Summary
Prokaryotes contain cytoskeletal proteins such as the tubulin-like FtsZ, which forms the Z ring at the cell center for cytokinesis [1], and the actin-like MreB, which forms a helix along the long axis of the cell and is required for shape maintenance [2]. Using time-lapse analysis of Escherichia coli cells expressing FtsZ-GFP, we found that FtsZ outside of the Z ring also localized in a helix-like pattern and moved very rapidly within this pattern. The movement occurred independently of the presence of Z rings and was most easily detectable in cells lacking Z rings. Moreover, we observed oscillation waves of FtsZ-GFP in the helix-like pattern, particularly in elongated cells, and the period of this oscillation was similar to that of the Min proteins. The MreB helix was not required for the rapid movement of FtsZ or the oscillation of MinD. The results suggest that FtsZ not only forms the Z ring but also is part of a highly dynamic, potentially helical cytoskeleton in bacterial cells.
Results and Discussion
FtsZ is essential for bacterial cell division [1, 3]. Despite its structural similarity to tubulins, FtsZ behavior is more akin to that of actin because FtsZ assembles into a cytokinetic ring, called the Z ring, at the cell midpoint between segregating daughter chromosomes [4, 5]. In Escherichia coli, midcell placement of the Z ring depends in part on the action of three Min proteins, MinC, MinD, and MinE, which migrate in an oscillatory pattern from one cell pole to the other over tens of seconds [6–8]. The localization pattern of the Min proteins is helical, suggesting that their oscillation follows a helical path [9].
Despite being in excess of the concentration needed for Z ring assembly [10], FtsZ does not form Z rings in E. coli cells until the time of chromosome replication termination and does not do so in newborn cells [11]. Once formed, FtsZ in Z rings has a turnover rate of about 30 s, with 30% of FtsZ present in the Z ring and 70% in the cytoplasm [12]. The connection between these different populations of FtsZ is unknown, as is the mechanism for triggering assembly at the correct cellular location. We hypothesized that, just as microtubules form multiple structures in the eukaryotic cell, non-ring FtsZ might have an organized structure as well. This hypothesis was also prompted by the helical organization previously observed for FtsZ outside of Z rings under specialized conditions, such as FtsZ overproduction in E. coli [5] or sporulation in B. subtilis [13].
FtsZ Moves Rapidly within a Helix-like Structure Apart from the Z Ring
To monitor the localization and movement of FtsZ, we used a green fluorescent protein (GFP) fusion to FtsZ, FtsZ-GFP, which is generally not capable of fully replacing native FtsZ for function. Nevertheless, FtsZ-GFP can localize like FtsZ and, when expressed at levels equivalent to or lower than those of native FtsZ, it acts as an accurate tracer for FtsZ localization and dynamics [5, 12–14]. E. coli EC448 expresses a single copy of FtsZ-GFP under IPTG control as well as wild-type FtsZ under control of its native promoter. EC488 is isogenic with EC448 except that wild-type chromosomal ftsZ is replaced by the ftsZ84(ts) allele. Under typical IPTG induction conditions with these strains and their derivatives, quantitative immunoblotting indicated that approximately 30%–40% of the FtsZ in the cells was FtsZ-GFP (data not shown), and cell division and length were normal (Figures 1A–1C and data not shown). In most cases, fluorescence microscopy of immobilized live cells under these conditions revealed typical medial Z rings (e.g., Figures 1A and 1C, left cell) that were much brighter than the surrounding fluorescence.
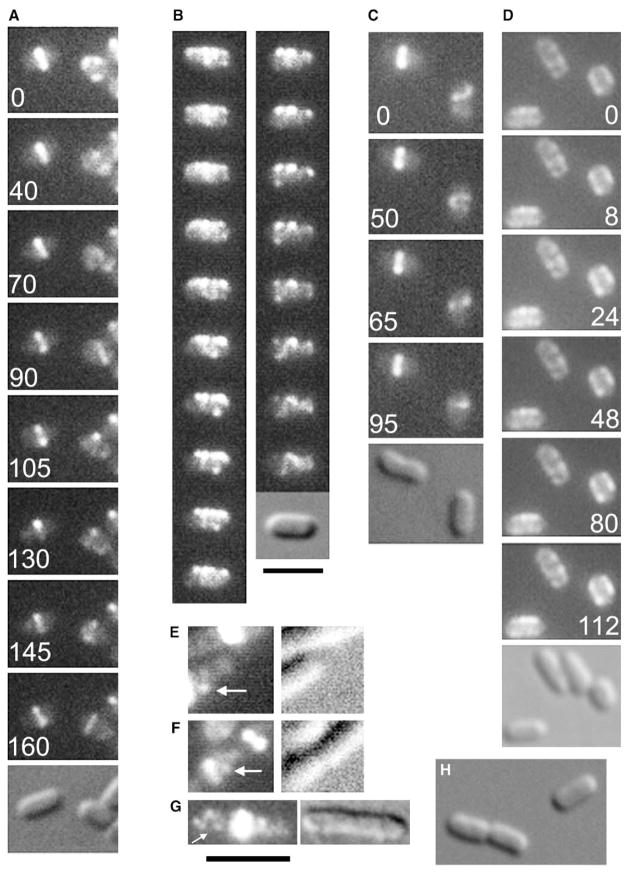
Figure 1. Non-Ring FtsZ Moves Rapidly in Helix-like Patterns.
(A–C) Time-lapse series of EC488 (A and B) or EC448 (C) cells grown in 40 μM IPTG at 30°C (B) or 37°C (A and C) to express FtsZ-GFP; for each series, fluorescence images are shown with a DIC image shown last. In (A) and (C), the cell on the left has a stable Z ring, whereas the cell on the right has not yet assembled a Z ring and exhibits the rapidly moving helix-like patterns. (D) A time-lapse series of WM2024 cells expressing GFP-MreB shows that the MreB coils do not move appreciably over the time course. Cells were prepared as in (A–C). For (B), the time interval between images was approximately 5 s; elapsed times in s are shown for (A), (C), and (D). (E–G) Fixed wild-type TX3772 cells were examined by immunofluorescence with FtsZ antibody; shown are fluorescence (left panels) and corresponding DIC image (right panels). Arrows highlight what appear to be helix-like structures. Very bright foci are Z rings purposely overexposed in order to show the faint non-ring fluorescence patterns. (H) Uninduced EC448 cells. Scale bars represent 3 μm for (A–D), (H) (under [B]) and for (E–G) (under [G]).
A closer look at patterns of fluorescence in newborn EC448 cells lacking distinct Z rings revealed the presence of fluorescence patterns were suggestive of a partial helix (Figure 1C, right cell). Unlike the previously reported cases of FtsZ helices, the FtsZ-GFP in these helix-like patterns was strikingly mobile, and cells exhibiting this behavior were actively dividing and within the normal 1.5–3 μm length range of the uninduced control cells (Figure 1H) or the MC4100 parent (data not shown). Most predivisional cells of EC448 or EC488 that were grown at the permissive temperature and were in the process of assembling a Z ring displayed mobile helices (Figure 1A–1C; Movies 1 and 2 in the Supplemental Data available with this article online). In the time courses in Figures 1A and 1C, the cell on the left has a clear Z ring at the beginning of the septation process, and a predivisional cell on the right has no Z ring. Although the FtsZ-GFP fluorescence in the cells with the Z ring did not fluctuate significantly in intensity or position, the fluorescence pattern was highly changeable in the predivisional cells. Merging pseudocolored images from different time points to show the positional displacement of fluorescence over time further demonstrated the degree of FtsZ-GFP mobility (Figures 2C and 2D).
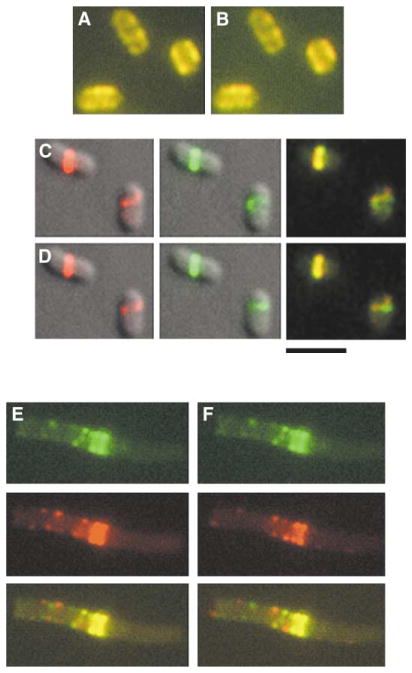
Figure 2. Difference Analysis of FtsZ-GFP Movement between Time Points.
(A and B) In RGB mode, the 8 s panel from the GFP-MreB time-lapse series in Figure 1D was pseudocolored red and merged with the 24 s panel or the 112 s panel (pseudocolored green) to give rise to the merged images in (A) and (B), respectively.
(C) The 0 and 50 s panels from the FtsZ-GFP time-lapse series in Figure 1C were pseudocolored red (left panel) and green (middle panel), respectively, and overlaid with the DIC image. The red and green images were then merged (right panel).
(D) The 65 and 95 s panels from the FtsZ-GFP time-lapse series in Figure 1C were pseudocolored red (left panel) and green (middle panel), respectively, and overlaid with the DIC image. The red and green images were then merged (right panel). The cell with the Z ring on the left serves as a useful image alignment control.
(E) Figure 3A panels 3 and 16, representing an interval of four complete FtsZ oscillation cycles, were pseudocolored green (top) and red (middle), respectively, and merged (bottom).
(F) Same as (E), except with Panels 3 and 10, representing an interval of two complete FtsZ oscillation cycles. Yellow pixels in the merged images represent overlapping red and green pixels and indicate no gross changes in localization between the two time points. Green and red pixels in the merged images, on the other hand, indicate positions where fluorescence localization changed over time. Note that panels (A) and (B) feature mainly yellow pixels, suggesting little movement over time, whereas green and red dots in panels (E) and (F) indicate areas of low fluorescence overlap between the two time points. The central ring or helical structure serves as a useful alignment control in the merged image. The scale bar represents 3 μm.
Although relatively long integration times were necessary for obtaining sufficient fluorescence signal for most of the above observations, shorter exposures sometimes also allowed visualization of FtsZ-GFP helical mobility. What became clear from these shorter exposures was that the movement of FtsZ-GFP was extremely rapid, with clear changes of position occurring within several seconds. This can be appreciated in the time- lapse panels, such as Figure 1B (5 s intervals). As with the oscillation of Min proteins, the movement of FtsZ-GFP did not require protein synthesis or even net growth because mobile FtsZ-GFP helical patterns were easily detected in stationary-phase cells, e.g., WM1135 (Movie 3) or in cells treated with chloramphenicol (data not shown).
To address potential artifacts caused by FtsZ-GFP, we also visualized FtsZ in fixed wild-type TX3772 cells by immunofluorescence. Z rings were easily the most prominent source of fluorescence intensity, but approximately 10% of the cells exhibited helix-like patterns of fluorescence, although the patterns were not as sharp as those in live cells with FtsZ-GFP (Figures 1E–1G, arrows). To address whether the observed movement was caused solely by subtle changes in focal planes during the time courses, we followed a time course of cells expressing GFP-MreB (WM2024), which forms a helical pattern in E. coli cells most easily observed by 3D image reconstruction [9]. Although the coiled pattern was reminiscent of that made by FtsZ-GFP, little if any movement was detected during a 2 min time period (Figures 1D, 2A, and 2B; Movie 4), suggesting that GFP-MreB localization is relatively static in E. coli and that the majority of the FtsZ-GFP movement seen in other time-lapse images was not a result of focal-plane changes.
FtsZ-GFP Moves in Slow Oscillatory Waves Similar to Those of MinD
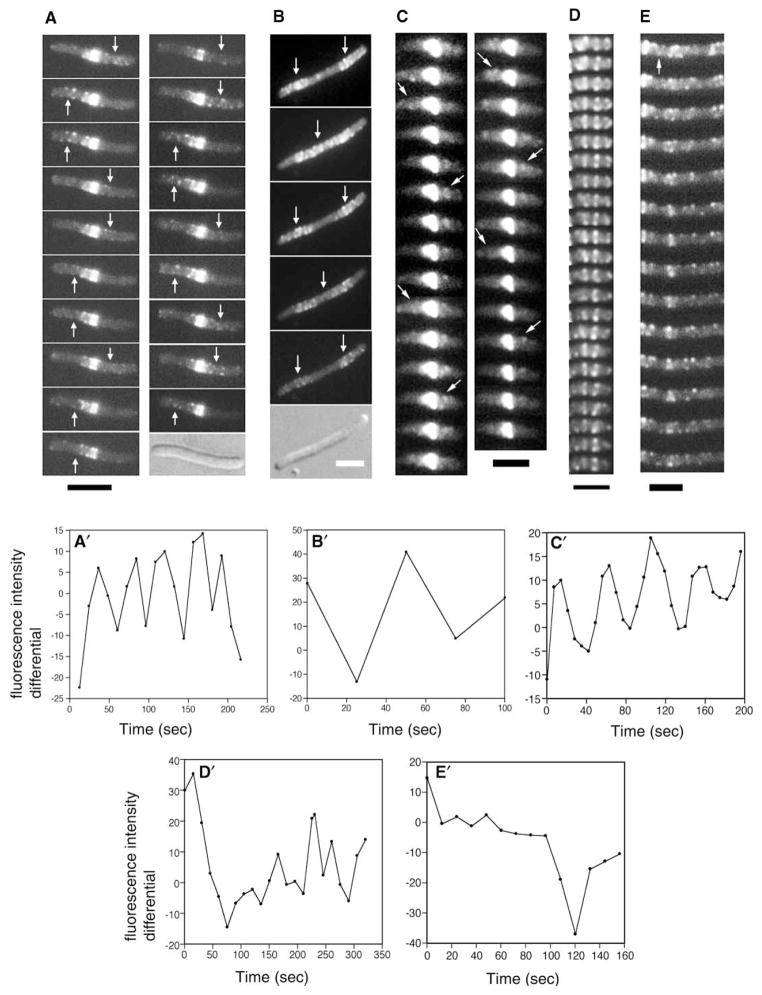
Analysis of live cells expressing FtsZ-GFP over longer time intervals indicated that, in addition to the rapid traffic of FtsZ within the helices, there was a longer-range back-and-forth movement of a large fraction of total fluorescence. This bulk oscillatory movement of fluorescence was sometimes observed in cells of normal length but was difficult to detect because of the low fluorescence signal compared with that of the Z ring and the small oscillation length (data not shown). However, increasing the cell length by thermoinactivation of the chromosomal ftsZ(ts) allele (EC488), coexpression of SulA (WM2012), or inactivation of the later cell division protein FtsI with cephalexin in otherwise wild-type EC448 facilitated the direct observation of these oscillation waves in many cells. These waves also exhibited a helix-like pattern and generally ranged from 30 to 60 s for a complete long-range cycle (Figures 3A–3C and 3A′–3C′; Movie 5). The maximum oscillation distance was approximately 8–9 μm; cells longer than this tended to have two oscillation waves (Figure 3B). It should be noted that for the experiment in Figure 3A, the FtsZ-GFP fusion protein may have partially complemented the thermoinactivated ftsZ84(ts) allele. However, inhibition of FtsZ itself was not required for the oscillation to be visualized (Figures 3C and 3C′), although inactivation of FtsI by cephalexin can destabilize Z rings indirectly [15]. The observation of this FtsZ oscillation in many cells further argues against the notion that the observed FtsZ movement is an artifact of the GFP fusion or focal-plane changes.
Figure 3. FtsZ Moves in MinD-like Oscillation Waves.
(A) Time-lapse images of EC488 cells grown at 42°C to thermoinactivate the chromosomal copy of ftsZ84(ts) and supplemented with 40 μM IPTG to express FtsZ-GFP. Intervals between panels are 12–15 s. Arrows highlight the FtsZ-GFP oscillation.
(B) Time-lapse images (approximately 25 s intervals) of a typical WM2012 filamentous cell grown for 2 hr at 37°C with 0.1% arabinose to induce synthesis of SulA + 60 μM IPTG to induce synthesis of FtsZ-GFP. Note that no Z rings were visible because of the action of SulA, yet the fluorescence still oscillated (arrows).
(C) Time-lapse images of EC448 treated with 40 μg/ml cephalexin for 0.5 hr to inhibit late septation events. The Z rings were overexposed so that the dim fluorescence moving in an oscillatory manner (arrows) could be detected.
(D) Time course of a typical segment of a WM1994 (ΔminCDE::kan in EC448) cell expressing FtsZ-GFP at 30°C with 40 μM IPTG. A cell segment was used because most Δmin cells are short filaments. Time intervals between frames were approximately 12 s.
(E) Same as (D), except that a typical segment of a WM1993 cell (ΔminCDE in EC488, with ftsZ84 on the chromosome) was used. The arrow highlights a transient helix-like segment. Cells containing Δmin and ftsZ84 are highly filamentous even at 30°C because of a synthetic effect [24].
(A′–E′) Plots of differential peak fluorescence intensities between two adjacent cell segments (see Experimental Procedures); panels correspond to (A)–(E), respectively. The cell segments used for the plot were the right half minus the left half of cells in (A), (C), (D), and (E), and the right segment minus the middle of the filament in (B). Scale bars represent 3 μm.
The above oscillation behaviors are strikingly similar to those of the Min proteins, which have a similar oscillation cycle, length, and localization pattern. To address whether the FtsZ-GFP oscillation was Min-dependent, we examined the FtsZ-GFP localization pattern in ΔminCDE cells lacking the Min proteins (WM1994). Such cells are significantly longer on average because of the division of polar septa at the expense of medial septa [16], facilitating the detection of potential oscillatory waves of FtsZ-GFP. Analysis of more than 20 min− cells via time-lapse movies (Movie 6, Figure 3D′, and data not shown) indicated that no obvious bulk oscillatory movement of FtsZ-GFP from one part of the cell to another occurred with visible periodicity; nevertheless, the rapid traffic within the helix-like pattern remained (Figure 3D and Movie 6). Cells containing ftsZ84(ts) as well as ΔminCDE (WM1993) were also examined and were found to exhibit similar FtsZ-GFP movement with-out obvious oscillations (Figures 3E and 3E′; Movie 7).
We can conclude from these data that the Min proteins, which move in a helical pattern, are not required for the rapid mobility of FtsZ or its helix-like pattern. The apparent lack of regular periodicity of FtsZ-GFP movement in Min− cells compared to Min+ cells (Figures 3A′–3E′) suggests that the 30–60 s FtsZ oscillations are Min dependent. However, because some periodic patterns were observed between different cell segments (Figures 3D′, 3E, and 3E′ and data not shown), we cannot rule out the possibility that a more subtle Min-independent FtsZ-GFP oscillation that cannot easily be detected may be occurring.
MreB Is Not Required for Either FtsZ or MinD Mobility
We next were interested in identifying the source of the FtsZ helix-like localization patterns that were often observed. Overlays of pairs of time-lapse images show that oscillating FtsZ-GFP fluorescence often appeared not to revisit the same spots, which might represent the edges of a helix, over short time intervals (Figures 2E, 2F, and 3E; data not shown). These differences could be explained by subtle changes in the focal plane. However, given that such changes were not evident in the GFP-MreB time course (Figures 1D, 2A, and 2B), an equally likely possibility was that the FtsZ helix-like pattern itself might be able to shift translationally. Deconvolution microscopy would normally be useful for visualizing the differences in helix-like localization but in this case did not help because meaningful deconvolution of optical sections cannot work when the fluorescence moves while sections are being obtained.
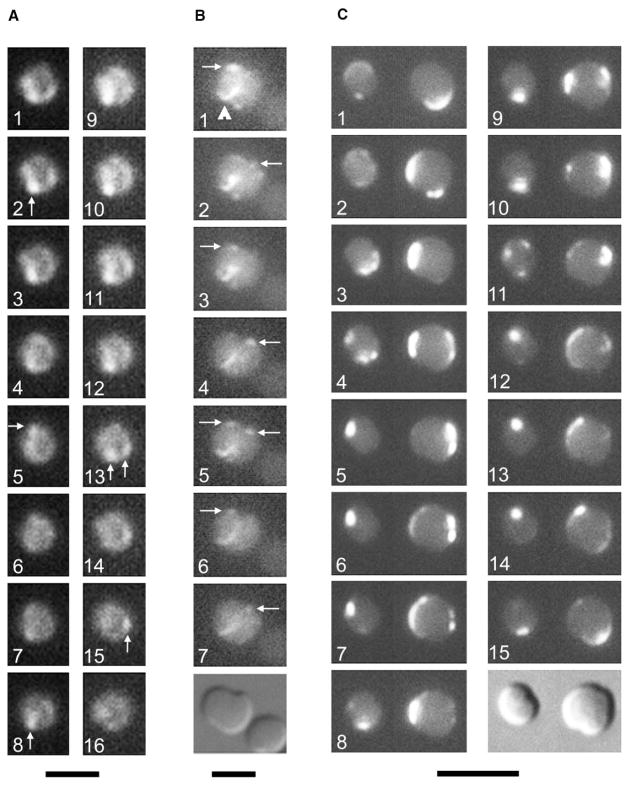
To further explore the identity of the helix, we asked whether the rapid mobility of FtsZ was dependent on MreB. The MreB helix, at least in B. subtilis, has a slow turnover rate [2, 17]. This helix is probably used to direct cell wall biosynthesis and may be important as a track for chromosome segregation. The failure of a GFP-MreB fusion to display movement during time-lapse experiments (see above) is consistent with the idea that MreB is a relatively stable helical structure. We expressed FtsZ-GFP in ΔmreB::cat mutants, which grow and divide as round cells because they lack MreB-directed cylindrical shape. FtsZ does not need MreB for localization because many FtsZ structures were present at sites of cytokinesis in ΔmreB::cat cells (Figure 4B, arrowhead, and data not shown). These structures, similar to those observed in rodA round-cell mutants [18], were often spiral shaped.
Figure 4. The MreB Helix Is Not Required for the Rapid Movement of FtsZ or MinD.
(A and B). Time courses of FtsZ-GFP movement in cells lacking MreB (WM2002). For panel (A), images were taken 12–15 s apart. For panel (B), times elapsed were 0, 60, 120, 160, 180, 200, and 240 s, respectively, with a DIC image at the end. Arrows highlight the most visible fluorescent foci, which appeared to move in an oscillatory manner perpendicular to the division plane (most obvious in [B]). The arrowhead in (B) points to a FtsZ arc at the medial constriction site.
(C) Time course of GFP-MinD movement in two WM1928 cells (ΔmreB::cat) at 8 s intervals, with a DIC image at the end. Note the complex localization patterns, including the assembly at multiple foci in panel 11. The scale bar for (A) represents 2 μm; that in (B) represents 3 μm; and that in (C) represents 5 μm.
Accurate imaging of FtsZ-GFP movement was more problematic in round cells because of their increased focal depth. Nevertheless, it was apparent that the protein retained its ability to move rapidly between localized areas of fluorescence (Figures 4A and 4B; Movie 8). In some cases, FtsZ-GFP appeared to move back and forth in a manner perpendicular to the incipient division plane (Figure 4B, arrows) and in a fashion similar to that of GFP-MinD in round rodA mutant cells [18]. However, the fluorescence intensity was sufficiently low to make it difficult to state conclusively that FtsZ-GFP was exhibiting oscillatory movement in these cells. The oscillation period and localization pattern of GFP-MinD in round cells is known to be highly variable, even within the same cell [18], and therefore could not serve as a guide for a predicted oscillation period for FtsZ-GFP in round cells. Nevertheless, these results indicate that the rapid mobility of FtsZ-GFP does not require MreB. Interestingly, the movement of Min proteins also did not require MreB because GFP-MinD displayed clear oscillation patterns similar to those in rodA mutant cells; these patterns featured directional drift and the formation of multiple simultaneous assembly points (Figure 4C), as observed previously [18].
Implications and Conclusions
Our results indicate that FtsZ exists in two states in bacterial cells. One, the cytokinetic Z ring, is well characterized. However, once FtsZ disassembles from the Z ring, it does not appear to diffuse evenly in the cytoplasm; such organized redistribution was first suggested by our previous time-lapse studies with 3D-reconstructed images [14] but may have been overlooked because such reconstruction cannot be used to localize fluorescence undergoing extremely rapid movement. We propose that upon disassembly of the Z ring, FtsZ in daughter cells is redistributed to and stored in a highly dynamic helical FtsZ cytoskeleton, which may be analogous to cytoplasmic microtubules in eukaryotes, until the next Z ring is assembled. Although a potential FtsZ helix was harder to detect in normal cells with Z rings, probably because non-ring fluorescence was depleted, we speculate that such a putative helix persists and creates the reservoir for rapid FtsZ turnover of monomers observed by FRAP [12]. It is possible that the FtsZ helices observed during sporulation of B. subtilis [13] reflect a similar cytoskeletal organization with much slower FtsZ mobility [19]. We speculate that the high mobility of the FtsZ cytoskeleton in E. coli may serve to constantly scan the cell surface for potential division sites more efficiently than if it were dispersed randomly throughout the cytoplasm.
The driving force for the rapid mobility of FtsZ is not known. However, our results suggest that this seemingly random motion may be given global direction by the Min proteins, which promote the bulk transport of FtsZ away from cell areas, such as the cell poles, that would be deleterious for formation of the Z ring. The Min proteins interact with a septal component [20] and help to disassemble the Z ring [21]. The apparent ability of the Min proteins to stimulate bulk movement of FtsZ suggests that FtsZ may be assembled in oligomers in the putative helices, with the intensity of fluorescence possibly being a measure of the extent of the assembly. It is intriguing that FtsZ oscillatory movement may be driven by a Min-directed Brownian ratchet [22].
At first glance, the helix-like path apparently traced by FtsZ in E. coli cells might suggest that FtsZ moves along the MreB helix in wild-type cells. However, our results suggest that FtsZ traffic follows an MreB-independent path. The strongest evidence is that FtsZ mobility does not require MreB. Additionally, MreB helices appear to be relatively static, whereas the helical pathway for FtsZ appears to fluctuate over time. The potentially helical organization of FtsZ also hints that the Z ring itself may be part of a FtsZ helix [23], which makes sense if FtsZ can rapidly interconvert between the two states. This model predicts that Z ring assembly may be triggered upon a change in assembly equilibrium within a preexisting FtsZ helix. One important question to address in the future is whether this putative FtsZ helix is self-formed or if FtsZ is traveling down a helical pathway determined by another factor. It will also be interesting to determine whether other proteins that localize to specific cellular addresses in bacterial cells use helical pathways along the membrane to be transported efficiently to their destination.
Supplementary Material
Acknowledgments
We thank Stacie Meaux for initial construction of the ΔmreB strain, Brian Corbin for preliminary data for GFP-MinD oscillation, Brett Geissler for the immunofluorescence images, David Ehrhardt, Eugenia Mileykovskaya, and members of the Margolin lab for helpful discussion, and David S. Weiss for EC448 and EC488. The research was supported by National Institutes of Health grant GM-61074 to W.M.
Footnotes
Supplemental Experimental Procedures and a table showing strains used in this study are available online at http://www.current-biology.com/cgi/content/full/14/13/1167/DC1/.
References
- 1.Addinall SG, Holland B. The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J Mol Biol. 2002;318:219–236. doi: 10.1016/S0022-2836(02)00024-4. [DOI] [PubMed] [Google Scholar]
- 2.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 3.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2002;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hale CA, Meinhardt H, de Boer PA. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 2001;20:1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu X, Shih YL, Zhang Y, Rothfield LI. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc Natl Acad Sci USA. 2001;98:980–985. doi: 10.1073/pnas.031549298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 11.Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM, Nanninga N. Timing of FtsZ assembly in Escherichia coli. J Bacteriol. 1999;181:5167–5175. doi: 10.1128/jb.181.17.5167-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 14.Sun Q, Margolin W. FtsZ dynamics during the cell division cycle of live Escherichia coli. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Begg K, Nikolaichik Y, Crossland N, Donachie WD. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol. 1998;180:881–884. doi: 10.1128/jb.180.4.881-884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carballido-Lopez R, Errington J. The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- 18.Corbin BD, Yu XC, Margolin W. Exploring intra-cellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 2002;21:1988–2008. doi: 10.1093/emboj/21.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolin W. Bacterial sporulation: FtsZ rings do the twist. Curr Biol. 2002;12:391–392. doi: 10.1016/s0960-9822(02)00882-5. [DOI] [PubMed] [Google Scholar]
- 20.Johnson JE, Lackner LL, de Boer PA. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J Bacteriol. 2002;184:2951–2962. doi: 10.1128/JB.184.11.2951-2962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichoff S, Lutkenhaus J. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J Bacteriol. 2001;183:6630–6635. doi: 10.1128/JB.183.22.6630-6635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oster G. Brownian ratchets: Darwin’s motors. Nature. 2002;417:25. doi: 10.1038/417025a. [DOI] [PubMed] [Google Scholar]
- 23.Bramhill D, Thompson CM. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc Natl Acad Sci USA. 1994;91:5813–5817. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu XC, Margolin W. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J Bacteriol. 2000;182:6203–6213. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.