Abstract
In mitochondria of Saccharomyces cerevisiae, a single aminoacyl-tRNA synthetase (aaRS), MST1, aminoacylates two isoacceptor tRNAs, tRNA1Thr and tRNA2Thr, that harbor anticodon loops of different size and sequence. As a result of this promiscuity, reassignment of the CUN codon box from leucine to threonine is facilitated. However, the mechanism by which a single aaRS binds distinct anticodon loops with high specificity is not well understood. Herein, we present the crystal structure of MST1 in complex with the canonical tRNA2Thr and non-hydrolyzable analog of threonyl adenylate. Our structure reveals that the dimeric arrangement of MST1 is essential for binding the 5′-phosphate, the second base pair of the acceptor stem, the first two base pairs of the anticodon stem and the first nucleotide of the variable arm. Further, in contrast to the bacterial ortholog that ‘reads’ the entire anticodon sequence, MST1 recognizes bases in the second and third position and the nucleotide upstream of the anticodon sequence. We speculate that a flexible loop linking strands β4 and β5 may be allosteric regulator that establishes cross-subunit communication between the aminoacylation and tRNA-binding sites. We also propose that structural features of the anticodon-binding domain in MST1 permit binding of the enlarged anticodon loop of tRNA1Thr.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) ensure accurate decoding of the genetic code through their ability to select and pair each proteinogenic amino acid with a specific set of isoacceptor transfer RNAs (tRNAs) (1). AaRSs activate the carboxyl group of the amino acid with ATP and then promote its coupling to the 3′-end of tRNA. The final reaction product, aminoacyl-tRNA (aa-tRNA), serves as an obligate substrate for the translating ribosome where the final steps of the decoding process and peptide bond synthesis take place. Because protein translation factors and the ribosome are not capable of identifying and editing misacylated tRNA species, it is presumed that aaRSs dictate the accuracy of protein synthesis. This notion is supported by the fact that the error rate of the aminoacylation reaction (∼10−4 per decoding event) closely reflects the overall error rate translation (2).
AaRSs achieve a relatively high level of accuracy through substrate selection as well as by their ability to edit reaction products. Selection of the amino acid is defined by the configuration of the active site of the enzyme. Although the catalytic groove can exclude a number of chemical structures, the activation of incorrect amino acids is still promoted by almost every aaRS, albeit at the very low rate of 10−4 to 10−5 (2). The misactivated amino acids are hydrolyzed either in the active site before the transfer onto the tRNA (i.e. pre-transfer editing) or in a distinct editing site after the coupling reaction is complete (i.e. post-transfer editing) (3–5). Whereas the selection of the amino acids is constrained within the relatively small cavities of the catalytic and editing sites, the tRNA selection requires engagement of larger parts of the enzyme surface. In general, the catalytic- and anticodon-binding domains recognize elements in the acceptor and anticodon arms of the cognate tRNA, respectively, and it is thus not surprising that the frequency of errors stemming from charging the non-cognate tRNAs are 10−6 or lower (2). With the exception of a few aaRSs (6), most aaRSs recognize their cognate tRNA(s) through specific interactions with the anticodon loop and the anticodon sequence. In fact, the strict recognition of the second and third position in the anticodon sequence provided initial evidence for the ‘wobble’ hypothesis and, subsequently, led to the belief that the genetic code is universal and frozen (7,8). Multiple studies, however, revealed codon reassignment events in mitochondrial and nuclear genomes in all domains of life (9–11), suggesting that the genetic code is, in fact, evolving.
For instance, in mitochondria, the mutated tRNATrp facilitates the reassignment of the UGA codon to tryptophan (Trp) (11). In bacteria, archaea and eukaryotes, the recoding of the nuclear opal stop codon into the 21st amino acid, selenocysteine (Sec), is mediated by a distinct tRNASec (9,12). Further, several methanogenic archaea rewire the amber stop codon into a signal for incorporation of the 22nd amino acid, pyrrolysine (Pyl) (13,14), whereas the anomalous recognition of the AUA codon by tRNAMet alters the meaning of this particular triplet from isoleucine (Ile) to methionine (Met) in the mitochondria of several yeast species (11). Perhaps the most peculiar reassignment event was observed in the mitochondria of budding yeast: Saccharomyces, Vanderwaltozyma and Nakaseomyces. Protein sequencing and mass spectroscopy analysis of these yeasts confirmed that the CUN box (N designates A, G, U or C) was reassigned to encode threonine (Thr) instead of the canonical leucine (Leu). In S. cerevisiae alone, the Leu -> Thr reassignment occurred in at least six mitochondrial proteins (15). Moreover, it was shown that the canonical tRNA2Thr is no longer present in these yeasts and that the codon reassignment event is facilitated by the anticodon sequence UAG, present in the enlarged 8-nucleotide (nt) long anticodon loop encoded in the unusual tRNA1Thr. However, the yeast mitochondria also express a canonical tRNA2Thr that reads the ACN codons. It was believed that because of the major differences in anticodon loops and sequences, two mitochondrial ThrRSs must be charging tRNA1Thr and tRNA2Thr. However, our biochemical and biophysical studies have firmly established that a single ThrRS, designated as MST1, binds to and acts on both mitochondrial tRNAThr species equally well. Furthermore, the phylogenetic, mutational and kinetic studies demonstrated that tRNA1Thr evolved from tRNAHis and that it co-evolved with MST1 (16).
MST1 is a class II aaRS devoid of the editing domain. Its structure closely resembles that of the bacterial orthologs and differs significantly from the archaeal ThrRS (17). In spite of the sequence divergence, MST1 primarily recognizes the anticodon loops of the substrate tRNAs. Whereas the size of the anticodon loop is critical for recognition of tRNA1Thr, it is the anticodon sequence that drives productive binding of tRNA2Thr (17). Further, distinct pockets within the anticodon-binding domain of MST1 might be responsible for coordinating different anticodon loops (17). However, in spite of the earlier studies, the exact mechanism by which a single ThrRS is capable of recognizing distinct anticodon loops of the two isoacceptor tRNAs remained unclear. Herein, we determined the crystal structure of MST1 in complex with the canonical tRNA2Thr and a non-hydrolyzable analog of threonyl adenylate (TAM). Our structure shows that the MST1 dimer binds two tRNAs and that the dimeric arrangement is essential for binding to the 5′-phosphate, and the acceptor and anticodon stems. The enzyme ‘reads’ the nucleotide immediately upstream of the anticodon sequence, the second and third position of the anticodon triplet, the first two base pairs of the anticodon stem and the second base pair of the acceptor arm. Structural analysis reveals putative relay elements that communicate between the tRNA-binding and catalytic sites as well as differences in the tRNA-recognition mechanism used by the yeast mitochondrial and bacterial enzymes.
MATERIALS AND METHODS
Expression and purification of MST1
The S. cerevisiae MST1 gene was cloned into a pET28a expression vector (Novagen) with an N-terminal His6-tag. The recombinant protein was overexpressed for 18 h at +15°C in the Rosetta pLysS (Novagen) E. coli expression strain following the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside to the culture. The recombinant protein was captured from the cell lysate using a Ni2+ affinity column (GE Healthcare) following a standard purification protocol. The affinity column eluate was then subjected to a HiLoad 26/600 Superdex 200 pg size exclusion column (GE Healthcare) equilibrated with 20 mM Tris HCl pH 7.5, 300 mM NaCl and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP). The purified protein was concentrated, flash-frozen in liquid nitrogen and stored at −80°C prior to use.
In vitro transcription and purification of tRNA2Thr
The tRNA2Thr gene was cloned into the pUC18 vector and transformed into XL1-Blue competent cells (Stratagene). The gene was amplified using polymerase chain reaction (PCR). The tRNA was synthesized by in vitro T7 RNA polymerase run-off transcription. The transcription reaction was performed at +37°C for 3 h in 40 mM Tris, pH 8.1, 22 mM MgCl2, 5 mM DTT, 2 mM spermidine, 50 μg/ml BSA, 20 mM GMP, 4 mM nucleotides (ATP, GTP, CTP and UTP), the PCR product of the tRNA2Thr gene (60 μg/ml) and 8 μM T7 RNA polymerase. The filtered reaction was loaded onto a Resource-Q column (GE Healthcare) and tRNA2Thr was purified using a linear gradient of NaCl (0.5–0.65 M) in 20 mM Tris HCl, pH 8.1. Following elution, the tRNA2Thr was further purified on a S200 Superdex size-exclusion column (GE Healthcare) equilibrated with 20 mM Tris, pH 8.1 and 150 mM NaCl. The pure tRNA2Thr was concentrated and flash frozen in liquid nitrogen and stored at −80°C.
Reconstitution and crystallization of the MST1-tRNA2Thr complex in the presence of the threonyl sulfamoyl adenylate (TAM)
MST1 and tRNA2Thr were mixed to a molar ratio of one tRNA molecule per MST1 dimer and the complex was concentrated to 3.6 mg/ml. TCEP, MnCl2 and TAM were added to a final concentration of 1 mM prior to crystallization trials. Crystals of the ternary complex were obtained by the sitting-drop vapor-diffusion method. Equal volumes of the solution containing the MST1-tRNA2Thr-TAM complex and the well buffer were mixed, and the drops were incubated at +12°C. The rod-like crystals of the ternary complex grew in the buffer containing 0.2 M lithium sulfate, 0.1 M Tris-HCl, pH 7.0 and 2.0 M ammonium sulfate. Crystals were mounted directly from the mother liquor and frozen in liquid nitrogen without additional cryoprotection. The diffraction data were collected on the Pilatus detector at GM/CA@APS (23ID-D) beam line at the Advanced Photon Source, Argonne National Laboratory.
Structure determination
The diffraction data were processed in HKL2000 (18). The crystal structure of the MST1-tRNA2Thr-TAM ternary complex was determined by molecular replacement in Phaser (19) using the crystal structure of apo MST1 (PDB ID: 3UGQ) as a search model. The structure of the bacterial tRNAThr (PDB ID: 1QF6) was used as the initial model of tRNA. The structure refinement was performed in Phenix (20), the model building was done in Coot (21) and structure superimpositioning was done using CEalign (22,23) in PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC). All figures were produced in PyMOL.
Aminoacylation assays
Aminoacylation experiments were performed as described (16) in the presence of 100 mM Na-HEPES pH 7.2, 30 mM KCl, 10 mM MgCl2, 2 mM ATP, 25 μM [3H] Thr (100 μCi/ml), 0.1–1.6 μM tRNA transcripts and 100–1000 nM MST1.
RESULTS
MST1 binds two tRNA2Thr molecules
Previously, we determined the crystal structures of apo MST1 (PDB ID: 3UGQ) and MST1 complexed with threonyl- (TAM) and seryl- (SAM) adenylate analogues (PDB ID: 3UH0 and 4EO4, respectively) to elucidate the mechanism MST1 employs to recognize amino acid and ATP substrates in the active site (17,24). Additionally, our kinetic and mutagenesis results suggested elements in MST1 that could be important for the recognition of tRNA substrates (17). Our results revealed that MST1, like other threonyl-tRNA synthetases, recognizes the anticodon loop of its substrate tRNAs as a major recognition element – despite the difference in size and sequence of the tRNA1Thr and tRNA2Thr anticodon loops (Supplementary Figure S1). Based on our results, we have proposed that the sequence of the anticodon loop is critical for recognition of the canonical tRNA2Thr, whereas the size of the corresponding element in tRNA1Thr dictates its binding to the enzyme (17). We also suggested that different residues in the anticodon-binding domain of MST1 define the specificity for one isoacceptor tRNA over the other. In spite of these efforts, our predictions were limited and could not fully explain why MST1 can act on two isoacceptor tRNAs that carry different anticodon sequences.
As part of our efforts to determine the mechanism at the structural level by which MST1 recognizes its substrate tRNAs, we determined the crystal structure of MST1 in complex with tRNA2Thr and TAM to 2.3 Å resolution (PDB ID: 4YYE). The ternary complex crystals belonged to the triclinic (P1) space group and the asymmetric unit contained one MST1 dimer complexed with two tRNA2Thr molecules. The crystals were phased by molecular replacement using the crystal structure of apo MST1 (PDB ID: 3UGQ) as a search model. Inspection of the initial unbiased Fo-Fc electron density difference map revealed strong positive peaks corresponding to two tRNA molecules, as well as a TAM analog and a Zn2+ ion in each active site. With the exception of the disordered region spanning residues 213–222 in MST1, the model is well ordered, of excellent geometry (Table 1), and agrees exceptionally well with the final 2mFo-DFc electron density map. The structure, which comprises 836 amino acids, 148 ribonucleotides, 2 TAM molecules, 2 Zn2+ ions, 4 SO4 ions and 636 water molecules, refined to a final Rwork / Rfree of 16.0/20.2% (Table 1).
Table 1. Data collection and structure refinement statistics.
| Crystal | MST1-tRNA2Thr-TAM |
|---|---|
| Space group | P1 |
| Cell dimensions, Å | a = 69.0, b = 77.2, c = 86.2 |
| α = 63.9, β = 75.0, γ = 89.39 | |
| Data collection | |
| Resolution limit, Å | 2.3 |
| Unique reflections | 58 583 |
| Completeness (overall/last shell),% | 86.2/89.1 |
| Rsym (overall/last shell),% | 10.3/31.2 |
| I/ σI (overall/last shell) | 13.3/11.7 |
| Redundancy (overall/last shell) | 2.3/2.0 |
| Refinement | |
| Average B-factor, Å2 | |
| macromolecule (protein, RNA) | 29.1 |
| solvent (water, ions) | 32.7 |
| ligands | 60.2 |
| Number of atoms | |
| macromolecule (protein, RNA) | 9944 |
| ligand | 22 |
| solvent molecules (water, ions) | 636 |
| Rwork (|F| > 0σ),% | 16 |
| Rfree (|F| > 0σ),% | 20 |
| Rms deviations from ideality | |
| Bond lengths, Å | 0.008 |
| Bond angles, ° | 1.10 |
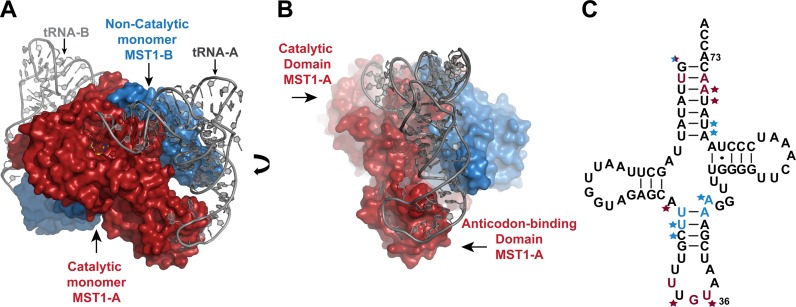
The MST1 dimer in the ternary complex crystal adopts a similar structure to that of apo MST1 and the MST1-TAM complex (PDB ID: 3UGQ and 3UH0, respectively). The enzyme forms a stoichiometric complex with both tRNA-binding sites fully occupied (Figure 1). The anticodon-binding domain intimately interacts with the anticodon loop and ‘reads’ the anticodon sequence. On the other hand, the catalytic domain binds the acceptor-TΨC arm and presumably positions the 3′-end of tRNA into the catalytic crevice. Although predominantly bound to one tRNA2Thr molecule, each MST1 subunit establishes cross-subunit interactions with the tRNA molecule that is bound to the second subunit. For instance, monomer A (MST1-A) interacts with both the anticodon loop and the acceptor arm of tRNA-A, but it also binds the 5′-phosphate, and the acceptor and anticodon stems of tRNA-B, which in turn interacts more intimately with subunit B (MST1-B) (Figure 1C). In this particular arrangement, MST1-A serves as the ‘catalytic’ subunit and MST1-B is the ‘non-catalytic’ subunit for tRNA-A, whereas the opposite is true for tRNA-B. The extent of interactions between a given tRNA and ‘catalytic’ and ‘non-catalytic’ MST1 subunits is quite different. To illustrate, the binding of tRNA-A to MST1-A buries 1,372 Å2 of the solvent accessible surface area, which is significantly larger than the surface area buried due to interactions between tRNA-A and MST1-B (526 Å2). For the sake of simplicity, we shall further discuss interactions between the MST1 dimer and tRNA-A only.
Figure 1.
MST1 dimer binds to two molecules of tRNA2Thr and each monomer interacts with both tRNAs. (A) Surface representation of MST1 dimer (subunits colored red and blue) in complex with tRNA2Thr (grey). MST1 monomer A (MST1-A), shown in red, binds to the acceptor stem and anticodon loop of tRNA molecule A (tRNA-A). MST1 monomer B (MST1-B), shown in blue, makes cross subunit contacts with the acceptor and anticodon stems of tRNA-A. (B) The ternary complex is shown in the view rotated clockwise ∼90° around the vertical axis. The catalytic and anticodon-binding domains of MST1-A are highlighted. Also, interactions between MST1-B and the anticodon stem of tRNA-A are shown. (C) Nucleotides in tRNA2Thr that interact with MST1 are highlighted in the cloverleaf diagram. Bases that are specifically recognized by MST1-A and MST1-B are colored in red and blue, respectively. Stars indicate interactions between the sugar-phosphate backbone and the enzyme that do not specifically recognize the base of the nucleotide.
The acceptor arm of tRNA2Thr interacts with both enzyme subunits
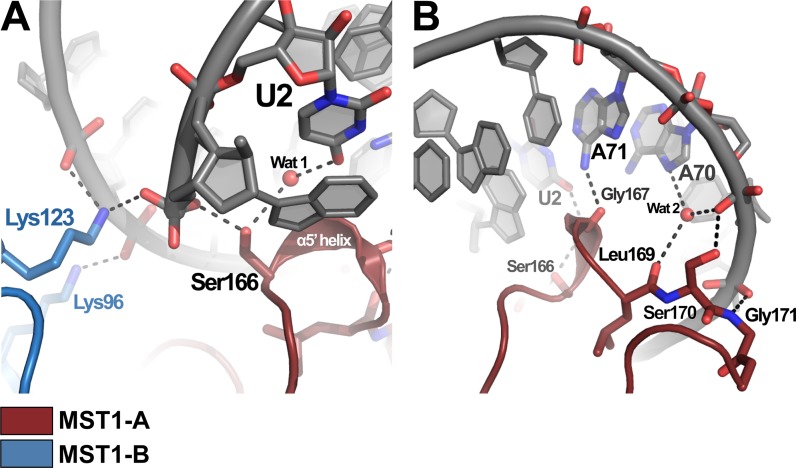
One of the main features of the binary aaRS-tRNA complexes are the base- and shape-specific interactions between the enzyme and the acceptor arm of the elongator tRNA. In our crystal, the catalytic domain of the MST1-A subunit interacts with the acceptor arm of tRNA-A using primarily residues 166–169 in the newly formed helical turn, α5′. The side-chain hydroxyl of Ser166 coordinates the 5′ phosphate of the tRNA as well as a water molecule (Wat1), which, in turn, forms a H-bond with the O4 atom of U2 (Figure 2A). The second base-specific interaction in this region is formed between the carbonyl oxygen of Gly167 and the N6 atom of A71 (Figure 2B). Residues 169–171 stabilize the complex through interactions with the sugar-phosphate backbone of the acceptor arm (Figure 2B). In particular, the backbone carbonyl of Leu169 coordinates a water molecule (Wat 2) that hydrogen bonds to N7 and a non-bridging oxygen of A70. The side chain hydroxyl of Ser170 and the backbone amide of Gly171 hydrogen bond to non-bridging oxygens of A70 and U69, respectively. These observations and the conservation of the U2:A71 pair indicate that the second base pair in the acceptor arm is an important recognition element in the yeast mitochondrial tRNAThr. To test this, we changed the second base pair from UA to CG (as in bacterial tRNAThr) and performed aminoacylation analyses. Whereas the U2C/A71G mutations almost completely abolished the activity of tRNA1Thr (Supplementary Figure S2A), they decreased the aminoacylation efficiency of tRNA2Thr by only 2-fold (Supplementary Figure S2B, Supplementary Table S1). This suggests that whereas U2:A71 is a major identity element for tRNA1Thr, it is less important for tRNA2Thr, presumably because other major identity elements in the anticodon of tRNA2Thr effectively compensate the recognition of U2:A71 by MST1. In our crystal structure, we did not observe electron density for the nucleotides C75 or A76, suggesting that the 3′ end of the tRNA remains flexible even after binding to MST1 (Supplementary Figure S3).
Figure 2.
Both monomers of MST1 interact with the acceptor stem of tRNA2Thr. (A) Both MST1 monomers create the binding pocket for the 5′ phosphate in tRNA2Thr. Lys123 from monomer B (blue) and Ser166 from monomer A (red) both coordinate the 5′ phosphate of tRNA bound to MST1-A. (B) Ser166 and Gly167 (MST1-A) recognize the second base pair in the acceptor stem of tRNA-A. Residues 169–171 make additional contacts with non-bridging oxygens in the sugar-phosphate backbone.
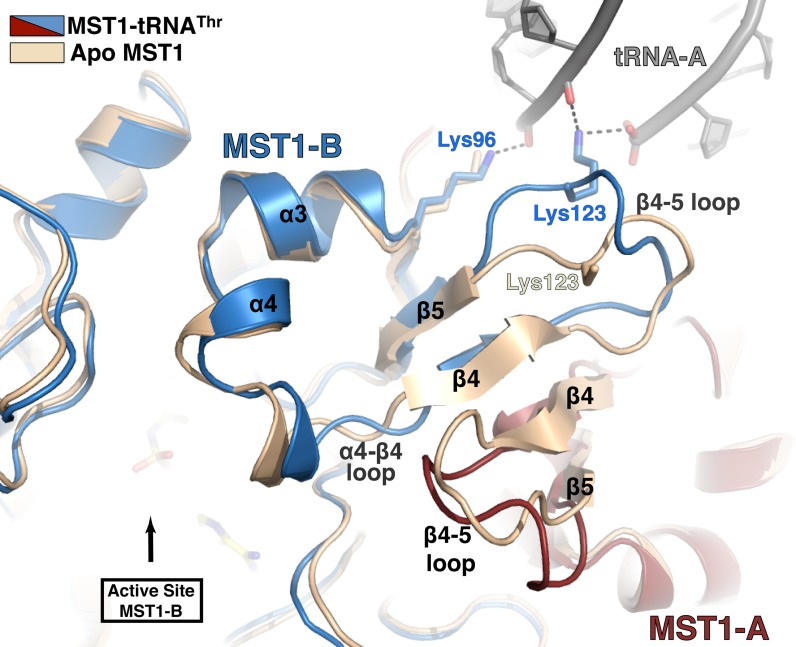
Besides binding to MST1-A, the acceptor stem of tRNA-A also makes contacts with the ‘non-catalytic’ MST1-B. The side chains of Lys96 and Lys123 from MST1-B are within the H-bonding distance from backbone phosphates of A66 and U67 (Figures 2A and 3). Particularly important is the side-chain NH2 of Lys123, which also coordinates the 5′-phosphate of tRNA-A and together with Ser166 from MST1-A forms the 5′-phosphate-binding groove (Figure 2A). Interestingly, Lys123 resides in the flexible loop linking strands β4 and β5. Because of its spatial relationship with the active site and the structural rearrangement it undergoes upon complex formation, we suggest that the β4-β5 loop may be a relay that signals to both catalytic sites of the MST1 dimer that the tRNA is bound to the enzyme (Figure 3).
Figure 3.
Cross-subunit interactions with the acceptor stem induce a conformational rearrangement in the active-site lid. Superimpositioning of the apo-MST1 structure (PDB ID: 3UGQ; beige) onto the MST1-tRNA complex (red, blue and grey) reveals structural rearrangements in the catalytic domain upon tRNA binding. Residues Lys96 and Lys123 from the MST1-B monomer (blue) interact with the sugar-phosphate backbone of the acceptor stem of the tRNA primarily bound to the MST1-A monomer (red). These interactions presumably induce to a conformational change in flexible loops β4-β5 and α4-β4 that result in movement of the helix α4 and thus widening of the active-site lid to accommodate the 3′ end of tRNA. This is a possible mechanism of allosteric regulation that allows the active site of one monomer to ‘sense’ when tRNA is bound to the other monomer.
In conclusion, the acceptor arm of the yeast mitochondrial tRNA2Thr is recognized by both subunits of the MST1 dimer. Whereas the ‘catalytic’ subunit establishes the identity of the threonine tRNA, the ‘non-catalytic’ subunit stabilizes the complex through interactions with the sugar-phosphate backbone of the acceptor arm. Lastly, both subunits are required for formation of the 5′-phosphate binding pocket(s).
The dimeric structure of MST1 is required for recognition of both the anticodon loop and anticodon stem of tRNA2Thr
Most aaRSs recognize the anticodon sequence in the cognate tRNAs. The accumulated biochemical and structural evidence demonstrates that the anticodon loop of tRNA binds to the anticodon-binding domain of the aaRS, where sequence-specific interactions are established. It is therefore not surprising that the anticodon loop, including the anticodon sequence it harbors, often serves as the major identity element in the elongator tRNA that is recognized by the cognate aaRS. Our previous biochemical studies suggested that MST1 employs this principle for substrate recognition as well (17). What was surprising, however, was the observation that MST1 specifically recognized two mitochondrial tRNAThr isoacceptors through interactions with their anticodon loops even though they differed both in size and sequence. Our crystal structure of MST1 in complex with the canonical tRNA2Thr provides the first glimpse at the mechanism of the anticodon stem and loop recognition by MST1. The structure shows that, as expected, the anticodon loop of tRNA-A binds to the anticodon-binding domain of MST1-A. On the other hand, its anticodon stem interacts in a cross-dimer fashion with the catalytic domain of MST1-B.
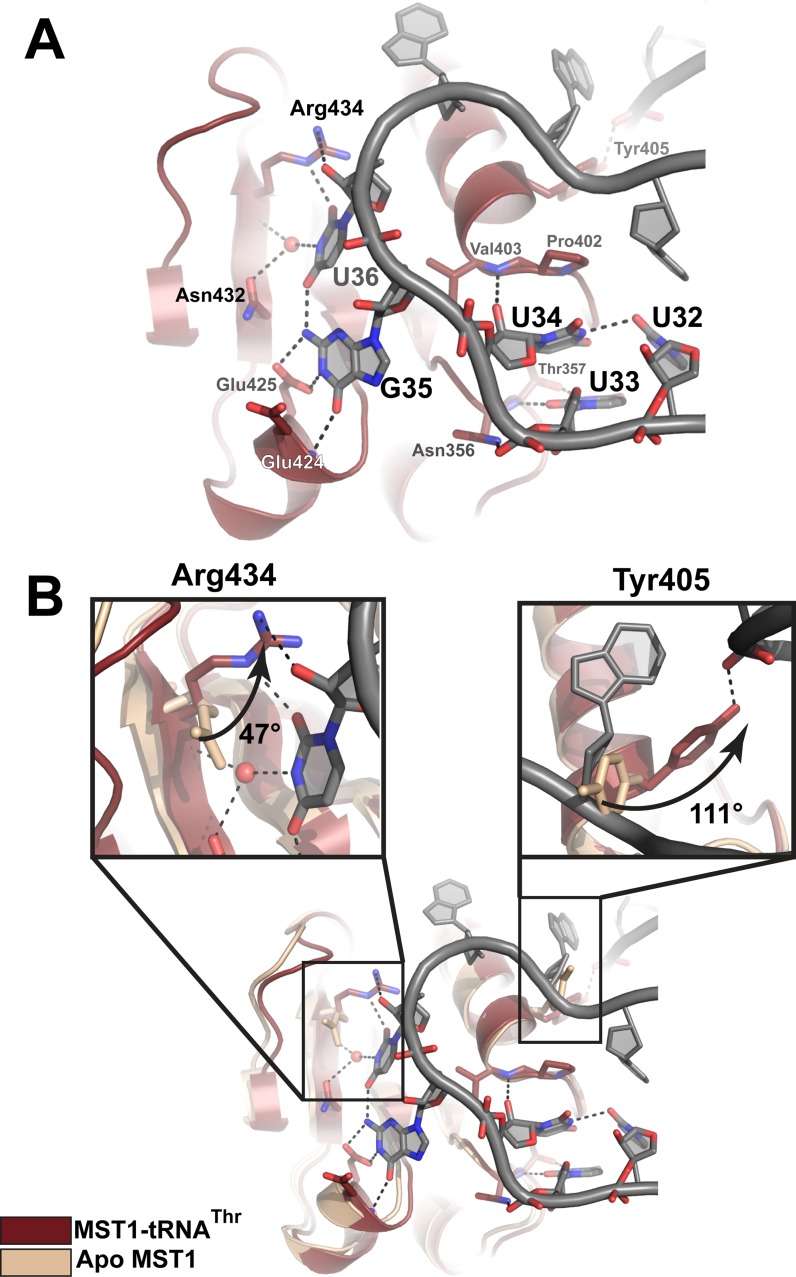
In our crystal, the anticodon-binding domain of MST1-A binds to the anticodon loop of tRNA2Thr (Figure 1). Not only does MST1 ‘read’ the second and third position of the anticodon sequence, but it also recognizes the base of the nucleotide located upstream of the anticodon triplet (Figure 4A). The side-chain hydroxyl and the backbone amide of Thr357 form H-bonds with the N3 and O2 atoms of the U33 base, respectively. In addition, the phosphate connecting U33 and U34 is held in place by H-bonds with the side chain of Asn356. The base of U34, the first nucleotide of the anticodon sequence, is not specifically recognized. Instead, its pyrimidine ring stacks over the pyrrolidine ring of Pro402, which is located in the loop that links strand β16 with helix α10. The base configuration of U34 is stabilized by a H-bond between its N3 imino moiety and the O2 keto group of U32, whereas the interaction with the backbone amide of Val403 anchors the ribose ring in place. Further, the Watson–Crick face of G35 is recognized by the backbone amide of Glu424 and the side-chain carboxyl of Glu425. The recognition of U36, however, is a bit more elaborate. For instance, the O4 and N3 atoms of U36 form H-bonds with the N2 amino of G35 and a water molecule, respectively. The water molecule is held in place through its contacts with the side chain of Asn432 and the backbone amide of Arg434. Lastly, the guanidinium group of Arg434 stabilizes the orientation of U36 through contacts with the O2 atom of the base and the 2′OH group of ribose. Interactions with the distal part of the anticodon loop are comprised of Van der Waals contacts and non-specific polar contacts between the enzyme and the sugar-phosphate backbone.
Figure 4.
Recognition of the anticodon sequence and structural rearrangements in the anticodon-binding domain of MST1 upon complex formation. (A) The first position in the anticodon sequence, U34, is not specifically recognized by MST1. Instead, Thr357 recognizes U33, the base upstream of the anticodon. The second base, G35, is recognized by the backbone amide of Glu424 and Glu425, whereas the third position, U36, is recognized by Asn435, the backbone atoms of Arg434, and a water molecule. The backbone atoms of MST1 and tRNA are red and grey, respectively. (B) Superimpositioning of apo MST1 (PDB ID 3UGQ; beige) onto the MST1-tRNA complex (red and grey) reveals structural rearrangements in the anticodon-binding domain upon tRNA binding. The first inset highlights the movement of Arg434 that is important for recognition of U36. The side chain of Arg434 rotates ∼47° and translates ∼1.4 Å toward the tRNA. The second inset shows rotation (∼111°) of Tyr405. This movement positions the side-chain hydroxyl within the H-bonding distance from the sugar-phosphate backbone of A26 in the anticodon stem.
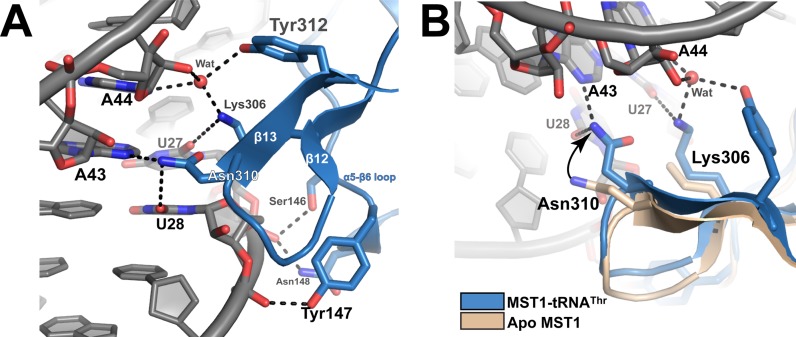
The anticodon stem of tRNA-A establishes cross-subunit contacts with the catalytic domain of MST1-B (Figure 1B). In the bacterial complex, the cross-dimer interactions with the anticodon stem of tRNAThr were sequence non-specific (25). In a sharp contrast, our structure reveals that MST1-B specifically recognizes the first two base pairs of the anticodon stem (i.e. U27:A43 and U28:A42) and the first base in the variable arm (i.e. A44) in tRNA2Thr (Figure 5A). The anticodon stem of tRNA-A sits atop α5-β6 loop (residues 142–150) and β12-β13 turn (residues 302–313). There, Lys306 forms a H-bond with the O2 atom in the base of U27, whereas the side chain of Asn310 makes contacts with the O2 and N3 atoms of U28 and A43, respectively. Also, the side-chain NH2 group of Lys306 and the side-chain OH of Tyr312 coordinate a water molecule that, in turn, makes polar contacts with the 2′OH and the N3 imino of A44. The sugar-phosphate backbone flanking U27:A43, U28:A42 and A44 is stabilized through contacts with side chains of Ser146, Tyr147 and Asn148.
Figure 5.
Cross dimer interactions between MST1-B and the anticodon stem of tRNA-A stabilize the ternary complex. (A) The catalytic domain of MST1-B establishes sequence-specific contacts with the anticodon stem of tRNA-A. The side chains of Lys306 and Tyr312 participate in recognition of A44. Lys306 is also involved in ‘reading’ the identity of U27, whereas Asn310 makes base specific contacts with U28 and A43. (B) The structural overlay of apo (PDB ID: 3UGQ; beige) and tRNA-bound MST1 (blue and grey) reveals that residues of the catalytic domain of MST1-B that ‘read’ the sequence of the anticodon stem undergo a conformational change upon ternary complex formation. Asn310 moves 3.3 Å from its position in the apo structure to interact with U28 and A43. Lys306 is partially disordered in apo MST1, but it is ordered in the ternary complex in which it interacts with a water molecule and the base of U27.
In summary, our structure shows that the homodimeric structure of MST1 is an essential prerequisite for productive binding to both the anticodon stem and loop of tRNA2Thr. The anticodon-binding domain of the catalytic MST1 subunit ‘reads’ the second and third position of the anticodon sequence as well as the nucleotide located immediately upstream of the anticodon sequence. The non-catalytic subunit, on the other hand, recognizes the first two base pairs of the anticodon stem, the first nucleotide of the variable arm and the shape of the flanking sugar-phosphate backbone.
Structural rearrangements in MST1 upon tRNA2Thr binding
We have previously reported that the active site of apo MST1 is dynamic and able to adopt both open and closed conformations (17). The crystal structure of the tRNA-bound MST1 provides a platform for examining structural rearrangements in the enzyme induced by formation of the productive complex with tRNA and aminoacyl adenylate. The structural comparison reveals that the tRNA binding induces conformational changes in the catalytic domain of both subunits of the MST1 dimer and that there are minor structural adjustments in the anticodon-binding domain.
The superimpositioning of 832 residues of apo-MST1 in the closed conformation (PDB ID: 3UGQ) onto the corresponding residues of MST1 bound to tRNA2Thr and TAM yields a low root mean square deviation (r.m.s.d.) value of 1.17Å. Although the structural overlay of 296 residues of the catalytic domain yielded similarly low r.m.s.d. value of 1.11 Å, we observed marked differences in several segments of the catalytic domain. Particularly, the flexible loops and helices comprising the dimer interface and the lid of the active site rearrange upon tRNA binding. The loop connecting strands β4 and β5 moves ∼3.6 Å away from the dimer interface and closer to the tRNA (Figure 3). Consequently, the side chain of Lys123 is brought in the proximity of the 5′ phosphate and the backbone between A66 and U67. Further, helices α3 and α4, which form the lid of the active site, are repositioned in the ternary complex. Also, loop α4-β4 of the catalytic monomer moves closer to the β4-β5 loop in the non-catalytic monomer upon complex formation thereby creating a larger cleft near the active site that presumably serves to accommodate the CCA-end of the tRNA. Lastly, residues that interact with the first two base pairs of the anticodon stem, Lys306 and Asn310, undergo structural adjustments that facilitate productive interactions with the tRNA (Figure 5B). The side chain of Asn310 rotates moves 3.3 Å from the position occupied in the apo MST1 and the side chain of Lys306 is fully ordered in the complex structure. These rearrangements facilitate interactions of Lys306 and Asn310 with U27 and A28:U43, respectively.
By contrast, the overall tertiary structure of the anticodon-binding domain does not alter upon complex formation. The superimpositioning of 120 residues of the anticodon-binding domain from the apo and tRNA-bound MST1 yields a very low r.m.s.d. value of 0.55 Å. However, residues that recognize the anticodon sequence of tRNA2Thr undergo structural changes (Figure 4B). The backbone atoms of Arg434 translate ∼1.4 Å toward the tRNA while the side chain rotates approximately 47° to coordinate the O2 atom of the U36 base. Likewise, the side chain of Tyr405 rotates ∼111° thereby positioning its hydroxyl moiety to H-bond a non-bridging oxygen of A26. Further, β-strands of the anticodon-binding domain shift when in complex with the tRNA and the domain adopts a more closed conformation when compared to the apo enzyme.
In summary, several surface loops of the catalytic domain are repositioned upon tRNA binding and, consequently, a larger cleft for binding the CCA-end is formed near the active site. On the other hand, the anticodon-binding domain largely adopts a very similar structure to the one observed in the apo enzyme. Only residues that directly interact with the anticodon loop exhibit altered conformation.
MST1 and E. coli ThrRS employ distinct tRNA recognition mechanisms
The superimpositioning of 392 analogous residues from the E. coli and yeast complexes yields an r.m.s.d value of 1.95 Å, suggesting that the overall complex architecture is conserved from bacteria to yeast (Figure 6). However, a closer inspection reveals a series of structural differences between the two complexes that may be of functional significance (25).
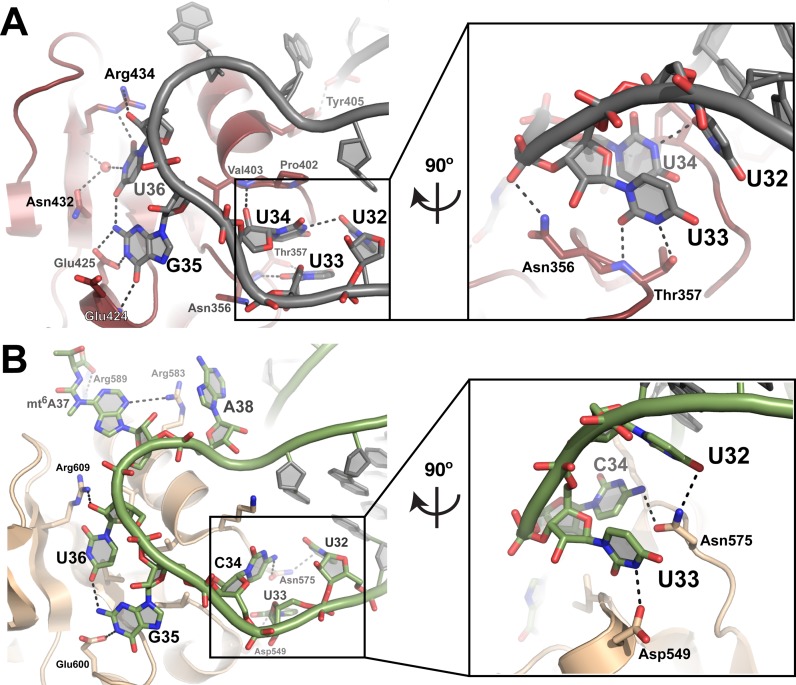
Figure 6.
The bacterial and yeast mitochondrial ThrRS employ distinct mechanisms to recognize the anticodon loop of the cognate tRNA. (A) Anticodon binding domain of MST1 (red) bound to the anticodon loop of tRNA2Thr (grey). Inset shows the coordination of U34 by U32. U33 is coordinated by Asn356 and Thr357. (B) Interactions between E. coli ThrRS (wheat) and tRNAThr (green) (based on PDB ID:1QF6). Inset shows the coordination of U32 and C34 by Asn575 as well as interactions between U33 and Asp549. G35 and U36 are coordinated in a similar manner in both the yeast and bacterial systems. Bases of nucleotides in positions 37 and 38 are coordinated by Arg583 and Arg589 in E. coli, but are not recognized by MST1.
The complex formation with MST1 buries ∼15% of the solvent accessible surface area of tRNA2Thr, whereas ∼20% of the bacterial tRNAThr surface is engaged in binding to ThrRS. This difference is due to interactions the acceptor arm of the bacterial tRNAThr establishes with the N-terminal editing domain, which is absent in MST1. The lack of this domain not only affects the editing function, but also the subtleties of the tRNA recognition mechanism of MST1. For instance, the editing and catalytic domains of the bacterial ThrRS form contacts with the minor- and major-groove sides of the acceptor stem, respectively. In particular, residues 201–214 of the bacterial N2 sub-domain of the editing domain form a hairpin motif that recognizes the sequence of the first two base pairs of the acceptor stem. As we have seen above, the second base pair in the yeast tRNA2Thr is recognized by the α5′ helix in the catalytic domain of MTS1. Hence, although conserved, α5′ helix plays a minor role in substrate recognition in the bacterial system. Instead, its role is performed by a segment of the editing domain, which is absent in MST1. Further, our analysis revealed differences in the mode of recognition of the 3′ and 5′ ends of the tRNAs. In the bacterial structure, the conserved Arg375, located in the loop in motif 2, forms base-specific contacts with C74. The side chain of the corresponding residue in MST1, Arg174, is only partially ordered in our crystal and does not make any contacts with tRNA. In addition, multiple residues of the bacterial catalytic domain coordinate either ribose hydroxyls or the base atoms of A76. By contrast, A76 is disordered in the binary MST1-tRNA2Thr complex. Lastly, while side chains from both MST1 monomers form the 5′-phosphate-binding pocket (Figure 2A), only Ser367, aided by water molecules, forms the corresponding groove in the bacterial ortholog.
Marked disparities are also present in the mode by which the anticodon stem and anticodon loop of the cognate tRNAs are coordinated. The bacterial ThrRS only interacts with the sugar-phosphate backbone between the second and third base pairs of the anticodon stem, but MST1 specifically ‘reads’ the sequence of the first two base pairs in the anticodon stem and the first base of the variable arm (Figure 5). Next, while the second and third positions of the anticodon sequence are recognized by a conserved mechanism, the identity of the first position is established by interactions that are distinct between the two complexes. In the bacterial complex, the side chain of Asn575 interacts with the base of C34. To the contrary, the analogous residue in MST1, Asn400, does not interact with the base of U34. Instead, its backbone atoms interact with a nucleotide upstream of the anticodon sequence U32. The base of U32 along with Pro402 and Val403 interact with U34. Finally, residues of the anticodon-binding domain of the bacterial ThrRS interact closely with the backbone atoms of nucleotides 32–37, and the side chain of Arg583 stacks over the base of A38 (Figure 6). In the MST1-tRNA2Thr complex, an analogous Lys408 residue does not interact with tRNA and A38, but it is rather oriented toward the anticodon stem where it coordinates a water molecule.
DISCUSSION
The ability of aaRSs to aminoacylate cognate tRNAs is of vital importance for the maintenance of both the genetic code and cellular proteome, and for the integrity of biological processes. With the notable exception of SerRS (26), AlaRS (27), LeuRS (28) and PylRS (14), other aaRSs establish identity of the cognate tRNA primarily through sequence-specific interactions with the anticodon sequence. Surprisingly, a single ThrRS in the mitochondria of certain yeast species, designated as MST1, aminoacylates equally well two isoacceptor threonine tRNAs, tRNA1Thr and tRNA2Thr, which harbor anticodon loops of different lengths and unrelated anticodon sequences (Supplementary Figure S1). Even more puzzling is the fact that the anticodon loop serves as the major recognition element for both tRNAs. This is in a startling contrast to the other known case of isoacceptor ‘promiscuitiy’ in which the mammalian mitochondrial SerRS is able to bind distinct TΨC-loops in two serine tRNAs (29,30). The question was, therefore, raised as to how MST1 achieves the tRNA specificity while exhibiting substrate promiscuity? In other words, how is this aaRS capable of binding to completely different anticodon loops and sequences with high specificity? In our earlier studies we suggested that the anticodon-binding domain of MST1 could employ different residues/surfaces for binding to these anticodon loops, but the structural evidence was lacking. Herein, in an effort to define the tRNA-recognition mechanism(s) of MST1, we determined the crystal structure of MST1 bound to the canonical tRNA2Thr and compared it to the homologous structure derived from the bacterial system.
The asymmetric unit of our crystal contained one MST1 dimer bound to two tRNA molecules in the arrangement very similar to that observed in the bacterial ThrRS-tRNAThr complex structure (25). The low r.m.s.d. value, which was calculated after superimpositioining the backbone atoms of the bacterial ThrRS onto the yeast MST1, also implies that the two complexes are similar. However, due to the missing N-terminal editing domain, MST1 establishes fewer contacts with the cognate tRNA. This is illustrated by the fact that only 15% of the tRNA2Thr surface participates in the complex formation compared to ∼20% of the bacterial tRNAThr. Also, while primarily interacting with one monomer (i.e. MST1-A), each tRNA forms a significant number of specific cross-subunit interactions with the other MST1 monomer (i.e. MST1-B). This implies that the dimeric structure of MST1 is critical for binding to both the acceptor and anticodon arms. Both E. coli ThrRS and S. cerevisiae MST1 recognize the second base pair of the acceptor arm of the tRNA, but the recognition patterns are distinct. In E. coli tRNAThr, the second base pair is C2:G71 and mutating it to U2:A71 decreased the aminoacylation activity over 100-fold (31). On the other hand, our data show that MST1 clearly prefers U2:A71 over C2:G71 (Supplementary Figure S2, Supplementary Table S1). Further, while the catalytic and editing domains of the bacterial ThrRS dimer ‘read’ the first two base pairs of the acceptor arm, the 5'-phosphate binding pocket is composed of residues from one subunit only. By contrast, the second base pair of the acceptor arm of tRNA-A is recognized by a newly formed helix α5′ in MST1-A, but its 5'-phosphate interacts with residues from both MST1 subunits. Of particular importance for the 5'-end recognition is the side chain of Lys123, which is located in the flexible β4-β5 loop that undergoes a conformational change upon tRNA binding. Because of its proximity to the catalytic sites, the β4-β5 loop could serve as a sensor that ‘informs’ the active site that the complex with tRNA is formed and it could represent an element that allosterically regulates the enzyme activity. The structural flexibility of the catalytic domain thus may be important for binding to the acceptor arm of mitochondrial tRNA1Thr and tRNA2Thr and could play a role in modulating the enzymatic activity of MST1. Further studies on probing this mechanism are warranted.
As in case of the acceptor arm, recognition of the anticodon stem is dependent on the dimeric structure of MST1. MST1-A only binds the anticodon stem of tRNA-A via an interaction between residue Tyr405 and A26, while base specific interactions are formed with the catalytic domain of MST1-B only. Through cross-subunit interactions, MST1-B ‘reads’ the sequence of the first two base pairs of the anticodon stem of tRNA2Thr, U27:A43 and U28:A42, as well as that of the first nucleotide in the variable arm, A44. This is in a striking contrast to the bacterial ThrRS that forms only non-specific contacts with the sugar-phosphate backbone of the anticodon arm. Hence, although MST1 makes fewer contacts with the cognate tRNA when compared with the bacterial ThrRS, more of those contacts are sequence-specific. Also, the binding of tRNA2Thr to MST1 is completely dependent on the dimeric structure of the enzyme, perhaps more so than the bacterial tRNAThr.
The crystal structures of MST1- tRNA2Thr and E. coli ThrRS-tRNAThr provide the opportunity to compare how respective anticodon sequences are recognized. In both structures, the anticodon-binding domain of monomer A binds the anticodon sequence of tRNA-A with minor structural adjustments to the enzyme. Our analysis shows that MST1, just like bacterial ThrRS, makes specific contacts with the second and third positions of the anticodon sequence. The conservation of the recognition mechanism ends here. Whereas the E. coli ThrRS ‘reads’ the first position of the anticodon, MST1 recognizes the base of the nucleotide located immediately upstream (i.e. U33) of the anticodon and not the first position. The base of the first nucleotide in the anticodon of tRNA2Thr is positioned through stacking with Pro402, and hydrogen bonds with U32 and the backbone carbonyl of Asn400. In addition, the bacterial ThrRS makes extensive contacts with the sugar-phosphate backbone of the distal part of the anticodon loop (nucleotides 32–38). By contrast, only a few interactions with A38 are present in the MST1- tRNA2Thr complex. Thus, it could well be that differences in a segment of the anticodon-binding domain that interacts with the distal part of the anticodon loop permit binding of the enlarged loop of tRNA1Thr.
We also correlated our earlier predictions based on mutagenesis and kinetic assays (17) with interactions observed in the complex structure. We have previously suggested that side chains of Thr357, Gln362, Tyr405, Lys408 and Ser409 are important for recognition of the canonical tRNA2Thr. The side chain of Gln362 does not specifically interact with the tRNA yet it stabilizes multiple residues (including Thr357) in the α10-β16 loop. Supporting our prediction of the importance of Thr357, the backbone amino group and the side chain hydroxyl hydrogen bond to the Watson–Crick face of U33. In our structure, Tyr405 rotates away from the tRNA anticodon loop and instead positions the phosphate of A26 in the acceptor stem. Our previous data suggested important roles for Lys408 and Ser409, however, neither of these residues interacts with the tRNA directly. Lys408 is positioned between A37 and A38 and is partially disordered, while the role of Ser409 remains unclear. Ser409 is located in α11 and may play a role in the stabilization of this helix and thus the positioning of Tyr405.
In summary, although adopting similar architectures, the structures of the yeast mitochondrial and bacterial ThrRS-tRNAThr complexes reveal multiple differences in the mode of tRNA recognition. MST1 forms fewer contacts with tRNA2Thr when compared to the bacterial ThrRS, but it establishes a greater number of sequence-specific contacts. In addition, the dimeric structure of MST1 is an absolute pre-requisite for binding to the acceptor and anticodon arms. This stands in contrast to the bacterial system in which cross-dimer interactions are fewer and less specific. Moreover, the structural flexibility in the catalytic domain of MST1 subunits is vital for recognition of the acceptor stem. Perhaps the most important is the flexible β4-β5 loop of the catalytic domain, which might play a role of a sensor (or relay) that signals to the catalytic site that the substrate tRNA is bound to the enzyme. Additionally, our analysis shows that MST1 and bacterial ThrRS most likely employ distinct mechanisms for anticodon sequence recognition. Surprisingly, a segment of the anticodon-binding domain of MST1 that interacts with the distal part of the anticodon loop establishes a limited number of interactions with the tRNAThr. Therefore, we speculate that this particular element in MST1 could play a critical role in accommodating an enlarged anticodon loop of tRNA1Thr. Lastly, the second base pair of the acceptor stem plays a more important role in recognition of tRNA1Thr compared with tRNA2Thr. To completely unravel the recognition mechanism of tRNA1Thr, however, a high-resolution structural study on MST1 bound to tRNA1Thr is needed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Jana Ognjenović, Ruslan Sanishvili and staff of the GM/CA@APS beam line for help during data collection and processing. The coordinates and structure factors for the MST1-tRNA2Thr complex are deposited in PDB with the accession code 4YYE.
FUNDING
National Institute of General Medical Sciences of the National Institutes of Health [R01 GM097042 to M.S. and R01 GM115431 to J.L.]; U.S. DOE [under Contract No. DE-AC02-06CH11357 to Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory]; GM/CA@APS (23ID-D and 23BM) sector has been funded from the NCI (ACB-12002) and NIGMS (AGM-12006). Funding for open access charge: National Institute of General Medical Sciences of the National Institutes of Health [R01 GM097042 to M.S.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ibba M., Söll D. Aminoacyl-tRNA Synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Jakubowski H., Goldman E. Editing of errors in selection of amino acids for protein synthesis. Microbiol. Rev. 1992;56:412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo M., Schimmel P. Structural analyses clarify the complex control of mistranslation by tRNA synthetases. Curr. Opin. Struct. Biol. 2012;22:119–126. doi: 10.1016/j.sbi.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascarenhas A.P., An S., Rosen A.E., Martinis S.A., Musier-Forsyth K. Protein Engineering. Berlin Heidelberg: Springer Science +Business Media; 2009. pp. 155–203. [Google Scholar]
- 5.Reynolds N.M., Lazazzera B.A., Ibba M. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 2010;8:849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 6.Giegé R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crick F.H.C., Barnett L., Brenner S., Watts-Tobin R.J. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 8.Crick F.H.C. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 9.Ambrogelly A., Palioura S., Söll D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 10.Moura G.R., Paredes J.A., Santos M.A.S. Development of the genetic code: Insights from a fungal codon reassignment. FEBS Lett. 2010;584:334–341. doi: 10.1016/j.febslet.2009.11.066. [DOI] [PubMed] [Google Scholar]
- 11.Sengupta S., Yang X., Higgs P.G. The mechanisms of codon reassignments in mitochondrial genetic codes. J. Mol. Evol. 2007;64:662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schön A., Böck A., Ott G., Söll D. The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res. 1989;17:7159–7165. doi: 10.1093/nar/17.18.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krzycki J.A. The direct genetic encoding of pyrrolysine. Curr. Opin. Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Nozawa K., O'Donoghue P., Gundllapalli S., Araiso Y., Ishitani R., Umehara T., Söll D., Nureki O. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009;457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebald W., Wachter E., Tzagoloff A. Identification of amino acid substitutions in the dicyclohexylcarbodiimide-binding subunit of the mitochondrial ATPase complex from oligomycin-resistant mutants of saccharomyces cerevisiae. Eur. J. Biochem. 1979;100:599–607. doi: 10.1111/j.1432-1033.1979.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 16.Su D., Lieberman A., Lang B.F., Simonović M., Söll D., Ling J. An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine. Nucleic Acids Res. 2011;39:4866–4874. doi: 10.1093/nar/gkr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling J., Peterson K.M., Simonovic I., Cho C., Söll D., Simonović M. Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3281–3286. doi: 10.1073/pnas.1200109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z., Minor W. Methods in Enzymology. NY: Elsevier BV; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 19.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Cryst. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P., Cowtan K. Coot : model-building tools for molecular graphics. Acta Crystallogr. D Biol. Cryst. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Cryst. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 23.Bramucci E., Paiardini A., Bossa F., Pascarella S. PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinformatics. 2012;13:S2. doi: 10.1186/1471-2105-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling J., Peterson K.M., Simonovic I., Söll D., Simonović M. The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase. J. Biol. Chem. 2012;287:28518–28525. doi: 10.1074/jbc.M112.372920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sankaranarayanan R., Dock-Bregeon A.-C., Romby P., Caillet J., Springer M., Rees B., Ehresmann C., Ehresmann B., Moras D. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 26.Biou V., Yaremchuk A., Tukalo M., Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser) Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 27.Musier-Forsyth K., Usman N., Scaringe S., Doudna J., Green R., Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu M., Asahara H., Tamura K., Hasegawa T., Himeno H. The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNAs by their aminoacyl-tRNA synthetases. J. Mol. Evol. 1992;35:436–443. doi: 10.1007/BF00171822. [DOI] [PubMed] [Google Scholar]
- 29.Chimnaronk S., Gravers Jeppesen M., Suzuki T., Nyborg J., Watanabe K. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005;24:3369–3379. doi: 10.1038/sj.emboj.7600811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimada N., Suzuki T., Watanabe K. Dual mode recognition of two isoacceptor trnas by mammalian mitochondrial seryl-tRNA synthetase. J. Biol. Chem. 2001;276:46770–46778. doi: 10.1074/jbc.M105150200. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa T., Miyano M., Himeno H., Sano Y., Kimura K., Shimizu M. Identity determinants of E. coli threonine tRNA. Biochem. Biophys. Res. Commun. 1992;184:478–484. doi: 10.1016/0006-291x(92)91219-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.