Abstract
During exocytosis, fusion pores form the first aqueous connection that allows escape of neurotransmitters and hormones from secretory vesicles. Although it is well established that SNARE proteins catalyze fusion, the structure and composition of fusion pores remain unknown. Here, we exploited the rigid framework and defined size of nanodiscs to interrogate the properties of reconstituted fusion pores, using the neurotransmitter glutamate as a content-mixing marker. Efficient Ca2+-stimulated bilayer fusion, and glutamate release, occurred with approximately two molecules of mouse synaptobrevin 2 reconstituted into ~6-nm nanodiscs. The transmembrane domains of SNARE proteins assumed distinct roles in lipid mixing versus content release and were exposed to polar solvent during fusion. Additionally, tryptophan substitutions at specific positions in these transmembrane domains decreased glutamate flux. Together, these findings indicate that the fusion pore is a hybrid structure composed of both lipids and proteins.
Membrane fusion and fission reactions underlie the compartmentalization that is a hallmark of all eukaryotic cells. Fusion underlies many processes ranging from fertilization and viral entry to hormone signaling and synaptic transmission1,2. Defects in membrane-fusion pathways are detrimental to cellular homeostasis and are associated with numerous human diseases3-5. In most cases, intracellular fusion reactions are directly mediated by soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs), which form the core of a conserved membrane fusion machine. In presynaptic nerve terminals, the SNARE complex that mediates synaptic vesicle (SV) exocytosis is composed of the vesicular SNARE (v-SNARE) synaptobrevin 2 (syb2; also referred to as VAMP2) and the target-membrane SNAREs (t-SNAREs) syntaxin 1A and SNAP-25B. These proteins assemble into four-helix bundles, to which SNAP-25B contributes two helices, and syb2 and syx1A contribute one helix each6. v- and t-SNAREs have been proposed to progressively zipper from their N-terminal domains toward their C-terminal domains, pulling the bilayers together and providing sufficient energy for fusion. SNARE-complex assembly is strictly regulated by a large number of regulatory factors7, including synaptotagmin 1 (syt1), a Ca2+ sensor that triggers rapid neuronal exocytosis8–10.
Although the central role of SNAREs in membrane fusion is well established, the structure of the fusion pore remains enigmatic. Two distinct hypotheses have been proposed11-13. In the lipid-stalk-fusion hypothesis, the outer leaflets of the membranes destined to fuse merge, initially forming a hemifusion stalk that resolves to a hemifusion diaphragm; the fusion pore then forms in the diaphragm and is purely lipidic14. This model is supported by molecular dynamic simulations and by physical chemistry experiments showing that membrane fusion, under specific conditions, can occur in the absence of proteins15-18. Alternatively, a proteinaceous fusion-pore model has also been proposed, in which the fusion pore is lined by the transmembrane domains (TMDs) of syntaxin1A and synaptobrevin 2 (ref. 11). This model is supported by experiments showing that mutations in the transmembrane domains of SNARE proteins alter the flux of hormones through fusion pores in a predictable manner19-21.
A major limitation in the study of fusion pores concerns their ephemeral nature. For example, in endocrine cells, the duration of the initial open state of the fusion pore is on the order of milliseconds; the pore then either closes (kiss-and-run exocytosis) or dilates to result in full fusion11,22. The transient nature of fusion pores has severely limited biochemical efforts to probe their composition and structure. Here, we attempt to address this critical question by using the rigid framework of nanodiscs, which prevent the dilation of fusion pores. We developed an assay in which nanodiscs bearing v-SNAREs fuse with small unilamellar vesicles (SUVs) containing t-SNAREs, in a manner accelerated by Ca2+ and synaptotagmin-1. We also made use of a bona fide neurotransmitter, glutamate, to monitor flux through the reconstituted pores via an optical sensor, iGluSnFR23. We found that efficient Ca2+-stimulated bilayer fusion, and glutamate release, required only two molecules of synaptobrevin 2 (syb2) and occurred when we used 6-nm nanodiscs. In addition, we determined that the transmembrane domains of SNAREs are exposed to solvent during fusion. Collectively, these data reveal that the fusion pore is formed by a combination of lipids and SNARE transmembrane domains.
RESULTS
Reconstitution of Syb2 into 6- and 13-nm nanodiscs
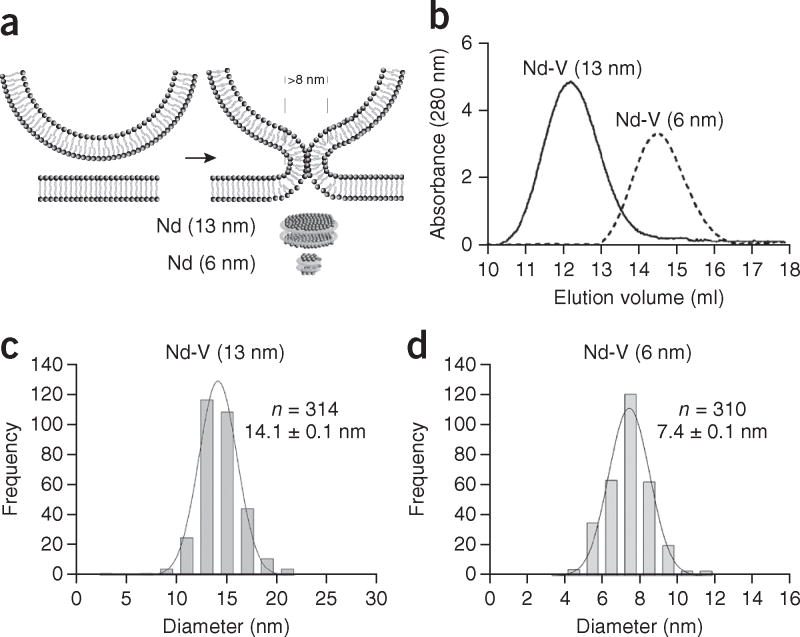
According to the lipid-stalk-fusion hypothesis, the fusion pore is purely lipidic. Given that each bilayer has a thickness of 4–5 nm (refs. 24,25), the fusion pore formed in nanodiscs would require a diameter >8 nm (Fig. 1a). Importantly, the size of nanodiscs can be controlled by the use of different membrane scaffold proteins (MSPs)26. In principle, the lipid-stalk-fusion model could be tested by use of discs that are too small to allow for the formation of a purely lipidic pore. If pores are solely lipidic, they should not form between liposomes and 6-nm nanodiscs, whereas robust membrane fusion and content release should be observed with 13-nm nanodiscs. Notably, an earlier study has reported bilayer fusion and content release in systems with SNAREs alone and 13-nm nanodiscs27.
Figure 1.
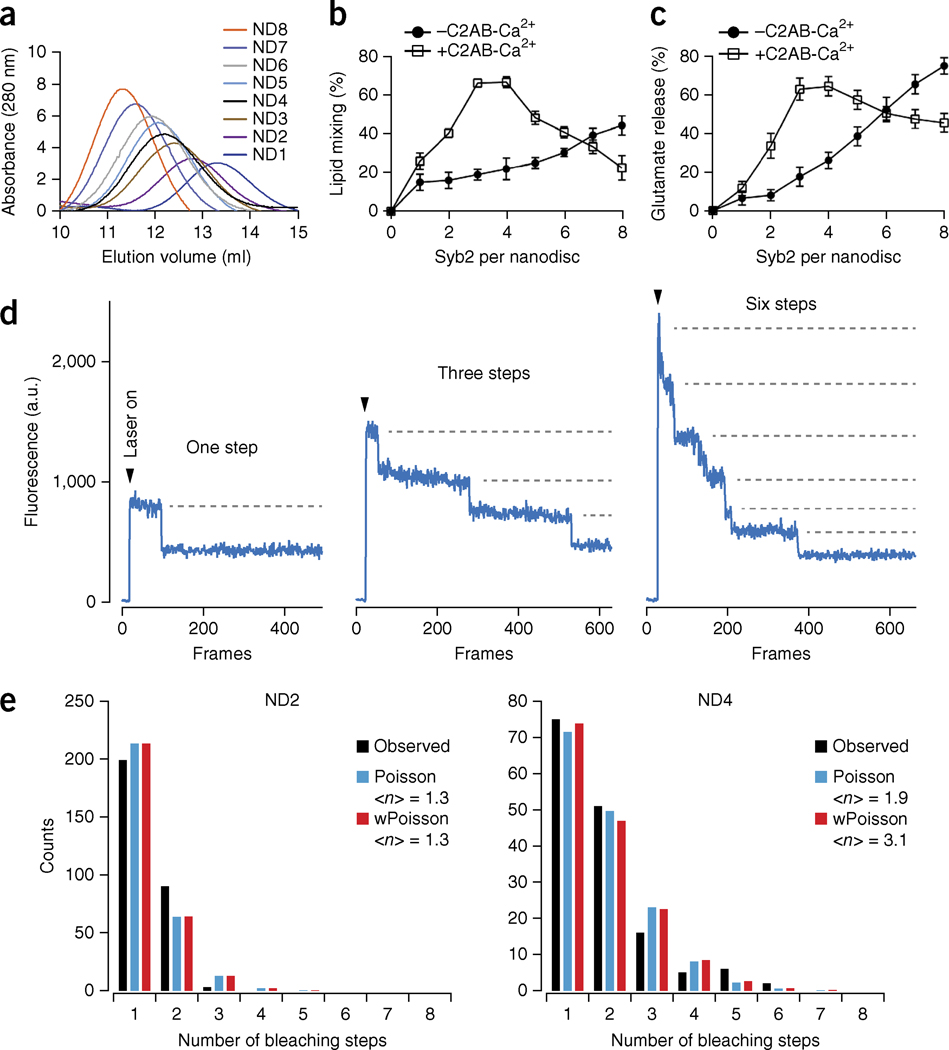
Reconstitution of syb2 into 6- and 13-nm nanodiscs. (a) Illustration of a lipidic fusion pore and the relative sizes of the two kinds of nanodiscs used in this study. (b) SEC of 6- and 13-nm nanodiscs. (c,d) Diameter distributions of 6-nm nanodiscs (d) and 13-nm nanodiscs (c) determined by AFM imaging. Data represent means ± s.d. (n = number of particles analyzed).
To conduct these experiments, we reconstituted syb2 into nanodiscs, using MSP1D1ΔH4–H6 (which yields 6-nm discs)28 and MSPE3D1 (which yields 13-nm discs)26. Size-exclusion chromatography (SEC) analysis of these v-SNARE–bearing nanodiscs (Nd-V) revealed sizes of 90 ± 10 and 230 ± 20 kDa (Fig. 1b), respectively, for these two distinct scaffolding proteins. Taking into account the size of the empty 6- and 13-nm nanodiscs (Supplementary Fig. 1a), we estimated that three and four molecules, respectively, of syb2 were reconstituted into the small and large discs (discussed further below). Using atomic force microscopy (AFM) imaging, we determined the nanodisc diameters to be 7.4 and 14 nm (Fig. 1c,d and Supplementary Fig. 1b), respectively; moreover, the size distributions were tightly clustered. Because scanning AFM imaging tends to overestimate the sizes of particles29, and because the scaffolding proteins contribute to the measured diameter, the bilayer diameter in the small discs is likely to be 6 nm or less, in agreement with results from previous studies26,28. In addition, we also reconstituted t-SNARE heterodimers into nanodiscs (Supplementary Fig. 1c,d). However, because SEC analysis revealed a broad elution profile indicative of a polydisperse sample, we did not use these discs in the following experiments; instead, we focused on Nd-V.
Bilayer fusion between nanodiscs and proteoliposomes
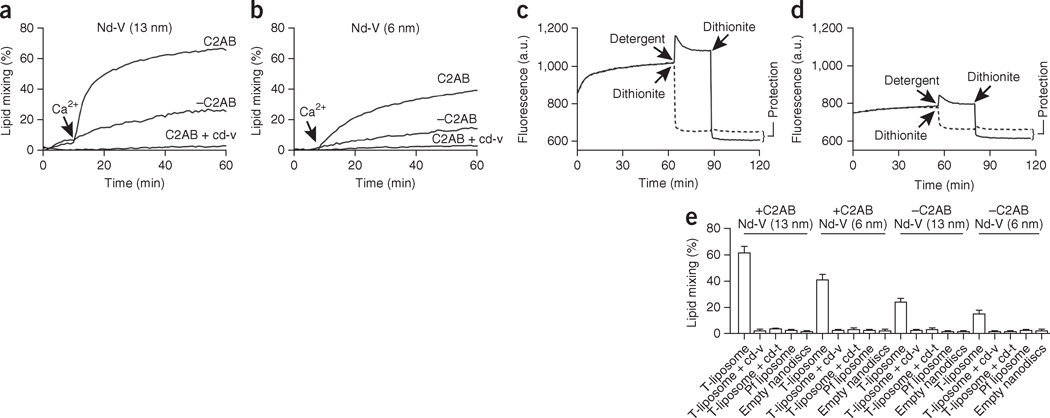
We first used a lipid-mixing assay to test whether 6-nm and 13-nm nanodiscs could fuse with t-SNARE SUVs (Fig. 2 and Supplementary Fig. 2a). We reconstituted nanodiscs with syb2, nitro-2-1,3-benzoxadiazol-4-yl (NBD)-phosphatidylethanolamine (PE) and rhodamine-PE (1.5 mol% each), probes that are widely used to study SNARE-catalyzed fusion8,27,30,31. In this system, fusion between Nd-V and t-SNARE liposomes resulted in the dilution of the NBD-rhodamine FRET pair and consequent dequenching of the NBD fluorescence, as we observed when using both 6- and 13-nm nanodiscs (Fig. 2a,b). In the presence of the cytoplasmic domain of syt1 (comprising tandem Ca2+-binding C2 domains tethered together by a short linker, designated C2AB), fusion was inhibited; then, upon addition of Ca2+, fusion was strongly stimulated. Lipid mixing was undetectable when we used protein-free (pf) liposomes or empty nanodiscs, and it was blocked by addition of the cytoplasmic domain of syb2 (cd-v) or the cytoplasmic domain of t-SNAREs (cd-t) (Fig. 2e), thus demonstrating that lipid mixing was mediated by trans-SNARE pairing.
Figure 2.
Bilayer fusion between v-SNARE nanodiscs (Nd-V) and t-SNARE vesicles. (a,b) Time courses of lipid mixing with 13-nm Nd-V (a) and 6-nm Nd-V (b), in the presence or absence of C2AB and cd-V; Ca2+ was added as indicated. (c,d) Dithionite quenching after fusion between t-SNARE vesicles and 13-nm Nd-V (c) or 6-nm Nd-V (d). A.u., arbitrary units. (e) Nd-SUV lipid mixing assays performed with t-SNARE liposomes, pf liposomes or empty nanodiscs; in each case, assays were run in the presence or absence of cd-v, cd-t and C2AB, and in the presence of 1 mM Ca2+. Data represent means ± s.d. (n = 3 technical replicates).
Because lipid mixing can be the result of hemifusion, rather than full fusion between nanodiscs and liposomes, we used dithionite to determine whether both leaflets fused in the nanodisc-SUV fusion assays8,27; lipid mixing in both leaflets is indicative of full fusion. After initiating lipid mixing, we added dithionite, which irreversibly quenches the fluorescence of NBD. Because dithionite does not readily cross lipid bilayers, the NBD in the inner leaflet would be protected if: (i) full fusion were to occur, and (ii) fusion pores were to close after full fusion (Supplementary Fig. 2a). Notably, a previous study has reported that all fusion pores that form between nanodiscs and SUVs ultimately close, probably because of the extreme curvature of the fusion product27. Protection from dithionite quenching, after fusion, was clearly observable in samples with either 6-nm or 13-nm nanodiscs but not in detergent-solubilized samples (Fig. 2c,d). Coflotation assays and AFM imaging revealed that nanodiscs remained bound to t-SNARE liposomes (Supplementary Fig. 2d,e). Therefore, full fusion occurred, and this was followed by closure of fusion pores via a mechanism that did not involve dissociation of the Nd-V. We observed no protection of the NBD from dithionite when we used empty nanodiscs (Supplementary Fig. 2b) or when we added dithionite at the beginning of the reactions to allow the reducing agent to enter the lumen of SUVs via nascent pores (Supplementary Fig. 2c).
Reconstitution of glutamate release with nanodiscs
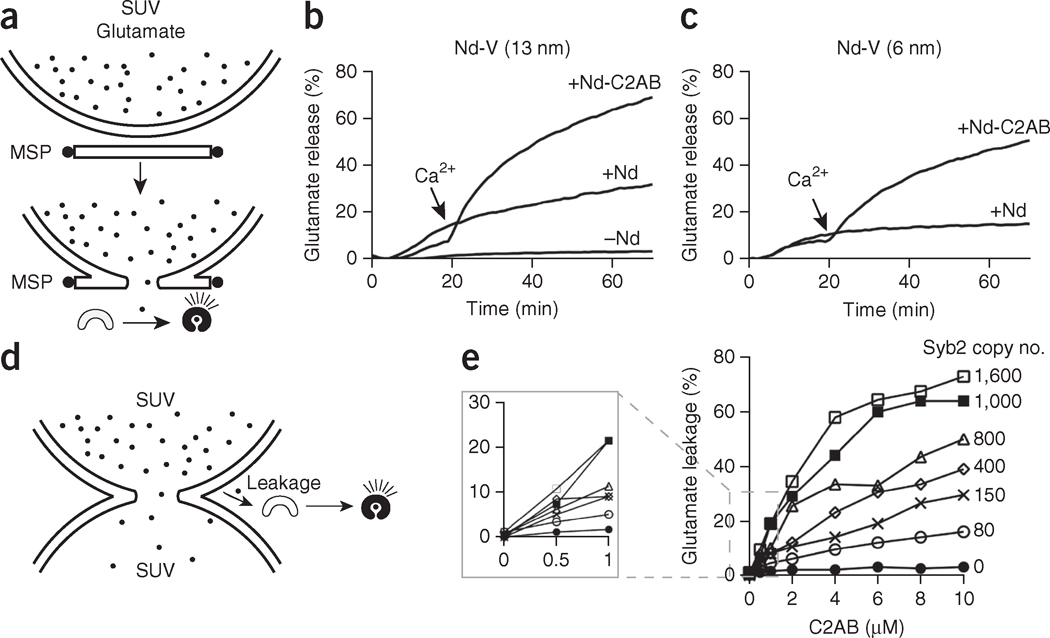
The fusion pore formed in nanodiscs can reseal and thus might not remain open long enough for neurotransmitter release, at least under some experimental conditions27. To address this, we tested whether a bona fide neurotransmitter, glutamate, could be released through fusion pores formed in the nanodisc-SUV fusion assay. We encapsulated glutamate into t-SNARE liposomes, which we then incubated with Nd-V in the presence of the optical sensor for glutamate iGluSnFR23. Glutamate flux through the fusion pore resulted in a robust increase in the iGluSnFR fluorescence signal (Fig. 3a) in systems with either 6- or 13-nm Nd-V (Fig. 3b,c), and release was strongly stimulated by addition of C2AB plus Ca2+. We also measured glutamate release under a wide range of nanodisc concentrations (Supplementary Fig. 3). The data revealed a Hill coefficient of ~0.9, which indicated that multiple nanodiscs do not work together, or cooperate, to destabilize vesicles and cause glutamate release. We conclude that Ca2+-stimulated bilayer fusion and glutamate release can be reconstituted with 6-nm nanodiscs. Finally, we confirmed the findings obtained in the presence of C2AB by also reconstituting full-length syt1, and we obtained similar results (Supplementary Fig. 4).
Figure 3.
Reconstitution of glutamate release through nanodiscs. (a) Illustration summarizing the glutamate release assay. During fusion between Nd-V and t-SNARE vesicles, glutamate is released through the fusion pore, thus resulting in increases in the fluorescence signal of the glutamate sensor (iGluSnFR). (b,c) Time courses of glutamate release with 13-nm Nd-V (b) and 6-nm Nd-V (c). (d) Illustration summarizing the membrane leakage assay using v- and t-SNARE vesicles. Glutamate leakage during fusion between v-SNARE and t-SNARE liposomes is detected via iGluSnFR. (e) Glutamate leakage as a function of Ca2+-C2AB concentration and v-SNARE copy number. Data represent means ± s.d. (n = 3 technical replicates).
Previous studies have shown that membrane fusion can be accompanied by leakage of content-mixing markers32,33, so we set out to determine whether the Ca2+-stimulated release of glutamate indeed occurred through fusion pores or whether there was substantial leakage. To address this, we used SUV-SUV fusion assays and took advantage of the glutamate-iGluSnFR reporter system. By placing the sensor in the medium, outside of the liposomes, leakage could readily be measured. This approach sharply contrasts with the common use of self-quenching soluble fluorescent dyes as content release markers, because these reporters do not discriminate between flux through pores (from the lumen of one SUV to another) and leakage (diffusion into the medium).
We incubated t-SNARE liposomes, containing glutamate, with syb2 liposomes in the presence of Ca2+ and C2AB (Fig. 3d). Glutamate leakage during fusion between t- and v-SNARE liposomes was readily monitored via iGluSnFR in the medium. We observed that high levels of syb2 (~1,600 syb2 per vesicle), in conjunction with a high concentration of Ca2+-C2AB (10 μM), led to excessive glutamate leakage (~70%). However, when we used low levels of syb2 (~80 syb2 per vesicle) and Ca2+-C2AB (1 μM), glutamate leakage was only ~5% (Fig. 3e). Hence, in the subsequent nanodisc-SUV fusion experiments we used 1 μM C2AB and always used eight or fewer copies of syb2 per disc, to ensure <5% glutamate leakage. Because glutamate release in the nanodisc-SUV system was ~65–70%, neurotransmitter flux in this system mainly occurred through fusion pores; we return to this point further below, in experiments using mutant and modified SNAREs.
Uncoupling lipid mixing from content mixing
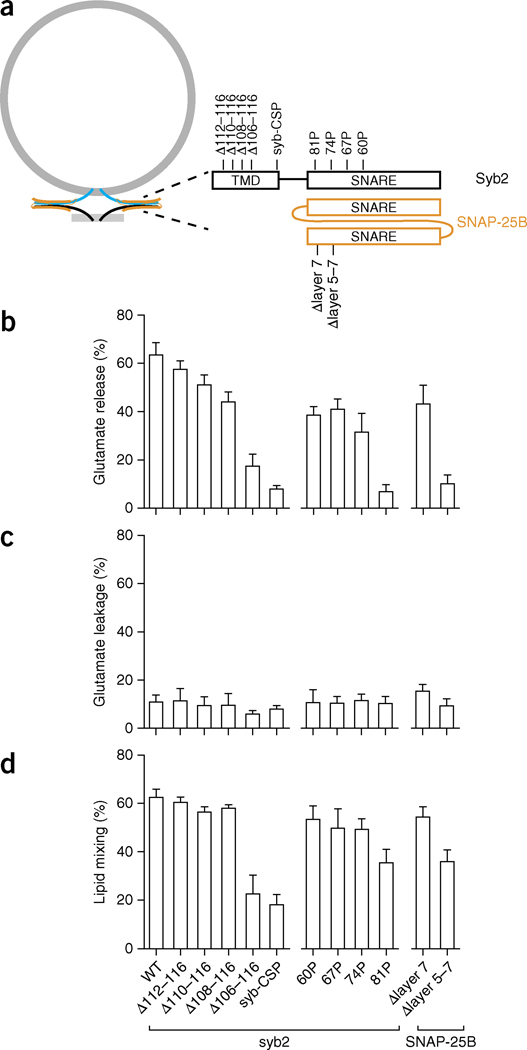
In the next series of experiments, we took advantage of the nanodisc-SUV fusion system to clarify whether alterations in SNARE proteins could differentially affect lipid mixing versus content release. We perturbed the TMD of syb2 and the SNARE motifs of both syb2 and SNAP-25B (Fig. 4a), and studied how these alterations affected lipid mixing, cargo leakage and cargo flux (Fig. 4b–d).
Figure 4.
Distinct structural elements of SNAREs differentially affect bilayer fusion, membrane leakage and glutamate release. (a) Illustration of the alterations in syb2 and SNAP-25B used to dissociate their functions. Constructs: syb-CSP, cytoplasmic domain of syb2 fused to the cysteine-rich segment of the SV protein CSP (cysteine string protein); Δlayer 5–7, deletion from layers 5 to 7 in the second SNARE motif of SNAP-25B; Δlayer 7, deletion of layer 7 in the same motif of SNAP-25B. (b–d) Characterization of SNARE mutants through glutamate release (b), glutamate leakage (c) and lipid mixing assays (d). Data represent means ± s.d. (n = 3 technical replicates). WT, wild type.
Previous studies have proposed that SNARE proteins progressively zipper together, in discrete stages, thereby forming four-helix bundles34. Hence, we used proline mutations, which perturb helices and often break helical continuity35,36, to disrupt the SNARE motif in specific positions. We introduced proline substitutions in the second half of the motif within syb2, at positions 60, 67, 74 and 81 (Fig. 4a). All of these mutants gave rise to similar levels of membrane leakage (Fig. 4c) but yielded unexpected effects on glutamate release and lipid mixing activity (Fig. 4b,d). The 60, 67 and 74P mutations resulted in modest, but significant (P < 0.05 by two-tailed Student’s t test; n = 3 technical replicates) and equal, reductions in lipid-mixing activity. These same mutations had larger effects on glutamate release, but again all three mutants appeared to be equivalent. However, a proline at position 81 profoundly inhibited content release compared to lipid-mixing activity. These experiments demonstrate that mutations in SNAREs can uncouple their functions in lipid mixing and content release and that the extreme C-terminal end of the SNARE motif has a particularly important role in content release37, perhaps by holding the fusion pore open for a long enough time to allow efficient escape of cargo. These findings are further supported by the observations that deletion of layers 5–7 (Δlayer 5–7) in SNAP-25B had no effect on membrane leakage, and had only a modest effect on lipid mixing, but strongly inhibited glutamate flux through the pore. Again, the membrane-proximal region of the SNARE motif appears to be particularly important for content release.
Next, we fused the cytoplasmic domain of syb2 to the cysteine-rich segment of the SV protein CSP (cysteine string protein) and anchored the fusion protein onto the headgroup of PE on the nanodisc surface, through a maleimide-thiol reaction. The resulting mutant (designated syb-CSP) exhibited 30% of the lipid mixing and 10% of the glutamate release activity exhibited by the wild-type protein (Fig. 4b,d). These results suggest that the TMD of syb2 might also have distinct functions during lipid mixing versus glutamate release. We explored this issue further by preparing a set of syb2 mutants that contained truncations in their TMDs. Stepwise truncations from residue 116 to 108 of syb2 gradually decreased glutamate release yet had no effect on lipid mixing, in agreement with previous work showing that a deletion in the TMD of a yeast v-SNARE can result in hemifusion rather than full fusion38. In contrast, further truncation by only two additional residues (to position 106) diminished both lipid mixing and glutamate release by 50% (Fig. 4b,d). Analogously to the results for the proline mutations detailed above, membrane leakage was not significantly affected by these deletions in the SNARE TMDs. In summary, the integrity of the syb2 TMD appears to have a key role in opening the fusion pore39 or keeping it open, thus yielding efficient glutamate release, but is less important for lipid mixing.
Two Syb2 molecules are sufficient for Ca2+-stimulated fusion
Six-nanometer nanodiscs are too small to accommodate the two lipid bilayers (each 4–5 nm thick) needed to form a purely lipidic fusion pore, yet we observed membrane fusion and content release. These findings suggest that the fusion pore might not proceed through the classical lipid-stalk-fusion model formed by only phospholipids14. We therefore conducted experiments to determine whether the pore is formed, in part, by the transmembrane domains of SNARE proteins, as posited by the proteinaceous-fusion-pore model11. To address this issue, we reconstituted increasing copy numbers of syb2 into nanodiscs and determined the effects of copy number on bilayer fusion and glutamate release (Fig. 5). We generated nanodiscs (13 nm) containing one (ND1) to eight (ND8) molecules of syb2 (Supplementary Fig. 5a) and confirmed the copy number by SEC (Fig. 5a) and single-molecule photobleaching experiments (Fig. 5d,e and Supplementary Fig. 5e–h).
Figure 5.
Two molecules of syb2 mediate efficient membrane fusion and content release. (a) SEC profiles of nanodiscs containing 1 to 8 molecules of syb2. (b,c) Lipid mixing (b) and glutamate release (c), in the absence or presence of Ca2+-C2AB, plotted versus the syb2 copy number per nanodisc. Data represent means ± s.d. (n = 3 technical replicates). (d) Examples of single-molecule photobleaching traces. (e) Histograms of the number of observed photobleaching steps fit to either a Poisson distribution or a binomially weighted (w) Poisson distribution. <n>, average number of syb2 molecules per nanodisc.
In the absence of Ca2+-C2AB, glutamate release fell from 70% to 5%, when the copy number of syb2 per nanodisc was reduced from eight to one (Fig. 5c). However, the lipid-mixing signal decreased only ~50% (Fig. 5b). These results are consistent with those from a previous study showing that content mixing, but not lipid mixing, is highly sensitive to the number of the SNARE complexes in the absence of additional regulatory proteins27.
In contrast, in the presence of Ca2+-C2AB, varying the number of syb2 molecules per nanodisc resulted in similar relative changes in levels of both lipid mixing and glutamate release (Fig. 5b,c). We achieved optimal fusion by using nanodiscs containing three or four molecules of syb2. As shown previously, high copy numbers of syb2 resulted in inhibition by Ca2+-C2AB31, perhaps by interfering with the ability of Ca2+-C2AB to bind to the membrane surface. A key finding was that two molecules of syb2 resulted in relatively efficient lipid mixing and glutamate release (~40% each; maximal efficiency ~70%). Because single-molecule photobleaching experiments revealed that 99% of Nd2 nanodiscs contained only one or two molecules of syb2, the fusion observed in the presence of Nd2 was not the result of a subpopulation of nanodiscs (<1%) containing more than two copies of syb2 (Fig. 5e). In line with this conclusion, we also prepared 6-nm nanodiscs that contained two molecules of syb2 and again observed efficient lipid mixing and glutamate release (Supplementary Fig. 5b–d). Because two molecules of syb2 are too few to form a proteinaceous channel, we posit that the fusion pore reconstituted in the nanodisc-SUV system is composed of both lipids and SNAREs. However, direct experiments showing that the TMDs of SNAREs physically line the pore were needed to confirm this conclusion; these experiments are described below.
TMDs of SNAREs line the fusion pore
If the fusion pore consists of both lipids and SNAREs, the SNARE TMDs should be transiently accessible to the medium when the fusion pore opens. To test this hypothesis, we performed scanning cysteine-accessibility assays on the TMDs of both syntaxin1A and syb2 (Fig. 6d–f). We initially attempted these experiments in cells. However, because of the short open lifetime of fusion pores, in conjunction with the fact that minute number of SNAREs are involved in fusion at any given time, these experiments were not successful. We therefore turned to the nanodisc-SUV system, because fusion pores cannot dilate, and efficient fusion can be observed with a relatively low copy number of SNARE proteins.
Figure 6.
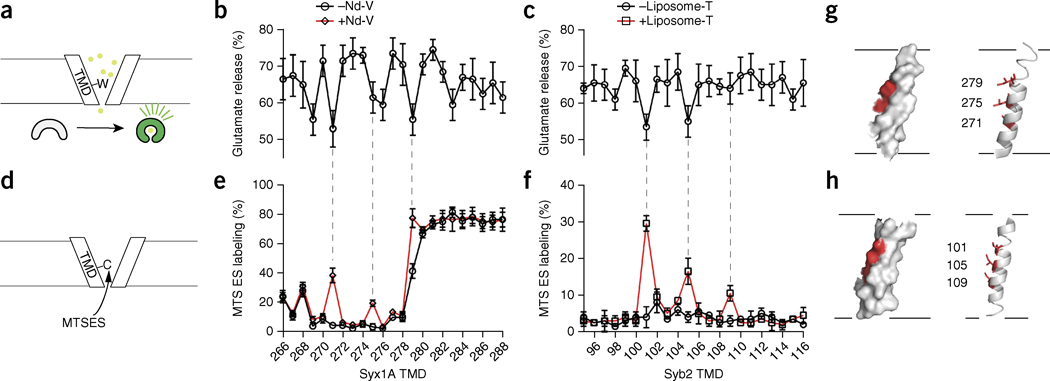
SNARE TMDs are present in the fusion pore. (a) Illustration of the scanning tryptophan assay. (b,c) Glutamate release efficiency mediated by tryptophan-mutant forms of syx1A (b) and syb2 (c). Data represent means ± s.d. (n = 5 technical replicates). (d) Illustration of the substituted cysteine-accessibility assay to probe the TMDs of syb2 and syx1A during fusion. The accessibility of single-cysteine substitutions in syx1A (e) and syb2 (f), with (red trace) or without (black trace) fusion, as measured by the degree of MTSES labeling. Data represent means ± s.d. (n = 3 technical replicates). (g,h) Solvent-exposed residues (red) mapped onto the structure of syx1A (g) and syb2 (h).
We used MTSES, a membrane-impermeant thiol-reactive probe, to determine the exposure of single cysteine residues—substituted in the TMDs of SNAREs—to the aqueous medium upon fusion of nanodiscs with SUVs. These experiments revealed that specific residues within these TMDs were in fact labeled in a fusion-dependent manner (Fig. 6e,f); we observed only low levels of labeling in control experiments lacking a fusion partner (i.e., in the absence of nanodiscs or t-SNARE liposomes). Hence, the TMDs of SNARE are present within fusion pores and are transiently exposed to the medium during fusion. In the crystal structure of the cis-SNARE complex, all the TMDs form α-helices that coalesce into four-helix bundles40. When mapped onto these structures, the MTSES-labeled residues are all located on the same side of the syx1A and syb2 TMDs (Fig. 6g,h); therefore, this face of the helix lines the pore.
To further corroborate this observation, we carried out tryptophan-scanning mutagenesis of the TMDs (Fig. 6a–c). As described in cell-based work, the bulky side chains of tryptophan residues can, in principle, decrease transmitter flux by partially occluding a partially or completely proteinaceous fusion pore19-21. Under the conditions used, wild-type SNAREs, as well as most of the tryptophan mutants, gave rise to glutamate release efficiencies of ~65–75%. However, when we changed specific residues to tryptophan, we observed a significant (P <0.05 by two-tailed Student’s t test; n = 5 technical replicates) further decrease of up to ~20% (Fig. 6b,c). Four of these positions (271 and 279 of syx1A, 101 and 105 of syb2) correlated with the MTSES-labeling experiments, thus corroborating the conclusion that these residues line the fusion pore during exocytosis. Two additional positions (269 and 283 in syx1A), when changed to tryptophan, also gave rise to diminished glutamate flux but did not exhibit fusion-dependent MTSES labeling. There are a number of plausible explanations for this latter observation, including the possibility that these two tryptophan mutations shortened the fusion-pore open time such that it could not be labeled chemically. Nonetheless, these experiments directly demonstrate that the TMDs of SNAREs partially line the nascent fusion pore.
DISCUSSION
Although progress has been made regarding the identification and reconstitution of the machinery that mediates neuronal exocytosis, the structure and composition of the central intermediate structure in the process—the fusion pore—remains unknown. This lack of information is largely because of the transient nature of fusion pores, which remain open for milliseconds in neuroendocrine cells, or probably even a shorter time during SV exocytosis, before either rapidly dilating to yield full fusion or closing again. Here, we took advantage of the rigid framework of nanodiscs, which prevent fusion-pore dilation, in conjunction with biochemical approaches, to address the composition of reconstituted fusion pores. Our results are difficult to reconcile with either purely lipidic or purely proteinaceous fusion pores; instead, our findings indicate that the pore is lined by a combination of lipids and the TMDs of SNARE proteins.
Because each lipid bilayer is 4–5 nm thick, a 1-nm-wide fusion pore would require a nanodisc bilayer of at least 9 nm diameter, according to the purely lipidic fusion-pore model. Consequently, we initially thought that fusion would not occur between 6-nm nanodiscs and liposomes. Unexpectedly, our results showed that Ca2+-stimulated bilayer fusion, and glutamate release, could be reconstituted through use of 6-nm nanodiscs, thus suggesting that the fusion pore might not be formed via the simple merger of two lipid bilayers. The lipid-mixing signals reported here did not arise from lipid transfer between the proximal leaflets (hemifusion), because dithionite quenching experiments demonstrated that lipid mixing occurred in both the outer and inner leaflets; hence, the bilayers indeed fused in this system. Notably, previous studies have shown that placing additional amino acids at the C terminus of the TMD of syb2 significantly decreases catecholamine release from cultured chromaffin cells41 and that mutations in the TMDs of syx1A and syb2 alter hormone flux through fusion pores in PC12 cells and chromaffin cells, respectively19-21. These findings are also consistent with the idea that SNARE TMDs line the exocytotic fusion pore.
To monitor the release of a fluid-phase marker from pores formed in the nanodisc-SUV system, we used a new approach based on glutamate, a bona fide neurotransmitter, in conjunction with the fluorescent glutamate sensor iGluSnFR23. In a previous study, high concentrations of Ca2+ were used as the content marker27, but high Ca2+ concentrations can directly affect the fusion of lipid bilayers42,43 and can confound experiments that include the Ca2+ sensor syt1. Glutamate is an ideal cargo because the flux of this transmitter mimics the release of glutamate from presynaptic boutons, without cause for concern about possible effects on the physical chemistry of the reconstituted membranes used in these in vitro studies. This approach may also facilitate comparisons between cell-based experiments with reconstitution approaches. However, one concern regarding the use of content-release markers is that membrane fusion can be accompanied by membrane leakage, as shown during the fusion of yeast vacuoles32. At present, this caveat has not been carefully examined in reconstituted v-SUV–t-SUV fusion studies because the readout in previous studies has usually been the dequenching of soluble fluorescent dyes upon dilution44; these reporters did not distinguish between membrane fusion and membrane leakage. Using the glutamate-iGluSnFR reporter system, we were able to determine the relationship between bilayer fusion, glutamate release and membrane leakage during v-SUV–t-SUV fusion. We found that the extent of leakage depended on the concentration of syb2 and C2AB; at a low copy number of syb2, and at low C2AB concentration, we observed only low levels of membrane leakage (5%) (Fig. 3e). Hence, we used these conditions for most of the experiments reported here. In addition, we found that membrane leakage was not affected by proline mutations that disrupt full zippering of the SNARE motif or by deletions in the syb2 TMD (Fig. 4). Thus, leakage can be readily uncoupled from flux through pores.
The data reported here indicate that the TMDs of SNAREs affect the dynamics of fusion pores by either increasing the probability of opening or increasing the open lifetime of pores to allow more efficient escape of content. The latter possibility could arise if a few residues in the TMDs of syb2 and syx 1A, near the interface of the membranes destined to fuse, were to physically interact with each other to hold pores open longer40. It should also be noted that SNAREs might oligo-merize45-47, and at high copy numbers they could potentially form largely or purely proteinaceous channels11-13. Transient fusion pores, formed by the reversible assembly of SNAREs into oligomers, is an appealing idea, because different stoichiometries of SNAREs might determine the size of the pore, which can range from 0.5 nm for small vesicles in the posterior pituitary to 2 nm in beige mouse mast cells48,49. In this light, we note that the number of SNAREs estimated to drive fusion range from 2 to 15 in different kinds of cells50-53.
We also observed that relatively efficient (~40%) glutamate release could be achieved with only two copies of syb2 in the presence of Ca2+-C2AB, thus supporting previous optical experiments showing that two copies of syb2 are sufficient for Ca2+-triggered SV exocytosis from cultured neurons50. Because a proteinaceous channel requires at least three transmembrane domains, the fusion pore, reconstituted with two molecules of syb2, is likely to be composed of both lipids and SNARE TMDs. In summary, purely proteinaceous channels can, in principle, form at high copy numbers of SNAREs, but at low copy numbers, the fusion pore is composed of both lipids and proteins.
To directly determine whether SNARE TMDs transiently line fusion pores, and to probe their conformation during fusion, we performed scanning cysteine-accessibility experiments, which revealed that residues in the TMDs are in fact exposed to solvent. These findings provide the most direct support to date for the idea that SNARE TMDs compose at least part of the fusion pore. Furthermore, systematic mutagenesis revealed positions in SNARE TMDs that, when substituted with bulky tryptophan residues, resulted in decreases in glutamate release. Similar tryptophan-scanning experiments, performed on secretory cells, have also revealed reductions in hormone flux when the same residues, 101 and 105 in syb2, were mutated21. In the case of syx1A, our tryptophan-scanning data (indicating that mutations at positions 271, 275 and 279 reduced flux) differ somewhat from the cell-based findings (indicating that mutations at positions 269, 276 and 283 reduced flux)19. This discrepancy might be due to different stoichiometries of SNAREs in cells versus our reconstituted system, as alluded to above. Future experiments, in which glutamate flux is measured at the single-event level, will be required to address these issues.
ONLINE METHODS
Reagents
Nitrilotriacetic acid (Ni2+-NTA)-chelating Sepharose and Superdex 200 were obtained from GE Healthcare. N-dodecylphosphocholine (DPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), 1,2-dioleoyl-sn-glycero- 3-phospho-l-serine (PS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate) (PI(4,5)P2), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) (biotin-PE), 1,2-dipalmitoyl-sn-glycero-3-phospho-ethanolamine-N-(7-nitro- 2-1,3-benzoxadiazol-4-yl) (NBD-PE) and N-(lissamine rhodamine B sulfonyl)-1,2- dipalmitoyl-sn-glycero-3-phosphoethanolamine (rhodamine-PE) were obtained from Avanti Polar Lipids. 2-Sulfonatoethyl methanethiosulfonate sodium salt (MTSES) was purchased from Biotium. All other chemicals were from Sigma.
Recombinant proteins
Syb2 and t-SNARE heterodimers were purified as previously described8,31, except that n-dodecyl β-d-maltoside (DDM) and N-dodecylphosphocholine (DPC), respectively, were used in place of n-octyl glucoside (OG). Purification of full length (residues 1–421) and the cytoplasmic domain (C2AB; residues 96–421) of syt1 was carried out as described previously54. Purification of the glutamate sensor (iGluSnFR) and the membrane scaffold proteins (MSPE3D1 and MSP1D1ΔH4–H6) was carried out as described previously23,26,28.
Proteoliposome reconstitution
SNARE proteins were mixed together with lipids (15% PE, 25% PS and 60% PC) in reconstitution buffer (25 mM HEPES, pH 7.5, 100 mM KCl, and 1 mM DTT) plus 0.02% DDM. To encapsulate glutamate into proteoliposomes, 50 mM glutamate was included in the mixture. For fusion assays involving full-length syt1, 2% PI(4,5)P2 was included in the t-SNARE liposomes. Detergent was removed with BioBeads under gentle shaking (overnight, 4 °C). t-SNARE liposomes were then isolated by flotation8, and this was followed by dialysis against reconstitution buffer (overnight, 4 °C). Trypsin digestion experiments revealed that ~50–70% of SNARE proteins were correctly oriented (data not shown).
Nanodisc reconstitution
Nanodisc reconstitution was performed as previously described27,55. A typical reconstitution experiment involved mixing syb2, MSP and lipids (15% PE, 40% PS and 45% PC) in reconstitution buffer containing 0.02% DDM. To prepare nanodiscs for lipid mixing experiments, the lipid composition was: 12% PE, 40% PS, 45% PC, 1.5% NBD-PE and 1.5% rhodamine-PE. For 13-nm nanodiscs, the ratio of MSP/lipid was 2:120, whereas for 6-nm nanodiscs, a ratio of 2:30 was used. To prepare nanodiscs containing different copy numbers of syb2, the following MSP/syb2 ratios were used: 2:0.2 (ND1), 2:0.4 (ND2), 2:1 (ND3), 2:2 (ND4), 2:4 (ND5), 2:6 (ND6), 2:8 (ND7) and 2:10 (ND8). Detergent was slowly removed with BioBeads (1/3 volume, Sigma) with gentle shaking (overnight, 4 °C). The reconstituted discs were centrifuged (20 min at 100,000g) and purified with Ni2+-NTA beads; this was followed by gel filtration with a Superdex 200 10/300 GL column equilibrated in reconstitution buffer plus 5% glycerol.
Lipid mixing assay
Lipid mixing assays were carried out as previously described8,31 with nanodiscs (0.2 μM) and t-SNARE liposomes (0.5 μM) in the absence or presence of C2AB (1 μM). The reaction mixtures were incubated at 37 °C for 10 min in reconstitution buffer plus 0.2 mM EGTA, and this was followed by addition of Ca2+ (1 mM final concentration). The NBD signal was monitored for an additional 1 h. After each run, 20 μl 2.5% of DDM was added to each reaction to calculate the maximal fluorescence signal, and data were collected for another 25 min. The maximal fluorescence signal after addition of detergent, minus the fluorescence signal at the beginning of the assay, was used to normalize the percentage of lipid mixing. Data were obtained from three independent trials.
Glutamate release assay
Glutamate release assays were performed with the glutamate sensor iGluSnFR (1 μM)23, nanodiscs (0.2 μM) and glutamate encapsulated t-SNARE liposomes (0.5 μM) in the absence or presence of C2AB (1 μM). The reaction mixtures were incubated at 37 °C for 20 min in reconstitution buffer plus 0.2 mM EGTA; this was followed by addition of Ca2+ (1 mM final concentration), and the iGluSnFR fluorescence signal was monitored for an additional 1 h. After each run, 20 μl 2.5% of DDM was added to each reaction, and data were collected for another 25 min. Data were obtained from three independent trials.
Scanning cysteine-accessibility method
Nd-V (0.2 μM) and t-SNARE liposomes (0.5 μM) were incubated with MTSES (0.1 mM) at 37 °C for 1 h, in the presence of Ca2+ (1 mM) and C2ABC277A (0.1 μM). The reaction mixture was diluted 20-fold into 25 mM HEPES, pH 7.5, plus 5 mM cysteine; this was followed by centrifugation at 150,000g at 4 °C for 40 min in a Beckman Optima MAX-E (Beckman Coulter) tabletop ultracentrifuge. The pellets were washed with 25 mM HEPES, pH 7.5, and resuspended in labeling buffer (25 mM HEPES, pH 7.5, 6 M urea and 0.5% DDM); samples were then incubated with 5-iodoacetamidofluorescein (5-IAF, 0.1 mM) at 37 °C for 1 h and subjected to SDS-PAGE; this was followed by fluorescence scanning and Coomassie blue staining. MTSES labeling efficiency was determined by protection from 5-IAF labeling in comparison with controls that were not treated with MTSES. Data were obtained from three independent trials.
AFM imaging
Purified v-SNARE nanodiscs and t-SNARE liposomes were suspended in HBS (100 mM NaCl and 50 mM HEPES, pH 7.5) containing 1 mM CaCl2, and 45 μl of the sample was deposited onto freshly-cleaved mica (10 mm diameter discs). After a 5-min adsorption period, the sample was rinsed with the same buffer solution to remove unabsorbed material. AFM imaging was carried out with a Dimension FastScan Bio AFM. All samples were imaged in tapping mode (in fluid) with silicon nitride probes (FastScan D, Bruker AFM Probes). These cantilevers had a spring constant of ~0.25 N/m and a drive frequency of ~85 kHz (10–20% below the resonance frequency). The applied imaging force was kept as low as possible (AS/A0~0.85). Images were captured at a scan rate of 20 Hz, with 512 scan lines per area. Data analysis was performed with Gwyddion 2.40. Nanodisc diameter was determined by drawing cross-sections of the imaged structures.
Single-molecule photobleaching
Syb2-61C/103A was incubated with Cy5 or DY650 maleimide at a molar ratio of 1:5 (room temperature, 3h). Labeled protein was purified with a Zeba desalting column (Life Technologies) equilibrated in reconstitution buffer containing 0.02% DDM. The labeling efficiencies were ~0.6 and 0.7 for Cy5 and DY650, respectively. For single-molecule photobleaching experiments, the lipid composition for nanodiscs was: 1% biotin-PE, 14% PE, 40% PS and 45% PC. Purified nanodiscs containing syb2-DY650 were immobilized on a biotin/streptavidin-coated surface within arrays of zero-mode waveguide nanoholes (~200 nm diameter, Pacific Biosciences) and imaged on an inverted microscope (Olympus IX71) with a 100× oil immersion objective (Olympus; NA, 1.49). Imaging buffer was 10 mM Na2HPO4, pH 7, 2 mM KH2PO4, 200 mM NaCl, 2 mg/ml BSA, 1 mM TCEP, 2.5 mM PCA, 250 nM PCD, and 1 mM Trolox. Concentrations of syb2-DY650 were chosen so that approximately 1% of the nanoholes in each imaged array contained fluorescent dye, such that each nanohole could be reasonably assumed to contain at most one nanodisc. DY650 was excited by epi-illumination from a 637-nm laser (Coherent OBIS). Each zero-mode waveguide nanohole limits the effective excitation volume to attoliters at the optical surface such that only fluorescence from surface-immobilized dye is observed. Fluorescence from arrays of ~1,000 nanoholes was imaged with a 512 × 512 EMCCD camera (Andor iXon Ultra 9899) equipped with a dichroic and emission filter (Semrock Brightline FF560/659-Di01 and FF01-577/690, respectively).
Bleaching step distributions were initially described by a Poisson distribution, under the assumption that incorporation of syb2 molecules into nanodiscs was both random and independent.
where k is the number of bleach steps, and λ is the average number of fluorescently labeled syb2 molecules per nanodisc. If each syb2 molecule has a probability p < 1 of having a fluorescent label, the observed number of bleaching steps k for nanodiscs containing n syb2 molecules will be described by a binomial distribution:
Thus, to account for incomplete fluorescent dye labeling of syb2, we further fitted bleaching step distributions to a weighted Poisson distribution in which the probability of observing k bleaching steps is given by:
where λ is the average number of syb2 molecules per nanodisc (labeled or not), p is the probability that each syb2 molecule has a fluorescent label, and N is the maximum possible number of syb2 molecules per nanodisc (in practice any N large enough such that Poisson (N, λ) is negligible). The summation includes the probabilities that k observed bleach steps came from n ≥ k molecules with fractional labeling p weighted by the Poisson probability of having n molecules.
Supplementary Material
Acknowledgments
We thank G. Wagner for providing the MSP1D1ΔH4–H6 plasmid. This study was supported by a grant from the US National Institutes of Health (MH061876 to E.R.C.). H.B. is supported by a postdoctoral fellowship from the Human Frontier Science Program. B.C. and M.G.-O. are supported by funding from the US National Institutes of Health (R01 GM084140 to B.C.). P.J. is supported by Kidney Research UK. J.M.E. is supported by the Biotechnology and Biological Sciences Research Council (BB/J018236/1 to J.M.E.) and Kidney Research UK. E.R.C. is supported as an Investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
M.G.-O. performed the single-molecule experiments in the laboratory of B.C. P.J. and J.M.E. carried out the AFM experiments; H.B. performed nanodisc reconstitutions, fusion assays and cysteine-accessibility assays; and H.B. and E.R.C. conceived the project and designed the experiments. H.B. and E.R.C. wrote the paper, and all other authors edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Any Supplementarsy Information and Source Data files are available in the online version of the paper.
References
- 1.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothman JE. The principle of membrane fusion in the cell (Nobel lecture) Angew Chem Int Edn Engl. 2014;53:12676–12694. doi: 10.1002/anie.201402380. [DOI] [PubMed] [Google Scholar]
- 3.Bolger AP, et al. Neurohormonal activation and the chronic heart failure syndrome in adults with congenital heart disease. Circulation. 2002;106:92–99. doi: 10.1161/01.cir.0000020009.30736.3f. [DOI] [PubMed] [Google Scholar]
- 4.Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 6.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 7.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhalla A, Chicka MC, Tucker WC, Chapman ER. Ca2+-synaptotagmin directly regulates t-SNARE function during reconstituted membrane fusion. Nat Struct Mol Biol. 2006;13:323–330. doi: 10.1038/nsmb1076. [DOI] [PubMed] [Google Scholar]
- 9.Chicka MC, Hui E, Liu H, Chapman ER. Synaptotagmin arrests the SNARE complex before triggering fast, efficient membrane fusion in response to Ca2+ Nat Struct Mol Biol. 2008;15:827–835. doi: 10.1038/nsmb.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai Y, et al. Fusion pore formation and expansion induced by Ca2+ and synaptotagmin 1. Proc Natl Acad Sci USA. 2013;110:1333–1338. doi: 10.1073/pnas.1218818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson MB, Chapman ER. The fusion pores of Ca2+-triggered exocytosis. Nat Struct Mol Biol. 2008;15:684–689. doi: 10.1038/nsmb.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindau M, Almers W. Structure and function of fusion pores in exocytosis and ectoplasmic membrane fusion. Curr Opin Cell Biol. 1995;7:509–517. doi: 10.1016/0955-0674(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 13.Fang Q, Lindau M. How could SNARE proteins open a fusion pore? Physiology (Bethesda) 2014;29:278–285. doi: 10.1152/physiol.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlovsky Y, Kozlov MM. Stalk model of membrane fusion: solution of energy crisis. Biophys J. 2002;82:882–895. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinreb G, Lentz BR. Analysis of membrane fusion as a two-state sequential process: evaluation of the stalk model. Biophys J. 2007;92:4012–4029. doi: 10.1529/biophysj.106.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty H, Tarafdar PK, Bruno MJ, Sengupta T, Lentz BR. Activation thermodynamics of poly(ethylene glycol)-mediated model membrane fusion support mechanistic models of stalk and pore formation. Biophys J. 2012;102:2751–2760. doi: 10.1016/j.bpj.2012.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chernomordik LV, Melikyan GB, Chizmadzhev YA. Biomembrane fusion: a new concept derived from model studies using two interacting planar lipid bilayers. Biochim Biophys Acta. 1987;906:309–352. doi: 10.1016/0304-4157(87)90016-5. [DOI] [PubMed] [Google Scholar]
- 19.Han X, Wang CT, Bai J, Chapman ER, Jackson MB. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 2004;304:289–292. doi: 10.1126/science.1095801. [DOI] [PubMed] [Google Scholar]
- 20.Han X, Jackson MB. Electrostatic interactions between the syntaxin membrane anchor and neurotransmitter passing through the fusion pore. Biophys J. 2005;88:L20–L22. doi: 10.1529/biophysj.104.056739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang CW, et al. A structural role for the synaptobrevin 2 transmembrane domain in dense-core vesicle fusion pores. J Neurosci. 2015;35:5772–5780. doi: 10.1523/JNEUROSCI.3983-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alabi AA, Tsien RW. Perspectives on kiss-and-run: role in exocytosis, endocytosis, and neurotransmission. Annu Rev Physiol. 2013;75:393–422. doi: 10.1146/annurev-physiol-020911-153305. [DOI] [PubMed] [Google Scholar]
- 23.Marvin JS, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods. 2013;10:162–170. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hristova K, White SH. Determination of the hydrocarbon core structure of fluid dioleoylphosphocholine (DOPC) bilayers by x-ray diffraction using specific bromination of the double-bonds: effect of hydration. Biophys J. 1998;74:2419–2433. doi: 10.1016/S0006-3495(98)77950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 26.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 27.Shi L, et al. SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science. 2012;335:1355–1359. doi: 10.1126/science.1214984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun S, et al. Receptor binding enables botulinum neurotoxin B to sense low pH for translocation channel assembly. Cell Host Microbe. 2011;10:237–247. doi: 10.1016/j.chom.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 31.Tucker WC, Weber T, Chapman ER. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 2004;304:435–438. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- 32.Starai VJ, Jun Y, Wickner W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc Natl Acad Sci USA. 2007;104:13551–13558. doi: 10.1073/pnas.0704741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nickel W, et al. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc Natl Acad Sci USA. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 36.McNew JA, et al. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sørensen JB, et al. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Zhang F, Su Z, McNew JA, Shin YK. Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 39.Grote E, Baba M, Ohsumi Y, Novick PJ. Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J Cell Biol. 2000;151:453–466. doi: 10.1083/jcb.151.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngatchou AN, et al. Role of the synaptobrevin C terminus in fusion pore formation. Proc Natl Acad Sci USA. 2010;107:18463–18468. doi: 10.1073/pnas.1006727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kachar B, Fuller N, Rand RP. Morphological responses to calcium-induced interaction of phosphatidylserine-containing vesicles. Biophys J. 1986;50:779–788. doi: 10.1016/S0006-3495(86)83518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hui SW, Nir S, Stewart TP, Boni LT, Huang SK. Kinetic measurements of fusion of phosphatidylserine-containing vesicles by electron microscopy and fluorometry. Biochim Biophys Acta. 1988;941:130–140. doi: 10.1016/0005-2736(88)90173-3. [DOI] [PubMed] [Google Scholar]
- 44.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hohl TM, et al. Arrangement of subunits in 20 S particles consisting of NSF, SNAPs, and SNARE complexes. Mol Cell. 1998;2:539–548. doi: 10.1016/s1097-2765(00)80153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rickman C, Hu K, Carroll J, Davletov B. Self-assembly of SNARE fusion proteins into star-shaped oligomers. Biochem J. 2005;388:75–79. doi: 10.1042/BJ20041818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy R, Laage R, Langosch D. Synaptobrevin transmembrane domain dimerization-revisited. Biochemistry. 2004;43:4964–4970. doi: 10.1021/bi0362875. [DOI] [PubMed] [Google Scholar]
- 48.Klyachko VA, Jackson MB. Capacitance steps and fusion pores of small and large-dense-core vesicles in nerve terminals. Nature. 2002;418:89–92. doi: 10.1038/nature00852. [DOI] [PubMed] [Google Scholar]
- 49.Breckenridge LJ, Almers W. Final steps in exocytosis observed in a cell with giant secretory granules. Proc Natl Acad Sci USA. 1987;84:1945–1949. doi: 10.1073/pnas.84.7.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha R, Ahmed S, Jahn R, Klingauf J. Two synaptobrevin molecules are sufficient for vesicle fusion in central nervous system synapses. Proc Natl Acad Sci USA. 2011;108:14318–14323. doi: 10.1073/pnas.1101818108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hua Y, Scheller RH. Three SNARE complexes cooperate to mediate membrane fusion. Proc Natl Acad Sci USA. 2001;98:8065–8070. doi: 10.1073/pnas.131214798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohrmann R, de Wit H, Verhage M, Neher E, Sørensen JB. Fast vesicle fusion in living cells requires at least three SNARE complexes. Science. 2010;330:502–505. doi: 10.1126/science.1193134. [DOI] [PubMed] [Google Scholar]
- 53.Montecucco C, Schiavo G, Pantano S. SNARE complexes and neuroexocytosis: how many, how close? Trends Biochem Sci. 2005;30:367–372. doi: 10.1016/j.tibs.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Liu H, Gu Y, Chapman ER. Reconstituted synaptotagmin I mediates vesicle docking, priming, and fusion. J Cell Biol. 2011;195:1159–1170. doi: 10.1083/jcb.201104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi L, et al. Preparation and characterization of SNARE-containing nanodiscs and direct study of cargo release through fusion pores. Nat Protoc. 2013;8:935–948. doi: 10.1038/nprot.2013.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.