SUMMARY
Multiple levels of interkingdom signaling have been implicated in maintaining the ecological balance between Candida albicans and commensal streptococci to assure a state of oral health. To better understand the molecular mechanisms involved in the initial streptococcal response to the presence of C. albicans that can initiate oral surface colonization and biofilm formation, hypha-forming cells were incubated with Streptococcus gordonii cells for 30 minutes to assess the streptococcal transcriptome response. A genome wide microarray analysis and quantitative PCR validation of S. gordonii transcripts identified a number of genes, the majority of which were involved in metabolic functions that were differentially expressed in the presence of hyphae. The fruR, fruB and fruA genes encoding the transcriptional regulator, fructose-1-phosphate kinase, and fructose-specific permease, respectively, of the phosphoenolpyruvate-dependent fructose phosphotransferase system, were consistently up-regulated. An S. gordonii mutant in which these genes were deleted by allelic replacement, formed an architecturally-distinct, less robust biofilm with C. albicans than did parental strain cells. Complementing the mutant with plasmid borne fruR, fruB and fruA genes caused phenotype reversion, indicating that the genes in this operon played a role in dual species biofilm formation. This genome wide analysis of the S. gordonii transcriptional response to C. albicans has identified several genes that have potential roles in interkingdom signaling and responses.
Keywords: interkingdom signaling, polymicrobial communities, mRNA, PTS, coaggregation
INTRODUCTION
Establishment of multispecies oral biofilms involves specific physicochemical adhesin-receptor interactions that can facilitate cell attachment and coaggregation as well as intercellular communication between microorganisms (Jenkinson, 2011; Wright et al., 2013). These interactions can be mediated by diffusible signals and metabolic interactions that affect the microenvironment within a developing biofilm (Kolenbrander et al., 2010; Nobbs & Jenkinson, 2015). Recent studies have provided insights into the many levels of cross talk between commensal oral streptococcal species and Candida albicans cells that cohabit the same niche in the oral cavity (Morales & Hogan, 2010; Xu et al., 2014a; Bertolini et al., 2015; Jack et al., 2015). Balances in growth between these two genera (Diaz et al., 2012) are thought to be essential in preventing Candida opportunistic infections and maintaining oral health. Contrarily, there are synergistic interactions between streptococci and C. albicans that have been shown to enhance fungal (Xu et al., 2014b) or bacterial (Falsetta et al., 2014) pathogenicity.
Studies of Candida-streptococcal interactions suggest that oral Streptococcus gordonii, a model commensal oral streptococcal species, releases diffusible signals and metabolic byproducts that influence C. albicans hypha formation and dual species biofilm development (Bamford et al., 2009). On the other hand, proteomic (Peters et al., 2010) and transcriptomic studies (Sztajer et al., 2014) have shown that the presence of C. albicans can result in changes in cumulative protein and transcript levels in Gram-positive bacterial species after several hours of co-cultivation.
To better understand the molecular mechanisms involved in the initiation of C. albicans-streptococcal biofilm formation, the present studies focused on the streptococcal short-term response to the presence of C. albicans under hyphae-forming conditions. Microarray analysis of the S. gordonii transcriptome identified a number of genes that were differentially expressed in response to hyphae. There was consistent up-regulation of the fruR, fruB and fruA genes which constitute the operon expressing a fructose-specific phosphotransferase. This operon has previously been shown to influence S. gordonii biofilm formation (Loo et al., 2003). Accordingly, the role of these genes in early dual species biofilm formation was explored utilizing a S. gordonii ΔfruRBA mutant strain in which the operon was deleted by allelic replacement. The results indicate that the fruRBA operon, and a range of other metabolic genes, are differentially expressed in S. gordonii in response to the presence of C. albicans hyphae. Furthermore, the fruR, fruB and fruA genes are implicated in the initiation and architecture of C. albicans-streptococcal biofilms.
METHODS
Interaction of strain DL1 with C. albicans hyphae
S. gordonii Challis DL1 cells were grown to mid-log phase (approximately 108 cells ml−1) in TY medium (5 g l−1 Tryptone, 5 g l−1 yeast extract, 23 mM K2HPO4, pH 7.5) supplemented with 5 g l−1 glucose (TY-G) under stationary conditions at 37°C under 5% CO2. Candida albicans SC5314 cells were grown in YPD medium (1% yeast extract, 2% mycological peptone, 2% glucose) aerobically with shaking at 225 rpm, for 16 h at 30°C (OD600 ~2.6). The C. albicans cells were harvested by centrifugation (1,370 x g for 5 min), washed once in complete salts-biotin (CSB) nutrient-limited medium (7.5 mM ammonium sulfate, 15 mM KH2PO4, 0.1 mM MgSO4.7H2O, 3.6 μg ml−1 biotin, 0.05 mg ml−1 CaCl2, pH 6.5), and suspended in fresh CSB at OD600 = 1.0 (~1 x107 cells ml−1). The culture was incubated aerobically with 225 rpm-shaking for 16 h at 37°C to induce starvation (Holmes & Shepherd, 1988). C. albicans hyphae formation was then triggered by the addition of one-tenth volume 10% glucose (CSB-Glc) to the C. albicans culture, and incubation with shaking was continued for 3 h at 37°C, at which time the pH was measured. Under these conditions at least 80% of the cells were forming hyphal filaments (germ tubes) (Jenkinson et al., 1990).
Equal volumes of the S. gordonii and C. albicans cultures, or S. gordonii with final pH-adjusted (see above) fresh CSB-Glc medium as control, were mixed, centrifuged (3,800 x g for 5 min) and the cell pellets were incubated without shaking for 30 min at 37° C aerobically (in 5% CO2) to allow the S. gordonii cells to interact with the C. albicans hyphae in close proximity. The S. gordonii-CSB-Glc control adjusted to the pH of the C. albicans hyphae-forming culture took into account any effects of centrifugation, and the shift in both nutritional status and pH for S. gordonii when mixed with C. albicans. At the end of this co-incubation, total RNA was extracted from the cell pellets. For microarray transcriptome analyses a minimum of two biologically independent S. gordonii + C. albicans hyphae interaction experiments were performed.
To verify the specificity of S. gordonii gene responses to live hyphae, two equivalent volumes of hyphae were harvested by centrifugation (3800 x g) and one aliquot was suspended in PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4 containing 0.137 M NaCl and 2.7 mM KCl) and heated for 60 min at 100°C to induce killing. Killed or live hyphae suspensions, as well as a suspension of an equivalent number of cells of the irrelevant bacterium Enterococcus faecalis strain OG1X as a control, were each resuspended with an equivalent number of S. gordonii cells in pH-adjusted CSB, re-centrifuged and co-incubated as above. Two independent biological replicate experiments were performed.
RNA isolation and cDNA synthesis
RNA was prepared from the S. gordonii + C. albicans hyphae cell mixtures or S. gordonii + CSB-Glc control with a FastPrep120 Instrument (MP Biomedicals, Santa Ana, CA, USA) and RNAPro Blue RNA Extraction Kit (MP Biomedicals) for prokaryotic cells according to the manufacturer’s directions. The extracted RNA was digested with DNase (Roche Applied Science, Indianapolis, IN, USA) and purified with QIAgen RNA mini spin columns (QIAgen, Germantown, MD, USA) according to the manufacturer’s directions. RNA integrity was confirmed by comparison of the ratios of absorbance at 260/280 nm and 260/230 nm as quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA, USA). For real-time PCR experiments cDNA was reverse transcribed from the purified RNA using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA, USA) according to the manufacturer’s directions. For microarray experiments cDNA was reverse transcribed using Superscript III (Life Technologies, Grand Island, NY, USA), end-labeled via amino-allyl-labeling (Life Technologies, Grand Island, NY, USA) and Cye-dye coupled (GE Healthcare/Amersham, Piscataway, NY, USA) to make microarray probes as previously described (Vickerman et al., 2007). Probes were purified over QIAgen PCR purification columns (QIAgen) to remove excess dye before use.
Microarray hybridization
For microarray experiments, cDNA probes were hybridized to aminosilane-coated glass slides spotted with unique 70-mers representing each open reading frame from the S. gordonii strain CH1 genome sequence (Vickerman et al., 2007; Genbank number CP000725.1) in replicates of six (obtained from the Pathogen Functional Genomics Resource Center; NIDCR Oral Microbe Microarray Initiative). Each slide contained a total of 500 Arabidopsis thaliana 70-mers as controls. An in silico analysis of the A. thaliana and 2,076 S. gordonii probes via a BLAST-N algorithm (Altschul et al., 1990) returned no significant similar sequences in the C. albicans SC5314 genome (AACQ00000000). Control experiments using cDNA prepared from pure C. albicans hyphal cells showed no specific hybridization of C. albicans cDNA to any of the S. gordonii spotted oligonucleotides.
Slides were blocked during pre-hybridization at 42°C in 5x SSC blocking buffer (1x SSC is 0.015 M sodium citrate, pH 7.4, containing 0.15 M NaCl, 0.1% SDS, and 1% bovine serum albumin), then washed successively with water and isopropanol before drying via centrifugation prior to hybridization. To accommodate potential increases in cDNA quantity based upon contributions from the C. albicans hyphae, two-fold more cDNA from the dual species interaction samples was mixed with S. gordonii + CSB-Glc control samples for each interaction comparison. To account for biases during the labeling process, flip-dyes, in which the fluorescent labeling was reversed for each sample, were used (Quackenbush, 2002). Cy3-labeled S. gordonii CSB-Glc cDNA was combined with Cy5-labeled dual species cDNA in appropriate ratios; similarly, Cy5-labeled S. gordonii-CSB-Glc and Cy3-labeled dual species cDNAs were combined. The dye-labeled cDNA mixtures were dried, suspended in 50 μl hybridization buffer (40% v/v formamide, 5x SSC, 0.1% SDS, 0.6 mg ml−1 Salmon sperm DNA), heated for 10 min at 95°C, placed on ice, and individual mixtures added to each microarray slide under a Lifterslip (Erie Scientific Company, Portsmouth, NH, USA). Each slide was incubated for 16 h at 42°C in a sealed hybridization chamber (Corning Life Sciences, Tewksbury, MA, USA), then washed successively in 0.1% SDS-2x SSC, 0.1% SDS-0.1x SSC, and 0.1x SSC, and dried by centrifugation.
Microarray analysis
The Axon 4200A (Axon Instruments, Foster City, CA, USA) was used to obtain digital images of the microarrays. Data were analyzed using the TM4 Microarray Software Suite, consisting of Spotfinder, Midas, and MeV (http://www.tm4.org/) (Saeed et al., 2003). The fluorescence signals from the flip-dye hybridizations were measured using Spotfinder. Normalization and regularization of the data were performed using Midas (Lowess normalization, block mode, smooth parameter of 0.33, Cy3 reference; standard deviation regularization, block mode, Cy3 reference; flip-dye consistency checking and combination, using the data trim option of the standard deviation mode and keeping all data within 2.0 standard deviations; replicate spots were averaged to be used in the determination of the final ratio for each spot). Using MeV, with 100 permutations and a P value cutoff of 0.05 the final expression ratios were generated and used to determine the fold change for hybridization of each probe; fold changes were ranked from the greatest increase to the greatest decrease. Experimental cDNA probes from S. gordonii cells incubated with hyphae that hybridized to the oligonucleotide spots with average fold changes 1.6-times greater than or 1.6-times less than fold changes of S. gordonii-CSB-Glc controls in two biologically independent experiments were considered to be potentially up- or down- regulated, respectively.
Data deposit
Microarray data have been deposited in NCBI’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are available through GEO Series accession number: GSE68752. The files are linked to Genbank entry CP000725.1. Currently, a new annotation of the S. gordonii Challis genome is in progress (NC_009785) using the annotation method of Tatusova et al. (2014). A comparison of original and updated locus tags is included in supplemental Table S2.
Real-time PCR and data analysis
Qualitative real-time PCR (qPCR) assays were performed to confirm the expression of selected S. gordonii DL1 genes that appeared to be up- or down-regulated in response to the presence of hyphae, based upon the microarray results. Primers for specific genes were designed using Primer3, available online at http://primer3.ut.ee/ (Koressaar & Remm, 2007; Untergrasser et al. 2012) to generate 100- or 150-bp amplicons as required (Table S1). Specificity of the primers was confirmed by searching for nucleotide sequence similarity in the S. gordonii genome (CP00725.1). The qPCR was performed using the Applied Biosystems 7500 Real-Time PCR System (Life Technologies) and carried out in 96-well microtiter plates using Power®SYBR® Green Master Mix (Life Technologies) according to the manufacturer’s directions. A minimum of 3 replicates were assayed for each sample. Data were analyzed using SDS Software v1.4 (Life Technologies). Expression values were determined using the 2−ΔΔCt method (Livak & Schmittgen, 2001).
Construction of fruRBA deletion and complemented strains
A derivative of the parental strain Challis DL1 with a deletion of the fruRBA chromosomal region was generated by a PCR ligation mutagenesis approach (Figure S1). Successive overlapping PCR products were made with primers listed in Table S1 using strain DL1 chromosomal template to amplify the DNA regions that flank the 5-prime and 3-prime ends of the fru operon with an intervening 1084-bp aad9 gene encoding spectinomycin resistance. The resulting amplicon was a dsDNA fragment in which 3,659 bp of the fruRBA operon, including the putative −10 promoter region and predicted catabolite repressible element binding site (Loo et al., 2003), were replaced with 1,084-bp DNA region encoding aad9 with its native promoter and terminator from plasmid pFW5 (Podbielski et al., 1996). This DNA fragment was transformed directly into serum-competent strain DL1 cells and transformant colonies, which arose via allelic replacement (Fig. S1), were selected by growth on agar medium supplemented with 100 μg ml−1 spectinomycin. After confirmation of the expected chromosomal sequence by nucleotide sequence analysis, the resulting strain with a deletion of the fruRBA region was designated strain UB2683.
To complement the mutation, a 3,984-bp amplicon was made from strain DL1 chromosomal template with primers BamFruCompF and BamFruCompR (Table S1) by PCR using Elongase enzyme (Lifetech). The resulting DNA encoding the complete fruRBA operon (Fig. S1) was digested with BamH1 and ligated into an engineered BamH1 site (Vickerman et al 2003) of the streptococcal replicative plasmid pVA749 (Macrina et al., 1981) and transformed into the parental strain. The sequence fidelity of the cloned fragment was confirmed by nucleotide sequence analysis and the resulting streptococcal plasmid designated p49MfruRBA was transformed into strain UB2683. Putative transformants carrying the plasmid were selected on Todd Hewitt agar medium with erythromycin (5 μg ml−1) and a representative UB2683/p49MfruRBA transformant was chosen for further characterization and biofilm studies.
Preparation of saliva
Pooled human saliva, collected from a minimum of six volunteers as approved by the National Research Ethics Committee South Central Oxford C (Number 08/H0606/87+5), was treated with 0.25 M dithiothreitol, and clarified by centrifugation, as previously described (Dutton et al., 2014). The resulting supernatant was diluted to 10% with sterile water, filter sterilized with 0.45 micron pore size membranes, stored at −20°C, and used to coat glass cover slips as substrata in biofilm growth experiments.
Single species biofilms
For mono-strain studies to determine the abilities of the S. gordonii parent or mutant cells to form biofilms on saliva-coated substrata, cells were grown in one of four different biofilm media: TY-G, TY medium with 5 g l−1 fructose (TY-F), and Yeast Nitrogen Base, Tryptone, KH2PO4-K2HPO4 pH 7.0 (YPT) with either 1 g l−1 glucose (YPT-G) or 1 g l−1 fructose (YPT-F). Biofilms were grown on 19-mm glass cover slips contained within 12-well tissue culture plates. Sterile cover slips were incubated with 10% sterile saliva for 16 h at 4°C. Saliva was removed and 0.9 ml of either TY-G, TY-F, YPT-G or YPT-F medium was added. Streptococcal strains were grown for 16 h at 37°C in Brain Heart Infusion-Yeast Extract (BHY) medium, inoculated (1:10 dilution) into TY-G, TY-F, YPT-G or YPT-F medium, and incubated until OD600 = 0.6 was obtained. Culture aliquots (0.1 ml) were added to wells containing 0.9 ml of the corresponding fresh media and incubated in a candle jar for 16 h at 37°C. Cover slips were removed into a fresh 12 well plate, washed twice with PBS, stained with 0.5% crystal violet, washed 3 times with PBS, air dried, and visualized by transmitted light microscopy (Leica DMLB). Alternatively, for biomass studies, following staining with crystal violet, cover slips were transferred to a fresh 12 well plate, and washed three times with PBS. The stain was then released by the addition of 10% acetic acid; the absorbance of the eluate was measured at 595 nm (Jakubovics et al., 2005).
Dual species biofilms
Biofilms were grown in 35-mm diameter plastic culture dishes with 14-mm No. 1.0 base glass cover slip windows (Mat Tek). Cover slip windows were coated with 2 ml sterile saliva for 16 h at 4°C. Saliva was removed by aspiration and 1.8 ml YPT-G and 0.2 ml C. albicans cell suspensions (2 × 106 in YPT) was added to each dish. Dishes were incubated for 1 h at 37°C, with gentle shaking at 50 rpm in a humid environment, and then the unattached C. albicans cells were removed. For control biofilms (C. albicans only), fresh YPT-G medium was added. For dual species biofilms, and streptococcal biofilm controls, overnight cultures of S. gordonii cells were suspended at OD600 = 0.5 in YPT-G and 2 ml aliquots were added to the C. albicans-inoculated plates to initiate dual species biofilm formation, or to saliva only-coated cover slip windows (S. gordonii biofilm controls). After incubation for 30 min at 37°C, to allow streptococcal cell attachment, unattached cells were removed by aspiration, fresh YPT-G medium was added (2 ml) and incubation was continued aerobically for up to 5 h at 37°C. At the end of the experiment, the biofilm-coated cover slip windows were removed, rinsed with YPT, and either stained with crystal violet for examination by light microscopy, or stained with fluorescein isothiocyanate (FITC), rinsed, and examined by confocal laser scanning microscopy (CLSM) using a Leica SP5-AOBS confocal microscope attached to a Leica DM I6000 inverted epifluorescence microscope as previously described (Dutton et al., 2014). Velocity® software was used to generate 3D images.
RESULTS
DL1 transcriptional changes in response to co-incubation with hyphae
S. gordonii genes found to be differentially expressed in S. gordonii DL1 cells co-incubated for 30 minutes with hyphae as compared to genes in strain DL1 cells incubated with control CSB-Glc medium are shown in Table 1. All genes listed exhibited an average fold change difference of at least 1.6 in two independent biological replicate experiments and met the significance cutoff of 95% confidence. Complete transcriptome data from these experiments have been deposited in the GEO database.
Table 1.
S. gordonii genes differentially expressed by microarray analysis
| Locusa | Descriptionb | Symbol | Predicted Functional Category/Rolec | Fold Changed | P Valuee |
|---|---|---|---|---|---|
| Up-regulated | |||||
| SGO_1112 | 1-phosphofructokinase | fruB | Energy metabolism/Glycolysis_gluconeogenesis | 4.0 | 0.002 |
| SGO_1111 | Phosphotransferase system repressor | fruR | Energy metabolism/Biosynthesis and degradation of polysaccharides | 3.4 | 0.002 |
| SGO_1113 | PTS system, fructose specific IIABC components | fruA | Regulatory functions/Other | 3.4 | 0.001 |
| SGO_0702 | hypothetical protein | Hypothetical | 3.1 | 0.002 | |
| SGO_1109 | aspartate carbamoyltransferase catalytic subunit | pyrB | Purines, pyrimidine, nucleosides, and nucleotides/Pyrimidine ribonucleotide biosynthesis | 2.0 | 0.048 |
| SGO_1190 | hypothetical protein | Hypothetical | 1.6 | 0.029 | |
| SGO_1244 | hypothetical protein | Hypothetical | 1.6 | 0.026 | |
| SGO_1686 | hypothetical protein | Hypothetical | 1.6 | 0.021 | |
| SGO_1695 | enoyl-acyl carrier protein (ACP) reductase | Fatty acid and phospholipid metabolism/Biosynthesis | 1.6 | 0.027 | |
| SGO_1696 | hypothetical protein | Hypothetical | 1.6 | 0.014 | |
| Down-regulatedf | |||||
| SGO_1592 | ornithine carbamoyltransferase | arcB | Energy metabolism/Amino acids and amines | −2.2 | 0.034 |
| SGO_0383 | proton/sodium- glutamate symport protein | Transport and binding proteins/Amino acids, peptides and amines | −2.0 | 0.002 | |
| SGO_1301 | hypothetical protein | Hypothetical | −2.0 | 0.003 | |
| SGO_0440 | L-iditol 2-dehydrogenase | Energy metabolism/Sugars | −2.0 | 0.019 | |
| SGO_1593 | arginine deiminase | arcA | Energy metabolism/Amino acids and amines | −1.9 | 0.009 |
| SGO_1757 | glucosamine-fructose-6-phosphate aminotransferase | glmS | Central intermediary metabolism/Amino sugars | −1.9 | 0.008 |
| SGO_1300 | NADPH-dependent reductase | Energy metabolism/Electron transport | −1.8 | 0.005 | |
| SGO_0985 | amino acid permease protein SP0711 | Transport and binding proteins/Amino acids, peptides and amines | −1.8 | 0.01 | |
| SGO_1216 | putative bacteriocin transport accessory protein | Transport and binding proteins/Unknown substrate | −1.8 | 0.007 | |
| SGO_0184 | urease cluster protein | Energy metabolism/Electron transport | −1.8 | 0.008 | |
| SGO_0384 | putative carboxylate-amine/thiol ligase | Energy metabolism/Other | −1.8 | 0.008 | |
| SGO_0631 | alpha-glycerophosphate oxidase | Energy metabolism/Other | −1.8 | 0.007 | |
| SGO_1720 | Aminotransferase, class-V | Central intermediary metabolism_Amino acid biosynthesis/Other_Aspartate family | −1.8 | 0.011 | |
| SGO_0630 | glycerol uptake facilitator protein | Transport and binding proteins/Other | −1.7 | 0.012 | |
| SGO_0278 | peptide methionine sulfoxide reductase | msrA | Central intermediary metabolism/Other | −1.7 | 0.01 |
| SGO_0982 | amino acid transport protein | Transport and binding proteins/Amino acids, peptides and amines | −1.6 | 0.03 | |
| SGO_1283 | pyridine nucleotide-disulfide oxidoreductase | Energy metabolism/Electron transport | −1.6 | 0.043 | |
Locus tag in S. gordonii genome sequence for CP000725.1 (Vickerman et al., 2007).
Locus description in NCBI database annotation for CP000725.1.
Predicted Functional Category/Role (Tanenbaum et al., 2010)
Average fold change, based upon cDNA probes hybridizing to a minimum of 24 up to a maximum of 48 independent 70-oligomer spots for each locus, of DL1 cells co-incubated with hyphae compared to baseline values of DL1-CSB-Glc pH-matched controls in two biologically independent experiments.
P value is based upon the rank product algorithm (Breitling et al., 2004) of the MEV software.
A negative sign added to the absolute fold change value for the downregulated genes indicates the value for DL1 cells incubated with hyphae is less than the DL1-CSB-Glc value.
Microarray analysis indicated a consistent 3- to 4- fold upregulation of the fruRBA operon (Table 1) in strain DL1 in response to a 30-minute incubation with C. albicans hyphae. Results from each microarray experiment were validated by independently-synthesized cDNA by quantitative PCR (Table 2). All three genes in the fruRBA operon were up-regulated in strain DL1 cells that interacted with hyphae compared to control DL1 cells incubated with pH-adjusted CSB-Glc. Furthermore, a substantial portion of this up-regulation could be specifically attributed to live hyphae (Table 3) since the response elicited from coincubation with hyphae was higher than the response to coincubation with equivalent numbers of heat-killed hyphae or the unrelated bacterium E. faecalis.
Table 2.
Quantitative PCR validation of gene expression during S. gordonii and hyphal C. albicans interactions.
| Locus | Gene Name | Relative Expression (2−(ΔΔCt))a
|
|---|---|---|
| Avg. Expression | ||
| Potentially Up-regulated by Microarray analysis
| ||
| SGO_1111 | fruR | 6.5 ± 1.98 |
| SGO_1112 | fruB | 9.05 ± 0.64 |
| SGO_1113 | fruA | 3.75 ± 0.92 |
| SGO_0701 | hup | 0.95 ± 0.21 |
| SGO_0702 | Hypothetical | 1.95 ± 0.35 |
| Potentially Down-regulated by Microarray analysis | ||
| SGO_1593 | arcA | 0.6 ± 0.28 |
| SGO_1592 | arcB | 0.6 ± 0.28 |
| SGO_1757 | glmS | 0.5 ± 0.14 |
Values were normalized to 150- or 100-bp amplicons of gyrA as appropriate. Expression values shown are those of DL1 cells in the hyphal interaction compared to baseline values (set at 1) of DL1 cells in pH-matched CSB-Glc. Relative expression values greater than 1 indicate up-regulation; values less than 1 indicate down-regulation. N=6.
Table 3.
Specificity of the S. gordonii fruRBA response to C. albicans hyphae
| Co-incubations in CSB | Average Relative Expression (2(−ΔΔCt))a |
|---|---|
| fruR | |
| DL1 + hyphae | 9.6 ± 4.9 |
| DL1 + heat-killed hyphae | 2.8 ± 1.5 |
| DL1 + E. faecalis OG1X | 1.6 ± 0.8 |
| fruB | |
| DL1 + hyphae | 12.1 ± 1.3 |
| DL1 + heat-killed hyphae | 3.7 ± 0.3*b |
| DL1 + E. faecalis OG1X | 0.8 ± 0.5* |
| fruA | |
| DL1 + hyphae | 18.9 ± 4.0 |
| DL1 + heat-killed hyphae | 3.1 ± 2.0* |
| DL1 + E. faecalis OG1X | 3.8 ± 2.5* |
All values normalized to 150-bp amplicons of gyrA and compared to the baseline expression of DL1 cells alone.
values with asterisks differ significantly from DL1 + hyphae (p<0.05). Significance patterns are consistent with 5-prime to 3-prime net directionality of prokaryotic transcript decay (Alifano et al. 1994). N ≥ 12.
The microarray data also showed a consistent upregulation of locus SGO_0702 which encodes a small hypothetical protein of unknown function (Table 1). This locus overlapped the convergent locus SGO_0701 which encodes a putative DNA-binding histone-like HU protein. Due to the small size of these open reading frames (276 and 237 bp for SGO_0701 and SGO_0702, respectively) the microarray probes for the two loci overlapped each other by 64 of 70 nucleotides. Nevertheless, the unidirectionality of the cDNA probes used in the microarray analysis allowed specific detection of transcripts arising from SGO_0702, which were up-regulated, versus the transcripts from the convergent SGO_0701 which was not differentially expressed. Two independent qPCR reactions were designed to validate these results (Table 2). The primers for SGO_0701 were completely within the open reading frame and indicated no significant upregulation of this gene in response to hyphae. In contrast, qPCR results for SGO_0702 showed an increase in relative expression of this gene in strain DL1 cells that interacted with hyphae. However, due to the small size of the two loci and the limits of primer design, primer 702-left was within SGO_0702 but primer 702-right overlapped the 3-prime end of SGO_0701. Since qPCR requires bidirectional amplification of the transcript, it cannot be ruled out that some of the upregulation of SGO_0702 detected by qPCR is due to a contribution of read through of the transcript from SGO_0701. Overall, the increased expression of SGO_702 taken together with the microarray results suggests that there is increased transcription of this locus in S. gordonii DL1 cells that interacted with hyphae under the conditions tested. Other genes that were up-regulated in the microarray analyses were several additional hypothetical proteins as well as SGO_1109 involved in pyrimidine synthesis and SGO_1695 involved in fatty acid biosynthesis pathways (Table 1).
The genes that showed the largest negative fold changes in response to hyphae encoded proteins that generally fell into three predicted functional categories (Tanenbaum et al., 2010): energy metabolism (SGO_0184, SGO_0440, SGO_0631, SGO_1283, SGO_1300, SGO_1592, SGO_1593, SGO_0384), central intermediary metabolism (SGO_0278, SGO_1720, SGO_1757), and transport and binding proteins (SGO_0383, SGO_0630, SGO_0982, SGO_0985, SGO_1216). The shifts in expression generally relate to genes involved in amino acid metabolism and transport. Accordingly, representative genes arcA and arcB, involved in arginine catabolism, and glmS, involved in the formation of glutamate from glutamine were chosen for validation of decreased expression in DL1 in response to hyphae by real time PCR (Table 2).
The role of fruRBA in strain DL1 biofilm formation
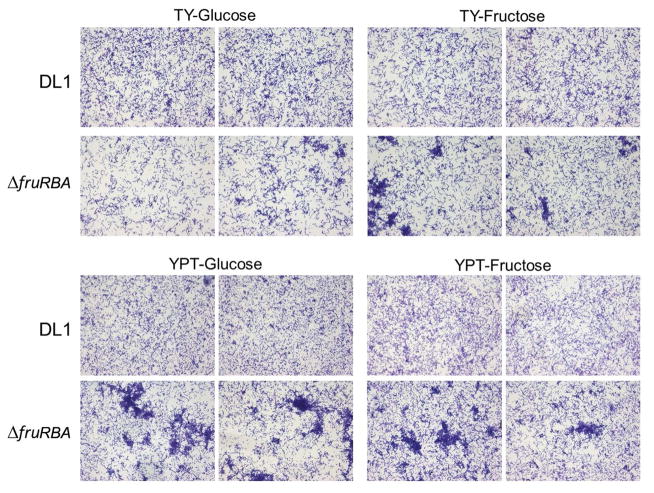
The doubling times of planktonic cultures of the parent strain DL1 and a ΔfruRBA mutant strain were similar in TY or YPT media independent of either glucose or fructose as a carbohydrate source. In biofilm experiments, cultures were grown to the same OD600 as starting inocula, in the respective medium to avoid any nutritional shift effects. The biofilms formed over a 16 h period by the ΔfruRBA mutant were less dense than those of the parental strain DL1 in TY-G medium (Fig. 1). Although there were no statistically significant differences in biofilm mass based upon crystal violet staining assay, there were distinct differences in the architectural patterns of cells present within the biofilms. The mutant cells tended to form aggregates and these aggregates were even more visually evident in the TY-F medium biofilms (Fig. 1). In YPT-G medium, the ΔfruRBA mutant formed denser aggregates, whereas parental cells were more evenly dispersed across the substratum. Aggregates of the ΔfruRBA mutant were also visible in YPT-F medium biofilms (Fig. 1).
Figure 1.
Biofilms formed on saliva-coated glass cover slips. Sixteen hour biofilms of S. gordonii DL1 or ΔfruRBA grown in TY-G or TY-F media (top panels), and in YPT-G or YPT-F media (bottom panels). Two representative views of transmitted light microscopy fields of crystal violet -stained biofilms formed in each medium are shown for each strain.
Role of fruRBA in dual species biofilms
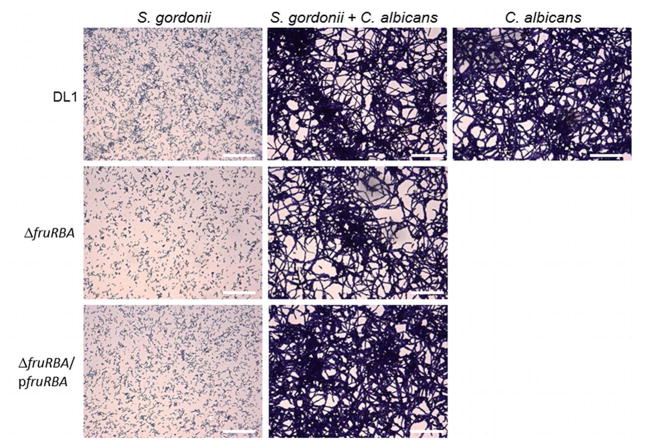
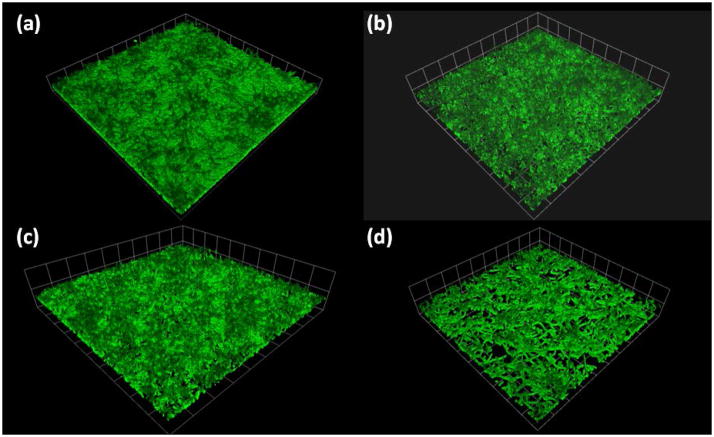
Because the aggregation differences between parent and mutant cells were most visually evident in YPT-G medium, this medium was chosen for additional dual species biofilm studies. Visual examination of light microscopy images (Fig. 2) indicated that biofilms formed by C. albicans were enhanced in the presence of S. gordonii DL1, covering more of the substratum surface than those biofilms formed by C. albicans alone. In contrast, the dual species biofilm formed by C. albicans with strain UB2683, the ΔfruRBA mutant, covered less of the substratum surface and had fewer intertwining hyphal filaments than dual species biofilms formed with the parental strain DL1. Complementing the ΔfruRBA mutation with plasmid-borne fruRBA genes in strain UB2683/p49MfruRBA resulted in a reversion of the dual-species biofilm architecture to one more similar to that seen with strain DL1. CLSM images of dual-species biofilms suggested that overall there were fewer ΔfruRBA mutant cells associated with C. albicans hyphal filaments compared with S. gordonii DL1-C. albicans biofilms (Fig. 3). In the ΔfruRBA mutant-C. albicans biofilms, many more hyphal filaments were visibly exposed (Fig. 3b) and they extended further into the environment. Consequently the ΔfruRBA mutant dual-species biofilms were of greater maximum height (see legend to Fig. 3) than the other biofilms in which the hyphal filaments tended to be more closely retained within the mixed species communities (Fig. 3). The complemented mutant produced a dual-species biofilm similar to wild type (Fig. 3c). Thus, the fruRBA operon is necessary for normal biofilm formation of S. gordonii with C. albicans, and is one of the operons up-regulated early in S. gordonii under co-culture conditions.
Figure 2.
Images of single- or dual-species biofilm formation in YPT-G medium by S. gordonii cells on saliva-coated cover slips with or without attached C. albicans SC5314 cells. Representative light microscopic fields of crystal violet-stained biofilms are shown. The middle panels show S. gordonii biofilms of the parent (top), ΔfruRBA mutant (middle) and fruRBA complemented (bottom) strains (DL1, UB2683, and UB2683/p49MfruRBA, respectively) grown from cells that bound during a 30-min incubation period to substrata with attached C. albicans cells. The flanking panels show corresponding monospecies biofilms of S. gordonii (left panels) grown from cells that bound saliva-coated substrata over 30-min incubation, and C. albicans biofilm (right panel) grown from cells that attached during 1-h incubation. Magnification Bar = 50 μm.
Figure 3.
CLSM images of dual-species C. albicans SC5314-S. gordonii biofilms formed on saliva-coated glass cover slips in YPT-G medium. Biofilms were grown as described in Methods for 6 h at 37°C and fluorescently stained with FITC. For each of the panels (A–D) a series of x-y sections were collected to generate 3D reconstructions of representative CLSM images with Volocity® software. Heights were measured across ten CLSM image stacks from two independent experiments ± SD. Panels: (a), C. albicans-S.gordonii DL1, height 81.4 ± 7.50 μm; (b), C. albcians-S. gordonii ΔfruRBA, height 113 ± 10.4 μm*; (c), C. albicans-S. gordonii ΔfruRBA/pfruRBA, height 70.6 ± 6.40 μm; (d), C. albicans, height 86.8 ± 11.6. *P<0.001 when heights were compared using the t test. Note that in (b) there is more substratum vacant and more hyphae are visible compared with the other dual-species biofilm panels (a) and (c).
DISCUSSION
Oral streptococci and C albicans can co-habit the same niche on human oral cavity surfaces. Dual species interaction studies have identified phenotypic responses and molecular mechanisms that mediate their interactions in oral biofilms. Communication and cellular interactions among members of multispecies oral biofilms can influence the relative abundance of specific microorganism, and consequently influence the relative health or pathogenic potential of the oral microbiome. Such microbial interactions may be mediated by physical-chemical properties, interactions between cell-associated surface molecules, or via extracellular products such as peptides or diffusible fatty acids (Xu et al., 2014a).
To better understand the intercellular communication from the streptococcal perspective, the initial response of S. gordonii cells exposed to C. albicans forming hyphal filaments was examined by transcriptome analysis. A 30 minute co-incubation period was chosen since S. gordonii gene expression is known to respond to environmental signals during this timeframe (Vickerman et al., 2007). Hyphal cells were employed in these interaction studies since this morphological form predominates in C. albicans biofilms. During this co-incubation, there was no evident up-regulation of genes encoding the streptococcal cell -wall anchored surface proteins that have previously been shown to mediate S. gordonii-C. albicans binding. For example, S. gordonii CshA protein domains (Holmes et al., 1996) and SspB protein (Silverman et al., 2011) have been shown to interact with the C. albicans cell surface. However, expression of neither cshA nor sspB was up-regulated during the 30 min co-incubation indicating that adhesion mediated by these proteins probably does not result from early specific transcriptional responses to C. albicans hyphal cells. Although the products of these genes play important role in inter-microbial interactions, their expression may not be specifically induced by the presence of hyphae, or their expression may be affected at a later time point in dual-species growth.
Genome-wide transcriptional studies of S. mutans after 4 to 24 hour of biofilm growth with C. albicans demonstrated up-regulation of genes involved in the alternative competence pathway (Sztajer et al., 2014). This effect was only noted after eight hours of dual species biofilm growth, by which time multiple environmental signals and responses would have occurred affecting multiple gene pathways. Likewise, a role for S. gordonii competence genes has been demonstrated in determining the architecture of dual-species biofilms but again, this was seen phenotypically after several hours of co-cultivation (Jack et al., 2015). The competence pathway of S. gordonii differs from that of S. mutans and is more similar to that of Streptococcus pneumoniae with a response that is essentially completed 45 min after induction (Vickerman et al., 2007). In the present studies there were no differentially-expressed competence peptide-responsive genes seen in S. gordonii cells co-incubated with C. albicans in the 30 min time frame. Taken collectively, the various studies suggest that competence-related responses play a role after the initiation of the dual species biofilms.
In general, the S. gordonii genes that initially responded to the presence of hyphae, when compared to pH-matched medium controls, were those involved in carbohydrate and amino acid metabolism. The majority of former were up-regulated whereas the latter were down-regulated. In silico analyses using various online sequence comparison programs such as Fuzznuc and Clustal (Rice et al., 2000) did not detect DNA sequences conserved among the upstream promoter regions of all the up- or down-regulated genes that would be suggestive of potential signal factor binding sites (data not shown). Fold changes in gene expression were modest, but genes were detected with differential expression that was reproducible and statistically significant. Several loci for hypothetical proteins were responsive to hyphae, but their functions are unknown and the potential roles of those genes cannot yet be determined. The most significantly up-regulated hyphal-responsive S. gordonii genes were those in the fruRBA operon. The fruR gene encodes a phosphotransferase system (PTS) repressor, fruB encodes a 1-phosphofructokinase, and fruA encodes the fructose specific permease of the phosphoenolpyruvate-dependent sugar phosphotransferase system. The phosphotransferase system enzymes in streptococci comprise the main mechanisms for transporting carbohydrates across the membrane into the cell for energy (Pessione, 2012). These carbohydrate transporters show some redundancy in their sugar specificity (Deutsher et al., 2014; Ajdic & Pham, 2007). The genes in the S. gordonii fructose PTS operon were originally identified and characterized (Loo et al., 2003) based upon their sequence and functional similarities to the S. mutans fruRKI genes. However, since S. gordonii is more closely related phylogenetically to S. pneumoniae, which was used as the basis for annotation of S. gordonii genome sequence (Vickerman et al., 2007), and the S. pneumoniae homologs are designated fruRBA, in the present paper we refer to these S. gordonii genes with the fruRBA nomenclature used in the NCBI genome database.
Co-incubation of DL1 cells with heat-killed vs. live hyphae in fresh pH-matched medium confirmed that as-yet-unidentified heat-stable component(s) associated with the living hyphae themselves (e.g. carbohydrates, small peptides) contribute to eliciting the fruRBA operon response. Reporter gene studies have shown that fruB is induced by fructose, sucrose, xylitol and human serum, and that expression of this operon is subject to glucose catabolite repression (Loo et al., 2003). The surfaces of C. albicans cells are covered with carbohydrate-containing compounds such as chitin, mannans and β-glucans (Dongari-Bagtzoglou et al., 2009). These can be hydrolyzed into carbohydrate moieties via various streptococcal and fungal enzymes e.g. N-acetylglucosaminidase, glucosidase, glucanase; and so upregulation of the fruRBA operon in response to hyphae is consistent with bacterial sensing alternative carbohydrate availability.
Another intriguing gene that was up-regulated in response to hyphae is the gene encoding the enoyl-acyl carrier protein. Additional fatty acid metabolism genes were up-regulated in response to hyphae but they did not meet our stringent statistical cutoff to be included in Table 1 (data are in the GEO files). Recent studies suggest that diffusible fatty acid signals made by several oral streptococcal species (Vilchez et al., 2010) may be involved in influencing or inhibiting hyphae formation, but it is not known at present if S. gordonii produces these. Future studies may provide more insights into the signals and cognate responses of these two microorganisms at various stages of S. gordonii-C. albicans biofilm initiation and growth.
Small peptides are a common mechanism of streptococcal intercellular signaling and quorum sensing (Kleerebezem et al., 1997). Although there was no down-regulation among genes expressing known S. gordonii quorum sensing peptides, a putative bacteriocin accessory protein, SGO_1216, was down-regulated 1.8-fold, consistent with the expression of its homolog in S. mutans which was ca. 2- to 5-fold down-regulated in 6-, 10- and 24-h dual species biofilm growth with C. albicans (Sztajer et al., 2014). The functional specificity of this transporter remains to be determined, as does its role in Streptococcus-C. albicans interactions. Nevertheless, this result is intriguing because of the shared down-regulation response of these homologs in two different oral streptococcal species in response to C. albicans.
The majority of the S. gordonii genes that were down-regulated in an initial response to the environmental presence of C. albicans hyphae appeared to indicate metabolic changes. Two main sources of energy in S. gordonii and other lactic acid bacteria are sugar phosphorylation and arginine deimination (Pessione, 2012). Indeed, many of the down-regulated genes encode putative functions in the arginine metabolic pathways (Kloosterman & Kuipers, 2011). Glutamine is centrally involved in both arginine and pyrimidine metabolism pathways. It can be converted to glutamate and involved in the arginine biosynthesis pathways via ornithine and citrulline, or it can be converted to carbamoyl-phosphate with roles in energy production and pyrimidine synthesis (Kloosterman & Kuipers, 2011). The gene encoding GlmS involved in the formation of glutamate from glutamine also has a role in arginine and pyrimidine synthetic pathways (Kloosterman & Kuipers, 2011). Similarly, three genes of the arginine deiminase pathway were down-regulated in response to hyphae. This pathway involves arginine conversion by ArcA to citrulline and ammonia. Citrulline is converted to ornithine or carbamoyl phosphate by Arc B. Carbamoyl phosphate is involved in pyrimidine metabolism, or via ArcC can yield ATP, as well as ammonia to neutralize potential acidic pH. Changes in expression of metabolic genes in response to the initial presentation of the numerous sugars available from C. albicans would suggest a switch to carbohydrate utilization, consistent with metabolic pathway-utilization gene or protein changes seen in other Gram-positive bacteria co-cultivated with C. albicans (Peters et al., 2010; Sztajer et al., 2014). Overall, our transcriptome analyses were not exhaustive and genes that were differentially expressed below our stringent threshold cutoff levels may also be biologically important. It would be expected that gene expression patterns would change as dual species growth progresses. Nevertheless, the present studies have identified genes that were consistently and reproducibly affected by initial exposure of S. gordonii to hyphae, thereby preparing groundwork for future genetic and functional investigations.
Because genes in the fruRBA operon showed the most significant up-regulation in response to the presence of hyphae, we further investigated the potential role of these genes in dual-species biofilm formation. Previous studies have shown that S. gordonii strains with mutations in genes in the fruRBA operon formed less robust biofilms on abiotic glass, saliva-coated (Loo et al., 2003); or fibronectin-coated (Bizzini et al., 2006) substrata compared to those of parental cells. Nonpolar mutations in fruB and fruA resulted in significant reductions of biofilm formation on glass or experimental salivary pellicle, whereas nonpolar disruption of fruR had no effect (Loo et al., 2003). The mechanism underlying the reduction in streptococcal biofilm formation is unknown and was speculated to involve additional roles of FruB in two-component signal transduction by acting as a kinase for a different regulatory system via cross-regulation (Deutscher et al., 2014). This possibility is consistent with studies suggesting that PTS’s may have global regulatory roles in response to environmental cues (Lengeler & Jahreis, 2009; Pereira et al., 2012). In the present studies, deletion of all three genes of the fruRBA operon resulted in changes to the biofilm architecture and surface coverage patterns on saliva-coated glass cover slips. The streptococcal fruRBA mutant cells formed more macroscopic aggregates thereby leaving more of the substratum uncovered. Such aggregation of cells may be a community mechanism to keep cells in closer proximity in order to optimize signaling and shared extracellular products that may favor nutrient processing and availability (Rickard et al., 2003).
The experimental design of the dual-species biofilm experiments only allowed streptococcal cells that attached in 30 min to a hyphae-coated substratum to contribute to the mature biofilm formation. Based upon the subsequent growth pattern, the mutant cells tended to attach in clusters suggesting that during the initial attachment phase the cells remained nearer each other rather than being more randomly distributed across the hyphal filament substratum. The similar chain lengths of planktonic parent and mutant cells in the various growth media (data not shown) suggested that this pattern was due to independent cell attachment rather than cell clusters due to cell chaining differences. The difference in parent and mutant biofilms formed during the 30 min attachment period, a time during which the microarray studies indicated that parental cells up-regulated expression of the fruRBA operon, support the idea that these genes play a role in the cellular adhesion patterns during initial interactions with the C. albicans cells. Alternatively, it is possible that the phenotypic variation in biofilms of the fruRBA mutant strain were due to compensatory expression of other genes in the mutant strain. An examination of the transcriptome of the S. gordonii fruRBA mutant either alone or in the presence of hyphae has not been performed. Nevertheless, complementing the mutant strain with plasmid-borne fruRBA resulted in a reversion of the biofilm phenotype, confirming a role of the fruRBA genes in dual species biofilm formation.
Overall, these studies provide the first genome-wide insights into the initial responses of S. gordonii to C. albicans hyphae under the conditions described here. The major streptococcal transcriptional effects involve responses to the changed microenvironment and carbohydrate availability. They provide clues to the metabolic responses that result from cross talk between these two microorganisms. In a recent study (Dutton et al., 2015), when S. gordonii and transitioning hyphae were co-cultured for a minimum of one hour in YPT-G medium rather than the shorter time period and CSB medium used to detect transcriptional responses in the present study, induction of the S. gordonii fruRBA operon was not detected. This is consistent with RNA sequencing findings which demonstrate that both streptococcal and C. albicans gene expression profiles and encoded gene products are influenced by culture conditions as well as growth stage and biofilm development (Dutton et al., 2015, Sztajer et al., 2014). Such limitations are inherent in in vitro models. Examination of multiple conditions will be necessary to gain a more complete understanding of the complex biological processes involved in dual species communication that may go on to influence biofilm development.
Supplementary Material
Map of DNA used for construction of ΔfruRBA mutant and complement strains. The chromosomal region of strain DL1 is shown with large arrows showing the positions and directions of the fruRBA genes. Software searches identified putative promoters (P) and transcriptional terminators (lollipops) in this region. The ΔG values for putative transcriptional terminators are shown as calculated with mfold (Zuker, 2003; http://mfold.rna.albany.edu). Positions of the Fru.F1/FruR1 and FruF2/FruR2 amplicons are shown above their corresponding genomic locations and include the upstream S. gordonii gene locus SGO_1110 and the downstream locus SGO_1114, respectively. Numbers directly below the ends of the amplicons (1153026 and 1158017) correspond to the nucleotide numbers in the S. gordonii Challis genome sequence (Genbank CP000725.1). The lower diagram shows the linear DNA fragment used to transform strain DL1 to construct strain UB2683, the ΔfruRBA deletion mutant in which the upstream and downstream amplicons flank an aad9 determinant with its own promoter and terminator. The upper panel shows the position of the amplicon made with primers BamFruCompF and BamFruCompR that was cloned into a replicative plasmid to generate p49MfruRBA used to complement strain UB2683.
Acknowledgments
We thank Lindsay Dutton for technical assistance and for CLSM image analysis. This work was supported by PHS-1-R01-DE016690. Microarray slides were obtained through a NIDCR Oral Microbe Microarray Initiative award.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Table S1. Oligonucleotide primers used.
Table S2. Comparison of S. gordonii CH1 genome loci annotation for CP000725.1 and NC_009785.
Figure S1. S. gordonii fruRBA chromosomal region and DNA used for strain construction.
References
- Ajdić D, Pham VT. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol. 2007;189:5049–5059. doi: 10.1128/JB.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifano P, Bruni CB, Carlomagno MS. Control of mRNA processing and decay in prokaryotes. Genetica. 1994;94:157–172. doi: 10.1007/BF01443430. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini MM, Xu H, Sobue T, Nobile CJ, Del Bel Cury AA, Dongari-Bagtzoglou A. Candida-streptococcal mucosal biofilms display distinct structural and virulence characteristics depending on growth conditions and hyphal morphotypes. Mol Oral Microbiol. 2015 Mar 7; doi: 10.1111/omi.12095. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzini A, Beggah-Moller S, Moreillon P, Entenza JM. Lack of in vitro biofilm formation does not attenuate the virulence of Streptococcus gordonii in experimental endocarditis. FEMS Immuno Med Microbiol. 2006;48:419–423. doi: 10.1111/j.1574-695X.2006.00168.x. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Aké FM, Derkaoui M, et al. The bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Xie Z, Sobue T, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80:620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS ONE. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton LC, Nobbs AH, Jepson K, et al. O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. MBio. 2014;5:e00911. doi: 10.1128/mBio.00911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton LC, Paszkiewicz KH, Silverman RJ, et al. Transcriptional landscape of trans-kingdom communication between Candida albicans and Streptococcus gordonii. 2015 doi: 10.1111/omi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82:1968–1981. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AR, McNab R, Jenkinson HF. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect Immun. 1996;64:4680–4685. doi: 10.1128/iai.64.11.4680-4685.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AR, Shepherd MG. Nutritional factors determine germ tube formation in Candida albicans. J Med Vet Mycol. 1988;26:127–131. [PubMed] [Google Scholar]
- Jack AA, Daniels DE, Jepson MA, et al. The Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics NS, Strömberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol Microbiol. 2005;55:1591–1605. doi: 10.1111/j.1365-2958.2005.04495.x. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF. Beyond the oral microbiome. Environ Microbiol. 2011;13:3077–3087. doi: 10.1111/j.1462-2920.2011.02573.x. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Lala HC, Shepherd MG. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun. 1990;58:1429–1436. doi: 10.1128/iai.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M, Quadri LEN, Kuipers OP, de Vos WM. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- Kloosterman TG, Kuipers OP. Regulation of arginine acquisition and virulence gene expression in the human pathogen Streptococcus pneumoniae by transcription regulators ArgR1 and AhrC. J Biol Chem. 2011;286:44594–605. doi: 10.1074/jbc.M111.295832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Lengeler JW, Jahreis K. Bacterial PEP-dependent carbohydrate:phophotransferase systems couple sensing and global control mechanisms. Contrib Microbiol. 2009;16:65–87. doi: 10.1159/000219373. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Δ-ΔC(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J Bacteriol. 2003;185:6241–6254. doi: 10.1128/JB.185.21.6241-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina FL, Tobian JA, Jones KR, Evans RP. Molecular cloning in the streptococci. In: Hollaender A, DeMoss R, Kaplan S, Konisky J, Savage D, Wolfe R, editors. Genetic Engineering of Microorganisms for Chemicals. New York: Plenum; 1981. pp. 195–210. [Google Scholar]
- Morales DH, Hogan DA. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010;6:e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Jenkinson HF. Interkingdom networking within the oral microbiome. Microbes Infect. 2015 Mar 21; doi: 10.1016/j.micinf.2015.03.008. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Santos AJM, Bejerano-Sagie M, Correia PB, Marques JC, Xavier KB. Phosphoenolpyruvate phosphotransferase system regulates detection and processing of the quorum sensing signal autoinducer-2. Mol Microbiol. 2012;84:93–104. doi: 10.1111/j.1365-2958.2012.08010.x. [DOI] [PubMed] [Google Scholar]
- Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol. 2012;2:86. doi: 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME. Microbial interactions and differential protein expression in Staphylococcus aureus-Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol. 2010;59:493–503. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lutticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nature Genetics Supplement. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztajer H, Szafranski SP, Tomasch J, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014;8:2256–2271. doi: 10.1038/ismej.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum DM, Goll J, Murphy S, et al. The JCVI standard operating procedure for annotating prokaryotic metagenomic shotgun sequencing data. Stand Genomic Sci. 2010;2:229–237. doi: 10.4056/sigs.651139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42 (Database issue):D553–9. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergrasser A, Cutcutache I, Koressaar T, et al. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman MM, Wang M, Baker LJ. An amino acid change near the carboxyl terminus of the Streptococcus gordonii regulatory protein Rgg affects its abilities to bind DNA and influence expression of the glucosyltransferase gene gtfG. Microbiology. 2003;149:399–406. doi: 10.1099/mic.0.25983-0. [DOI] [PubMed] [Google Scholar]
- Vickerman MM, Iobst S, Jesionowski AM, Gill SR. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol. 2007;189:7799–7807. doi: 10.1128/JB.01023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez R, Lemme A, Ballhausen B, et al. Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF) Chembiochem. 2010;11:1552–1562. doi: 10.1002/cbic.201000086. [DOI] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in the building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in health and disease. Mol Oral Microbiol. 2014a;29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sobue T, Thompson A, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014b;16:214–231. doi: 10.1111/cmi.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map of DNA used for construction of ΔfruRBA mutant and complement strains. The chromosomal region of strain DL1 is shown with large arrows showing the positions and directions of the fruRBA genes. Software searches identified putative promoters (P) and transcriptional terminators (lollipops) in this region. The ΔG values for putative transcriptional terminators are shown as calculated with mfold (Zuker, 2003; http://mfold.rna.albany.edu). Positions of the Fru.F1/FruR1 and FruF2/FruR2 amplicons are shown above their corresponding genomic locations and include the upstream S. gordonii gene locus SGO_1110 and the downstream locus SGO_1114, respectively. Numbers directly below the ends of the amplicons (1153026 and 1158017) correspond to the nucleotide numbers in the S. gordonii Challis genome sequence (Genbank CP000725.1). The lower diagram shows the linear DNA fragment used to transform strain DL1 to construct strain UB2683, the ΔfruRBA deletion mutant in which the upstream and downstream amplicons flank an aad9 determinant with its own promoter and terminator. The upper panel shows the position of the amplicon made with primers BamFruCompF and BamFruCompR that was cloned into a replicative plasmid to generate p49MfruRBA used to complement strain UB2683.