Abstract
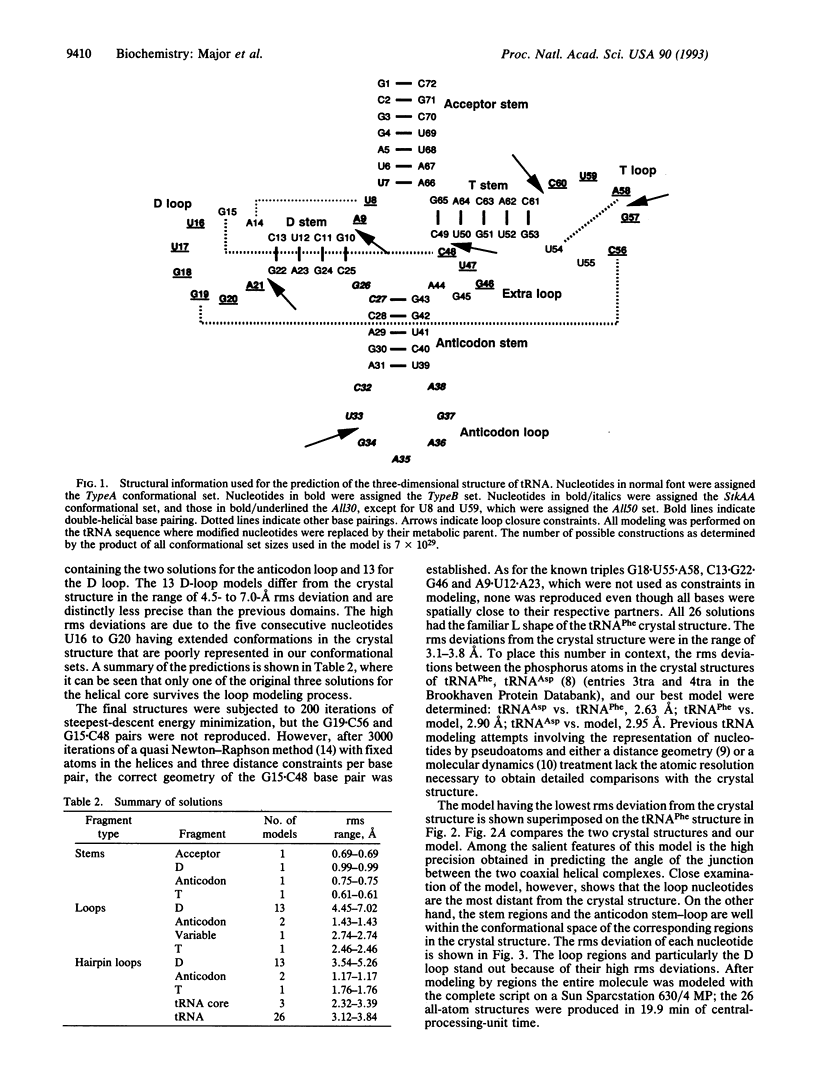
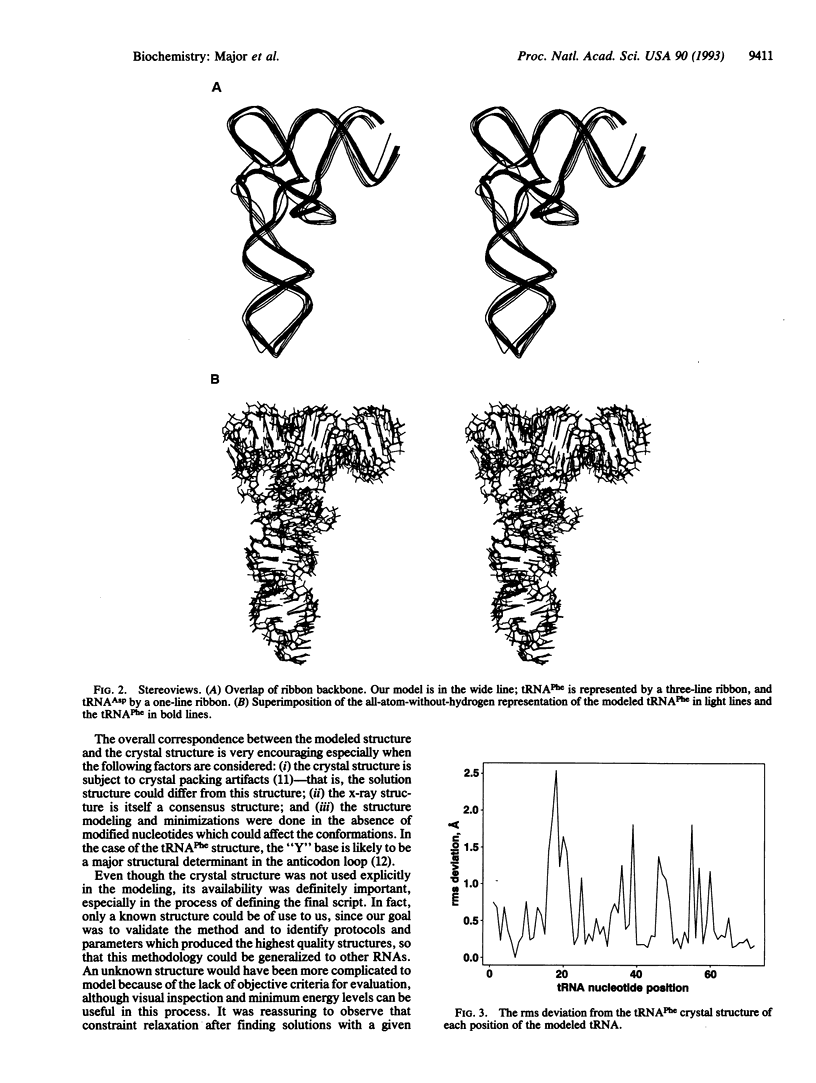
The three-dimensional structure of yeast tRNA(Phe) was reproduced at atomic resolution with the automated RNA modeling program MC-SYM, which is based on a constraint-satisfaction algorithm. Structural constraints used in the modeling were derived from the secondary structure, four tertiary base pairs, and other information available prior to the determination of the x-ray crystal structure of the tRNA. The program generated 26 solutions (models), all of which had the familiar "L" form of tRNA and root-mean-square deviations from the crystal structure in the range of 3.1-3.8 A. The interaction between uridine-8 and adenosine-14 was crucial in the modeling procedure, since only this among the tertiary pairs is necessary and sufficient to reproduce the L form of tRNA. Other tertiary interactions were critical in reducing the number of solutions proposed by the program.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gautheret D., Major F., Cedergren R. Modeling the three-dimensional structure of RNA using discrete nucleotide conformational sets. J Mol Biol. 1993 Feb 20;229(4):1049–1064. doi: 10.1006/jmbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Power A., Hertz G. Z., Putz E. J., Stormo G. D. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res. 1992 Nov 11;20(21):5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U. A note on crystal packing and global helix structure in short A-DNA duplexes. J Biomol Struct Dyn. 1991 Feb;8(4):801–811. doi: 10.1080/07391102.1991.10507846. [DOI] [PubMed] [Google Scholar]
- Houssier C., Grosjean H. Temperature jump relaxation studies on the interactions between transfer RNAs with complementary anticodons. The effect of modified bases adjacent to the anticodon triplet. J Biomol Struct Dyn. 1985 Oct;3(2):387–408. doi: 10.1080/07391102.1985.10508425. [DOI] [PubMed] [Google Scholar]
- Hubbard J. M., Hearst J. E. Predicting the three-dimensional folding of transfer RNA with a computer modeling protocol. Biochemistry. 1991 Jun 4;30(22):5458–5465. doi: 10.1021/bi00236a019. [DOI] [PubMed] [Google Scholar]
- Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969 Nov 22;224(5221):759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- Major F., Turcotte M., Gautheret D., Lapalme G., Fillion E., Cedergren R. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science. 1991 Sep 13;253(5025):1255–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- Malhotra A., Tan R. K., Harvey S. C. Prediction of the three-dimensional structure of Escherichia coli 30S ribosomal subunit: a molecular mechanics approach. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1950–1954. doi: 10.1073/pnas.87.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Ninio J., Favre A., Yaniv M. Molecular model for transfer RNA. Nature. 1969 Sep 27;223(5213):1333–1335. doi: 10.1038/2231333a0. [DOI] [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. Folding of circularly permuted transfer RNAs. Science. 1991 Nov 29;254(5036):1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Seeman N. C., Wang A. H., Suddath F. L., Rich A. Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res. 1975 Dec;2(12):2329–2341. doi: 10.1093/nar/2.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr A. 1988 Mar 1;44(Pt 2):112–123. [PubMed] [Google Scholar]


