Introduction
Pegylated interferon-α (peginterferon-α or PEG-IFN-α) is an antiviral and immunoregulatory drug that has served as the foundation for the treatment of hepatitis C virus (HCV) infection for more than two decades. Currently, there are two PEG-IFNs on the market, in combination with the broad-spectrum antiviral, ribavirin (RBV), for the treatment of chronic HCV: PEG-IFN-α 2a and PEG-IFN-α 2b. The two drugs are differentiated by the size and nature of their covalently attached polyethylene glycol (PEG) moiety. Although PEG-IFN-α 2a and 2b exhibit differences in pharmacokinetics and dosing regimens, the consensus is that the two drugs are clinically equivalent [1].
HCV infection affects 170–200 million people worldwide and is the leading cause of cirrhosis and hepatocellular carcinoma [2]. Although the prevalence of HCV appears to be decreasing in most developed nations, mortality due to liver diseases secondary to HCV infection is expected to continue to rise over the next 20 years [3]. Although a minority of patients with acute HCV infections are able to clear the virus spontaneously, 55–85% of patients develop chronic HCV infection, defined as detectable HCV RNA for longer than 6 months [4]. Chronic infection, once established, rarely resolves spontaneously [5]. The primary goal of treatment is achievement of sustained virologic response (SVR), characterized by undetectable serum viral RNA 24 weeks after the end of therapy, which is considered clinically as cure of infection and is associated with lower morbidity and mortality [6]. HCV is divided into seven recognized genotypes (1–7), which differ at 30–35% of nucleotide sites [7]. HCV genotypes are differentially distributed globally, with genotype 1 being most prevalent in North America, South America, and Europe [8]. Further, the HCV genotype is highly associated with response to PEG-IFN-α/RBV combination therapy, and the rates of treatment-induced SVR are lowest for patients infected with HCV genotypes 1 and 4 [9].
Before 2011, the standard-of-care for patients with chronic HCV was a combination of PEG-IFN-α and RBV therapy (either PEG-IFN-α 2a or 2b) administered for either 48 weeks, for HCV genotypes 1, 4, 5, and 6, or 24 weeks, for genotypes 2 and 3. However, these combination therapies yield SVR rates of only 40–50% in HCV genotype 1 patients [10]. In contrast, SVR rates for patients with genotypes 2 and 3 are ~ 70–80%, although these rates apply to selected populations without the comorbidities that often accompany HCV [10–14]. Further, combination therapy is costly, is associated with several moderate-to-severe side effects (influenza-like symptoms, depression, thrombocytopenia, and hemolytic anemia) [15], and is contraindicated in many patients (hepatic decompensation, portal hypertension, hypersplenism, severe psychiatric depression, major systems impairment, and pregnancy) [16]. Positive predictive factors for achieving SVR include young age, female sex, and low pretreatment HCV-RNA levels [17]. Conversely, SVR is less likely in HCV patients with high baseline HCV-RNA levels (>800 000 IU/ml), steatosis, insulin resistance, coinfection with HIV, and more advanced liver fibrosis [5,17].
The strongest pretreatment predictor of SVR rate is variations in IFNL3 (formerly known as IL28B), which is located on chromosome 19. The gene encodes IFN-λ 3, a member of the type 3 IFN-λ family that exhibits antiviral, antiproliferative, and immunomodulatory activities [18]. The strongest and most commonly tested polymorphisms are rs12979860 and rs8099917, which are located 3 and 8 kb upstream of IFNL3, respectively, and exhibit strong linkage disequilibrium [19]. As described in detail below, patients with the favorable rs12979860 CC or rs8099917 TT genotype have a greater than twofold increased likelihood of achieving treatment-induced SVR. The Clinical Pharmacogenetics Implementation Consortium, which advises clinicians on how to utilize genetic information in treatment decisions, has produced detailed guidelines on the basis of IFNL3 genotyping for PEG-IFN-α/RBV-based treatment regimens [20]. IFNL3 genotype can inform decisions on whether to initiate PEG-IFN-α/RBV therapy given the risk for adverse events and treatment failure. Once the decision to initiate therapy is made, the IFNL3 genotype can be used to inform decisions on both treatment composition [e.g. whether to prescribe PEG-IFN-α/RBV dual therapy or triple therapies that include direct-acting antiviral agents (DAAs)] and duration (e.g. whether to shorten treatment duration in certain patients). The current consensus is that the rs1297860 genotype is the single-nucleotide polymorphism (SNP) most likely to predict treatment response if a single SNP has to be selected for diagnostic purposes, although tests for both rs12979860 and rs8099917 genotyping are commercially available [21,22].
Recently, the advent of DAAs has allowed for even higher SVR rates in HCV genotype 1 patients. DAAs directly target specific stages of the HCV life cycle. Although PEG-IFN and RBV remained essential components of therapy with the first waves of these medications, they are no longer part of the recommended regimens. The first regimens were called triple therapies and included one DAA in combination with PEG-IFN and RBV. These regimens significantly improved SVR rates and allowed for shorter treatment regimes in many patients [23]. In 2011, the American Association for the Study of Liver Diseases updated its practice guidelines for chronic HCV genotype 1 patients to triple therapies that include either of the ‘first-generation’ protease inhibitor DAAs, boceprevir (BOC) or telaprevir (TVR) [23]. These triple therapies have demonstrated SVR rates of 65–75% [24–26]. The ‘second-generation’ protease inhibitor simeprevir was approved by the Food and Drug Administration (FDA) in 2013 and raised the SVR rates to ~ 80% [27]. Sofosbuvir, which was recently approved by the FDA, has shown great potential to improve SVR rates even further, either as part of triple therapies in combination with PEG-IFN-α/RBV [28–30] or as part of IFN-free therapies with either simeprevir or ledipasvir [31,32]. Finally, trials of the recently FDA-approved IFN-free regime consisting of paritaprevir (ABT-450) boosted by ritonavir, ombitasvir, dasabuvir, and ribavirin have demonstrated SVR rates greater than 95% in both treatment-naive patients and previous nonresponders with HCV genotype 1 and no cirrhosis [33,34]. As a result, the American Association for the Study of Liver Diseases recently revised its practice guidelines for HCV genotype 1 patients to include only IFN-free regimes, which boast of improved tolerability as well as efficacy [35].
Pharmacokinetics
Attachment of a PEG moiety (pegylation) was a major advancement that led to improvement in the pharmacokinetics of IFNs. Compared with administration of unmodified IFN-α three times a week, PEG-IFNs allow for a once-weekly dosing administration and avoid large fluctuating serum concentrations. PEG-IFN-α 2a has a branched 40 kDa PEG chain covalently attached to lysine residues and circulates as an intact molecule, whereas PEG-IFN-α 2b has a linear 12 kDa PEG chain covalently attached through an unstable urethane bond that is hydrolyzed after injection. The size and nature of the PEG moiety attached causes differences in the pharmacokinetics and dosing regimens of the drugs [36].
The absorption half-life of unmodified IFN-α is 2.3 h, whereas those of PEG-IFN-α 2a and 2b are ~ 50 and 4.6 h, respectively [37]. In addition to its longer half-life, PEG-IFN-α 2a is more highly localized than PEG-IFN-α 2b, having a smaller volume of distribution (0.99 l/kg), with highest concentrations occurring in the liver [37]. Patients administered a single dose of PEG-IFN-α 2a reached a mean maximum serum concentration of 14.2 μg/l in a mean time of 78 h after administration [38]. Following multiple doses (180 μg weekly), the mean maximum serum concentration was 25.6 μg/l, which was reached in a mean time of 45 h. In contrast, maximum serum concentrations for PEG-IFN-α 2b were achieved between 15 and 44 h after administration and sustained for 48–72 h. PEG-IFN-α 2a had a smaller peak-to-trough ratio of 1.5–2, as compared with a ratio of greater than 10 after multiple doses. Thus, PEG-IFN-α 2a exhibited less fluctuation in serum concentration during the 1-week dosing interval [37].
The longer half-life and limited distribution of PEG-IFN-α 2a allows for a fixed, weekly dose. Conversely, weekly dosage of PEG-IFN-α 2b must be adjusted according to body weight [1]. Because of the relatively short serum half-life of PEG-IFN α 2b, a significant number of patients may have undetectable levels of the drug by the end of the weekly dose interval, which may cause viral rebound [39,40]. It has been suggested that a shorter dosage interval, such as a twice-weekly regimen, may be necessary for PEG-IFN-α 2b, although this has not yet been formally tested [38].
PEG-IFN-α 2a is cleared by both the liver and the kidney; ~ 30% of PEG-IFN-α 2b is cleared by the kidney, with the rest cleared by the liver or degraded after interacting with cellular IFN receptors [37]. Compared with standard IFN-α, PEG-IFN-α 2a and 2b have significantly reduced renal clearance [41].
RBV coadministration does not affect the pharmacokinetics of PEG-IFNs, nor do PEG-IFNs affect that of RBV [42]. For patients administered PEG-IFN-α 2b/RBV combination therapy, the mean peak plasma RBV concentrations at week 1 were 741, 799, and 1101 ng/ml for daily RBV doses of 600, 800, and 1000–1200 mg, respectively. At week 4, these dosing regimens produced mean peak plasma RBV concentrations of 1770, 2297, and 2750 ng/ml, respectively, accounting for five-fold RBV accumulation. Regardless of dosing group, apparent clearance of RBV was consistently 23–26 l/h [37].
Pharmacodynamics
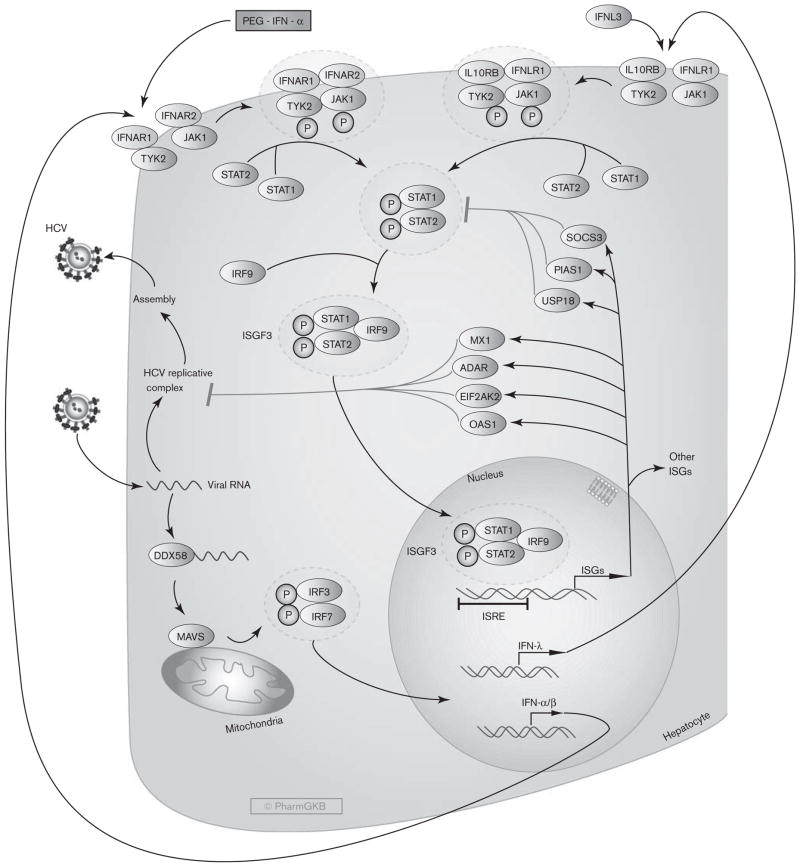
IFN-α acts both directly and indirectly as an antiviral, although the exact mechanisms by which PEG-IFN-α inhibits HCV replication are still not well understood [1]. IFN-α exerts its effects by inducing a non-virus-specific immune response in cells through up-regulation of many IFN-stimulated genes (ISGs) that encode antiviral effector proteins. As shown in Fig. 1, when HCV RNA is detected by the host innate immune system, it is recognized by several pathogen recognition receptors such as retinoic acid inducible gene-1, encoded by the DDX58 gene. Retinoic acid inducible gene-1 activates the adapter mitochondrial antiviral signaling protein, which induces phosphorylation of IFN regulatory factor 3 (IRF3) and IRF7. The two proteins heterodimerize, then translocate to the nucleus and induce the expression and secretion of various IFNs, including IFN-α, IFN-β, and IFN-λ.
Fig. 1.
Stylized depiction of the pharmacodynamics of Peginterferon-α in hepatocytes. A fully interactive version is available at PharmGKB (http://www.pharmgkb.org/pathway/PA166126086).
Circulating IFN-α binds to two unique cell surface receptor subunits, IFN-α R1 and IFN-α R2. Once bound to IFN-α R1 and IFN-α R2, a heterodimer is formed and activates Janus-activated kinase (JAK) and tyrosine kinase, which in turn phosphorylate the cytoplasmic signal transducers and activators of transcription (STAT) proteins, STAT1 and STAT2. Phosphorylated STAT1 and STAT2 dimerize and bind IRF9, forming the interferon-stimulated gene factor 3 (ISGF3) complex. ISGF3 then translocates to the nucleus, where it binds to interferon-stimulated response elements. Upon binding DNA, ISGF3 induces transcription of ISG mRNAs encoding a number of effector proteins. Microarray analyses have shown that IFN-α induces hundreds of ISGs, many of which are related to antiviral activity and others are involved in lipid metabolism, apoptosis, protein degradation, and inflammatory responses [43]. Among the major ISGs believed to play important roles in inhibiting HCV replication are 2′,5′ oligoadenylate synthetase 1, eukaryotic translation initiation factor 2-alpha kinase 2 (formerly known as PKR), adenosine deaminase RNA-specific, and MX dynamin-like GTPase 1. Conversely, antiviral signaling is controlled by anti-inflammatory ISGs, such as ubiquitin-specific peptidase 18, suppressor of cytokine signaling 3, and protein inhibitor of activated STAT1, which act as part of a negative feedback loop that restricts both the extent and duration of the IFN response by direct or indirect inhibition of STAT signaling [44,45].
In addition to its antiviral activity, IFN-α exerts immunologic effects through interaction with the adaptive and innate immune responses to promote memory T-cell proliferation, prevent T-cell apoptosis, and stimulate natural-killer cell activation and dendritic-cell maturation [46]. Finally, IFN-α has been shown to increase the production of major histocompatibility complex class I and class II molecules [47]. However, these immunological properties are likely not directly involved in HCV clearance by IFN-based therapies, which are thought to exert their effects primarily through their antiviral activity [48].
Virological decline, often referred to as the ‘viral kinetics of response’, is an important on-treatment predictor of achieving an SVR during PEG-IFN-α/RBV therapy. Rapid virological clearance (RVR) and early virological response (EVR), defined as undetectable HCV RNA after 4 weeks and a 2 log decline in HCV RNA after 12 weeks, respectively, have both been shown to be associated with SVR [23,49]. Indeed, SVR rates for patients who achieve RVR are greater than 70%, and the treatment regime can be shortened in these patients [50,51].
The pharmacodynamics of PEG-IFN-α 2a and 2b appears to be similar. Pharmacodynamic comparisons have shown that the two PEG-IFN-α forms have similar biological activity, as evidenced by induction patterns of 2′,5′-oligoadenylate synthetase, neopterin, and the major histocompatibility complex class I molecule β2-micro-globulin [1,52]. However, one study found that PEG-IFN-α 2b exhibited greater activation of ISGs in a group of 31 patients with HCV genotype 1 infection [53].
Similar to IFN-α, IFN-λ activates the JAK-STAT pathway, which upregulates a number of ISGs by binding to the IFN-stimulated response element, and establishes an antiviral state in cells. IFN-λ has been shown in experimental models to inhibit HCV replication, and when coadministered with IFN-α, this antiviral effect was additive [54,55]. Although IFN-α and IFN-λ increase the expression of a similar set of ISGs through the same downstream pathways to exert their effects, they signal through unique receptors: IFN-λ binds to the IL10R–IL28R receptor complex, which is highly expressed only in hepatocytes [56]. Further, the kinetics of response differ between the two IFNs: IFN-λ exhibits more rapid phosphorylation of STAT, yet more delayed and prolonged expression of various ISGs compared with IFN-α [55]. Because of the more limited distribution of the IFN-λ receptor, among other factors, it has been suggested that a pegylated form of IFN-λ may be better tolerated than PEG-IFN-α [57,58].
The mechanism of action of RBV against HCV remains unclear. It has been suggested that RBV acts directly to inhibit HCV replication or inhibits the host enzyme inosine monophosphate dehydrogenase [58]. Conversely, RBV may act indirectly against HCV viral replication by causing rapid and lethal mutations of virions or depletion of intracellular guanosine triphosphate, which is required for viral RNA synthesis [59,60]. Finally, it has been suggested that RBV may exert its effect through immunomodulatory activity [5].
Pharmacogenomics
IFNL3 variation and PEG-IFN-α/RBV treatment response
Marked variation in HCV clearance and response to PEG-IFN-α among different ethnic groups and among patients infected with the same HCV genotype have long pointed to host genetic factors as determinants of treatment success [61]. In 2009, several genomewide association studies independently showed that several SNPs on chromosome 19, near IFNL3, are strongly associated with response to PEG-IFN-α/RBV therapy in individuals infected with HCV genotype 1 or 4. These studies demonstrated that HCV genotype 1 patients with favorable variants in the IFNL3 locus (rs12979860 CC or rs8099917 TT) were two to three times more likely to achieve SVR with PEG-IFN-α/RBV combination therapy and also had higher rates of spontaneous HCV clearance in the absence of treatment, as compared with patients with the unfavorable T or C genotype, respectively [62–66]. These findings were later validated in several candidate gene studies [67–70]. The association between the IFNL3 genotype and SVR has been confirmed by independent studies in various populations from Asia, Europe, and Latin America [22,71–77]. Thus, IFNL3 variation constitutes the strongest pretreatment indicator of PEG-IFN-α/RBV response for HCV genotype 1 patients, even when accounting for known clinical predictive factors [62,65]. Once PEG-IFN-α/RBV therapy is initiated, the favorable rs12979860 CC genotype is also associated with significantly higher rates of viral clearance at the 2, 4, 12, and 48-week treatment points, indicating better viral kinetics, as well as increased rates of RVR, EVR, and end-of-treatment response, and a reduced virological relapse rate [78].
Most studies of patients infected with HCV genotypes 2 and 3 have found a relatively weak correlation between the IFNL3 genotype and treatment response, but many have failed to achieve statistical significance [65,79–83], likely because SVR rates are generally high among HCV genotype 2 and 3 patients [22]. Further, as variation in SVR rates among the different IFNL3 genotypes is generally smaller in patients infected with HCV genotypes 2 and 3, IFNL3 genotyping may be of little value in predicting SVR. However, significant associations between the IFNL3 genotype and RVR have been demonstrated in studies on HCV genotype 2 and 3 patients [84–87]. The IFNL3 genotype may be more useful in predicting RVR in patients with HCV genotypes 2 and 3, which can inform decisions to shorten treatment duration and minimize treatment-associated adverse effects [88].
Role of the IFNL3 genotype in HCV-HIV coinfected and liver transplant patients
Coinfection with HCV is relatively common (15–40%) among patients with HIV infection, likely because of common routes of transmission [89]. As the progression of HCV-related liver disease is accelerated in coinfected patients [90,91], HCV has become an important cause of morbidity and mortality among HIV-infected individuals [92]. Unfortunately, HIV–HCV-coinfected patients are also less likely to respond to PEG-IFN-α/RBV therapy compared with HCV monoinfected patients [89]. Thus, predictors of response to PEG-IFN-α/RBV treatment are useful for identifying the best candidates for current therapies. The association between the IFNL3 genotype and PEG-IFN-α/RBV therapy outcome has been consistently demonstrated across studies in subgroup analyses of HIV patients coinfected with HCV genotypes 1 and 4, but not in patients with HCV genotypes 3 and 4 [79,84]. Several studies have shown an association between the favorable IFNL3 genotypes (rs12979860 CC or rs8099917 TT) and higher SVR rates [79,84,93–95], although one study that compared the two SNPs found that rs12979860 was a better predictor of response [96]. IFNL3 genotyping may also be useful in patients coinfected with HIV and HCV genotype 1 or 4 who had undergone previously failed PEG-IFN-α/RBV therapy, to identify nonresponders who would likely benefit from retreatment [84,97].
In addition, the IFNL3 genotype has been demonstrated to be an important predictor of reinfection in HCV patients who undergo liver transplantation. In several independent cohorts, the favorable rs12979860 CC and rs8099917 TT genotypes of both donor and recipient have been shown to be associated with higher SVR rates [98–101]. However, most studies that identified correlations were performed in HCV genotype 1 patients and there is little evidence of associations among other genotypes [22].
Distribution of IFNL3 alleles among ethnic groups
The favorable IFNL3 rs12979860 polymorphism is differentially distributed among ethnic groups, with African-Americans having the lowest frequency (allele frequency 0.39), Eastern and South-Eastern Asians having the highest frequency (nearing 0.9), and Caucasians and Hispanics having an intermediate frequency (0.63 and 0.55, respectively) [62,64,78,102]. The SNPs rs12979860 and rs8099917 exhibit strong linkage disequilibrium, except in patients of African ancestry, with partial linkage disequilibrium in Caucasians and near-complete linkage disequilibrium in Asians [62,65]. The varying frequencies of favorable alleles help explain differences in SVR rates among different ethnic populations [62,103]. For example, it is estimated that approximately half of the difference in SVR rates among European-Americans and African-Americans is due to the rs12979860 C allele [62]. Still, in African-American patients with the favorable rs1297860 CC genotype, viral kinetics have been shown to be slower and SVR rates lower than in European-Americans of the same genotype. This may suggest that ethnicity remains an independent predictor of outcome and points to the existence of currently unknown gene variants that influence treatment response, particularly in African-Americans [78].
Proposed mechanism of action of IFNL3 variations on treatment response
The biological implications of IFNL3 variations – that is, the actual mechanisms by which the IFNL3 genotype affects response to PEG-IFN-α and RBV – remain unclear. Although the effects of IFNL3 polymorphisms on IFN-λ production and intrahepatic ISG expression are still controversial, it has been suggested that ISG expression patterns may in part explain the difference in SVR rates. Interestingly, patients with the unfavorable IFNL3 genotype (rs1297860 CT/TT) were found to have higher intrahepatic ISG expression at baseline, and this is associated with failure to respond to PEG-IFN-α/RBV treatment [104,105]. One explanation for this is that patients with the unfavorable genotype exhibit continual, ineffectual intrahepatic expression of ISGs. At the same time, IFN-signaling inhibitors such as SOC3 and protein inhibitor of activated STAT 1 are also upregulated; this preactivated ISG response may be insufficient to clear the virus, but it may cause reduced sensitivity to PEG-IFN-α therapy through negative regulation of JAK-STAT signaling by IFN-signaling inhibitors [104,106]. Thus, even when PEG-IFN-α is administered, the cell may fail to induce strong enough ISG expression and be unable to clear the virus [106].
Conversely, patients with the favorable IFNL3 genotype may induce a weaker response to HCV RNA, and thus less IFN expression occurs, resulting in lower ISG levels at baseline and higher viral loads. When IFN-α is administered, there may be less inhibition of IFN signaling by negative regulatory molecules, causing the cell to be more sensitive to IFNs, and a stronger ISG induction is possible. Ultimately, this unrestrained IFN signal transduction and strong ISG stimulation may result in more effective clearance of the virus. Indeed, patients with the favorable IFNL3 genotype are observed to eliminate HCV more efficiently at each time point of treatment, indicating better viral kinetics [107]. Yet, despite known association with treatment response and a proposed mechanism for its effect, it remains unknown whether the favorable IFNL3 SNPs actually exert biological effects or are simply in linkage disequilibrium with other functional polymorphisms.
IFN4 (ss469415590) and PEG-IFN-α/RBV treatment response
Most recently, studies of the genetic region upstream of IFNL3 have uncovered a new transiently induced region that encompasses a dinucleotide variant, rs368234815 TT/ΔG (originally designated ss469415590). IFNL3 is in high linkage disequilibrium with rs12979860 and has been suggested as a possible causal variant [108–110]. ss469415590[ΔG] is a frame-shift variant that creates a novel gene, IFNL4, encoding the IFN-λ4 protein. IFN-λ4 is similar but not identical to IFN-λ3 [109]. Like the existing three members of the IFN-λ family, IFN-λ4 exhibits antiviral activity in vitro and binds the IL10RB/IFNLR1 receptor complex to activate the JAK-STAT pathway. Patients with the ss469415590 [ΔG] allele express the full protein, whereas the TT allele causes a frame shift that leads to an abrogation of the IFN-λ4 protein [109]. Interestingly, the TT allele is strongly associated with spontaneous or treatment-induced SVR, indicating that disruption of IFNL4 is favorable [109,110]. The apparent associations between IFN-λ4 production and a reduced likelihood of spontaneous or PEG-IFN-α/RBV therapy-induced clearance of HCV suggests that this novel IFN may be responsible for the observed clinical phenotypes, although the mechanism remains unclear [109]. In contrast to most IFNs, the IFN-λ4 protein is poorly secreted, and it has been hypothesized that it may impair HCV clearance by blocking other IFN-λs from binding the IL10RB/IFNLR1 receptor. Of note, ss469415590, but not rs12979860, has been shown in peripheral blood mono-nuclear cells to be involved in the stimulation of IFN-α and IFN-λ-inducible protein 10 mRNA, high plasma levels of which are associated with treatment failure [110].
As ss469415590 is strongly correlated with rs12979860 of IFNL3, the novel variant provides no additional information for predicting treatment response in Caucasian patients. Conversely, IFNL4 is only moderately correlated with rs8099917 in Caucasian patients, suggesting that it may serve as a better predictor of treatment outcome [111,112]. Likewise, the IFNL4 genotype may be a better predictor of treatment response in patients of African ancestry because ss469415590 shows only weak correlation with rs8099917 and moderate correlation with rs12979860 in these populations [109]. Taken together, the identification and characterization of the ss469415590 TT/ΔG functional variant represents a new step toward elucidating the genetic mechanisms of HCV clearance and the role of IFN polymorphisms in predicting HCV treatment response.
IFNL3 in the age of direct-acting antivirals
With the incorporation of DAAs into the HCV treatment arsenal, the question emerges of whether the IFNL3 genotype will remain relevant in predicting treatment outcomes to triple-acting therapies that include these new agents. Initial data suggest that the IFNL3 genotype may remain useful, but less so than for traditional PEG-IFN-α/RBV dual therapy [113,114]. To date, the most well-studied triple therapies are those that combine PEG-IFN-α and RBV with the ‘first-generation’ protease inhibitors BOC and TVR. In treatment-naive patients, SVR rates for patients treated with triple therapies that included BOC and TVR, respectively, were significantly higher for those with the favorable rs12979860 CC genotype (80 and 90%) as compared with those with the CT (71 and 71%) and TT (59 and 73%) genotypes [25,115,116]. Further, patients exhibited better viral kinetics of response and a lower risk of selection of resistance-associated HCV variants [116, 117]. In patients previously treated with PEG-IFN-α/RBV therapy, the IFNL3 genotype was not significantly associated with SVR in those who had relapsed or in partial responders, but previous null responders with the favorable rs12979860 CC genotype exhibited higher SVR rates compared with those with unfavorable genotypes [118].
With regard to ‘second-generation’ protease inhibitors, the effect of the IFNL3 genotype on treatment response has been shown to be weaker, but it is expected to remain useful in predicting response, especially in treatment-naive patients. For example, the IFNL3 genotype has been shown to be significantly associated with SVR at week 12 in both treatment-naive and treatment-experienced HCV genotype 1 patients undergoing triple therapy with the NS3 protease inhibitor simeprevir [119]. Similarly, rs12979860 was identified as an independent pretreatment predictor of SVR in treatment-naive patients with HCV genotypes 1, 4, 5, and 6 treated with the NS5B polymerase inhibitor sofosbuvir in combination with PEG-IFN-α/RBV [29]. It has also been suggested that stratification of patients according to IFNL3 genotype in ongoing clinical trials of new treatments will be useful in individualizing future triple therapies and optimizing outcomes [17].
Finally, IFNL3 appears to marginally affect treatment response in newly developed IFN-free therapies [120]. Although the IFNL3 genotype may not be relevant in predicting SVR, it has been demonstrated that IFNL3 variations are associated with viral kinetics during IFN-free treatments [120,121]. However, studies with larger cohorts are needed to confirm these associations.
Other host genes and PEG-IFN-α/RBV response
Apart from the well-established associations described above, numerous studies have demonstrated that polymorphisms in other genes may be associated with treatment response and adverse reactions to PEG-IFN-α/RBV therapy. One of the most significant drug-related toxicities for combination therapy is RBV-induced hemolytic anemia, which causes ~15% of patients to reduce RBV dose or discontinue treatment [122]. In 2010, a genome-wide association study identified a polymorphism of the ITPA gene (rs6051702 A), encoding inosine triphosphate pyrophosphohydrolase (ITPA), shown to protect against ribavirin-induced hemolytic anemia in HCV genotype 1 patients treated with PEG-IFN-α/RBV [123]. Subsequent analysis identified SNPs on two functional variants carrying the rs6051702 C allele that also protect against RBV-induced anemia in a European-American HCV genotype 1 population [124]. Further, these results have been replicated in a Japanese population and extended to HCV genotypes 2, 3, and 6, as well as to patients treated with PEG-IFN-α/RBV plus TVR triple therapy [125–130]. Although these variants have established associations with ITPA deficiency, the mechanism by which this condition protects against RBV-induced anemia remains unclear.
A genetic variant of the LDLR gene, which encodes the low-density lipoprotein-cholesterol receptor, has been shown to predict SVR in chronic HCV patients treated with PEG-IFN-α/RBV therapy [131]. The LDLR and IFNL3 variants appear to have additive effects in predicting SVR [132]. Thus, consideration of both LDLR and IFNL3 is more accurate in predicting SVR than use of either genotype singly. Finally, it has been shown that 25-OH vitamin D deficiency is associated with non-response to PEG-IFN-α/RBV combination therapy, suggesting that variants of the vitamin D receptor gene (VDR) may play a role in treatment outcomes [133,134]. It was recently demonstrated that a common nonsynonymous SNP in the VDR gene (rs2228570) predicts treatment response to PEG-IFN-α/RBV therapy in HCV genotype 1 and 4 patients, with minor T-allele carriers having an increased likelihood of achieving an SVR [135].
Conclusion
Treatment of HCV genotype 1 infection has made remarkable progress over the last two decades, allowing SVR rates to climb to new highs for a chronic infection that was traditionally difficult to cure. In treatment-naive patients, an ~ 30% increase in SVR has been possible with the addition of DAAs to PEG-IFN-α/RBV combination therapies. Pharmacogenetics offers another useful tool to optimize individual treatment regimens by allowing clinicians to make more informed decisions on treatment options, to modify dose, and to decide the duration of treatment regimens and avoid certain adverse reactions associated with therapies. In particular, the IFNL3 genotype has been established as a robust predictor of treatment outcomes to PEG-IFN-α/RBV-based therapies. In the era of new DAAs, IFNL3 variation may remain useful, although the effects will be attenuated. However, newer therapies are extremely expensive and thus PEG-IFN-α/RBV-based regimens will likely remain the mainstay of HCV treatment in developing nations. As developed nations like the USA adopt newer, more effective treatments, studies will be needed to confirm the initial associations found between the IFNL3 genotype and treatment response. Although likely theories have been suggested, it is still not certain how the IFNL3 genotype affects treatment outcomes, and further research will be needed to elucidate the exact mechanism. An improved mechanistic understanding of PEG-IFN-α and genetic determinants of SVR will help identify novel biomarkers for spontaneous clearance, determine new targets to personalize treatments for HCV infection, and ultimately allow for further improvement of treatment response rates. As SVR rates climb to new highs of greater than 95% with the advent of IFN-free regimens, which have now replaced IFN-based therapies in developed nations, the future of genotype 1 HCV treatment is bright indeed.
Acknowledgments
The authors thank Feng Liu for assistance with the graphics and Julia Barbarino for assistance with the manuscript. PharmGKB is supported by the NIH/NIGMS R24 GM61374.
Footnotes
Conflicts of interest
R.B.A. and T.E.K. are stockholders in Personalis Inc. For the remaining authors, there are no conflicts of interest.
References
- 1.Foster GR. Pegylated interferons for the treatment of chronic hepatitis C: pharmacological and clinical differences between peginterferon-alpha-2a and peginterferon-alpha-2b. Drugs. 2010;70:147–165. doi: 10.2165/11531990-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2. [Accessed 28 April 2015];Hepatitis C WHO fact sheet. Available at: http://www.who.int/mediacentre/factsheets/fs164/en/
- 3.Razavi H, Elkhoury AC, Elbasha E, Estes C, Pasini K, Poynard T, Kumar R. Chronic hepatitis C virus (HCV) disease burden and cost in the United States. Hepatology. 2013;57:2164–2170. doi: 10.1002/hep.26218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 5.Hoofnagle JH, Seeff LB. Peginterferon and ribavirin for chronic hepatitis C. N Engl J Med. 2006;355:2444–2451. doi: 10.1056/NEJMct061675. [DOI] [PubMed] [Google Scholar]
- 6.van der Meer AJ, Veldt BJ, Feld JJ, Wedemeyer H, Dufour JF, Lammert F, et al. Association between sustained virological response and all-cause mortality among patients with chronic hepatitis C and advanced hepatic fibrosis. JAMA. 2012;308:2584–2593. doi: 10.1001/jama.2012.144878. [DOI] [PubMed] [Google Scholar]
- 7.Simmonds P, Alberti A, Alter HJ, Bonino F, Bradley DW, Brechot C, et al. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994;19:1321–1324. [PubMed] [Google Scholar]
- 8.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghany MG, Strader DB, Thomas DL, Seeff LB American Association for the Study of Liver Diseases. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosen HR. Clinical practice. Chronic hepatitis C infection. N Engl J Med. 2011;364:2429–2438. doi: 10.1056/NEJMcp1006613. [DOI] [PubMed] [Google Scholar]
- 11.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 12.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 13.Hadziyannis SJ, Sette H, Jr, Morgan TR, Balan V, Diago M, Marcellin P, et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 14.Strader DB. Understudied populations with hepatitis C. Hepatology. 2002;36:S226–S236. doi: 10.1053/jhep.2002.36991. [DOI] [PubMed] [Google Scholar]
- 15.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 16. [Accessed 8 February 2015];World Hepatitis C: global alert and response. Available at: http://www.who.int/csr/disease/hepatitis/whocdscsrlyo2003/en/index5.html.
- 17.Kamal SM. Pharmacogenetics of hepatitis C: transition from interferon-based therapies to direct-acting antiviral agents. Hepat Med. 2014;6:61–77. doi: 10.2147/HMER.S41127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lotsch J, Hofmann WP, Schlecker C, Zeuzem S, Geisslinger G, Ultsch A, Doehring A. Single and combined IL28B, ITPA and SLC28A3 host genetic markers modulating response to anti-hepatitis C therapy. Pharmacogenomics. 2011;12:1729–1740. doi: 10.2217/pgs.11.99. [DOI] [PubMed] [Google Scholar]
- 20.Muir AJ, Gong L, Johnson SG, Lee MT, Williams MS, Klein TE, et al. Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon-alpha-based regimens. Clin Pharmacol Ther. 2014;95:141–146. doi: 10.1038/clpt.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afdhal NH, McHutchison JG, Zeuzem S, Mangia A, Pawlotsky JM, Murray JS, et al. Hepatitis C pharmacogenetics: state of the art in 2010. Hepatology. 2011;53:336–345. doi: 10.1002/hep.24052. [DOI] [PubMed] [Google Scholar]
- 22.Kawaguchi-Suzuki M, Frye RF. The role of pharmacogenetics in the treatment of chronic hepatitis C infection. Pharmacotherapy. 2014;34:185–201. doi: 10.1002/phar.1349. [DOI] [PubMed] [Google Scholar]
- 23.Ghany MG, Nelson DR, Strader DB, Thomas DL, Seeff LB American Association for Study of Liver Diseases. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 2011;54:1433–1444. doi: 10.1002/hep.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405–2416. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 26.Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, et al. Telaprevir for retreatment of HCV infection. N Engl J Med. 2011;364:2417–2428. doi: 10.1056/NEJMoa1013086. [DOI] [PubMed] [Google Scholar]
- 27.Dieterich D, Rockstroh JK, Orkin C, Gutierrez F, Klein MB, Reynes J, et al. Simeprevir (TMC435) with pegylated interferon/ribavirin in patients coinfected with HCV genotype 1 and HIV-1: a phase 3 study. Clin Infect Dis. 2014;59:1579–1587. doi: 10.1093/cid/ciu675. [DOI] [PubMed] [Google Scholar]
- 28.Ilyas JA, Vierling JM. An overview of emerging therapies for the treatment of chronic hepatitis C. Med Clin North Am. 2014;98:17–38. doi: 10.1016/j.mcna.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Lawitz E, Gane EJ. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;369:678–679. doi: 10.1056/NEJMc1307641. [DOI] [PubMed] [Google Scholar]
- 30.Kowdley KV, Lawitz E, Crespo I, Hassanein T, Davis MN, DeMicco M, et al. Sofosbuvir with pegylated interferon alfa-2a and ribavirin for treatment-naive patients with hepatitis C genotype-1 infection (ATOMIC): an open-label, randomised, multicentre phase 2 trial. Lancet. 2013;381:2100–2107. doi: 10.1016/S0140-6736(13)60247-0. [DOI] [PubMed] [Google Scholar]
- 31.Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Subramanian GM, et al. Efficacy of nucleotide polymerase inhibitor sofosbuvir plus the NS5A inhibitor ledipasvir or the NS5B non-nucleoside inhibitor GS-9669 against HCV genotype 1 infection. Gastroenterology. 2014;146:736–743. e1. doi: 10.1053/j.gastro.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Lawitz E, Poordad FF, Pang PS, Hyland RH, Ding X, Mo H, et al. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383:515–523. doi: 10.1016/S0140-6736(13)62121-2. [DOI] [PubMed] [Google Scholar]
- 33.Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, et al. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med. 2014;370:1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 34.Zeuzem S, Jacobson IM, Baykal T, Marinho RT, Poordad F, Bourliere M, et al. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med. 2014;370:1604–1614. doi: 10.1056/NEJMoa1401561. [DOI] [PubMed] [Google Scholar]
- 35. [Accessed 10 February 2015];Recommendations for testing, managing, and treating hepatitis C. Available at: http://www.hcvguidelines.org.
- 36.Bruno R, Sacchi P, Cima S, Maiocchi L, Novati S, Filice G, Fagiuoli S. Comparison of peginterferon pharmacokinetic and pharmacodynamic profiles. J Viral Hepat. 2012;19(Suppl 1):33–36. doi: 10.1111/j.1365-2893.2011.01519.x. [DOI] [PubMed] [Google Scholar]
- 37.Zeuzem S, Welsch C, Herrmann E. Pharmacokinetics of peginterferons. Semin Liver Dis. 2003;23(Suppl 1):23–28. doi: 10.1055/s-2003-41631. [DOI] [PubMed] [Google Scholar]
- 38.Noureddin M, Ghany MG. Pharmacokinetics and pharmacodynamics of peginterferon and ribavirin: implications for clinical efficacy in the treatment of chronic hepatitis C. Gastroenterol Clin North Am. 2010;39:649–658. doi: 10.1016/j.gtc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruno R, Sacchi P, Ciappina V, Zochetti C, Patruno S, Maiocchi L, Filice G. Viral dynamics and pharmacokinetics of peginterferon alpha-2a and peginterferon alpha-2b in naive patients with chronic hepatitis C: a randomized, controlled study. Antivir Ther. 2004;9:491–497. [PubMed] [Google Scholar]
- 40.Di Bisceglie AM, Ghalib RH, Hamzeh FM, Rustgi VK. Early virologic response after peginterferon alpha-2a plus ribavirin or peginterferon alpha-2b plus ribavirin treatment in patients with chronic hepatitis C. J Viral Hepat. 2007;14:721–729. doi: 10.1111/j.1365-2893.2007.00862.x. [DOI] [PubMed] [Google Scholar]
- 41.Rajender Reddy K, Modi MW, Pedder S. Use of peginterferon alfa-2a (40 kD) (Pegasys) for the treatment of hepatitis C. Adv Drug Deliv Rev. 2002;54:571–586. doi: 10.1016/s0169-409x(02)00028-5. [DOI] [PubMed] [Google Scholar]
- 42.Glue P, Rouzier-Panis R, Raffanel C, Sabo R, Gupta SK, Salfi M, et al. A dose-ranging study of pegylated interferon alfa-2b and ribavirin in chronic hepatitis C. The Hepatitis C Intervention Therapy Group. Hepatology. 2000;32:647–653. doi: 10.1053/jhep.2000.16661. [DOI] [PubMed] [Google Scholar]
- 43.De Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 44.Mahlakoiv T, Ritz D, Mordstein M, DeDiego ML, Enjuanes L, Muller MA, et al. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J Gen Virol. 2012;93:2601–2605. doi: 10.1099/vir.0.046284-0. [DOI] [PubMed] [Google Scholar]
- 45.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 46.Tilg H. New insights into the mechanisms of interferon alfa: an immunoregulatory and anti-inflammatory cytokine. Gastroenterology. 1997;112:1017–1021. doi: 10.1053/gast.1997.v112.pm9041265. [DOI] [PubMed] [Google Scholar]
- 47.Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawlotsky JM. Hepatitis C virus: standard-of-care treatment. Adv Pharmacol. 2013;67:169–215. doi: 10.1016/B978-0-12-405880-4.00005-6. [DOI] [PubMed] [Google Scholar]
- 49.European Association for the Study of the L. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245–264. doi: 10.1016/j.jhep.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 50.Mangia A, Minerva N, Bacca D, Cozzolongo R, Ricci GL, Carretta V, et al. Individualized treatment duration for hepatitis C genotype 1 patients: A randomized controlled trial. Hepatology. 2008;47:43–50. doi: 10.1002/hep.22061. [DOI] [PubMed] [Google Scholar]
- 51.Chen TM, Huang PT, Lin CH, Tsai MH, Lin LF, Liu CC, et al. Feasibility of individualized treatment for hepatitis C patients in the real world. J Gastroenterol Hepatol. 2010;25:61–69. doi: 10.1111/j.1440-1746.2009.05946.x. [DOI] [PubMed] [Google Scholar]
- 52.Bruno R, Sacchi P, Scagnolari C, Torriani F, Maiocchi L, Patruno S, et al. Pharmacodynamics of peginterferon alpha-2a and peginterferon alpha-2b in interferon-naive patients with chronic hepatitis C: a randomized, controlled study. Aliment Pharmacol Ther. 2007;26:369–376. doi: 10.1111/j.1365-2036.2007.03392.x. [DOI] [PubMed] [Google Scholar]
- 53.Silva M, Poo J, Wagner F, Jackson M, Cutler D, Grace M, et al. A randomised trial to compare the pharmacokinetic, pharmacodynamic, and antiviral effects of peginterferon alfa-2b and peginterferon alfa-2a in patients with chronic hepatitis C (COMPARE) J Hepatol. 2006;45:204–213. doi: 10.1016/j.jhep.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 56.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Jilg N, Shao RX, Lin W, Fusco DN, Zhao H, et al. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J Hepatol. 2011;55:289–298. doi: 10.1016/j.jhep.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52:822–832. doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 59.Maag D, Castro C, Hong Z, Cameron CE. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J Biol Chem. 2001;276:46094–46098. doi: 10.1074/jbc.C100349200. [DOI] [PubMed] [Google Scholar]
- 60.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 61.Hayes CN, Imamura M, Aikata H, Chayama K. Genetics of IL28B and HCV – response to infection and treatment. Nat Rev Gastroenterol Hepatol. 2012;9:406–417. doi: 10.1038/nrgastro.2012.101. [DOI] [PubMed] [Google Scholar]
- 62.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 63.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 64.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. 1345.e1–7. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 66.Ruiz-Extremera A, Munoz-Gamez JA, Salmeron-Ruiz MA, de Rueda PM, Quiles-Perez R, Gila-Medina A, et al. Genetic variation in interleukin 28B with respect to vertical transmission of hepatitis C virus and spontaneous clearance in HCV-infected children. Hepatology. 2011;53:1830–1838. doi: 10.1002/hep.24298. [DOI] [PubMed] [Google Scholar]
- 67.Stattermayer AF, Stauber R, Hofer H, Rutter K, Beinhardt S, Scherzer TM, et al. Impact of IL28B genotype on the early and sustained virologic response in treatment-naive patients with chronic hepatitis C. Clin Gastroenterol Hepatol. 2011;9:344–350. e2. doi: 10.1016/j.cgh.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 68.Montes-Cano MA, Garcia-Lozano JR, Abad-Molina C, Romero-Gomez M, Barroso N, Aguilar-Reina J, et al. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology. 2010;52:33–37. doi: 10.1002/hep.23624. [DOI] [PubMed] [Google Scholar]
- 69.Abe H, Ochi H, Maekawa T, Hayes CN, Tsuge M, Miki D, et al. Common variation of IL28 affects gamma-GTP levels and inflammation of the liver in chronically infected hepatitis C virus patients. J Hepatol. 2010;53:439–443. doi: 10.1016/j.jhep.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 70.McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 72.Ridruejo E, Solano A, Marciano S, Galdame O, Adrover R, Cocozzella D, et al. Genetic variation in interleukin-28B predicts SVR in hepatitis C genotype 1 Argentine patients treated with PEG IFN and ribavirin. Ann Hepatol. 2011;10:452–457. [PubMed] [Google Scholar]
- 73.Liu CH, Liang CC, Liu CJ, Tseng TC, Lin CL, Yang SS, et al. Interleukin 28B genetic polymorphisms and viral factors help identify HCV genotype-1 patients who benefit from 24-week pegylated interferon plus ribavirin therapy. Antivir Ther. 2012;17:477–484. doi: 10.3851/IMP2026. [DOI] [PubMed] [Google Scholar]
- 74.Lyoo K, Song MJ, Hur W, Choi JE, Hong SW, Kim CW, et al. Polymorphism near the IL28B gene in Korean hepatitis C virus-infected patients treated with peg-interferon plus ribavirin. J Clin Virol. 2011;52:363–366. doi: 10.1016/j.jcv.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 75.Chen JY, Lin CY, Wang CM, Lin YT, Kuo SN, Shiu CF, et al. IL28B genetic variations are associated with high sustained virological response (SVR) of interferon-alpha plus ribavirin therapy in Taiwanese chronic HCV infection. Genes Immun. 2011;12:300–309. doi: 10.1038/gene.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sporea I, Popescu A, Curescu M, Sirli R, Dan I, Goldis A, et al. The correlation of Il28B genotype with sustained virologic response in Romanian patients with chronic hepatitis C. Hepat Mon. 2011;11:975–979. doi: 10.5812/kowsar.1735143x.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liao XW, Ling Y, Li XH, Han Y, Zhang SY, Gu LL, et al. Association of genetic variation in IL28B with hepatitis C treatment-induced viral clearance in the Chinese Han population. Antivir Ther. 2011;16:141–147. doi: 10.3851/IMP1703. [DOI] [PubMed] [Google Scholar]
- 78.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. e118. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 79.Cariani E, Villa E, Rota C, Critelli R, Trenti T. Translating pharmacogenetics into clinical practice: interleukin (IL)28B and inosine triphosphatase (ITPA) polymophisms in hepatitis C virus (HCV) infection. Clin Chem Lab Med. 2011;49:1247–1256. doi: 10.1515/CCLM.2011.618. [DOI] [PubMed] [Google Scholar]
- 80.Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821, 827, 827.e1. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
- 81.Sarrazin C, Susser S, Doehring A, Lange CM, Muller T, Schlecker C, et al. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415–421. doi: 10.1016/j.jhep.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 82.Sakamoto N, Nakagawa M, Tanaka Y, Sekine-Osajima Y, Ueyama M, Kurosaki M, et al. Association of IL28B variants with response to pegylated-interferon alpha plus ribavirin combination therapy reveals intersubgenotypic differences between genotypes 2a and 2b. J Med Virol. 2011;83:871–878. doi: 10.1002/jmv.22038. [DOI] [PubMed] [Google Scholar]
- 83.Kawaoka T, Hayes CN, Ohishi W, Ochi H, Maekawa T, Abe H, et al. Predictive value of the IL28B polymorphism on the effect of interferon therapy in chronic hepatitis C patients with genotypes 2a and 2b. J Hepatol. 2011;54:408–414. doi: 10.1016/j.jhep.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 84.Clark PJ, Thompson AJ. Host genomics and HCV treatment response. J Gastroenterol Hepatol. 2012;27:212–222. doi: 10.1111/j.1440-1746.2011.06918.x. [DOI] [PubMed] [Google Scholar]
- 85.Yu ML, Huang CF, Huang JF, Chang NC, Yang JF, Lin ZY, et al. Role of interleukin-28B polymorphisms in the treatment of hepatitis C virus genotype 2 infection in Asian patients. Hepatology. 2011;53:7–13. doi: 10.1002/hep.23976. [DOI] [PubMed] [Google Scholar]
- 86.Scherzer TM, Hofer H, Staettermayer AF, Rutter K, Beinhardt S, Steindl-Munda P, et al. Early virologic response and IL28B polymorphisms in patients with chronic hepatitis C genotype 3 treated with peginterferon alfa-2a and ribavirin. J Hepatol. 2011;54:866–871. doi: 10.1016/j.jhep.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 87.Moghaddam A, Melum E, Reinton N, Ring-Larsen H, Verbaan H, Bjoro K, Dalgard O. IL28B genetic variation and treatment response in patients with hepatitis C virus genotype 3 infection. Hepatology. 2011;53:746–754. doi: 10.1002/hep.24154. [DOI] [PubMed] [Google Scholar]
- 88.Matsuura K, Watanabe T, Tanaka Y. Role of IL28B for chronic hepatitis C treatment toward personalized medicine. J Gastroenterol Hepatol. 2014;29:241–249. doi: 10.1111/jgh.12475. [DOI] [PubMed] [Google Scholar]
- 89.Soriano V, Puoti M, Sulkowski M, Cargnel A, Benhamou Y, Peters M, et al. Care of patients coinfected with HIV and hepatitis C virus: 2007 updated recommendations from the HCV-HIV International Panel. AIDS. 2007;21:1073–1089. doi: 10.1097/QAD.0b013e3281084e4d. [DOI] [PubMed] [Google Scholar]
- 90.Graham CS, Baden LR, Yu E, Mrus JM, Carnie J, Heeren T, Koziel MJ. Influence of human immunodeficiency virus infection on the course of hepatitis C virus infection: a meta-analysis. Clin Infect Dis. 2001;33:562–569. doi: 10.1086/321909. [DOI] [PubMed] [Google Scholar]
- 91.Martin-Carbonero L, Benhamou Y, Puoti M, Berenguer J, Mallolas J, Quereda C, et al. Incidence and predictors of severe liver fibrosis in human immunodeficiency virus-infected patients with chronic hepatitis C: a European collaborative study. Clin Infect Dis. 2004;38:128–133. doi: 10.1086/380130. [DOI] [PubMed] [Google Scholar]
- 92.Weber R, Sabin CA, Friis-Moller N, Reiss P, El-Sadr WM, Kirk O, et al. Liver-related deaths in persons infected with the human immunodeficiency virus: the D:A:D study. Arch Intern Med. 2006;166:1632–1641. doi: 10.1001/archinte.166.15.1632. [DOI] [PubMed] [Google Scholar]
- 93.Rallon NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, et al. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23–F29. doi: 10.1097/QAD.0b013e3283391d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pineda JA, Caruz A, Rivero A, Neukam K, Salas I, Camacho A, et al. Prediction of response to pegylated interferon plus ribavirin by IL28B gene variation in patients coinfected with HIV and hepatitis C virus. Clin Infect Dis. 2010;51:788–795. doi: 10.1086/656235. [DOI] [PubMed] [Google Scholar]
- 95.Aparicio E, Parera M, Franco S, Perez-Alvarez N, Tural C, Clotet B, Martinez MA. IL28B SNP rs8099917 is strongly associated with pegylated interferon-alpha and ribavirin therapy treatment failure in HCV/HIV-1 coinfected patients. PLoS One. 2010;5:e13771. doi: 10.1371/journal.pone.0013771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.De Castellarnau M, Aparicio E, Parera M, Franco S, Tural C, Clotet B, Martinez MA. Deciphering the interleukin 28B variants that better predict response to pegylated interferon-alpha and ribavirin therapy in HCV/HIV-1 coinfected patients. PLoS One. 2012;7:e31016. doi: 10.1371/journal.pone.0031016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Labarga P, Barreiro P, Mira JA, Vispo E, Rallon N, Neukam K, et al. Impact of IL28B polymorphisms on response to peginterferon and ribavirin in HIV-hepatitis C virus-coinfected patients with prior nonresponse or relapse. AIDS. 2011;25:1131–1133. doi: 10.1097/QAD.0b013e3283471d83. [DOI] [PubMed] [Google Scholar]
- 98.Coto-Llerena M, Perez-Del-Pulgar S, Crespo G, Carrion JA, Martinez SM, Sanchez-Tapias JM, et al. Donor and recipient IL28B polymorphisms in HCV-infected patients undergoing antiviral therapy before and after liver transplantation. Am J Transplant. 2011;11:1051–1057. doi: 10.1111/j.1600-6143.2011.03491.x. [DOI] [PubMed] [Google Scholar]
- 99.Eurich D, Boas-Knoop S, Bahra M, Neuhaus R, Somasundaram R, Neuhaus P, et al. Role of IL28B polymorphism in the development of hepatitis C virus-induced hepatocellular carcinoma, graft fibrosis, and posttransplant antiviral therapy. Transplantation. 2012;93:644–649. doi: 10.1097/TP.0b013e318244f774. [DOI] [PubMed] [Google Scholar]
- 100.Duarte-Rojo A, Veldt BJ, Goldstein DD, Tillman HL, Watt KD, Heimbach JK, et al. The course of posttransplant hepatitis C infection: comparative impact of donor and recipient source of the favorable IL28B genotype and other variables. Transplantation. 2012;94:197–203. doi: 10.1097/TP.0b013e3182547551. [DOI] [PubMed] [Google Scholar]
- 101.Eurich D, Boas-Knoop S, Ruehl M, Schulz M, Carrillo ED, Berg T, et al. Relationship between the interleukin-28b gene polymorphism and the histological severity of hepatitis C virus-induced graft inflammation and the response to antiviral therapy after liver transplantation. Liver Transpl. 2011;17:289–298. doi: 10.1002/lt.22235. [DOI] [PubMed] [Google Scholar]
- 102.Kobayashi M, Suzuki F, Akuta N, Sezaki H, Suzuki Y, Hosaka T, et al. Association of two polymorphisms of the IL28B gene with viral factors and treatment response in 1,518 patients infected with hepatitis C virus. J Gastroenterol. 2012;47:596–605. doi: 10.1007/s00535-012-0531-1. [DOI] [PubMed] [Google Scholar]
- 103.Satapathy SK, Lingisetty CS, Proper S, Chaudhari S, Williams S. Equally poor outcomes to pegylated interferon-based therapy in African Americans and Hispanics with chronic hepatitis C infection. J Clin Gastroenterol. 2010;44:140–145. doi: 10.1097/MCG.0b013e3181ba9992. [DOI] [PubMed] [Google Scholar]
- 104.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci USA. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 106.Asahina Y, Tsuchiya K, Muraoka M, Tanaka K, Suzuki Y, Tamaki N, et al. Association of gene expression involving innate immunity and genetic variation in interleukin 28B with antiviral response. Hepatology. 2012;55:20–29. doi: 10.1002/hep.24623. [DOI] [PubMed] [Google Scholar]
- 107.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-lambda4: the paradoxical new member of the interferon lambda family. J Interferon Cytokine Res. 2014;34:829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stattermayer AF, Strassl R, Maieron A, Rutter K, Stauber R, Strasser M, et al. Polymorphisms of interferon-lambda4 and IL28B – effects on treatment response to interferon/ribavirin in patients with chronic hepatitis C. Aliment Pharmacol Ther. 2014;39:104–111. doi: 10.1111/apt.12547. [DOI] [PubMed] [Google Scholar]
- 112.Covolo L, Bibert S, Donato F, Bochud PY, Lagging M, Negro F, Fattovich G. The novel ss469415590 variant predicts virological response to therapy in patients with chronic hepatitis C virus type 1 infection. Aliment Pharmacol Ther. 2014;39:322–330. doi: 10.1111/apt.12568. [DOI] [PubMed] [Google Scholar]
- 113.Chayama K, Hayes CN, Abe H, Miki D, Ochi H, Karino Y, et al. IL28B but not ITPA polymorphism is predictive of response to pegylated interferon, ribavirin, and telaprevir triple therapy in patients with genotype 1 hepatitis C. J Infect Dis. 2011;204:84–93. doi: 10.1093/infdis/jir210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, et al. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52:421–429. doi: 10.1002/hep.23690. [DOI] [PubMed] [Google Scholar]
- 115.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Poordad F, Bronowicki JP, Gordon SC, Zeuzem S, Jacobson IM, Sulkowski MS, et al. Factors that predict response of patients with hepatitis C virus infection to boceprevir. Gastroenterology. 2012;143:608–618. e1–5. doi: 10.1053/j.gastro.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 117.Pol S, Aerssens J, Zeuzem S, Andreone P, Lawitz EJ, Roberts S, et al. Limited impact of IL28B genotype on response rates in telaprevir-treated patients with prior treatment failure. J Hepatol. 2013;58:883–889. doi: 10.1016/j.jhep.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 118.McHutchison JG, Manns MP, Muir AJ, Terrault NA, Jacobson IM, Afdhal NH, et al. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362:1292–1303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 119.Jacobson IM, Dore GJ, Foster GR, Fried MW, Radu M, Rafalsky VV, et al. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet. 2014;384:403–413. doi: 10.1016/S0140-6736(14)60494-3. [DOI] [PubMed] [Google Scholar]
- 120.Chu TW, Kulkarni R, Gane EJ, Roberts SK, Stedman C, Angus PW, et al. Effect of IL28B genotype on early viral kinetics during interferon-free treatment of patients with chronic hepatitis C. Gastroenterology. 2012;142:790–795. doi: 10.1053/j.gastro.2011.12.057. [DOI] [PubMed] [Google Scholar]
- 121.Zeuzem S, Soriano V, Asselah T, Bronowicki JP, Lohse AW, Mullhaupt B, et al. Faldaprevir and deleobuvir for HCV genotype 1 infection. N Engl J Med. 2013;369:630–639. doi: 10.1056/NEJMoa1213557. [DOI] [PubMed] [Google Scholar]
- 122.Fowell AJ, Nash KL. Telaprevir: a new hope in the treatment of chronic hepatitis C? Adv Ther. 2010;27:512–522. doi: 10.1007/s12325-010-0047-0. [DOI] [PubMed] [Google Scholar]
- 123.Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 124.De Franceschi L, Fattovich G, Turrini F, Ayi K, Brugnara C, Manzato F, et al. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31:997–1004. doi: 10.1053/he.2000.5789. [DOI] [PubMed] [Google Scholar]
- 125.Ochi H, Maekawa T, Abe H, Hayashida Y, Nakano R, Kubo M, et al. ITPA polymorphism affects ribavirin-induced anemia and outcomes of therapy – a genome-wide study of Japanese HCV virus patients. Gastroenterology. 2010;139:1190–1197. doi: 10.1053/j.gastro.2010.06.071. [DOI] [PubMed] [Google Scholar]
- 126.Thompson AJ, Fellay J, Patel K, Tillmann HL, Naggie S, Ge D, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181–1189. doi: 10.1053/j.gastro.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eskesen AN, Melum E, Moghaddam A, Bjoro K, Verbaan H, Ring-Larsen H, Dalgard O. Genetic variants at the ITPA locus protect against ribavirin-induced hemolytic anemia and dose reduction in an HCV G2/G3 cohort. Eur J Gastroenterol Hepatol. 2012;24:890–896. doi: 10.1097/MEG.0b013e3283546efd. [DOI] [PubMed] [Google Scholar]
- 128.Seto WK, Tsang OT, Liu K, Chan JM, Wong DK, Fung J, et al. Role of IL28B and inosine triphosphatase polymorphisms in the treatment of chronic hepatitis C virus genotype 6 infection. J Viral Hepat. 2013;20:470–477. doi: 10.1111/jvh.12047. [DOI] [PubMed] [Google Scholar]
- 129.Suzuki F, Suzuki Y, Akuta N, Sezaki H, Hirakawa M, Kawamura Y, et al. Influence of ITPA polymorphisms on decreases of hemoglobin during treatment with pegylated interferon, ribavirin, and telaprevir. Hepatology. 2011;53:415–421. doi: 10.1002/hep.24058. [DOI] [PubMed] [Google Scholar]
- 130.Ogawa E, Furusyo N, Nakamuta M, Kajiwara E, Nomura H, Dohmen K, et al. Clinical milestones for the prediction of severe anemia by chronic hepatitis C patients receiving telaprevir-based triple therapy. J Hepatol. 2013;59:667–674. doi: 10.1016/j.jhep.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 131.Hennig BJ, Hellier S, Frodsham AJ, Zhang L, Klenerman P, Knapp S, et al. Association of low-density lipoprotein receptor polymorphisms and outcome of hepatitis C infection. Genes Immun. 2002;3:359–367. doi: 10.1038/sj.gene.6363883. [DOI] [PubMed] [Google Scholar]
- 132.Pineda JA, Caruz A, Di Lello FA, Camacho A, Mesa P, Neukam K, et al. Low-density lipoprotein receptor genotyping enhances the predictive value of IL28B genotype in HIV/hepatitis C virus-coinfected patients. AIDS. 2011;25:1415–1420. doi: 10.1097/QAD.0b013e328348a7ac. [DOI] [PubMed] [Google Scholar]
- 133.Lange CM, Bojunga J, Ramos-Lopez E, von Wagner M, Hassler A, Vermehren J, et al. Vitamin D deficiency and a CYP27B1–1260 promoter polymorphism are associated with chronic hepatitis C and poor response to interferon-alfa based therapy. J Hepatol. 2011;54:887–893. doi: 10.1016/j.jhep.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 134.Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 135.Garcia-Martin E, Agundez JA, Maestro ML, Suarez A, Vidaurreta M, Martinez C, et al. Influence of vitamin D-related gene polymorphisms (CYP27B and VDR) on the response to interferon/ribavirin therapy in chronic hepatitis C. PLoS One. 2013;8:e74764. doi: 10.1371/journal.pone.0074764. [DOI] [PMC free article] [PubMed] [Google Scholar]