Abstract
The reversible acetylation of specific lysine residues on core histones regulates gene transcription in eukaryotes. Since the discovery of GCN5 as the first transcription-regulating histone acetyltransferase (HAT), a variety of HATs have now been identified and shown to acetylate different sites on histones as well as on non-histone proteins, including transcription regulators. In general, purified recombinant HATs expressed in bacteria or in insect cells are able to acetylate free histones and sometimes other substrates in vitro. However, such activity is often restricted to certain substrates and/or is very weak on physiological substrates, such as nucleosomes. Moreover, it does not reflect the actual scenario inside the cell, where HATs generally associate with other proteins to form stable multisubunit complexes. Importantly, these peripheral proteins significantly influence the functions of the catalytic HAT subunit by regulating its intrinsic catalytic activity and/or by modulating its target substrate selectivity. In this chapter, we describe detailed methods for the rapid (two step) and efficient purification of large, multiprotein HAT complexes from nuclear extracts of mammalian epitope-tagged cell lines, including protocols for the generation and large-scale suspension culture of these cell lines. These methods have been used to purify and characterize different human GCN5 HAT complexes that retain activity toward their physiological substrates in vitro.
Keywords: Transcription, Histone acetyltransferase, Protein complexes, GCN5, Affinity purification, FLAG tag, S-Sepharose, Cell line
1. Introduction
RNA polymerase II-mediated transcription in eukaryotes is a tightly regulated multistep process involving many types of regulatory proteins, such as transcriptional activators and repressors, different cofactors, and general transcription factors (1). The picture is further complicated as nuclear genomic DNA is packed into chromatin by both histone and non-histone proteins, rendering DNA inaccessible to transcription regulators (2). To overcome this barrier, eukaryotes have evolved to use either ATP-dependent enzymes to move/evict the nucleosomes or chromatin-modifying enzymes to modify DNA or histone proteins (3). Indeed, posttranslational modifications of nucleosomal histones have been correlated with changes in chromatin structure and transcription regulation (4, 5). Although several different histone modifications have now been identified and shown to play a role in transcription, lysine acetylation was the first to be correlated with transcription activation (6) and is the best characterized.
Mechanistically, histone tail acetylation was first shown to facilitate binding of transcriptional activators to nucleosomal DNA (7), consistent with findings that active chromosomal domains are generally associated with hyperacetylated histones (8) while inactive or heterochromatin domains are associated with hypoacetylated histones (9, 10). However, a more direct link between histone acetylation and transcription came originally from the identification of the first histone acetyltransferase (HAT) Gcn5 and the first histone deacetylase (HDAC) Rpd3, two factors that were previously genetically defined as a coactivator and a corepressor of gene transcription, respectively (11, 12). Thus, the steady-state acetylation level of histone proteins is accomplished by an intricate balance between HATs and HDACs that is important for proper cellular function (13, 14). Acetylation affects high-order folding of chromatin fibers, loosens the contacts between the DNA and the nucleosomes, and alters the interactions between histones and non-histone proteins. Acetylated lysine residues on histone tails are also “marks” that are recognized by bromodomain-containing proteins, which include components of the transcription machinery and ATP-dependent nucleosome-remodeling enzymes that displace modified nucleosomes from promoters (15, 16). Not surprisingly, histone acetylation has been connected with changes in chromatin that occur not only during transcription, but also during DNA replication and repair in vivo (17–20). Although histone proteins are the primary targets of HAT activity, HAT enzymes also acetylate a growing number of non-histone substrates, including many transcription regulators (21, 22). In this respect, a direct proof that histone tail acetylation (rather than other non-histone substrate acetylation) is causally linked to gene activation was provided only relatively recently. This was achieved via reconstitution of transcription activation on chromatin in vitro with purified components, including a recombinant coactivator-HAT (p300) and recombinant nucleosomal templates bearing substitutions of the acetylated lysine residues on the tails of core histones (23, 24).
Based on their catalytic domains, HATs have been grouped into several families, including the Gcn5-related N-acetyltransferases (GNATs) family and the MYST (MOZ, Ybf2/Sas3, Sas2, and Tip60)-related family (25). As the prototypical HAT, Gcn5 has been the focus of intense study over the last decade. Although purified, recombinant Gcn5 displays HAT activity on free histones, it fails to acetylate the more physiological, nucleosomal histone substrates in vitro. This led to the discovery that Gcn5 exists as multisubunit complexes inside the cell – e.g., SAGA/STAGA [Spt3-Taf-Ada-Gcn5 Acetylase] complexes that (1) can acetylate nucleosomes, (2) are recruited to promoters by DNA-binding activators, and (3) have HAT activity-dependent transcription coactivator functions on nucleosomal genes/promoters in vitro and in vivo (26–33). Importantly, the functions and specificity of the GCN5 HAT – and most other HATs – depend largely on the context of other subunits within those complexes (15, 26). Moreover, HAT-associated subunits within these complexes have additional roles in transcription coactivation, as shown, for instance, for specific GCN5-associated subunits within SAGA/STAGA complexes, which directly interact with components of the general/basal transcription machinery (34) or have additional catalytic activities targeting non-histone proteins (35).
Several multiprotein HAT complexes have now been identified and characterized in different model systems and shown to be evolutionarily conserved from yeast to human (15). However, depending on the organism and the experimental design, different laboratories utilize different strategies to purify these multiprotein complexes. A successful purification scheme to purify multisubunit HAT complexes generally contains at least one highly specific and stringent Affinity separation step that retains only the target protein and the stably associated subunits, but not proteins that interact only weakly or nonspecifically. Among all the purification strategies, immunoprecipitation/co-immunoprecipitation (IP/co-IP) with specific antibodies and Affinity chromatography using epitope tags are the most commonly used methods (31, 33, 36, 37). In some cases, protein complexes can also be purified via their Affinity for specific peptides/substrates, such as histone tails. For example, peptide pull-down assay has been shown to purify histone methyltransferase complexes (38). Conventional biochemical purification (gel filtration and ion-exchange chromatography) is, per se, generally not the method of choice, although it can be used as one additional step to complement the Affinity step (as described below). A summary of these individual methods with their strengths and weaknesses is given in Table 1.
Table 1.
Commonly used strategies for protein complex purification
| Approach | Strength | Weakness |
|---|---|---|
| Anti-epitope tag immunoaffinity |
|
|
| Regular IP/Co-IP |
|
|
| Peptide affinity pull down |
|
|
| Ion-exchange and gel filtration chromatography |
|
|
IP/co-IP experiments are most frequently used to test endogenous association of two or more proteins. Therefore, they are also suitable techniques to capture endogenous protein complexes. Typically, an antibody recognizing a particular component of the complex is incubated with cell lysates followed by immobilization on protein A or protein G resins. The bound protein complex is then washed extensively to remove the contaminants for downstream applications. However, there are some limitations with these methods. First, the antibody of interest may not be commercially available or may not work in IP. Second, the antibody has to be highly specific with minimal cross-reactivity against other nonspecific proteins, which might give rise to false-positive results. Therefore, re-IP against another component of the complex is sometimes performed to validate the interaction.
Another powerful method is immunoAffinity purification with specific monoclonal antibodies recognizing an epitope tag carried by one of the subunits of the complex (39, 40). Typically, the protein of interest (bait) is fused to an epitope tag (e.g., FLAG, HA) and then overexpressed in the cell. To retrieve the bait protein and its cognate protein complexes, an antibody directed against the tag is used. As a result, many different subunits of the same complex can be tagged and purified by the same procedure. A variety of expression vectors containing the tags and the corresponding antibody-immobilized resins are commercially available. The complexes can be further eluted (in an active form) from the antibody resin by competition with an excess of epitope peptides.
Often a highly pure protein complex is derived from more than one purification step. This is because no matter how stringent the purification condition is, it always carries trace amount of contaminants that are specific to that particular method. A second purification helps remove the contaminants and also concentrates the sample for downstream applications. Besides the aforementioned antibody-based purification methods, conventional size-exclusion and ion-exchange chromatography are also used to fractionate protein complexes and are most useful in conjunction with an Affinity method (31, 33, 39–41).
Technically, there are a number of parameters involving protein complex purification. These include binding affinities between components of the complex, cellular expression level of each subunit, wash stringency, etc. Therefore, it is important to validate the complex of interest by a combination of methods. In this chapter, we describe a straightforward, two-step purification method that has been successful in purifying different human GCN5 HAT-containing complexes (STAGA and ATAC) for their subunit characterization and for analysis of their catalytic activities on physiological, nucleosomal histone and non-histone substrates (31, 33, 42).
2. Materials
2.1. Cell Culture
2.1.1. Equipment and Supplies
Tissue culture incubators (both a humidified incubator with 5% CO2 and a nonhumidified/non-CO2, “reach-in” incubator for large spinner flasks).
Carbon dioxide (CO2) gas.
Dulbecco’s modified Eagle’s medium (DMEM; Cellgro).
Minimum essential medium (S-MEM; Gibco).
Penicillin–streptomycin solution (Cellgro).
Fetal bovine serum (FBS).
Bovine calf serum (BCS).
Trypsin-ethylenediaminetetraacetic acid (trypsin-EDTA) solution.
Phosphate-buffer saline (PBS).
Plastic tissue culture dish.
LipoD293™ DNA In Vitro Transfection Reagent (SignaGen Laboratories).
Geneticin/G418 (Gibco).
Conical tubes (15 and 50 ml).
pIRES vector (Clontech).
FLAG antibody (Sigma).
Spinner flasks and magnetic stirrers for cell suspension culture (many different types/providers work well).
Trypan blue stain 0.4% (Gibco).
Dounce homogenizer.
2.1.2. Buffers
Lysis buffer: 50 mM HEPES, pH 7.9, 250 mM NaCl, 0.1% (v/v) IGEPAL CA-630, 0.2 mM EDTA, 0.2 mM PMSF, 2 mM β-mercaptoethanol.
2.2. Nuclear Extracts’ Preparation from Mammalian Cells
2.2.1. Buffers
Hypotonic buffer: 10 mM Tris–HCl, pH 7.9, 10 mM KCl, 1.5 mM MgCl2. Autoclave and store at 4°C. To complete the buffer before use, take the required amount and add to final concentration 10 mM β-mercaptoethanol and 0.2 mM PMSF.
2× BC-O: 40 mM Tris–HCl, pH 7.9, 40% (v/v) glycerol, 0.4 mM EDTA. Autoclave and store at 4°C. This is used to make all 1× BC buffers.
BC-500: 20 mM Tris–HCl, pH 7.9, 20% (v/v) glycerol, 0.2 mM EDTA, 500 mM KCl. To complete the buffer before use, take the required amount and add to final concentration 10 mM β-mercaptoethanol and 0.2 mM PMSF. (Working BC buffers are made fresh by diluting 2× BC-O with autoclaved deionized water and appropriate salt).
2.3. Purification of Multisubunit HAT Complexes
2.3.1. Buffers
BC-330: 20 mM Tris–HCl, pH 7.9, 20% glycerol, 0.2 mM EDTA, 330 mM KCl. To complete the buffer before use, take the required amount and add to final concentration 10 mM β-mercaptoethanol, 0.2 mM PMSF, and 0.05% (v/v) IGEPAL (CA–630).
BC-100: 20 mM Tris–HCl, pH 7.9, 20% glycerol, 0.2 mM EDTA, 100 mM KCl. To complete the buffer before use, take the required amount and add to final concentration 10 mM β-mercaptoethanol, 0.2 mM PMSF, and 0.05% IGEPAL (CA–630).
BC-400: 20 mM Tris–HCl, pH 7.9, 20% glycerol, 0.2 mM EDTA, 400 mM NaCl. To complete the buffer before use, take the required amount and add to final concentration 10 mM β-mercaptoethanol, 0.2 mM PMSF, and 0.05% IGEPAL (CA–630).
1× SDS loading buffer: 60 mM Tris–HCl, pH 6.8, 10% glycerol, 2% (w/v) SDS, 0.1% (w/v) bromophenol blue.
2.3.2. Other Materials and Reagents
Thin, round gel-loading tips.
Protein A–Agarose (Pierce).
Anti-FLAG® M2 Affinity Gel/Resin (Sigma).
FLAG® peptide (Sigma).
S-Sepharose Fast Flow (Pharmacia).
3. Methods
3.1. Cell Culture and Cell Line Generation
Culture HEK293 cells (or other mammalian cell line of interest) in a 6-cm culture dish in DMEM supplemented with 10% FBS and penicillin–streptomycin solution at 37°C with 5% CO2.
When the cells reach ~95% confluence, decant the medium from the dish and wash the cells three times with PBS. After the last wash, decant the PBS completely and add 0.3 ml trypsin-EDTA into the dish. Leave the cells in the incubator for 1 min to allow detaching (from the dish and from each other). Resuspend the cells in 6 ml prewarmed DMEM supplemented with 10% FBS and penicillin–streptomycin.
Distribute the cells into two 6-cm culture dishes evenly (each with a total volume ~3 ml). Wait overnight for the cells to attach (see Note 1).
Transfect one dish with 1 µg pFH-GeneX-IRESneo vector containing your gene X of interest with a FLAG (F) and/or HA (H) epitopes in frame at the N or C terminus (31). Gene X can be the catalytic HAT subunit (e.g., GCN5) or another subunit of the multisubunit HAT complex. Transfect the other dish with control empty vector (pFH-IRESneo) and handle the control cells in parallel afterward (see Note 2).
Change medium 24 h after transfection.
24 h later, trypsinize the cells from the dish and transfer them into a 50-ml conical tube containing 45 ml prewarmed DMEM supplemented with 10% FBS and penicillin–streptomycin. Gently resuspend the cells by pipetting up and down. Distribute the cells evenly into two 15-cm dishes (~23 ml per dish).
24 h later, add geneticin/G418 to a concentration of 500 µg/ml to select for stably transfected clones. Select for 2–3 weeks by changing medium every other day to refresh the selection drug and remove the dead cells.
Pick single geneticin/G418-resistent colony and subculture in 3.5-cm culture dish for expansion. Choose several colonies.
Duplicate each clone into two dishes. Use one dish to check for the expression of your FLAG-tagged protein and the other dish for expansion.
To check for expression, when the cells reach 95–100% confluence, decant the medium and wash the cells three times with 1–2 ml of PBS. After the last wash, use a pipettman to remove all the remaining PBS completely.
Add 150 µl of ice-cold lysis buffer to the dish and keep it on ice for 10 min.
Use a policeman to scrap the cells off the dish and pipet the lysate into a microcentrifuge tube.
Spin the lysate on a countertop microcentrifuge at 13,000 rpm/10 min/4°C. Take an aliquot of the supernatant and check for positive clones by SDS-PAGE and western blotting with the FLAG antibody.
Choose a cell clone that expresses your FLAG -tagged protein and adapt the cells from monolayer to suspension. To adapt the cells, trypsinize the cells from the dish and resuspend them gently in SMEM supplemented with 20% FBS in a spinner flask (see Note 3). The density of suspended cells should be 3–4 × 105 cells/ml (see Note 4).
24 h later, check the viability of the cells by staining an aliquot (2–3 ml) of the cells with trypan blue and examine under a microscope (see Note 5).
When the cells reach 7–8 × 105 cells/ml, remove half of the culture medium (i.e., half of the cells) and replace with the same amount of fresh SMEM plus 10% FBS (see Note 6).
Repeat steps 13 and 14 by replacing the medium with different amounts of serum. In repeat 1, replace the medium with SMEM plus 5% FBS and 5% BCS; in repeat 2, replace with SMEM plus 20% BCS; in repeat 3, replace with SMEM plus 10% BCS; in repeat 4, replace with SMEM plus 5% BCS. The cells are ready for expansion after they are adapted to 5% BCS.
Expand the cells to the desired amount (~16 l) for nuclear extracts (NE)’s preparation. Grow the cells to a density of 1 × 106 cells/ml before harvest (see Note 7).
3.2. Preparation of Nuclear Extracts
All procedures should be performed on ice or in the cold room and finished within the same day.
Pour the suspended cells into 1-l bottles. Use a Beckman Coulter J6-HC centrifuge to spin down the cells at 3,000 rpm/10 min/4°C with a JS-4.2 rotor and swinging buckets. Decant the medium carefully. Repeat the process until all the cells in the spinner are collected (see Note 8).
Transfer cells into six 50-ml conical tubes. Use a Beckman Coulter J6-HC centrifuge to spin down the cells at 2,000 rpm/5 min/4°C (JS-4.2 rotor with swinging buckets).
Remove the medium and resuspend the cells gently with ice-cold PBS. Transfer the cell pellet into four 50-ml conical tubes.
Spin down the cells at 2,000 rpm/3 min/4°C, as above.
Remove the supernatant and wash the cells again with ice-cold PBS. Transfer the cell pellet into two 50-ml conical tubes.
Spin down the cells at 2,000 rpm/3 min/4°C.
Measure packed cell volume (PCV).
Remove the supernatant and resuspend the cells with 30–40 ml of ice-cold, complete hypotonic buffer by gently inverting the cells until completely resuspended.
Spin down the cells at 2,000 rpm/3 min/4°C. Use a pipette to remove the supernatant carefully (see Note 9).
Add 1/3 PCV ice-cold, complete hypotonic buffer and resuspend cells carefully by gently inverting the tube.
Swell cells on ice for 10 min.
Pour the cells into a 40 ml ice-cold Dounce homogenizer.
Homogenize the cells on ice using a “B” pestle. To homogenize the cells, slowly and carefully twist the pestle up and down for 15 strokes (see Note 10).
Use a microscope to check the cell breakage and release of nuclei by staining ~5 µl of the homogenate with 10–20 µl of trypan blue on a petri dish (see Note 11).
If there is less than 90% cell breakage, give two more strokes and check again. Repeat this process until more than 90% of the cells have released their nuclei (see Note 12).
Pour the nuclei from the Dounce homogenizer into a 50-ml conical tube and spin the nuclei at 3,500 rpm/20 min/4°C (Beckman JS-4.2 rotor with swinging buckets); measure nuclear pellet volume (NPV) (see Note 13).
Transfer the supernatant (cytoplasmic fraction) to another 50-ml conical tube and measure the volume (cytoplasmic fraction volume (CFV) (see Note 14).
Add 1/4 CFV of 100% glycerol, 1/29 CFV of 3 M KCl, and 1/1,000 CFV of 250 mM EDTA to the cytoplasmic fraction. Gently mix by inverting the tube. Keep it on ice.
Extract the nuclei by adding 2 NPV of ice-cold, complete BC-500 to the nuclear pellet (NP). Resuspend the pellet by pipetting up and down several times with a pipette aid (see Note 15).
Rotate for 1 h at 4°C (see Note 16).
Transfer the nuclei/nuclear extracts mixture and the cytoplasmic fraction into different centrifuge tubes for Sorvall SS-34 rotor.
Use a Sorvall RC-5B centrifuge (or equivalent) to spin at 14,000 rpm/30 min/4°C.
Transfer the supernatant from the nuclei/nuclear extracts mixture tube into a 50-ml conical tube and label nuclear extracts.
Scrap off the nuclear pellet into a 50-ml conical tube and label nuclear pellet.
Transfer the supernatant from the cytoplasmic fraction tube into a 50-ml conical tube and label cytoplasmic extracts (CEs). Discard the pellet.
Keep a small aliquot of NE, NP, and CE.
Snap freeze all these fractions by liquid nitrogen and store at −80°C (see Note 17).
3.3. Purification of FLAG-Tagged Complexes from Nuclear Extracts
3.3.1. First-Step Purification with Anti-FLAG® M2 Affinity Resin
Thaw nuclear extracts on ice (from both control parental cells and cell line expressing your FLAG-tagged protein).
Transfer 15 ml nuclear extracts into a centrifuge tube for Sorvall SS-34 rotor. Add 10% (v/v) of NP-40 (IGEPAL CA–630) to a final concentration of 0.05%. Mix gently by inverting.
Spin at 14,000 rpm/20 min/4°C.
Transfer ~14 ml of nuclear extracts into a 15-ml conical tube. Avoid lipid on the top and pellet at the bottom.
Aliquot 30 µl M2 resin (60 µl 50% slurry) and protein A resin for each 14 ml nuclear extracts (see Note 18).
Wash the resin three times with ice-cold/complete BC-330. After last wash, keep the resin as 50% slurry in BC-330 (see Note 19).
Preclear nuclear extracts by adding 30 µl of preequilibrated protein A resin into each tube. Rotate for 30 min/4°C.
Use a Beckman Coulter J6-HC centrifuge to spin down the resin at 2,000 rpm/1 min/4°C (JS-4.2 rotor with swinging buckets).
Transfer the nuclear extracts into a new 15-ml conical tube. Avoid taking the protein A resin at the bottom.
Add 30 µl of preequilibrated M2 resin into the precleared nuclear extracts (see Note 20).
Rotate for 3 h/4°C (see Note 21).
Spin down the resin at 2,000 rpm/1 min/4°C.
Transfer the supernatant (unbound) into a new 15-ml conical tube and label M2-1×. Snap freeze in liquid nitrogen and store at −80°C (see Note 22).
Wash the resin with 15 ml of ice-cold/complete BC-330 by gently inverting the tube.
Spin down the resin at 2,000 rpm/1 min/4°C. Carefully remove the wash without disturbing the resin.
Repeat the wash step twice.
After the third wash, add 1 ml of ice-cold/complete BC-330 to the 15-ml conical tube. Gently pipet up and down to resuspend the resin.
Transfer the resin into a microcentrifuge tube.
Use a countertop microcentrifuge (Eppendorf or equivalent) to spin down the resin at 2,000 rpm/1 min/4°C. Carefully remove the wash without disturbing the resin.
Repeat the wash step twice.
Adjust salt concentration to 100 mM by washing the resin twice with 1 ml ice-cold/complete BC-100.
Remove the final wash completely (see Note 23).
To elute, add 35 µl of 0.3 mg/ml FLAG peptide solution to the resin and shake it at 900 rpm/30 min/20°C (see Note 24).
Spin down the resin at 3,000 rpm/10 s/4°C.
Use a gel-loading tip to transfer the eluent into a new microcentrifuge tube and label “Eluate 1.” Be careful not to take the resin. Keep eluate 1 on ice.
Repeat the elution step twice. Label the eluates as “Eluate 2” and “Eluate 3.”
Snap freeze all the eluates in liquid nitrogen and store at −80°C (see Note 25).
Analyze 5–8 µl of each eluate (including the control eluates of mock purifications from parental untagged cells) by SDS-PAGE and silver staining.
3.3.2. Second-Step Concentration and Purification on S-Sepharose Resin
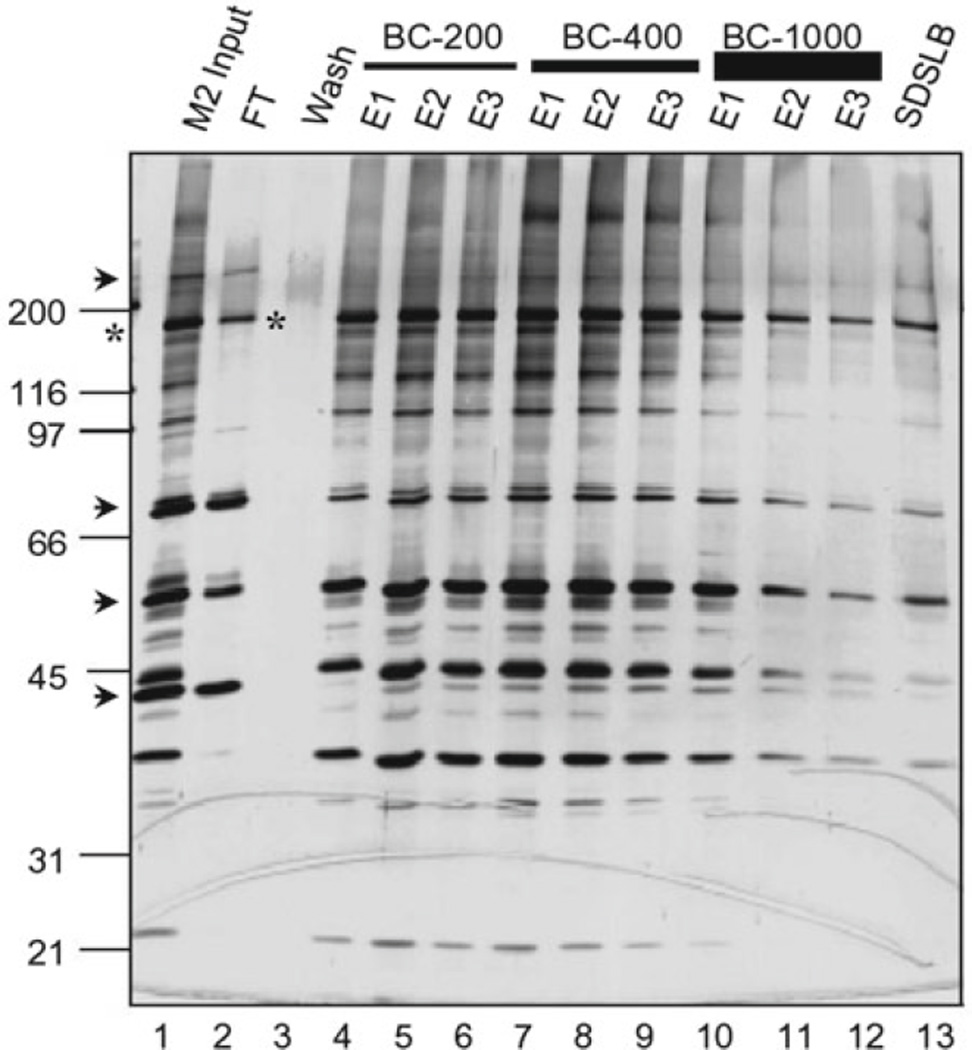
Generally, single-step FLAG/M2-purified HAT complexes are suitable for downstream applications, such as acetylation of histone and non-histone substrates. However, except for the first eluates of the initial M2 Affinity round of purification, subsequent eluates may be too diluted; in addition, proteins that bind nonspecifically to the anti-FLAG/M2 resin (e.g., Hsp70) are still present. These contaminants can be detected by SDS-PAGE and silver staining in the control lane containing the mock purification sample from parental cells that do not express any tagged protein (step 28 above). A simple way to concentrate the pool of diluted complexes and to remove most of the contaminants (including the excess FLAG peptide) is to batch adsorb FLAG-tagged complexes onto a small amount of the negatively charged S-Sepharose (or SP-Sepharose) ion-exchange resin. Most contaminants do not bind tightly to S-Sepharose as illustrated in Fig. 1, which shows a representative S-Sepharose purification of GCN5 HAT-containing ATAC complexes isolated from cells expressing the FLAG-tagged YEATS2 subunit (33).
Thaw M2-purified FLAG-tagged HAT complexes on ice (process the control/mock-purified eluates in parallel).
Pool M2 eluates together (~1 ml total) into one microcentrifuge tube (see Note 26).
Spin on a countertop centrifuge at 13,000 rpm/10 min/4°C.
Transfer 800 µl of the supernatant into a new microcentrifuge tube (see Note 27).
Adjust final salt concentration to 60 mM KCl by adding 533 µl of ice-cold/complete 1× BC-0 to a total of 1,333 µl. Gently mix by inverting the tube.
Keep 20 µl as “input.”
Aliquot 20 µl of S-Sepharose resin (40 µl of 50% slurry) into a microcentrifuge tube and wash three times with ice-cold/complete BC-60. After last wash, keep the resin as 50% slurry in BC-60.
Add 20 µl of preequilibrated S-Sepharose resin (40 µl slurry) to the pooled M2 eluates.
Rotate for 3 h/4°C.
Spin down the resin at 2,000 rpm/1 min/4°C.
Transfer the supernatant into a new microcentrifuge tube and label “S-Sepharose flow through.”
Wash the resin with 1 ml of ice-cold/complete BC-100 by gently inverting the tube.
Spin down the resin at 2,000 rpm/1 min/4°C. Keep the wash and label “wash 1.”
Repeat the wash step three times and label “wash 2, wash 3, wash 4.”
Remove the final wash completely.
To elute, add 25 µl of ice-cold/complete BC-400 to the resin (see Note 28). Gently flick the microcentrifuge tube several times with a finger.
Spin down the resin at 3,000 rpm/10 s/4°C.
Use a gel-loading tip to transfer the eluate into a new microcentrifuge tube and label “S-Sepharose eluate 1.”
Repeat the elution step up to four times and label accordingly.
Add 20 µl of 1× SDS loading buffer to the resin.
Analyze 2–4 µl of each eluate by SDS-PAGE and silver staining. Snap freeze the remaining eluates in liquid nitrogen and store at −80°C.
Fig. 1.
S-Sepharose fractionation of anti-FLAG/M2-purified ATAC complexes. Shown is a silver-stained, SDS-PAGE gel containing different S-Sepharose elution fractions. M2-purified, FLAG-tagged ATAC complexes (M2 input, lane 1) were adsorbed onto S-Sepharose resin in BC-60, washed with BC-100 (Wash, lane 3), and eluted successively with three resin volumes (E1–E3) of the indicated BC buffers containing from 200 mM NaCl (BC–200) to 1,000 mM NaCl (BC-1000) and finally with SDS loading buffer (SDSLB). FT unbound “flow through” fraction. Proteins that bind nonspecifically to M2 agarose (arrow-heads) and contaminate the M2-purified complexes (M2 input) do not bind to S-Sepharose (i.e., present in FT fraction, lane 2). Excess, “free,” FLAG-tagged YEATS2 subunit (asterisks) does not bind to S-Sepharose either (lane 2).
Acknowledgments
The authors would like to thank Dr. Jennifer Liu for advice. This work was supported by grants R01CA100464 and MCB0448488 from NIH and NSF, respectively.
Footnotes
Mammalian cells double approximately every 24 h. Therefore, by the next day, the cells should reach ~95% confluence again.
It is important to select appropriate transfection reagent and perform transfection according to manufacturer’s protocol. To transfect HEK293 cells, we use LipoD293™ at 1:3 ratio (DNA [µg]:transfection reagent [µl]). To obtain good transfection efficiency and low cytotoxicity, transfection conditions should be tested by varying DNA and transfection reagent ratios. Optimal DNA (µg):transfection reagent (µl) ratio generally lies between 1:0.5–1:5. It may be optimal to transfect the cells again the next day if transfection efficiency is not good.
It is important not to use geneticin/G418 during the adaptation process; otherwise, the cells may die.
Depending on the cell type, one 10-cm dish generally equals to 1–2 × 107 cells. Assuming that one 10-cm dish contains 1 × 107 cells, three 10-cm dishes would be 3 × 107 cells. Therefore, cells from three confluent 10-cm dishes can be resuspended into 100 ml of SMEM plus serum at the density of 3 × 105 cells/ml.
Trypan blue goes inside the dead cells and stains them blue. On the contrary, the live ones are not stained and therefore have a shiny contour.
The cell density should be reduced in half but no less than 3 × 105 cells/ml. The cells do not grow if density drops below 3 × 105 cells/ml.
Depending on the cell type, serum concentration, and medium, the density of cells may vary. 16 l of HEK293 cells at a density of 1 × 106 cells/ml typically generate about 30–40 ml nuclear extracts.
Do not decant the medium completely, as this may result in cell loss. Leave approximately 200 ml of medium each time.
The cells swell and increase volume after adding hypotonic buffer.
The time to complete 1 stroke should be ~30 s. Homogenizing the cells too quickly may shear the nuclei. In addition, if the pestle is not twisted well, the cell membrane may not break.
Intact cells appear large and shiny while released nuclei are intensely stained by Trypan blue and tend to aggregate.
Avoid excessive homogenization, which leads to nuclei breakage and release of DNA/chromatin.
Typically, NPV is 15–18 ml for 16 l of HEK293 cell culture.
Typically, CFV is 10–12 ml for 16 l of HEK293 cell culture.
Nuclear pellet is very dense and viscous. Pipetting up and down for several minutes may be required. It is important to dissociate the clumps as much as possible to increase the surface area for optimal nuclear extraction.
Final salt/KCl concentration should be ~333 mM, which should be verified with a conductivity meter.
The protein concentration of the nuclear extracts and the cytoplasmic extracts should be 7–8 mg/ml by using the Bradford protein quantification method with BSA as a standard.
Optimal resin/NE volume ratio is ~1/500 for our FLAG-tagged GCN5 complex cell lines, but varies depending on specific tagged proteins and complexes and must be determined empirically. Avoid the use of too much resin, which increases the “sticking” of nonspecific contaminants.
330 mM is the salt concentration of the nuclear extracts.
Resin may stick to the wall of the tip and microcentrifuge tube. Use the nuclear extracts to pipet up and down several times to wash the remaining resin into the 15-ml conical tube.
If incubation time is too short, FLAG-tagged complexes may not bind efficiently to the M2 resin; if incubation time is too long, nonspecific binding and protein degradation may occur.
The unbound fraction may be used up to three times to purify the remaining FLAG-tagged complexes. However, the yield decreases proportionally.
To completely remove the final wash, place a gel-loading tip against the bottom of the tube and carefully remove all remaining buffer. A small amount of resin may stick to the tip. Carefully circle the tip against the wall of the microcentrifuge tube to collect the tip-bound resin.
Dilute FLAG peptide in ice-cold/complete BC-100 at a concentration of 0.3 mg/ml.
Optional: Repeat the purification (steps 10 – 27) with the unbound protein extracts (step 13) up to twice more.
It is important to check each elution of FLAG-purified complexes by silver staining to decide which eluates are worth pooling.
Minor M2 resin carryover may be at the bottom. Be careful not to take the resin.
BC-400 contains 400 mM of NaCl instead of KCl. This is because loading 400 mM KCl into an SDS-PAGE gel may cause precipitation in the well.
References
- 1.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 4.Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 5.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 8.Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 10.Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 11.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 12.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 13.Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions. Curr Opin Cell Biol. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- 14.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 15.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 16.Weake VM, Swanson SK, Mushegian A, Florens L, Washburn MP, Abmayr SM, Workman JL. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes & Dev. 2009;23:2818–2823. doi: 10.1101/gad.1846409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carruthers LM, Hansen JC. The core histone N termini function independently of linker histones during chromatin condensation. J Biol Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe AP, Hansen JC. Nuclear visions: functional flexibility from structural instability. Cell. 2001;104:631–634. doi: 10.1016/s0092-8674(02)01453-8. [DOI] [PubMed] [Google Scholar]
- 20.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 21.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–689. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol Cell. 2002;9:811–821. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 24.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14:942–948. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 27.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 28.Candau R, Zhou JX, Allis CD, Berger SL. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 30.Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 31.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 33.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Vorontchikhina M, Wang YL, Faiola F, Martinez E. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol. 2008;28:108–121. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol. 2009;29:1649–1660. doi: 10.1128/MCB.01076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Shogren-Knaak MA. The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J Biol Chen. 2009;284:9411–9417. doi: 10.1074/jbc.M809617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Q, Lieberman PM, Boyer TG, Berk AJ. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes & Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- 40.Chiang CM, Ge H, Wang Z, Hoffmann A, Roeder RG. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, 3rd, Washburn MP, Abmayr SM, Workman JL. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, Farina A, Martinez E. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25:10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]