Abstract
We determined the effects of DNA damage caused by ultraviolet radiation on gene expression in Leptospira interrogans using DNA microarrays. These data were integrated with DNA binding in vivo of LexA1, a regulator of the DNA damage response, assessed by chromatin immunoprecipitation and massively parallel DNA sequencing (ChIP-seq). In response to DNA damage, Leptospira induced expression of genes involved in DNA metabolism, in mobile genetic elements and defective prophages. The DNA repair genes involved in removal of photo-damage (e.g. nucleotide excision repair uvrABC, recombinases recBCD and resolvases ruvABC) were not induced. Genes involved in various metabolic pathways were down regulated, including genes involved in cell growth, RNA metabolism and the tricarboxylic acid cycle. From ChIP-seq data, we observed 24 LexA1 binding sites located throughout chromosome 1 and one binding site in chromosome 2. Expression of many, but not all, genes near those sites was increased following DNA damage. Binding sites were found as far as 550 bp upstream from the start codon, or 1 kb into the coding sequence. Our findings indicate that there is a shift in gene expression following DNA damage that represses genes involved in cell growth and virulence, and induces genes involved in mutagenesis and recombination.
INTRODUCTION
Coping with DNA lesions is essential for many types of pathogenic bacteria to survive in the host and in the environment. During infection, treatment with antibiotics and oxidative bursts from macrophages can cause substantial damage in bacterial DNA (1). While in the environment, bacteria often encounter agents (2,3) such as ultraviolet (UV) light and heavy metals which can damage their DNA (2,3). DNA damage can cause the replication fork to stall, triggering the SOS response, a complex system that orchestrates DNA repair and tolerance of remaining lesions. The controller of the SOS response is LexA, a transcriptional repressor from the S24 family of peptidases that binds to operators called SOS boxes. These binding sites are usually located in promoter regions, inhibiting transcription of associated genes (4). When DNA is damaged, single-stranded DNA (ssDNA) can be formed by stalled replication forks or enzymatic processing of broken DNA ends (2). RecA protein binds to ssDNA to become activated and then stimulate the autocleavage of S24 peptidases, including LexA (5,6). Cleaved LexA releases from the promoter regions of controlled genes, allowing derepression of transcription (4,7). Some genes of the SOS regulon are associated with DNA repair (8–10). Other genes are involved in bacterial adaptation to the environment by regulating mutagenesis. Expression of virulence factors, including Shiga-like toxins and the type III secretion system and associated factors in enterohemorrhagic Escherichia coli, are also induced during the SOS response (11–13). This makes the SOS response a key component of bacterial virulence and adaptation (14,15).
Every year the Spirochaete L. interrogans kills ∼48 000 people worldwide (16). It colonizes the kidney of rats and other mammals, is shed in the urine, and is able to survive for weeks in water and mud. Humans are usually infected through abrasions on skin or mucous membranes (17). Once in the bloodstream, the bacteria can target eyes, kidneys, liver and lungs, causing a variety of illnesses (18,19) which range from mild flu-like symptoms to thrombocytopenia, uveitis, renal and liver failure, and pulmonary hemorrhage (20).
We previously found that L. interrogans serovar Copenhageni, the most frequent causative of human Leptospirosis in Brazil, activates an SOS response following DNA damage (21). This bacterium has two lexA genes: lexA1, homologous to L. interrogans serovar Lai lexA (22) and lexA2, which is exclusive to the serovar Copenhageni serovar (21). LexA1 is probably the main regulator of the SOS response. In addition to both lexA genes and their operons, initial experiments identified recA, recN and dinP as LexA1-bound and induced by DNA damage in L. interrogans (21). LexA1 binds to the upstream sequence of both lexA1 and lexA2. In contrast, LexA2 is only known to bind to its own promoter, through a different motif than that used by LexA1. As is the case for the horizontally acquired lexA2, from Pseudomonas aeroginosa (23), leptospiral lexA2 has probably restricted activity outside its own genetic island.
Pathogenic leptospires impose several difficulties to genetic manipulation, especially serovar Copenhageni, as even random transposon insertion techniques showed limited success (24). Consequently, global analyses that can be performed in wild-type cells are extremely important to study complex responses in this bacterium. In this work, we performed an integrated approach that combines different omics data, discovering LexA1 binding sites and their relationship with global transcriptional modulation after DNA damage induction in L. interrogans. Gene expression profiling data were integrated with global DNA binding site identification in order to investigate the mechanisms employed by Leptospira to deal with DNA insults. Transcriptional profiling analysis in response to DNA damage indicated that L. interrogans turns down virulence gene expression and increases chromosome instability during genotoxic stress. We also expanded the known SOS network from 7 to over 25 genes, with little overlap with the LexA regulon from other organisms. We found that the LexA1 binding site is a 16 base pair palindromic motif different from the previously proposed binding site (21,22). Based on our new findings, we were able to reconcile apparent inconsistences in previous leptospiral SOS studies. These binding sites were not restricted to promoter regions, as LexA1 showed binding in intra- and intergenic regions. We compared LexA1 binding data to the changes in gene expression following DNA damage and found that expression of most, but not all, LexA1-bound transcriptional units increased following DNA damage.
MATERIALS AND METHODS
Growth conditions and treatments
L. interrogans serovar Copenhageni Fiocruz L1–130 was cultivated at 30°C in supplemented EMJH medium (Difco) without agitation (25). The strain was frequently re-isolated from experimentally infected hamsters to maintain its virulence, as described (26). We used a protocol for irradiation with short wavelength ultraviolet light (UV-C) that has been described elsewhere (21). Briefly, cultures of exponentially growing bacteria were either exposed (IR) or not exposed (NI) to a UV-C germicidal lamp (254 nm), at a dose known to kill 50–60% of cells (5 J.m−2). After treatment, the same volume of fresh medium was added to both cultures to stimulate cellular growth and they were incubated at 30°C for 12 h. For ciprofloxacin treatment, 5 μg/ml antibiotic was added to the cultures, which were incubated for 4 h or 12 h. Next, cells were harvested by centrifugation (6000xg, 15 min) and pellets were immediately frozen in liquid nitrogen and stored at −80°C until use. All procedures were performed in the dark and repeated for four different biological samples.
Production of polyclonal antibodies
The mouse polyclonal antiserum against LexA1 used in this study is the same employed previously (21). Briefly, five Balb/C mice were inoculated intraperitoneously with 10 μg of recombinant protein, using Al(OH)3 as adjuvant. Seven days after the fourth weekly boost, blood was retrieved from the retro obital plexus, incubated at 37°C, and spun to collect serum.
Chromatin immunoprecipitation combined with massively parallel DNA sequencing (ChIP-seq)
Cross-linked lysate preparation and immunoprecipitation were performed as described by Lin & Grossman (27), with some modifications. Cells in the fifth day of growth in liquid medium, in exponential phase, were harvested by centrifugation (6000xg, 15 min), washed and resuspended in formaldehyde (2% final concentration). After 40 or 60 min incubation at room temperature, glycine was added and cross-linked cells were lysed. DNA present in lysates was sheared by sonication, yielding fragments between 200 to 600 bp, as visualized in a BioAnalyzer 2100 (Agilent). Aliquots from lysates (‘total’ samples) were saved for later analysis. Immunoprecipitations were performed with 1:100 final dilution of anti-LexA1 mouse serum (21), followed by incubation with Protein G Sepharose resin (GE Healthcare). DNA from elution (‘IP’) and total samples were purified using a QIAquick PCR Purification Kit (Qiagen) and used for PCR or high-throughput sequencing.
High-throughput sequencing analysis
Both pairs of IP and total samples (40 and 60 min of cross-linking) were selected for 200–400 bp fragments and sequenced on a HiSeq 2500 (Illumina) platform. At least 17 million 40-nt reads were obtained for each sample (Supplementary Table S1). These reads were aligned to both L. interrogans serovar Copenhageni chromosomes (accession numbers NC_005823.1 and NC_005824.1) using Bowtie2 (28), allowing no mismatches. The percentage of reads aligned to the reference genome ranged from 88 to 94%. The number of reads at each chromosomal position was normalized to the total number of reads of the sample. The reads were extended in silico to the estimated average length of 250 bp. The peak-calling tool from CisGenome (29) was used to define enriched regions in each IP sample, using the total sample as background control. The parameters were: –e 250 –b 5 –c 2 –maxgap 25 –minlen 50 –bw 2. Only regions with at least 2-fold enrichment, present in both IP samples and absent from total samples were selected. Aligned reads and peak-calling data were deposited into GEO (accession number GSE73688). Graphs shown are for the 60-min cross-link sample pair, since it presented more number of reads and it was representative of both treatments. The de novo motif search was performed in MEME (30) with 100 bp of sequence center on the peak from each region, allowing any number of motif occurrences.
DNA microarrays analysis
RNA from UV-C irradiated and non-irradiated samples was purified using an RNEasy mini-kit with RNA Protect, as described by the manufacturer (Qiagen). RNA quality was assessed by BioAnalyzer (Agilent), with RNA integrity numbers (RIN) > 9. Complementary DNA (cDNA) synthesis and labeling were performed with FairPlay III Microarray Labeling Kit following manufacturer's instructions (Agilent), but using only Cy-3. Samples were hybridized to custom Agilent gene expression slides for L. interrogans serovar Copenhageni, containing eight arrays of 15 K spots. Images were captured by Agilent DNA Microarray Scanner Bundle and extracted with Agilent Feature Extraction software version 9.5.3. Microarray intensity data were converted into log2 and normalized by Quantile. Student's t-test analysis was used to identify probes differentially expressed at 12 h post-irradiation compared to non-irradiated samples and probes representing the same gene were collapsed by taking the probe with the lowest P-value. Expression data were deposited in GEO (accession number GSE73687).
Gene set enrichment analysis (GSEA)
Gene Set Enrichment Analysis (GSEA), available at http://www.broadinstitute.org/gsea (31), was used to determine statistically differentially expressed functional groups. Gene categories were derived from Uniprot (http://www.uniprot.org) and KEGG (http://www.kegg.jp) annotations. Analyses were run through 1000 permutations, with weighted statistics. We considered as enriched categories with normalized enrichment score (NER) > 1 and normalized P-value < 0.1.
Quantitative PCR (qPCR)
Supplementary Table S2 lists all oligos used in this work. All qPCR experiments were repeated three times for each biological sample. For validation of ChIP conditions, the amount of DNA corresponding to the rrs gene (not bound by LexA1) and recA promoter (bound by LexA1) amplified in both IP and total samples was quantified. Fold enrichment was calculated by the ratio (recAIP/rrsIP)/(recAtotal/rrstotal). ChIP conditions used in this work resulted in 3.5- to 6-fold enrichment of recA promoter. For validation of microarray, fold change was calculated by the 2−ΔΔCt method, with the rrs (16S) coding region as normalizer. P-values were calculated by two-tailed Student's t-test.
Electrophoretic mobility shift assay (EMSA)
These assays were performed as described previously (21), using LexA1 binding regions from several targets (see Supplementary Table S2 for PCR oligos) as competitors for LexA1 binding to a 32P-labeled DNA fragment corresponding to the recA promoter region. These assays use leptospiral extract, whose specificity was validated elsewhere (21). Briefly, 40 μg of leptospiral extract was incubated with 1.55 fmol labeled probe and 1 μg poly[d(A-T)] as nonspecific competitor. After 20 min on ice, competitors were added in 50-fold excess and incubated for another 20 min. Electrophoresis and detection were performed essentially as described previously (21). Band densitometry was calculated by ImageJ, available at http://imagej.nih.gov/ij/.
RESULTS
Overview
We assessed the gene expression profile of the pathogenic L. interrogans after UV-C irradiation. We also determined the (detectable) chromosomal binding sites occupied by LexA1 using chromatin immunoprecipitation assay coupled to massively parallel DNA sequencing (ChIP-seq) to determine the LexA1 chromosomal binding sites. This combination of LexA1 binding sites and gene expression analyses resulted in a more extensive view of the gene network that controls prophage, motility, repair, tolerance and adaptation responses to UV-C-induced DNA damage in L. interrogans.
Global transcriptional profiling during UV-C DNA damage response
Previously, we found that a UV-C dose of 5 J m−2 kills approximately 50% of a leptospiral culture, and causes an increase in expression of recA, recN, dinP and both lexA genes (21). The upregulation of these SOS response genes starts about 8 h after treatment with UV-C and reaches a summit after 12 h, which coincides with the 8–12 h doubling time of pathogenic leptospiras in the growth medium used (18).
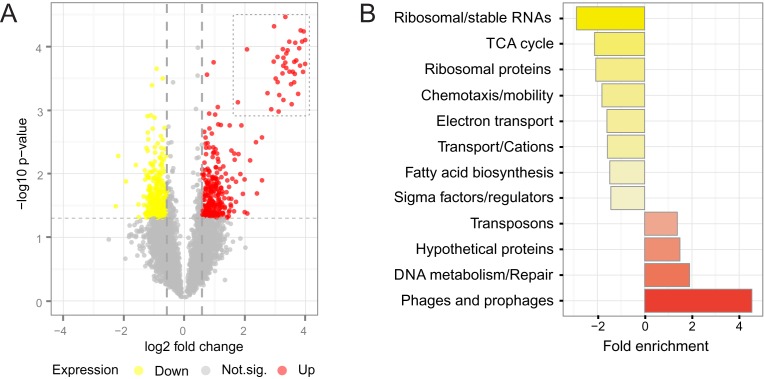
Here, we used DNA microarrays to measure the genome-wide changes in gene expression (mRNA levels) 12 h after exposure of L. interrogans to UV-C light. Exposure of bacteria to UV-C induced changes in expression of 568 genes (15% of total genes) at 12 h post-irradiation (Figure 1A). Among them, 299 genes were up regulated and 269 genes were down regulated. Relative expression of 37 genes was independently confirmed by analysis of mRNA levels using qPCR (rs = 0.71, P-value < 0.001, Supplementary Figure S1).
Figure 1.
DNA damage response in L. interrogans. (A) Volcano plot of all microarray data, comparing signals from non-irradiated and irradiated cultures, 12 h after treatment. Red indicates up regulation (upper right corner) and yellow, down-regulation of genes (upper left corner). Horizontal and vertical dotted lines mark the t-test P-value cutoff of 0.05 and fold change cutoff of 1.5. The dotted square englobes highly expressed prophage genes. (B) Genes displaying altered expression upon DNA damage were ranked and analyzed through gene set enrichment analyses (GSEA). Red bars to the right represent induced categories, and yellow bars to the left, repressed.
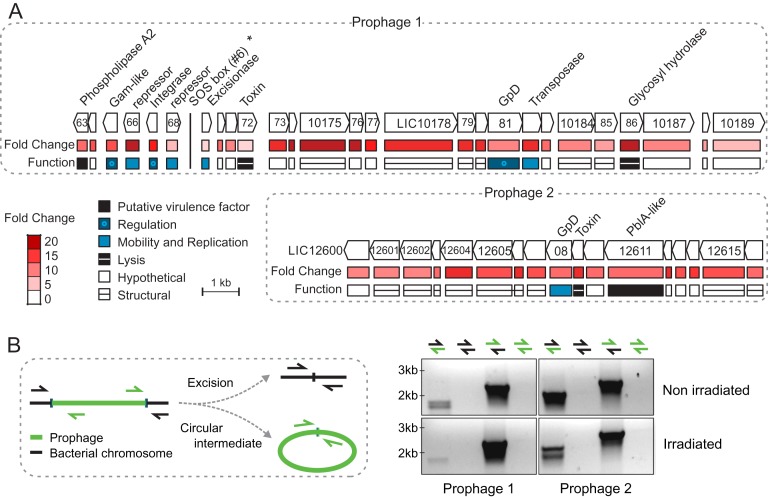
We performed GSEA (GSEA, http://www.broadinstitute.org/ gsea/) to assess the major functional categories affected by UV-C stress. Prophages and transposons were both induced (Figure 1B). Nine transposases, from at least four different families (IS1533, IS1501, IS3 and ISlin1), were induced ∼2-fold. In addition, of the top 50 up regulated genes after UV-C irradiation, 43 were phage-related genes (Figure 2A and Supplementary Table S3). The majority mapped in two genomic islands enriched in prophage genes: a gene cluster ranging from LIC10136 to LIC10189, previously described in serovar Lai (32) (which will be referred to as Prophage 1), and a previously undescribed cluster (referred here as Prophage 2) from LIC12600 to LIC12616 (Figure 2A). Prophage 1 lacks several replication and DNA-packaging genes, but encodes the only phospholipase A2 (PLA2, LIC10163) in the L. interrogans genome, and also a putative integrase and excisionase pair (LIC10167 and LIC10169). Prophage 1 encodes two documented cI/Cro repressors, one of them shown to be functional (32). However, neither of them harbors the S24 domain typically found in those repressors which is needed for RecA-stimulated auto-cleavage. Prophage 2 encodes a putative plasmid maintenance system toxin (LIC12609) and a phage tail protein (LIC12611) structurally similar to pblA, used by Streptococcus mitis to bind to platelets (33). Despite the up regulation of integrase and excisionase post-irradiation (Figure 2A), we did not detect excision of either Prophage 1 or Prophage 2 from the chromosome in UV-C treated or non-treated cells (Figure 2B). Since increased levels of excisionase in relation to integrase favor excision (34), lower expression of the excisionase (LIC10169; 2.8-fold) in relation to integrase (LIC10167; 15.3-fold) may be responsible for the stability of this region in the chromosome. It is also possible that these proteins and/or the phage attachment sites are not functional.
Figure 2.
(A) Two prophages heavily induced 12 h after UV-C irradiation. The Prophage 1 cluster begins at LIC10163 and the prophage 2 at LIC12600. Names of annotated genes are indicated, and below are the function and fold change, as assessed by microarray data. The vertical line indicates the LexA1 binding site correspondent to peak #6 (asterisk; see next section). (B) Phage excision test. Genomic DNA from cultures non-irradiated and irradiated with UV-C were used as template for PCR with primers flanking the bacteria-prophage interface (mixed-color arrows) and regions only available if the prophage would excise and form a circular intermediate (same-color arrows). Products predicted size for them would be around 2 kb.
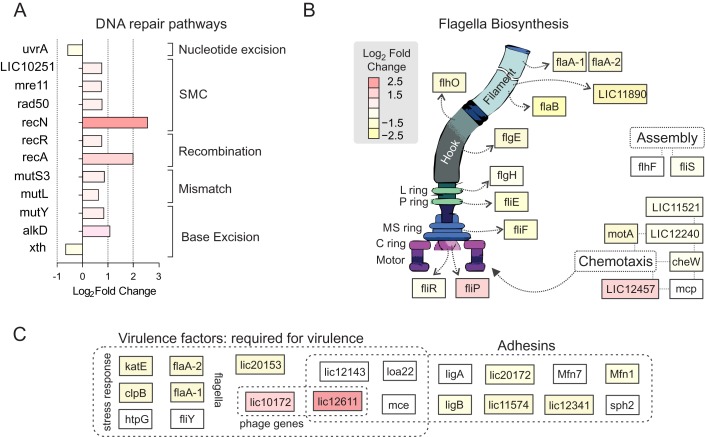
The second most overrepresented functional category among the up regulated genes was DNA metabolism (Figure 3A). Some genes involved in repair of different types of DNA damage were induced, including mutY and alkD (LIC12552), responsible for removal of oxidative and alkylation lesions, respectively. Post-replicative mismatch repair genes mutS3 and mutL were also up regulated, as well as structural maintenance of the chromosome (SMC) genes recN, rad50, mre11 and the recombination repair components recA and recR (Figure 3A). SMC proteins interact with DNA free ends and are involved in chromosomal rearrangements and recombination (35,36). Compared to other bacteria, though, relatively few DNA repair genes were up regulated after UV-C damage. None of the canonical DNA repair genes involved in removal of UV-C-induced DNA damage (components of nucleotide excision repair uvrABC, recombinases recBCD and resolvases ruvABC) were induced. Instead, the uvrA gene was slightly down regulated (Figure 3A and Supplementary Table S3). This suggests that DNA lesions, as cyclobutane pyrimidine dimers and pyrimidine-(6–4)-pyrimidone photoproducts, are mostly repaired by homologous recombination rather than by direct DNA removal.
Figure 3.
Alteration in expression of genes from specific functional categories during genotoxic stress. (A) Genes associated with DNA repair. Four repair pathways and structural maintenance of the chromosome (SMC) were represented by differentially expressed genes. (B) Genes involved in flagella biosynthesis, assembly and chemotaxis. Structures formed by the gene products are indicated by open arrows. (C) Level of expression of genes required for virulence and adhesion. See Supplementary Table S4 for specific functions and references. Differential expression of all genes shown is indicated by colors.
In contrast to several other bacteria, leptospires down regulated genes in several functional categories associated with cell growth, virulence, RNA metabolism and motility after genotoxic stress (Figure 1B). This repression may be due to down regulation of the major sigma factor rpoD (Supplementary Table S3), which drives expression of genes related to translation (ribosome proteins and ribosome RNA) and central metabolism (TCA cycle, electron transport and fatty acid biosynthesis). This down regulation seems to result in lower levels of the corresponding proteins, at least in ciprofloxacin-induced SOS response (37) – further discussed below. Additionally, several genes involved in chemotaxis, flagella biosynthesis and flagellar function were down regulated, representing one-third of flagella structural genes (Figure 3B). This includes flaA-1 and flaA-2 flagellins, which are essential leptospiral virulence factors (38).
There are currently 10 genes, including flaA-1 and flaA-2, considered as the virulence factors in L. interrogans. They are required for pathogenesis, as knockout strains lose capacity to cause disease (38–45). In addition, at least 10 putative virulence-associated factors are involved in adhesion and toxicity (46–52). From these 20 genes, 7 were down regulated after DNA damage (Figure 3C and Supplementary Table S4 for more information), including the stress response genes katE and clpB, and LIC10153, which is required for colonization of carriers. While the phage-related LIC10172 and LIC12611 are up regulated, expression of the other virulence genes was not altered (Figure 3C).
Genome-wide mapping of LexA1 binding sites
To better understand regulation of the UV-C response in an organism where genetic manipulation is limited, we identified L. interrogans chromosomal regions that were bound by LexA1 using ChIP-seq experiments. The second repressor LexA2 is highly divergent from LexA1 and probably acts only in its own transcriptional unit (21). Nonetheless, we employed a LexA1-specific antibody, previously characterized (21). L. interrogans cells were grown to exponential phase in defined EMJH-enriched medium, crosslinked, LexA1 was immunoprecipitated and associated DNA was analyzed by deep sequencing (Materials and Methods).
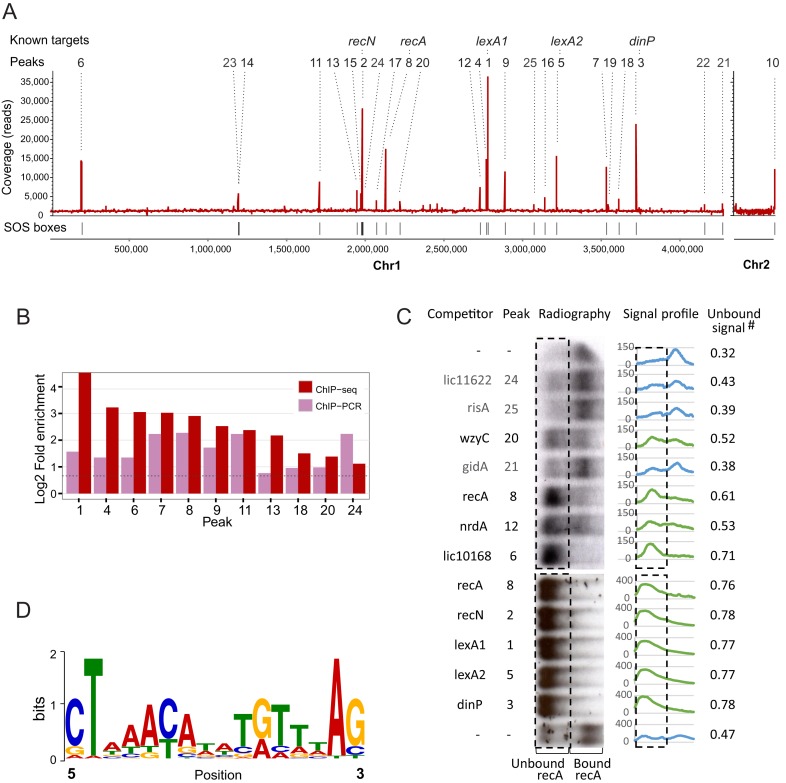
We found that LexA1 was associated with 24 loci distributed throughout chromosome 1, and to one site near the origin of replication of chromosome 2 (Figure 4A). Peaks were ranked from #1 to #25 according to the fold enrichment of sequenced reads in relation to the background. Five of the peak-associated transcriptional units were previously found to be bound by LexA1 (lexA1, recN, dinP, lexA2/LIC12653 and recA) and to be induced by DNA damage (21). We verified binding in vivo to 11 sites using ChIP-qPCR (Figure 4B and Supplementary Table S5).
Figure 4.
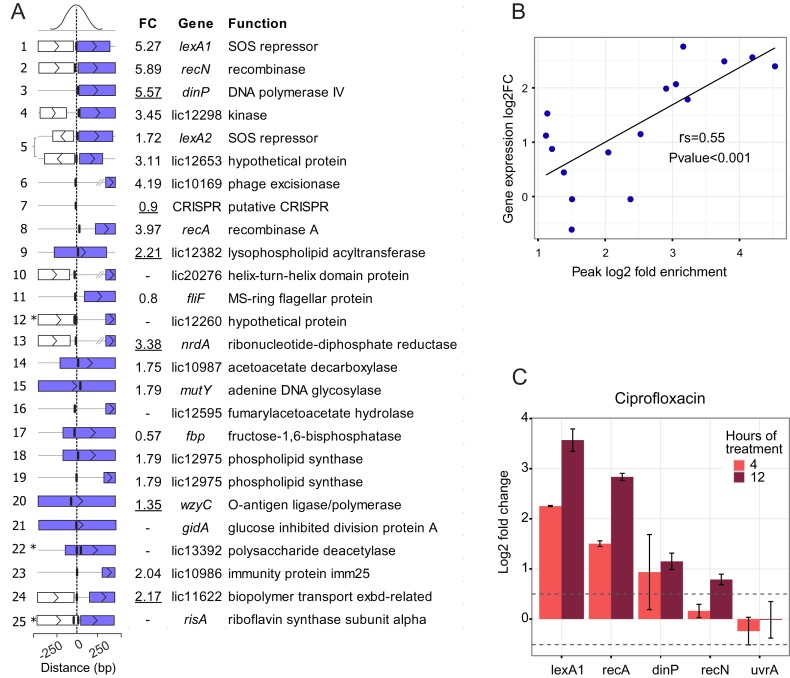
Overview of the results from LexA1 ChIP-seq in L. interrogans serovar Copenhageni. (A) ChIP-seq coverage map, plotted on the y-axis versus chromosomal position on x-axis. All 25 identified peaks are indicated by numbers, ranked from the most to the least enriched in relation to background, as assessed by CisGenome. Identified peaks are indicated above, and peaks associated with previously known targets of LexA1 are named. (B) Validation of ChIP-seq results by ChIP-qPCR of independent samples. Fold enrichment of ChIP-qPCR was calculated using the total sample, and 16S coding sequence as background controls. (C) Binding reactions between labeled recA promoter DNA and LexA1 were challenged by addition of 50-fold excess of unlabeled double-stranded oligonucleotides correspondent to LexA1 binding sites as competitors. The resulting radiographies are tilted 90°, so the rectangles enclose the region where the unbound, outcompeted labeled recA promoter region is expected to be. Signal profiles show the amount of signal present in each lane, and unbound signal is the fraction of total signal that falls within the rectangle. The green profiles correspond to the ones with more than 50% of unbound labeled recA (i.e. LexA1 binds to the tested region), while the blue indicate the ones where the added fragment did not compete. The first and last lanes show binding reactions without competitors added. (D) Sequence logo of the position-specific weight matrix generated by MEME, using the peak sequences. This motif has a P-value of 1.7e-5.
We tested the ability of LexA1 to bind these regions in vivo using gel EMSAs and competitive binding. Purified LexA1 binds to a DNA fragment containing the recA promoter region (Figure 4C and Supplementary Figure S2). To this gel shift assay, we added DNA fragments, each with a different regulatory region that was identified as a possible LexA1 target in vivo. If the fragment was able to compete LexA1 away from the recA promoter region, we inferred that the fragment contained a LexA1 binding site. This competition worked with 9 of the 12 regions identified in vivo. Together, our results indicate that there are at least 15 LexA1 binding sites in L. interrogans. These binding sites include the previously identified sites (recA, recN, lexA1, lexA2, dinP) and 10 additional sites (Figure 4). The regions unable to displace the recA promoter fragment from LexA1 corresponded to peaks 21, 24 and 25 (Figure 4B). These sites may be true binding regions of LexA1, but not strong enough to compete with the binding site in the recA promoter region. Alternatively, LexA1 might need another factor to bind these regions, or the binding detected in vivo might be spurious.
Determination of LexA1 SOS box
The DNA sequences corresponding to each peak of LexA1 binding were used in a motif search using MEME (30). This analysis indicated that the LexA1 binding site is a 16 nt palindrome whose consensus was roughly CTnnACnnnnGTnnAG (Figure 4D). This motif was found at least once per sequence. There were two binding sites less than 15 bp apart for peaks #12 (LIC12660 promoter region), #22 (LIC13392) and #25 (risA) (Supplementary Table S3). This consensus sequence is different from the previously proposed SOS box for leptospira (TTTGN5CAAA). However, the newly proposed consensus is present in all sequences shown to bind to LexA1 in serovar Copenhageni (21) (see also Supplementary Figure S2) and closely overlaps the assigned SOS box upstream of recA in serovar Lai (22). This apparent discrepancy in the new consensus compared to that previously proposed is discussed below (see Discussion).
Almost every peak of LexA1 binding was located upstream of a gene, in potential regulatory regions. Exceptions were peak #5, which was located in the intergenic region between LIC12653 and lexA2 (which are transcribed from opposite strands, see Supplementary Figure S3A) and peaks #18 and #19, located respectively upstream and inside the 5′ end of the LIC12975 coding region (Figure 5A). As already mentioned above, upstream regions of five transcripts were previously shown to be bound by LexA1. The predicted biological function of the remaining genes include: DNA metabolism (mutY, nrdA and LIC20276), phage excision (excisionase) and resistance (CRISPR), synthesis and modification of the membrane and peptideoglycan (LIC13392 and wzyC), central metabolism (LIC12298, LIC12382, LIC10987, LIC12595, fbp, LIC12975 and risA), motility (fliF), biopolymer transport (LIC11622), tRNA modification (gidA) and hypothetical proteins (LIC12260, LIC10986 and LIC11622).
Figure 5.
(A) Genes directly associated with LexA1 binding sites. Peaks are ordered from the most to the least enriched, with the distance from the center of the peak to the next coding region graphically represented. Asterisks indicate peaks containing two closely localized binding sites. Purple rectangles represent the controlled genes, while white ones indicate other ORFs in the vicinity; vertical lines indicate the location of LexA1 binding sites. Differential expression (FC, linear fold change) of genes during the SOS response, as assessed by microarrays or qPCR (underlined values), is shown. (B) Correlation between ChIP-seq peak fold enrichment and log2 fold change of the nearest gene after UV-C damage. Both correlation coefficient (rs) and P-value were computed by Spearman's method. (C) Expression of some core SOS (lexA1, recA, dinP and recN) and not SOS induced (uvrA) genes after 4 or 12h of treatment with 5 μg/ml ciprofloxacin. Values are log2 fold change, calculated by the RT-qPCR 2−ΔΔCt method with six replicates. Error bars represent the standard deviation, and dotted lines comprise fold changes of less than 1.5.
DNA damage-inducible LexA1-bound genes
The expression data were integrated with ChIP-seq results, revealing that 14 LexA1 binding sites were associated with 15 transcriptional units that were up regulated in response to DNA damage (Figure 5A and Supplementary Table S5). Despite the caveats and difficulties of inferring quantitative binding from crosslinking and immunoprecipitation results, there was a significant positive correlation between the fold-change in expression of these genes and the fold enrichment of the LexA binding sites associated with them (Figure 5B). The correlation indicates that stronger binding sites were probably more strongly repressed in normal conditions, and that the amount of derepression was greater than that of genes downstream from weaker binding sites. This finding is in agreement with LexA1 dissociation from binding sites inducing expression of the correspondent genes (53).
Although the majority of binding sites associated with induced genes was located close to putative promoter regions, others were not. In some cases, the binding sites were more than 300 bp upstream of a nearby gene. Some of these genes included nrdA and the putative phage excisionase. In the latter, the LexA1 binding site was more than 500 bp from the coding sequence, 100 bp upstream the putative promoter (Figure 5A). Expression of nrdA could not be determined by microarray, but quantitative RT-PCR confirmed up regulation after UV-C treatment. RT-PCR was also used to confirm the transcriptional response of many of the other genes potentially regulated by LexA1 (Figure 5A, underlined, and Supplementary Table S5).
Seven of the SOS boxes were present inside a coding sequence, namely LIC12382, LIC10987, mutY, fbp, LIC12975, wzyC and gidA (Figure 5A and Supplementary Figure S3). Apart from gidA and fbp, these genes were up regulated after UV-C irradiation, suggesting LexA1 may be able to control gene expression even when bound far from the promoter (Figure 5A). Quantitative RT-PCR performed for wzyC, not represented in the microarrays, showed it is also induced, despite binding of LexA1 more than 1 kb downstream its putative promoter (Supplementary Figure S3). This binding region was also able to compete with recA promoter for binding to LexA1 (Figure 4B) indicating that it is a bona fide binding site. WzyC is responsible for the last step in lipopolysaccharide formation, which is critical for gram-negative pathogen infections.
LexA1-bound genes showing decreased or no differential expression after DNA damage
Expression of the fbp gene, which possesses an SOS box within its coding sequence (Supplementary Figure S3), was repressed during the SOS response, mirroring the expression trend of genes involved in central metabolism. This finding indicated that either LexA1 might be functioning as a transcriptional activator or does not have a regulatory role in this site. LIC20276 and LIC12260, both uninduced after DNA damage, contain a LexA1-binding site, respectively, 140 and 80 bp upstream of their putative promoter, more than 300 bp from the start codon (Supplementary Table S5). There were also five binding sites situated near putative promoters upstream of genes whose expression did not significantly change following DNA damage (Figure 5A). These genes included fliF, coding for the flagella MS ring. These genes may require a combination of stimuli to be induced, due to the binding of additional regulators.
Finally, peak #7 was located in a putative CRISPR (clustered regularly interspaced short palindromic repeats) region (Supplementary Figure S3), an adaptive immunity against phage infection (54). One kilobase downstream of the peak was a complete set of CRISPR-associated (cas) genes, none of which had UV-C-mediated differences in expression (Supplementary Table S3). As usual, this cas cassette was accompanied by an RNA leader with a set of spacers and repeats used to guide the machinery to specific foreign DNA (55–57).
Data available on L. interrogans protein expression during treatment with three different antibiotics—doxycycline, penicillin and ciprofloxacin (37)—show ciprofloxacin elicits a similar response to UV-C. Several components from the prophages were enriched, as well as mismatch repair proteins. As observed for UV-C, pathways involved in cell growth (TCA cycle, fatty acid biosynthesis, glycolysis/gluconeogenesis) and motility (chemotaxis) were repressed (37). To confirm the induction of SOS response, we treated leptospires with 5 μg/ml ciprofloxacin, resulting in 70% survival (data not shown). The mRNA levels from four core SOS genes, namely lexA1, recA, recN and dinP, were measured by RT-qPCR after 4 and 12 h of continuous ciprofloxacin treatment (Figure 5C). The SOS regulators lexA1 and recA were induced after 4 h, while it took 8 h after UV-C irradiation to trigger their induction (21). Levels of dinP and recN also increased. On the other hand, uvrA remained uninduced, once more excluding leptospiral uvrA as a DNA damage inducible gene under such stressing conditions.
DISCUSSION
Although the core SOS regulon is most commonly focused on DNA repair, bacteria can also use it to better adapt to their environment, regulating mutagenesis and expression of virulence factors (11–13). We previously showed that LexA1 is capable of binding to five promoter regions, including the lexA1 and lexA2 promoters (21). Here, by combining ChIP-seq and global gene expression profiling, we expanded this set to 25 genomic loci, associated with at least 25 transcriptional units, the majority of which show increased expression levels after UV-C treatment. The DNA damage induction also caused an enormous rearrangement in the expression landscape, changing mRNA levels of over 500 genes. Several, but not all, LexA1-bound genes were up regulated following DNA damage induction. The remaining genes with expression alteration in response to UV-C might be under indirect SOS control (as a secondary consequence of SOS genes regulation), or might be affected by RecA-independent pathways.
LexA1 binds to 16-nt SOS boxes in both inter- and intragenic sequences
ChIP-seq is a powerful tool to identify DNA targets of transcriptional regulators. Using it in conjunction with de novo motif search, it enabled us to identify in vivo binding sites of LexA1 and its regulon. The resulting SOS box, CTnnACnnnnGTnnAG, differed from the first LexA binding site identified in leptospires by DNAse I footprinting in the recA promoter from the closely related serovar Lai (TTTCGTATACAAA) (22). Despite seeming contradictory at first, the newly identified SOS box actually reconciles both studies on leptospiral SOS box and overlaps with the previously proposed binding site in the recA promoter (Supplementary Figure S2E). The study from Cuñé et al. (22) did not observe LexA binding to its own promoter in vitro, disrupting the paradigm of LexA self-regulation. However, our previous work showed binding of LexA1 not only to its own but also to other gene regulatory regions. In attempt to reconcile both works, it was proposed that the leptospiral LexA protein would display relaxed specificity, allowing it to recognize degenerate SOS boxes (21). In the present work, we identified more binding site sequences, improving the power of motif discovery by computational methods. The ChIP-seq-derived motif overlaps the recA DNAse I-protected region, and it is present in all previously identified sequences able to bind LexA1 (see Supplementary Figure S2). In addition, it is absent from the lexA upstream sequence unable to bind to LexA in serovar Lai (22).
Several LexA1 binding sites were found in regions not suspected to be near promoters, including some far into an open reading frame, including LIC10169 (far upstream), LIC12382, mutY and wzyC (into the open reading frame). There are examples of regulatory protein binding sites that are not in the promoter region. Examples include the pyrimidine catabolism regulator RutR in E. coli (58) and SigF in Mycobacterium tuberculosis (59). In addition, E. coli LexA is capable of binding to an ectopic SOS box inside the coding region of lacZ even in condition of high transcription (60). On the other hand, LexA is able to regulate genes binding to sites 700 bp upstream of the transcriptional start site, as in the case of the hox operon in the cyanobacterium Synechocystis (61). As with other distal binding sites that contribute to LexA-dependent regulation (62,63), LexA1 binding to these distal sites might be involved in DNA looping.
DNA repair, mutagenesis and mobile elements induction in response to DNA damage
In most bacteria, the SOS response induces proteins involved in DNA repair by the nucleotide excision repair pathway (UvrABC) and homologous recombination, represented by the RecABC and RuvABC complexes. This is not the case for L. interrogans serovar Copenhageni, as only recA, recN and mutY are regulated by LexA1. The low number of DNA repair genes activated during the SOS response is not a consequence of genome reduction, as leptospires are significantly enriched in DNA repair-related genes (64). Many of DNA repair genes are up regulated when cells are subjected to stresses imposed by the host (64). Indeed, such results indicate the existence of additional regulators that control DNA repair genes.
The down regulation of uvrA after irradiation and the up regulation of LexA1-bound dinP (the error-prone DNA polymerase IV) and nrdA (ribonucleotide reductase) indicate mutagenic mechanisms might have a special role during the DNA damage response. Pol IV is important for replication through damaged sites, but it is potentially mutagenic, since it can incorporate any base at specific lesion sites and lacks proofreading activity (4,65,66). In E. coli, nrdA expression increases up to 3-fold after UV-C irradiation, causing a 10-fold increase in the dNTP pool. This increase generates a ‘dNTP mutator’ phenotype, characterized by a decrease in fidelity of the replicative polymerase (67). In addition, the structural maintenance of the chromosome and recombination systems also had increased expression. Homologous recombination stabilizes and restores blocked replication forks, in some situations assisted by error-prone DNA polymerases that bypass the damage with promotion of mutagenesis (4).
Several transposons (IS elements) and two defective prophages were induced in L. interrogans after DNA damage. Both prophage regions are under tight regulation and may have roles in specific phases of pathogenesis. They are activated after UV-C or ciprofloxacin treatment (37), and are repressed in leptospires inside the peritoneum of rats (68). Moreover, genome comparison of the culture-attenuated strain 56601 of L. interrogans serovar Lai with its virulent, isogenic parent identified the corresponding prophage components LIC10172 and LIC12611 (LA0202 and LA1056, respectively) as putative virulence factors (46). The up regulation of these two genes was on sharp contrast to the constant or decreased expression of virulence genes we detected after DNA damage.
Possible consequences of leptospiral DNA damage response
While expression of genes in mobile genetic elements and some components of DNA repair systems increased in response to genotoxic stress, genes related to almost every aspect of cell growth and mobility were repressed (Figures 1 and 3). This profile of increased expression in such specific DNA repair genes combined with decreased cell growth and motility is similar to the response of L. interrogans to ciprofloxacin (37,69). We indeed showed induction of four SOS genes in response to this antibiotic, although in early time points than UV-C treatment. This difference is probably a consequence from the different lesions both agents force into DNA: UV-C-induced pyrimidine dimers would only trigger SOS after replication forks arrive at a lesion location, while ciprofloxacin inhibition of DNA gyrase would immediately affect active replisomes (4,70). Although the link between quinolones such as ciprofloxacin and the SOS response is well known, each organism has a specific profile of pathways induced or repressed after ciprofloxacin treatment. For example, in Pseudomonas aeroginosa, all non-direct SOS genes involved in several aspects of DNA metabolism and TCA cycle were down regulated (56), while Staphyloccocus aureus induces the TCA cycle after ciprofloxacin treatment (55), among other differences.
It was also recently shown that SOS-inducing antibiotics have only temporary effects on L. interrogans chronically infecting mice (71). After treatment with ciprofloxacin, the reduced population infecting the kidneys regrows to previous levels, whereas treatment with a non-SOS inducing antibiotic successfully controlled the infection. The early SOS induction in response to ciprofloxacin, shown in the present study, may have a role in this phenomenon. The SOS system and DNA damage response could also be inducing persistence in Leptospira. The slow-growing, less virulent bacteria may be more successful in surviving the antibiotic treatment, and after the response shuts off, they would be able to grow and recolonize the niche.
DNA damage responses may be acting to protect the cell from oxidative damage. The down regulation of TCA cycle enzymes, as well as the electron transport chain, could decrease both production and consumption of NADH+. In addition, some genes from the oxidative damage repair of DNA are active, possibly protecting the genetic material from hydroxyl radical-induced damages. Slowing down the production and consumption of NADH+, through an isocitrate dehydrogenase (icdA) deletion, helps E. coli survive treatment with bactericidal antibiotics in a RecA-dependent way (71–73).
In this work, we integrated powerful techniques for assessment of transcription factor targets and transcriptional profiling to provide an unprecedented view of the DNA damage response in L. interrogans, and the possible role of LexA1 in its regulation. General DNA damage stress response seems to be preferred rather than photolesion-specific repair. The global expression profile shifts from cell growth and virulence toward DNA repair, mutagenesis and mobile genetic elements induction. Some of those effects are related to the LexA1 regulon, which involves several aspects of cell biology. LexA1 binding sites could be found from −500 bp relative to the start codon of DNA damage-induced genes to deep into their coding sequence. Our findings provide support to novel experimental approaches to understand the stress response and its implications during disease development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We would like to thank Carlos Moreira Filho, Silvia Y Bando and Fernanda Bertonha for assistance in microarray samples preparation and hybridization, to Enéas Carvalho for providing the microarrays slides, to Frederico Gueiros-Filho for discussion and Christopher M. Johnson for critical reading of the manuscript.
FUNDING
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP); Fundação Butantan; National Institute of General Medical Sciences of the National Institutes of Health [Award number GM41934 to A.D.G., in part]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. Funding for open access charge: Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Conflict of interest statement. None declared.
REFERENCES
- 1.Schlosser-Silverman E., Elgrably-Weiss M., Rosenshine I., Kohen R., Altuvia S. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 2000;182:5225–5230. doi: 10.1128/jb.182.18.5225-5230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baharoglu Z., Mazel D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 2014;38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 3.Erill I., Campoy S., Barbé J. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev. 2007;31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA repair and mutagenesis. 2nd edn. Washington: ASM Press; 2006. [Google Scholar]
- 5.Hewat E.A., Ruigrok R.W., DiCapua E. Activation of recA protein: the pitch of the helical complex with single-stranded DNA. EMBO J. 1991;10:2695–2698. doi: 10.1002/j.1460-2075.1991.tb07813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Little J.W. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. U.S.A. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butala M., Zgur-Bertok D., Busby S.J. The bacterial LexA transcriptional repressor. Cell Mol. Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Rocha R.P., de Miranda Paquola A.C., do Valle Marques M., Menck C.F.M., Galhardo R.S. Characterization of the SOS regulon of Caulobacter Crescentus. J. Bacteriol. 2008;190:1209–1218. doi: 10.1128/JB.01419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truglio J.J., Croteau D.L., Van Houten B., Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 10.Goranov A.I., Kuester-Schoeck E., Wang J.D., Grossman A.D. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J. Bacteriol. 2006;188:5595–5605. doi: 10.1128/JB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toshima H., Yoshimura A., Arikawa K., Hidaka A., Ogasawara J., Hase A., Masaki H., Nishikawa Y. Enhancement of Shiga toxin production in Enterohemorrhagic Escherichia coli serotype O157:H7 by DNase colicins. Appl. Environ. Microbiol. 2007;73:7582–7588. doi: 10.1128/AEM.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Veen S., van Schalkwijk S., Molenaar D., de Vos W.M., Abee T., Wells-Bennik M.H.J. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology. 2010;156:374–384. doi: 10.1099/mic.0.035196-0. [DOI] [PubMed] [Google Scholar]
- 13.Mellies J.L., Haack K.R., Galligan D.C. SOS regulation of the type III secretion system of enteropathogenic Escherichia coli. J. Bacteriol. 2007;189:2863–2872. doi: 10.1128/JB.01859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ubeda C., Maiques E., Tormo M.A., Campoy S., Lasa I., Barbe J., Novick R.P., Penades J.R. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol. Microbiol. 2007;65:41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 15.Guerin E., Cambray G., Sanchez-Alberola N., Campoy S., Erill I., Da Re S., Gonzalez-Zorn B., Barbe J., Ploy M.C., Mazel D. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 16.Abela-Ridder B., Bertherat E., Durski K. Prince Mahidol Award Conference2013. Bangkok, Thailand: Prince Mahidol Award Conference; 2013. Global burden of Human Leptospirosis and cross-sectoral interventions for its prevention and control. [Google Scholar]
- 17.Faine S. In: Bacterial Infection of Humans: Epidemiology and Control. 3rd edn. Evans AS, Brachman PS, editors. Nova Iorque: Springer; 1998. pp. 395–420. [Google Scholar]
- 18.Faine S., Adler B., Bolim C., Perolat P. Leptospira and leptospirosis. 2nd edn. Melbourne, Australia: MediSci; 1999. p. 296. [Google Scholar]
- 19.Ko A.I., Galvao Reis M., Ribeiro Dourado C.M., Johnson W.D., Jr, Riley L.W. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet. 1999;354:820–825. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- 20.Levett P.N. Leptospirosis. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonseca L.S., da Silva J.B., Milanez J.S., Monteiro-Vitorello C.B., Momo L., de Morais Z.M., Vasconcellos S.A., Marques M.V., Ho P.L., da Costa R.M.A. Leptospira interrogans serovar Copenhageni Harbors Two lexA Genes Involved in SOS Response. PLoS One. 2013;8:e76419. doi: 10.1371/journal.pone.0076419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuñé J., Cullen P.A., Mazon G., Campoy S., Adler B., Barbe J. The Leptospira interrogans lexA gene is not autoregulated. J. Bacteriol. 2005;187:5841–5845. doi: 10.1128/JB.187.16.5841-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abella M., Campoy S., Erill I., Rojo F., Barbe J. Cohabitation of two different LexA regulons in Pseudomonas putida. J. Bacteriol. 2007;189:8855–8862. doi: 10.1128/JB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray G.L., Morel V., Cerqueira G.M., Croda J., Srikram A., Henry R., Ko A.I., Dellagostin O.A., Bulach D.M., Sermswan R.W., et al. Genome-Wide Transposon Mutagenesis in Pathogenic Leptospira Species. Infect. Immun. 2009;77:810–816. doi: 10.1128/IAI.01293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellinghausen H.C., McCullough W.G. Albumin fatty acid broth for Leptospira, modified by Johnson and Harris. J. Bacteriol. 1967;94:27–31. [Google Scholar]
- 26.da Silva J.B., Carvalho E., Covarrubias A.E., Ching A.T.C., Mattaraia V.G.M., Paiva D., de Franco M., Fávaro R.D., Pereira M.M., Vasconcellos S., et al. Induction of TNF-alfa and CXCL-2 mRNAs in different organs of mice infected with pathogenic Leptospira. Microbiol. Pathog. 2012;52:206–216. doi: 10.1016/j.micpath.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Lin D.C.-H., Grossman A.D. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 28.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji H., Jiang H., Ma W., Wong W.H. Using CisGenome to analyze ChIP-chip and ChIP-seq data. Curr. Protoc. Bioinformatics. 2011;33:2.13.1–2.13.45. doi: 10.1002/0471250953.bi0213s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin J.-H., Zhang Q., Zhang Z.-M., Zhong Y., Yang Y., Hu B.-Y., Zhao G.-P., Guo X.-K. Identification of a novel prophage-like gene cluster actively expressed in both virulent and avirulent strains of Leptospira interrogans serovar lai. Infect. Immun. 2008;76:2411–2419. doi: 10.1128/IAI.01730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bensing B.A., Siboo I.R., Sullam P.M. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect. Immun. 2001;69:6186–6192. doi: 10.1128/IAI.69.10.6186-6192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franz B., Landy A. The Holliday junction intermediates of lambda integrative and excisive recombination respond differently to the bending proteins integration host factor and excisionase. EMBO J. 1995;14:397–406. doi: 10.1002/j.1460-2075.1995.tb07014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cromie G.A., Connelly J.C., Leach D.R.F. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita E., van der Linden E., Sanchez H., Wyman C. RAD50, an SMC family member with multiple roles in DNA break repair: how does ATP affect function. Chromosome Res. 2009;17:277–288. doi: 10.1007/s10577-008-9018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt A., Beck M., Malmström J., Lam H., Claassen M., Campbell D., Aebersold R. Absolute quantification of microbial proteomes at different states by directed mass spectrometry. Mol. Syst. Biol. 2011;7:510. doi: 10.1038/msb.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert A., Picardeau M., Haake D.A., Sermswan R.W., Srikram A., Adler B., Murray G.A. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of the flagellum sheath. Infect. Immun. 2012;80:2019–2025. doi: 10.1128/IAI.00131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King A.M., Pretre G., Bartpho T., Sermswan R.W., Toma C., Suzuki T., Eshghi A., Picardeau M., Adler B., Murray G.L. High-temperature protein G is an essential virulence factor of Leptospira interrogans. Infect. Immun. 2014;82:1123–1131. doi: 10.1128/IAI.01546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eshghi A., Lourdault K., Murray G.L., Bartpho T., Sermswan R.W., Picardeau M., Adler B., Snarr B., Zuerner R.L., Cameron C.E. Leptospira interrogans catalase is required for resistance to H2O2 and for virulence. Infect. Immun. 2012;80:3892–3899. doi: 10.1128/IAI.00466-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao S., Sun A., Ojcius D., Wu S., Zhao J., Yan J. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 2009;9:253–262. doi: 10.1186/1471-2180-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lourdault K., Cerqueira G.M., Wunder E.A., Jr, Picardeau M. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 2011;79:3711–3717. doi: 10.1128/IAI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray G.L., Srikram A., Henry R., Hartskeerl R.A., Sermswan R.W., Adler B. Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol. Microbiol. 2010;78:701–709. doi: 10.1111/j.1365-2958.2010.07360.x. [DOI] [PubMed] [Google Scholar]
- 44.Ristow P., Bourhy P., McBride F.v.W.d.C., Figueira C.P., Huerre M., Ave P., Girons I.S., Ko A.I., Picardeau M. The OmpA-like protein Loa22 is essential for Leptospiral virulence. PLoS Pathog. 2007;3:e97. doi: 10.1371/journal.ppat.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong Y., Chang X., Cao X.-J., Zhang Y., Zheng H., Zhu Y., Cai C., Cui Z., Zhang Y., Li Y.-Y., et al. Comparative proteogenomic analysis of the Leptospira interrogans virulence-attenuated strain IPAV against the pathogenic strain 56601. Cell Res. 2011;21:1210–1229. doi: 10.1038/cr.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehmann J.S., Fouts D.E., Haft D.H., Cannella A.P., Ricaldi J.N., Brinkac L., Harkins D., Durkin S., Sanka R., Sutton G., et al. Pathogenomic inference of virulence-associated genes in Leptospira interrogans. PLoS Negl. Trop. Dis. 2013;7:e2468. doi: 10.1371/journal.pntd.0002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toma C., Murray G.L., Nohara T., Mizuyama M., Koizumi N., Adler B., Suzuki T. Leptospiral outer membrane protein LMB216 is involved in enhancement of phagocytic uptake by macrophages. Cell. Microbiol. 2014;16:1366–1377. doi: 10.1111/cmi.12296. [DOI] [PubMed] [Google Scholar]
- 48.Ching A.T., Favaro R.D., Lima S.S., Chaves Ade A., de Lima M.A., Nader H.B., Abreu P.A., Ho P.L. Leptospira interrogans shotgun phage display identified LigB as a heparin-binding protein. Biochem. Biophys. Res. Commun. 2012;427:774–779. doi: 10.1016/j.bbrc.2012.09.137. [DOI] [PubMed] [Google Scholar]
- 49.Evangelista K.V., Hahn B., Wunder E.A., Jr, Ko A.I., Haake D.A., Coburn J. Identification of Cell-Binding Adhesins of Leptospira interrogans. PLoS Negl. Trop. Dis. 2014;8:e3215. doi: 10.1371/journal.pntd.0003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Figueira C.P., Croda J., Choy H.A., Haake D.A., Reis M.G., Ko A.I., Picardeau M. Heterologous expression of pathogen-specific genes ligA and ligB in the saprophyte Leptospira biflexa confers enhanced adhesion to cultured cells and fibronectin. BMC Microbiol. 2011;11:129–129. doi: 10.1186/1471-2180-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcsisin R.A., Bartpho T., Bulach D.M., Srikram A., Sermswan R.W., Adler B., Murray G.L. Use of a high-throughput screen to identify Leptospira mutants unable to colonise the carrier host or cause disease in the acute model of infection. J. Med. Microbiol. 2013 doi: 10.1099/jmm.0.058586-0. [DOI] [PubMed] [Google Scholar]
- 52.Pinne M., Matsunaga J., Haake D. Leptospiral outer membrane protein microarray, a novel approach to identification of host ligand-binding proteins. J. Bacteriol. 2012;194:6074–6087. doi: 10.1128/JB.01119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butala M., Klose D., Hodnik V., Rems A., Podlesek Z., Klare J.P., Anderluh G., Busby S.J.W., Steinhoff H.-J., Zgur-Bertok D. Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res. 2011:1–12. doi: 10.1093/nar/gkr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cirz R.T., Jones M.B., Gingles N.A., Minogue T.D., Jarrahi B., Peterson S.N., Romesberg F.E. Complete and SOS-mediated response of staphylococcus aureus to the antibiotic Ciprofloxacin. J. Bacteriol. 2007;189:531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cirz R.T., O'Neill B.M., Hammond J.A., Head S.R., Romesberg F.E. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic Ciprofloxacin. J. Bacteriol. 2006;188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bisognano C., Kelley W.L., Estoppey T., Francois P., Schrenzel J., Li D., Lew D.P., Hooper D.C., Cheung A.L., Vaudaux P. A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 2004;279:9064–9071. doi: 10.1074/jbc.M309836200. [DOI] [PubMed] [Google Scholar]
- 58.Shimada T., Ishihama A., Busby S.J.W., Grainger D.C. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008;36:3950–3955. doi: 10.1093/nar/gkn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartkoorn R.C., Sala C., Uplekar S., Busso P., Rougemont J., Cole S.T. Genome-wide definition of the SigF Regulon in Mycobacterium tuberculosis. J. Bacteriol. 2012;194:2001–2009. doi: 10.1128/JB.06692-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wade J.T., Reppas N.B., Church G.M., Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutekunst K., Phunpruch S., Schwarz C., Schuchardt S., Schulz-Friedrich R., Appel J. LexA regulates the bidirectional hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 as a transcription activator. Mol. Microbiol. 2005;58:810–823. doi: 10.1111/j.1365-2958.2005.04867.x. [DOI] [PubMed] [Google Scholar]
- 62.Smollett K.L., Smith K.M., Kahramanoglou C., Arnvig K.B., Buxton R.S., Davis E.O. Global analysis of the regulon of the transcriptional repressor LexA, a key component of the SOS response in Mycobacterium tuberculosis. J. Biol. Chem. 2012;287:22004–22014. doi: 10.1074/jbc.M112.357715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haydel S.E., Clark-Curtiss J.E. The Mycobacterium tuberculosis TrcR response regulator represses transcription of the intracellularly expressed Rv1057 gene, encoding a seven-bladed β-propeller. J. Bacteriol. 2006;188:150–159. doi: 10.1128/JB.188.1.150-159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martins-Pinheiro M., Schons-Fonseca L., da Silva J., Domingos R., Momo L., Simões A., Ho P., da Costa R.A. Genomic survey and expression analysis of DNA repair genes in the genus Leptospira. Mol. Genet. Genomics. 2015 doi: 10.1007/s00438-015-1135-2. doi:10.1007/s00438-015-1135-2. [DOI] [PubMed] [Google Scholar]
- 65.Ohmori H., Friedberg E.C., Fuchs R.P.P., Goodman M.F., Hanaoka F., Hinkle D., Kunkel T.A., Lawrence C.W., Livneh Z., Nohmi T., et al. The Y-Family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 66.Goodman M.F., Woodgate R. Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gon S., Napolitano R., Rocha W., Coulon S., Fuchs R.P. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caimano M.J., Sivasankaran S.K., Allard A., Hurley D., Hokamp K., Grassmann A.A., Hinton J.C.D., Nally J.E. A model system for studying the transcriptomic and physiological changes associated with mammalian host-adaptation by Leptospira interrogans Serovar Copenhageni. PLoS Pathog. 2014;10:e1004004. doi: 10.1371/journal.ppat.1004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malmstrom J., Beck M., Schmidt A., Lange V., Deutsch E.W., Aebersold R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hiasa H., Shea M.E. DNA Gyrase-mediated wrapping of the DNA strand is required for the replication fork arrest by the DNA Gyrase-Quinolone-DNA ternary complex. J. Biol. Chem. 2000;275:34780–34786. doi: 10.1074/jbc.M001608200. [DOI] [PubMed] [Google Scholar]
- 71.Ratet G., Veyrier F.J., Fanton d'Andon M., Kammerscheit X., Nicola M.-A., Picardeau M., Boneca I.G., Werts C. Live imaging of bioluminescent Leptospira interrogans in mice reveals renal colonization as a stealth escape from the blood defenses and antibiotics. PLoS Negl. Trop. Dis. 2014;8:e3359. doi: 10.1371/journal.pntd.0003359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 73.Bos J., Zhang Q., Vyawahare S., Rogers E., Rosenberg S.M., Austin R.H. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc. Natl. Acad. Sci. U.S.A. 2015;112:178–183. doi: 10.1073/pnas.1420702111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.