Abstract
Signal Transducers and Activators of Transcription (STATs) are principal transcription factors downstream of cytokine receptors. Although STAT5A is expressed in most tissues it remains to be understood why its premier, non-redundant functions are restricted to prolactin-induced mammary gland development and function. We report that the ubiquitously expressed Stat5a/b locus is subject to additional lineage-specific transcriptional control in mammary epithelium. Genome-wide surveys of epigenetic status and transcription factor occupancy uncovered a putative mammary-specific enhancer within the intergenic sequences separating the two Stat5 genes. This region exhibited several hallmarks of genomic enhancers, including DNaseI hypersensitivity, H3K27 acetylation and binding by GR, NFIB, ELF5 and MED1. Mammary-specific STAT5 binding was obtained at two canonical STAT5 binding motifs. CRISPR/Cas9-mediated genome editing was used to delete these sites in mice and determine their biological function. Mutant animals exhibited an 80% reduction of Stat5 levels in mammary epithelium and a concomitant reduction of STAT5-dependent gene expression. Transcriptome analysis identified a class of mammary-restricted genes that was particularly dependent on high STAT5 levels as a result of the intergenic enhancer. Taken together, the mammary-specific enhancer enables a positive feedback circuit that contributes to the remarkable abundance of STAT5 and, in turn, to the efficacy of STAT5-dependent mammary physiology.
INTRODUCTION
Cells receive a wide range of extracellular signals, which are transmitted through receptors and transcription factors to elicit genomic responses. The six members of the family of Signal Transducers and Activators of Transcription (STATs) are the principal transcription factors conveying signals of most, if not all, cytokines and peptide hormones, such as interleukins, growth hormone and prolactin (1–3). STATs thereby permit cells to adapt and respond to a wide range of extracellular cues. While STATs in general modulate the biology of existing cells, it remains to be determined why the establishment of two unique lineages, mammary alveolar cells (4,5) and T cells (6) is dependent on STAT5A/B (referred to as STAT5), the most promiscuous members of the STAT family. Mammary alveoli are distinct from other cells in that their formation, proliferation and function are exclusively controlled by prolactin, the key activator of STAT5.
The Stat5 locus encodes two genes, Stat5a and Stat5b that are positioned in a head-to-head orientation and separated by ∼10 kb (7). Although STAT5A and STAT5B are thought to be functionally redundant, differences in the phenotypes in mice lacking one or the other suggest paralog specific functions. Alternatively, and more likely, cell-specific abundance of either isoform could contribute to the observed phenotypes (8). Inactivation of the entire Stat5a/b locus in mice has revealed its importance in the establishment of functional mammary alveoli (4,9) as well as T cells (6,10). In other cell types, such as hepatocytes and muscle, STAT5 serves a more modest and modulating role (11–14).
Mammary alveolar epithelium is a cyclical organ that is generated de novo with each pregnancy, with the sole purpose to produce large quantities of milk to nourish offspring. Proliferation and differentiation of alveoli during pregnancy are controlled by prolactin (PRL) mainly through STAT5A (15). In contrast, body growth (16,17) and development and expansion of T cells (16,18) are controlled mainly through STAT5B. Loss-of-function mutations in human Stat5b also result in short stature and immunological dysfunction (16). The unique and differential contribution of STAT5A and STAT5B to mammary epithelium and T cells is likely the consequence of their relative abundance in these cell types. While STAT5A and STAT5B are present at low levels in most cell types, high levels of STAT5A are found specifically in mammary tissue and high STAT5B levels in immune cells. The relative abundance of these two isoforms is reflected by distinct defects in mice carrying mutations in either gene (8). Genetic studies in mice have also demonstrated the presence of differentiation-specific gene classes that respond to different STAT5 concentrations (8). Notably, expression of several mammary-specific genes encoding milk proteins is greatly reduced in mice carrying only two Stat5b alleles (Stat5a-null) (15). This leads to a compromised first lactation, which, however, is compensated for after subsequent pregnancies as a result of increased expression of Stat5b (19). The presence of exceptional high levels of STAT5A in mammary tissue suggests that the corresponding gene is tightly regulated. However, the underlying mechanism is not understood.
While studies using mouse genetics have suggested similar, if not identical, functions between STAT5A and STAT5B, a wealth of in vitro studies have eluded to unique and distinct functions of these two isoforms. Isoform-specific knockdown studies in cell lines have led to the identification of specific target genes and non-canonical STAT5 binding sites (20). Esr1 and Esr2 were among differential target genes activated by a constitutively active PRLR (21) and it has also been shown that STAT5A and STAT5B can bind to unique targets with different kinetics (22). Differences in serine phosphorylation between the two proteins have been described but their biological significance is not clear (23). While in most cases individual GAS motifs are associated with regulatory sequences and are recognized by STAT5 dimers, in a limited number of genes STAT5 tetramers bind juxtaposed GAS motifs (24). Biological significance for such tetramers comes from mouse studies in which the tetramerization sequence in the two STAT5 isoforms had been disrupted (25).
Genome-wide analyses of histone modifications and transcription factor binding have provided tools to predict the presence of regulatory elements (26) and RNA-guided genome editing (27) can be used to validate their existence. This study was designed to explore the possibility that transcription of the Stat5 locus is under control of a mammary-specific enhancer, which would ensure high levels of STAT5 needed for functional development of mammary epithelium during pregnancy and the activation of differentiation-associated genes. We used DNaseI hypsersensitivity, H3K27ac patterns and transcription factor binding to identify putative enhancers and CRISPR/Cas9-mediated mutations to investigate their biological significance.
MATERIALS AND METHODS
BioGPS gene expression analyses
To determine the Stat5a and Stat5b gene expression profiles across tissues, microarray expression data of Stat5a and Stat5b in 49 different mouse tissues from GeneAtlas MOE430 gcrma dataset were downloaded using the BioGPS Gene Expression Database (http://biogps.org). The gcRMA-processed values of the Stat5a and Stat5b transcripts from seven representative tissues were analyzed by GraphPad Prism software.
DNase I hypersensitive sites sequencing (DNase-Seq)
DNase-Seq in mammary gland was done following published method with several modifications (28). Briefly, L1 mammary gland was isolated and snap frozen in liquid nitrogen. Frozen tissue was grinded into powder and homogenized with buffer A (15 mM Tris–HCl pH 8.0, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.5 mM EGTA, 0.15 mM Spermine, 0.5 mM Spermidine, 0.5 mM DTT, 1 mM PMSF with proteinase inhibitors). After cells were lysed in buffer A supplemented with 0.2% NP40, nuclei were collected, counted and re-suspended in DNase buffer. 10 U DNase I (New England Biolab) was used to digest 10 million nuclei at 37°C for 5 min followed by proteinase K digestion. Genomic DNA was then purified. Approximately, 50–100 bp DNA fragments were selected for further library construction and sequencing.
Chromatin immunoprecipitation coupled by parallel sequencing (ChIP-seq)
Frozen-stored mammary tissues collected from lactation day 1 (L1) were ground into powder with mortar and pestle and then crosslinked with 1% formaldehyde (Sigma–Aldrich) for 10 min. After adding 0.125 M glycine to stop crosslinking, nuclei were isolated with Farnham Lysis Buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40, supplemented with PMSF and proteinase inhibitor cocktails). The chromatin was fragmented to 200–500 bp using sonicator 3000 (25 cycles; 30 s pulse/30 s rest, Misonix Sonicators) and further lysed in RIPA buffer. One milligram of chromatin was immunoprecipitated with Dynabeads Protein A (Novex) coated with anti-H3K4me3 (Millipore, 17-614), anti-H3K27ac (Abcam, ab4729), anti-RNA Polymerase II (Abcam, ab5408), anti-MED1 (Bethyl Laboratory, A300-793A), or anti-STAT5A (Santa Cruz, sc-1081). After serial bead washes, ChIP DNA was reverse-crosslinked at 65°C overnight in the presence of 1% SDS and 1 mg/ml of Proteinase K (Roche), and DNA was purified with QIAquick PCR Purification Kit (Qiagen). The DNA fragments were blunt-ended using End-it DNA End-Repair Kit (Epicentre Biotechnology), ligated to the Illumina Indexed DNA adaptors, and sequenced with HiSeq 2000 (Illumina).
Processing sequence data
ChIP-Seq and DNase-Seq data were mapped to the reference genome mm9 using Bowtie2 aligner (29) and visualized using HOMER (http://homer.salk.edu/homer/) and IGV (30). GAS motifs were searched with IGV browser. For visualization, the total reads number of mapped result in each sample was normalized to 10 million and background signals of <2 were eliminated. RNA-Seq data was mapped to the reference genome mm9 using STAR aligner (31). To compare sample expression levels, cufflinks program (32) was used to generate FPKM values. Transcripts with FPKM >1 were kept for further analysis. Fold change and variance were calculated using DESeq2 package in R (33). Transcripts with over 2-fold change and significant pairwise variance (P < 0.05) are classified as differentially expressed genes. NGS data sets have been deposited at GEO (GSE 72724).
GSEA analysis was performed as described (34). Briefly, unabridged RNA-Seq datasets were used to compute a ranked list of genes that were differentially expressed in GAS2 site mutant relative to wild type controls. Enrichment was calculated against a user generated gene set that included 199 genes (see Results section). Enrichment score curves and member ranks were generated by the GSEA software package (Broad Institute, Cambridge, MA, USA). Volcano plot was generated with DataGraph (Visual Data Tools, Inc.). Heatmap was generated with gplots package in R.
Statistical analyses
Data were presented as SEM in each group and were evaluated with a two-tailed, unpaired Student's t-test using GraphPad Prism. Statistical significance was obtained by comparing all groups to Wild Type. Asterisks denote significant differences (P < 0.05) between mutants and Wild type controls.
CRISPR/Cas9 plasmid and sgRNA preparation
The CRISPR sgRNA constructs were designed based on their proximity to the mutation sites and their off-target scores (calculated by the online tool at crispr.mit.edu). Specifically, the 5′-ctaTTCCCAGAAacaaagga-3′ and 5′-CTGAAgagtgttagagacca-3′ sequences were separately cloned into the pDR274 vector (Addgene #42250), and injectable RNAs were in vitro transcribed using the MEGAshortscript T7 kit (Life Technologies). Cas9 mRNA was in vitro transcribed from plasmid MLM3613 (Addgene #42251) using the mMESSAGE mMACHINE T7 kit (Life Technologies).
Generation of CRISPR/Cas9 targeted mice
Mice used in this study were handled and housed in accordance with NIH guidelines. Animal experiments were approved by the NIDDK Animal Care and Use Committee. Zygote preparation and microinjection were performed as previously described (35). Superovulated B6CBAF1/J female mice (JAX) were mated with B6CBAF1/J males, and fertilized eggs were collected from their oviducts. For microinjection, 100 ng/μl of Cas9 mRNA and 50 ng/μl of each sgRNA in nuclease-free microinjection buffer (10 mM Tris, pH 7.5, 0.1 mM EDTA) were microinjected into the cytoplasm of fertilized eggs. Injected zygotes were cultured overnight in M16 medium at 37°C in 5% CO2. The next morning, those embryos that had reached the 2-cell stage were implanted into oviducts of pseudopregnant fosters (Swiss Webster, Taconic Farm). Of 157 microinjected mouse eggs, 46 live pups were born and genotyped using PCR and DNA sequencing. Seven mosaic founder mice with mutations outside the GAS sites were not pursued in this study due to ambiguous allelic overlapping in chromatograms (19).
Genotyping founder mice and breeding to homozygosity
Pups were genotyped by PCR of genomic DNA from mouse tail tips and subsequent Sanger sequencing. Genomic DNA was purified using the Wizard Genomic DNA Purification Kit (Promega, A1120) according to the manufacturer's instructions. Fifty nanograms of genomic DNA were PCR-amplified in a 50 μl reaction using DreamTaq Green PCR Master Mix (Life Technologies, K1071) with primers flanking the GAS sites. PCR forward primer: 5′-TGTGTGCCTAGAGTACCTGACC-3′; reverse primer: 5′-GATCATGAAGCCAGAATAGGC-3′. Primers used for Sanger sequencing: forward: 5′-TGGTCTCTCATCTGGGTAGG-3′; reverse: 5′-GTGTGTTCTCTGTAAATGC-3′.
Of the 46 F0 mice, three mice carrying different mutations were bred to C57BL/6 wild-type mice to generate F1 heterozygote mice. Heterozygous F1 mice were then inter-bred to generate mice homozygous for the various GAS mutations. This study focused on three mouse lines carrying unique GAS mutations. All mice were genotyped by PCR amplification of genomic DNA purified from mouse tails followed by Sanger sequencing as previously above.
Isolation of RNA and quantitative RT-PCR
RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized from total RNA using Superscript II (Invitrogen) and quantitative PCR was performed with gene-specific primers.
Stat5a (213 bp): The primers span exons 16–18. Forward: 5′-ATTACACTCCTGTACTTGCGA-3′; Reverse: 5′-GGTCAAACTCGCCATCTTGG-3′.
Stat5b: (313 bp) The primers span from exon 16 to exon 19. Forward: 5′-TCCCCTGTGAGCCCGCAAC-3′; Reverse: 5′-GGTGAGGTCTGGTCATGACT-3′
Csn2 (334 bp): The primers span exon 7. Forward: 5′-ACTCCAGCATCCAGTCACAGC-3′; Reverse: 5′-AGATGAGTCTGAGGAAAAGCC-3′
Csn1s2b (182 bp): The primers span from exon 6 to exon 11. Forward: 5′-CATTCCCCAAATGCAATCTG-3′; Reverse: 5′-TTACAAACTGTGGCCAGGTC-3′
Wap (156 bp) The primers span from exon 2 to exon 4. Forward: 5′-CCAGAATGCCATGTGCTGTC-3′; Reverse: 5′-CGGTCGCTGGAGCATTCTAT-3′
Socs2: (186 bp) The primers span exon 3. Forward: 5′-TCAGCTGGACCGACTAACCT-3′; Reverse: 5′-CAGGTGAACAGTCCCATTCC-3′
Gapdh: (199 bp) The primers span exon 6 to exon 7. Forward: 5′-TTGTCAAGCTCATTTCCTGGT-3′; Reverse: 5′-TTACTCCTTGGAGGCCATGTA-3′
Real-time PCR was carried out using the BioRad CFX96 Real-Time PCR Detection System (185-5196; BioRad). Individual PCRs were performed in triplicate using the reference gene Gapdh for normalization. The threshold cycle (Cq) was determined by default settings. Relative gene expression (as a fold change) was calculated with the 2−ΔΔCq method. Each run was completed with a melting curve analyses to confirm the specificity of the amplification and the absence of primer dimers.
RNA-sequencing (RNA-seq)
Total RNA was isolated from mouse mammary tissues at L1 using RNeasy Plus Mini Kit (Qiagen). One microgram of total RNA was purified to Poly (A) RNA, synthesized to cDNA, and prepared to RNA-seq library using a TruSeq RNA Sample Preparation Kit v2 (Illumina) according to the manufacturer's protocol. The quantity and the quality of libraries were assessed using Quant-iT Picogreen dsDNA reagent (Invitrogen) and Agilent High Sensitivity DNA kit (Agilent Biotechnology) and sequenced with HiSeq 2000 system (Illumina).
RESULTS
Cell-specific features of the Stat5 locus
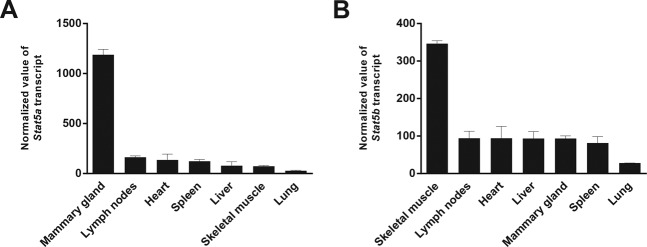
Although Stat5a mRNA is found in most, if not all, cell types, its levels are highest in lactating mammary tissue, exceeding expression in other tissues, such as lung, by up to 40-fold (http://biogps.org/#goto=genereport&id=20850) (Figure 1). These microarray-based data sets were complemented with RNA-seq experiments from liver and lactating mammary tissue. In mammary tissue 56 Stat5a FPKM and 20 Stat5b FPKM were detected (GSE37646). This contrasts two Stat5a and four Stat5b FPKM in liver. In fact, this relative abundance facilitated the discovery of STAT5A, which was originally cloned from sheep mammary tissue (36) and subsequently together with STAT5B from mouse mammary mRNA (37). It is also notable that, in contrast to most other tissues (e.g. muscle, spleen) Stat5a is more abundant than Stat5b in mammary epithelium. The high expression levels in mammary epithelium suggest the possibility that the Stat5 locus (Figure 2) is subject to cell-specific transcriptional control.
Figure 1.
Stat5 gene expression levels across tissues. Stat5a and Stat5b mRNA levels in different tissues were based on microarray data sets and were obtained from GNF Mouse GeneAtlas V3 (GSE10246) using open access BioGPS transcriptome database. The normalized values of the Stat5a and Stat5b transcripts from seven representative tissues were graphically plotted using GraphPad Prism software. Stat5a was predominantly expressed in mammary tissue whereas Stat5b was highly expressed in skeletal muscles compared to other tissues.
Figure 2.
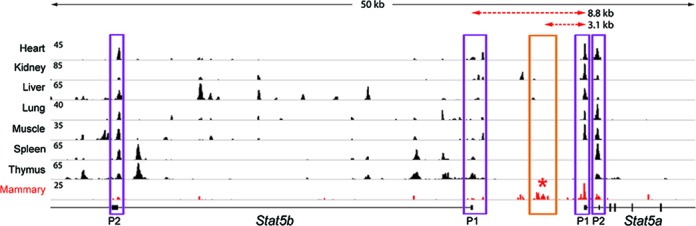
Mammary-specific DNaseI hypersensitive sites in the Stat5 locus. DNase-seq data sets from seven published studies (GSE37074) and from mammary tissue at day 1 of lactation (L1) were analyzed. The Stat5a gene has two promoters, which display differential DHS. While DHS in spleen and thymus is restricted to P2, both P1 and P2 are hypersensitive in heart, lung, liver, kidney and mammary glands. DHS over the distal promoter (P1) of the Stat5b gene was strongest in spleen and thymus, which express high levels of Stat5b. DHS sites were detected in the intergenic region ∼3.5 kb upstream of the Stat5a gene in mammary tissue but not in other cell types. This region coincides with mammary-specific STAT5 binding (Figure 3). The promoters and the intergenic region are marked by rectangles.
DNaseI hypersensitivity is commonly used to identify cis-regulatory DNA elements. To discover putative mammary-specific regulatory elements, DNase-seq was performed on mammary tissue at day 1 of lactation (L1) and the resulting hypersensitivity maps were compared to ENCODE DNase-seq data sets (GSE37074) from seven different tissues. The Stat5a gene features two distinct promoters with distinct transcriptional start sites (TSS) (Figure 2). DHS is evident at both P1 and P2 within heart, kidney, liver, lung, muscle and lactating mammary, but is restricted to P2 within spleen and thymus. The Stat5b gene also contains two promoters that are separated by ∼30 kb, with P2 displaying stronger DHS than P1 in all tissues. Notably, a mammary-specific DHS area was detected in the intergenic region ∼3.5 kb upstream of the Stat5a gene (Figure 2), suggesting the presence of relevant cell-specific regulatory elements. Although tissue culture cells are frequently used to explore molecular mechanisms underlying cell-specific gene regulation, mammary cell lines are suboptimal as expression of bona fide mammary-specific genes, i.e. milk protein genes, is generally not detected. Notably, DHS sites associated with mammary-specific regulatory elements are not present in the mammary cell line 3134 (38) (Supplementary Figure S1) further supporting their biological relevance in the intact gland.
ChIP-seq experiments from L1 mammary tissue were used to collect additional cues for a putative mammary-specific regulatory region (Figure 3A). H3K4me3 marks coincided with Stat5a and Stat5b promoter regions (37). The presence of H3K27ac marks was used as another indicator of putative regulatory sequences. In addition to covering promoter sequences, the mammary-specific DHS region 3.5 kb upstream of the Stat5a TSS was also enriched for H3K27ac marks (Figure 3A). A search for putative transcription factor binding sites revealed two GAS motifs in this region and strong binding of STAT5A was observed in mammary tissue from lactating mice (Figure 3A). Binding of the transcriptional co-activator MED1 coincided with STAT5 occupancy. The H3K27ac pattern was mutually exclusive to that of STAT5 binding (Figure 3B), suggesting the exclusion of modified histones from STAT5 binding sites. RNA Polymerase II (Pol II) loading was observed preferentially on the Stat5a gene, which reflects its preferential expression in mammary tissue. Pol II binding was also detected in the region associated with mammary-specific H3K27ac, the DHS and STAT5 binding (Figure 3), further indicating the presence of a bona fide enhancer. In contrast to mammary tissue no H3K27ac and no STAT5 binding were detected in T cells (GSE36890 and GSE60353) and liver (GSE31578 and GSE31039) further supporting that this region encodes mammary-specific regulatory elements. Notably, only the center peak of the area occupied by STAT5 coincided with GAS motifs. An imperfect GAS motif with five matching base pairs was identified in the peak to the right but no related motif was identified in the left peak. However, several NFIB motifs were identified in the left peak and a glucocorticoid receptor (GR) motif was located in the right peak suggesting that STAT5 could bind indirectly through these transcription factors. Indeed, NFIB and GR binding has been identified in these regions (Supplementary Figure S2).
Figure 3.
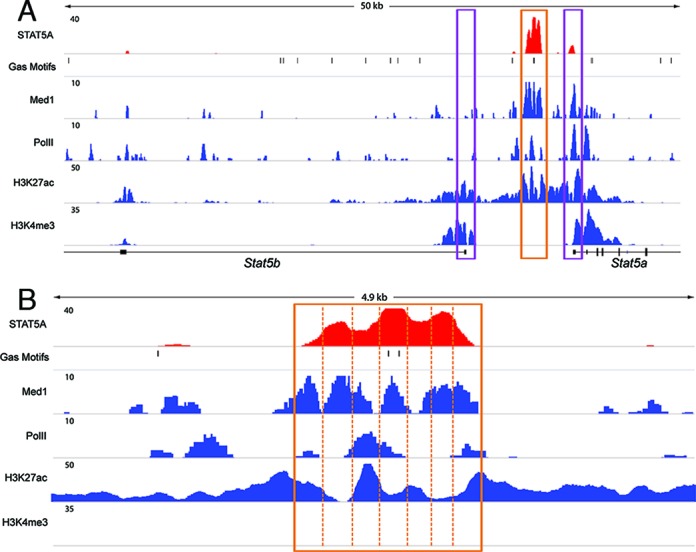
Mammary specific H3K27ac and STAT5 binding in the Stat5 locus. H3K4me3 marks feature promoter sequences of the Stat5a and Stat5b genes in mammary tissue (GSE60353). (A) H3K27ac marks in the Stat5a/b intergenic region were detected in mammary tissue but not in T cells (GSE60353). STAT5A occupancy to Stat5a/b intergenic sequences was only detected in mammary tissue (GSE40930) and not in T cells (GSE36890). MED1 binding coincided with STAT5 occupancy. RNA Pol II binding also coincided with these sequences. (B) An enlarged image of the putative mammary-specific enhancer. Mammary specific STAT5A (GSE40930) binding coincides with two GAS motifs. H3K27ac marks and Pol II loading are detected in the STAT5 binding region. The three mammary STAT5A binding peaks coincide with sequences devoid of H3K27ac marks.
Since STAT5 is present in most, if not all, cell types, its binding to the Stat5a/b intergenic region specifically in mammary tissue might be the results of a combinatorial effect with other, possibly more cell-restricted, transcription factors. Additional ChIP-seq experiments were conducted at day 13 of pregnancy, a stage when mammary enhancers are established, and binding of the GR, ELF5 and NFIB coincided with STAT5 binding in the intergenic region (Supplementary Figure S2). ELF5 is preferentially expressed in mammary epithelium and required for mammary development during pregnancy (39), glucocorticoids act synergistically with prolactin to activate milk protein genes (40) and NFIB co-regulates mammary-specific genes (41). These findings are in agreement that a combination of common and cell-enriched transcription factors facilitates mammary-restricted STAT5 binding.
CRISPR/Cas9-mediated genome editing induced a wide array of mutations
CRISPR/Cas9 methodology was employed to generate mice lacking the two STAT5-binding sites within the putative intergenic Stat5a/b enhancer. Heterozygous mutations were found in approximately 72% of founder mice, of which 67% were mosaic. Founder mice had various combinations of mutations within the intergenic enhancer (Figure 4). Three unique germline mutations were bred to homozygosity and further studied (Figure 5). Line A contained a 5 bp deletion, inactivating GAS1, line B contained a 39bp deletion spanning GAS2 and line C contained a 103 bp deletion straddling GAS1 and GAS2 without any additional alterations. To determine whether the CRISPR/Cas9 gene editing procedure had yielded additional mutations, the entire 10 kb Stat5a/b intergenic region from line C, including the promoters and first exons of Stat5a and Stat5b was sequenced. No additional mutations were detected (data not shown). Mice carrying the Stat5 enhancer mutations were overtly normal, fertile and dams were able to raise litters.
Figure 4.
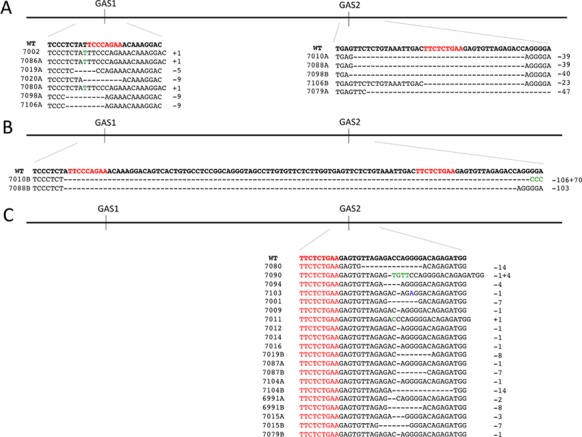
Characterization of the F0 mice generated by CRISPR/Cas9 mutagenesis to mutate GAS motifs in the Stat5a/b intergenic region. (A) Alleles with mutations in either the GAS1 or GAS2 sites. (B) Alleles with mutations spanning both GAS1 and GAS2 sites. (C) Alleles with mutations starting after the GAS sites. GAS sites are indicated in red, deleted nucleotides are indicated as dashes, inserted nucleotides are indicated in green, and transitions are indicated in blue.
Figure 5.
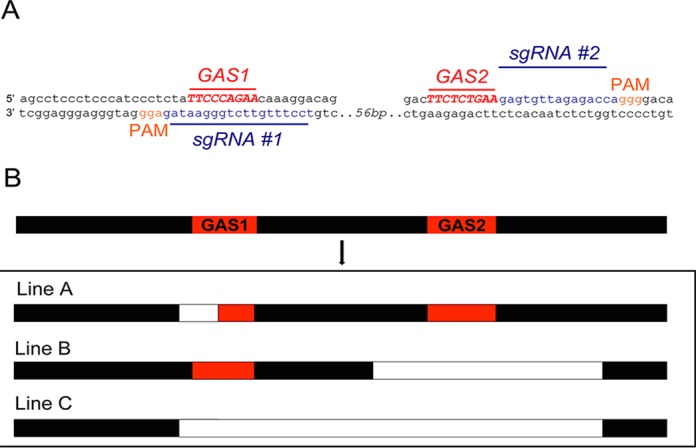
Genetic mapping of the two GAS motifs in the intergenic region of the Stat5a/b locus and the experimental design in using CRISPR/Cas9 mutagenesis to establish mice with GAS mutations. (A) Scheme of Cas9 cleavage at the sgRNA sites. (B) Comparison of the deleted sequences in the CRISPR/Cas9 mice used in this study. Line A represents mice with a GAS1 only deletion (5 bp), line B represents mice with GAS2 only deletions (39 bp) and the deletion in line C spans both GAS sites (103 bp). GAS sites are shown in red, deleted regions are shown in white color.
A mammary-specific STAT5-dependent enhancer controls Stat5 expression and downstream targets
To determine whether the intergenic enhancer regulates expression of the adjacent Stat5 genes we compared Stat5 mRNA levels in wild type and enhancer mutant mice at day one of lactation (L1) (Figure 6A). Stat5a and Stat5b mRNA levels were reduced by ∼40–50% in line A, by ∼60% and ∼40% in line B and by ∼80% and ∼65% in line C. RNA-seq data from lines B and C validated the reduction of Stat5a and Stat5b levels (Supplementary Table S2). Taken together, these data confirm that the two GAS sites within the intergenic enhancer significantly contribute to Stat5a and Stat5b expression during lactation. They also indicate that Stat5a is more sensitive to GAS site inactivation than Stat5b, possibly due to their greater proximity to the Stat5a promoter. Indication that reduced Stat5 expression is the consequence of loss of GAS sites comes from line A, which harbors a 5bp deletion that inactivates only the GAS1 motif and does not disturb flanking sequences. Stat5 expression was reduced the most in line C suggesting a contribution of not only the second GAS site but potentially of other transcription factors (Supplementary Figure S2). Notably, a glucocorticoid receptor motif was identified between the two GAS sites and coincides with binding (Supplementary Figure S2). This finding points to a complex enhancer and the total number of interacting transcription factors remains to be determined.
Figure 6.
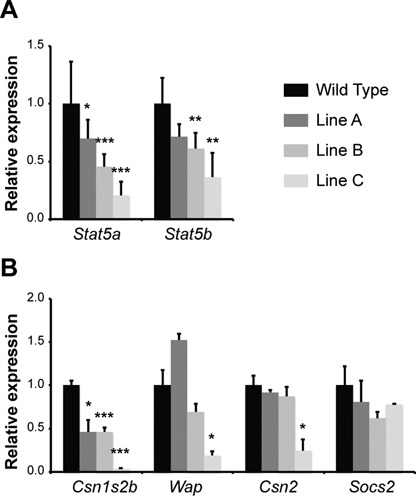
Relative expression levels at day one of lactation analyzed by qRT-PCR. The mean Cq values were normalized against the reference gene Gapdh and converted to a ratio relative to wild type. (A) Relative expression of Stat5a and Stat5b in the various GAS mutant mice. (B) Relative expression of STAT5 target genes in the various GAS mutant mice. Line A represents mice with a GAS1 deletion, line B represents mice with a GAS2 deletion, line C represents mice with both GAS1 and GAS2 deletions. A two-tailed unpaired t-test was used for statistical analysis (*P < 0.05). Data represent the mean of six biological replicates for line B and line C and four biological replicates for wild type and line A.
To further validate the mammary-specificity of the enhancer, we analyzed STAT5 levels in lymphoid cells and liver tissue (Supplementary Figures S3 and S4). Stat5a and Stat5b mRNA levels in B and T cells as well as total STAT5 levels in these cells and in T regulatory cells were equivalent in wild type and mutant mice. Moreover, B and T cell numbers were not affected by loss of the Stat5 enhancer and expression of the lymphoid-specific Il2rα gene was normal. Stat5a and Stat5b mRNA levels in liver tissue were also not altered in mutant mice (Supplementary Figure S4). These experiments substantiate the mammary-specificity of the Stat5 intergenic enhancer.
We have previously shown that progressive deletion of individual Stat5a or Stat5b alleles has deleterious effects on mammary physiology (8). These studies had also established that the presence of just one out of four Stat5 alleles (low concentrations of STAT5) is sufficient to promote alveolar cell proliferation during pregnancy whereas at least two Stat5 alleles (high STAT5 concentrations) were required for functional differentiation (8). We therefore considered that deletion of the GAS sites within the intergenic enhancer would have a similar effect as germline deletion of two or three Stat5 alleles. To test this hypothesis, we first measured steady-state mRNA levels of a subset of well-characterized mammary-specific, STAT5-dependent genes in mammary tissue from the three mutants (Figure 6B). All three of the milk protein genes that were analyzed, Csn1s2b, Wap, and Csn2, exhibited significant reductions in mRNA levels within mutant tissue from line C (Figure 6). Notably, Csn1s2b mRNA levels were already reduced by ∼50% in lines A and B, lacking site GAS1 and GAS2, respectively. The Csn1s2b gene was virtually silent in line C, which agrees with RNA-seq data (Supplementary Table S2). Wap and Csn2 mRNA levels were reduced by ∼80% only in line C that lacks both GAS motifs. Expression of Socs2, a known STAT5 target that is expressed across cell types, was similar in the enhancer mutant mouse lines (Figure 6B). These results clearly show a hierarchy among STAT5 target genes, with some being very sensitive to reduced STAT5 levels while others are rather indolent and are expressed even at very low STAT5 levels. Mutant dams were able to raise their litters suggesting that under laboratory conditions mammary tissue retains functionality in the presence of reduced Stat5 levels. This is in agreement with our earlier genetic studies on mice carrying selected Stat5 alleles (8).
To assess the genome-wide impact of deleting the Stat5a/b intergenic enhancer, we initially compared transcriptomes of WT and GAS2 mutant mammary tissue (line B) at parturition. Overall, expression of ∼200 genes was significantly reduced in mutant tissue, and 155 genes whose expression was up-regulated (Figure 7A). Among the former were Stat5a and Csn1s2b, thereby confirming the results of our PCR studies, as well as additional mammary-specific genes that are highly expressed during lactation, including Wap, Lalba and Saa1 (Figure 7B and C). Because many of these genes are subject to the prolactin/STAT5 axis (8), we performed gene set enrichment analysis (GSEA) to determine if STAT5-dependent genes were globally affected in GAS2 mutants. To that end, we first mined the data from our previous studies involving mice lacking individual Stat5 alleles to generate a set of ∼200 genes whose expression is STAT5-dependent and largely restricted to mammary tissue (Supplementary Table S1). Next, we ran this set against the transcriptome analysis and found that, indeed, STAT5-dependent genes were heavily enriched in WT controls or, stated otherwise, that they tended to be reduced in GAS2 mutants (Figure 7B and C).
Figure 7.
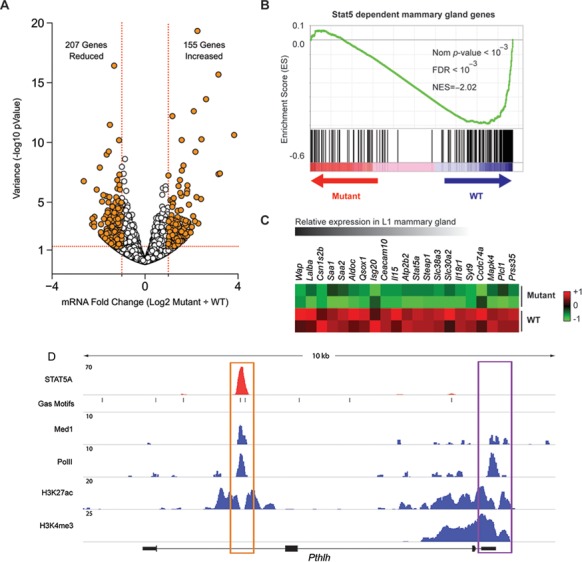
Impaired STAT5 target gene expression in Stat5 mutant mammary tissue. RNA-seq was performed on L1 mammary tissues from mice lacking the GAS2 site (mutant) and wild type controls (WT). RNA-Seq data from Stat5 mutants were acquired from biological duplicates (see Supplementary Figure S11 for quality control). (A) Volcano plot shows the fold change (log2 transformed) and variance for all transcripts relative to normal controls. Differentially expressed transcripts are highlighted in orange. Those with greater abundance in mutant (upper right) or WT (upper left) are summed. Dotted red lines indicate 2-fold changes and 0.05 P-value. (B) GSEA for STAT5 dependent mammary gland genes comparing WT and mutant samples. The analysis shows skewed distribution toward WT (Nom P-value, normalized P-value; FDR, false discovery rate; NES, normalized enrichment score). (C) Heatmap depicting relative expression change of representative STAT5 dependent genes in WT and mutant mammary tissues. Genes are arranged according to relative expression level in WT. (D) ChIP-seq data depict binding of STAT5, MED1 and Pol II in the Pthlh gene. GAS motifs (STAT5 binding sites) are shown. ChIP-seq data also demonstrate the location of H3K27ac (enhancer) and H3K4me3 (promoter) marks. STAT5 binding has been identified to a putative intronic enhancer that coincides with H3K27ac. Expression of Pthlh is reduced in Stat5 enhancer mutant mammary tissue (Supplementary Figure S12 and Table S2).
These results were complemented with additional RNA-seq data from line C that carries a deletion spanning both GAS sites (Supplementary Table S2). In agreement with qRT-PCR data, Stat5a levels were reduced approximately 6-fold and Stat5b levels 3-fold. In addition to well-known milk protein genes, several members of the solute carrier proteins (Slc) emerged as STAT5 targets. These include Slc30a2 and Slc30a4, zinc transporters linked to mammary function (42–44). Elf5 levels were reduced by 2-fold, reaffirming its status as a STAT5 target. Stat1 levels were unaltered. Collectively, these data establish that intergenic enhancer driven auto-regulation significantly contributes to maintaining the high levels of STAT5 necessary for maximal mammary-specific gene expression during pregnancy.
To validate that reduced expression of STAT5 target genes in mutant tissue is the result of diminished STAT5 binding to key regulatory sequences, we conducted a STAT5 ChIP experiment on the Wap gene, whose expression is reduced by >80% in line C (Supplementary Table S2). STAT5 binding to a putative Wap gene enhancer was reduced by >60% (Supplementary Figure S5). STAT5 binding to the mutant intergenic Stat5 enhancer was reduced by >90%. To confirm that the genes with reduced expression in the intergenic enhancer mutants are bound by STAT5, we searched for coincident STAT5 and MED1 binding, Pol II loading and H3K27ac. As predicted by the GSEA analysis, STAT5 binding was observed at putative cis-regulatory elements for many of these genes (Supplementary Figures S6–S10). One emblematic example of this trend is the Csn1s2b gene, which exhibited robust STAT5 binding at its promoter, the proximal upstream region and within intron 9 (Supplementary Figure S6). In these, and many other instances, such as Lalba (Supplementary Figure S7) STAT5 binding occurred at GAS motifs and coincided with MED1 binding and H3K27ac, suggesting that these are bona fide enhancer elements.
Beyond those that have been previously described, such as Csn1s2b and Wap, the integrated transcriptomic, DNA binding and epigenomic analyses identified novel STAT5 dependent genes that have been linked to mammary gland development or could potentially impact mammary function. Notably, the parathyroid hormone related hormone gene (Pthlh), which is essential for the formation of embryonic mammary buds and mammary gland development (45), contains an intronic STAT5-driven enhancer (Figure 7D). The Pthlh gene as well as several solute carrier (SLC) proteins, including the sodium phosphate cotransporter Slc34a (Supplementary Figure S8) and the Ca2+ transporting membrane protein gene Atp2b2 (Supplementary Figure S9) exhibited robust STAT5 binding that coincided with MED1 binding, Pol II occupancy and the presence of H3K27ac marks. Expression of both Slc34a2 and Atp2b2 was reduced more than 90% in mutant tissue (Supplementary Table S2). Surprisingly, we also identified Il15 as a direct STAT5 target (Supplementary Figure S10), whose expression was reduced in mammary tissues from the intergenic enhancer mice (Figure 7C) by >95% (Supplementary Table S2). Since lymph nodes had been removed from mammary tissue prior to the preparation of RNA, it is likely that the Il15 signal is inherent to epithelial cells. This gene encodes a secreted cytokine that has been extensively studied within the immune system, particularly in terms of lymphoid cell homeostasis and cancer surveillance.
DISCUSSION
Transcription factors are central to the regulation of common and cell-specific gene networks that control diverse programs ranging from cell specification to modulating cell physiology. However, it remains to be understood how communal transcription factors also control cell- and differentiation-specific transcription programs. This study has uncovered the presence of a mammary-specific autoregulatory enhancer within the Stat5 locus that ensures extraordinary high Stat5a levels required to implement the activation of mammary-specific differentiation programs. STAT5 is a transcription factor that is activated by a range of cytokines, including prolactin, growth hormone and interleukins. Although STAT5 modulates the biology of many cell types, its presence is essential during pregnancy for the proliferation and differentiation of mammary epithelium.
STAT5-induced transcriptional regulation
The extraordinary high expression levels of mammary-specific genes are controlled by prolactin through the transcription factor STAT5 (46–49) and during lactation more than 95% of mRNA in secreting mammary epithelium is encoded by less than 10 genes (8). Importantly, activation of these genes during pregnancy is induced up to several thousand-fold and is largely the result of a temporally defined STAT5 recruitment to putative enhancer sequences (50). The current dogma suggests that levels of STAT5, like those of the other STAT members, remains constant in a given cell type regardless of its differentiation status and that key to its function is the degree of tyrosine phosphorylation (51). Although tyrosine phosphorylation of STAT5 is essential for its activity and biology, our study demonstrates for the first time that an increase of STAT5 levels in a defined cell lineage progressing through a developmental program is important to attain a specific differentiation status. High STAT5 levels in mammary tissue were only obtained in the presence of the cell-specific intergenic enhancer bound STAT5 itself. Notably, already the ablation of individual GAS sites resulted in reduced levels of STAT5. It is possible that the autoregulation of the Stat5 locus is dependent on STAT5 tetramers (25) although there is no experimental evidence to support this notion. The Stat5 intergenic enhancer is not only bound by STAT5 itself but also by additional factors, including GR, NFIB, MED1 and the mammary-enriched transcription factor ELF5, suggesting that mammary-specificity is attained through a combinatorial effect. Since the two GAS motifs were the distinct DNA binding motifs in this region, we propose that STAT5 provides a platform for the other factors.
Exploiting autoregulatory control as a means to boost expression in specific cell types is a concept adopted also by other transcription factors and possibly more prevalent as recognized. Levels of GATA1, which is central to erythropoiesis, increase as the erythroid lineage differentiates into the proerythroblast stage. Similarly levels of Gata2, Gata3 and Runx1 in hematopoietic compartments exceed those in other cell types. Gata1 is under autoregulation in hematopoietc cells as reported recently (52) with GATA1 binding to an upstream enhancer. Autoregulatory loops can be augmented as shown for PU.1, where differential activity is established through association with cell type specific transcription factors on distinct autoregulatory elements in myeloid and lymphoid B cells (53). Based on our genome-wide studies the molecular explanation for the mammary-specificity of the STAT5 autoregulatory loop is likely to be found in the co-binding of additional transcription factors, including MED1, GR, NFIB and the mammary-enriched transcription factor ELF5.
STAT5 autoregulation controls differentiation specific programs
Differentiation of mammary alveolar epithelium during pregnancy is characterized by the sequential activation of mammary-specific genes (54), with those expressed just prior to parturition being most sensitive to STAT5 levels. Stat5 autoregulation likely enables these genes to acquire maximum activity during lactation. Among them are several members of the solute carrier (SLC) group of membrane transport proteins, some of which are required for mammary epithelial differentiation (42,44) and connexins (gjb), which participate in the establishment of functional mammary tissue (41,55). Notably, also the transcriptional co-activator CIDEA (56) that participates in lipid secretion and mammary development and the critical Pthlh gene (45) are controlled by STAT5. In contrast to these differentiation-specific genes, expression of common STAT5 targets, such as Socs2 and Cish, is rather insensitive to changing STAT5 levels (57).
While our study has identified a mammary-specific enhancer that enables high STAT5 levels in milk secreting epithelium, an enhancer responsible for exceptionally high STAT5B levels in T cells is still elusive. Although a strong lymphoid-specific DHS site at the Stat5b proximal promoter and T cell-specific STAT5 binding (ENCODE data) to the first intron provide clues to the elusive lymphoid enhancer, their biological relevance needs to be validated. It can already be hypothesized that cell-specificity of STAT5-driven enhancers is achieved through the additional recruitment of transcription factors enriched in respective cell types, such as NFIB and ELF5 in mammary tissue.
Supplementary Material
Acknowledgments
We acknowledge Dr Harold Smith from the NIDDK genomics core for NGS and Dr Chengyu Liu from the NHLBI transgenic core for generating the CRISPR/Cas9 based mouse mutants. Sumin Oh is a graduate student of the Individual Graduate partnership Program between NIH/NIDDK and Dankook University (Cheonan, Republic of Korea).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
IPR of the NIDDK/NIH and NIAMS/NIH. Funding for open access charge: Intramural research program of the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Villarino A.V., Kanno Y., Ferdinand J.R., O'Shea J.J. Mechanisms of Jak/STAT signaling in immunity and disease. J. Immunol. 2015;194:21–27. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Shea J.J., Lahesmaa R., Vahedi G., Laurence A., Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat. Rev. Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hennighausen L., Robinson G.W. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–721. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C.X., Robinson G.W., Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004;24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyoshi K., Shillingford J.M., Smith G.H., Grimm S.L., Wagner K.U., Oka T., Rosen J.M., Robinson G.W., Hennighausen L. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J. Cell Biol. 2001;155:531–542. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Z., Cui Y., Watford W.T., Bream J.H., Yamaoka K., Hissong B.D., Li D., Durum S.K., Jiang Q., Bhandoola A., et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyoshi K., Cui Y., Riedlinger G., Robinson P., Lehoczky J., Zon L., Oka T., Dewar K., Hennighausen L. Structure of the mouse Stat 3/5 locus: evolution from Drosophila to zebrafish to mouse. Genomics. 2001;71:150–155. doi: 10.1006/geno.2000.6433. [DOI] [PubMed] [Google Scholar]
- 8.Yamaji D., Kang K., Robinson G.W., Hennighausen L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res. 2013;41:1622–1636. doi: 10.1093/nar/gks1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyoshi K., Meyer B., Gruss P., Cui Y., Renou J.P., Morgan F.V., Smith G.H., Reichenstein M., Shani M., Hennighausen L., et al. Mammary epithelial cells are not able to undergo pregnancy-dependent differentiation in the absence of the helix-loop-helix inhibitor Id2. Mol. Endocrinol. 2002;16:2892–2901. doi: 10.1210/me.2002-0128. [DOI] [PubMed] [Google Scholar]
- 10.Yao Z., Kanno Y., Kerenyi M., Stephens G., Durant L., Watford W.T., Laurence A., Robinson G.W., Shevach E.M., Moriggl R., et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klover P., Chen W., Zhu B.M., Hennighausen L. Skeletal muscle growth and fiber composition in mice are regulated through the transcription factors STAT5a/b: linking growth hormone to the androgen receptor. FASEB J. 2009;23:3140–3148. doi: 10.1096/fj.08-128215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klover P., Hennighausen L. Postnatal body growth is dependent on the transcription factors signal transducers and activators of transcription 5a/b in muscle: a role for autocrine/paracrine insulin-like growth factor I. Endocrinology. 2007;148:1489–1497. doi: 10.1210/en.2006-1431. [DOI] [PubMed] [Google Scholar]
- 13.Zhu B.M., McLaughlin S.K., Na R., Liu J., Cui Y., Martin C., Kimura A., Robinson G.W., Andrews N.C., Hennighausen L. Hematopoietic-specific Stat5-null mice display microcytic hypochromic anemia associated with reduced transferrin receptor gene expression. Blood. 2008;112:2071–2080. doi: 10.1182/blood-2007-12-127480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y., Hosui A., Sun R., Shen K., Gavrilova O., Chen W., Cam M.C., Gao B., Robinson G.W., Hennighausen L. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration. Hepatology. 2007;46:504–513. doi: 10.1002/hep.21713. [DOI] [PubMed] [Google Scholar]
- 15.Liu X., Robinson G.W., Wagner K.U., Garrett L., Wynshaw-Boris A., Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 16.Kofoed E.M., Hwa V., Little B., Woods K.A., Buckway C.K., Tsubaki J., Pratt K.L., Bezrodnik L., Jasper H., Tepper A., et al. Growth hormone insensitivity associated with a STAT5b mutation. N. Engl. J. Med. 2003;349:1139–1147. doi: 10.1056/NEJMoa022926. [DOI] [PubMed] [Google Scholar]
- 17.Udy G.B., Towers R.P., Snell R.G., Wilkins R.J., Park S.H., Ram P.A., Waxman D.J., Davey H.W. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imada K., Bloom E.T., Nakajima H., Horvath-Arcidiacono J.A., Udy G.B., Davey H.W., Leonard W.J. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X., Gallego M.I., Smith G.H., Robinson G.W., Hennighausen L. Functional rescue of Stat5a-null mammary tissue through the activation of compensating signals including Stat5b. Cell Growth Differ. 1998;9:795–803. [PubMed] [Google Scholar]
- 20.Basham B., Sathe M., Grein J., McClanahan T., D'Andrea A., Lees E., Rascle A. In vivo identification of novel STAT5 target genes. Nucleic acids Res. 2008;36:3802–3818. doi: 10.1093/nar/gkn271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frasor J., Barkai U., Zhong L., Fazleabas A.T., Gibori G. PRL-induced ERalpha gene expression is mediated by Janus kinase 2 (Jak2) while signal transducer and activator of transcription 5b (Stat5b) phosphorylation involves Jak2 and a second tyrosine kinase. Mol. Endocrinol. 2001;15:1941–1952. doi: 10.1210/mend.15.11.0722. [DOI] [PubMed] [Google Scholar]
- 22.Nelson E.A., Walker S.R., Alvarez J.V., Frank D.A. Isolation of unique STAT5 targets by chromatin immunoprecipitation-based gene identification. J. Biol. Chem. 2004;279:54724–54730. doi: 10.1074/jbc.M408464200. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita H., Xu J., Erwin R.A., Farrar W.L., Kirken R.A., Rui H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J. Biol. Chem. 1998;273:30218–30224. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- 24.Soldaini E., John S., Moro S., Bollenbacher J., Schindler U., Leonard W.J. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell. Biol. 2000;20:389–401. doi: 10.1128/mcb.20.1.389-401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J.X., Li P., Liu D., Jin H.T., He J., Ata Ur Rasheed M., Rochman Y., Wang L., Cui K., Liu C., et al. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity. 2012;36:586–599. doi: 10.1016/j.immuni.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 27.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 28.He H.H., Meyer C.A., Hu S.S., Chen M.W., Zang C., Liu Y., Rao P.K., Fei T., Xu H., Long H., et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H., Wang H., Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 2014;9:1956–1968. doi: 10.1038/nprot.2014.134. [DOI] [PubMed] [Google Scholar]
- 36.Wakao H., Gouilleux F., Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Robinson G.W., Gouilleux F., Groner B., Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Nat. Acad. Sci. U.S.A. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X., Chatterjee R., John S., Bravo H., Sathyanarayana B.K., Biddie S.C., FitzGerald P.C., Stamatoyannopoulos J.A., Hager G.L., Vinson C. Contribution of nucleosome binding preferences and co-occurring DNA sequences to transcription factor binding. BMC Genomics. 2013;14:428. doi: 10.1186/1471-2164-14-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J., Chehab R., Tkalcevic J., Naylor M.J., Harris J., Wilson T.J., Tsao S., Tellis I., Zavarsek S., Xu D., et al. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 2005;24:635–644. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pittius C.W., Sankaran L., Topper Y.J., Hennighausen L. Comparison of the regulation of the whey acidic protein gene with that of a hybrid gene containing the whey acidic protein gene promoter in transgenic mice. Mol. Endocrinol. 1988;2:1027–1032. doi: 10.1210/mend-2-11-1027. [DOI] [PubMed] [Google Scholar]
- 41.Robinson G.W., Kang K., Yoo K.H., Tang Y., Zhu B.M., Yamaji D., Colditz V., Jang S.J., Gronostajski R.M., Hennighausen L. Coregulation of genetic programs by the transcription factors NFIB and STAT5. Mol. Endocrinol. 2014;28:758–767. doi: 10.1210/me.2012-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang L., Gitschier J. A novel gene involved in zinc transport is deficient in the lethal milk mouse. Nat. Genet. 1997;17:292–297. doi: 10.1038/ng1197-292. [DOI] [PubMed] [Google Scholar]
- 43.McCormick N.H., Kelleher S.L. ZnT4 provides zinc to zinc-dependent proteins in the trans-Golgi network critical for cell function and Zn export in mammary epithelial cells. Am. J. Physiol. Cell Physiol. 2012;303:C291–C297. doi: 10.1152/ajpcell.00443.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S., Hennigar S.R., Alam S., Nishida K., Kelleher S.L. Essential role for ZnT2-mediated zinc transport in mammary gland development and function during lactation. J. Biol. Chem. 2015;290:13064–13078. doi: 10.1074/jbc.M115.637439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wysolmerski J.J., Philbrick W.M., Dunbar M.E., Lanske B., Kronenberg H., Broadus A.E. Rescue of the parathyroid hormone-related protein knockout mouse demonstrates that parathyroid hormone-related protein is essential for mammary gland development. Development. 1998;125:1285–1294. doi: 10.1242/dev.125.7.1285. [DOI] [PubMed] [Google Scholar]
- 46.Burdon T.G., Maitland K.A., Clark A.J., Wallace R., Watson C.J. Regulation of the sheep beta-lactoglobulin gene by lactogenic hormones is mediated by a transcription factor that binds an interferon-gamma activation site-related element. Mol. Endocrinol. 1994;8:1528–1536. doi: 10.1210/mend.8.11.7877621. [DOI] [PubMed] [Google Scholar]
- 47.Archibald A.L., McClenaghan M., Hornsey V., Simons J.P., Clark A.J. High-level expression of biologically active human alpha 1-antitrypsin in the milk of transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5178–5182. doi: 10.1073/pnas.87.13.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenberg N.M., Anderson J.W., Hsueh A.J., Nishimori K., Reeves J.J., deAvila D.M., Ward D.N., Rosen J.M. Expression of biologically active heterodimeric bovine follicle-stimulating hormone in milk of transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8327–8331. doi: 10.1073/pnas.88.19.8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pittius C.W., Hennighausen L., Lee E., Westphal H., Nicols E., Vitale J., Gordon K. A milk protein gene promoter directs the expression of human tissue plasminogen activator cDNA to the mammary gland in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 1988;85:5874–5878. doi: 10.1073/pnas.85.16.5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang K., Yamaji D., Yoo K.H., Robinson G.W., Hennighausen L. Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Mol. Cell. Biol. 2014;34:464–473. doi: 10.1128/MCB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X., Robinson G.W., Hennighausen L. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol. Endocrinol. 1996;10:1496–1506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- 52.Moriguchi T., Suzuki M., Yu L., Takai J., Ohneda K., Yamamoto M. Progenitor stage-specific activity of a cis-acting double GATA motif for Gata1 gene expression. Mol. Cell. Biol. 2015;35:805–815. doi: 10.1128/MCB.01011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leddin M., Perrod C., Hoogenkamp M., Ghani S., Assi S., Heinz S., Wilson N.K., Follows G., Schonheit J., Vockentanz L., et al. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood. 2011;117:2827–2838. doi: 10.1182/blood-2010-08-302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hennighausen L., Robinson G.W. Information networks in the mammary gland. Nat. Rev. Mol. Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 55.Bry C., Maass K., Miyoshi K., Willecke K., Ott T., Robinson G.W., Hennighausen L. Loss of connexin 26 in mammary epithelium during early but not during late pregnancy results in unscheduled apoptosis and impaired development. Dev. Biol. 2004;267:418–429. doi: 10.1016/j.ydbio.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 56.Wang W., Lv N., Zhang S., Shui G., Qian H., Zhang J., Chen Y., Ye J., Xie Y., Shen Y., et al. Cidea is an essential transcriptional coactivator regulating mammary gland secretion of milk lipids. Nat. Med. 2012;18:235–243. doi: 10.1038/nm.2614. [DOI] [PubMed] [Google Scholar]
- 57.Yamaji D., Na R., Feuermann Y., Pechhold S., Chen W., Robinson G.W., Hennighausen L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009;23:2382–2387. doi: 10.1101/gad.1840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.