ABSTRACT
Purpose:
The aim of this study was to identify possible protein biomarkers and/or candidates for therapeutic targets in tissues of patients with SCCP, infected by HPV, applying one dimensional electrophoresis (1DE), followed by direct mass spectrometry (MS) analysis.
Materials and Methods:
Tissues from 10 HPV positive patients with SCCP and from 10 patients with HPV negative non-tumorous penile foreskins were analyzed applying 1D electrophoresis, followed by analysis with direct mass spectrometry (MS).
Results:
Sixty-three different proteins were identified in the first group and 50 in the second group. Recognition was possible for 28 proteins exclusively detected in Group 1 and 21 proteins presented only in Group 2.
Conclusion:
Some proteins in the first group are directly involved in the development of other types of cancer, and therefore, suitable for analysis. Complement C3 protein is a strong candidate for evaluating SCCP patients.
Key words: Carcinoma, Squamous Cell, Penis, Human papillomavirus
INTRODUCTION
Cancer of the penis is a rare neoplasm with a high incidence in developing countries. This fact clearly indicates the disease's association with local economic conditions (1). Penile cancer has a low overall incidence, representing approximately 0.4% of malignancies in the Unites States. In Brazil, despite the high incidence in some regions, this disease accounts for about 2.1% of malignancies (2, 3). A recent Brazilian epidemiologic study on penile carcinoma revealed the profile of these patients (4).
The etiology of penile cancer has not been fully elucidated. However, its incidence varies according to the practice of circumcision, personal hygiene, presence of phimosis, human papillomavirus (HPV) infection, and tobacco use (5–9). The mechanism of tumor induction and promotion related to HPV infection is not completely understood. It is believed that the incorporation of viral DNA to the human genome leads to hyper-expression of viral genes E6 and E7 and inactivates the host cell's tumor suppressor gene products p53 and pRb (10).
The presence and extent of inguinal metastases are the most important prognostic factor related to the survival of patients with penile carcinoma. At the time of its initial presentation, 50% of patients with SCCP have inguinal lymphadenopathy; however, only half of these actually show metastatic lymph node involvement. Furthermore, 20% of patients with clinically negative inguinal lymph nodes have micro-metastases that will only be diagnosed by histopathologic examination of surgical specimens obtained from lymphadenectomy, a procedure associated with a significant morbidity (1, 4). Therefore, SCCP remains a challenge for the urologist, because there is no consensus for an appropriate therapy for all forms of disease presentation. The possibility of using reliable biomarkers to predict disease prognosis and to establish procedures less aggressive for patients at low risk for metastasis becomes necessary. In this sense, the development of more accurate molecular diagnostic methods and prognostic value tumor markers is essential.
Proteomics is the large-scale identification of proteins. Proteomics technologies are currently under development and several methodological approaches can be applied depending on the objectives. The great advantage of proteomics over genomics or transcriptomics studies is that the real functional molecules of the cell are being studied. Therefore, in this study, the aim was to identify possible protein biomarkers and/ or candidates for therapeutic targets in tissues of patients with SCCP, infected by HPV, applying one dimensional electrophoresis (1DE), followed by direct mass spectrometry (MS) analysis.
MATERIALS AND METHODS
Patients and controls
Between January 2009 and December 2011, 20 patients treated at three health institutions in the state of Rio de Janeiro were recruited and divided into two groups for prospective tissue proteomic analysis. Group 1 was composed of 10 patients with positive HPV malignant SCCP treated at the Brazilian National Cancer Institute (INCA) and Mario Kröeff Hospital. Group 2 (control group) was composed of 10 patients with HPV negative non-tumorous penile foreskins collected at Santa Veronica Hospital after circumcision procedures. HPV typing was performed as previously published (11, 12) and reported (13).
Pathological material was reviewed in both groups and all tumors were histologically classified based on Broders’ system. Only two pathologists were responsible for reviewing the specimens. The clinical and pathological staging for Group 1 was done according to the 2002 TNM classification system. Patients’ treatment varied according to primary tumor presentation. The distribution of T and N categories is shown in Table-1. The criterion for performing a radical inguinal lymphadenectomy (RIL) in all cases was the stage, grade and/or presence of lymphovascular invasion. Two patients with stage T1 underwent lymphadenectomy. The first presented unilateral inguinal lymphadenopathy and the second presented grade 2 tumor. In our services, we only do not perform RIL in patients with stage T1N0M0 grade 1 tumor without lymphovascular invasion ages ranged from 38 to 90 years (mean, 63.56) for Group 1 and 23 to 83 years (mean, 60) for Group 2. The pathological variables studied were histological type, grade of tumor differentiation, corpus spongiosum and/or infiltration of the corpora cavernosa, urethral in-filtration, and inguinal lymph nodes involvement. Patient recruitment did not take into account any criteria of poor prognosis, and the tissue specimens were randomly selected respecting the number of pre-established patients for the study. All patients involved in the current study gave their informed consent. This study was approved by the Brazilian National Cancer Institute Ethical Board (registrations # 38/05 and 67/07). Because this was a pilot study and unprecedented in literature the number of patients was pre-established in both groups in the design of work, aiming preliminary results for further investigation. The only exclusion criterion was positivity for HPV in the control group. Our study aimed qualitative detection of proteins in the 2 groups not being our objective to quantify the identified proteins. All tests in tumor samples from patients revealed the presence of HPV. Because of the rarity of HPV-negative patients, a second study with HPV-negative patients will be necessary.
Table 1. Histopathologic findings, pathologic staging and treatment option for patients from group 1.
| Pts | Histology | Grade | Stage TNM | HPV type | Surgery |
|---|---|---|---|---|---|
| 1 | Squamous cell carcinoma | G2 | T4N2Mx | MY-/16+ | Total Amputation + Bilateral RIL |
| 2 | Squamous cell carcinoma | G1 | T2N3Mx | MY-/18+ | Partial Amputation + Bilateral RIL |
| 3 | Squamous cell carcinoma | G2 | T2N0Mx | 16+;45+ | Partial Amputation + Bilateral RIL |
| 4 | Squamous cell carcinoma | G2 | T2N0Mx | MY-/18+ | Partial Amputation + Bilateral RIL |
| 5 | Squamous cell carcinoma | G1 | T1N1Mx | MY-/45+ | Partial Amputation + Bilateral RIL |
| 6 | Squamous cell carcinoma | G2 | T1N0Mx | 45+ | Partial Amputation + Bilateral RIL |
| 7 | Squamous cell carcinoma | G1 | T2N0Mx | MY-/16+ 45+ | Partial Amputation + Bilateral RIL |
| 8 | Squamous cell carcinoma | G2 | T2N1Mx | MY+/45+ | Partial Amputation + Bilateral RIL |
| 9 | Squamous cell carcinoma | G2 | T2N1Mx | MY+/16+ | Partial Amputation + Bilateral RIL |
| 10 | Squamous cell carcinoma | G2 | T2N0Mx | MY-/45+ | Partial Amputation + Bilateral RIL |
RIL = Radical Inguinal Lymphadenectomy; MY-MY09/11 Consensus Primers
Tissue protein extraction and quantification
Tissues were macerated in 200μL of lysis buffer (7 M urea, 2 M Thiourea, 4% CHAPS and 1% DTT) with the addition of 0.2-mM PMSF. This mixture was stirred for 1 hour at room temperature and then centrifuged at 14.000g for 15 minutes. The supernatant was collected and stored at −80°C (14) until experimentation.
The protein extracts were quantified by 2D Quant Kit (GE Healthcare, Cat #. 80-6483-56), according to the manufacturer's instructions. Measurement was performed at 650 nm in Elisa Spectra Max 190 device from Molecular Devices. The analysis of quantification was performed by the program SOFT® Pro 4.3 max, Life Sciences Edition.
Gel 1D
After quantification, two protein pools were formed with 10 SCCP tissues and with 10 control tissues, separately. Each pool contained 3.3μg of proteins from each sample, a total of 33μg. The SCCP and control pools were applied on a 12% SDS-PAGE gel. Proteins were separated in Tris-Glycine buffer (25-mM Tris and 250-mM Glycine pH 8,3) and 0.1% SDS at 80 V and 50 mA (15). The proteins were visualized with Coomassie blue G-250. The gels were scanned on Image ScannerTM (GE Healthcare) using the program Labscan™ (GE Healthcare) for protein lanes reading.
Mass spectrometry analysis
The lanes were fractioned in approximately 2-5 mm slices. The bands in the slices were destained in a solution of 25-mM ammonium bicarbonate (NH4HCO3) pH 8.8/50% and acetonitrile (ACN) overnight on a shaker, at room temperature. To reduce proteins, the gel was incubated with 10mM DTT in 25-mM NH4HCO3 at 56°C for 1 hour. The supernatant was discarded and the gel was washed in a solution of 25-mM NH4HCO3 twice. After protein disulfide bonds were reduced, cysteines were alkylated with iodoacetamide 55 mM for 45 minutes at room temperature in the dark. The supernatant was discarded and the gel was washed with 25-mM NH4HCO3 solution in 50% ACN. The supernatant was removed again and gel slices were dehydrated with 100% ACN for 5 minutes and posteriorly in a vacuum centrifuge. Proteins were digested with trypsin (Promega) 10ng/μL dilution, overnight, at 37°C. After digestion with trypsin, peptides were extracted from gels by adding a solution containing 0.1% formic acid/50% ACN for 30 minutes. This solution was transferred to another tube and the procedure was repeated twice. The samples were completely dried in a vacuum centrifuge. The pellets were resuspended in water and purified through Ziptip Perfect Pure C18 (Eppendorf, cat # 0030.008.405) and then dried in a vacuum centrifuge.
For mass spectrometry analysis, the peptides were resuspended in 20μL of acetonitrile 3% and acid formic 0.1% solution. The peptides were analyzed by mass spectrometer ESI-Q/TOF Micro (Waters) linked to a nanoACQUITYUPLC® (Waters). The peptides was loaded on symmetric C18 trap column (Waters) followed by fraction in a nanoEase BEH 130 C18 100 mm × 100μm column (Waters) at a flow rate of 0.5μL/min and eluted with a linear acetonitrile gradient (from 10 to 50%) of 0.1% formic acid. Spectrometer analysis was performed on positive mode. Acquisition parameters on mass spectrometer was: cone voltage 30 V, capillary voltage 3500 V, source temperature 80°C, scanning a mass-to-charge ratio (m/z) MS mode 400-2000 and MS/MS mode 50-2000. The three ions with more intensity with charge states of +2, +3, or +4 were selected for MS/MS fragmentation. The reference ion used was the monocharged ion m/z 588.8692 of phosphoric acid. The data acquisition was performed by MassLynx 4.0 software (Micromass/Waters) and the process data by proteinLynx Global Service (PLGS 2.4, Waters).
Proteins were identified by correlation of tandem mass spectra to the NCBInr proteins database, using Mascot online (Matrix Science, London, UK - http://www.matrixscience.com/cgi/se-arch_form.pl?FORMVER=2&SEARCH=MIS) with restricted taxonomy Homo sapiens. The NCBI (National Center for Biotechnology Information) protein database is an on line collection of sequences from several sources, including translations from annotated coding regions in GenBanK, RefSeq and TPA, as well as records from Swiss-Prot, PIR, PRF, and PDB; “nr” refers to non-redundant protein sequences. The NCBI is a division of the National Library of Medicine (NLM) at the National Institutes of Health (NIH), USA. The parameters were as follows: MS and MS/MS tolerance of 0.1 Da, tryptic specificity allowing for one missed cleavage, fixed modification of carbamidomethylation of cysteine residues, and variable modification of oxidation of methionine, phosphorylation of tyrosine, serine and threonine residues and propionamide. Positive protein identification was accepted with at least two peptides with a Mascot peptide score ≥35.
RESULTS
The pathological features of the primary tumor and inguinal lymph nodes, the type of treatment instituted, and the HPV type (13) for each patient from Group 1 are described in Table-1.
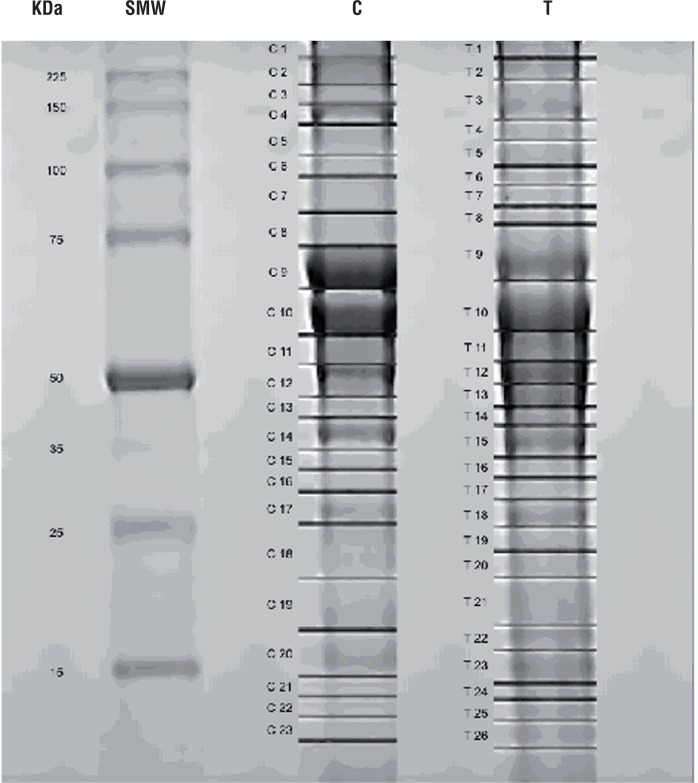
The protein extracts obtained from Groups 1 and 2 were separated by electrophoresis in a 12% SDS-PAGE gel. The protein bands of each group were identified and compared for differences. Twenty-six protein spots from Group 1 and 21 from Group 2 were identified, sliced out from the gel and analyzed through mass spectrometry (Figure-1). Sixty-three different proteins were identified in Group 1 and 50 in Group 2. After a comparative analysis of both groups, it was possible to recognize 28 proteins exclusively detected in Group 1 and 21 proteins presented only in Group 2 (Tables 2 and 3).
Figure 1. 1DE analysis of tissue samples from SCCP HPV patients and control group. Each pool contained 3.3µg of proteins from each sample, a total of 33µg. The SCCP and control pools were applied on a 12% SDS-PAGE gel. The gel was stained with Coomassie blue G. The markers and numbers in gel represent the sections that were excised for mass spectrometry analysis.
T=penile cancer SCCP; C=control group; SMW=Standard Molecular Weight
Table 2. Proteins identified in pool of patients with SCCP (Group 1).
| Gel Slice | NCBI ID | Proteins | Score | Representative Peptides | KEGG | Function | Cell Compartment |
|---|---|---|---|---|---|---|---|
| T1 | gi|179212 | Na+ K+ ATPase alpha subunit | 41 | R.SPDFTNENPLETR.N | hsa:476 | ATP biosynthesis | Cell membrane |
| T2;3 | gi|189036 | Nonmuscle myosin heavy chain (Myosin 9) | 78 | K.ALELDSNLYR.I K.HSQAVEELAEQLEQTKR.V | hsa:4627 | Cell communication | Cytosol |
| T2 | gi|1296662 | Plectin | 74 | R.SQVEEELFSVR.V K.VLALPEPSPAAPTLR.S | hsa:5339 | Apoptosis | Cytosol, cytoskeletal |
| T2 | gi|93141049 | Collagen alpha-1 (XII) chain short isoform precursor | 60 | K.ALALGALQNIR.Y R.WYSPVDGTRPSESIVVPGNTR.M | hsa:1303 | Cell adhesion | Extracellular matrix |
| T5 | gi|4507677 | Endoplasmin | 44 | K.SILFVPTSAPR.G | hsa:7184 | Proteins processing | Cytosol |
| T6 | gi|223170 | Fibrinogen gamma | 58 | K.EGFGHLSPTGTTEFWLGNEK.I K.MLEEIMKYEASILTHDSSIR.Y K.AIQLTYNPDESSKPNMIDAATLK.S | hsa:2266 | Blood coagulation | Secreted |
| T9 | gi|5729877 | Heat Shock cognate 71-kDa protein isoform 1 | 41 | K.DAGTIAGLNVLR.I(U) | hsa:3312 | Stress response, transcription regulation | Cytosol, nucleus, cell surface |
| T11 | gi|2982019 | Chain B, Fab Fragment of engineered Human Monoclonal Antibody A5b7 | 47 | K.GPSVFPLAPCSR.S/ R.STSESTAALGCLVK.D/ E.VQLLESGGGLVQPGGSLR.L | Unknown | Cell defense | Secreted |
| T11;12 | gi|178375 | Aldehyde dehydrogenase | 44 | K.LPEWAADEPVEK.T/ R.SLEEAIQFINQR.E | hsa:218 | Glycolysis, metabolism of amino acids and xenobiotics | Cytosol |
| T13 | gi|4503571 | Alpha-enolase isoform 1 | R.GNPTVEVDLFTSK.G/ R.YISPDQLADLYK.S/ K.WIGMDVAASEFFR.S + Oxidation (M) / K.VNQIGSVTESLQACK.L / K.YNQLLRIEEELGSK.A/ | hsa:2023 | Glicolysis; gliconeogenese | Cytosol | |
| 90 | K.LAMQEFMILPVGAANFR.E + 2 Oxidation (M) /R.EIFDSRGNPTVEVDLFTSK.G/ K.DATNVGDEGGFAPNILENKEGLELLK.T | ||||||
| T13 | gi|1710248 | Protein disulfide isomerase-related protein 5 | 55 | R.TGEAIVDAALSALR.Q/ K.LAAVDATVNQVLASR.Y | hsa:10130 | Proteins processing | Endoplasmatic reticulum |
| T13 | gi|31170 | Chain A, Crystal Structure Of Human Beta Enolase Enob | 44 | K.VNQIGSVTESIQACK.L | hsa:2027 | Glicolysis | Cytosol, fosfopiruvat hydratase complex |
| T13 | gi|40068518 | 6-phosphogluconate dehydrogenase, decarboxylating | 42 | K.IISYAQGFMLLR.Q + Oxidation (M) / K.GILFVGSGVSGGEEGAR.Y | hsa:5226 | Pentose pathways | Cytosol |
| T14 | gi|4505763 | Phosphoglycerate kinase 1 | 39 | K.ITLPVDFVTADKFDENAK.T | hsa:5230 | Glicolysis | Cytosol |
| T14 | gi|306882 | Haptoglobin precursor | 36 | K.VTSIQDWVQK.T/ K.SPVGVQPILNEHTFCAGMSK.Y + Oxidation (M) | hsa:3240 | Defense | Secreted |
| T15 | gi|35222 | 70-kDa heat shock protein | 43 | R.TTPSYVAFTDTER.L (U) | hsa:3312 | Regulation of cell cycle; cellular membrane organization | Cytosol; plasma membrane |
| T16 | gi|63252913 | Macrophage-capping protein | 44 | R.QAALQVAEGFISR.M | hsa:822 | Actin filament organization | Cytosol |
| T17 | gi|5174391 | Alcohol dehydrogenase [NADP+] | 44 | K.GLVQALGLSNFNSR.Q/ R.GLEVTAYSPLGSSDR.A | hsa:10327 | Glicolysis, glicerolipids metabolism | Cytosol |
| T17 | gi|31397 | Fibronectin precursor | 40 | R.VPGTSTSATLTGLTR.G | hsa:2335 | Angiogenesis, cell adhesion, platelet activation and degranulation | Secreted, extracellular matrix |
| T18 | gi|31645 | Glyceraldehyde- 3-phosphate dehydrogenase | 46 | R.GALQNIIPASTGAAK.A/ R.VPTANVSVVDLTCR.L/ K.LISWYDNEFGYSNR.V/ K.LTGMAFRVPTANVSVVDLTCR.L + Oxidation (M) / K.IKWGDAGAEYVVESTGVFTTMEK.A + Oxidation (M) / K.VIHDNFGIVEGLMTTVHAITATQK.T + Oxidation (M) | hsa:2597 | Glycolysis | Cytosol, plasma membrane |
| T19;20 | gi|809185 | Chain A, The Effect Of Metal Binding On The Structure Of Annexin V And Implications For Membrane Binding | 52 | RSEIDLFNIRK/ KGLGTDEESILTLLTSRS/ KWGTDEEKFITIFGTRS/ RGTVTDFPGFDERADAETLRK | hsa:308 | Blood coagulation | Cytosol |
| T19 | gi|2906146 | Malate dehydrogenase precursor | 46 | K.IFGVTTLDIVR.A/ K.VDFPQDQLTALTGR.I | hsa:4191 | Citric acid cycle | Mitochondria |
| T19 | gi|4826643 | Annexin A3 | 37 | K.MLISILTER.S +Oxidation (M)/ K.GAGTNEDALIEILTTR.T | hsa:306 | Defense response | Phagocytic vesicle |
| T19 | gi|4929769 | Glyoxalase domain-containing protein 4 (CGI-150 protein) | 36 | K.ILTPLVSLDTPGK.A(U) | hsa:51031 | Unknown | Mitochondria |
| T20 | gi|4502599 | Carbonyl reductase [NADPH] 1 | 46 | R.LFSGDVVLTAR.D/ R.VVNVSSIMSVR.A/ R.GQAAVQQLQAEGLSPR.F/ K.VADPTPFHIQAEVTMK.T + Oxidation | hsa:873 | Lipid metabolism -arachidonic acid | Cytosol |
| T23 | gi|9844110 | cAMP-specific phosphodiesterase 4D | 42 | K.LSPVISPR.N | hsa:5144 | Smooth muscle contraction; regulation of receptor activity | Cytosol |
| T24 | gi|2204207 | Glutathione S-transferase | 75 | M.PPYTVVYFPVR.G -.MPPYTVVYFPVR.G + Oxidation (M) M.PPYTVVYFPVRGR.C K.EEVVTVETWQEGSLK.A K.FQDGDLTLYQSNTILR.H K.ALPGQLKPFETLLSQNQGGK.T K.YISLIYTNYEAGKDDYVK.A | hsa:2940 | Amino acids metabolism | Cytosol |
| T26 | gi|181250 | Cyclophilin | 20 | K.TVDNFVALATGEK.G | hsa:5480 | Signal transduction | Cytosol |
Table 3. Proteins identified in pool of patients with non-tumor tissue (Group 2).
| Gel Slice | NCBI ID | Proteins | Score | Representative Peptides | KEGG | Function | Cell Compartment |
|---|---|---|---|---|---|---|---|
| C1 | gi|78101267 | Chain A, Human Complement Component C3 | 57 | K.TIYTPGSTVLYR.I R.IPIEDGSGEVVLSR.K R.LVAYYTLIGASGQR.E | hsa:718 | Inflammatory response; innate immunity; lipid metabolism | Secreted |
| C2 | gi|28243 | Filamin A | 49 | R.IANLQTDLSDGLR.L R.SAGQGEVLVYVEDPAGHQEEAK.V K.LDVQFSGLTK.G K.SPFSVAVSPSLDLSK.I R.EGPYSISVLYGDEEVPR.S R.FGGEHVPNSPFQVTALAGDQPSVQPPLR.S | hsa:2316 | Actin coupler | Cytosol |
| C3 | gi|2104553 | Myosin heavy chain (MHY11)(5′partial) | 59 | K.HAQAVEELTEQLEQFKR.A R.ALEEALEAKEELER.T K.IAQLEEQVEQEAREK.Q | hsa:4629 | Vascular muscle contraction; cell communication | Extracellular matrix |
| C7 | gi|34228 | Prelamin-A/C isoform 1 precursor; lamin A protein; progerin | 56 | R.VAVEEVDEEGKFVR.L | hsa:4000 | Intermediate filament | Nucleus |
| C7 | gi|386758 | GRP78 (Glucose-regulated protein 78) precursor | 45 | R.ITPSYVAFTPEGER.L R.IINEPTAAAIAYGLDKR.E | hsa:3309 | Anti-apoptosis | Endoplasmic reticulum |
| C7 | gi|23200154 | Chain A, NMR Structures Of The C-Terminal Globular Domain Of Human Lamin AC | 43 | R.VAVEEVDEEGKFVR.L | hsa:4000 | Structural molecule activity | Nucleus |
| C8 | gi|762885 | Plakoglobin | 72 | K.SAIVHLINYQDDAELATR.A R.ALMGSPQLVAAVVR.T Oxidation (M) R.LVQNCLWTLR.N R.NEGTATYAAAVLFR.I | hsa:3728 | Cell adhesion, cell migration | Cytosol; plasmatic membrane |
| C8 | gi|642534 | Lumican | 42 | K.SLEYLDLSFNQIAR.L | hsa:4060 | Collagen fibrils organization | Extracellular matrix |
| C8 | gi|110590597 | Chain A, Apo-Human Serum Transferrin (Non-Glycosylated) | 36 | K.FYLGYEYVTAIR.N | hsa:7018 | Mineral absorption | Secreted |
| C10 | gi|35505 | Pyruvate kinase | 39 | R.TATESFASDPILYRPVAVALDTKGPEIR.T R.EAEAAIYHLQLFEELRR.L | hsa:5315 | Glycolysis | Cytosol; plasmatic membrane |
| C11;13; 15; 16 | gi|3411130 | Mutant Desmin | 48 | R.FLEQQNAALAAEVNR.L | hsa:1674 | Cytoskeleton structural protein activity | Intermediate filament C |
| C13 | gi|340219 | Vimentin | 60 | K.ILLAELEQLK.G K.ILLAELEQLKGQGK.S K.LQEEMLQREEAENTLQSFR.Q Oxidation (M) R.KVESLQEEIAFLK.K R.QVQSLTCEVDALKGTNESLER.Q R.EYQDLLNVK.M K.MALDIEIATYR.K Oxidation (M) R.ISLPLPNFSSLNLR.E | hsa:7431 | Apoptosis; cell mobility | Cytosol |
| C13 | gi|704416 | Elongation factor Tu | 45 | K.LLDAVDTYIPVPAR.D | hsa:7284 | Oxidative phosphorylation | Mitochondria |
| C15 | gi|34234 | Laminin-binding protein | 75 | R.AIVAIENPADVSVISSR.N R.FTPGTFTNQIQAAFREPR.L | hsa:3921 | Ribosome | Cytosol |
| C16 | gi|47519616 | Tropomyosin beta chain isoform 2 | 73 | R.IQLVEEELDR.A R.IQLVEEELDRAQER.L R.LATALQKLEEAEK.A | hsa:7169 | Muscle contraction | Cytosol |
| C17 | gi|31645 | Glyceraldehyde-3-phosphate dehydrogenase | 72 | K.VIHDNFGIVEGLMTTVHAITATQK.T Oxidation (M) R.DGRGALQNIIPASTGAAK.A R.GALQNIIPASTGAAK.A R.VPTANVSVVDLTCR.L K.LISWYDNEFGYSNR.V | hsa:2597 | Glycolysis | Cytosol; plasmatic membrane |
| C19 | gi|4505773 | Prohibitin | 91 | R.IFTSIGEDYDER.V R.FDAGELITQR.E | hsa:5245 | DNA synthesis | Mitochondrial membrane |
| C19 | gi|66473265 | Beta globin chain | 50 | K.VNVDEVGGEALGR.L R.LLVVYPWTKR.F | hsa:5245 | Oxygen transport | Hemoglobin |
| C21 | gi|494066 | Chain A, Three- Dimensional Structure Of Class Pi Glutathione S-Transferase From Human Placenta In Complex With S-Hexylglutathione At2.8 Angstroms Resolution | 36 | .PPYTVVYFPVRGR.C K.FQDGDLTLYQSNTILR.H | hsa:2940 | Amino acids metabolism | Cytosol |
DISCUSSION
A large number of proteins were identified in both Groups 1 and 2. Some of these proteins found in Group 1 are also directly involved in the development of other types of cancers and therefore, suitable for analysis.
The major stress-inducible heat shock protein, Hsp70, that is a chaperone protein abundantly and preferentially expressed in tumors, was detected in Group 1. Owing to the ability of Hsp70 to protect cells from a wide range of apoptotic and necrotic stimuli, it has been assumed that Hsp70 may confer survival advantage to tumor cell lines. Nylandsted et al. (16) demonstrated that the depletion of Hsp70 by an adenovirus expressing antisense Hsp70 resulted in a massive cell death of tumorigenic cell lines of breast, colon, prostate, and liver. The authors advocate that Hsp70 is a prerequisite for the survival of human cancer cells. Similarly, Aghdassi et al. (17) demonstrated that the depletion of Hsp70 by short interfering RNA treatment induced apoptosis in pancreatic adenocarcinoma.
Plectin is a cytolinker protein of the plakin family. Plakins connect intermediate filaments to desmosomes and hemidesmosomes, stabilize cells mechanically, regulate cytoskeleton dynamics, and serve as a scaffolding platform for signaling molecules. Niwa et al. (18) reported that Plectin misexpression leads to displacement of the centrosome, therefore contributing to genomic instability and cancer development. Nevertheless, plectin is not expressed by most normal tissues, with the exception of the skin and genitourinary tract. Interestingly, we have detected plectin solely in Group 1. Complement plays a central part of the innate immune system, providing a highly effective means for destruction of invading microorganisms: clearance of immune complexes; and elimination of dead, apoptotic, and tumor cells. During the evolution of a cancer cell, neo-antigens are produced. These elements distinguish cancer cells from their normal counterparts and may well be recognized by the immune system, eliminating many or most tumors (19, 20). Although most in vivo observations support that many cancers activate the autologous complement system, it is also well-known that the efficiency of complement-mediated tumor cytotoxicity is hampered by various protective mechanisms (21). In this work, human complement C3 was detected only in Group 2. A possible explanation for these findings lies on the theory that patients with malignancies have a poorer immune response. Our result corroborates the study of Ornellas et al. (22), in which the authors have demonstrated that human complement fragments C3 and C4A/B were downregulated in plasma of patients with SCCP. In the present series, all patients from Group 1 were HPV positive and this could explain the absence of complement C3 because viral proteins counteract the immune response (23).
Enolase is a key glycolytic enzyme that has been used as a diagnostic marker to identify human lung cancers (24). Higher α-enolase plasma levels were also identified in patients with renal cell carcinoma (25). In cancer cells, enolase is overexpressed and localizes on their surface, where it acts as a key protein in tumor metastasis, promoting cellular metabolism in anaerobic conditions and driving tumor invasion through plasminogen activation and extracellular matrix degradation. It also displays a characteristic pattern of acetylation, methylation, and phosphorylation that regulates protein functions and immunogenicity. In the present study, alfa and beta enolase isoforms were identified exclusively in Group 1. This finding may suggest that in the future, enolase can be used as a possible clinical biomarker. Nevertheless, further studies are needed to corroborate these findings and to determine the usefulness of this protein in clinical scope.
Prohibitin is a potential tumor suppressor, which was originally identified because of its anti-proliferative activities. The human prohibitin gene was identified and cloned in 1991, as a result of a search for potential tumor suppressors, on the basis of its anti-proliferative activities (26). Furthermore, prohibitin is capable of inhibiting cell proliferation by repressing the transcriptional activity mediated by E2F which regulates many genes involved in the transition G1/S and DNA synthesis (27). In addition to transcriptional repression, prohibitin can induce p53-mediated transcription, indicating that prohibitin may have dual functions in modulating transcription (28).
In a study conducted by Joshi et al. (29), the authors supported this theory by demonstrating that prohibitin can differentially regulate the Yin-Yang 1 and caspase 7 gene promoter activities. Additional functions related to prohibitin were linked to cell apoptosis (30). In this series, prohibitin was exclusively presented in Group 2, supporting its potential tumor suppressor activity. The critical functions of prohibitin in growth control and transcriptional regulation clearly indicate the need for further investigations to elucidate its importance in SCCP development.
To our knowledge this is the first study that analyzed penile tumors through proteomics technologies. Unfortunately, as all samples in our analysis were typed as HPV +, it was not possible to perform a comparison concerning HPV status in the group with tumor. As the selected patients were positive for HPV DNA, this fact can cause false negative for complement proteins. The variability could have been better analyzed if there were compared to patients with cancer of the penis, whose tests did not reveal the presence of HPV. The proteomic consequences of HPV infection in penile carcinoma are not known. Analysis of differentially expressed proteins by HPV status revealed enrichment of proteins involved in epithelial cell development, keratinization and extracellular matrix organization in HPV- oropharyngeal carcinoma (OPC), whereas enrichment of proteins in DNA initiation and replication and cell cycle control was found for HPV+ (OPC) (31). Due to the rarity of penile tumors and the high percentage of HPV positive in our samples (8, 13) it is difficult to compare the tumors according to HPV status. However, a second study is underway to compare our results and identify the presence or absence of complement in tissue of SCCP patients negative for HPV.
CONCLUSIONS
We identified a large number of proteins in patients with penile cancer and in the control group. Some of these proteins, found in the first group, are also directly involved in the development of other types of cancers and therefore, suitable for analysis. Further studies are needed to corroborate these findings and to determine the usefulness of each discussed protein in the clinical scope of SCCP patients. Remarkably, this work reinforces that the C3 complement protein is a strong biomarker candidate for evaluating SCCP patients. Further studies should be conducted comparing samples positive for HPV with other HPV negative.
ACKNOWLEGMENTS
This work was supported by FAPERJ (APQ1 E-26/110.812/2009 and E-26/111.336/2013) and Programa de Oncobiologia (Brazil).
REFERENCES
- 1.Solsona E, Algaba F, Horenblas S, Pizzocaro G, Windahl T. European Association of Urology. EAU Guidelines on Penile Cancer. Eur Urol. 2004;46:1–8. doi: 10.1016/j.eururo.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Brunini R. Resultados. Ministério da Saúde - Campanha Nacional de Combate ao Câncer; Rio de Janeiro, RJ: 1982. Câncer no Brasil: Dados histopatológicos: 1976-80; pp. 118–118. [Google Scholar]
- 3.Parkin DM, Muir CS. Cancer Incidence in Five Continents. Comparability and quality of data. IARC Sci Publ. 1992;120:45–173. [PubMed] [Google Scholar]
- 4.Koifman L, Vides AJ, Koifman N, Carvalho JP, Ornellas AA. Epidemiological aspects of penile cancer in Rio de Janeiro: evaluation of 230 cases. Int Braz J Urol. 2011;37:231–240. doi: 10.1590/s1677-55382011000200010. discussion 240-3. [DOI] [PubMed] [Google Scholar]
- 5.Barrasso R, De Brux J, Croissant O, Orth G. High prevalence of papillomavirus-associated penile intraepithelial neoplasia in sexual partners of women with cervical intraepithelial neoplasia. N Engl J Med. 1987;317:916–923. doi: 10.1056/NEJM198710083171502. [DOI] [PubMed] [Google Scholar]
- 6.Maiche AG. Epidemiological aspects of cancer of the penis in Finland. Eur J Cancer Prev. 1992;1:153–158. doi: 10.1097/00008469-199202000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Maden C, Sherman KJ, Beckmann AM, Hislop TG, Teh CZ, Ashley RL, et al. History of circumcision, medical conditions, and sexual activity and risk of penile cancer. J Natl Cancer Inst. 1993;85:19–24. doi: 10.1093/jnci/85.1.19. [DOI] [PubMed] [Google Scholar]
- 8.Scheiner MA, Campos MM, Ornellas AA, Chin EW, Ornellas MH, Andrada-Serpa MJ. Human papillomavirus and penile cancers in Rio de Janeiro, Brazil: HPV typing and clinical features. Int Braz J Urol. 2008;34:467–474. doi: 10.1590/s1677-55382008000400009. discussion 475-6. [DOI] [PubMed] [Google Scholar]
- 9.McCance DJ, Kalache A, Ashdown K, Andrade L, Menezes F, Smith P, et al. Human papillomavirus types 16 and 18 in carcinomas of the penis from Brazil. Int J Cancer. 1986;37:55–59. doi: 10.1002/ijc.2910370110. [DOI] [PubMed] [Google Scholar]
- 10.Peclat de Paula AA, Neto JCA, Cruz AD, Freitas R de., Jr Carcinoma epidermóide do pênis: considerações epidemiológicas, histopatológicas, infuência viral e tratamento cirúrgico. Rev Bras Cancer. 2005;51:243–252. [Google Scholar]
- 11.Silva KC, Rosa ML, Moyse N, Afonso LA, Oliveira LH, Cavalcanti SM. Risk factors associated with human papillomavirus infection in two populations from Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2009;104:885–891. doi: 10.1590/s0074-02762009000600011. [DOI] [PubMed] [Google Scholar]
- 12.Melgaço FG, Rosa ML, Augusto EF, Haimuri JG, Jacintho C, Santos LS, et al. Human papillomavirus genotypes distribution in cervical samples from women living with human immunodeficiency virus. Arch Gynecol Obstet. 2011;283:809–817. doi: 10.1007/s00404-010-1443-z. [DOI] [PubMed] [Google Scholar]
- 13.Afonso LA, Moyses N, Alves G, Ornellas AA, Passos MR, Oliveira Ldo H, et al. Prevalence of human papillomavirus and Epstein-Barr virus DNA in penile cancer cases from Brazil. Mem Inst Oswaldo Cruz. 2012;107:18–23. doi: 10.1590/s0074-02762012000100003. [DOI] [PubMed] [Google Scholar]
- 14.Ericsson C, Franzén B, Nistér M. Frozen tissue biobanks. Tissue handling, cryopreservation, extraction, and use for proteomic analysis. Acta Oncol. 2006;45:643–661. doi: 10.1080/02841860600818047. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;15(227):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Nylandsted J, Brand K, Jäättelä M. Heat shock protein 70 is required for the survival of cancer cells. Ann N Y Acad Sci. 2000;926:122–125. doi: 10.1111/j.1749-6632.2000.tb05605.x. [DOI] [PubMed] [Google Scholar]
- 17.Aghdassi A, Phillips P, Dudeja V, Dhaulakhandi D, Sharif R, Dawra R, et al. Heat shock protein 70 increases tumorigenicity and inhibits apoptosis in pancreatic adenocarcinoma. Cancer Res. 2007;15(67):616–625. doi: 10.1158/0008-5472.CAN-06-1567. [DOI] [PubMed] [Google Scholar]
- 18.Niwa T, Saito H, Imajoh-ohmi S, Kaminishi M, Seto Y, Miki Y, et al. BRCA2 interacts with the cytoskeletal linker protein plectin to form a complex controlling centrosome localization. Cancer Sci. 2009;100:2115–2125. doi: 10.1111/j.1349-7006.2009.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 20.Smyth MJ, Godfrey DI, Trapani JA. A fresh look at tumor immunosurveillance and immunotherapy. Nat Immunol. 2001;2:293–299. doi: 10.1038/86297. [DOI] [PubMed] [Google Scholar]
- 21.Jurianz K, Ziegler S, Garcia-Schüler H, Kraus S, Bohana-Kashtan O, Fishelson Z, et al. Complement resistance of tumor cells: basal and induced mechanisms. Mol Immunol. 1999;36:929–939. doi: 10.1016/s0161-5890(99)00115-7. [DOI] [PubMed] [Google Scholar]
- 22.Ornellas P, Ornellas AA, Chinello C, Gianazza E, Mainini V, Cazzaniga M, et al. Downregulation of C3 and C4A/B complement factor fragments in plasma from patients with squamous cell carcinoma of the penis. Int Braz J Urol. 2012;38:739–749. doi: 10.1590/1677-553820133806739. [DOI] [PubMed] [Google Scholar]
- 23.Campo MS, Graham SV, Cortese MS, Ashrafi GH, Araibi EH, Dornan ES, et al. HPV-16 E5 down-regulates expression of surface HLA class I and reduces recognition by CD8 T cells. Virology. 2010;407:137–142. doi: 10.1016/j.virol.2010.07.044. [DOI] [PubMed] [Google Scholar]
- 24.He P, Naka T, Serada S, Fujimoto M, Tanaka T, Hashimoto S, et al. Proteomics-based identification of alpha-enolase as a tumor antigen in non-small lung cancer. Cancer Sci. 2007;98:1234–1240. doi: 10.1111/j.1349-7006.2007.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko N, Gotoh A, Okamura N, Matsuo E, Terao S, Watanabe M, et al. Potential tumor markers of renal cell carcinoma: α-enolase for postoperative follow up, and galectin-1 and galectin-3 for primary detection. Int J Urol. 2013;20:530–535. doi: 10.1111/j.1442-2042.2012.03206.x. [DOI] [PubMed] [Google Scholar]
- 26.Nuell MJ, Stewart DA, Walker L, Friedman V, Wood CM, Owens GA, et al. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblastos and HeLa cells. Mol Cell Biol. 1991;11:1372–1381. doi: 10.1128/mcb.11.3.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501–3510. doi: 10.1038/sj.onc.1202684. [DOI] [PubMed] [Google Scholar]
- 28.Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853–47861. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- 29.Joshi B, Rastogi S, Morris M, Carastro LM, DeCook C, Seto E, et al. Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J. 2007;401:155–166. doi: 10.1042/BJ20060364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowdhury I, Xu W, Stiles JK, Zeleznik A, Yao X, Matthews R, et al. Apoptosis of rat granulosa cells after staurosporine and sérum withdrawal is suppressed by adenovirus-directed overexpression of prohibitin. Endocrinology. 2007;148:206–217. doi: 10.1210/en.2006-0187. [DOI] [PubMed] [Google Scholar]
- 31.Slebos RJ, Jehmlich N, Brown B, Yin Z, Chung CH, Yarbrough WG, et al. Proteomic analysis of oropharyngeal carcinomas reveals novel HPV-associated biological pathways. Int J Cancer. 2013;132:568–579. doi: 10.1002/ijc.27699. [DOI] [PMC free article] [PubMed] [Google Scholar]