Abstract
Cell cycle regulation and the maintenance of genome integrity are crucial for the development and virulence of the pathogenic plant fungus Fusarium graminearum. To identify transcription factors (TFs) related to these processes, four DNA-damaging agents were applied to screen a F. graminearum TF mutant library. Sixteen TFs were identified to be likely involved in DNA damage responses. Fhs1 is a fungal specific Zn(II)2Cys6 TF that localises exclusively to nuclei. fhs1 deletion mutants were hypersensitive to hydroxyurea and defective in mitotic cell division. Moreover, deletion of FHS1 resulted in defects in perithecia production and virulence and led to the accumulation of DNA damage. Our genetic evidence demonstrated that the FHS1-associated signalling pathway for DNA damage response is independent of the ATM or ATR pathways. This study identified sixteen genes involved in the DNA damage response and is the first to characterise the novel transcription factor gene FHS1, which is involved in the DNA damage response. The results provide new insights into mechanisms underlying DNA damage responses in fungi, including F. graminearum.
DNA functions as the carrier of genetic information, and therefore genome integrity and stability are essential for all organisms. However, DNA is easily subjected to damage from both intrinsic and extrinsic sources, including single- and double-strand breaks, base damage, and DNA-protein crosslinks1. If left unrepaired or incorrectly repaired, damaged DNA will result in cell cycle arrest, cell death, loss of genetic information, and genomic instability2.
To address the fundamental problem of genomic erosion, DNA damage response (DDR) systems have evolved to detect DNA damage and promote the repair of damaged DNA. DDR ensures the maintenance of genome integrity and stability by regulating DNA repair mechanisms. Moreover, DNA replication, gene transcription, DNA repair, and cell cycle checkpoints should be interlinked to assure cell survival and the normal regulation of cellular functions following DNA damage3,4.
Despite significant advances in agricultural biotechnology and fungal genetics, fungal threats to public health and food security are still increasing5. Fusarium graminearum is a disastrous pathogenic plant fungus and is the major causative agent of Fusarium head blight (FHB) on major cereal crops worldwide6. F. graminearum causes yield and quality losses due to sterility of florets and formation of discoloured, withered and light test weight kernels7. In addition, the fungal infection results in the accumulation of mycotoxins, such as zearalenone and deoxynivalenol, that are harmful to animals and humans8. Because several previously proposed strategies to control FHB, including biological and chemical measures, have not been effective in preventing disease outbreaks, FHB epidemics continue to occur throughout the world7.
Various forward and reverse genetic approaches have been applied in F. graminearum; however, most of the previous studies have focused on signal transduction pathways or orthologous genes, which tend to be highly conserved in eukaryotes7,9,10. Characterisation of novel genes or mechanisms involved in central transduction pathways or tolerance to specific environmental stresses would be novel molecular targets for the development of new chemical/biological pesticides or molecular breeding. Massive genetic approaches applying forward and reverse genetic tools are necessary for mining those genes or mechanisms.
In a previous study, we constructed a F. graminearum transcription factor (TF) gene deletion mutant library and obtained a comprehensive phenotypic dataset, which can be accessed at FgTFPD (http://ftfd.snu.ac.kr/FgTFPD)11. The results identified TFs involved in sexual and asexual reproduction, secondary metabolism, stress responses, and virulence. Because TFs are involved in most biological processes, screening the TF mutant library with specific stress conditions will shed light on novel mechanisms for these processes. As mentioned above, all organisms encounter DNA damage and have evolved their own DNA repair mechanisms. Therefore, the identification of TFs related to DNA damage responses could uncover novel mechanisms in F. graminearum.
The objectives for this study were to 1) construct a phenotypic dataset for DNA damage responses from the F. graminearum TF mutant library and 2) understand the molecular mechanisms underlying the DNA damage responses of a novel TF in F. graminearum. We screened the previously generated F. graminearum TF mutant library using four different DNA-damaging agents and discovered that the fhs1 mutant was highly sensitive to hydroxyurea (HU). To elucidate the regulatory mechanisms for the hypersensitivity of the fhs1 mutant to HU, we performed genetic studies and analysed the transcriptome. This study is the first to characterise a novel transcription factor in DNA damage responses and will provide new insights into mechanisms underlying the DNA damage responses in F. graminearum as well as other filamentous fungi.
Results
Identification of transcription factors involved in DNA damage responses
To identify the transcription factors involved in DNA damage responses, we screened 675 previously generated11 transcription factor gene deletion mutants using four chemical reagents that cause different types of DNA damage (Supplementary Table S1). Experimental conditions were established such that the F. graminearum wild-type strain exhibited approximately one half of the radial growth observed on complete medium (CM) without any DNA-damaging agent: methyl methanesulfonate (MMS, 0.1 μl/ml), hydroxyurea (HU, 10 mM), bleomycin (BLM, 10 mU/ml), and camptothecin (CPT, 0.4 μM).
Sixteen mutants exhibited altered sensitivity to at least one DNA-damaging agent (Table 1 and Fig. 1). Some of the screening results were consistent with previous studies. RFX1 functions in DNA damage repair in F. graminearum12. Homologues of ARS2 (FGSG_01106)13, MSN2 (FGSG_09368)14, SFP1 (FGSG_15767)15, CTR9 (FGSG_06948)16, CUL-4A (FGSG_05520)17, MYO1 (FGSG_16982)18, and RAD18 (FGSG_04258)19 are all required for DNA damage responses and/or cytokinesis in model organisms.
Table 1. Putative transcription factors involved in DNA damage responses.
| Locus ID | DNA-damaging agents |
Description of the gene product | Species | Homologue | Reference | |||
|---|---|---|---|---|---|---|---|---|
| MMS | HU | BLM | CPT | |||||
| FGSG_01106 | S | S | Related to arsenite-resistance protein 2 | Human | ARS2 | 13 | ||
| FGSG_09368 | S | S | Related to C2H2 zinc finger protein | S. cerevisiae | MSN2 | 14 | ||
| FGSG_15767 | S | S | S | S | Related to zinc finger protein SFP1 | S. cerevisiae | SFP1 | 15 |
| FGSG_09565 | S | R | Probable siderophore regulation protein (GATA factor) | A. nidulans | SREA | 23 | ||
| FGSG_01182 | S | S | S | S | Conserved hypothetical protein | S. cerevisiae | DBP4 | 20 |
| FGSG_05304 | S | S | S | S | Conserved hypothetical protein | S. cerevisiae | DBP3 | 21 |
| FGSG_01488 | S | S | Conserved hypothetical protein | N/A | N/A | N/A | ||
| FGSG_06948 | S | S | Related to tetratricopeptide repeat protein tpr1 | S. cerevisiae | CTR9 | 16 | ||
| FGSG_05520 | S | S | S | S | Related to cullin homologue 4A | Human | CUL-4A | 17 |
| FGSG_07420 | S | S | S | S | Related to cephalosporin C regulator 1 (cpcR1 gene) | F. graminearum | RFX1 | 12 |
| FGSG_16982 | S | S | Related to myosin 2 | S. cerevisiae | MYO1 | 18 | ||
| FGSG_04258 | S | Probable DNA repair protein UVS-2 | S. cerevisiae | RAD18 | 19 | |||
| FGSG_01176 | S | Conserved hypothetical protein | F. graminearum | FHS1 | This study | |||
| FGSG_00404 | R | R | S | Conserved hypothetical protein | N/A | N/A | N/A | |
| FGSG_00574 | S | Related to purine utilisation positive regulator | S. cerevisiae | PPR1 | 47 | |||
| FGSG_00573 | S | Related to GAL4-like transcriptional activator | S. cerevisiae | PPR1 | 47 | |||
MMS, methyl methanesulfonate; HU, hydroxyurea; BLM, bleomycin; CPT, camptothecin; S, more sensitive than wild-type; R, more resistant than wild-type; N/A, not applicable.
Figure 1. Sixteen transcription factor gene deletion mutants exhibited altered sensitivity to DNA-damaging agents.
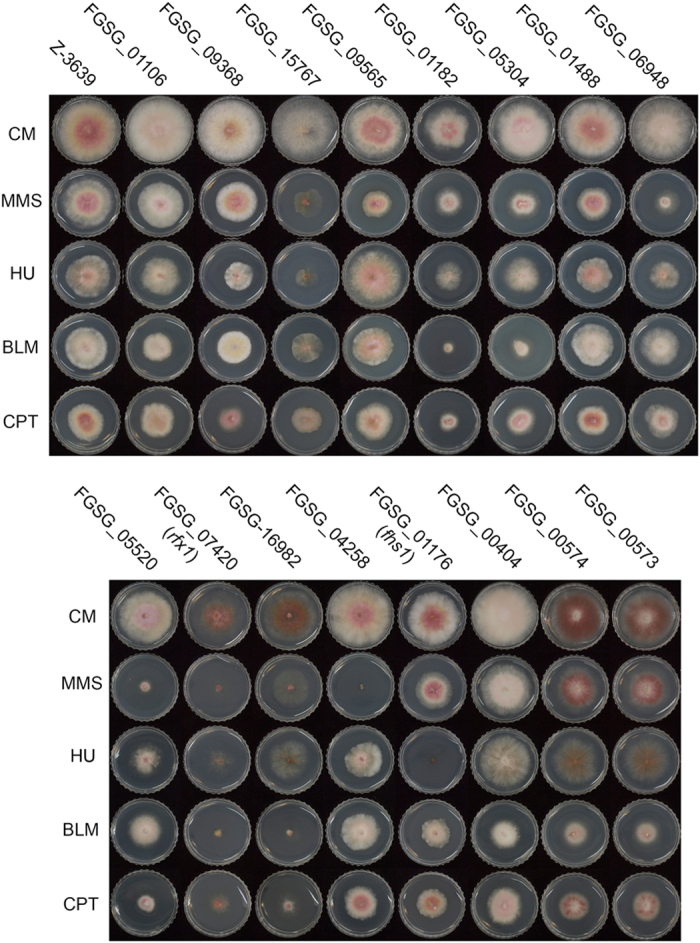
Pictures were taken 3 days after inoculation on CM with or without DNA-damaging agent (0.1 μl/ml MMS [methyl methanesulfonate], 10 mM HU [hydroxyurea], 10 mU/ml BLM [bleomycin], and 0.4 μM CPT [camptothecin]). Each locus ID is denoted above the corresponding deletion mutant.
Homologues of DBP3 (FGSG_05304) and DBP4 (FGSG_01182) encode the third and fourth subunits of DNA polymerase ε, respectively20,21, and both FGSG_00573 and FGSG_00574 are predicted to encode Ppr1, which regulates de novo pyrimidine biosynthesis in yeast22. These four genes are potentially involved in DNA damage responses, because they are closely linked to DNA replication and nucleotide synthesis1.
FGSG_09565 encodes a homologue of the GATA transcription factor SreA, which is involved in siderophore biosynthesis in A. nidulans23. A putative transcription factor containing an HMG box DNA-binding domain (FGSG_01488) and two genes (FGSG_01176 and FGSG_00404) with a Zn(II)2Cys6 fungal-type DNA-binding domain (IPR001138) have not been functionally characterised in other fungi. Of these, we selected one mutant (FGSG_01176) that was specifically sensitive to HU for further study and designated the gene name F. graminearum Hydroxyurea Sensitive 1 (FHS1).
Identification of the novel transcription factor gene FHS1 in F. graminearum
FHS1 (locus ID: FGSG_01176) encodes 492 amino acids containing the Zn(II)2Cys6 fungal-type DNA-binding domain (IPR001138) and a nuclear localisation signal (NLS; SHKKSR). Fhs1 homologues are highly conserved in the Pezizomycotina subdivision of Ascomycota, particularly in the class Sordariomycetes, including Neurospora crassa, Podospora anserina, and Magnaporthe oryzae (Supplementary Fig. S1a,b). No homologue has been functionally characterised to date.
Effect of HU on fhs1 mutants
To confirm the function of FHS1, targeted gene deletion and complementation experiments were performed. Both genetic manipulations were confirmed using Southern blot analyses (Supplementary Fig. S1c,d). The fhs1 deletion mutant exhibited a slight growth defect on CM compared with wild-type and was highly sensitive to HU supplementation in accordance with our screening data (Fig. 2a). The complemented strain fully abolished the high HU sensitivity of the fhs1 mutant, confirming the role of FHS1 in HU resistance.
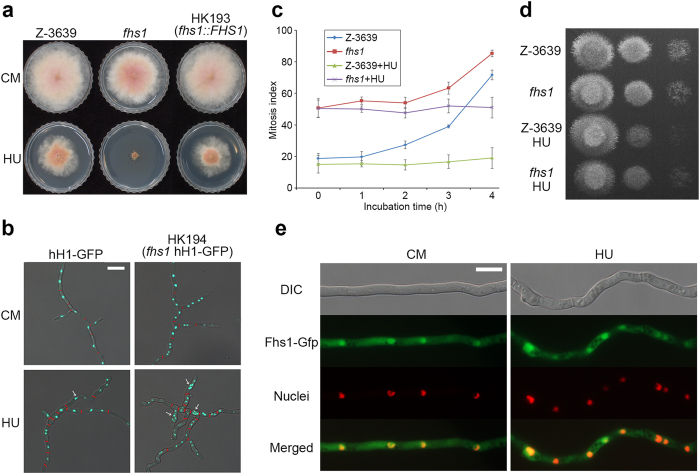
Figure 2. Fungal specific transcription factor Fhs1 involved in hydroxyurea sensitivity.
(a) Mycelia growth of F. graminearum strains on CM and CM supplemented with 10 mM HU. Pictures were taken 4 days after inoculation. (b) Nuclei and septa formation in hyphae. Conidia of both strains were inoculated into CM and CM supplemented with 10 mM HU for 24 h. Histone H1 was tagged with Gfp to visualise nuclei in both wild-type and fhs1 mutant strains. Septa confirmed by Calcofluor white staining are indicated with red bars. DIC and fluorescent protein images were merged. Scale bar = 20 μm. (c) Mitosis assay. Germlings bearing cells with two or more nuclei for 4-h incubation with and without 100 mM HU were scored. One hundred conidia were assessed for each strain with three biological replicates. (d) Serial dilutions of all strains were point-inoculated onto CM after 4-h incubation with or without HU treatment (106, 105, and 104 conidia/ml). (e) Subcellular localisation of Fhs1. Fhs1 was fused with Gfp, and histone H1 was fused with Rfp. Mycelia were harvested for microscopic observation 12 h after conidia inoculation in liquid CM supplemented with or without 5 mM HU. The yellow colour in the merged images indicates colocalisation. DIC, differential interference contrast. Scale bar = 10 μm.
To visualise the nuclei of hyphae, the HK194 strain (Δfhs1 hH1-GFP) was generated from an outcross between the mat1g (Δmat1 hH1-GFP) and fhs1 strains (Supplementary Table S2). Septa visualised using Calcofluor white staining were marked with red bars. Compared with the strain hH1-GFP carrying an FHS1 wild-type allele, the fhs1 mutant often produced meandering hyphae (Fig. 2b). Moreover, each hyphal cell of the fhs1 mutant tended to contain more nuclei (2.9 nuclei per cell) compared with the hH1-GFP strain (1.4 nuclei per cell). When HU was exogenously amended, hyphae of the hH1-GFP strain with wild-type FHS1 began to meander and the number of nuclei per cell increased as in the fhs1 mutant without HU supplementation (2.6 nuclei per cell). The phenotypic defect became more severe when HU was added to the fhs1 mutant (4.9 nuclei per cell).
Because the lack of FHS1 seemed to cause defects in nuclear division (Fig. 2c), we further performed a mitosis assay. Conidia of fhs1 mutants bore more nuclei than wild type. When conidia were incubated with a high concentration of HU (100 mM), nuclear division was arrested in the wild-type and fhs1 stains. After 4 h of HU treatment, both wild-type and fhs1 mutant strains resumed mycelial growth (Fig. 2d).
Subcellular localisation of Fhs1
To examine Fhs1 localisation, the FHS1-GFP fusion construct under the control of its native promoter was transformed for genetic complementation (Supplementary Fig. S1b). To compare Fhs1-Gfp with nuclei, the HK224 (∆fhs1::FHS1-GFP-HYG hH1-RFP-GEN) strain was generated from an outcross between the mat1r and HK193 strains. Fhs1-Gfp in the HK224 strain accumulated in the nuclei (Fig. 2e). localisation patterns were similar regardless of the exogenous HU treatment.
Sexual development and virulence test
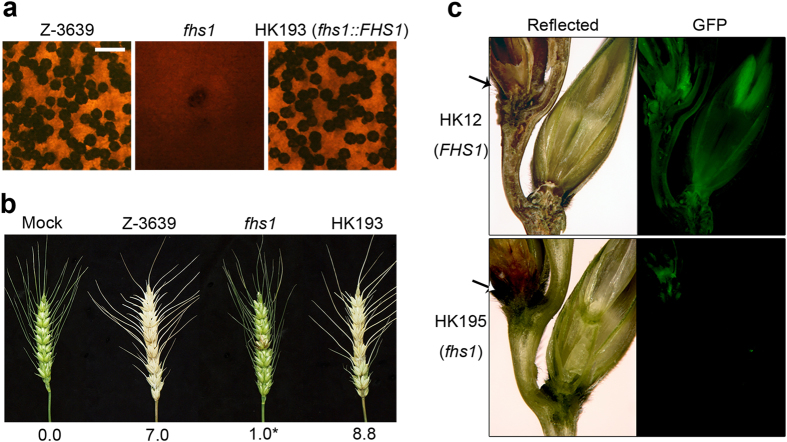
The wild-type and complemented strains produced normal perithecia bearing mature ascospores 7 days after sexual induction (Fig. 3a). By contrast, vigorous mycelial growth was observed in the sexually induced fhs1 mutant culture and only a few immature perithecia were produced. Perithecia of the fhs1 mutant neither matured nor produced ascospores.
Figure 3. Sexual development and virulence of F. graminearum strains.
(a) Sexual reproduction on carrot agar. Pictures were taken 7 days after sexual induction. Scale bar = 500 μm. (b) Virulence on wheat heads. A centre spikelet of each wheat head was injected with 10 μl of conidia suspension. Disease index (diseased spikelets per wheat head) is denoted below the picture. Asterisks indicate that the data differed significantly (P < 0.05) based on Tukey′s test. The pictures were taken 14 days after inoculation. Mock, negative control mock-inoculated with 0.01% Tween 20. (c) Sections of infected wheat heads. Wheat spikelets were inoculated with conidia suspensions of cytosolic Gfp-expressing F. graminearum strains. Infected wheat heads were dissected 6 days after inoculation and examined using fluorescence microscopy. Spreading of the Gfp signal represents spreading of hyphae from the points of inoculation. Arrows mark the inoculated spikelets. Reflected, reflected light.
The virulence of the fungal strains was examined by point inoculation of wheat spikelets. Fourteen days after inoculation, the wild-type and complemented strains colonised the injected spikelet and adjacent spikelets, causing normal head blight symptoms (Fig. 3b). The fhs1 mutant infected the inoculated spikelet; however, it lost its ability to spread into the neighbouring spikelets.
To reveal the mechanism underlying the virulence of the fhs1 deletion mutant, we generated an fhs1 deletion strain constitutively expressing Gfp to observe mycelial movement during plant infection through an outcross between the KM19 (Δmat1 GFP-HYG) and fhs1 mutant strains. The resulting HK195 (Δfhs1 GFP-HYG) strain showed the same phenotype as the parental strain fhs1 mutant and exhibited constitutive expression of cytosolic Gfp. Approximately 6 days after inoculation, the strain (HK12) carrying the wild-type allele of FHS1 readily colonised the injected spikelet and began to infect adjacent spikelets through rachis nodes (Fig. 3c). The fhs1 mutant successfully colonised the injected spikelets as in the wild-type, but the hyphae of the fhs1 mutant could not spread into the adjacent spikelet from the inoculated one. Hyphal growth through rachis was rarely observed in fhs1 mutants.
Accumulation of DNA damage in cells
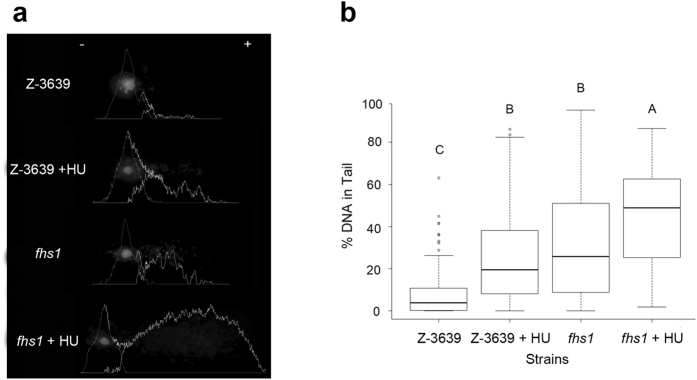
The alkaline comet assay was performed to analyse DNA single-strand breaks, double-strand breaks, and alkali-labile sites of DNA in each cell. The head of the comet shows undamaged DNA, whereas the tail of comet signals the damaged DNA in each cell. Therefore, cells with more DNA damage exhibit a higher percentage of DNA in the tail. We tested the wild-type and fhs1 strains with and without HU treatment (Fig. 4). HU induced DNA damage in F. graminearum, increasing the comet tail in HU treated cells. DNA damage levels in HU-untreated fhs1 cells and HU-treated wild-type cells were similar (P < 0.01) and both increased relative to the HU-untreated wild-type. This result suggests that deletion of FHS1 triggered DNA damage in the absence of a DNA-damaging agent. HU-treated fhs1 cells exhibited significant increases in the percentage of DNA in the tail compared with HU-treated wild-type cells, demonstrating that DNA in fhs1 mutants is more easily damaged when exposed to HU than the wild-type strain (P < 0.01).
Figure 4. Detection of increased DNA damage.
(a) Photographs of comets and analysis of the alkaline comet assay. DNA fragments generated from DNA single-strand breaks, double-strand breaks, and alkali-labile sites in the DNA migrate toward the anode, creating the comet assay. Between the comets of fhs1 and HU-treated wild-type, fhs1 cells produced longer tails. Graphs depict image analysis quantifying the DNA contents of the head and tail. (b) Box-plot of average percentages of DNA in comet tails. fhs1 cells contained higher percentages of DNA in the comet tail than wild-type but similar to HU-treated wild-type cells (P < 0.01). HU-treated fhs1 cells contained significantly higher percentages of DNA in the comet tail than wild-type and HU-treated wild-type cells (P < 0.01).
Transcriptome analyses
To reveal the molecular mechanisms underlying the increased DNA damage level of the fhs1 mutant, we analysed the transcriptomes of the wild-type and fhs1 mutant strains using RNA-seq (Supplementary Dataset 1). Genes with transcript levels exhibiting a 3-fold or greater difference between the wild-type and fhs1 mutant strains were categorised based on their predicted functions (Table 2). Of the total predicted 13,820 genes, 898 (6.5%) were up-regulated and 522 (3.8%) were down-regulated in the fhs1 strain, respectively. High proportions of genes included in “Cell rescue, defense and virulence”, “Metabolism”, “Cellular transport”, “Interaction with the environment”, and “Energy” were highly up-regulated or down-regulated in the fhs1 mutant compared with the other groups, except genes encoding “Unclassified proteins”.
Table 2. Changes in transcription levels between fhs1 and wild-type strains by functional category.
| FunCat ID | FunCat category name | No. of genes inthe genome | No. of genes up-regulated over 3-fold | No. of genes down-regulated over 3-fold |
|---|---|---|---|---|
| 01 | Metabolism | 2322 | 121 (5.21%) | 79 (3.40%) |
| 02 | Energy | 503 | 22 (4.37%) | 15 (2.98%) |
| 10 | Cell cycle and DNA procession | 659 | 6 (0.91%) | 3 (0.46%) |
| 10.01.05.01 | DNA repair | 156 | 2 (1.28%) | 1 (0.64%) |
| 11 | Transcription | 718 | 4 (0.56%) | 6 (0.84%) |
| 12 | Protein synthesis | 370 | 0 (0.00%) | 3 (0.81%) |
| 14 | Protein fate | 920 | 15 (1.63%) | 7 (0.76%) |
| 16 | Protein with binding function or cofactor requirement | 1714 | 53 (3.09%) | 33 (1.93%) |
| 18 | Regulation of metabolism and protein function | 242 | 2 (0.83%) | 1 (0.41%) |
| 20 | Cellular transport, transport facilities and transport routes | 1390 | 63 (4.53%) | 51 (3.67%) |
| 30 | Cellular communication/signal transduction mechanism | 312 | 5 (1.60%) | 3 (0.96%) |
| 32 | Cell rescue, defence and virulence | 856 | 67 (7.83%) | 34 (3.97%) |
| 34 | Interaction with the environment | 606 | 27 (4.46%) | 24 (3.96%) |
| 40 | Cell fate | 240 | 2 (0.83%) | 2 (0.83%) |
| 41 | Development | 55 | 2 (3.64%)) | 2 (3.64%) |
| 42 | Biogenesis of cellular components | 617 | 18 (2.92%) | 7 (1.13%) |
| 99 | Unclassified proteins | 9074 | 716 (7.89%) | 398 (4.39%) |
| – | Total | 13820 | 898 (6.50%) | 522 (3.78%) |
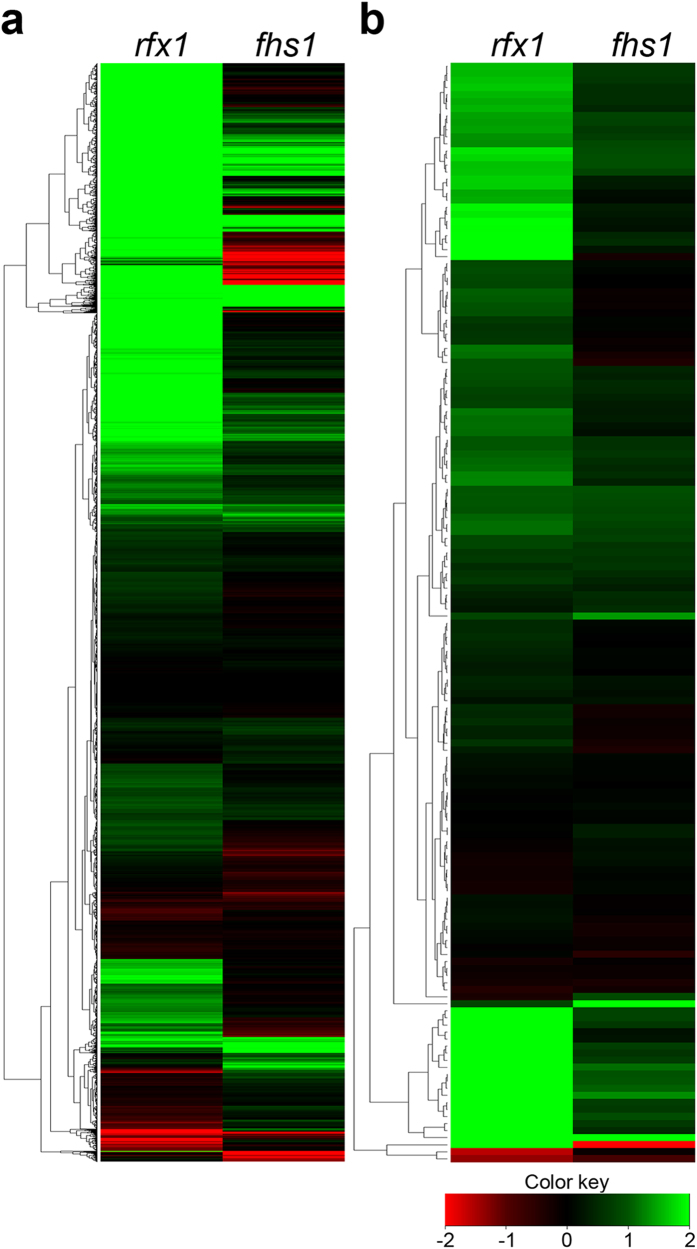
Because RFX1 deletion also increased DNA damage levels compared with wild-type12, we compared the transcriptomes of rfx1 and fhs1 mutants (Fig. 5). Approximately one third of total genes (5,300/13,820) were up-regulated and a small portion (146) was down-regulated in the rfx1 mutant, whereas only 898 and 522 genes were up- and down-regulated, respectively, in the fhs1 mutant (Fig. 5a). Genes altered in transcript levels were also markedly different between the two mutants. In particular, the transcript levels of DNA repair-related genes were rarely changed in the fhs1 mutant, whereas 22.4% of these genes (35/156) were up-regulated when RFX1 was deleted (Fig. 5b). In the fhs1 mutant, only two genes related to DNA repair (FGSG_05924 and FGSG_16955) were significantly up-regulated (P < 0.01) (Supplementary Dataset 2).
Figure 5. Comparison of the transcriptomes of the rfx1 and fhs1 mutants using cluster analysis.
(a) Heat map depicting the results of cluster analysis of all genes. (b) Heat map depicting the results of cluster analysis of genes involved in “DNA repair”. Green represents higher expression, red represents lower expression, and rows represent transcriptional units.
We further examined 18 genes whose homologues in A. nidulans24 are involved in HU sensitivity to characterise the genetic relationships between FHS1 and those genes (Supplementary Dataset 3). None of the transcript levels of the homologues including ATR, ATM, CHK1, and CHK2 were altered in the fhs1 mutant.
Characterisation of central regulators for DNA damage responses
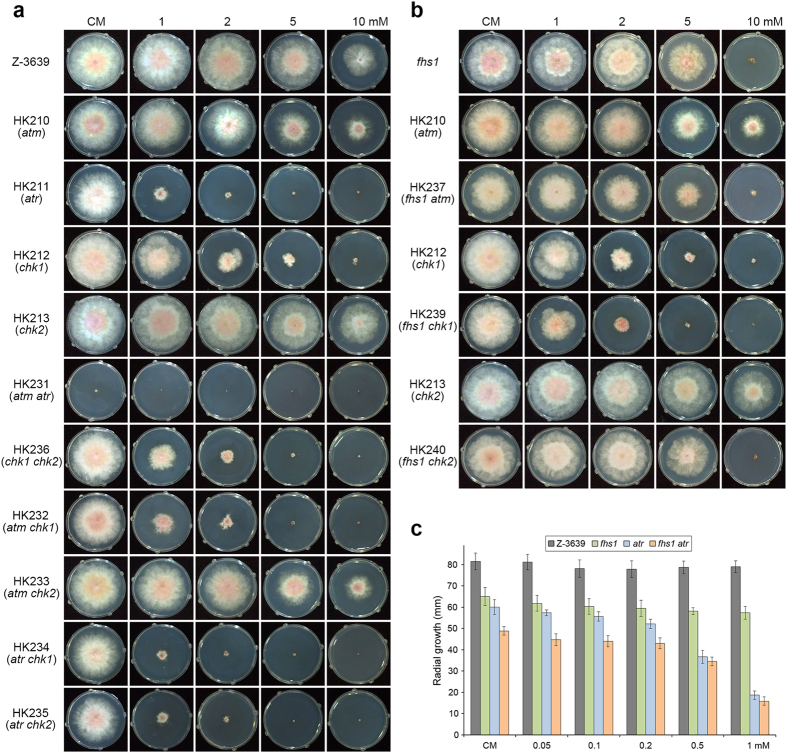
To determine whether central regulators of DNA damage responses had functionally conserved roles in F. graminearum, we identified ATM (FGSG_16493), ATR (FGSG_13318), CHK1 (FGSG_01506), and CHK2 (FGSG_07121) (Supplementary Dataset 3) and individually deleted genes from the wild-type strain Z-3639 (Supplementary Fig. S2). Outcrossing was used to generate the strains described in Supplementary Table S2. For example, to generate the double deletion mutants, the ∆mat2 strain was outcrossed with HK210 (atm deletion mutant) and HK213 (chk2 deletion mutant) to select for HK228 (mat2 atm) and HK230 (mat2 chk2), respectively (Supplementary Table S2). Finally, each double deletion mutant HK231 (atm atr) and HK236 (chk1 chk2) was obtained from the outcrosses HK228 × HK211 and HK230 × HK212, respectively. HK238 (fhs1 atr) and HK239 (fhs1 chk1) were generated by transformation due to ascospore lethality.
Three single deletion mutants (atm, atr, and chk1) were more sensitive to HU compared with the wild-type strain. In particular, atr and chk1 mutants rarely grew on CM supplemented with 1 and 2 mM HU, respectively. Both double deletion mutants HK231 (atm atr) and HK236 (chk1 chk2) exhibited increased sensitivity to HU compared with the corresponding single deletion mutants. Moreover, the double deletion of ATM and CHK1 had a synergistic effect on HU sensitivity. Taken together, we concluded that the cooperative ATR-CHK1 and ATM-CHK2 signalling pathways for DNA damage responses induced by HU are conserved in F. graminearum.
Genetic relationships between FHS1 and central regulators for DNA damage responses
Radial growth of the double deletion mutants, HK237 (fhs1 atm) and HK238 (fhs1 atr), both harbouring FHS1 deletions, was markedly reduced on CM with and without HU supplementation compared with the corresponding single deletion mutants (Fig. 6b,c). And deletion of FHS1 with ATM and CHK1 exhibited a synergistic effect on HU sensitivity. Transcript levels of FHS1 were not altered when central regulator genes were deleted under HU treated condition (Supplementary Fig. S3). These results suggest that the FHS1-dependent signalling pathway for mitotic cell division and DNA damage response is distinct and independent of central regulators, such as ATM and ATR.
Figure 6. Mycelial growth of F. graminearum strains on CM and CM supplemented with HU.
(a) Mycelial growth of ATM and ATR pathway gene deletion mutants. (b) Mycelial growth of double deletion mutants carrying the FHS1 deletion. Pictures were taken 3 days after inoculation on CM. (c) Radial growth of double deletion mutants carrying the FHS1 deletion on CM and CM supplemented with 0.05–1 mM HU. Data were obtained 4 days after inoculation.
Discussion
The increase in applicable molecular tools in F. graminearum has enabled large scale genetic approaches and in-depth biochemical studies to dissect the molecular mechanisms underlying fungal development and virulence25. The construction of a transcription factor (TF) mutant library shed light on fungal genetics, because TFs are thought to be involved in most biological processes11. In addition, chemical genetics utilizing our TF mutant library led us to unveil hidden functions of TFs and new mechanisms for specific cellular processes. A novel regulatory mechanism for sodium tolerance was recently reported from a genetic screen of sodium/lithium-sensitive TF mutants26. In the current study, we successfully identified 16 TFs involved in DNA damage responses through genetic screens using four DNA-damaging agents. Further in-depth functional analyses revealed that the novel gene FHS1 is involved in mitotic cell cycle arrest and DNA damage response in F. graminearum.
The results of genetic screens generated in this study provide new insight into the DNA damage responses in filamentous fungi, including F. graminearum. We identified that twelve genes (homologues for ARS2, MSN2, SFP1, DBP3, DBP4, CTR9, CUL-4A, RFX1, MYO1, RAD18, and PPR1) were involved in DNA damage responses, as was previously shown in model organisms, demonstrating that their roles are also conserved in F. graminearum and possibly in other filamentous fungi (Table 1). SreA (the homologue of FGSG_09565) is a repressor of siderophore biosynthesis, which is required for iron uptake in A. nidulans23. Because intracellular iron levels may result in DNA damage27, further genetic studies on the role of SreA in DNA damage response are necessary. Moreover, we discovered three novel genes involved in DNA damage responses, two of which (FGSG_01176 and FGSG_00404) are fungal specific TFs included in the Zn(II)2Cys6 family. Unveiling the up- and downstream pathways for these genes will provide novel fungal specific molecular mechanisms for DNA damage responses.
HU is a specific inhibitor of ribonucleotide reductase (RNR), which is responsible for the synthesis of dNTPs, and therefore stalls DNA replication28. Defective cell cycle processes and checkpoint regulation results in hypersensitivity to HU29. FHS1 deletion also resulted in hypersensitivity to HU. Mitosis and alkaline comet assays showed that FHS1 deletion resulted in defects in the mitotic cell division and subsequent genomic instability (Figs 2c and 4). Moreover, fhs1 mutants did not exhibit normal DNA damage responses against accumulated DNA damages (Supplementary Dataset 2 and Fig. 5). DNA repair defect of fhs1 mutant seems to be reason for increased sensitivity to HU than a checkpoint defect since HU successfully arrested mitotic cell division in the wild-type and fhs1 strains (Fig. 2c). Taken together, we concluded that FHS1 is required for proper mitotic cell division and DNA damage responses.
Two phosphoinositide 3-kinase-related kinases, Atm and Atr, are highly conserved key factors for cell cycle regulation and DNA damage responses in eukaryotic cells. The downstream effectors Chk1 and Chk2 are directly phosphorylated by Atm and Atr, respectively, for further signal transduction1. Whereas the ATM pathway mainly responds to DNA damage, such as double-strand breaks, Atr regulates cell cycle and genes required for DNA damage response caused by disturbed replication forks, such as with HU treatment30,31. Because our genetic evidence revealed that FHS1 is required for mitotic cell division and DNA damage responses, we tried to characterise genetic relationships among ATM, ATR, CHK1, CHK2, and FHS1 in F. graminearum (Fig. 6). Our genetic evidence revealed that the ATR-CHK1 and ATM-CHK2 pathways and their cross talk are conserved in F. graminearum. However, FHS1-associated regulatory mechanisms appear to be distinct from and independent of both ATR and ATM pathways. We also could not find distinct genetic correlation between FHS1 and 14 homologous genes responsible for A. nidulans S-phase checkpoint mutations (Supplementary Dataset 3).
Hypersensitivity to HU and fungal development of double mutations between FHS1 and ATR were synergistic. Defects in vegetative growth and HU sensitivity were markedly increased in the fhs1 atr double deletion mutants compared with single deletion mutants (Fig. 6b,c). Moreover, outcrosses between fhs1 and atr or chk1 to generate double deletion mutants resulted in ascospore lethality, suggesting that they have overlapping roles in fertility. Taken together, the biological roles of the FHS1-dependent pathway and the ATR pathway for DNA damage response and fungal development are not wholly distinct.
Various fungal developmental processes, including sexual and asexual reproduction and virulence, necessarily involve the reprograming of cell cycle regulation for unique morphogenesis. F. graminearum produces both sexual (ascospores) and asexual (conidia) spores. Ascospores are produced and forcibly discharged from the perithecia for primary inocula32. Perithecia production is a complex cellular differentiation process under polygenic control33. Conidia function as secondary inocula and are produced from specialized conidiogenous cells34. Moreover, the fungus differentiates hyphal cells to appressoria-like structures for successful plant infection35. Therefore, disruption of the cell cycle usually results in severe pleiotropic defects in F. graminearum12. In addition, accumulation of DNA damages triggered by plant defense responses such as increased levels of reactive oxygen species or reactive nitrogen species might be the reason for avirulent feature of fhs1 mutant36. Fhs1-dependent regulations for mitosis and DNA damage responses are specifically related to sexual development and virulence but not closely related to asexual development or vegetative growth.
In summary, we identified 16 TF genes involved in cell cycle regulation and/or DNA damage responses in F. graminearum. FHS1 is related to DNA damage response and genome integrity in F. graminearum. Moreover, disruption of FHS1 function resulted in DNA damage in cells and multiple defects in virulence and sexual development. Because FHS1 participates in novel signalling pathways for cell cycle regulation and DNA damage response, further study will focus on the characterisation of regulons of Fhs1 using chromatin immunoprecipitation-sequencing coupled with transcriptome analysis.
Methods
Fungal strains and media
The wild-type strain Z-3639 and transgenic strains derived from this strain were used in this study (Supplementary Table S2). F. graminearum transcription factor gene deletion mutants were obtained from a previous study11. All strains were stored as conidia and mycelia in 30% glycerol solution at −80 °C. Conidia production was induced in carboxymethyl cellulose medium (CMC) or on yeast malt agar (YMA) as previously described37,38. All of the other media used in this study were prepared as described in the Fusarium laboratory manual32.
The DNA-damaging agents used in the study were MMS, HU, BLM, and CPT. The concentration of each DNA-damaging agent at which the wild-type strain exhibited approximately half of the radial growth observed on CM without any DNA-damaging agent was established (Supplementary Table S1). CM supplemented with each DNA-damaging agent was used to screen 657 transcription factor gene deletion mutants. Statistical and clustering analyses were conducted using R statistical software packages39.
Nucleic acid manipulation, PCR primers and conditions
Genomic DNA was extracted from lyophilised mycelia according to the Fusarium laboratory manual32. Restriction endonuclease digestion, agarose gel electrophoresis, Southern blotting, and hybridisation with 32P-labeled probes were performed following standard protocols40. Total RNA was isolated from mycelia ground in liquid nitrogen using the Easy-Spin Total RNA Extraction Kit (iNtRON Biotech, Seongnam, Korea).
PCR and quantitative real-time (qRT)-PCR primers used in this study were synthesised by an oligonucleotide synthesis facility (Bionics, Seoul, Korea) (Supplementary Table S3). General PCR procedures were performed in accordance with the manufacturer’s instructions (TaKaRa Bio Inc., Otsu, Japan).
Genetic manipulations and fungal transformations
The double-joint (DJ) PCR strategy was applied to construct fusion PCR products for targeted gene deletion and complementation41. For FHS1 deletion, the 5′ and 3′ flanking regions of FHS1 were amplified from the genomic DNA of the wild-type strain using the primer pairs FHS1-5F/FHS1-5R and FHS1-3F/FHS1-3R, respectively. A geneticin resistance gene cassette (GEN) under the control of the A. nidulans trpC promoter and terminator was amplified from pII99 using a primer pair Gen-for/Gen-Rev. Three amplicons (5′ flanking region, GEN, and 3′ flanking region) were mixed at 1:3:1 molar ratio and fused by a second round of DJ PCR. Finally, the fusion constructs were amplified with the nested primers using the second round PCR product as template.
For complementation, the 5′ flanking region that included the FHS1 open reading frame with its own promoter and 3′ flanking region were amplified from genomic DNA of the wild-type strain using the primer pairs FHS1-5F/FHS1-5R GFP and FHS1-3F GFP/FHS1-3R, respectively. The GFP-HYG construct was amplified from pIGPAPA using pIGPAPA-sGFP/HYG-F1 primers. Three amplicons were then fused in a second round of DJ PCR. Finally, the fusion constructs for transformation were amplified with the nested primers using the second round PCR product as a template.
Fungal transformation was performed as previously described42. Conidia were harvested 3–4 days after inoculation in CMC medium, and freshly harvested conidia were inoculated into 50 ml YPG (1% yeast extract, 1% peptone, and 2% glucose) for 12 h at 25 °C. To generate protoplasts, mycelia were harvested through sterilised filter paper and incubated in 35 ml NH4Cl containing 10 mg/ml Driselase (Sigma-Aldrich, St. Louis, MO, USA) for 3 h at 30 °C with shaking (50 rpm). Polyethylene glycol (PEG)-mediated fungal transformation was applied for deletion and complementation. Fungal transformants carrying GEN or HYG were regenerated in the regeneration medium and overlaid with 1% water agar containing geneticin (150 μg/ml; Sigma-Aldrich, St. Louis, MO, USA) or hygromycin B (150 μg/ml; Calbiochem, La Jolla, CA, USA), respectively. After 2–5 days incubation, antibiotic-resistant colonies were selected for further study.
Sexual crosses and virulence tests
For selfing, cultures were grown on carrot agar plates for 5 days. Sexual reproduction was induced by removing aerial mycelia with sterile 2.5% Tween 60 solution32. For outcrosses, heterothallic ∆mat1 strain was fertilised with 1 ml of a conidial suspension (105 conidia/ml) from fertilising parents. All cultures were incubated at 25 °C for perithecial development for an additional 10 days under near UV light (wavelength: 365 nm, HKiv Import & Export Co., Ltd., Xiamen, China).
For virulence tests, point inoculation on wheat cultivar Eunpamil’s heads was performed as previously described42. The conidia used for inoculation were harvested from CMC cultures and suspended in 0.01% Tween 20 solution at 106 spores/ml. After inoculation with 10 μl conidia suspension in the middle of the spikelet, inoculated plants were incubated in a high humidity chamber at 25 °C for 3 days and transferred to a greenhouse for an additional 11 days to assess disease symptoms. Five replicated inoculations per strain and three independent mutant strains were used in the experiment.
Microscopic observation
To observe localisation of Fhs1 and histone H1 (hH1) in the nuclei, the mat1r strain (Δmat1::GEN hH1-RFP-GEN) was fertilised with the HK193 (Δfhs1::FHS1-GFP-HYG) strain. Ascospores carrying both FHS1-GFP-HYG and hH1-RFP-GEN were selected using antibiotic resistance and confirmed using PCR. Localisation was observed in cultures from CM with and without 5 mM HU supplementation. Chitin staining was conducted by adding Calcofluor white stock solution (10 mg/ml) to mycelia samples on slide glasses. Microscopic observation was performed with a DE/Axio Imager A1 microscope (Carl Zeiss, Oberkochen, Germany) using filter set 38HE (excitation 470/40; emission 525/50) for Gfp, filter set 15 (excitation 546/12; emission 590) for Rfp, and filter set 49 (excitation 356; emission 445/50) for Calcofluor white.
Quantitative real-time PCR (qRT)-PCR
Freshly harvested conidia were inoculated into 50 ml liquid CM and incubated for 24 h at 25 °C with shaking. Mycelia were then harvested and inoculated into CM or CM supplemented with 10 mM HU for an additional 2 h. Total RNA was isolated from mycelia that were ground in liquid nitrogen using an Easy-Spin total RNA extraction kit (iNtRON Biotech), and each first-strand cDNA was synthesised using SuperScriptIII reverse transcriptase (Invitrogen, Carlsbad, CA, USA). qRT-PCR was performed with SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) and a 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with corresponding primer pairs (Supplementary Table S3). The ubiquitin C-terminal hydrolase gene UBH (FGSG_01231.3) was used as a reference gene. We compared the cycle threshold (2−∆∆CT) to measure the transcript levels of target genes in different conditions. PCR was performed three times with two replicates per run.
Alkaline comet assay
The alkaline comet assay was performed as previously described with some modifications43. Freshly harvested conidia were incubated in 50 ml YPG at 25 °C for 12 h with shaking. Mycelia were harvested through sterilised filter paper and incubated in 35 ml of 1 M NH4Cl containing 10 mg/ml Driselase with or without 10 mM HU supplementation at 30 °C to generate protoplasts. Protoplasts were collected by centrifugation after 4 h and resuspended in 1 M NH4Cl (2 × 104 cells/ml).
Low-gelling-temperature agarose (1%, Sigma-Aldrich) was melted in 1 M NH4Cl and kept at 40 °C. To improve agarose adhesion, we scored the edges of dust-free frosted-end microscope slides using a diamond-tipped pen. To prepare agarose-precoated slides, the slides were dipped into molten 1% agarose, and one side was wiped clean and air-dried. Protoplast suspension (0.4 ml) was mixed with 1.2 ml low-gelling-temperature agarose at 40 °C in a 5 ml plastic disposable tube. Then, 1.2 ml agarose mixtures were poured onto a pre-coated slide and allowed to gel. Slides were submerged in alkaline lysis conditions to detect DNA single-strand breaks, double-strand breaks, and alkali-labile sites in the DNA, in accordance with a standard protocol43. After overnight lysis in the dark, the slides were washed three times with alkaline rinse solution for 20 min each to ensure removal of salt and detergent. Each slide was submerged in fresh alkaline rinse solution in an electrophoresis chamber and electrophoresed for 25 min at a voltage of 0.6 V/cm. Slides were washed with distilled water and stained in ethidium bromide solution, rinsed again with distilled water, and dried at room temperature. The dried agarose was rehydrated for fluorescence microscopy. The pictures for comet analysis were taken on an Axio Imager A1 microscope with a CCD camera and the 550-nm/605-nm filter set. The percentage of DNA in the tail was analysed for individual cells using the CometScore image analysis software (TriTek Corp., Sumerduck, VA).
RNA-seq analysis
RNA-seq analysis was performed as previously described34. Wild-type and fhs1 conidia were harvested from CMC cultures and inoculated into CM at 25 °C for 24 h with shaking. Total RNA was extracted as stated above. RNA-seq libraries were constructed using the Illumina TruSeqTM RNA sample prep kit with no modifications to the standard low-throughput protocol. Samples were run on an Illumina HiSeq2000 instrument using the reagents provided in the Illumina TruSeq paired-end (PE) cluster kit V3-cBot-HS and the TruSeq SBS kit v3-HS (200 cycles).
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus44 and are accessible through GEO Series accession number GSE73891 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73891). Genome-wide transcript levels were quantified in reads per kilobase of exon per million mapped sequence reads (RPKM)45. RPKM values of 0 were changed to 1 to calculate the fold change of the transcript level. Genes for which differential transcript levels were detected were functionally characterised using the Munich Information Centre for Protein Sequences FunCat functional classification and annotation system46. Three replicates of RNA-seq were performed for each sample.
Additional Information
How to cite this article: Son, H. et al. A novel transcription factor gene FHS1 is involved in the DNA damage response in Fusarium graminearum. Sci. Rep. 6, 21572; doi: 10.1038/srep21572 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (2008–0061897 and 2013R1A6A3A04059121) and the Cooperative Research Program for Agricultural Science and Technology Development (Project PJ01085602), Rural Development Administration, Republic of Korea.
Footnotes
Author Contributions H.S., M.F., K.M. and Y.W.L. conceived and designed the experiments. H.S., M.F., Y.L. and K.M. performed experiments. J.Y.L. analysed computational work. J.C.K. and G.J.C. did the pathogenicity experiments. H.S., M.F. and Y.W.L. wrote the manuscript. All authors read, corrected and approved the final manuscript.
References
- Zhou B.-B. S. & Elledge S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000). [DOI] [PubMed] [Google Scholar]
- Jackson S. P. & Bartek J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. & Jackson S. P. Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547–551 (2002). [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G., Zotter A. & Vermeulen W. DNA damage response. Cold Spring Harb. Perspect. Biol. 3, a000745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M. C. et al. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R. et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R. S. & Kistler H. C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525 (2004). [DOI] [PubMed] [Google Scholar]
- Desjardins A. E. & Proctor R. H. Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 119, 47–50 (2007). [DOI] [PubMed] [Google Scholar]
- Trail F. For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiol. 149, 103–110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Z. et al. Recent advances in genes involved in secondary metabolite synthesis, hyphal development, energy metabolism and pathogenicity in Fusarium graminearum (teleomorph Gibberella zeae). Biotechnol. Adv. 32, 390–402 (2014). [DOI] [PubMed] [Google Scholar]
- Son H. et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 7, e1002310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K. et al. Transcription factor RFX1 is crucial for maintenance of genome integrity in Fusarium graminearum. Eukaryot. Cell 13, 427–436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriyama M., Kobayashi Y., Saito M., Ishikawa F. & Yonehara S. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 29, 4729–4741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A. P. & McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93, 5777–5782 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. & Norris D. The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics 150, 1419–1428 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como C. J., Chang H. & Arndt K. T. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol. Cell. Biol. 15, 1835–1846 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiyanov P., Nag A. & Raychaudhuri P. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274, 35309–35312 (1999). [DOI] [PubMed] [Google Scholar]
- Luo J. et al. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae. J. Cell Biol. 165, 843–855 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita H., Soshi T. & Inoue H. The Neurospora uvs-2 gene encodes a protein which has homology to yeast RAD18, with unique zinc finger motifs. Mol. Gen. Genet. 238, 225–233 (1993). [DOI] [PubMed] [Google Scholar]
- Ohya T., Maki S., Kawasaki Y. & Sugino A. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase ε in Saccharomyces cerevisiae. Nucleic Acids Res. 28, 3846–3852 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H. et al. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 19, 4867–4872 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn P. J. & Reece R. J. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol. Cell. Biol. 19, 882–888 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., Zadra I., Stöffler G. & Angermayr K. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 274, 4613–4619 (1999). [DOI] [PubMed] [Google Scholar]
- Arnaud M. B. et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 40, D653–D659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L.-J. & Tang W.-H. The omics era of Fusarium graminearum: opportunities and challenges. New Phytol. 207, 1–3 (2015). [DOI] [PubMed] [Google Scholar]
- Son H., Park A. R., Lim J. Y. & Lee Y.-W. Fss1 is involved in the regulation of an ENA5 homologue for sodium and lithium tolerance in Fusarium graminearum. Environ. Microbiol. 17, 2048–2063 (2015). [DOI] [PubMed] [Google Scholar]
- Keyer K. & Imlay J. A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA 93, 13635–13640 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koç A., Wheeler L. J., Mathews C. K. & Merrill G. F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279, 223–230 (2004). [DOI] [PubMed] [Google Scholar]
- Ye X. S., Fincher R. R., Tang A., O’Donnell K. & Osmani S. A. Two S-phase checkpoint systems, one involving the function of both BIME and Tyr15 phosphorylation of p34cdc2, inhibit NIMA and prevent premature mitosis. EMBO J. 15, 3599–3610 (1996). [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem. Sci. 31, 402–410 (2006). [DOI] [PubMed] [Google Scholar]
- Nyberg K A., Michelson R J., Putnam C. W. & Weinert T. A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617–656 (2002). [DOI] [PubMed] [Google Scholar]
- Leslie J. F. & Summerell B. A. The Fusarium laboratory manual, 1st edn. (Blackwell, 2006). [Google Scholar]
- Trail F. & Common R. Perithecial development by Gibberella zeae: a light microscopy study. Mycologia 92, 130–138 (2000). [Google Scholar]
- Son H. et al. AbaA regulates conidiogenesis in the ascomycete fungus Fusarium graminearum. PLoS One 8, e72915 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen C. et al. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 102, 16892–16897 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna B., Fodor J., Harrach B. D., Pogány M. & Király Z. The Janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 59, 37–43 (2012). [DOI] [PubMed] [Google Scholar]
- Cappellini R. A. & Peterson J. L. Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia 57, 962–966 (1965). [Google Scholar]
- Harris S. D. Morphogenesis in germinating Fusarium graminearum macroconidia. Mycologia 97, 880–887 (2005). [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2006). [Google Scholar]
- Sambrook J. & Russell D. W. Molecular cloning: a laboratory manual, 4th edn. (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- Yu J.-H. et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981 (2004). [DOI] [PubMed] [Google Scholar]
- Son H., Lee J., Park A. R. & Lee Y.-W. ATP citrate lyase is required for normal sexual and asexual development in Gibberella zeae. Fungal Genet. Biol. 48, 408–417 (2011). [DOI] [PubMed] [Google Scholar]
- Olive P. L. & Banath J. P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protocols 1, 23–29 (2006). [DOI] [PubMed] [Google Scholar]
- Barrett T. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 41, D991–D995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Ruepp A. et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32, 5539–5545 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljelund P., Losson R., Kammerer B. & Lacroute F. Yeast regulatory gene PPR1: II. Chromosomal localization, meiotic map, suppressibility, dominance/recessivity and dosage effect. J. Mol. Biol. 180, 251–265 (1984). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.