Abstract
Copy number variations are a common cause of intellectual disability (ID). Determining the contribution of copy number variants (CNVs), particularly gains, to disease remains challenging. Here, we report four males with ID with sub-microscopic duplications at Xp11.2 and review the few cases with overlapping duplications reported to date. We established the extent of the duplicated regions in each case encompassing a minimum of three known disease genes TSPYL2, KDM5C and IQSEC2 with one case also duplicating the known disease gene HUWE1. Patients with a duplication encompassing TSPYL2, KDM5C and IQSEC2 without gains of nearby SMC1A and HUWE1 genes have not been reported thus far. All cases presented with ID and significant deficits of speech development. Some patients also manifested behavioral disturbances such as hyperactivity and attention-deficit/hyperactivity disorder. Lymphoblastic cell lines from patients show markedly elevated levels of TSPYL2, KDM5C and SMC1A, transcripts consistent with the extent of their CNVs. The duplicated region in our patients contains several genes known to escape X-inactivation, including KDM5C, IQSEC2 and SMC1A. In silico analysis of expression data in selected gene expression omnibus series indicates that dosage of these genes, especially IQSEC2, is similar in males and females despite the fact they escape from X-inactivation in females. Taken together, the data suggest that gains in Xp11.22 including IQSEC2 cause ID and are associated with hyperactivity and attention-deficit/hyperactivity disorder, and are likely to be dosage-sensitive in males.
Introduction
Genomic microarrays are the first tier diagnostic tool in developmental disorders and intellectual disability (ID).1, 2, 3 Current genomic microarrays can detect tens to hundreds of copy number variations (CNVs) in a single individual, ranging anywhere from 1 to 10 kb upto several megabases. Increasingly, it is not the detection of CNVs that is difficult, but the interpretation of their clinical significance that is the greatest challenge.4 The estimates of the contribution of CNVs to disease are continually being refined, with variations dependent upon the specific cohorts and disorders in question. For example, Cooper et al5 show that 25.7% of ID/DD children harbor an event of >400 kb compared with 11.5% of controls, suggesting that an estimated 14.2% of ID/DD is due to CNV >400 kb. Similarly, large CNVs (>400 kb) are estimated to be causative between 15 and 20% of cases with ID,2, 5, 6, 7 and as high as 25% of cases with ID and multiple congenital abnormalities.8 With increasingly large patient cohorts being harnessed, morbidity mapping of CNVs has identified a noticeable burden of smaller CNVs. Many CNVs of 250 kb and smaller in size are also associated with ID5 and developmental delay.9 The evidence is less clear for smaller CNVs (<50 kb) in autism spectrum disorder.10
Typically, CNVs involve regions containing several to dozens of genes, often with multiple candidates in the smallest region of overlap between similar cases. Hence, determining the dosage-sensitive genes that underlie the phenotypes is not trivial. The prediction of clinical consequences of copy number gains remains particularly challenging. Deletions are seen in excess on autosomes compared with duplications and show on average greater penetrance.5, 9, 11, 12 In contrast, on the X-chromosome, duplications are either as prevalent or more common than deletions. When considering cases with ID, approximately 10% of all potentially causative CNVs are X-linked,13, 14 with ~7% being duplications, ~3% being deletions,14 and 80–90% maternally inherited.8
Clinical consequences of duplications in females can vary depending upon the degree of skewing of X-inactivation, as well as variations in cell-specific patterns of gene regulation.15 In addition, there are some genes on the X-chromosome that escape X-inactivation further complicating the assessment of dosage sensitivity. Despite these complexities, increased dosage of X-chromosome genes has been implicated in ID and autism. These include several at Xq28; namely methyl CpG binding protein, MECP2 (MIM 300260),16, 17 and GDP dissociation inhibitor 1, GDI1 (MIM 300104).18 Interestingly, not all genes from the duplicated region(s) are genomic dosage-sensitive, while they are mRNA expression-sensitive, for example, HCFC1 included within MECP2 duplications.19 Another frequently identified gain is a non-recurrent microduplication at Xp11.22 implicating HECT, UBA and WWE domain containing 1, E3 ubiquitn protein ligase, HUWE1 (MIM 300706).20, 21 Here, we report four novel cases of male patients with sub-microscopic duplications at Xp11.2, all including gains of testis-specific protein Y-encoded Like 2, TSPYL2, lysine specific demethylase, KDM5C, and IQ motif and Sec7 domain 2, IQSEC2. Together with published cases, we implicate copy number gains in a known ID gene, IQ motif and Sec7 domain 2, IQSEC2 (MIM 300522)22 that escapes X-inactivation in females as the cause of ID and behavioral disturbances.
Materials and methods
Clinical descriptions of the patients and families
The research protocols were approved by the appropriate institution review boards and informed consent was obtained from the parents of affected patients. Genomic DNA from patients and healthy controls was isolated from peripheral blood according to standard procedures and stored at 4°C. Variants and phenotypes for patients in this study have been submitted to Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER)30 (http://decipher.sanger.ac.uk/), with specific patient IDs indicated in section for each patient below.
Patient 1
This patient (Decipher ID 306072) was a 3-year-old male referred for language delay and behavioral problems. The patient was born at term of an uncomplicated gestation with a birth weight of 3150g (25th–50th centile). Parents were both healthy and cognitively normal. He started walking at 19 months of age. He was referred to genetics service for diagnostic assessment of developmental delay. His speech development was delayed with no words until he was approximately 3 years of age and slow progression of language with only four to five words by the age of 4 years. At the age of 4 years, his weight was 23 kg (>95th centile), length 108 cm (90th centile) and head circumference 50.4 cm (between average and -2SD). He has downward corners of the mouth. When examined at the age of 4 years, the patient displayed poor socialization, although with some recent improvement. It is unclear whether there will be additional psychiatric disturbances due to young age of the patient.
Patient 2
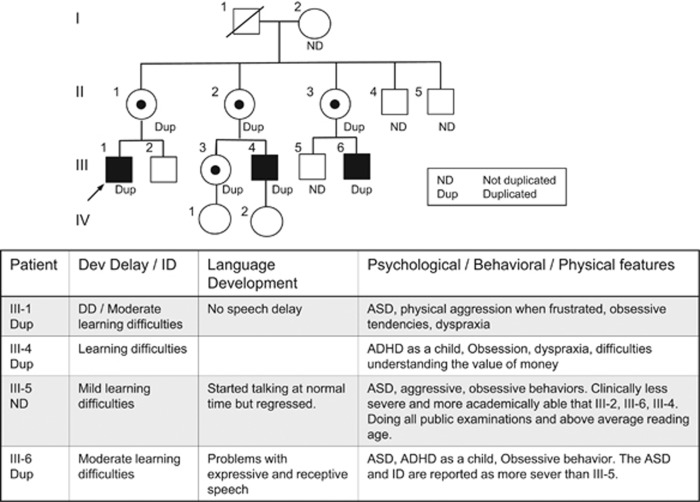
The proband (Decipher ID 27212) was a 14-year-old male who presented with ID and autism spectrum disorder. The pregnancy was complicated by hypertension and labor was induced at 39 weeks gestation because of preeclampsia. He was delivered by forceps delivery for fetal distress, with a birth weight of 3104 g (25–50th centile) and head circumference of 35 cm (50–75th centile). There was neonatal jaundice persisting for 10.5 weeks. He was a poor feeder and was slow to gain weight. He was an unhappy baby who screamed a lot and did not like being touched. He sat late, and first walked at the normal time, but was clumsy and was diagnosed with dyspraxia at the age of 8 years. He had learning difficulties and was transferred to a special school for moderate learning difficulties at the age of 11 years. At the age of 19 years, he required assistance with daily living activities and was reported to have poor social skills. He had challenging behavior with physical aggression when frustrated and had obsessions about articular items, which he collected. He avoided eye contact, but was non-dysmorphic with no neurocutaneous signs. This family has a history of mild-to-moderate learning difficulties and autistic traits in three maternal male cousins, suggesting an X-linked inheritance (Figure 1). The mother and her two sisters were normal with no ID or behavioral problems.
Figure 1.
Pedigree of Family 2. Pedigree of a family identified with submicroscopic duplication at Xp11.22. All tested individuals are marked with ND if the duplication was not present and DUP if the duplication was identified. Open symbols represent normal individuals, and filled squares represent affected males. The proband is indicated with an arrow. Individual generations are numbered with Roman numerals (I, II, III). III-5 does not carry the duplication.
Patient 3
This patient (Decipher ID 308362) was a 5-year and 8-month-old male referred for global developmental delay and behavioral problems. He was born at term following a normal pregnancy to healthy parents. The developmental delay across his postnatal history included absence of crawling at 1 year of age, and walking independently at 2 years. His speech development was also delayed with no words until he was approximately 3 years of age. On examination at the age of 5 years and 8 months, he speaks fluently in conversational speech, but has a significant ongoing issue with pronunciation. He has significant behavioral difficulties and requires the assistance in a special education class. His height was 102.5 cm, his weight was 16.3 kg and his head circumference was 50.5 cm, on the 5th, 10th and 40th percentiles, respectively.
Patient 4
This patient (Decipher ID 249420) was born at 38 weeks gestation following a period of intrauterine growth retardation and oligohydramnios detected by ultrasound at 35 weeks. His weight was 2550 g (3rd centile), length was 46 cm (3rd–10th centile) and head circumference 31 cm (1 cm smaller than the 3rd centile). Developmental delay was first noted at age 6 months. Review at age 9 years revealed severe developmental delay. The patient could sit and shuffle on his bottom but was non-ambulant. He was able to smile and shake hands but has no words and little receptive language. He has a head circumference of 48 cm (1.5 cm smaller than the 2nd centile), weight 25 kg (25th centile) and length 120 cm (3rd centile) at 9 years of age. He had finely arched eyebrows, a short nose and down-turned corners of the mouth. His hands and feet measure on the 3rd centile and he had single palmar creases. He had a small penis with undescended testes. Owing to gastroesophageal reflux, the patient had a fundoplication with percutaneous enterogastrostomy tube insertion. The patient had myoclonic seizures with no recognizable electroclinical syndrome and an EEG that showed an abnormal background with constant generalized sharp slow discharges around 1 Hz with frontal dominance. The seizures commenced at age 5 years and vary from several seizures per day to a seizure every month. Most seizures occur shortly after falling asleep and sodium valproate improved seizure control. At the age of 9 years, the seizures occurred with a frequency of approximately one per month.
Array CGH and FISH
Genomic DNA from patient and control samples was extracted from blood collected using EDTA as the anticoagulant. High-resolution chromosomal microarray and analysis was conducted by contributing cytogenetic diagnostic laboratories including 4 × 180 K Agilent array (Santa Clara, CA, USA) for patient 1 and Affymetrix Cytoscan HD array (750 K) (Santa Clara, CA, USA) for patients 2, 3 and 4. Annotation of the CNVs identified was based on the UCSC Genome Browser, National Centre for Biotechnology Information, Genome Reference Consortium Human Build (GRCh37/ hg19), February 2009. FISH analysis on patient 2 and family used BAC probes RP11-236P24 (Xp11.2) and RP13-130F17 (Xq12) (BlueGnome/Illumina, Melbourne, VIC, Australia).
Quantitative real-time polymerase chain reaction
For cDNA expression analysis, total RNA was extracted from lymphoblastoid cell lines (LCL) for male (n=4) and female (n=5) pooled controls and patients using TRIzol (Life Technologies, Melbourne, VIC, Australia) and RNeasy Mini Kit (Qiagen, Melbourne, VIC, Australia) and treated with DNase I (Qiagen) following the manufacturer's instruction. LCL could not be sourced for patient 1 or 2. Routinely, 2 μg of total RNA was primed with random hexanucleotides and then subjected to reverse transcription with SuperScript III reverse transcriptase. Pooled male and female cDNAs were made from equal amounts of each male control RNAs (n=4) and female control RNAs (n=5). Validation of the reaction was carried out by PCR using primers specific to the ubiquitously expressed EsteraseD. cDNAs were amplified using TaqMan Gene Expression Assays (Life Technologies) using the standard curve method for TSPYL2 (Hs00223173_m1), KDM5C (Hs01011846_m1) and SMC1A (Hs00196849_m1). Probes of target genes were FAM-labeled. The probe for the housekeeping gene HPRT (4326321E, Applied Biosystems, Melbourne, VIC, Australia) was VIC-labeled and enabled normalization of expression in each well. Data analysis was carried out with StepOne Software (Life Technologies).
In silico analysis of the distribution gene expression in males and females
In silico analysis of gene expression was performed on the publicly accessible datasets deposited in Gene Expression Omnibus (GEO) by The National Center for Biotechnology Information (NCBI) using the GEO2R analysis tool available from GEO. We selected studies conducted on the Affymetrix Human Exon array ST 1.0 platform in which the gender of participants was listed. Box-and-whisker plots were constructed to analyze the distribution of gene expression for TSPYL2, KDM5C, IQSEC2, SMC1A and HUWE1 based on gender within each dataset. As the level of IQSEC2 expression in human LCLs is negligible, we included data from studies listed on the GEO database examining gene expression in the human brain.
Results
Microduplication of IQSEC2 and/or KDM5C contribute to ID
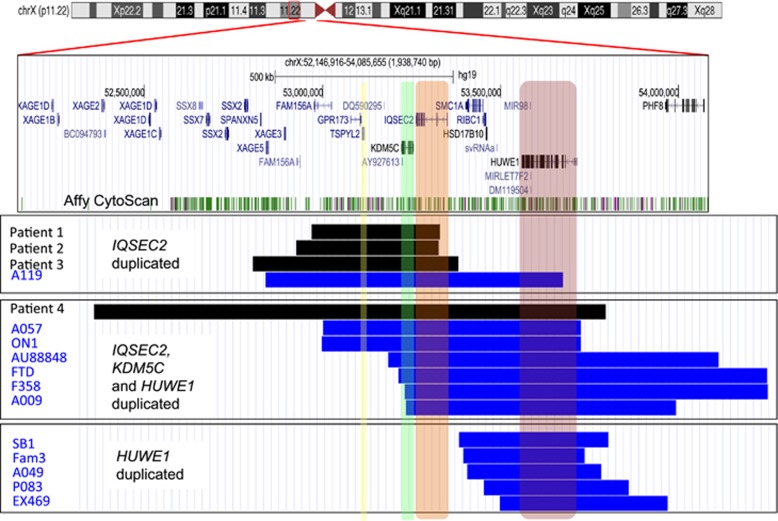
All four cases identified in this study have gains of two known ID genes, KDM5C,17 IQSEC2,22 and gain of TSPYL2, encoding a protein that interacts with the known ID CASK gene.23 All gains are described using genomic co-ordinates consistent with Human Feb. 2009, Build 37 hg19 Assembly. Patient 1 had a 361 kb chromosome X duplication extending from chrX.hg19:g.52,954,520_53,315,542dup duplicating the short isoform of IQSEC2 and extends to FAM156A (Figure 2). This duplication is maternally inherited. In patient 2, a series of investigations by array CGH were undertaken to identify two submicroscopic duplications, both on the X chromosome, a 400 kb chromosome X duplication at Xp11.22 and a 220 kb duplication at Xq12. The 400 kb duplication extends from chrX.hg19:g.52,911,287_53,315,010dup and duplicating the short isoform of IQSEC2 extending to FAM156A (Figure 2). This duplication is maternally inherited. However, this gain is not present in the maternal grandmother who had normal CGH array and FISH results, suggesting she is mosaic for the abnormality or the duplication came from the deceased maternal grandfather, who was reported as normal. Hence, we cannot determine whether the maternal grandfather was either a mosaic or an unaffected carrier. The 400 kb duplication segregates through the family with the affected status (Figure 1). III-5, who displays a very mild phenotype, did not have the CNV gain. The second gain was identified at Xq12 from chrX.hg19:g.65,304,358_65,524,712dup. FISH indicates that the microduplication of Xp11.22 is inserted in the Xq arm and the maternally inherited Xq duplication is inserted into the Xp arm and may indicate a small pericentric inversion in these patients. Patient 3 has a maternally inherited 579 kb X chromosome duplication that extends from chrX.hg19:g.52,789,239_53,368,927dup and includes the region duplicated in patient 1 extending proximal of IQSEC2 and distal of SSX2 (Figure 1). Patient 4 has a de novo 1441 kb X chromosome duplication that extends from chrX.hg19:g.52,341,517_53,782,896dup and involves gains of TSPYL2, KDM5C and IQSEC2 seen in the previous three cases, and extends to include gains of HUWE1 and a region distal of XAGE1D (Figure 1). No other known disease-causing CNVs were identified in these patients.
Figure 2.
Submicroscopic duplications in Xp11.22 in patients with ID. From the top: A red box outlines the region of interest on the X-chromosome in the UCSC genome browser. UCSC genes, including RefSeq, GenBank, CCDS, Rfam, tRNAs and comparative Genomics, are shown below the Affymetrix Cytoscan HD array probe sets. Below are three boxed panels each showing cases of submicroscopic duplications in Xp11.22 clustered depending upon the genes duplicated. The top boxed panel shows cases with TSPYL2 (yellow highlight), KDM5C (green highlight) and IQSEC2 (orange highlight) duplicated, the middle boxed panel shows cases with gains of KDM5C, IQSEC2 and HUWE1, while the bottom panel shows cases with HUWE1 (red highlight) duplicated. Patient 1 to 4 are shown as black bars to differentiate from the patients with duplications reported by Froyen et al20,21, 43 shown as bright blue bars for male patients. Patient identification is given to the left of each bar. Schematic was constructed using the Human Feb. 2009 (hg19) Assembly (UCSC).
Behavioral disturbances are a frequent clinical feature in patients with microduplication of IQSEC2
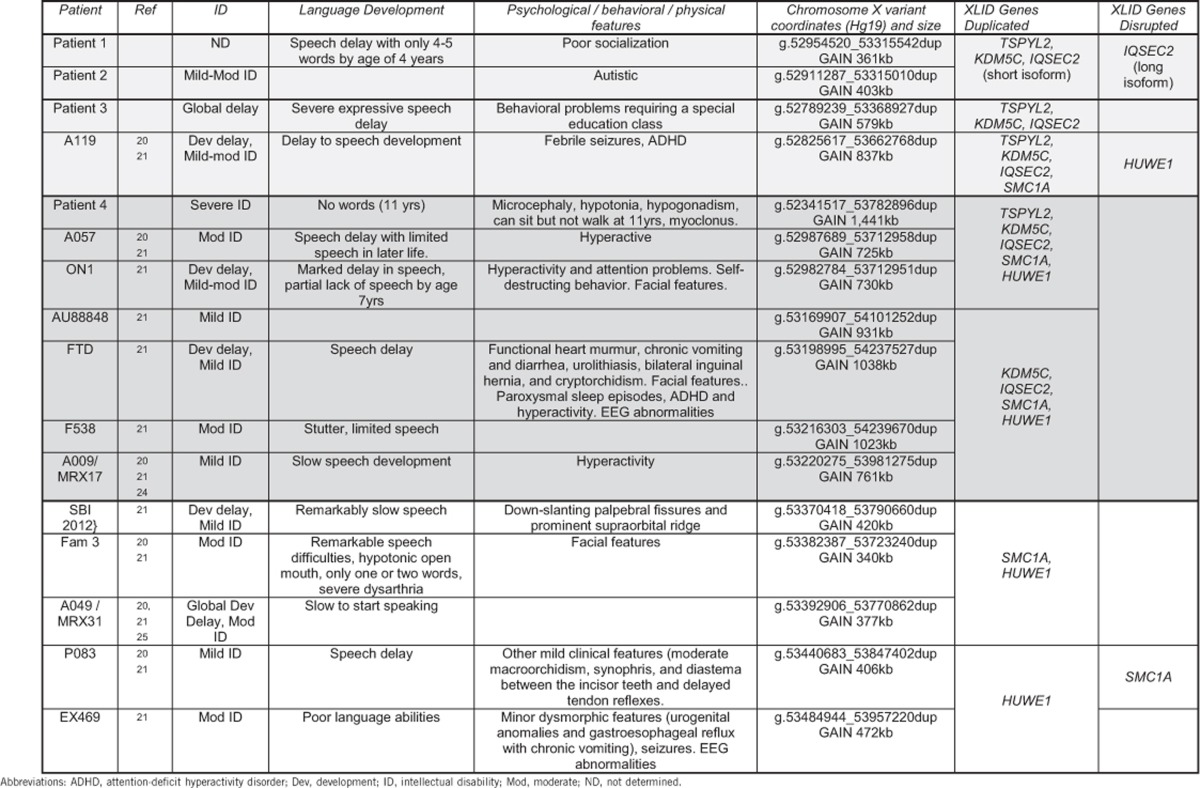
To investigate the potential impact of increased dosage of IQSEC2 on the clinical features in our patients, we reviewed published patients with gains in this chromosomal region.20, 21 All cases with gains in this region are shown on Figure 1, clustered depending upon which known ID genes are involved in the duplication. Four cases, including patients 1, 2 and 3 of this study, duplicate TSPYL2, KDM5C and IQSEC2, but do not duplicate or only partially duplicate (disrupt) the HUWE1 gene (Top boxed panel, Figure 1). There are seven cases, including patient 4, with copy number gains encompassing KDM5C, IQSEC2 and HUWE1 (middle boxed panel, Figure 1). However, only three of these, including patient 4, have gains in TSPYL2 (middle boxed panel, Figure 1). Lastly, there are five cases of gains to HUWE1, without gains to TSPYL2, KDM5C or IQSEC2 (bottom panel, Figure 1).
When considering the clinical findings of these patients, we noted that a consistent feature includes disturbances to both intellectual function and global development as well as significant deficits to speech development (Table 1). These features are present in patients with different size gains involving different known ID genes in this region and do not segregate exclusively with any single duplicated gene. On the other hand, clinical features that arise in cases with specific gene content duplicated can be assigned to two broad categories. When HUWE1 is duplicated, with or without a change to the dosage of TSPYL2, KDM5C or IQSEC2, physical features including dysmorphism (often mild) are frequently observed (Table 1). In contrast, patients with a duplication encompassing IQSEC2, KDM5C and TSPYL2 (with or without gains of HUWE1) frequently present with behavioral disturbances of hyperactivity or ADHD.
Table 1. Clinical features of patients with submicroscopic duplications involving XLID genes at Xp11.22.
TSPYL2, KDM5C and SMC1A mRNA expression testing
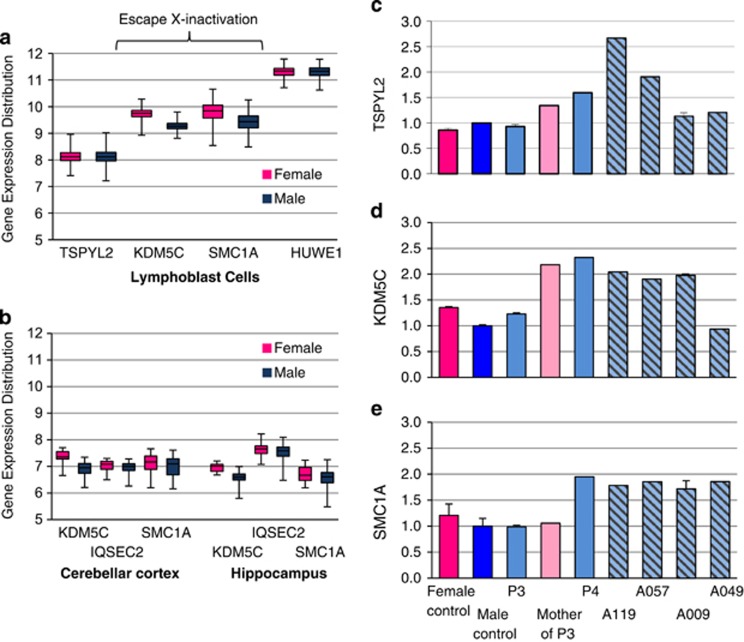
The region duplicated in our patients contains several genes known to escape X-inactivation, including KDM5C, IQSEC2 and SMC1A (TSPYL2 is subject to X-inactivation). We interrogated data from NCBI GEO database (selecting only Affymetrix Human Exon array ST 1.0) using GEO2R analysis tool to determine the normal distribution of expression of these genes in males and females. We analyzed two data series (i) GEO series GSE49531 ‘Utilizing patient LCLs from participants in the genetics of microangiopathic brain injury study' and (ii) GEO series GSE46706 ‘Expression data generated from post-mortem human brain tissue originating from neurologically and neuropathologically control individuals'.26 We report data on KDM5C and SMC1A from both GEO series, but given the negligible expression in LCLs, for IQSEC2, we report the data from the GEO series arising only for brain tissue. Our analysis from LCLs includes genes TSPYL2 and HUWE1 as relevant examples of X-chromosome genes that do not escape X-inactivation. In each data set, we stratified gene expression based on the gender and examined the normal distribution within each gender and between genders. In LCLs, KDM5C and SMC1A that escape X-inactivation to some degree, both have higher levels of gene expression in females compared with males. In comparison, both TSPYL2 and HUWE1 had similar levels of expression in both genders as expected because of X-inactivation status (Figure 3a). Our analysis indicates that KDM5C is consistently expressed at higher (but modest) levels in females compared with males in the tissues analyzed. In contrast, both SMC1A and in particular IQSEC2 expression in males was very similar to the expression levels (and distribution) in females in each tissue analyzed (Figure 3b).
Figure 3.
Expression distribution of XLID genes involved in submicroscopic duplications of Xp11.2. Distribution of XLID gene expression in each gender: Data from NCBI GEO database of Affymetrix Human Exon array ST 1.0 expression was analyzed using GEO2R. Distribution of normal expression in males (blue box plot) and females (pink box plot) is shown for TSPYL2, KDM5C, SMCIA and HUWE1 in LCLs (a), and KDM5C, IQSEC2 and SMC1A in two regions of the brain–Cerebellar cortex (left panel) and Hippocampus (right panel) (b). Data sets included GEO series GSE49531 ‘Gene expression data from LCLs from participants in genetics of microangiopathic brain injury' (females, n=515 and males, n=368). Series GSE46706 ‘Expression data generated from post-mortem human brain tissue originating from neurologically and neuropathologically control individuals' (females, n=23 and males, n=73). All data sets can be found at NCBI GEO gene expression omnibus site, searching with the specific GEO series number. Changes to expression of TSPYL2 (c) and XLID genes KDM5C (d) and SMC1A (e): cDNA was prepared from total RNA extracted from LCL male (n=4) and female (n=5) pooled controls and patients 3 and 4, and the mother of patient 3. Transcript levels measured by qRT-PCR were normalized to housekeeping gene HPRT and shown as relative difference to male pool set at 1.0. Error bars indicate standard error of the mean from replicate cDNA samples. Female control is shown in bright pink, male control in dark blue, male patients in pale blue, mother of patient in pale pink and patients reported in Froyen et al20 in pale blue background with diagonal lines.
Next, we examined the impact of the submicroscopic duplication on the expression of genes involved in the gain, and show marked elevation of KDM5C and SMC1A expression in LCLs from patient 4. In the case of patient 3, levels of KDM5C mRNA were elevated in the mother, with very modest gains in KDM5C in patient 3 (Figure 3c). Interestingly, TSPYL2 expression was not elevated in patient 3 and showed only modest gains in the mother of patient 3 and in patient 4. These modest increases in KDM5C expression in females seen in the analysis of the GEO series data was also observed in our control LCLs (Figure 3c). We extended our expression analysis to available patients reported by Froyen et al20, 21 and show elevated expression of TSPYL2, KDM5C and SMC1A in male patients A119, A057, with A009 having elevated expression of KDM5C and SMC1A and male patient A049 having elevated expression for SMC1A but not TSPYL2 or KDM5C, consistent with the extent of the duplication in each case (Figure 3c). Taken together, these data suggest that dosage of these genes, in particular IQSEC2, is similar in males and females despite their escape from X-inactivation in females.
Discussion
We report four novel patients and review published cases with submicroscopic copy number gains on Xp11.22, and we implicate IQSEC2 as a dosage-sensitive gene contributing to ID and behavioral disturbances. Our study identifies four male patients all with ID and/or developmental delay, speech delay and when not severely developmentally delayed, behavioral problems. Although not recurrent, the CNVs in our cases range from 361 kb to 1441 kb and encompass a number of genes, including known ID genes. In all four cases, we identified CNV gains across the region encompassing the known ID genes KDM5C and IQSEC2, and the TSPYL2 gene known to be linked to the ID CASK gene.23 Copy number gains of these genes without the inclusion of nearby SMC1A or HUWE1 have not been reported. In one of the four patients reported in this study, the CNV gain extended to include SMC1A and the known dosage-sensitive HUWE1 gene.20, 21
The region of Xp11.22 contains a cluster of known X-linked ID (XLID) genes, including KDM5C, IQSEC2, SMC1A and HUWE1. In brief, the mutations in KDM5C cause Claes-Jensen type syndromic XLID (MIM 300534)17, 27, 28, 29, 30 characterized by moderate-to-severe ID, speech abnormalities and other clinical findings such as seizures and aggressive behavior in some individuals. Mutations in IQSEC2 were originally identified in cases of non-syndromic XLID.22 In these families, seizures and autism spectrum disorder were infrequent clinical findings, with more consistent comorbidity in additional de novo cases,31, 32, 33, 34 including two cases of small intragenic duplications, predicted to cause termination mutations.33 In the case of SMC1A, mutations in this gene are responsible for an estimated 4–6% of patients with Cornelia de Lange syndrome type 2.35, 36 This is a clinically heterogeneous developmental disorder characterized by malformations affecting multiple systems, including dysmorphic facial features, distal limb defects, cleft palate growth retardation and developmental delay.35, 36, 37, 38, 39, 40, 41 Mutations in HUWE1 cause XLID with moderate-to-severe ID often coupled with disturbances to speech development.20, 42, 43 Of the ID genes in this cluster, HUWE1 is the only one known to be dosage-sensitive, with twofold overexpression in males leading to mild-to-profound cognitive impairment in syndromic XLID, Turner type.20, 21
When assessing the potential contribution of CNV gains to our patients' clinical phenotype, we need to keep in mind that several of the involved genes are known to escape X-inactivation in the human setting, including KDM5C, IQSEC2 and SMC1A.44,45 When more than one X-chromosome is present (ie, in females), one X-chromosome is inactivated or silenced, resulting in similar dosage of X-chromosome genes between the males and females. However, it is known that some genes do escape this silencing, a phenomenon more prevalent in humans than other species, although the reason for this remains unknown. Moreover, the extent of escape from X-inactivation may be underappreciated with a substantial number of genes recently identified as subject to some degree of escape from X-inactivation.45,46 As the expression from the inactive X chromosome is limited, the total expression level in females is usually less than twice that in males.47, 48 Even so, a modest increased gene dosage that escapes X-inactivation may contribute to disease, such as the extra dosage in X-aneuploidy in Klinefelter syndrome (47, XXY).49 Hence, genes that escape X-inactivation are potential candidates for dosage-mediated phenotypic disruptions.48, 50 Our analysis shows that KDM5C expression from the X-chromosome was consistently higher in females than males in both LCLs and brain tissue. Although KDM5C from the X-chromosome is uniformly expressed at higher levels in females compared with males in LCLs, the expression in males is compensated (and exceeds those in females) by expression from the Y-chromosome.47 Hence, it is unclear whether variable levels of dosage are tolerated between the two genders, or whether additional levels are required by males. In the case of IQSEC2, our analysis indicates that dosage of this gene is similar in males and females, despite escaping X-inactivation in females. This suggests that the expression of IQSEC2 in females is regulated to achieve a similar dosage to that of males. It is attractive to speculate that females may have adequate capacity to regulate IQSEC2 expression even in the case of increased gene dosage. As males normally only have one copy of IQSEC2, the capacity to regulate increased dosage of this gene may be lacking. In this setting, an increased dosage of IQSEC2 in males due to CNV gain may be pathogenic.
The clinical findings of the cases reported in this study along with published cases of CNV gains implicate the region containing multiple XLID genes at Xp11.2 as a hotspot for dosage-sensitive genes impacting on development of cognitive ability and speech. Interestingly, the CNV gains that include TSPYL2, KDM5C and in particular IQSEC2 are associated with behavioral disturbances, including hyperactivity and ADHD. It remains to be established whether one or more of these genes are dosage-sensitive in males. Given the function of KDM5C protein as a versatile epigenetic regulator of chromatin, it is not obvious how increased dosage in males would contribute to the behavioral aspects seen in our cases. In the case of TSPYL2, the interaction of this protein with the known ID gene, CASK, makes this an interesting potential candidate for dosage sensitivity in our patients. The interaction of TSPYL2 with CASK allows the formation of a complex with the transcription factor Tbr-1,23 which regulates the expression of genes including NMDA receptor subunits encoded by Grin2b.23 Recent findings modelling loss-of-function in mice indicate that TSPYL2 may contribute to cognitive variability via regulation of expression of Grin2a and Grin2b.51 Stimulation of the synapse with NMDA leads to a reduction in the protein levels of TSPYL2, but not other members of the regulatory complex, CASK and Trb-1.23 This downregulation contributes to regulating the transcriptional activity of Trb-1-CASK-TSPYL2 protein complex, and hence the expression of Grin2b. In this setting, we can speculate that overexpression of TSPYL2 due to copy number gain may disturb regulation in response to synaptic signalling. In the case of IQSEC2, we know this gene encodes for a guanine-nucleotide exchange factor for the ARF family of small GTPase proteins. IQSEC2 protein is enriched in the post-synaptic density at excitatory synapses52, 53 and associates with PSD95, a PDZ domain-containing synaptic protein involved in NMDA receptor clustering. IQSEC2 transcripts are localized in both hippocampal neuronal cell bodies and dendritic processes53 suggesting that local translation of IQSEC2 mRNA in an activity-dependent manner could contribute to synaptic plasticity. When we consider patients with ADHD, there is a significant enrichment of CNVs affecting the metabotrophic glutamate receptor genes GRM5, GRM8, GRM7 and GRM1, indicating abnormal glutamate signaling in the cause of ADHD.54 Similarly, IQSEC2 is linked to glutamate signaling, via the ionotrophic glutamate receptor (NMDA) through the interaction with DLG proteins (eg, PSD95).53 Hence, it is attractive to speculate that increased dosage of IQSEC2 may lead to an inappropriate response to glutamate signaling with similar phenotypic outcomes as those seen with CNV gains to GRM1.
Acknowledgments
We thank the patients, their families and physicians for their participation in this study. The Molecular Neurogenetics research program in the Department of Paediatrics, University of Adelaide, Australia was funded by the Australian National Health and Medical Research Council (NHMRC) (Grant No 1006586). CS is supported by Australian Research Council (Future Fellowship FT120100086). JG is supported by NHMRC senior research fellowship 1041920. This study makes use of data generated by the DECIPHER Consortium. A full list of centers that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
The authors declare no conflict of interest.
References
- 1Mefford HC, Cooper GM, Zerr T et al: A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res 2009; 19: 1579–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Miller DT, Adam MP, Aradhya S et al: Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010; 86: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Vissers LE, de Ligt J, Gilissen C et al: A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–1112. [DOI] [PubMed] [Google Scholar]
- 4Hehir-Kwa JY, Pfundt R, Veltman JA, de Leeuw N: Pathogenic or not? Assessing the clinical relevance of copy number variants. Clin Genet 2013; 84: 415–421. [DOI] [PubMed] [Google Scholar]
- 5Cooper GM, Coe BP, Girirajan S et al: A copy number variation morbidity map of developmental delay. Nat Genet 2011; 43: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Hochstenbach R, van Binsbergen E, Engelen J et al: Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet 2009; 52: 161–169. [DOI] [PubMed] [Google Scholar]
- 7Qiao Y, Mercier E, Dastan J et al: Copy number variants (CNVs) analysis in a deeply phenotyped cohort of individuals with disability (ID). BMC Med Genet 2014; 15: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Vulto-van Silfhout AT, Hehir-Kwa JY, van Bon BW et al: Clinical significance of de novo and inherited copy-number variation. Hum Mutat 2013; 34: 1679–1687. [DOI] [PubMed] [Google Scholar]
- 9Coe BP, Witherspoon K, Rosenfeld JA et al: Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 2014; 46: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Girirajan S, Dennis MY, Baker C et al: Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet 2013; 92: 221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Grayton HM, Fernandes C, Rujescu D, Collier DA: Copy number variations in neurodevelopmental disorders. Prog Neurobiol 2012; 99: 81–91. [DOI] [PubMed] [Google Scholar]
- 12Rosenfeld JA, Kim KH, Angle B et al: Further evidence of contrasting phenotypes caused by reciprocal deletions and duplications: duplication of NSD1 causes growth retardation and microcephaly. Mol Syndromol 2013; 3: 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Froyen G, Van Esch H, Bauters M et al: Detection of genomic copy number changes in patients with idiopathic mental retardation by high-resolution X-array-CGH: important role for increased gene dosage of XLMR genes. Hum Mutat 2007; 28: 1034–1042. [DOI] [PubMed] [Google Scholar]
- 14Whibley AC, Plagnol V, Tarpey PS et al: Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet 2010; 87: 173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Wu H, Luo J, Yu H et al: Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron 2014; 81: 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Van Esch H, Bauters M, Ignatius J et al: Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet 2005; 77: 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Jensen LR, Amende M, Gurok U et al: Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 2005; 76: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Pinto JC, Pereira V, Marques SL, Amorim A, Alvarez L, Prata MJ: Mirandese language and genetic differentiation in Iberia: a study using X chromosome markers. Ann Hum Biol 2014; 42: 1–6. [DOI] [PubMed] [Google Scholar]
- 19Huang L, Jolly LA, Willis-Owen S et al: A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am J Hum Genet 2012; 91: 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Froyen G, Corbett M, Vandewalle J et al: Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet 2008; 82: 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Froyen G, Belet S, Martinez F et al: Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am J Hum Genet 2012; 91: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Shoubridge C, Tarpey PS, Abidi F et al: Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat Genet 42: 486–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Wang GS, Hong CJ, Yen TY et al: Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron 2004; 42: 113–128. [DOI] [PubMed] [Google Scholar]
- 24Gedeon A, Kerr B, Mulley J, Turner G: Pericentromeric genes for non-specific X-linked mental retardation (MRX). Am J Med Genet 1994; 51: 553–564. [DOI] [PubMed] [Google Scholar]
- 25Donnelly AJ, Partington MW, Ryan AK, Mulley JC: Regional localisation of two non-specific X-linked mental retardation genes (MRX30 and MRX31). Am J Med Genet 1996; 64: 113–120. [DOI] [PubMed] [Google Scholar]
- 26Trabzuni D, Thomson PC United Kingdom Brain Expression Consortium: Analysis of gene expression data using a linear mixed model/finite mixture model approach: application to regional differences in the human brain. Bioinformatics 2014; 30: 1555–1561. [DOI] [PubMed] [Google Scholar]
- 27Tzschach A, Lenzner S, Moser B et al: Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum Mutat 2006; 27: 389. [DOI] [PubMed] [Google Scholar]
- 28Abidi FE, Holloway L, Moore CA et al: Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. J Med Genet 2008; 45: 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Adegbola AA, Gonzales ML, Chess A, LaSalle JM, Cox GF: A novel hypomorphic MECP2 point mutation is associated with a neuropsychiatric phenotype. Hum Genet 2009; 124: 615–623. [DOI] [PubMed] [Google Scholar]
- 30Rujirabanjerd S, Nelson J, Tarpey PS et al: Identification and characterization of two novel JARID1C mutations: suggestion of an emerging genotype-phenotype correlation. Eur J Hum Genet 2010; 18: 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Gandomi SK, Farwell Gonzalez KD, Parra M et al: Diagnostic exome sequencing identifies two novel IQSEC2 mutations associated with X-linked intellectual disability with seizures: implications for genetic counseling and clinical diagnosis. J Genet Couns 2014; 23: 289–298. [DOI] [PubMed] [Google Scholar]
- 32Rauch A, Wieczorek D, Graf E et al: Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- 33Tran Mau-Them F, Willems M, Albrecht B et al: Expanding the phenotype of IQSEC2 mutations: truncating mutations in severe intellectual disability. Eur J Hum Genet 2013; 22: 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Allen AS, Berkovic SF, Cossette P et alEpi4K Consortium, Epilepsy Phenome/Genome Project: De novo mutations in epileptic encephalopathies. Nature 2013; 501: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Musio A, Selicorni A, Focarelli ML et al: X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet 2006; 38: 528–530. [DOI] [PubMed] [Google Scholar]
- 36Hoppman-Chaney N, Jang JS, Jen J, Babovic-Vuksanovic D, Hodge JC: In-frame multi-exon deletion of SMC1A in a severely affected female with Cornelia de Lange Syndrome. Am J Med Genet A 2012; 158A: 193–198. [DOI] [PubMed] [Google Scholar]
- 37Borck G, Zarhrate M, Bonnefont JP, Munnich A, Cormier-Daire V, Colleaux L: Incidence and clinical features of X-linked Cornelia de Lange syndrome due to SMC1L1 mutations. Hum Mutat 2007; 28: 205–206. [DOI] [PubMed] [Google Scholar]
- 38Deardorff MA, Kaur M, Yaeger D et al: Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 2007; 80: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Liu J, Feldman R, Zhang Z et al: SMC1A expression and mechanism of pathogenicity in probands with X-Linked Cornelia de Lange syndrome. Hum Mutat 2009; 30: 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Limongelli G, Russo S, Digilio MC et al: Hypertrophic cardiomyopathy in a girl with Cornelia de Lange syndrome due to mutation in SMC1A. Am J Med Genet A 2010; 152A: 2127–2129. [DOI] [PubMed] [Google Scholar]
- 41Gervasini C, Picinelli C, Azzollini J et al: Genomic imbalances in patients with a clinical presentation in the spectrum of Cornelia de Lange syndrome. BMC Med Genet 2013; 14: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Nava C, Lamari F, Heron D et al: Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry 2012; 2: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Isrie M, Froyen G, Devriendt K et al: Sporadic male patients with intellectual disability: contribution of X-chromosome copy number variants. Eur J Med Genet 2012; 55: 577–585. [DOI] [PubMed] [Google Scholar]
- 44Tsuchiya KD, Greally JM, Yi Y, Noel KP, Truong JP, Disteche CM: Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res 2004; 14: 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ: Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet 2015; 24: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Zhang Y, Castillo-Morales A, Jiang M et al: Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol Biol Evol 2013; 30: 2588–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT: Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet 2008; 4: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Berletch JB, Yang F, Xu J, Carrel L, Disteche CM: Genes that escape from X inactivation. Hum Genet 2011; 130: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Geschwind DH, Boone KB, Miller BL, Swerdloff RS: Neurobehavioral phenotype of Klinefelter syndrome. Ment Retard Dev Disabil Res Rev 2000; 6: 107–116. [DOI] [PubMed] [Google Scholar]
- 50Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L: A review of trisomy X (47,XXX). Orphanet J Rare Dis 2010; 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Tsang KH, Lai SK, Li Q et al: The nucleosome assembly protein TSPYL2 regulates the expression of NMDA receptor subunits GluN2A and GluN2B. Sci Rep 2014; 4: 3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Murphy JA, Jensen ON, Walikonis RS: BRAG1, a Sec7 domain-containing protein, is a component of the postsynaptic density of excitatory synapses. Brain Res 2006; 1120: 35–45. [DOI] [PubMed] [Google Scholar]
- 53Sakagami H, Sanda M, Fukaya M et al: IQ-ArfGEF/BRAG1 is a guanine nucleotide exchange factor for Arf6 that interacts with PSD-95 at postsynaptic density of excitatory synapses. Neurosci Res 2008; 60: 199–212. [DOI] [PubMed] [Google Scholar]
- 54Elia J, Glessner JT, Wang K et al: Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet 2012; 44: 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]