Abstract
For decades, odour-baited traps have been used for control of tsetse flies (Diptera; Glossinidae), vectors of African trypanosomes. However, differential responses to known attractants have been reported in different Glossina species, hindering establishment of a universal vector control tool. Availability of full genome sequences of five Glossina species offers an opportunity to compare their chemosensory repertoire and enhance our understanding of their biology in relation to chemosensation. Here, we identified and annotated the major chemosensory gene families in Glossina. We identified a total of 118, 115, 124, and 123 chemosensory genes in Glossina austeni, G. brevipalpis, G. f. fuscipes, G. pallidipes, respectively, relative to 127 reported in G. m. morsitans. Our results show that tsetse fly genomes have fewer chemosensory genes when compared to other dipterans such as Musca domestica (n>393), Drosophila melanogaster (n = 246) and Anopheles gambiae (n>247). We also found that Glossina chemosensory genes are dispersed across distantly located scaffolds in their respective genomes, in contrast to other insects like D. melanogaster whose genes occur in clusters. Further, Glossina appears to be devoid of sugar receptors and to have expanded CO2 associated receptors, potentially reflecting Glossina's obligate hematophagy and the need to detect hosts that may be out of sight. We also identified, in all species, homologs of Ir84a; a Drosophila-specific ionotropic receptor that promotes male courtship suggesting that this is a conserved trait in tsetse flies. Notably, our selection analysis revealed that a total of four gene loci (Gr21a, GluRIIA, Gr28b, and Obp83a) were under positive selection, which confers fitness advantage to species. These findings provide a platform for studies to further define the language of communication of tsetse with their environment, and influence development of novel approaches for control.
Author Summary
Chemical sensing is crucial to survival of tsetse flies; the sole cyclical vectors of African trypanosomes that cause the neglected zoonotic tropical disease sleeping sickness in humans. For many years, vector control has been used to mitigate trypanosome infections among rural populations of sub-Saharan Africa. Nevertheless, development of an all-inclusive strategy to control tsetse flies using odour-baited traps has been limited by disparate responses to the odors exhibited by various tsetse species. In this study, proteins that are putatively involved in chemical sensing were identified and compared among five tsetse species and their close relatives with an aim of enhancing our knowledge on tsetse olfaction. Our findings suggest that the chemosensory genes are conserved across tsetse fly species despite their documented differential responses in odours. We found no species-specific sequence variations among the five species to suggest that differential response to odours is due to loss or gain of genes. It could therefore be hypothesized that the observed differences emerge during the downstream processing of odour molecules involving post translational modification of the chemosensory proteins. We thus recommend functional studies on the identified proteins to determine their roles and molecular interactions.
Introduction
Tsetse flies (Glossina spp.) are the sole cyclical vectors of African trypanosomes that cause the devastating Human African Trypanosomiasis (HAT, sleeping sickness) and Animal African Trypanosomiasis (AAT, nagana) across sub-Saharan Africa [1]. It is estimated that approximately 70 million people and 50 million cattle inhabiting tsetse-fly infested areas are at risk of contracting trypanosomiasis [2,3], and that nagana accounts for up to $ 4.75 billion annual losses [4]. Currently, there are no prophylactic drugs or vaccines against HAT. Moreover, the available chemotherapeutic remedies are not ideal due to their toxicity, difficulty in administration and growing resistance [4–6].
It has long been known that comprehensive and sustainable control of trypanosomiasis requires a vector control component [7]. Efforts to suppress tsetse populations include trapping, which rely on traps baited with various host derived odours [8–10]. Differences in response to the available baits have been observed among tsetse species and/or between males and female flies [11,12]. For example the palpalis/riverine species are thought to be attracted to kairomones released by monitor lizards, but unresponsive to odours that are highly attractive to Savannah species [13]. This differentiation of responses to odours is shown by the varied host preference in different sub-groups [14,15].
Chemoreception in tsetse and other insects is mediated by a group of peri-receptor and surface proteins/receptors encoded by different gene families [16] including: odorant binding proteins (OBPs), chemosensory proteins (CSPs), sensory neuron membrane proteins (SNMPs), gustatory receptors (GRs), ionotropic receptors (IRs) and odorant receptors (ORs). Genes encoding various chemosensory proteins are expressed at different olfactory receptor neurons (ORNs) located mainly on the surface of antennae and in fewer numbers on the maxillary palpi [17,18].
The OBPs and CSPs that recognize and solubilize hydrophobic odor molecules, shuttling them to the dendritic membrane [19,20], are characterized by the presence of a signal peptide and α-helices joined by disulphide bonds [21]. OBPs (~150 aa) are highly diverse proteins thought to bind to a wide range of odorants including pheromones. In Drosophila, four different sub-groups of OBPs have been described based on the number of conserved cysteine residues that participate in formation of their tertiary structures. These include (i) Classic OBPs that harbor six highly conserved cysteines and three disulphide bridges, (ii) Classic-Dimer OBPs that have two of the six-cysteine signatures, (iii) Minus-C OBPs which have lost two conserved cysteine residues and (iv) Plus-C OBPs which have additional conserved cysteine residues and a conserved proline [22]. On the other hand, CSPs are characterized by four conserved cysteines and an average length of 130 aa [19]. The latter have been implicated in non-olfactory functions in Drosophila [23]. Expression of OBPs and CSPs has been linked to host seeking by adult female in G. m. morsitans [24,25]. A third class of proteins that play a role in olfaction is the SNMPs which belong to the CD36 super family that act as scavenger proteins in humans [25–27]. An earlier study by Xa and colleagues demonstrated involvement of SNMP1 in chemoreception as a requirement for pheromone detection by Drosophila [28].
Insect ORs are highly diverse and are characterized by a reversed N-terminal topology and presence of a seven trans-membrane domain [29]. Specific ORs combine with Orco (Or83b), a non-conventional co-receptor, to form functional ion channels that confer specificity to a variety of semiochemicals [29,30]. Fewer ORs were identified in G. m. morsitans relative to D. melanogaster genome, but with an expansion of a gene critical role in recognition of male the pheromone, cis-vaccenyl acetate (cVA) (OR67d) [31]. Insect GRs are responsible for distinguishing between odor tastes and contact pheromones [16,32]. Fewer GRs were also identified in tsetse than in D. melanogaster and other Diptera [28]. No receptors for sugar were identified in G. m. morsitans, probably due to the hematophagous feeding behavior of the insect [31].
Another class of divergent insect chemosensory receptors is the ionotropic receptors; IRs [33,34]. The IRs, like ORs, function in complexes formed by up to three subunits and one or two of co-receptors (Ir25a and Ir8a) [33,35]. However, unlike ORs, IRs are expressed by coeloconic olfactory neurons [33], and show responses to a variety of odours including acids, aldehydes, amines and humidity [36]. Between two and three heterodimers in IRs, similar to those observed in ORs, are required to form functional complexes involved in distinct odor perception [33,37]. Antennal IRs are not similar to ionotropic glutamate receptors (iGluRs), but have higher specificity to volatiles than ORs [33]. Characterization of IRs has not been reported among Glossina species to date. Insect chemosensory genes are divergent and evolve through duplication, pseudogenisation and/or deletion incidences [38]. Functional olfactory genes have been reported to be under natural selection in other organisms including humans [39] and Drosophila [40]. Positive selection confers a fitness advantage to a given species relative to the rest of the population and/or increases its genetic diversity [36]. On the other hand, negative (purifying) selection is known to remove deleterious alleles [41].
Understanding molecular factors that underpin the differences observed among species of tsetse, in response to odours is key to success of vector control and management of this vector-borne disease. Availability of the complete genome sequences of five Glossina genomes presents fortuity for comparing molecular properties of proteins that mediate olfaction at species level. Recent characterization of major chemosensory protein gene families (OBPs and CSPs) [24,25] and identification of genes encoding GRs and ORs in G. m. morsitans [31,42] formed a basis to compare genes in different tsetse species. We hypothesize that differences in responses to odours observed among tsetse species are mediated by differences in their chemosensory repertoire. Genes annotated in four newly sequenced tsetse species were compared with their homologs in G. m. morsitans and close dipterans (Ceratitis capitata, D. melanogaster, M. domestica and An. gambiae). The choice of insects used in comparative analysis was informed by their evolutionary grouping under tree of life [43]. Results obtained from this study will form a prototype for undertaking functional studies on tsetse chemosensory proteins to identify their role in tsetse speciation and differential host-selection. Further, the findings will provide insight for improvement of existing vector control tools and development of novel strategies.
Methods
Identification and Annotation of Chemosensory Genes
Genome sequences of G. austeni, G. brevipalpis, G. f. fuscipes and G. pallidipes, their associated gene sets (transcripts, peptides) and gene loci feature files were retrieved from VectorBase database, Release VB-2014-12 [44]. Chemosensory gene sequences from D. melanogaster, An. gambiae, and M. domestica were sourced from FlyBase [45], Uniprot [46], and [47] (through Hugh Robertson of University of Illinois), respectively. The OBP sequences for C. capitata were obtained from GenBank [48] using the published Accession numbers [49]. BLASTp algorithm with an e-value cutoff of ≤ 1.0e-5 was used to identify homologs to chemosensory genes annotated in G. m. morsitans [24,31] and/or in D. melanogaster [16]. Presence of definitive domain(s) in CSPs (OS-D-like), OBPs (PBP/GOBP), ORs (7tm-6), GRs (7tm-7) and IRs (Lig-Chan, ANF, NMDA) was confirmed through Delta Blast searches against the NCBI’s Conserved Domain Database [50]. Where applicable, gene loci that showed incomplete domains and/or had incomplete sequences were manually curated using Artemis genome viewer tool [51]. For curation, flanking regions of the gene loci (in respective scaffolds) were interrogated for Open Reading Frames (ORF) using NCBI’s ORF- Finder [52]. Results of ORF-Finder were used to manually curate the gene models observing rules of intron-exon junction and the subsequent sequences re-blasted against NCBI’s non-redundant database to confirm homology before inclusion into the genes list. Genes with incomplete or no conserved functional domains were considered putative pseudogenes.
The identified Glossina genes were renamed after their closest Drosophila homologs for easier comparison. Abbreviations (Ga–G. austeni, Gbr-G. brevipalpis, Gff-G. f fuscipes, Gmm-G. m. morsitans and Gpd-G. pallidipes) of the species names were used as prefixes to the specific gene name to identify them. The G. m. morsitans OBPs without homologs in D. melanogaster were named as described by Liu and colleagues [24].
Comparative Phylogenetic Analysis of Glossina Chemosensory Genes
Multiple sequence alignments for each class of the chemosensory genes were generated using MUSCLE v3.6 [53] with default settings. Resulting alignmentswere manually edited using standalone Jalview v2 [54] (S2 Dataset), then converted into Phylip format using ClustalX v2.1 [55]. The best substitution model for the alignment was determined using ProtTest server v3.2.1 [56]. Phylogeny inference for the aligned sequences were deduced using a Maximum-likelihood approach as implemented in RAxML v8.2.0 [57] with 1000 bootstrap iterations. Obtained phylogenetic trees were viewed and rendered using Fig Tree viewer v1.4.1. Based on their relationship to other species in the tree of life [43], D. melanogaster and An. gambiae were used as out groups.
Selection Analysis
Codon alignment of Glossina orthologs was done using Prank v 140603 [58] and their corresponding phylogenetic trees constructed using RAxML v8.2.0 [57]. Signatures of natural selection on orthologs were evaluated by calculating ratios of nonsynonymous to synonymous substitutions (dN/dS) in codeml in PAML package v4 [59]. Three site models including M1a (Nearly neutral), M2a (Positive Selection) and M8 (beta & w) were evaluated against their null models to test for selection using log-likelihood ratio (LRT). In case of duplicates, copies of gene loci were separated in order to assess the levels of selection across intra-species paralogs. Corresponding p-value was calculated to test for significance of selection. A p-value < = 0.05 was used to consider a gene to be under positive selection. Similarly, selection analysis was carried out using HyPhy package [60] hosted on Datamonkey web server [41]. In this case, neighbor joining trees were constructed within the package and an appropriate model of nucleotide evolution was determined for each alignment, prior to analysis. Two algorithms; Mixed effects model of Evolution (MEME) [61] and PARRIS [62] were used to identify sites under episodic selection taking recombination events into account. A p-value < = 0.05 was implemented to estimate the rate of false positives (type I error) in which neutrally evolving sites may be erroneously reported to be under selection.
Accession Numbers
Accession numbers of Glossina spp. annotated chemosensory proteins and those used in comparative analysis. Glossina ids were retrieved from Vectrobase alongside those of Anopheles gambiae and Musca domestica. Uniprot accession ids are provided for Drosophila melanogaster while those of Ceratitis capitata are from Genebank.
Odorant binding proteins
Glossina austeni
GAUT003576-PA,GAUT045923-PA,GAUT045912-PA,GAUT045925-PA,GAUT045144-PA,GAUT048147-PA,GAUT018078-PA,GAUT030435-PA,GAUT041055-PA,GAUT039149-PA,GAUT028974-PA,GAUT051622-PA,GAUT040992-PA,GAUT029308-PA,GAUT028968-PA,GAUT026721-PA,GAUT019500-PA,GAUT029664-PA,GAUT019501-PA,GAUT019501-PA,GAUT030010-PA,GAUT030009-PA,GAUT030008-PA,GAUT044447-PA,GAUT043978-PA,GAUT051640-PA,GAUT051645-PA,GAUT051620-PA
Glossina brevipalpis
GBRI030526-PA,GBRI036202-PA,GBRI035551-PA,GBRI035552-PA,GBRI035549-PA,GBRI010734-PA,GBRI012886-PA,GBRI045128-PA,GBRI026688-PA,GBRI016471-PA,GBRI016436-PA,GBRI010929-PA,GBRI040269-PA,GBRI036199-PA,GBRI041963-PA,GBRI013864-PA,GBRI031755-PA,GBRI031753-PA,GBRI031754-PA,GBRI031756-PA,GBRI031703-PA,GBRI031705-PA,GBRI031704-PA,GBRI023685-PA,GBRI009351-PA,GBRI012898-PA,GBRI012882-PA
Glossina fuscipes fuscipes
GFUI025618-PA,GFUI007906-PA,GFUI000760-PA,GFUI000759-PA,GFUI000757-PA,GFUI048313-PA,GFUI004675-PA,GFUI008988-PA,GFUI008564-PA,GFUI009068-PA,GFUI007894-PA,GFUI026749-PA,GFUI040667-PA,GFUI048612-PA,GFUI048613-PA,GFUI048614-PA,GFUI049167-PA,GFUI004156-PA,GFUI004155-PA,GFUI027466-PA,GFUI045274-PA,GFUI035804-PA,GFUI035776-PA
Glossina morsitans morsitans
GMOY008038-PA,GMOY009475-PA,GMOY001927-PA,GMOY005386-PA,GMOY004772-PA,GMOY010761-PA,GMOY001365-PA,GMOY003305-PA,GMOY009271-PA,GMOY006265-PA,GMOY011399-PA,GMOY006479-PA,GMOY006480-PA,GMOY005796-PA,GMOY005084-PA,GMOY010839-PA,GMOY003312-PA,GMOY004392-PA,GMOY007472-PA,GMOY005479-PA,GMOY012018-RB,GMOY012323-PA,GMOY012193-PA,GMOY012195-PA,GMOY012218-PA,GMOY012239-PA,GMOY012253-PA,GMOY012276-PA,GMOY012356-PA,GMOY012357-PA,GMOY005610-PA
Glossina pallidipes
GPAI017685-PA,GPAI006440-PA,GPAI032191-PA,GPAI032193-PA,GPAI032197-PA,GPAI018668-PA,GPAI045033-PA,GPAI017770-PA,GPAI004501-PA,GPAI008752-PA,GPAI008777-PA,GPAI018009-PA,GPAI008860-PA,GPAI009631-PA,GPAI013560-PA,GPAI013557-PA,GPAI013558-PA,GPAI013555-PA,GPAI031702-PA,GPAI031704-PA,GPAI031703-PA,GPAI005408-PA,GPAI041909-PA,GPAI045017-PA,GPAI045022-PA,GPAI045024-PA
D. melanogaster
O02372,Q27377,P54192,Q9V8Y9,Q9V8Y2,Q8MMF9,P54193,Q9VAJ4,Q9VAI6,Q8SY61,Q9V931,Q23970,Q9VR94,P54195,Q8MKJ4,Q9V938,P54194,P54191,P54185,Q8MKK0,A1ZBQ4,Q9W372,Q9VR95,Q9VNL2,Q7JVM1,Q9VAI7,Q7KE33,Q9VWM0,Q7KE32,Q7K084,Q9VR96,D1FYT3,Q4V3N1,Q7K088,Q8MVX6,D1FYH5,Q9VNL1,A1Z8I9,A1Z8E4,A1ZBP9,Q9VHQ9,A1ZBQ3,A1ZBP7,Q9W209,A1Z9Q5,A1Z9Q6,Q7KUQ3,Q86BF9,A1Z9Q2,Q9VDE1,A1Z8E3,A1Z9Q4,Q8T6R8,E2DBU7,E2DCD5,A9QK61
M. domestica
MDOA007276-PA,MDOA004728-PA,MDOA013142-PA,MDOA009850-PA,MDOA014153-PA,MDOA012315-PB,MDOA007587-PA,MDOA000539-PA,MDOA009520-PA,MDOA000889-PA,MDOA012315-PA,MDOA010320-PA,MDOA006902-PA,MDOA005410-PA,MDOA012373-PA,MDOA013526-PA,MDOA013340-PA,MDOA011898-PA,MDOA012293-PA,MDOA005617-PA,MDOA014993-PA,MDOA002810-PA,MDOA009465-PA,MDOA003634-PA,MDOA011594-PA,MDOA004718-PA,MDOA005255-PA,MDOA012958-PA,MDOA012814-PA,MDOA008946-PA,MDOA008603-PA,MDOA008774-PA,MDOA003332-PA,MDOA003832-PA,MDOA001753-PA,MDOA003303-PA,MDOA010146-PA,MDOA011279-PA,MDOA015523-PA,MDOA011147-PA,MDOA011314-PA,MDOA003913-PA,MDOA012317-PA,MDOA005286-PA,MDOA013466-PA,MDOA003735-PA,MDOA012772-PA,MDOA008740-PA,MDOA000714-PA,MDOA002286-PA,MDOA013644-PA,MDOA000734-PA,MDOA002802-PA,MDOA009637-PA,MDOA013698-PA,MDOA004040-PA,MDOA007337-PA,MDOA010340-PA,MDOA001064-PA,MDOA003787-PA,MDOA014452-PA,MDOA010806-PA,MDOA004094-PA,MDOA004456-PA,MDOA008804-PA,MDOA003429-PA,MDOA003694-PA,MDOA004433-PA,MDOA001399-PA,MDOA002259-PA,MDOA009815-PA,MDOA004406-PA,MDOA003400-PA,MDOA004070-PA,MDOA014188-PD,MDOA014188-PG,MDOA004116-PA,MDOA001908-PA
Ceratitis capitata
XM_004521128.1,XM_004524969.1,XM_004524970.1,XM_004524978.1,XM_00425083.1,XM_004254959.1,XM_004517746.1,XM_004518409.1,XM_004523388.1,XM_004523387.1,XM_004529312.1,XM_0045211129.1,XM_004521127.1
Chemosensory receptors
Glossina austeni
GAUT014421-PA, GAUT038415-PA,GAUT027332-PA,GAUT027343-PA,GAUT046063-PA
Glossina brevipalpis
GBRI045129-PA, GBRI011414-PA,GBRI020682-PA,GBRI020713-PA
Glossina fuscipes fuscipes
GFUI014924-PA, GFUI040903-PA,GFUI003186-PA,GFUI003196-PA,GFUI039843-PA
Glossina morsitans morsitans
GMOY010026-PA, GMOY010882-PA,GMOY012164-PA,GMOY010874-PA,GMOY009731-PA
Glossina pallidipes
GPAI012674-PA, GPAI011776-PA,GPAI029774-PA,GPAI029784-PA,GPAI031814-PA
Drosophila melanogaster
Q8MLP9, Q9W0X2, D5A7M1, Q27377
Musca domestica
MDOA006615-PA, MDOA008546-PA, MDOA001428-PA,MDOA000806-PA,MDOA008937-PA
An.gambaie
AGAP008058-PA, AGAP008055-PA, AGAP008059-PA,AGAP008062-PA,AGAP008052-PA,AGAP008051-PA,AGAP008054-PA
Sensory neuron membrane proteins
Glossina austeni
GAUT049266-PA, GAUT008732-PA
Glossina breviplapis
GBRI029848-PA, GBRI009197-PA
Glossina fuscipes fuscipes
GFUI000887-PA, GFUI009502-PA
Glossina morsitans morsitans
GMOY002994-PA, GMOY006180-PA
D. melanogaster
M. domestica
MDOA006272-PB,MDOA006435-PA
An.gambaie
AGAP002451-PA,AGAP005716-PAhttp://www.uniprot.org/uniprot/E1JI63
Gustatory receptors
Glossina austeni
GAUT050702-PA,GAUT041339-PA,GAUT018372-PA,GAUT018371-PA,GAUT037007-PA,GAUT018378-PA,GAUT030746-PA,GAUT018082-PA,GAUT016799-PA,GAUT032734-PA,GAUT042077-PA,GAUT025297-PA,GAUT018813-PA
Glossina brevipalpis
GBRI008315-PA,GBRI004163-PA,GBRI016968-PA,GBRI016977-PA,GBRI039848-PA,GBRI043822-PA,GBRI043906-PA,GBRI014933-PA
Glossina fuscipes fuscipes
GFUI005702-PA,GFUI034303-PA,GFUI041369-PA,GFUI018032-PA,GFUI027606-PA,GFUI026404-PA,GFUI051944-PA,GFUI022205-PA,GFUI025370-PA,GFUI036605-PA,GFUI041074-PA
Glossina morsitans morsitans
GMOY008001-PA,GMOY003231-PA,GMOY004207-PA,GMOY007472-PA,GMOY011615-PA,GMOY006209-PA,GMOY011510-PA,GMOY011903-PA,GMOY005361-PA
Glossina pallidipes
GPAI014620-PA,GPAI045887-PA,GPAI035388-PA,GPAI037163-PA,GPAI019874-PA,GPAI039461-PA,GPAI004494-PA,GPAI040289-PA,GPAI040385-PA,GPAI007341-PA,GPAI024994-PA,GPAI040381-PA,GPAI043562-PA
D. melanogaster
Q9W497,Q9VSH2,P83293,Q9W367,P58950,P58952,P58953,P58954,Q9V4K2,P58962,P83295,Q9VZJ6,P83297,Q9W0M2,Q9VD76,P83296,Q9VTN0,Q9VYZ2,P84181,Q8IRL8,Q9VJF2,Q9W2B2,P58955,Q8INZ2,Q8IN58,Q8INM9,Q9VEU0,Q9VB26,Q8IMN5,Q8IMN6,Q9VB30,Q0E9G8,H0RNL7,D3PK93,E1JJC5,Q8MLS6,Q7KV53,Q8IN22,M9PAZ2,M9PGM7,A1Z881,M9PBP0,Q9W1V0,B4PH96,B4PH99,B4PHA1,B4PX40,B4PHA0,B4PH98,B4PZC5,B4PH97,B6ZDW0
M. domestica
MDOA000140-PA,MDOA000302-PA,MDOA000316-PA,MDOA000580-PA,MDOA000804-PA,MDOA000952-PA,MDOA001249-PA,MDOA002394-PA,MDOA002976-PA,MDOA002995-PA,MDOA003120-PA,MDOA003761-PA,MDOA003814-PA,MDOA004047-PA,MDOA004843-PA,MDOA004883-PA,MDOA005532-PA,MDOA006053-PA,MDOA006078-PA,MDOA006341-PA,MDOA006396-PA,MDOA006542-PA,MDOA007003-PA,MDOA007173-PA,MDOA007349-PA,MDOA007502-PA,MDOA008622-PA,MDOA008716-PA,MDOA008860-PA,MDOA008965-PA,MDOA009078-PA,MDOA009179-PA,MDOA009364-PA,MDOA009614-PA,MDOA009686-PA,MDOA009754-PA,MDOA009880-PA,MDOA011018-PA,MDOA011119-PA,MDOA011281-PA,MDOA012391-PA,MDOA012949-PA,MDOA013669-PA,MDOA014425-PA,MDOA014604-PA,MDOA015305-PA,MDOA002641-PA,MDOA002364-PA,MDOA014947-PA,MDOA015347-PA
An.gambiae
AGAP004716-PA,AGAP004727-PA,AGAP005047-PA,AGAP005495-PA,AGAP005514-PA,AGAP006143-RD,AGAP006399-PA,AGAP006450-PA,AGAP006713-RA,AGAP006716-PA,AGAP006717-PA,AGAP006874-PA,AGAP006875-PA,AGAP006876-PA,AGAP006877-RB,AGAP006917-PA,AGAP001915-PA,AGAP002633-PA,AGAP002635-RA,AGAP001125-PA,AGAP003098-PA,AGAP003256-PA,AGAP003255-PA,AGAP003254-PA,AGAP003253-PA,AGAP003260-PA,AGAP003259-RA,AGAP004114-PA,AGAP001171-PA,AGAP001172-PA,AGAP001173-PA,AGAP001170-PA,AGAP001169-RA,AGAP004313-PA,AGAP002275-PA,AGAP011915-PA,AGAP007757-PA,AGAP009256-RA,AGAP009802-PA,AGAP009803-PA,AGAP009804-PA,AGAP009805-RA,AGAP009853-PA,AGAP009854-PA,AGAP009856-PA,AGAP009857-PA,AGAP009858-PA,AGAP009999-PA,AGAP009855-PA,AGAP007756-PA,AGAP012713-PA,AGAP001114-PA,AGAP001117-RA,AGAP001119-PA,AGAP001122-PA,AGAP001123-PA,AGAP001121-PA,AGAP001120-PA,AGAP001115-PA,AGAP001137-PA,AGAP010195-PA,
Odorant receptors
Glossina austeni
GAUT014395-PA,GAUT050371-PA,GAUT004311-PA,GAUT045920-PA,GAUT028888-PA,GAUT021583-PA,GAUT000836-PA,GAUT050213-PA,GAUT050213-PA,GAUT022268-PA,GAUT044021-PA,GAUT022034-PA,GAUT028238-PA,GAUT011101-PA,GAUT016620-PA,GAUT005608-PA,GAUT042364-PA,GAUT042360-PA,GAUT018044-PA,GAUT003629-PA,GAUT038273-PA,GAUT018383-PA,GAUT032244-PA,GAUT021320-PA,GAUT051820-PA,GAUT021321-PA,GAUT035779-PA,GAUT050214-PA,GAUT003281-PA,GAUT005460-PA,GAUT006649-PA,GAUT040462-PA,GAUT036655-PA,GAUT005363-PA,GAUT034813-PA
Glossina brevipalpis
GBRI045111-PA,GBRI018062-PA,GBRI036522-PA,GBRI035583-PA,GBRI036342-PA,GBRI002464-PA,GBRI016989-PA,GBRI044639-PA,GBRI034666-PA,GBRI009897-PA,GBRI026647-PA,GBRI008361-PA,GBRI028428-PA,GBRI026891-PA,GBRI015995-PA,GBRI011898-PA,GBRI011904-PA,GBRI011358-PA,GBRI031244-PA,GBRI031534-PA,GBRI002179-PA,GBRI026158-PA,GBRI017432-PA,GBRI017598-PA,GBRI040021-PA,GBRI044640-PA,GBRI018811-PA,GBRI027004-PA,GBRI041284-PA,GBRI030235-PA,GBRI005734-PA,GBRI013056-PA,GBRI012762-PA,GBRI030714-PA
Glossina fuscipes fuscipes
GFUI014938-PA,GFUI043297-PA,GFUI032492-PA,GFUI028755-PA,GFUI007794-PA,GFUI003104-PA,GFUI003105-PA,GFUI003499-PA,GFUI028213-PA,GFUI008162-PA,GFUI032116-PA,GFUI005658-PA,GFUI037305-PA,GFUI034469-PA,GFUI045476-PA,GFUI009257-PA,GFUI038138-PA,GFUI038147-PA,GFUI042981-PA,GFUI027054-PA,GFUI051694-PA,GFUI007388-PA,GFUI043789-PA,GFUI036188-PA,GFUI022534-PA,GFUI022472-PA,GFUI003500-PA,GFUI053522-PA,GFUI022126-PA,GFUI047908-PA,GFUI049134-PA,GFUI037003-PA,GFUI024278-PA,GFUI012941-PA,GFUI035140-PA
Glossina morsitans morsitans
GMOY008038-PA,GMOY009475-PA,GMOY001927-PA,GMOY005386-PA,GMOY004772-PA,GMOY010761-PA,GMOY001365-PA,GMOY003305-PA,GMOY009271-PA,GMOY006265-PA,GMOY011399-PA,GMOY006479-PA,GMOY006480-PA,GMOY005796-PA,GMOY005084-PA,GMOY010839-PA,GMOY003312-PA,GMOY004392-PA,GMOY007472-PA,GMOY005479-PA,GMOY012018-RB,GMOY012323-PA,GMOY012193-PA,GMOY012195-PA,GMOY012218-PA,GMOY012239-PA,GMOY012253-PA,GMOY012276-PA,GMOY012356-PA,GMOY012357-PA,GMOY005610-PA
Glossina pallidipes
GPAI034871-PA,GPAI027642-PA,GPAI015219-PA,GPAI004010-PA,GPAI034198-PA,GPAI039623-PA,GPAI039631-PA,GPAI031316-PA,GPAI031326-PA,GPAI029610-PA,GPAI041951-PA,GPAI026906-PA,GPAI014680-PA,GPAI009882-PA,GPAI009200-PA,GPAI039539-PA,GPAI001497-PA,GPAI004557-PA,GPAI045424-PA,GPAI045426-PA,GPAI039747-PA,GPAI017649-PA,GPAI041241-PA,GPAI037164-PA,GPAI033169-PA,GPAI012943-PA,GPAI012945-PA,GPAI046202-PA,GPAI002749-PA,GPAI042230-PA,GPAI031315-PA,GPAI024118-PA,GPAI001626-PA,GPAI040919-PA,GPAI002024-PA,GPAI004056-PA,GPAI027550-PA,GPAI009882-PA,GPAI035133-PA
D. melanogaster
Q9VPT1,Q9VZL7,P81909,P81910,O46077,Q9V3Q2,P81915,P81917,P81921,Q9VNB5,Q9I816,Q9VXL0,Q9VYZ1,Q9W5G6,P81912,P81911,P81913,Q9VLE5,P81916,P81914,Q9V9I2,Q9V589,P81919,P81922,P81918,Q9V3N2,Q9V9I4,Q9V6A9,Q9V6H2,Q9V568,Q9V8Y7,Q9W1P8,P81923,P82982,Q9VT90,Q9VT92,Q9VT08,Q9VT20,Q9VVF3,Q9W3I5,Q9VHQ7,Q9VHE6,Q9VHS4,Q9VFN2,P82986,Q9VAZ3,Q9W2U9,Q8IRZ5,Q9VZW8,Q9VNB3,Q9VHQ6,Q9VCS9,Q9VCS8,E1JIA4,M9NFD3,E2E626,E2E5L1,E2E5L0,E2E510,B4NY14
M. domestica
MDOA000926-PA,MDOA000137-PA,MDOA000385-PA,MDOA000464-PA,MDOA001095-PA,MDOA001330-PA,MDOA001508-PA,MDOA001711-PA,MDOA001967-PA,MDOA002017-PA,MDOA002113-PA,MDOA002222-PA,MDOA002540-PA,MDOA002654-PA,MDOA002736-PA,MDOA002822-PA,MDOA002922-PA,MDOA003091-PA,MDOA003495-PA,MDOA003512-PA,MDOA003540-PA,MDOA003948-PA,MDOA004405-PA,MDOA004757-PA,MDOA004936-PA,MDOA004949-PA,MDOA005313-PA,MDOA005821-PA,MDOA005976-PA,MDOA006361-PA,MDOA006570-PA,MDOA006773-PA,MDOA006970-PA,MDOA007213-PA,MDOA007232-PA,MDOA007549-PA,MDOA007555-PA,MDOA007822-PA,MDOA007881-PA,MDOA008272-PA,MDOA008672-PA,MDOA008787-PA,MDOA009136-PA,MDOA009183-PA,MDOA009203-PA,MDOA009938-PA,MDOA010127-PA,MDOA010179-PA,MDOA010267-PA,MDOA010394-PA,MDOA010396-PA,MDOA010576-PA,MDOA011183-PA,MDOA011663-PA,MDOA011814-PA,MDOA011954-PA,MDOA012084-PA,MDOA012436-PA,MDOA012443-PA,MDOA012722-PA,MDOA012767-PA,MDOA012864-PA,MDOA012897-PA,MDOA012955-PA,MDOA013188-PA,MDOA013204-PA,MDOA013213-PA,MDOA013229-PA,MDOA013697-PA,MDOA014353-PA,MDOA014482-PA,MDOA014540-PA,MDOA014647-PA,MDOA014744-PA,MDOA014843-PA,MDOA014864-PA,MDOA014904-PA,MDOA015346-PA,MDOA015469-PA,MDOA015496-PA,MDOA015498-PA,MDOA005448-PA,MDOA007080-PA,MDOA007097-PA,MDOA010057-PA,MDOA013717-PA
Ionotropic & ionotropic glutatmate receptors
Glossina austeni
GAUT036857-PA,GAUT010844-PA,GAUT032862-PA,GAUT032862-PA,GAUT018821-PA,GAUT036856-PA,GAUT051652-PA,GAUT029664-PA,GAUT011688-PA,GAUT019628-PA,GAUT028361-PA,GAUT017831-PA,GAUT035430-PA,GAUT051179-PA,GAUT013397-PA,GAUT013397-PA,GAUT003875-PA,GAUT051343-PA,GAUT037856-PA,GAUT038749-PA,GAUT002274-PA,GAUT026102-PA,GAUT023024-PA,GAUT005991-PA,GAUT026111-PA,GAUT031582-PA,GAUT008471-PA,GAUT032864-PA
Glossina brevipalpis
GBRI004368-PA,GBRI037007-PA,GBRI006509-PA,GBRI004366-PA,GBRI004366-PA,GBRI013356-PA,GBRI037007-PA,GBRI012928-PA,GBRI001929-PA,GBRI023337-PA,GBRI000712-PA,GBRI039411-PA,GBRI033584-PA,GBRI012051-PA,GBRI033291-PA,GBRI016181-PA,GBRI016181-PA,GBRI012020-PA,GBRI018928-PA,GBRI009997-PA,GBRI002787-PA,GBRI010267-PA,GBRI006799-PA,GBRI006802-PA,GBRI006799-PA,GBRI029815-PA,GBRI013857-PA,GBRI040612-PA
Glossina fuscipes fuscipes
GFUI019198-PA,GFUI016186-PA,GFUI018591-PA,GFUI019200-PA,GFUI019200-PA,GFUI031610-PA,GFUI041857-PA,GFUI035802-PA,GFUI017944-PA,GFUI008852-PA,GFUI031962-PA,GFUI025996-PA,GFUI041337-PA,GFUI028023-PA,GFUI019558-PA,GFUI029180-PA,GFUI029178-PA,GFUI043801-PA,GFUI005590-PA,GFUI004860-PA,GFUI020203-PA,GFUI000065-PA,GFUI009601-PA,GFUI000460-PA,GFUI000063-PA,GFUI045184-PA,GFUI050910-PA
Glossina morsitans morsitans
GMOY004222,GMOY012186,GMOY006490,GMOY007988,GMOY001514,GMOY012037,GMOY006751,GMOY001810,GMOY004959,GMOY005753,GMOY007825,GMOY000804,GMOY012048,GMOY012127,GMOY008789,GMOY008540,GMOY012136,GMOY006890,GMOY009209,GMOY002585,GMOY004997,GMOY009750,GMOY004578
Glossina pallidipes
GPAI011564-PA,GPAI006854-PA,GPAI010111-PA,GPAI011561-PA,GPAI011561-PA,GPAI019869-PA,GPAI006854-PA,GPAI045043-PA,GPAI016226-PA,GPAI011331-PA,GPAI007758-PA,GPAI004624-PA,GPAI022505-PA,GPAI032358-PA,GPAI017485-PA,GPAI036018-PA,GPAI036018-PA,GPAI025294-PA,GPAI027894-PA,GPAI044391-PA,GPAI022870-PA,GPAI042411-PA,GPAI006139-PA,GPAI006142-PA,GPAI029067-PA,GPAI010422-PA,GPAI006139-PA,GPAI006944-PA
D. melanogaster
Q9W365,Q9W3P2,A1Z882,E9NA96,A1Z8N9,B7Z069,Q9VCM4,A1ZBM8,M9PCT4,A1ZBM7,Q2MGM0,B7YZQ4,B7Z0P2,A1Z8P2,Q9VCM0,Q9W191,Q9VDH6,Q9VYN4,Q9VVU7,Q9V9T2,Q8IN10,A1ZA17,Q8IN09,B7Z0Y1,A8JNV9,Q9W155,Q9VTH3,B7Z0X5,Q9VDN3,A8JUR3,A1ZBM9,B7Z0X6,Q9VTT6,A1Z6D6,Q9VVL1,Q8IMY8,A1ZAY9,Q8IN08,X2JCB2,A1ZA14,Q9W3P0,Q9W3P4,Q9VRL4,A1ZA16,A1ZBG7,Q9VPI2,Q9V9N1,Q9VHL4,Q8IPB8,Q9VVL2,Q9VCM1,Q9VRI8,A1ZA15,A1Z9Y5,M9PGG3,Q9VFV0,Q8IQE2,B7YZQ6,Q9VIA5,Q9VT09,E9NA95,E9NA98,E9NA99,E7E521
M. domestica
MDOA007071-PA,MDOA010874-PA,MDOA004663-PA,MDOA002700-PA,MDOA000608-PA,MDOA010444-PA,MDOA014373-PA,MDOA014396-PA,MDOA009431-PA,MDOA011131-PA,MDOA003227-PA,MDOA000640-PA,MDOA003336-PA,MDOA015494-PA,MDOA004232-PA,MDOA015201-PA,MDOA010345-PA,MDOA012059-PA,MDOA009027-PA,MDOA008354-PA,MDOA001109-PA,MDOA006236-PA,MDOA009699-PA,MDOA007005-PA,MDOA002252-PA,MDOA001895-PA,MDOA012195-PA,MDOA013121-PA,MDOA007819-PA,MDOA012117-PA,MDOA008579-PA,MDOA010627-PA,MDOA002571-PA,MDOA012119-PA,MDOA005930-PA,MDOA008763-PA,MDOA003307-PA,MDOA011360-PA,MDOA009489-PA,MDOA000887-PA,MDOA005477-PA,MDOA003828-PA,MDOA007088-PA,MDOA011259-PA,MDOA015382-PA,MDOA009859-PA,MDOA008038-PA,MDOA008826-PA,MDOA008185-PA,MDOA014666-PA,MDOA011062-PA,MDOA000255-PA,MDOA010951-PA,MDOA012239-PA,MDOA013187-PA,MDOA005137-PA,MDOA007608-PA,MDOA010321-PA,MDOA000493-PA,MDOA003357-PA,MDOA008271-PA,MDOA010652-PA,MDOA004469-PA,MDOA012545-PA,MDOA007828-PA,MDOA005225-PA,MDOA008360-PA,MDOA012330-PA,MDOA011161-PA,MDOA014865-PA,MDOA014404-PA,MDOA008618-PA,MDOA005214-PA,MDOA002045-PA,MDOA006466-PA,MDOA005355-PA,MDOA007990-PA,MDOA012546-PA,MDOA000971-PA,MDOA005099-PA,MDOA005808-PA,MDOA003734-PA,MDOA009668-PA,MDOA006290-PA,MDOA012758-PA,MDOA006255-PA,MDOA002539-PA,MDOA002539-PB,MDOA014635-PA,MDOA004067-PA,MDOA003912-PA,MDOA005542-PA,MDOA004606-PA,MDOA013782-PA,MDOA011463-PA,MDOA011711-PA,MDOA004225-PA,MDOA013355-PA,MDOA000458-PA,MDOA003685-PA,MDOA007697-PA,MDOA001982-PA,MDOA008624-PA,MDOA001178-PA,MDOA009650-PA,MDOA011682-PA,MDOA002092-PA,MDOA002232-PA,MDOA001533-PA,MDOA013906-PA,MDOA007071-PA,
An.gambiae
AGAP004923-PA,AGAP004969-PA,AGAP005466-RA,AGAP005527-PA,AGAP005677-PA,AGAP005678-PA,AGAP005679-PA,AGAP006407-PA,AGAP006440-PA,AGAP006691-PA,AGAP007498-PA,AGAP001811-PA,AGAP001812-PA,AGAP013085-PA,AGAP013436-PA,AGAP013242-PA,AGAP013363-PA,AGAP013285-PA,AGAP002763-PA,AGAP013416-PA,AGAP002797-RB,AGAP002904-PA,AGAP013473-PA,AGAP003531-PA,AGAP012951-PA,AGAP013425-PA,AGAP004021-PA,AGAP001478-PA,AGAP004432-PA,AGAP012969-PA,AGAP004475-PA,AGAP013520-PA,AGAP013172-PA,AGAP013409-PA,AGAP000714-PA,AGAP013154-PA,AGAP000803-PA,AGAP000801-RB,AGAP000798-PA,AGAP000140-PA,AGAP000256-PA,AGAP000293-PA,AGAP010411-PA,AGAP011943-PA,AGAP011968-PA,AGAP007951-PA,AGAP008511-PA,AGAP008759-PA,AGAP009014-PA,AGAP010272-PA,AGAP012429-PA,AGAP012447-PA
Results
Annotation and Genomic Arrangement of Glossina Chemosensory Genes
The numbers of chemosensory gene families identified and annotated in this study are summarized in Table 1 and their metadata in S1 Dataset.
Table 1. Summary of putative chemosensory genes annotated in Glossina species.
G. austeni, G. brevipalpis, G. f. fuscipes, G. m. morsitans and G. pallidipes against those of selected dipterans.
| Species | CSPs† | GRs | IRs/IGluRs | OBPs | ORs | SNMPs | Reference(s) |
|---|---|---|---|---|---|---|---|
| G. austeni | 5 | 14 | 28 | 29 | 40 (5) | 2 | This study |
| G. brevipalpis | 4 | 11 | 28 | 28 | 42 (5) | 2 | This study |
| G. f. fuscipes | 5 | 14 | 31 (2) | 30 (3) | 42 (6) | 2 | This study |
| G. pallidipes | 5 | 14 | 30 (1) | 30 (2) | 42 (3) | 2 | This study |
| G. m. morsitans | 5 | 14 | 30 (2) | 30 (3) | 46 (3) | 2 | 24,25,31 |
| An. gambiae | 8 | 76 | 48 | 82 | 79 | 2 | 34,68 |
| D. melanogaster | 4 | 60 (13) | 66(9) | 52 | 62 (2) | 2 | 22,64 |
| M. domestica | 5 | 103 | 110 | >87 | 86 | 2 | 47 |
† CSPs—chemosensory specific proteins, GRs—gustatory receptors, IRs/IGluRs- ionotropic receptors/ionotropic glutamate receptors, OBPs- odorant binding proteins, ORs- odorant receptors, SNMPs- sensory neuron membrane proteins.
Number of genes in parentheses represents putative pseudogenes i.e. either incomplete genes or genes missing functional domain.
Overall, the results presented in Table 1 show that the five tsetse species have fewer chemosensory genes compared to the other dipterans used in this study. Majority of the chemosensory proteins identified in this study (S1 Dataset) contained their respective definitive domains (7tm_7 superfamily in GRs, 7tm_6 in ORs, PBP, ANF- receptor and Lig_Chan in IRs, PBP-GOBP in OBPs, OS-D in CSPs and CD36 in SNMPs). However, a few genes were missing the domain signatures. These included Obp73a in all tsetse species, Obp56h in G. austeni, Obp20, Or85e and Gr33a in G. brevipalpis, SNMP1 and Or56a in G. f. fuscipes, and Or67d3 in G. pallidipes. The GRs and ORs in G. austeni, G. brevipalpis, G. f. fuscipes, and G. pallidipes were 269–480 aa and 295–508 aa long, respectively. Similarly, CSPs and OBPs were 108–178 aa and 108–257 aa long, respectively. The SNMPs and IRs had longer sequences than other gene families, being 384–540 aa and 407–1070 aa long, respectively.
Our analysis revealed a general genome-wide dispersion of the chemosensory genes in all the tsetse species analyzed (S1 Dataset). Fourteen loci were duplicated. The loci included one CSP (Ejbp3; that have two copies namely Ejbp3A and Ejbp3B), three GRs (Gr21a; with three copies namely Gr2a1, Gr2a2 and Gr21a3, Gr28b; two to three copies per genome Gr28bB, Gr28bC, and/or Gr28bD and Gr59f; with two copies: Gr59f1-2). Two OBPs (Obp83a which has four copies; Obp83a1-4, Obp56e; with two copies Obp56e1 and Obp56e2, and eight ORs (Or7a with three copies: Or7a1-3, Or45a with three copies: Or45a1-3, Or67d with five copies: Or67d1-5 and Or56a with two copies: Or56a1 and Or56a2, Or43a, Or46a, Or63a, and Or67c with two copies each). All four copies of Obp83a homolog were in tandem in all the five tsetse genomes, and represented evidence of structural gene variation and rearrangement (S1 Fig, panel A). One of the Obp83a copies was located on the reverse strand. In contrast, duplicated ORs including three copies of Or45a, two copies of Or7a and four to six copies of Or67d homologs were located in different scaffolds (S1 Dataset).
Comparative Analyses of Glossina Chemosensory Gene Families
Sequence alignment of Obp56i and Obp19 from selected dipterans showed variation of amino acids between the third and fourth conserved cysteine residues (labeled C3 and C4 in S2 Fig). Glossina Obp56i and Obp19 showed sequence deletions between C3 and C4. In contrast, their homologs from D. melanogaster and M. domestica showed amino acid conservation around the same regions.
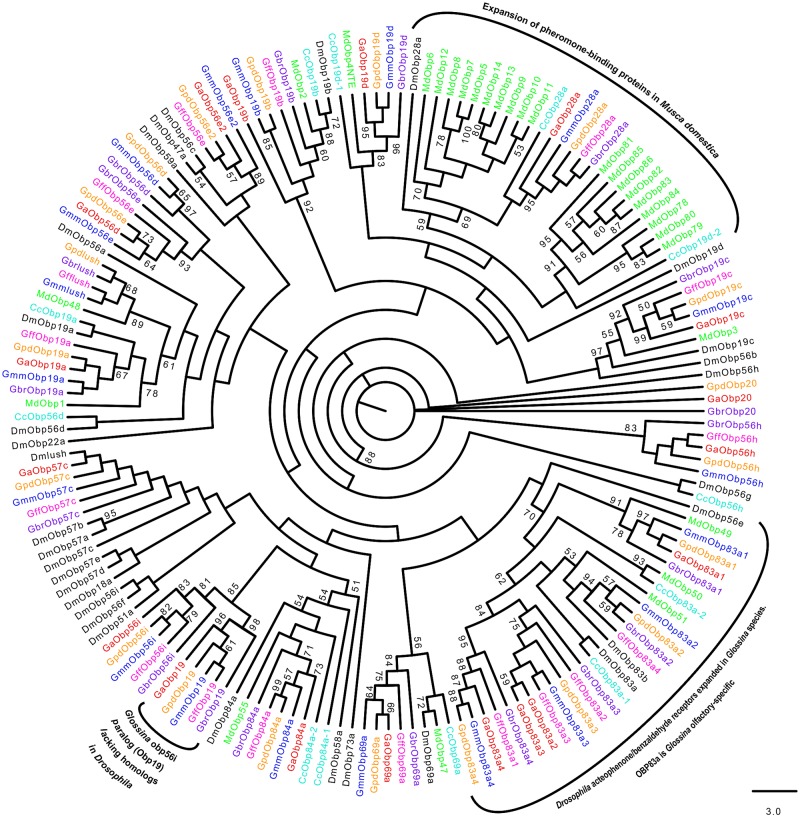
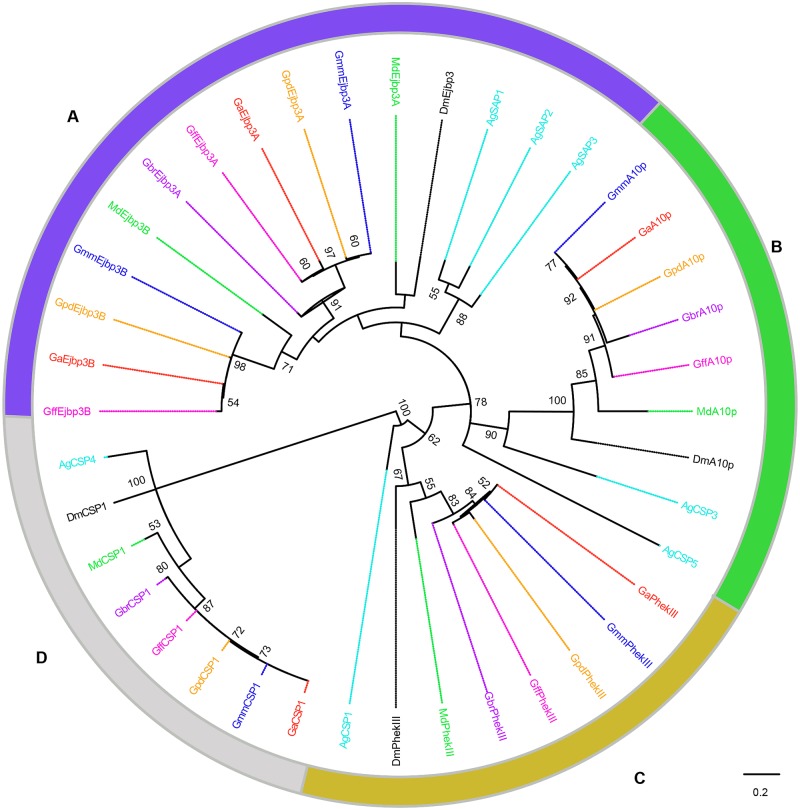
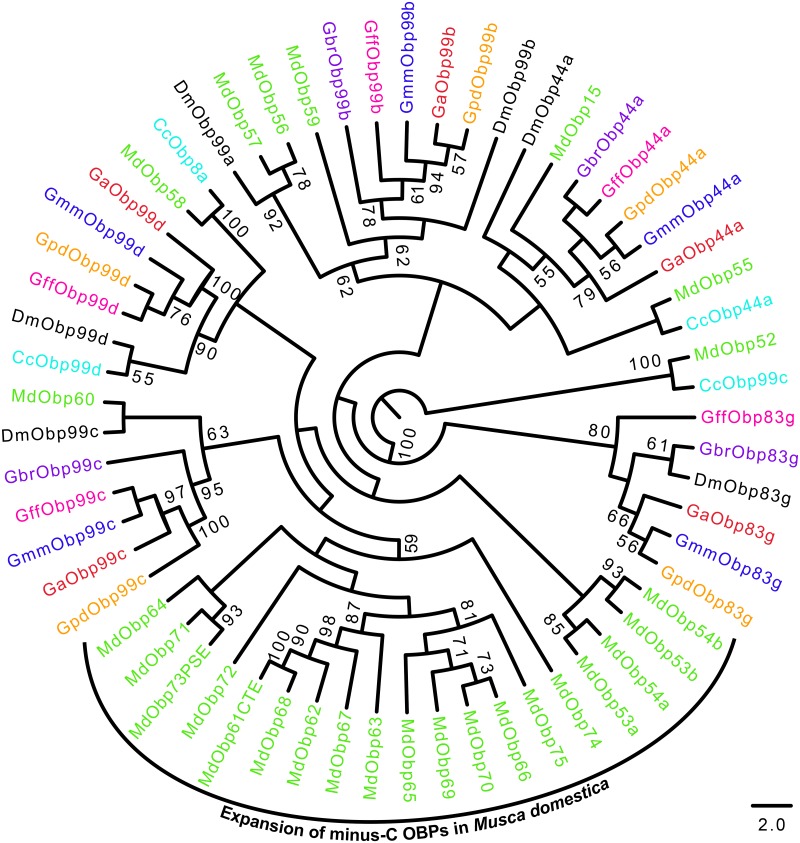
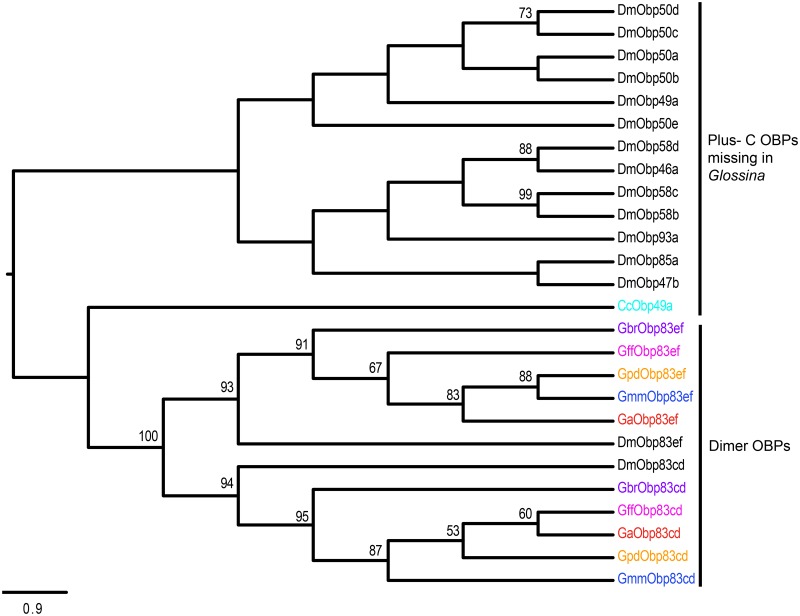
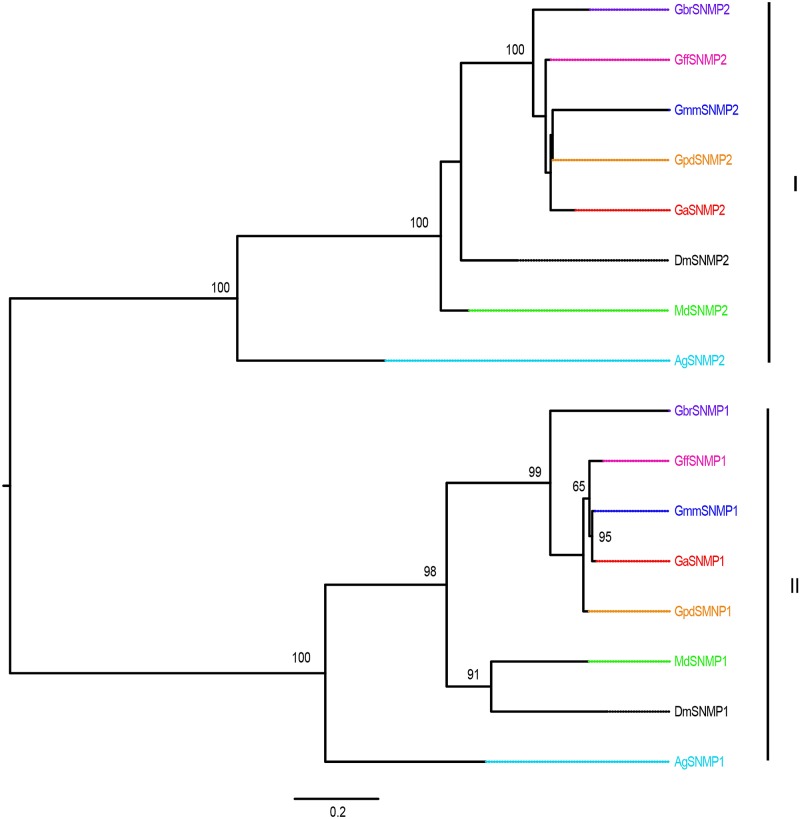
Multiple alignments of the OBPs and CSPs revealed high conservation of conserved cysteine residues (for formation of disulphide bridges) and hydrophobic amino acid residues (for formation ligand-binding sites) (See S2 Dataset). Phylogenetic relationships predicted among the OBPs and CSPs identified in Glossina species against those in C. capitata, D. melanogaster and M. domestica are shown in Figs 1–4. About 68.9% (n = 29) of the Glossina OBPs were grouped into the Classic subfamily (Fig 1) (with six conserved cysteines) while six OBPs in each of the tsetse species were identified into the Minus-C subfamily (with less than the conventional six cysteines) (Fig 2). We did not identify any Plus-C /Atypical subfamily members in any of the Glossina species studied (Fig 3). Expansions of Obp56e (two copies) and Obp83a (four copies) classic subfamily were observed in all tsetse species (Fig 1), while M. domestica and C. capitata had three and two copies of gene encoding Obp83a respectively. The Obp28a, and Obp19d, were among the list of OBP genes highly expanded in M. domestica (Fig 1). There were four distinct clades (A–D), of the CSPs (Fig 4). All tsetse species except G. brevipalpis had two copies of ejaculatory–bulb specific protein 3 (Ejbp3). G. brevipalpis on the other hand, had a single copy of Ejbp3 similar to M. domestica (Clade A, Fig 4). Further, orthologs of SNMP1 and SNMP2 reported in D. melanogaster, Ae. aegypti and various Lepidoptera species [63] were present in all tsetse species. Two SNMP sub-clades with one-to-one orthology across all insects were identified (Fig 5).
Fig 1. Phylogeny of classic odorant binding proteins.
Insect classic OBPs are characterized by six conserved cysteine residues. Different symbols depict OBPs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Ceratitis capitata (sky blue*) and Musca domestica (lime green*). The symbol * represents the name of the specific OBP. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship was inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations.
Fig 4. Phylogeny of chemosensory proteins.
Clade A shows duplication of ejaculatory bulb protein 3 (Ejbp3 in four tsetse species). Clade B shows expansion of A10p—like homologs in An. gambiae while clades C and D depicts conservation of Pherokine-3 and CSP1 across the species compared, respectively. Different symbols depict CSPs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Anopheles gambiae (sky blue*) and Musca domestica (lime green*). The symbol * represents the name of the specific CSP. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship was inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations.
Fig 2. Phylogeny of Minus-C odorant binding proteins.
The minus-C OBPs have less than six conserved cysteine residues (Missing C1or C2 and/or C5). Different symbols depict OBPs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (Dm*), Ceratitis capitata (sky blue*) and Musca domestica (lime green*). The symbol * represents the name of the specific OBP. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship was inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations.
Fig 3. Phylogeny of Plus-C and Classic-Dimer odorant binding proteins.
The Plus-C OBPs are characterized by having more than six cysteines and a conserved proline residue. The Classic-dimers have two conserved domains of classic sub-family. Different symbols depict OBPs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Ceratitis capitata (sky blue*) and Musca domestica (lime green*). The symbol * represents the name of the specific OBP. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship was inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations.
Fig 5. Phylogeny of sensory neuron membrane proteins.
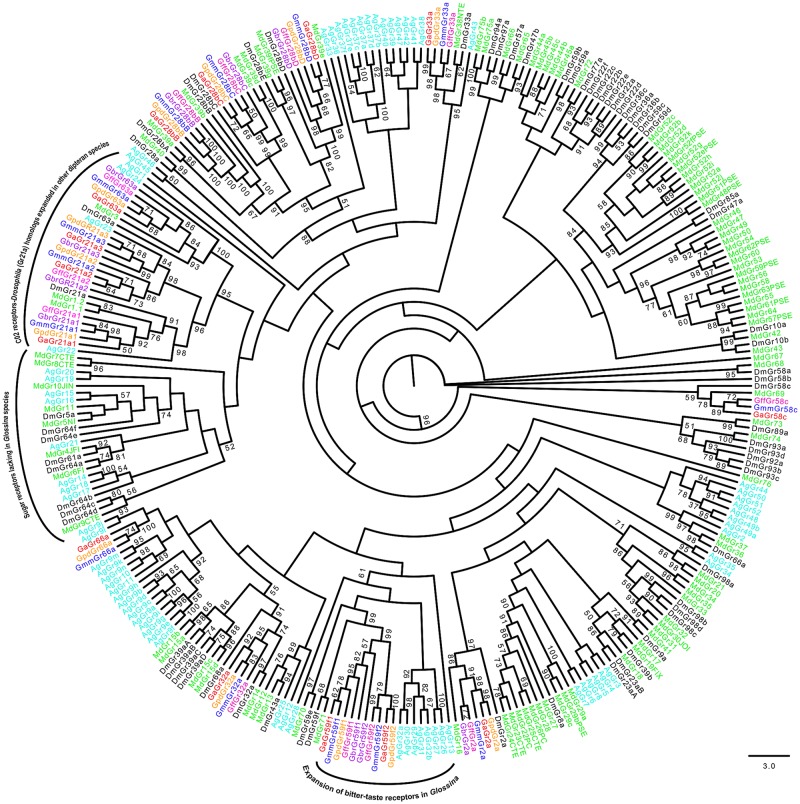
Both clades I and II show one to one orthology of the specific SNMP from different insect species. Different symbols depict SNMPs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Anopheles gambiae (sky blue*) and Musca domestica (lime green*). The symbol * represents the name of the specific SNMP. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship was inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations. Phylogenetic relationships of GRs identified in Glossina genes and their homologs in An. gambiae, D. melanogaster and M. domestica are shown in Fig 6. In all the tsetse species, there was expansion of Gr21a, associated with CO2 detection in fruit fly and mosquitoes [64,65]. Similarly, expansion of CO2 receptors was noted in An. gambiae which has expanded Gr63a, a protein co-expressed with Gr21a and involved in CO2 detection [65]. No homologs to sugar receptors in D. melanogaster [66] were identified in any of the five Glossina species (Fig 6). Similarly, D. melanogaster Gr43a, implicated in internal fructose sensing [67] was absent in all tsetse species.
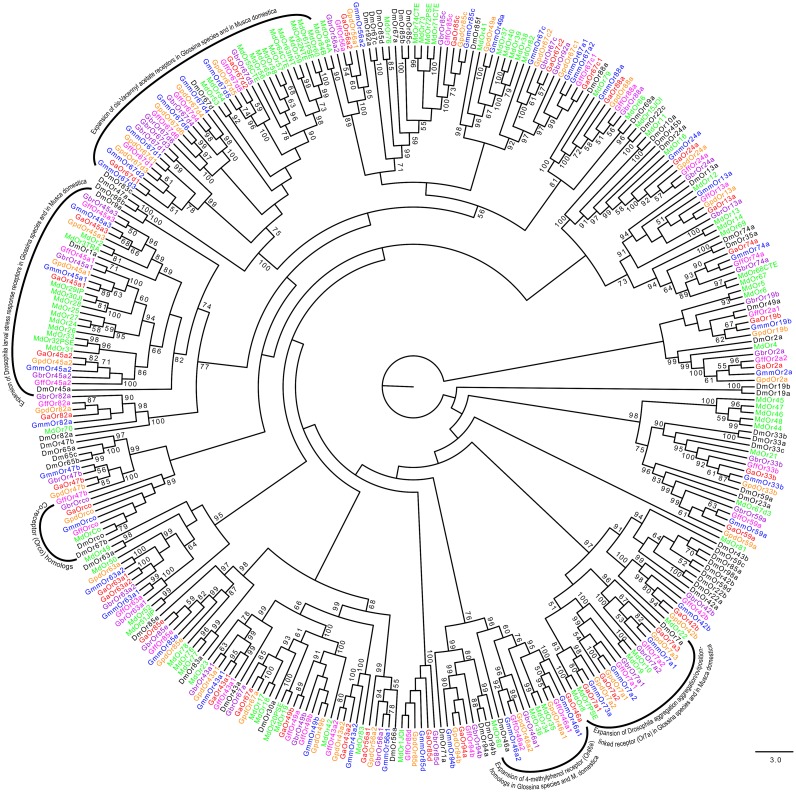
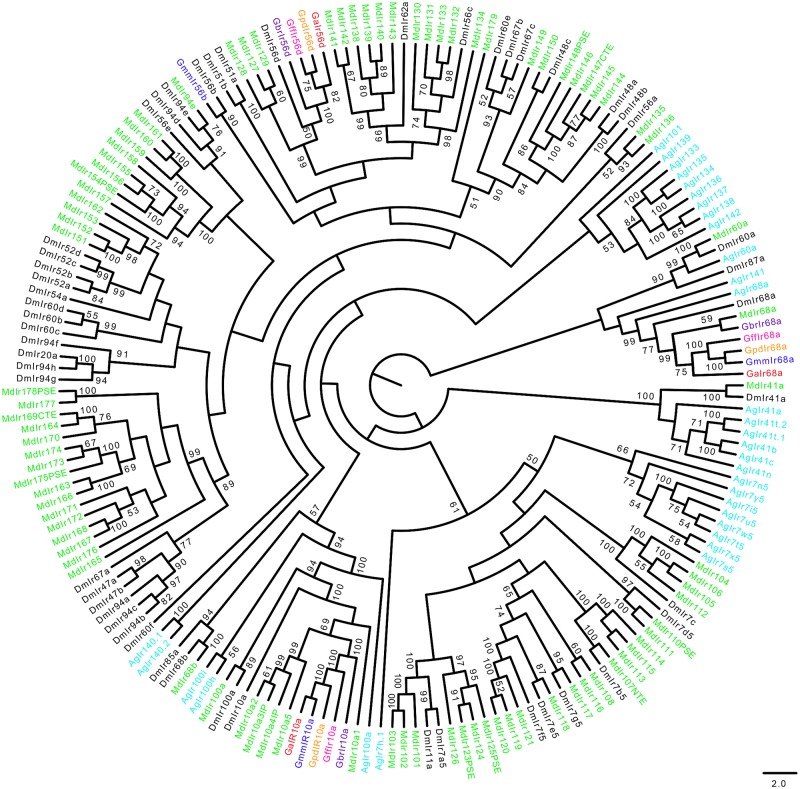
A single copy of the co-receptor (Orco) ortholog was identified in all tsetse species (Fig 7). There were 75–85% amino acid identity between Orco in all tsetse and those of its homologs in M. domestica, D. melanogaster and An. gambiae. Phylogenetic analysis was resolved into distinct clades among Glossina species, D. melanogaster, M. domestica and An. gambiae ORs [68] (Fig 7). Three paralogs of Or45a which is responds to stress in Drosophila larvae [69], were identified in all tsetse species (Fig 7). Expansion of Or7a and Or46a was also noted in Glossina spp. and M. domestica (Fig 7). A clade containing Drosophila cis- Vacennyl acetate receptor; Or67d homologs shows its expansion in tsetse flies. Four Glossina species (G. austeni, G. brevipalpis, G. f. fuscipes and G. pallidipes) had a total of five Or67d paralogs compared to six copies reported in G. m. morsitans [31]. Other genes that showed expansion in Glossina species include Or67c and Or43a (Fig 7).
Fig 7. Phylogeny of odorant receptors.
Expansion of cis-Vaccenyl acetate receptor (Or67d), 4-Methylphenol receptor (Or46a) and aggregation-linked receptor (Or7a) is observed in Glossina species and Musca domestica relative to Drosophila. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship was inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations. Different symbols and colors were used to depict ORs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*) and Musca domestica (lime green*). The symbol * represents the name of the specific OR.
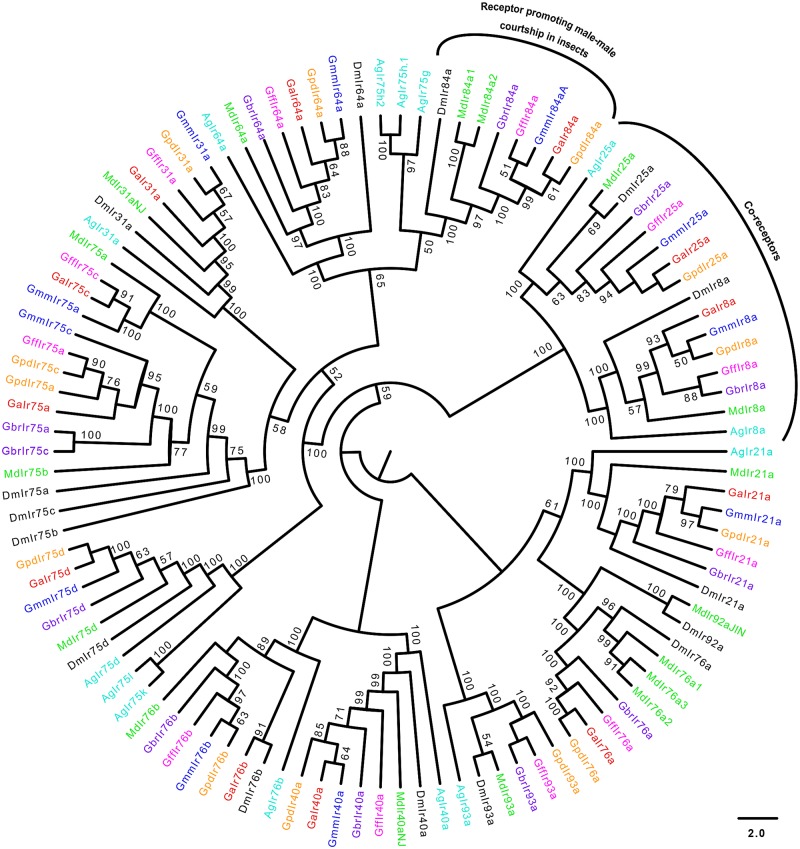
Similar numbers of IRs/iGluRs were identified in all tsetse species (Table 1). The homolog of a Drosophila Ir93a was not identified in G. austeni. Phylogeny reconstruction of IRs and iGluRs yielded highly supported clades (Figs 8–10). A total of 13 Glossina IR homologs clustered with their antennal Drosophila orthologs (Ir40a, Ir25a, Ir8aa, Ir93a, Ir21a, Ir76a, Ir76b, Ir31a, Ir75c, Ir75a, Ir75d, Ir64a and Ir84a) (Fig 8). Further, Drosophila-specific Ir84a and was found to have homologs in all Glossina species studied.
Fig 8. Phylogeny of antennal ionotropic receptors.
Antennal IRs are primarily expressed at the antenna of the insect. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations. Different symbols and colors were used to depict IRs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Musca domestica (lime green*) and Anopheles gambiae (sky blue*). The symbol * represents the name of the specific IR.
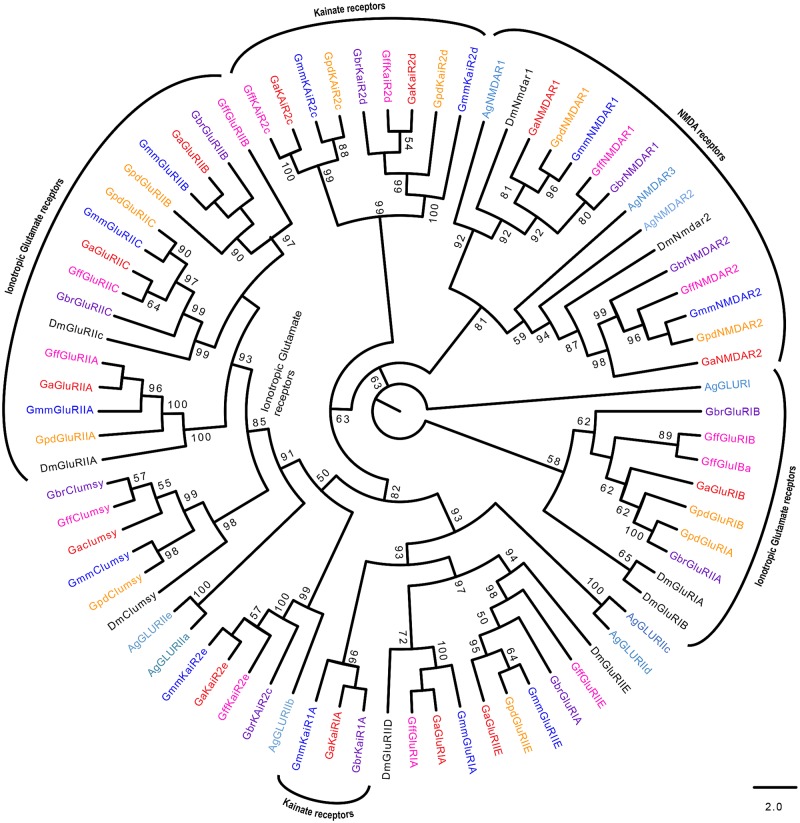
Fig 10. Phylogeny of ionotropic glutamate receptors and kainate receptors.
Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations. Different symbols and colors were used to depict IGluRs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Musca domestica (lime green*) and Anopheles gambiae (sky blue*). The symbol * represents the name of the specific IGluR.
Only three of the Glossina predicted Irs clustered with the divergent IRs (Fig 9). These include Ir68a, Ir10a and Ir56d in Glossina species except G. m. morsitans which had a homolog of Ir56b. Although the alignment of IRs and iGluRs show similar modular arrangements (S3 and S4 Figs, respectively), iGluRs have an extra conserved arginine residue which most IRs lack. Phylogeny of the iGluRs (Fig 10) depicts their high conservation across Diptera.
Fig 9. Phylogeny of divergent ionotropic receptors.
Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations. Different symbols and colors were used to depict IRs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Musca domestica (lime green*) and Anopheles gambiae (sky blue*). The symbol * represents the name of the specific IR.
Selection Analysis
The M8 (beta & w) codeml model was found to have better representation of the data relative to M1a and M2a models, hence its adoption in calculation of LRT values. Nevertheless, some of the dN/dS (w1M8) values were too high to be considered reliable (S1 Table); such values result from low counts of synonymous substitutions compared nonsynonymous substitutions. In addition, majority (67.02%) of the alignments were seen to have a significant p-value under the M8-M8a model. Different levels of selection were noted for majority of the intra-species paralogs (S1 Table). For instance whereas Ejbp3B showed significant selection, Ejbp3A did not show significant selection signatures. Similarly, Or45a2 and Or45a3 showed significant selection while Or45a1 did not show significant selection. Other genes with similar pattern of selection pressures are shown in S1 Table. On the contrary, only a small subset (13.64%) of gene loci was significantly identified to be under selection in the HyPhy package (S2 Table). Only four gene loci (Gr21a, Gr28b, Obp83a and GluRIIA) that were identified by the two packages could be conclusively indicated to be under selection (Table 2). Various factors such as the low number of sequences per gene loci and lack of divergence within sequences have been indicated to introduce false positives (type I error) and lack meaningful inference [70,71].
Table 2. Summary of four Glossina chemosensory gene loci identified to have signatures of positive selection.
Selection analysis was performed using HyPhy package using MEME and PARRIS and compared with PAML –codeml using the M8-M8a model. lnL M8 is the likelihood of the experimental model (M8).
| Gene id | lnL M8 | lnL M8a | ΔLRT | p –value | w1M8 | Sites by MEME | Singleton (S) /Duplicate (D) | Number of codons analyzed | ΔLRT MEME |
|---|---|---|---|---|---|---|---|---|---|
| Obp83a | -529.17 | -533.174 | 7.943 | 0.0048 | 1.075 | 29 | D | 498 | 57.927 |
| Gr21a | -1642.19 | -1656.36 | 28.346 | 1.014E-7 | 1.18653 | 39 | D | 621 | 21.87 |
| GluRIIA | -1431.52 | -1387.04 | 8.34 | 0.00387 | 1.4264 | 2 | S | 1807 | 12.58 |
| Gr28b | -1557.62 | -1566.29 | 17.34 | 4.85E-5 | 1.75 | 44 | D | 569 | 6.98 |
lnL M8a is the likelihood of the null model (M8a), ΔLRT is the Likelihood Ratio Test = 2*(lnL M8- lnL M8a), w1M8 is the ratio of non-synonymous to synonymous mutations (dN/dS) predicted under M8 model/and p-value is the statistical measure of significance.
Discussion
Identification and annotation of chemosensory gene families in four tsetse genomes (G. austeni, G. brevipalpis, G. f. fuscipes and G. pallidipes), which are representatives of all tsetse fly sub-genera, has provided a comprehensive gene repertoire necessary for undertaking comparative functional genomics.
Results of this study show a general conservation of chemosensory gene families in terms of sequence length, gene structure, and gene copy numbers across the five tsetse species. This included the previously described G. morsitans morsitans [31,24]. Specifically, high levels of conservation were observed in OBPs and CSPs; genes involved in trafficking of hydrophobic molecules across the sensillum lymph of insects [72], suggesting a safeguarded role in odorant binding. The two protein families are characterized by six and four conserved cysteine residues, respectively, with CSPs being more conserved [73]. This supports earlier observations by Sanchez-Gracia et al., (2009) that the CSPs family is more conserved compared to OBPs family. The majority of OBPs identified across the Glossina genus fall under the Classic subfamily with six conserved cysteine residues (Fig 1). This is consistent with what has been reported in genomes of related insect species such as Drosophila and the Mediterranean fly [14,49], suggesting that classical OBPs have conserved functions in all insects. Expansion of Obp83a (previously named Obp8-10,12 in G. m. morsitans [24] was noted in all tsetse species. Liu and colleagues [24] suggested that Obp83a1 could be olfactory-specific as it is expressed highly in starved females. Therefore, the expansion of Obp83a across the five tsetse species studied so far implicates its participation in host seeking, with the duplication indicating the investment made by tsetse in finding food. Co-localization of the four copies under the same scaffold (S1 Fig) suggests that they are recently duplicated paralogs that perhaps could be co-regulated. On the other hand, the presence of two Glossina odorant receptor paralogs (copies of Or45a and Or7a) in distantly located scaffolds may indicate the involvement of transposition in emergence. Notably, gene transposition has been reported earlier in three Drosophila species (D. melanogaster, D. yakuba and D. simulans) [74–76], adding credence to the occurrence of transposition as a mode of gene emergence in insects.
The complete loss of genes and/or distortion in their gene structure observed in G. brevipalpis could be attributed to evolutionary events given that it is the most ancient among the Glossina species studied [77]. This correlates an assumption made by Gooding and colleagues [78] who proposed that the oldest subgenus would exhibit more genetic differences, assuming a constant rate of evolution. Of two GRs (Gr32a and Gr68a), that are known to respond to pheromones, only Gr32a was present in all five Glossina species. This is not surprising as Gr68a also participates in sound reception [66]. Absence of Gr68a in tsetse could imply that tsetse flies rely on a different receptor other than Gr68a for sound reception, or that the insects rely entirely on their tympanal organ for this function [79]. Additionally, absence of Gr68a has been reported to reduce male-male courtship in Drosophila and perhaps may play the same role in tsetse flies [32]. Glossina IRs/iGluRs shows conservation of copy numbers. Notably, the Ir84a have homologs in all tsetse species studied here. Ir84a is a candidate receptor for phenylacetyaldehyde and has been reported to promote male courtship in Drosophila [80]. Presence of Ir84a in Glossina support male courtship to be conserved across tsetse species. On the other hand, the absence of Ir93a in G. austeni whose ligand is unknown [81] could potentially encode a defective response to either aldehydes, amines or carboxylic acids, which are primarily recognized by IRs [37].
Based on the number of chemosensory genes identified across Glossina, it is apparent that all tsetse fly species have a reduced chemosensory repertoire compared to D. melanogaster and M. domestica. This is in agreement with findings reported in G. m morsitans [31,42]. Noteworthy is the absence of all sugar receptors (Gr64a-f and Gr5a) in all tsetse species studied here (Fig 6). This is presumably due to the obligate hematophagous nature of both sexes in tsetse flies. Sugar receptors are present in M. domestica, D. melanogaster and An. gambiae, which feed on nectar as primary or secondary source of nutrients. Also, tsetse species lack homologs to Gr43a, which has been attributed to internal fructose sensing in Drosophila [67]. Gr43a mutants show an abolished preference of fructose but no difference in response to other sugars [82]. All tsetse species showed expansion of Gr21a homologs that mediates CO2 recognition confirming that tsetse flies are attracted to their vertebrate hosts through this volatile gas [83]. Similar to M. domestica [47], expansions of Or45a and Or67d that mediate stress response [84] and cVA reception [85], respectively, in Drosophila, were noted in all tsetse species. Or45a in Drosophila is expressed only in larvae [69] where it serves as a receptor for octyl-acetate that trigger a repellency effect [84]. Though the significance of expansion of Or45a in tsetse is yet to be understood, the receptor may potentially play roles in recognizing some undesirable cues present in tsetse’s uterus during larval development. Further, expansion of Or67d in the majority of the insect species compared in this study may point to its importance in enhancing their pheromone perception, hence mate selection [85]. Other ORs that showed expansion in tsetse include Or67c whose role is yet to be determined and Or43a, linked to benzaldehyde perception in Drosophila [86]. Among the annotated Glossina OBPs, Obp19 (a gene without homologs in Drosophila) was seen to have homologs in hemipterans, Lygus lineoralis and Microplitis demolitor and not in any of the close dipterans such as M. domestica or Stomoxys calcitrans. Moreover, Obp19 showed close phylogenetic relationship with Obp56i from all the Glossina species. This could imply that Obp19 is a recent paralog of Obp56i that assumes similar function to that of its homologs in hemipterans. Close phylogenetic relationship observed among Glossina OBPs and genes related to pheromone binding protein receptor proteins (PBPRPs) from other insects including (i.e. Obp19d, Obp28a, Obp69a, Obp83a and Obp84a) is similar to what was reported in C. capitata [87]. This implies that the role of PBRPs is well conserved in tsetse flies, as in other insect species.
Fig 6. Phylogeny of gustatory receptors.
Gustatory receptors responsible for CO2 detection show expansion in Glossina species and Musca domestica relative to Drosophila. On the contrary, all receptors responsible for sugar detection are found to be absent in Glossina. Different symbols depict GRs from the different species at the terminal nodes: Glossina austeni (red*), Glossina brevipalpis (purple*), Glossina fuscipes fuscipes (pink*), Glossina morsitans morsitans (dark blue*), Glossina pallidipes (light orange*), Drosophila melanogaster (black*), Anopheles gambiae (sky blue*) and Musca domestica (lime green*). The symbol * represents the name of the specific GR. Sequence alignment was performed using MuSCLE v3.8.31 and phylogeny relationship was inferred using RAxML v8 with best fitting Wheelan and Goldman (WAG) model and 1000 bootstrap iterations.
Three of four gene loci (Table 2), showing strongest indication of positive selection are evolving under duplication suggesting a rapid rate in their evolution as earlier reported in ants [20] and in Drosophila [88]. The three genes are potentially involved in host seeking and/or taste discrimination in tsetse species and could therefore serve as targets for behavior manipulation as control measure. The Gr21a has three copies in all the five Glossina species and is believed to play a role in detection of CO2; a tsetse volatile cue from vertebrate hosts [83]. Expansion of CO2 receptors is also noted in the malaria vector, An. gambiae (Fig 6). This highlights the importance of CO2 in host location. Similar to Gr21a, Obp83a has four copies in each of the five Glossina species characterized so far and has been reported to be highly expressed in adult females 48 hours post feeding [24] suggesting its role in host finding. The only singleton found to be under significant selection is GluRIIA, but its role in tsetse is unknown. However, the homolog of GluRIIA in Drosophila has been implicated in postsynaptic signaling at the neuromuscular junction [89]. Though few genes were found to harbor signatures of natural selection, it is evident that those identified are inclined towards host seeking and perhaps are responsible for diverse host preference observed across different species [13,90,91]. The discrepancy in the number of gene loci identified to be under positive selection by PAML and HyPhy package could be due to few sequences available for the analysis. In addition to forces of natural selection, the observed behavioral differences exhibited by tsetse species could be as a result of unraveled diversity in their signal transduction machinery and/or post translational modification in their respective chemosensory proteins. Two different odor transduction mechanisms have been proposed in insects [92]. They include (I) the receptor-mediated (ion-channel) mechanism which does not rely on G-protein signaling pathway [93] and (II) the G-protein cascade approach in which binding of semiochemicals to ORs is thought to activate the cyclic-nucleotide pathway [94,95]. To date, little is known about the interaction between the tsetse’s specific ORs and their corresponding ligands and their downstream processing in the fly’s central nervous system (CNS). Receptor-ligand interaction marks the beginning of odor processing that leads to a behavioral response. Post-translational modification is known to permit change of the amino acid properties as a reaction towards physiological needs of an organism [96]. For instance, phosphorylation has been attributed to elasticity of ion channels involved in signaling [97]. Thus, it is important to study the downstream processes involved in odor processing across tsetse species to identify any underlying differences responsible for their behavior towards hosts. Additionally, tsetse species may have developed an adaptation to specific odours based on learning. This type of learning has been reported to influence host selection in tsetse [91]. It is therefore possible that learning could play a role in differentially recognizing odours observed across different tsetse species.
In general, tsetse species have a conserved chemosensory gene repertoire with genes sparsely distributed across their genomes. This study did not find significant gene loss/gain between species, except G. brevipalpis, the presumed ancestral species. A few of the chemosensory genes in tsetse are rapidly evolving through duplication and among them, genes potentially associated with host finding are under strong positive selection pressure, presumably to confer adaptation to host odours. These genes among others could form potential molecular targets for control. The power to detect genes under natural selection and its influence on shaping olfaction in tsetse flies was limited by the number of sequences available. More gene sequences may yield better results in future. This study highlights the need to undertake functional studies on chemosensory genes of tsetse and to study the down-stream odor signaling pathway to enhance our understanding on differential behavior observed across tsetse species and how it can be used in improving current control strategies. Knowledge of differential host responses of sympatric tsetse species will aid in development of an integrated universal and cost-effective control strategy for vectors of trypanosomiasis.
Supporting Information
Metadata for each protein chemosensory family is contained in a separate sheet. CSPs—sheet 1, SNMPs—sheet 2, GRs –sheet3, OBPs –sheet 4, ORs –sheet 5 and IRs –sheet 6. For every sequence, the following data is provided in columns A-G of each sheet: Gene name, VectorBase identifier, scaffold where it is located, number of exons, strand orientation, length of the amino acid sequence and the associated scaffold coordinates.
(XLS)
Multiple sequence alignments of five Glossina species, Drosophila melanogaster, Anopheles gambaie, Musca domestica and Ceratitis capitata used in construction of phylogenies of the chemosensory gene.
(ZIP)
Screenshots illustrating gene structure and tandem arrangement of selected chemosensory genes. Four copies of Obp83a (part A) thought to be olfactory specific in GlossinaTwo Or7a homologs (part B) and two Or56a homologs (part C).
(PDF)
Variation of amino acids between conserved cysteine(s) C3 and C4 in Obp56i and Obp19 from Glossina. Their homologs in M. domestica and D. melanogaster appear more conserved around the same region.
(PDF)
(PDF)
(PDF)
lnL M8 is the likelihood of the experimental model (M8), lnL M8a is the likelihood of the null model (M8a), ΔLRT is the Likelihood Ratio Test = 2*(lnL M8- lnL M8a), w1M8 is the ratio of Non-synonymous to synonymous mutations (dN/dS) predicted under M8 model/and p-value is the statistical measure of significance.
(PDF)
(PDF)
Acknowledgments
We are grateful to Dr. Jing-Jiang Zhou Department of Biological Chemistry and Crop Protection Rothamsted Research, BBSRC and Dr. George Obiero of icipe, Kenya for providing set of chemosensory sequences identified in G. m. morsitans and Dr Hugh Robertson for providing the gene set identified in house fly. Dr. Henry Kariithi of Kenya Agricultural and Livestock Research Organization who helped in editing the manuscript and Collins Omogo of icipe, who helped with editing of figures.
Data Availability
The data used in this Manuscript are available in the Vectorbase database (https://www.vectorbase.org/) and in the supporting files provided. All accession numbers are in the Methods section of the manuscript.
Funding Statement
This study was funded by the German Academic Exchange Service (DAAD) through icipe’s African Regional Postgraduate Program for Insect Science (ARPPIS) that awarded a study fellowship to RM. Research reported in this publication was also supported by the Fogarty International Center of the National Institutes of Health under Award Number R03TW009444. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aksoy S. Control of tsetse flies and trypanosomes using molecular genetics. Vet Parasitol. 2003. 125–145. [DOI] [PubMed] [Google Scholar]
- 2.Simarro PP, Diarra A, Postigo JAR, Franco JR, Jannin JG. The human african trypanosomiasis control and surveillance programme of the world health organization 2000–2009: The way forward. PLoS Negl Trop Dis. 2011;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FAO. Food and Agriculture Organization of the United Nations. FISHSTAT. Global Aquaculture Production. 2014.
- 4.FAO. The state of food and agriculture, 2013. Lancet. 2013.
- 5.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010;375: 148–159. 10.1016/S0140-6736(09)60829-1 [DOI] [PubMed] [Google Scholar]
- 6.Barrett MP, Boykin DW, Brun R, Tidwell RR. Human African trypanosomiasis: pharmacological re-engagement with a neglected disease. Br J Pharmacol. 2007;152: 1155–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hocking KS, Lamerton JF, Lewis EA. Tsetse-fly control and eradication. Bull World Health Organ. 1963;28: 811–823. [PMC free article] [PubMed] [Google Scholar]
- 8.Dransfield RD, Brightwell R, Kyorku C, Williams B. Control of tsetse fly (Diptera: Glossinidae) populations using traps at Nguruman, south-west Kenya. Bull Entom Res 1990. 265. [Google Scholar]
- 9.Hall DR, Beevor PS, Cork A, Nesbitt BF, Vale GA. 1-Octen-3-ol. A potent olfactory stimulant and attractant for tsetse isolated from cattle odours. International Journal of Tropical Insect Science. 1984. 335–339. [Google Scholar]
- 10.Gikonyo NK, Hassanali A, Njagi PGN, Gitu PM, Midiwo JO. Odor composition of preferred (buffalo and ox) and nonpreferred (waterbuck) hosts of some savanna tsetse flies. J Chem Ecol. 2002;28: 969–981. [DOI] [PubMed] [Google Scholar]
- 11.Gikonyo NK, Hassanali A, Njagi PGN, Saini RK. Responses of Glossina morsitans morsitans to blends of electroantennographically active compounds in the odors of its preferred (buffalo and ox) and nonpreferred (waterbuck) hosts. J Chem Ecol. 2003;29: 2331–2345. [DOI] [PubMed] [Google Scholar]
- 12.Mireji PO, Mabveni AM, Dube BN, Ogembo JG, Matoka CM, Mangwiro TNC. Field Responses of Tsetse Flies (Glossinidae) and Other Diptera to Oils in Formulations of Deltamethrin. Int J Tropl Insect Sci. 2003. 317–323. [Google Scholar]
- 13.Omolo MO, Hassanali A, Mpiana S, Esterhuizen J, Lindh J, Lehane MJ, et al. Prospects for developing odour baits to control Glossina fuscipes spp., the major vector of human African trypanosomiasis. PLoS Negl Trop Dis. 2009;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Späth J. Feeding patterns of three sympatric tsetse species (Glossina spp.) (Diptera: Glossinidae) in the preforest zone of Cote d’Ivoire. Acta Trop. 2000;75: 109–118. [DOI] [PubMed] [Google Scholar]
- 15.Muturi CN, Ouma JO, Malele II, Ngure RM, Rutto JJ, Mithöfer KM, et al. Tracking the feeding patterns of tsetse flies (glossina genus) by analysis of bloodmeals using mitochondrial cytochromes genes. PLoS One. 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vieira FG, Sánchez-Gracia A, Rozas J. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 2007;8: R235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mamidala P, Wijeratne AJ, Wijeratne S, Poland T, Qazi SS, Doucet D, et al. Identification of Odor-Processing Genes in the Emerald Ash Borer, Agrilus planipennis. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson MN, Grosse-Wilde E, Keeling CI, Bengtsson JM, Yuen MMS, Li M, et al. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics. 2013;14: 198 10.1186/1471-2164-14-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leal WS. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Ann Rev Entomol. 2011. 120928130709004. [DOI] [PubMed] [Google Scholar]
- 20.Kulmuni J, Havukainen H. Insights into the Evolution of the CSP Gene Family through the Integration of Evolutionary Analysis and Comparative Protein Modeling. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozaki K, Utoguchi A, Yamada A, Yoshikawa H. Identification and genomic structure of chemosensory proteins (CSP) and odorant binding proteins (OBP) genes expressed in foreleg tarsi of the swallowtail butterfly Papilio xuthus. Insect Biochem Mol Biol. 2008;38: 969–976. 10.1016/j.ibmb.2008.07.010 [DOI] [PubMed] [Google Scholar]
- 22.Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-Wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002. pp. 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mameli M, Tuccini A, Mazza M, Petacchi R, Pelosi P. Soluble proteins in chemosensory organs of phasmids. Insect Biochem Mol Biol. 1996;26: 875–882. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Lehane S, He X, Lehane M, Hertz-Fowler C, Berriman M, et al. Characterisations of odorant-binding proteins in the tsetse fly Glossina morsitans morsitans. Cell Mol Life Sci. 2010;67: 919–929. 10.1007/s00018-009-0221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu R, He X, Lehane S, Lehane M, Hertz-Fowler C, Berriman M, et al. Expression of chemosensory proteins in the tsetse fly Glossina morsitans morsitans is related to female host-seeking behaviour. Insect Mol Biol. 2012;21: 41–8. 10.1111/j.1365-2583.2011.01114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronderos DS, Smith DP. Diverse signaling mechanisms mediate volatile odorant detection in Drosophila. Fly (Austin). 2009;3(4).290–297. [DOI] [PubMed] [Google Scholar]
- 27.Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450: 289–293. [DOI] [PubMed] [Google Scholar]
- 28.Jin X, Ha TS, Smith DP. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc Natl Acad Sci USA 2008;105(31).10996–11001. 10.1073/pnas.0803309105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4: 240–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annu Rev Entomol. 2006;51: 113–135. [DOI] [PubMed] [Google Scholar]
- 31.Obiero GFO, Mireji PO, Nyanjom SRG, Christoffels A, Robertson HM, Masiga DK. Odorant and gustatory receptors in the tsetse fly Glossina morsitans morsitans. PLoS Negl Trop Dis. 2014;8: e2663 10.1371/journal.pntd.0002663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montell C. A taste of the Drosophila gustatory receptors. CurrOpinion in Neurobiol. 2009. 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benton R, Vannice KS, Gomez-diaz C, Vosshall LB. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell. Elsevier Inc.; 2009;136: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69: 44–60. 10.1016/j.neuron.2010.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25: 8359–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rytz R, Croset V, Benton R. Ionotropic Receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 2013;43: 888–897. 10.1016/j.ibmb.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 38.Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Human Gen. 2006. 505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4: 0446–0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardiner A, Barker D, Butlin RK, Jordan WC, Ritchie MG. Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol Ecol. 2008;17: 1648–57. 10.1111/j.1365-294X.2008.03713.x [DOI] [PubMed] [Google Scholar]
- 41.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2010;26: 2455–2457. 10.1093/bioinformatics/btq429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Attardo GM, Abila PP, Auma JE, Baumann AA, Benoit JB, Brelsfoard CL, et al. Genome Sequence of the Tsetse Fly (Glossina morsitans): Vector of African Trypanosomiasis. Science (80). 2014;344: 380–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim J-W, Lambkin C, et al. Episodic radiations in the fly tree of life. Proc Natl Acad Sci U S A. 2011;108: 5690–5695. 10.1073/pnas.1012675108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner R V., Butler R, et al. VectorBase: A data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelbart WM, Rindone WP, Chillemi J, Russo S, Crosby M, Mathews B, et al. FlyBase: The Drosophila database. Nucleic Acids Research. 1996. pp. 53–56. 8594600 [Google Scholar]
- 46.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32: D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott JG, Warren WC, Beukeboom LW, Bopp D, Clark AG, Giers SD, et al. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 2014;15: 466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siciliano P, Scolari F, Gomulski LM, Falchetto M, Gabrieli P, et al. Sniffing Out Chemosensory Genes from the Mediterranean Fruit Fly, Ceratitis capitata.PLOS ONE; 2014;9 1: 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. Domain enhanced lookup time accelerated BLAST. Biology Direct. 2012. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16: 944–945. [DOI] [PubMed] [Google Scholar]
- 52.Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF-FINDER: A vector for high-throughput gene identification. Gene. 2002;282: 33–41. [DOI] [PubMed] [Google Scholar]
- 53.Edgar RC, Drive RM, Valley M. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009;25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chenna R, Sugawara H, Koike T, Lopez R, Gibson T, Higgins D, et al. , Multiple sequence alignment with clustal series of programs. Nucleic Acid Res. 2003, 31(13):3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 2005;21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 57.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30: 1312–3. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Löytynoja A. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 2014;1079: 155–170. 10.1007/978-1-62703-646-7_10 [DOI] [PubMed] [Google Scholar]
- 59.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 60.Kosakovsky Pond SL, Frost SDW, Muse S V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005;21: 676–679. [DOI] [PubMed] [Google Scholar]
- 61.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheffler K, Martin DP, Seoighe C. Robust inference of positive selection from recombining coding sequences. Bioinformatics 2006;22: 2493–2499. [DOI] [PubMed] [Google Scholar]
- 63.Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, Staples J, et al. The insect SNMP gene family. Insect Biochem Mol Biol. 2009;39: 448–456. 10.1016/j.ibmb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 64.Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO 2 reception in Drosophila.Proc Natl Acad Sci USA 2007;104(9):3574–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445: 86–90. [DOI] [PubMed] [Google Scholar]
- 66.Isono K, Morita H, Bickmeyer U, Wegener A. Molecular and cellular designs of insect taste receptor system.Front Cell Neurosci 2010;4: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyamoto T, Slone J, Song X, Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012;151: 1113–1125. 10.1016/j.cell.2012.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fox AN, Pitts RJ, Robertson HM, Carlson JR, Zwiebel LJ. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci U S A. 2001;98: 14693–14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30: 505–533. [DOI] [PubMed] [Google Scholar]
- 70.Poon AFY, Frost SDW, Pond SLK. Detecting signatures of selection from DNA sequences using datamonkey. Methods Mol Biol. 2009;537: 163–183. 10.1007/978-1-59745-251-9_8 [DOI] [PubMed] [Google Scholar]
- 71.Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dyer NA, Lawton SP, Ravel S, Choi KS, Lehane MJ, Robinson AS, et al. Molecular Phylogenetics and Evolution Molecular phylogenetics of tsetse flies (Diptera: Glossinidae) based on mitochondrial (COI, 16S, ND2) and nuclear ribosomal DNA sequences, with an emphasis on the palpalis group. 2008;49: 227–239. [DOI] [PubMed] [Google Scholar]
- 73.Angeli S, Ceron F, Scaloni A, Monti M, Monteforti G, Minnocci A, et al. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur J Biochem. 1999;262: 745–754. [DOI] [PubMed] [Google Scholar]
- 74.Heger A, Ponting CP. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res. 2007;17: 1837–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ponting CP, Mott R, Bork P, Copley RR. Novel protein domains and repeats in Drosophila melanogaster: insights into structure, function, and evolution. Genome Res. 2001;11: 1996–2008. [DOI] [PubMed] [Google Scholar]
- 76.Inohara N, Nuez G. ML—A conserved domain involved in innate immunity and lipid metabolism. Trends in Biochemical Sciences. 2002. 219–221. [DOI] [PubMed] [Google Scholar]
- 77.Rio RVM, Symula RE, Wang J, Lohs C, Wu Y neng, Snyder AK, et al. Insight into the Transmission Biology and Species-Specific Functional Capabilities of Tsetse (Diptera: Glossinidae) Obligate Symbiont Wigglesworthia. MBio. 2012;3: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gooding RH, Krafsur ES. Tsetse genetics: contributions to biology, systematics, and control of tsetse flies. Annu Rev Entomol. 2005;50: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tuck EJ, Windmill JFC, Robert D. Hearing in tsetse flies? Morphology and mechanics of a putative auditory organ. Bull Entomol Res. 2009;99: 107–119. 10.1017/S0007485308006160 [DOI] [PubMed] [Google Scholar]
- 80.Grosjean Y, Rytz R, Farine J-P, Abuin L, Cortot J, Jefferis GSXE, et al. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011. 236–240. [DOI] [PubMed] [Google Scholar]
- 81.Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GSXE, et al. Complementary Function and Integrated Wiring of the Evolutionarily Distinct Drosophila Olfactory Subsystems. Journal of Neuroscience. 2011. 13357–13375. 10.1523/JNEUROSCI.2360-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mishra D, Miyamoto T, Rezenom YH, Broussard A, Yavuz A, Slone J, et al. The molecular basis of sugar sensing in Drosophila larvae. Curr Biol. 2013;23: 1466–1471. 10.1016/j.cub.2013.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Torr SJ, Mangwiro TNC, Hall DR. The effects of host physiology on the attraction of tsetse (Diptera: Glossinidae) and Stomoxys (Diptera: Muscidae) to cattle. Bull Entomol Res. 2006;96: 71–84. [DOI] [PubMed] [Google Scholar]
- 84.Bellmann D, Richardt A, Freyberger R, Nuwal N, Schwärzel M, Fiala A, et al. Optogenetically Induced Olfactory Stimulation in Drosophila Larvae Reveals the Neuronal Basis of Odor-Aversion behavior. Front Behav Neurosci. 2010;4: 27 10.3389/fnbeh.2010.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang L, Anderson DJ. Identification of an aggression-promoting pheromone and itw receptor neurons in Drosophila. Nature. 2010;463: 227–231. 10.1038/nature08678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rollmann SM, Wang P, Date P, West SA, Mackay TFC, Anholt RRH. Odorant Receptor Polymorphisms and Natural Variation in Olfactory Behavior in Drosophila melanogaster. Genetics. 2010. pp. 687–697. 10.1534/genetics.110.119446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Siciliano P, He XL, Woodcock C, Pickett JA, Field LM, Birkett MA, et al. Identi fi cation of pheromone components and their binding af fi nity to the odorant binding protein CcapOBP83a-2 of the Mediterranean fruit fl y, Ceratitis capitata. 2014;48: 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Almeida FC, Sánchez-Gracia A, Campos JL, Rozas J. Family size evolution in Drosophila chemosensory gene families: a comparative analysis with a critical appraisal of methods. Genome Biol Evol. 2014;6: 1669–82. 10.1093/gbe/evu130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morimoto T, Nobechi M, Komatsu A, Miyakawa H, Nose A. Subunit-specific and homeostatic regulation of glutamate receptor localization by CaMKII in Drosophila neuromuscular junctions. Neuroscience. 2010;165: 1284–1292. 10.1016/j.neuroscience.2009.11.059 [DOI] [PubMed] [Google Scholar]
- 90.Torr SJ, Chamisa A, Vale GA, Lehane MJ, Lindh JM. Responses of tsetse flies, Glossina morsitans morsitans and Glossina pallidipes, to baits of various size. Med Vet Entomol. 2011;25: 365–369. 10.1111/j.1365-2915.2011.00947.x [DOI] [PubMed] [Google Scholar]
- 91.Bouyer J, Pruvot M, Bengaly Z, Guerin PM, Lancelot R. Learning influences host choice in tsetse. Biol Lett. 2007;3: 113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dionne VE, Dubin AE. Transduction diversity in olfaction. J Exp Biol. 1994;194: 1–21. [DOI] [PubMed] [Google Scholar]
- 93.Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452: 1002–1006. 10.1038/nature06850 [DOI] [PubMed] [Google Scholar]
- 94.Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452: 1007–1011. 10.1038/nature06861 [DOI] [PubMed] [Google Scholar]
- 95.Gomez-Diaz C, Martin F, Alcorta E. The cAMP transduction cascade mediates olfactory reception in Drosophila melanogaster. Behav Genet. 2004;34: 395–406. [DOI] [PubMed] [Google Scholar]
- 96.Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdisciplinary Rev: Systems Biol and Med. 2012. 565–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levitan IB. Modulation of ion channels by protein phosphorylation and dephosphorylation. Annu Rev Physiol. 1994;56: 193–212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metadata for each protein chemosensory family is contained in a separate sheet. CSPs—sheet 1, SNMPs—sheet 2, GRs –sheet3, OBPs –sheet 4, ORs –sheet 5 and IRs –sheet 6. For every sequence, the following data is provided in columns A-G of each sheet: Gene name, VectorBase identifier, scaffold where it is located, number of exons, strand orientation, length of the amino acid sequence and the associated scaffold coordinates.
(XLS)
Multiple sequence alignments of five Glossina species, Drosophila melanogaster, Anopheles gambaie, Musca domestica and Ceratitis capitata used in construction of phylogenies of the chemosensory gene.
(ZIP)
Screenshots illustrating gene structure and tandem arrangement of selected chemosensory genes. Four copies of Obp83a (part A) thought to be olfactory specific in GlossinaTwo Or7a homologs (part B) and two Or56a homologs (part C).
(PDF)
Variation of amino acids between conserved cysteine(s) C3 and C4 in Obp56i and Obp19 from Glossina. Their homologs in M. domestica and D. melanogaster appear more conserved around the same region.
(PDF)
(PDF)
(PDF)
lnL M8 is the likelihood of the experimental model (M8), lnL M8a is the likelihood of the null model (M8a), ΔLRT is the Likelihood Ratio Test = 2*(lnL M8- lnL M8a), w1M8 is the ratio of Non-synonymous to synonymous mutations (dN/dS) predicted under M8 model/and p-value is the statistical measure of significance.
(PDF)
(PDF)
Data Availability Statement
The data used in this Manuscript are available in the Vectorbase database (https://www.vectorbase.org/) and in the supporting files provided. All accession numbers are in the Methods section of the manuscript.