Abstract
Formation of the FtsZ ring (Z ring) in Escherichia coli is the first step in the assembly of the divisome, a protein machine required for cell division. Although the biochemical functions of most divisome proteins are unknown, several, including ZipA, FtsA and FtsK, have overlapping roles in ensuring that the Z ring assembles at the cytoplasmic membrane, and that it is active. As shown previously, a single amino acid change in FtsA, R286W, also called FtsA*, bypasses the requirement for either ZipA or FtsK in cell division. In this study, the properties of FtsA* were investigated further, with the eventual goal of understanding the molecular mechanism behind the bypass. Compared to wild-type FtsA, the presence of FtsA* resulted in a modest but significant decrease in the mean length of cells in the population, accelerated the reassembly of Z rings, and suppressed the cell-division block caused by excessively high levels of FtsZ. These effects were not mediated by Z-ring remodelling, because FtsA* did not alter the kinetics of FtsZ turnover within the Z ring, as measured by fluorescence recovery after photobleaching. FtsA* was also unable to permit normal cell division at below normal levels of FtsZ, or after thermoinactivation of ftsZ84(ts). However, turnover of FtsA* in the ring was somewhat faster than that of wild-type FtsA, and overexpressed FtsA* did not inhibit cell division as efficiently as wild-type FtsA. Finally, FtsA* interacted more strongly with FtsZ compared with FtsA in a yeast two-hybrid system. These results suggest that FtsA* interacts with FtsZ in a markedly different way compared with FtsA.
INTRODUCTION
The Escherichia coli cell division machine, called the divisome, is composed of at least 13 known essential proteins (Vicente et al., 2006). Inactivating any of these components causes a block in divisome assembly, resulting in the formation of filamentous cells that eventually lyse. Although specific roles for all 13 proteins have not been established, functions have recently been attributed to many of them (Errington, 1996; Weiss, 2004); those without defined roles are thought to function in either divisome formation or septum generation.
Based on structural and biochemical similarities, FtsZ is a prokaryotic homologue of tubulin (Lowe & Amos, 1998). Upon segregation of newly duplicated chromosomes, FtsZ polymerizes into a ring structure at midcell called the Z ring (Bi & Lutkenhaus, 1991). Although seemingly quite static, the Z ring is actually highly dynamic, exhibiting rapid turnover of FtsZ monomers from the cytoplasmic pool (Stricker et al., 2002). The Z ring acts as a scaffold for the assembly of other divisome proteins, and divisome assembly does not proceed in its absence (Goehring & Beckwith, 2005).
Two other essential divisome proteins of E. coli, ZipA and FtsA, are dependent on FtsZ for their localization to the Z ring, and help to stabilize the ring. When either ZipA or FtsA is inactivated, many Z rings form, but are often missing at potential division sites between nucleoids. Inactivating both ZipA and FtsA abolishes Z-ring formation (Pichoff & Lutkenhaus, 2002), suggesting that they have overlapping functions. Indeed, both ZipA and FtsA bind to the membrane and to FtsZ, and part of their essential function is to anchor the Z ring to the membrane (Hale & de Boer, 1997, 1999; Liu et al., 1999; Pichoff & Lutkenhaus, 2002). In Bacillus subtilis, which lacks a ZipA homologue, FtsA is also not essential for the formation of Z rings (Harry, 2001). However, most Z rings formed in the absence of FtsA are non-functional, indicating that, as in E. coli, B. subtilis FtsA is required for proper recruitment of downstream proteins by the Z ring (Jensen et al., 2005).
Structure–function studies of FtsA have helped to highlight its general roles in cell division, although the mechanisms are not well understood. Subdomain 1c of FtsA (van Den Ent & Lowe, 2000) is required for recruitment of downstream divisome proteins and maturation of the divisome (Corbin et al., 2004; Rico et al., 2004). At the opposite end of the FtsA molecule, the S12–S13 loop of subdomain 2b is not essential for FtsA function or recruitment of downstream divisome proteins, but instead has an unknown role in regulating Z ring assembly (Rico et al., 2004).
A gain-of-function mutation in the same loop, R286W (also known as ftsA*), can fully compensate for the loss of ZipA, normally an essential protein, and can partially compensate for the loss of another essential division protein, FtsK (Geissler & Margolin, 2005; Geissler et al., 2003). Furthermore, FtsA* confers resistance to overproduction of MinC, which normally disassembles Z rings (Pichoff & Lutkenhaus, 2001). The ability of FtsA* to antagonize a known destabilizer of Z rings and to mimic a stabilizer such as ZipA (Raychaudhuri, 1999) suggests that, compared to FtsA, FtsA* enhances the structural integrity of the Z ring. FtsA* also suppresses the toxic effects of high levels of ZipA (Geissler et al., 2003), which may act to destabilize the Z ring. FtsA* is otherwise fully functional, because it can replace FtsA with little effect on cell viability.
These special properties of FtsA* prompted us to investigate further its effects on FtsZ and the Z ring. In this work, we discovered that FtsA* accelerates assembly of the Z ring, resulting in cell division at significantly shorter cell lengths. We then investigated whether this enhanced Z ring assembly was caused specifically by an altered interaction between FtsA and FtsZ. FtsA* did not change the kinetics of FtsZ turnover within the Z ring, or affect the partitioning between FtsZ within and outside the ring. However, compared with wild-type FtsA, FtsA* suppressed the toxicity caused by perturbing the FtsZ : FtsA ratio, and interacted more strongly with FtsZ in a yeast two-hybrid system. These results suggest that FtsA* has an altered interaction with FtsZ.
METHODS
Bacterial strains, plasmids and media
E. coli strains and plasmids used in this study are listed in Table 1. Bacteria were grown in either Luria–Bertani (LB) (0.5 % NaCl) or M9 minimal medium supplemented with Casamino acids (CA) (0.2 %) and glycerol (0.1 %, v/v) or glucose (0.4 %). If required, antibiotics were added at the following concentrations: 20 μg chloramphenicol (Cm) ml−1, 25 μg kanamycin ml−1, 100 or 25 μg ampicillin ml−1, or 10 μg tetracycline ml−1. L-Arabinose was added at the concentrations indicated to induce expression from PBAD promoters (Guzman et al., 1995). IPTG was used to induce expression of green fluorescent protein (GFP) fusion constructs. Unless stated otherwise, phage P1 transduction, viability plating, anti-FtsZ immunofluorescence microscopy (IFM), differential interference contrast (DIC) microscopy, cell fixation, and staining with 4′,6-diamidino-2-phenylindole (DAPI) were done essentially as described previously (Geissler et al., 2003). ImageJ software was used to calculate the percentage of ring-bound FtsZ–GFP versus cytoplasmic, non-ring FtsZ–GFP, by determining the fluorescence intensity of the entire cell and comparing it to the fluorescence found within the midcell Z ring (Anderson et al., 2004).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| General E. coli strains | ||
| WM1074 | TX3772 | Laboratory collection |
| WM1659 | TX3772 ftsA* | Geissler et al. (2003) |
| WM1657 | WM1659 ΔzipA : : kan | Geissler et al. (2003) |
| WM1032 | TX3772 ΔminCDE : : kan | Laboratory collection |
| WM1940 | WM1659 ΔminCDE : : kan | This study |
| WM2195 | WM1657 fadR : : Tn10 (ΔminCDE) | This study |
| WM2176 | W3110 | Laboratory collection |
| WM2177 | W3110 ftsA* leu : : Tn10 | This study |
| WM2230 | ttk : : kan (FB21358) | F. Blattner† |
| WM2775 | WM1074 ttk : : kan | This study |
| WM2776 | WM1659 ttk : : kan | This study |
| WM1597 (SSR996) | sulAp–gfp, nadA57 : : Tn10 | McCool et al. (2004) |
| WM2739 | WM1074+sulAp–gfp | This study |
| WM2740 | WM1659+sulAp–gfp | This study |
| WM1488 | EC436 (λInch trc99p–gfp–ftsI) | Weiss et al. (1999) |
| ftsZ84 and FtsZ depletion | ||
| E. coli strains | ||
| WM1109 | TX3772 ftsZ84 | Laboratory collection |
| WM1985 | WM1109+pET28 | This study |
| WM1986 | WM1109+pET-FtsA | This study |
| WM1987 | WM1109+pET-FtsA* | This study |
| WM2757 | WM1985+trc99p–gfp–ftsI from WM1488 | This study |
| WM2758 | WM1986+trc99p–gfp–ftsI from WM1488 | This study |
| WM2759 | WM1987+trc99p–gfp–ftsI from WM1488 | This study |
| WM747 | WX7/λGL100 (ftsZ0, lacp–ftsZ) | Laboratory collection |
| WM2637 | WM747+pBAD33 | This study |
| WM2638 | WM747+pBAD-FtsA | This study |
| WM2639 | WM747+pBAD-FtsA* | This study |
| ftsZ–gfp E. coli strains | ||
| WM1085 (EC448) | MC4100 ftsZ–gfp | Weiss et al. (1999) |
| WM2026 | WM1074 ftsZ–gfp | This study |
| WM2027 | WM1659 ftsZ–gfp | This study |
| WM2720 | WM1940 ftsZ–gfp | This study |
| WM2721 | WM2195 ftsZ–gfp | This study |
| WM2722 | WM1032 ftsZ–gfp | This study |
| WM2724 | WM2176 ftsZ–gfp | This study |
| WM2725 | WM2177 ftsZ–gfp | This study |
| Plasmids | ||
| pWM1261 | pET28a expression vector | Novagen |
| pWM1260 | pET-FtsA | Geissler et al. (2003) |
| pWM1690 | pET-FtsA* | Geissler et al. (2003) |
| pWM1728 | pBAD33 expression vector | Guzman et al. (1995) |
| pWM1727 | pBAD33-FtsA | Geissler et al. (2003) |
| pWM1726 | pBAD33-FtsA* | Geissler et al. (2003) |
| pMK4 | pMK4, Ptac-ftsZ in IncP plasmid | Ma et al. (1996) |
| pJSB101 | pJSB2-FtsZ D45A | H. Erickson‡ |
| pWM971 | pET11a-FtsZ | H. Erickson‡ |
| pWM976 | pBAD33-FtsZ-GFP | Laboratory collection |
| pWM2463 | pBAD33-FtsZ | This study |
| pWM2248 | pZA′Q | Geissler & Margolin (2005) |
| pDSW209 | Ptrc-gfp pBR322 derivative | Weiss et al. (1999) |
| pWM2760 | pDSW209-FtsA | This study |
| pWM2761 | pDSW209-FtsA* | This study |
| pWM1891 | pGADT7 | Clontech |
| pWM1892 | pGBKT7 | Clontech |
| pWM2770 | pGAD-FtsZ | This study |
| pWM1896 | pGAD-FtsA | This study |
| pWM1898 | pGAD-FtsA* | This study |
| pWM2771 | pGAD-ZipA | This study |
| pWM1902 | pGBK-FtsZ | This study |
| pWM1897 | pGBK-FtsA | This study |
| pWM1899 | pGBK-FtsA* | This study |
| pWM2772 | pGBK-ZipA | This study |
University of Wisconsin.
Duke University.
Plasmid construction
Plasmids used in this study were constructed using standard molecular biology protocols (Sambrook et al., 1989). To construct plasmids expressing either GFP–FtsA or GFP–FtsA*, ftsA or ftsA* were amplified by PCR from strain WM1074 or WM1659 genomic DNA, respectively, using Roche Hi-Fidelity DNA polymerase and oligonucleotide primers WM822 and WM833. These products and pDSW209 were digested with XbaI and PstI, then ligated together, yielding pGFP-FtsA (WM2760) or pGFP-FtsA* (WM2761). Constructs used for yeast two-hybrid assays were constructed by either PCR-amplifying ftsA or ftsA* from WM1074 or WM1659 genomic DNA; ftsZ was removed from pWM971 by digestion with NdeI/BamHI. pGAD-T7 and pGBK-T7 (Clontech) and the PCR products were digested with NdeI and BamHI, then ligated together to make pWM1896, pWM1897, pWM1898, pWM1899, pWM1902 and pWM2770. pBAD-FtsZ was made by replacing ftsZ–gfp in pWM976 with ftsZ from pJSB101 (a gift from Harold Erickson, Duke University) by digestion with KpnI/HindIII and subsequent ligation.
Temperature-shift experiments with ftsZ84(ts)
We performed temperature-shift experiments on cultures of the ftsZ84(ts) strains WM1985, WM1986 and WM1987, essentially as described by Addinall et al. (1996). Briefly, 2 ml cultures were shaken at the permissive temperature of 30 °C to mid-exponential phase (OD600 ~0.25), and 1 ml was either fixed with (final concentrations) 0.04 % paraformaldehyde, 2.6 % glutaraldehyde, 32.25 mM Na3PO4, or diluted 1 : 4 into fresh medium pre-warmed to 42 °C. These cultures were incubated for 2 min at 42 °C, then split for fixation or dilution (1 : 2) into fresh medium kept at 30 °C. These cultures were further incubated at 30 °C for 5 min, at which point a 1 ml aliquot was fixed for IFM. Strains WM2757, WM2758 and WM2759, which are WM1985, WM1986 and WM1987, respectively, containing an IPTG-inducible gfp–ftsI fusion, were grown for 90 min with 2.5 μM IPTG, before shifting to 42 °C. These cultures were allowed to recover for 30 min before an aliquot was taken for fixation, and then for another 30 min before a second aliquot was fixed for IFM. We followed the IFM staining procedure used elsewhere (Addinall et al., 1996), for the samples collected during this experiment.
Cell length and Z-ring measurements
Cultures of strain WM1074 or WM1659 were grown in LB at 32 °C until early exponential or stationary phase. An aliquot from each sample at each time point was immobilized in 2 % LB agarose, and observed microscopically; cell lengths of over 100 cells for each strain were measured with Object Image (Norbert Vischer). To measure the number of c.f.u. during steady-state growth, WM1074 and WM1659 were grown in LB at 32 °C to OD600 0.61; the cultures were then diluted 105-fold and plated (0.1 ml) on LB plates in quadruplicate, and the total number of colonies was counted.
To determine the timing of Z-ring assembly in strain WM1074 or WM1659, cultures were grown in LB at 30 °C to early exponential phase, at which point an aliquot was fixed for IFM. The inter-nucleoid distance was calculated by measuring the space between nucleoids in cells containing a clear Z ring, using the pixel-measuring tool in Adobe Photoshop 6.0.
FtsZ depletion
Cultures of strains WM2637, WM2638 and WM2639 were grown overnight in LB+Cm+20 μM IPTG at 30 °C, and diluted 1 : 100 into fresh medium containing 20 μM IPTG. At early exponential growth, 1 ml of each culture was spun down and washed two times with LB, then resuspended in 500 μl LB. Subsequently, 100 μl of the washed cells was added to 1.9 ml fresh LB+Cm containing various concentrations of IPTG (1.0, 5, 10, 12.5, 15, 17.5 or 20 μM) or 2 % glucose, and allowed to grow for 3 h at 30 °C. Aliquots were then fixed for IFM or added to 1 % SDS for SDS-PAGE. For immunoblotting, 10 μg total protein, as determined by the bicinchonic acid (BCA) assay (Pierce), was separated by SDS-PAGE and transferred to nitrocellulose membranes. Affinity-purified antisera against FtsZ or GFP and secondary antibody (goat anti-rabbit-conjugated horseradish peroxidase) were used to detect FtsZ or GFP–FtsA (or FtsA*). After incubating with antibodies, membranes were developed using standard ECL reagents (Sigma).
Fluorescence recovery after photobleaching (FRAP)
Photobleaching experiments were carried out essentially as described by Ghosh & Young (2005). Briefly, strains expressing FtsZ–GFP or GFP–FtsA (FtsA*) were grown overnight at 30 °C in M9+ CA+glycerol, and subcultured 1 : 100 into fresh medium containing 40 μM IPTG to induce FtsZ–GFP, or 2.5 μM IPTG for GFP–FtsA or FtsA*. Cultures were allowed to grow at 30 °C until they reached mid-exponential phase, at which point, 3 μl culture was applied to a 1 % agarose pad containing M9+CA+glycerol attached to a cover slip, inverted, and placed in the well of a glass-bottomed dish (Difco).
Image acquisition and FRAP experiments were performed on a Zeiss LSM510-Meta inverted microscope using a Plan-Apochromat 63x/1.4 numerical aperture (NA) oil objective. Ten iterations of × 100 laser power were used to photobleach selected regions of interest. To reduce non-specific photobleaching during the course of the experiment, ~300 ms exposures were taken using 2–4 % transmitted light. Measurements of fluorescence intensity were obtained using LSM AIM software (Carl Zeiss). Microsoft Excel was used to correct for background fluorescence and to calculate the half-time of recovery t½ (Stenoien et al., 2001). Microsoft Excel was also used for statistical analysis of the data using the t test data analysis tool.
Yeast two-hybrid assays
Protocols for yeast transformation and liquid β-galactosidase assays were obtained from the Yeast Protocols Handbook (Clontech). Briefly, we transformed Saccharomyces cerevisiae Y190 with our constructs expressing ftsA, ftsA* or ftsZ. Overnight cultures were diluted into fresh SC-Leu-Trp and grown at 30 °C to OD600 0.5–1.1, and processed for liquid β-galactosidase assays, using ONPG as a substrate for the reactions. Values shown are the means±SD of at least five separate assays from two separate transformations.
RESULTS
Cells expressing ftsA* are shorter than wild-type cells
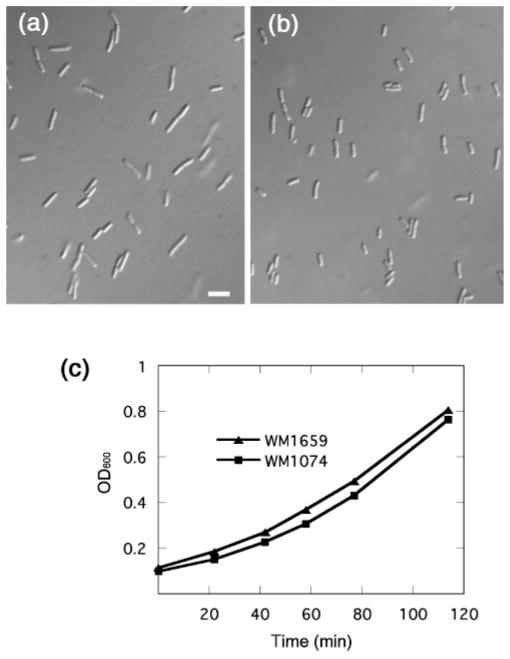
In our previous report of ftsA* being able to bypass the requirement for zipA, we focused on the ability of ftsA* to permit cell division under conditions normally not favourable for division, and found that otherwise wild-type cells with ftsA* mostly seemed to divide normally. However, upon closer inspection, we noticed that during exponential growth at 32 °C, cells of strain WM1659 (ftsA replaced with ftsA*) were 27 % shorter on average (3.3 μm) than cells of the isogenic parental strain WM1074 that contains ftsA (4.5 μm). Representative fields of these cells are shown in Fig. 1(a, b). The growth rates of the two strains were essentially identical (Fig. 1c), ruling out the possibility that WM1074 cells were larger merely because they were growing faster. Moreover, in stationary phase, WM1659 cells were also 12 % shorter than WM1074 cells.
Fig. 1.
FtsA* cells are shorter than normal. Representative fields are shown of cells of strain WM1074 (a) and its isogenic ftsA* derivative WM1659 (b), at the same magnification. Bar, 5 μm. Both strains were grown at 32 °C in LB to exponential phase, and samples were briefly spun to concentrate them five-fold prior to placing on microscope slides. Growth curves of the strains cultured at 32 °C in LB are shown in (c).
Because ftsA* decreased the mean cell length in the population, it might also have been expected to decrease the length of newborn cells. This can be seen in liquid cultures (Fig. 1a, b), but growth and division of cells were also examined in time-lapse on an agar pad under the microscope. We found that newly formed daughter cells of strain WM1659 were indeed 23 % shorter than those of wild-type strain WM1074, and mother cells were 27 % shorter at the time of visible septation (data not shown). Once again, this difference was not a result of different rates of elongation, as cells of both strains grew at a similar rate. This size difference was more pronounced at faster growth rates. For example, when grown in LB at 37 °C, WM1659 cells were 35 % shorter than WM1074 cells (data not shown).
To confirm these findings, we measured the number of viable cells in populations of strains WM1074 and WM1659 growing under the steady-state conditions used to measure the cell lengths above. Measurement of c.f.u. of cultures grown to identical cell densities revealed 11 % higher cell counts for WM1659 than for WM1074, consistent with the smaller cells of WM1659. We conclude that the presence of ftsA* caused the cells to divide at a significantly smaller size than the ftsA+ parent.
Influence of ftsA* on placement of Z rings between nucleoids
The ability of FtsA* to stimulate cell division at a smaller cell size led us to investigate further the effects of FtsA* on wild-type Z-ring formation. The combined effects of the Min system and nucleoid occlusion normally prevent Z-ring assembly at the cell poles and over the nucleoid, respectively (Rothfield et al., 2005; Yu & Margolin, 1999). Positioning the Z ring between nucleoids ensures that incompletely segregated chromosomes will not be severed by an ingrowing septum, and that daughter cells will contain an equal amount of genetic material.
We investigated whether the shorter ftsA* cells contained a smaller space between nucleoids for Z-ring assembly. The rationale for this idea was that if ftsA* potentially accelerated Z-ring assembly, such that midcell rings formed in cells that were shorter than normal, then those cells may have been at an earlier stage in chromosome segregation. WM1074 and WM1659 cells were grown in rich medium (LB) at 30 °C, and we measured the distance between the nucleoids in cells containing a Z ring at midcell. Of the 174 cells of strain WM1074 examined, 91 % contained Z rings, and the mean distance between nucleoids in these cells with rings was 0.35±0.06 μm. In comparison, of the 178 cells of strain WM1659 examined, 79 % contained Z rings, and the mean distance between nucleoids in these cells with rings was 0.20±0.02 μm, which is 43 % less. This modest but consistent decrease in inter-nucleoid distance supports the idea that FtsA* promotes Z-ring formation earlier in the cell cycle relative to chromosome segregation. We also measured the percentage of cells with visibly segregated nucleoids. Out of 259 cells of WM1074, 205 (79 %) had clearly separated nucleoids, whereas only 130 out of 218 cells of WM1659 (60 %) had separated nucleoids. These results support the idea that the shorter cells of WM1659 were, on average, at an earlier stage in their chromosome segregation cycle than cells of WM1074.
Although the growth rate of ftsA* cells is similar to that of wild-type cells, the smaller space between nucleoids prompted us to investigate whether there might be an increase in chromosome guillotining in ftsA* cells. A chromosomal sulAp–GFP reporter regulated by DNA damage (McCool et al., 2004) was introduced into wild-type WM1074 or ftsA* mutant WM1659 cells. Whereas only ~1 % of WM1074+sulAp–GFP (WM2739) cells exhibited significant GFP fluorescence when grown in rich or minimal media, only ~2 % of WM1659+sulAp–GFP (WM2740) cells exhibited similar GFP fluorescence (data not shown). Therefore, despite having less space between nucleoids in cells with FtsA*, Z rings did not significantly induce chromosome guillotining or DNA damage compared to wild-type cells. Moreover, removing the nucleoid occlusion protein SlmA by introducing a ttk : : kan insertion did not significantly affect Z-ring formation or viability in wild-type (WM2775) or ftsA* (WM2776) cells growing in rich medium (data not shown). This supports the idea that sufficient space remains between nucleoids in ftsA* cells to restrict Z-ring assembly to that location, and suggests that the Min system is sufficient to keep rings precisely at the midpoint of these smaller cells.
FtsA* accelerates reassembly of the Z ring and the divisome
Z rings formed in strains containing the thermosensitive ftsZ84(ts) allele disassemble within 2 min after shifting to the non-permissive temperature of 42 °C (Addinall et al., 1996). Two minutes after returning the culture to the permissive temperature (30 °C), Z rings are largely reassembled. When an additional copy of the zipA gene, which encodes a proposed Z-ring stabilizing protein, is introduced in an ftsZ84(ts) strain, the thermosensitivity is suppressed, allowing Z rings to form at 42 °C (Raychaudhuri, 1999). Based on its ability to permit the complete removal of ZipA, we tested whether FtsA* could also relieve this temperature sensitivity, restore Z-ring formation, and increase viability.
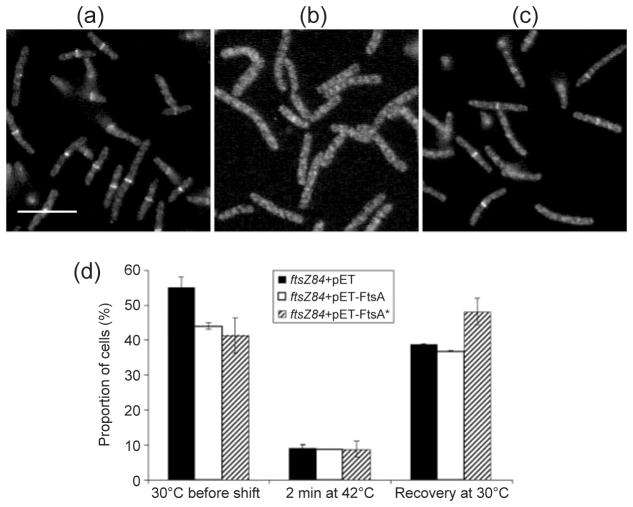
An ftsZ84(ts) strain containing either empty plasmid vector (pET28), pET-FtsA or pET-FtsA* (WM1985, WM1986 or WM1987) was subjected to a series of temperature shifts. Aliquots of each strain were immediately fixed for IFM after each shift, as described in Methods. Uninduced basal expression levels of ftsA or ftsA* from the pET plasmid derivatives, like expression from pBAD-FtsA or FtsA* previously reported (Geissler et al., 2003), complements an ftsA mutant, indicating that the levels produced in this system are physiological. IFM with antibodies directed against FtsZ showed that ftsZ84+pET28, ftsZ84+pET-FtsA or ftsZ84+pET-FtsA* cultures grown at 30 °C prior to the shift contained Z rings in 55.0, 44.0 or 41.3 % of the cells, respectively (Fig. 2a, d). These differences may have resulted from the slight toxicity of FtsA made from pET-FtsA, and the decrease in the size of the cells caused by expression of FtsA* (see below).
Fig. 2.
FtsA* promotes more rapid Z-ring formation in ftsZ84(ts) cells after thermoinactivation and recovery. (a–c) Representative fluorescence micrographs of cells with ftsZ84 +pET28a vector (WM1985) grown at 30 °C (a), shifted to 42 °C for 2 min (b), then shifted back to 30 °C for 5 min (c). Samples were taken after each shift, fixed, and processed for IFM using anti-FtsZ specific antiserum. Cultures of ftsZ84+pET-FtsA (WM1986) and ftsZ84+pET-FtsA* (WM1987) were similarly processed for IFM (data not shown). (d) Percentages of ftsZ84 cells containing pET vector (black columns), pET-FtsA (white columns) or pET-FtsA* (hatched columns) containing a clear Z ring after each step of the temperature-shift experiment. Bar, 5 μm.
After the shift to 42 °C, distinct Z rings remained in ~9 % of cells for all three strains (Fig. 2b, d). These results confirm that FtsZ84 assembly into Z rings was thermoinactivated, and indicate that FtsA* from pET-FtsA* failed to enhance Z-ring assembly at the non-permissive temperature. We transformed pBAD-FtsA* into the ftsZ84(ts) strain and induced expression of ftsA* with arabinose, to test whether increased levels of FtsA* could allow division of ftsZ84, but we were unable to suppress the thermosensitivity significantly at any concentration of inducer (data not shown). This suggests that, at least at the levels produced in our system from pET-FtsA* or pBAD-FtsA*, FtsA* does not directly mimic doubled gene dosage of ZipA.
Upon shifting the cultures back to 30 °C for 5 min, Z rings were found in 48.1 % of ftsZ84+pET-FtsA* cells, compared to 38.8 % of ftsZ84+pET28 or or 36.8 % of ftsZ84+pET-FtsA cells (Fig. 2c, d). Despite the apparent ability of FtsA* to alter Z-ring assembly kinetics, the viability of ftsZ84+pET-FtsA*, ftsZ84+pET28, or ftsZ84+pET-FtsA was indistinguishable when grown at 30, 36, 39 or 42 °C (data not shown).
To confirm this result, we monitored the localization of another divisome component, FtsI, following a similar series of temperature shifts. We used GFP–FtsI, which localizes to Z rings later in the divisome assembly pathway, and thus should be a good marker for a nearly completely assembled divisome (Weiss et al., 1999). After a 10 min shift to 42 °C, followed by 30 min of recovery at 30 °C, GFP–FtsI fluorescence was localized to midcell in 52.6 % of ftsZ84 gfp-ftsI+pET-FtsA* (WM2759) cells, compared to 27.3 % of ftsZ84 gfp-ftsI+pET-FtsA (WM2758) or 38.2 % of ftsZ84 gfp-ftsI+pET28 (WM2757) cells (data not shown). Importantly, the percentage of cells with GFP–FtsI rings levelled out at ~45 % for all three strains after 60 min at 30 °C, indicating that divisome formation was not significantly altered by pET28 or pET-FtsA (data not shown). These results suggest that under pseudo-synchronized conditions in an ftsZ84(ts) mutant, FtsA* enhances both the kinetics of Z-ring assembly and the subsequent formation of a nearly complete divisome.
FtsA* suppresses toxicity of excess FtsZ by maintaining Z rings
An ~10-fold or greater excess of FtsZ inhibits cell division, causing cell filamentation (Ward & Lutkenhaus, 1985). The cause of this effect is unknown, although concomitant increases in FtsA levels restore the FtsZ : FtsA ratio and suppress the filamentation. Because FtsA* can suppress filamentation caused by excess ZipA, we investigated whether the toxic effects of excess FtsZ were also suppressed.
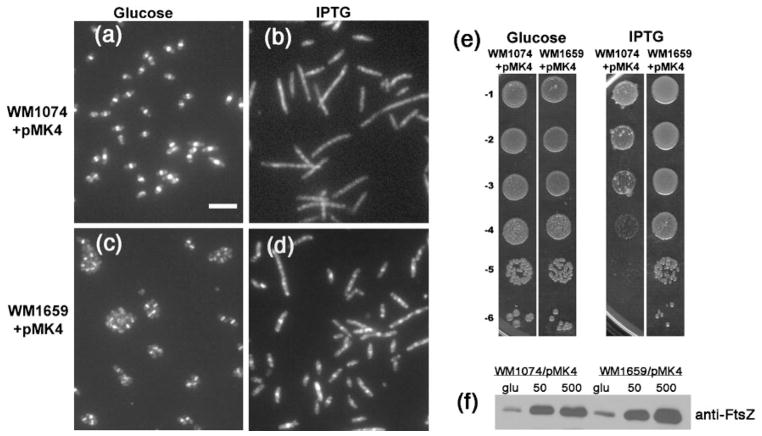
We transformed strains WM1074 and WM1659 with plasmids expressing ftsZ under the control of either the IPTG-inducible lac promoter (pMK4) or the arabinose-controlled PBAD promoter (pBAD-FtsZ). Dilution-plating experiments showed that WM1659 (ftsA*)+pMK4 was ~103-fold more viable than WM1074 (WT)+pMK4, when grown on medium containing 500 μM IPTG at 30 °C (Fig. 3e). In support of this decreased viability, WM1074+pMK4 cultures grown with 500 μM IPTG contained 25 % more filamentous cells (defined as longer than 5 μm) than did cultures of WM1659+pMK4, 88.4 versus 63.8 % (Fig. 3a–d). These filamentous cells likely arose because they were unable to form Z rings, as suggested by the IFM images showing few Z rings in filamentous cells (Fig. 3b, d; compare with Fig. 3a, c). Importantly, whereas very long filamentous cells (>20 μm) were only rarely present in the WM1659 population (Fig. 3d), they were often observed in the WM1074 population (Fig. 3b). Immunoblotting with anti-FtsZ showed similar increased amounts of FtsZ overproduction in both strains (about three times) grown in 50 μM IPTG versus glucose (Fig. 3f), indicating that FtsZ was overproduced to similar levels at this IPTG concentration. It was harder to interpret the FtsZ levels at the higher concentrations of IPTG (500 μM) used for the micrographs; a continued increase in FtsZ levels was observed for WM1659+pMK4, but not for WM1074+pMK4, probably because many cells of the latter were dead or dying.
Fig. 3.
FtsA* suppresses the toxicity caused by excess FtsZ. (a–d) Micrographs showing anti-FtsZ IFM of WM1074+pMK4 (a, b) or WM1659+pMK4 (c, d) grown at 30 °C for 4 h in LB containing either glucose (a, c) or 500 μM IPTG (b, d). Bar, 5 μm. (e) Viability plating of WM1074+pMK4 or WM1659+pMK4 on LB containing glucose (left panels) or 500 μM IPTG (right panels). (f) Immunoblot of extracts from cells described in (a–d), grown in glucose (glu), 50 μM IPTG (50) or 500 μM IPTG (500), and probed with anti-FtsZ. An equivalent amount of cells, as measured by optical density, was loaded into each lane. The FtsZ band density in the WM1074/pMK4 (500 μM IPTG) lane was low, probably because many of the cells had already been lysed by the high levels of FtsZ.
In support of these results, WM1074+pBAD-FtsZ formed filaments three to four cell-equivalents long, with irregular diffuse Z rings (>95 % of cells) when grown in 0.01 % arabinose, whereas cells of WM1659+pBAD-FtsZ under the same induction conditions were mostly normal in length, and ~90 % of these cells contained a single regular midcell Z ring (data not shown). When the concentration of arabinose was increased to 0.1 %, both strains formed non-dividing filaments, suggesting that the endogenous level of FtsA* cannot provide resistance to very high levels of FtsZ.
FtsA* cannot stimulate division in cells with decreased levels of FtsZ
In addition to testing the effect of ftsA* in cells with increased concentrations of FtsZ, we also examined the effects of lower than normal FtsZ levels. We were prompted to do this because in B. subtilis, ZapA promotes assembly of the Z ring, and becomes essential for division when the level of FtsZ is decreased (Gueiros-Filho & Losick, 2002). To determine if FtsA* could compensate for lower than normal levels of FtsZ, we used an E. coli strain which has the chromosomal ftsZ gene inactivated, and a copy of ftsZ under Plac control integrated at the phage lambda attachment site (WM747) to decrease the level of FtsZ. In medium supplemented with 20 μM IPTG, this strain produced near wild-type levels of FtsZ, and mostly divided normally (data not shown). At IPTG concentrations <20 μM, WM747 formed long non-septate filaments and became less viable on plates, indicating that these levels of FtsZ were not sufficient for cell division. We transformed pBAD33, pBAD-FtsA or pBAD-FtsA* into WM747 and examined the viability, cell morphology and ability to form Z rings at various concentrations of IPTG. All three strains (WM 2527, WM2528 and WM2529) had fewer Z rings, and displayed similar cell filamentation and viability at several IPTG concentrations in the range 0–20 μM (data not shown). FtsZ levels were the same in all strains at a given IPTG concentration (data not shown). Therefore, whereas FtsA* could protect against Z-ring disassembly by high levels of FtsZ, it could not help cells divide if FtsZ levels were below a minimum amount.
FtsA* does not affect turnover and partitioning of FtsZ into the Z ring
The potential increase in Z-ring stability provided by FtsA* prompted us to investigate whether FtsZ subunit turnover within the ring was altered in cells containing ftsA* versus wild-type ftsA. Using FRAP, Anderson et al. (2004) have found that removing proposed Z-ring-stabilizing proteins in B. subtilis only slightly increases the t½ of FtsZ–GFP. The same study has also shown that removing minCDE increases the t½ of FtsZ–GFP in E. coli, and those authors suggest that the Min system is implicated in directly affecting Z-ring stability.
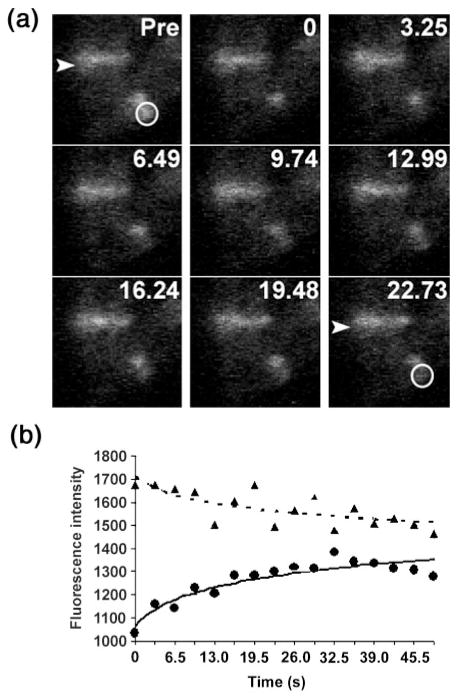
We used FRAP to determine whether FtsZ–GFP turnover was influenced by the presence of ftsA*. We measured the t½ of FtsZ–GFP in two wild-type E. coli parental strains and those transduced with ftsA*. One such measurement is shown in Fig. 4. Table 2 shows that the presence of ftsA* had no significant effect on FtsZ–GFP recovery, in either TX3772 or W3110 strain backgrounds. We measured FtsZ–GFP turnover in a ΔminCDE strain (WM2722) as a positive control for altered dynamics, and confirmed the previous finding that deleting minCDE increased the t½ of FtsZ–GFP modestly but significantly. The mean t½ of WM2720 (ftsA* ΔminCDE) was also statistically distinguishable from that of wild-type cells (P=0.017). As shown in Table 2, ftsA* slightly increased FtsZ–GFP turnover in the absence of minCDE (WM2720), although the difference between WM2720 and WM2722 was not statistically significant. The small variation between WM2720 (ftsA* ΔminCDE) and WM2027 (ftsA*) may have been a result of the FtsA*-mediated immunity of FtsZ to MinC. Deleting zipA in the presence of ftsA* (WM2721) had little effect on FtsZ turnover (Table 2), further supporting the previous observation that FtsZ-associated proteins have little effect on FtsZ turnover, but perhaps modulate initial assembly of the Z ring. Although similar, our mean values for t½ differed slightly from those obtained previously. This may be a result of different strain backgrounds, different levels of FtsZ–GFP, and/or a different microscope and software setup used to obtain images.
Fig. 4.
FRAP analysis. (a) Micrographs from one FRAP experiment with FtsZ–GFP expressed at low levels in an otherwise wild-type strain (WM2026). Three cells with Z rings are visible: two cells aligned vertically (upper left), and one horizontally (lower right) that was used for bleaching. Each panel represents a sequential time point in seconds, with the first prior to photobleaching (Pre). The arrowhead highlights one of the unbleached Z rings used as an internal control for background photobleaching during the experiment. The white circle shows the photobleached portion of another Z ring, which recovered during the time-course. (b) The fluorescence intensities (arbitrary units) for the two Z rings are plotted versus time. The FRAP data from this and all other time-courses are summarized in Table 2. ●, Bleached; ▲, no bleaching.
Table 2.
Summary of FtsZ–GFP and GFP–FtsA (FtsA*) FRAP measurements
| Strain | Average t½ | SD | Rings† | Different from wild-type‡ |
|---|---|---|---|---|
| FtsZ–GFP§ | ||||
| WM1074 (WM2026) | 11.2 | 3.5 | 37 | – |
| WM1074 ftsA* (WM2027) | 12.0 | 3.6 | 39 | No |
| WM1074 min− (WM2722) | 15.3 | 6.0 | 12 | Yes (P=0.0046) |
| WM1074 min− ftsA* (WM2720) | 13.4 | 2.8 | 19 | Yes (P=0.017) |
| WM1074 min− ftsA* zipA− (WM2721) | 12.7 | 1.4 | 10 | No |
| W3110 (WM2724) | 11.3 | 4.1 | 16 | – |
| W3110 ftsA* (WM2725) | 11.1 | 2.8 | 18 | No |
| GFP–FtsA (FtsA*)|| | ||||
| WM1074+pWM2760 | 16.3 | 5.2 | 31 | – |
| WM1074 ftsA*+pWM2761 | 11.9 | 4.7 | 27 | Yes (P=0.0015) |
Number of rings bleached for each strain.
As determined by the t test. Comparing WM1074 to other WM1074 mutant strains, W3110 to W3110 ftsA*, or GFP–FtsA* to GFP–FtsA.
Strains were grown in M9+CA+Gly+40 μM IPTG for ~5 h at 30 °C.
Strains were grown in M9+CA+Gly+2.5 μM IPTG for ~5 h at 30 °C.
In addition to FtsZ turnover, we measured the relative percentage of FtsZ found within Z rings during exponential growth, using similar strains that synthesized both endogenous FtsZ and low levels of FtsZ–GFP. As ftsA* functions to increase stability of the Z ring, one prediction is that an increased percentage of FtsZ might be found incorporated into the ring. However, we found no significant difference between wild-type and ftsA* strains in the percentage of FtsZ–GFP at midcell compared to the rest of the cell. Both contained ~40 % of the total fluorescence within the ring (data not shown). These results suggest that the positive effects of ftsA* on Z-ring integrity do not result from higher FtsZ concentration in the ring under these conditions.
FtsA* affects FtsA dynamics and is less toxic in excess than wild-type FtsA
We have previously found that in a zipA+ strain, additional FtsZ, FtsA* and FtsQ reduce viability and produce misshapen cells, whereas additional FtsZ, wild-type FtsA and FtsQ produce minicells, but have much less effect on overall morphology (Geissler et al., 2003). Moreover, it is known that excess FtsA inhibits cell division. These results led us to examine whether extra FtsA* alone was toxic. Previously, we have determined that very high induction of either FtsA or FtsA* from pBAD-FtsA or pBAD-FtsA* with arabinose blocks cell division and produces non-viable filaments (Geissler et al., 2003). Nevertheless, we wished to test whether moderate overproduction of FtsA or FtsA* had differential effects.
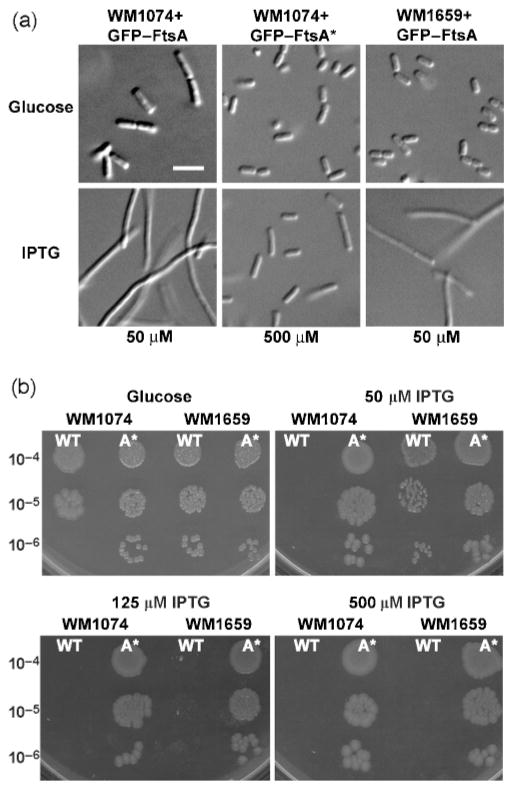
We constructed GFP fusions to both FtsA and FtsA* under the control of the weakened Ptrc promoter from pDSW209 (Weiss et al., 1999), so that the effects and localization of FtsA could be monitored simultaneously, and examined the effects of their overproduction at a range of IPTG concentrations. When grown in LB, wild-type cells expressing GFP–FtsA formed long filaments even at low concentrations (50 μM) of IPTG, whereas strains expressing GFP–FtsA* at the same or higher (50 or 500 μM) IPTG concentrations were normal in length (Fig. 5a). As expected (Ma et al., 1996), GFP–FtsA or GFP–FtsA* fusions localized to Z rings, but were not fully functional, as judged by complementation of an ftsA12(ts) mutant (data not shown).
Fig. 5.
Overproduction of GFP–FtsA* is less toxic to cells than that of GFP–FtsA. (a) Micrographs of WM1074+GFP–FtsA (left column), WM1074+GFP–FtsA* (middle column) or WM1659+GFP–FtsA (right column) grown in LB containing either glucose (top row), 50 μM IPTG (bottom-left and bottom-right images) or 500 μM IPTG (bottom-middle image). Bar, 5 μm. (b) Dilution plating of WM1074 or WM1659 containing either GFP–FtsA (wild-type, WT) or GFP–FtsA* (A*) grown on glucose, or 50 μM, 125 μM or 500 μM IPTG. Serial dilutions are shown to the left of the panels.
Dilution-plating experiments showed that GFP–FtsA was very toxic to wild-type cells, even at a low level of induction (50 μM), whereas GFP–FtsA* was not toxic to wild-type cells, even with 500 μM IPTG (Fig. 5b). When GFP–FtsA was introduced into strain WM1659, the chromosomal ftsA* provided some resistance to GFP–FtsA at 50 μM IPTG (Fig. 5a, b), but addition of higher amounts of inducer resulted in toxicity similar to that of the wild-type strain. In contrast, cells were mostly normal when production of GFP–FtsA or GFP–FtsA* was suppressed by growth in glucose (Fig. 5a, b). Probing immunoblots with anti-GFP antibody confirmed that additional IPTG increased the amounts of both fusion proteins to similar levels (data not shown). This toxicity is consistent with our findings that excess FtsA lacking the GFP tag negatively affects Z-ring assembly (D. Shiomi & W. Margolin, unpublished results), which suggests that the GFP tag is not required for the toxicity.
We used FRAP to monitor the turnover of GFP–FtsA or FtsA* within the ring. Despite its toxicity in LB, low levels of IPTG (2.5 μM) in minimal medium only slightly increased cell length, but did not significantly affect the growth rate or localization of WM1074+pGFP-FtsA, and had no effect on WM1659+pGFP-FtsA*. Despite significant variability and modest recovery of fluorescence, the calculated t½ of GFP–FtsA* (11.9±4.7 s) was shorter than that of GFP–FtsA (16.3±5.2 s) (Table 2). This difference, which was statistically significant (P=0.002), might reflect an increase in FtsA*–FtsZ interaction relative to FtsA–FtsZ.
FtsA* interacts more strongly with FtsZ than does FtsA in a yeast two-hybrid system
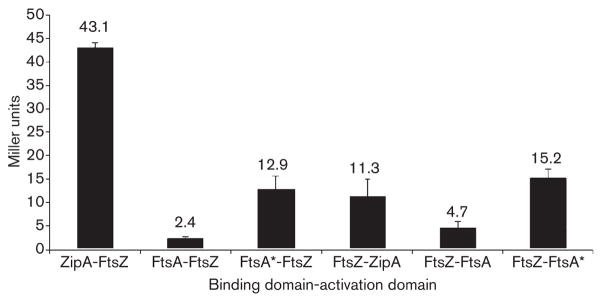
To test if FtsA* affects protein–protein interactions with FtsZ, we measured the interactions between ZipA, FtsA, FtsA* and FtsZ using a standard yeast two-hybrid reporter system. As a positive control for interaction with FtsZ, we monitored β-galactosidase activity in liquid cultures of yeast expressing both ZipA and FtsZ, which are known to interact strongly in E. coli, and showed a significant interaction in yeast as expected (Fig. 6). Interestingly, we found that yeast reporter strains containing plasmids expressing FtsA* and FtsZ exhibited 5.4 and 3.2 times more β-galactosidase activity, respectively, than did strains expressing wild-type FtsA and FtsZ (Fig. 6), depending on which protein was fused to the activation or DNA-binding domain. Yeast expressing fusions to FtsA, FtsA* or FtsZ and an unfused DNA-binding or activation domain did not show detectable β-galactosidase activity (data not shown), supporting the idea that significant β-galactosidase activity reflected real interactions. The increased activity in strains expressing FtsZ and FtsA* versus FtsA suggests that FtsZ has a higher affinity for FtsA* than for FtsA.
Fig. 6.
Yeast two-hybrid assays of FtsZ interactions with FtsA or FtsA*. S. cerevisiae Y190 containing plasmids producing the indicated proteins fused to the GAL4 activation domain were assayed for β-galactosidase activity (Miller units) in liquid culture, as described in Methods. The values shown are the means of at least five separate assays of two different transformations, with the SEM indicated by error bars.
DISCUSSION
The isolation of FtsA* as a suppressor of a zipA deletion suggests that FtsA*, like ZipA, enhances the integrity of the Z ring. This idea is supported by the resistance of Z rings in ftsA* mutants to high levels of MinC, which normally disassembles Z rings and blocks cell division (Hu et al., 1999). We have also recently reported that FtsA* can partially bypass the removal of the essential gene ftsK (Geissler & Margolin, 2005), which is consistent with FtsA* enhancing the integrity of the Z ring to compensate for the loss of another divisome protein. In this study, we investigated in more detail the role of FtsA* in promoting Z-ring integrity. We monitored its effects on FtsZ during various points of the Z-ring assembly–disassembly cycle (Romberg & Levin, 2003), including the initial assembly of FtsZ into Z rings, maintenance of the Z-ring structure, and the partitioning and exchange of FtsZ subunits between the Z ring and the cytoplasm.
Replacing ftsA with ftsA* shortened the mean cell length by 27 % during growth at 32 °C, and shortened newborn cells, suggesting that FtsA* promotes cell division at shorter than normal cell lengths. The smaller distances between segregating nucleoids in these shorter cells remained sufficient to permit normal Z-ring function without significant chromosome guillotining. The decrease in cell length we observed is similar to the 29 % decrease found previously when levels of FtsZ and wild-type FtsA were increased up to sevenfold with plasmid pZAQ′ (Begg et al., 1998). We should emphasize, however, that the level of FtsZ in the ftsA* strains was equal to that of its parental strain (Geissler et al., 2003; data not shown). This indicates that ftsA* promotes Z-ring formation without increasing total levels of FtsZ.
One possible mechanism for this would be for FtsA* to increase the fraction of FtsZ within the Z ring. However, our measurements indicate that there is a similar fraction of FtsZ within the Z ring in wild-type and ftsA* strains. Another potential mechanism for ftsA* to enhance Z-ring activity is to alter the kinetics of FtsZ-subunit turnover within the ring. Previous FRAP analysis has indicated that FtsZ cycles rapidly into the Z ring, with a t½ of ~10 s, indicating that the seemingly static Z ring is being constantly remodelled. Such remodelling may be important for its stability, because the thermosensitive FtsZ84 protein, which has a defective GTPase, turns over about threefold more slowly (Anderson et al., 2004). However, our FRAP studies indicate that FtsA* does not significantly alter the kinetics of FtsZ-subunit exchange. Inactivation of other known Z-ring-stabilizing or -destabilizing proteins, such as EzrA, ZapA or MinC, also has little effect on subunit exchange (Anderson et al., 2004), suggesting that most regulators of FtsZ assembly do not regulate FtsZ turnover as part of their mechanisms.
The t½ of GFP–FtsA was somewhat longer than that observed for FtsZ–GFP. Although the effect was not dramatic, it suggests that either the two proteins do not always exist as a complex during turnover within the ring, or the GFP tag alters the dynamics of the proteins; as FRAP in live cells is generally performed with fluorescent protein tags, there is no way to disprove the latter possibility. On the other hand, the t½ of GFP–FtsA* is nearly identical to that of FtsZ–GFP, suggesting that FtsA* and FtsZ may interact during turnover. This idea is supported by the yeast two-hybrid data, which indicate that FtsA* interacts more efficiently with FtsZ compared with wild-type FtsA. As one function of FtsA is to anchor FtsZ to the cytoplasmic membrane, an increased binding affinity between FtsA* and FtsZ might enhance the ability of FtsA* to promote assembly of FtsZ protofilaments into a compact Z ring, and strengthen the membrane binding of the Z ring.
The acceleration of Z-ring reassembly after ftsZ84(ts) thermoinactivation is completely consistent with the idea that FtsA* enhances the integrity of the Z ring structure, perhaps by increasing the cooperativity of assembly. Because FtsA* can replace ZipA, which probably bundles FtsZ protofilaments in vivo, one idea is that FtsA* also bundles FtsZ protofilaments, although there is no experimental proof for such an activity. Other evidence presented herein, however, suggests that FtsA* does not merely duplicate the activities of ZipA. For example, unlike ZipA, FtsA* does not permit ftsZ84(ts) cells to divide normally under the conditions tested. In addition, although FtsA* can stimulate FtsZ activity, it is not able to compensate for lower than normal levels of FtsZ.
Overproduction of FtsA or FtsZ alone is normally toxic, because it perturbs the normal FtsZ : FtsA ratio. Here, we have shown that FtsA* is significantly less toxic than FtsA when overproduced. One potential explanation for this is that FtsA* does not interact as well with FtsZ as does FtsA. However, the yeast two-hybrid data indicate the converse, as does the ability of FtsA* to replace ZipA, which normally binds strongly to FtsZ. Therefore, a more likely explanation is that FtsA* provides resistance to any destabilization of Z rings. Too much FtsZ relative to FtsA, or vice versa, causes destabilization, and hence FtsA* would counter this effect; too much FtsA* itself would also counter the effect of lowering the FtsZ : FtsA ratio, although if produced in sufficient quantity, FtsA* itself becomes toxic. Interestingly, while overproduction of FtsA and FtsZ in the correct ratio from pZAQ enhances cell division, overproduction of FtsA* and FtsZ in the correct ratio from pZA*Q is toxic, forming filaments with multiple constrictions (Geissler et al., 2003). This suggests that FtsZ structures become hyperstabilized in the presence of FtsA* and in the absence of any antagonistic effects from destabilizers, and is consistent with the regulatory balance that controls FtsZ assembly (Romberg & Levin, 2003). Further work needs to be done to understand how FtsA interacts with FtsZ, and why the FtsZ : FtsA stoichiometry is so important. Nevertheless, the ftsA* allele has already provided important insights into divisome function, particularly showing that ZipA must recruit downstream divisome proteins via an indirect effect on the Z ring, and not directly. Continued dissection of the mechanism by which FtsA* confers its unique properties on FtsA should help to define the roles of FtsA in cell division.
Acknowledgments
We thank Harold Erickson, Duke University, for providing pJSB101, and Susan Rosenberg, Baylor College of Medicine, for providing SSR966. We are grateful to Andrew Morris and James Broughman, University of Texas – Houston, and Zuzana Berkova and Shuo Dong, Baylor College of Medicine, for assistance with FRAP experiments and calculations. This work was supported by National Institutes of Health grant R01-GM61074.
Abbreviations
- Cm
chloramphenicol
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- IFM
immunofluorescence microscopy
- t½
half-time of recovery
References
- Addinall SG, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol. 2004;186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K, Nikolaichik Y, Crossland N, Donachie WD. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol. 1998;180:881–884. doi: 10.1128/jb.180.4.881-884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in E. coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Corbin BD, Geissler B, Sadasivam M, Margolin W. A Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Determination of cell fate in Bacillus subtilis. Trends Genet. 1996;12:31–34. doi: 10.1016/0168-9525(96)81386-2. [DOI] [PubMed] [Google Scholar]
- Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AS, Young KD. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 2005;187:1913–1922. doi: 10.1128/JB.187.6.1913-1922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry EJ. Bacterial cell division: regulating Z-ring formation. Mol Microbiol. 2001;40:795–803. doi: 10.1046/j.1365-2958.2001.02370.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SO, Thompson LS, Harry EJ. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-ring assembly. J Bacteriol. 2005;187:6536–6544. doi: 10.1128/JB.187.18.6536-6544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci U S A. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Microbiol. 2004;53:1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J Bacteriol. 2001;183:6630–6635. doi: 10.1128/JB.183.22.6630-6635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico AI, Garcia-Ovalle M, Mingorance J, Vicente M. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol Microbiol. 2004;53:1359–1371. doi: 10.1111/j.1365-2958.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL. Spatial control of bacterial division-site placement. Nat Rev Microbiol. 2005;3:959–968. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O’Malley BW, Mancini MA. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat Cell Biol. 2001;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Den Ent F, Lowe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Rico AI, Martinez-Arteaga R, Mingorance J. Septum enlightenment: assembly of bacterial division proteins. J Bacteriol. 2006;188:19–27. doi: 10.1128/JB.188.1.19-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JE, Lutkenhaus J. Overproduction of FtsZ induces minicells in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Weiss DS. Bacterial cell division and the septal ring. Mol Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]