Abstract
Synthetic genetics is a subdiscipline of synthetic biology that aims to develop artificial genetic polymers (also referred to as xeno-nucleic acids or XNAs) that can replicate in vitro and eventually in model cellular organisms. This field of science combines organic chemistry with polymerase engineering to create alternative forms of DNA that can store genetic information and evolve in response to external stimuli. Practitioners of synthetic genetics postulate that XNA could be used to safeguard synthetic biology organisms by storing genetic information in orthogonal chromosomes. XNA polymers are also under active investigation as a source of nuclease resistant affinity reagents (aptamers) and catalysts (xenozymes) with practical applications in disease diagnosis and treatment. In this review, we provide a structural perspective on known antiparallel duplex structures in which at least one strand of the Watson–Crick duplex is composed entirely of XNA. Currently, only a handful of XNA structures have been archived in the Protein Data Bank as compared to the more than 100 000 structures that are now available. Given the growing interest in xenobiology projects, we chose to compare the structural features of XNA polymers and discuss their potential to access new regions of nucleic acid fold space.
INTRODUCTION
Elucidating the three-dimensional (3D) structures of proteins and nucleic acids with atomic-level resolution—a celebrated rarity less than half a century ago (the first crystal structure of an enzyme (1) and a DNA oligonucleotide (2) were determined in 1965 and 1979, respectively)—now occurs at an astonishing rate of hundreds per month. The coordinates of over 100 000 structures of proteins, along with some 1600 structures of DNA and 1100 structures of RNA can be downloaded at the website of the Research Collaboratory for Structural Biology (www.rcsb.org) (3). Single crystal X-ray crystallography and solution nuclear magnetic resonance (NMR) have revealed the rich variety of DNA structural motifs (4,5) and RNA's conformational repertoire, from simple duplexes and hairpin loops to the awe-inspiring complexity of the ribosome (6–8). Compared to the flurry of structures available for natural biopolymers, the structural information on artificial genetic polymers, generally referred to as xeno-nucleic acids or XNAs, is only beginning to emerge and includes about a dozen base-pairing systems (Figures 1 and 2).
Figure 1.
Representative structures illustrate the structural diversity and plasticity of natural and artificial nucleic acid (XNA) backbones. Structures are shown in alphabetic order. (A) Natural genetic polymers: B-form DNA (black), DNA:RNA hybrid and A-form RNA (gray). (B) Representative structures of XNA heteroduplexes with RNA or DNA. The RNA strand is shown in gray, the DNA strand in black and the orientation of the XNA strand is indicated. (C) XNA homoduplexes. Homo-XNA duplexes adopt a variety of structures. (D) Representative XNA-only heteroduplexes. FAF:FAF stands for FANA(F)-ANA(A)-FANA(F) XNA:XNA heteroduplex. Alt and chim indicate the alternated or chimeric order of FANA-segments in the duplex sequences respectively. The depicted duplexes have the following PDB ID codes in the Protein Data Bank (http://www.rcsb.org): B-DNA (3BSE); DNA:RNA (1EFS); A-RNA (3ND4); ANA(purple):RNA (2KP3); CeNA(blue):RNA (3KNC); FANA(violet):RNA (2KP4); HNA(yellow):RNA (2BJ6); LNA(cyan):RNA (1H0Q); PNA(orange):DNA (1PDT); PNA(orange):RNA (176D); CeNA:CeNA (blue, 2H0N); hDNA:hDNA (sky blue, 2H9S); FRNA:FRNA (magenta, 3P4A); GNA:GNA (red, 2XC6); HNA:HNA (yellow, 481D); LNA:LNA (cyan, 2×2Q); PNA:PNA (orange, 2K4G), TNA:TNA (green, coordinates not deposited in the PDB; see also Table 1); dXyNA:dXyNA (brown, coordinates not deposited in the PDB; see also Table 1); XyNA:XyNA (light green, 2N4J); FAF:FAF (FANA in violet, ANA in purple, 2LSC), FRNA:FANA (alt) (FRNA in magenta, FANA in violet, 2M8A); FRNA:FANA (chim) (FRNA in magenta, FANA in violet, 2M84).
Figure 2.
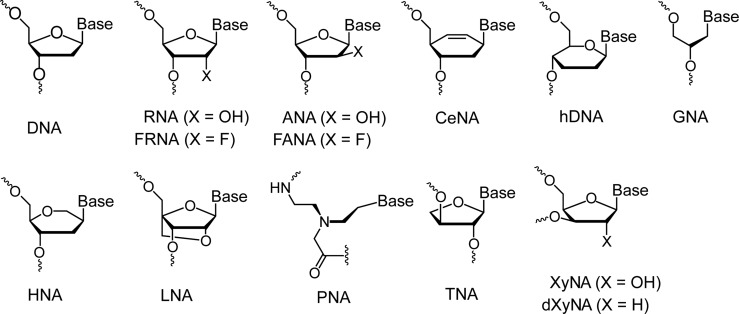
XNA backbone repeating units. Chemical structures of the natural and artificial (XNA) nucleic acid repeating units in alphabetic order. The variety of existing nucleic acid backbones gives rise to tremendous potential for structural diversity with applications that span diverse fields.
Chemical modification of the natural DNA and RNA framework was motivated by two main objectives. The first concerns medicinal chemistry and the generation of analogs with tailor-made properties that render them suitable for antisense, siRNA and microRNA targeting, aptamer and ribozyme applications (9). The second aims at gaining new insight into the chemical etiology of ribofuranosyl nucleic acids (DNA and RNA) as nature's choice for an information-carrying biopolymer (10,11). The available large assortment of chemical modifications has paved the road to nucleic acid chemical biology (12) and provided a solid basis for systematically evaluating the consequences of chemical modifications on structure and pairing stability (13,14). In the latter realm, the first crystal structures of fully modified DNA, (N3′→P5′ phosphoramidate DNA (15), and RNA, 2′-O-(2-methoxyethyl)-RNA (16), demonstrated the impact of the anomeric effect on DNA and RNA backbone conformation as well as the role of hydration and conformational preorganization in nucleic acid pairing stability.
In the world of nucleic acid polymers, seemingly minor differences in chemical structure can produce dramatic changes in the dynamics and stability of antiparallel Watson–Crick duplexes. Consider the case of natural DNA and RNA, for example, DNA commonly adopts a B-type helix, but the 2′-hydroxyl group of the ribose sugar locks RNA in an A-form helical structure (17). The ability of RNA to preorganize into a preferred helical geometry causes RNA to dominate the DNA conformation in RNA:DNA duplexes (18,19). However, one should always keep in mind that nucleic acid helices are highly polymorphic and even RNA:DNA duplexes can adopt a range of conformations, from canonical A-form to shapes that match neither A- nor B-form (20). More recent work on 2′-deoxy-2′-fluororibonucleic acid (2′-F RNA or FRNA) demonstrated that simple replacement of the ribose 2′-OH group by fluorine alters strand hydration, thermodynamic stability and in vitro and in vivo siRNA activity, without affecting the standard A-form geometry (21). The increased thermal stability is enthalpy-based; thanks to the higher Watson–Crick H-bonding strength between base pairs in FRNA compared to RNA and DNA duplexes (22,23). Additional stability is contributed by C2′(F)H…O4′ nonconventional H-bonds as a result of significant polarization effects provoked by the fluorine substituent (24). Owing to the enhanced pairing stability, FRNA is widely used as an RNA analog.
XNA artificial pairing systems, that may or may not pair with DNA and/or RNA, have opened the door to an emerging area of synthetic biology called synthetic genetics (25,26). Researchers working in this area are developing polymerases that copy genetic information back and forth between DNA and XNA and hope to one day create enzymes that will directly copy information between XNA itself (27,28). While such systems remain in their infancy, many hypothesize that XNA polymers, by virtue of their backbone structure, will provide access to vast new regions of nucleic acid fold space. This possibility, coupled with enhanced nuclease resistance, provides substantial motivation for advancing the use of XNA polymers in synthetic biology and molecular medicine (25,26).
While this review focuses on the structural features of XNA duplexes in which at least one strand of the Watson–Crick duplex is composed entirely of XNA, numerous other studies have examined the role of modified bases and backbones in the context of chemical biology. Notable accomplishments include expansion of the genetic alphabet to include unnatural bases that are capable of Darwinian evolution (29,30), formation of size and strand expanded helices (31–33) and the development of semi-synthetic organisms that carry modified bases within the DNA of actively dividing cells (34,35). Since a thorough discussion of these modifications is beyond the scope of this review, we direct readers interested in learning more about modified nucleotides and their interactions with polymerases to several excellent reviews on this topic (36–38).
This review provides a current analysis of the structural properties of known XNA duplex structures that are predominantly available in the Protein Data Bank (Table 1). We perform a comparative conformational analysis of XNA self-pairing and cross-pairing with natural nucleic acids and discuss their potential as genetic information carriers. Not surprisingly, the geometries of XNA duplexes cover a wide conformational range relative to standard DNA and RNA. It is our hope that this review will encourage others to pursue XNA research and further advance the structural knowledge of XNA polymers.
Table 1. Selected structural parameters for XNA homoduplexes and XNA heteroduplexes with DNA or RNA.
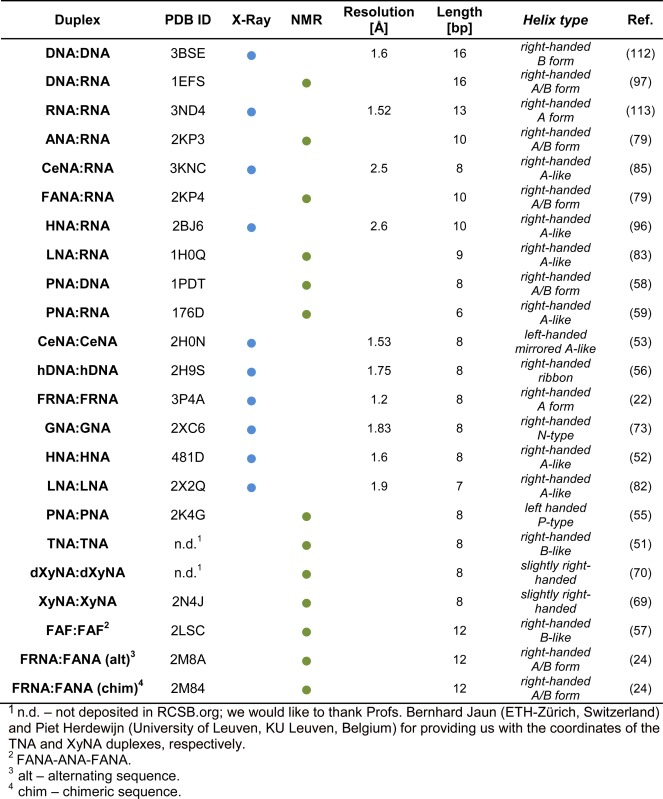
|
METHODS TO STUDY THE 3D STRUCTURE AND DYNAMICS OF NUCLEIC ACID POLYMERS
Single crystal X-ray crystallography is the most powerful approach in the structural biologist's toolbox for determining the 3D structures of nucleic acids, proteins and their complexes with molecular weights ranging from a few kilodalton up to megadalton (39). Crystallography has no inherent size limits and larger complex structures that are out of reach for solution NMR spectroscopy can be analysed by X-ray diffraction. In addition, molecules that are too small for single-particle cryo-electron microscopy are compatible with X-ray crystallography. It is therefore the method of choice for studying larger XNA aptamer structures and XNA-polymerase complexes. A further advantage of using X-ray crystallography is the ability to visualize oligonucleotide–ion interactions and hydration in high-resolution structures. However, the need for single crystals that diffract X-rays to high resolution is a major limitation of crystallography. Unfortunately, many oligonucleotides fail to produce crystals of sufficient quality and as a result, extensive attempts are necessary to screen length and sequence of oligomeric fragments to obtain such crystals. This effort can be time-consuming and requires milligram amounts of highly pure material. For XNAs, the latter condition is frequently expensive and an established synthetic route and purification protocols are prerequisites for embarking on the structural analysis of an artificial genetic polymer.
A second major hurdle on the road to a crystallographic 3D model of XNA concerns the generation of phase information necessary to compute an initial electron density map (39). If the XNA under investigation deviates conformationally from DNA or RNA, a common approach for solving the phase problem, the molecular replacement technique (40), cannot be applied. This is because molecular replacement typically fails if the root mean square deviation between model and actual structure amounts to just a few angstrom. In such cases, the multiple or single wavelength anomalous dispersion (MAD or SAD, respectively) approaches have to be used to determine a structure (41). The success of MAD or SAD is dependent on the presence of anomalously scattering atoms in the crystal. Their positions can then be identified from anomalous Patterson maps. Anomalously scattering atoms incorporated into nucleotides (e.g. 5-bromo-uridine or 2′-SeMe uridine) or amino acids (e.g. selenomethionine), or heavy atoms/anomalous scatterers present in the crystallization solution, such as cobalt hexamine, strontium and barium, are often used for overcoming the phase problem with nucleic acid crystals (42). It is important to keep in mind that nucleotide building blocks for solid phase synthesis of derivatized DNA (e.g. 5-bromo-dC) or RNA (e.g. 5-bromo- or 2′-SeMe-U) are not commercially available for XNA, or the approach cannot be used for a particular XNA (e.g. the RNA 2′-SeMe modification). The crystal structure of homo-DNA constitutes a case in point: octamers with incorporated 5-bromo-U or 5-bromo-C in place of T or C, respectively, did not crystallize and solving the phase problem required incorporation of selenium in the form of phosphoroselenoate into the oligonucleotide backbone (43).
In principle it is possible to phase oligonucleotide crystal structures using the anomalous signals of phosphorus (44) (available for any XNA with phosphate in the backbone) or sulfur (45) (using phosphorothioate or phosphorodithioate modification). However, neither the P-SAD (44,46) nor the S-SAD (47) approach has been met with much success in nucleic acid crystallography thus far. If the resolution of the diffraction data is >>1 Å, so-called direct methods to obtain phase information can also be used by exploiting known phase relationships between certain groups of reflections (48).
Another proven method for structural analysis is NMR spectroscopy (49). Because NMR sensitivity is inversely proportional to molecular size, the current limitation of NMR studies of nonisotope enriched samples is about 50 nucleotides (nt) for high-resolution structural characterization (50). Isotopic labeling can increase the size range of samples amenable to NMR structural studies; however, for XNA studies this significantly increases the costs associated with sample preparation. In the context of structural studies, synthetic restrictions have historically limited the size of the molecules to relatively short XNA duplexes of about 8–10 nucleotides and as a result present an excellent opportunity for NMR structural characterization. To date, approximately one-half of the 3D structures of XNA duplexes and their associated heteroduplexes with natural genetic polymers (DNA and RNA) have been determined by NMR. The limited chemical shift dispersion and nuclear Overhauser effect (NOE)-restraint network, which can complicate the assignment and structure calculations of natural polymers, are overcome by the small size and spectral variability of XNA duplexes. Commonly explored modifications, such as a fluorine substitution in 2′-deoxy-2′-fluoroarabinonucleic acid (FANA), can increase the chemical shift dispersion pattern, which allows for facile resonance assignment and NMR-based structure determination. Fluoro-substituted XNA molecules also allow for direct 19F detection using this abundant and sensitive NMR-active isotope. Moreover, their increased backbone stability and resistance to naturally occurring exo- and endo-nucleases (26) make XNA amenable to long NMR experiments over a wide range of temperatures.
A major advantage of NMR spectroscopy is the possibility to study artificial genetic polymers under near-physiological conditions. Beyond traditional structural studies, NMR can also be used to provide information on conformational plasticity and dynamics of XNA molecules, thereby facilitating comparative studies of duplex stability, thermodynamic properties, base pair lifetimes and direct assessment of XNA duplex conformations through measurement of various NMR parameters. Both techniques combined, X-ray and NMR, provide a powerful framework to explore the XNA fold space and to gain detailed insight into the structure and dynamics of artificial nucleic acid pairing systems.
STRUCTURAL PLASTICITY OF ARTIFICIAL AND NATURAL NUCLEIC ACIDS
Artificial genetic polymers can self-assemble into antiparallel Watson–Crick duplexes that adopt a wide range of helical geometries (Figure 1). Natural genetic polymers are known to form right-handed antiparallel helices. RNA self-pairing and cross-paring with DNA leads to the wider A-form helical conformation that is characterized by a compact rise, inclined base pairs and a ribbon-like arrangement of strands. By contrast, DNA self-pairing leads to a B-form helix that is narrower than the A-form helix and is characterized by tightly wound strands and base pairs that are oriented roughly perpendicular to the helical axis (Figure 1a). In contrast to natural helical conformations observed for DNA and RNA, XNAs adopt a variety of structural conformations. The 3D structures of XNA homoduplexes (Figure 1c) range from those that are similar to the standard A-form RNA helix, like α-L-threofuranosyl-(3′→2′) nucleic acid (TNA) (51) and 2′,3′-dideoxy-1′,5′-anhydro-D-arabino-hexitol nucleic acid (HNA) (52) to more diverse structures, like the left-handed antiparallel, mirrored A-type helix of cyclohexene nucleic acid (CeNA) (53), the P-helix formed by peptide nucleic acid (PNA) (54,55), or the slowly writhing, more ladder-like (4′→6′) linked oligo-2′,3′-dideoxy-β-D-glucopyranose nucleic acid (homo-DNA or hDNA) (56). One interesting XNA structure is that of a self-complementary chimeric FANA-ANA-FNA strand that self-assembles into an antiparallel right-handed duplex in which the central and flanking regions undergo ANA:ANA and FANA:FANA self-pairing (57). Heteroduplexes of XNAs with native genetic polymers also display a range of helical shapes, with variations in diameter, helical rise, inclination and twist (Figure 1b). Interestingly, the morphology of these helices, in most cases, is right-handed and antiparallel, even for PNA:DNA (58) and PNA:RNA (59), and resembles double-stranded RNA more closely than DNA.
THE BUILDING BLOCKS OF ARTIFICIAL GENETIC POLYMERS
One of the founding ideas behind the field of synthetic genetics was to produce chemically stable, nuclease-resistant oligonucleotides that are capable of cross-pairing with DNA and RNA. The first XNA synthesized and published in 1991 was PNA (60). PNA has a backbone that is composed of repeating N-(2-aminoethyl)-glycine units linked by amide bonds (Figure 2). Adenine, thymine, cytosine and guanine bases are attached to the backbone by a methylene bridge (CH2) and a carbonyl group (C = O). Unlike DNA and RNA, PNA lacks a sugar moiety and a repeating negatively charged phosphodiester moiety. Another acyclic nucleic acid polymer is glycerol nucleic acid (GNA), which is composed of a repeating three-carbon sugar linked by phosphodiester bonds. (Figure 2) (61). GNA can exist as two enantiomers (R) or (S), as indicated in Figure 2. (S)-GNA and (R)-GNA strands can self-assemble into homochiral antiparallel right-handed and left-handed, respectively, duplexes that are held together by Watson–Crick base pairs. Interestingly, the (S)-enantiomer of GNA is capable of cross-pairing with RNA, but not DNA (61). Another group of XNA molecules contains a 6-membered pyranose ring instead of the more common 5-membered furanose ring found in DNA and RNA. Representative members of this group include hDNA (62), HNA (63) and CeNA (53). TNA, explored in the context of a chemical etiology of the nucleic acids, has a threose backbone that is one atom (or one bond) shorter compared to DNA and RNA (64). TNA and GNA were originally dismissed as artificial base-pairing systems due to the perceived notion that a 6-atom backbone repeat unit was required to form stable duplexes with DNA and RNA. However, we now know that this dogma was incorrect as both TNA and GNA are capable of base pairing with natural genetic polymers. XNA molecules with more subtle chemical differences relative to the native counterparts include arabinonucleic acid (ANA) (65), in which ribose is replaced by arabinose, FANA (66) and locked nucleic acid (LNA) that features the bicyclic β-D-2′-O-4′-C-methylene ribofuranose sugar (67) (Figure 2). With the exception of PNA, all of these artificial pairing systems feature phosphate groups in the backbone.
XNA PAIRING MODES
Examples of XNA homo- or heterostructures in which at least one strand is composed entirely of XNA are shown in Figure 1. Although many XNAs pair with natural DNA and RNA, some display orthogonal base-pairing properties that are distinct from the natural DNA and RNA. For example, (R)-GNA and hDNA have strong self-pairing properties, but do not cross-pair with natural DNA and RNA (61,68). Similarly, the recently published xylonucleic acid (XyNA) and deoxy-xylonucleic acids (dXyNA) also constitute an autonomous pairing system, with the caveat that stable triplexes can be formed with both, DNA and RNA poly-A strands (69–71). In the crystal structure of a homochiral DNA (hDNA) octamer duplex, the two strands form Watson–Crick base pairs, with one adenine per strand being extruded from the duplex and forming a reverse-Hoogsteen base pair with a thymine base from an adjacent duplex (56). Interestingly, hDNA duplexes are more stable compared to DNA duplexes of the same sequence, whereby the increased stability is entropy-based (68). Another hallmark of the hDNA duplex is the strong inclination and concomitant cross-strand base stacking. A recent fascinating finding is that D-β-hDNA and L-β-hDNA can form heterochiral duplexes that are of higher stability than the corresponding homochiral duplexes (72). In the GNA family, the (S)- and (R)- enantiomers do not pair with each other and a structure for (R)-GNA has not yet been reported. Nevertheless, the structure of (S)-GNA provides a number of interesting observations that include cross-strand base stacking as opposed to the more common intrastrand base stacking and a strong hydrophobic effect from the 2′-methylene group (73–75). PNA is capable of forming a stable antiparallel duplex and can also cross-pair efficiently with DNA and RNA. Three types of PNA duplex structures have been reported in the literature (55,58,59) with PNA:PNA duplexes adopting both left- and right-handed helical structures (76). PNA:DNA and PNA:RNA duplexes have higher thermal stability when compared to natural DNA or RNA duplexes with the same sequence (77). Like PNA, TNA is capable of stable self-pairing and cross-pairs with RNA and DNA (38); however TNA:RNA duplexes are more stable than TNA:DNA duplexes (78). Although the structure of an all-TNA duplex was reported in 2008 (28), no structures of TNA:DNA or TNA:RNA heteroduplexes have been reported. ANA and FANA can form stable duplexes with RNA, whereby their conformation mimics that of DNA in DNA:RNA hybrids. FANA has a higher affinity towards RNA than ANA, probably because of formation of a favorable intrastrand pseudo-hydrogen bond (F2′ to H8 from the 3′-adjacent purine), which contrasts with the unfavorable 2′-OH…nucleobase steric contributions and lack of favorable electrostatic contacts in the case of ANA (79,80). Solution NMR structures of FANA and ANA hybrids with RNA have been published (79). In addition, crystal structures of A- and B-form DNA duplexes with incorporated FANA and ANA nucleotides revealed that the FANA conformation is more compatible with an A-type duplex form than that adopted by ANA (81), consistent with FANA's favorable pairing with RNA. For LNA, structures have been reported for a homoduplex and a heteroduplex with RNA and both are consistent with A-like helical geometry (82,83). Introduction of LNA nucleotides into RNA results in increased melting temperatures of the oligoribonucleotide (84). The LNA:DNA duplex has been analysed by NMR and the resulting data suggest it is similar to a DNA:RNA hybrid, but no detailed structure has been determined (83). For CeNA, crystal structures are available for the CeNA homoduplex (53) and a CeNA:RNA hybrid duplex (85). CeNA forms an antiparallel right-handed duplex with standard Watson–Crick base pair geometry (Figure 1b). The XNA molecules highlighted here demonstrate that fold space diversity of artificial nucleic acids is increased relative to natural DNA and RNA by the adoption of multiple base-pairing modes. The diversity stems from the extended conformational range afforded by different backbone chemistries, Watson–Crick (most XNAs) and reverse-Hoogsteen base pairing (purine–purine pairs in hDNA duplexes (68)), and homochiral (virtually the rule) and heterochiral pairing (hDNA (72)) modes.
GEOMETRIC PARAMETERS OF XNA DUPLEXES
Helical parameters of natural A- and B-form helices
Natural DNA and RNA adopt dynamic helices with local conformational plasticity that is determined to a large extent by their sequence, which is necessary for their biological function (86,87). Most of the available structures show that these systems adopt either an A- or a B-form antiparallel right-handed helical conformation that is stabilized by base stacking interactions together with hydrogen bonding, electrostatics and solvation (88,89). DNA sequences that contain alternating patterns of purine and pyrimidine repeats (i.e. [CG]n) can form left-handed antiparallel duplexes in which phosphate groups of each strand follow a zig-zag pattern (Z-DNA) (86). The potential for Z-DNA structure correlates with regions of human DNA that are under active transcription (90).
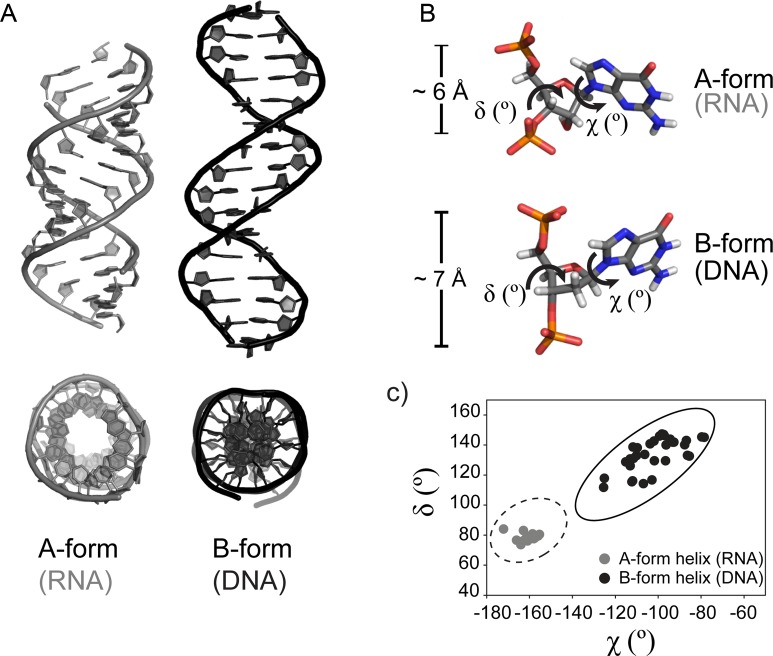
RNA adopts an A-form helix (Figure 3a), which is characterized by a wide and shallow minor groove of about 11 Å width and a deep and narrow major groove of about 3 Å width, calculated as the shortest distance between phosphates across the groove minus the sum of van der Waals radii of the two phosphates. Approximately 11 base pairs constitute a single A-form helical turn. In contrast, DNA is mostly observed as a B-form duplex (Figure 3a). This helix is more compact, featuring limited core space and deep major and minor grooves of ca. 7.5–8.5 Å depth. The major groove is wider (ca. 12 Å), exposing a different pattern of hydrogen bond donor and acceptor groups, as compared to the A-form helix, which is recognized by different sets of enzymes (91). Approximately 10 base pairs form one turn of a standard B-form helix. Normally, a given calculated structure will not match all of the classical criteria of either an A- or a B-form helix; however, the average values will fall into specific ranges. The conformational differences between natural A- and B-form helices have been characterized in detail (92).
Figure 3.
Geometric parameters of natural A- and B-form helices. (A) Natural genetic polymers (RNA and DNA) adopt different helical geometries. Left: canonical A-form helix (RNA, PDB code 3ND4). Right: canonical B-form helix (DNA, PDB code 3BSE). A view from the top is presented underneath each structure. (B) Differences in sugar puckering and P–P distances between A- and B-form helices. A close-up view of a single nucleotide in a canonical A-helix (top panel, RNA) and a B-helix (bottom panel, DNA) color coded by atom type. The χ and δ torsion angles and the distances between two adjacent phosphates in 5′-3′ direction (PiPi+1) are indicated. (C) (χ,δ) angle covariance matrix for natural A- and B-form helices. Each dot represents a (χ,δ) angle covariance for a single nucleotide from the structures listed above. The values for χ and δ torsion angles of A- and B-form helices cluster in distinct regions of the plot and allow a simple measure of structural diversity. Example values were generated using PYMOL software, based on the PDB code 3ND4 (gray circles, A-form, RNA) and PDB code 3BSE (black circles, B-form, DNA) structures. The ellipses show published value ranges for the (χ,δ) angle covariance matrices of an A-DNA (dashed line) and a B-DNA (solid line) (92).
The distinctive morphologies of A- and B-form duplexes originate from different pucker conformations of the sugar ring, reflected in their pseudorotation phase angles and correlate with nucleotide torsion angle values (Figure 3). The C3′-endo ribose conformation in the A-form duplex places intrastrand backbone phosphates closer together and at an approximate distance of 6 Å. The C2′-endo sugar conformation in the B-form duplex forces them further apart, to a distance of ca. 7 Å. Due to the high conformational plasticity of nucleic acids, a single torsion angle value does not allow discrimination between A- and B-structures (93). However, clustering of pairs of parameters for χ (the glycosidic torsion angle between sugar and base) and δ (backbone torsion angle associated with the sugar ring) has the discriminative power to identify helical morphology (92,94). Surveys with large sets of 3D structures show, that the (χ,δ) value pairs of A- and B-form helices occupy distinct areas of conformational space (Figure 3c). In case of an A-form helix, χ values cluster between -170° and -140° and δ values in the range between 65° and 100°. In a B-form helix, χ values lie between -130° and -70° and δ angles between 90° and 160°. Pseudorotation phase angles P, describing the sugar pucker and (χ,δ) covariance matrices offer the means for deeper insights into the 3D morphology of duplexes and allow structural comparisons between natural nucleic acids and XNAs.
Sugar pseudorotation phase angles in natural and XNA duplexes
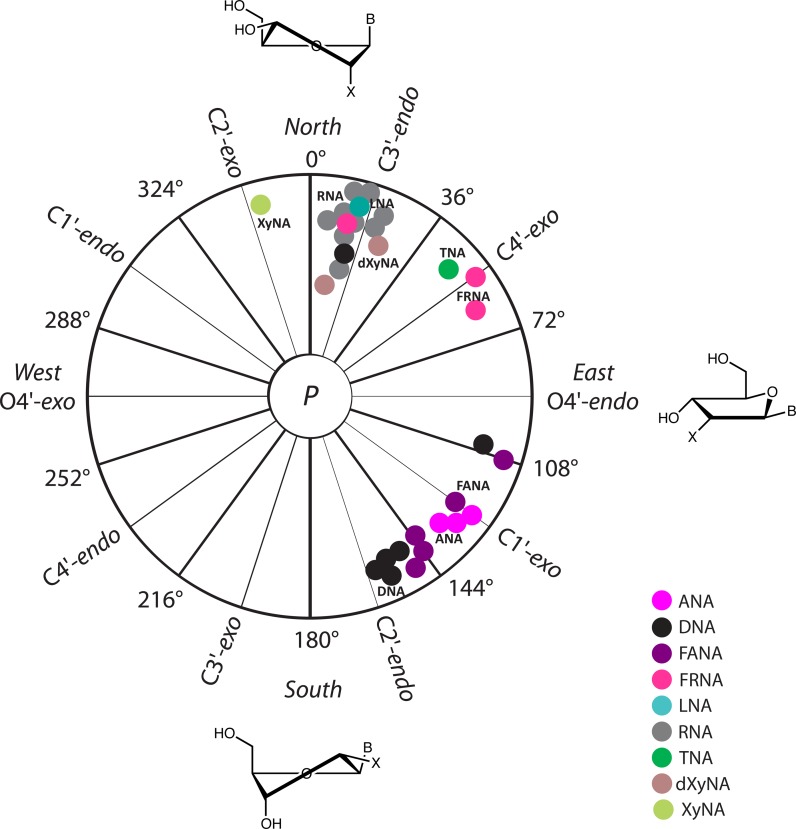
The conformation of the sugar ring can be described by the pseudorotation phase angle parameter P. A standard conformation (P = 0°) is defined with a maximally positive C1′-C2′-C3′-C4′ torsion angle with the P value ranging from 0° to 360°. Conformations in a region within the upper half of the circle with P = 0±45° are denoted Northern, N, and those in a region within the lower half of the circle with P = 180±45° are denoted Southern, S. For comparative purposes, we calculated the pseudorotation phase angles for all XNA duplexes that contain furanose (5-membered) sugar rings. As evident from the data shown in Figure 4, riboses in RNA adopt a C3′-endo conformation regardless of the pairing partner (DNA or XNA). An identical picture emerges for LNA that adopts the same sugar conformation as RNA. In DNA, the 2′-deoxyribose sugar moieties adopt a C2′-endo conformation. However, in duplexes that feature DNA paired with PNA, the DNA strand adapts to the PNA strand and the 2′-deoxyribose sugars display a C3′-endo or O4′-endo conformation. In ANA and FANA oligonucleotides, the average sugar pucker is of the C1′-exo type, but FANA's extends to the Eastern range whereas ANA puckers appear limited more to the Southeast (Figure 4). Crystallographic data indicated that FANA nucleotides in an A-form duplex environment can adopt a Northeastern pucker (81). TNA is unique in terms of its sugar conformation as the tetrose adopts a C4′-exo pucker. For HNA and CeNA, the sugar conformation lies in the Northern range. Like with DNA and RNA, the Western half of the pseudorotation phase cycle remains unpopulated as structural restrictions prevent XNAs from adopting such puckers.
Figure 4.
Pseudorotation phase angles P for XNA duplexes. Angles were only calculated for oligonucleotides with 5-membered sugar moieties, using the program PROSIT (http://cactus.nci.nih.gov/prosit/) (114). All angles are averages, whereby terminal residues were excluded. Angles were not calculated for HNA, CeNA and homo-DNA that all contain 6-membered sugar moieties, but HNA and CeNA are compatible with an N-type sugar pucker. Different colors represent the various sugar chemistries: purple (ANA), black (DNA), violet (FANA), magenta (FRNA), cyan (LNA), gray (RNA), green (TNA), brown (dXyNA) and light green (XyNA). The sugar puckers for RNA and DNA opposite XNAs are as follows: C3′-endo (RNA and LNA), C4′-exo (TNA), C1′-exo (FANA, ANA), O4′-endo (DNA opposite PNA; NMR solution structure with PDB code 1PDT), C3′-endo or C2′-endo (DNA opposite PNA; crystal structure with PDB code 1NR8) and C2′-endo (DNA and PS-DNA). Different gray and black spots for DNA and RNA respectively, represent average puckers in DNA and RNA strands opposite various XNAs.
Characteristic geometries of XNA structures
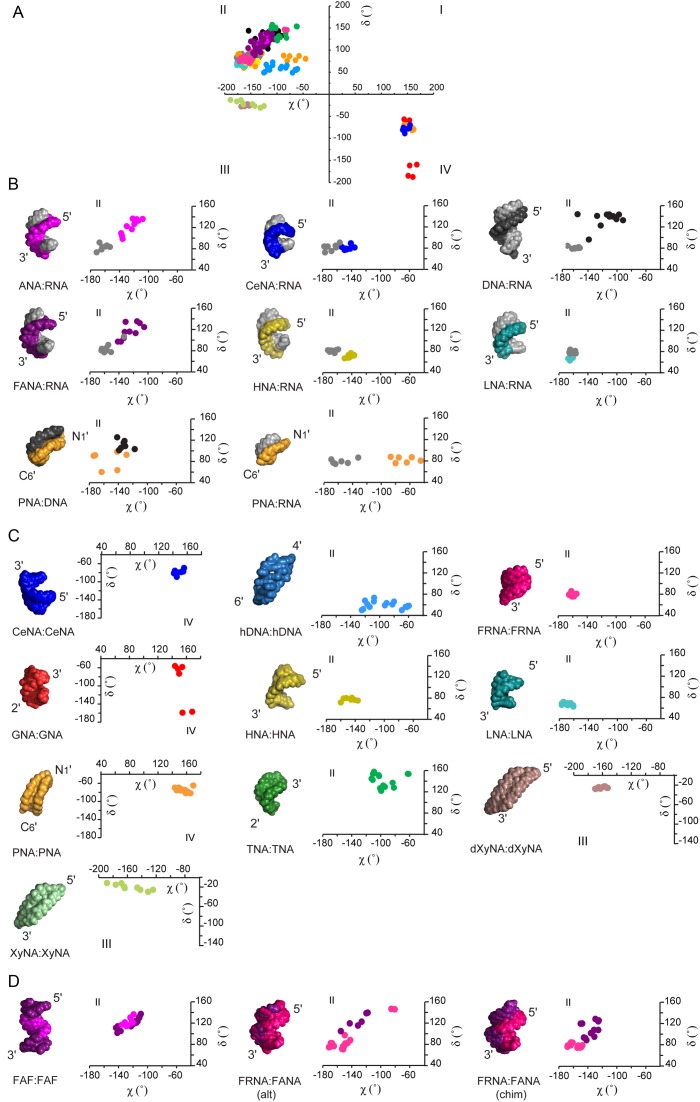
Heteroduplexes between naturally occurring nucleic acids (DNA:RNA hybrids) and most XNA homo- and heteroduplexes form right-handed antiparallel helical structures, which is reflected in their (χ,δ) covariance plots (Figure 5). The values of pairs of (χ,δ) torsion angles and their equivalents in XNAs predominantly cluster in the second quadrant of Cartesian space (Figure 5a), which is also occupied by (χ,δ) angles of canonical A- and B-form duplexes (Figure 3c). The exceptions are formed by double-stranded PNA, CeNA and GNA helices that cluster in the fourth quadrant of Cartesian space, and XyNA and dXyNA helices occupying the third quadrant.
Figure 5.
(χ,δ) angle covariance matrices reflect the structural diversity and plasticity of xeno-nucleic acid (XNA) backbones. (A) The values for χ and δ torsion angles of natural and artificial nucleic acid helices cluster in quadrants II, III and IV of the Cartesian coordinate system. Each dot represents a (χ,δ) covariance for a single nucleotide in the sequence. (B) The 3D structure and a close-up view of the (χ,δ) covariance plot for DNA and RNA heteroduplexes of natural and artificial nucleic acids in alphabetic order. The RNA strand is shown in gray and the DNA strand is shown in black. The Cartesian quadrant of the plot is indicated on top. From left to right: ANA (purple):RNA (PDB code 2KP3); CeNA (blue):RNA (PDB code 3KNC); DNA:RNA (PDB code 1EFS); FANA (violet):RNA (PDB code 2KP4), HNA (yellow):RNA (PDB code 2BJ6); LNA (cyan):RNA (PDB code 1H0Q); PNA (orange):DNA (PDB code 1PDT); PNA (orange):RNA (PDB code 176D). (C) The 3D structure and a close-up view of the (χ,δ) covariance plot for XNA homoduplexes. The Cartesian quadrant of the plot is indicated. From left to right CeNA:CeNA (blue, PDB code 2H0N); hDNA:hDNA (sky blue, PDB code 2H9S); FRNA:FRNA (magenta, PDB code 3P4A); GNA:GNA (red, PDB code 2XC6); HNA:HNA (yellow, PDB code 481D); LNA:LNA (cyan, PDB code 2×2Q); PNA:PNA (orange, PDB code 2K4G); TNA:TNA (green, coordinates not deposited); dXyNA:dXyNA (brown, coordinates not deposited); XyNA:XyNA (light green, PDB code 2N4J). (D) The 3D structure and a close-up view of the (χ,δ) covariance plot for representative XNA-only heteroduplexes. The Cartesian quadrant of the plot is indicated on top of the plot. From left to right: FANA(F)-ANA(A)-FANA(F) heteroduplex (FANA in purple, ANA in violet, PDB code 2LSC), FRNA:FANA (alt) (FRNA in magenta, FANA in violet, PDB code 2M8A); FRNA:FANA (chim) (FRNA in magenta, FANA in violet, PDB code 2M84). Alt and chim indicate the alternated or chimeric order of FANA-segments in the duplex sequences respectively. Glycosidic torsion angles between sugar and base (χ) and the backbone torsion angle associated with the sugar ring (δ) were extracted from the structures, shown above, using PyMol software (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.) and defined as torsion angles between the following atoms in RNA or DNA. χ (pyrimidines): O4′-C1′-N1-C2; χ (purines): O4′-C1′-N9-C4; δ: C5′-C4′-C3′-O3′. In the case of XNAs atoms at the equivalent positions were used for calculations.
Crystal structures indicate that achiral PNA can form right-handed and left-handed P-type helices (54). In the P-type helix, an antiparallel double helix form uniquely attributed to PNA, the strands are held together by standard Watson–Crick base pairs and are wound more loosely relative to DNA. This difference leads to a helical structure with a larger diameter than DNA. To date only left-handed P-type helices have been detected by NMR in solution (55). The (χ,δ) value pairs of left-handed PNA fall into the fourth quadrant of Cartesian space. The structure of the antiparallel duplex formed by another left-handed XNA molecule, CeNA, exhibits geometric parameters that closely resemble a left-handed RNA. Thus, it has been characterized as belonging to a mirrored A-type family (53). The duplex is held together by normal Watson–Crick base pairs. The CeNA (χ,δ) value pairs cluster in the fourth quadrant of Cartesian space, and are positioned diagonally across those for an A-type helix. The backbone of GNA contains only three carbons and a stereo center derived from propylene glycol. Previous crystallographic studies reveal that (S)-GNA can form two different helical types—a more elongated M-type helix, containing metallo base pairs (74), and a condensed N-type structure that contains brominated base pairs (73,75), presented in Figure 5c. Overall, GNA helices differ significantly from canonical A- and B-forms. Although the structures share some similarities with RNA-helices, they may be better described as a helical ribbon loosely wrapped around the helix axis. The GNA N-type duplex retains the canonical Watson–Crick base-pairing pattern; however, its backbone torsion angle δ is found in alternating gauche and anti conformations and its (χ,δ) pairs are found in the fourth quadrant of Cartesian space.
(χ,δ) angles of dXyNA and XyNA occupy the third quadrant of Cartesian space. Their recently described homoduplexes (69,70) do not form cylindrical helices in solution, but adopt slightly right-handed ladder-like, extended structures with almost indistinguishable major and minor grooves and highly inclined bases. The chiral inversion in the 3′-carbon center leads to changes in the backbone dihedral angles, including the backbone torsion angle δ, which is close to -20°. Though XyNA and dXyNA structures are predominantly stabilized by zipper-like interstrand stacking interactions, the canonical Watson–Crick base-pairing is retained.
To date, all structurally characterized XNAs that form antiparallel right-handed duplexes do so through canonical Watson–Crick base-pairing interactions. Their (χ,δ) pairs cluster in the second quadrant of Cartesian space (Figure 5a). However, a closer look reveals considerable differences in their backbone morphologies (Figure 5c). For example, other parameters such as helical twist, roll, number of base pairs per turn (ca. 14–15) and a wide major groove, place double-stranded HNA and double-stranded LNA more in the structural vicinity of large-diameter P-helices similar to that adopted by PNA (52,82). These backbones seem to have an A-form like morphology, with the (χ,δ) values occupying the A-DNA segment of the covariance space. Double-stranded TNA, on the other hand, is generally considered structurally close to an A-type helix, as judged from the average values for base-pair parameters, intrastrand phosphate distance, a wide and shallow minor groove and stable cross-pairing with RNA (51). Interestingly, given the short length of the oligonucleotide and high curvature of the structure, no reliable information could be extracted for the width of the TNA major groove. Moreover, its backbone torsion angle pairs fall into the B-DNA sector of the (χ,δ) covariance angle space, indicating a need for further structural studies of a longer TNA oligonucleotide.
hDNA represents an outlier among members of the family of right-handed antiparallel XNA homoduplexes (56). Although hDNA displays stable self-pairing in a canonical Watson–Crick fashion, the structure of the octamer duplex virtually lacks intrastrand stacking, but instead features extensive overlaps between adjacent bases from opposite strands. hDNA also lacks a major groove but features a shallow minor groove, and therefore resembles a flat ribbon. Its helical geometry is distinct from the cylindrical helices of A-form and B-form natural nucleic acids and this is reflected in (χ,δ) correlations that occupy a distinct area of the second quadrant of Cartesian space (Figure 5c).
Based on the currently known structures of artificial oligonucleotides paired with RNA or DNA (Figure 5b), XNA heteroduplexes adopt predominantly antiparallel, right-handed helices. They are held together by standard Watson–Crick base pairs and are predominantly stabilized by intrastrand and partial interstrand stacking interactions. Stacking interactions were proposed to play the major role in nucleic acid duplex stability and the ability of XNAs to form helices with natural partners (88,95). Although helices display a distinct pitch and curvature, they feature ca. 11–12 base pairs per turn, and χ/δ covariance plots indicate that the backbones of XNA:RNA or XNA:DNA heteroduplexes adopt an architecture that is either closely related to the A-form, as in the case of HNA:RNA (96), LNA:RNA (83), CeNA:RNA (85) and PNA:RNA (59), or between the A- and B-forms, as seen in the structures of DNA:RNA (97), ANA:RNA (79), FANA:RNA (79) and PNA:DNA (98). Interestingly, the (χ,δ) value pairs of natural nucleic acids in the heteroduplexes fall into the covariance areas usually occupied by (χ,δ) angles of the respective RNA or DNA homoduplexes. Thus, it seems that RNA and DNA even in XNA heteroduplexes may prefer backbone morphologies that resemble those in their respective canonical structures. Still, the wide and loosely packed A-form helix may have more structural plasticity to accommodate increased variability in the nucleic acid backbone, which in general forces the XNA heteroduplex backbones to adapt to it.
POTENTIAL FUNCTIONAL AND BIOLOGICAL APPLICATIONS OF XNA
Recent advances in polymerase engineering have made it possible to copy genetic information back and forth between DNA and certain XNA polymers (25,26). Such accomplishments build on years of basic research into the molecular recognition properties of DNA and RNA polymerases with modified substrates. While early studies focused mainly on close structural analogs of DNA and RNA (99), recent work has been extended to include XNAs with more diverse chemical compositions. In the area of TNA, for example, enzyme-screening assays were used to identify polymerases that could copy DNA on a chemically synthesized TNA template and other polymerases that could synthesize TNA on a DNA template (100–103). In the case of FANA, Damha et al. were the first to show that natural polymerases could synthesize FANA on a DNA template and DNA on a FANA template (104). Related work by Wengel et al. has shown that LNA can be enzymatically incorporated into growing DNA strands (105). These, and many other studies, helped raise awareness for the importance of developing engineered polymerases for synthetic biology (106).
Recognizing the limitations of natural polymerases as tools for synthetic biology, directed evolution methods have been developed to create XNA polymerases that function with increased efficiency, fidelity and processivity. Such efforts have produced a number of polymerases that can ‘transcribe’ DNA into XNA and ‘reverse transcribe’ XNA back into DNA (27,28). By inserting a selective binding step into the replication cycle, researchers have isolated the first examples of XNA aptamers—molecules that function as synthetic antibodies by folding into shapes with specific ligand binding activity (26,107). The list of XNA aptamers generated to date includes a TNA aptamer with affinity to human α-thrombin, two HNA aptamers, one that binds the HIV trans-activating response RNA element and another that binds protein hen-egg lysozyme and an FANA aptamer to HIV-RT (26,107,108). More recently, this concept of functional activity was further extended to include XNAs with rudimentary catalytic activity (109). Together, these examples demonstrate that XNAs, like DNA and RNA, are dynamic molecules that can fold into complex 3D structures that complement the shape and charge of a target molecule.
In addition to functional activity, XNA polymers have shown promise in nanotechnology and materials engineering (26). GNA, which has a phosphodiester backbone with a single stereocenter was used to construct two identical nanostructures with mirror-image symmetry (110). While DNA is also capable of accessing similar structures, the chemical simplicity of GNA relative to DNA makes the design of hierarchical assemblies with mirror-image structures more accessible by chemical synthesis. Because XNAs have diverse helical geometries, one could easily speculate that future work in this area will lead to new folding conformations with structures that are not accessible to DNA. Likewise, XNA has the potential to expand the range of mechanical properties available for molecular engineering and structural nanotechnology, as demonstrated by Maher et al. in a recent study on the bending and flexibility of several DNA analogs (111).
These applications along with those that have yet to be developed will be aided by structural studies that help elucidate the molecular underpinnings of XNA structure and function. By improving our understanding of the structural properties of XNA, it should be possible to design new XNA molecules for nanotechnology, enhance the functional properties of in vitro selected aptamers and catalysts and develop computational programs for rationally designing new XNA sensors and other ligand responsive elements. In addition, there is a pressing need to explore the structural properties of engineered polymerases that are used to ‘transcribe’ and ‘reverse transcribe’ genetic information. In combination, XNA and their requisite polymerases, have broad applications from information storage to the development of orthogonal cells with engineered pathways that function independent of the natural genetic system (35).
CONCLUSIONS AND FUTURE OUTLOOKS
From a small sampling of known XNA structures, it is clear that XNA can self-assemble into antiparallel Watson–Crick duplexes with helical geometries that extend beyond the structural space occupied by DNA and RNA. Advances in nucleic acid chemistry and structural biology indicate that the total number of XNA polymers will continue to increase. These advances suggest that future XNA duplexes will greatly expand our understanding of the limits of Watson–Crick base pairing and nucleic acid helicity. One question appearing on the horizon concerns how these molecules will fold into complex 3D-structures with specific ligand binding or chemical catalysis properties? One could speculate based on the structural heterogeneity of known XNA duplexes that highly structured XNA molecules represent an untapped source of nucleic acid fold space. While many technical hurdles will need to be solved before we can get our first glimpse into the structural diversity of XNA fold space, the current pace of polymerase engineering and XNA evolution suggests that these structures may be accessible in the near future. As we begin to move in this direction, nucleic acid chemists, structural biologists and polymerase engineers will need to work in close collaboration to help bring these new technologies to the forefront of mainstream molecular biology.
Acknowledgments
Dedicated to Professor Albert Eschenmoser, ETH-Zürich, on the occasion of his 90th birthday.
FUNDING
Defense Advanced Research Projects Agency (DARPA) Fold F(x) Program [DARPA-14–13-FOLD-PA-004 to J.C.C., M.E. and W.V.H]. The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blake C.C., Koenig D.F., Mair G.A., North A.C., Phillips D.C., Sarma V.R. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206:757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- 2.Wang A.H., Quigley G.J., Kolpak F.J., Crawford J.L., van Boom J.H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 3.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egli M., Pallan P.S. The many twists and turns of DNA: template, telomere, tool, and target. Curr. Opin. Struct. Biol. 2010;20:262–275. doi: 10.1016/j.sbi.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neidle S. Principles of Nucleic Acid Structure. London: Academic Press, Elsevier Inc; 2008. [Google Scholar]
- 6.Mortimer S.A., Kidwell M.A., Doudna J.A. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 7.Yusupova G., Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 8.Klostermeier D., Hammann C., editors. RNA Structure and Folding. Berlin; Boston: De Gruyter; 2013. [Google Scholar]
- 9.Egli M., Herdewijn P., editors. Chemistry and Biology of Artificial Nucleic Acids. Weinheim: Wiley-VCH Publishers; 2012. [Google Scholar]
- 10.Eschenmoser A. Chemical etiology of nucleic acid structure. Science. 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- 11.Eschenmoser A. Etiology of potentially primordial biomolecular structures: from vitamin B12 to the nucleic acids and an inquiry into the chemistry of life's origin: a retrospective. Angew. Chem. Int. Ed. 2011;50:12412–12472. doi: 10.1002/anie.201103672. [DOI] [PubMed] [Google Scholar]
- 12.Blackburn G.M., Gait M.J., Loakes D., Williams D.M., editors. Nucleic Acids in Chemistry and Biology. Cambridge: Royal Society of Chemistry; 2006. [Google Scholar]
- 13.Egli M., Pallan P.S. Insights from crystallographic studies into the structural and pairing properties of nucleic acid analogs and chemically modified DNA and RNA oligonucleotides. Annu. Rev. Biophys. Biomol. Struct. 2007;36:281–305. doi: 10.1146/annurev.biophys.36.040306.132556. [DOI] [PubMed] [Google Scholar]
- 14.Egli M., Pallan P.S. Crystallographic studies of chemically modified nucleic acids: a backward glance. Chem. Biodiv. 2010;7:60–89. doi: 10.1002/cbdv.200900177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tereshko V., Gryaznov S., Egli M. Consequences of replacing the DNA 3′-oxygen by an amino group: high-resolution crystal structure of a fully modified N3’→P5’ phosphoramidate DNA dodecamer duplex. J. Am. Chem. Soc. 1998;120:269–283. [Google Scholar]
- 16.Teplova M., Minasov G., Tereshko V., Inamati G.B., Cook P.D., Manoharan M., Egli M. Crystal structure and improved antisense properties of 2 ‘-O-(2-methoxyethyl)-RNA. Nat. Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 17.Saenger W. Principles of Nucleic Acid Structure. NY: Springer; 1984. [Google Scholar]
- 18.Ban C., Ramakrishnan B., Sundaralingam M. A single 2′-hydroxyl group converts B-DNA to A-DNA. Crystal structure of the DNA-RNA chimeric decamer duplex d(CCGGC)r(G)d(CCGG) with a novel intermolecular G-C base-paired quadruplet. J. Mol. Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 19.Egli M., Usman N., Zhang S.G., Rich A. Crystal structure of an Okazaki fragment at 2-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 1992;89:534–538. doi: 10.1073/pnas.89.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowotny M., Gaidamakov S.A., Crouch R.J., Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Manoharan M., Akinc A., Pandey R.K., Qin J., Hadwiger P., John M., Mills K., Charisse K., Maier M.A., Nechev L., et al. Unique gene-silencing and structural properties of 2 ‘-fluoro-modified siRNAs. Angew. Chem. Int. Ed. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pallan P.S., Greene E.M., Jicman P.A., Pandey R.K., Manoharan M., Rozners E., Egli M. Unexpected origins of the enhanced pairing affinity of 2′-fluoro-modified RNA. Nucleic Acids Res. 2011;39:3482–3495. doi: 10.1093/nar/gkq1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patra A., Paolillo M., Charisse K., Manoharan M., Rozners E., Egli M. 2 ‘-Fluoro RNA shows increased Watson-Crick H-bonding strength and stacking relative to RNA: evidence from NMR and thermodynamic data. Angew. Chem. Int. Ed. 2012;51:11863–11866. doi: 10.1002/anie.201204946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Pintado N., Deleavey G.F., Portella G., Campos-Olivas R., Orozco M., Damha M.J., Gonzalez C. Backbone FC-H…O hydrogen bonds in 2′ F-substituted nucleic acids. Angew. Chem. Int. Ed. 2013;52:12065–12068. doi: 10.1002/anie.201305710. [DOI] [PubMed] [Google Scholar]
- 25.Chaput J.C., Yu H., Zhang S. The emerging world of synthetic genetics. Chem. Biol. 2012;19:1360–1371. doi: 10.1016/j.chembiol.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Pinheiro V.B., Holliger P. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol. 2014;32:321–328. doi: 10.1016/j.tibtech.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S.Y., McLaughlin S.H., et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H., Zhang S., Dunn M., Chaput J.C. An efficient and faithful in vitro replication system for threose nucleic acid. J. Am. Chem. Soc. 2013;135:3583–3591. doi: 10.1021/ja3118703. [DOI] [PubMed] [Google Scholar]
- 29.Sefah K., Yang Z., Bradley K.M., Hoshika S., Jimenez E., Zhang L., Zhu G., Shanker S., Yu F., Turek D., et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimoto M., Yamashige R., Matsunaga K.-I., Yokoyama S., Hirao I. Generation of high affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotech. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 31.Liu H., Gao J., Lynch S.R., Saito Y.D., Maynard L., Kool E.T. A four-base paired genetic helix with expanded size. Science. 2003;302:868–871. doi: 10.1126/science.1088334. [DOI] [PubMed] [Google Scholar]
- 32.Chaput J.C., Switzer C. A DNA pentaplex incorporating nucleobase quintets. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10614–10619. doi: 10.1073/pnas.96.19.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang M., Heuberger B., Chaput J.C., Switzer C., Feigon J. Solution structure of a parallel-stranded oligoisoguanine DNA pentaplex formed by d(T(iG)(4) T) in the presence of Cs(+) ions. Angew. Chem. Int. Ed. 2012;51:7952–7955. doi: 10.1002/anie.201203459. [DOI] [PubMed] [Google Scholar]
- 34.Malyshev D.A., Dhami K., Lavergne T., Chen T., Dai N., Foster J.M., Correa I.R., Romesberg F.E. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marliere P., Patrouix J., Doring V., Herdewijn P., Tricot S., Cruveiller S., Bouzon M., Mutzel R. Chemical evolution of a bacterium's genome. Angew. Chem. Int. Ed. 2011;50:7109–7114. doi: 10.1002/anie.201100535. [DOI] [PubMed] [Google Scholar]
- 36.Malyshev D.A., Romesberg F.E. The expanded genetic alphabet. Angew. Chem. Int. Ed. 2015;54:11930–11944. doi: 10.1002/anie.201502890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Appella D.H. Non-natural nucleic acids for synthetic biology. Curr. Opin. Chem. Biol. 2009;13:687–696. doi: 10.1016/j.cbpa.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brundo Y., Liu D.R. Recent progress toward the templated synthesis and directed evolution of sequence-defined synthetic polymers. Chem. Biol. 2009;16:265–276. doi: 10.1016/j.chembiol.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rupp B. Biomolecular Crystallography. Principles, Practice, and Applications to Structural Biology. Garland Science Taylor & Francis Group; 2009. [Google Scholar]
- 40.Taylor G. The phase problem. Acta Cryst. D. 2003;59:1881–1890. doi: 10.1107/s0907444903017815. [DOI] [PubMed] [Google Scholar]
- 41.Ealick S.E. Advances in multiple wavelength anomalous diffraction crystallography. Curr. Opin. Chem. Biol. 2000;4:495–499. doi: 10.1016/s1367-5931(00)00122-8. [DOI] [PubMed] [Google Scholar]
- 42.Ennifar E.E. Nucleic Acid Crystallography: Methods and Protocols. NY: Humana Press, Springer Science and Business Media; 2015. [Google Scholar]
- 43.Egli M., Lubini P., Pallan P.S. The long and winding road to the structure of homo-DNA. Chem. Soc. Rev. 2007;36:31–45. doi: 10.1039/b606807c. [DOI] [PubMed] [Google Scholar]
- 44.Luo Z., Dauter M., Dauter Z. Phosphates in the Z-DNA dodecamer are flexible, but their P-SAD signal is sufficient for structure solution. Acta Cryst. D. 2014;70:1790–1800. doi: 10.1107/S1399004714004684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller-Dieckmann C., Panjikar S., Schmidt A., Mueller S., Kuper J., Geerlof A., Wilmanns M., Singh R.K., Tucker P.A., Weiss M.S. On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths. Acta Cryst. D. 2007;63:366–380. doi: 10.1107/S0907444906055624. [DOI] [PubMed] [Google Scholar]
- 46.Raiber E.A., Murat P., Chirgadze D.Y., Beraldi D., Luisi B.F., Balasubramanian S. 5-Formylcytosine alters the structure of the DNA double helix. Nat. Struct. Mol. Biol. 2015;22:44–49. doi: 10.1038/nsmb.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q., Dahmane T., Zhang Z., Assur Z., Brasch J., Shapiro L., Mancia F., Hendrickson W.A. Structures from anomalous diffraction of native biological macromolecules. Science. 2012;336:1033–1037. doi: 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egli M., Tereshko V., Teplova M., Minasov G., Joachimiak A., Sanishvili R., Weeks C.M., Miller R., Maier M.A., An H.Y., et al. X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers (Nucleic Acid Sci.) 1998;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 49.Evans J.N.S. Biomolecular NMR Spectroscopy. NY; Tokyo: Oxford University Press; 1995. [Google Scholar]
- 50.Furtig B., Richter C., Wohnert J., Schwalbe H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 51.Ebert M.O., Mang C., Krishnamurthy R., Eschenmoser A., Jaun B. The structure of a TNA-TNA complex in solution: NMR study of the octamer duplex derived from alpha-(L)-threofuranosyl-(3′-2′)-CGAATTCG. J. Am. Chem. Soc. 2008;130:15105–15115. doi: 10.1021/ja8041959. [DOI] [PubMed] [Google Scholar]
- 52.Declercq R., Van Aerschot A., Read R.J., Herdewijn P., Van Meervelt L. Crystal structure of double helical hexitol nucleic acids. J. Am. Chem. Soc. 2002;124:928–933. doi: 10.1021/ja016570w. [DOI] [PubMed] [Google Scholar]
- 53.Robeyns K., Herdewijn P., Van Meervelt L. Structure of the fully modified left-handed cyclohexene nucleic acid sequence GTGTACAC. J. Am. Chem. Soc. 2008;130:1979–1984. doi: 10.1021/ja077313f. [DOI] [PubMed] [Google Scholar]
- 54.Yeh J.I., Pohl E., Truan D., He W., Sheldrick G.M., Du S., Achim C. The crystal structure of non-modified and bipyridine-modified PNA duplexes. Chemistry. 2010;16:11867–11875. doi: 10.1002/chem.201000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He W., Hatcher E., Balaeff A., Beratan D.N., Gil R.R., Madrid M., Achim C. Solution structure of a peptide nucleic acid duplex from NMR data: features and limitations. J. Am. Chem. Soc. 2008;130:13264–13273. doi: 10.1021/ja800652h. [DOI] [PubMed] [Google Scholar]
- 56.Egli M., Pallan P.S., Pattanayek R., Wilds C.J., Lubini P., Minasov G., Dobler M., Leumann C.J., Eschenmoser A. Crystal structure of homo-DNA and nature's choice of pentose over hexose in the genetic system. J. Am. Chem. Soc. 2006;128:10847–10856. doi: 10.1021/ja062548x. [DOI] [PubMed] [Google Scholar]
- 57.Martin-Pintado N., Yahyaee-Anzahaee M., Campos-Olivas R., Noronha A.M., Wilds C.J., Damha M.J., Gonzalez C. The solution structure of double helical arabino nucleic acids (ANA and 2′ F-ANA): effect of arabinoses in duplex-hairpin interconversion. Nucleic Acids Res. 2012;40:9329–9339. doi: 10.1093/nar/gks672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eriksson M., Nielsen P.E. Solution structure of a peptide nucleic acid-DNA duplex. Nat. Struct. Biol. 1996;3:410–413. doi: 10.1038/nsb0596-410. [DOI] [PubMed] [Google Scholar]
- 59.Brown S.C., Thomson S.A., Veal J.M., Davis D.G. NMR solution structure of a peptide nucleic acid complexed with RNA. Science. 1994;265:777–780. doi: 10.1126/science.7519361. [DOI] [PubMed] [Google Scholar]
- 60.Nielsen P.E., Egholm M., Berg R.H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L., Peritz A., Meggers E. A simple glycol nucleic acid. J. Am. Chem. Soc. 2005;127:4174–4175. doi: 10.1021/ja042564z. [DOI] [PubMed] [Google Scholar]
- 62.Böhringer M., Roth H.-J., Hunziker J., Göbel M., Krishnan R., Giger A., Schweizer B., Schreiber J., Leumann C., Eschenmoser A. Why pentose and not hexose nucleic acids? Part II. Preparation of oligonucleotides containing 2′ 3′ -dideoxy-β-D-glucopyranosyl building blocks. Helv. Chim. Acta. 1992;75:1416–1477. [Google Scholar]
- 63.Lescrinier E., Esnouf R., Schraml J., Busson R., Heus H.A., Hilbers C.W., Herdewijn P. Solution structure of a HNA–RNA hybrid. Chem. Biol. 7:719–731. doi: 10.1016/s1074-5521(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 64.Schoning K., Scholz P., Guntha S., Wu X., Krishnamurthy R., Eschenmoser A. Chemical etiology of nucleic acid structure: the alpha-threofuranosyl-(3′→2′) oligonucleotide system. Science. 2000;290:1347–1351. doi: 10.1126/science.290.5495.1347. [DOI] [PubMed] [Google Scholar]
- 65.Noronha A.M., Wilds C.J., Lok C.N., Viazovkina K., Arion D., Parniak M.A., Damha M.J. Synthesis and biophysical properties of arabinonucleic acids (ANA): circular dichroic spectra, melting temperatures, and ribonuclease H susceptibility of ANA.RNA hybrid duplexes. Biochemistry. 2000;39:7050–7062. doi: 10.1021/bi000280v. [DOI] [PubMed] [Google Scholar]
- 66.Wilds C.J., Damha M.J. 2′-Deoxy-2′-fluoro-beta-D-arabinonucleosides and oligonucleotides (2′ F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koshkin A.A., Singh S.K., Nielsen P., Rajwanshi V.K., Kumar R., Meldgaard M., Olsen C.E., Wengel J. LNA (Locked Nucleic Acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 68.Hunziker J., Roth H.J., Böhringer M., Giger A., Diedrichsen U., Göbel M., Krishnan R., Jaun B., Leumann C., Eschenmoser A. Why pentose and not hexose nucleic acids? Part III. Oligo(2′,3′-dideoxy-β-D-glucopyranosyl)nucleotides (‘homo-DNA’): base-pairing properties. Helv. Chim. Acta. 1993;76:259–352. [Google Scholar]
- 69.Maiti M., Maiti M., Knies C., Dumbre S., Lescrinier E., Rosemeyer H., Ceulemans A., Herdewijn P. Xylonucleic acid: synthesis, structure, and orthogonal pairing properties. Nucleic Acids Res. 2015;43:7189–7200. doi: 10.1093/nar/gkv719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maiti M., Siegmund V., Abramov M., Lescrinier E., Rosemeyer H., Froeyen M., Ramaswamy A., Ceulemans A., Marx A., Herdewijn P. Solution structure and conformational dynamics of deoxyxylonucleic acids (dXNA): an orthogonal nucleic acid candidate. Chemistry. 2012;18:869–879. doi: 10.1002/chem.201102509. [DOI] [PubMed] [Google Scholar]
- 71.Poopeiko N.E., Juhl M., Vester B., Sorensen M.D., Wengel J. Xylo-configured oligonucleotides (XNA, xylo nucleic acid): synthesis of conformationally restricted derivatives and hybridization towards DNA and RNA complements. Bioorg. Med. Chem. Lett. 2003;13:2285–2290. doi: 10.1016/s0960-894x(03)00441-4. [DOI] [PubMed] [Google Scholar]
- 72.D'Alonzo D., Amato J., Schepers G., Froeyen M., Van Aerschot A., Herdewijn P., Guaragna A. Enantiomeric selection properties of beta-homoDNA: enhanced pairing for heterochiral complexes. Angew. Chem. Int. Ed. 2013;52:6662–6665. doi: 10.1002/anie.201301659. [DOI] [PubMed] [Google Scholar]
- 73.Johnson A.T., Schlegel M.K., Meggers E., Essen L.O., Wiest O. On the structure and dynamics of duplex GNA. J. Org. Chem. 2011;76:7964–7974. doi: 10.1021/jo201469b. [DOI] [PubMed] [Google Scholar]
- 74.Schlegel M.K., Essen L.O., Meggers E. Duplex structure of a minimal nucleic acid. J. Am. Chem. Soc. 2008;130:8158–8159. doi: 10.1021/ja802788g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schlegel M.K., Essen L.O., Meggers E. Atomic resolution duplex structure of the simplified nucleic acid GNA. Chem. Commun. (Camb) 2010;46:1094–1096. doi: 10.1039/b916851f. [DOI] [PubMed] [Google Scholar]
- 76.Rasmussen H., Kastrup J.S., Nielsen J.N., Nielsen J.M., Nielsen P.E. Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 A resolution. Nat. Struct. Biol. 1997;4:98–101. doi: 10.1038/nsb0297-98. [DOI] [PubMed] [Google Scholar]
- 77.Egholm M., Buchardt O., Christensen L., Behrens C., Freier S.M., Driver D.A., Berg R.H., Kim S.K., Norden B., Nielsen P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 78.Pallan P.S., Wilds C.J., Wawrzak Z., Krishnamurthy R., Eschenmoser A., Egli M. Why does TNA cross-pair more strongly with RNA than with DNA? an answer from X-ray analysis. Angew. Chem. Int. Ed. 2003;42:5893–5895. doi: 10.1002/anie.200352553. [DOI] [PubMed] [Google Scholar]
- 79.Watts J.K., Martin-Pintado N., Gomez-Pinto I., Schwartzentruber J., Portella G., Orozco M., Gonzalez C., Damha M.J. Differential stability of 2′ F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 2010;38:2498–2511. doi: 10.1093/nar/gkp1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anzahaee M.Y., Watts J.K., Alla N.R., Nicholson A.W., Damha M.J. Energetically important C-H…F-C pseudohydrogen bonding in water: evidence and application to rational design of oligonucleotides with high binding affinity. J. Am. Chem. Soc. 2011;133:728–731. doi: 10.1021/ja109817p. [DOI] [PubMed] [Google Scholar]
- 81.Li F., Sarkhel S., Wilds C.J., Wawrzak Z., Prakash T.P., Manoharan M., Egli M. 2′-Fluoroarabino- and arabinonucleic acid show different conformations, resulting in deviating RNA affinities and processing of their heteroduplexes with RNA by RNase H. Biochemistry. 2006;45:4141–4152. doi: 10.1021/bi052322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eichert A., Behling K., Betzel C., Erdmann V.A., Furste J.P., Forster C. The crystal structure of an ‘All Locked’ nucleic acid duplex. Nucleic Acids Res. 2010;38:6729–6736. doi: 10.1093/nar/gkq505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nielsen K.E., Rasmussen J., Kumar R., Wengel J., Jacobsen J.P., Petersen M. NMR studies of fully modified locked nucleic acid (LNA) hybrids: solution structure of an LNA:RNA hybrid and characterization of an LNA:DNA hybrid. Bioconjug. Chem. 2004;15:449–457. doi: 10.1021/bc034145h. [DOI] [PubMed] [Google Scholar]
- 84.Wahlestedt C., Salmi P., Good L., Kela J., Johnsson T., Hokfelt T., Broberger C., Porreca F., Lai J., Ren K., et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ovaere M., Herdewijn P., Van Meervelt L. The crystal structure of the CeNA:RNA hybrid ce(GCGTAGCG):r(CGCUACGC) Chemistry. 2011;17:7823–7830. doi: 10.1002/chem.201003594. [DOI] [PubMed] [Google Scholar]
- 86.Lescrinier E., Froeyen M., Herdewijn P. Difference in conformational diversity between nucleic acids with a six-membered ‘sugar’ unit and natural ‘furanose’ nucleic acids. Nucleic Acids Res. 2003;31:2975–2989. doi: 10.1093/nar/gkg407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vesnaver G., Breslauer K.J. The contribution of DNA single-stranded order to the thermodynamics of duplex formation. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3569–3573. doi: 10.1073/pnas.88.9.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yakovchuk P., Protozanova E., Frank-Kamenetskii M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Auffinger P., Hashem Y. Nucleic acid solvation: from outside to insight. Curr. Opin. Struct. Biol. 2007;17:325–333. doi: 10.1016/j.sbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 90.Champ P.C., Maurice S., Vargason J.M., Camp T., Ho P.S. Distributions of Z-DNA and nuclear factor I in human chromosome 22: a model for coupled transcriptional regulation. Nucleic Acids Res. 2004;32:6501–6510. doi: 10.1093/nar/gkh988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu X.J., Shakked Z., Olson W.K. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 93.Chandrasekaran R., Arnott S. The structure of B-DNA in oriented fibers. J. Biomol. Struct. Dyn. 1996;13:1015–1027. doi: 10.1080/07391102.1996.10508916. [DOI] [PubMed] [Google Scholar]
- 94.Schneider B., Neidle S., Berman H.M. Conformations of the sugar-phosphate backbone in helical DNA crystal structures. Biopolymers. 1997;42:113–124. doi: 10.1002/(sici)1097-0282(199707)42:1<113::aid-bip10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 95.Ebert M.O., Jaun B. Oligonucleotides with sugars other than ribo- and 2′-deoxyribofuranose in the backbone: the solution structures determined by NMR in the context of the ‘Etiology of nucleic acids’ project of Albert Eschenmoser. Chem. Biodiv. 2010;7:2103–2128. doi: 10.1002/cbdv.201000096. [DOI] [PubMed] [Google Scholar]
- 96.Maier T., Przylas I., Strater N., Herdewijn P., Saenger W. Reinforced HNA backbone hydration in the crystal structure of a decameric HNA/RNA hybrid. J. Am. Chem. Soc. 2005;127:2937–2943. doi: 10.1021/ja045843v. [DOI] [PubMed] [Google Scholar]
- 97.Hantz E., Larue V., Ladam P., Le Moyec L., Gouyette C., Dinh T.H. Solution conformation of an RNA-DNA hybrid duplex containing a pyrimidine RNA strand and a purine DNA strand. Int. J. Biol. Macromol. 2001;28:273–284. doi: 10.1016/s0141-8130(01)00123-4. [DOI] [PubMed] [Google Scholar]
- 98.Eriksson M., Nielsen P.E. PNA-nucleic acid complexes. Structure, stability and dynamics. Q. Rev. Biophys. 1996;29:369–394. doi: 10.1017/s0033583500005886. [DOI] [PubMed] [Google Scholar]
- 99.Lauridsen L.H., Rothnagel J.A., Veedu R.N. Enzymatic recognition of 2′-modified ribonucleoside 5′-triphosphates: towards the evolution of verstatile aptamers. ChemBioChem. 2012;13:19–25. doi: 10.1002/cbic.201100648. [DOI] [PubMed] [Google Scholar]
- 100.Chaput J.C., Ichida J.K., Szostak J.W. DNA polymerase-mediated DNA synthesis on a TNA template. J. Am. Chem. Soc. 2003;125:856–857. doi: 10.1021/ja028589k. [DOI] [PubMed] [Google Scholar]
- 101.Chaput J.C., Szostak J.W. TNA synthesis by DNA polymerases. J. Am. Chem. Soc. 2003;125:9274–9275. doi: 10.1021/ja035917n. [DOI] [PubMed] [Google Scholar]
- 102.Kempeneers V., Vastmans K., Rozenski J., Herdewijn P. Recognition of threosyl nucleotides by DNA and RNA polymerases. Nucleic Acids Res. 2003;31:6221–6226. doi: 10.1093/nar/gkg833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Horhota A., Zou K., Ichida J.K., Yu B., McLaughlin L.W., Szostak J.W., Chaput J.C. Kinetic analysis of an efficient DNA-dependent TNA polymerase. J. Am. Chem. Soc. 2005;127:7427–7434. doi: 10.1021/ja0428255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng C.G., Damha M.J. Polymerase-directed synthesis of 2′-deoxy-2′-fluoro-b-D-arabinonucleic acids. J. Am. Chem. Soc. 2007;129:5310–5311. doi: 10.1021/ja069100g. [DOI] [PubMed] [Google Scholar]
- 105.Veedu R.N., Vester B., Wengel J. Enzymatic incorporation of LNA nucleotides into DNA strands. ChemBioChem. 2007;8:490–492. doi: 10.1002/cbic.200600501. [DOI] [PubMed] [Google Scholar]
- 106.Chaput J.C. Replicating an expanded genetic alphabet in cells. ChemBioChem. 2014;15:1869–1871. doi: 10.1002/cbic.201402289. [DOI] [PubMed] [Google Scholar]
- 107.Yu H., Zhang S., Chaput J.C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012;4:183–187. doi: 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- 108.Ferreira-Bravo I.A., Cozens C., Holliger P., DeStefano J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase. Nucleic Acids Res. 2015;43:9578–9599. doi: 10.1093/nar/gkv1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taylor A.I., Pinheiro V.B., Smola M.J., Morgunov A.S., Peak-Chew S., Cozens C., Weeks K.M., Herdewijn P., Holliger P. Catalysts from synthetic genetic polymers. Nature. 2015;518:427–430. doi: 10.1038/nature13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang S., McCullum E.O., Chaput J.C. Synthesis of two mirror image 4-helix junctions derived from glycerol nucleic acid. J. Am. Chem. Soc. 2008;130:5846–5847. doi: 10.1021/ja800079j. [DOI] [PubMed] [Google Scholar]
- 111.Peters J.P., Yelgaonkar S.P., Srivatsan S.G., Tor Y., Maher L.J. Mechanical properties of DNA-like polymers. Nucleic Acids Res. 2013;41:10593–10604. doi: 10.1093/nar/gkt808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Narayana N., Weiss M.A. Crystallographic analysis of a sex-specific enhancer element: sequence-dependent DNA structure, hydration, and dynamics. J. Mol. Biol. 2009;385:469–490. doi: 10.1016/j.jmb.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mooers B.H., Singh A. The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate. RNA. 2011;17:1870–1883. doi: 10.1261/rna.2880311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sun G., Voigt J.H., Marquez V.E., Nicklaus M.C. Prosit, an online service to calculate pseudorotational parameters of nucleosides and nucleotides. Nucleos. Nucleot. Nucleic Acids. 2005;24:1029–1032. doi: 10.1081/ncn-200059757. [DOI] [PubMed] [Google Scholar]