Abstract
MiR399f plays a crucial role in maintaining phosphate homeostasis in Arabidopsis thaliana. Under phosphate starvation conditions, AtMYB2, which plays a role in plant salt and drought stress responses, directly regulates the expression of miR399f. In this study, we found that miR399f also participates in plant responses to abscisic acid (ABA), and to abiotic stresses including salt and drought. Salt and ABA treatment induced the expression of miR399f, as confirmed by histochemical analysis of promoter-GUS fusions. Transgenic Arabidopsis plants overexpressing miR399f (miR399f-OE) exhibited enhanced tolerance to salt stress and exogenous ABA, but hypersensitivity to drought. Our in silico analysis identified ABF3 and CSP41b as putative target genes of miR399f, and expression analysis revealed that mRNA levels of ABF3 and CSP41b decreased remarkably in miR399f-OE plants under salt stress and in response to treatment with ABA. Moreover, we showed that activation of stress-responsive gene expression in response to salt stress and ABA treatment was impaired in miR399f-OE plants. Thus, these results suggested that in addition to phosphate starvation signaling, miR399f might also modulates plant responses to salt, ABA, and drought, by regulating the expression of newly discovered target genes such as ABF3 and CSP41b.
Keywords: ABA, abiotic stress, arabidopsis, drought, microRNA, salt
INTRODUCTION
During growth and development, plants encounter a wide array of environmental stresses that trigger physiological and genetic responses (Chinnusamy and Zhu, 2009; Cushman and Bohnert, 2000). In addition, primary stresses lead to secondary stresses such as oxidative stress and thermal shock. Accordingly, plants have evolved various response mechanisms that help them adapt or acclimate to the stresses (Yamaguchi-Shinozaki and Shinozaki, 2006). A large proportion of plant genes are regulated by biotic (e.g., bacterial pathogens, virus, fungi, insects, and nematodes) (Brotman et al., 2012; Fagard et al., 2007) and abiotic stresses (e.g., drought, soil salinity, extreme temperatures, and heavy metals) (Chao et al., 2005; Si et al., 2009). Various cellular processes, such as RNA processing and post-transcriptional or even post-translational modifications, participate in regulation of the expression of genes in response to biotic and abiotic stresses.
MicroRNAs (miRNAs) can repress gene expression at the post-transcriptional level in plants (Bartel, 2004; Mallory and Vaucheret, 2006). MiRNAs are generally generated via a multistep process associated with the activation of DCL1 (DICER-LIKE 1), HEN1, and HYL1 (Jones-Rhoades et al., 2006). The binding of miRNAs to the mRNAs of target genes leads to the degradation and/or translational inhibition of the targets (Guo et al., 2005; Voinnet, 2009).
Recent studies have shown that plants respond to environmental stresses by modulating gene expression at post-transcriptional levels via miRNAs. MiRNAs act in a wide variety of metabolic and biological processes during plant hormone signaling (Liu and Chen, 2009), abiotic stress responses (Lu and Huang, 2008; Sunkar et al., 2007), and immune responses (Katiyar-Agarwal and Jin, 2010; Lu et al., 2008; Voinnet, 2008). Similarly, numerous studies have revealed the involvement of specific miRNAs in plant responses to biotic and abiotic stresses. For example, the expression of miR169 is induced in response to drought (Li et al., 2008), cold (Zhou et al., 2008), salt (Zhao et al., 2009), nitrogen deficiency (Zhao et al., 2011), and UV-B radiation (Zhou et al., 2007). MiR393 is involved in nitrate signaling (Vidal et al., 2010), drought stress, and auxin signaling (Chen and Xiong, 2012). The expression of miR398 is suppressed in response to salt stress, abscisic acid (ABA) treatment (Jia et al., 2009), oxidative stress (Sunkar et al., 2006), high light (Siré et al., 2009), and biotic stress (Jagadeeswaran et al., 2009; Naya et al., 2014). MiR394 is involved in the regulation of plant responses to salt and drought stresses (Song et al., 2013). MiR395 is highly expressed in response to sulfate deficiency (Liang et al., 2010) as well as salt and dehydration (Kim et al., 2010). MiR159, miR397, and miR402 are up-regulated, but miR389 is down-regulated, in response to ABA (Reyes and Chua, 2007; Sunkar and Zhu, 2004). MiR399 expression is strongly induced by phosphate deficiency, and regulates phosphate uptake and root-to-shoot phosphate trans-location as a systemic signaling molecule (Bari et al., 2006; Fujii et al., 2005; Pant et al., 2008). MiR156, miR778, miR827, and miR2111 are highly expressed under phosphate-deficiency conditions, whereas miR169, miR395, and miR398 are repressed by phosphate deficiency (Buhtz et al., 2008; Hsieh et al., 2009; Pant et al., 2009). Numerous studies have tested whether miRNAs function as linkers between nutrient homeostasis and hormone signaling or abiotic stress.
Previously we reported that miR399f, which acts in phosphate homeostasis, is directly activated by AtMYB2 (Baek et al., 2013). AtMYB2 regulates the expression of genes that respond to salt and drought stresses (Abe et al., 2003; Yoo et al., 2005). In this study, we revealed that miR399f plays an important role in plant responses to abiotic stresses, including salt and drought stresses. The transcription of miR399f is up-regulated by salt stress and exogenous ABA. Furthermore, we showed that transgenic Arabidopsis overexpressing miR399f displayed tolerance to salt stress and ABA treatment, but were hypersensitive to drought. Moreover, we identified candidate target genes of miR399f that function in plant abiotic stress signaling. Our study reports a novel biological function of miR399f and its possible regulatory mechanism in plant responses to abiotic stresses.
MATERIALS AND METHODS
Plant materials and stress treatments
Seedlings of Arabidopsis thaliana ecotype Colombia (Col-0) plant were sown and grown on 1/2 Murashige and Skoog (MS) medium containing 1.5% sucrose, and 0.6% agar, pH 5.7 for 10 days, and then were treated with different stresses for indicated times (Figs. 1 and 3). Plants were grown in a growth chamber with a cycle of 16 h light (approximately 100 µE m−2 s−1) and 8 h dark at 22°C.
Fig. 1.
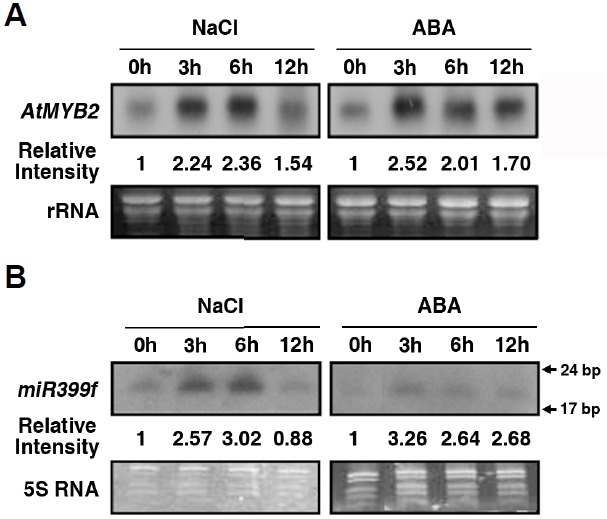
Expression of AtMYB2 and miR399f under salt stress and ABA treatment. For RNA gel blot and small RNA blot analysis, total RNAs were extracted from ten-day-old seedlings treated with 100 mM NaCl or 100 μM ABA for the indicated times. (A) RNA gel blot analysis of AtMYB2 expression. (B) Small RNA blot analysis of miR399f expression. The rRNA and 5S RNA are shown as loading controls. “Relative Intensity” indicates the signal intensity relative to that at time 0 of treatment.
Fig. 3.
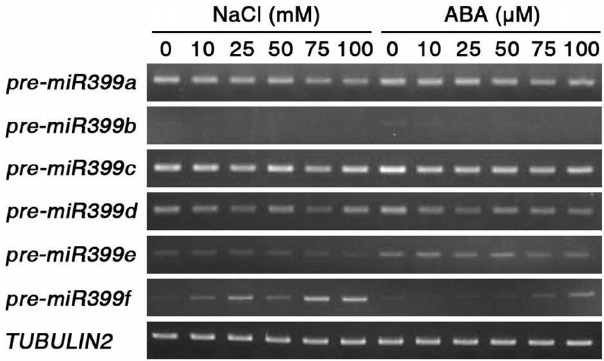
Expression of precursors of miR399 family members in response to salt stress and ABA treatment. Total RNA extracted from ten-day-old seedlings after treatment with different concentrations of 100 mM NaCl or 100 μM ABA for 3 h. TUBULIN2 was used as a loading control.
Dr. Zhu kindly provided the seeds of wild type (Col-gl) and transgenic plants overexpressing miR399f (Fujii et al., 2005). These plants were grown on 1/2 MS medium containing 1.5% sucrose, and 1.2% agar, pH 5.7 for 4 days, and then transferred to media containing various concentrations of NaCl (0 to 150 mM) and further grown for 8 days. To test cotyledon greening under ABA treatment conditions, seeds of these plants were grown on 1/2 MS medium without or with ABA (0 to 1 μM) for 5 days.
For drought treatments, 3-week-old plants were treated with natural drought (water was withheld). After 12 days without watering, the drought-treated plants were re-watered, and recovery was checked after 1 day. Drought experiments were repeated five times and at least 7 plants for each individual line were used in each repeated experiment and one representative picture was shown.
Measurement of water loss
The rate of water loss by the leaves was measured. The shoots of 4-week-old plants were detached from the root, and weighed immediately. The shoots were placed in a covered plate at room temperature and weighed at various time intervals. The loss of fresh weight was calculated on the basis of the initial weight of the plant. At least three biological replicates for each sample were used for water loss assays.
Small RNA and RNA gel blot analyses
RNA was extracted from seedlings using Plant RNA Reagent (Invitrogen, USA) following the supplier’s instructions. For detection of miR399f, 20 μg total RNA was resolved on 15% polyacrylamide gels containing 7 M urea and transferred electrophoretically to nylon membranes (EMD Millipore, USA) using semi-dry transfer (GE Healthcare, USA). Probes complementary to miR399f were 5′-end labeled with γ32P-ATP using Optikinase (Usb, USA). For small RNA blotting, blots were prehybridized for at least 1 h and hybridized for 24 h using PerfectHyb Plus Hybridization Buffer (Sigma, USA) at 37°C.
For detection of AtMYB2 mRNA, 15 μg total RNA was separated on formaldehyde agarose gels and transferred to nylon membranes (EMD Millipore, USA). For hybridization, blots were pre-hybridized for at least 1 h and hybridized for 18 h at 65°C. Blots were washed three times (2× SSC and 0.1% SDS for 20 min, 0.5× SSC and 0.1% SDS for 20 min, 0.1× SSC and 0.1% SDS for 20 min) at 50°C. Ethidium bromide staining was used for RNA loading controls. The relative intensity of detected bands was measured with Image J program.
RT-PCR and quantitative Real-time PCR analysis
For RT-PCR, total RNA was isolated using an RNaeasy Kit (Qiagen, USA) according to the manufacturer’s instructions. Total RNA was treated with DNase I (Qiagen, USA) to remove genomic DNA contamination. The first-strand cDNA was synthesized using 2 μg total RNA with a cDNA synthesis kit (Invitrogen, USA), and subjected to RT-PCR analysis for examination of gene expression.
Quantitative real-time PCR (qRT-PCR) was used to assay gene expression levels with a CFX384TM Real-Time PCR Detection System (Bio-Rad, USA) following a standard protocol. The QuantiSpeed SYBR kit (PhileKorea, Korea) was used for 20 μl PCR reactions as follows: 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 5 s and 60°C for 30 s. The relative expression levels of all samples were automatically calculated and analyzed three times by CFX Manager software (Bio-Rad, USA). The specific primers used in RT-PCR and qRT-PCR analysis are described in Supplementary Table 2. TUBULIN2 primers were used for RNA normalization.
Histological staining of GUS activity
Transgenic plants of PromiR399f:GUS or vector-only controls (Baek et al., 2013) were grown on 1/2 MS medium for 10 days, and then treated 100 mM NaCl and 100 μM ABA for 3 h. For GUS histological staining, seedlings from treated transgenic plants were incubated at 37°C for 6 h in the dark, in staining buffer (0.5 M Tris, pH 7.0, 10% Triton X-100) with 1 mM X-Gluc (5-bromo-4-chloro-3-indolyl β-D-glucuronide; Park et al., 2013). Chlorophyll was removed using an ethanol series: 20%, 35%, and 50% ethanol at room temperature for 30 min each.
In silico analysis
Plant microRNA database (PMRD; http://bioinformatics.cau.edu.cn/PMRD/) and Web microRNA designer (WMD3; http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) were used to search for putative target genes of miR399f.
RESULTS
Expression of AtMYB2 and miR399f in response to salt stress and ABA treatment
Previously we showed that AtMYB2, a transcription factor that functions in ABA and salt stress signaling in Arabidopsis, also acts in phosphate starvation signaling by regulating miR399f transcription (Baek et al., 2013; Fujii et al., 2005). To test whether miR399f participates in plant responses to ABA and salt stress signaling, we analyzed patterns of AtMYB2 and miR399f expression in ten-day-old Arabidopsis seedlings treated with 100 mM NaCl and 100 μM ABA for various times (Fig. 1). Consistent with previous reports (Urao et al., 1993), the expression of AtMYB2 increased strongly in response to NaCl and ABA treatment (Fig. 1A). The transcript level of AtMYB2 increased within 3 h of NaCl and ABA treatment and stayed high up to 6 h of treatment. Similarly, miR399f was strongly induced by 100 mM NaCl or 100 μM ABA (Fig. 1B). Thus, miR399f and AtMYB2 showed similar expression patterns. These results suggested that miR399f, which plays an important role in phosphate homeostasis, might also participate in salt stress and ABA signaling and moreover that the regulation of miR399f expression during salt stress and ABA treatment involves AtMYB2.
To further investigate the regulation of miR399f expression by salt and ABA treatment, we performed histochemical staining for β-glucuronidase (GUS) expression in one-week-old transgenic Arabidopsis plants harboring the miR399f promoter fused to the GUS reporter gene (PromiR399f:GUS) and grown in media containing NaCl or ABA (Fig. 2). Transgenic plants harboring the empty GUS vector were used as a control. Strong promoter activity of miR399f was detected in vascular tissues of rosette leaves of seedlings grown in the presence of salt or ABA, whereas very weak GUS staining was observed in PromiR399f:GUS seedlings grown under normal conditions. GUS expression in PromiR399:GUS plants was mainly observed in vascular tissues under salt and ABA stress condition, but we could also detected weaker expression of GUS in leaf tissues (Supplementary Fig. S1). It is possible that miR399f generated from vascular tissue can move to neighboring leaf tissues and regulate its target gene expression, because cell-to-cell movement of miRNA has been previously reported (Lin et al., 2008). These results confirmed that miR399f expression was influenced by salt and ABA.
Fig. 2.
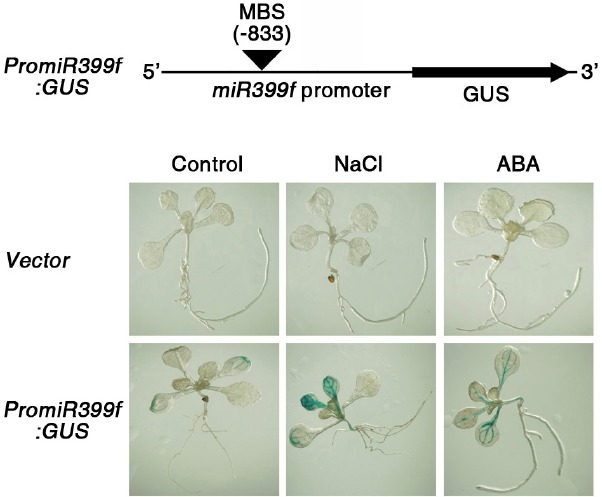
Promoter activity of miR399f under salt and ABA treatment. Histochemical analysis of GUS activity was conducted using ten-day-old PromiR399f:GUS and vector control transgenic seedlings treated with 100 mM NaCl or 100 μM ABA for 3 h.
Expression of miR399 family members in response to salt and ABA treatments
In Arabidopsis thaliana, the miR399 family consists of six members, miR399a to miR399f. Pi starvation conditions induce the expression of miR399 family members, which regulate the expression of an ubiquitin-conjugating enzyme 24 (UBC24) gene involved in phosphate homeostasis (Aung et al., 2006). To investigate the responses of the miR399 family members to salt and ABA stresses, we analyzed transcript levels of miR399 family genes by RT-PCR in ten-day-old seedlings that had been treated with different concentrations of NaCl and ABA for 3 h (Fig. 3). With increasing concentrations of NaCl and ABA, the expression of the miR399f precursor remarkably increased. In contrast to miR399f, NaCl and ABA treatments did not affect the expression of other miR399 family members, such as miR399a, miR399b, miR399c, miR399d, and miR399e. This result suggested that the expression of miR399f, but not other miR399 family members, is induced by not only Pi starvation, but also by NaCl stress and ABA.
miR399f-overexpressing plants have different responses to NaCl, ABA, and drought
We obtained two Arabidopsis transgenic lines overexpressing miR399f (kindly provided by Dr. Jian-Kang Zhu); one line showed relatively weak expression of miR399f (#14) and the other line showed strong expression (#15) (Fujii et al., 2005; Supplementary Fig. S2). To determine the effect of miR399f overexpression on plant responses to salt stress, we performed root growth assays on Murashige and Skoog (MS) medium containing various concentrations of NaCl (Figs. 4A and 4B). Four-day-old seedlings of wild type (WT) and miR399f-overexpressing plants (miR399f-OE #14 and #15) grown on MS medium were transferred to MS medium containing different concentrations of NaCl and their primary root lengths were measured. The primary root elongation of WT seedlings was strongly suppressed under 100 mM NaCl conditions, in which root length was approximately 41% that of WT seedlings grown under normal conditions. However, the root elongation of miR399f-OE #15 seedlings showed more resistance to NaCl treatment. At 100, 125, and 150 mM NaCl, the root length of miR399f-OE #15 seedlings was 1.52-, 1.75-, and 4.83-fold that of WT plants grown under the same conditions, respectively (Fig. 4B). By contrast, the root elongation of miR399f-OE #14 was similar to that of the WT plants under NaCl stress conditions.
Fig. 4.
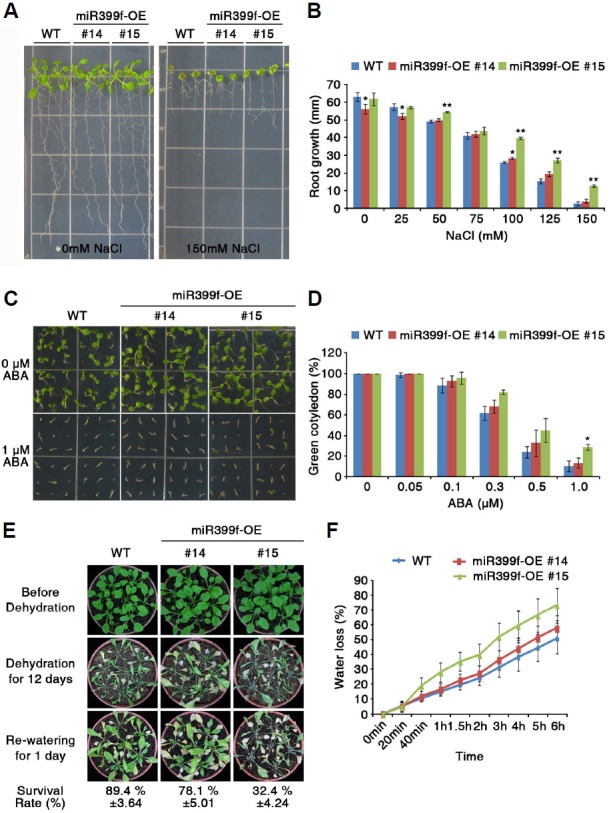
Responses of miR399f-overexpressing plants to salt, ABA and drought. (A) WT and two independent lines overexpressing miR399f (miR399f-OE #14 and #15) were grown on 1/2 MS agar medium for 4 days, and then transferred to 1/2 MS agar without NaCl (0 mM NaCl) or with NaCl (150 mM NaCl), and incubated for 8 days. (B) Comparison of root elongation at different concentrations of NaCl in WT and miR399f-OE plants. Bars represent the means ± standard error of three replicates with 16 seedlings per replicate. Asterisks represent significant differences from the WT (*; p-value ≤ 0.05, Student’s t-test). (C) Seeds of WT and miR399f-OE plants were germinated on 1/2 MS agar medium without ABA (0 μM ABA) or with ABA (1 μM ABA) for 5 days. (D) Comparison of cotyledon greening at different concentrations of ABA in WT and miR399-OE plants. Bars represent the means ± standard error of three replicates with 30 seeds per replicate. Asterisks represent significant differences from the WT (*; p-value ≤ 0.05, Student’s t-test). (E) Photographs show plants before and after dehydration stress. WT and miR399f-OE plants were grown in soil with sufficient water for 3 weeks, and then the water were withheld for 12 days. Plants were re-watered for 1 day before the photograph was taken. (F) Water loss from detached leaves of 4-week-old plants measured at room temperature. Bars represent the means ± standard error of three replicates with 5 seedlings per replicate.
We further examined the response of miR399f-OE plants to ABA treatment by measuring the number of green cotyledons after seed germination (Figs. 4C and 4D). The seeds of WT and miR399f-OE plants were germinated on MS media containing 0 to 1 µM ABA and grown for 5 days. The seeds of WT and miR399f-OE plants showed different responses to ABA treatment. At 0.3 µM ABA, approximately 82% of miR399f-OE #15 seedlings developed green cotyledons, compared to approximately 62% of WT seedlings. The difference was more obvious at higher concentrations of ABA. At 1 µM ABA, the number of miR399f-OE #15 seedlings with green cotyledons was 2.78-fold higher than that of WT seedlings.
In contrast to their tolerance of NaCl and ABA, miR399f-OE plants were more sensitive to drought than WT plants. Three-week-old WT and miR399f-OE plants were subjected to drought stress for 12 days and then re-watered. Under drought conditions, most WT and miR399f-OE plants withered, but one day after re-watering, WT plants resumed growth, whereas miR399f-OE plants had not fully recovered (Fig. 4E). To further confirm the response to drought stress, we examined water loss of WT and miR399f-OE plants, using detached rosette leaves of 4-week-old plants and placing them on petri dishes at room temperature. Water loss proceeded more quickly from leaves of miR399f-OE plants than from WT leaves (Fig. 4F). At 6 h after detachment, the leaves of miR399f-OE plants lost almost 74% of their water, while WT leaves lost only 51%. Together, these results indicated that miR399f-overexpressing plants were resistant to NaCl and ABA, but hypersensitive to drought stress. Moreover, these findings support the idea that miR399f plays a role in plant responses to multiple abiotic stresses.
CSP41b and ABF3 are putative target genes of miR399f in salt stress and ABA signaling
The post-transcriptional activity of UBC24 is partially inhibited upon an increase in miR399f expression during the phosphate-deficiency response (Fujii et al., 2005). To understand the mode of action of miR399f in plant responses to salt, ABA, and drought, we attempted to identify the putative target genes of miR399f by in silico analysis using the Plant MicroRNA Database (PMRD) and Web MicroRNA Designer (WMD3) (Supplementary Table 1). In silico analysis revealed five putative miR399f target genes, BASS2 (BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 2), CSP41b, ABF3 (ABA-RESPONSIVE ELEMENT-BINDING TRANSCRIPTION FACTOR3), At1g04985, and At3g26730, in addition to UBC24. If miR399f exerts its role by mRNA cleavage, levels of the putative target mRNA should decrease in the miR399f-overexpressing plants. To determine whether miR399f mediates the mRNA cleavage of these target genes, we analyzed the transcript levels of the five putative target genes by quantitative real time-PCR (qRT-PCR) in WT and miR399f-OE #15 plants grown in the presence of NaCl and ABA (Fig. 5A and B, Supplementary Fig. S3). We performed this study with miR399f-OE #15 line, because miR399f-OE #15 plants expressed more miR399f than miR399f-OE #14 line and both lines showed similar phenotype under stress conditions (Fig. 4). Indeed, two candidate target genes, CSP41b and ABF3, were down-regulated in miR399f-OE plants under salt stress conditions. In addition, the mRNA level of CSP41b decreased in miR399f-OE plants in the presence of ABA. However, the other three putative target genes (BASS2, At1g04985, and At3g26730) showed no significant difference in mRNA levels between WT and miR399f-OE plants (Supplementary Fig. S3). ABF3 is involved in salt stress, ABA, and drought stress signaling and regulates the expression of abiotic stress-responsive genes (Finkelstein et al., 2005; Yoshida et al., 2010). CSP41b encodes a chloroplast RNA binding protein and to date has not been reported to be involved in abiotic stress. The qRT-PCR results showed that the expression of CSP41b gradually decreased in response to NaCl and ABA treatment in WT plants, suggesting a possible role for CSP41b in stress signaling (Figs. 5A and 5B). In silico analysis suggested that sites for cleavage by miR399f are located at the 3rd exon of CSP41b and 5th exon of ABF3 (Fig. 5C), although there are five mismatches, G-A, G-G, G-U, A-A and U-C, or three mismatches, C-U, G-G and C-U, between miR399f and the CSP41b and ABF3 mRNAs, respectively (Supplementary Table 1). Our results suggested that CSP41b and ABF3 are candidate target genes regulated by miR399f during salt stress and/or ABA signaling. Moreover, the results also suggested that the regulatory mechanism of ABF3 expression might be different in NaCl- and ABA-responsive signaling pathways.
Fig. 5.
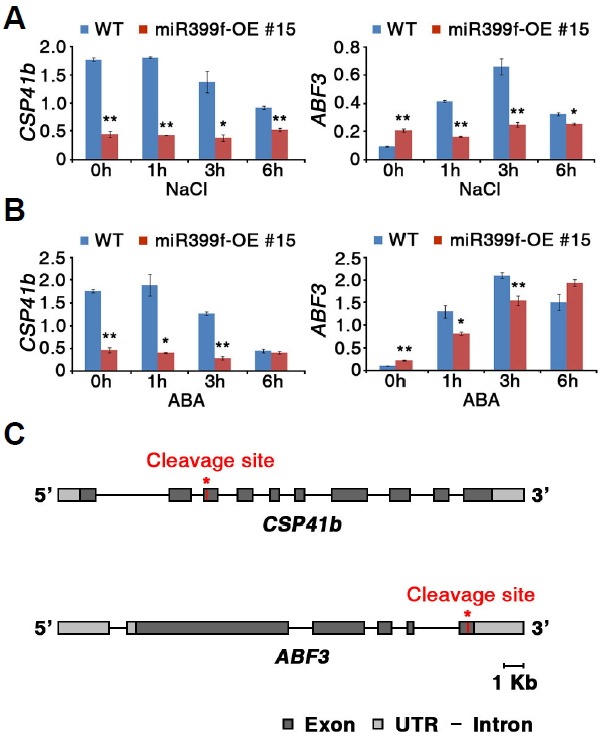
Expression of putative target genes of miR399f. (A, B) Analysis by qRT-PCR of mRNA levels of CSP41b and ABF3 in ten-day-old WT and miR399f-OE seedlings grown in the presence of NaCl (A) or ABA (B). Total RNA was extracted from ten-day-old seedlings after 100 mM NaCl or 100 μM ABA treatment for the indicated times. TUBULIN2 was used for normalization. Bars represent the means ± standard error of three biological replicates with two technical replicates each. Asterisks represent significant differences from the WT (*; 0.01< p-value ≤ 0.05, **; p-value ≤ 0.01, Student’s t-test). (C) Schematic diagram of the CSP41b and ABF3 genes. Red asterisks indicate the predicted cleavage sites of miR399f.
Expression of stress-responsive genes in miR399f-overexpressing plants in response to NaCl or ABA treatment
The expression of stress-responsive genes is remarkably impaired in the abf3 mutant (Yoshida et al., 2010). The down-regulation of ABF3 expression in miR399f-OE plants prompted us to test the expression of stress-responsive genes in Arabidopsis transgenic plants overexpressing miR399f. To investigate whether overexpression of miR399f affects the expression of stress-responsive genes, we analyzed the mRNA levels of stress-responsive genes such as RD29A, RD29B, RD22, DREB1A, DREB2A, and P5CS1 in WT and miR399f-OE #15 plants treated with NaCl or ABA. In WT plants, expression of these genes was highly induced by treatment with NaCl or ABA (Fig. 6). In miR399f-OE #15 plants, however, induction of stress-responsive gene expression by NaCl and ABA treatments was significantly suppressed. This result was consistent with the observations in abf3 mutants (Yoshida et al., 2010). The suppression was more obvious for RD29A, RD29B, DREB1A, and DREB2A expression in response to NaCl treatment (Fig. 6A), and for RD29B and RD22 in response to exogenous ABA (Fig. 6B). These results suggested that miR399f contributes to the regulation of stress-responsive gene expression in response to salt, ABA, and drought. Taken together, our results suggested the existence of a novel regulatory mechanism mediated by miR399f in plant responses to abiotic stresses.
Fig. 6.
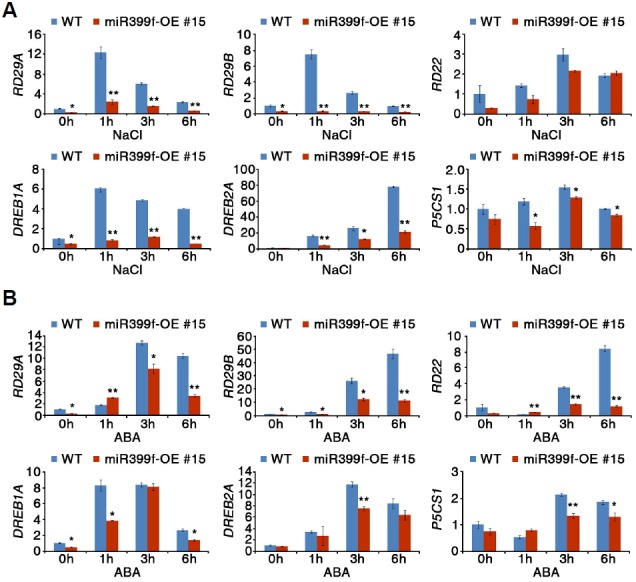
Expression of stress-responsive genes in miR399f-OE plants in response to salt stress and ABA. The mRNA levels of stress-responsive genes in WT and miR399f-OE in the presence of NaCl (A) or ABA (B) were determined by qRT-PCR using total RNA extracted from ten-day-old seedlings after treatment with 100 mM NaCl or 100 μM ABA for the indicated times. TUBULIN2 was used for normalization. Bars represent the means ± standard error of three biological replicates with two technical replicates each. Asterisks represent significant differences from the WT (*; 0.01< p-value ≤ 0.05, **; p-value ≤ 0.01, Student’s t-test).
DISCUSSION
Abiotic stresses affect various physiological processes in plant development, such as seedling growth and seed germination. Exposure to different abiotic stresses can lead to similar responses in plants. Moreover, different kinds of stresses can trigger responses through the induction of similar types of miRNAs (Sunkar and Zhu, 2004). This suggests that plants share common signaling pathways that act in different abiotic stress responses. The relevant miRNAs are either up- or down-regulated after exposure to stress treatments that influence plant growth and developmental processes (Lu and Huang, 2008). The role of miR399 has been established mainly in the plant response to phosphate starvation (Bari et al., 2006). However, little is known about the potential biological function of miR399 in responses to other abiotic stresses. In this report, we uncovered a possible role of miR399f in plant responses to osmotic stresses, including salt, drought, and ABA. The miR399 family consists of six members, miR399a to miR399f, which share similar mature sequences. A previous report indicated that phosphate starvation induces the expression of miR399 family genes (Bari et al., 2006). Our data showed that the expression of miR399f was remarkably induced by NaCl and ABA treatment. These results suggested that the members of miR399 family might have distinct roles in plant responses to various abiotic stresses.
Several miRNAs function in the ABA-mediated stress response (Chen et al., 2012; Jia et al., 2009; Reyes and Chua, 2007). Some studies suggested a genetic connection between miRNAs and ABA-mediated stress responses (Kim et al., 2010; Song et al., 2013). Consistent with our results indicating the involvement of Arabidopsis miR399f in salt stress and ABA responses, miRNA array analysis revealed that poplar miR399 is induced in response to ABA and NaCl (Jia et al., 2009). These findings indicate that some stress-responsive miRNAs might be similarly regulated across different species. These miRNAs potentially play vital roles in the morphological and metabolic adaptation of plants to salinity and ABA-mediated stress responses, and a genotype-specific expression model might explain the distinct stress tolerances among species.
Plant responses to various stresses are associated with multiple transcriptional cascades mediated by miRNAs (Lu and Huang, 2008; Sunkar et al., 2007). Identification of direct down-stream target genes regulated by miRNAs in each of these cascades is crucial for understanding miRNA-mediated plant responses to stresses. Through our in silico prediction and subsequent gene expression analysis, we identified two candidate downstream target genes for miR399f, namely CSP41b and ABF3, the expression levels of which significantly decreased in miR399f-OE plants under salt and ABA treatment conditions. However, the expression patterns of CSP41b and ABF3 genes were different in WT under the stress conditions. Unlike CSP41b, ABF3 expression was decreased after 6 h of stress treatment. These results suggested that the mode of miR399f action on the regulation of CSP41b and ABF3 expression may be different. One possibility would be that miR399f regulate ABF3 expression at the late stage to turn-off the ABF3-mediated stress signaling. In the future, we are going to verify the specific regulatory mechanism for miR399f-mediated CSP41b and ABF3 gene expression.
While the role of CSP41b is not yet clear, it is known that ABF3 is a transcription factor involved in transcriptional cascades in response to salt, ABA and drought. Mutation of ABF3 enhances the tolerance of plant to salt stress and ABA treatment, but significantly reduces their capacity to survive drought stress (Finkelstein et al., 2005; Yoshida et al., 2010). Consistent with the phenotype of the abf3 mutant under abiotic stresses, miR399f-OE plants displayed increased tolerance to salt stress and exogenous ABA, but hypersensitivity to drought stress (Fig. 4). These results indicate that miR399f acts as a positive regulator of plant tolerance of salt stress and ABA, but a negative regulator for plant response to drought stress. Recent work reported that miR168a- and miR394a-overexpressing plants were hypersensitive to salt stress, but resistant to drought stress (Li et al., 2012; Song et al., 2013). These results support our findings that same miRNA can have different, stress-specific roles in plant responses. Our findings point to a need to explore the role of CSP41b and its regulation by miR399f for better understanding of the function of miR399f in plant stress signaling.
Overall, the present study identified a biological function of miR399f. Genetic and physiological studies revealed that over-expression of miR399f resulted in salt and ABA tolerance in Arabidopsis by overcoming the arrest of root growth and seed germination under salt and ABA treatment. By contrast, miR399f-OE plants displayed a hypersensitive phenotype under drought conditions (Fig. 4). In miR399f-OE plants, the expression of several stress-responsive genes, such as RD29A, RD29B, DREB1A, and DREB2A was suppressed. Even though our results suggested that miR399f might function in abiotic stress signaling via ABF3 and CSP41b, but still we cannot rule out the possibility of the involvement of other genes in miR399f-mediated signaling. That’s the reason why we tested a number of stress-responsive genes in our experiment. Moreover, we showed that RD29B containing ABRE element in its promoter was down regulated in miR399f-OE plants (Fig. 6). This impaired induction of stress-responsive genes in miR399f-OE plant might contribute, at least in part, to its hypersensitive phenotype under drought conditions. Further characterization of the putative components might reveal the association between miR399f and salt stress or ABA signals.
In summary, we provide the first evidence of the involvement of miR399f in osmotic stress signaling, including responses to salt, ABA, and drought, and of its putative downstream target genes in Arabidopsis. Our results suggested that the regulation of CSP41b and/or ABF3 expression by miR399f would be important for the maintenance of a phenotype favorable for the adaptive responses to salt stress, ABA, and drought stresses. Furthermore, miR399f and its target genes have distinct roles in plant responses to different types of environmental stresses.
Acknowledgments
This work was supported by a grant from National Research Foundation funded by the Gyeongsang National University Fund for Professors on Sabbatical Leave (2015), Korean Government (MSIP; 2013R1A2A1A01005170) and Next-Generation BioGreen21 Program (SSAC, grant#: PJ01105101 and PJ01105102), Rural Development Administration Republic of Korea; and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2013R1A1A2011511) to D. Baek.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006;141:1000–1011. doi: 10.1104/pp.106.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Kim M.C., Chun H.J., Kang S., Park H.C., Shin G., Park J., Shen M., Hong H., Kim W.Y., et al. Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol. 2013;161:362–373. doi: 10.1104/pp.112.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brotman Y., Lisec J., Méret M., Chet I., Willmitzer L., Viterbo A. Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology. 2012;158:139–146. doi: 10.1099/mic.0.052621-0. [DOI] [PubMed] [Google Scholar]
- Buhtz A., Springer F., Chappell L., Baulcombe D.C., Kehr J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 2008;53:739–749. doi: 10.1111/j.1365-313X.2007.03368.x. [DOI] [PubMed] [Google Scholar]
- Chao D.Y., Luo Y.H., Shi M., Luo D., Lin H.X. Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Res. 2005;15:796–810. doi: 10.1038/sj.cr.7290349. [DOI] [PubMed] [Google Scholar]
- Chen H., Li Z., Xiong L. A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett. 2012;586:1742–1747. doi: 10.1016/j.febslet.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman J.C., Bohnert H.J. Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. 2000;3:117–124. doi: 10.1016/s1369-5266(99)00052-7. [DOI] [PubMed] [Google Scholar]
- Fagard M., Dellagi A., Roux C., Périno C., Rigault M., Boucher V., Shevchik V.E., Expert D. Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Mol. Plant Microbe. Interact. 2007;20:794–805. doi: 10.1094/MPMI-20-7-0794. [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S.S., Lynch T.J., Thomas T.L., Rock C.D. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- Fujii H., Chiou T.J., Lin S.I., Aung K., Zhu J.K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Guo H.S., Xie Q., Fei J.F., Chua N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh L.C., Lin S.I., Shih A.C., Chen J.W., Lin W.Y., Tseng C.Y., Li W.H., Chiou T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G., Saini A., Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta. 2009;229:1009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- Jia X., Wang W.X., Ren L., Chen Q.J., Mendu V., Willcut B., Dinkins R., Tang X., Tang G. Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol. Biol. 2009;71:51–59. doi: 10.1007/s11103-009-9508-8. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. Micro-RNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Jin H. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Lee H.J., Jung H.J., Maruyama K., Suzuki N., Kang H. Overexpression of microRNA395c or 395e affects differently the seed germination of Arabidopsis thaliana under stress conditions. Planta. 2010;232:1447–1454. doi: 10.1007/s00425-010-1267-x. [DOI] [PubMed] [Google Scholar]
- Li W.X., Oono Y., Zhu J., He X.J., Wu J.M., Iida K., Lu X.Y., Cui X., Jin H., Zhu J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Yang F., Yu D. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010;62:1046–1057. doi: 10.1111/j.1365-313X.2010.04216.x. [DOI] [PubMed] [Google Scholar]
- Lin S.I., Chiang S.F., Lin W.Y., Chen J.W., Tseng C.Y., Wu P.C., Chiou T.J. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 2008;147:732–746. doi: 10.1104/pp.108.116269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Chen Y.Q. Insights into the mechanism of plant development: interactions of miRNAs pathway with phytohormone response. Biochem. Biophys. Res. Commun. 2009;384:1–5. doi: 10.1016/j.bbrc.2009.04.028. [DOI] [PubMed] [Google Scholar]
- Lu X.Y., Huang X.L. Plant miRNAs and abiotic stress responses. Biochem. Biophys. Res. Commun. 2008;368:458–462. doi: 10.1016/j.bbrc.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Lu Y.D., Gan Q.H., Chi X.Y., Qin S. Roles of micro-RNA in plant defense and virus offense interaction. Plant Cell Rep. 2008;27:1571–1579. doi: 10.1007/s00299-008-0584-z. [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- Naya L., Paul S., Valdés-López O., Mendoza-Soto A.B., Nova-Franco B., Sosa-Valencia G., Reyes J.L., Hernández G. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS One. 2014;9:e84416. doi: 10.1371/journal.pone.0084416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B.D., Buhtz A., Kehr J., Scheible W.R. Micro-RNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008;53:731–738. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B.D., Musialak-Lange M., Nuc P., May P., Buhtz A., Kehr J., Walther D., Scheible W.R. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.Y., Kim S.A., Lee S.J., Kim S.Y. ATHB17 is a positive regulator of abscisic acid response during early seedling growth. Mol. Cells. 2013;35:125–133. doi: 10.1007/s10059-013-2245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.L., Chua N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Si Y., Zhang C., Meng S., Dane F. Gene expression changes in response to drought stress in Citrullus colocynthis. Plant Cell Rep. 2009;28:997–1009. doi: 10.1007/s00299-009-0703-5. [DOI] [PubMed] [Google Scholar]
- Siré C., Moreno A.B., Garcia-Chapa M., López-Moya J.J., San Segundo B. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS Lett. 2009;583:1039–1044. doi: 10.1016/j.febslet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- Song J.B., Gao S., Sun D., Li H., Shu X.X., Yang Z.M. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 2013;13:210. doi: 10.1186/1471-2229-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Chinnusamy V., Zhu J., Zhu J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 1993;5:1529–1539. doi: 10.1105/tpc.5.11.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E.A., Araus V., Lu C., Parry G., Green P.J., Coruzzi G.M., Gutiérrez R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Post-transcriptional RNA silencing in plant-microbe interactions: a touch of robustness and versatility. Curr. Opin. Plant Biol. 2008;11:464–470. doi: 10.1016/j.pbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant micro-RNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoo J.H., Park C.Y., Kim J.C., Heo W.D., Cheong M.S., Park H.C., Kim M.C., Moon B.C., Choi M.S., Kang Y.H., et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in arabidopsis. J. Biol. Chem. 2005;280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Zhao B., Ge L., Liang R., Li W., Ruan K., Lin H., Jin Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009;8:10–29. doi: 10.1186/1471-2199-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Ding H., Zhu J.K., Zhang F., Li W.X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011;190:906–915. doi: 10.1111/j.1469-8137.2011.03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang G., Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang G., Sutoh K., Zhu J.K., Zhang W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta. 2008;1779:780–788. doi: 10.1016/j.bbagrm.2008.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


