Abstract
Macrophages are essential for the progression and maintenance of many cancers, but their role during the earliest stages of tumor formation is unclear. To test this, we used a previously described transgenic mouse model of wound-induced skin tumorigenesis, in which expression of constitutively active MEK1 in differentiating epidermal cells results in chronic inflammation (InvEE mice). Upon wounding, the number of epidermal and dermal monocytes and macrophages increased in wildtype and InvEE skin, but the increase was greater, more rapid, and more sustained in InvEE skin. Macrophage ablation reduced tumor incidence. Furthermore, bioluminescent imaging in live mice to monitor macrophage flux at wound sites revealed that macrophage accumulation was predictive of tumor formation; wounds with the greatest number of macrophages at day 5 went on to develop tumors. Gene expression profiling of flow-sorted monocytes, macrophages, and T cells from InvEE and wildtype skin showed that as wound healing progressed, InvEE macrophages altered their phenotype. Throughout wound healing and after wound closure, InvEE macrophages demonstrated sustained upregulation of several markers implicated in alternative macrophage activation including arginase-1 (ARG1) and mannose receptor (CD206). Notably, inhibition of ARG1 activity significantly reduced tumor formation and epidermal proliferation in vivo, whereas addition of L-arginase to cultured keratinocytes stimulated proliferation. We conclude that macrophages play a key role in early, inflammation-mediated skin tumorigenesis, with mechanistic evidence suggesting that ARG1 secretion drives tumor development by stimulating epidermal cell proliferation. These findings highlight the importance of cancer immunotherapies aiming to polarize tumor-associated macrophages towards an antitumor phenotype.
Keywords: macrophages, skin cancer, arginase, wound healing, keratinocyte proliferation
INTRODUCTION
The notion that the local microenvironment is actively involved in tumor development has gained wide acceptance in the recent years. Interactions between cancer cells and surrounding non-malignant cells are crucial determinants of cancer development (1-3). It has been suggested that the construction of a pre-cancerous niche is required for initiated cancer cells to survive and evolve into a tumor (4). This niche is typically shaped by resident or recruited stromal cells, bone marrow derived cells and signals or secreted factors (cytokines, chemokines, exosomes) from all these populations (5).
We have previously described a mouse model of wound-induced, inflammation-mediated skin tumorigenesis (6, 7). In this model non-dividing cells in the suprabasal layers of the epidermis express constitutively active MEK1 (InvEE transgenics), resulting in epidermal hyperproliferation and a chronic inflammatory skin infiltrate. Between 40-60 % of InvEE transgenics develop benign tumors (papillomas and keratoacanthomas) at the site of a full-thickness circular wound, several days after the wound has healed (6). In this model MEK1 expressing, non-dividing cells stimulate keratinocytes in the basal layer to divide and form the proliferative compartment of the tumor (6). MEK1 expressing suprabasal cells recruit an inflammatory infiltrate via secretion of pro-inflammatory cytokines including IL-1α (7), which in turn induces expression of CD26 on stromal fibroblasts (8).
The presence of an immune infiltrate in InvEE skin is essential for wound-induced tumor formation, since treatment with the broad-spectrum anti-inflammatory drug Dexamethasone protects against tumor formation (6). Depletion of circulating γδ T cells, but not αβ T cells or B cells, reduces the incidence of tumors and leads to a reduction in the number of skin macrophages (6). Depletion of skin macrophages by treatment of the wound site with clodronate liposomes also decreases tumor incidence (6).
The identification of macrophages as key contributors to InvEE tumor formation is consistent with studies in a wide variety of other tumor types (9). Macrophages are highly specialized to support established neoplastic lesions, by promoting immunosuppression, proliferation, angiogenesis, and metastases (10). Correlations between persistent, macrophage-mediated chronic inflammation and cancer have been noted (11, 12). However, little is known about the role of macrophages in the earliest stages of tumor development. Initiating events, such as tissue or DNA damage caused by the release of cytotoxic factors through inflammatory cells (12), are difficult to identify as they may occur long before overt tumor development and in most experimental systems the precise site of the tumor cannot be predicted. Our model allows the observation of events that ultimately lead to tumor formation in a well-defined pre-cancerous niche, the wound.
MATERIALS AND METHODS
Animals
Mice from the InvEE founder line 3376A (7) were maintained on an F1 (CBA × C57BL/6) background as heterozygotes. Transgene-negative littermates were used as controls. PU.1-YFP mice (13), kindly donated by Claus Nerlov (The Weatherall Institute of Molecular Medicine, University of Oxford, UK), were crossed with InvEE F1 mice and maintained on an F1 background as homozygotes for PU.1-YFP. Homozygote CD11b-DTR mice from a BL/6 background (Jackson Laboratory) were crossed with CBA mice and used as bone marrow donors only. Animal procedures were subject to ethical approval from Cancer Research UK and King’s College London and performed under a UK Government Home Office license.
Animal procedures
Wounding was performed as previously described (6). A single 8 mm (diameter) wound was made into the back skin with a punch biopsy (Stiefel). The following drugs were administered before or immediately after wounding: Dexamethasone (400 μg/day; s.c., Organon), Diphtheria Toxin (10 ng/g, i.p, 3x/week, Sigma Aldrich), nor-NOHA (40 mg/kg/day, Bachem, in an s.c. implanted Alzet® osmotic pump, model 1002), Lucigenin (20 mg/kg, i.p., 3x/week, Sigma Aldrich) and Luminol (100 mg/kg, i.p., 3x/week, Sigma Aldrich). Bone marrow transplantations were performed between MHC-matched mice as described previously (6, 14). Sublethally irradiated (2 × 5 Gy, separated by 3 h rest) 6- to 12-week-old InvEE females were reconstituted within 24 h by i.v. injection with 200 μL saline containing 2–5 × 106 bone marrow cells freshly isolated from tibia and femur of male CD11b-DTR donor mice. Chimerism was detected by Y-chromosome in situ hybridization. Bioluminescent imaging was performed on anaesthetized mice previously wounded and i.p. injected with either 200 μL of Luminol or Lucigenin. Sequential bioluminescence imaging was performed on an IVIS Lumina II (Perkin Elmer) at 5-10 minute intervals. Each imaging step comprised a 1 min acquisition time, f/stop = 1, binning = 16 (large), fovD and 0 sec delay. The imaging procedure was repeated approximately 9x in 2-3 day intervals to monitor wound healing.
Histology and immunohistochemistry
The following antibodies were used: Alexa 647/488-conjugated anti-F4/80, anti-CD11b (all eBioscience), anti-GFP (Invitrogen, for YFP expression), anti-CD68 (Abcam), anti-CD206 (Serotec), anti-K14 (Covance), uncoupled NOS2-rabbit polyclonal (Santa Cruz), anti-Ki67 (rabbit polyclonal, Novocastra, NCL-Ki67p), anti-K14 (LL002, in house) and species-specific antibodies conjugated with Alexa 488, 555, or 647/633 (Molecular Probes). For EdU stainings the Click-iT® EdU Alexa Fluor® 488 Imaging Kit (Molecular Probes) was used. Frozen sections were fixed at −20 °C in 4% paraformaldehyde or acetone for 15 min. Dewaxed paraffin sections were subjected to heat-mediated antigen retrieval (citrate buffer, pH 6). Sections were blocked in 1% BSA, 10% goat or donkey serum before antibody labeling and counter-stained with DAPI. Images were captured with a bright field (Nikon), Nikon A1 Upright confocal microscope or Leica SP5 confocal microscope and cells were counted manually using ImageJ 1.49 software (NIH). Epidermal Ki67+ cells were counted and stated relatively to the length of the epidermis (mm) in a 20X magnification field. Quantification was performed on both wound edges of each animal (if available).
Cell quantifications and flow cytometry
Frozen skin sections were stained with antibodies and scanned on the Ariol scanning system at 20x magnification. Regions were drawn at both edges of the wound with a minimum area of at least 50,000 μm2, which is equivalent to three 40x fields on a microscope (field of view of 0.4 mm). Immune cells were counted manually and extrapolated to number of cells/mm2. At least 2 regions per wound and animal were analyzed and a minimum of 3 mice used per data point. For quantifications using flow cytometry a 1.5 x 2 cm piece of back skin was harvested; dermal and epidermal cells were isolated as described previously (8) and labelled with the following antibodies: CD45-APC Cy7, CD3-PerCP (both BD), CD11b-PacBlue (Invitrogen), CD11b-AF594, MHC class II-brilliant violet 421 (all Biolegend), F4/80-PE, F4/80-alexa 647, Gr1-alexa 660 (RB6-8C5) and Ly6C-eF780 (all eBioscience). Labeled cells were sorted on a BD FACSARIA II cell sorter. DAPI or LIVE/DEAD Fixable Violet Dead Cell Stain (Life Technologies) was used to exclude dead cells.
Cell culture and proliferation assay
Primary mouse keratinocytes were isolated from InvEE and WT back skin and cultured feeder-free in calcium-free DMEM medium supplemented with 10% chelated fetal calf serum, hydrocortisone (0.5 μg/mL), insulin (5 μg/mL), cholera toxin (8.4 ng/mL), 1% Sodium Pyruvate (100 mM), Glutamine (450 μg/mL), penicillin (100 U/mL), streptomycin (100 μg/mL) and epidermal growth factor (10 ng/mL) in culture flasks (15). Cells were passaged 4-7 times before they were plated in μClear 96-well plates, treated with L-Arginase (Sigma Aldrich) every 10 hours for 2 days and subsequently labelled with EdU (1 hour pulse), fixed (4% PFA) and labelled with antibodies to K14 (Covance), EdU (Molecular Probes) and DAPI. Plates were analyzed using the Operetta High Content Imaging System (PerkinElmer) and single cell population distributions were quantified based on standard algorithms for nuclear labelling and cell shape.
Western blots
Skin lysates were run on 4–15% Precast polyacrylamide gel (Biorad, 4561085), transferred to PVDF membrane (Biorad, 1704156), and blocked for 1h at RT in 5% skimmed milk and 0.1% Tween-20 in PBS. Membranes were incubated with primary antibodies overnight at 4 °C and with secondary antibodies for 1h at RT. Membranes were washed in 0.05% Tween-20 in PBS. Protein bands were visualized using Clarity Western ECL Substrate (Biorad, 1705061) and the Chemidoc Touch Imaging System (Biorad). Quantification of protein levels was performed using Biorad software. The following antibodies and were used: goat anti-arginase I Antibody (V-20) (Santa Cruz; 1:400), donkey anti-goat igG HRP (Santa Cruz, 1:10000), mouse anti-alpha tubulin (Sigma-Aldrich, 1:5000), donkey anti-mouse HRP (Jackson Immuno Research Inc., 1:10000). Bovine L-Arginase (Sigma) was used as positive control.
Arginase activity and NO analysis
Arginase activity in skin lysates and plasma samples was assessed by measuring urea production using the Quantichrome Arginase Assay Kit (Bioassay Systems) according to manufacturer's protocol. Serum samples were filtered through a 10 kDa molecular weight cut-off filter before measurement to deplete urea that could interfere with the assay, and diluted 1:50 in dH2O. Skin lysate samples were diluted 1:40. To determine the concentration of nitrate and nitrite, the supernatant of cultivated and Arginase-treated keratinocytes was collected and analyzed. To normalize the samples, protein concentration in cell lysates was measured using a BCA Protein Assay Kit (Thermo Scientific).
Quantitative real-time PCR
For RNA analysis of specific cell populations, cells were FACS sorted into RNA later reagent (Amgen) and pelleted at 7,000 g for 10 minutes, RLT lysis buffer was added and cells were homogenized using a 1 mL syringe and a 21-gauge needle by passing them through the needle at least 5-10 times. RNA was extracted using the RNeasy micro kit® (Qiagen) according to manufacturer’s instructions. RNA concentration and quality was assessed using an Agilent 2100 Bioanalyzer and cDNA was generated using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative PCR was performed on a 7900HT Fast RT-PCR system (Applied Biosystems) at CR-UK CI or on a CFX 384 Touch RT-PCR detection system (Bio-Rad) at KCL. For each sample expression ratios were normalized to the reference primer GAPDH. Full primer sequences are listed in Supplementary Table S1.
Statistics
Statistical analyses were performed using GraphPad Prism (GraphPad Software). Statistical differences were determined using either student’s t-tests or Wilcoxon tests for paired, nonparametric data. A p-value of <0.05 was considered significant.
RESULTS
Elevated numbers of macrophages in InvEE skin
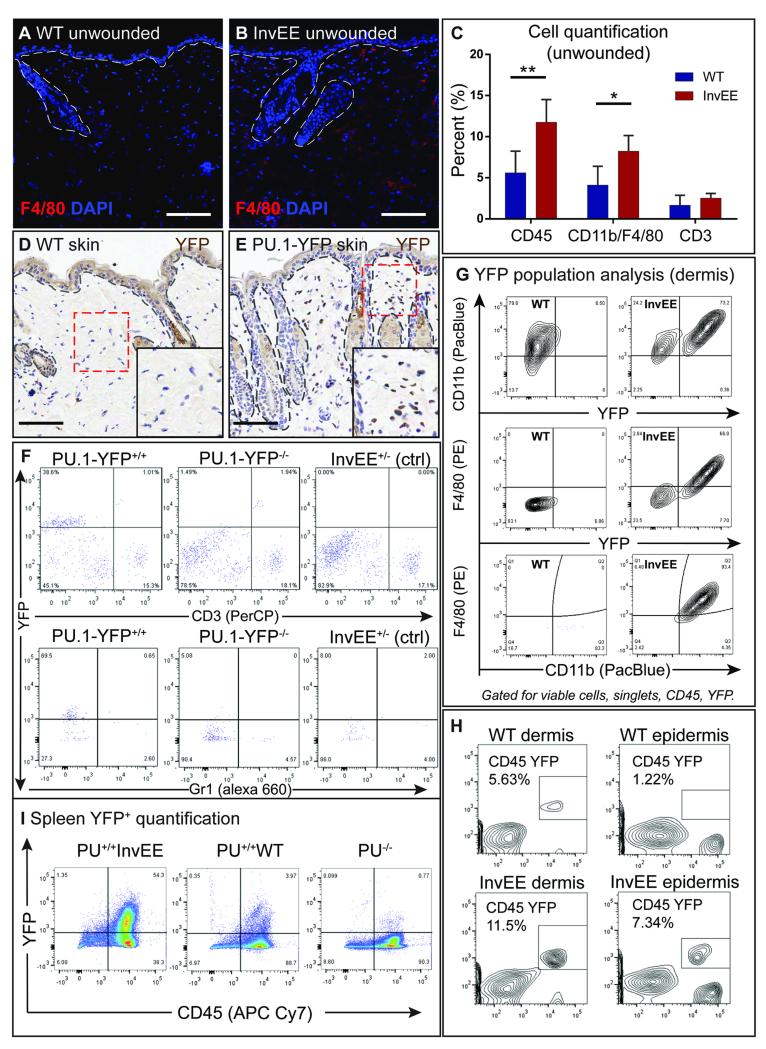
Immunofluorescent staining has previously shown that the immune infiltrate in unwounded InvEE skin consists primarily of CD11b+ monocytes, F4/80+ macrophages and CD3+ T cells (Fig. 1A-C, CD45: p = 0.0067, CD11b/F4/80 p = 0.0141) (6, 7). To facilitate analysis of myeloid cells we used a PU.1-YFP reporter mouse (13) (Fig. 1D-E). This strain expresses yellow fluorescent protein (YFP) attached to the C terminus of the PU.1 transcription factor. PU.1 is an ets family transcription factor required for the development of multiple lineages of the immune system (16). Differentiation into the myeloid lineage requires PU.1 expression, with high expression levels being linked to macrophage differentiation (17). In tissues with small numbers of earlier hematopoietic progenitors PU.1 expression can therefore be used as a marker for myeloid cells, in particular of the monocyte and macrophage lineage. YFP signal strength is correlated with PU.1 expression levels (13), and cells that express low levels of PU.1 (such as B cells or certain subtypes of T cells (18)) cannot be detected on the basis of YFP expression. The fusion of YFP to PU.1 does not affect PU.1 function, as mice homozygous for the allele are viable and do not show any detectable hematopoietic defects (13).
Fig. 1.
The inflammatory infiltrate in unwounded InvEE mice. (A-B) Immunofluorescent staining of back skin sections of unwounded InvEE and WT mice stained with a F4/80 antibody and DAPI. (C) Flow cytometric quantification (% of total counts) of leukocytes (CD45), monocytes (CD11b) and macrophages (F4/80) and T cells (CD3) in unwounded PU.1-YFPxInvEE−/− (WT) and PU.1-YFPxInvEE+/− (InvEE) skin. Significant differences (*p < 0.05, **p < 0.01; Student’s t-test) are indicated by asterisk. Data are presented as the mean ± SEM (WT n = 5, InvEE n = 5). (D-E) Immunohistochemical staining with an antibody against nuclear YFP in unwounded skin sections of (D) WT and (E) PU.1-YFP skin. High-magnification images of red boxed area shown in the lower, right image corner. (F) Flow cytometric analysis of YFP+ cell populations in PU.1-YFP+/+, PU.1-YFP−/− (WT) and PU.1−/−InvEE+/− (control) mice. Populations were analyzed for co-expression of T cell (CD3) or granulocyte (Gr-1) markers. Cells gated for viable cells, singlets and CD45+. (G) Flow cytometric analysis of YFP+ populations in the dermis and co-labelling with CD11b and F4/80. Cells gated for viable cells, singlets and CD45+. Cells in the third panel also gated for YFP. (H) Flow cytometric analysis of YFP+CD45+ populations in WT and InvEE dermis and epidermis. (I) Flow cytometric quantification of YFP+ populations in PU.1-YFPxInvEE, PU.1-YFPxWT and PU.1-YFP−/− control spleen. Dashed lines denote basement membrane of IFE & hair follicles. Scale bars = 100 μm.
When tested by flow cytometry and immunostaining, the majority of YFP+ cells isolated from skin were positive for the pan leukocyte marker CD45 and common monocyte and murine macrophage markers such as CD11b, F4/80 and CD68 (Fig. 2 E-F). No YFP+ cells co-expressed the T cell marker CD3 or the granulocyte/monocyte marker Gr-1 (this clone reacts predominantly with Ly6G but can also stain cells with highest Ly6C expression) (Fig. 1F) and few expressed markers of inflammatory monocytes (Ly6Cint/high MHC-IIlow) (Fig. S1A).
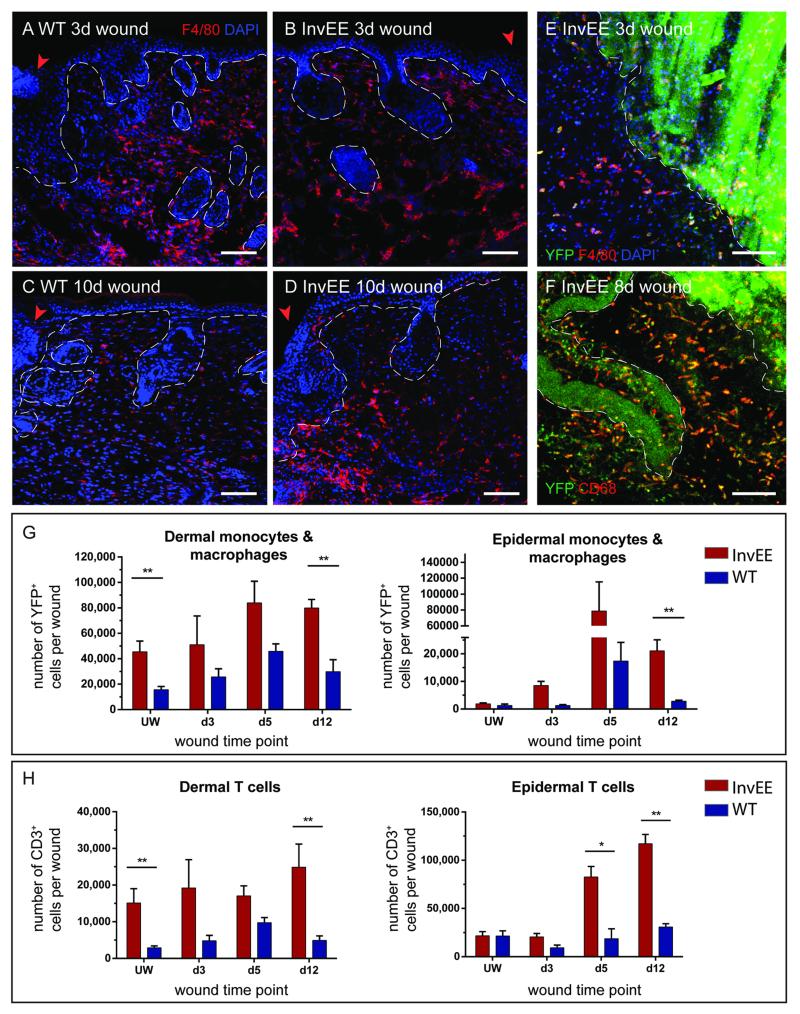
Fig. 2.
Cell quantifications in wounded skin. (A-D) Immunofluorescent stainings of InvEE and WT backskin sections from (A, B) 3 and (C, D) 10 day old wounds with a F4/80 antibody and DAPI. (E-F) Horizontal flat mounts of PU.1-YFPxInvEE skin 3 and 8 days after wounding, immunostained with F4/80, CD68 and DAPI. (G) Quantification of YFP+ cells in the dermis and epidermis of PU.1-YFPxInvEE+/− (InvEE) and PU.1-YFPxInvEE−/− (WT) skin during different wound healing time points and in unwounded skin. Cells were gated for viable cells, singlets and CD45+ YFP+ populations. Significant differences (*p < 0.05, **p < 0.01; Student’s t-test) are indicated by asterisk. Data are presented as the mean ± SEM (n = 6 biological replicates per time-point and group. (H) Quantification of CD3+ cells in the dermis and epidermis of InvEE and WT skin during different wound healing time points and in unwounded skin. Cells were gated for viable cells, singlets and CD45+ CD3+ populations. Significant differences (*p < 0.05, **p < 0.01; Student’s t-test) are indicated by asterisk. Data are presented as the mean ± SEM (n = 4 biological replicates per time-point and group. Arrowhead denotes the wound edge. Dashed line in A-D represents the basement membrane of IFE and hair follicles, in E-F the wound edge. Scale bars = 100 μm.
We next generated PU.1-YFPxInvEE double transgenic mice and analyzed PU.1high expressing monocytes and macrophages in the dermis and epidermis. In unwounded skin, a mean of 93.3% InvEE (PU.1-YFP+/+InvEE+/−) and 63.8% WT (PU.1-YFP+/+InvEE−/−) YFP+ F4/80+ cells were Ly6Clo and MHC IIlow/high mature macrophages (Fig. S1A). We conclude that the YFP signal in PU.1-YFP skin is primarily attributable to F4/80+ monocytes and macrophages (Fig. 1G).
There was a significant increase in YFP+ leukocyte infiltration in unwounded InvEE skin compared to WT (Fig. 1H, Fig. S1A). In addition, an increase in YFP+ cells in spleens of unwounded InvEE mice was evident (Fig. 1I), correlating with the previously reported splenomegaly (6).
To induce tumors in PU.1-YFP+/+InvEE+/− mice we used 8 mm punch biopsies to make a full thickness wound in the back skin of each mouse. This typically results in 40-60% of mice developing a tumor at the site of the wound (6). We then determined the number of F4/80+ cells in immunostained skin sections (Fig. 2A-D, S2A) and FACS sorted YFP+ cells (Fig. 2G, S1B-C) at intervals following wounding InvEE (n = 19) and WT (n = 17) mice.
YFP+ monocytes and macrophages were more abundant in InvEE than WT skin at all time points examined (Fig. 2G, Fig. S1), both in epidermis and dermis. The number of Ly6Chigh MHC-IIlow inflammatory monocytes was slightly increased during early wounding healing stages in InvEE skin (Fig. S1B). Ly6Chigh MHC-IIlow cells represented 28.3% (InvEE) and 10.1% (WT) of YFP+ F4/80+ cells 5 days after wounding, but did not show substantial differences at 10 days, with 24.5% (InvEE) and 24.1% (WT), respectively (Fig. S1B-C). Five days after wounding, 65.42% of InvEE and 74.72% of WT YFP+ F4/80+ populations consisted of Ly6Clo and MHC IIlow/high mature macrophages (Fig. S1B) and ten days after wounding this number was maintained at 60.01% in InvEE and decreased to 45.93% in WT skin (Fig. S1C). The peak in macrophage infiltration was at day 5 after wounding (Fig. 2G), but it was notable that at day 12 macrophage numbers remained elevated in InvEE epidermis and dermis while declining in WT (Fig. 2G, UW p = 0.0097, d12 dermis p = 0.0045, d12 epidermis p =0.0007).
The number of dermal CD3+ T lymphocytes was also significantly elevated in unwounded InvEE skin (Fig. 2H, p = 0.0048), correlating with previously published results (7). At day 12 after wounding there were significantly more CD3+ T cells in InvEE than in WT dermis (p = 0.0076). In contrast, the number of epidermal T cells increased at day 5 and 12 after wounding in InvEE epidermis but remained relatively unchanged in WT epidermis (Fig. 2H, day 5: p = 0.0126, day 12: p = 0.0019).
Depletion of monocytes and macrophages reduces tumor incidence
To investigate whether macrophages are required for wound-induced tumor formation, we used the CD11b-DTR mouse model (19). This transgenic strain allows specific ablation of CD11b+ cells (monocytes and macrophages) on administration of 10 ng/g Diphtheria Toxin (DTx) three times a week (20), without any depletion of neutrophils (19). To avoid previously reported toxicity to the liver and lungs (20), we irradiated InvEE mice (6-12 weeks old) and transplanted them with bone marrow from CD11b-DTR donor mice at 5-6 weeks of age. Five weeks after bone marrow reconstitution, InvEE mice were treated with DTx to ablate CD11b+ cells in the transplanted bone marrow. Three days after the start of treatment full thickness wounds were made in the back skin and treatment with DTx (n = 14) or saline (control, n = 10) was continued. To confirm successful chimerism, male bone marrow was transplanted into female recipients and the spleen of recipient mice were subjected to Y probe in situ hybridization (Fig. S2B).
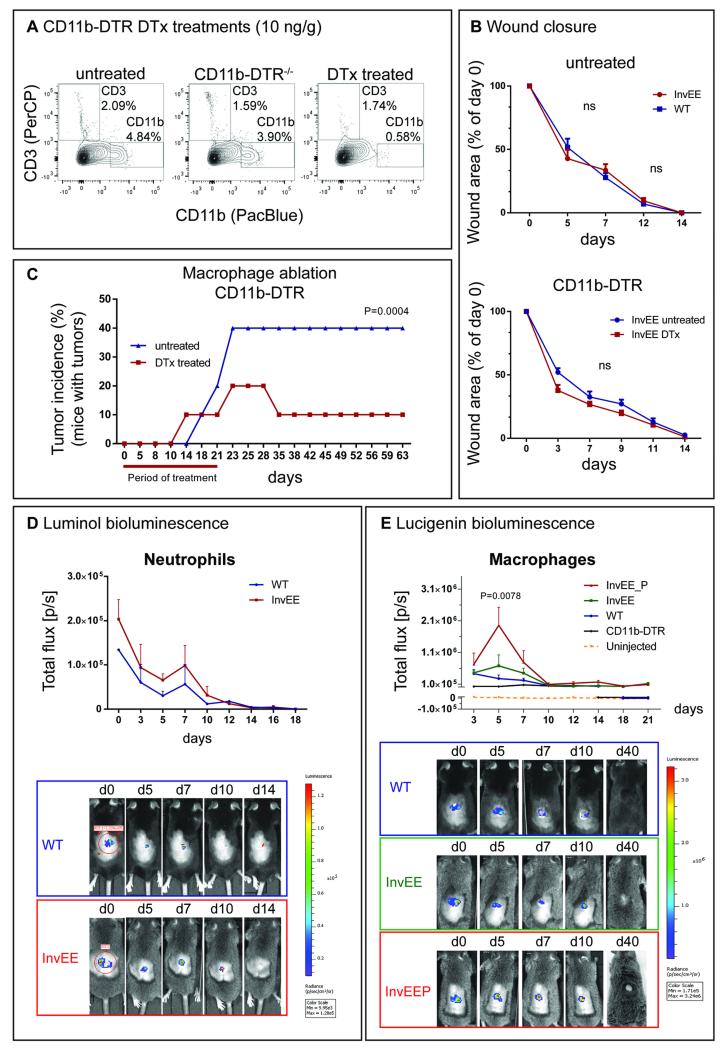
Analysis of the skin of treated animals 2 days after the start of treatment and 12 days after wounding showed successful CD11b+ cell depletion, while CD3+ cell numbers remained unchanged (Fig. 3A, Fig. S2C-E, p = 0.0285 for CD45, p = 0.00005 for CD11b and p = 0.00005 for F4/80). Ablation of monocytes and macrophages had no effect on wound closure time (Fig. 3B, CD11b-DTR), concurring with previous findings (21). Similarly, no difference in wound closure timing was observed between untreated InvEE and WT skin (Fig. 3B, untreated), with wound closure in both cases typically occurring at day 14 (±1d).
Fig. 3.
Macrophage ablation and tumor prediction. (A) Flow cytometric analysis of CD3+ and CD11b+ cell populations in WT (CD11b-DTR−/−), untreated and DTx treated CD11b-DTR+/+ mice after administration of 2 x 10 ng/g DTx. For quantifications see Fig. S2C. Cells gated for singlets and viable cells. n = 3 biological replicates. (B) Wound closure duration in untreated InvEE and WT mice and in untreated and DTx treated InvEE chimeras that received CD11b-DTR bone marrow. (C) Tumor incidence rates (% of mice with 1 tumor) in InvEE CD11b-DTR bone marrow chimeras treated with DTx (n=14) and saline (untreated, n=10) for 3 days before and 21 days after wounding. Wilcoxon test: p = 0.0004. (D) Luminol bioluminescent signal intensities in WT (n = 3) and InvEE (n = 4) mice during different wound healing time-points, starting 4 hours after wounding/ injection (d0). (E) Lucigenin bioluminescent signal intensities in uninjected (n = 3), DTx-treated CD11b-DTR (n = 3), WT (n = 9), InvEE mice which do not develop tumors (InvEE, n = 7) and InvEE mice which develop tumors (InvEEP, n = 7) during different wound healing time points. Wilcoxon-test: p = 0.0078 (WT vs InvEEP), p = 0.0391 (InvEE vs InvEEP), p = 0.0078 (CD11b-DTR vs InvEEP), p = 0.0234 (InvEE vs WT).
Treatment of InvEE mice with DTx for 21 days after wounding revealed a significant reduction in tumor incidence on macrophage ablation (Fig. 3C, p = 0.0004). We conclude that macrophages are required for wound-induced tumor formation.
Increase in macrophage populations at the wound site predicts tumor risk
To determine whether the abundance of macrophages in InvEE wounds was predictive of whether a wound would develop a tumor, we set up a Lucigenin-based bioluminescence assay to monitor macrophage influx at sites of wounded InvEE (n = 9) and WT skin (n = 7). Lucigenin detects superoxide (O2−) production by NADPH oxidase (Phox) in phagocytic cells (22). This non-invasive in vivo imaging approach allows detection and tracking of macrophages in the same animal from prior to wounding up to wound-induced tumor formation (23). Because Phox is also enzymatically active in other phagocytes, such as neutrophils (24), we used Luminol as a second reagent to monitor neutrophil behavior. Luminol interacts with myeloperoxidase (MPO), an enzyme that is predominantly active in neutrophils and monocytes in the acute phases of inflammation (25). Lucigenin signal contamination by neutrophils is unlikely since they translocate the enzyme to the phagosome after assembly, whereas macrophages present it on the plasma membrane (26). Due to its structure and the two positive charges, Lucigenin is barely membrane-permeable (23). It can therefore be assumed that the imaging agent is selective in its detection of the macrophage population.
After intraperitoneal (i.p.) injection of 100 mg/kg of Luminol into wounded InvEE (n=4) and WT (n=2) mice, bioluminescence was found within and in close proximity to the wound site, demonstrating peak intensity 4 hours after wounding (Fig. 3D, d0). Signal strength then dropped profoundly and after day 7 only a weak residual signal could be detected in either InvEE or WT mice, confirming that there were few neutrophils present at that stage (Fig. 1F). In line with these results, there was no difference between Luminol signal intensity in InvEE and WT mice, suggesting that neutrophil populations are of similar size (Fig. 3D). Likewise, no correlation could be found between InvEE tumor risk and Luminol signal intensity. Signal saturation typically occurred 10-15 minutes after Luminol injection (Fig. S2F).
Following i.p. injection of 20 mg/kg of Lucigenin 4 hours after wounding (d0) we detected a robust bioluminescent signal in all wounds (Fig. 3E). Signal saturation typically occurred 30 minutes after injection in InvEE mice and 10 minutes after injection in WT mice, which were selected as measuring points (Fig. S2G). Macrophage-ablated CD11b-DTR mice (Fig. 3E, CD11b-DTR, p = 0.0078) showed little signal above baseline (Fig. 3E, uninjected). In WT mice with an intact immune system the initial signal decreased progressively until by day 10 it was the same as in macrophage-depleted mice (Fig. 3E, WT). Importantly, in those InvEE mice that did not develop a tumor, the Lucigenin signal was constant for approximately 5 days and declined thereafter. In contrast, in those mice that went on to develop a tumor signal intensity increased substantially, with a peak 5 days after wounding (Fig. 3E, InvEEP, p = 0.0078), decreasing to WT level at day 10. We conclude that an elevated number of macrophages recruited to the wound correlates with an elevated tumor risk (Fig. S2H).
Macrophage polarization in InvEE wounds
Classically, two distinct functional classifications have been defined for macrophages based on their activation states: classically activated (M1) macrophages have the role of inflammatory, cytotoxic effector cells in Th1 immune responses, while alternatively activated (M2) macrophages are involved in immunosuppression, scavenging of debris and tissue repair (27). The current consensus is that these classifications represent extreme states in a spectrum of activation profiles and cannot fully reproduce the complex nature of most macrophages in vivo (28). In response to signals from the microenvironment macrophages show remarkable plasticity and while they transiently adopt M1- or M2-‘like’ activation profiles (29), respective subtypes and functions may vary based on external signals received (28). Reportedly, alternatively activated macrophages express high levels of scavenger mannose (CD206, also known as MRC1) and galactose receptors, release anti-inflammatory cytokines (such as IL-10, IL-1ra or TGF-β) and downregulate pro-inflammatory cytokines in different settings (such as TNF, IL-1β, IL-12, IL-6 and CCL3)(30). In some alternatively activated phenotypes expression of MHC-II and/or Arginase is also upregulated (30). Classically polarized macrophages typically express pro-inflammatory cytokines such as TNF, IL-1, IL-6 (also present in some M2-like subtypes (31)), IL-12 and IL-23. The antimicrobial functions of M1 macrophages are linked to upregulation of inducible nitric oxide synthase (iNOS, also known as NOS2), which converts Arginine into nitric oxide (NO)(32).
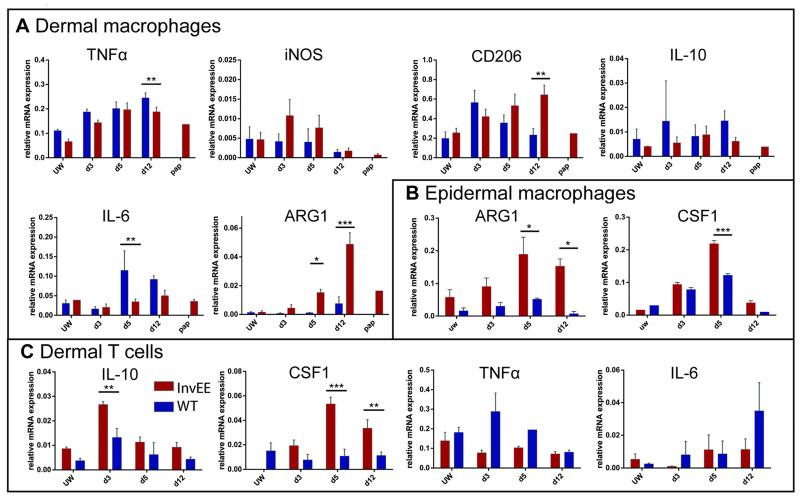
To characterize their polarization status, YFP+ F4/80+ monocytes and macrophages were isolated from PU.1-YFPxInvEE and PU.1-YFPxWT mice and FACS sorted (Fig. S3A). The cells were collected from dermis and epidermis of unwounded skin and at different times after wounding. Specific gene expression was analyzed using qRT-PCR (Fig. 4A-B). Most macrophage polarization markers, including IL-10, were not altered between InvEE and WT macrophages (Fig. 4A). A conventional marker of classical activation, iNOS, was expressed at similar levels in both InvEE and WT mice, with a slight increase in the first days of wound healing (Fig. 4A), an observation that was supported by immunofluorescent labelling of skin sections (Fig. S4B). Additional markers of alternative macrophage activation that were elevated on wounding to the same extent in WT and InvEE mice were Ym1, a murine M2 marker, TGF-β, VEGF and MCP1 (also known as CCL2) (Fig. S3B). Pro-inflammatory cytokines such as IL-6 and TNFα were significantly decreased during mid- and late InvEE wound healing stages compared to WT (Fig. 4A, p = 0.0094 for IL-6 at day 5; p = 0.0051 for TNFα at day 12).
Fig. 4.
Gene expression profiling of wound macrophages & monocytes and T cells. (A) Analysis of mRNA expression in sorted YFP+F4/80+ dermal wound macrophages/monocytes during different wound healing time points, in unwounded skin (UW) and papillomas (pap). (B) Analysis of mRNA expression in sorted YFP+F4/80+ epidermal wound macrophages/monocytes during different wound healing time-points and in unwounded skin (UW). (C) Analysis of mRNA expression in sorted CD3+F4/80− dermal wound T cells during different wound healing time points and in unwounded skin (UW). Data are presented as the mean ± SEM (n = 4 for InvEE, n = 4 for WT per time point). Ct values normalized to GAPDH. Significant differences (*p < 0.05, **p < 0.01; Student’s t-test) are indicated by asterisk.
Although some markers did not differ significantly between WT and InvEE skin, mannose receptor (Cd206) expression steadily increased in dermal InvEE macrophages from 3 days after wounding with significant differences at day 12 while it peaked in WT mice at around day 3 and then decreased again (Fig. 4A, p = 0.0030). The gene expression data were supported by immunofluorescent labelling of skin sections (Fig. S4).
Similarly, Arg1 expression increased during the late wound healing stage in InvEE dermis and epidermis while it showed only a slight upregulation during WT wound healing (Fig. 4A; p = 0.0231 for day 5, p = 3.44E-08 for day 12 in the dermis) (Fig. 4B; p = 0.0146 for day 5 and p = 0.0105 for day 12 in the epidermis). Arginase-expressing macrophages were also elevated in the blood of InvEE mice following wounding (Fig. S5A). Additionally, integrin α6+ basal keratinocytes expressed only low levels of ARG1 in healthy InvEE skin and an insignificant increase after wound healing (Fig. S5B).
Dermal T cells also contributed to the wound environment. In early and later stages of InvEE wound healing they exhibited increased IL-10 and decreased Tnfα expression relative to WT macrophages, suggesting a Th2 phenotype (Fig. 4C, p = 0.0022 for IL-10 at day 3). Most strikingly, T cells also upregulated Csf1 expression throughout InvEE wound healing, indicating a vital role as macrophage attractors (Fig. 4C, p = 0.0003 at day 5 and p = 0.0035 at day 12).
We conclude that InvEE macrophages undergo marked phenotypic changes during wound healing response. They do not, however, correspond with the full phenotypes of M2a, M2b or M2c macrophages (30).
In vivo inhibition of ARG1 in wounds reduces tumor formation
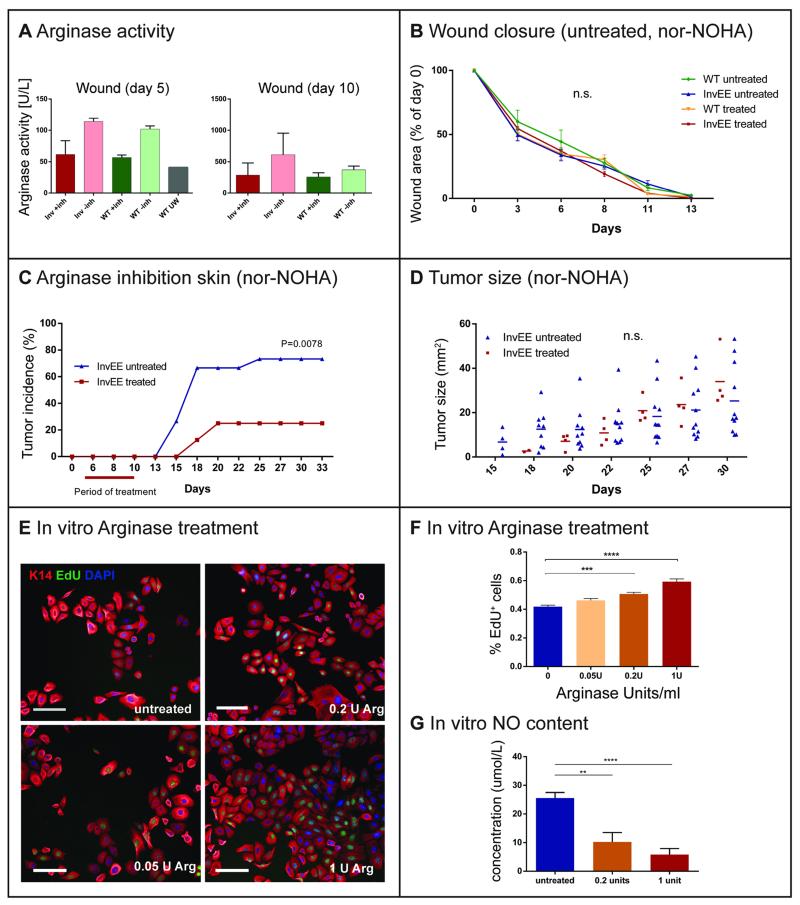
Given the strong expression of ARG1 in wound-recruited macrophages, we examined its effect on wound healing and tumor development in InvEE transgenics. Human and murine ARG1 activity can be blocked by the selective inhibitor N(omega)-hydroxy-nor-l-arginine (nor-NOHA) (33), which was delivered using a subcutaneously implanted osmotic pump. Pumps were inserted in close proximity to the wound 5 days after wounding (Fig. S5C), at the time when ARG1 expression typically started to increase in InvEE animals (Fig. 4A-B). Continuous dosing at a rate of 0.25 μl per hour ensured constant compound levels. Successful ARG1 inhibition was confirmed in blood plasma and wounded skin samples taken 5 days after implantation (Fig. 5A, Fig. S5D, n = 4). Although the abundance of ARG1 was greatly increased in InvEE skin relative to WT, both at the mRNA (Fig. 4A-B) and protein level (Fig. S5E), enzyme activity normalized to the amount of protein was the same (Fig. 5A).
Fig. 5.
In vivo Arginase inhibition and regulation of keratinocyte proliferation. (A) Arginase activity assay from isolated wound tissue (measured through urea production in Units per liter) in Nor-NOHA (+inh) or saline (-inh) treated mice, 5 and 10 days after wounding. Data are presented as the mean ± SEM (InvEE n = 4, WT n = 4). (B) Time to completed wound closure in Nor-NOHA and saline treated WT (n = 5 per group) and InvEE (n = 8 per group) mice, days after wounding. (C) Effects of the Arginase-inhibitor nor-NOHA (40 mg/kg/d) on InvEE tumor incidence following a 5d treatment, starting 5d after wounding (n = 21), compared to saline (n = 20). Wilcoxon-test: p = 0.0078. (D) Effects of nor-NOHA on tumor sizes (mm2) in InvEE mice, days after wounding. (E) Immunofluorescent staining of murine keratinocytes cultured feeder-free and substituted for 48 hours with saline (untreated), 0.05, 0.2 and 1 Unit of bovine L-Arginase. Cells were stained with antibodies against K14, Edu and DAPI. Scale bar = 100 μm. (F) Percentage of Edu+ proliferating keratinocytes after 48h Arginase treatment in different concentrations (Units per mL). Data are presented as the mean ± SEM (n = 4 independent experiments, 12 replicates). (G) Measurement of in vitro nitric oxide (NO) production by keratinocytes following treatment with 0, 0.2 and 1 units of L-Arginase. Data are presented as the mean ± SEM (n = 2 independent experiments, 12 replicates). Significant differences (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; Student’s t-test) are indicated by asterisk.
When InvEE and WT animals were treated with nor-NOHA 5 days after wounding, the rate of wound healing was unaffected (Fig. 5B). When monitored for 33 days, inhibitor-treated InvEE mice developed significantly fewer papillomas (Fig. 5C, p = 0.0078, n = 21) than the control group, which had received saline (n = 20) even though pump-administered nor-NOHA lost its Arginase inhibiting activity 10 days after implantation (Fig. 5A, n = 4). In InvEE mice treated with saline tumors occasionally appeared at the site where an incision had been made to implant the osmotic pump (Fig. S5C), whereas inhibitor-treated mice were protected in these areas. Those papillomas that did develop in nor-NOHA-treated mice had a similar size to those of untreated animals (Fig. 5D). We conclude that increased ARG1 activity in the late stages of wound healing is required for wound-induced papilloma formation.
Arginase activity stimulates keratinocyte proliferation
To examine whether ARG1 activity affected epidermal proliferation in vivo, we quantitated the number of Ki67+ keratinocytes in skin sections (Fig. S6). In InvEE mice, the numbers of Ki67+ keratinocytes were elevated in undamaged skin, as reported previously (6, 7) and throughout all wound healing stages, compared to WT (Fig. S6A). Nor-NOHA treatment of InvEE mice led to a significant decline in Ki67+ keratinocytes 10 days after wounding, compared with samples from saline-treated InvEE mice (Fig. S6B).
To examine whether ARG1 activity can directly influence keratinocyte proliferation, we added L-Arginase to cultures of primary mouse keratinocytes (passage 4-7). The presence of active Arginase during a 48 hour incubation period promoted keratinocyte proliferation in a concentration-dependent manner, as evaluated by cellular EdU incorporation into the DNA. While the addition of 0.05 Units/mL of L-Arginase did not lead to a significant change in EdU incorporation, concentrations of 1 Unit and 0.2 Units per mL were stimulatory (Fig. 5E-F, p < 0.0001 for 0 vs 1 U/mL, p < 0.0001 for 0 vs 0.2 U/mL, n = 3). This correlates well with the observation that 200 Units per liter correspond to enzyme levels in a murine wound environment (Fig. 5A). In addition to stimulating EdU incorporation, ARG1 treatment increased total cell numbers significantly (Fig. 5E, Fig. S5F, p = 0.0073 for 0.2 vs 0 U/mL, p = 0.0432 for 1 vs 0 U/mL).
We next investigated the metabolic mechanism behind the growth-promoting effect of Arginase. It has long been known that the main enzyme substrate Arginine is metabolized by two opposing pathways: nitric oxide synthase (NOS) and Arginase (34), which have contrasting biological effects. NOS activity creates a cytotoxic wound environment via nitric oxide (NO) production (35), whereas the Arginase pathway promotes cell growth and proliferation by increasing the availability of ornithine, which is the precursor for the synthesis of polyamines and collagen (34). If Arginase out-competes NOS for the substrate Arginine, the additional effect is a reduction in NO production, which has been shown to promote keratinocyte and fibroblast proliferation (36, 37). When we measured the NO content of keratinocyte conditioned medium, we found that concentrations in the wells of highly proliferating, Arginase-treated cells (1 Unit/mL and 0.2 Units/mL) were significantly lower than in untreated wells (Fig. 5G, p < 0.0001, n = 2). In line with a study published by Krischel et al. (37), we found that keratinocytes proliferated in the presence of low NO concentrations (5-10 μmol/L) in wells that had been treated with 0.2 or 1 Unit of bovine Arginase (Fig. 5G). Medium that was conditioned by untreated cells showed a higher NO concentration (35-50 μmol/L) (Fig. 5G), which was previously reported to induce differentiation (37).
We conclude that the most likely explanation for the anti-tumorigenic effect of ARG1 inhibition is that ARG1 expression by macrophages stimulates keratinocyte proliferation by reducing NO production.
DISCUSSION
The role of macrophages in tumor maintenance and progression is well established in many tumor types, but whether they also contribute to early events in tumor promotion and, if so, how, is obscure. Here we show that in a mouse model of chronic epidermal hyperproliferation and skin inflammation the level of macrophage accumulation 5 days after wounding is predictive of which wounds will convert into tumors. This correlates with a peak in expression of the alternative macrophage activation marker, Arginase 1. Arginase 1 stimulates keratinocytes to proliferate by reducing the availability of Nitric Oxide (NO) (Fig. 5E-F) (37) and pharmacological inhibition of Arginase prevents tumor formation.
Increased ARG1 expression during normal murine and human skin wound healing has been reported previously (38, 39) (Fig. 4A-B). However, InvEE macrophages differ from WT in that the level of ARG1 following wounding is considerably higher and expression remains elevated for at least 12 days, whereas in WT macrophages it declines after day 5. In addition to ARG1 directly stimulating keratinocyte proliferation it is likely to be immunosuppressive, dampening the effects on T lymphocyte activation and proliferation, as observed in other tumor models (40, 41). However, the primary focus of this study was on its growth-promoting potential. As a consequence of sustained and elevated ARG1 expression, anti-inflammatory, proliferative stimuli are intensified and extended after wound re-epithelialization in InvEE skin. In psoriasis, ARG1 overexpression in keratinocytes leads to a substantial increase in proliferation (36) and ARG1 has also been reported to be expressed in wound-activated fibroblasts (42). However, in the InvEE model macrophages are the predominant source of ARG1, which is only detectable at low levels in keratinocytes (Fig. S5B) and fibroblasts (Esther Hoste, unpublished data) before and after wounding.
By monitoring macrophages at different times after wounding, we demonstrated that at day 5 they switch their phenotype from pro-inflammatory to an alternative activation state. While they upregulate Arginase-1 and other common M2 markers such as CD206, increases in markers associated with M2c alternative macrophage activation (CD163, CD206, TGFβ) were not statistically significant. Thus InvEE wound macrophages do not display all the typical markers of M2 polarization. The markers they do express mostly correlate with tissue repair macrophages (43), which downregulate IL-10 and upregulate CD206 and exhibit highly context-dependent iNOS expression. Nevertheless, upregulation of ARG1, CD206, TGFβ and VEGF is also typically found in tumor-associated macrophages in established lesions (27, 44). Some of the phenotypic variation evident during gene expression profiling could reflect the contribution of Ly6Chigh MHC IIlow inflammatory monocytes to the YFP+ F4/80+ populations, constituting around 10-28% of sorted cells during intermediate and late wound healing stages.
Macrophages are not the only immune cells that accumulate in InvEE skin, as increased numbers of CD4+ T cells, γδ T cells, mast cells and neutrophils are also present (5-7, 41) and deletion of bone marrow derived γδ T cells leads to a reduction in the abundance of macrophages in InvEE skin (6). We speculate that T cells attract macrophages at least in part by releasing CSF-1, which could also promote an M2-like skewing of macrophage functions (45). Wound-recruited T cells in InvEE mice could further exert an immunosuppressive effect on the microenvironment via IL-10 expression. In mice, IL-10 is typically an indicator of Th2 polarization in T lymphocytes and downregulates the Th1 response (46).
The overall picture that emerges from our studies of wound-induced tumor formation is of tumorigenesis via collaboration between multiple cell types and signaling molecules, including upregulation of IL-1α in differentiating keratinocytes driving increased expression of the dipeptidyl peptidase CD26 in fibroblasts (6, 8), recruitment of macrophages and other immune cells and a pivotal role of macrophages in both creating an immunosuppressive environment and stimulating keratinocytes to proliferate.
There are interesting parallels between the role of macrophages in our model and in other tumor settings. In mouse models of Lewis lung carcinoma and melanoma, expression of ARG1 mRNA by tumor-associated macrophages promotes tumor growth, possibly via the production of polyamines or an influence on the nitrogen metabolism (an exact mechanism was not presented).(47) In a murine model of glioma, carcinogenesis is initiated through a persistent CNS wound environment that recruits local macrophages and stimulates prolonged inflammation and growth factor secretion, thereby activating local neural stem cells to hyperproliferate (48). Macrophage-dependent tumor initiation is observed in colitis-associated cancer in mice (49) and is dependent on NF-κB signalling in macrophages (49). In the InvEE model ablation of MyD88, an adapter protein for NF-κB signalling, in radiosensitive leukocytes also prevents tumour formation (50). The importance of macrophages in gastric cancer progression and invasion is now well established (51), and infection with H. pylori, a major risk factor for gastric cancer, causes prolonged macrophage infiltration and activation (52). In good agreement with this observation, we have recently established a tumor-promoting role for flagellated bacteria in the wounds of InvEE mice (50). Thus, a common characteristic of macrophage-dependent epithelial cancers is prolonged exposure to a pathogenic agent and/or repeated injury, resulting first in acute and then sustained chronic inflammation and an aberrant regenerative response of recruited macrophages (53). These studies highlight the growing importance of cancer immunotherapies, where attempts to reeducate macrophages have already shown some success in glioma (54) and ovarian cancer (55).
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by the European Union FP7 programme (HEALING), Cancer Research UK (at the CR-UK Cambridge Research Institute), and grants for F.M.W. from the Medical Research Council and Wellcome Trust. A.R is the recipient of a MRC PhD studentship. The authors acknowledge the use of Core Facilities provided by the financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London and King’s College Hospital NHS Foundation Trust. Input from Beate M Lichtenberger, Gernot Walko and Alexander Schreiner is gratefully acknowledged. The authors also thank Claus Nerlov for providing the PU.1-YFP strain, KCL and CR-UK CI core staff for technical support, and the Nikon Imaging Centre at KCL for expert assistance.
Footnotes
The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Developmental cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–71. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 3.Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–31. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 4.Barcellos-Hoff MH, Lyden D, Wang TC. The evolution of the cancer niche during multistage carcinogenesis. Nat Rev Cancer. 2013;13:511–8. doi: 10.1038/nrc3536. [DOI] [PubMed] [Google Scholar]
- 5.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170–80. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- 6.Arwert EN, Lal R, Quist S, Rosewell I, van Rooijen N, Watt FM. Tumor formation initiated by nondividing epidermal cells via an inflammatory infiltrate. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1007404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hobbs RM, Silva-Vargas V, Groves R, Watt FM. Expression of activated MEK1 in differentiating epidermal cells is sufficient to generate hyperproliferative and inflammatory skin lesions. J Invest Dermatol. 2004;123:503–15. doi: 10.1111/j.0022-202X.2004.23225.x. [DOI] [PubMed] [Google Scholar]
- 8.Arwert EN, Mentink RA, Driskell RR, Hoste E, Goldie SJ, Quist S, et al. Upregulation of CD26 expression in epithelial cells and stromal cells during wound-induced skin tumour formation. Oncogene. 2012;31:992–1000. doi: 10.1038/onc.2011.298. [DOI] [PubMed] [Google Scholar]
- 9.Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9:259–70. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–8. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 11.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7:211–7. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–56. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 14.Cui YZ, Hisha H, Yang GX, Fan TX, Jin T, Li Q, et al. Optimal protocol for total body irradiation for allogeneic bone marrow transplantation in mice. Bone Marrow Transplant. 2002;30:843–9. doi: 10.1038/sj.bmt.1703766. [DOI] [PubMed] [Google Scholar]
- 15.Lichtenberger BM, Tan PK, Niederleithner H, Ferrara N, Petzelbauer P, Sibilia M. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140:268–79. doi: 10.1016/j.cell.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 16.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–41. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 17.Back J, Allman D, Chan S, Kastner P. Visualizing PU.1 activity during hematopoiesis. Exp Hematol. 2005;33:395–402. doi: 10.1016/j.exphem.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Carotta S, Wu L, Nutt SL. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev. 2010;238:63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 19.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115:56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoneman V, Braganza D, Figg N, Mercer J, Lang R, Goddard M, et al. Monocyte/macrophage suppression in CD11b diphtheria toxin receptor transgenic mice differentially affects atherogenesis and established plaques. Circ Res. 2007;100:884–93. doi: 10.1161/01.RES.0000260802.75766.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin P, D'Souza D, Martin J, Grose R, Cooper L, Maki R, et al. Wound healing in the PU.1 null mouse--tissue repair is not dependent on inflammatory cells. Curr Biol. 2003;13:1122–8. doi: 10.1016/s0960-9822(03)00396-8. [DOI] [PubMed] [Google Scholar]
- 22.Rembish SJ, Trush MA. Further evidence that lucigenin-derived chemiluminescence monitors mitochondrial superoxide generation in rat alveolar macrophages. Free Radic Biol Med. 1994;17:117–26. doi: 10.1016/0891-5849(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 23.Tseng JC, Kung AL. In vivo imaging of inflammatory phagocytes. Chemistry & biology. 2012;19:1199–209. doi: 10.1016/j.chembiol.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiological reviews. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 25.Gross S, Gammon ST, Moss BL, Rauch D, Harding J, Heinecke JW, et al. Bioluminescence imaging of myeloperoxidase activity in vivo. Nat Med. 2009;15:455–61. doi: 10.1038/nm.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson A, Jesaitis AJ, Lundqvist H, Magnusson KE, Sjolin C, Karlsson A, et al. Different subcellular localization of cytochrome b and the dormant NADPH-oxidase in neutrophils and macrophages: effect on the production of reactive oxygen species during phagocytosis. Cell Immunol. 1995;161:61–71. doi: 10.1006/cimm.1995.1009. [DOI] [PubMed] [Google Scholar]
- 27.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–55. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 28.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–96. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 29.Davis MJ, Tsang TM, Qiu Y, Dayrit JK, Freij JB, Huffnagle GB, et al. Macrophage M1/M2 polarization dynamically adapts to changes in cytokine microenvironments in Cryptococcus neoformans infection. mBio. 2013;4:e00264–13. doi: 10.1128/mBio.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–86. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 31.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–12. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 32.Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004;75:639–53. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 33.Di Costanzo L, Ilies M, Thorn KJ, Christianson DW. Inhibition of human arginase I by substrate and product analogues. Archives of biochemistry and biophysics. 2010;496:101–8. doi: 10.1016/j.abb.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shearer JD, Richards JR, Mills CD, Caldwell MD. Differential regulation of macrophage arginine metabolism: a proposed role in wound healing. The American journal of physiology. 1997;272:E181–90. doi: 10.1152/ajpendo.1997.272.2.E181. [DOI] [PubMed] [Google Scholar]
- 35.Shi HP, Most D, Efron DT, Tantry U, Fischel MH, Barbul A. The role of iNOS in wound healing. Surgery. 2001;130:225–9. doi: 10.1067/msy.2001.115837. [DOI] [PubMed] [Google Scholar]
- 36.Bruch-Gerharz D, Schnorr O, Suschek C, Beck KF, Pfeilschifter J, Ruzicka T, et al. Arginase 1 overexpression in psoriasis: limitation of inducible nitric oxide synthase activity as a molecular mechanism for keratinocyte hyperproliferation. Am J Pathol. 2003;162:203–11. doi: 10.1016/S0002-9440(10)63811-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krischel V, Bruch-Gerharz D, Suschek C, Kroncke KD, Ruzicka T, Kolb-Bachofen V. Biphasic effect of exogenous nitric oxide on proliferation and differentiation in skin derived keratinocytes but not fibroblasts. J Invest Dermatol. 1998;111:286–91. doi: 10.1046/j.1523-1747.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- 38.Campbell L, Saville CR, Murray PJ, Cruickshank SM, Hardman MJ. Local arginase 1 activity is required for cutaneous wound healing. J Invest Dermatol. 2013;133:2461–70. doi: 10.1038/jid.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kampfer H, Pfeilschifter J, Frank S. Expression and activity of arginase isoenzymes during normal and diabetes-impaired skin repair. J Invest Dermatol. 2003;121:1544–51. doi: 10.1046/j.1523-1747.2003.12610.x. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–49. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 41.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–8. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 42.Witte MB, Barbul A, Schick MA, Vogt N, Becker HD. Upregulation of arginase expression in wound-derived fibroblasts. J Surg Res. 2002;105:35–42. doi: 10.1006/jsre.2002.6443. [DOI] [PubMed] [Google Scholar]
- 43.Novak ML, Koh TJ. Macrophage phenotypes during tissue repair. J Leukoc Biol. 2013;93:875–81. doi: 10.1189/jlb.1012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 46.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170:2081–95. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–63. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seyfried TN. Perspectives on brain tumor formation involving macrophages, glia, and neural stem cells. Perspectives in biology and medicine. 2001;44:263–82. doi: 10.1353/pbm.2001.0035. [DOI] [PubMed] [Google Scholar]
- 49.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Hoste E, Arwert EN, Lal R, South EP, Salas-Alanis JC, Murrell DF, et al. Innate sensing of microbial products promotes wound-induced skin cancer (ACCEPTED) Nat Commun. 2014 doi: 10.1038/ncomms6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardoso AP, Pinto ML, Pinto AT, Oliveira MI, Pinto MT, Goncalves R, et al. Macrophages stimulate gastric and colorectal cancer invasion through EGFR Y(1086), c-Src, Erk1/2 and Akt phosphorylation and smallGTPase activity. Oncogene. 2014;33:2123–33. doi: 10.1038/onc.2013.154. [DOI] [PubMed] [Google Scholar]
- 52.Schumacher MA, Donnelly JM, Engevik AC, Xiao C, Yang L, Kenny S, et al. Gastric Sonic Hedgehog acts as a macrophage chemoattractant during the immune response to Helicobacter pylori. Gastroenterology. 2012;142:1150–9 e6. doi: 10.1053/j.gastro.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 54.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–72. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagemann T, Lawrence T, McNeish I, Charles KA, Kulbe H, Thompson RG, et al. “Re-educating” tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–8. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.