Abstract
Microbial bioprocessing of lignocellulose to bioethanol still poses challenges in terms of substrate catabolism. The most important challenge is to overcome substrate recalcitrance and to thus reduce the number of steps needed to biorefine lignocellulose. Conventionally, conversion involves chemical pretreatment of lignocellulose, followed by hydrolysis of biomass to monomer sugars that are subsequently fermented into bioethanol. Consolidated bioprocessing (CBP) has been suggested as an efficient and economical method of manufacturing bioethanol from lignocellulose. CBP integrates the hydrolysis and fermentation steps into a single process, thereby significantly reducing the amount of steps in the biorefining process. Filamentous fungi are remarkable organisms that are naturally specialised in deconstructing plant biomass and thus they have tremendous potential as components of CBP. The fungus Fusarium oxysporum has potential for CBP of lignocellulose to bioethanol. Here we discuss the complexity and potential of CBP, the bottlenecks in the process, and the potential influence of fungal genetic diversity, substrate complexity and new technologies on the efficacy of CPB of lignocellulose, with a focus on F. oxysporum.
Keywords: Bioethanol, Fungi, Fusarium, Consolidated bioprocessing, Lignocellulose
Background
Energy is the single most important commodity in the world today. In many ways, a nation’s success is largely dependent on their level of energy security. Obtaining a secure, renewable, environmentally-benign and cheap supply of energy is of crucial global importance. The world is highly dependent on fossil fuels for energy. The transportation sector in particular is almost entirely reliant on petroleum-based fuels. Energy use is also inexorably linked to climate change. In OECD countries, the transportation sector is responsible for 23 % of worldwide carbon dioxide emissions and over 70 % of global carbon monoxide emissions (International Energy Agency 2014). The substantial release of these greenhouse gases into the atmosphere has drastically quickened the rate of global warming and subsequent climate change. This rate of pollution shows no sign of abating and expectations are that there will be over 2 billion vehicles worldwide by 2030 (International Energy Agency 2015). This growth will put enormous stress on global ecosystems and the global climate (Balat 2011). The increasing demand and increases in atmospheric CO2 concentrations means that the continued use of fossil fuels at the current levels is clearly unsustainable. Therefore, the only way to stem this energy crisis is to reduce fossil fuel dependence and move to alternative fuels. With these factors in mind, the need for a cheaper, ‘greener’ and more self-sufficient energy source is of global importance.
For the past few decades, industry and governments have diversified research in an attempt to discover, develop and commercialise new technologies for alternative transportation fuels, including biofuels derived from plant biomass. Generous subsidies were given in many developed nations in order to stimulate the production of biofuel crops. But there are huge concerns regarding the increasing diversion of starch- or sucrose-rich crop materials and land from food to biofuel production. This has shifted attention to the use of lignocellulose-derived bioethanol as a biofuel (Morales et al. 2015; Himmel and Bayer 2009; Bentsen et al. 2014; Limayem and Ricke 2012; Banerjee et al. 2010). Lignocellulose is the structural backbone of plant material, mainly composed of cellulose and hemicelluloses linked by lignin. It is one of the most abundant materials on this planet but is generally not used as a human foodstuff.
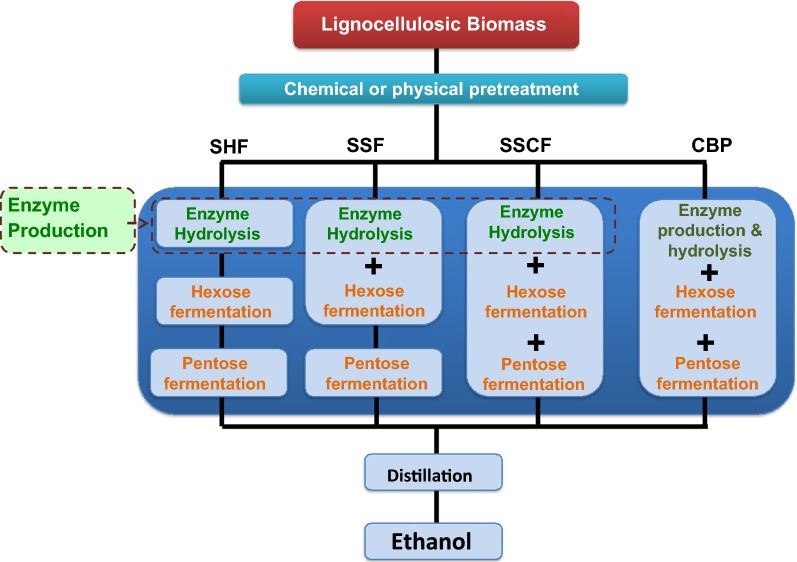
Despite having such advantages, lignocellulosic bioethanol cannot compete price-wise with starch or sucrose -based bioethanol (Balan et al. 2013; Guo et al. 2015). It’s tough molecular design has evolved over 400 million years and keeps plants upright and protects them from other organisms (Sanderson 2011). Converting lignocellulose into biofuel is costly; it requires several pretreatments, the enzymatic hydrolysis to release the sugars and the fermentation of sugars to bioethanol (Fig. 1). Several strategies can be used to convert lignocellulose to bioethanol. These include pretreatment followed by simultaneous saccharification and fermentation, simultaneous saccharification and co-fermentation and consolidated bioprocessing (CBP) (Li et al. 2014; Brethauer and Studer 2014; Lynd et al. 2008; Mosier et al. 2005; Xu et al. 2009). But, these processes are not yet commercially adopted. The current practice involves separate hydrolysis and fermentation, distinct steps in the process including the production of enzymes, biomass hydrolysis and subsequent fermentation of hexose and pentose sugars. However, the accumulation of high glucose content can inhibit some enzymes, principally glucosidases. Simultaneous saccharification and fermentation would circumvent this problem by preventing a build-up of glucose in the reactor (Olofsson et al. 2008). CBP is the ideal process, wherein the same microorganism is able to produce the enzymes, hydrolyse biomass and convert sugars into ethanol (Xu et al. 2009).
Fig. 1.
The steps involved in the bioprocessing to lignocellulose to ethanol. Following chemical pretreatment to break down the tough, recalcitrant material in lignocellulosic biomass, it is more susceptible to enzymatic attack because of the exposure of the underlying carbohydrates (i.e., cellulose and hemicelluloses). Thereafter, there are four possible routes to ethanol production; SHF separate hydrolysis and fermentation, SSF simultaneous saccharification and fermentation, SSCF simultaneous saccharification and co-fermentation, CBP consolidated bioprocessing. Microbial enzymes produced by bacteria, fungi and other micro-organisms are used to convert the exposed cellulose and hemicellulose sugar polymers to simple sugars which can then be efficiently fermented; in all cases except CBP, these enzymes need to be added (the CBP organism(s) both saccharify and ferment the substrate). Microbial fermentation is the final phase in the bioconversion process. By this stage, the hydrolysate contains a mixture of hexose and pentose sugars such as glucose, xylose, mannose, fructose, galactose and arabinose, which are all fermentable by micro-organisms to produce bioethanol
Lignocellulosic bioethanol and current bottlenecks
Lignocellulosic material is the most abundant source of biomass on earth and includes wood, grasses, agricultural residues or any non-food-plant sources. The fermentable sugars are derived via the hydrolysis of the cellulose and hemicellulose components, which account for approximately 60 % of lignocellulosic materials, while lignin accounts for 15–25 % (Dionisi et al. 2015; Menon and Rao 2012; Wyman 1994). Lignocellulose offers several benefits over sugar and starch as a substrate for bioethanol production (Morales et al. 2015; Solomon et al. 2007). Most importantly, when lignocellulose is derived from non-food parts of plants, and as long as land use is not diverted to lignocellulose production, it does not interfere with food security (if the land use pattern is not changed).
Low carbon biofuels from commercial-scale cellulosic ethanol has become a reality in recent times. Numerous cellulosic ethanol refineries have now come online worldwide with several more in the pipeline (European Biofuels Technology Platform 2015). To date, the largest cellulosic ethanol industrial-scale refinery is the Beta Renewables/Novozyme funded plant situated at Crescentino in north-western Italy which commenced operations in October 2013. The facility is entirely self-sufficient, using the lignin and biogas by-products to power the plant which generates 75 million litres annually of cellulosic ethanol, enough fuel for more than 50,000 cars. A sister plant in Strazke, Slovak Republic is currently under construction while in Fuyang, China a biorefinery four times the capacity of the Crescentino plant is under development. In the US, there are over 200 corn-based ethanol plants in operation (Gnansounou 2010). Many of these bioethanol plants are evolving to become cellulosic ethanol production facilities utilising cheaper agricultural residues and non-food substrates. In October 2014 the Spanish renewable energy giant, Abengoa officially opened its 25 million gallons per year ethanol commercial biorefinery at Hugoton, Kansas. A similar-scaled plant financed through a collaborative venture between chemical group Du Pont and Murex LLC is also nearing completion in Nevada, Iowa. Encouraging yields ranging from 68 to 83 gallons per tonne of biomass have recently been reported by several bioenergy groups such as Abengoa Bioenergy, Iogen Energy and Poet, LLC from their respective pilot cellulosic ethanol plants (Guo et al. 2015). With the inevitable upsurge in oil price back to pre-2014 levels an unavoidable reality allied with advancements in the relevant technology, industrial-scale lignocellulosic bioethanol will continue to spread worldwide in the near future.
The nature of lignocellulose leads to several bottlenecks in improving the commercial viability of second generation bioethanol. These include the substrate recalcitrance to digestion, the lack of fermentability of some of the sugars released via hydrolysis, the substrate heterogeneity and the transportation costs. The transportation cost can be reduced by building bioethanol refineries close to biomass production sites (Eggeman and Elander 2005). The heterogeneity in both the chemistry and structure of lignocellulose means that it is often difficult to optimise the production process in terms of pretreatment, enzymatic hydrolysis and fermentation to suit the various types of lignocellulosic biomass. The pretreatment of lignocellulosic biomass to reveal the cellulose and hemicellulose is very expensive and contributes up to 33 % of the overall costs of producing lignocellulosic bioethanol (Behera et al. 2014; Tomás-Pejó et al. 2009). Hence the focus on the genetic manipulation of the lignin biosynthetic pathway in plants in order to improve bioethanol yields from lignocellulosic materials (Fu et al. 2011; Hopkins et al. 2007). Traditional breeding can also be used in order to breed plant genotypes with altered composition/degradability, as demonstrated for wheat (Ali et al. 2012a). The micro-organisms used in the fermentation process to date are incompetent at efficiently co-fermenting the variety of sugars released from lignocellulosic materials. Many microbial strains used in bioethanol production, including Saccharomyces cerevisiae, can successfully ferment glucose and other hexose sugars but struggle to adequately ferment pentose sugars such as xylose and arabinose (Gírio et al. 2010; Hahn-hägerdal et al. 2007; Hector et al. 2008). Xylose is the second most common sugar found in plants and the inability of current microbial strains to successfully ferment xylose is an immensely limiting factor. While several yeast strains have been genetically modified for improved xylose fermentation (Zha et al. 2014; Lee et al. 2012; Chu and Lee 2007; Eliasson et al. 2000; Ho et al. 1998; Kuyper et al. 2005; Laluce et al. 2012), a naturally-occurring yeast or fungal strain equipped with an independent xylose transporter has yet to be found (Hector et al. 2008). In the race towards making the lignocellulosic bioethanol economically viable, some researchers are using directed mutagenesis to improve the activity of enzymes in microbes currently used for bioconversion; others are trying to build the ultimate microbe in the laboratory that exhibits high efficiency in the processes of both saccharification and fermentation leading to CBP; a few research teams are hunting for new microbes with enhanced hydrolytic capacities; others are exploring the possibility of incorporating advance non-biological steps into the process (Lynd et al. 2008).
Consolidated bioprocessing (CBP)
CBP employs microbes to perform all the four biologically-mediated transformations viz. the production of saccharolytic enzymes, the hydrolysis of carbohydrate components present in biomass to simple sugars, the fermentation of hexose sugars and the fermentation of pentose sugars in a single step (Fig. 1) (Lynd et al. 2005; Amore and Faraco 2012; Schuster and Chinn 2013). Thus, it offers the potential for lowering the cost and enhancing the efficiency of bioethanol production, as compared to independent hydrolysis and fermentation steps. Independent hydrolysis and fermentation can lead to end-product inhibition by sugars and acetates produced via hydrolysis (Wright et al. 1988). These inhibitors disrupt the fermentation capabilities of the ethanologen and result in substantial yield reductions. CBP might eliminate problems attributed with glucose accumulation and other such inhibitors (Olofsson et al. 2008; Dashtban et al. 2009). Another advantage of using CBP is that glucose does not need to be separated from the lignin fraction following the hydrolysis step, thus minimising sugar loss (Olofsson et al. 2008).
Increasing evidence suggests that CBP may be feasible (Ali et al. 2012b, 2013, 2014; Xu et al. 2009; Xiros and Christakopoulos 2009; Bokinsky et al. 2011; Hyeon et al. 2011; Okamoto et al. 2014). Ever since the concept of CBP was proposed in 1996, CBP research has focused on the development of new and even more effective CBP microorganisms, which has been a key challenge (Lynd et al. 2005). Bacteria and yeast have been the primary candidates for CBP research and some progress has been made in this regard (Lynd et al. 2005; Amore and Faraco 2012; den Haan et al. 2015). Fungi have not been widely proposed as CBP microorganisms, but there are a few recent reports of researchers developing strains of the fungi Fusarium oxysporum and Trichoderma reesei with enhanced CBP potential (Ali et al. 2012b, 2013, 2014; Xiros and Christakopoulos 2009; Xu et al. 2009; Huang et al. 2014).
There are several drawbacks associated with CBP. Microbial growth, enzymatic hydrolysis and the fermentation phases are carried out synchronically and it is very difficult to find culture conditions that are optimal for all these processes. Traditionally, proponents of CBP processes have identified two approaches capable of producing industrially-viable microbial agents for CBP. These are: (i) engineering a cellulase producer, such as Clostridium thermocellum, to be ethanologenic, and (ii) engineering an ethanologen, such as S. cerevisiae or Zymomonas mobilis, to be cellulolytic (Xu et al. 2009). Efforts have so far been focused mainly on the second approach. The bacteria Z. mobilis (Linger et al. 2010; Luo and Bao 2015), Escherichia coli (Ingram et al. 1999; Tao et al. 2001; Ko et al. 2013) and Klebsiella oxytoca (Tran et al. 2011; Wood et al. 2005), and the yeasts S. Cerevisiae (Hong et al. 2014; Xu et al. 2014; van Zyl et al. 2007), Pachysolen tannophilus (Slininger et al. 1987), Pichia stipitis and Candida shehatae (Prior et al. 1989) have been modified in order to improve their performance in CBP. But there are various difficulties and challenges in the conversion of a candidate microorganism using gene transfer technology. These include the adverse effects of the co-expression of multiple heterologous genes on cell performance, the modulation of simultaneous co-expression of multiple genes at the transcription level and improper folding of some secretory proteins (Xu et al. 2009). For instance several studies have found that the heterologous expression of cellobiohydrolases I and II in S. cerevisiae resulted in poor levels of conversion of crystalline cellulose degradation (den Haan et al. 2007; Hong et al. 2003; Chow et al. 1994). Thus, the search for a native CBP agent is very much essential and their study will also increase the arsenal of genes available for genetic manipulation of microbes currently being developed for CBP.
The potential of filamentous fungi as CBP agents
Despite recent advances in engineering cellulases to be more efficient and less costly, the complete saccharification of lignocellulose still requires a very long time for digestion and high loadings of enzyme (30–50 mg enzyme g−1 of crystalline cellulose) (Xu et al. 2009). Thus, a biorefinery consuming thousands of tons of biomass per day will require many tons of cellulase enzymes to operate. Only fungi appear to be able to produce sufficient amounts of cellulase to meet this need. For example, T. reesei is reported to be able to produce more than 100 g cellulase enzyme per litre of culture broth (Cherry and Fidantsef 2003; Vitikainen et al. 2010). In comparison, the most productive cellulolytic bacteria produce only a few grams per litre (Xu et al. 2009). The ability to produce and secrete enzymes of complex structure, such as the cellobiohydrolase I, requires a robust secretion system, including the endoplasmic reticulum and the golgi complex within the cytosol of the cell (Xu et al. 2009). Bacteria do not normally have such systems and it may be difficult or impossible to engineer these organisms to produce cellulolytic enzymes in sufficient quantities for a biorefinery (Xu et al. 2009).
Microorganisms used in the CBP of lignocellulose to bioethanol must display high levels of alcohol/sugar tolerance, thermotolerance and tolerance to other inhibitors that may be produced as a result of either pretreatment or the CBP process itself. An ability to utilise multiple sugars is also a prerequisite. The proposed use of filamentous fungi in the CBP process goes a long way towards satisfying these requirements. T. reesei is considered to be a one of the best CBP fungi due to its capacity to produce and secrete lignocellulolytic enzymes. The extensive knowledge of its physiology and cellulolytic machinery (Kubicek et al. 2009; Schmoll et al. 2010; Silva-Rocha et al. 2014) and the availability of a range of tools for its genetic manipulation enhance its preference as a CBP organism (Kück and Hoff 2010). Yet, T. reesei also presents some challenges that must be addressed before it can become an efficient CBP organism. Its main limitations are the low ethanol yield and rate of production, low ethanol tolerance, and difficulties during fermentation associated with its cell morphology (Xu et al. 2009). Another restriction is the fact that genes encoding enzymes crucial for glycolysis are repressed in the absence of oxygen (Bonaccorsi et al. 2006), which limits its growth without oxygen. Xu et al. (2009) demonstrated that T. reesei can survive, but not thrive, and convert soluble sugars to ethanol under anaerobic or microaerobic conditions. In addition to T. reesei, there are alternative fungi presenting a significant potential to become a CBP organism. These include Fusarium oxysporum (Ali et al. 2012b; Christakopoulos et al. 1989), Mucor indicus (Karimi et al. 2006), Monilia sp. (Gong et al. 1981), Rhizopus oryzae (Karimi et al. 2006; Battaglia et al. 2011), Paecilomyces sp. (Wu 1989), Aspergillus oryzae (Skory et al. 1997), Neocallimastix patriciarum (Wang et al. 2011) and Neurospora crassa (Dogaris et al. 2009). These can all produce bioethanol from cellulosic material. Most of these filamentous fungi possess the ability to assimilate and metabolise numerous sugars, both hexose and pentose types (Taherzadeh and Karimi 2007). Furthermore, these fungi have a greater degree of thermotolerance than many bacteria and can grow at 37 °C, which is closer to the optimal temperature of enzymatic hydrolysis (40–50 °C) (Millati et al. 2005). This advantage was considered to improve theoretical ethanol yields by as much as 15 % compared to S. cerevisae (enzymatic hydrolysis followed by fermentation) when using M. indicus and R. oryzae in CBP of dilute-acid pretreated rice straw (Karimi et al. 2006). Bioethanol production from cellulosic biomass by various filamentous fungi has been summarised in Table 1.
Table 1.
The potential of filamentous fungi as CBP agents
| Organism | Number of strains tested | Theoretical ethanol yield from biomass (%)a | Reference | |||
|---|---|---|---|---|---|---|
| Cellulose | Other lignocellulose | Glucose | Xylose | |||
| Aspergillus awamori | 2 | 1.5–1.1 | – | 28.2–23.5 | 5.4–3.5 | Skory et al. (1997) |
| A. foetidus | 1 | 1.1 | – | 20.3 | 13.3 | Skory et al. (1997) |
| A. niger | 1 | 0.7 | – | 22.3 | 4.7 | Skory et al. (1997) |
| A. oryzae | 4 | 3.1–2.3 | – | 95.6–62.3 | 18.4–10.1 | Skory et al. (1997) |
| A. sojae | 6 | 1.5–2.7 | – | 56.4–31.3 | 21.1–8.2 | Skory et al. (1997) |
| A. tamari | 5 | 2.3–0.3 | – | 72.9–38.4 | 13.7–11.3 | Skory et al. (1997) |
| Fusarium oxysporum | 3 | 89.2 (alkali-treated) | – | 80 | 48 | Christakopoulos et al. (1989) |
| F. oxysporum | 1 | – | 83 (ball milled wheat straw); 67 (alkali treated wheat straw) | – | – | Christakopoulos et al. (1991a, b) |
| F. oxysporum | 17 | 19.7 (untreated wheat straw); 80.2 (alkali treated wheat straw) | Ali et al. (2012b) | |||
| Flammulina velutipes | 1 | – | 57.2 (sweet sorghums) | – | – | Mizuno et al. (2009) |
| Fomitopsis palustris | 1 | 88.2 (cellobiose) | – | 90.2 | – | Okamoto et al. (2011b) |
| Gloeophyllum trabeum | 1 | 10.4 (pretreated corn fiber) | – | – | Rasmussen et al. (2010) | |
| Mucor indicus | 1 | 61 (avicel) | 68 (rice straw) | – | – | Karimi et al. (2006) |
| Monilia sp. | 1 | 60 | – | – | – | Gong et al. (1981) |
| Mucor hiemalis | 1 | – | 80 (pretreated sweet sorghum bagasse) | – | – | Goshadrou et al. (2011) |
| Neurospora crassa | 1 | 60 | – | – | – | Rao et al. (1983) |
| N. crassa | 1 | 100 (avicel) 91 (alkali-treated) | 54 (alkali treated sugarcane bagasse) | 96.9 | 64.2 | Deshpande et al. (1986) |
| N. crassa | 1 | – | 11.76 (sorghum bagasse) | – | – | Dogaris et al. (2009) |
| Phanerochaete chrysosporium | 1 | – | 6.8 (corn fiber) | – | – | Shrestha et al. (2010) |
| Phlebia sp. | 1 | 65.7 (alkali treated sugarcane bagasse) | Khuong et al. (2014) | |||
| Paecilomyces sp. | 1 | 18 (wheat bran and brewers spent grain mix) | 61.8 | 82 | Zerva et al. (2014) | |
| Paecilomyces sp. | 1 | 78.4 | – | – | 78 | Wu (1989) |
| Rhizopus javanicus | 3 | 5–1.9 | – | 92.1–48.6 | 45.8–6.6 | Skory et al. (1997) |
| R. oryzae | 6 | 5.4–.7 | – | 99.5–58.4 | 42.3–3.9 | Skory et al. (1997) |
| R. oryzae | 1 | 76 (avicel) | 74 (rice straw) | – | – | Karimi et al. (2006) |
| Trichoderma harzianum | 1 | 7.8 | – | 19.6 | 2.3 | Stevenson and Weimer (2002) |
| T. reesei | 1 | – | 14.5 (corn fiber) | – | – | Shrestha et al. (2010) |
| Trametes hirsuta | 1 | – | 78.8 (wheat bran), 57.4 (rice straw) | 96 | 86.2 | Okamoto et al. (2011a) |
aMaximum theoretical yield was calculated based on the fact that 0.51 g ethanol and 0.49 g of CO2 are yielded from 1 g of glucose. The % theoretical yield was calculated based on the sugar content of lignocellulosic material. When sugar content was not mentioned in the original reference, it is calculated based on average composition of lignocellulose which can theoretically produce 0.336 g ethanol g−1 of biomass (Szczodrak 1988)
Fusarium oxysporum as a potent CBP agent
Fusarium oxysporum is a filamentous soil-borne fungus that is more widely regarded as a phytopathogen responsible for vascular wilt disease in a variety of different plant species or as a mycotoxin-producing contaminant of human and animal food (Di Pietro et al. 2003). The majority of crops cultivated worldwide are hosts to a pathogenic form of F. oxysporum. It is among the handful of microbial species that are known to have the enzymatic systems needed to break down cellulose and hemicellulose and to ferment the released hexoses and pentose sugars to bioethanol in a single step (Ali et al. 2012b; Christakopoulos et al. 1989; Panagiotou et al. 2005b; Singh et al. 1992; Suihko 1983). These and other associated characteristics render it relatively efficient for the CBP of lignocellulose to bioethanol. This ability stems from the relatively high levels of cellulases and xylanases produced by F. oxysporum (Christakopoulos et al. 1989, 1996a, b; Panagiotou et al. 2003). Fusarium oxysporum also exhibits high levels of tolerance to sugars, ethanol and inhibitors such as acetate (Hennessy et al. 2013; Singh and Kumar 1991). Fusarium oxysporum-mediated fermentation of glucose is unaffected until ethanol concentrations reach 4.5–5.0 % in the reactor (Enari and Suihko 1983). Acetate is a major inhibitory compound produced by microbes during the ethanol fermentation process, but growing cells of F. oxysporum are capable of reducing acetate to ethanol (Enari and Suihko 1983).
Fusarium oxysporum requires an aerobic growth phase (for initial fungal growth) followed by an oxygen-limited fermentation phase in order to produce ethanol from glucose (Panagiotou et al. 2005a). To optimise the performance of F. oxysporum during the fermentation phase, a limited oxygen supply of 18–20 % is required (Panagiotou et al. 2005d). Such level of oxygen will facilitate growth and the breakdown of acetate by growing cells (Enari and Suihko 1983). As shown in Table 1, the highest reported bioethanol yield from any unprocessed lignocellulosic material by a filamentous fungus to date was achieved by F. oxysporum strain 11C, producing up to 80 mg bioethanol g−1 wheat straw/bran (Ali et al. 2012b). With alkali-treated straw as the substrate, the yield increased to 326 mg bioethanol g−1 wheat straw/bran, a theoretical yield of 80.2 %. Although still below industrial exploitable yields, this yield is higher than that reported for other fungi grown on pre-treated agricultural waste (Deshpande et al. 1986; Mizuno et al. 2009; Karimi et al. 2006; Okamoto et al. 2011a, b; Goshadrou et al. 2011). Interestingly, the ability of F. oxysporum to produce ethanol during CBP was found not be reflective of either cellulose or alcohol dehydrogenase (ADH) activity (Ali et al. 2012b). Explorative work by Ali et al. (2014) identified a large consortium of F. oxysporum genes (<210) that were upregulated in F. oxysporum strain 11C and may have conferred this strain with an enhanced capacity for bioethanol production from lignocellulosic residues. These genes were identified by comparing the transcriptome of F. oxysporum strain 11C with that of a low efficacy F. oxysporum strain. Many of the genes identified and their encoded proteins could be assigned to various categories such as carbohydrate metabolism, energy, protein and sugar transport and detoxification. Furthermore, a consortium of novel genes were discovered that had no direct link to saccharification or fermentation but were shown to be activated during CBP, highlighting the complexity of the process (Ali et al. 2014). Another promising strain, F. oxysporum strain F3, has been reported to directly convert alkali-treated and ball-milled wheat straw to bioethanol, producing 67.8 and 83 % of theoretical yield, respectively (Christakopoulos et al. 1991a, b). The same strain has been reported to yield 109 g bioethanol kg−1 of dry alkali- pretreated brewer’s spent grain under microaerobic conditions, corresponding to 60 % of the theoretical yield (Xiros and Christakopoulos 2009). Further studies were done with this particular F. oxysporum strain to saccharify and co-ferment wheat straw with S. cerevisiae (Panagiotou et al. 2011); yields of bioethanol were very promising (40 g kg−1 of pretreated wheat straw).
The CBP metabolic pathway of F. oxysporum
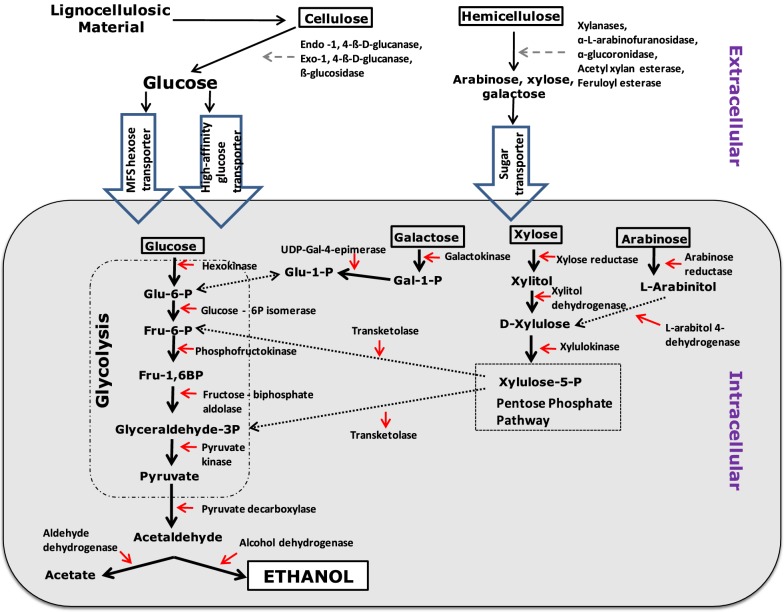
During CBP the first metabolic step for any microbial agent is the production of cellulases and hemicellulases followed by extracellular secretion of these enzymes for the purpose of substrate hydrolysis. Celluloytic and hemicelluloytic enzymes produced by F. oxysporum are used to breakdown cellulose and hemicellulose to their simple sugar derivatives which can then be imported into the cell by sugar transporters. Glucose enters glycolysis on immediate arrival inside the cell. Conversely, the hemicellulosic sugars (xylose, galactose, arabinose) must enter alternative metabolic pathways where they are enzymatically converted to glycolytic intermediates, allowing them to enter glycolysis and undergo subsequent fermentation to bioethanol (Fig. 2). Cellulase is a multienzyme complex consisting of endo-l,4-β-d-glucanase, exo-l,4-β-d-glucanase and β-glucosidase. Synergistic activity of these enzymes is required for the complete hydrolysis of insoluble cellulose (Halliwell 1961). Fusarium oxysporum has been shown to produce all three of these enzymes (Ali et al. 2012b; Christakopoulos et al. 1989; Kumar et al. 1991). Cellulase production by F. oxysporum is a function of the level of the soluble hydrolysis products (glucose and cellobiose) that can be utilised by the fungal cells; this in turn is dependent on the rate of adsorption of endoglucanase onto the cellulose and rate of hydrolysis of this substrate (Targonski and Szajer 1979; Singh et al. 1991).
Fig. 2.
The metabolic pathway involved in the bioconversion of lignocellulosic material to bioethanol by Fusarium oxysporum. Glu-6-P Glucose-6-Phosphate, Fru-6-P Fructose-6-Phosphate, Fru-1, 6BP fructose-1, 6-biphosphate, glyceraldehyde-3P glyceraldehyde-3-phosphate, Glu-1-P glucose-1-phosphate, Gal-1-P galactose-1-phosphate, Xylulose-5-P xylulose-5-phosphate
Three endoglucanases and one β-glucosidase from F. oxysporum strain F3 has been purified and fully characterised (Christakopoulos et al. 1994, 1995a, b, c). The significantly higher β-glucosidase activity of this particular strain prevented the inhibitory effect of cellobiose on cellulase activity (Panagiotou et al. 2005b) and a low molecular mass endoglucanase may have played an important role in cellulose degradation as it can penetrate through the cellulose fibre easily (Christakopoulos et al. 1995c). Interestingly, in the case of F. oxysporum strain 11C, the capacity to produce ethanol during fungal CBP was not linked to cellulase activity (Ali et al. 2012b). Alternatively, a large consortium of up-regulated genes encoding proteins assigned to various categories, including carbohydrate metabolism, energy, protein and sugar transport and detoxification were identified as likely factors in F. oxysporum-mediated CBP (Ali et al. 2014).
The variable structure and organisation of hemicellulose necessitates the concerted action of many enzymes for its complete degradation. The catalytic modules of hemicellulases are either glycoside hydrolases (GHs) that hydrolyse glycosidic bonds, or carbohydrate esterases (CEs), which hydrolyse ester linkages of acetate or ferulic acid side groups (Shallom and Shoham 2003). Shallom and Shoham (2003) described thirteen different enzymes required for the complete hydrolysis of hemicellulose; various hemicullases from F. oxysporum has been extensively studied and this strain can produce most of the enzymes required for the complete hydrolysis of hemicellulose (Singh et al. 1995; Christakopoulos et al. 1996a; Cheilas et al. 2000; Panagiotou et al. 2003; Anasontzis et al. 2011; Ali et al. 2012b, 2013).
Sugar transport across the cell membrane is the first step in the metabolism of sugars and this occurs through facilitated diffusion (Jeffries 1983; Kim et al. 2003). In yeast there are twenty different glucose transporter genes and expression of these transporters are regulated by the glucose concentration (Ali et al. 2013; Ozcan and Johnston 1995). Brandao and Loureiro-Dias (1990) observed that sugar transport of F. oxysporum was under the same regulatory mechanism as that of yeast and other eukaryotic microorganisms. It has been observed that the rate of sugar transport determines the rate of anaerobic fermentation and ethanol production/tolerance in yeast (Leão and Uden 1985; Alexandre et al. 2001) and Fusarium (Ali et al. 2013; Hennessy et al. 2013). Indeed, manipulation of a Hxt gene encoding for a high affinity glucose transporter in F. oxysporum was found to have a profound effect on the overall activity and productivity of the fungus during CBP, with Hxt overexpression mutants achieving the highest yield to date of a fungus from pre-treated agricultural waste (Ali et al. 2013). Transport of d-xylose has been found to be related to d-glucose transport, since the influx of d-xylose or d-arabinose is more rapid in the presence of glucose under anaerobic conditions than under aerobic conditions (Jeffries 1983). Concomitantly, it was also observed that overexpression of the Hxt gene in F. oxysporum increased not only glucose uptake rates but likewise xylose uptake rates resulting in enhanced fermentation rates (Ali et al. 2013). In regards to tolerance, transcriptional studies have revealed the functional role filamentous fungal sugar transporters play in times of alcohol stress (Alexandre et al. 2001; Chandler et al. 2004; Reyes et al. 2011; Hennessy et al. 2013).
In yeast and fungi, glucose is metabolised to ethanol via the Embden-Meyerhof-Parnas (EMP) pathway (Rose and Harrison 1970). Theoretically, 0.51 g ethanol and 0.49 g of CO2 are yielded from 1 g of glucose. However, the real ethanol and CO2 yields are 0.46 g and 0.44 g respectively, since 0.10 g of glucose is metabolised for biomass production (Singh and Kumar 1991). In the case of F. oxysporum, the highest bioethanol yield to date from glucose was recorded as 0.409 gg−1 (Panagiotou et al. 2005d). Panagiotou et al. (2005b) has analysed the intracellular metabolic profile of F. oxysporum strain F3 under aerobic and anaerobic conditions. He observed that there was a high glycolytic flux under anaerobic growth conditions, characterised by a high efflux of glyceraldehyde-3-phosphate (G3P) and fructose-6-phosphate from the pentose phosphate pathway (PPP) to the EMP pathway, resulting in the highest bioethanol production under these conditions. The amino acid profile of fermenting F. oxysporum cells clearly suggested that the TCA cycle was primarily active under aerobic cultivation. On the other hand, the presence of high levels of γ-amino-n-butyric acid (GABA) under anaerobic conditions suggested a functional GABA bypass and a possible block in the TCA cycle under these conditions (Panagiotou et al. 2005d). An accumulation of sedoheptulose-7-P was observed indicating a barrier in the PPP that affects the production of NADPH and results in the production of acetate. Furthermore, the presence of high levels of GABA under anaerobic conditions suggest a functional GABA bypass and a possible block in the TCA cycle, which may also contribute to acetate production (Panagiotou et al. 2005d). It was interesting to observe that F. oxysporum does not co-metabolise xylose with glucose during anaerobic growth and starts only after glucose exhaustion (Panagiotou et al. 2005c). Under anaerobic conditions xylitol is the main product of xylose metabolism (Panagiotou et al. 2005c). Therefore, it can be predicted that a metabolic engineering approach to block the acetate and xylitol pathway will dramatically boost bioethanol yielded via CBP by F. oxysporum.
A roadmap for enhanced CBP activity
Fusarium oxysporum is known for its slow rate of growth which is a major drawback for use as a CBP ethanologenic agent as the microorganism takes longer to reach the required critical mass before the transition to anaerobic conditions for fermentation. Quickening the maximum specific growth rate of F. oxysporum has been successfully achieved recently through the overexpression of both phosphoglucomutase and transaldolase (Anasontzis and Christakopoulos 2014). Further improvements to speed up the growth rate of F. oxysporum would be a major breakthrough in the quest towards designing the ultimate CBP microbial agent. As F. oxysporum and other CBP agents are already adept at fermenting the cellulosic fraction of plant biomass, the greatest gains in bioethanol yield will likely come from increased catabolism of the under-utilised hemicellulosic fraction. Therefore, the tactical co-expression of hemicelluloytic genes in a designer strain of F. oxysporum such as the gene encoding for a α-l-arabinofuranosidase (AbfB) (Ali et al. 2014) involved in degradation of hemicellulose to its simple sugar derivatives with the previously highlighted Hxt gene (involved in sugar transport) (Ali et al. 2013) could drastically improve hemicellulosic fermentation rates. Development of a designer strain with the ability to metabolise all hemicellulosic sugars would represent a significant breakthrough for the lignocellulosic bioethanol industry.
During CBP, F. oxysporum produces large amounts of acetic acid, an unwanted by-product of the fermentation process that impedes the bioethanol productivity of the fungus. Higher transcript levels of a gene encoding acetyl-CoA hydrolase has been observed in F. oxysporum strain 11C before which is likely associated with the increased acetic acid production of the strain (Ali et al. 2012b, 2014). The targeted disruption of this gene and other genes that are involved in by-product formation could serve to further enhance F. oxysporum’s CBP efficacy to minimize the production of undesirable by-products such as acetic acid during lignocelluloytic bioconversion.
Fusarium oxysporum is a species with high morphological and physiological variation and its ubiquitous presence in ecological activities worldwide indicate a highly diverse and important role in nature. Though the fungus is highly successful as a saprophyte in soil, as stated earlier, most of the interest in it arises from its ability to cause detrimental diseases on various economically important crops. Both plant pathogenic and non-pathogenic isolates could provide a rich source of industrial enzyme sources for bioethanol production (King et al. 2011). Unlike the pathogenic strains, very little is known about the genetic and molecular variation among the non-pathogenic F. oxysporum strains and these could potentially serve as a rich source of plant-degrading enzymes. Ali et al. (2012b, 2013) observed significant inter-strain divergence in regards to the capacity of different F. oxysporum strains to produce alcohol from wheat straw and identified 210 transcripts encoding proteins assigned to various categories, including carbohydrate metabolism, energy, protein and sugar transport and detoxification that were overexpressed in a high as compared to low efficacy F. oxysporum strains during CBP. The level of diversity recorded in the bioethanol production capacity among the isolates means that a targeted screening of populations of selected isolates could greatly improve bioprocessing yields, in terms of providing both new host strains and candidate genes for the bioethanol industry.
Conclusion
Under the present predicted scenario of a food and fuel crisis, coupled with global warming, cellulosic bioethanol shows promise as an alternative to petroleum. Filamentous fungi such as F. oxysporum could be commercially competitive CBP agents, or components of their enzymatic arsenal could contribute to the development of a ‘designer’ CBP agent. CBP needs much robust microbial agents in order to bring this process to the level of industrial expectations. The most efficient CBP of lignocellulose will most likely be achieved using a consortium of enzymes contributed by several microbes that are either working in unison or as donors of genes pyramided into one or more ‘designer’ organisms. Approach for such a designer agent should include (i) increasing the ethanol yield, (ii) eliminating by-products, (iii) improving the tolerance to ethanol, and (iv) introduction of new metabolic pathways for assimilating lignocellulose sugars. Understanding all these components and the complexity of the networks involved will be important in selecting such a consortia or designing such an organism. A functional genomic or proteomic approach could help unfold the networks involved and would open up many more avenues for the improvement of CBP agents. Then synthetic biology can provide new tools to rewire the cell components (promoters, regulators, terminators, enzymes, operons, transporters, etc.) in order to reach the desired features for the production of economically viable biofuels. Although some successful examples were already reported in bacteria and yeast, a crucial remaining challenge is to apply these approaches in fungi, which have a tremendous potential since they are the effective producers of critical cellulases.
Acknowledgements
Shahin Ali was supported by the Irish Department of Agriculture, Fisheries and Food Research Stimulus Fund (RSF 07 513) and Brian Nugent was supported by the Earth and Natural Sciences (ENS) Doctoral Studies Programme. The ENS programme is funded by the Higher Education Authority (HEA) through the Programme for Research at Third Level Education, Cycle 5 (PRTLI-5) and is co-funded by the European Regional Development Fund (ERDF). The authors thank Bredagh Moran and Brian Fagan for technical assistance.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Shahin S. Ali and Brian Nugent contributed equally to this work
Contributor Information
Shahin S. Ali, Email: shahinsharif.ali@gmail.com
Brian Nugent, Email: briannugent88@gmail.com.
Ewen Mullins, Email: ewen.mullins@teagasc.ie.
Fiona M. Doohan, Email: fiona.doohan@ucd.ie
References
- Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 2001;498:98–103. doi: 10.1016/S0014-5793(01)02503-0. [DOI] [PubMed] [Google Scholar]
- Ali SS, Khan M, Mullins E, Doohan FM. Identification of Fusarium oxysporum genes associated with lignocellulose bioconversion competency. Bioenergy Res. 2014;7:110–119. doi: 10.1007/s12155-013-9353-0. [DOI] [Google Scholar]
- Ali SS, Khan M, Mullins E, Doohan FM. The effect of wheat genotype on ethanol production from straw and the implications for multifunctional crop breeding. Biomass Bioenergy. 2012;42:1–9. doi: 10.1016/j.biombioe.2012.03.020. [DOI] [Google Scholar]
- Ali SS, Khan M, Mullins E, Fagan B, Doohan FM. Exploiting the inter-strain divergence of Fusarium oxysporum for microbial bioprocessing of lignocellulose to bioethanol. AMB Express. 2012;2:16. doi: 10.1186/2191-0855-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SS, Nugent B, Mullins E, Fagan B, Doohan FM. Insights from the fungus Fusarium oxysporum point to high affinity glucose transporters as targets for enhancing ethanol production from lignocellulose. PLoS ONE. 2013;8:e54701. doi: 10.1371/journal.pone.0054701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amore A, Faraco V. Potential of fungi as category I consolidated BioProcessing organisms for cellulosic ethanol production. Renew Sustain Energy Rev. 2012;16:3286–3301. doi: 10.1016/j.rser.2012.02.050. [DOI] [Google Scholar]
- Anasontzis GE, Christakopoulos P. Challenges in ethanol production with Fusarium oxysporum through consolidated bioprocessing. Bioengineered. 2014;5:1–3. doi: 10.4161/bioe.36328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anasontzis GE, Zerva A, Stathopoulou PM, Haralampidis K, Diallinas G, Karagouni AD, Hatzinikolaou DG. Homologous overexpression of xylanase in Fusarium oxysporum increases ethanol productivity during consolidated bioprocessing (CBP) of lignocellulosics. J Biotechnol. 2011;152:16–23. doi: 10.1016/j.jbiotec.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Balat M. Production of bioethanol from lignocellulosic materials via the biochemical pathway: a review. Energ Convers Manage. 2011;52:858–875. doi: 10.1016/j.enconman.2010.08.013. [DOI] [Google Scholar]
- Balan V, Chiaramonti D, Kumar S. Review of US and EU initiatives toward development, demonstration, and commercialization of lignocellulosic biofuels. Biofuel Bioprod Bior. 2013;7:732–759. doi: 10.1002/bbb.1436. [DOI] [Google Scholar]
- Banerjee S, Mudliar S, Sen R, Giri B, Satpute D, Chakrabarti T, Pandey RA. Commercializing lignocellulosic bioethanol: Technology bottlenecks and possible remedies. Biofuel Bioprod Bior. 2010;4:77–93. doi: 10.1002/bbb.188. [DOI] [Google Scholar]
- Battaglia E, Benoit I, van den Brink J, Wiebenga A, Coutinho PM, Henrissat B, de Vries RP. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genom. 2011;12:38. doi: 10.1186/1471-2164-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S, Arora R, Nandhagopal N, Kumar S. Importance of chemical pretreatment for bioconversion of lignocellulosic biomass. Renew Sustain Energy Rev. 2014;36:91–106. doi: 10.1016/j.rser.2014.04.047. [DOI] [Google Scholar]
- Bentsen NS, Felby C, Thorsen BJ. Agricultural residue production and potentials for energy and materials services. Prog Energy Combust Sci. 2014;40:59–73. doi: 10.1016/j.pecs.2013.09.003. [DOI] [Google Scholar]
- Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Soon Lee T, Tullman-Ercek D, Voigt CA, Simmons BA, Keasling JD. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci USA. 2011;108:19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi ED, Ferreira AJS, Chambergo FS, Ramos ASP, Mantovani MC, Simon Farah JP, Sorio CS, Gombert AK, Tonso A, El-Dorry H. Transcriptional response of the obligatory aerobe Trichoderma reesei to hypoxia and transient anoxia: implications for energy production and survival in the absence of oxygen. Biochemistry. 2006;45:3912–3924. doi: 10.1021/bi052045o. [DOI] [PubMed] [Google Scholar]
- Brandao RL, Loureiro-Dias MC. Regulation of sugar transport systems in Fusarium oxysporum var. lini. Appl Environ Microbiol. 1990;56:2417–2420. doi: 10.1128/aem.56.8.2417-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brethauer S, Studer MH. Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ Sci. 2014;7:1446. doi: 10.1039/c3ee41753k. [DOI] [Google Scholar]
- Chandler M, Stanley G, Rogers P, Chambers P. A genomic approach to defining the ethanol stress response in the yeast Saccharomyces cerevisiae. Ann Microbiol. 2004;54:427–454. [Google Scholar]
- Cheilas T, Stoupis T, Christakopoulos P, Katapodis P, Mamma D, Hatzinikolaou DG, Kekos D, Macris BJ. Hemicellulolytic activity of Fusarium oxysporum grown on sugar beet pulp. Production of extracellular arabinanase. Process Biochem. 2000;35:557–561. doi: 10.1016/S0032-9592(99)00103-X. [DOI] [Google Scholar]
- Cherry JR, Fidantsef AL. Directed evolution of industrial enzymes: an update. Curr Opin Biotechnol. 2003;14:438–443. doi: 10.1016/S0958-1669(03)00099-5. [DOI] [PubMed] [Google Scholar]
- Christakopoulos P, Goodenough PW, Kekos D, Macris BJ, Claeyssens M, Bhat MK. Purification and characterisation of an extracellular β-glucosidase with transglycosylation and exo-glucosidase activities from Fusarium oxysporum. Eur J Biochem. 1994;224:379–385. doi: 10.1111/j.1432-1033.1994.00379.x. [DOI] [PubMed] [Google Scholar]
- Christakopoulos P, Kekos D, Kolisis FN, Macris BJ. Controlling simultaneous production of endoglucanase and β-glucosidase by Fusarium oxysporum in submerged culture. Biotechnol Lett. 1995;17(8):883–888. doi: 10.1007/BF00129023. [DOI] [Google Scholar]
- Christakopoulos P, Kekos D, Macris BJ, Claeyssens M, Bhat MK. Purification and characterization of a less randomly acting endo-1,4-β-d-glucanase from the culture filtrates of Fusarium oxysporum. Arch Biochem Biophys. 1995;316:428–433. doi: 10.1006/abbi.1995.1057. [DOI] [PubMed] [Google Scholar]
- Christakopoulos P, Kekos D, Macris BJ, Claeyssens M, Bhat MK. Purification and mode of action of a low molecular mass endo-1,4-β-glucanase from Fusarium oxysporum. J Biotechnol. 1995;39(1):85–93. doi: 10.1016/0168-1656(94)00147-5. [DOI] [Google Scholar]
- Christakopoulos P, Koullas D, Kekos D, Koukios E, Macris B. Direct conversion of straw to ethanol by Fusarium oxysporum: effect of cellulose crystallinity. Enzyme Microb Technol. 1991;13:272–274. doi: 10.1016/0141-0229(91)90141-V. [DOI] [Google Scholar]
- Christakopoulos P, Koullas D, Kekos D, Koukios E, Macris B. Direct ethanol conversion of pretreated straw by Fusarium oxysporum. Bioresour Technol. 1991;35:297–300. doi: 10.1016/0960-8524(91)90128-7. [DOI] [Google Scholar]
- Christakopoulos P, Macris BJ, Kekos D. Direct fermentation of cellulose to ethanol by Fusarium oxysporum. Enzyme Microb Technol. 1989;11:236–239. doi: 10.1016/0141-0229(89)90098-7. [DOI] [Google Scholar]
- Christakopoulos P, Mamma D, Nerinckx W, Kekos D, Macris B, Claeyssens M. Production and partial characterization of xylanase from Fusarium oxysporum. Bioresour Technol. 1996;58:115–119. doi: 10.1016/S0960-8524(96)00091-0. [DOI] [Google Scholar]
- Christakopoulos P, Nerinckx W, Kekos D, Macris B, Claeyssens M. Purification and characterization of two low molecular mass alkaline xylanases from Fusarium oxysporum F3. J Biotechnol. 1996;51:181–189. doi: 10.1016/0168-1656(96)01619-7. [DOI] [PubMed] [Google Scholar]
- Chu BCH, Lee H. Genetic improvement of Saccharomyces cerevisiae for xylose fermentation. Biotechnol Adv. 2007;25:425–441. doi: 10.1016/j.biotechadv.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Chow CM, Yague E, Raguz S, Wood DA, Thurston CF. The cel3 gene of Agaricus bisporus codes for a modular cellulase and is transcriptionally regulated by the carbon source. Appl Environ Microbiol. 1994;60:2779–2785. doi: 10.1128/aem.60.8.2779-2785.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashtban M, Schraft H, Qin W. Fungal bioconversion of lignocellulosic residues; Opportunities and perspectives. Int J Biol Sci. 2009;5:578–595. doi: 10.7150/ijbs.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan R, Mcbride JE, La Grange DC, Lynd LR, van Zyl WH. Functional expression of cellobiohydrolases in Saccharomyces cerevisiae towards one-step conversion of cellulose to ethanol. Enzyme Microb Technol. 2007;40:1291–1299. doi: 10.1016/j.enzmictec.2006.09.022. [DOI] [Google Scholar]
- den Haan R, van Rensburg E, Rose SH, van Gorgens JF, van Zyl WH. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing. Curr Opin Biotechnol. 2015;33:32–38. doi: 10.1016/j.copbio.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Deshpande V, Keskar S, Mishra C, Rao M. Direct conversion of cellulose/hemicellulose to ethanol by Neurospora crassa. Enzyme Microb Technol. 1986;8:149–152. doi: 10.1016/0141-0229(86)90103-1. [DOI] [Google Scholar]
- Di Pietro A, Madrid MP, Caracuel Z, Delgado-Jarana J, Roncero MIG. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol Plant Pathol. 2003;4:315–325. doi: 10.1046/j.1364-3703.2003.00180.x. [DOI] [PubMed] [Google Scholar]
- Dionisi D, Anderson JA, Aulenta F, McCue A, Paton G. The potential of microbial processes for lignocellulosic biomass conversion to ethanol: a review. J Chem Technol Biotechnol. 2015;90:366–383. doi: 10.1002/jctb.4544. [DOI] [Google Scholar]
- Dogaris I, Vakontios G, Kalogeris E, Mamma D, Kekos D. Induction of cellulases and hemicellulases from Neurospora crassa under solid-state cultivation for bioconversion of sorghum bagasse into ethanol. Ind Crops Prod. 2009;29:404–411. doi: 10.1016/j.indcrop.2008.07.008. [DOI] [Google Scholar]
- Eggeman T, Elander RT. Process and economic analysis of pretreatment technologies. Bioresour Technol. 2005;96:2019–2025. doi: 10.1016/j.biortech.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol. 2000;66:3381–3386. doi: 10.1128/AEM.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari T, Suihko ML. Ethanol production by fermentation of pentoses and hexoses from cellulosic materials. Crit Rev Biotechnol. 1983;1:229–240. doi: 10.3109/07388558309077980. [DOI] [Google Scholar]
- European Biofuels Technology Platform. 2015. Newsletter 21, January 2015. www.biofuelstp.eu/newsletters.
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M, Chen F, Foston M, Ragauskas A, Bouton J. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA. 2011;108:3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gírio FM, Fonseca C, Carvalheiro F, Duarte LC, Marques S, Bogel-Lukasik R. Hemicelluloses for fuel ethanol: a review. Bioresour Technol. 2010;101:4775–4800. doi: 10.1016/j.biortech.2010.01.088. [DOI] [PubMed] [Google Scholar]
- Gnansounou E. Production and use of lignocellulosic bioethanol in Europe: current situation and perspectives. Bioresour Technol. 2010;101:4842–4850. doi: 10.1016/j.biortech.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Gong CS, Maun CM, Tsao GT. Direct fermentation of cellulose to ethanol by a cellulolytic filamentous fungus Monilia sp. Biotechnol Lett. 1981;3:77–82. doi: 10.1007/BF00145114. [DOI] [Google Scholar]
- Goshadrou A, Karimi K, Taherzadeh MJ. Bioethanol production from sweet sorghum bagasse by Mucor hiemalis. Ind Crops Prod. 2011;34:1219–1225. doi: 10.1016/j.indcrop.2011.04.018. [DOI] [Google Scholar]
- Guo M, Song W, Buhain J. Bioenergy and biofuels: history, status, and perspective. Renew Sustain Energy Rev. 2015;42:712–725. doi: 10.1016/j.rser.2014.10.013. [DOI] [Google Scholar]
- Halliwell G. The action of cellulolytic enzymes from Myrothecium verrucaria. Biochem J. 1961;79:185–192. doi: 10.1042/bj0790185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn-hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Hector RE, Qureshi N, Hughes SR, Cotta MA. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl Microbiol Biotechnol. 2008;80:675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- Hennessy RC, Doohan F, Mullins E. Generating phenotypic diversity in a fungal biocatalyst to investigate alcohol stress tolerance encountered during microbial cellulosic biofuel production. PLoS One. 2013;8:e77501. doi: 10.1371/journal.pone.0077501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel ME, Bayer EA. Lignocellulose conversion to biofuels: current challenges, global perspectives. Curr Opin Biotechnol. 2009;20:316–317. doi: 10.1016/j.copbio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Ho NWY, Chen Z, Brainard AP. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Tamaki H, Yamamoto K, Kumagai H. Cloning of a gene encoding thermostable cellobiohydrolase from Thermoascus aurantiacus and its expression in yeast. Appl Microbiol Biotechnol. 2003;63:42–50. doi: 10.1007/s00253-003-1379-3. [DOI] [PubMed] [Google Scholar]
- Hong J, Yang H, Zhang K, Liu C, Zou S, Zhang M. Development of a cellulolytic Saccharomyces cerevisiae strain with enhanced cellobiohydrolase activity. World J Microbiol Biotechnol. 2014;30:2985–2993. doi: 10.1007/s11274-014-1726-9. [DOI] [PubMed] [Google Scholar]
- Hopkins DW, Webster EA, Boerjan W, Pilate G, Halpin C. Genetically modified lignin below ground. Nat Biotechnol. 2007;25:168–169. doi: 10.1038/nbt0207-168. [DOI] [PubMed] [Google Scholar]
- Huang J, Chen D, Wei Y, Wang Q, Li Z, Chen Y, Huang R. Direct ethanol production from lignocellulosic sugars and sugarcane bagasse by a recombinant Trichoderma reesei strain hj48. Sci World J. 2014 doi: 10.1155/2014/798683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon JE, Jeon WJ, Whang SY, Han SO. Production of minicellulosomes for the enhanced hydrolysis of cellulosic substrates by recombinant Corynebacterium glutamicum. Enzyme Microb Technol. 2011;48:371–377. doi: 10.1016/j.enzmictec.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Ingram L, Aldrich H, Borges A, Causey T, Martinez A, Morales F, Saleh A, Underwood S, Yomano L, York S. Enteric bacterial catalysts for fuel ethanol production. Biotechnol Prog. 1999;15:855–866. doi: 10.1021/bp9901062. [DOI] [PubMed] [Google Scholar]
- International Energy Agency. 2014. CO2 Emissions from fuel combustion highlights. doi: 10.1787/co2_fuel-2014-en.
- International Energy Agency. 2015. Energy technology perspectives: mobilising innovation to accelerate climate action. Edition: 2015.
- Jeffries TW. Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng Biotechnol. 1983;27:1–32. doi: 10.1007/BFb0009101. [DOI] [PubMed] [Google Scholar]
- Karimi K, Emtiazi G, Taherzadeh MJ. Ethanol production from dilute-acid pretreated rice straw by simultaneous saccharification and fermentation with Mucor indicus, Rhizopus oryzae and Saccharomyces cerevisiae. Enzyme Microb Technol. 2006;40:138–144. doi: 10.1016/j.enzmictec.2005.10.046. [DOI] [Google Scholar]
- Khuong LD, Kondo R, De Leon R, Kim Anh T, Shimizu K, Kamei I. Bioethanol production from alkaline-pretreated sugarcane bagasse by consolidated bioprocessing using Phlebia sp. MG-60. Int Biodeterior Biodegradation. 2014;88:62–68. doi: 10.1016/j.ibiod.2013.12.008. [DOI] [Google Scholar]
- Kim J-H, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BC, Waxman KD, Nenni NV, Walker LP, Bergstrom GC, Gibson DM. Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol Biofuels. 2011;4:4. doi: 10.1186/1754-6834-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko KC, Lee JH, Han Y, Choi JH, Song JJ. A novel multifunctional cellulolytic enzyme screened from metagenomic resources representing ruminal bacteria. Biochem Biophys Res Commun. 2013;441:567–572. doi: 10.1016/j.bbrc.2013.10.120. [DOI] [PubMed] [Google Scholar]
- Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels. 2009;2:19. doi: 10.1186/1754-6834-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U, Hoff B. New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol. 2010;86:51–62. doi: 10.1007/s00253-009-2416-7. [DOI] [PubMed] [Google Scholar]
- Kumar P, Singh A, Schügerl K. Formation of acetic acid from cellulosic materials by Fusarium oxysporum. Appl Microbiol Biotechnol. 1991;34:570–572. doi: 10.1007/BF00167900. [DOI] [Google Scholar]
- Kuyper M, Hartog MMP, Toirkens MJ, Almering MJH, Winkler AA, Dijken JP, Pronk JT. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005;5:399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Laluce C, Schenberg ACG, Gallardo JCM, Coradello LFC, Pombeiro-Sponchiado SR. Advances and developments in strategies to improve strains of Saccharomyces cerevisiae and processes to obtain the lignocellulosic ethanol: a review. Appl Biochem Biotechnol. 2012;166:1908–1926. doi: 10.1007/s12010-012-9619-6. [DOI] [PubMed] [Google Scholar]
- Leão C, Uden N. Effects of ethanol and other alkanols on the temperature relations of glucose transport and fermentation in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1985;22:359–363. doi: 10.1007/BF00582420. [DOI] [Google Scholar]
- Lee SM, Jellison T, Alper HS. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 2012;78:5708–5716. doi: 10.1128/AEM.01419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin J, Zhou P, Wu K, Liu H, Xiong C, Gong Y, Xiao W, Liu Z. One-pot simultaneous saccharification and fermentation: a preliminary study of a novel configuration for cellulosic ethanol production. Bioresour Technol. 2014;161:171–178. doi: 10.1016/j.biortech.2014.02.130. [DOI] [PubMed] [Google Scholar]
- Limayem A, Ricke SC. Lignocellulosic biomass for bioethanol production: current perspectives, potential issues and future prospects. Prog Energy Combust Sci. 2012;38:449–467. doi: 10.1016/j.pecs.2012.03.002. [DOI] [Google Scholar]
- Linger JG, Adney WS, Darzins A. Heterologous expression and extracellular secretion of cellulolytic enzymes by Zymomonas mobilis. Appl Environ Microbiol. 2010;76:6360–6369. doi: 10.1128/AEM.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Bao J. Secretive expression of heterologous β-glucosidase in Zymomonas mobilis. Bioresour Bioprocess. 2015;2:29. doi: 10.1186/s40643-015-0053-9. [DOI] [Google Scholar]
- Lynd LR, Laser MS, Bransby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J. How biotech can transform biofuels. Nat Biotechnol. 2008;26:169–172. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- Lynd LR, Van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Menon V, Rao M. Trends in bioconversion of lignocellulose: biofuels, platform chemicals and biorefinery concept. Prog Energy Combust Sci. 2012;38:522–550. doi: 10.1016/j.pecs.2012.02.002. [DOI] [Google Scholar]
- Millati R, Edebo L, Taherzadeh MJ. Performance of Rhizopus, Rhizomucor and Mucor in ethanol production from glucose, xylose and wood hydrolyzates. Enzyme Microb Technol. 2005;36:294–300. doi: 10.1016/j.enzmictec.2004.09.007. [DOI] [Google Scholar]
- Mizuno R, Ichinose H, Honda M, Takabatake K, Sotome I, Takai T, Maehara T, Okadome H, Isobe S, Gau M, Kaneko S. Use of whole crop sorghums as a raw material in consolidated bioprocessing bioethanol production using Flammulina velutipes. Biosci Biotechnol Biochem. 2009;73:1671–1673. doi: 10.1271/bbb.90099. [DOI] [PubMed] [Google Scholar]
- Morales M, Quintero J, Conejeros R, Aroca G. Life cycle assessment of lignocellulosic bioethanol: environmental impacts and energy balance. Renew Sustain Energy Rev. 2015;42:1349–1361. doi: 10.1016/j.rser.2014.10.097. [DOI] [Google Scholar]
- Mosier N, Wyman C, Dale B, Elander R, Lee Y, Holtzapple M, Ladisch M. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol. 2005;96:673–686. doi: 10.1016/j.biortech.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Nitta Y, Maekawa N, Yanase H. Direct ethanol production from starch, wheat bran and rice straw by the white rot fungus Trametes hirsuta. Enzyme Microb Technol. 2011;48:273–277. doi: 10.1016/j.enzmictec.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Sugita Y, Nishikori N, Nitta Y, Yanase H. Characterization of two acidic ß-glucosidases and ethanol fermentation in the brown rot fungus Fomitopsis palustris. Enzyme Microb Technol. 2011;48:359–364. doi: 10.1016/j.enzmictec.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Uchii A, Kanawaku R, Yanase H. Bioconversion of xylose, hexoses and biomass to ethanol by a new isolate of the white rot basidiomycete Trametes versicolor. Springerplus. 2014;3:121. doi: 10.1186/2193-1801-3-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson K, Bertilsson M, Lidén G. A short review on SSF—an interesting process option for ethanol production from lignocellulosic feedstocks. Biotechnol Biofuels. 2008;1:7. doi: 10.1186/1754-6834-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/MCB.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotou G, Christakopoulos P, Olsson L. The influence of different cultivation conditions on the metabolome of Fusarium oxysporum. J Biotechnol. 2005;118:304–315. doi: 10.1016/j.jbiotec.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Panagiotou G, Christakopoulos P, Olsson L. Simultaneous saccharification and fermentation of cellulose by Fusarium oxysporum F3–growth characteristics and metabolite profiling. Enz Microb Technol. 2005;36:693–699. doi: 10.1016/j.enzmictec.2004.12.029. [DOI] [Google Scholar]
- Panagiotou G, Christakopoulos P, Villas-Boas SG, Olsson L. Fermentation performance and intracellular metabolite profiling of Fusarium oxysporum cultivated on a glucose-xylose mixture. Enz Microb Technol. 2005;36:100–106. doi: 10.1016/j.enzmictec.2004.07.009. [DOI] [Google Scholar]
- Panagiotou G, Kekos D, Macris BJ, Christakopoulos P. Production of cellulolytic and xylanolytic enzymes by Fusarium oxysporum grown on corn stover in solid state fermentation. Ind Crops Prod. 2003;18:37–45. doi: 10.1016/S0926-6690(03)00018-9. [DOI] [Google Scholar]
- Panagiotou G, Villas-Bôas SG, Christakopoulos P, Nielsen J, Olsson L. Intracellular metabolite profiling of Fusarium oxysporum converting glucose to ethanol. J Biotechnol. 2005;115:425–434. doi: 10.1016/j.jbiotec.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Panagiotou G, Topakas E, Moukouli M, Christakopoulos P, Olsson L. Studying the ability of Fusarium oxysporum and recombinant Saccharomyces cerevisiae to efficiently cooperate in decomposition and ethanolic fermentation of wheat straw. Biomass Bioenergy. 2011;35:3727–3732. doi: 10.1016/j.biombioe.2011.05.005. [DOI] [Google Scholar]
- Prior B, Kilian S, Du Preez J. Fermentation of d-xylose by the yeasts Candida shehatae and Pichia stipitis: prospects and problems. Process Biochem. 1989;24:21–32. [Google Scholar]
- Rao M, Deshpande V, Keskar S, Srinivasan MC. Cellulase and ethanol production from cellulose by Neurospora crassa. Enzyme Microb Technol. 1983;5:133–136. doi: 10.1016/0141-0229(83)90048-0. [DOI] [Google Scholar]
- Rasmussen ML, Shrestha P, Khanal SK, Pometto Iii AL, van Leeuwen J. Sequential saccharification of corn fiber and ethanol production by the brown rot fungus Gloeophyllum trabeum. Bioresour Technol. 2010;101:3526–3533. doi: 10.1016/j.biortech.2009.12.115. [DOI] [PubMed] [Google Scholar]
- Reyes LH, Almario MP, Kao KC. Genomic library screens for genes involved in n-butanol tolerance in Escherichia coli. PLoS One. 2011;6:e17678. doi: 10.1371/journal.pone.0017678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Harrison J. The yeasts. vol. 2 and 3. Orlando: Academic Press; 1970.
- Sanderson K. Lignocellulose: a chewy problem. Nature. 2011;474:S12–S14. doi: 10.1038/474S012a. [DOI] [PubMed] [Google Scholar]
- Schmoll M, Esquivel-Naranjo EU, Herrera-Estrella A. Trichoderma in the light of day—Physiology and development. Fungal Genet Biol. 2010;47:909–916. doi: 10.1016/j.fgb.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster BG, Chinn MS. Consolidated bioprocessing of lignocellulosic feedstocks for ethanol fuel production. Bioenergy Res. 2013;6:416–435. doi: 10.1007/s12155-012-9278-z. [DOI] [Google Scholar]
- Shallom D, Shoham Y. Microbial hemicellulases. Curr Opin Microbiol. 2003;6:219–228. doi: 10.1016/S1369-5274(03)00056-0. [DOI] [PubMed] [Google Scholar]
- Shrestha P, Khanal SK, Pometto AL, Hans van Leeuwen J. Ethanol production via in situ fungal saccharification and fermentation of mild alkali and steam pretreated corn fiber. Bioresour Technol. 2010;101:8698–8705. doi: 10.1016/j.biortech.2010.06.089. [DOI] [PubMed] [Google Scholar]
- Silva-Rocha R, Castro LDS, Antoniêto ACC, Guazzaroni ME, Persinoti GF, Silva RN. Deciphering the cis-regulatory elements for XYR1 and CRE1 regulators in Trichoderma reesei. PLoS ONE. 2014;9:e99366. doi: 10.1371/journal.pone.0099366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kuhad RC, Kumar M. Xylanase production by a hyperxylanolytic mutant of Fusarium oxysporum. Enzyme Microb Technol. 1995;17:551–553. doi: 10.1016/0141-0229(94)00074-2. [DOI] [Google Scholar]
- Singh A, Kumar PK. Fusarium oxysporum: status in bioethanol production. Crit Rev Biotechnol. 1991;11:129–147. doi: 10.3109/07388559109040619. [DOI] [PubMed] [Google Scholar]
- Singh A, Kumar PKR, Schugerl K. Adsorption and reuse of cellulases during saccharification of cellulosic materials. J Biotechnol. 1991;18:205–212. doi: 10.1016/0168-1656(91)90248-T. [DOI] [Google Scholar]
- Singh A, Kumar PKR, Schiigerl K. Bioconversion of cellulosic materials to ethanol by filamentous fungi. Adv Biochem Eng Biotechnol. 1992;45:29–55. doi: 10.1007/BFb0008753. [DOI] [Google Scholar]
- Skory CD, Freer SN, Bothast RJ. Screening for ethanol-producing fillamentous fungi. Biotecnol letter. 1997;19:203–206. doi: 10.1023/A:1018337003433. [DOI] [Google Scholar]
- Slininger PJ, Bolen PL, Kurtzman CP. Pachysolen tannophilus: properties and process considerations for ethanol production from d-xylose. Enzyme Microb Technol. 1987;9:5–15. doi: 10.1016/0141-0229(87)90043-3. [DOI] [Google Scholar]
- Solomon BD, Barnes JR, Halvorsen KE. Grain and cellulosic ethanol: history, economics, and energy policy. Biomass Bioenergy. 2007;31:416–425. doi: 10.1016/j.biombioe.2007.01.023. [DOI] [Google Scholar]
- Stevenson DM, Weimer PJ. Isolation and characterization of a Trichodermastrain capable of fermenting cellulose to ethanol. Appl Microbiol Biotechnol. 2002;59:721–726. doi: 10.1007/s00253-002-1027-3. [DOI] [PubMed] [Google Scholar]
- Suihko ML. The fermentation of different carbon sources by Fusarium oxysporum. Biotechnol Lett. 1983;5:721–724. doi: 10.1007/BF01386491. [DOI] [Google Scholar]
- Szczodrak J. The enzymatic hydrolysis and fermentation of pretreated wheat straw to ethanol. Biotechnol Bioeng. 1988;32(6):771–776. doi: 10.1002/bit.260320608. [DOI] [PubMed] [Google Scholar]
- Taherzadeh MJ, Karimi K. Acid-based hydrolysis processes for ethanol from lignocellulosic materials: a review. Bioresources. 2007;2:472–499. [Google Scholar]
- Tao H, Gonzalez R, Martinez A, Rodriguez M, Ingram L, Preston J, Shanmugam K. Engineering a homo-ethanol pathway in Escherichia coli: increased glycolytic flux and levels of expression of glycolytic genes during xylose fermentation. J Bacteriol. 2001;183:2979–2988. doi: 10.1128/JB.183.10.2979-2988.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targonski Z, Szajer C. The dynamics of cellulase synthesis in Fusarium cultures: influence of the cellulose structure. Biotechnol Lett. 1979;1:75–80. doi: 10.1007/BF01398312. [DOI] [Google Scholar]
- Tomás-Pejó E, Oliva J, González A, Ballesteros I, Ballesteros M. Bioethanol production from wheat straw by the thermotolerant yeast Kluyveromyces marxianus CECT 10875 in a simultaneous saccharification and fermentation fed-batch process. Fuel. 2009;88:2142–2147. doi: 10.1016/j.fuel.2009.01.014. [DOI] [Google Scholar]
- Tran DT, Lin CW, Lai CY, Wu CH. Ethanol production from lignocelluloses by native strain Klebsiella oxytoca THLC0409. Waste Biomass Valorization. 2011;2:389–396. doi: 10.1007/s12649-011-9082-6. [DOI] [Google Scholar]
- Vitikainen M, Arvas M, Pakula T, Oja M, Penttilä M, Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genom. 2010;11:441. doi: 10.1186/1471-2164-11-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zyl WH, Lynd LR, Den Haan R, McBride JE. Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol. 2007;108:205–235. doi: 10.1007/10_2007_061. [DOI] [PubMed] [Google Scholar]
- Wang TY, Chen HL, Lu MYJ, Chen YC, Sung HM, Mao CT, Cho HY, Ke HM, Hwa TY, Ruan SK. Functional characterization of cellulases identified from the cow rumen fungus Neocallimastix patriciarum W5 by transcriptomic and secretomic analyses. Biotechnol Biofuels. 2011;4:24. doi: 10.1186/1754-6834-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood BE, Yomano LP, York SW, Ingram LO. Development of industrial-medium-required elimination of the 2,3-butanediol fermentation pathway to maintain ethanol yield in an ethanologenic strain of Klebsiella oxytoca. Biotechnol Prog. 2005;21:1366–1372. doi: 10.1021/bp050100e. [DOI] [PubMed] [Google Scholar]
- Wright JD, Wyman CE, Grohmann K. Simultaneous saccharification and fermentation of lignocellulose. Appl Biochem Biotechnol. 1988;18:75–90. doi: 10.1007/BF02930818. [DOI] [Google Scholar]
- Wu JF (1989) Process for producing ethanol from plant biomass using the fungus Paecilomyces sp. US Patent 4,840,903.
- Wyman CE. Ethanol from lignocellulosic biomass: technology, economics, and opportunities. Bioresource Technol. 1994;50:3–15. doi: 10.1016/0960-8524(94)90214-3. [DOI] [Google Scholar]
- Xiros C, Christakopoulos P. Enhanced ethanol production from brewer’s spent grain by a Fusarium oxysporum consolidated system. Biotechnol Biofuels. 2009;2:4. doi: 10.1186/1754-6834-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Singh A, Himmel ME. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr Opin Biotechnol. 2009;20:364–371. doi: 10.1016/j.copbio.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Xu L, Shen Y, Hou J, Peng B, Tang H, Bao X. Secretory pathway engineering enhances secretion of cellobiohydrolase I from Trichoderma reesei in Saccharomyces cerevisiae. J Biosci Bioeng. 2014;117:45–52. doi: 10.1016/j.jbiosc.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Zerva A, Savvides AL, Katsifas EA, Karagouni AD, Hatzinikolaou DG. Evaluation of Paecilomyces variotii potential in bioethanol production from lignocellulose through consolidated bioprocessing. Bioresour Technol. 2014;162:294–299. doi: 10.1016/j.biortech.2014.03.137. [DOI] [PubMed] [Google Scholar]
- Zha J, Shen M, Hu M, Song H, Yuan Y. Enhanced expression of genes involved in initial xylose metabolism and the oxidative pentose phosphate pathway in the improved xylose-utilising Saccharomyces cerevisiae through evolutionary engineering. J Ind Microbiol Biotechnol. 2014;41:27–39. doi: 10.1007/s10295-013-1350-y. [DOI] [PubMed] [Google Scholar]