Abstract
Using Nanodiscs, we quantitate the heterotropic interaction between two different drugs mediated by monomeric CYP3A4 incorporated into a native-like membrane environment. The mechanism of this interaction is deciphered by global analysis of multiple turnover experiments performed under identical conditions using the pure substrates progesterone (PGS) and carbamazepine (CBZ) and their mixtures. Activation of CBZ epoxidation and simultaneous inhibition of PGS hydroxylation are measured and quantitated through differences in their respective affinities towards both a remote allosteric site and the productive catalytic site near the heme iron. Preferred binding of PGS at the allosteric site and higher preference of CBZ binding at the productive site give rise to a non-trivial drug-drug interaction. Molecular dynamics simulations indicate functionally important conformational changes caused by PGS binding at the allosteric site and by two CBZ molecules positioned inside the substrate binding pocket. Structural changes involving Phe-213, Phe-219, and Phe-241 are suggested to be responsible for the observed synergetic effects and positive allosteric interactions between these two substrates. Such a mechanism is likely of general relevance to the mutual heterotropic effects caused by biologically active compounds which exhibit different patterns of interaction with the distinct allosteric and productive sites of CYP3A4, as well as other xenobiotic metabolizing cytochromes P450 that are also involved in drug-drug interactions. Importantly, this work demonstrates that a monomeric CYP3A4 can display the full spectrum of activation and cooperative effects that are observed in hepatic membranes.
Cytochromes P450 are a key class of enzymes involved in the metabolism of xenobiotics, with the CYP3A4 isoform in human liver responsible for many critical biotransformations. Given their involvement in the metabolism of numerous drugs, these enzymes constitute a major site of drug-drug interactions. CYP3A4 can bind multiple copies of many compounds, and the enzyme often exhibits cooperativity with multiple substrates. Very often, these interactions are detected as inhibition of CYP3A4-mediated metabolism of test substrates by the presence of an effector or another compound (1), which can result in a dangerously high concentration of an administered therapeutic. Some compounds can accelerate CYP3A4-mediated metabolism in vitro, e.g., α-naphthoflavone (ANF)1 (2), quinidine (3), steroids (4), and a number of potential drug candidate molecules (5). This metabolic interaction between two different compounds is termed "heterotropic" and, like the homotropic case, is manifested through the simultaneous binding of multiple molecules to the enzyme. Despite the importance of CYP3A4 to human pharmacology, the detailed mechanisms of such heterotropic effects are not completely understood. Given different systems and protocols employed by various research groups, it has been difficult to directly compare the reported results, leading to confusion and conflicting claims. For instance, ANF was stated to be both an inhibitor (6, 7) and an activator of CYP3A4 (8–10). Further complications arise when one considers the possibility of oligomeric complexes of multiple P450s (10).
The anticonvulsant drug carbamazepine (CBZ) has been reported to increase the clearance of warfarin (11), and this positive heterotropic effect has been attributed to induction of CYP3A4 (12). Direct interaction of CBZ with the sedative drug midazolam inside the substrate binding pocket has been suggested to be the molecular basis underlying the observed changes in stereo-specificity of midazolam hydroxylation (13). The accelerated metabolism of steroids (including oral contraceptives) during CBZ administration has been also attributed to the induction of CYP3A4 (14–16). On the other hand, CBZ has been reported to act as an inhibitor of hydroxylation of both steroids and midazolam in vitro (13, 17), while 10,11-epoxidation of CBZ by CYP3A4 is activated in the presence of various endogenous steroids (4, 17, 18) as well as other effectors, e.g., ANF, artemisinin, and quinidine (19).
In order to understand the mechanism of heterotropic cooperativity in monomeric CYP3A4 we initiated a series of experimental studies using a pair of substrates with substantially different physical properties, namely, the clinically relevant steroid progesterone (PGS) and the anti-epileptic drug CBZ. Both drugs are known CYP3A4 substrates, and their mutual metabolic effects have been reported in several previous studies, although no mechanistic interpretation was provided for the observed heterotropic interactions (17, 19). In addition, both drugs can act as allosteric effectors, activating the metabolism of other substrates (PGS and other steroids activate CBZ epoxidation (19)) or changing the regio-selectivity of hydroxylation for other substrates catalyzed by CYP3A4 (13). However, their relative affinities with respect to the productive binding site inside the substrate binding pocket near the heme iron (P-site) and to the nonproductive allosteric binding site (A-site) are anticipated to be distinct due to the differences in their molecular shape and solubility. Importantly, heterotropic effects could be most pronounced in the mixture of two substrates with preferential affinities to the productive site (P-type substrate) for one, and to the allosteric site (A-type substrate) for the other. In such a case, preferential binding of the A-type substrate to the allosteric site is expected to favor binding and metabolism of the P-type substrate inside the binding pocket, if its affinity to the latter is higher.
In addition to the experimental investigations focused on determining the fundamental catalytic properties of CYP3A4 in its multiple substrate-bound states, we also performed a series of molecular dynamics (MD) simulations of the CYP3A4 monomer in a membrane environment, with CBZ and PGS bound to the enzyme. The allosteric site of CYP3A4 located near the membrane interface was probed with PGS and CBZ in independent simulations, in order to study the dynamics of the two substrates at this non-productive binding site. The dynamics of CBZ near the productive site of CYP3A4 were probed using both one and two copies of this substrate in the active site and provided critical structural insight into the mechanism of heterotropic cooperativity of CYP3A4.
Experimental Procedures
Protein expression and purification
Expression and purification of membrane scaffold protein (MSP), cytochrome P450 CYP3A4 and rat P450 reductase, as well as preparation of CYP3A4 in POPC Nanodiscs (ND) was executed following previously described protocols (20–22). Cytochrome P450 CYP3A4 was expressed from the NF-14 construct in the PCWori+ vector with a C-terminal pentahistidine tag generously provided by Dr. F. P. Guengerich (Vanderbilt University, Nashville, TN). Cytochrome P450 reductase (CPR) was expressed using the rat CPR/pOR262 plasmid, a generous gift from Dr. Todd D. Porter (University of Kentucky, Lexington, KY). Incorporation of CPR into preformed and purified CYP3A4-Nanodiscs was made by direct addition of CPR at 1:4 CYP3A4/CPR molar ratio, as described (23). All experiments were performed at 37° C using POPC Nanodisc system analogous to our earlier detailed mechanistic studies (24, 25) in order to allow direct comparison of results. This reconstitution system provides a stable well characterized and monodisperse preparation of CYP3A4 incorporated into the model lipid bilayer effectively mimicking the native membrane.
UV-Vis spectroscopy
Substrate titration experiments were performed at 1 µM CYP3A4 in Nanodiscs using a Cary 300 spectrophotometer (Varian, Lake Forest, CA) at 37°C. For the mixed titration experiments the mixtures of steroid substrate PGS with CBZ in methanol were prepared and added to the CYP3A4-Nanodisc solution, thereby maintaining the constant substrate ratios. The final concentration of methanol was less than 1.5%.
NADPH oxidation and product formation
CYP3A4 incorporated Nanodiscs with CPR in a 1:4 molar ratio, and substrate were preincubated for 5 minutes at 37°C, in a 1 ml reaction volume in 100 mM HEPES buffer (pH 7.4), 10 mM MgCl2, 0.1 mM dithiothreitol. The concentration of CYP3A4 was in the range from 60 to 100 nM. The reaction was initiated with the addition of 200 nmol of NADPH. NADPH consumption was monitored for 5 min and calculated from the absorption changes at 340 nm using the extinction coefficient 6.22 mM−1 cm−1. A short reaction time was used to avoid covalent modification of CYP3A4 by CBZ (26). At the end of the incubation period, 0.5 ml of the sample was removed from the cuvette, mixed with 2 ml of dichloromethane and used for product analysis. Cortexolone (2.5 nmol per sample) was added as an internal standard. The samples were thoroughly mixed; after phase separation, the organic layer was isolated and the solvent was removed under a stream of nitrogen. The dried sample was dissolved in 70 ul of methanol and 40 ul was injected onto Ace 3 C18 HPLC column, 2.1 × 150 mm (MAC-MOD Analytical, Chadds Ford,PA). The mobile phase contained 15% acetonitrile and 15% methanol in water; products of PGS hydroxylation and CBZ epoxidation were separated in linear gradient of acetonitrile and methanol rising from 15% to 37% each over 35 min at flow rate 0.2 ml/min. The calibration and method validation was performed using commercially available metabolites of PGS and CBZ. The chromatograms were processed with Millennium software (Waters).
Global Analysis for deconvoluting apparent cooperative effects in P450
Global analysis of homotropic cooperativity for metabolism of one substrate was performed, as previously described (24), by simultaneously fitting the experimental data sets to the four state linear equilibrium binding scheme:
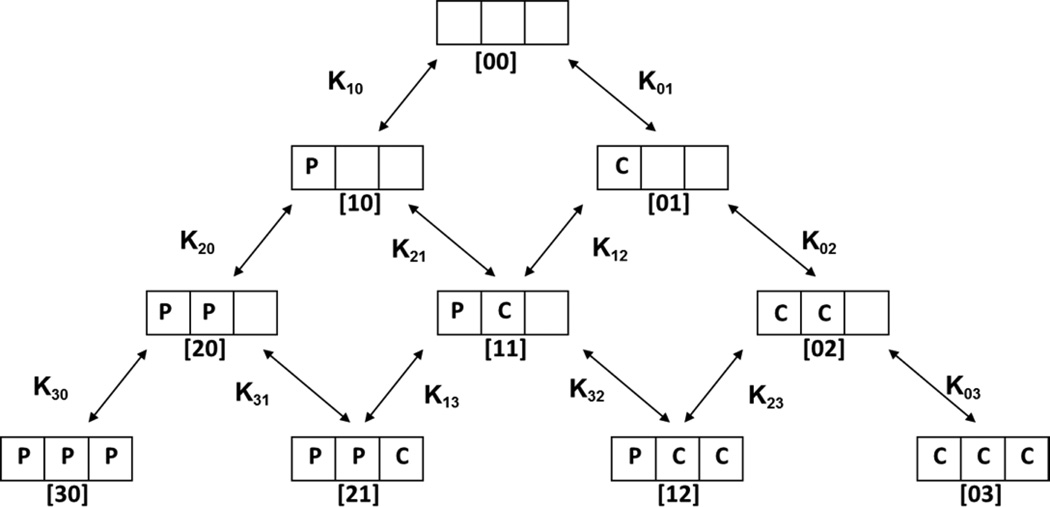
Here E is the concentration of substrate-free CYP3A4 (designated [00] in Scheme 1), S is the concentration of the free substrate, and ESi are the concentrations of the binding intermediates, i.e., complexes of CYP3A4 with i molecules of substrate bound (i = 1, 2, 3). These binding intermediates are designated as [10], [20], [30] for one substrate, and [01], [02], [03] for another (Scheme 1), with stoichiometric dissociation constants Ki0 and Koi (i=1, 2, 3). Binding of up to three molecules of these substrates to one CYP3A4 monomer was described in previous studies (27–29), and attempts to fit the data with only two binding sites proved unsuccessful.
Scheme 1.
Binding equilibria for a CYP3A4 monomer with three binding sites. Two types of substrates designated P (PGS) and C (CBZ) can bind to the same or similar binding sites, so that up to three substrate molecules can bind to one CYP3A4 monomer. The fractional populations of all ten binding intermediates are determined by the twelve equilibrium constants Kij. Three additional equations determined by the microscopic reversibility in the closed cycles (K10K21 = K01K12, K20K31 = K21K13, and K12K32 = K02K23) reduce the number of independent constants from twelve to nine. See text for further description.
The fractions of the enzyme-substrate complexes were expressed using the standard binding polynomials (30),
with the functional properties at different substrate concentrations represented as the linear combination of the fractional contributions from binding intermediates. For example, the fraction of the high-spin CYP3A4 in Type I titrations, YS, is calculated as the weighted sum of the signals from the cytochrome P450 molecules with 0, 1, 2, or 3 substrate molecules bound, having a0, a1, a2, and a3 fractions of high-spin state correspondingly:
The set of such equations for the spectral titration, NADPH consumption, and product formation have been used for the simultaneous fitting of the experimental data obtained under the same conditions, using the same set of dissociation constants, which then corresponds to a total of twelve parameters. The fitting program was written in MATLAB using the Nelder-Mead simplex minimization algorithm implemented in the MATLAB subroutine “fminsearch.m.”
Heterotropic interactions were analyzed using the system of stoichiometric equilibria shown in the Scheme 1. A complete description of ten states with different combinations of two substrates which can bind at three binding site requires twelve binding constants, although only nine of them are independent variables (Scheme 1 and legend). The functional properties of these binding intermediates cannot be individually measured for the pure states with exception of [00] and possibly [30] and [03], if substrate saturation can be reached. However, the ten-state system can be efficiently separated into three parts by the choice of experimental conditions. The homotropic paths [00] → [10] → [20] → [30] and [00] → [01] → [02] → [03] can be measured separately with only one substrate, as described above and in our previous publications (25). Subsequently, the full sets of obtained parameters can be used for the experiments with the mixture of two substrates, if all experiments are performed under identical conditions. In this case only three equilibrium dissociation constants K11, K12, K21, and other functional properties for three mixed intermediates [11], [21], and [12] need to be resolved from the similar set of experiments, with the number of unknown parameters the same as for the homotropic problem, and can be successfully treated using global analysis (24, 25, 31). This was done using the same approach as outlined above for the global analysis of homotropic experiments. Cooperative free energies ΔΔG for the mixed intermediates [11], [21], and [12] were calculated from ratios of corresponding stoichiometric binding constants obtained in fitting: ΔΔG11 =RT ln(K21/K20), ΔΔG21 =RT ln(K31/K30), and ΔΔG12 =RT ln(K23/K03), where RT=0.617 kcal/mol at T=310 K.
Computational Procedures
CYP3A4 model and initial configurations
A membrane-bound model of the globular domain of CYP3A4 (PDB entry 1TQN (32)), including the transmembrane helix formed by residues 1 to 27, was adopted from the last frame of one of the membrane binding simulations reported previously (33). The system is composed of CYP3A4 bound to a solvated POPC membrane. This system was then minimized for 1000 steps and equilibrated for 100 ps while restraining the heavy atoms of the protein and the lipids within 3.5 Å of the protein with a force constant k = 1 kcal mol−1 Å−2. Following this step, the system was simulated without restraints for 100 ns.
Molecular docking and ligand-bound simulations
In order to study the binding mode and dynamics of PGS and CBZ bound to CYP3A4, we first employed molecular docking using Autodock 4.2 (34, 35) to probe potential binding sites for these drugs in the enzyme. Taking advantage of the above-described MD simulation of membrane-bound CYP3A4, a large number of snapshots taken from the 100 ns trajectory were included in our docking as described in the following (in contrast to a single structure used in most docking studies). This approach allows us to take into the account the dynamics of the protein in the presence of the membrane and probe in our docking cavities and putative binding sites that only transiently arise during the simulations. The snapshots of membrane-bound CYP3A4 were taken from the MD trajectory at 1 ns intervals, resulting in 100 different snapshots, which were then used for docking of either PGS or CBZ.
Protein-ligand interactions were modeled with the Lamarckian genetic algorithm (34) with a grid box spanning the peripheral binding site region (near the F-F’ and G-G’ loops in CYP3A4) for both PGS and CBZ, and around the active site for CBZ. The top 10 docking poses for each snapshot and for each ligand (1000 pose for PGS and 1000 poses for CBZ) were collected for further analysis. The resulting docked structures were then clustered based on their root mean square deviation (RMSD), using an RMSD cutoff of 2 Å, resulting in a total of five clusters for each ligand. For PGS in the peripheral binding site, the docked poses in which the substrate adopts a similar orientation to the crystal structure of substrate-bound CYP3A4 (36) were chosen as starting positions for the MD simulations of ligand-bound systems. For CBZ in the peripheral site, the docking poses with the highest docking scores from the pool of the first four clusters were selected as starting positions for ligand-bound MD simulations. In the case of CBZ in the active site, the best docked pose (highest docking score) in each cluster was selected as initial positions for MD simulations. Two of the initial positions of CBZ were discarded based on the distance between the sites of metabolism of the substrate (carbons C10 and C11) and the heme iron (> 6 Å), resulting in only three initial orientations for a single CBZ molecule in the active site. The resulting systems were minimized for 1000 steps and equilibrated for 100 ps while harmonically restraining (k = 1 kcal mol−1 Å−2) the heavy atoms of the protein backbone and the ligand. This step was then followed by unrestrained, production simulations. Simulations of substrates in the allosteric site were run each for 30–100 ns for CBZ, and 100 ns for PGS. For a single CBZ in the active site, the production simulations were each 10–20 ns.
In order to generate models for two CBZ molecules bound to CYP3A4, the general molecular docking approach was repeated employing snapshots from the single CBZ simulations where the distance between CBZ C10/C11 and the heme iron was < 6 Å, resulting in 30 frames. In this case, each snapshot also included the first CBZ molecule, in addition to the protein. The resulting docked poses of the two CBZ molecules were clustered based on RMSD, with an RMSD cutoff of 2 Å. The best docked pose for each cluster was then selected as the initial configuration for additional MD simulations, resulting in four independent systems each with two CBZ bound.
Molecular Dynamics (MD) Simulation Conditions and Protocols
MD Simulations were performed using NAMD2 (37), utilizing the CHARMM27 force field with cMAP (38) corrections for the protein and CHARMM36 (39) for lipids. Parameters for PGS were derived from available testosterone and cholesterol CHARMM parameters (40, 41). Parameters for CBZ were obtained by analogy (42, 43) from the CHARMM General Force Field (39), and were further optimized employing the Force Field Toolkit (44), implemented in VMD (45). The TIP3P model was used for water (46). All simulations were performed as an NPT ensemble at 1.0 atm and 310 K, and with a time step of 2 fs. Constant pressure was maintained using the Nosé-Hoover Langevin piston method (47, 48), and constant temperature was maintained by Langevin dynamics with a damping coefficient of 0.5 ps−1 applied to all atoms. Non-bonded interactions were cut off after 12 Å with a smoothing function applied after 10 Å. Bond distances involving hydrogen atoms were constrained using the SHAKE algorithm (49). The particle mesh Ewald (PME) method (50) was used for long-range electrostatic calculations with a grid density greater than 1 Å−3.
Results
In order to understand the mechanism of activation of CYP3A4 mediated metabolism of one or both substrates in the mixture, we collected the spin shift titration data and steady-state kinetics of substrate turnover with PGS and CBZ separately, as well as in the mixed titration experiments as described in our earlier work on CYP3A4 heterotropic cooperativity (25, 31). All experiments have been performed under identical conditions allowing us to use the same set of binding constants in the analysis of multiple experiments.
Kinetics of PGS and CBZ oxidation
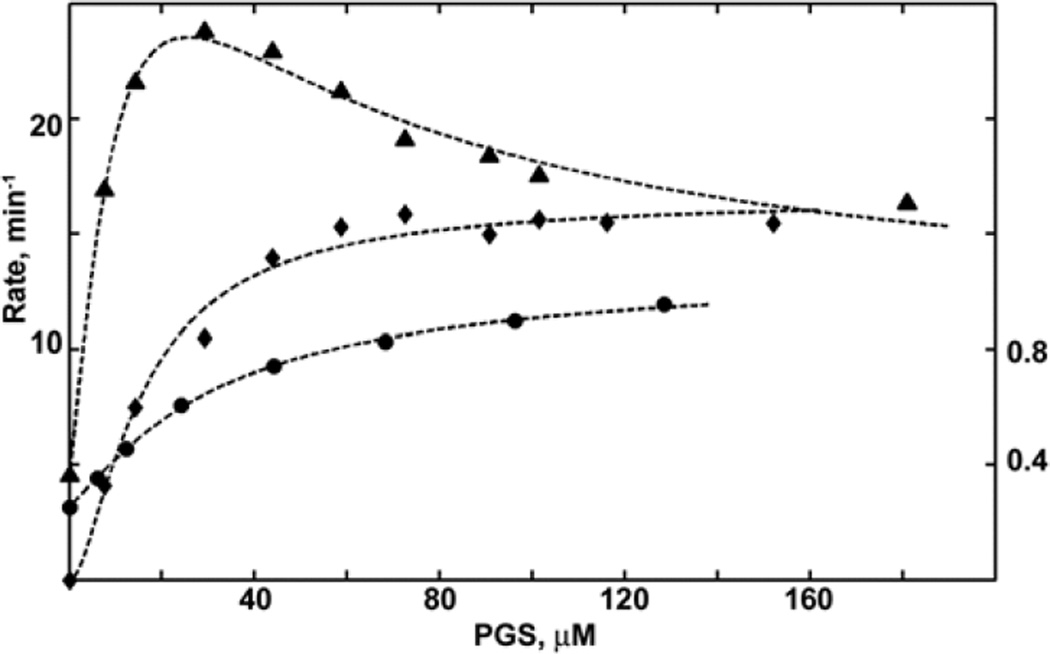
The results of the spectral titration of CYP3A4 with PGS are shown in Figure 1 together with the rate of steady-state NADPH oxidation and the rate of product formation at various substrate concentrations. The spin shift titration and the rate of hydroxylation reveal pronounced apparent cooperativity with a sigmoidal concentration dependence. The rate of PGS hydroxylation reaches 16.6 min−1 at saturation, in agreement with earlier results (8, 51). Fitting with the Hill equation shows nH = 1.7, also similar to earlier reports for CYP3A4 with PGS (51). The rate of NADPH consumption reaches a maximum value of 240 min−1 at 30 µM PGS and then slowly decreases at higher PGS concentrations down to 160 min−1. These observations are in a good agreement with our results obtained earlier for testosterone (TST) and ANF metabolism by CYP3A4 reconstituted in Nanodiscs (24, 25, 31). Stepwise dissociation constants K1, K2, K3 were determined to be 7, 14, and 36 µM. This corresponds to an overall tighter binding but a similar progressive pattern as that resolved earlier in our work with TST (which had values of 19, 37, and 56 µM respectively (24)). The only difference between the parameters resolved for TST and PGS is a moderate spin shift (37%) obtained for the second binding event with PGS (Table 1) as compared with an almost completely high spin state obtained for TST (24). The apparent Km for PGS hydroxylation was estimated from the fitting curve as the concentration of substrate at which v = Vmax/2 for the purpose of a proper choice of molar stoichiometry of PGS and CBZ in the mixed substrate experiments. For PGS the value of Km estimated from the fit is 16 µM (Figure 1).
Figure 1.
Results of the global analysis for PGS hydroxylation by CYP3A4. Spin shift titration (circles, right axis), steady state rate of NADPH oxidation (triangles, left axis) and product formation rates (diamonds, left axis) measured at various substrate concentrations are globally fitted to the stepwise binding scheme as described in Materials and Methods. The absolute values of experimentally measured rates and percentage of high-spin shift were normalized for fitting (NADPH rate divided by 10, and fraction of high spin multiplied by 10) to ensure the same statistical weight of contributions from three different experimental data sets to the globally fitted parameters which are shown in the Table 1.
Table 1.
Parameters derived from single substrate global analyses. Confidence intervals are shown in parentheses below each fitted parameter.
| PGS Kd (µM) | 7 (4.4 – 10.4) High Spin Fraction |
14 (11 – 18) NADPH Rate (nmol/nmol/min) |
36 (27 – 48) Product Forming Rate (nmol/nmol/min) |
|---|---|---|---|
| [00] | 21 - |
44 - |
0 - |
| [10] | 22 (25 – 40) |
171 (126 – 220) |
0 (0.01) |
| [20] | 37 (26 – 47) |
388 (343 – 450) |
14.7 (13.8 – 15.8) |
| [30] | 92 (88 – 97) |
108 (100 – 114) |
16.6 (16.3 – 16.9) |
| CBZ Kd (µM) | 100 (73 – 134) High Spin Fraction |
210 (113 – 340) NADPH Rate (nmol/nmol/min) |
570 (330 – 900) Product Forming Rate (nmol/nmol/min) |
| [00] | 21 - |
41 - |
0 - |
| [01] | 22 (12 – 31) |
195 (155 – 227) |
0 (.01) |
| [02] | 22 (8 – 33) |
124 (90 – 163) |
1.7 (0.9 – 2.4) |
| [03] | 59 (54 – 64) |
56 (46 – 66) |
3.0 (2.7 – 3.4) |
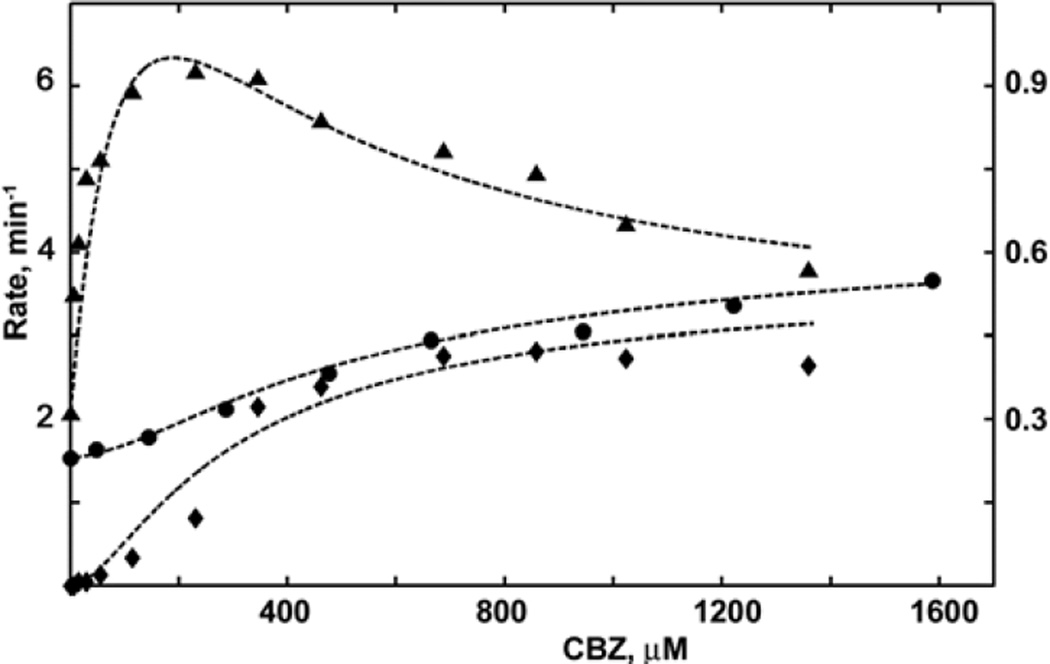
The results obtained with pure CBZ as a substrate are shown in Figure 2. The NADPH oxidation rate reaches a maximal value of 123 min−1 at 200 µM CBZ and then decreases at higher concentrations of the substrate. The same effect is observed with PGS, although the absolute rate of NADPH consumption with CBZ is two times lower at maximum. In contrast, the rate of product formation is low at 200–300 µM CBZ and reaches its maximal value only at concentrations of 500 µM and higher. The spin shift dependence on concentration is much smaller than with the steroids, as mentioned earlier, but also reveals the same sigmoidal shape with the lag at low CBZ concentrations. Moderate positive cooperativity with Hill coefficients from 1.4 to 2.1 was also reported in a recent study of CBZ metabolism by recombinant wild-type CYP3A4 and several mutants (29).
Figure 2.
Results of the global analysis for CBZ epoxidation by CYP3A4. Spin shift titration (circles, right axis), steady state rate of NADPH oxidation (triangles, left axis) and product formation rates (diamonds, left axis) measured at various substrate concentrations are globally fitted to the stepwise binding scheme as described in Materials and Methods. The absolute values of experimentally measured rates and percentage of high-spin shift were normalized for fitting (NADPH rate divided by 10, and fraction of high spin multiplied by 10) to ensure the same statistical weight of contributions from three different experimental data sets to the globally fitted parameters which are shown in the Table 1.
All data for CBZ shown in Figure 2 were simultaneously fitted using the same three-site binding scheme used for the steroid substrates (24). The results of this fit are plotted in Figure 2 as smooth curves, and calculated parameters are shown in Table 1. According to the fitted parameters, the product formation rate is negligible when only one CBZ molecule is bound to CYP3A4. The absence of the product from the first binding intermediate was also reported for CBZ epoxidation by CYP3A4 in human liver microsomes (52, 53) and in insect cells microsomes (17, 54). Binding of the second substrate molecule triggers productive metabolism with an estimated rate of 1.7 min−1, which increases up to 3 min−1 after the third binding event. This pattern is different from the one observed with TST and PGS, where the maximal hydroxylation rate is already achieved after the second substrate binding, and the third substrate binding resulted only in better coupling but not faster catalysis. A similar difference is seen in the high spin fraction of the [20] intermediate, where the CYP3A4 with two PGS molecules bound exhibits a substantial spin shift, while binding of two CBZ molecules still does not cause any spin shift.
Asymmetric heterotropic effect in PGS/CBZ mixtures
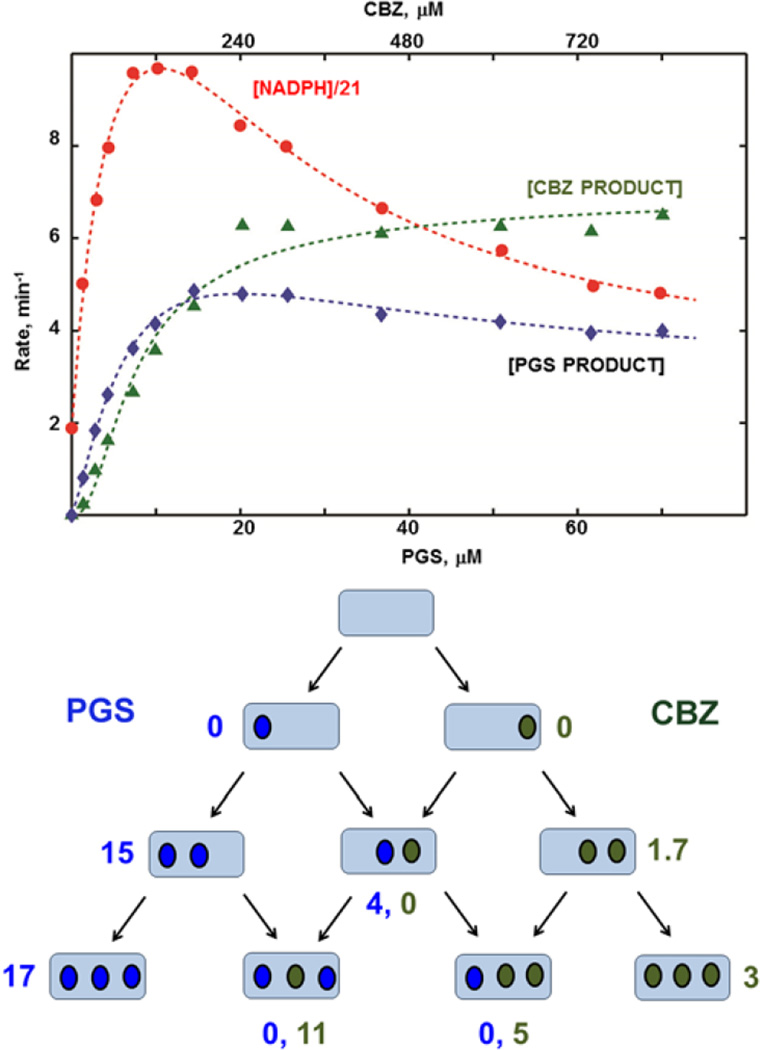
The results of experiments with the mixture of CBZ and PGS are shown in Figure 3. The molar ratio of two substrates CBZ/PGS 12:1 was selected based on the apparent averaged Km values determined from the midpoints of the fitting curves for product formation rates for pure substrates in Figures 1 and 2. The absolute rate of NADPH consumption at maximum, and the shape of its dependence on the substrate concentration, are very similar to those observed with pure CBZ (Figure 2). However, the maximum rate is reached already at 10 µM PGS + 120 µM CBZ, i.e., at significantly lower concentrations as compared to when only a single substrate is present. In addition, significantly different values of the product formation rates are observed for both PGS and CBZ. For PGS the maximum product formation rate 5 min−1 is reached already at 18–20 µM PGS, with the subsequent decrease to 4 min−1 at 60 µM and higher PGS concentrations. This is four times slower than the maximum rate observed with pure PGS, indicating substantial inhibition of PGS hydroxylation in the presence of CBZ. At the same time, the rate of CBZ epoxidation reaches a maximum value of 6 min−1 at 240 µM CBZ and then remains approximately the same up to 800 µM CBZ concentration. This rate is two times higher than the 3 min−1 observed with pure CBZ, meaning significant activation of CBZ metabolism in the presence of PGS. Note that all rate values are calculated per total CYP3A4, which metabolizes both substrates in the mixture, so the actual activation for CBZ and inhibition for PGS is even more pronounced. Importantly, this activation is seen with monomeric CYP3A4.
Figure 3.
(Top) NADPH consumption rate and product forming rates for PGS and CBZ observed using the mixture of PGS and CBZ at 1:12 molar ratio. PGS concentrations are shown at the bottom, and corresponding CBZ concentrations on the top. (Bottom) Rates (min−1) of product formation for all ten binding intermediates with results for PGS hydroxylation shown in blue, and for CBZ epoxidation in green.
The global fitting of these experimental results has been performed using fixed parameters for homotropic parts of Scheme 1, which were determined in separate experiments (Table 1). Parameters evaluated for the mixed intermediates are shown in Table 2. As evident from the results shown in Figure 3, PGS hydroxylation is observed at the relatively slow rate of 4 min−1 only from the [11] intermediate, where one PGS and one CBZ molecule are bound simultaneously to the CYP3A4 monomer. In addition, formation of [11] is favorable by the additional ΔΔGcoop of approximately 1 kcal/mol, resulting in a five-fold higher population of this intermediate as compared with [20] and [02]. This fact partly explains the steep increase in the rates of NADPH oxidation and product formation at relatively low concentrations of substrates seen in Figure 3.
Table 2.
Parameters derived from the global analysis of the mixed substrate experiments. Confidence intervals are shown in parentheses below each fitted parameter.
| ΔΔGcoop (kcal/mol) |
−1 (−1.2 – −0.8) NADPH Rate (nmol/nmol/min) |
0.7 (0.55 – 0.9) PGS Product Rate (nmol/nmol/min) |
0.3 (0.2 – 0.45) CBZ Product Rate (nmol/nmol/min) |
|---|---|---|---|
| [11] | 410 (387 – 434) |
4 (3.7 – 4.2) |
0 (0.01) |
| [21] | 120 (106 – 134) |
0 (0.02) |
11 (9.8 – 12.2) |
| [12] | 128 (82 – 166) |
0 (0.01) |
4.5 (2.0 – 6.0) |
Neither of the [21] and [12] intermediates shows any measurable product formation for PGS, indicating predominant positioning of CBZ at the productive site near the heme iron in these intermediates. The product formation rates for CBZ obtained for these intermediates (11 min−1 and 4.5 min−1, respectively) are significantly higher than 3 min−1 for CBZ alone, revealing these faster rates as the main source of activation of CBZ epoxidation in the presence of PGS. Interestingly, no CBZ product is formed in the [11] intermediate. This may be explained either by preferential PGS positioning at the productive site in this intermediate, or low efficiency of productive CBZ catalysis in [11], if both PGS and CBZ have access to the catalytically active iron-oxo intermediate. Taking into account the very high rate of NADPH oxidation (410 min−1) and the low rate of PGS hydroxylation (4 min−1) obtained for the [11] intermediate (Table 2), the second explanation seems to be more consistent with all experimental data, although no definite conclusion can be made without site-specific structural information, which is currently unavailable. As a result, CBZ epoxidation is significantly accelerated in the presence of PGS, while PGS hydroxylation is strongly inhibited. This asymmetric heterotropic effect can be explained only by an uneven distribution of two substrates between the productive binding site close to the heme iron and the unproductive peripheral binding sites.
Dynamics of PGS and CBZ in the allosteric site of CYP3A4
In order to explore the effects of multiple ligand binding and gain insight into the structural basis of heterotropic cooperativity in CYP3A4 we performed several MD simulations of the enzyme in the presence of PGS or CBZ at different binding sites. It has been suggested that the mechanism of cooperativity of CYP3A4 is regulated by the binding of a ligand to a peripheral binding site (10, 36, 55–57). The binding of a substrate in the periphery of CYP3A4 might trigger a conformational change necessary to facilitate the cooperative behavior of the enzyme. In this regard, experimental structural studies have identified a peripheral binding site in the proximity of the F-G region of the enzyme (36, 55), with X-ray crystal structures revealing PGS binding at this site. In the membrane-bound form of CYP3A4, the peripheral binding site is located in a region that interacts with the membrane (33, 58). Given the partitioning of steroids in the membrane (59) in a region located between the head group and lipid tails, it is likely that a substrate can be recruited by the enzyme and bind to this peripheral site.
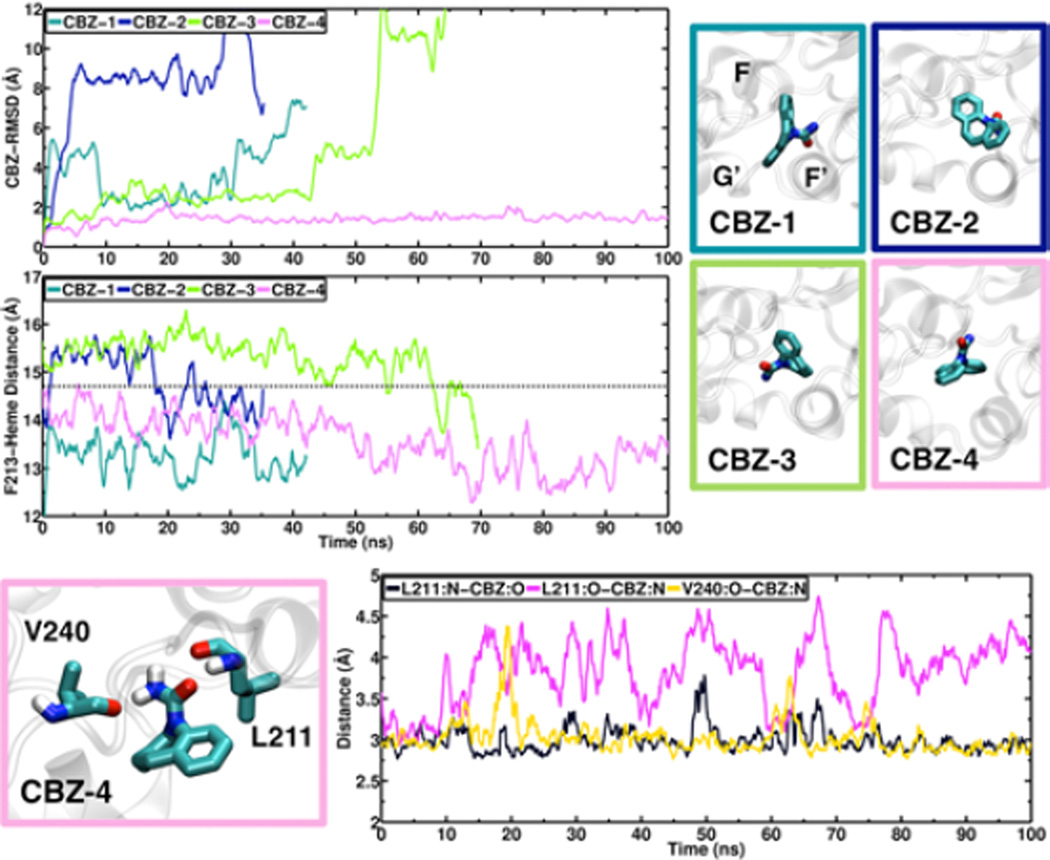
To study the dynamics of PGS in the peripheral binding site of CYP3A4 in the presence of a lipid bilayer, we performed three independent MD simulations starting from docked poses where the ligand has a similar conformation to the available crystal structure of PGS-bound CYP3A4 (36) (Figure 4). Similarly, we executed four independent simulations with CBZ docked in the peripheral binding site. Since there is no structural information about CBZ binding to this allosteric site, we tested four different docked poses for CBZ, one in each simulation (Figure 5). In the resulting docked poses, PGS and CBZ are packed in the vicinity of the hydrophobic side chains in the peripheral binding site. During the simulations, PGS maintains its contacts with hydrophobic side chains located in this region, particularly with Phe-219 and Phe-220, while forming additional transient contacts with Leu-211, Asp-217, and Arg-243. In the case of CBZ, the substrate detached from the enzyme in three out of the four simulations, as quantified by its RMSD evolution (Figure 5). In the single simulation where CBZ remains bound to the enzyme, it is stabilized by interactions between its carbonyl oxygen and the backbone amide group of Leu-211, and between its amine nitrogen and backbone carbonyl oxygen atoms of Leu-211 and Val-240 (Figure 5).
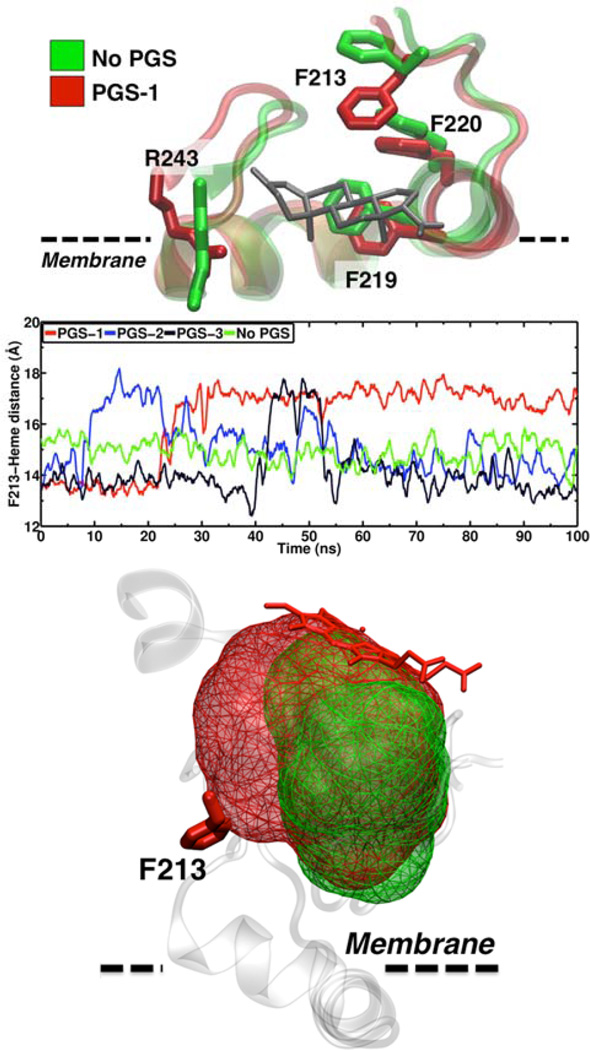
Figure 4.
Dynamics of PGS in the peripheral binding site of CYP3A4. (Top) Representative snapshots of the conformation of the F-G region of CYP3A4 for both substrate-free (green) and PGS-bound simulations (red). Side-chains of residues interacting with PGS are shown in a stick representation in their respective color. PGS is shown in a gray stick representation. (Middle) Time series of the center of mass (COM) distance between the Phe-213 side chain and the heme. The average distance for the substrate-free simulation over 100 ns is 14.7 Å. (Bottom) Change in the active site volume resulting from the displacement of the Phe-213 out of the active site. The Phe-213 side chain and the heme moiety are shown in red stick representation. The volume was calculated using the program “mdpocket” [27].
Figure 5.
Dynamics of CBZ in the peripheral binding site of CBZ. (Top left) Snapshots of the four different orientations of CBZ in the peripheral binding site probed with MD simulations. (Top right) Time series of the RMSD of CBZ and the COM distance between the Phe-213 side chain and the heme for each simulation. The average distance for the 100 ns substrate-free simulation is 14.7 Å (dashed line). (Bottom left) Snapshot of CBZ interactions with residues Leu-211 and Val-240, obtained from system CBZ-4. CBZ and residues Leu-211 and Val-240 are shown in stick representation. (Bottom right) Time series of the distance between interacting elements of CBZ and Leu-211 and Val-240.
Interestingly, the presence of PGS in the peripheral binding site induces a change in the orientation of the Phe-213 side chain, located in the F-F’ loop (Figure 4). In the absence of the substrate, Phe-213 is located closer to the productive binding site (active site), with its side chain pointing towards the heme and away from the membrane. In this orientation, the average distance between Phe-213 and the heme is 14.7 Å. In the simulations where PGS is bound to CYP3A4, a transient change in the orientation of Phe-213 is observed. In these cases, the Phe-213 side chain flips out of the active site and packs against PGS, resulting in an increase of ~4 Å in the distance between its side chain and the heme. In contrast, in the CBZ simulations Phe-213 remains pointing towards the active site, with the distances between the side chain and heme ~14 Å, similar to the substrate-free simulation, even in the simulation where CBZ remains bound to CYP3A4 (Figure 5). This local conformational change induced by PGS favors an increase in the volume of the CYP3A4 active site, from 675 Å3 in the absence of the substrate to 990 Å3 when PGS is present in the peripheral binding site. The relevance of Phe-213 in cooperativity has been observed experimentally, where mutation of this residue results in reduced cooperativity of CYP3A4 (60) and decreased stimulation by ANF (8). In the context of the membrane, our simulations suggest that Phe-213 is important to modulate cooperativity by allowing a volume increase in the active site, therefore facilitating the binding of other substrates in the productive site of CYP3A4.
Dynamics of CBZ in the active site
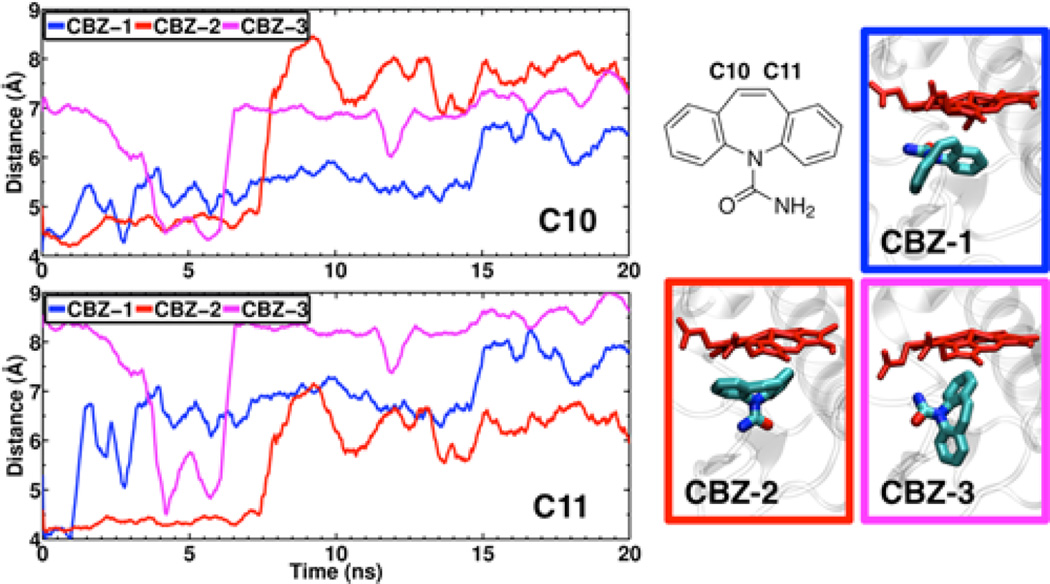
The active site of CYP3A4 is capable of accommodating more than one substrate molecule, as suggested by X-ray structures where two ketoconazole (61) or ritonavir (62, 63) molecules were found in the active site. To study the dynamics of CBZ in the active site, we have performed several independent simulations where one or two CBZ molecules are located in the active site. Although several binding modes of CBZ based on molecular docking have been previously proposed (64), none of the previous studies took into account the dynamics of the binding site and the substrate. Therefore, in order the take into account different potential orientations of the substrate in the active site, we have performed three independent simulations, each starting from one of the three most representative docked poses of a single CBZ molecule in the active site (Figure 6). During these simulations, CBZ is significantly displaced from its initial position in the active site, as shown by the distances between the heme iron and carbons C10 and C11 of CBZ (Figure 6). For example, in system CBZ-3, the molecule transiently approaches the heme (heme iron and C10 or C11 distances < 5 Å), while in systems CBZ-1 and CBZ-3 the molecule gradually drifts away from its initial position and from the heme. Together, these simulations suggest that it may be difficult for a single CBZ molecule to find a stable position and orientation that would facilitate its oxidation.
Figure 6.
Dynamics of single CBZ in active site of CYP3A4. (Right) Snapshots of the three different orientation of CBZ in the active site probed with MD simulations. The structure of CBZ is also shown, indication its sites of metabolism, carbons C10 and C11. CBZ is shown in stick representation. The heme moiety is show in red stick representation. (Left) Time series of the distance between carbons C10 and C11 of CBZ and the heme iron atom.
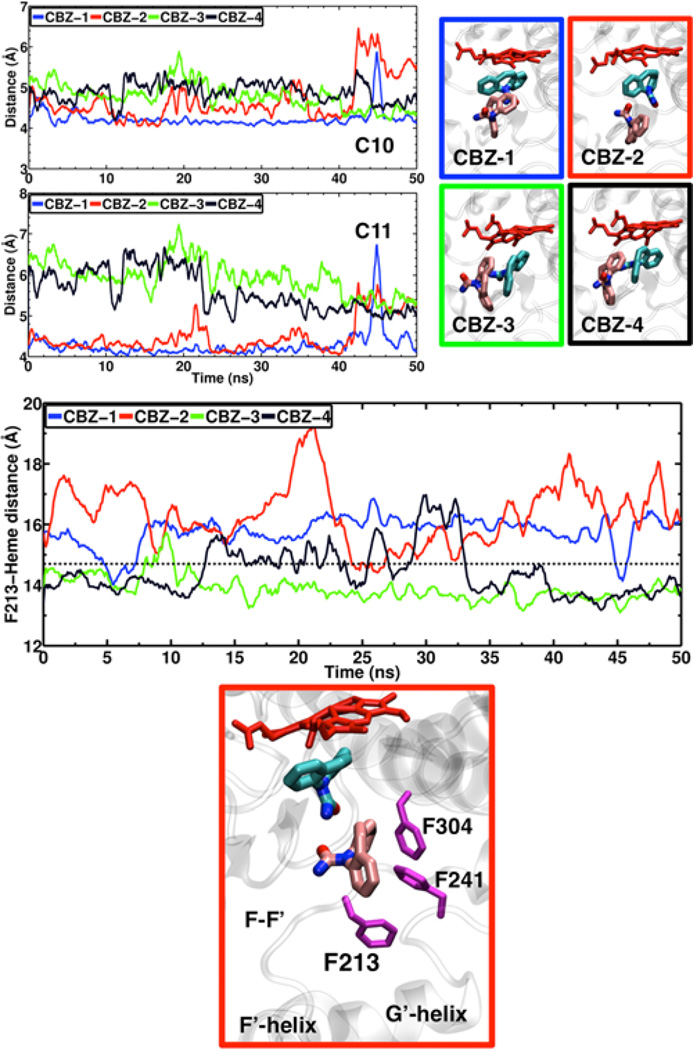
Starting from the snapshots obtained from single-CBZ simulations, and employing molecular docking, we have also generated membrane-bound models of CYP3A4 with two CBZ molecules in the active site. For this condition, we tested four different initial configurations. In these systems, the CBZ molecule that is closer to the heme remains in its position during the simulations, with C10 and C11-heme iron distances of ~5 Å (Figure 7). This observation suggests that the presence of a second CBZ molecule could favor the epoxidation of the first molecule by keeping it in the proximity of the heme. Moreover, the simulations reveal that some of the tested two-CBZ configurations favor the transient motion of Phe-213 out of the active site (Figure 7). This is particularly evident in system CBZ-2, where the distance between the Phe-213 side chain and the heme remains above the average of 14.7 Å observed in the substrate-free simulation. In this case, the Phe-213 side chain is displaced out of the active site, allowing the second CBZ molecule to occupy its position and contact Phe-241 and Phe-304. The observed displacement of Phe-213 in this case suggests that the binding of two or more substrates in the active site of CYP3A4 might be facilitated by the binding of another substrate, such as PGS in our case, to the peripheral binding site. Therefore, these simulations provide some structural insight into the role of the different substrates in the mechanism of cooperativity of CYP3A4.
Figure 7.
Two CBZ molecules in active site of CYP3A4. (Top right) Snapshots of the four different configurations of two CBZ molecules in the active site probed with MD simulations. CBZ molecules are shown in stick representation in cyan (first molecule) and pink (second molecule) colors. (Top left) Time series of the distance between carbons C10 and C11 of the first CBZ and the heme iron atom. (Middle) Time series of the COM distance between the Phe-213 side chain and the heme. The average distance for the 100 ns substrate-free simulation is 14.7 Å (dashed line). (Bottom) Snapshot showing a representative configuration from system CBZ-2 where Phe-213 is displaced out of the active site. Phe-213, Phe-241 and Phe-304 are shown in magenta stick representation.
Discussion
In this manuscript we provide a detailed analysis of heterotropic interactions between two substrates and monomeric CYP3A4 incorporated in POPC Nanodiscs. Using a reconstituted system with CPR we measured the steady-state rates of NADPH oxidation and product formation with CBZ and PGS as a function of substrate concentration both in homotropic (with individual substrates) and heterotropic (with substrate mixture at a constant molar ratio) titration experiments. In addition we performed Type I spin shift titration experiments with pure substrates and their mixture under identical conditions. Simultaneous analysis of multiple experimental data sets allows for deciphering the stepwise substrate binding constants and fractional contributions from the binding intermediates with one, two, or three bound substrate molecules to the overall experimentally measured metabolic parameters of CYP3A4 in Nanodiscs (results shown in Tables 1 and 2). The resolved parameters provide a detailed picture of cooperative properties of CYP3A4, similar to the mechanism of homotropic cooperativity with TST as a substrate, as described in the earlier work (24).
The direct manifestation of heterotropic interactions between two different substrates bound to one CYP3A4 monomer is determined by the contribution of three mixed intermediates, i.e., [11], [21], and [12] in Scheme 1 to the overall observed functional properties of the enzyme. This contribution depends both on the functional properties of these intermediates, as well as on their populations. The first factor depends on the probability of a particular substrate to occupy the productive site, which can be different for a given intermediate. For instance, in [11] one of the substrates may be predominantly bound near the heme iron, while another may be bound at the remote non-productive site. In addition, the rate of product formation depends on the efficiency of catalytic step, or coupling at this configuration, which also can vary significantly, even if both substrates occupy the productive site with the same probability. The second factor, the relative population of a given binding intermediate, depends on the corresponding stoichiometric binding constants, including possible allosteric or cooperative interactions between substrate molecules, and, obviously, on the absolute concentrations of both substrates. Clearly, if simultaneous binding of two types of substrates to one CYP3A4 molecule is strongly unfavorable, then the overall functional heterotropic effects will be low, because the relative population of the mixed intermediates will be also low, and their contribution will be small. In the opposite scenario, when the population of mixed intermediates is high due to a positive cooperative interaction between the two substrates, the behavior of the system will be determined by their functional properties, because they will dominate in the mixture. The experimental protocol and global analysis approach described in this study allow one to determine these binding constants and properties of the mixed binding intermediates and to understand the molecular mechanisms of heterotropic cooperativity in CYP3A4.
Comparison of the properties of homotropic binding intermediates [20] and [02] with those of [11] reveals a strong asymmetry in the latter case. While the rate of PGS hydroxylation is 14.7 min−1 at NADPH consumption rate of 388 min−1 for [20], it drops to 4 min−1 in [11] with NADPH consumption even faster (410 min−1) and yet no CBZ product is produced (Figure 3). This drop in the coupling ratio suggests a looser packing of both substrates when both substrates are in the binding cavity or a much weaker allosteric effect of CBZ on the PGS hydroxylation if CBZ binds at the remote non-productive binding site (Figure 5). Although no site-specific structural information is currently available to allow one to distinguish between these two cases, useful information can be obtained from molecular modeling. For example, flexible docking of CBZ and TST in CYP3A4 (26) suggested that the optimal positioning of TST in productive mode near the heme iron is consistent with CBZ positioning in the substrate binding pocket between TST and the G–G’ loop near Cys-239. This result provides a reasonable explanation for the observed preferential PGS hydroxylation in the [11] intermediate. In addition, CBZ was found to bind covalently inside the substrate binding cavity after prolonged incubation in functionally reconstituted mixture (26), with concomitant moderate inactivation of the enzyme with respect to TST hydroxylation. Analogously, PGS hydroxylation in the [11] intermediate is also slower than in [20] by a factor of 4 (Tables 1 and 2).
The reversal of substrate selectivity and activation of CBZ epoxidation observed in the intermediates [12] and [21] indicates that PGS binding at the allosteric site changes the shape of the active site and favors placement of CBZ in the vicinity of the heme iron. The absence of PGS hydroxylation products from these intermediates strongly suggests PGS binding at the remote, non-productive site, and minimal, if any at all, occupation of the heme active site. At the same time, the rate of CBZ epoxidation is 1.5–3.5 times faster in [12] and [21], highlighting the positive heterotropic effect of PGS on CBZ metabolism. NADPH oxidation rates measured for [12] and [21] are approximately the same, and both are 2.5 times faster than for [03], indicating essentially no changes in the coupling ratio due to the replacement of one or two CBZ molecules by PGS.
Dramatic acceleration of CBZ epoxidation in the presence of PGS is consistent with results reported by Houston and collaborators (4), where 2–6 fold faster CBZ metabolism by CYP3A4 in the presence of high excess PGS or other steroids was observed using human liver microsomes and human hepatocytes. Because the rate observed in the presence of 100 µM of PGS was directly compared to the rate of CBZ epoxidation observed at the same low concentration of this substrate, 5 µM (4), activation can be attributed to the intermediate [21], in which two PGS and one CBZ molecules are bound to CYP3A4. Our results for the state [21] with the CBZ metabolism rate of 11 min−1 and four-fold acceleration compared to the state [03] are in agreement with these earlier results (4, 19) and provide the specific molecular assignment of the main binding intermediate responsible for the acceleration of CBZ metabolism in the presence of steroids. Importantly, we also observe this significant heterotropic activation of CBZ epoxidation with PGS as an effector in monomeric CYP3A4 in Nanodiscs, which allows ruling out effects of oligomerization.
Overall, the results summarized in Table 2 show distinct patterns of metabolism of CBZ and PGS by CYP3A4 in the mixed intermediates, as compared to the homotropic experiments shown in Table 1. As discussed above, this difference is explained by preferential distribution of these two substrates between the productive and non-productive sites, while binding to the latter results in significant allosteric effects, as confirmed by significant activation of CBZ epoxidation in [21] and [12]. In both these intermediates the absence of the PGS product implies dominant binding of this substrate at non-productive sites. MD simulations reported in this study suggest a structural mechanism explaining the observed synergy between PGS, found to bind between F-F’ and G-G’ helices, and activation of CBZ metabolism. As seen in Figure 4, PGS binding at this peripheral site results in the movement of Phe-213 side chain out of the binding pocket and concomitant increase of the volume of the substrate binding cavity. Moreover, MD simulation of two CBZ molecules inside the substrate binding pocket (Figure 7) reveals the same outward movement of Phe-213 to provide the necessary space for the second CBZ molecule to bind. This striking similarity of conformational changes involving Phe-213 and its neighboring residues in response to PGS binding at the allosteric site and those involved in accommodating two CBZ molecules inside the substrate binding cavity in a productive position provides a possible molecular mechanism of allosteric communication between the remote and productive binding sites in CYP3A4.
The experimental and simulation results described herein provide a basis for the general picture of cooperative interactions between two different substrates in monomeric CYP3A4. Up to three substrate molecules of moderate size can bind simultaneously to one CYP3A4 molecule in the Nanodisc bilayer. However, at any given moment only one substrate molecule can be positioned at the nearest vicinity to the catalytically active iron-oxygen intermediate for the oxidative transformation. This position may be termed the productive site (P-site). The remote site which is not productive and spectroscopically silent at the spectral binding experiments was suggested to have allosteric properties (A-site), because substrate binding to this site can result in perturbation of the functional properties of cytochrome P450, such as substrate positioning, regiospecificity of catalysis (13, 65–67), rate of autoxidation (22), and overall coupling (24, 25). Based on the functional properties of site-specific mutants (8, 51, 68), the X-ray structure of PGS-bound CYP3A4 (36) and fluorescent probes of its location (10, 55, 69), the remote site is thought to be located at the surface of the protein in the region between the F-F’ and G-G’ loops at the membrane interface (58).
Many of the substrates can interact with both the productive and allosteric sites with or without pronounced cooperativity in binding. However, the observable properties, i.e., spectroscopic response and the product turnover rate, strongly depend on the specific positioning of the bound substrate. Based to differential binding affinities to the productive and remote allosteric sites, all substrates of CYP3A4 can be roughly separated into two classes, P-type and A-type. The presence of two substrate molecules inside the substrate binding pocket can place one near the heme iron for metabolism, and the second in an unfavorable position to interact with the iron-oxo intermediate itself while in direct steric contact with the first. Examples of the latter type of binding can be seen in the X-ray structures of CYP3A4 with ketoconazole (61) and analogues of ritonavir (62, 63), and also in P450eryF with androstendione (70), CYP158A2 with flavioline (71), and in CYP2B4 and CYP2B6 structures (72, 73). In addition, dramatic acceleration of hydroxylation of small hydrocarbons and benzene by CYP102A1 has been demonstrated in the presence of a “decoy” effector molecule of perfluorodecanoic acid, which filled the active site cavity and stabilized binding of small hydrocarbon substrates near the active iron-oxygen intermediate (74, 75). Cooperativity seems to be a much more common property for cytochromes P450 than it was realized in the past. Biochemical analyses have indicated homotropic cooperativity in substrate binding by CYP392E10 from spider mite Tetranychus urticae (76) and heterotropic cooperativity in DDT metabolism by the insect P450 (CYP6M2 in A. gambiae) (77), similarly to what is reported for CYP3A4 and other human cytochromes P450, such as CYP11A1(78) and CYP46A1 (79).
In the mixture of A-type and P-type substrates at a 1:1 ratio, normalized to the concentration of each needed for half-maximal spectral response or activity, the actual sites occupied by the two different substrates are likely to be asymmetric, with an A-type substrate showing preference for A-site association, and P-type substrate preferentially binding inside the substrate binding cavity. The magnitude of such competition for the active site between P-type and A-type substrates will also depend on the details of their mutual packing in the substrate binding pocket and the preferential positioning of one close to the iron-oxygen catalytic intermediate.
The presence of an allosteric site at the membrane interface suggests a potentially significant variation between experimentally determined drug-drug interactions with P450 enzymes when either incorporated in the lipid bilayer or in detergent solubilized systems (78). This may be an important factor for the proper choice of an in vitro system for P450 activity studies in reconstituted systems. For evaluation of potential drug-drug interactions between two or more substrates, the presence of the bilayer may be critically important. For analysis of unknown mixtures of several P450s using substrate cocktails (80) the results may turn out different when compared to a single P450 tested using only a single substrate, as other compounds may also serve as allosteric effectors. In cases where there are pronounced interactions between P450 monomers resulting in formation of P450 oligomers, it has been suggested that this protein – protein interface may involve the allosteric A-site (10). If correct, interactions with A-type substrates could alter the monomer-oligomer equilibrium and the functional properties of oligomers. A role of oligomerization in observed cooperativity of CYP3A4 was reviewed by Davydov and Halpert (81, 82) and updated in their recent papers (10, 57). As these authors pointed out (10, 57), a significant fraction of P450 enzymes present in oligomers cannot interact with CPR and therefore may be not functional. Interactions with substrates and effectors can result in redistribution of cytochromes P450 between functional and non-functional forms and thus play an important regulatory role. Further elucidations of mechanistic behavior of oligomeric CYP3A4 and other P450 enzymes outlined in (10, 57) need to be based on the detailed understanding of modulation of the functional properties of monomeric CYP3A4 provided by oligomerization. Our results clearly demonstrate that even monomeric CYP3A4 in a membrane can display complex activation and allosteric responses.
In conclusion, we describe the results of experimental and computational studies of heterotropic interactions between two substrates of CYP3A4. Different properties of PGS and CBZ with respect to their binding affinity to productive and non-productive allosteric sites give rise to strongly asymmetric mutual perturbation of their metabolic turnover catalyzed by monomeric CYP3A4, with strong activation of CBZ epoxidation and simultaneous inhibition of PGS hydroxylation in the mixed titration experiments. Using a global analysis, we have deconvoluted the partial contributions of the mixed binding intermediates into the overall observed rates of product formation and found a 1.5–3.5 fold acceleration of CBZ metabolism upon binding of one or two PGS molecules to the same CYP3A4 monomer. Importantly, our results obtained using CYP3A4 monomer in Nanodiscs are in very good agreement with the earlier results of Houston et al. (4, 52) obtained with the same substrates using human liver microsomes and human hepatocytes, despite recent claim that such heterotropic activation will not be reproducible in monomeric systems (10). Results of MD simulations of PGS bound in the suggested allosteric remote site and one or two CBZ molecules bound inside the substrate binding cavity of CYP3A4 provide detailed insight into the possible conformational changes caused by these binding events. These conformational changes involving the side chains of several phenylalanine residues at the F-F’ and G-G’ loops are likely to give rise to the allosteric interactions between PGS binding outside of the binding cavity and CBZ binding inside the substrate binding pocket and explain the positive heterotropic cooperativity observed experimentally. Such a mechanism is expected to manifest itself with other substrates metabolized by CYP3A4, especially if their relative binding affinities with respect to the allosteric and to productive sites are significantly different.
Moreover, a similar mechanism may also apply to other membrane-bound eukaryotic cytochromes P450s, especially those with the broad substrate specificities, because of the general similarity of their protein folds (83–85) and/or of their positioning in the membrane (58), so that the F-F’ and G-G’ region is contacting the membrane surface and forms a pocket potentially suitable for accommodation of medium-sized hydrophobic molecules, such as endogenous steroids.
Three site models for CYP3A4 have been suggested previously (27, 28) in order to account for the rich variety of heterotropic effects observed in biochemical results before the solution of the first X-ray structure of a eukaryotic cytochrome P450 and without any knowledge on the mode of incorporation of P450 into the membrane. We now have a more detailed picture of the possible modes of interactions of human cytochromes P450s with the membrane based on a large array of biophysical data and computational modeling (33, 58, 83, 86). However, the high resolution structural information on the possible substrate binding sites for cytochromes P450 incorporated in the membrane is still not available. Current efforts to identify the location of allosteric site(s) and possible molecular mechanisms of cooperative interactions between non-productive remote binding events and productive substrate binding to CYP3A4 based on combination of biochemical and biophysical data with computational modeling suggest the mechanism involving several residues at the F-F’ and G-G’ loops, as illustrated in Figures 4 and 7, in agreement with earlier mutational studies (8, 51, 68). Further studies employing site-specific structural methods together with specific mutations probing this site will bring better understanding of this mechanism in CYP3A4 and possibly other drug metabolizing cytochromes P450s.
Acknowledgments
Funding Source Statement:
This work was supported by National Institutes of Health, grant R01-GM33775 to S. G. S., grant R01-GM101048 to S.G.S. and E.T., and grants R01-GM086749, U54-GM087519, and P41-GM104601 to E.T.. All simulations have been performed using XSEDE resources (grant number MCA06N060).
Abbreviations
- ANF
α-naphthoflavone
- CBZ
carbamazepine
- COM
center of mass
- CPR
cytochrome P450 reductase
- MD
molecular dynamics
- MSP
membrane scaffold protein
- PGS
progesterone
- RMSD
root mean square deviation
- TST
testosterone
References
- 1.Evers R, Dallas S, Dickmann LJ, Fahmi OA, Kenny JR, Kraynov E, Nguyen T, Patel AH, Slatter JG, Zhang L. Critical review of preclinical approaches to investigate cytochrome p450-mediated therapeutic protein drug-drug interactions and recommendations for best practices: a white paper. Drug Metab Dispos. 2013;41:1598–1609. doi: 10.1124/dmd.113.052225. [DOI] [PubMed] [Google Scholar]
- 2.Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Activation of CYP3A4: evidence for the simultaneous binding of two substrates in a cytochrome P450 active site. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 3.Ngui JS, Chen Q, Shou M, Wang RW, Stearns RA, Baillie TA, Tang W. In vitro stimulation of warfarin metabolism by quinidine: increases in the formation of 4'- and 10-hydroxywarfarin. Drug Metab Dispos. 2001;29:877–886. [PubMed] [Google Scholar]
- 4.Henshall J, Galetin A, Harrison A, Houston JB. Comparative analysis of CYP3A heteroactivation by steroid hormones and flavonoids in different in vitro systems and potential in vivo implications. Drug Metab Dispos. 2008;36:1332–1340. doi: 10.1124/dmd.108.021279. [DOI] [PubMed] [Google Scholar]
- 5.Blobaum AL, Bridges TM, Byers FW, Turlington ML, Mattmann ME, Morrison RD, Mackie C, Lavreysen H, Bartolome JM, MacDonald GJ, Steckler T, Jones CK, Niswender CM, Conn PJ, Lindsley CW, Stauffer SR, Daniels JS. Heterotropic activation of the midazolam hydroxylase activity of CYP3A by a positive allosteric modulator of mGlu5: in vitro to in vivo translation and potential impact on clinically relevant drug-drug interactions. Drug Metab. Dispos. 2013;41:2066–2075. doi: 10.1124/dmd.113.052662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emoto C, Iwasaki K. Enzymic characteristics of CYP3A5 and CYP3A4: A comparison of in vitro kinetic and drug-drug interaction patterns. Xenobiotica. 2006;36:219–233. doi: 10.1080/00498250500489968. [DOI] [PubMed] [Google Scholar]
- 7.Shou M, Dai R, Cui D, Korzekwa KR, Baillie TA, Rushmore TH. A kinetic model for the metabolic interaction of two substrates at the active site of cytochrome P450 3A4. J. Biol. Chem. 2001;276:2256–2262. doi: 10.1074/jbc.M008799200. [DOI] [PubMed] [Google Scholar]
- 8.Domanski TL, He Y-A, Khan KK, Roussel F, Wang Q, Halpert JR. Phenylalanine and Tryptophan Scanning Mutagenesis of CYP3A4 Substrate Recognition Site Residues and Effect on Substrate Oxidation and Cooperativity. Biochemistry. 2001;40:10150–10160. doi: 10.1021/bi010758a. [DOI] [PubMed] [Google Scholar]
- 9.Woods CM, Fernandez C, Kunze KL, Atkins WM. Allosteric activation of cytochrome P450 3A4 by alpha-naphthoflavone: branch point regulation revealed by isotope dilution analysis. Biochemistry. 2011;50:10041–10051. doi: 10.1021/bi2013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davydov DR, Davydova NY, Sineva EV, Kufareva I, Halpert JR. Pivotal role of P450-P450 interactions in CYP3A4 allostery: the case of alpha-naphthoflavone. Biochem. J. 2013;453:219–230. doi: 10.1042/BJ20130398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walzer M, Bekersky I, Blum RA, Tolbert D. Pharmacokinetic drug interactions between clobazam and drugs metabolized by cytochrome P450 isoenzymes. Pharmacotherapy. 2012;32:340–353. doi: 10.1002/j.1875-9114.2012.01028.x. [DOI] [PubMed] [Google Scholar]
- 12.Parrish RH, Pazdur DE, O'Donnell PJ. Effect of carbamazepine initiation and discontinuation on antithrombotic control in a patient receiving warfarin: case report and review of the literature. Pharmacotherapy. 2006;26:1650–1653. doi: 10.1592/phco.26.11.1650. [DOI] [PubMed] [Google Scholar]
- 13.Roberts AG, Yang J, Halpert JR, Nelson SD, Thummel KT, Atkins WM. The structural basis for homotropic and heterotropic cooperativity of midazolam metabolism by human cytochrome P450 3A4. Biochemistry. 2011;50:10804–10818. doi: 10.1021/bi200924t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konishi H, Tanaka K, Minouchi T, Yamaji A. Urinary 6beta-hydroxycortisol/17-hydroxycorticosteroids ratio as a measure of hepatic CYP3A4 capacity after enzyme induction. Ann Clin Biochem. 2004;41:335–337. doi: 10.1258/0004563041201527. [DOI] [PubMed] [Google Scholar]
- 15.Ohno Y, Hisaka A, Ueno M, Suzuki H. General framework for the prediction of oral drug interactions caused by CYP3A4 induction from in vivo information. Clin Pharmacokinet. 2008;47:669–680. doi: 10.2165/00003088-200847100-00004. [DOI] [PubMed] [Google Scholar]
- 16.Spina E, Pisani F, Perucca E. Clinically significant pharmacokinetic drug interactions with carbamazepine. An update. Clin Pharmacokinet. 1996;31:198–214. doi: 10.2165/00003088-199631030-00004. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura H, Torimoto N, Ishii I, Ariyoshi N, Nakasa H, Ohmori S, Kitada M. CYP3A4 and CYP3A7-mediated carbamazepine 10,11-epoxidation are activated by differential endogenous steroids. Drug Metab. Dispos. 2003;31:432–438. doi: 10.1124/dmd.31.4.432. [DOI] [PubMed] [Google Scholar]
- 18.Kerr BM, Thummel KE, Wurden CJ, Klein SM, Kroetz DL, Gonzalez FJ, Levy RH. Human liver carbamazepine metabolism. Role of CYP3A4 and CYP2C8 in 10,11-epoxide formation. Biochem Pharmacol. 1994;47:1969–1979. doi: 10.1016/0006-2952(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 19.Egnell A-C, Houston JB, Boyer CS. Predictive models of CYP3A4 heteroactivation: in vitro-in vivo scaling and pharmacophore modeling. J. Pharmacol. Exp. Ther. 2005;312:926–937. doi: 10.1124/jpet.104.078519. [DOI] [PubMed] [Google Scholar]
- 20.Denisov IG, Grinkova YV, Baas BJ, Sligar SG. The ferrous-dioxygen intermediate in human cytochrome P450 3A4: Substrate dependence of formation of decay kinetics. J. Biol. Chem. 2006;281:23313–23318. doi: 10.1074/jbc.M605511200. [DOI] [PubMed] [Google Scholar]
- 21.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 22.Denisov IG, Grinkova YV, McLean MA, Sligar SG. The one-electron autoxidation of human cytochrome P450 3A4. J. Biol. Chem. 2007;282:26865–26873. doi: 10.1074/jbc.M704747200. [DOI] [PubMed] [Google Scholar]
- 23.Grinkova YV, Denisov IG, Sligar SG. Functional reconstitution of monomeric CYP3A4 with multiple cytochrome P450 reductase molecules in Nanodiscs. Biochem. Biophys. Res. Commun. 2010;398:194–198. doi: 10.1016/j.bbrc.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: Linkages in substrate binding, spin state, uncoupling, and product formation. J. Biol. Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 25.Frank DJ, Denisov IG, Sligar SG. Analysis of heterotropic cooperativity in cytochrome P450 3A4 using alpha-naphthoflavone and testosterone. J Biol Chem. 2011;286:5540–5545. doi: 10.1074/jbc.M110.182055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang P, Liao M, Wester MR, Leeder JS, Pearce RE, Correia MA. CYP3A4-mediated carbamazepine (CBZ) metabolism: formation of a covalent CBZ-CYP3A4 adduct and alteration of the enzyme kinetic profile. Drug Metab. Dispos. 2008;36:490–499. doi: 10.1124/dmd.107.016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domanski TL, He Y-A, Harlow GR, Halpert JR. Dual role of human cytochrome P450 3A4 residue Phe-304 in substrate specificity and cooperativity. J. Pharmacol. Exp. Ther. 2000;293:585–591. [PubMed] [Google Scholar]
- 28.Hosea NA, Miller GP, Guengerich FP. Elucidation of Distinct Ligand Binding Sites for Cytochrome P450 3A4. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 29.Muller CS, Knehans T, Davydov DR, Bounds PL, von Mandach U, Halpert JR, Caflish A, Koppenol WH. Concurrent Cooperativity and Substrate Inhibition in the Epoxidation of Carbamazepine by Cytochrome P450 3A4 Active Site Mutants Inspired by Molecular Dynamics Simulations. Biochemistry. 2015;54:711–721. doi: 10.1021/bi5011656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Cera E. Thermodynamic Theory of Site-Specific Binding Processes in Biological Macromolecules. Cambridge: Cambridge University Press; 1995. [Google Scholar]
- 31.Frank DJ, Denisov IG, Sligar SG. Mixing apples and oranges: Analysis of heterotropic cooperativity in cytochrome P450 3A4. Arch. Biochem. Biophys. 2009;488:146–152. doi: 10.1016/j.abb.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J. Biol. Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 33.Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J Am Chem Soc. 2013;135:8542–8551. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 35.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock and AutoDockTools: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 37.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 39.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fishelovitch D, Shaik S, Wolfson HJ, Nussinov R. Theoretical Characterization of Substrate Access/Exit Channels in the Human Cytochrome P450 3A4 Enzyme: Involvement of Phenylalanine Residues in the Gating Mechanism. J. Phys. Chem. B. 2009;113:13018–13025. doi: 10.1021/jp810386z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim JB, Rogaski B, Klauda JB. Update of the Cholesterol Force Field Parameters in CHARMM. J. Phys. Chem. B. 2012;116:203–210. doi: 10.1021/jp207925m. [DOI] [PubMed] [Google Scholar]
- 42.Vanommeslaeghe K, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanommeslaeghe K, Raman EP, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J. Chem. Inf. Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. Rapid parameterization of small molecules using the force field toolkit. J. Comput. Chem. 2013;34:2757–2770. doi: 10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VDM: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. plates, 27–28. [DOI] [PubMed] [Google Scholar]
- 46.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 47.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 48.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 49.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 50.Darden T, York D, Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 51.Harlow GR, Halpert JR. Analysis of human cytochrome P450 3A4 cooperativity: construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Egnell A-C, Houston B, Boyer S. In vivo CYP3A4 heteroactivation is a possible mechanism for the drug interaction between felbamate and carbamazepine. J. Pharmacol. Exp. Ther. 2003;305:1251–1262. doi: 10.1124/jpet.102.047530. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura H, Nakasa H, Ishii I, Ariyoshi N, Igarashi T, Ohmori S, Kitada M. Effects of endogenous steroids on CYP3A4-mediated drug metabolism by human liver microsomes. Drug Metab Dispos. 2002;30:534–540. doi: 10.1124/dmd.30.5.534. [DOI] [PubMed] [Google Scholar]
- 54.Maekawa K, Yoshimura T, Saito Y, Fujimura Y, Aohara F, Emoto C, Iwasaki K, Hanioka N, Narimatsu S, Niwa T, Sawada J. Functional characterization of CYP3A4.16: catalytic activities toward midazolam and carbamazepine. Xenobiotica. 2009;39:140–147. doi: 10.1080/00498250802617746. [DOI] [PubMed] [Google Scholar]
- 55.Davydov DR, Rumfeldt JA, Sineva EV, Fernando H, Davydova NY, Halpert JR. Peripheral ligand-binding site in cytochrome P450 3A4 located with fluorescence resonance energy transfer (FRET) J. Biol. Chem. 2012;287:6797–6809. doi: 10.1074/jbc.M111.325654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Denisov IG, Sligar SG. A novel type of allosteric regulation: functional cooperativity in monomeric proteins. Arch Biochem Biophys. 2012;519:91–102. doi: 10.1016/j.abb.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davydov DR, Davydova NY, Sineva EV, Halpert JR. Interactions among Cytochromes P450 in Microsomal Membranes: Oligomerization of Cytochromes P450 3A4, 3A5 and 2E1 and its Functional Consequences. J Biol Chem. 2015;290:3850–3864. doi: 10.1074/jbc.M114.615443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berka K, Paloncyova M, Anzenbacher P, Otyepka M. Behavior of Human Cytochromes P450 on Lipid Membranes. J Phys Chem B. 2013;117:11556–11564. doi: 10.1021/jp4059559. [DOI] [PubMed] [Google Scholar]
- 59.Vijayan R, Biggin PC. A steroid in a lipid bilayer: localization, orientation, and energetics. Biophys. J. 2008;95:L45–L47. doi: 10.1529/biophysj.108.140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernando H, Rumfeldt JA, Davydova NY, Halpert JR, Davydov DR. Multiple substrate-binding sites are retained in cytochrome P450 3A4 mutants with decreased cooperativity. Xenobiotica. 2011;41:281–289. doi: 10.3109/00498254.2010.538748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ekroos M, Sjoegren T. Structural basis for ligand promiscuity in cytochrome P 450 3A4. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sevrioukova IF, Poulos TL. Dissecting cytochrome P450 3A4-ligand interactions using ritonavir analogues. Biochemistry. 2013;52:4474–4481. doi: 10.1021/bi4005396. [DOI] [PubMed] [Google Scholar]
- 63.Sevrioukova IF, Poulos TL. Ritonavir analogues as a probe for deciphering the cytochrome P450 3A4 inhibitory mechanism. Curr Top Med Chem. 2014;14:1348–1355. doi: 10.2174/1568026614666140506120647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuki H, Honma T, Hata M, Hoshino T. Prediction of sites of metabolism in a substrate molecule, instanced by carbamazepine oxidation by CYP3A4. Bioorg. Med. Chem. 2012;20:775–783. doi: 10.1016/j.bmc.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Jiang Y, Ortiz de Montellano PR. Cooperative effects on radical recombination in CYP3A4-catalyzed oxidation of the radical clock beta-thujone. Chembiochem. 2009;10:650–653. doi: 10.1002/cbic.200800772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Atkins WM, Isoherranen N, Paine MF, Thummel KE. Evidence of CYP3A allosterism in vivo: analysis of interaction between fluconazole and midazolam. Clin Pharmacol Ther. 2012;91:442–449. doi: 10.1038/clpt.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrelson JP, Atkins WM, Nelson SD. Multiple-ligand binding in CYP2A6: probing mechanisms of cytochrome P450 cooperativity by assessing substrate dynamics. Biochemistry. 2008;47:2978–2988. doi: 10.1021/bi702020y. [DOI] [PubMed] [Google Scholar]
- 68.Harlow GR, Halpert JR. Alanine-scanning mutagenesis of a putative substrate recognition site in human cytochrome P450 3A4. Role of residues 210 and 211 in flavonoid activation and substrate specificity. J. Biol. Chem. 1997;272:5396–5402. doi: 10.1074/jbc.272.9.5396. [DOI] [PubMed] [Google Scholar]
- 69.Tsalkova TN, Davydova NY, Halpert JR, Davydov DR. Mechanism of Interactions of alpha -Naphthoflavone with Cytochrome P450 3A4 Explored with an Engineered Enzyme Bearing a Fluorescent Probe. Biochemistry. 2007;46:106–119. doi: 10.1021/bi061944p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cupp-Vickery J, Anderson R, Hatziris Z. Crystal structures of ligand complexes of P450eryF exhibiting homotropic cooperativity. Proc. Natl. Acad. Sci. USA. 2000;97:3050–3055. doi: 10.1073/pnas.050406897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR. Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J. Biol. Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 72.Jang HH, Davydov DR, Lee GY, Yun CH, Halpert JR. The role of cytochrome P450 2b6 and 2b4 substrate access channel residues predicted based on crystal structures of the amlodipine complexes. Arch Biochem Biophys. 2014;545:100–107. doi: 10.1016/j.abb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shah MB, Wilderman PR, Pascual J, Zhang Q, Stout CD, Halpert JR. Conformational adaptation of human cytochrome P450 2B6 and rabbit cytochrome P450 2B4 revealed upon binding multiple amlodipine molecules. Biochemistry. 2012;51:7225–7238. doi: 10.1021/bi300894z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shoji O, Kunimatsu T, Kawakami N, Watanabe Y. Highly selective hydroxylation of benzene to phenol by wild-type cytochrome P450BM3 assisted by decoy molecules. Angew. Chem. Int. Ed. 2013;52:6606–6610. doi: 10.1002/anie.201300282. [DOI] [PubMed] [Google Scholar]
- 75.Kawakami N, Shoji O, Watanabe Y. Direct hydroxylation of primary carbons in small alkanes by wild-type cytochrome P450BM3 containing perfluorocarboxylic acids as decoy molecules. Chem. Sci. 2013;4:2344–2348. [Google Scholar]
- 76.Demaeght P, Dermauw W, Tsakireli D, Khajehali J, Nauen R, Tirry L, Vontas J, Lummen P, Van Leeuwen T. Molecular analysis of resistance to acaricidal spirocyclic tetronic acids in Tetranychus urticae: CYP392E10 metabolizes spirodiclofen, but not its corresponding enol. Insect Biochem Mol Biol. 2013;43:544–554. doi: 10.1016/j.ibmb.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 77.Mitchell SN, Stevenson BJ, Muller P, Wilding CS, Egyir-Yawson A, Field SG, Hemingway J, Paine MJ, Ranson H, Donnelly MJ. Identification and validation of a gene causing cross-resistance between insecticide classes in Anopheles gambiae from Ghana. Proc Natl Acad Sci U S A. 2012;109:6147–6152. doi: 10.1073/pnas.1203452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mast N, Linger M, Pikuleva IA. Inhibition and stimulation of activity of purified recombinant CYP11A1 by therapeutic agents. Mol. Cell. Endocrinol. 2013;371:100–106. doi: 10.1016/j.mce.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mast N, Li Y, Linger M, Clark M, Wiseman J, Pikuleva IA. Pharmacologic Stimulation of Cytochrome P450 46A1 and Cerebral Cholesterol Turnover in Mice. J. Biol. Chem. 2014;289:3529–3538. doi: 10.1074/jbc.M113.532846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dixit V, Hariparsad N, Desai P, Unadkat JD. In vitro LC-MS cocktail assays to simultaneously determine human cytochrome P450 activities. Biopharm Drug Dispos. 2007;28:257–262. doi: 10.1002/bdd.552. [DOI] [PubMed] [Google Scholar]
- 81.Davydov DR. Microsomal monooxygenase as a multienzyme system: the role of P450-P450 interactions. Expert Opin. Drug Metab. Toxicol. 2011;7:543–558. doi: 10.1517/17425255.2011.562194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davydov DR, Halpert JR. Allosteric P450 mechanisms: multiple binding sites, multiple conformers or both? Expert Opin Drug Metab Toxicol. 2008;4:1523–1535. doi: 10.1517/17425250802500028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Denisov IG, Shih AY, Sligar SG. Structural differences between soluble and membrane bound cytochrome P450s. J Inorg Biochem. 2012;108:150–158. doi: 10.1016/j.jinorgbio.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnson EF, Stout CD. Structural diversity of eukaryotic membrane cytochrome P450s. J. Biol. Chem. 2013;288:17082–17090. doi: 10.1074/jbc.R113.452805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poulos TL, Johnson EF. Structures of Cytochrome P450 Enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Function, Genetics. 3rd. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 87–114. [Google Scholar]
- 86.Mast N, Liao WL, Pikuleva IA, Turko IV. Combined use of mass spectrometry and heterologous expression for identification of membrane-interacting peptides in cytochrome P450 46A1 and NADPH-cytochrome P450 oxidoreductase. Arch Biochem Biophys. 2009;483:81–89. doi: 10.1016/j.abb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]