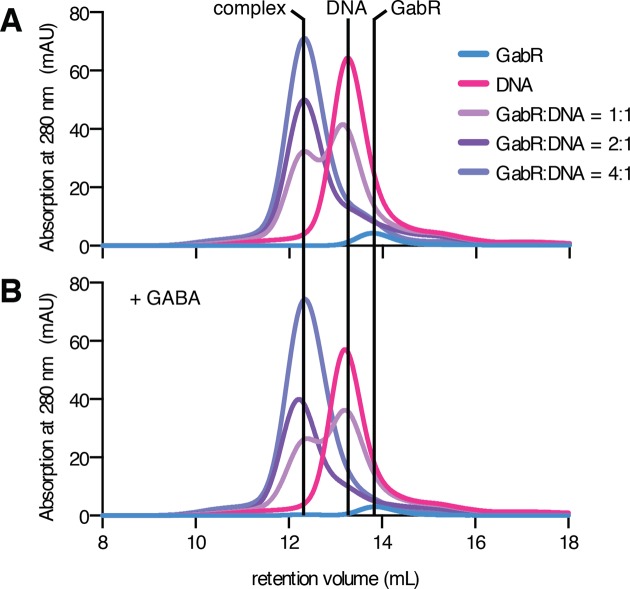
To distinguish between the two proposed GabR–DNA binding models (Supplementary Figure S1), we determined the stoichiometry of the GabR–DNA complex with SEC (Figure 2) and multi-angle laser light scattering (MALLS) (Supplementary Figure S2) to distinguish between different models for transcriptional regulation. The molecular weight of PLP-bound GabR determined by SEC-MALLS (109.9 ± 0.7 kDa) was consistent with sedimentation velocity experiments (12) and confirmed that GabR is a dimer in solution. Incubation of GabR with a 53 bp DNA fragment (MW 31.1 ± 1.5 kDa) encompassing the GabR binding sequence (Figure 1) in increasing molar ratios of GabR:DNA, demonstrates that a 2:1 molar ratio was required to completely shift all DNA into a GabR–DNA complex that had lower retention volume than either DNA or GabR alone. Higher molar ratios of GabR:DNA had no additive effect (Figure 2A). The molecular weight of the complex determined by MALLS (137.6 ± 3.3 kDa) was consistent with a 2:1 binding stoichiometry of GabR to DNA. SEC experiments were also conducted in the presence of GABA, which induces a change in the complex leading to transcriptional activation (14). Binding of GABA had no effect on the stoichiometry of the DNA–GabR complex (Figure 2B). Taken together, these MALLS and SEC data show that one PLP-bound GabR dimer interacts with one DNA binding fragment containing two wHTH binding sequences and that this stoichiometry was unchanged in the presence of GABA. These observations are consistent with recent isothermal calorimetry measurements (19) and exclude the model for transcriptional regulation that involves the binding of a two dimers (12).
Figure 2.
One GabR dimer binds to DNA to form the activating and repressing complex. Size exclusion chromatography elution profiles monitored by absorbance at 280 nm of GabR, DNA and of samples containing GabR + DNA pre-incubated at molar ratios of 1:1, 2:1 and 4:1. (A) and (B) show the elution profiles in the absence and presence of the cofactor GABA, respectively.
The shape of the GabR dimer in solution is consistent with the crystal structure
Mutations in DNA bases within either of the two direct repeat sequences substantially reduced the binding affinity between GabR and DNA in vitro (13). This observation together with the 2:1 stoichiometry of the complex described above, point to a mode of binding where each wHTH domains on the GabR dimer binds to a different ATACCA sequence on a single 53 bp DNA strand. However, in the dimeric GabR crystal structure the wHTH domains are not located in a position that would allow their simultaneous interaction with a linear strand of DNA. Given that there is a long unstructured linking peptide and a small interface between the dimeric core of the structure and the wHTH domains (∼400 Å2), it has been postulated that the wHTH domains may readily dissociate from the dimeric core to facilitate simultaneous binding of both wHTH domains to DNA (12). This spontaneous dissociation of the wHTH domains would lead to a substantially different shape of the dimer in solution.
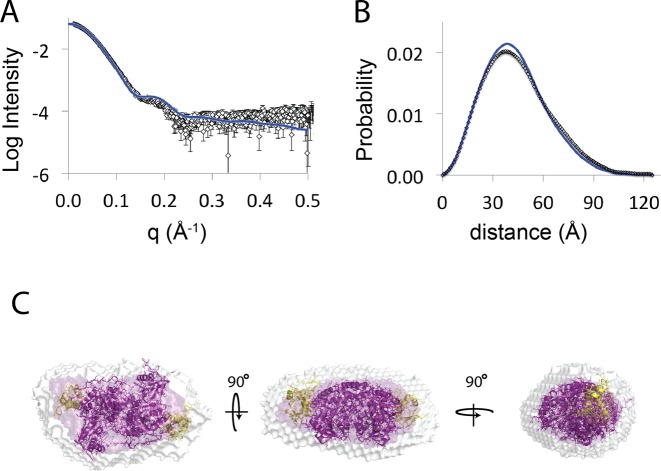
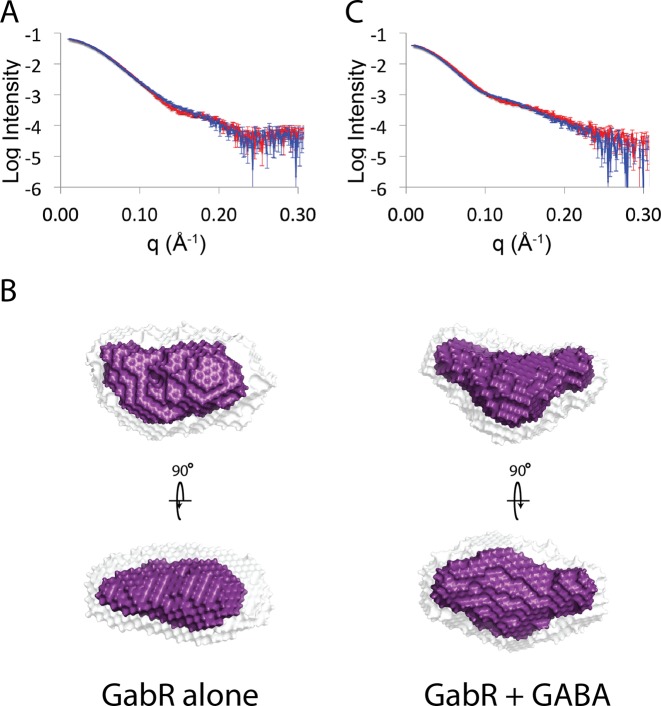
To investigate this further, we determined the shape of the PLP-bound GabR dimer in solution by collecting SAXS data (Supplementary Figure S3) directly from the elution of a SEC column (SEC-SAXS) to exclude the possibility of aggregates (20,21). Data were processed with the ATSAS suite according to the materials and methods (22). Estimated molecular weights from SAXS data were consistent with a dimeric protein (Supplementary Table S1) and the radius of gyration (Rg) of GabR in solution was 34.6 ± 0.1 Å, which is similar to that calculated from the crystal structure (Rg = 33.3 Å). The theoretical scattering and interatomic distance distribution (P(r)) plots calculated from the crystal structure of the GabR dimer (PDBID: 4N0B) (12) using the program crysol, (23) was also similar to the experimental scattering data (Figure 3A and B). The maximum dimensions (Dmax) from SAXS data were also consistent with the crystal structure, indicating that extended conformations, which would result from the dissociation of wHTH domains from the dimeric core, were not detectable in solution. A total of 20 ab initio shape restorations, independently produced very similar shapes as indicated by a low normalized spatial distribution (NSD = 0.517 ± 0.004) (24) and averaged aligned models were consistent with the shape of the dimeric GabR crystal structure (Figure 3C). These data indicate that the shape of GabR in solution is consistent with the crystal structure and there is no evidence for the spontaneous dissociation of wHTH domains that were proposed as a requirement for DNA binding.
Figure 3.
Comparison between the small-angle X-ray scattering data and the crystal structure of GabR. (A) Buffer subtracted X-ray scattering data from the GabR protein is shown as white triangles with errors depicted as twice the standard error of the mean. The theoretical scattering calculated from the dimeric GabR crystal structure (PDBID:4NOB) is shown as a blue line. (B) Plot of the interatomic distance distributions calculated from scattering profiles in (A). (C) Average shape (purple) and aligned shapes (white) from 20 independent ab initio shape restorations. The crystal structure of the GabR dimer depicted in cartoon representation is docked into the average shape restoration. The dimeric core is colored in purple and the DNA binding HTH domains are highlighted in yellow.
DNA undergoes a conformational change to facilitate binding to the GabR dimer
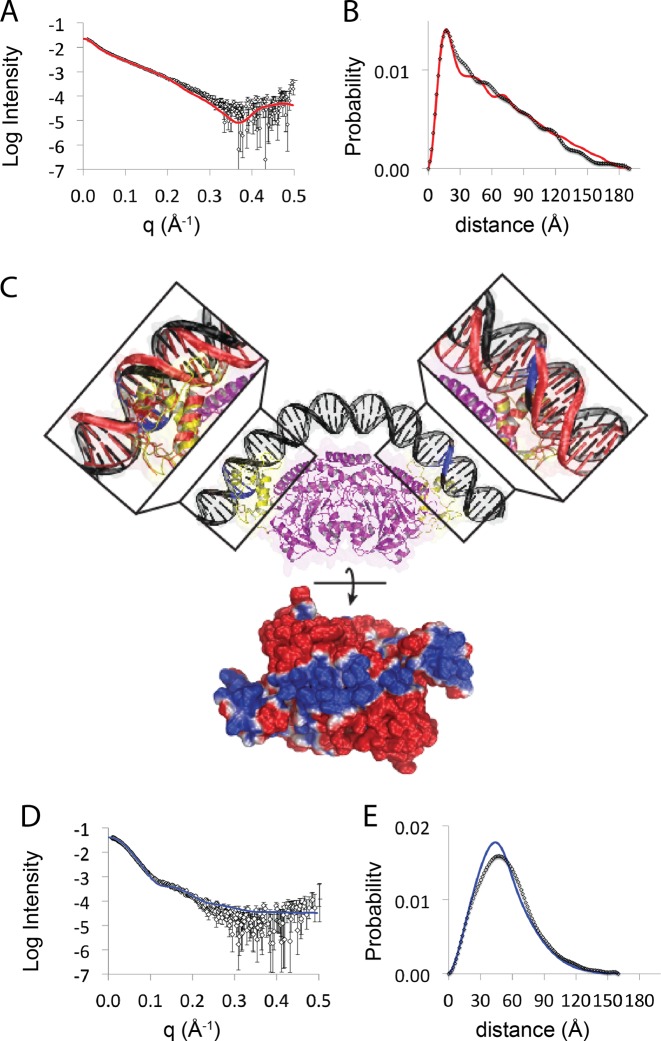
We next analyzed the shapes of the 53 bp DNA fragment and the GabR–DNA complex in solution to obtain further clues for the mode of interaction. The DNA alone had a Rg of 49.7 ± 0.3 Å. This was significantly smaller than the Rg calculated from a 3D model of an ideal 53 bp DNA duplex, with a periodicity of 10.5 bp per turn (Rg ≈ 52 Å). Differences can also been seen in a P(r) plots and theoretical scattering and calculated from the idealized model of DNA, which systematically deviates from experiment at higher diffraction angles (Figure 4A and B). This suggests that the GabR DNA binding sequence in solution samples conformational states that are more compact than an ideal straight DNA duplex of the same length.
Figure 4.
Small-angle X-ray scattering data and model showing a conformational change in DNA upon binding to GabR. (A) Buffer subtracted X-ray scattering data from the 53 bp DNA fragment (white triangles) overlayed with theoretical scattering curves calculated from an atomic model of the DNA fragment as an ideal rigid rod with a periodicity of 10.5 bp per turn (red line) or in a highly bent conformation as shown in (C) (blue line). (B) Interatomic distance distribution profiles from the data and models shown in (A). (C) Top: structural model of the GabR–DNA complex. The bent DNA is shown in black with the tandem cytosine bases that are necessary for binding to the HTH highlighted in blue. The dimeric core of GabR is shown in purple and the wHTH domains are shown in yellow. The insets show the crystal structures of the HTH domain of FadR in complex with a short strand of DNA (PDBID:1H9T) (red) superimposed on the HTH domains of GabR. Bottom: electrostatic potential surface map of the protein rotated by 90° around the X-axis; positive potentials are shown in blue and negative potentials are shown in red. (D) Buffer subtracted X-ray scattering data from the GabR–DNA complex (white triangles) overlayed with the theoretical scattering curve (red line) calculated from an atomic model of the GabR–DNA shown in (C). (E) Interatomic distance distribution profiles calculated from (D). Errors are depicted as twice the standard error of the mean.
Surprisingly, the GabR–DNA complex was more compact than the DNA alone with a substantially smaller Rg of 41.2 ± 0.2 Å and a reduction in the maximal dimensions by around 30 Å, (Supplementary Table S1 and Supplementary Figure S4). This suggests that GabR induces a conformational change in DNA that results in the DNA becoming more compact. Short DNA duplexes can be highly bendable (25) and the propensity of DNA to be bent on average is sequence dependent (26). Inspection of the GabR binding region reveals the presence of three repeated adenine-thymine tracts (A-tracts) that are in phase with each other and between the repeated wHTH binding sites (Figure 1). This arrangement of A-tracts is known to produce curvature (27). Further analysis of the sequence with an anisotropic bendability model (26) reveals that these tracts correspond to rigid segments that alternate with regions of high bendability (Figure 1). The DNA conformation predicted on the basis of this sequence (Figure 1B) shows a slight bend that brings the wHTH binding sequences toward each other. Alternative models generate 3D structures curve in the same direction albeit to different extents, supporting the general model of a curved DNA with wHTH binding sequences facing each other in the correct orientation for GabR binding (Supplementary Figure S5). Thus, one possibility is that the GabR DNA binding sequence bends upon interaction with GabR.
We therefore generated a model of bent DNA bound to the GabR dimer. To achieve this, we utilized a structure of a homologous wHTH binding domain from the acyl-CoA-responsive transcription factor FadR in complex with a short strand of DNA (RCSB ID:1H9T) (28). This structure was superimposed onto both of the wHTH domains of the dimeric GabR crystal structure (Figure 4C—insets). We then structurally superimposed the ATACCA sequence from the ideal linear model of the 53-base GabR DNA binding fragment onto the short DNA strands on one of the FadR–DNA crystal structures. This provided a model defining the orientation of the GabR binding fragment relative to the first wHTH domain. We then bent the DNA model evenly over the base pairs in between two ATACCA repeat sequences toward the second FadR–DNA crystal structure until the bent DNA model aligned with both FadR–DNA crystal structures. Remarkably, the bent DNA model aligned well with the short strands of DNA on both FadR crystal structures with the second wHTH around 3 bp from the second ATACCA repeat sequence (Figure 4C—top and Supplementary Figure S6). This indicates that the distance between the GabR binding motifs and the bent DNA model is complementary with the location of the wHTH domains on the dimeric GabR crystal structure. To determine whether this model was consistent with experimental scattering data, we used the model coordinates to calculate a theoretical X-ray scattering profile. The Rg of the model (Rg = 39.7 Å) is similar to the Rg from SAXS data of the GabR–DNA complex and the theoretical and experimental scattering profiles overlayed reasonably well up to mid q scattering angles (q < 0.2 Å−1) (Figure 4D) and the corresponding interatomic distance distribution profiles are also similar (Figure 4E), indicating that our theoretical model is a reasonable description of the shape of the GabR–DNA complex in solution. Interestingly, a comparison of P(r) plots calculated from the model of bent DNA alone and SAXS data, indicate that the 53 bp DNA fragment is not already highly bent in the absence of GabR (Figure 4B). Combined, these data suggest that GabR stabilizes DNA in a bent conformation, and point to an alternative mechanism of interaction between the GabR dimer and DNA that allows for binding without a dramatic conformational change in the protein. Notably however, since the ATACCA repeat sequences run in the same direction, if the wHTH domains are oriented symmetrically as in the crystal structure, one of the wHTH domains will be in the opposite direction to its corresponding direct repeat sequence.
The bendability of the bridging sequence between the ATACCA repeats is a determinant of complex stability
To test the structural model described above, we next investigated whether the sequence-dependent bendability of the bridging DNA sequence between the repeated wHTH binding sites, affects the binding strength between GabR and DNA. We used biolayer interferometry to obtain estimates of the dissociation constant (KD) for the complex formed between GabR in solution and wild-type or mutant DNA duplexes immobilized on the sensor surface (Figure 5 and Supplementary Figure S7). This allowed us to determine whether mutations in the bridging DNA sequence that alter the predicted bendability, would affect the KD of the interaction. The dissociation constant for the complex with wild-type DNA (KD = 27.4 ± 8.5 nM with respect to the concentration of the GabR dimer) was similar to the values determined previously using a fluorescence polarization assay (KD = 70.2 ± 3.7 nM with respect to the concentration of the GabR monomer) (12) and isothermal titration calorimetry (KD = 36.4 ± 21.6 nM) (19). In the presence of GABA the complex appeared to be more stable than in its absence (KD = 13 ± 0.9 nM).
Figure 5.
Mutations in the repeat sequences or bridging region affect complex stability. (A) Comparison of 3D models of wild-type and mutant DNA sequences defined in Supplementary Figure S8 and used for biolayer interferometry. DNA structural models were ‘consensus’ scale for an anisotropic bendability model in an online DNA curvature analysis tool (www.lfd.uci.edu/∼gohlke/dnacurve/). The repeated wHTH binding sites are rendered in purple (B) Dissociation constants obtained for wild type and mutant DNA sequences from the biolayer interferometry traces. (C) Kinetic scheme of GabR–DNA complex formation.
Next we confirmed the importance of the base-readout interactions between the wHTH domains and the two ATACCA for complex formation. Mutations of the CC dinucleotide to GG in either of the two ATACCA sequences, previously shown to be important for DNA binding (13), resulted in a pronounced reduction of the GabR–DNA complex stability (KD = 6.4 ± 0.9 μM and 7.1 ± 1.9 μM for mutations in the first and second repeat, respectively). As expected, mutation of both direct repeat sequences abolished GabR binding in the concentration range used in the experiment (up to 10 μM).
To quantify the contribution of shape recognition to the stability of the GabR–DNA complex, we introduced a range of DNA mutants in the region that bridges the ATACCA sequences. These were designed to perturb the bendability of the DNA according to a sequence-dependent anisotropic bendability model (26). The predicted 3D structural models of the mutants are shown in Figure 5A (see Supplementary Figure S8 for the sequence of all mutants and S9 for the corresponding bendability plots). To test for the possibility of base recognition in the bridging sequence, a control mutant with a GG to CC substitution located in the middle of the linker preserves the sequence-dependent DNA bendability and intrinsic curvature of the wild-type sequence and is known to retain GabR binding (13). Three other DNA mutants are predicted to alter bendability by deviating from the curvature of the wild-type sequence by different degrees. First, single nucleotide mutations that convert the four regions of high bendability to regions of intermediate bendability were chosen to generate a structure that bends in the same direction as wild type albeit to a lesser extent (‘rigid/curved’). Second, a highly bendable bridging sequence that was designed by introducing mutations into the three rigid regions; this mutant linker can bend equally well in all directions but is straight on average (‘bendable/straight’). Third, we created a mutant that bends in the opposite direction to the wild type sequence by shifting the register of the pattern of bendable and rigid regions by four nucleotides with respect to ATACCA repeat sequences; this shift was achieved by moving the CATC tetranucleotide sequence from the 3′ end of the bridging sequence to the 5′ end (‘inverted curvature’).
As expected the mutated control DNA showed essentially the same affinity (KD = 44 ± 21 nM) as wild-type DNA. All other mutants that were predicted to alter the bendability of the DNA duplex reduced its affinity to GabR. The reduction in affinity of the ‘bendable/straight’ mutant compared to the wild type sequence was pronounced (KD = 1.16 ± 0.65 μM) whereas the effect of the ‘rigid/curved’ mutations was relatively mild (KD = 101 ± 16 nM). The affinity of the ‘inverted curvature’ mutant were similar to those observed for DNA with mutations in either of the two direct repeat sequences suggesting that the complex does not proceed beyond the singly-bound state (KD = 11.7 ± 5.9 μM). Thus, a greater predicted deviation in DNA shape from wild-type resulted in a greater reduction in affinity between GabR and DNA. This supports the model of DNA bending to facilitate a stable interaction with GabR and indicates that shape recognition is equally important to sequence recognition for this interaction to occur.
The bridging sequence between the direct repeats consists of the putative −35 region of the gabT promoter with the sequence TTTTCA, which contains one of the three rigid segments of the linker (Figure 1B). Restoring the −35 region to its consensus sequence in B. subtilis (TTGACA) renders this segment highly bendable with a concomitant reduction of the predicted curvature (Supplementary Figure S9). The dissociation constant of this mutant (KD = 114 ± 23 nM) was ∼4× higher than of wild-type but considerably lower than for the ‘bendable/straight’ mutant in line with its intermediate deviation from wild-type bendability and curvature. Thus, the deviation from the consensus sequence may be required to preserve the DNA shape for recognition by the GabR dimer.
It has been shown that local DNA unwinding at nucleotides between individual recognition elements provides one mechanism for establishing the correct spatial orientation for binding of multimeric transcription factors (29). The bridging sequence between the ATACCA sites contains an AT-rich region (TATAAT) that is prone to DNA untwisting and we asked whether untwisting at this sequence was involved in GabR binding (30). The mutation of an AT dinucleotide to CG removes the propensity of this stretch of DNA to untwist while preserving the bendability and curvature of the wild-type sequence (Supplementary Figure S9). This mutation had no discernible effect on GabR–DNA complex formation (KD = 24.6 ± 11.9 nM) suggesting that changes in the twist, at least in this sequence, are not important for complex formation.
Kinetic analysis of the binding and dissociation traces with a 1:1 interaction model yielded estimates of the association and dissociation rate constants, kon and koff (Supplementary Table S2 and Supplementary Figure S10). The association rate constants for all bendability mutants were within a factor of two of wild-type (kon = 0.14 ± 0.04 μM−1 s−1). The differences in KD observed between constructs were therefore largely due to differences in koff. As expected, control mutant and the ‘no untwisting’ mutant, which had a bendability profile similar to that of wild-type DNA had a koff within error of the wild-type value (koff = 0.0067 ± 0.0033 s−1) while the dissociation rate constants for the ‘bendable/straight’ mutant (koff = 0.076 ± 0.018 s−1) was one order of magnitude higher and those for the ‘inverted curvature’ and both single repeat mutants were two orders of magnitude higher (Supplementary Table S2) than for wild-type DNA. These kinetic data are consistent with the sequential binding of the GabR wHTH domains to DNA, consisting of the rate limiting binding of one wHTH domain followed the stabilization of the complex via a rapid binding of the second wHTH domain.
GabR undergoes a conformational change during switch from repressor to activator
GabR activates expression of the gabTD operon upon binding to its effector molecule GABA (14). To determine whether GabR undergoes a structural rearrangement in response to binding to GABA, we compared the shape of GabR alone and in complex with DNA in solution, in the presence and absence of GABA (Supplementary Figure S3 and Supplementary Table S1). There was a detectable increase in the Rg of GabR in the presence of GABA (Rg = 35.4 ± 0.1 Å) and differences in scattering at mid-Q angles pointing to a redistribution of mass (Figure 6A). This is also reflected in 24 independent ab initio shape restorations, which were self consistent (NSD = 0.507 ± 0.03 Å), and different to reconstructions generated from SAXS data of GabR in the absence of GABA (Figure 6B). Similarly, when bound to DNA, there was also a significant increase in Rg in the presence of GABA (Rg = 42.6 ± 0.2 Å) and a detectable difference in scattering (Figure 6C). These data indicate that the GABA-induced transition to the activator state may involve a conformational rearrangement of the protein.
Figure 6.
GABA induces a small conformational change in GabR. (A) Buffer subtracted scattering of GabR in the absence (red) and presence (blue) of GABA. (B) Average shape (purple) and aligned shapes (white) from 20 independent ab initio shape restorations for GabR in the absence (left) and presence (right) of GABA. (C) Buffer subtracted scattering of the GabR–DNA complex also in the absence (red) and presence (blue) of GABA. Errors are plotted as twice the standard error of the mean.
DISCUSSION
Our structural and mutational analysis of GabR–DNA complex formation allows us to arrive at the following conclusions: (i) binding of one GabR dimer to a DNA fragment from the gabRTD regulatory region containing the two wHTH binding sequences separated by a 29 bp bridging sequence, leads to a compaction of the DNA fragment that is consistent with a DNA structural change. (ii) Simultaneous binding of both wHTH domains to the ATACCA repeat sequences is essential for complex stability because the affinity of a single wHTH domain for the ATACCA sequence is low. (iii) Mutations in the linker region predicted to alter its sequence-dependent bendability and intrinsic curvature lead to weaker complexes. We thus propose a structural model for the GabR–DNA complex, which does not require large-scale rearrangements of the wHTH domains. Instead, DNA bends around the dimeric core to allow simultaneous contacts between the wHTH domains located at opposing ends of the GabR dimer and the ATACCA repeat sequences (Figure 4C).
Orientation of the wHTH domains
In this model the wHTH domains could either unusually contact the ATACCA motif in different orientations (as shown in Figure 4C) or in the same orientation if upon DNA binding one of the wHTH domains flips around and associates with the dimeric core in an alternative orientation to that seen in the the crystal structure. Our data cannot distinguish between either of these possibilities. For the wHTH domains to flip upon DNA binding, the binding energy between DNA and the second wHTH domain has to be larger than the energy required to reorientate the wHTH domains. The latter can be approximated by the interaction strength between the wHTH domains and the linking peptide to the GabR dimeric core. This interaction comprises of an interface with a relatively small surface area (12) and hence may well be disrupted by the formation of the second wHTH–DNA interaction. This is because once the first ATACCA repeat sequence is bound, the effective concentration of the second ATACCA sequence at the second wHTH domain is large and subsequently there binding energy between these structures.
GabR binding mechanism and dissection of DNA base and shape recognition
Specific protein–DNA interactions involve both base readout (the formation of hydrogen bonds between amino acid side-chains and a specific sequence of DNA bases) and shape readout mechanisms (recognition of local or global DNA structure) but the relative contributions of these mechanisms to binding are often difficult to dissect. Here we used mutational analysis to separate the roles of these mechanisms for GabR binding. Overall it was observed that both the integrity of the ATACCA repeats as well as preserving the bendability profile in between were critical for complex formation. The energetics of shape recognition were directly altered with mutations that are predicted to affect the bendability of the gabRTD regulatory region. Mutations that disfavor bending in the correct direction (‘inverted curvature’) are just as detrimental to complex stability as mutations in either of the two ATACCA repeats. This suggests that the ‘inverted curvature’ mutant can bind to only one wHTH domain at a time, presumably because the energy required for bending at the rigid segments in the direction required for double binding is too high. Interestingly, mutants with high isotropic bendability also lead to a decrease in complex stability. This is likely to be due to a higher entropic cost associated with confining more flexible DNA to the conformation in the bound state. Thus, the free energy of the complex formed with the bendable mutants (‘bendable/straight’, ‘consensus restored’) is higher than that for wild-type DNA. Taken together the observations suggest that GabR binding to DNA involves a combination of base and shape readout mechanisms, which is emerging as a general concept for protein–DNA interactions (6).
We propose the following kinetic model for GabR binding to DNA (Figure 5C): the first step is governed by the binding of one of the wHTH domains to one of the ATACCA repeats in the DNA. This process is expected to be independent of the properties of the bridging sequence, which is reflected in the similarity of the association rate constants between wild-type and all bendability mutants. In the second step the DNA samples various conformations until the second repeat is oriented correctly to allow binding of the second wHTH domain. With the wild-type DNA sequence the second step is fast, because the effective concentration of the second ATACCA sequence relative to the unbound wHTH domain is high. But this is dependent on the mechanical properties of the bridging DNA sequence, which has evolved to readily form a shape that is complementary to the structure of GabR. The final complex is stable but possibly dynamic with brief excursions into the singly-bound intermediate. Electrostatic interactions and local shape recognition mechanisms between residues on the positive ridge of GabR and the DNA bridging region may further contribute to complex stability. Analysis of the bridging sequence with DNAshape, a tool for the prediction of DNA structural features (31), reveals a narrowing of the minor groove in two locations (Supplementary Figure S11). While our structural model does not specify the precise alignment between DNA bases and surface amino acids, it is possible that insertion of lysine or arginine residues into the narrow minor groove may contribute to the shape readout of the bridging sequence (8). Consistent with this speculation, weakly binding mutants show a shift of the regions of minor groove narrowing away from the positive ridge (‘inverted curvature’) or a reduction in minor groove narrowing (‘bendable/straight’).
General implications for DNA shape recognition
These observations reinforce and extend previous discoveries that show the importance of DNA bending for protein binding. For example, the papillomavirus E2 protein is a dimeric protein that binds to two binding sites on DNA separated by a four-nucleotide linker with intrinsic curvature (32). The crystal structure of this protein–DNA complex shows that the DNA bends at the linker sequence to facilitate binding (33). Mutations in this four-nucleotide linker sequence that abolish its intrinsic curvature also lead to complexes with lower affinity (34). Here we provide mutational analysis showing that shape readout is not restricted to the cognate binding site and a few adjacent nucleotides. Rather, this readout mode can involve regions of several tens of base pairs, which instead of being inert, contribute to the stability of the protein–DNA complex. This observation is consistent with recent computational analyses of protein-binding microarrays or chromatin immunoprecipitation data, suggesting that transcription factor binding to E-box binding sites depends on the structural features of flanking regions (35,36). Indeed, prediction of transcription factor binding sites is improved when using binding data that provides sequence information of the flanking regions (37).
This discovery is likely to have broader relevance to other DNA binding proteins including in eukaryotic systems. Recent data shows that the mammalian transcription factor GATA3 (38) and members of the FOXP subfamily of Forkhead transcription factors (39,40) can bind to DNA in a bridging mode whereby the DNA-binding domains of the dimeric transcription factor contact two sites that are distal from each other. While this bridging mode is thought to be involved in long-range regulation via DNA looping or linking of chromosomes, these types of proteins could in principle also bind to sites with a spacing similar to that observed here. The contribution of DNA bendability in these cases may be less pronounced than observed for GabR but would nevertheless contribute to distinguishing sites of different affinity and thus impact on transcription factor site searches. This finding has implications for the prediction of TF binding sites whereby algorithms need to take into account not only the sequence and shape of the DNA at the binding site but also properties of bridging regions between TF binding sites.
Supplementary Material
Acknowledgments
We would like to acknowledge the beamline scientists and other support staff SAXS/WAXS beamline at the at Australian Synchrotron, Victoria, Australia where SAXS data was collected. We would also like to thank Xin Wang for his technical assistance and Horace R. Drew for useful discussions on DNA shape.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Australian Research Council [DP130102219]; Ramaciotti Foundation (Establishment Grant) [ES2014/010] and HFSP grant [RGY0084/2014]; Australian Research Council Discovery Early Career Research Award [DE140100262 to L.K.L.]; Australian Research Council Future Fellowship [FT100100411 to T.B.]; NIH [1R15GM113229-01 to D.L.]. Funding for open access charge: Australian Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev. Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 2.Brennan R.G., Matthews B.W. The helix-turn-helix DNA binding motif. J. Biol. Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 3.Ellenberger T.E., Brandl C.J., Struhl K., Harrison S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 4.Moscou M.J., Bogdanove A.J. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 5.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 6.Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordân R., Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abe N., Dror I., Yang L., Slattery M., Zhou T., Bussemaker H.J., Rohs R., Mann R.S. Deconvolving the recognition of DNA shape from sequence. Cell. 2015;161:307–318. doi: 10.1016/j.cell.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Z., Wang Q., Liu Z., Zhang M., Machado A.C., Chiu T.-P.P., Feng C., Zhang Q., Yu L., Qi L., et al. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 2015;6:7642. doi: 10.1038/ncomms8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding P., McFarland K.A., Jin S., Tong G., Duan B., Yang A., Hughes T.R., Liu J., Dove S.L., Navarre W.W., et al. A novel AT-rich DNA recognition mechanism for bacterial Xenogeneic silencer MvaT. PLoS Pathog. 2015;11:e1004967. doi: 10.1371/journal.ppat.1004967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edayathumangalam R., Wu R., Garcia R., Wang Y., Wang W., Kreinbring C.A., Bach A., Liao J., Stone T.A., Terwilliger T.C., et al. Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17820–17825. doi: 10.1073/pnas.1315887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belitsky B.R. Bacillus subtilis GabR, a protein with DNA-binding and aminotransferase domains, is a PLP-dependent transcriptional regulator. J. Mol. Biol. 2004;340:655–664. doi: 10.1016/j.jmb.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Belitsky B.R., Sonenshein A.L. GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis. Mol. Microbiol. 2002;45:569–583. doi: 10.1046/j.1365-2958.2002.03036.x. [DOI] [PubMed] [Google Scholar]
- 15.van Dijk M., Bonvin A.M.J.J. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2009;37:W235–W239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda K., Kato S., Ito T., Shiraki S., Kawase Y., Goto M., Kawashima S., Hemmi H., Fukada H., Yoshimura T. Role of the aminotransferase domain in Bacillus subtilis GabR, a pyridoxal 5′-phosphate-dependent transcriptional regulator. Mol. Microbiol. 2014;95:245–257. doi: 10.1111/mmi.12861. [DOI] [PubMed] [Google Scholar]
- 20.David G., Pérez J. Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. Appl. Crystallogr. 2009;42:892–900. [Google Scholar]
- 21.Hynson R.M.G., Duff A.P., Kirby N., Mudie S., Lee L.K. Differential ultracentrifugation coupled to small-angle X-ray scattering on macromolecular complexes. J. Appl. Crystallogr. 2015;48:769–775. [Google Scholar]
- 22.Konarev P.V., Petoukhov M.V., Volkov V.V., Svergun D.I. ATSAS2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svergun D., Barberato C., Koch M.H.J. CRYSOL– a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 24.Kozin M.B., Svergun D.I. Automated matching of high-and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- 25.Vafabakhsh R., Ha T. Extreme bendability of DNA less than 100 pbase pairs long revealed by single-molecule cyclization. Science. 2012;337:1097–1101. doi: 10.1126/science.1224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munteanu M.G., Vlahovicek K., Parthasarathy S., Simon I., Pongor S. Rod models of DNA: sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem. Sci. 1998;23:341–347. doi: 10.1016/s0968-0004(98)01265-1. [DOI] [PubMed] [Google Scholar]
- 27.Hizver J., Rozenberg H., Frolow F., Rabinovich D., Shakked Z. DNA bending by an adenine–thymine tract and its role in gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Aalten D.M., DiRusso C.C., Knudsen J. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001;20:2041–2050. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., Zhang X., Dantas Machado A.C., Ding Y., Chen Z., Qin P.Z., Rohs R., Chen L. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Res. 2013;41:8368–8376. doi: 10.1093/nar/gkt584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calladine C.R., Drew H.R., Luisi B.F., Travers A.A. Understanding DNA. 3rd edn. Waltham: Elsevier Inc; 2004. [Google Scholar]
- 31.Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013;41:W56–W62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozenberg H., Rabinovich D., Frolow F., Hegde R.S., Shakked Z. Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15194–15199. doi: 10.1073/pnas.95.26.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegde R.S., Grossman S.R., Laimins L.A., Sigler P.B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 34.Hines C.S., Meghoo C., Shetty S., Biburger M., Brenowitz M., Hegde R.S. DNA structure and flexibility in the sequence-specific binding of papillomavirus E2 proteins. J. Mol. Biol. 1998;276:809–818. doi: 10.1006/jmbi.1997.1578. [DOI] [PubMed] [Google Scholar]
- 35.Hadžić T., Park D., Abruzzi K.C., Yang L., Trigg J.S., Rohs R., Rosbash M., Taghert P.H. Genome-wide features of neuroendocrine regulation in Drosophila by the basic helix-loop-helix transcription factor DIMMED. Nucleic Acids Res. 2015;43:2199–2215. doi: 10.1093/nar/gku1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordân R., Shen N., Dror I., Zhou T., Horton J., Rohs R., Bulyk M.L. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou T., Shen N., Yang L., Abe N., Horton J., Mann R.S., Bussemaker H.J., Gordân R., Rohs R. Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl. Acad. Sci. U.S.A. 2015;112:4654–4659. doi: 10.1073/pnas.1422023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y., Bates D.L., Dey R., Chen P.-H., Machado A.C.D., Laird-Offringa I.A., Rohs R., Chen L. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep. 2012;2:1197–1206. doi: 10.1016/j.celrep.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandukwala H.S., Wu Y., Feuerer M., Chen Y., Barboza B., Ghosh S., Stroud J.C., Benoist C., Mathis D., Rao A., et al. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011;34:479–491. doi: 10.1016/j.immuni.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stroud J.C., Wu Y., Bates D.L., Han A., Nowick K., Paabo S., Tong H., Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.