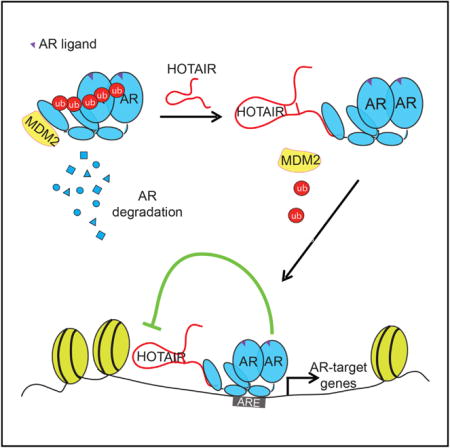
SUMMARY
Understanding the mechanisms of androgen receptor (AR) activation in the milieu of low androgen is critical to effective treatment of castration-resistant prostate cancer (CRPC). Here, we report HOTAIR as an androgen-repressed lncRNA, and, as such, it is markedly upregulated following androgen deprivation therapies and in CRPC. We further demonstrate a distinct mode of lncRNA-mediated gene regulation, wherein HOTAIR binds to the AR protein to block its interaction with the E3 ubiquitin ligase MDM2, thereby preventing AR ubiquitination and protein degradation. Consequently, HOTAIR expression is sufficient to induce androgen-independent AR activation and drive the AR-mediated transcriptional program in the absence of androgen. Functionally, HOTAIR overexpression increases, whereas HOTAIR knockdown decreases, prostate cancer cell growth and invasion. Taken together, our results provide compelling evidence of lncRNAs as drivers of androgen-independent AR activity and CRPC progression, and they support the potential of lncRNAs as therapeutic targets.
Graphical abstract
INTRODUCTION
Long noncoding RNAs (lncRNAs) are RNA transcripts that are more than 200 nt long but have little protein-coding potential. Within the last few years, thousands of lncRNAs have been identified and their function in biological processes begun to be understood (Iyer et al., 2015; Rinn and Chang, 2012). For example, lncRNA-p21 has been shown to act in trans or in cis to regulate target gene expression (Dimitrova et al., 2014). LncRNAs also regulate chromatin states by association with chromatin-modifying proteins, such as the polycomb repressive complexes (PRC2) (Gupta et al., 2010a; Khalil et al., 2009). The lncRNA HOTAIR has been shown to bind PRC2 through its 5′ region but to bind the LSD1/REST complex with its 3′ domain, thereby serving as scaffolds that coordinate co-targeting of these proteins to selected chromatin regions (Tsai et al., 2010). In addition, emerging evidence suggests that lncRNAs are able to regulate cellular processes through post-translational mechanisms. For instance, lncRNA FAL1 associates with BMI to regulate its stability and target gene transcription (Hu et al., 2014), whereas lncRNA HOTAIR interacts with E3 ubiquitin ligases through their RNA-binding domains to accelerate substrate ubiquitination and protein degradation (Yoon et al., 2013). Through these regulatory mechanisms, lncRNAs are believed to play critical roles in human development and diseases.
Prostate cancer (PCa) is a frequently diagnosed disease in American men. Androgen ablation therapies (ADTs) are the mainline treatment for patients with advanced PCa. Despite the high initial response rates, remissions following ADT are temporary due to the emergence of castration-resistant prostate cancer (CRPC). Studies of CRPC have revealed that resumption of androgen receptor (AR)-dependent transcriptional activity is a critical event in nearly all cases (Grasso et al., 2012). Identifying new molecular mechanisms underlying aberrant AR activation in the milieu of low androgen holds great promise to improve the treatment of CRPC. Integrative genomic analysis of microarray datasets has revealed many lncRNAs that were dys-regulated in various cancer types when compared to corresponding benign samples (Du et al., 2013). Transcriptome sequencing across PCa cohorts has identified a list of lncRNAs that stratify benign, localized, and metastatic PCa samples (Iyer et al., 2015; Prensner et al., 2011). These cancer-expressed lncRNAs, like protein-coding genes, may serve as markers of the disease and may play essential roles in cancer progression. In particular, SChLAP1, an lncRNA that is overexpressed in a subset of PCa, has been shown to promote PCa invasiveness and tumor metastasis by antagonizing the tumor-suppressive SWI/SNF chromatin-modifying complex (Prensner et al., 2013). Yang et al. (2013) reported two lncRNAs, PRNCR1 and PCGEM1, that are highly overexpressed in aggressive PCa and bind successively to the AR protein to enhance the AR-mediated gene activation program and induce PCa growth. Yet, a subsequent study reported conflicting results (Prensner et al., 2014). Thus, lncRNAs that interfere with the critical AR pathway remain to be characterized.
HOTAIR, a 2.2-kb-long transcript localized to the boundaries of the HOXC gene cluster, is highly expressed and correlated with metastasis in a variety of cancer types, including breast (Gupta et al., 2010a; Sørensen et al., 2013), colorectal (Kogo et al., 2011), lung (Nakagawa et al., 2013), and pancreatic (Kim et al., 2013) cancers. HOTAIR promotes breast cancer metastasis at least in part by associating with the PRC2 complex to induce genome-wide retargeting of PRC2 to hundreds of genes involved in tumor metastasis (Gupta et al., 2010a). Integrative genomic analyses have revealed HOTAIR among hundreds of lncRNAs that are deregulated in various cancers (Du et al., 2013). HOTAIR, however, has not been well characterized in PCa. Through lncRNA profiling, a recent study showed that HOTAIR was inhibited by genistein, a phytoestrogen, and may have tumor-suppressive roles (Chiyomaru et al., 2013). This study, however, was carried out solely in the AR-negative DU145 and PC3 cell lines that do not represent clinical CRPC. In our study, we investigated whether the lncRNA HOTAIR interacts with the AR protein, modulates the AR-mediated transcriptional program, and contributes to CRPC.
RESULTS
HOTAIR Is Repressed by Androgen and Upregulated in CRPC
To identify lncRNAs that are potentially regulated by AR, we re-analyzed chromatin immunoprecipitation sequencing (ChIP-seq) data that we previously published in lymph node carcinoma of prostate (LNCaP) cells (Yu et al., 2010). Of 8,403 lncRNAs annotated in Ensemble, 3,418 contained at least one AR-binding event within 50 kb upstream of intragenic regions (Figure 1A; Table S1). Among these, we nominated HOTAIR, an lncRNA that has been studied in multiple cancer types, but has not been well characterized in PCa, for further investigation. ChIP-seq showed three strong AR-binding events at 46 kb upstream and 66 and 137 kb downstream of the HOTAIR gene (Figure S1A). As AR is known to regulate target genes through strong binding to enhancers and weak binding at promoters mediated by chromatin looping, we asked whether AR binds to the HOTAIR promoter. Motif analysis of the HOTAIR promoter revealed three highly scored androgen response element (ARE) motifs (Figure S1B). We then designed three primer pairs flanking these three AREs as well as a primer set targeting a distal region as a negative control. ChIP-PCR confirmed strong enrichment of known AR target enhancers of PSA, TMPRSS2, FKBP5, as well as the HOTAIR promoter, but not the distal region, by AR antibody relative to the IgG control (Figure 1B). Moreover, ChIP-qPCR in LNCaP cells demonstrated that androgen greatly increased AR binding to the HOTAIR promoter as well as known AR target enhancers, but not to the distal HOTAIR region (Figure S1C).
Figure 1. HOTAIR Is Repressed by Androgen and Is Thus Upregulated in CRPC.
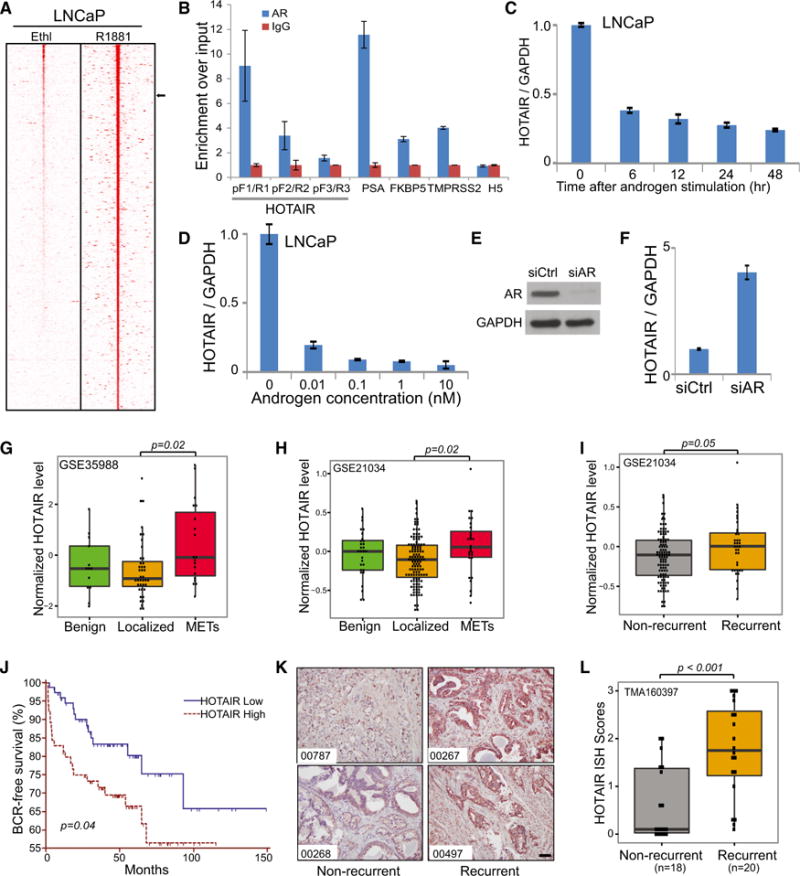
(A) Heatmap showing AR ChIP-seq reads around lncRNAs. Previously published AR ChIP-seq data from ethanol or R1881 (10 nM)-stimulated LNCaP cells were re-analyzed for enrichment around lncRNAs (Table S1). Normalized AR ChIP-seq reads around (±50 kb) lncRNAs are shown. Black arrow on the right denotes HOTAIR.
(B) AR occupies the HOTAIR promoter. AR and IgG ChIP were performed in LNCaP cells and ChIP-qPCR was carried out using primers flanking the HOTAIR promoter regions (see Figure S1B for detail), well-established AR target gene enhancers, and the 3′ end of the HOTAIR gene as a negative control (H5).
(C and D) Androgen inhibits HOTAIR expression. The qRT-PCR analysis of HOTAIR expression in LNCaP cells, stimulated either by 1 nM androgen R1881 over a time course (C) or by different doses of R1881 (D) for 48 hr, is shown. Data were normalized to GAPDH. Data shown are mean ± SEM and representative of at least two independent experiments.
(E and F) AR depletion restores HOTAIR expression. LNCaP cells were treated with control and AR-targeting small interfering RNA (siRNA), confirmed for AR knockdown (E) and subjected to qRT-PCR analysis of HOTAIR expression (F).
(G–I) HOTAIR is upregulated in metastatic prostate tumors. HOTAIR expression in publicly available datasets (GEO: GSE35988 and GSE21034) was comparatively plotted in benign prostate, localized PCa, and metastatic PCa (G and H) or in localized PCa with or without biochemical recurrence (I). Significant p values are indicated.
(J) High HOTAIR level is associated with poor clinical outcome. Kaplan-Meier analysis of PCa outcome using the GEO: GSE21034 dataset is shown. Localized PCa cases were stratified based on their HOTAIR expression level and analyzed for biochemical recurrence. The p values for Kaplan-Meier curves were determined using a log-rank test.
(K and L) The lncRNA ISH was performed using a tissue microarray of recurrent versus non-recurrent PCa. ISH HOTAIR staining in two representative cases of recurrent and non-recurrent PCa is shown (K). Scale bar, 100 μm. HOTAIR ISH staining intensity was scored in all cases, compared between two groups (p < 0.001 by t test), and visualized by boxplot (L).
Next, we asked whether AR binding at HOTAIR regulatory regions results in modulation of gene expression. Our qRT-PCR analysis of LNCaP cells stimulated with R1881 showed that HOTAIR expression is highly sensitive to androgen; HOTAIR was inhibited by androgen rapidly and significantly up to 10-fold, while PSA showed a 14-fold increase (Figures 1C, 1D, and S1D). A similar decrease in HOTAIR expression following androgen treatment also was observed in another independent PCa cell line, vertebral cancer of prostate (VCaP) (Figures S1E and S1F). To demonstrate that this regulation is mediated by AR, we carried out AR knockdown through RNA interference and found that HOTAIR expression is markedly restored in AR-depleted LNCaP cells (Figures 1E and 1F). To further support that androgen inhibits HOTAIR through modulation of active transcription, we performed RNA PolII ChIP-qPCR and indeed observed a decrease in RNA PolII occupancy at HOTAIR and several known AR-repressed genes, but an increase at AR-induced genes such as PSA and KLK2 (Figure S1G). Concordantly, ChIP-seq revealed slightly decreased enhancer activity (indicated by H3K27Ac) around the HOTAIR gene upon androgen stimulation (Figure S1A). By contrast, RNA decay assays showed no significant difference in the rates of HOTAIR lncRNA degradation in ethanol- and androgen-treated cells, precluding RNA stability as a mechanism to HOTAIR repression by androgen (Figure S1H).
Due to its androgen-regulated nature, we hypothesized that HOTAIR may be differentially expressed during PCa progression. We performed qRT-PCR analysis of a panel of PCa cell lines and observed that HOTAIR was drastically upregulated in CRPC cell line models, such as LNCaP-abl and C4-2B (Figure S1I). To confirm this in primary specimens, we re-analyzed two publicly available datasets and found that HOTAIR was significantly upregulated in metastatic CRPC when compared to localized prostate tumors (Figures 1G and 1H). Next, we stratified localized PCa into two groups based on biochemical recurrence and found that HOTAIR was significantly upregulated in patients with recurrent PCa (Figure 1I). Further, Kaplan-Meier analysis revealed that high HOTAIR level was significantly associated with poor clinical outcome (Figure 1J). To further validate these findings in independent patient cohorts, we conducted RNA in situ hybridization (ISH) in PCa tissue microarray (Figure 1K). Our data demonstrated that HOTAIR staining was indeed significantly stronger in recurrent PCa compared to non-recurrent PCa (Figure 1L). Therefore, HOTAIR is an androgen-repressed lncRNA and is upregulated in aggressive PCa and in CRPC tumors.
The lncRNA HOTAIR Directly Interacts with the AR Protein
Since both HOTAIR and AR are upregulated in late-stage PCa, we sought to determine whether HOTAIR might interact with AR protein. We performed in vitro transcription to synthesize biotinylated RNA from sense and antisense HOTAIR 1–360 bp, the region that interacts with EZH2, which was then incubated with nuclear extracts from LNCaP cells to allow protein interactions, precipitated using streptavidin beads, and analyzed by SDS-PAGE gel. Silver staining revealed that HOTAIR, but not the antisense fragment, precipitated a protein of 110 kDa (Figure S2A). To investigate whether this band represents the AR protein, we carried out western blot analysis and demonstrated that biotinylated HOTAIR RNA, but not the antisense transcript or no-probe control, retrieved AR protein (Figure 2A).
Figure 2. HOTAIR Directly Interacts with the AR Protein.
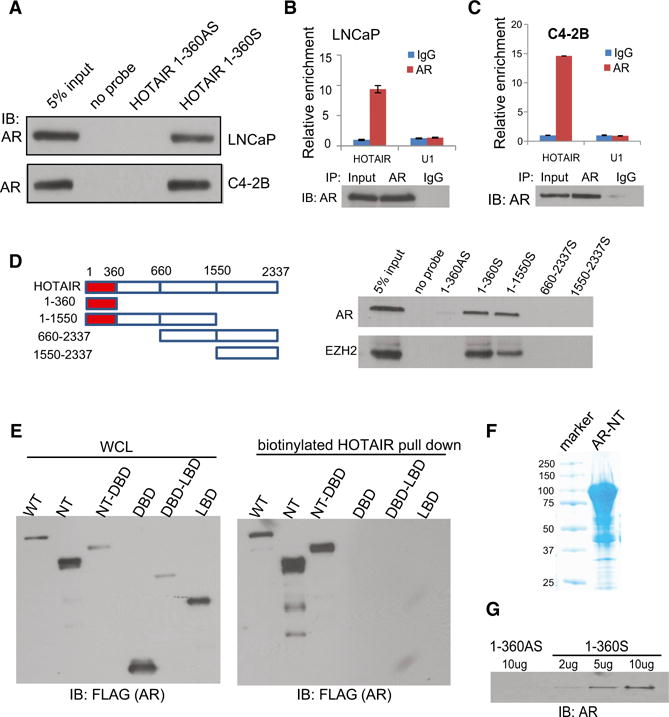
(A) Western blot showing AR protein interaction with HOTAIR RNA. RNA pull-down assay was performed in LNCaP and C4-2B cells using biotin-labeled HOTAIR RNA probe transcribed in vitro from the first 360 bp of the sense HOTAIR gene and detected using western blots. No probe and HOTAIR 1–360AS probe were used as negative controls.
(B and C) The qRT-PCR showing HOTAIR enrichment by an anti-AR antibody. LNCaP (B) and C4-2B (C) cells were subjected to RIP assay using an anti-AR antibody or IgG. IP-enriched RNA was then analyzed by qRT-PCR using U1 RNA as a negative control. (Bottom) Western blots confirm AR protein IP. Error bars are mean ± SEM.
(D) The 5′ end of HOTAIR transcript binds AR. RNA pull-down was carried out in LNCaP cells with various HOTAIR deletion probes (top), and the retrieved protein extract was analyzed by western blotting (bottom).
(E) NTD of the AR protein interacts with HOTAIR. FLAG-tagged AR deletion constructs were transfected into 293T cells and the expression confirmed by western blot analysis (left). RNA pull-down using biotinylated HOTAIR 1–360S probe retrieved the NTD-containing AR proteins (right).
(F and G) Coomassie blue staining verifying purified recombinant MBP-AR-NT protein synthesized in vitro (F) and western blot of AR-NT proteins retrieved by increasing doses of HOTAIR probes in in vitro binding assay (G). HOTAIR 1–360AS probe was used as a negative control.
To further validate this interaction between HOTAIR and AR, we carried out RNA immunoprecipitation (RIP) of LNCaP cell nuclear extracts. A qPCR analysis of antibody-enriched RNA revealed that AR antibody (PG-21 targeting the N terminus of AR, EMD Millipore) pulled down significantly more HOTAIR lncRNA than the IgG control (Figure 2B). By contrast, no differential enrichment of U1 RNA was observed. This was validated using a second AR antibody targeting the C terminus of AR (C-19, SC-815x from Santa Cruz Biotechnology) (Figure S2B). In addition, RIP experiments showed that AR antibodies were able to significantly enrich HOTAIR in hormone-deprived cells, albeit with slightly reduced fold enrichment (Figures S2C and S2D). The amount of HOTAIR immunoprecipitated by the anti-AR antibodies was comparable to that enriched by EZH2 antibody, supporting a strong interaction between HOTAIR transcript and the AR protein (Figure S2E). Moreover, RIP analysis of C4-2B cell lysate confirmed such interaction and further showed that more HOTAIR transcript was enriched by anti-AR antibody in C4-2B than in LNCaP cells, supporting increased HOTAIR expression and regulatory activities in CRPC cells (Figure 2C).
Next, we sought to determine which region of the HOTAIR transcript mediates its interaction with the AR protein. We generated a series of HOTAIR deletion probes, which were biotinylated and utilized for RNA pull-down assay. We first confirmed that all of these probes were able to retrieve the HOTAIR gene itself with RNA pull-down efficiency around 15-fold when compared with the antisense control (Figure S2F). Next, RNA pull-down followed by western blot analysis showed that RNA probes containing 1–360 nt of the HOTAIR gene, but not others, were capable of pulling down AR, suggesting that the first 360 bp of HOTAIR are necessary and sufficient to bind the AR protein (Figure 2D). As a positive control, the 5′ end of HOTAIR also pulled down the EZH2 protein as previously reported (Tsai et al., 2010). On the other hand, we were interested in knowing which AR domain modulates its interaction with HOTAIR. To test this, we generated a series of FLAG-tagged AR domain constructs, which were subsequently transfected into 293T cells (Figure S2G). RNA pull-down experiments revealed that AR constructs containing the N-terminal domain (NTD), but not the DNA-binding domain (DBD) and ligand-binding domain (LBD), interact with HOTAIR (Figure 2E). To further demonstrate that this is a direct interaction, we synthesized the AR-NTD protein through in vitro translation (Figure 2F). In vitro binding assay that incubates AR-NTD with increasing amounts of HOTAIR 1–360-bp probes showed that HOTAIR sense probe pulled down the AR protein in a dose-dependent manner, whereas the antisense probe did not (Figure 2G). To determine whether this interaction crosstalks with the well-known AR protein interaction with DNA through its DBD, we performed electrophoretic mobility shift assay (EMSA) and found that HOTAIR probe, and thus its interaction with AR-NTD, did not disrupt the interaction between AR-DBD and biotinylated ARE, whereas ARE oligo fully blocked the gel shift as expected (Figure S2H). Therefore, our data strongly suggest that the NTD of the AR protein directly interacts with the 5′ end of the HOTAIR lncRNA.
HOTAIR Stabilizes AR Protein by Blocking Its Interaction with MDM2
To investigate the consequences of HOTAIR and AR interaction, we overexpressed HOTAIR in LNCaP and LAPC4 cells through lentivirual transduction followed by antibiotic selection of stable clones (Figure S3A). Immunoblotting analysis showed that AR protein level increased following HOTAIR overexpression in both cell lines (Figure 3A). However, qRT-PCR analysis revealed a lack of change of AR at the mRNA level (Figure S3B). Since HOTAIR lncRNA previously has been shown to interact with nuclear proteins, we performed nuclear and cytoplasmic extraction of LNCaP and LAPC4 cell lysates and confirmed that AR primarily localized in the nuclei, which is significantly increased by HOTAIR overexpression (Figures 3B and S3C). To test whether HOTAIR stabilizes AR protein, we treated LNCaP and LAPC4 cells with cycloheximide (CHX), an inhibitor of protein biosynthesis. Western blot analysis showed that the half-life of AR protein was about 4 hr in control cells, which was increased to about 16 hr in both LNCaP and LAPC4 cells with HOTAIR overexpression (Figures 3C and S3D). On the other hand, HOTAIR knockdown dramatically accelerated AR protein degradation in C4-2B cells, which could be rescued by concurrent overexpression of small hairpin RNA (shRNA)-resistant HOTAIR (Figure S3E). To further demonstrate that HOTAIR hinders AR protein degradation, we treated control and HOTAIR-overexpressing LNCaP cells with proteasome inhibitor MG132. Western blot analysis confirmed that MG132 dramatically increased AR protein level, confirming successful blockade of proteasome-mediated protein degradation (Figure 3D). Further, while HOTAIR overexpression significantly increased AR protein level in the control-treated cells, it had a much smaller effect on the cells pretreated with MG132, supporting a mechanism dependent on proteasome-mediated AR degradation.
Figure 3. HOTAIR Stabilizes AR and Blocks MDM2-Mediated Ubiquitination.
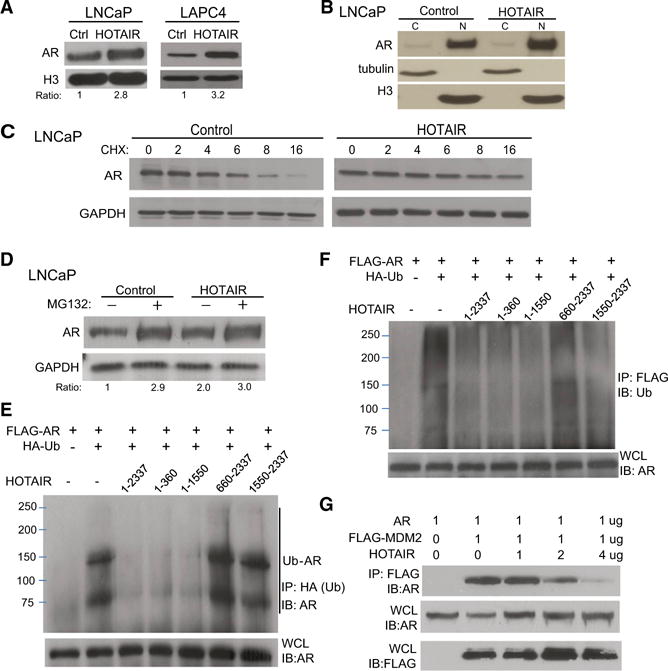
(A) Western blot shows total AR protein level in control and HOTAIR-overexpressing LNCaP and LAPC4 cells.
(B) Cytoplasmic and nuclear AR protein levels in control and HOTAIR-overexpressing LNCaP cells. Tubulin and H3 were used as cytoplasmic and nuclear protein-loading controls, respectively.
(C) HOTAIR overexpression stabilizes AR protein. Control, HOTAIR-overexpressing LNCaP cells were treated with 10 μM CHX for 6 hr and then collected at the indicated times for western blotting.
(D) Effects of proteasome inhibitor MG132 on HOTAIR-mediated AR protein level. LNCaP cells with control or HOTAIR overexpression were treated with control or 10 μM MG132 for 6 hr and subjected to western blot analysis.
(E and F) HOTAIR regulation of AR ubiquitination. The FLAG-AR and HA-Ub constructs were co-transfected into 293T cells with different regions of the HOTAIR transcript. (E) Whole-cell lysates (WCLs) were confirmed for AR expression (bottom) and subjected to IP using an anti-HA (Ub) antibody, followed by western blot using an anti-AR antibody (top). In a reverse coIP (F), WCLs were subjected to IP using an anti-FLAG (AR) antibody, followed by western blot using an anti-Ubiquitin antibody.
(G) CoIP showing AR and MDM2 protein interaction. The 293T cells were co-transfected with AR and different amounts of FLAG-MDM2 and HOTAIR. Cell lysates were confirmed for the expression of AR and MDM2 using anti-AR and anti-FLAG (MDM2) antibodies, respectively. CoIP using anti-FLAG (MDM2) followed by anti-AR western blot detected a reduced amount of AR pulled down following HOTAIR overexpression.
AR previously has been shown to degrade through the ubiquitination pathway (Lin et al., 2002). To examine whether HOTAIR affects AR ubiquitination, we performed co-transfection of FLAG-AR, HA-ubiquitin, HOTAIR or various HOTAIR deletion constructs into 293T cells. Western blot analysis of whole-cell lysate (WCL) confirmed AR protein expression in all experimental conditions. Cell lysates were then subjected to immunoprecipitation (IP) by an anti-HA antibody followed by western blotting using an anti-AR antibody. Our results showed that the presence of ubiquitin led to AR protein ubiquitination, which was blocked by the co-expression of HOTAIR that contained the first 360 bp, but not others (Figure 3E). Concordantly, reverse coIP using an anti-FLAG (AR) antibody followed by western blotting using an anti-ubiquitin antibody confirmed that HOTAIR transcripts prevent AR from binding to ubiquitin proteins (Figure 3F).
Previous studies have shown that MDM2, an E3 ligase, interacts with AR through its NTD domain, leading to AR protein ubiquitination (Lin et al., 2002). We thus hypothesized that HOTAIR binding with the AR protein masks the NTD and, thus, prevents AR from interacting with MDM2. To test this, we co-transfected 293T cells with AR, FLAG-tagged MDM2, and increasing doses of HOTAIR. Western blot analysis of WCL confirmed AR and FLAG (MDM2) expression. FLAG IP followed by western blot of AR proteins demonstrated that MDM2 physically interacts with the AR protein, which, however, is inhibited by concomitant HOTAIR expression in a dose-dependent manner (Figure 3G). To confirm this mechanism in a physiological setting, we performed IP using an anti-AR antibody in LNCaP cells with control and HOTAIR overexpression and in C4-2B cells with control and HOTAIR knockdown. Immunoblotting using an anti-MDM2 antibody demonstrated decreased and increased MDM2 pull-down, respectively, following HOTAIR overexpression in LNCaP cells and HOTAIR knockdown in C4-2B cells (Figure S3F). Taken together, our data suggest that HOTAIR stabilizes AR protein by binding to its NTD, thus blocking MDM2-AR interaction and decreasing AR ubiquitination and protein degradation.
HOTAIR Expression Enhances AR Transcriptional Activity
Next, to determine how HOTAIR affects the genomic action of AR, we first extracted nuclear and chromatin-bound protein fractions. Western blot analysis showed that HOTAIR expression dramatically increased both nuclear and chromatin-bound AR protein (Figure 4A). To examine whether this leads to an increase in AR-binding events, we performed AR ChIP-seq in LNCaP cells with either control or HOTAIR overexpression. Remarkably, ChIP-seq analysis revealed that the amount of AR-binding events was nearly doubled in LNCaP cells with HOTAIR overexpression (Figure 4B). In contrast to the marked gain of new AR-binding sites (ARBSs), very few were lost probably due to variability between replicate experiments. Moreover, all ARBSs, including gained, lost, and conserved ones, showed increased enrichment following HOTAIR overexpression (Figure 4C). A genome browser view of several known AR-bound enhancers confirmed highly enriched and sharp AR-binding peaks at target sites in HOTAIR-overexpressing cells (Figures 4D, 4E, S4A, and S4B).
Figure 4. HOTAIR Expression Enhances AR Transcriptional Activity.
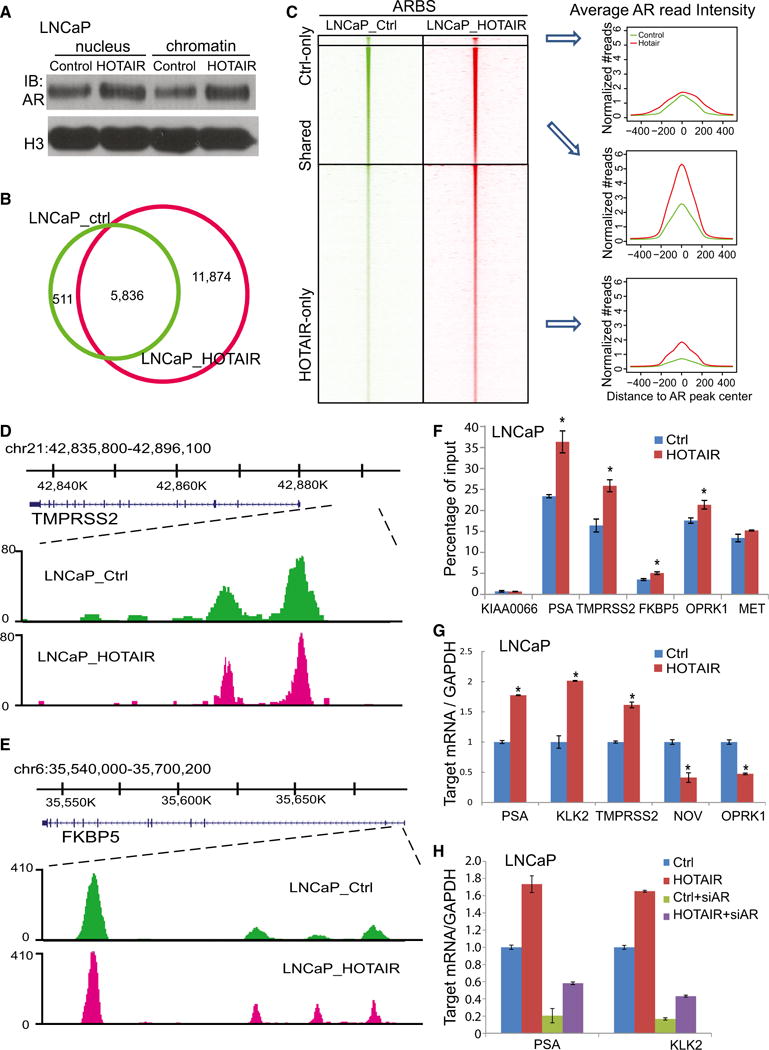
(A) Nuclear and chromatin-bound fractions of AR protein in control and HOTAIR-overexpressing LNCaP cells. H3 was used as a loading control.
(B) Overlap of AR-binding sites (ARBSs) detected by ChIP-seq in LNCaP cells with control (LNCaP_ctrl) or HOTAIR overexpression (LNCaP_HOTAIR) is shown.
(C) Heatmap depicting AR ChIP-seq read intensity around (±5 kb) ARBSs detected in control only, HOTAIR-overexpressing LNCaP cells only, or both. Average AR ChIP-seq read intensity around ARBSs (±500 bp) is shown (right).
(D and E) Genome browser view of ChIP-seq AR-binding events around known AR downstream genes TMPRSS2 (D) and FKBP5 (E) is shown.
(F) ChIP-qPCR analysis of known AR target genes. AR ChIP was performed in LNCaP_ctrl and LNCaP_HOTAIR cells followed by qPCR using primers targeting enhancers of several AR downstream genes. KIAA0066 was used as a negative control. Data shown are mean ± SEM.
(G) The qRT-PCR analysis of AR downstream genes in LNCaP cells with control, HOTAIR overexpression. Gene expression was normalized to GAPDH. Data shown are mean ± SEM.
(H) The qRT-PCR analysis of AR downstream genes in LNCaP_ctrl and LNCaP_HOTAIR cells with control or AR knockdown. Gene expression was normalized to GAPDH. Data shown are mean ± SEM.
To validate these findings, we performed AR ChIP-qPCR on several known AR target genes using site-specific primers. Our data confirmed that HOTAIR overexpression indeed increased AR recruitment to enhancers of PSA, TMPRSS2, FKBP5, OPRK1, and MET (Zhao et al., 2012; Figure 4F). Consistent with this increase in AR occupancy, qRT-PCR demonstrated that HOTAIR increased the expression of AR-induced genes, whereas it reduced that of AR-repressed genes in both LNCaP and LAPC4 cells, supporting overall augmentation of AR transcriptional activity (Figures 4G and S4C). Further, HOTAIR-induced AR target gene expression was fully abolished by AR knockdown via RNA interference (Figure 4H). Together, these results suggest that HOTAIR enhances AR chromatin-binding and transcriptional activity.
HOTAIR Drives Androgen-Independent AR Chromatin Targeting
We sought to determine whether HOTAIR overexpression is able to activate AR even in the absence of androgen. Western blot analysis showed a dramatically increased amount of nuclear AR in HOTAIR-expressing, but hormone-deprived, LNCaP cells (Figure 5A). Concordantly, AR immunofluorescence staining in hormone-deprived or -stimulated control and HOTAIR-expressing LNCaP cells revealed that, similar to androgen stimulation, HOTAIR overexpression resulted in evident AR nuclear staining in the absence of androgen (Figure 5B). ChIP-seq showed that, as expected, very few ARBSs were detected in the control, hormone-deprived cells. Remarkably, HOTAIR overexpression increased AR-binding events from 112 to 5,566 sites, strongly supporting androgen-independent AR activation (Figures 5C and 5D). A genome browser view confirmed that HOTAIR overexpression resulted in substantial AR-binding events at the enhancers of known AR target genes, such as TMPRSS2, FKBP5, PSA, and KLK2 (Figures 5E, 5F, S5A, and S5B). Moreover, ChIP-qPCR confirmed that HOTAIR overexpression is sufficient to dramatically induce AR binding to target genes even under androgen-depleted conditions (Figure 5G).
Figure 5. HOTAIR Overexpression Leads to Androgen-Independent AR Chromatin Targeting.
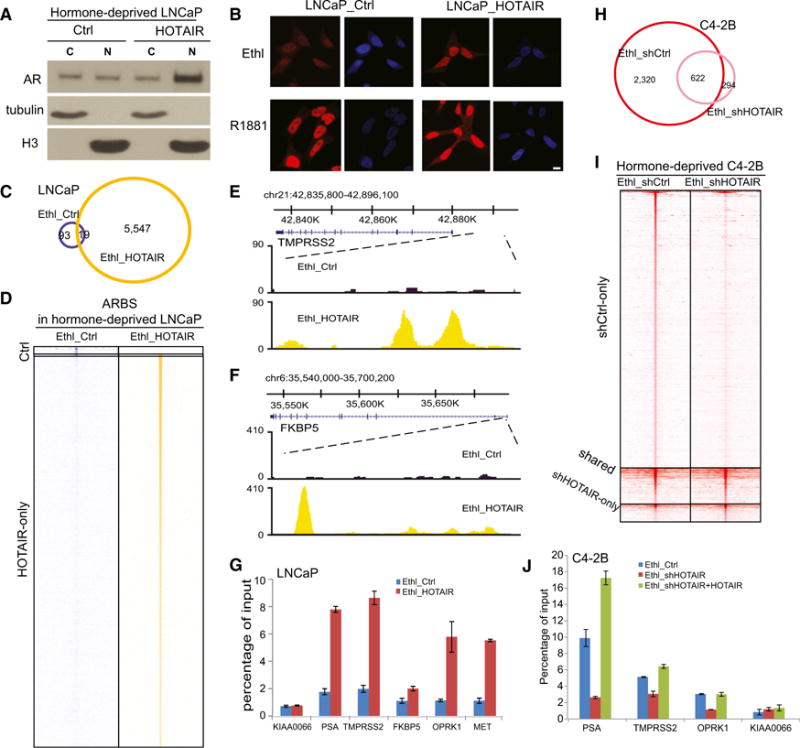
(A) Cytoplasmic and nuclear AR protein levels in hormone-deprived, control and HOTAIR-overexpressing LNCaP cells. Tubulin and H3 were used as cytoplasmic and nuclear protein-loading controls, respectively.
(B) Immunofluorescence staining of AR. LNCaP_ctrl and LNCaP_HOTAIR cells were hormone-starved for 3 days and treated with ethanol or 1 nM R1881 for 24 hr before being subjected to immunostaining using an anti-AR antibody. Scale bar, 10 μm.
(C) Overlap of ARBSs detected in hormone-deprived LNCaP_ctrl and LNCaP_HOTAIR cells is shown.
(D) Heatmap depicts AR ChIP-seq read intensity around (±5 kb) ARBSs shown in (C).
(E and F) Genome browser view of ChIP-seq AR-binding events around AR target genes TMPRSS2 (E) and FKBP5 (F) is shown.
(G) ChIP-qPCR of AR binding on known target genes. AR ChIP was performed in hormone-deprived LNCaP_ctrl and LNCaP_HOTAIR cells. KIAA0066 was the negative control gene. Data shown are mean ± SEM.
(H) Overlap of ARBSs detected in C4-2B cells with control or HOTAIR knockdown is shown.
(I) Heatmap depicts AR ChIP-seq read intensity around (±5 kb) ARBSs shown in (H).
(J) ChIP-qPCR of AR binding on known target genes. AR ChIP was performed in hormone-deprived C4-2B_ctrl, C4-2B shHOTAIR, and C4-2B shHOTAIR + HOTAIR WT cells. KIAA0066 was the negative control gene. Data shown are mean ± SEM.
Next, we asked whether HOTAIR expression is required for AR transcriptional activity in CRPC cells. Since we have shown that HOTAIR is upregulated in CRPC cell lines, we carried out HOTAIR knockdown in C4-2B cells (Figure S5C). ChIP-seq showed that, unlike the androgen-dependent LNCaP cells, C4-2B cells harbor substantial AR-binding events even in the absence of androgen, being consistent with androgen-independent AR activation (Figure 5H). Most importantly, HOTAIR knockdown abolished a majority of these AR-binding events, suggesting that HOTAIR is required for androgen-independent AR activation in CRPC cells. Analysis of ChIP-seq read counts around these ARBSs confirmed a significant decrease at most ARBSs (Figure 5I). Further, ChIP-qPCR verified that HOTAIR knockdown in C4-2B cells decreased AR binding at known AR target genes, which could be rescued by the overexpression of shRNA-resistant HOTAIR gene (Figure 5J). Taken together, our results support that HOTAIR expression is critical in inducing and maintaining androgen-independent AR activation and genomic targeting.
HOTAIR Induces AR Transcriptional Program in the Absence of Androgen
To understand how the HOTAIR-mediated AR program relates to that induced by androgen, we first compared the ARBSs induced by HOTAIR (under androgen-deprived conditions) with those by androgen in LNCaP cells. Interestingly, we found that more than 70% of HOTAIR-induced ARBSs overlapped with androgen-induced ones, supporting that, instead of reprogramming AR, HOTAIR primarily acts to increase AR activity (Figure 6A). A heatmap view of AR ChIP-seq reads around all ARBSs further confirmed that, under androgen-deprived conditions, HOTAIR overexpression induced an AR-binding profile that highly resembles that stimulated by androgen (Figure 6B). Moreover, HOTAIR expression and androgen stimulation showed synergistic effects in maximizing AR binding.
Figure 6. HOTAIR Drives AR Transcriptional Program in the Absence of Androgen.
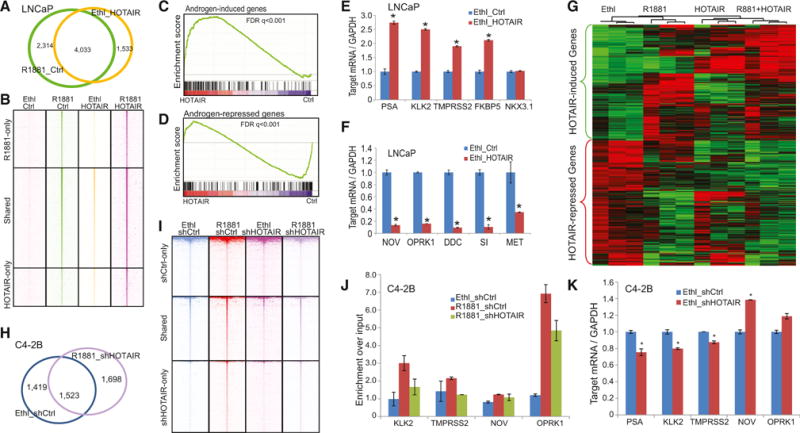
(A) Overlap of ARBSs detected in hormone-deprived LNCaP cells with either R1881 or HOTAIR overexpression is shown.
(B) Heatmap depicting AR ChIP-seq read intensity around (±5 kb) ARBSs shown in (A). AR ChIP-seq was carried out in LNCaP cells treated with R1881, HOTAIR overexpression, or both. ChIP-seq enrichment was normalized to total number of reads for comparative visualization.
(C and D) GSEA depicting HOTAIR regulation of AR-mediated gene program. GSEA was carried out to determine the enrichment of AR-induced (C) and AR-repressed (D) gene sets in expression dataset profiling hormone-deprived LNCaP cells with control or HOTAIR overexpression.
(E and F) The qRT-PCR analysis of AR-induced (E) and AR-repressed (F) genes in hormone-deprived LNCaP with control or HOTAIR overexpression. Data were normalized to GAPDH. Error bars are mean ± SEM.
(G) Heatmap view of HOTAIR-regulated genes in triplicate microarray experiments profiling hormone-deprived LNCaP_ctrl and LNCaP_HOTAIR cells stimulated with ethanol or androgen. Each row represents one gene, and each column corresponds to one expression microarray. Differentially expressed genes were identified by limma R package with p value cutoff <0.01.
(H) Overlap of ARBSs detected in C4-2B cells with hormone-deprivation or HOTAIR knockdown is shown.
(I) Heatmap depicting AR ChIP-seq read intensity around (±5 kb) ARBSs shown in (H). AR ChIP-seq was carried out in C4-2B cells treated with R1881, HOTAIR knockdown, or both. ChIP-seq enrichment was normalized to the total number of reads for comparative visualization.
(J) AR ChIP was performed in C4-2B_shCtrl or C4-2B-shHOTAIR cells in the presence or absence of androgen. ChIP-qPCR was carried out using primers targeting enhancers of different AR downstream genes. KIAA0066 was the negative control. Error bars are mean ± SEM.
(K) The qRT-PCR analysis of AR downstream genes in C4-2B_shCtrl and C4-2B-shHOTAIR cells cultured in hormone starvation medium. Data were normalized to GAPDH. Error bars are mean ± SEM.
To determine how these genomic binding events relate to target gene expression, we performed expression microarrays of hormone-deprived LNCaP cells with or without HOTAIR overexpression. To investigate how HOTAIR modulates the expression of androgen-regulated genes, we carried out gene set enrichment analysis (GSEA). Androgen-induced and -repressed gene sets were obtained from our previously published data in LNCaP cells stimulated with control or androgen (Zhao et al., 2012). Importantly, GSEA demonstrated that androgen-induced genes were significantly enriched for upregulation by HOTAIR, whereas androgen-repressed genes were downregulated (Figures 6C and 6D). Moreover, qRT-PCR analysis confirmed that androgen-induced genes, such as PSA and KLK2, were significantly increased, while androgen-repressed genes, including NOV and OPRK1, decreased following HOTAIR overexpression (Figures 6E and 6F).
To further characterize HOTAIR downstream genes, we carried out triplicate microarray experiments and identified 512 and 560 genes, respectively, that were significantly (p < 0.01) induced and repressed by HOTAIR in hormone-deprived LNCaP cells. To obtain some understanding of the function of these genes, we conducted gene ontology (GO) analysis and found that HOTAIR-induced genes were strongly enriched for cellular processes such as cell cycle, mitosis, and nuclear division (Table S2). HOTAIR-repressed genes, on the other hand, were involved in DNA binding, nucleus, and zinc-finger proteins (Table S3). Next, to determine how HOTAIR correlates with androgen signaling, we performed clustering analysis of triplicate microarray experiments profiling control and HOTAIR-overexpressing LNCaP cells before and after androgen stimulation. A heatmap analysis using Treeview demonstrated that a majority of HOTAIR-increased genes also were induced by androgen while HOTAIR-decreased genes were inhibited by androgen (Figure 6G). Moreover, these genes exhibited maximal regulation in LNCaP cells with HOTAIR overexpression and androgen stimulation.
To further confirm our findings, we treated control and HOTAIR-knockdown C4-2B with androgen and performed AR ChIP-seq. We observed that HOTAIR depletion decreased ARBSs in C4-2B cells under both androgen-deprived and -stimulated conditions. Interestingly, ARBSs found in androgen-stimulated, but HOTAIR-depleted, C4-2B cells largely overlapped with those found in androgen-deprived control cells, suggesting that HOTAIR knockdown counteracted the effect of androgen stimulation (Figure 6H). HOTAIR is required for AR-binding events in the milieu of low androgen, and HOTAIR collaborates with androgen in maximizing AR binding to chromatin (Figure 6I). Furthermore, ChIP-qPCR analysis confirmed that HOTAIR depletion abolished androgen-stimulated AR activation, while qRT-PCR assays validated that HOTAIR knockdown hampered AR transcriptional regulation (Figures 6J and 6K). Therefore, using two independent prostate cancer models, we demonstrated that HOTAIR overexpression induces an AR transcriptional program similar to that activated by androgen.
HOTAIR Promotes CRPC Progression
As androgen-independent AR activation is a main driver to CRPC, we next investigated the roles of HOTAIR in CRPC progression. Cell proliferation assays showed that HOTAIR overexpression significantly increased LNCaP cell growth, while HOTAIR knockdown in C4-2B cells markedly reduced cell growth, being consistent with its role in enhancing the AR transcriptional program (Figure 7A). As HOTAIR is able to drive androgen-independent AR activation, we performed cell proliferation assays in hormone-deprived PCa cells. Our results demonstrated that HOTAIR overexpression indeed increased LNCaP cell growth in the absence of androgen, whereas HOTAIR knockdown abolished androgen-independent C4-2B cell growth (Figure 7B). Besides cell growth, androgen signaling also is known to induce cell invasion. Boyden chamber assay showed that ectopic HOTAIR overexpression substantially increased LNCaP cell invasion (Figures 7C and 7D). On the other hand, HOTAIR depletion in C4-2B cells dramatically reduced cell invasion (Figures 7E and 7F). Therefore, being consistent with its role in enhancing AR chromatin-targeting and transcriptional activities, HOTAIR increases PCa cell growth and invasion.
Figure 7. HOTAIR Promotes CRPC Progression.
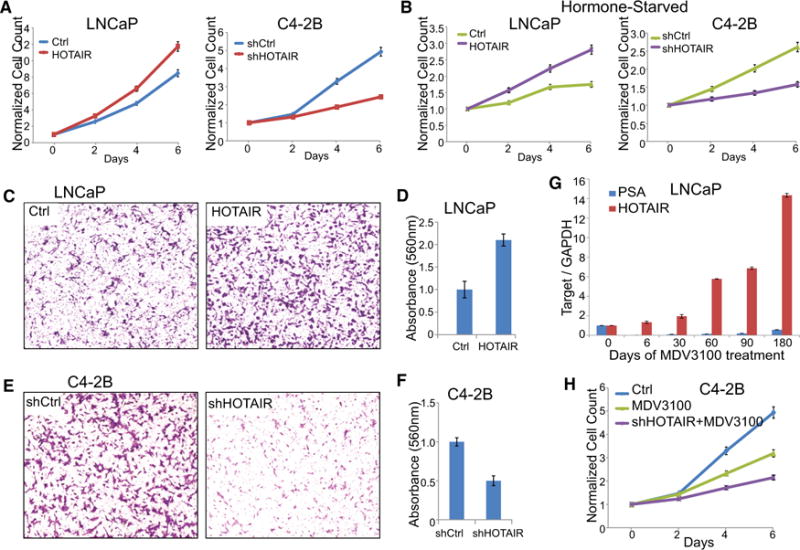
(A) HOTAIR regulation of androgen-dependent cell growth. Cell growth was assayed in LNCaP cells with control or HOTAIR overexpression and C4-2B cells with control or HOTAIR knockdown. Cells were grown in androgen-containing medium. Data shown are mean ± SEM of triplicate wells and are representative of at least two replicate experiments.
(B) HOTAIR regulation of androgen-independent growth of LNCaP and C4-2B cells grown in hormone-deprived condition is shown.
(C and D) HOTAIR overexpression increases LNCaP cell invasion. Cell invasion was monitored by Boyden chamber assay in LNCaP_Ctrl and LNCaP_HOTAIR cells. Representative images of invaded cells are shown (C), and the total number of invaded cells was quantified using colorimetry with absorbance at 560 nm (D). (E and F) HOTAIR depletion diminished C4-2B cell invasion as examined using Boyden chamber assay. Representative images of invaded cells are shown (E), and the total number of invaded cells was quantified using colorimetry with absorbance at 560 nm (F).
(G) The qRT-PCR showing PSA and HOTAIR expression in LNCaP cells following MDV3100 treatment. Gene expression was normalized to GAPDH. Error bars are mean ± SEM.
(H) Cell growth of C4-2B cells treated with MDV3100 alone or in combination with HOTAIR depletion is shown.
Next, we attempted to investigate the role of HOTAIR in resistance to androgen-deprivation therapies. We generated enzalutamide-resistant cells by culturing LNCaP cells in the presence of 1 μM MDV3100 for several months. Our qRT-PCR analysis confirmed that PSA level was drastically decreased by MDV3100 initially and slowly recovered after 2 months of continuous drug treatment (Figure 7G). Interestingly, HOTAIR expression was gradually increased over time following the MDV3100 treatment and markedly upregulated to as much as 14-fold. Together with the role of HOTAIR in enhancing the androgen-independent AR program, these data suggest that HOTAIR upregulation may account at least partially for enzalutamide resistance and that HOTAIR knockdown may inhibit CRPC cell proliferation. Indeed, cell growth assays showed that HOTAIR knockdown further decreased C4-2B cell growth in the presence of enzalutamide (Figure 7H).
DISCUSSION
LncRNAs have been shown to regulate gene transcription and protein function through various mechanisms. For example, by binding to the PRC2 proteins, HOTAIR acts to guide and recruit PRC2 to target genes related with tumor metastasis in breast cancer (Gupta et al., 2010b). HOTAIR also can function as a scaffold that, by simultaneously binding to the EZH2 and LSD1 proteins through its 5′ end and 3′ end, respectively, direct these proteins to co-occupy the same genomic regions (Tsai et al., 2010). Through this scaffold function, HOTAIR also has been reported to link E3 ubiquitin ligases with their substrates to accelerate proteolysis (Yoon et al., 2013). Findings from our study revealed a distinct mode of action of lncRNAs in gene regulation. Here we propose a working model wherein the lncRNA HOTAIR binds the NTD of the AR protein, which consequently blocks recruitment of the E3 ubiquitin ligase MDM2 that interacts with AR through the same domain. Through this competitive mechanism, HOTAIR expression inhibits AR ubiquitination, prevents AR protein degradation, and, thus, increases AR protein stability. Our study demonstrates that the lncRNA is able to mask a protein-interacting domain, thereby preventing co-regulator protein binding.
In this paper, we show that HOTAIR is an androgen-repressed lncRNA that enhances the AR-activated gene expression program by directly binding to AR to protect it from ubiquitin-mediated degradation. In the absence of androgen, HOTAIR overexpression induced an AR-binding profile that largely overlapped with that induced by androgen, probably due to a shared mechanism of action, as androgen-induced ligand binding is well known to stabilize the AR protein (Zhou et al., 1995). In addition, a study by Steven Balk’s laboratory previously found that AR itself is an androgen-repressed gene that is upregulated in CRPC to drive aberrant AR activity (Cai et al., 2011). Unlike androgen-induced genes, the androgen-repressed genes, such as AR, are able to escape from AR-mediated repression in CRPC cells and, thus, remain upregulated. HOTAIR may become upregulated in CRPC cells through similar mechanisms and further stabilize AR and enhance its activity.
Being highly consistent with its positive regulation of AR signaling, HOTAIR overexpression in androgen-dependent LNCaP cells was found to increase cell growth even in the absence of androgen. On the other hand, HOTAIR knockdown in CRPC cell line C4-2B markedly decreased cell proliferation, supporting that HOTAIR expression is required for CRPC cell growth. Importantly, our results demonstrated that HOTAIR is remarkably upregulated in enzalutamide-resistant PCa cells and its depletion decreased CRPC cell growth in the presence of enzalutamide. The lncRNA HOTAIR thus may be a biomarker of enzalutamide resistance and a promising target for therapeutic intervention in CRPC.
EXPERIMENTAL PROCEDURES
Patient Specimens
Specimens were collected by the Northwestern University Prostate Cancer Specialized Program of Research Excellence Tissue Core (SPORE). Tissue samples were collected in a de-identified manner with informed consent of the patients and prior Institutional Review Board approval.
Plasmids and Reagents
The HOTAIR gene was amplified by RT-PCR and then cloned into a lentiviral vector pCDH-MSCV-mcs-EF1-GFP-T2A-Pu (SBI) at EcoR1 and Not1 sites using the Cold Fusion kit (SBI). To generate HOTAIR overexpression stable cells, cells were transduced with control or HOTAIR overexpression lentivirus particles and selected with puromycin for 3 days. HOTAIR knockdown construct was created by inserting oligo (CCGGGAACGGGAGTACAGAGAGAATCTCGAGATTCTCTCTGTACTCCCGTTCTTTTTG) into the pLKO.1 vector. Lentiviral particles were generated by transfecting 293T cells with packaging vectors, psPAX2 and pMD2.g. All primers were designed using Primer 3 and synthesized by Integrated DNA Technologies (Table S4). The qRT-PCR was performed using Bullseye EvaGreen qPCR SYBR Green Mastermix (MIDSCI) on an Applied Biosystems StepOne Plus. A detailed list of other plasmids and reagents used in this study is provided in the Supplemental Experimental Procedures.
RNA Pull-Down Assay
Biotin-labeled HOTAIR probe and cell protein extract were incubated for 60 min before being incubated with streptavidin agarose beads (Invitrogen) at room temperature (RT) for 1 hr. After PBS wash, the retrieved protein was subjected to western blot analysis. For detecting direct binding of AR with HOTAIR, AR-NT recombinant protein was incubated with biotin-labeled RNA probe to proceed with the IP process.
RIP
Briefly, cells were resuspended in radio-immunoprecipitation assay (RIPA) buffer and incubated with 20 μl protein beads and 1 μg antibody complex overnight at 4°C. After incubation with RNase-free DNase I (Promega) at 37°C for 15 min and proteinase K at 45°C for 45 min, RNA samples were extracted with 1 ml Trizol and subjected to RT-PCR analysis.
LncRNA ISH
ISH was used to detect HOTAIR in clinical specimens. Biotin-labeled anti-sense HOTAIR LNA probe/5Biosg/G+C+C+TTGCTCCCTT+G+CCTGCATTTCT+C+T+G was synthesized by EXIQON. ISH was performed on paraffin-embedded PCa tissue microarray. Detailed procedure is provided in the Supplemental Experimental Procedures.
Gene Expression, ChIP-Seq, and Data Analysis
Total RNAs were isolated using TRIzol reagent (Invitrogen). The integrity of the RNA was monitored using Bioanalyzer 2100. Microarray profiling was performed using HumanHT-12 v 4.0 Expression BeadChip (Illumina). Bead-level data were processed using GenomeStudio (Illumina), and the expression values were quantile-normalized using the limma package in Bioconductor (Ritchie et al., 2015). Differentially expressed genes were identified based on p < 0.01. GO term enrichment was analyzed using DAVID 6.7 (Huang da et al., 2009). Heatmap view of differentially expressed genes was created by Cluster and Java Treeview (de Hoon et al., 2004; Saldanha, 2004).
ChIP was carried out as previously described (Yu et al., 2010). ChIP DNA was prepared into libraries according to standard protocols using Bioo Scientific’s DNA Sample Kit (514101). Libraries were sequenced using Illumina Hi-Seq platforms. Sequence reads were aligned to the Human Reference Genome (assembly hg19) using Burrows-Wheeler Alignment Tool (bwa) version 0.6.1 (Li and Durbin, 2009). Peak identification and overlapping were performed using the Hypergeometric Optimization of Motif Enrichment suite (Heinz et al., 2010). Heatmaps and intensity plots of peaks were generated by Perk script, R, and/or Java Treeview.
Supplementary Material
Highlights.
The lncRNA HOTAIR is repressed by androgen and, thus, upregulated in CRPC
HOTAIR inhibits AR degradation by blocking its binding to E3 ubiquitin ligase MDM2
HOTAIR increases AR chromatin targeting and enhances the AR-mediated gene program
HOTAIR drives androgen-independent AR activation and promotes CRPC
Acknowledgments
We thank Dr. Ximing J. Yang (Northwestern University) for assistance on RNA ISH and Dr. Nicholas Erho (GenomeDx Biosciences) for data mining. Tissue microarrays were provided by the Northwestern University prostate cancer SPORE (P50 CA090386). J.K. and Y.A.Y were supported by the NIH T32 CA080621 and T32 CA09560, respectively. This work was supported in part by the NIH R01CA172384 (to J.Y.), U.S. Department of Defense W81XWH-14-1-0023 (to J.Y.), and the Research Scholar Award RSG-12-085-01 (to J.Y.) from the American Cancer Society.
Footnotes
ACCESSION NUMBERS
The accession number for the microarray and short-read sequencing data reported in this paper is GEO: GSE61270.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.08.069.
AUTHOR CONTRIBUTIONS
A.Z., J.K., K.F., and Y.A.Y performed the experiments. J.C.Z. analyzed microarray and ChIP-seq data. J.Y. and A.Z. wrote the manuscript. D.C. provided critical discussions and Y.Y.M. helped with RIP and RNA pull-down protocols. J.Y. designed and supervised the project. All authors discussed the results and commented on the manuscript.
References
- Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, Chen S, Nelson PS, Liu XS, Brown M, Balk SP. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S, Saini S, Chang I, Tanaka Y, Enokida H, et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS ONE. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long non-coding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010a;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010b;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open Source Clustering Software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T, Satoh K. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Sahu A, Iyer MK, Malik R, Chandler B, Asangani IA, Poliakov A, Vergara IA, Alshalalfa M, Jenkins RB, et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5:1434–1438. doi: 10.18632/oncotarget.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sørensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M, Larsen MJ, Kruse TA. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142:529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JC, Yu J, Runkle C, Wu L, Hu M, Wu D, Liu JS, Wang Q, Qin ZS, Yu J. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome Res. 2012;22:322–331. doi: 10.1101/gr.131508.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZX, Lane MV, Kemppainen JA, French FS, Wilson EM. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol Endocrinol. 1995;9:208–218. doi: 10.1210/mend.9.2.7776971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


