Abstract
Background:
Candida albicans (C. albicans) is a major cause of candidaemia in people with impaired immunity. Blood culture is a “gold standard” for candidaemia detection but is time-consuming and relatively insensitive. We established a real-time PCR assay for C. albicans detection in blood by LightCycler PCR and melting curve analysis.
Methods:
Five milliliter blood samples from healthy volunteers were spiked with 100-106 C. albicans cells to determine the detection limit of our method. DNA was extracted from whole blood using glass beads and the QIAamp DNA Blood Mini Kit (Qiagen, Hilden Germany). DNA from C. albicans isolates were amplified with primers and inserted into Escherichia coli (E. coli) DH5α.1 cells with the TA cloning vector (Invitrogen). The plasmid was used for standardization and optimization. A quantitative PCR assay with the LightCycler amplification and detection system based on fluorescence resonance energy transfer (FRET) with two different specific probes was established. To assess the precision and reproducibility of real-time PCR the intra-assay precision was determined in six consecutive assays.
Results:
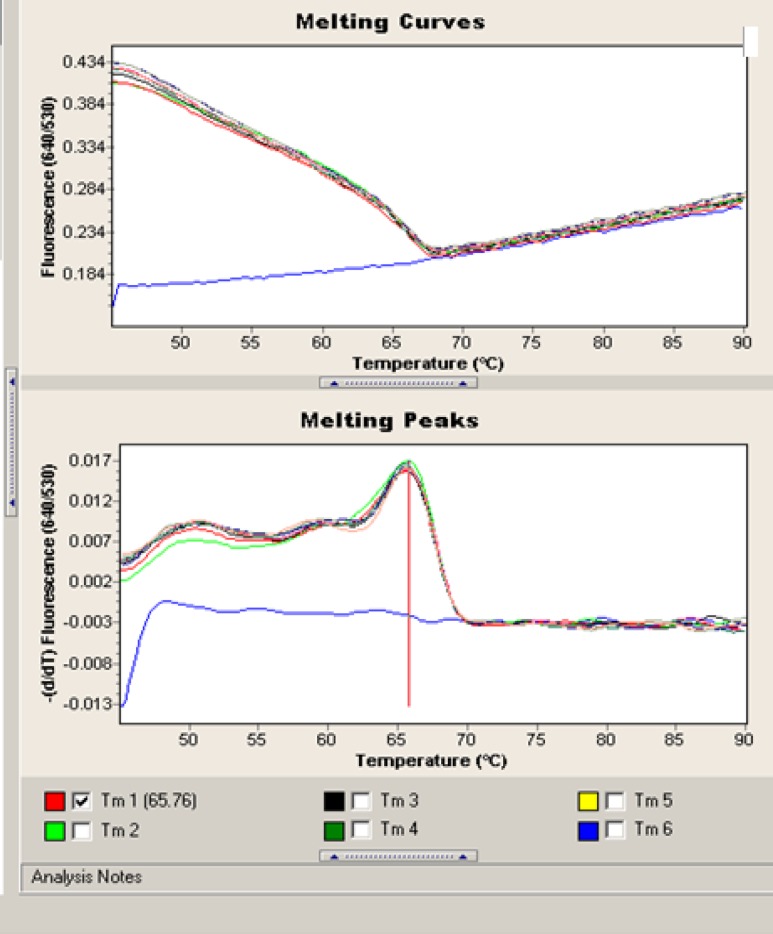
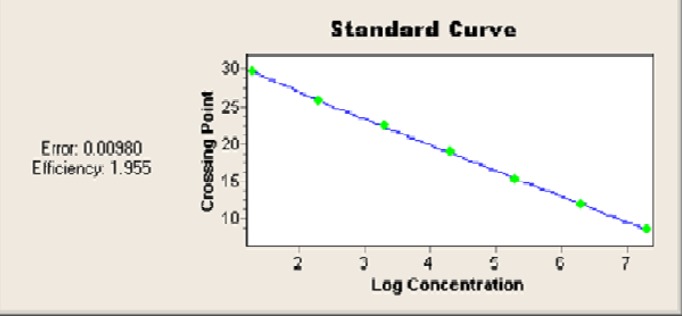
No cross-reactivity of the hybridization probes with the DNA of non-C. albicans species or human genomic DNA was observed, which confirmed its 100% specificity. The minimum limit detected was one C. albicans cell or 100 CFU/ml (10 fg) per PCR reaction. The real-time PCR efficiency rate for Candida was high (E = 1.95). Melting curve analysis of C. albicans showed a specific melting peak temperature of 65.76 °C.
Conclusion:
The real-time PCR assay we developed is highly specific and sufficiently sensitive to detect the fungal load for early diagnosis of invasive candidiasis.
Key Words: Invasive candidiasis, Real-time PCR, Candida albicans
Introduction
Invasive fungal infections have proved to be major causes of morbidity and mortality in people with impaired immunity, particularly neutropenic patients with hematological malignancies, and recipients of allogeneic bone marrow transplants (1). Invasive candidiasis, a common cause of nosocomial infections, is a main cause of infections in patients receiving bone marrow transplants and those with leukemia or other cancers, and is also associated with high rates of morbidity and mortality (2-4).
Candida species represent the fourth most-frequent cause of bloodstream infections in critically ill patients (5). However, diagnosis remains difficult and, more importantly, rapid identification of Candida infection is necessary for early, effective, and appropriate antifungal therapy (6-9). In hospitals, Candida spp. represent 8-9% of all nosocomial bloodstream infections, and the risk is higher in patients in intensive care units (ICU) and cancer patients (10, 11). As many as half of the cases are not diagnosed ante mortem (12, 13).
In North America, non-Candida albicans (C. albicans) spp. are currently more prevalent than C. albicans; C. glabrata, and C. krusei, which are less susceptible to fluconazole than C. albicans, account for 28% of all candidaemias (14). Mortality rates are high, ranging from 40 to 54% (15, 16).
Conventional identification methods from positive blood cultures need at least one extra day for the pure culture and germ tube test. Germ tube negatives require two to four additional days for chlamydospore formation, and assimilation tests can take five days or longer. Identifying certain Candida spp. might take even longer, which can delay appropriate antifungal treatment (17, 18). The current “gold standard” for identification of candidaemia is blood culture. However, not only is this method time-consuming, but its sensitivity for early detection of infection has been reported to be as low as 50% (19, 20). Non-culture-based methods such as antigen, antibody, and metabolite detection, which can be less time-consuming, have been investigated. None of them is accepted as ideal, and few have been produced commercially (21, 22).
Nucleic acid detection systems, such as DNA detection by PCR, have been developed to assist in the rapid diagnosis of infections, allowing for the initiation of species-oriented therapies as early as 6 h after the onset of sepsis (23).
The quantitative PCR test is based on fluorescence resonance energy transfer (FRET) with the LightCycler (Roche Diagnostics, Mannheim, Germany). This technology combines rapid glass capillary thermo cycling with online fluorescence detection of the PCR amplicon.
In this study we optimized a real-time PCR/FRET assay for detection of C. albicans DNA in blood samples.
Materials and Methods
Bacterial Strains, Plasmid and mice
Three standard strains of fungi (C. albicans, CBS 562; C. glabrata, CBS 138; and C. dubliniensis, CBS 7987) were obtained from the CBS (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands) and cultured on Sabouraud-glucose-agar for 72 h at 30 °C. Serial dilutions of fungal cells were adjusted with saline to a McFarland standard of 0.5.
To determine the detection limit of Candida, 5 mL of EDTA anti-coagulated whole-blood samples from normal volunteers were spiked with serial dilutions of C. albicans cells (100-106 cells per ml of blood). DNA was extracted from whole blood as described by Loffler et al. (24)with minor modifications (25). Briefly, erythrocytes were hypotonically lysed in Red Cell Lysis Buffer (RCLB:10 mM Tris pH = 7.6, 5 mM MgCl2, 10 mM NaCl) for 10 min at 37 °C. The pellets were obtained by centrifugation at 5000 g for 10 min and vortexed with glass beads (425-600 µm, Sigma-Aldrich Corp, St. Louis, MO USA) for 3 min. The supernatants were treated with 180 U of lyticase (Sigma-Aldrich Corp, St. Louis, MO USA) at 37 oC for 30 min. Two hundred µl of the treated supernatants were extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden Germany) following the manufacturer’s instructions. To test for possible contamination in the DNA extractions, aliquots of saline, or DNA from healthy volunteers were analyzed simultaneously. To exclude the presence of cross-contamination in the amplifications, sterile saline and no DNA template samples were processed in parallel with all experimental samples.
To determine the sensitivity of the assay, we created a plasmid containing a Candida DNA amplicon. The 500-bp amplicon was purified using Micro Spin S-400 HR columns, ligated into the TA cloning vector (Invitrogen, CA USA) as described in the manufacturer’s instructions, and transfected into Escherichia coli (E.coli) DH5α cells. Colonies containing the inserts were identified by color screening on LB medium supplemented with ampicillin (200 mg/L). The colorless colonies were selected and successful cloning of the PCR product was confirmed by real-time PCR. The plasmid DNA was extracted and purified with the Quick Plasmid Miniprep Kit (Qiagen, Hilden Germany).
The plasmid DNA concentrations were spectrophotometrically determined at 260 nm by SmartSpec Plus (Bio-Rad Laboratories, Hercules, California) and serially diluted ten-fold in TE buffer (pH, 8.0) from 100-106 plasmid copies/μL for use in real-time PCR. The standard curve was established with the logarithm of the standard sample concentrations as the Y axes and the Ct values as the X axes.
To determine the specificity of the oligonucleotide hybridization based on the FRET technique, a panel of archived DNAs, including those from filamentous fungi such as A. fumigatus, and healthy human fibroblasts, was analyzed.
A quantitative PCR assay with the LightCycler (Roche Diagnostics, Mannheim, Germany) amplification and detection system based on FRET with two different specific probes was established. PCR was performed in glass capillaries, which ensures rapid equilibration between the air and the reaction components because of the high surface area to volume ratio of the capillaries.
The primers bind to conserved regions of the fungal 18s rRNA gene. Two different probes were designed that hybridized to internal sequences of the C. albicans 18s RNA amplicon. One probe was labeled with LightCycler Red 640 phosphate, and the other with fluorescein (Table 1). Amplicons were detected using the FastStart LightCycler DNA Master Hybridization Probes Kit (Roche Diagnostics, Mannheim, Germany) as described by the manufacturer. During FRET, fluorescein is excited by the light source of the LightCycler instrument. The excitation energy is transferred to the acceptor phosphate, LightCycler Red 640, and the emitted fluorescence is measured after being annealed by the photohybrids of the instrument. The PCR mixture contained Taq polymerase, LightCycler hybridization reaction buffer, a deoxynucleoside triphosphate mixture (with dUTP instead of dTTP), 3 mM MgCl2, and 12.5 pmol of primers. Samples were amplified in parallel by performing 45 repeated cycles of denaturation (5 s at 95 °C), annealing (15 s at 62 °C), and enzymatic chain extension (25 s at 72 °C). To enhance specificity, TaqStart Antibody (Clontech, Palo Alto, CA) was used.
Table 1.
Primers and probes
| Primers (Eurofins MWG Operon, Ebensburg, Germany) |
| CAN-F (5´-ATT GGA GGG CAA GTC TGG TG) CAN-R (5´-ATC CCT TAG TCG GCA TAG-3´) |
| Probes (Tibmolbiol, Berlin, Germany) |
| LC 640 5′ TGG CGA ACC AGG ACT TTT ACT TTG A- phosphate 5′AGC CTT TCC TTC TGG GTA GCC ATT-LC fluorescein |
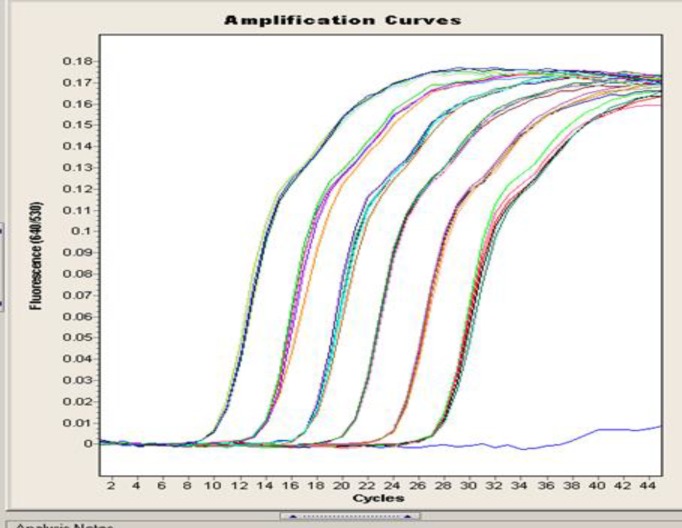
Quantification was performed by monitoring the time point at which the logarithmic linear phase could be distinguished from the background (crossing point). Serially-diluted samples of genomic fungal DNA obtained from C. albicans cultures (100-106 CFU, corresponding to 10 fg-10 ng of DNA) were used as external standards in each run (Fig. 2). The cycle numbers of the logarithmic linear phases were plotted against the logarithms of the template DNA concentrations. The concentrations of fungal DNA in the clinical samples were calculated by comparing the cycle numbers of the logarithmic linear phases of the samples with the cycle numbers of the external standards. After each LightCycler run, products were electrophoresed in 2% agarose gels containing ethidium bromide (EtBr) in TAE buffer (40 mM Tris-acetate, 1 mM EDTA) and visualized under UV light. A 100-bp DNA ladder was used as a standard (Life Technologies, Karlsruhe, Germany).
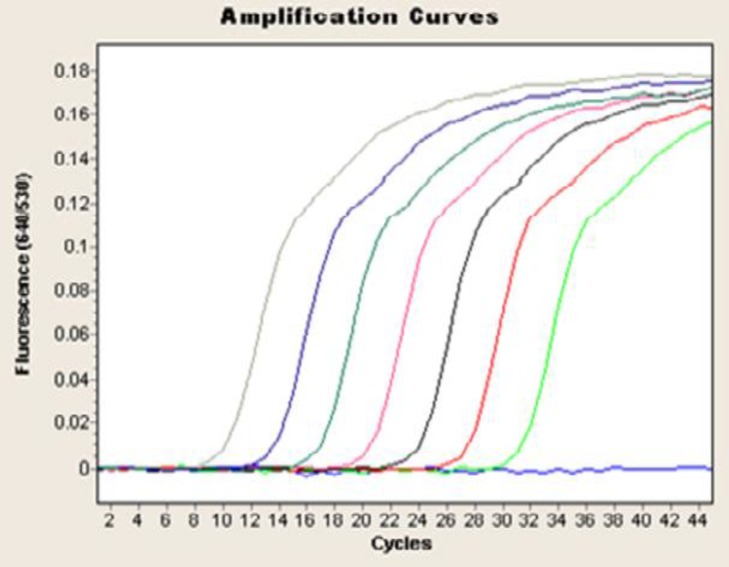
Fig.2.
Quantification of serially-diluted 100-106 C. albicans cells using LightCycler-based PCR
Results
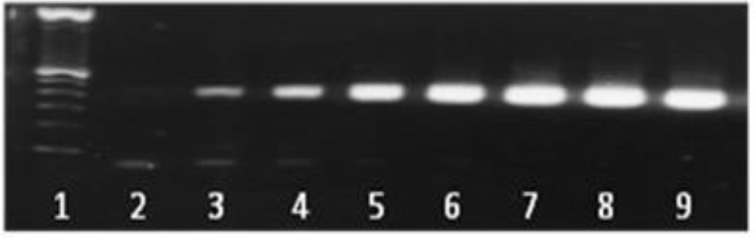
Candida DNAs were efficiently amplified with the LightCycler instrument. All serially-diluted samples containing at least 100 CFU/ml produced single bands of 500 bp; the bands represent the fungus-specific amplicons (Fig. 1). For sensitivity testing, blood from healthy volunteers was spiked with Candida cells (100-106 cells per ml of blood, in serial dilutions). Using the LightCycler FRET technique, we demonstrated a sensitivity of 1 CFU/ml of blood for Candida cells (Fig. 2).
Fig. 1.
Agarose gel electrophoresis of serially diluted 100-106 C. albicans cells (100-106 CFU, corresponding to 10 ng to 10 fg-10 ng of DNA) showing a single, specific band at 500 bp. Lanes: 1: 100-bp ladder; 2: negative control (double-distilled H2O); 3: 100 CFU (10 fg); 4: 101 CFU (100 fg); 5: 102 CFU (1 pg); 6: 103 CFU (10 pg); 7: 104 CFU (100 pg); 8: 105 CFU (1 ng); 9: 106 CFU (10 ng). Amplifications were performed in a conventional thermocycler
All PCRs were negative with other fungal and human genomic DNAs, and the probes successfully hybridized the DNA extracted from C. albicans, demonstrating 100% specificity. Melting curve analysis of C. albicans showed a specific melting peak temperature of 65.76 °C (Fig. 3).
Fig.3.
Melting peaks obtained from C. albicans with the corresponding species-specific biprobes. The values on the y axis are the first negative derivative of the change in fluorescence (dF) divided by the change in temperature (dT) (dF/dT
The linear range of the assay was 100-106 Candida cells. The real-time PCR efficiency rate (E) was 1.95 (Fig. 4).
Fig.4.
The standard curve demonstrates the linear range of the assay
The inter- and intra-assay reproducibility of the PCR was assessed by serial dilution of standard C. albicans DNA for six consecutive assays in parallel and on two following days. Results were highly similar, reflecting a high level of reproducibility (Fig. 5).
Fig.5.
Intra-assay: Samples were serially diluted from 100 -106 CFU per ml of blood with standard C.albican's DNA for six consecutive time points and amplified by real-time PCR
Discussion
We detected DNA from C. albicans, the most common Candida species, by LightCycler PCR and melting curve analysis. This study represents a step forward in detection, speed, and differentiation in PCR-based diagnostic methods for C. albicans. Standardized PCR assays have not yet become routine in clinical settings. Traditional techniques such as cultures and direct examination of tissues are practical but not sufficiently sensitive for the early diagnosis of fungal infections. In addition, the sensitivities of Candida mannan, (1, 3)-beta-D-glucan (BDG), and anti-mannan antibodies, singly or in combination, for early diagnosis of invasive candidiasis are not high (26). Real-time PCR represents an advance compared to the detection of molds by culture methods, which lack sensitivity and specificity and require more time than real-time PCR to allow diagnoses.
About 80% of systemic fungal infections are caused by Candida species. Approximately 60% of these are caused by C. albicans, but in recent years, nearly 50% of all cases of candidaemia are caused by non-albicans species such as C. tropicalis, C. parapsilosis, C. glabrata, and C. krusei (27-29).
It was essential to design a method that would be able to identify C. albicans without the need for a time-consuming post-amplification handling step. The amplification and post-amplification analyses performed in closed glass capillaries minimized the risk of carryover contamination. Using two fluorescently labeled probes on the LightCycler-FRET detection system enabled us to detect C. albicans DNA with no cross-hybridization with DNA from other fungal pathogens or human genomic DNA. This finding, combined with our previous study (30), supports the diagnostic value of the LightCycler-FRET detection system in the sensitive and specific diagnosis of C. albicans.
The method used in this study allowed detection of one cell (100 CFU/ ml) equivalent (10 fg) of C. albicans genomic DNA in whole blood. The limit of detection in the Gurtner et al. study was at least 1 CFU per PCR, corresponding to 5 to 10 CFU/ml blood (31). Fricke et al. detected at least two genome equivalents per assay in spiked serum samples for C. albicans, which is highly sensitive and comparable to other described methods (32).
The detection limits for PCR-ELISA and LightCycler PCR were reported to be 5 and 20 CFU/ml, respectively (33, 34). In this study we used the 18S rDNA gene, which is highly conserved in fungi, as the primary target for the detection of Candida in PCR-based diagnoses. Klingspor and Jalal used the 18S rDNA region for identification of Candida, although Hsu et al. used the 28S rDNA region. These assays established a highly sensitive and specific method for the identification and quantification of Candida species in blood (35, 36).
We also used a melting temperature (Tm) analysis that allowed species differentiation based on Tms. In a Fricke et al. study the melting temperature for C. albicans was 66.9 C (37). Due to the emergence of non-albicans Candida strains, eg. C. krusei and C. glabrata, which are resistant to common antifungal drugs such as fluconazole, the ability to identify the infecting species with the amplification/detection procedure will allow physicians to choose appropriate therapies for patients (38).
These assays are valuable tools for the detection of Candida DNA from blood in a variety of clinical settings. Accordingly, a study to validate our findings for detection of invasive candidiasis in patients at high risk for the infection is underway and more results will be available soon.
In conclusion, we adapted the LightCycler PCR assay technique to a procedure that was standardized, rapid, accurate, and reproducible, and combines rapid in vitro amplification with real-time species identification and quantification of the fungal load.
References
- 1.Denning DW. Invasive aspergillosis. Clinic Infect Dis. 1998 Apr;26(4):781–803. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 2.Horn R, Wong B, Kiehn TE, Armstrong D. Fungemia in a cancer hospital: Changing frequency, earlier onset, and results of therapy. Rev Infect Dis. 1985 Sep-Oct;7(5):646–55. doi: 10.1093/clinids/7.5.646. [DOI] [PubMed] [Google Scholar]
- 3.Whimbey E, Kiehn TE, Brannon P, Blevins A, Armstrong D. Bacteremia and fungemia in patients with neoplastic disease. Am J Med. 1987 Apr;82(4):723–30. doi: 10.1016/0002-9343(87)90007-6. [DOI] [PubMed] [Google Scholar]
- 4.Pittet D, Wenzel RP. Nosocomial bloodstream infections. Secular trends in rates, mortality, and contribution to total hospital deaths. Arch Intern Med. 1995 Jun;155(11):1177–84. doi: 10.1001/archinte.155.11.1177. [DOI] [PubMed] [Google Scholar]
- 5.Nishikawa A, Sugita T, Shinoda T. Rapid identification of Debaryomyces hansenii/Candida famata by polymerase chain reaction. Med Mycol. 1999 Apr;37(2):101–4. [PubMed] [Google Scholar]
- 6.Foster N, Symes C, Barton R, Hobson R. Rapid identification of Candida glabrata in Candida bloodstream infections. J Med Microbiol. 2007 Dec;56(Pt 12):1639–43. doi: 10.1099/jmm.0.47406-0. [DOI] [PubMed] [Google Scholar]
- 7.Cuenca-Estrella M, Rodriguez D, Almirante B, Morgan J, Planes AM, Almela M, et al. In vitro susceptibilities of bloodstream isolates of Candida species to six antifungal agents: results from a population-based active surveillance programme, Barcelona, Spain, 2002–2003. J Antimicrob Chemother. 2005 Feb;55(2):194–9. doi: 10.1093/jac/dkh548. [DOI] [PubMed] [Google Scholar]
- 8.Hong Nguyen M, Peacock Jr JE, Morris AJ, Tanner DC, Nguyen ML, Snydman DR, et al. The changing face of candidemia: emergence of non- Candida albicans species and antifungal resistance. Am J Med. 1996 Jun;100(6):617–23. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 9.Pfaller M, Jones R, Doern G, Sader H, Messer S, Houston A, et al. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997-1998. Antimicrob Agents Chemother. 2000 Mar;44(3):747–51. doi: 10.1128/aac.44.3.747-751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: A three-year analysis. Clin Infect Dis. 1999 Aug;29(2):239–44. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 11.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004 Aug;39(3):309–17. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 12.Chandrasekar P, Weinmann A, Shearer C. Autopsy-identified infections among bone marrow transplant recipients: a clinico-pathologic study of 56 patients. Bone Marrow Transplantation Team. Bone Marrow Transplant. 1995 Nov;16(5):675–81. [PubMed] [Google Scholar]
- 13.Einsele H, Hebart H, Roller G, Löffler J, Rothenhofer I, Müller C, et al. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997 Jun;35(6):1353–60. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, et al. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis. 2009 Jun;48(12):1695–703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- 15.Alonso-Valle H, Acha O, Garcia-Palomo J, Farinas-Alvarez C, Fernandez-Mazarrasa C, Farinas M. Candidemia in a tertiary care hospital: Epidemiology and factors influencing mortality. Eur J Clin Microbiol Infect Dis. 2003 Apr;22(4):254–7. doi: 10.1007/s10096-003-0890-x. [DOI] [PubMed] [Google Scholar]
- 16.Pappas PG, Rex JH, Lee J, Hamill RJ, Larsen RA, Powderly W, et al. A prospective observational study of candidemia: Epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis. 2003 Sep;37(5):634–43. doi: 10.1086/376906. [DOI] [PubMed] [Google Scholar]
- 17.Dooley D, Beckius ML, Jeffrey BS. Misidentification of clinical yeast isolates by using the updated Vitek Yeast Biochemical Card. J Clin Microbiol. 1994 Dec;32(12):2889–92. doi: 10.1128/jcm.32.12.2889-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maaroufi Y, De Bruyne J-M, Duchateau V, Georgala A, Crokaert F. Early Detection and Identification of Commonly Encountered Candida Species from Simulated Blood Cultures by Using a Real-Time PCR-Based Assay. J Mol Diagn. 2004 May;6(2):108–14. doi: 10.1016/S1525-1578(10)60498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez LJ, Rex JH, Anaissie EJ. Update on invasive candidiasis. Adv Pharmacol. 1996;37:349–400. doi: 10.1016/s1054-3589(08)60955-2. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad S, Khan Z, Mustafa AS, Khan ZU. Seminested PCR for diagnosis of candidemia: comparison with culture, antigen detection, and biochemical methods for species identification. J Clin Microbiol. 2002 Jul;40(7):2483–9. doi: 10.1128/JCM.40.7.2483-2489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maertens J, Marchetti O, Herbrecht R, Cornely O, Flückiger U, Frere P, et al. European guidelines for antifungal management in leukemia and hematopoietic stem cell transplant recipients: summary of the ECIL 3—2009 update. Bone Marrow Transplant. 2011 May;46(5):709–18. doi: 10.1038/bmt.2010.175. [DOI] [PubMed] [Google Scholar]
- 22.Karageorgopoulos DE, Vouloumanou EK, Ntziora F, Michalopoulos A, Rafailidis PI, Falagas ME. β-D-glucan assay for the diagnosis of invasive fungal infections: A meta-analysis. Clin Infect Dis. 2011 Mar;52(6):750–70. doi: 10.1093/cid/ciq206. [DOI] [PubMed] [Google Scholar]
- 23.McMullan R, Metwally L, Coyle P, Hedderwick S, McCloskey B, O'Neill H, et al. A prospective clinical trial of a real-time polymerase chain reaction assay for the diagnosis of candidemia in nonneutropenic, critically ill adults. Clin Infect Dis. 2008 Mar;46(6):890–6. doi: 10.1086/528690. [DOI] [PubMed] [Google Scholar]
- 24.Löffler J, Hebart H, Schumacher U, Reitze H, Einsele H. Comparison of different methods for extraction of DNA of fungal pathogens from cultures and blood. J Clin Microbiol. 1997 Dec;35(12):3311–2. doi: 10.1128/jcm.35.12.3311-3312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgener-Kairuz P, Zuber J, Jaunin P, Buchman T, Bille J, Rossier M. Rapid detection and identification of Candida albicans and Torulopsis (Candida) glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol-alpha-demethylase (L1A1) gene fragment. J Clin Microbiol. 1994 Aug;32(8):1902–7. doi: 10.1128/jcm.32.8.1902-1907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alam FF, Mustafa AS, Khan ZU. Comparative evaluation of (1, 3)-β-D-glucan, mannan and anti-mannan antibodies, and Candida species-specific snPCR in patients with candidemia. BMC Infect Dis. 2007 Sep;7:103. doi: 10.1186/1471-2334-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diekema D, Messer S, Brueggemann A, Coffman S, Doern G, Herwaldt L, et al. Epidemiology of candidemia: 3-year results from the emerging infections and the epidemiology of Iowa organisms study. Journal of Clinical Microbiology. 2002;40(4):1298–302. doi: 10.1128/JCM.40.4.1298-1302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis SL, Vazquez JA, McKinnon PS. Epidemiology, risk factors, and outcomes of Candida albicans versus non-albicans candidemia in nonneutropenic patients. Ann pharmacother. 2007 Apr;41(4):568–73. doi: 10.1345/aph.1H516. [DOI] [PubMed] [Google Scholar]
- 29.Passos XS, Costa CR, Araujo CR, Nascimento ES, e Souza LKH, Fernandes OdFL, et al. Species distribution and antifungal susceptibility patterns of Candida spp. bloodstream isolates from a Brazilian tertiary care hospital. Mycopathologia. 2007 Mar;163(3):145–51. doi: 10.1007/s11046-007-0094-5. [DOI] [PubMed] [Google Scholar]
- 30.Nabili M, Shokohi T, Janbabaie G, Hashemi-Soteh M, Ali-Moghaddam K, Aghili S. Detection of invasive aspergillosis in bone marrow transplant recipients using real-time PCR. J Glob Infect Dis. 2013 Apr;5(2):68–75. doi: 10.4103/0974-777X.112296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schabereiter-Gurtner C, Selitsch B, Rotter ML, Hirschl AM, Willinger B. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J Clin Microbiol. 2007 Mar;45(3):906–14. doi: 10.1128/JCM.01344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White PL, Shetty A, Barnes RA. Detection of seven Candida species using the Light-Cycler system. J Med Microbiol. 2003 Mar;52(Pt 3):229–38. doi: 10.1099/jmm.0.05049-0. [DOI] [PubMed] [Google Scholar]
- 33.Loeffler J, Hebart H, Sepe S, Schumacher U, Klingebiel T, Einsele H. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med Mycol. 1998 Oct;36(5):275–9. doi: 10.1080/02681219880000441. [DOI] [PubMed] [Google Scholar]
- 34.Maaroufi Y, Heymans C, De Bruyne JM, Duchateau V, Rodriguez-Villalobos H, Aoun M, et al. Rapid detection of Candida albicans in clinical blood samples by using a TaqMan-based PCR assay. J Clin Microbiol. 2003 Jul;41(7):3293–8. doi: 10.1128/JCM.41.7.3293-3298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klingspor L, Jalal S. Molecular detection and identification of Candida and Aspergillus spp. from clinical samples using real‐time PCR. Clin Microbiol Infect. 2006 Aug;12(8):745–53. doi: 10.1111/j.1469-0691.2006.01498.x. [DOI] [PubMed] [Google Scholar]
- 36.Hsu M-C, Chen K-W, Lo HJ, Chen YC, Liao MH, Lin YH, et al. Species identification of medically important fungi by use of real-time LightCycler PCR. J Med Microbiol. 2003 Dec;52(12):1071–6. doi: 10.1099/jmm.0.05302-0. [DOI] [PubMed] [Google Scholar]
- 37.Fricke S, Fricke C, Schimmelpfennig C, Oelkrug C, Schönfelder U, Blatz R, et al. A real‐time PCR assay for the differentiation of Candida species. J Appl Microbiol. 2010 Oct;109(4):1150–8. doi: 10.1111/j.1365-2672.2010.04736.x. [DOI] [PubMed] [Google Scholar]
- 38.Vanden BH, Dromer F, Improvisi I, Lozano-Chiu M, Rex J, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl 1):119–28. [PubMed] [Google Scholar]