Abstract
Biotin is a water-soluble vitamin and serves as a coenzyme for five carboxylases in humans. Biotin is also covalently attached to distinct lysine residues in histones, affecting chromatin structure and mediating gene regulation. This review describes mammalian biotin metabolism, biotin analysis, markers of biotin status, and biological functions of biotin. Proteins such as holocarboxylase synthetase, biotinidase, and the biotin transporters SMVT and MCT1 play crucial roles in biotin homeostasis, and these roles are reviewed here. Possible effects of inadequate biotin intake, drug interactions, and inborn errors of metabolism are discussed, including putative effects on birth defects.
Keywords: Biotin, biotinidase, deficiency, holocarboxylase synthetase, metabolism, transport
1. History
Boas was the first to demonstrate the requirement for the water-soluble vitamin biotin in mammals [1]. Subsequently, biotin was isolated [2], its chemical structure was identified [3], and its chemical synthesis was accomplished [4].
2. Biosynthesis and catabolism of biotin
Mammals cannot synthesize biotin but depend on dietary intake from microbial and plant sources. The route of microbial biosynthesis of biotin was largely elaborated by Eisenberg and coworkers in studies of Escherichia coli. In this pathway, dethiobiotin is formed from the oleate metabolite pimelyl-CoA and carbamyl phosphate [5]. Sulfur is incorporated into dethiobiotin in a synthase-dependent step, generating biotin [6].
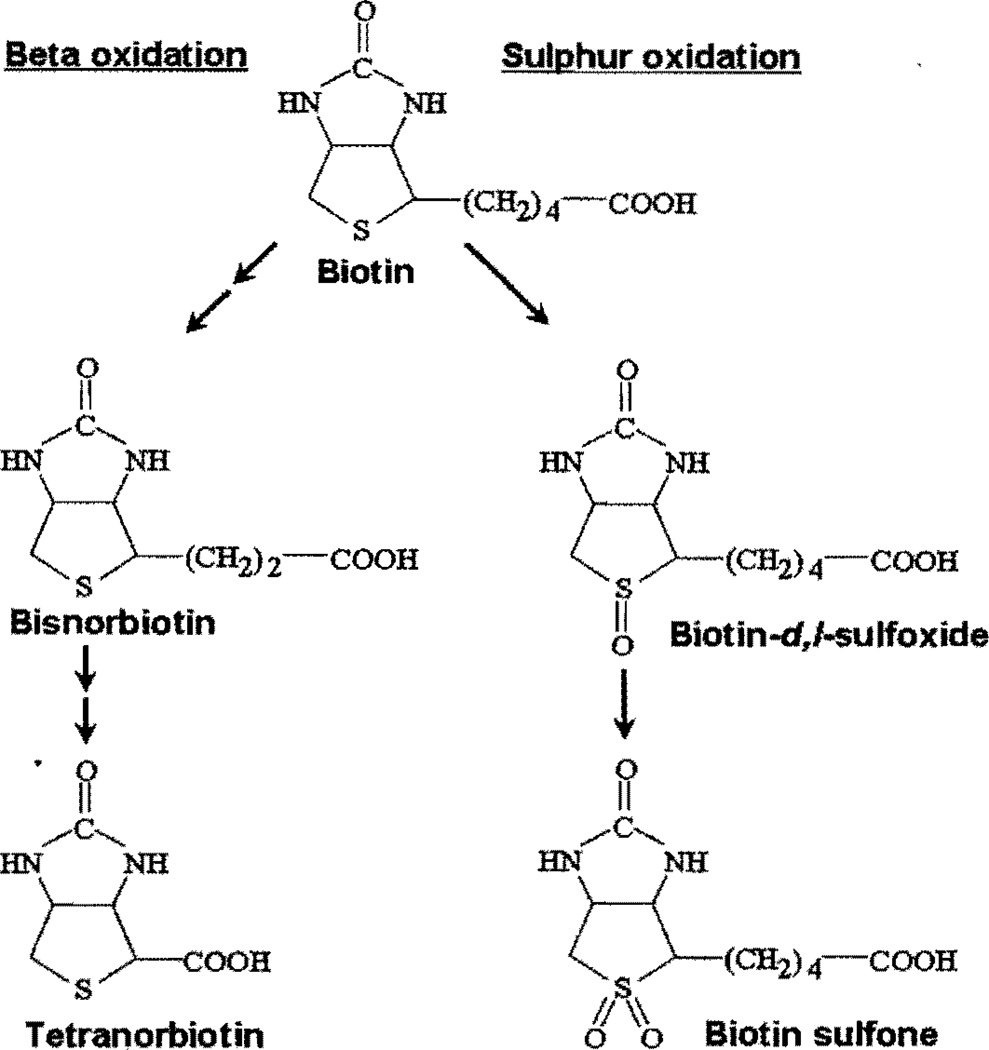
Early studies of biotin catabolism were primarily conducted by using microbes as model organisms but largely apply to mammals as well [7–9]. McCormick and Wright identified two pathways of biotin catabolism (Fig. 1). In one pathway, biotin is catabolized by β-oxidation of the valeric acid side chain [7]. This pathway leads to the formation of bisnorbiotin, tetranorbiotin, and intermediates known to result from β-oxidation of fatty acids (i.e. α,β-dehydro-, β-hydroxy, and β-keto-intermediates). Spontaneous decarboxylation of β-ketobiotin and β-ketobisnorbiotin yields bisnorbiotin methyl ketone and tetranorbiotin methyl ketone [7,9]. After degradation of biotin to tetranorbiotin, microorganisms cleave and degrade the heterocyclic ring [7]; degradation of the heterocyclic ring is quantitatively minor in mammals [8], In a second pathway of biotin catabolism, the sulfur in the heterocyclic ring is oxidized to produce biotin−/−sulfoxide, biotin-d-sulfoxide, and biotin sulfone [7,9]. Sulfur oxidation in the biotin molecule occurs in the smooth endoplasmic reticulum in a reaction that depends on nicotinamide adenine dinucleotide phosphate [10]. Biotin is also catabolized by a combination of both β-oxidation and sulfur oxidation, producing compounds such as bisnorbiotin sulfone [7,9].
Fig. 1.
Pathways of biotin catabolism.
3. Biological functions of biotin
Biotin has long been recognized for its role as a covalently bound coenzyme for carboxylases [11]. More recently, evidence emerged that biotin also plays unique roles in cell signaling, epigenetic regulation of genes, and chromatin structure [12].
3.1. Biotin-dependent carboxylases
In mammals, biotin serves as a covalently bound coenzyme for acetyl-CoA carboxylases 1 and 2 (E.C. 6.4.1.2), pyruvate carboxylase (E.C. 6.4.1.1), propionyl-CoA carboxylase (E.C. 6.4.1.3), and 3-methylcrotonyl-CoA carboxylase (E.C. 6.4.1.4) [11,13]. The attachment of biotin to the ε-amino group of a specific lysine residue in apocarboxylases is catalyzed by holocarboxylase synthetase (HCS) (E.C. 6.3.4,10); biotinylation of carboxylases requires ATP and proceeds in the following two steps [14]:
ATP + biotin + HCS → Biotin-AMP-HCS + pyrophosphate
-
Biotin-AMP-HCS + apocarboxylase → holocarboxylase + AMP + HCS
(Net) ATP + biotin + apocarboxylase → holocarboxylase + & AMP + pyrophosphate
Biotin-dependent carboxylases mediate the covalent binding of bicarbonate to organic acids by the following carboxylation sequence [15]. First, bicarbonate and ATP form carboxy phosphate, releasing ADP. Second, carboxy phosphate reacts with the 1'-N of the biotinyl moiety in holocarboxylases (“biotinyt carboxylase”) to form 1'-N-carboxybiotinyl carboxylase and to release inorganic phosphate, Pi. Third, 1'-N-carboxybiotinyl carboxylase incorporates carboxylate into an acceptor, i.e. a specific organic acid for each of the carboxylases.
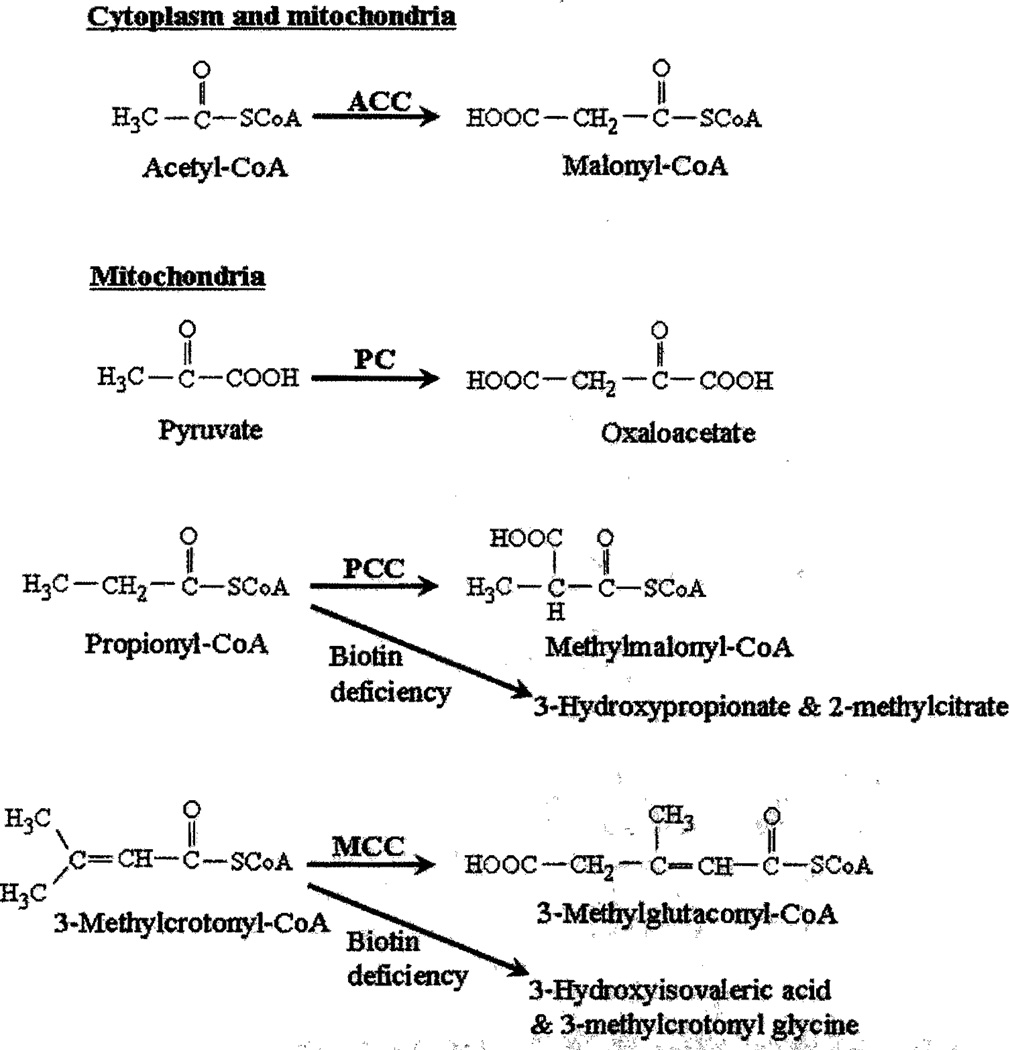
Two isoforms of acetyl-CoA carboxylase have been identified: the cytoplasmic acetyl-CoA carboxylase 1 and the mitochondrial acetyl-CoA carboxylase 2 [13]. Both acetyl-CoA carboxylase 1 and 2 catalyze the binding of bicarbonate to acetyl-CoA, generating malonyl-CoA (Fig. 2). Acetyl-CoA carboxylase 1 is a key enzyme in fatty acid synthesis in the cytoplasm. In contrast, acetyl-CoA carboxylase 2 participates in the regulation of fatty acid oxidation in mitochondria. This effect of acetyl-CoA carboxylase 2 is mediated by malonyl-CoA, which is an inhibitor of fatty acid transport into mitochondria.
Fig. 2.
Biotin-dependent carboxylases. ACC, acetyl-CoA carboxylase; MCC, 3-methylcrotonyl-CoA carboxylase; PC, pyruvate carboxylase; PCC, propionyl-CoA carboxylase.
Pyruvate carboxylase localizes to mitochondria and is a key enzyme in gluconeogenesis (Fig. 2). The mitochondrial propionyl-CoA carboxylase and 3-methylcrotonyl-CoA carboxylase comprise biotin-containing alpha subunits and biotin-free beta subunits. Propionyl-CoA carboxylase catalyzes an essential step in the metabolism of isoleucine, valine, methionine, threonine, the cholesterol side chain, and odd-chain fatty acids. β-Methylcrotonyl-CoA carboxylase catalyzes an essential step in leucine metabolism. Additional carboxylases have been identified in some microbes but are not further discussed here [11].
Proteolytic degradation of holocarboxylases leads to the formation of biotinyl peptides. These peptides are further degraded by biotinidase (BTD) (E.C. 3.5.1.12) to release biotin which is recycled in holocarboxylase synthesis [16].
3.2. Epigenetics
3.2.1. Chromatin and gene regulation
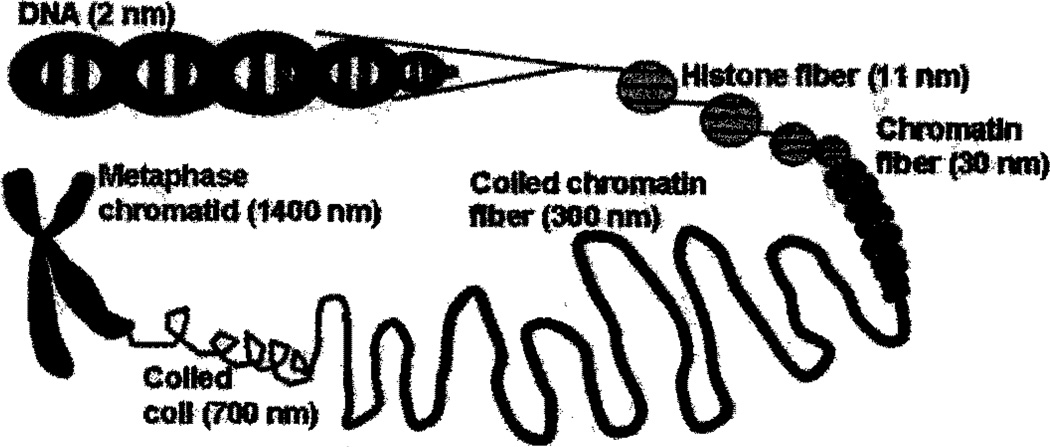
Chromatin comprises of DNA and DNA-binding proteins, i.e. histories and nonhistone proteins (Fig. 3). Histories play a crucial role in the folding of DNA in chromatin [17]. Five major classes of histones have been identified in mammals: linker histone H1, and core histones H2A, H2B, H3, and H4. Histones consist of a globular domain and a more flexible N-terminal tail. DNA and histones form repetitive nucleoprotein units, the nucleosomes [17]. Each nucleosome (“nucleosomal core particle”) consists of 147 base pairs of DNA wrapped around an octamer of core histones (one H3-H3-H4-H4 tetramer and two H2A–H2B dimers).
Fig. 3.
The structure of chromatin. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The N-terminal tails of core histones protrude from the nucleosomal surface; covalent modifications of these tails affect the structure of chromatin and form the basis for gene regulation [18,19]. Amino acid residues in histone tails are modified by covalent acetylation, methylation, phosphorylation, ubiquitination, and poly(ADP-ribosylation) [17–19]. The various modifications of histones have distinct functions. For example, trimethylation of K4 in histone H3 is associated with transcriptional activation of surrounding DNA, whereas dimethylation of K9 is associated with transcriptional silencing [18,19]. Covalent modifications of histones are reversible [19].
3.2.2. Histone biotinylation
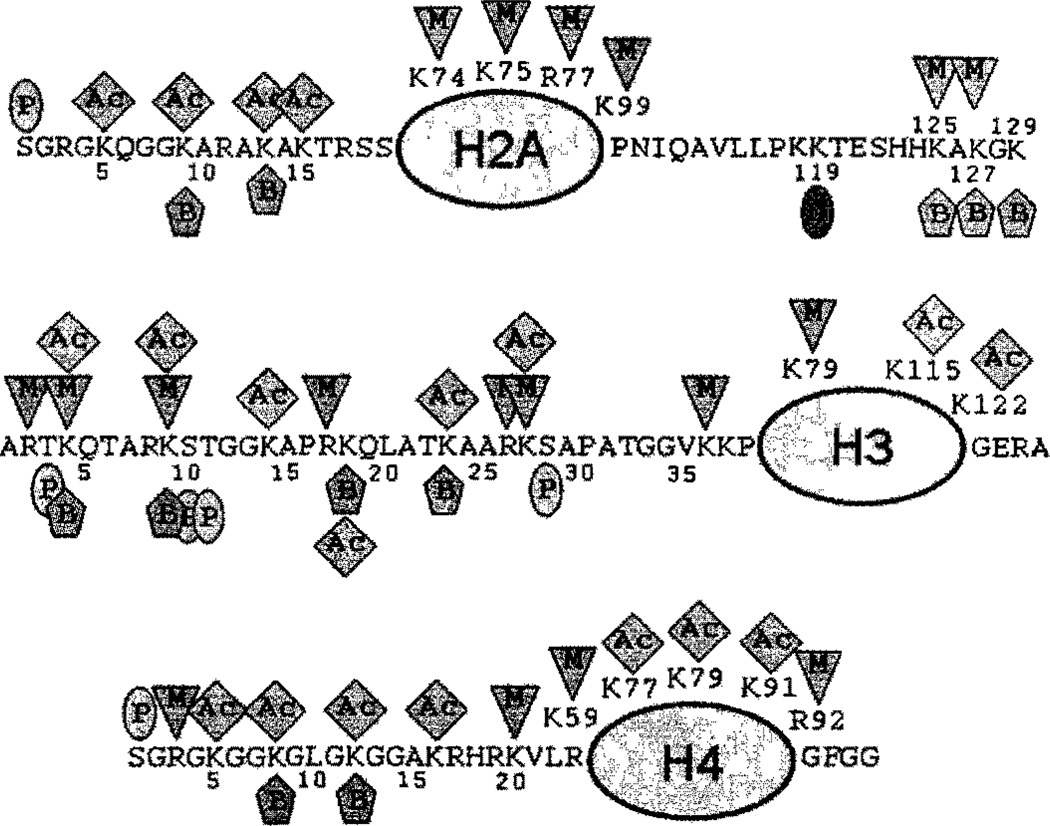
Evidence has been provided that biotin is attached to histones (DNA-binding proteins) via an amide bond [20,21]. The following 11 biotinylation sites have been identified in human histones: lysine (K)-9, K13, K125, K127, and K129 in histone H2A [22]; K4, K9, K18, and perhaps K23 in histone H3 [23,24]; and K8 and K12 in histone H4 [25] (Fig. 4).
Fig. 4.
Modification sites in histories H2A, H3, and H4. Ac, acetate; B, biotin; M, methyl; P, phosphate; U, ubiquitin, [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Functions of histone biotinylation in chromatin biology are emerging. For example, biotinylation of K12 in histone H4 plays roles in gene repression, DNA repair, heterochromatin structures, and repression of transposons, thereby promoting genomic stability [26–30]. Importantly, biotinylation of histones depends on dietary biotin supply [31,32].
Originally, it was believed that biotinylation of histones is catalyzed by BTD. Hymes et al. have proposed a reaction mechanism by which cleavage of biocytin (biotin-ε-lysine) by BTD leads to the formation of a biotinyl-thioester intermediate at or near the active site of BTD [20,33]. In a next step, the biotinyl moiety is transferred from the thioester to the ε-amino group of lysines in histories. The substrate (biocytin) for biotinylation of histories is generated in the breakdown of biotin-dependent carboxylases [34,35].
Subsequent studies by Narang et al. provided evidence that HCS can also biotinylate histones [36]. Notwithstanding the ability of BTD to catalyze biotinylation of histones, studies by Camporeale et al. suggest that HCS is more important than BTD for biotinylation of histones [27]. Importantly, knockdown of HCS or BTD decreases histone biotinylation, causes abnormal gene expression patterns, and causes phenotypes such as decreased life span and heat resistance in Drosophila melanogaster [27]. Effects of BTD knockdown are likely due to impaired biotin recycling, causing biotin deficiency.
3.2.3. Debiotinylation of histones
Biotinylation of histones is a reversible modification but the mechanisms mediating debiotinylation of histones are largely unknown. Recent studies suggest that BTD might catalyze both biotinylation and debiotinylation of histones [37]. Variables such as the microenvironment (substrate availability) in chromatin, protein-BTD interactions, and posttranslational modifications and alternative splicing of BTD might theoretically determine whether BTD acts as a biotinyl histone transferase or histone debiotinylase [38,39]. An assay for analysis of histone debiotinylases is available [40].
3.3. Gene expression
About 40 years ago, evidence emerged suggesting that biotin may affect gene expression [41]; these pioneering studies provided evidence that expression of rat liver glucokinase (E.C. 2.7.1.2) depends on biotin. Since then, more than 2,000 biotin-dependent genes have been identified in human lymphoid and liver cells [12]. These genes are not randomly distributed in the human genome but can be assigned to gene clusters, based on signaling pathways, chromosomal location, cellular localization of gene products, biological function, and molecular function [12]. Evidence has been provided that bisnorbiotin also affects gene expression, suggesting that biotin catabolites might have biotin-like activities in humans [12]. Effects of biotin on gene expression are mediated by a variety of cell signals, including biotinyl-AMP, cGMP, NF-κB, Sp1, and Sp3, and receptor tyrosine kinases [12,42].
Evidence has been provided that biotin also affects gene expression at the post-transcriptional level. For example, the expression of asialoglycoprotein receptor in HepG2 hepatocarcinoma cells and propionyl-CoA carboxylase in rat hepatocytes depends on biotin; these effects are not caused by alterations in the abundance of mRNA coding for these proteins [12].
4. Methods of biotin analysis
4.1. Microbial growth assays
Microorganisms such as Lactobacillus plantarum, Lactobacillus casei, Ochromonas danica, Escherichia coli C162, Saccharomyces cerevisiae, and Kloeckera brevis depend on biotin for growth and are commonly used in microbial growth assays [43]. These test organisms show variable growth responses to biotin and biotin precursors, catabolites, and analogs. For some microbes (e.g. Ochromonas danica, Lacto-bacillus plantarum), acid or enzymatic hydrolysis of samples is required to release biotin from protein. In contrast, for other organisms (e.g. Kloeckera brevis) the detectable biotin decreases with enzymatic hydrolysis.
4.2. Avidin-binding assays
The proteins avidin and streptavidin are widely used in biotin analysis because they bind biotin with exceptional strength and specificity; the dissociation constant of the avidin–biotin complex is 1.3 × 10−15 M [44]. Avidin-binding assays generally measure the ability of biotin to compete with [3H] biotin or [14C] biotin for binding to avidin; to prevent binding of enzyme-conjugated avidin to biotinylated protein adhered to plastic; or to prevent the binding of biotinylated enzyme to avidin [43]. Streptavidin has greater specificity for biotin than avidin and, therefore, is typically the protein of choice in (strept) avidin-binding assays [44].
Avidin-binding assays are subject to the following potential pitfalls. First, avidin binds biotin catabolites less tightly compared with biotin [43]. Hence, avidin-binding assays may underestimate the true concentration of biotin plus catabolites if calibrated by using biotin. Second, compounds structurally similar to biotin (e.g. lipoic acid, urea, hexanoic acid) may bind to avidin and cause artificially large readings for “apparent biotin.” Taken together, avidin-binding compounds in biological samples need to be resolved by chromatography prior to analysis of individual chromatographic fractions against authentic standards. Appropriate analytical procedures have been reviewed elsewhere [43].
4.3. 4'-Hydroxyazobenzene-2-carboxylic acid dye assay
This is a simple assay to quantify biotin at concentrations that exceed those typically found in biological samples. In the absence of biotin, 4'-hydroxyazobenzene-2-carboxylic acid forms noncovalent complexes with avidin at its biotin-binding sites to produce a characteristic absorption band at 500 nm [43]. The addition of biotin to this complex results in displacement of 4'-hydroxyazobenzene-2-carboxytic acid from the binding sites. As 4'-hydroxyazobenzene-2-carboxylic acid is displaced, the absorbance of the complex decreases proportionally.
5. Digestion, absorption, storage, and excretion
5.1. Digestion
A large fraction of dietary biotin is covalently linked to lysine residues in proteins [16,45]. Gastrointestinal proteases and peptidases digest biotin-containing proteins to release biocytin (biotinyl-ε-lysine) and biotin-containing peptides [16]. BTD releases free biotin from biocytin and biotinylated peptides; BTD activity can be found in pancreatic fluids and other intestinal secretions, intestinal flora, and brush-border membranes [16,46,47]. The primary site(s) for hydrolysis of biotinylated peptides is unknown. Small quantities of biotmylated peptides might be absorbed without prior hydrolysis [48].
5.2. Absorption
Biotin transport across the apical membrane in the brush-border membrane occurs through a sodium-dependent, electroneutral mechanism, whereas the transport across the basolateral membrane is sodium-independent and electrogenic [49–52]. The apparent Michaelis–Menten constant of the biotin transporter in mouse jejunum is 22 µmol/L [53]. The intestinal, sodium-dependent uptake of biotin is mediated by the sodium-dependent multivitamin transporter SMVT, which also has affinity for pantothenic acid and lipoic acid [54–57]. Passive diffusion across cell membranes may contribute to biotin uptake if extracellular biotin concentrations exceed 25 µmol/L; carrier-mediated uptake predominates at biotin concentrations of less than 5 µmol/L [47]. Intestinal biotin transporter activity depends on dietary biotin supply [58], and is regulated by protein kinase C in concert with Ca2+/catmodutin-mediated pathways [52] and transcription factors KLF-4 and AP-2 [59,60]. A mechanism has been proposed by which biotinylation of histones at the SMVT promoter locus regulates expression of the SMVT gene and, hence, biotin transport rates [31]. Doses of biotin that exceed the normal dietary intake 600 times are 100% bioavailable [61].
5.3. Uptake, storage, and distribution in liver and peripheral tissues
SMVT not only mediates intestinal absorption of biotin, but also is crucial for biotin uptake into liver and peripheral tissues and for renal reabsorption [52,57,62,63]. Evidence has been provided that monocarboxylate transporter 1 might account for biotin uptake in some cell lineages such lymphoid cells and keratinocytes [64,65].
A relatively large fraction of intravenously administered biotin accumulates in rat liver, consistent with a role for this organ in biotin storage [66]. Depletion and repletion experiments of biotin-dependent carboxylases in rat liver provided evidence that mitochondrial acetyl-CoA carboxylase 2 may serve as a reservoir for biotin [67]. Note that the central nervous system (CNS) retains most of its biotin during phases of depletion at the expense of other tissues such as liver [68]. In rat CNS, biotin is enriched in specific regions such as the cerebellar motor system and brainstem auditory nuclei that correlates to the associated neurological symptoms associated with biotin deficiency [69].
5.4. Cellular compartments
Biotin is distributed unequally across cellular compartments [66]. The vast majority of biotin in rat liver localizes to mitochondria and cytoplasm, whereas only a small fraction localizes to microsomes [66]. The relative enrichment of biotin in mitochondria and cytoplasm is consistent with the role of biotin as a coenzyme for carboxylases in these compartments. A quantitatively small but qualitatively important fraction of biotin localizes to the cell nucleus, i.e. about 0.7% of total biotin in human lymphoid cells can be recovered from the nuclear fraction [21]. The relative abundance of nuclear biotin increases to about 1% of total biotin in response to proliferation, consistent with a role for histone in cell proliferation [21,70].
5.5. Urinary and biliary excretion
Healthy adults excrete ~100 nmol of biotin and catabolites per day into urine [9] (Table 1). Biotin accounts for approximately half of the total; the catabolites bisnorbiotin, biotin-d,l-sulfoxides, bisnorbiotin methyl ketone, biotin sulfone, and tetranorbiotin−/−sulfoxide account for most of the balance [9]. If physiological or pharmacological doses of biotin are administered parenterally to humans, rats, or pigs, 43–75% of the dose is excreted into urine [8,61]. Renal epithelia reclaim biotin that is filtered in the glomeruli in an SMVT-mediated process [72]. The biliary excretion of biotin and catabolites is quantitatively minor. Less than 2% of an intravenous dose of [14C] biotin was recovered in rat bile but more than 60% of the dose was excreted in urine [73].
Table 1.
| Compound | Serum (pmol/L) |
Urine (nmol/24 h) |
|---|---|---|
| Biotin | 244 ± 61 | 35 ± 14 |
| Bisnorbiotin | 189 ± 135 | 68 ± 48 |
| Biotin-d,l-sulfoxide | 15 ± 33 | 5±6 |
| Bisnorbiotin methyl ketone | NDa | 9±9 |
| Biotin sulfone | NDa | 5±5 |
| Total biotinyl compounds | 464 ± 178b | 122 ± 66 |
Means ± SD are reported (n = 15 for serum; n = 6 for urine).
ND, not determined. Bisnorbiotin methyl ketone and biotin sulfone had not been identified at the time when this study of serum was conducted and, hence, quantification of these “unknowns” was based on using biotin as a standard.
Including three unidentified biotin catabolites.
6. Biotin status
6.1. Direct measures
The urinary excretion of biotin and biotin catabolites decreases rapidly and substantially in biotin-deficient individuals, suggesting that the urinary excretion is an early and sensitive indicator of biotin deficiency [74]. A decreased urinary excretion of biotin, together with an increase in the ratios of bisnorbiotin and biotin-d,l-sulfoxide to biotin, reflect increased biotin catabolism as observed in women smokers [75]. In contrast, serum concentrations of biotin, bisnorbiotin, and biotin-d,l-sulfoxide do not decrease in biotin-deficient individuals [74] and in patients on biotin-free total parenteral nutrition [76] during reasonable periods of observation. Hence, serum concentrations are not good indicators of marginal biotin deficiency.
6.2. Indirect measures
Activities of biotin-dependent carboxylases in lymphocytes are reliable markers for assessing the biotin status in humans [77,78]. Some investigators used a modified approach and calculated a “carboxylase activation index,” representing the ratio of carboxylase activities in cells incubated with excess supplemental biotin to cells without supplemental biotin [76]. High values for the activation index suggest that a substantial fraction of a given carboxylase was present as apo-enzyme, indicating biotin deficiency.
Reduced carboxylase activities in biotin deficiency affects intermediary metabolism (Fig. 2). Reduced activity of β-methylcrotonyl-CoA carboxylase impairs leucine catabolistm. As a consequence, β-methylcrotonyl-CoA is shunted to alternative pathways, leading to an increased formation of 3-hydroxyisovaleric acid and 3-methylcrotonyl glycine. Biotin deficiency studies in humans suggest that the urinary excretion of 3-hydroxyisovaleric acid is an early and sensitive indicator of biotin status [75,79].
Reduced activity of propionyl-CoA carboxylase causes a metabolic block in propionic acid metabolism. Consequently, propionic acid is shunted to alternative metabolic pathways. In these pathways, 3-hydroxypropionic add and 2-methylcitric acid are formed. However, recent studies in biotin-deficient individuals suggest that the urinary excretion of 3-hydroxypropionic acid and 2-methylcitric acid is not a good indicator of marginal biotin deficiency [80]. Theoretically, propionic acid might be consumed in the synthesis of odd-chain fatty acids [81]. In severe biotin deficiency, the activity of pyruvate carboxylase may be reduced, leading to the accumulation of lactate [32].
7. Biotin deficiency
7.1. Clinical findings of frank biotin deficiency
Signs of frank biotin deficiency have been described in patients with BTD deficiency [35], in severely malnourished children in developing countries [82], and in individuals consuming large amounts of raw egg white which contains the protein avidin [83]. Binding of biotin to avidin in the gastrointestinal tract prevents absorption of biotin. Clinical findings of frank biotin deficiency include periorificial dermatitis, conjunctivitis, alopecia, ataxia, hypotonia, ketolactic acidosis/organic aciduria, seizures, skin infection, and developmental delay in infants and children [35,45].
7.2. Immune system, cell proliferation, and stress resistance
Biotin deficiency has adverse effects on cellular and humoral immune functions. For example, children with hereditary abnormalities of biotin metabolism developed Candida dermatitis and presented with absent delayed-hypersensitivity skin-tests responses, IgA deficiency, and subnormal percentages of T lymphocytes in peripheral blood [84]. In rodents, biotin deficiency decreases antibody synthesis [85], decreases the number of spleen cells and the percentage of B lymphocytes in spleen [86], and impairs thymocyte maturation [87]. Decreased rates of cell proliferation may cause some of the effects of biotin on immune function [88–90].
Biotin deficiency is linked also to cell stress, enhancing the nuclear translocation of the transcription factor NF-κB in human lymphoid cells [91]. NF-κB mediates activation of anti-apoptotic genes; this is associated with enhanced survival of biotin-deficient cells in response to cell death signals compared with biotin-sufficient controls [91]. Stress-resistant Drosophila can be selected by feeding biotin-deficient diets for multiple generations [32].
7.3. Lipid metabolism
Consistent with roles of biotin-dependent acetyl-CoA carboxylases 1 and 2, and propionyl-CoA carboxylase in lipid metabolism, biotin deficiency causes alterations of the fatty acid profile in liver, skin, and serum of several animal species [92]. Biotin deficiency is associated with increased abundance of odd-chain fatty acids, suggesting that odd-chain fatty acid accumulation may be a marker for reduced propionyl-CoA carboxylase activity in biotin deficiency. Biotin deficiency does not affect the fatty acid composition in brain tissue to the same extent as in liver [92].
Biotin deficiency also causes abnormalities in fatty acid composition in humans. In patients who developed biotin deficiency during parenteral alimentation, the percentage of odd-chain fatty acids (15:0, 17:0) in serum increased for each of the four major lipid classes, i.e. cholesterol esters, phospholipids, triglycerides, and free fatty acids [92]. However, the relative changes in these four classes of lipids have not always been consistent among studies [92].
7.4. Teratogenic effects of biotin deficiency
Mock and coworkers suggested that about half of the pregnant women in the U.S. are marginally biotin deficient despite a normal dietary biotin intake [93–95]. If there was a link between marginal biotin deficiency and fetal malformations in humans, the findings by Mock and coworkers would have important implications for health policies and intake recommendations. As of today, this link is somewhat uncertain. While animal studies have clearly demonstrated that biotin deficiency is teratogenic, the severity of deficiency in these animal studies typically exceeded what was observed in pregnant women. Notwithstanding these limitations, the teratogenic effects of biotin deficiency in animal models warrant a brief summary [92,96,97]. In some strains of mice, biotin deficiency during pregnancy causes substantial increases in fetal malformations and mortality [92,97]. The most common fetal malformations in biotin-deficient rats include cleft palate, micrognathia, and micromelia. Evidence has been provided that biotin catabolism to bisnorbiotin is increased in pregnant women compared with nonpregnant controls [93,98]. Mock and coworkers reported that the activity of propionyl-CoA carboxylase is reduced by ~90% in the fetus at term in response to feeding an egg-white that causes only a 50% reduction in maternal hepatic propionyl-CoA carboxylase activity [99].
7.5. Biotin homeostasis in the CNS
Disturbances in biotin homeostasis in the CNS cause encephalopathies [12]. Factors leading to biotin imbalances in CNS include deficiencies of BTD, HCS, and perhaps biotin transporters [12] as described below. Afflicted patients typically respond to the administration of large doses of biotin with maintaining normal neurological function [12].
Moderate dietary biotin deficiency is typically not associated with neurological symptoms. This is consistent with the hypothesis that under conditions of moderate biotin deficiency the CNS maintains normal concentrations of biotin at the expense of other tissues. Indeed, evidence has been provided that biotin deficiency causes a >90% decrease of biotinylated carboxylases in rat liver, whereas brain carboxylases remain unchanged [68]. The tissue-specific response to biotin supply is probably due to tissue-specific changes in expression.
8. Inborn errors of biotin metabolism
8.1. BTD deficiency
Low activities of the enzyme BTD cause a failure to recycle biotin from degraded carboxylases, i.e. to release biotin from biocytin. Substantial amounts of biocytin are excreted into urine [100], eventually leading to biotin deficiency. Thus, clinical and biochemical features in children with BTD deficiency are similar to those described earlier for biotin deficiency [35].
Typically, symptoms of BTD deficiency appear at age 1 week to >1 year [35]. Wolf distinguishes between patients with profound BTD deficiency (less than 10% of normal serum BTD activity) and patients with partial deficiency (10–30% of normal BTD activity) [101]. Procedures for prenatal diagnosis of BTD activity in cultured amniotic fluid cells and for neonatal screening using blood samples have been proposed [35]. BTD activity is measured by quantitating the release of p-aminobenzoic acid from N-biotinyl-p-aminobenzoate [102]. The mean (±SD) normal activity of BTD is 5.8 ± 0.9 nmol p-aminobenzoate liberated per minute per milliliter of serum [102].
The combined incidence of profound and partial deficiency is 1 in 60,089 live births; an estimated 1 in 123 individuals is heterozygous for the disorder [101]. Mutations of the BTD gene have been well characterized at the molecular level [103–105]. Children with profound BTD deficiency are treated with 5–20 mg of biotin per day [35]. If identified early, symptomatic patients improve rapidly after biotin therapy is initiated; therapy must be continued throughout life [35].
8.2. Deficiencies of carboxylases and HCS
Afflicted individuals present with either isolated deficiencies of individual carboxylases or multiple deficiencies of biotin-dependent carboxylases because of defective HCS [106]. Patients with multiple carboxylase deficiency characteristically exhibit low activities of all five biotin-dependent carboxylases. Mutations of the HCS gene have been well characterized at the molecular level [107]. It has been estimated that the incidence of HCS deficiency is less than 1 in 1,000 live births in Japan [107]. Afflicted individuals typically respond well to administration of pharmacological doses of biotin, in particular if the mutation of the gene resides in the biotin-binding region of the protein [107]. Only a few cases of isolated carboxylase deficiencies have been reported [105,108–111].
8.3. Biotin transporter deficiency
Recently, a case of inborn biotin transporter deficiency has been identified [12]. The afflicted patient exhibited the typical signs of biotin deficiency despite normal biotin intake; transport rates of biotin in lymphoid tissues were substantially smaller compared with healthy controls. Clinical features are similar to those described for multiple carboxylase deficiency and BTD deficiency.
9. Biotin–drug interactions
9.1. Anticonvulsants
Biotin requirements may be increased during anticonvulsant therapy. The anticonvulsants primidone and carbamazepine inhibit biotin uptake into brush-border membrane vesicles from human intestine [45]. Long-term therapy with anticonvulsants increases both biotin catabolism and urinary excretion of 3-hydroxyisovaleric acid [45]. Phenobarbital, phenytoin, and carbamazepine displace biotin from BTD, conceivably affecting plasma transport, renal handling, or cellular uptake of biotin [45].
9.2. Lipoic acid
Lipoic acid competes with biotin for binding to SMVT [45], potentially decreasing the cellular uptake of biotin. Indeed, chronic administration of pharmacological doses of lipoic acid decreased the activities of pyruvate carboxylase and β-methylcrotonyl-CoA carboxylase in rat liver to 64–72% of controls [45].
10. Requirements and recommended intakes
10.1. Adequate intakes
The Food and Nutrition Board of the National Research Council has released recommendations for adequate intake of biotin, ranging from 5 µg/day in newborn infants to 35 µg/day in lactating women (21–143 nmol/day) [112]. These recommendations are based on estimated biotin intakes (not to be confused with “requirements” and “Recommended Dietary Allowances”) in a group of healthy people. Adequate intakes are set when requirements of a certain nutrient are unknown; they may serve as goals for the nutrient intake of individuals. Biotin supplements may contribute substantially to biotin intake. For example, 15–20% of individuals in the U.S. report consuming biotin-containing dietary supplements [112].
10.2. Factors that affect biotin requirements
Pregnancy may be associated with an increased demand for biotin. Recent studies provide evidence for marginal biotin deficiency in human gestation as judged by increased urinary excretion of 3-hydroxyisovaleric acid [98]. Pregnancy and smoking accelerate biotin catabolism in women [75,93].
Lactation may generate an increased demand for biotin. At 8 days postpartum, biotin in human milk was ~8 nmol/L and accounted for 44% of total avidin-binding substances; bisnorbiotin and biotin-d,l-sulfoxides accounted for 48% and 8%, respectively [113]. By 6 weeks postpartum, the biotin concentration had increased to ~30 nmol/L and accounted for about 70% of total avidin-binding substances; bisnorbiotin and biotin-d,l-sulfoxides accounted for about 20% and less than 10%, respectively.
11. Intake and food sources
The majority of biotin in meats and cereals appears to be protein bound [45]. Most studies of biotin content in foods depended on using bioassays. Biotin is widely distributed in natural foodstuffs. Foods relatively rich in biotin include egg yolk, liver, and some vegetables. The dietary biotin intake in Western populations is about 35–70 µg/day (143–287 nmol/day) [45]. Infants consuming 800 mL of mature breast milk per day ingest ~6 µg (24 nmol) of biotin [113]. It remains unclear whether biotin synthesis by gut microorganisms contributes importantly to the total biotin absorbed [45].
12. Excess and toxicity
Empirically, ingestion of pharmacological doses of biotin is considered safe. For example, lifelong treatment of BTD deficiency patients with biotin doses that exceed the normal dietary intake by 300 times does not produce frank signs of toxicity [35]. Likewise, no signs of biotin overdose were reported after acute oral and intravenous administration of doses that exceeded the dietary biotin intake by up to 600 times [61]. Biotin supplementation affects the expression of numerous genes. It is unknown whether any of these alterations are undesirable.
Acknowledgments
This work was supported in part by the University of Nebraska Agricultural Research Division with funds provided through the Hatch Act. Additional support was provided by NIH Grants DK063945, DK077816, and ES015206, USDA Grant 2006-35200-17138, and NSF EPSCoR Grant EPS-0701892.
Abbreviations
- BTD
biotinidase
- HCS
holocarboxylase synthetase
- K
lysine
References
- 1.Boas MA. The effect of desiccation upon the nutritive properties of egg-white. Biochem. J. 1927;21:712–724. doi: 10.1042/bj0210712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kogl F, Tonnis B. Über das Bios-Problem. Darstellung von krystallisiertem Biotin aus Eigelb. Z. Physiol Chem. 1932;242:43–73. [Google Scholar]
- 3.du Vigneaud V, Melville DB, Folkers K, Wolf DE, Mozingo DE, Keresztesy JC, Harris SA. The structure of biotin: a study of desthiobiotin. J. Biol. Chem. 1942;146:475–485. [Google Scholar]
- 4.Harris SA, Wolf DE, Mozingo R, Folkers K. Synthetic biotin. Science. 1943;97:447–448. doi: 10.1126/science.97.2524.447. [DOI] [PubMed] [Google Scholar]
- 5.Hatakeyama K, Kobayashi M, Yukawa H. Analysis of biotin biosynthesis pathway in coryneform bacteria: Brevibacterium flavum. In: McCormick DB, Suttie JW, Wagner C, editors. Vitamins and Coenzymes, Part I. San Diego, CA: Academic Press; 1997. pp. 339–348. [DOI] [PubMed] [Google Scholar]
- 6.Flint DH, Allen RM. Purification and characterization of biotin synthases. In: McCormick DM, Suttie W, Wagner C, editors. Vitamins and Coenzymes, Part 1. San Diego, CA: Academic Press; 1997. pp. 349–356. [DOI] [PubMed] [Google Scholar]
- 7.McCormick DB, Wright LD. The metabolism of biotin and analogues. In: Florkin M, Stotz EH, editors. Metabolism of Vitamins and Trace Elements. Amsterdam, The Netherlands: Elsevier; 1971. pp. 81–110. [Google Scholar]
- 8.Lee HM, Wright LD, McCormick DB. Metabolism of carbonyl-labeled [14C] biotin in the rat. J. Nutr. 1972;102:1453–1464. doi: 10.1093/jn/102.11.1453. [DOI] [PubMed] [Google Scholar]
- 9.Zempleni J, McCormick DB, Mock DM. Identification of biotin sulfone, bisnorbiotin methyl ketone, and tetranorbiotin−/−sulf-oxide in human urine. Am. J. Clin. Nutr. 1997;65:508–511. doi: 10.1093/ajcn/65.2.508. [DOI] [PubMed] [Google Scholar]
- 10.Lee YC, Joiner-Hayes MG, McCormick DB. Microsomal oxidation of α-thiocarboxylic acids to sulfoxides. Biochem. Pharmacol. 1970;19:2825–2832. doi: 10.1016/0006-2952(70)90021-3. [DOI] [PubMed] [Google Scholar]
- 11.Knowles JR. The mechanism of biotin-dependent enzymes. Ann. Rev. Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- 12.Zempleni J. Uptake, localization, and noncarboxylase roles of biotin. Annu. Rev. Nutr. 2005;25:175–196. doi: 10.1146/annurev.nutr.25.121304.131724. [DOI] [PubMed] [Google Scholar]
- 13.Kim K-H, McCormick DB, Bier DM, Goodridge AG. Regulation of mammalian acetyl-coenzyme A carboxylase. Ann. Rev. Nutr. 1997;17:77–99. doi: 10.1146/annurev.nutr.17.1.77. [DOI] [PubMed] [Google Scholar]
- 14.Dakshinamurti K, Chauhan J. Biotin-binding proteins. In: Dakshinamurti K, editor. Vitamin Receptors: Vitamins as Ligands in Cell Communication. Cambridge, UK: Cambridge University Press; 1994. pp. 200–249. [Google Scholar]
- 15.Camporeale G, Zempleni J. Biotin. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. 9th. Washington, DC: International Life Sciences Institute; 2006. pp. 314–326. [Google Scholar]
- 16.Wolf B, Heard GS, McVoy JRS, Grier RE. Biotinidase deficiency. Ann. NY Acad. Sci. 1985;447:252–262. doi: 10.1111/j.1749-6632.1985.tb18443.x. [DOI] [PubMed] [Google Scholar]
- 17.Wolffe A. Chromatin. 3rd. San Diego, CA: Academic Press; 1998. [Google Scholar]
- 18.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Celt Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 20.Hymes J, Wolf B. Human biotinidase isn't just for recycling biotin. J. Nutr. 1999;129:485S–489S. doi: 10.1093/jn/129.2.485S. [DOI] [PubMed] [Google Scholar]
- 21.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur. J. Biochem. 2001;268:5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 22.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N-and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J. Nutr. Biochem. 2006;17:225–233. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–4259. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Rep. 2008;41:310–315. doi: 10.5483/bmbrep.2008.41.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur. J. Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 26.Kothapalli N, Sarath G, Zempleni J. Biotinylation of K12 in histone H4 decreases in response to DNA double strand breaks in human JAr choriocarcinoma cells. J. Nutr. 2005;135:2337–2342. doi: 10.1093/jn/135.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J. Nutr. 2006;136:2735–2742. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camporeale G, Zempleni J, Eissenberg JC. Susceptibility to heat stress and aberrant gene expression patterns in holocarboxylase synthetase-deficient Drosophila melanogaster are caused by decreased biotinylation of histones, not of carboxylases. J. Nutr. 2007;137:885–889. doi: 10.1093/jn/137.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J. Nutr. Biochem. 2007;18:760–768. doi: 10.1016/j.jnutbio.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chew YC, West JT, Kratzer SJ, Itvarsonn AM, Eissenberg JC, Dave BJ, Klinkebiel D, Christman JK, Zempleni J. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J. Nutr. 2008;138:2316–2322. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J. Nutr. Biochem. 2008;1:400–408. doi: 10.1016/j.jnutbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EM, Hoi JT, Eissenberg JC, Shoemaker ID, Neckameyer WS, Iivarsonn AM, Harshman LG, Schlegel VL, Zempleni J. Feeding Drosophila a biotin-deficient diet for multiple generations increases stress resistance and lifespan and alters gene expression and histone biotinylation patterns. J. Nutr. 2007;137:2006–2012. doi: 10.1093/jn/137.9.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase; assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem. Mot. Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 34.Pispa J. Animal biotinidase. Ann. Med. Exp. Biol. Fenniae. 1965;43:4–39. [PubMed] [Google Scholar]
- 35.Wolf B, Heard GS. Biotinidase deficiency. In: Barness L, Oski F, editors. Advances in Pediatrics. Chicago, IL: Medical Book Publishers; 1991. pp. 1–21. [PubMed] [Google Scholar]
- 36.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum. Mol, Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 37.Ballard TD, Wolff J, Griffin JB, Stanley JS, Calcar Sv, Zempleni J. Biotinidase catalyzes debiotinylation of histones. Eur. J. Nutr. 2002;41:78–84. doi: 10.1007/s003940200011. [DOI] [PubMed] [Google Scholar]
- 38.Cole H, Reynolds TR, Lockyer JM, Buck GA, Denson T, Spence JE, Hymes J, Wolf B. Human serum biotinidase cDNA cloning, sequence, and characterization. J. Biol. Chem. 1994;269:6566–6570. [PubMed] [Google Scholar]
- 39.Stanley CM, Hymes J, Wolf B. Identification of alternatively spliced human biotinidase mRNAs and putative localization of endogenous biotinidase. Mol. Genet. Metob. 2004;81:300–312. doi: 10.1016/j.ymgme.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Chew YC, Sarath G, Zempleni J. An avidin-based assay for quantification of histone debiotinylase activity in nuclear extracts from eukaryotic cells. J. Nutr. Biochem. 2007;18:475–481. doi: 10.1016/j.jnutbio.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dakshinamurti K, Cheah-Tan C. Liver glucokinase of the biotin deficient rat. Can. J. Biochem. 1968;46:75–80. doi: 10.1139/o68-012. [DOI] [PubMed] [Google Scholar]
- 42.Solorzano-Vargas RS, Pacheco-Alvarez D, Leon-Del-Rio A. Hotocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells. Proc. Natl. Acad. Sci. USA. 2002;99:5325–5330. doi: 10.1073/pnas.082097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zempleni J, Mock DM. Biotin. In: Song WO, Beecher GR, editors. Modern Analytical Methodologies on Fat and Water-Soluble Vitamins. New York, NY: Wiley; 2000. pp. 389–409. [Google Scholar]
- 44.Green NM. Avidin. Adv. Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 45.Zempleni J, Mock DM. Biotin biochemistry and human requirements. J. Nutr. Biochem. 1999;10:128–138. doi: 10.1016/s0955-2863(98)00095-3. [DOI] [PubMed] [Google Scholar]
- 46.Bowman BB, Rosenberg I. Biotin absorption by distal rat intestine. J. Nutr. 1987;117:2121–2126. doi: 10.1093/jn/117.12.2121. [DOI] [PubMed] [Google Scholar]
- 47.Bowman BB, Selhub J, Rosenberg IH. Intestinal absorption of biotin in the rat. J. Nutr. 1986;116:1266–1271. doi: 10.1093/jn/116.7.1266. [DOI] [PubMed] [Google Scholar]
- 48.Said HM, Thuy LP, Sweetman L, Schatzman B. Transport of the biotin dietary derivative biocytin (N-biotinyl-l-lysine) in rat small intestine. Gastraenterology. 1993;104:75–80. doi: 10.1016/0016-5085(93)90837-3. [DOI] [PubMed] [Google Scholar]
- 49.Said HM, Redha R, Nylander W. A carrier-mediated, Na+ gradient dependent transport for biotin in human intestinal brush border membrane vesicles. Am. J. Physiol. 1987;253:G631–G636. doi: 10.1152/ajpgi.1987.253.5.G631. [DOI] [PubMed] [Google Scholar]
- 50.Said HM, Redha R, Nylander W. Biotin transport in basolateral membrane vesicles of human intestine. Gastroenterology. 1988;94:1157–1163. doi: 10.1016/0016-5085(88)90007-8. [DOI] [PubMed] [Google Scholar]
- 51.Said HM, Derweesh I. Carrier-mediated mechanism for biotin transport in rabbit intestine–studies with brush-border membrane vesicles. Am. J. Physiol. 1991;261:R94–R97. doi: 10.1152/ajpregu.1991.261.1.R94. [DOI] [PubMed] [Google Scholar]
- 52.Said HM. Cellular uptake of biotin: mechanisms and regulation. J. Nutr. 1999;129:490S–493S. doi: 10.1093/jn/129.2.490S. [DOI] [PubMed] [Google Scholar]
- 53.Said HM, Redha R. A carrier-mediated system for transport of biotin in rat Intestine in vitro. Am. J. Physiol. 1987;252:G52–G55. doi: 10.1152/ajpgi.1987.252.1.G52. [DOI] [PubMed] [Google Scholar]
- 54.Prasad PD, Wang H, Kekuda R, Fujita T, Fei Y-J, Devoe LD, Leibach FH, Ganapathy V. Cloning and functional expression of a cDNA encoding a mammalian sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin, and lipoate. J. Biol. Chem. 1998;273:7501–7506. doi: 10.1074/jbc.273.13.7501. [DOI] [PubMed] [Google Scholar]
- 55.Prasad PD, Ramamoorthy S, Leibach FH, Ganapathy V. Characterization of a sodium-dependent vitamin transporter mediating the uptake of pantothenate, biotin and lipoate in human placental choriocarcinoma cells. Placenta. 1997;18:527–533. doi: 10.1016/0143-4004(77)90006-6. [DOI] [PubMed] [Google Scholar]
- 56.Prasad P, Wang H, Huang W, Fei YJ, Leibach FH, Devoe LD, Ganapathy V. Molecular and functional characterization of the intestinal Na+-dependent multivitamin transporter. Arch. Biochem. Biophys. 1999;366:95–106. doi: 10.1006/abbi.1999.1213. [DOI] [PubMed] [Google Scholar]
- 57.Said HM. Recent advances in carrier-mediated intestinal absorption of water-soluble vitamins. Annu. Rev. Physiol. 2004;66:419–446. doi: 10.1146/annurev.physiol.66.032102.144611. [DOI] [PubMed] [Google Scholar]
- 58.Said HM, Mock DM, Collins JC. Regulation of biotin intestinal transport in the rat: effect of biotin deficiency and supplementation. Am. J. Physiol. 1989;256:G306–G311. doi: 10.1152/ajpgi.1989.256.2.G306. [DOI] [PubMed] [Google Scholar]
- 59.Reidling JC, Nabokina SM, Said HM. Molecular mechanisms involved in the adaptive regulation of human intestinal biotin uptake: a study of the hSMVT system. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G275–G281. doi: 10.1152/ajpgi.00327.2006. [DOI] [PubMed] [Google Scholar]
- 60.Reidling JC, Said HM. Regulation of the human biotin transporter hSMVT promoter by KLF-4 and AP-2: confirmation of promoter activity in vivo. Am. J. Physiol. Cell Physiol. 2007;292:C1305–C1312. doi: 10.1152/ajpcell.00360.2006. [DOI] [PubMed] [Google Scholar]
- 61.Zempleni J, Mock DM. Bioavailability of biotin given orally to humans in pharmacologic doses. Am. J. Clin. Nutr. 1999;69:504–508. doi: 10.1093/ajcn/69.3.504. [DOI] [PubMed] [Google Scholar]
- 62.Said HM, Ortiz A, McCloud E, Dyer D, Moyer MP, Rubin S. Biotin uptake by human colonic epithelial NCM460 cells: a carrier-mediated process shared with pantothenic acid. Am. J. Physiol. 1998;275:C1365–C1371. doi: 10.1152/ajpcell.1998.275.5.C1365. [DOI] [PubMed] [Google Scholar]
- 63.Said HM, Mohammed ZM. Intestinal absorption of water-soluble vitamins: an update. Curr. Opin. Gastroenterol. 2006;22:140–146. doi: 10.1097/01.mog.0000203870.22706.52. [DOI] [PubMed] [Google Scholar]
- 64.Daberkow RL, White BR, Cederberg RA, Griffin JB, Zempleni J. Monocarboxylate transporter 1 mediates biotin uptake in human peripheral blood mononuctear cells. J. Nutr. 2003;133:2703–2706. doi: 10.1093/jn/133.9.2703. [DOI] [PubMed] [Google Scholar]
- 65.Grafe F, Wohlrab W, Neubett RH, Brandsch M. Transport of biotin in human keratinocytes. J. Invest. Dermatol. 2003;120:428–433. doi: 10.1046/j.1523-1747.2003.12058.x. [DOI] [PubMed] [Google Scholar]
- 66.Petrelli F, Moretti P, Paparelli M. Intracellular distribution of biotin-14C COOH in rat liver. Mol. Biol. Rep. 1979;4:247–252. doi: 10.1007/BF00777563. [DOI] [PubMed] [Google Scholar]
- 67.Shriver BJ, Roman-Shriver C, Allred JB. Depletion and repletion of biotinyl enzymes in liver of biotin-deficient rats: evidence of a biotin storage system. J. Nutr. 1993;123:1140–1149. doi: 10.1093/jn/123.6.1140. [DOI] [PubMed] [Google Scholar]
- 68.Pacheco-Alvarez D, Solorzano-Vargas RS, Gravel RA, Cervantes-Roldan R, Velazquez A, Leon-Del-Rio A. Paradoxical regulation of biotin utilization in brain and liver and implications for inherited multiple carboxylase deficiencies. J. Biol. Chem. 2005;279:52312–52318. doi: 10.1074/jbc.M407056200. [DOI] [PubMed] [Google Scholar]
- 69.McKay BE, Molineux ML, Turner RW. Biotin is endogenously expressed in select regions of the rat central nervous system. J. Comp. Neurol. 2004;473:86–96. doi: 10.1002/cne.20109. [DOI] [PubMed] [Google Scholar]
- 70.Dakshinamurti K, Bhagavan HN. Biotin. New York: New York Academy of Science; 1985. [Google Scholar]
- 71.Mock DM, Lankford GL, Mock NI. Biotin accounts for only half of the total avidin-binding substances in human serum. J. Nutr. 1995;125:941–946. doi: 10.1093/jn/125.4.941. [DOI] [PubMed] [Google Scholar]
- 72.Nabokina SM, Subramanian VS, Said HW. Comparative analysis of ontogenic changes in renal and intestinal biotin transport in the rat. Am. J. Physiol. Renal Physiol. 2003;284:F737–F742. doi: 10.1152/ajprenal.00364.2002. [DOI] [PubMed] [Google Scholar]
- 73.Zempleni J, Green GM, Spannagel AU, Mock DM. Biliary excretion of biotin and biotin metabolites is quantitatively minor in rats and pigs. J. Nutr. 1997;127:1496–1500. doi: 10.1093/jn/127.8.1496. [DOI] [PubMed] [Google Scholar]
- 74.Mock N, Malik M, Stumbo P, Bishop W, Mock D. Increased urinary excretion of 3-hydroxyisovaleric acid and decreased urinary excretion of biotin are sensitive early indicators of decreased status in experimental biotin deficiency. Am. J. Clin. Nutr. 1997;65:951–958. doi: 10.1093/ajcn/65.4.951. [DOI] [PubMed] [Google Scholar]
- 75.Sealey WM, Teague AM, Stratton SL, Mock DM. Smoking accelerates biotin catabolism in women. Am. J. Clin. Nutr. 2004;80:932–935. doi: 10.1093/ajcn/80.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Velazquez A, Zamudio S, Baez A, Murguia-Corral R, Rangel-Peniche B, Carrasco A. Indicators of biotin status: a study of patients on prolonged total parenteral nutrition. Eur. J. Clin. Nutr. 1990;44:11–16. [PubMed] [Google Scholar]
- 77.Mock DM, Mock NI. Lymphocyte propionyl-CoA carboxylase is an early and sensitive indicator of biotin deficiency in rats, but urinary excretion of 3-hydroxypropionic acid is not. J. Nutr. 2002;132:1945–1950. doi: 10.1093/jn/132.7.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mock DM, Henrich CL, Carnell N, Mock NI, Swift L. Lymphocyte propionyl-CoA carboxylase and accumulation of odd-chain fatty acid in plasma and erythrocytes are useful indicators of marginal biotin deficiency. J. Nutr. Biochem. 2002;13:462–470. doi: 10.1016/s0955-2863(02)00192-4. [DOI] [PubMed] [Google Scholar]
- 79.Mock DM, Henrich CL, Carnell N, Mock NI. Indicators of marginal biotin deficiency and repletion in humans: validation of 3-hydroxyisovaleric acid excretion and a leucine challenge. Am. J. Clin. Nutr. 2002;76:1061–1068. doi: 10.1093/ajcn/76.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mock DM, Henrich-Shell CL, Carnell N, Stumbo P, Mock HI. 3-Hydroxypropionic acid and methylcitric acid are not reliable indicators of marginal biotin deficiency in humans. J. Nutr. 2004;134:317–320. doi: 10.1093/jn/134.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mock DM, Johnson SB, Holman RT. Effects of biotin deficiency on serum fatty acid composition: evidence for abnormalities in humans. J. Nutr. 1988;118:342–348. doi: 10.1093/jn/118.3.342. [DOI] [PubMed] [Google Scholar]
- 82.Velazquez A, Martin-del-Campo C, Baez A, Zamudio S, Quiterio M, Aguilar JL, Perez-Ortiz B, Sanchez-Ardines M, Guzman-Hernandez J, Casanueva E. Biotin deficiency in protein-energy malnutrition. Eur. J. Clin. Nutr. 1989;43:169–173. [PubMed] [Google Scholar]
- 83.Spencer RP, Brody KR. Biotin transport by small intestine of rat, hamster, and other species. Am. J. Physiol. 1964;206:653–657. doi: 10.1152/ajplegacy.1964.206.3.653. [DOI] [PubMed] [Google Scholar]
- 84.Cowan MJ, Wara DW, Packman S, Yoshino M, Sweetman L, Nyhan W. Multiple biotin-dependent carboxylase deficiencies associated with defects in T-cell and B-cell immunity. Lancet. 1979;2:115–118. doi: 10.1016/s0140-6736(79)90002-3. [DOI] [PubMed] [Google Scholar]
- 85.Kumar M, Axelrod AE. Cellular antibody synthesis in thiamin, riboflavin, biotin and folic acid-deficient rats. Proc. Soc. Exp. Biol Med. 1978;157:421–423. doi: 10.3181/00379727-157-40068. [DOI] [PubMed] [Google Scholar]
- 86.Báez-Saldaña A, Díaz G, Esprnoza B, Ortega E. Biotin deficiency induces changes in subpopulations of spleen lymphocytes in mice. Am. J. Clin. Nutr. 1998;67:431–437. doi: 10.1093/ajcn/67.3.431. [DOI] [PubMed] [Google Scholar]
- 87.Baez-Saldana A, Ortega E. Biotin deficiency blocks thymocyte maturation, accelerates thymus involution, and decreases nose-rump length in mice. J. Nutr. 2004;134:1970–1977. doi: 10.1093/jn/134.8.1970. [DOI] [PubMed] [Google Scholar]
- 88.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J. Nutr. 2002;132:887–892. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 89.Crisp SERH, Camporeale G, White BR, Toombs CF, Griffin JB, Said HM, Zempleni J. Biotin supply affects rates of cell proliferation, biotinylation of carboxylases and histones, and expression of the gene encoding the sodium-dependent multivitamin transporter in JAr choriocarcinoma cells. Eur. J. Nutr. 2004;43:23–31. doi: 10.1007/s00394-004-0435-9. [DOI] [PubMed] [Google Scholar]
- 90.Dakshinamurti K, Chalifour LE, Bhullar RJ. Requirement for biotin and the function of biotin in cells in culture. In: Dakshinamurti K, Bhagavan HN, editors. Biotin. New York: New York Academy of Science; 1985. pp. 38–55. [DOI] [PubMed] [Google Scholar]
- 91.Rodríguez-Melendez R, Schwab LD, Zempleni J. Jurkat cells respond to biotin deficiency with increased nuclear translocation of NF-κB, mediating cell survival. Int. J. Vitam. Nutr. Res. 2004;74:209–216. doi: 10.1024/0300-9831.74.3.209. [DOI] [PubMed] [Google Scholar]
- 92.Zempleni J, Mock DM. Marginal biotin deficiency is teratogenic. Proc. Soc. Exp. Biol. Med. 2000;223:14–21. doi: 10.1046/j.1525-1373.2000.22303.x. [DOI] [PubMed] [Google Scholar]
- 93.Mock DM, Stadler DD. Conflicting indicators of biotin status from a cross-sectional study of normal pregnancy. J. Am. Coll. Nutr. 1997;16:252–257. doi: 10.1080/07315724.1997.10718682. [DOI] [PubMed] [Google Scholar]
- 94.Mock DM, Quirk JG, Mock NI. Marginal biotin deficiency during normal pregnancy. Am. J. Clin. Nutr. 2002;75:295–299. doi: 10.1093/ajcn/75.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stratton SL, Bogusiewicz A, Mock MM, Mock NI, Wells AM, Mock DM. Lymphocyte propionyl-CoA carboxylase and its activation by biotin are sensitive indicators of marginal biotin deficiency in humans. Am. J. Clin. Nutr. 2006;84:384–388. doi: 10.1093/ajcn/84.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mock DM. Marginal biotin deficiency is teratogenic in mice and perhaps humans: a review of biotin deficiency during human pregnancy and effects of biotin deficiency on gene expression and enzyme activities in mouse dam and fetus. J. Nutr. Biochem. 2005;16:435–437. doi: 10.1016/j.jnutbio.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 97.Mock DM, Mock NI, Stewart CW, LaBorde JB, Hansen DK. Marginal biotin deficiency is teratogenic in ICR mice. J. Nutr. 2003;133:2519–2525. doi: 10.1093/jn/133.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mock DM, Stadler D, Stratton S, Mock NI. Biotin status assessed longitudinally in pregnant women. J. Nutr. 1997;127:710–716. doi: 10.1093/jn/127.5.710. [DOI] [PubMed] [Google Scholar]
- 99.Seaiey WM, Stratton SL, Mock DM, Hansen DK. Marginal maternal biotin deficiency in CD-1 mice reduces fetal mass of biotin-dependent carboxylases. J. Nutr. 2005;135:973–977. doi: 10.1093/jn/135.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suormala T, Baumgartner ER, Bausch J, Holick W, Wick H. Quantitative determination of biocytin in urine of patients with biotinidase deficiency using high-performance liquid chromatography (HPLC) Clin. Chim. Acta. 1988;177:253–270. doi: 10.1016/0009-8981(88)90070-8. [DOI] [PubMed] [Google Scholar]
- 101.Wolf B. Worldwide survey of neonatal screening for biotinidase deficiency. J. Inherit. Metab. Dis. 1991;14:923–927. doi: 10.1007/BF01800475. [DOI] [PubMed] [Google Scholar]
- 102.Wolf B, Grier RE, Allen RJ, Goodman SI, Kien CL. Biotinidase deficiency: an enzymatic defect in late-onset multiple carboxylase deficiency. Clin. Chim. Acta. 1983;131:273–281. doi: 10.1016/0009-8981(83)90096-7. [DOI] [PubMed] [Google Scholar]
- 103.Moslinger D, Muhl A, Suormala T, Baumgartner R, Stockler-lpsiroglu S. Molecular characterisation and neuropsychological outcome of 21 patients with profound biotinidase deficiency detected by newborn screening and family studies. Eur. J. Pediatr. 2003;162:S46–S49. doi: 10.1007/s00431-003-1351-3. [DOI] [PubMed] [Google Scholar]
- 104.Laszlo A, Schuler EA, Sallay E, Endreffy E, Somogyi C, Varkonyi A, Havass Z, Jansen KP, Wolf B. Neonatal screening for biotinidase deficiency in Hungary: clinical, biochemical and molecular studies. J. Inherit Metab. Dis. 2003;26:693–698. doi: 10.1023/b:boli.0000005622.89660.59. [DOI] [PubMed] [Google Scholar]
- 105.Neto EC, Schulte J, Rubim R, Lewis E, DeMari J, Castithos C, Brites A, Giugliani R, Jensen KP, Wolf B. Newborn screening for biotinidase deficiency in Brazil: biochemical and molecular characterizations. Braz. J. Med. Biol. Res. 2004;37:295–299. doi: 10.1590/s0100-879x2004000300001. [DOI] [PubMed] [Google Scholar]
- 106.Wolf B, Feldman GL. The biotin-dependent carboxylase deficiencies. Am. J. Hum. Genet. 1982;34:699–716. [PMC free article] [PubMed] [Google Scholar]
- 107.Suzuki Y, Yang X, Aoki Y, Kure S, Matsubara Y. Mutations in the holocarboxylase synthetase gene HLCS. Hum. Mutat. 2005;26:285–290. doi: 10.1002/humu.20204. [DOI] [PubMed] [Google Scholar]
- 108.Perez B, Desviat LR, Rodriguez-Pombo P, Clavero S, Navarrete R, Perez-Cerda C, Ugarte M. Propionic acidemia: identification of twenty-four novel mutations in Europe and North America. Mol. Genet. Metab. 2003;78:59–67. doi: 10.1016/s1096-7192(02)00197-x. [DOI] [PubMed] [Google Scholar]
- 109.Robinson BH, Oei J, Saudubray JM, Marsac C, Barttett K, Quan F, Gravel R. The French and North American phenotypes of pyruvate carboxylase deficiency, correlation with biotin containing protein by 3H-biotin incorporation, 35S-streptavidin labeling, and northern blotting with a cloned cDNA probe. Am. J. Hum. Genet. 1987;40:50–59. [PMC free article] [PubMed] [Google Scholar]
- 110.Desviat LR, Perez-Cerda C, Perez B, Esparza-Gordillo J, Rodriguez-Pombo P, Penalva MA, Rodriguez De Cordoba S, Ugarte M. Functional analysis of MCCA and MCCB mutations causing methylcrotonylglycinuria. Mol. Genet. Metab. 2003;80:315–320. doi: 10.1016/S1096-7192(03)00130-6. [DOI] [PubMed] [Google Scholar]
- 111.Baumgartner MR, Dantas MF, Suormala T, Almashanu S, Giunta C, Friebel D, Gebhardt B, Fowler B, Hoffmann GF, Baumgartner ER, Valle D. Isolated 3-methylcrotonyl-CoA carboxylase deficiency: evidence for an allele-specific dominant negative effect and responsiveness to biotin therapy. Am. J. Hum. Genet. 2004;75:790–800. doi: 10.1086/425181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.National RC. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline. Washington, DC: National Academy Press; 1998. [PubMed] [Google Scholar]
- 113.Mock DM, Stratton SL, Mock NI. Concentrations of biotin metabolites in human milk. J. Pediatr. 1997;131:456–458. doi: 10.1016/s0022-3476(97)80077-7. [DOI] [PubMed] [Google Scholar]