Abstract
Chronic alcohol consumption can lead to the development of alcoholic fatty liver disease. The underlying pathogenic mechanisms however, have not been fully elucidated. Here, we review the current state of the art regarding the application of lipidomics to study alcohol’s effect on hepatic lipids. It is clear that alcohol has a profound effect on the hepatic lipidome, with documented changes in the major lipid categories (i.e. fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids and prenol lipids). Alcohol’s most striking effect is the marked change in the hepatic fatty acyl pool. This effect includes increased levels of 18-carbon fatty acyl chains incorporated into multiple lipid species, as well as a general shift toward increased unsaturation of fatty acyl moieties. In addition to our literature review, we also make several recommendations to consider when designing lipidomic studies into alcohol’s effects. These recommendations include integration of lipidomic data with other measures of lipid metabolism, inclusion of multiple experimental time points, and presentation of quantitative data. We believe rigorous analysis of the hepatic lipidome can yield new insight into the pathogenesis of alcohol-induced fatty liver. While the existing literature has been largely descriptive, the field is poised to apply lipidomics to yield a new level of understanding on alcohol’s effects on hepatic lipid metabolism.
Keywords: alcohol; fatty acyls; fatty liver; glycerolipids, glycerophospholipids; lipidomics; prenol lipids; sphingolipids; sterol lipids
Graphical abstract
General introduction
Alcohol abuse is associated with a significant global health burden, accounting for 6% of all deaths in the Americas, as well as ~9% of all disability-adjusted life years.1 A large part of this burden is associated with disease of the liver, of which there is a well-characterized disease spectrum beginning with alcohol-induced fatty liver, and then progressing to hepatitis, fibrosis and cirrhosis.2 As the initial step in this disease pathway, alcohol-induced fatty liver is of particular interest. Although this condition is commonly regarded as reversible with the cessation of alcohol consumption, it is well-recognized that the development of both non-alcoholic and alcohol-induced fatty liver confers an increased risk for the development of hepatitis, hepatic fibrosis and cirrhosis, as well as hepatocellular carcinoma.3,4,5,6,7,8 In this regard, an improved understanding of the pathogenesis of alcohol-induced fatty liver would be beneficial for the development of therapies that not only reverse lipid accumulation in the liver, but also counteract the progression of fatty liver to more severe forms of disease.
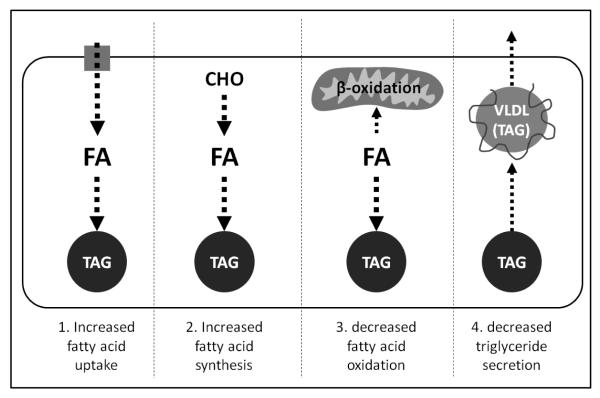
Fatty liver (steatosis) occurs when lipid - primarily triacylglycerol - accumulates in the liver, and is technically defined by a hepatic fat content in excess of 5-10% of liver weight.9 Theoretically, fatty liver can develop through multiple pathways,10 as summarized in Fig. 1. Many of these pathways have been implicated in the context of alcohol-induced fatty liver, including increased fatty acid uptake from the circulation into the liver, increased de novo lipogenesis, decreased fatty acid oxidation, and impaired VLDL secretion.11,12 The relative importance of these pathways and their contribution to alcohol-induced fatty liver remains unclear. Indeed, given the complexity and uncertainties surrounding the pathogenesis of alcohol-induced fatty liver, the ability to quantitatively and specifically measure multiple lipid species in the liver at the same time is advantageous. In this regard, new lipidomic techniques are available to capture the effect of alcohol on the hepatic lipidome and provide new insight into the pathogenesis of alcohol-induced fatty liver. In this review, we will introduce the basic principles of lipidomic analyses, consider recent attempts to characterize the hepatic lipidome, and discuss the current literature describing alcohol’s effects on the hepatic lipidome.
Fig. 1. Multiple possible pathways contribute to the development of alcohol-induced fatty liver.
This schematic highlights four pathways through which chronic alcohol consumption has been proposed to cause hepatic steatosis. 1) Alcohol stimulates the uptake of circulating fatty acids into the liver, which are then used as a substrate for triacylglycerol synthesis. 2) Alcohol stimulates de novo lipogenesis, with the newly synthesized fatty acids acting as a substrate for triacylglycerol synthesis. 3) Alcohol inhibits mitochondrial β-oxidation of hepatic fatty acids, leading to their accumulation and availability for synthesis of triacylglycerol. 4). Alcohol inhibits VLDL secretion from the liver, limiting the liver’s ability to secrete triacylglycerol and causing it to accumulate. FA: fatty acids, TAG: triacylglycerol; CHO: carbohydrate; VLDL: very low-density lipoprotein.
An introduction to lipidomics
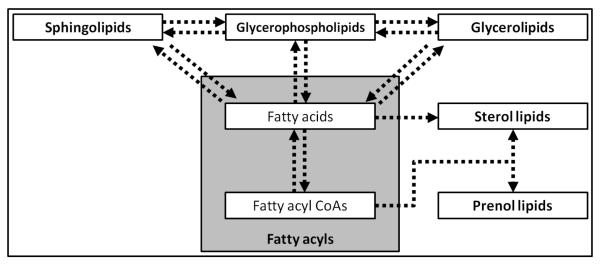
Lipids are a major class of biologically important molecules that can be broadly characterized as insoluble in water and soluble in organic solvents, thereby comprising small molecules that are hydrophobic or amphipathic. In this review, we strived to use the comprehensive classification system and nomenclature established by the LIPID MAPS consortium in accord with the International Lipids Classification and Nomenclature Committee, as outlined in Fig. 2.13,14 In simple terms, lipidomics refers to the comprehensive analysis of lipids in a given cell or tissue, and can be considered as a subdivision of analyses designed to determine the complete set of chemicals within cells (i.e. metabolomics). More broadly, lipidomics is a burgeoning scientific endeavor to identify and quantify the thousands of distinct lipids present in various cell types, and to establish lipid functions in health and disease. According to these definitions, when referring to the hepatic lipidome, we mean the complete spectrum of lipids within this tissue.
Fig. 2. Major mammalian lipid categories and their inter-relationships.
This scheme shows the major lipid categories as classified by the LIPID MAPS consortium including fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids and prenol lipids. This scheme also highlights the central importance of fatty acyls (i.e. fatty acids and fatty acyl CoAs) in the biosynthesis of other lipids (Adapted from Dennis et al., 2010, and Quehenberger et al., 2010).24,84
The complex analysis of lipids in biological samples was made possible by analytical advances in the field of mass spectrometry (MS).15 While gas chromatography MS (GC-MS) and tandem MS were developed in the second half of the 20th century and allowed the analysis of some lipids, the more recent introduction of electrospray ionization (ESI) allowed for a highly sensitive and specific analysis of molecules faster and more cheaply than before. In this regard, ESI-MS is currently thought to be the most sensitive, specific and direct technique for assessing the lipidome of biological samples.15 Parallel improvements in liquid chromatography (LC; e.g. ultra-high performance liquid chromatography), computing power, and the development of specific software applications have also greatly facilitated the production and analysis of lipidomic data.
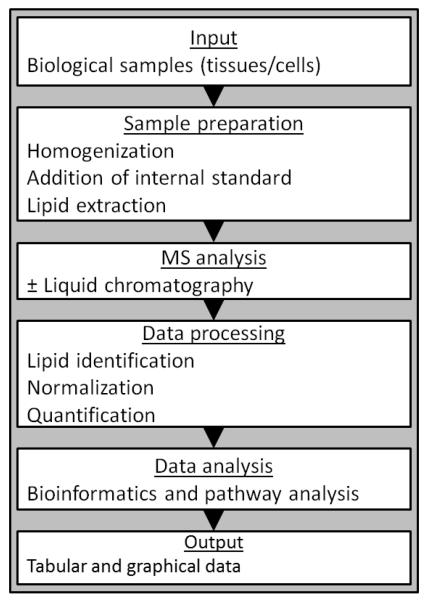
Most MS-based lipidomic analyses follow a standard workflow, as summarized in Fig. 3. While this workflow provides a general framework within which the profiling of lipids in biological samples can be conducted, there are two different analytical methods that can be used to identify and quantify lipids: LC-MS and shotgun lipidomics. LC-MS uses LC to separate lipids prior to MS, whereas shotgun lipidomics works by the direct infusion of lipid extracts into the MS. An in-depth discussion of the respective merits of LC-MS vs. shotgun lipidomics, and their technical refinements is beyond the scope of the current review, and the reader is referred to the following literature.16,17 In brief, the advantages of using LC-MS include higher sensitivity and quantification, especially for low abundance lipids. The use of LC to separate lipids prior to MS is particularly advantageous when analyzing complex biological samples such as the liver; furthermore, the use of LC allows retention time to be used as an additional analytic parameter. In accordance with the multiple categories of lipids and their numerous constituents, several different LC-MS protocols have been developed and optimized to measure specific lipids. An in-depth discussion of these different techniques is not possible in the current review; however, the reader is referred to the following publications that provide a detailed description of analytical protocols for measuring lipids in different categories.18,19,20,21,22,23,24 While less sensitive, shotgun lipidomics has the advantage of allowing large scale, high-throughput analysis of samples without the possibility of chromatographic anomalies. The direct infusion of samples also allows for a more complete analysis of low volume samples, though this is not necessarily a limiting factor when studying the liver because of its relatively large size. In shotgun lipidomics, a greater emphasis is placed on computational analysis for lipid identification and relative quantification.
Fig. 3. Representative workflow for the lipidomic analysis of biological samples.
This scheme shows the typical workflow for the analysis of biological samples using lipidomics (Adapted from Graessler et al., 2009).85
MS-based approaches are not the only techniques for generating lipidomic data. Nuclear magnetic resonance (NMR) spectroscopy can also be used for this purpose, applying a similar workflow to that of MS-based techniques (Fig. 3).25 Both approaches are comparable in terms of the time taken for sample analysis and data processing; however, NMR spectroscopy is less sensitive and the resonance overlay can limit the ability to identify and quantify molecules in complex samples.25,26
Irrespective of the analytical method chosen, the large quantity of data and its complexity makes the analysis and interpretation of lipidomic data a major challenge. Indeed, a strong bioinformatics component is essential if lipidomic datasets are to be effectively utilized, with limitations in data analysis and interpretation potential limiting factors in gathering the most information from these datasets.27,17. In terms of data analysis, several proprietary and open-source software platforms are available to facilitate the processing of MS data, particularly in terms of lipid identification, quantification and statistical analysis. Unsurprisingly, different data processing software packages are available depending on the analytic method used, for example Lipid Inspector is designed for shotgun lipidomic data, whereas Lipid Data Analyzer is designed for LC-MS data.17 Successful interpretation of lipidomic data requires accurate placement in a biological context, which is greatly facilitated by the construction of lipid pathways. These pathways annotate lipids along with accompanying enzymes involved in their synthesis and degradation, as well as potentially relevant binding proteins and other functional effectors (e.g. receptors). For example, the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg) contains a collection of lipid-specific pathways that can be used to create representations of specific pathways with integrated data.
There has been a recent proliferation in the number of published lipidomic studies, facilitated by rapid technological developments that have allowed improved data collection, analysis and interpretation. Although the complete characterization of the mammalian lipidome is a significant endeavor, progress has been made in the field. The LIPID MAPS structures database now contains over 37,000 biologically relevant lipids, and in 2009, 342 lipid species were identified in the yeast Saccharomyces cerevisiae, estimated to reflect 95% coverage of the complete yeast lipidome.28 More recently, changes in the lipidome of murine macrophages were characterized in response to stimulation with lipopolysaccharide; however this study only focused on 400 lipids out of a predicted total in the order of thousands.29 As discussed in the next section, the hepatic lipidome is beginning to be explored, allowing an improved understanding of the role lipids play in the normal functioning of this important organ.
The hepatic lipidome
Methodological strategies for the lipidomic analysis of liver tissue have been described; however, at this time there are fewer than 100 studies catalogued in the MEDLINE database that primarily report an analysis of the hepatic lipidome.30 It should also be noted that the majority of these studies describe the analysis of the hepatic lipidome in animal models, with few studies using human liver samples. In general, hepatic lipidomics studies can be split into two categories, technical papers describing methods to facilitate the analysis of hepatic lipids, and applied papers typically focusing on a particular type of liver disease.
Technical papers have reported methods for the analysis of constituents from the major lipid categories in the liver (i.e., fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids and prenol lipids), including both LC-MS and shotgun lipidomics.31,32. Regarding applied papers, there is an increasing number of studies that focus on the hepatic lipidome in various disease states, including non-alcoholic fatty liver disease,30,33,34 non-alcoholic steatohepatitis,35 liver cancer,35,36, and of course, alcoholic liver injury (discussed below). These papers can be divided generally into those attempting to identify a particular lipid biomarker of disease, or more mechanistic studies that are aimed at improving understanding disease pathogenesis though lipidomic data. As an example of the former, Kwan et al. set out to identify and validate biomarkers for hypertriglyceridemia in two different mouse models.37 The authors studied lipid levels in the liver and serum, and identified individual fatty acids and fatty acyl CoAs in the liver that formed a specific signature associated with hypertriglyceridemia. As an example of the latter, Fu et al. used lipidomic methods combined with proteomics to better understand the mechanisms underlying hepatic endoplasmic reticulum stress secondary to obesity.38 Using this approach, the authors concluded that abnormal lipid metabolism contributes to the development of endoplasmic reticulum stress in the livers of obese mice.
Attempts to characterize the hepatic lipidome have one important caveat: data reported from whole liver extracts reflect analytes from a heterogeneous cell population. There are at least four major cell types to consider in the liver: hepatocytes, hepatic stellate cells, Kupffer cells and endothelial cells. Thus, lipidomic analysis of liver samples reflects a composite lipidome of all the cells of the liver. This limitation becomes apparent when one considers experiments into specific liver diseases. For example, studies into hepatic steatosis would benefit from the specific analysis of hepatocytes – the primary site of triacylglycerol accumulation in the fatty liver; whereas studies into hepatic fibrosis might benefit from the specific analysis of hepatic stellate cells – which, when activated, are the primary contributors of extracellular matrix deposition in the fibrotic liver. Two approaches have been successfully applied to overcome this limitation. One approach involves MS imaging; this technique yields high resolution spatial localization of lipids in cross sections of the liver at the cellular level, and has been used successfully to visualize fatty acyls, glycerolipids, glycerophospholipids, sterol lipids and prenol lipids in the liver.39,40. Instead of imaging lipids in situ, a second approach is to isolate the particular cell type of interest prior to MS analysis. For example, lipidomic analyses of isolated hepatocytes, hepatic stellate cells and Kupffer cells have all been reported, whereas others have focused on specific organelles, including the analysis of purified mitochondria and hepatocyte lipid droplets.41,42,43,44,45
A further methodological approach that greatly improves the utility of lipidomic data obtained from liver samples is its coupling with other analyses. While differences in lipid levels can be informative, accompanying measures such as the concentration of related non-lipid metabolites or the expression level of related enzymes, allow the construction of integrated pathways that provide a more complete picture of lipid metabolism under different experimental conditions. Indeed, the literature contains several examples of researchers who have coupled their lipidomic analyses with a survey of gene expression at the mRNA level using quantitative PCR.34,46,47. At a more sophisticated level, others have combined lipidomics with broader metabolomic, proteomic and transcriptomic analyses to obtain a more complete picture of both metabolite, protein and gene expression levels, respectively.35,38,48,49,50,51,52 Indeed, this combined approach is well-suited to studies in the liver because of the abundance of tissue available. For example, the liver of a 3 month-old male mouse typically provides ~1 g of tissue available for analysis, whereas other organs such as the heart or kidneys typically weigh only ~0.15 g. This relatively large amount of tissue is advantageous because it allows multiple analyses from the same animal, thereby reducing the effect of biological variability between experimental animals in a given dataset. In our laboratory, we can routinely analyze the profile of lipids in a given liver, as well as perform separate analyses of RNA and protein expression levels using the same sample.47
The entire hepatic lipidome has yet to be characterized, although some authors have reported 80% coverage of the total lipid mass of the liver, including hundreds of its most abundant lipids.31 Despite the challenge that complete coverage of the hepatic lipidome represents, strategies for the comprehensive analysis of hepatic lipids, particularly in the context of hepatic steatosis, have been described.30 The next section reviews the current literature describing the effect of chronic alcohol consumption on the hepatic lipidome.
The effect of chronic alcohol consumption on the hepatic lipidome
At the time of writing, we identified eight publications from the MEDLINE database where the primary goal was to describe the metabolomic/lipidomic profile of liver tissue collected from control and alcohol-fed experimental animals.47,53,54,55,56,57,58,59 As summarized in Table 1, these publications encompass studies in different species, with different alcohol feeding protocols and methods of analysis. A brief summary of each of these reports is provided below, including information on the analytical method, experimental model, main findings and conclusions. Later, we will discuss patterns and trends that emerge from the available data, make some recommendations to be considered when designing studies into alcohol’s effect on the hepatic lipidome, and finish with an overall summary of the current state of the art with respect to the effect of chronic alcohol consumption on the hepatic lipidome. It should be noted that other researchers have analyzed alcohol’s effects on the lipid content of other tissues in addition to the liver. This work has largely focused on the identification of biomarkers for alcohol-induced liver injury in readily collected tissues such as plasma and urine.60,61 These papers will not be discussed further in this review because they do not specifically focus on hepatic lipids or the hepatic lipidome.
Table 1.
List of publications studying the hepatic lipidome in animal models of chronic alcohol consumption
| Author | Year | PMID | Model Species | Alcohol Feeding Protocol |
Tissue (s) Studied | Analytical Method | Lipid Category Analyzed |
|---|---|---|---|---|---|---|---|
| Fernando, H. | 2012 | 22884994 | Deer mice | LDeC | Liver | 1H NMR, 31P NMR | FA, GL, GP, ST |
| Jang, J-H | 2012 | 22532405 | Zebrafish | Water:EtOH* | Liver | 1H NMR, GC-MS | FA, GL, GP,ST |
| Zhao, Z. | 2011 | 21058963 | C57BL/6 mice | LDeC | Liver, skeletal muscle, heart, kidney, white adipose |
LC-MS | FA, GP, SP |
| Loftus, N. | 2011 | 21028815 | C57BL/6 mice, Wistar rats |
IG | Liver | LC-MS | FA,GL,GP,ST, PL |
| Clugston, R.D. | 2011 | 21856784 | C57BL/6 mice | LDeC | Liver, plasma | LC-MS | FA, SP |
| Fernando, H. | 2011 | 21736892 | Fischer 344 rats | LDeC | Liver, plasma | 1H NMR, 31P NMR | FA, GL, GP, ST |
| Fernando, H. | 2010 | 20682011 | Fischer 344 rats | LDeC | Liver, plasma | 1H NMR, 31P NMR | FA, GL, GP, ST |
| Bradford, B.U. | 2008 | 18674555 | C57BL/6mice | LDeC | Liver, urine |
1H NMR, direct infusion MS |
FA, PL |
Abbreviations: PMID: PubMed Identification number; FA, Fatty Acyls; GL, Glycerolipids; GP, Glycerophospholipids; SP, Sphingolipids; ST, sterol lipids; PL, prenol lipids; LDeC, Lieber-DeCarli liquid diet; IG: intrasgastric ethanol infusion
Jang et al., added ethanol to zebrafish system water
Metabolomic profiling of a modified alcohol liquid diet model for liver injury in the mouse uncovers new markers of disease
Bradford et al. studied the effect of alcohol on the hepatic metabolome in male mice consuming control and alcohol-containing Lieber-DeCarli liquid diets for up to 5 weeks.53 Data on lipid levels in the liver were obtained using direct infusion (shotgun) ESI-MS. While the majority of the data presented in this paper concerns non-lipid metabolites, it is included here because it was one of the first published studies to perform a metabolomic analysis of the alcohol-exposed liver. The lipid data reported in this paper focused on relative changes in fatty acyls, primarily fatty acids. It should be pointed out that the identification of lipids in this study was based primarily on mass, cross-referenced with known metabolite information available in the Human Metabolome Database and the LIPID MAPS database. This method of identification can be equivocal, for example a significant increase in a metabolite with the exact mass of 282.255 was observed, but the authors were unable to definitively ascertain if this related to vaccenic acid (C18:1 n-7) or oleic acid (C18:1 n-9).
1H and 31P NMR lipidome of ethanol-induced fatty liver
Here, we consider two papers published by Fernando and colleagues in 2010 and 2011.54,55 These papers describe changes in the hepatic lipidome of rats in response to consumption of Lieber-DeCarli liquid diets containing 5% ethanol for 1, 2 and 3 months. The 2010 paper focuses on changes in the hepatic lipidome at the 1 month time point, while the subsequent paper focuses on the 2 and 3 month time points. Both studies use the same analytical approach complimented by histology and basic biochemical measures. Methodologically, the authors used proton (1H) and phosphorous (32P) NMR spectroscopy to observe changes in hepatic lipids in response to alcohol. The main findings of both papers are in agreement, with both reporting increased levels of hepatic cholesterol and decreased phosphatidylcholine levels in response to alcohol. The extent of acyl chain saturation in all liver lipids was also observed to change in response to alcohol in both studies, such that there was decreased saturation of fatty acyls in alcohol-fed rats. These studies provide examples of the limitations of using NMR spectroscopy to study the hepatic lipidome. While the authors provided an excellent time series of changes in the hepatic lipidome in response to alcohol consumption, the lack of quantitative data made it impossible to discern progressive changes in lipid levels over time. Furthermore, NMR spectroscopy’s lack of specificity was apparent in the equivocal identification of some lipids. For example, while cholesterol could be positively identified through its unique composition, changes in metabolites with CH=CH groups could only be attributed to the subclass of unsaturated fatty acids. This limitation also extended into difficulties identifying lipid classes, for example the authors mentioned in their discussion that triacylglycerols could not be identified because of resonance overlap between the glycerol backbone of triacylglycerol and phospholipids.55 Similarly, the changes in fatty acyl chain composition could not be ascribed to changes in a specific lipid category.54 Nevertheless, these reports provide important reference data and allow comparisons to be made between studies using NMR spectroscopy vs. MS-based approaches.
Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study
Our group has also contributed to the literature regarding alcohol’s effects on the hepatic lipidome.47 In our study, we analyzed the effect of chronic alcohol consumption in mice using Lieber-DeCarli liquid diets (up to 6.4% alcohol for 5 weeks). Analytically, we used an LC-MS based approached that was targeted to specific fatty acyls and sphingolipids, allowing quantitative lipid levels to be obtained in the liver and plasma. With regard to fatty acyls, our main findings included the observation that alcohol feeding was associated with increased hepatic levels of fatty acids and decreased levels of CoAs. Whereas our analysis of sphingolipids revealed broad increases in ceramide levels, which was associated with increased levels of their precursor molecules sphingosine and sphinganine. Our study was different from the other alcohol-related papers discussed here in that we also performed an analysis of gene expression levels in the liver, allowing an improved mechanistic insight into alcohol’s effects on specific lipid pathways. For example, we correlated decreased expression of the enzyme Elovl5 with increased levels of the fatty acid C18:3. Given that the Elovl5 gene product catalyzes the elongation of the long-chain fatty acid C18:3 to C20:3, we interpreted this data to indicate that the accumulation of the C18:3 fatty acid could be explained by its decreased elongation to C20:3.47
Metabonomic investigation of liver profiles of nonpolar metabolites obtained from alcohol-dosed rats and mice using high mass accuracy MSn analysis
Loftus et al. studied alcohol’s effect on hepatic lipid levels in rats and mice using intragastric infusion of ethanol over a period of 4 weeks.56 Hepatic lipid levels were measured using LC-MS and the data included the relative quantification of lipids from all the major lipid categories (fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids and prenol lipids). The major findings reported in this paper include a relatively large decline in hepatic retinol and cholesterol levels, as well as consistently elevated levels of several fatty acid ethyl ester species (including ethyl arachidonate, oleate and linoleate). A strength of this paper was the analysis of liver tissue from alcohol-fed rats and mice. This allowed cross-species comparisons, showing that both species of rodent generally responded similarly to alcohol in terms of changes to the hepatic lipidome. While this paper had the broadest coverage of lipid categories compared to all the other papers reviewed here, it was limited by the fact the authors only reported relative changes in lipid levels. Indeed, in the discussion of their paper the authors proposed that further studies were needed to develop quantitative measures of lipids.
Ethanol-induced alterations in fatty acid-related lipids in serum and tissues in mice
Zhao et al. studied alcohol’s effects on lipids in a mouse model, using Lieber-DeCarli liquid diets (5% alcohol for 4 weeks).57 In this comprehensive study, the authors used LC-MS to study the effect of alcohol on the lipidomes of multiple tissues, including the liver, skeletal muscle, heart, kidneys, white adipose and serum. The study’s key findings include the observation that alcohol has a tissue specific effect on lipid levels, although some consistent patterns emerged between tissues. Strikingly, the authors observed that lipids containing 18 carbon fatty acyl species were increased in multiple tissues, including C18:0 sphingomyelin, C18:0 ceramide, and C18:0/C18:2 phosphatidylcholine. Similarly, lipids containing a C22:6 fatty acyl moiety also increased in many tissues. Cluster analysis of phospholipid changes in the different organs studied revealed significant clustering between the liver and serum relative to the changes observed in other tissues. This observation was taken to indicate that alcohol’s effects on lipid levels in the liver and serum are somewhat comparable, and opens the possibility of using changes in serum lipid levels as a proxy for changes in the liver, and therefore the development of serum biomarkers for alcohol toxicity. Strengths of this study include the analysis of multiple distinct lipids in several lipid categories (fatty acyls, glycerophospholipids, and sphingolipids), the analysis of different tissues, and the reporting of quantitative data.
Metabolic profiling of an alcoholic fatty liver in zebrafish (Danio rerio)
Jang et al. studied alcoholic fatty liver in zebrafish.59 This experimental model has previously been shown to be suitable for the study of alcoholic liver disease and has several unique advantages, particularly with respect to the economics of the research.62,63 The relatively short alcohol feeding protocol used in this study involved the direct addition of ethanol (1.2% v/v) into the fish’s water for 9 hours a day, repeated over a period of 7 days. The authors compiled a metabolomic profile of the liver of alcohol-exposed zebrafish, including lipids, combining data collected using 1H-NMR spectroscopy and GC-MS. Similar to the limitations experienced by Fernando et al.,58 the identification of individual lipids by 1H-NMR spectroscopy was limited and only changes in broad lipid categories were identified. For example, alcohol exposure was associated with increased hepatic phosphatidylcholine levels. The author’s GC-MS data allowed a more specific look at several distinct lipid species, including the alcohol-induced elevation of multiple fatty acids, including palmitic (C16:0), oleic (C18:1) and arachidonic acids (C20:4). The use of two analytic methods (1H-NMR spectroscopy and GC-MS) was a strength as it provided a broader array of analytic data, as well as a means to confirm data obtained by one method of analysis with the other. This study provides important data demonstrating that global metabolomic profiling of the zebrafish liver is possible and applicable for the study of alcoholic liver disease, highlighting the utility of this unique animal model.
Hepatic lipid profiling of deer mice fed ethanol using 1H and 31P NMR spectroscopy: a dose-dependent subchronic study
In addition to their studies in the rat model described above, Fernando et al. also used 1H and 32P NMR spectroscopy to study the hepatic lipidome of deer mice fed ethanol.58 These authors used Lieber-DeCarli liquid diets to feed deer mice alcohol for two months, at a final ethanol concentration of 1%, 2% or 3.5%. Moreover, they also compared the effect of alcohol feeding in the liver of wild-type animals, with deer mice carrying a natural genetic mutation in hepatic alcohol dehydrogenase (ADH), as previously described.64 The main findings of this study focused on the mutant ADH group fed 3.5% ethanol for 2 months, which had the most profound hepatic pathology compared to the other experimental groups. In this group, several changes were observed in the hepatic lipidome, including relatively increased levels of total cholesterol, cholesterol esters, fatty acid methyl esters, and triacylglycerols. The authors also reported decreased levels of free cholesterol and phospholipids in alcohol-fed animals. Although changes were observed in the hepatic lipidome, the author’s analysis did not generally extend beyond general lipid categories, thus little information on individual lipid species was available. Indeed, the authors concluded that there is a need for an alternative “approach to identify the individual lipid(s) at various stages of [alcohol liver disease]”.58 Indeed, this particular study can perhaps be best viewed as a proof of principle study, highlighting the utility of using NMR spectroscopy to probe the hepatic lipidome. Through the use of mutant ADH deer mouse, this study also highlights the benefits of using animal models with genetic mutations to gain further insight into the pathogenesis of alcohol-induced liver disease. Furthermore, through their use of low doses of alcohol, the authors were able to conclude that lipidomic approaches are sensitive enough to show changes in the lipid profile of the liver prior to the onset of frank fatty liver disease.
Emerging insights from the lipidomic analysis of the alcohol-exposed liver
The eight papers discussed in detail above analyze alcohol’s effect on hepatic lipids using distinct methodologies; including differences in the species studied, alcohol feeding protocol and/or analytical method. Such diversity in experimental approaches provides an opportunity to ask questions about what general trends emerge regarding alcohol’s effect on hepatic lipids. While it is well-established that chronic alcohol consumption is associated with an increase in hepatic triacylglycerol levels, lipidomic approaches allow us to dig deeper to yield new and potentially important information. Given the large amount of lipidomic data presented in these papers, it is not possible to provide a synthesis of all the reported results; however, the text below will discuss selected lipids where sufficient data is available to allow for comparisons between studies.
Fatty acyls are one of the more diverse lipid categories in the mammalian lipidome, and as shown in Fig. 1, fatty acids and fatty acyl CoAs are important building blocks that are incorporated into many other more complex lipids. The majority of data on this lipid category with regard to alcohol’s effects is primarily focused on the fatty acids. The studies by Bradford, Clugston, Jang, Loftus and Zhao et al. present data on specific fatty acids in the alcohol-exposed liver. A review of this data reveals several striking patterns. Most notably, fatty acid levels are increased by alcohol consumption. This finding is entirely consistent with historical data showing that alcohol is associated with increased hepatic fatty acid levels,65 though the use of lipidomic analyses allows a closer examination of individual fatty acid species. In particular, it is apparent that alcohol has a profound effect on fatty acids with an 18 carbon acyl chain. Specifically, alcohol increases multiple 18-carbon fatty acids with varying degrees of saturation (indicated in parentheses), as seen in the studies reported by Bradford (C18:0, C18:1 and C18:2), Clugston (C18:0, C18:1, C18:2 and C18:3), Loftus (C18:2 and C18:3), and Jang et al. (C18:0, C18:1 and C18:2).47,53,56,59 Other notable effects of alcohol on specific fatty acids include consistently reported increases in the level of docosahexaenoic acid (C22:6), which was significantly increased by alcohol in three of the studies reviewed.47,56,59 As a precursor for eicosanoid synthesis, arachidonic acid (C20:4) is also of particular interest; however, there is no clear consensus on alcohol’s effect on this important lipid. Bradford and Loftus et al. both reported that arachidonic acid was significantly decreased by alcohol in the mouse liver.53,56 Conversely, arachidonic acid levels were increased in the liver of rats and zebrafish.56,59 Thus, it appears alcohol may have a species-specific effect on arachidonic acid, or at least its response to alcohol is more complex than the other fatty acids mentioned above.
Fatty acyl ether esters (FAEEs) are an important lipid class within the category of fatty acyls. Only two of the lipidomic papers we reviewed reported FAEE data;47,56 however, they have been included in this synthesis for three reasons; first, FAEEs can be formed by the non-oxidative metabolism of ethanol and have been widely proposed to be biomarkers of alcohol consumption;66 second, the accumulation of FAEEs has been reported in multiple tissues (e.g. liver, heart, and lung), with increasing evidence suggesting they may be cytotoxic;67,68,69 and third, the observed pattern of FAEE accumulation in the alcohol-exposed liver is consistent with broader changes in the hepatic lipidome. Specifically, both Clugston and Loftus et al., reported significant increases in hepatic FAEE levels, with the largest increases observed for ethyl stearate (C18:0), oleate (C18:1) and linoleate (C18:2).47,56 Thus, similar to the accumulation of fatty acids described above, there is a propensity for FAEEs with an 18-carbon acyl chain to accumulate in the liver of alcohol-fed mice. It should be noted however, that increases in FAEEs were not limited to this group of lipids, with both Clugston and Loftus et al. reporting increased levels of ethyl palmitate (C16:0).47,56
Another group of lipids that have received wide attention in the literature are ceramides. It is becoming apparent that these sphingolipids are important in numerous cellular signaling pathways and their accumulation has been proposed to be cytotoxic.70,71 For example, it is thought that the accumulation of ceramide in the heart can cause cardiac dysfunction.72 There is also growing literature linking ceramides with hepatic steatosis and alcoholic liver injury.73,74 In terms of the effect that chronic alcohol consumption has on hepatic ceramide levels, Clugston et al. reported increased levels of total ceramide, with individual increases in multiple ceramides including N-stearoyl ceramide (C18:0 Cer) and N-oleoyl ceramide (C18:1 Cer).47 Zhao et al. also reported increased levels of N-stearoyl ceramide (C18:0 Cer) in the liver of alcohol-fed mice, as well as increased levels of N-palmitoyl ceramide (C16:0 Cer).57
Alcohol has a profound effect on the acyl composition of hepatic lipids. If we consider that fatty acids are important building blocks for the synthesis of many other complex lipids, then perhaps, unsurprisingly, the significant increases in 18-carbon fatty acids are reflected in many other lipid species that incorporate acyl chains derived from fatty acids. What is also apparent from several of the studies discussed here is the shift toward increasingly more unsaturated acyl chains. In absolute terms, Clugston et al. demonstrated that while the concentration of saturated fatty acids did not significantly increase in the liver of alcohol-fed mice, mono- and polyunsaturated fatty acids both significantly increased.47 This shift towards greater unsaturation is also reflected in the other studies that report specific levels of hepatic fatty acids, and in all three studies reported by Fernando et al. the authors reported an overall shift from lipids with saturated fatty acyl chains to those with unsaturated fatty acyl chains.54,55,58 Future studies into this general shift toward unsaturated fatty acyls, particularly those with 18-carbon acyl chains, are required to determine this shift’s possible impact on the initiation and progression of alcoholic liver injury. Given that oleic acid (C18:1) is known to be more steatogenic than palmitic acid, there could be a clear link between the accumulation of this lipid and the development of alcoholic fatty liver.75
It is well-established that chronic ethanol consumption is associated with increased hepatic triacylglycerol accumulation. Many of the studies reviewed here report increased hepatic triacylglycerol levels in alcohol consuming mice based on histology or simple biochemical assays; however, the use of lipidomic approaches to measure triacylglycerol composition has not yet provided much additional information. Those investigators using NMR spectroscopy all reported data on triacylglycerols, focusing on metabolites identified as comprising the glycerol backbone of triacylglycerols.54,55,58,59 These studies all reported an alcohol-associated increase in hepatic triacylglycerol levels, but were unable to provide further information, such as the fatty acyl composition or identification of individual triacylglycerol species. The MS-based approach used by Loftus et al. reported an alcohol-induced increase in specific triacylglycerols; however, changes in several possible isomers could have explained the effect.56 One of the reasons that triacylglycerols are not more widely studied is because of their complex molecular structure and the large number of unique triacylglycerol species present in the lipidome. For example, Murphy et al. highlighted the large number of triacylglycerols present in the mammalian lipidome, identifying >500 unique triacylglycerols in RAW 264.7 cells.76 Analyzing such a diverse class of lipids is complicated further by the presence of multiple isobaric species, for example triacylglycerols with the combined fatty acyl composition of C52:2 could reflect a molecule containing the fatty acyl moieties C16:0/C14:0/C22:2 or C16:0/C16:0/C20:2. Thus, the quantitative analysis of triacylglycerols requires the use of specialized MS protocols, meaning this class of lipids is under-studied in the context of alcohol’s effect on the hepatic lipidome.
Another lipid category that has only been studied at a superficial level in the livers of alcohol-fed animals is the glycerophospholipids. This is another complex category of lipids that contains multiple classes and sub-classes of lipids with their own high level of complexity and diversity. In their description of a LC-MS method to analyze glycerophospholipids, Retra et al. identified over 400 unique lipid species in the rat liver, highlighting the challenge that analysis of this lipid category represents.77 As described above for triacylglycerols, multiple isobaric species can also exist, making the identification of glycerophospholipids more difficult. For example, it is common to report the combined fatty acyl composition of phosphatidylcholines (e.g. C36:4), without providing additional information on the contributing fatty acyl groups.78 The papers we reviewed here reported alcohol’s effects on the following glycerophospholipids: glycerophosphocholines (phosphatidylcholine and lysophosphatidylcholine), glycerophosphethanolamines (phosphatidylethanolamine and lysophosphatidylethanolamine), glycerophosphoserines (phosphatidylserine) and glycerophosphoinositols (phosphatidylinositol).54,55,56,58,59. Similar to the triacylglycerols discussed above, the available data on the different glycerophospholipid classes only provides information on alcohol’s effect on the entire class, be it increased or decreased, with no information available on individual species within the class, or their fatty acyl composition.
Recommendations for future studies in the effect of alcohol on the hepatic lipidome
The research undertaken for this review has given us the opportunity to gain perspective on the advantages and limitations of using lipidomic approaches to study alcohol’s effect on hepatic lipids. Based on this analysis of the literature, we propose four recommendations that in our opinion are important considerations when designing studies into alcohol’s effect on the hepatic lipidome
1. Integrate lipidomic data with other metrics of lipid metabolism.
As discussed above, a comprehensive knowledge of lipid levels in a given tissue provides an important insight into the state of lipid metabolism within that tissue. It is important to enhance this level of understanding by providing complementary data on other important effectors in these pathways, such as enzymes, binding proteins, or receptors. An integrated approach fosters thinking in terms of metabolic pathways, and not just individual lipids in isolation. Complementary data can take the form of comprehensive metabolomic, proteomic or transcriptomic analyses, or more targeted protein and gene expression analyses similar to the approach we have used.47 This dual approach allows different datasets to be integrated, thereby providing a deeper level of understanding. As Zhao et al. concluded in their study of alcohol’s effect on tissue lipid levels, the “evaluation of specific enzymes and genes that are responsible for the observed lipidomic changes…could be useful in understanding the mechanism of early stage (alcoholic liver disease)”.57
2. Include multiple time points to capture dynamic changes in the hepatic lipidome
There are two related considerations relevant to the timing of data collection: first, that multiple experimental time points reveal dynamic changes in lipid levels and, second, that changes in lipids prior to the onset of diseases may reveal important information for understanding the pathogenic process. Our recent study into the effect of alcohol on the acyl composition of hepatic retinyl esters highlights the additional insight gained by studying multiple time points.78 Through the use of multiple time points – particularly those early in the alcohol feeding protocol – we revealed that while retinyl palmitate levels precipitously dropped shortly after the onset of alcohol consumption, there was a compensatory increase in retinyl oleate levels, such that total hepatic retinyl ester levels remained unchanged early on. Thus, through the use of multiple experimental time points, we established that alcohol has a dynamic effect on hepatic retinyl ester levels, providing a new understanding of alcohol’s known effects on hepatic retinoids.79 Fernando et al. studied the effect of chronic alcohol consumption for 1, 2 and 3 months in rats, revealing the dynamic effects of alcohol on broad lipid categories within the liver.54,55,58 These authors also highlighted the utility of using lipidomics at early time points, prior to the development of alcoholic fatty liver disease. While we recognize that the addition of extra experimental time points is not always practical, we do believe the extra information derived from such an experimental design is advantageous for a more meticulous understanding of alcohol’s effect on the lipidome.
3. Be quantitative
Only two of the eight studies we reviewed above provide quantitative data on hepatic lipid levels.47,57 While certain methodological approaches only generate relative data, we are of the opinion that quantitative data is more valuable because it provides an extra dimension of information that can highlight the biological relevance of the findings. Reporting relative changes in lipid levels clearly highlights what has changed in response to alcohol consumption, but provides no information on the relative abundance of lipids. For example, Bradford et al. highlighted that alcohol consumption increased hepatic levels of eicosadenoic acid (C20:2), yet this low abundance lipid accounts for less than 1% of fatty acids in the liver.53 On the other hand, the increase in oleic acid (C18:1) reported by several groups could be considered more biologically important since this lipid accounts for ~40% of hepatic fatty acids.47 This need for more quantitative studies is echoed by Loftus et al. who concluded that “the development of fully quantitative assays…will be required to better assess their value in understanding these models of alcohol-induced liver injury”.56 We do recognize however, that the presentation of relative data can be advantageous because it readily allows the reader to determine the direction of change of a given analyte. In this regard, our analysis of the hepatic lipidome included heat maps and bar charts showing relative changes in the lipids measured, but also supplied quantitative data in the supplemental material.47
4. Report ‘negative’ data
Seven of the eight studies reviewed here only presented data on lipids whose level had changed in response to alcohol. While this approach makes data presentation easier, the authors deny the reader a deeper understanding of the study’s findings. If a particular lipid is not presented in a table or figure should the reader assume that it was not measured, or that it was measured and found to be unchanged? The study by Zhao et al. shows that they were able to measure >50 unique lipids in the tissues of alcohol-fed mice, but only presented data on the 10 whose concentrations were altered in the liver.57 The presentation of ‘negative’ data is essential to understanding alcohol’s effects on the hepatic lipidome, and identifying pathways or classes of lipids that do not change are equally as informative as delineating pathways that do change. Similar to our recommendation regarding quantitative data, negative data could be published in the supplemental material to avoid cluttering the main manuscript.
The general recommendations listed above reflect our opinion on the important information that should be provided when studying the hepatic lipidome. An extra layer of complexity must be considered when we think about experimental models of chronic alcohol consumption. As highlighted in Table 1, the studies we have discussed used different alcohol feeding protocols in different species. A discussion of the advantages and limitations of these different approaches is beyond the scope of this review, and we do not make any specific recommendations in this regard. We do, however, refer the reader to the recent review by de la Hall et al. that critically evaluates different models of alcoholic liver disease, as well as the recent description of the National Institute of Alcohol Abuse and Alcoholism models of chronic and binge alcohol feeding.80,81 Suffice to say that methodological diversity is advantageous because it allows the identification of general phenomena that are not unique to a specific species or route of alcohol exposure, while a more uniform approach allows results from different research groups to be compared directly and integrated easily. In this respect, it would seem that the preferred model would be the one that best recapitulates alcohol’s effects in humans; however, to our knowledge no lipidomic studies into liver tissue from human alcoholics have been published.
A further important consideration when designing studies into the hepatic lipidome is what lipids to focus on. Ideally, methods that cover the entire hepatic lipidome would provide a complete snapshot of the entire lipid content of the liver. In practical terms, however, this complete coverage is not possible, therefore decisions on what lipids to study must be made a priori.
Given the current state of our knowledge, it is unsurprising that alcohol has an effect on hepatic fatty acids and triacylglycerol levels, but when we consider the pathogenesis of alcoholic fatty liver disease and its progression, what other lipids should be routinely included in lipidomic analyses of the alcohol-exposed liver? As discussed above, it is becoming apparent that ceramides can be lipotoxic and have been linked to alcoholic liver disease, thus their further study is warranted.73,73 Other toxic lipids that have been associated with liver disease include diacylglycerols, yet they are understudied in the context of alcoholism.82 Similarly, there is a growing literature linking endocannabinoid signaling and alcoholic liver disease, with our own lipidomic analysis reporting increased levels of anandamide in the alcohol-exposed liver.47,83 A particular group of lipids that is pertinent to our own research are the retinoids; there is a longstanding literature linking chronic alcohol consumption with the depletion of hepatic retinoid content, but it remains unclear what the disease-related significance of this is.78,79
When one considers the wide array of lipids that could be studied in the liver that may be important in the pathogenesis of alcohol-induced fatty liver, lipidomic analyses yield a double-edge sword. Lipidomics allows the identification of hundreds of lipids in the liver, with the hard task becoming sifting through the data and identifying pathologically significant changes. As discussed above, there is continual improvement in the bioinformatics resources available to investigators performing lipidomic studies. We believe these improvements, in combination with the above recommendations, should allow more meaningful data to be extracted from large lipidomic datasets.
Concluding remarks
Alcohol clearly has a profound effect on the hepatic lipidome, but it is also apparent that our knowledge of the large array of lipids normally present in the liver and their physiological significance is evolving constantly. As the methods for lipidomic analyses advance, we expect to learn more about the complex interplay of lipid metabolic pathways in the liver, and how these pathways are affected by chronic alcohol intake. This review establishes that alcohol has an effect on members of all the major lipid categories in the liver. The most striking change routinely observed is a shift towards unsaturation of the hepatic fatty acyl pool, with a propensity for significantly increased levels of mono- and poly-unsaturated 18 carbon fatty acyls. While the current literature describing alcohol’s effects on the hepatic lipidome is mostly descriptive, we anticipate that future studies will be more hypothesis driven and move toward an improved mechanistic understanding of the development of alcoholic fatty liver, especially when integrated with other relevant data.
Acknowledgments
This research was supported by the National Institutes of Health under award numbers: R21AA021336 (WSB), R01DK068437 (WSB) and K99AA022652 (RDC). MAG is supported by a Columbia University work exemption program grant.
References
- 1.Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet. 2009;373(9682):2223–33. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- 2.Mann RE, Smart RG, Govoni R. The epidemiology of alcoholic liver disease. Alcohol Res Health. 2003;27(3):209–19. [PMC free article] [PubMed] [Google Scholar]
- 3.Banciu T, Tudose N. Hepatic fibrosis in alcoholic steatosis. Morphologic aspects and evolutive tendencies. Med Interne. 1988;26(2):121–4. [PubMed] [Google Scholar]
- 4.Teli MR, Day CP, Burt AD, Bennett MK, James OF. Determinants of progression to cirrhosis or fibrosis in pure alcoholic fatty liver. Lancet. 1995;346(8981):987–90. doi: 10.1016/s0140-6736(95)91685-7. [DOI] [PubMed] [Google Scholar]
- 5.Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma--epidemiological trends and risk factors. Dig Dis. 2009;27(2):80–92. doi: 10.1159/000218339. [DOI] [PubMed] [Google Scholar]
- 6.Orman ES, Odena G, Bataller R. Alcoholic liver disease: pathogenesis, management, and novel targets for therapy. J Gastroenterol Hepatol. 2013;28(Suppl 1):77–84. doi: 10.1111/jgh.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pais R, Charlotte F, Fedchuk L, Bedossa P, Lebray P, Poynard T, Ratziu V. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J Hepatol. 2013;59(3):550–6. doi: 10.1016/j.jhep.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 8.Onnerhag K, Nilsson PM, Lindgren S. Increased risk of cirrhosis and hepatocellular cancer during long-term follow-up of patients with biopsy-proven NAFLD. Scand J Gastroenterol. 2014:1–8. doi: 10.3109/00365521.2014.934911. [DOI] [PubMed] [Google Scholar]
- 9.Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290(5):G852–8. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- 10.Hawkins RD, Kalant H. The metabolism of ethanol and its metabolic effects. Pharmacol Rev. 1972;24(1):67–157. [PubMed] [Google Scholar]
- 11.Moriya T, Naito H, Ito Y, Nakajima T. “Hypothesis of seven balances”: molecular mechanisms behind alcoholic liver diseases and association with PPARalpha. J Occup Health. 2009;51(5):391–403. doi: 10.1539/joh.k9001. [DOI] [PubMed] [Google Scholar]
- 12.Sozio MS, Liangpunsakul S, Crabb D. The role of lipid metabolism in the pathogenesis of alcoholic and nonalcoholic hepatic steatosis. Semin Liver Dis. 2010;30(4):378–90. doi: 10.1055/s-0030-1267538. [DOI] [PubMed] [Google Scholar]
- 13.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839–61. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50(Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44(6):1071–9. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31(1):134–78. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kofeler HC, Fauland A, Rechberger GN, Trotzmuller M. Mass spectrometry based lipidomics: an overview of technological platforms. Metabolites. 2012;2(1):19–38. doi: 10.3390/metabo2010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36(2):207–24. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Garrett TA, Guan Z, Raetz CR. Analysis of ubiquinones, dolichols, and dolichol diphosphate-oligosaccharides by liquid chromatography-electrospray ionization-mass spectrometry. Methods Enzymol. 2007;432:117–43. doi: 10.1016/S0076-6879(07)32005-3. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- 21.Krank J, Murphy RC, Barkley RM, Duchoslav E, McAnoy A. Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzymol. 2007;432:1–20. doi: 10.1016/S0076-6879(07)32001-6. [DOI] [PubMed] [Google Scholar]
- 22.McDonald JG, Thompson BM, McCrum EC, Russell DW. Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:145–70. doi: 10.1016/S0076-6879(07)32006-5. [DOI] [PubMed] [Google Scholar]
- 23.Haynes CA, Allegood JC, Sims K, Wang EW, Sullards MC, Merrill AH., Jr Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J Lipid Res. 2008;49(5):1113–25. doi: 10.1194/jlr.D800001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quehenberger O, Armando A, Dumlao D, Stephens DL, Dennis EA. Lipidomics analysis of essential fatty acids in macrophages. Prostaglandins Leukot Essent Fatty Acids. 2008;79(3-5):123–9. doi: 10.1016/j.plefa.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieterle F, Riefke B, Schlotterbeck G, Ross A, Senn H, Amberg A. NMR and MS methods for metabonomics. Methods Mol Biol. 2011;691:385–415. doi: 10.1007/978-1-60761-849-2_24. [DOI] [PubMed] [Google Scholar]
- 26.Barding GA, Jr, Beni S, Fukao T, Bailey-Serres J, Larive CK. Comparison of GC-MS and NMR for metabolite profiling of rice subjected to submergence stress. J Proteome Res. 2013;12(2):898–909. doi: 10.1021/pr300953k. [DOI] [PubMed] [Google Scholar]
- 27.Fahy E, Cotter D, Byrnes R, Sud M, Maer A, Li J, Nadeau D, Zhau Y, Subramaniam S. Bioinformatics for lipidomics. Methods Enzymol. 2007;432:247–73. doi: 10.1016/S0076-6879(07)32011-9. [DOI] [PubMed] [Google Scholar]
- 28.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(7):2136–41. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennis EA. Lipidomics joins the omics evolution. Proc Natl Acad Sci U S A. 2009;106(7):2089–90. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yetukuri L, Katajamaa M, Medina-Gomez G, Seppanen-Laakso T, Vidal-Puig A, Oresic M. Bioinformatics strategies for lipidomics analysis: characterization of obesity related hepatic steatosis. BMC Syst Biol. 2007;1:12. doi: 10.1186/1752-0509-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han X, Yang J, Cheng H, Ye H, Gross RW. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330(2):317–31. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Cifkova E, Holcapek M, Lisa M, Ovcacikova M, Lycka A, Lynen F, Sandra P. Nontargeted quantitation of lipid classes using hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry with single internal standard and response factor approach. Anal Chem. 2012;84(22):10064–70. doi: 10.1021/ac3024476. [DOI] [PubMed] [Google Scholar]
- 33.Gorden DL, Ivanova PT, Myers DS, McIntyre JO, VanSaun MN, Wright JK, Matrisian LM, Brown HA. Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; comparison to a murine model. PLoS One. 2011;6(8):e22775. doi: 10.1371/journal.pone.0022775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisinger K, Krautbauer S, Hebel T, Schmitz G, Aslanidis C, Liebisch G, Buechler C. Lipidomic analysis of the liver from high-fat diet induced obese mice identifies changes in multiple lipid classes. Exp Mol Pathol. 2014;97(1):37–43. doi: 10.1016/j.yexmp.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Muir K, Hazim A, He Y, Peyressatre M, Kim DY, Song X, Beretta L. Proteomic and lipidomic signatures of lipid metabolism in NASH-associated hepatocellular carcinoma. Cancer Res. 2013;73(15):4722–31. doi: 10.1158/0008-5472.CAN-12-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita Y, Sakaguchi T, Ikegami K, Goto-Inoue N, Hayasaka T, Hang VT, Tanaka H, Harada T, Shibasaki Y, Suzuki A, Fukumoto K, Inaba K, Murakami M, Setou M, Konno H. Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression. J Hepatol. 2013;59(2):292–9. doi: 10.1016/j.jhep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 37.Kwan HY, Hu YM, Chan CL, Cao HH, Cheng CY, Pan SY, Tse KW, Wu YC, Yu ZL, Fong WF. Lipidomics identification of metabolic biomarkers in chemically induced hypertriglyceridemic mice. J Proteome Res. 2013;12(3):1387–98. doi: 10.1021/pr3010327. [DOI] [PubMed] [Google Scholar]
- 38.Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473(7348):528–31. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debois D, Bralet MP, Le Naour F, Brunelle A, Laprevote O. In situ lipidomic analysis of nonalcoholic fatty liver by cluster TOF-SIMS imaging. Anal Chem. 2009;81(8):2823–31. doi: 10.1021/ac900045m. [DOI] [PubMed] [Google Scholar]
- 40.Thomas A, Charbonneau JL, Fournaise E, Chaurand P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal Chem. 2012;84(4):2048–54. doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- 41.Chitraju C, Trotzmuller M, Hartler J, Wolinski H, Thallinger GG, Lass A, Zechner R, Zimmermann R, Kofeler HC, Spener F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J Lipid Res. 2012;53(10):2141–52. doi: 10.1194/jlr.M028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leroux A, Ferrere G, Godie V, Cailleux F, Renoud ML, Gaudin F, Naveau S, Prevot S, Makhzami S, Perlemuter G, Cassard-Doulcier AM. Toxic lipids stored by Kupffer cells correlates with their pro-inflammatory phenotype at an early stage of steatohepatitis. J Hepatol. 2012;57(1):141–9. doi: 10.1016/j.jhep.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 43.Testerink N, Ajat M, Houweling M, Brouwers JF, Pully VV, van Manen HJ, Otto C, Helms JB, Vaandrager AB. Replacement of retinyl esters by polyunsaturated triacylglycerol species in lipid droplets of hepatic stellate cells during activation. PLoS One. 2012;7(4):e34945. doi: 10.1371/journal.pone.0034945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bird SS, Marur VR, Stavrovskaya IG, Kristal BS. Qualitative Characterization of the Rat Liver Mitochondrial Lipidome using LC-MS Profiling and High Energy Collisional Dissociation (HCD) All Ion Fragmentation. Metabolomics. 2013;9(1 Suppl):67–83. doi: 10.1007/s11306-012-0400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanninger J, Liebisch G, Eisinger K, Neumeier M, Aslanidis C, Voggenreiter L, Pohl R, Weiss TS, Krautbauer S, Buechler C. Adiponectin isoforms differentially affect gene expression and the lipidome of primary human hepatocytes. Metabolites. 2014;4(2):394–407. doi: 10.3390/metabo4020394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi Y, Jiang C, Cheng J, Krausz KW, Li T, Ferrell JM, Gonzalez FJ, Chiang JY. Bile acid signaling in lipid metabolism: Metabolomic and lipidomic analysis of lipid and bile acid markers linked to anti-obesity and anti-diabetes in mice. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbalip.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clugston RD, Jiang H, Lee MX, Piantedosi R, Yuen JJ, Ramakrishnan R, Lewis MJ, Gottesman ME, Huang LS, Goldberg IJ, Berk PD, Blaner WS. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: A targeted lipidomic and gene expression study. J Lipid Res. 2011 doi: 10.1194/jlr.M017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG, 2nd, Waters KM, Smith RD, Rice CM, Katze MG. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6(1):e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng Y, Meyer SA, Guan X, Escalon BL, Ai J, Wilbanks MS, Welti R, Garcia-Reyero N, Perkins EJ. Analysis of common and specific mechanisms of liver function affected by nitrotoluene compounds. PLoS One. 2011;6(2):e14662. doi: 10.1371/journal.pone.0014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen A, Tang Y, Davis V, Hsu FF, Kennedy SM, Song H, Turk J, Brunt EM, Newberry EP, Davidson NO. Liver fatty acid binding protein (L-Fabp) modulates murine stellate cell activation and diet induced nonalcoholic fatty liver disease. Hepatology. 2013 doi: 10.1002/hep.26318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernando H, Wiktorowicz JE, Soman KV, Kaphalia BS, Khan MF, Shakeel Ansari GA. Liver proteomics in progressive alcoholic steatosis. Toxicol Appl Pharmacol. 2013;266(3):470–80. doi: 10.1016/j.taap.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossmeisl M, Medrikova D, van Schothorst EM, Pavlisova J, Kuda O, Hensler M, Bardova K, Flachs P, Stankova B, Vecka M, Tvrzicka E, Zak A, Keijer J, Kopecky J. Omega-3 phospholipids from fish suppress hepatic steatosis by integrated inhibition of biosynthetic pathways in dietary obese mice. Biochim Biophys Acta. 2014;1841(2):267–78. doi: 10.1016/j.bbalip.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Bradford BU, O'Connell TM, Han J, Kosyk O, Shymonyak S, Ross PK, Winnike J, Kono H, Rusyn I. Metabolomic profiling of a modified alcohol liquid diet model for liver injury in the mouse uncovers new markers of disease. Toxicol Appl Pharmacol. 2008;232(2):236–43. doi: 10.1016/j.taap.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernando H, Kondraganti S, Bhopale KK, Volk DE, Neerathilingam M, Kaphalia BS, Luxon BA, Boor PJ, Shakeel Ansari GA. (1)H and (3)(1)P NMR lipidome of ethanol-induced fatty liver. Alcohol Clin Exp Res. 2010;34(11):1937–47. doi: 10.1111/j.1530-0277.2010.01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernando H, Bhopale KK, Kondraganti S, Kaphalia BS, Shakeel Ansari GA. Lipidomic changes in rat liver after long-term exposure to ethanol. Toxicol Appl Pharmacol. 2011;255(2):127–37. doi: 10.1016/j.taap.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loftus N, Barnes A, Ashton S, Michopoulos F, Theodoridis G, Wilson I, Ji C, Kaplowitz N. Metabonomic investigation of liver profiles of nonpolar metabolites obtained from alcohol-dosed rats and mice using high mass accuracy MSn analysis. J Proteome Res. 2011;10(2):705–13. doi: 10.1021/pr100885w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Z, Yu M, Crabb D, Xu Y, Liangpunsakul S. Ethanol-induced alterations in fatty acid-related lipids in serum and tissues in mice. Alcohol Clin Exp Res. 2011;35(2):229–34. doi: 10.1111/j.1530-0277.2010.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernando H, Bhopale KK, Boor PJ, Ansari GA, Kaphalia BS. Hepatic lipid profiling of deer mice fed ethanol using (1)H and (3)(1)P NMR spectroscopy: a dose-dependent subchronic study. Toxicol Appl Pharmacol. 2012;264(3):361–9. doi: 10.1016/j.taap.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jang ZH, Chung HC, Ahn YG, Kwon YK, Kim JS, Ryu JH, Ryu do H, Kim CH, Hwang GS. Metabolic profiling of an alcoholic fatty liver in zebrafish (Danio rerio) Mol Biosyst. 2012;8(7):2001–9. doi: 10.1039/c2mb25073j. [DOI] [PubMed] [Google Scholar]
- 60.Gika HG, Ji C, Theodoridis GA, Michopoulos F, Kaplowitz N, Wilson ID. Investigation of chronic alcohol consumption in rodents via ultra-high-performance liquid chromatography-mass spectrometry based metabolite profiling. J Chromatogr A. 2012;1259:128–37. doi: 10.1016/j.chroma.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manna SK, Patterson AD, Yang Q, Krausz KW, Li H, Idle JR, Fornace AJ, Jr, Gonzalez FJ. Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. J Proteome Res. 2010;9(8):4176–88. doi: 10.1021/pr100452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8(5):353–67. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 63.Passeri MJ, Cinaroglu A, Gao C, Sadler KC. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49(2):443–52. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shigeta Y, Nomura F, Leo MA, Iida S, Felder MR, Lieber CS. Alcohol dehydrogenase (ADH) independent ethanol metabolism in deermice lacking ADH. Pharmacol Biochem Behav. 1983;18(Suppl 1):195–9. doi: 10.1016/0091-3057(83)90171-5. [DOI] [PubMed] [Google Scholar]
- 65.Lieber CS, Schmid R. The effect of ethanol on fatty acid metabolism; stimulation of hepatic fatty acid synthesis in vitro. J Clin Invest. 1961;40:394–9. doi: 10.1172/JCI104266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaphalia BS, Cai P, Khan MF, Okorodudu AO, Ansari GA. Fatty acid ethyl esters: markers of alcohol abuse and alcoholism. Alcohol. 2004;34(2-3):151–8. doi: 10.1016/j.alcohol.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 67.Wu H, Cai P, Clemens DL, Jerrells TR, Ansari GA, Kaphalia BS. Metabolic basis of ethanol-induced cytotoxicity in recombinant HepG2 cells: role of nonoxidative metabolism. Toxicol Appl Pharmacol. 2006;216(2):238–47. doi: 10.1016/j.taap.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 68.Beckemeier ME, Bora PS. Fatty acid ethyl esters: potentially toxic products of myocardial ethanol metabolism. J Mol Cell Cardiol. 1998;30(11):2487–94. doi: 10.1006/jmcc.1998.0812. [DOI] [PubMed] [Google Scholar]
- 69.Hu C, Ge F, Hyodo E, Arai K, Iwata S, Lobdell H. t, Walewski JL, Zhou S, Clugston RD, Jiang H, Zizola CP, Bharadwaj KG, Blaner WS, Homma S, Schulze PC, Goldberg IJ, Berk PD. Chronic ethanol consumption increases cardiomyocyte fatty acid uptake and decreases ventricular contractile function in C57BL/6J mice. J Mol Cell Cardiol. 2013;59:30–40. doi: 10.1016/j.yjmcc.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arana L, Gangoiti P, Ouro A, Trueba M, Gomez-Munoz A. Ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 2010;9:15. doi: 10.1186/1476-511X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartke N, Hannun YA. Bioactive sphingolipids: metabolism and function. J Lipid Res. 2009;50(Suppl):S91–6. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park TS, Goldberg IJ. Sphingolipids, lipotoxic cardiomyopathy, and cardiac failure. Heart Fail Clin. 2012;8(4):633–41. doi: 10.1016/j.hfc.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bikman BT, Summers SA. Sphingolipids and hepatic steatosis. Adv Exp Med Biol. 2011;721:87–97. doi: 10.1007/978-1-4614-0650-1_6. [DOI] [PubMed] [Google Scholar]
- 74.Supakul R, Liangpunsakul S. Alcoholic-induced hepatic steatosis--role of ceramide and protein phosphatase 2A. Transl Res. 2011;158(2):77–81. doi: 10.1016/j.trsl.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ricchi M, Odoardi MR, Carulli L, Anzivino C, Ballestri S, Pinetti A, Fantoni LI, Marra F, Bertolotti M, Banni S, Lonardo A, Carulli N, Loria P. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol. 2009;24(5):830–40. doi: 10.1111/j.1440-1746.2008.05733.x. [DOI] [PubMed] [Google Scholar]
- 76.Murphy RC, James PF, McAnoy AM, Krank J, Duchoslav E, Barkley RM. Detection of the abundance of diacylglycerol and triacylglycerol molecular species in cells using neutral loss mass spectrometry. Anal Biochem. 2007;366(1):59–70. doi: 10.1016/j.ab.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Retra K, Bleijerveld OB, van Gestel RA, Tielens AG, van Hellemond JJ, Brouwers JF. A simple and universal method for the separation and identification of phospholipid molecular species. Rapid Commun Mass Spectrom. 2008;22(12):1853–62. doi: 10.1002/rcm.3562. [DOI] [PubMed] [Google Scholar]
- 78.Clugston RD, Jiang H, Lee MX, Berk PD, Goldberg IJ, Huang LS, Blaner WS. Altered hepatic retinyl ester concentration and acyl composition in response to alcohol consumption. Biochim Biophys Acta. 2013;1831(7):1276–86. [PubMed] [Google Scholar]
- 79.Clugston RD, Blaner WS. The adverse effects of alcohol on vitamin a metabolism. Nutrients. 2012;4(5):356–71. doi: 10.3390/nu4050356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de la MHP, Lieber CS, DeCarli LM, French SW, Lindros KO, Jarvelainen H, Bode C, Parlesak A, Bode JC. Models of alcoholic liver disease in rodents: a critical evaluation. Alcohol Clin Exp Res. 2001;25(5 Suppl ISBRA):254S–261S. doi: 10.1097/00000374-200105051-00041. [DOI] [PubMed] [Google Scholar]
- 81.Bertola A, Mathews S, Ki SH, Wang H, Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8(3):627–37. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Galbo T, Shulman GI. Lipid-induced hepatic insulin resistance. Aging (Albany NY) 2013;5(8):582–3. doi: 10.18632/aging.100585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeong WI, Osei-Hyiaman D, Park O, Liu J, Batkai S, Mukhopadhyay P, Horiguchi N, Harvey-White J, Marsicano G, Lutz B, Gao B, Kunos G. Paracrine activation of hepatic CB1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 2008;7(3):227–35. doi: 10.1016/j.cmet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 84.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, Merrill AH, Jr, Sullards MC, Wang E, Murphy RC, Raetz CR, Garrett TA, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. A mouse macrophage lipidome. J Biol Chem. 2010;285(51):39976–85. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One. 2009;4(7):e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]