Abstract
We developed a comprehensive resource for the genome-reduced bacterium Mycoplasma pneumoniae comprising 1748 consistently generated ‘-omics’ data sets, and used it to quantify the power of antisense non-coding RNAs (ncRNAs), lysine acetylation, and protein phosphorylation in predicting protein abundance (11%, 24% and 8%, respectively). These factors taken together are four times more predictive of the proteome abundance than of mRNA abundance. In bacteria, post-translational modifications (PTMs) and ncRNA transcription were both found to increase with decreasing genomic GC-content and genome size. Thus, the evolutionary forces constraining genome size and GC-content modify the relative contributions of the different regulatory layers to proteome homeostasis, and impact more genomic and genetic features than previously appreciated. Indeed, these scaling principles will enable us to develop more informed approaches when engineering minimal synthetic genomes.
INTRODUCTION
Recent molecular and systems biology studies in bacteria have revealed a surprisingly dynamic and complex regulation of gene expression, in some aspects even resembling that of eukaryotes (1,2). Consequently, the information flow from genome to RNA to protein, a central dogma in molecular biology (3,4), has been refined by newly identified regulatory layers and detailed regulatory mechanisms, including non-coding RNAs (ncRNAs; 1,5,6), post-translational modifications (PTMs; 2) and second messengers (7–9). Nevertheless, we still do not fully understand the contribution of these regulatory layers to protein abundances, nor the complex interplay that characterizes the physiological state of a cell.
Although considerable progress has been made to model some of the regulatory processes linking genomes to phenomes (10), dissecting their putative interactions and quantifying their contributions to the regulation of protein abundance remains difficult due to the paucity of available ‘-omics’ data sets. For example, recent work in humans resulted in a collection of ≈500 genes for which sufficient multi-omics data sets were available (11), but this only represents ≈5% of the genes with identified proteins (12,13). Furthermore, most ‘-omics’ data sets of model organisms have been derived in different laboratories under different conditions, thereby considerably hampering their integration.
To allow complex and systemic analyses within a single organism, we took various Mycoplasma pneumonia data sets that were previously generated under standardized conditions, integrated them into a single resource (MyMpn: http://mycoplasma.crg.eu/; 14) and added new screens for both ncRNAs and PTMs. In total, the 1748 data sets (1680 published, 68 new; Supplementary Table S1) include DNA methylomes (15), transcriptomes (1,16), proteomes (17), protein–protein interaction networks (18), PTMs (2), metabolomes (19) and a genome-wide essentiality map (20). For comparison purposes, we also generated some of the data sets using the related pathogen Mycoplasma genitalium (43 data sets; Supplementary Table S1; 10,15,21–24). These data feature biological replicates, synchronized experimental conditions (e.g. longitudinal data along the growth curve) and matching sample batches.
Our ‘-omics’ data generally cover a much larger fraction of genes or proteins than data for other model bacteria; for example, the M. pneumoniae interactome covers ≈90% of the tested soluble proteins (18), in contrast to the ≈77% in Escherichia coli (25). Overall, our data could link 99% (816 360 out of 816 394) of the base pairs of the M. pneumoniae genome to at least one data set (Figure 1 and Supplementary Figure S1), and represent the largest coordinated effort in ‘-omics’ profiling for a model bacterium (Figure 1B; Supplementary Table S2). While technical details of the databases have been previously described [14], here we summarize the data and illustrate the value of this unified resource by a number of integration approaches, to provide a quantitative view of the individual and combinatorial contributions of different regulatory layers to the fine-tuning of protein abundance. Our results indicate that antisense ncRNAs, lysine acetylation and phosphorylation correlate better with protein abundance than mRNA levels when combined or even evaluated individually in this genome-reduced bacterium. Comparative analyses of about 1600 bacteria reveal that both genomic guanine-cytosine (GC) content and genome size affect the abundances of ncRNAs, as well as lysine acetylation and phosphorylation. GC content and genome size were previously shown to be significantly correlated with each other (26). Our results suggest that genome-reduction in bacteria correlates with the prevalence of other genomic and genetic features and the respective wiring of different regulatory layers at transcriptomic and proteomic levels.
Figure 1.
A comprehensive resource for M. pneumoniae and comparisons with selected model bacteria. (A) -omics data and the number of data sets collected (see Supplementary Table S1 for more details); (B) Comparison of data coverage with the selected model bacteria (as of May 2013). The numbers for each selected model bacteria come from the individual study where the largest quantities of data sets were included; if two or more studies contain the same number of ‘-omics’ data sets, the one published most recently was chosen (Supplementary Table S2). (C) A graphical view of all available data sets for M. pneumoniae, where each circular layer represents an -omics datatype. Only data for the plus strand are shown; see Supplementary Figure S1 for a full-sized figure containing all data for both strands.
MATERIALS AND METHODS
Assembly of the 1748 ‘-omics’ data sets of M. pneumoniae
The 1748 multi-omics data sets from M. pneumoniae are summarized in Supplementary Table S1, of which 1680 were previously published. Additionally, 43 data sets were also generated for M. genitalium, including transcriptome profiling at two time points and proteome profiling at 12 time points from 0 to 96 h along the growth curve (Supplementary Table S1). These data have been used to manually annotate the genomes of M. pneumoniae and M. genitalium and the coding capacity of newly annotated genes; see Supplementary Tables S3 and S4 for all annotated genes of the two mycoplasmas. All data are freely accessible at MyMpn: http://mycoplasma.crg.eu (14).
Re-annotation of M. pneumoniae and M. genitalium by combining multi-omics data and manual inspection
We have identified non-coding genome regions, previously unannotated RNAs (ncMPNs), transcriptional start sites (TSSs), promoter sequences and 5′-untranslated regions (5′-UTRs) by analyzing newly-generated (this study) and previously published deep sequencing data (RNAseq; 27) and tiling array data of the M. pneumoniae transcriptome (1; Supplementary Figure S2). In order to validate the annotation of new genes and refine existing annotations, we integrated data concerning a new class of short RNAs, denominated tssRNAs, which precisely map to the TSSs of bacterial genes (27). The results for M. pneumoniae are shown in Supplementary Table S3. Briefly, if the TSS is downstream of the annotated translational start codon (TSC), the open reading frame (ORF) is annotated as shorter; ORFs with more than one TSS are indicated as having an alternative TSS. The transcripts with a 5′-UTR region longer than 40 base pairs are indicated by an ‘x’ in column I of Supplementary Table S3 (See legend for Supplementary Table S3). This analysis revealed 34 previously annotated protein-coding genes with the TSS downstream of the annotated translational start codon (TSC), 72 cases with an alternative TSS (of which 30 were found inside the annotated gene), 160 ORFs with a 5′-UTR longer than 40 nucleotides and 313 ncRNAs (ncMPNs; Supplementary Table S3).
In order to annotate putative coding genes, we translated all the identified transcripts into all three possible frames, and obtained 2392 putative new ORFs of at least 25 amino acids in length. Sequence comparison supports the transcriptome prediction of 22 new proteins (Supplementary Table S3). Mass spectrometry (MS) analysis using newly generated proteome data for this study detected 575 proteins with unique peptides (538 from the 689 annotated, and 37 novel) and 51 proteins (29 from the 689 annotated, and 22 of the predicted new ones) with shared peptides unable to be unequivocally assigned (Supplementary Table S3), covering 82% of the previously described M. pneumoniae proteome. The total number of detected proteins is similar to a previously published MS analysis of the M. pneumoniae proteome, where 557 out of 689 ORFs were found (28; Supplementary Table S3). Taking into consideration both studies, 706 proteins of the M. pneumoniae proteome have been identified, representing 96% of the total proteome. We detected all genes with significant expression levels (average log2 > 11 for tiling array and log2 > 5 for deep sequencing results) by MS, but found only 71% of the proteins with low expression levels or significantly shorter RNAs than expected from the formerly annotated ORFs. Considering that the number of totally or partially duplicated proteins is around 104 in the M. pneumoniae genome, we are approaching complete coverage. Most of the new coding ORFs (35 out of 38) are located in intragenic regions, showing that the same part of the genome encodes for different proteins. For instance, in the region of mpn199, two new proteins encoded by mpn199a (in antisense orientation versus mpn199) and mpn200a (in sense) have been identified. The newly identified proteins are usually very short, i.e. less than 50 amino acids (Supplementary Table S3).
Out of the 689 previously annotated ORFs, we identified by sequence comparison 11 proteins with putatively longer and shorter isoforms, 26 possibly longer proteins and 34 presumably shorter ones (Supplementary Table S3). We assigned molecular weights to 518 proteins based on SDS gel/MS analysis, confirming the existence of 12 out of the 26 predicted longer proteins (Supplementary Table S3). For five of the 26 putative larger proteins (MPN006, MPN148, MPN163, MPN388 and MPN664), we identified unique peptides corresponding to the extended sequences (Supplementary Table S3).
For six of the 30 ORFs that were found to have internal TSSs in the transcriptome analysis, we detected the expression of two proteins of different sizes: MPN310 (200/19 kDa), MPN130 (16.5/10 kDa), MPN410 (17.5/10 kDa), MPN073 (44/38 kDa), MPN196 (27/6.5 kDa) and MPN307 (33/20 kDa; Supplementary Table S3). In fact, the two isoforms of MPN310 have been previously described by Boonmee et al. (29).
Finally, we found two cases in which sequence analysis revealed proteins split into two different ORFs (mpn279 (LepA) and mpn520 (IleS)). Genome re-sequencing of these two ORFs revealed a frame shift that, when corrected, resulted in the correct functional proteins with peptides corresponding to the two split regions identified by MS (Supplementary Table S3).
In summary, the integration of transcriptomics and proteomics data with a library of the theoretical proteome of M. pneumoniae enabled us to identify 49 new ORFs (35 of them overlapping with annotated proteins), to change the length of 46 proteins (12 longer and 34 shorter), to correct two frame shifts and to identify two long and short isoforms for 6 proteins. It also revealed the existence of a significant number of small proteins (≈5%) of unknown function that are probably missing in the majority of bacterial genome annotations (20).
Re-annotation of M. genitalium genome
The same procedure was used to annotate the genome of M. genitalium. In total, we were able to refine the annotation for 12 previously annotated protein-coding genes (21), identify 23 new protein-coding genes and 494 new ncRNAs (Supplementary Table S4).
Orthologous groups and phylogenetic reconstruction of 16 completely sequenced mycoplasma genomes
The orthologous relationships between protein-coding genes of 16 completely sequenced mycoplasmas including M. pneumoniae and M. genitalium, and five additional closely related bacteria were reconstructed using the EGGNOG 3 (30) pipeline. The selected species and resulting orthologous groups are shown in Supplementary Tables S14 and S15. The one-to-one orthologous genes between M. pneumoniae and M. genitalium are defined as those that are in the same orthologous group, with each species having only one gene in that group.
The phylogenetic relationships among the 21 selected species were reconstructed based on the concatenation of 40 universal one-to-one marker genes (Supplementary Table S14) that were previously described (31). Briefly, for each of the one-to-one genes a multiple sequence alignment (MSA) was built using MUSCLE (32), with the maximum number of iterations set to 100, followed by GBLOCKS (33), with parameters ‘-b3 = 8 –b4 = 2 –n = y’ to remove poorly aligned positions. The resulting 40 MSAs were concatenated and RAxML (34) was used to reconstruct the phylogenetic relationships for the 21 species using the default JTT model with 100 bootstrap iterations. The resulting species tree can be found in Supplementary Table S14 and visualized using EvolView (35).
Characteristics of ncRNAs and their putative roles in gene regulation
ncRNAs are abundantly expressed in M. pneumoniae (Supplementary Figure S3). The majority (236 out of 311; 76%) of the ncRNAs are antisense to protein-coding genes, suggesting putative regulatory roles. As expected, we found that coding genes targeted by genome-encoded antisense ncRNAs had relatively lower mRNA abundances (P = 0.0039; Wilcoxon Rank Sum Test) than untargeted genes, suggesting putative transcriptional interference. Furthermore, we found that the protein/mRNA abundance ratios of targeted coding genes were also significantly lower compared to the untargeted ones (P = 0.006; Supplementary Figure S8); for example, the three most targeted genes (red dots in Supplementary Figure S8) showed lower than average protein/mRNA ratios, suggesting post-transcriptional regulation by ncRNAs. The respective regulatory effects of ncRNAs on the abundances of either the target mRNAs or corresponding proteins differ among the distinct classes of genes with different abundances (Supplementary Figure S9) or functional categories (Supplementary Figure S8).
Similar results were found in M. genitalium: most of the ncRNAs (447 out of 494; 90.4%) overlap with antisense protein-coding genes; genes that overlapped with antisense ncRNAs showed decreased mRNA and protein abundances as compared with those that did not (P < 0.05).
M. pneumoniae–specific and conserved genes (i.e. those also found in M. genitalium) have a similar likelihood of being targeted by ncRNAs, thus implying that there is no preference with regard to targeting conserved genes. Coding genes targeted by antisense ncRNAs are not random in M. pneumoniae: proteins involved in the translation machinery or the regulation of translation efficiency are often heavily targeted (Supplementary Table S6; Figure S10). For example, among the top 10 most targeted genes (ordered according to the percentage of gene length covered by the antisense ncRNA), two are related to the assembly and regulation of the 50S ribosome complex, rplC and yceC. rplC is a member of the large ribosome complex (Supplementary Figure S10A) while yceC is a putative regulator of its assembly (Supplementary Figure S10C), and their disruption could lead to fitness and lethal phenotypes respectively (20). In addition, Ygl3, a putative tRNA methyltransferase essential in M. pneumoniae (Supplementary Figure S10C), is capable of controlling translation efficiency by increasing the tRNA methylation in eukaryotes (36).
Generally, the ncRNAs are not conserved between the two mycoplasmas, and the extent to which the coding genes overlap with antisense ncRNAs between one-to-one orthologs varies significantly between the two species (R = −0.025, P = 0.592; Supplementary Table S6). However, the heavily targeted genes (i.e. those which have ≥50% of their lengths overlapping with antisense ncRNAs) in M. genitalium (Supplementary Table S6) fall in the same functional categories as those in M. pneumoniae (Supplementary Figure S10). For example, ribosomal proteins are preferentially targeted. The ribosomal proteins rpsP, rpsD and rpsF are targeted by ncRNAs with significant overlap in M. genitalium. Interestingly, genes of the small (30S; Supplementary Figure S10B) ribosome subunit are preferentially targeted in M. genitalium, while those of the large (50S; Supplementary Figure S10A) subunit are targeted in M. pneumoniae; this is somewhat similar to the regulation of cell cycle expression, where the level of selection is the complex and not the individual gene or protein (37).
Furthermore it appears that if only one subunit of a stoichiometrically well-balanced protein complex is targeted, the entire complex becomes low abundant regardless of the exact protein that has been suppressed by the ncRNA (Supplementary Figure S10).
Identification and comparative analyses of lysine acetylation in M. pneumoniae, M. genitalium and the larger pathogen Salmonella enterica, subsp. enterica serovar Typhimurium LT2
A method previously described in ref. (2) was used to identify lysine acetylation sites in the three bacteria. To maximize the identification, lysine-acetylated peptides were enriched and three technical replicates were performed for each bacterium. In total we identified 3045, 4156 and 2804 acetylated lysine residues in M. pneumoniae, M. genitalium and S. enterica, respectively. The total number of lysine sites and acetylated ones identified per protein are listed in Supplementary Tables S7–S9. Based on the overlap between the triplicated samples, we estimated the total number of acetylated sites to be as high as 3500, 4500 and 4000 for M. pneumoniae, M. genitalium and S. enterica, respectively (Supplementary Figure S11), indicating that we have captured most of the possible acetylated lysines in the two mycoplasmas (87% and 92% for M. pneumoniae and M. genitalium respectively) and the majority (70%) of them in S. enterica.
Conserved proteins are more likely to be acetylated (Supplementary Figure S4). Indeed, metabolic enzymes, chaperones and proteins involved in transcription, protein turnover and PTMs, were preferentially lysine-acetylated (for each protein the number of lysine acetylation sites was normalized by the total number of lysines identified by MS; Supplementary Tables S7–S9; Figure S12). Interestingly, we found that the enzymes involved in central carbon metabolism and production of acetyl-CoA were all frequently lysine-acetylated (Supplementary Figure S13). High levels of acetyl-coA are known to induce increased lysine acetylation in the mitochondria of mammalian cells (38). Acetyl-coA is a key indicator of cellular energy status and can regulate enzymatic activities (39), thus providing an evolutionary conserved feedback loop in central carbon metabolism (Supplementary Figure S13).
In addition, we found that the likelihood that a protein is lysine-acetylated in both M. pneumoniae and S. enterica increases with the number of species in which orthologs of this protein can be found (Supplementary Figure S4).
We found that both the total numbers and proportions of acetylated lysines per protein between one-to-one orthologs in the two mycoplasmas are significantly correlated (R2 = 0.376, P < 2.2e-16, Pearson correlation; Supplementary Figure S14A). However, when exact lysine sites were examined in the alignments of one-to-one orthologs of the two mycoplasmas (conserved and non-conserved lysine residues between one-to-one orthologs were identified by using MUSCLE (32) to align the two protein sequences), we found that the percentage of conserved and non-conserved lysine sites being acetylated in each species was similar (Supplementary Figure S14B). The results did not change even upon considering the so-called ‘neighboring effects’ (i.e. when the acetylated lysine was not conserved, an alternative lysine could frequently be found within the immediate neighboring amino acids of the original aligned site in other species (2)), suggesting fast evolution of the PTM sites and their putative species-specific regulatory roles. However, the conserved lysine sites are more likely to be acetylated in both mycoplasmas than is randomly expected (P = 0.00037; Supplementary Figure S15), indicating a selective functional advantage.
Collection of genomic and genetic features for 1600 complete prokaryotic genomes
M. pneumoniae is a rather unique bacterium with respect to its highly reduced genome and yet free-living lifestyle. In order to extrapolate our findings to other bacterial species with confidence, we needed to identify the evolutionary forces that were at work. We therefore also performed comparative analyses with different groups of bacterial species. Genome sequences and annotations for 1600 completely sequenced prokaryotic genomes (as of January 2013) were downloaded from the NCBI GenBank (40). The following features were then calculated for each genome: number of protein-coding genes, median and mean protein length, total number of amino acids, numbers of select amino acids such as K (lysine), Y (tyrosine), T (threonine) and S (serine), genome size, and genomic and coding GC-contents.
In order to identify possible transcription factors in each genome, protein sequences were searched against the PFAM (41) domain profiles version 18 using HMMER3 (42), with an e-value cutoff of 0.01. Then, resulting domain hits were cross-compared with a list of DNA-binding domains downloaded from DBD (43). A protein was marked as a putative transcription factor if it contained one or more DNA-binding domains.
Operon predictions were obtained from DOOR, the database of prokaryotic operons (44). tRNAs were predicted using tRNAscan-SE version 1.3.1 (45) with parameter –G (use general tRNA model) on the downloaded genome sequences.
Regular and partial correlations
Pearson correlation coefficients between mRNA and protein concentrations were calculated using a built-in function, cor.test() in R (46). To estimate the contribution of selected sequence features and -omics data sets on protein abundance, independent of its mRNA abundance, partial correlation coefficients (Pearson) were also calculated using the R function pcor.test() with default parameters. The pcor.test can be obtained from http://www.yilab.gatech.edu/pcor.R.
Multiple regression using MARS
Multivariate adaptive regression splines (MARS) was used to describe the individual as well as combined contribution of the selected features to protein abundance. MARS is a non-parametric regression technique and is implemented in the ‘earth’ package (47) in R (46).
RESULTS AND DISCUSSION
As a first use case, we derived high-quality annotations for the M. pneumoniae and M. genitalium genomes (Supplementary Figure S2) by integrating previously published transcriptomics (RNAseq (27) and tiling arrays (1)), with newly derived deep sequencing RNAseq (at 6 and 96 h), transcription start site (TSS) associated RNAs (tssRNAs; 27) and quantitative proteome data. We were able to refine annotated ORFs, annotate new protein-coding genes and ncRNAs, and assign TSSs for all of them (see Materials and Methods). Taken together, our current annotation for M. pneumoniae contains 694 ORFs (32 smORFs, 43 conventional RNAs (rRNAs and tRNAs) as well as 311 ncRNAs (195 new; Supplementary Table S3), while the annotation for M. genitalium contains 544 (23 new since the last annotation (21); 12 refined) protein-coding genes, 36 tRNAs, 3 rRNAs and 494 new ncRNAs (Supplementary Table S4).
The resulting large number of ncRNAs (311) in M. pneumoniae was quite striking as we found 30 times more per million base pairs (MB) when compared to E. coli (5,6; Supplementary Table S5). Many of the ncRNAs are abundantly expressed during all stages of the growth curve (Supplementary Figure S3). As many as 85% overlap with protein-coding genes, with 76% of these being on the opposite strand (Supplementary Table S6). We previously found that 5% of the newly annotated ncRNAs are essential for the growth of M. pneumoniae in rich-media (20). Although overexpression of 11 of these ncRNAs did not affect the transcriptome or proteome of M. pneumoniae (Llorens et al., in print), we observed in our data set that coding genes targeted by genome-encoded antisense ncRNAs have lower mRNA abundances (P = 0.0039; Wilcoxon Rank Sum Test) and protein/mRNA abundance ratios (P = 0.004) than the untargeted ones. This could be due to antisense ncRNAs functioning by the generated RNA (48) or their generation itself (20,49,50), or simply reflect that poorly transcribed genes could tolerate transcriptional noise in the opposite strand. Similar results were found for M. genitalium (Supplementary Table S6).
We have previously detected a high number of phosphorylation sites in M. pneumoniae, and, surprisingly, even more lysine acetylation sites (2). We have now also measured them comparatively with both M. genitalium and S. enterica (4857 Kbp, 4423 protein-coding genes (51); Materials and Methods). We not only found four times as many lysine acetylation sites as previously reported (3045 versus 759 sites (2); Supplementary Table S7) in M. pneumoniae, but also many more in the smaller M. genitalium (4156; Supplementary Table S8), and significantly less in the larger S. enterica (2804; Supplementary Table S9). Starting with the smallest genome, at least 82%, 58% and 20% of lysine-containing proteins, and 25%, 15% and 4.6% of all identified lysines are found acetylated in the three bacteria, respectively (Supplementary Table S9), implying that smaller genomes tend to have considerable higher rates of lysine acetylation. Consistent with our observations, recent studies on acetylome profiling identified 1070 (52) and 1355 (53) unique acetylation sites in E. coli and B. subtilis, respectively; these numbers are lower than those of the two mycoplasmas. We did identify more unique acetylation sites in S. enterica than in E. coli or B. subtilis, even though they have similar genome sizes; this is likely due to the more sensitive technology used in our study and our exhaustive sampling strategy. The higher acetylation rate in streamlined genomes is likely due to the fact that a higher proportion of proteins in these genomes are involved in essential processes such as translation, transcription and metabolism; these proteins are often conserved, and more likely to be acetylated (52; see also Supplementary Figure S4).
In M. pneumoniae both the total number of acetylated lysines per identified protein and the proportion of acetylated lysines out of all identified lysines (taken into account so that the protein abundance can be controlled for; see Materials and Methods) correlate positively with the protein abundance (Pearson Correlation R = 0.47 and 0.35, P < 2.2 × 10−16). As our identification of acetylated sites reached saturation in M. pneumoniae, i.e. the total number of identified acetylation sites did not increase with additional experiments (Supplementary Figure S11), the observed positive correlation cannot be a byproduct of sampling biases (e.g. abundant proteins have higher chance to be sampled) and may imply a functional role for acetylation in protein abundance. Indeed, previous studies suggested that lysine acetylation may play regulatory roles in protein stability (54). Similarly, a positive correlation between protein abundance and the number of phosphorylation sites was observed (R = 0.28 and 0.41, P < 1.1 × 10−8; using data from (2)). However, we cannot rule out that these results are related to the sensitivity of the mass spectrometer, where low abundant peptides are not detected or only very rarely detected, resulting in a depletion of acetylated sites in low abundant proteins.
So far we have shown that both the abundantly expressed ncRNAs and extensive PTMs correlate with (and may contribute to) protein abundances, but in order to quantify their predictive power to the latter, other factors have to be taken into account. For example, protein half-life in M. pneumoniae is generally longer than the generation-time (17), while mRNA half-life is rather short (on average 8 min; Llorens V., Yus E. & Serrano, L. in preparation), similar to other bacteria (e.g. on average ≈5 min in both E. coli (55) and B. subtilis (56)). We thus derived from our M. pneumoniae resource a total of 20 features that could possibly be predictive of protein abundance (11; see Supplementary Table S10 for a complete list) when the contribution of the mRNAs is controlled for (a method called ‘partial correlation’ (57); also see Materials and Methods). Among them, 10 were found to correlate significantly with protein abundance at a given mRNA abundance and thus were retained for subsequent analysis (Figure 2).
Figure 2.
Factors controlling protein abundance. (A) Correlations of individual factors with mRNA and protein (partial) abundances. Levels of significance: *** < 0.001, ** < 0.01, * < 0.05. Percentages higher than 10% in the last column are highlighted in red. (B) Combined contributions of the factors listed above in (A) to protein abundance variation using MARS (Multivariate adaptive regression splines) analysis. (C) A schematic view of the information flow from genome to RNA to protein and the additional regulatory layers. The widths of the dark-blue arrows correspond to the relative contributions to protein abundances as compared with mRNA abundances.
On average <11% of the variation in protein abundance could be explained by mRNA abundance under the same conditions (Figure 2); this is much lower than in larger bacteria (e.g. 30–50% for E. coli (58)). In M. pneumoniae, many factors could have an impact on protein abundance and are hence called ‘regulatory layers’ hereafter. The largest contributor is the extent of lysine acetylation, which explains as much as 24% of the protein abundance variation (Figure 2), followed by protein half-life (17.09%), the length of gene overlap with its antisense ncRNAs (10.51%), sequence features including the proportion of leucines (9.58%) and codon adaptation index (CAI; 9.57%), as well as phosphorylation (8.46%).
Considering redundancies among these factors (Supplementary Table S10), together they are capable of explaining 55.3% of the variance in protein abundance (Figure 2B, see also Supplementary Table S10). These results indicate the need to factor in sequence constraints and regulatory layers when drawing conclusions from transcriptomic readouts to the protein landscape of a cell. Some of the remaining variance could be attributed to technical limitations associated with the quantification of transcriptomes (≈4%) and proteomes (≈17%; see Materials and Methods). Notably, a substantial proportion of the protein abundance variation (23.6%) remains unexplained (Figure 2). This suggests the existence of additional, independent regulatory layers such as translational controls or second messengers for which data are currently not available in M. pneumoniae.
Although deriving comparable data for two closely related mycoplasmas validates our observations, the power of individual features to predict protein abundance can obviously vary in other bacteria and might be heavily constrained by genome-reduction. To extrapolate our findings and derive scaling laws (a scaling law is a functional relationship between two variables where one varies as a function of the other) for related features, we performed comparative analyses with different groups of bacteria with varying phylogenetic distances to M. pneumoniae and M. genitalium.
Unlike the number of protein-coding genes, which tightly correlates with genome size (R2 = 0.97; Supplementary Figure S5; using 1600 complete bacterial genomes from NCBI as of January 2013, Supplementary Table S11), the number of ncRNAs appears to follow a different principle, as the smaller bacterium M. genitalium expresses more ncRNAs (Supplementary Table S5). We recently found that the number of ncRNAs per million bases shows a strong negative correlation with genomic GC-content (R2 = 0.88) in 20 selected bacteria (unpublished results). As Sigma 70 factors in bacteria are known to recognize A/T rich regions, it is possible that in genomes with a low GC-content more regions promote transcription in an unspecific way, and therefore having more ncRNAs could provide a needed level of control.
The GC-content can also explain ≈87.7% and ≈40% of the variation in acetylation and phosphorylation substrates (lysine% and serine%; Figure 3A,B), respectively. The codons for these amino acids are AT-rich and occur frequently in low-GC genomes (Supplementary Table S12). Larger bacteria such as S. enterica and E. coli encode fewer lysine residues per protein, after normalizing for protein lengths (Supplementary Table S11); the same is true for tyrosines and serines (59). Both conserved and species-specific proteins are equally affected (Supplementary Figure S6).
Figure 3.
The percentage of post-translationally modifiable residuals (PTMRs) decreases with decreasing GC-content. Colored dots: selected model bacterial species; black dots: the other 1600 bacterial species. (A) The proportion of putative acetylation targets (lysine - K) in a genome decrease with an increasing genomic GC-content. (B) Proportion of putative phosphorylation targets as a function of genomic GC-content; shown is the major phosphorylation target serine – S.
These observations have several important implications. Firstly, in two species with comparable genome sizes but different GC-contents (e.g. E. coli and B. subtilis) the numbers of post-translationally modifiable residues (PTMRs) per protein are very different (Figure 3). Secondly, the incidences of the two types of PTMRs decrease differently. For instance, the genomic GC-content increases from 31.5% in M. genitalium to 51.9% in E. coli, and the frequency of lysines drops from 9.5% to 4.4% (2.15-fold), but that of tyrosines, threonines and serines remains largely unaffected (varying from 6.6% to 5.7%, or 1.15-fold). Due to the fact that the interactions between the two types of PTMs, i.e. decreased protein phosphorylation affects acetylation and vice versa, depend on the frequency of the modification (2), our results suggested that GC-content could be a key indicator in the dynamics of crosstalk among PTMs in prokaryotes.
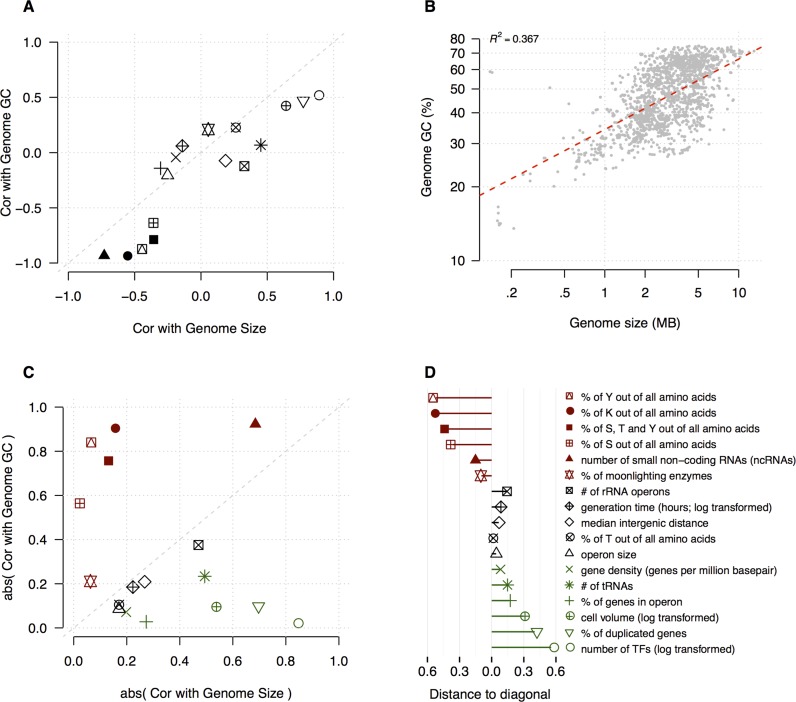
Many genomic and genetic properties such as the number of transcription factors or the percent of duplicated genes that correlate with GC-content, correlate similarly with genome size (Figure 4A), due to the correlation between the two factors (R2 = 0.367; Figure 4B). To dissect the relative predictive power of one factor independent of the other for selected genomic and genetic features, we again used partial correlation (57) to revisit existing and newly identified scaling laws. This way we were able to show that the number of transcription factors increases with genome size (60) regardless of GC-content (Figure 4). Similar results were found for operon size, proportion of genes in operons, gene density and proportion of duplicated genes (Figure 4C). Conversely, the correlations with the proportion of select amino acids (serine, tyrosine, lysine) could be attributed mostly to genome GC-content (Figure 4D). These results confirm that GC-content is a better predictor for features that have important regulatory roles while genome size appears to be more directly associated to the pool of proteins, i.e. functional capacity (Figure 4D).
Figure 4.
Dissecting the relative predictive powers of genome size and GC-content on selected genomic and genetic features that have been used to derive scaling laws. (A) Most of the selected features correlate with both genome size and GC-content in a similar way using regular Pearson Correlation; (B) Genome size and GC-content correlate significantly across 1600 bacteria. The dashed red-line represents the linear regression. (C) Separating the impact of one factor from the other using partial correlation. Genome size was found to have more predictive power than GC-content for some features (in dark-green), while for others, GC-content was found to be more predictive (in dark-red). A factor (i.e. genome size or genome GC) is defined as a major contributor if it has significantly more predictive power than the other. For this to be true one of the following conditions must be satisfied: (i) it (genome size or GC-content) correlates significantly with a genomic feature while the other factor (GC or genome size) does not, or (ii) both correlate significantly with a genomic feature, but one (GC or genome size) has an absolute correlation coefficient value that is twice as high or higher than the other (genome size or GC). (D) Distances of the features in (C) to the diagonal line showing the relative predictive power (absolute partial correlation coefficient value) of GC-content over genome size; the more to the left on the x-axis, the more predictive power of GC-content over genome size. Black data points in (C) and (D) are those for which the GC-content and genome size have similar predictive powers. See Supplementary Table S13 for the data.
In bacteria, the reduction of genome size has been attributed to a variety of factors, e.g. degenerative reduction because of parasitic life styles (e.g. pathogens) or adaptive streamlining because of environmental energetic constraints (61); either way, genome-reduction is often accompanied by decreasing genome GC-content (Supplementary Table S11). The decreasing complexity of traditional regulatory networks consisting of transcription factors that comes along with genome size reduction (Supplementary Figure S7; R2 = 0.8; see also (60)), appears to be counteracted by elevated nonconventional regulatory layers including ncRNAs, and PTMs. Thus, the evolutionary forces constraining genome size and GC-content modify the relative contributions of the regulatory mechanisms to proteome homeostasis, and impact more genomic and genetic features than previously appreciated.
Taken together, we make use of the richest resource that has been assembled so far for any bacterium, as measured by base pair coverage and diversity. This resource has a huge potential to boost systems biology research in M. pneumoniae and beyond. We were able to quantify the predictive power of different factors in estimating protein abundance, many of which follow simple scaling laws (see Supplementary Figure S16 for an overview of our data integration workflow), demonstrating that global principles can be derived from this genome-reduced bacterium.
Supplementary Material
Acknowledgments
We thank the CRG ultra sequencing facility (Heinz Himmelbauer) and CRG/UPF Proteomics Unit (Eduard Sabidó). Many thanks to Tony Ferrar for critical manuscript revision and language editing (http://theeditorsite.com).
Author contributions: P.B., L.S. and A.C.G. conceived the study; W.H.C., V.v.N. and M.L.S. assembled and analyzed the data and wrote the manuscript; P.B., L.S. and A.C.G. revised the manuscript; M.L.H. generated and helped with the analyses of the post-translational modification (PTM) data; J.A.H.W. developed the online database; E.Y., A.A. and G.R. generated and helped with the analyses of the transcriptome data; D.R.M. generated the orthologous groups and the species tree; C.P. and A.T. helped with the analyses of the PTM data; all authors have read and approved the manuscript.
Footnotes
Present address: Vera van Noort, Department of Microbial and Molecular Systems (M2S), KU Leuven, Kasteelpark Arenberg 20, 3001 Leuven, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007–2013)/ERC Grant Agreement No. 232913. We acknowledge support from the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’, SEV-2012–0208. This project has received funding from the European Union's Horizon 2020 research and innovation program under Grant Agreement No. 634942. Funding for open access charge: European Research council (ERC) advanced Grant Agreement No. 232913, the Fundación Marcelino Botin, the Spanish Ministerio de Economía y Competitividad BIO2007-61762, the ISCIII, Subdirección General de evaluación y fomento de la investigación PI10/01702 to the ICREA researcher L.S.
Conflict of interest statement. None declared.
REFERENCES
- 1.Guell M., van Noort V., Yus E., Chen W.H., Leigh-Bell J., Michalodimitrakis K., Yamada T., Arumugam M., Doerks T., Kuhner S., et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 2.van Noort V., Seebacher J., Bader S., Mohammed S., Vonkova I., Betts M.J., Kuhner S., Kumar R., Maier T., O'Flaherty M., et al. Cross-talk between phosphorylation and lysine acetylation in a genome-reduced bacterium. Mol. Syst. Biol. 2012;8 doi: 10.1038/msb.2012.4. 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G.-W., Xie X.S. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 5.Raghavan R., Groisman E.A., Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–1497. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D., Hong J.S., Qiu Y., Nagarajan H., Seo J.H., Cho B.K., Tsai S.F., Palsson B.O. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet. 2012;8:e1002867. doi: 10.1371/journal.pgen.1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duerig A., Abel S., Folcher M., Nicollier M., Schwede T., Amiot N., Giese B., Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd C.D., O'Toole G.A. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Ann. Rev. Cell Dev. Biol. 2012;28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonough K.A., Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat. Rev. Microbiol. 2012;10:27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karr J.R., Sanghvi J.C., Macklin D.N., Gutschow M.V., Jacobs J.M., Bolival B., Jr, Assad-Garcia N., Glass J.I., Covert M.W. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel C., de Sousa Abreu R., Ko D., Le S.-Y., Shapiro B.A., Burns S.C., Sandhu D., Boutz D.R., Marcotte E.M., Penalva L.O. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 2010;6 doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wodke J.A., Alibes A., Cozzuto L., Hermoso A., Yus E., Lluch-Senar M., Serrano L., Roma G. MyMpn: a database for the systems biology model organism Mycoplasma pneumoniae. Nucleic Acids Res. 2015;43:D618–D623. doi: 10.1093/nar/gku1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lluch-Senar M., Luong K., Llorens-Rico V., Delgado J., Fang G., Spittle K., Clark T.A., Schadt E., Turner S.W., Korlach J., et al. Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution. PLoS Genet. 2013;9:e1003191. doi: 10.1371/journal.pgen.1003191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vivancos A.P., Guell M., Dohm J.C., Serrano L., Himmelbauer H. Strand-specific deep sequencing of the transcriptome. Genome Res. 2010;20:989–999. doi: 10.1101/gr.094318.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier T., Schmidt A., Guell M., Kuhner S., Gavin A.C., Aebersold R., Serrano L. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol. Syst. Biol. 2011;7:511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhner S., van Noort V., Betts M.J., Leo-Macias A., Batisse C., Rode M., Yamada T., Maier T., Bader S., Beltran-Alvarez P., et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 19.Yus E., Maier T., Michalodimitrakis K., van Noort V., Yamada T., Chen W.H., Wodke J.A., Guell M., Martinez S., Bourgeois R., et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 20.Lluch-Senar M., Delgado J., Chen W.H., Llorens-Rico V., O'Reilly F.J., Wodke J.A., Unal E.B., Yus E., Martinez S., Nichols R.J., et al. Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome-reduced bacterium. Mol. Syst. Biol. 2015;11:780. doi: 10.15252/msb.20145558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser C.M., Gocayne J.D., White O., Adams M.D., Clayton R.A., Fleischmann R.D., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 22.Parraga-Nino N., Colome-Calls N., Canals F., Querol E., Ferrer-Navarro M. A Comprehensive Proteome of Mycoplasma genitalium. J. Proteome Res. 2012;11:3305–3316. doi: 10.1021/pr300084c. [DOI] [PubMed] [Google Scholar]
- 23.Suthers P.F., Dasika M.S., Kumar V.S., Denisov G., Glass J.I., Maranas C.D. A genome-scale metabolic reconstruction of Mycoplasma genitalium, iPS189. PLoS Comput. Biol. 2009;5:e1000285. doi: 10.1371/journal.pcbi.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glass J.I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M.R., Maruf M., Hutchison C.A. 3rd, Smith H.O., Venter J.C. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arifuzzaman M., Maeda M., Itoh A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H.C., Hirai A., et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCutcheon J.P., McDonald B.R., Moran N.A. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yus E., Guell M., Vivancos A.P., Chen W.H., Lluch-Senar M., Delgado J., Gavin A.C., Bork P., Serrano L. Transcription start site associated RNAs in bacteria. Mol. Syst. Biol. 2012;8:585. doi: 10.1038/msb.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaffe J.D., Berg H.C., Church G.M. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- 29.Boonmee A., Ruppert T., Herrmann R. The gene mpn310 (hmw2) from Mycoplasma pneumoniae encodes two proteins, HMW2 and HMW2-s, which differ in size but use the same reading frame. FEMS Microbiol. Lett. 2009;290:174–181. doi: 10.1111/j.1574-6968.2008.01422.x. [DOI] [PubMed] [Google Scholar]
- 30.Powell S., Szklarczyk D., Trachana K., Roth A., Kuhn M., Muller J., Arnold R., Rattei T., Letunic I., Doerks T., et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 32.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 34.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H., Gao S., Lercher M.J., Hu S., Chen W.H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–W572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 37.Jensen L.J., Jensen T.S., de Lichtenberg U., Brunak S., Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- 38.Kim S.C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson D.A., Karsch-Mizrachi I., Clark K., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 43.Wilson D., Charoensawan V., Kummerfeld S.K., Teichmann S.A. DBD–taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dam P., Olman V., Harris K., Su Z., Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 2007;35:288–298. doi: 10.1093/nar/gkl1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Team R.C. R: A language and environment for statistical computing. 2013. https://www.R-project.org/ https://www.R-project.org/ R Foundation for Statistical Computing.
- 47.Milborrow S. earth: Multivariate Adaptive Regression Splines. 2012. http://CRAN.R-project.org/package=earth http://CRAN.R-project.org/package=earth
- 48.Faghihi M.A., Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G.Z., Lercher M.J., Hurst L.D. Transcriptional coupling of neighboring genes and gene expression noise: evidence that gene orientation and noncoding transcripts are modulators of noise. Genome Biol. Evol. 2011;3:320–331. doi: 10.1093/gbe/evr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClelland M., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 52.Zhang K., Zheng S., Yang J.S., Chen Y., Cheng Z. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 2013;12:844–851. doi: 10.1021/pr300912q. [DOI] [PubMed] [Google Scholar]
- 53.Kosono S., Tamura M., Suzuki S., Kawamura Y., Yoshida A., Nishiyama M., Yoshida M. Changes in the Acetylome and Succinylome of Bacillus subtilis in Response to Carbon Source. PLoS One. 2015;10:e0131169. doi: 10.1371/journal.pone.0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caron C., Boyault C., Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays. 2005;27:408–415. doi: 10.1002/bies.20210. [DOI] [PubMed] [Google Scholar]
- 55.Selinger D.W., Saxena R.M., Cheung K.J., Church G.M., Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hambraeus G., von Wachenfeldt C., Hederstedt L. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics. 2003;269:706–714. doi: 10.1007/s00438-003-0883-6. [DOI] [PubMed] [Google Scholar]
- 57.Kim S.-H., Yi S. Understanding relationship between sequence and functional evolution in yeast proteins. Genetica. 2007;131:151–156. doi: 10.1007/s10709-006-9125-2. [DOI] [PubMed] [Google Scholar]
- 58.Taniguchi Y., Choi P.J., Li G.W., Chen H., Babu M., Hearn J., Emili A., Xie X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cozzone A.J. Protein phosphorylation in prokaryotes. Annu. Rev. Microbiol. 1988;42:97–125. doi: 10.1146/annurev.mi.42.100188.000525. [DOI] [PubMed] [Google Scholar]
- 60.van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 61.Giovannoni S.J., Tripp H.J., Givan S., Podar M., Vergin K.L., Baptista D., Bibbs L., Eads J., Richardson T.H., Noordewier M., et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.